Abstract
Background
Lead is a cardiotoxic metal with a variety of adverse health effects. In the absence of data on bone lead exposure, epigenetic biomarkers can serve as indicators of cumulative lead exposure and body burden. Herein, we leveraged novel epigenetic biomarkers of lead exposure to investigate their association with cardiovascular disease (CVD) incidence and mortality.
Methods and Results
Blood DNA methylation was measured using the Illumina MethylationEPIC BeadChip among 2231 participants of the Strong Heart Study (SHS) at baseline (1989–1991). Epigenetic biomarkers of lead levels in blood, patella, and tibia were estimated using previously identified cytosine‐guanine dinucleotide (CpG) sites. CVD incidence and mortality data were available through 2017. Median concentrations of lead epigenetic biomarkers were 13.8 μg/g, 21.3 μg/g, and 2.9 μg/dL in tibia, patella, and blood, respectively. In adjusted models, the hazard ratio (HR) (95% CI) of CVD mortality per doubling increase in lead epigenetic biomarkers were 1.42 (1.07–1.87) for tibia lead, 1.22 (0.93–1.60) for patella lead, and 1.57 (1.16–2.11) for blood lead. The corresponding HRs for incident CVD were 0.99 (0.83–1.19), 1.07 (0.89–1.29), and 1.06 (0.87–1.30). The association between the tibia lead epigenetic biomarker and CVD mortality was modified by sex (interaction P value: 0.014), with men at increased risk (HR, 1.42 [95% CI, 1.17–1.72]) compared with women (HR, 1.04 [95% CI, 0.89–1.22]).
Conclusions
Tibia and blood epigenetic biomarkers were associated with increased risk of CVD mortality, potentially reflecting the cardiovascular impact of cumulative and recent lead exposures. These findings support that epigenetic biomarkers of lead exposure may capture some of the disease risk associated with lead exposure.
Keywords: American Indian populations, DNA methylation, epigenetic biomarkers, lead
Subject Categories: Cardiovascular Disease, Epidemiology
Nonstandard Abbreviations and Acronyms
- DNAm
DNA methylation
- eBlood
DNA methylation–based biomarker of lead exposure in blood
- ePatella
DNA methylation–based biomarker of lead exposure in patella
- eTibia
DNA methylation–based biomarker of lead exposure in tibia
- NAS
Normative Aging Study
- SHS
Strong Heart Study
Clinical Perspective.
What Is New?
Higher levels of epigenetic biomarkers of lead exposure estimated in tibia and blood were associated with increased risk of cardiovascular disease mortality.
The association between the tibia lead epigenetic biomarker and cardiovascular disease mortality was modified by sex, with men at increased risk compared with women.
What Are the Clinical Implications?
In the absence of data on bone lead exposure, epigenetic biomarkers can serve as indicators of cumulative lead exposure.
Epigenetic lead biomarkers can capture some of the cardiovascular risk associated with lead exposure.
Lead (Pb) is a toxic metal associated with adverse cardiovascular, neurological, renal, hematological, immunological, reproductive, and developmental outcomes. 1 , 2 , 3 , 4 Before widespread bans in the 1970s, lead was included in gasoline, paint, water piping, and plumbing fixtures, resulting in extensive contamination of the air, soil, dust, and water. 5 Lead is still widely refined and processed in the United States, 6 where lead levels remain relatively high and individuals remain at risk of exposure. There is also evidence of racial and socioeconomic disparities concerning the burden of lead exposure, 7 , 8 , 9 with several racial and ethnic groups and low‐income populations facing increased exposures compared with other groups. Because of the persistence of lead in the environment and continuous new exposures, lead and its associated adverse health effects remain relevant today.
Despite its importance as a potential cardiovascular risk factor, large cohort studies of cardiovascular disease (CVD) often lack data on lead exposure. Traditionally, biomarkers of lead exposure have been measured in blood, urine, plasma, and bone. 10 Lead accumulates in bone with a half‐life of decades, and bone lead measures can be used to reflect cumulative lead exposure and long‐term health effects. 11 In blood, lead reflects both endogenous sources from bone and exogenous sources from the environment, with a half‐life of 1 to 2 months. Obtaining bone lead measures, however, can be challenging on a population scale, as the technology used requires exposure to radiation and is not widely available. 12
Genome‐wide DNA methylation (DNAm) data can serve as biomarkers of epigenetic signatures to estimate lead concentrations in tibia, patella, and blood, 12 by leveraging the knowledge that lead exposure induces sensitive and specific changes in whole blood DNAm. 13 These methylation‐based biomarkers were well correlated with lead concentrations in tibia and patella, 12 and in a separate analysis, increasing levels of the tibia DNAm biomarker was associated with increased odds of Parkinson disease status. 14 These results highlight the potential of methylation‐based biomarkers to provide estimates of lead exposure, and their relation to disease.
The Strong Heart Study (SHS), a study of CVD in American Indian adults across the Southwest and the Great Plains, 15 represents an opportunity to investigate the relationship of these epigenetic biomarkers with cardiovascular health. Lead exposure has been documented in American Indian communities, 16 , 17 where a legacy of environmental contamination remains a concern. Research has also identified that SHS communities have a high burden of CVD, 18 and that exposure to toxic metals, including cadmium and arsenic, contributes to this increased risk. 19 , 20 , 21 , 22 Because of the evidence highlighting the importance of metals on CVD, 23 , 24 there is further need to investigate the impact of lead in American Indian communities.
The objective of this study was to apply these recently developed epigenetic biomarkers of lead exposure and investigate their association with CVD incidence and mortality in the SHS. Given the accumulation of lead in bone, and consistent with previous findings, 25 we anticipated that epigenetic biomarkers of bone lead would be more strongly associated with cardiovascular outcomes than an epigenetic biomarker of blood lead. This work is a novel application of these epigenetic biomarkers to study cardiovascular outcomes.
METHODS
Study Population
The SHS is a prospective cohort of CVD and its risk factors among American Indians adults, funded by the National Heart, Lung, and Blood Institute and the National Institute of Environmental Health Sciences. 15 In 1989 to 1991, all adults aged 45 to 74 years across 13 tribes and communities in Arizona and Oklahoma, and random subsets in North Dakota and South Dakota, were eligible for recruitment. 15 The SHS protocol was approved by institutional review boards, participating tribes, and the respective area Indian Health Service institutional review boards. All participants provided informed consent. A total of 4549 adults were initially recruited. For this study, 1032 participants from one community were not included on their request. We also excluded 252 participants with CVD at baseline, 429 participants without sufficient urine for metal analyses, and 44 participants missing cardiovascular risk factors, resulting in 2792 participants eligible for analysis of blood DNAm. However, 445 participants had insufficient amounts of DNA for analysis, and 26 were further excluded in quality control, leaving a final sample size of 2321 participants included in this study. The data underlying this article cannot be shared publicly in an unrestricted manner because of limitations in the consent forms and in the agreements between the SHS tribal communities and the SHS investigators. The data can be shared to external investigators following the procedures established by the SHS, available at https://strongheartstudy.org/.
All participants provided sociodemographic and medical history information, including age, sex, education, study center of recruitment, smoking status (never, former, or current), body mass index, low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol, blood pressure, hypertension treatment (yes/no), and diabetes (yes/no) via baseline questionnaires, physical examinations, and laboratory analyses, as described previously. 26 Estimated glomerular filtration rate was calculated using age, sex, and plasma creatinine via the Chronic Kidney Disease Epidemiology Collaboration formula. 27 Diabetes status at baseline was defined as fasting glucose ≥126 mg/dL, 2‐hour plasma glucose ≥200 mg/dL, hemoglobin A1C level ≥6.5%, self‐reported history of diagnosis, or current use of diabetic medication.
DNA Methylation
Buffy coat from fasting blood samples was collected in 1989 to 1991 on recruitment, and biological specimens were stored at <−70 °C. DNA from white blood cells was extracted and stored at the MedStar Health Research Institute under a strict quality control system. In 2015, blood DNA was shipped to the analytical laboratory at the Texas Biomedical Research Institute for DNAm analysis. DNA was bisulfite converted with the EZ DNAm kit (Zymo Research, Irvine, CA), according to the manufacturer's instructions. Bisulfite‐converted DNA was measured using the Illumina MethylationEPIC BeadChip (850K; Illumina, San Diego, CA), which provides a measure of DNAm at a single‐nucleotide resolution at >850 000 CpGs. Samples were randomized across and within plates to remove potential batch artifacts and confounding effects, and replicate and across‐plate control samples were included on every plate. All the preprocessing was conducted using R version 3.6.1. 28 Data were read in 6 different batches (of ∼400 individuals each) and combined using the R package minfi (version 1.18.4). 29 CpGs with a P‐detection value of >0.01 in >5% of the individuals (6159 CpGs) were removed. Single sample normalization was conducted using the preprocess Noob function in minfi, 30 , 31 which includes a background correction with dye‐bias normalization for Illumina Infinium methylation arrays. Regression on correlated probes normalization was applied to account for probe type bias. 32
As a result of these preprocessing preliminary analyses, we had data from 2321 individuals and 860 079 CpGs. Cross‐hybridizing probes, sex chromosomes, and single‐nucleotide polymorphism probes with minor allele frequency >0.05 33 were removed from the analysis. The final number of CpGs for analysis was 790 026. Quality checks, data normalization, statistical preprocessing, and β‐value calculation, which ranges from 0 to 1 and represents the proportion of unconverted cytosines in bisulfite‐converted DNA at specific locations, were performed using the R package minfi. 30 We estimated Houseman cell proportions (CD8T cells, CD4T cells, natural killer cells, B cells, monocytes, and granulocytes) 34 using the R package minfi, to use them as adjustment variables in regression models. We detected and corrected for potential batch effects by sample plate, sample row, and DNA isolation time with the combat function (sva R package). 35 We annotated CpGs to the nearest gene, according to the Illumina Infinium MethylationEPIC Manifest File (version 1.0 B4). 30 , 36
Lead Epigenetic Biomarkers
Epigenetic biomarkers of lead exposure reflecting lead in patella (ePatella) and tibia (eTibia) were calculated according to Colicino et al (2019). 12 To generate these biomarkers, Colicino et al used blood DNAm and bone lead measures available in a subset of 348 elderly men from the NAS (Normative Aging Study), a prospective cohort study of aging in adult men established in 1963 by the US Department of Veterans Affairs. 37 Blood DNAm was obtained with the Illumina Infinium HumanMethylation450 BeadChip (450k) array, and probes overlapping the 450k array and the Infinium MethylationEPIC BeadChip platform (395 005 CpG sites) were included in their analysis. Lead levels in tibia and patella were measured noninvasively with K‐shell X‐ray fluorescence spectroscopy. 38 , 39
Epigenome‐wide robust linear regressions were performed to select the most significant CpG sites associated with log2‐transformed lead concentrations in each tissue separately, and then an elastic net regression approach, which takes into account the high‐dimensional nature of CpG data, 40 was used to create the lead epigenetic biomarkers. CpGs from the epigenome‐wide analysis were selected at P<0.0001. Elastic net with leave‐one‐out cross‐validation was performed on the training data set, composed of 80% of participants. Lead biomarkers were then validated in the remaining test data set of the NAS cohort. ePatella and eTibia biomarkers were calculated as the linear combination of regression coefficients, and DNAm β values were calculated from the test data set. Biomarkers were originally only estimated for lead in bone; however, we further extended the epigenetic biomarkers to estimate lead in blood (eBlood), to provide a marker of recent exposure, using the above approach in the NAS cohort (Data S1, Supplemental Methods). Lead levels in blood were measured using Zeeman background‐corrected flameless atomic absorption (graphite furnace). 41 , 42 Estimation and validation of the blood lead epigenetic biomarker can be found in Tables S1 and S2 and Figures S1 through S5. In the current analysis, we used the same significant CpG sites associated with lead levels in blood (74 of 75 CpG sites), patella (58 CpG sites), and tibia (138 CpG sites) in NAS to estimate 3 lead DNAm‐based biomarkers (eTibia, ePatella, and eBlood) in 2321 participants from the SHS. All CpG sites included in biomarker development in NAS, except one CpG used to calculate eBlood, were available for lead epigenetic biomarkers in SHS.
Cardiovascular Outcomes
Morbidity and mortality surveillance in the SHS is ongoing and has been previously described. 43 , 44 , 45 Briefly, all CVD outcomes and deaths are identified through coordination between Field Centers, the Coordinating Center, and Surveillance Reporting. For this study, all deaths and potential cardiovascular outcomes occurring through 2017 were reviewed by the Morbidity and Mortality Review Committee, which is composed of physicians with experience in reviewing medical records for the ascertainment of cardiovascular outcomes. 43 , 44 Incident CVD was defined as any definite or possible fatal or nonfatal coronary heart disease, stroke, or heart failure. Cardiovascular deaths were ascertained according to international diagnostic criteria, and events possibly meeting these criteria included the following: definite fatal myocardial infarction, definite sudden death attributable to coronary heart disease, definite fatal coronary heart disease, possible fatal coronary heart disease, definite fatal stroke, possible fatal stroke, definite fatal congestive heart failure, possible fatal congestive heart failure, and other fatal CVD.
Statistical Analysis
Distributions of eTibia, ePatella, and eBlood lead biomarkers were analyzed according to demographic and clinical covariates. Spearman correlations were performed among lead epigenetic biomarkers and urinary cadmium concentrations. We included urinary cadmium as this metal is moderately correlated with lead biomarkers in other studies. 46 Urinary lead biomarkers, unfortunately, are not available at the SHS examination 1 as the vials for sample collection were contaminated with lead. 47 Urinary cadmium concentrations were expressed in micrograms per gram of urine creatinine. The significance level in this analysis was P=0.05.
We used progressively adjusted multivariable Cox proportional hazards models to estimate the risk of incident CVD and CVD mortality according to each lead epigenetic biomarker. Lead epigenetic biomarkers were analyzed in tertiles, as continuous variables, and in a nonlinear manner using a restricted quadratic spline model with knots at the 10th, 50th, and 90th percentiles with the reference at the 10th percentile. Lead epigenetic biomarkers were log2 transformed to remove skewness (Figure S6). In all Cox models, center of recruitment was incorporated as a strata term, and age was used as the time metric. In progressively adjusted models, the first model (model 1) adjusted for sex, smoking status, body mass index, 5 genetic principal components (to account for population stratification 48 ), and Houseman cell proportions (CD8T cells, CD4T cells, natural killer cells, B cells, and monocytes). The second model (model 2) further adjusted for low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol, and diabetes status. Model 3 further adjusted for systolic blood pressure, hypertension treatment, and estimated glomerular filtration rate.
Effect modification of the relationship between lead epigenetic biomarkers and CVD incidence and mortality was investigated according to the following subgroups: age (44.0–49.9, 50.0–64.9, and 65.0–75.4 years), sex (male/female), study center (Arizona, Oklahoma, and North Dakota/South Dakota), smoking status (never/former/current), urinary cadmium (<0.71, 0.71–1.26, and >1.26 μg/g), and diabetes (yes/no). Cox proportional hazards models included interaction terms between the subgroup and an increase in an interquartile range change in the respective lead epigenetic biomarker (eTibia, ePatella, and eBlood). Finally, as these biomarkers were estimated in men, we examined the differences in methylation levels between men and women for individual probes included in the biomarkers (Table S3). t‐Tests were performed to examine statistically significant differences in these methylation levels. All analyses were conducted in R version 4.0.2. 28
RESULTS
Median (interquartile range) concentrations of lead epigenetic biomarkers were 21.3 (18.5–24.8) μg/g in ePatella, 13.8 (11.7–16.1) μg/g in eTibia, and 2.9 (2.6–3.4) μg/dL in eBlood. The concentrations of each lead epigenetic biomarker were largely similar across participant characteristics and medical covariates (Table 1). Correlations among lead epigenetic biomarkers and urinary cadmium, adjusted for urine creatinine, revealed slight but significant positive associations between eTibia lead and urinary cadmium (Spearman r=0.10; P<0.001), ePatella lead and urinary cadmium (r=0.07; P<0.001), and eTibia and eBlood lead (r=0.09; P<0.001) (Table 2). eTibia and ePatella lead biomarkers (r=−0.07; P<0.001) and ePatella and eBlood lead biomarkers (r=−0.13; P<0.001) were negatively correlated. The median (interquartile range) age at follow‐up among those who experienced a CVD event was 68.5 (62.4–74.9) years, whereas the median (interquartile range) age at follow‐up among those who did not experience a CVD event was 75.1 (68.7–80.5) years.
Table 1.
Lead Epigenetic Biomarkers by Participant Characteristics (N=2321)
| Variable | No. (%) | eTibia lead, μg/g | ePatella lead, μg/g | eBlood lead, μg/dL | Urinary cadmium, μg/g | |
|---|---|---|---|---|---|---|
| Sex | Men | 962 (41.5) | 13.2 (11.1–15.3) | 20.8 (18.1–24.5) | 2.9 (2.5–3.4) | 0.71 (0.47–1.10) |
| Women | 1359 (58.5) | 14.2 (12.2–16.6) | 21.6 (18.8–25.1) | 2.9 (2.6–3.4) | 1.16 (0.77–1.78) | |
| Age, y | 44.0–50.8 | 668 (28.8) | 14.0 (12.0–16.4) | 21.0 (18.6–24.3) | 3.0 (2.6–3.4) | 0.90 (0.56–1.31) |
| 50.9–59.5 | 1244 (53.6) | 13.8 (11.6–16.1) | 21.2 (18.3–24.9) | 2.9 (2.6–3.4) | 1.00 (0.64–1.55) | |
| 59.6–75.4 | 409 (17.6) | 13.7 (11.5–15.8) | 21.6 (19.0–25.2) | 2.9 (2.5–3.3) | 1.03 (0.64–1.59) | |
| Center | Arizona | 311 (13.4) | 13.9 (11.6–16.0) | 20.5 (17.8–23.3) | 2.9 (2.6–3.4) | 0.76 (0.53–1.19) |
| Oklahoma | 981 (42.3) | 13.9 (11.7–16.2) | 21.0 (18.3–24.5) | 3.0 (2.6–3.4) | 0.86 (0.55–1.33) | |
| ND/SD | 1029 (44.3) | 13.8 (11.7–16.1) | 21.7 (18.9–25.5) | 2.9 (2.5–3.4) | 1.11 (0.75–1.78) | |
| Smoking status | Never | 684 (29.5) | 13.8 (11.8–15.8) | 21.1 (18.3–24.8) | 3.0 (2.6–3.5) | 0.87 (0.55–1.36) |
| Former | 745 (32.1) | 13.8 (11.8–16.0) | 21.3 (18.4–25.1) | 2.9 (2.6–3.3) | 0.81 (0.55–1.25) | |
| Current | 892 (38.4) | 14.0 (11.7–16.5) | 21.3 (18.7–24.6) | 2.9 (2.6–3.4) | 1.17 (0.77–1.81) | |
| Urinary cadmium, μg/g | <0.71 | 758 (32.7) | 13.4 (11.3–15.5) | 20.8 (18.2–24.4) | 2.9 (2.6–3.4) | 0.51 (0.38–0.61) |
| 0.71–1.26 | 790 (34.0) | 13.8 (11.8–16.2) | 21.3 (18.4–25.0) | 3.0 (2.6–3.5) | 0.96 (0.83–1.10) | |
| >1.26 | 773 (33.3) | 14.1 (12.0–16.7) | 21.6 (18.8–25.2) | 2.9 (2.5–3.3) | 1.81 (1.50–2.33) | |
| BMI, kg/m2 | <25 | 406 (17.5) | 14.1 (12.0–16.3) | 21.6 (18.7–25.1) | 3.0 (2.6–3.5) | 1.21 (0.78–1.87) |
| 25–30 | 830 (35.8) | 13.6 (11.5–16.2) | 21.2 (18.5–24.8) | 3.0 (2.6–3.4) | 0.98 (0.62–1.51) | |
| ≥30 | 1085 (46.7) | 13.8 (11.7–16.0) | 21.2 (18.5–24.8) | 2.9 (2.5–3.3) | 0.87 (0.57–1.35) | |
| Diabetes | Yes | 966 (41.6) | 13.8 (11.5–15.9) | 21.2 (18.4–24.7) | 2.9 (2.5–3.4) | 0.89 (0.58–1.41) |
| No | 1355 (58.4) | 13.8 (11.8–16.3) | 21.3 (18.6–24.9) | 3.0 (2.6–3.4) | 1.02 (0.65–1.54) | |
| Hypertension treatment | Yes | 464 (20.0) | 14.0 (12.0–16.0) | 20.8 (18.2–24.6) | 3.0 (2.6–3.5) | 0.86 (0.56–1.33) |
| No | 1857 (80.0) | 13.7 (11.7–16.2) | 21.4 (18.6–24.9) | 2.9 (2.6–3.4) | 1.00 (0.64–1.54) | |
| SBP, mm Hg | <124 | 1146 (49.4) | 13.7 (11.6–16.0) | 21.2 (18.5–24.8) | 2.9 (2.6–3.4) | 1.01 (0.65–1.53) |
| ≥124 | 1175 (50.6) | 13.9 (11.8–16.3) | 21.3 (18.5–24.8) | 2.9 (2.6–3.4) | 0.92 (0.60–1.46) | |
| LDL‐C, mg/dL | <119 | 1159 (50.0) | 14.0 (11.8–16.3) | 21.4 (18.5–24.9) | 3.0 (2.6–3.4) | 0.97 (0.61–1.48) |
| ≥119 | 1162 (50.0) | 13.7 (11.5–16.0) | 21.1 (18.5–24.8) | 2.9 (2.5–3.3) | 0.96 (0.62–1.52) | |
| HDL‐C, mg/dL | <44 | 1140 (49.1) | 13.6 (11.4–15.9) | 21.0 (18.3–24.7) | 2.9 (2.5–3.4) | 0.91 (0.57–1.41) |
| ≥44 | 1181 (50.9) | 14.1 (12.0–16.3) | 21.5 (18.8–24.9) | 2.9 (2.6–3.4) | 1.03 (0.67–1.59) | |
| eGFR, mL/min per 1.73 m2 | <60 | 76 (3.3) | 14.7 (11.9–17.3) | 21.4 (18.9–25.6) | 2.9 (2.5–3.4) | 0.91 (0.53–1.51) |
| ≥60 | 2245 (96.7) | 13.8 (11.7–16.1) | 21.3 (18.5–24.8) | 2.9 (2.6–3.4) | 0.97 (0.62–1.50) | |
| CVD mortality | Yes | 452 (19.5) | 14.1 (11.7–16.3) | 21.4 (18.6–25.0) | 3.0 (2.6–3.4) | 1.00 (0.65–1.60) |
| No | 1869 (80.5) | 13.7 (11.7–16.1) | 21.2 (18.5–24.8) | 2.9 (2.6–3.4) | 0.96 (0.61–1.48) | |
| CVD incidence | Yes | 1023 (44.1) | 13.7 (11.4–16.0) | 21.3 (18.5–25.0) | 2.9 (2.5–3.3) | 0.99 (0.62–1.54) |
| No | 1298 (55.9) | 13.9 (11.9–16.2) | 21.2 (18.5–24.7) | 2.9 (2.6–3.4) | 0.95 (0.61–1.46) |
Data are given as median (25th–75th percentile), unless otherwise indicated. Urinary cadmium concentrations were expressed in micrograms per gram of urine creatinine. BMI indicates body mass index; CVD, cardiovascular disease; eBlood, DNA methylation–based biomarker of lead exposure in blood; eGFR, estimated glomerular filtration rate; ePatella, DNA methylation–based biomarker of lead exposure in patella; eTibia, DNA methylation–based biomarker of lead exposure in tibia; HDL‐C, high‐density lipoprotein cholesterol; LDL‐C, low‐density lipoprotein cholesterol; ND, North Dakota; SBP, systolic blood pressure; and SD, South Dakota.
Table 2.
Spearman Correlation Coefficients (P Value) of Lead Epigenetic Biomarkers (eTibia, ePatella, and eBlood Lead) and Urinary Cadmium Concentrations (N=2321)
| Variable | eTibia lead | ePatella lead | eBlood lead | Urinary cadmium |
|---|---|---|---|---|
| eTibia lead | 1.00 | |||
| ePatella lead | −0.07 (<0.001) | 1.00 | ||
| eBlood lead | 0.09 (<0.001) | −0.13 (<0.001) | 1.00 | |
| Urinary cadmium | 0.10 (<0.001) | 0.07 (<0.001) | −0.01 (0.62) | 1.00 |
Urinary cadmium concentrations were expressed in micrograms per gram of urine creatinine. eBlood indicates DNA methylation–based biomarker of lead exposure in blood; ePatella, DNA methylation–based biomarker of lead exposure in patella; and eTibia, DNA methylation–based biomarker of lead exposure in tibia.
For CVD mortality, the fully adjusted hazard ratio (HR) (95% CI) for a doubling increase in each lead epigenetic biomarker was 1.42 (1.07–1.87) for eTibia lead, 1.22 (0.93–1.60) for ePatella lead, and 1.57 (1.16–2.11) for eBlood lead (Table 3, model 3). Modeling the eBlood lead epigenetic biomarker in tertiles, the fully adjusted HR (95% CI) for CVD mortality comparing the highest with lowest tertile was 1.31 (1.03–1.67) in the partially adjusted model (model 2), and the association was similar after adjustment for systolic blood pressure, hypertension treatment, and estimated glomerular filtration rate (HR, 1.28 [95% CI, 1.00–1.64]; model 3). Flexible dose‐response models supported a linear relationship for the association of eTibia lead and eBlood lead epigenetic biomarkers (log2 transformed) with CVD mortality, whereas the ePatella lead biomarker showed a linear but not statistically significant relationship (Figure S7).
Table 3.
HRs (95% CIs) for CVD Mortality by Lead Epigenetic Biomarkers (N=2321)
| Epigenetic biomarker | Tertile 1 | Tertile 2 | Tertile 3 | Per double increase |
|---|---|---|---|---|
| eTibia lead, μg/g | <3.6 | 3.6–3.9 | >3.9 | |
| Model 1 | 1.00 (Reference) | 1.19 (0.94–1.50) | 1.16 (0.91–1.47) | 1.36 (1.03–1.79)* |
| Model 2 | 1.00 (Reference) | 1.22 (0.97–1.54) | 1.22 (0.95–1.55) | 1.47 (1.11–1.94)* |
| Model 3 | 1.00 (Reference) | 1.18 (0.94–1.49) | 1.19 (0.93–1.52) | 1.42 (1.07–1.87)* |
| ePatella lead, μg/g | <4.3 | 4.3–4.5 | >4.5 | |
| Model 1 | 1.00 (Reference) | 1.01 (0.80–1.28) | 1.08 (0.85–1.37) | 1.14 (0.87–1.48) |
| Model 2 | 1.00 (Reference) | 0.99 (0.79–1.25) | 1.13 (0.88–1.44) | 1.22 (0.93–1.59) |
| Model 3 | 1.00 (Reference) | 0.98 (0.78–1.24) | 1.12 (0.88–1.43) | 1.22 (0.93–1.60) |
| eBlood lead, μg/dL | <1.4 | 1.4–1.7 | >1.7 | |
| Model 1 | 1.00 (Reference) | 1.14 (0.90–1.45) | 1.23 (0.96–1.57) | 1.51 (1.12–2.04)* |
| Model 2 | 1.00 (Reference) | 1.20 (0.95–1.52) | 1.31 (1.03–1.67)* | 1.59 (1.19–2.15)* |
| Model 3 | 1.00 (Reference) | 1.17 (0.92–1.48) | 1.28 (1.00–1.64)* | 1.57 (1.16–2.11)* |
Model 1: adjusted for sex, smoking status (never, former, or current), body mass index (kg/m2), genetic principal components, and immune cell types (CD8+ cells, CD4+ cells, natural killer cells, B cells, and monocytes). Model 2: further adjusted for low‐density lipoprotein cholesterol (mg/dL), high‐density lipoprotein cholesterol (mg/dL), and diabetes status (yes/no). Model 3: further adjusted for blood pressure (mm Hg), hypertension treatment (yes/no), and estimated glomerular filtration rate (mL/min per 1.73 m2). All models included center of recruitment as a strata term, and age was accounted for in the follow‐up times of all models. Tertiles were calculated on log2‐transformed lead epigenetic biomarker concentrations. CVD indicates cardiovascular disease; eBlood, DNA methylation–based biomarker of lead exposure in blood; ePatella, DNA methylation–based biomarker of lead exposure in patella; eTibia, DNA methylation–based biomarker of lead exposure in tibia; and HR, hazard ratio.
Represents statistically significant associations.
None of the 3 lead epigenetic biomarkers was significantly associated with CVD incidence, either in models analyzing each epigenetic biomarker in tertiles or as continuous variables (Table 4) or in flexible dose‐response models (Figure S7), and with different levels of adjustment. In additional models treating cadmium as a confounder, we found that our analyses are robust to urine cadmium adjustment (Table S4).
Table 4.
HRs (95% CIs) for CVD Incidence by Lead Epigenetic Biomarkers (N=2321)
| Epigenetic biomarker | Tertile 1 | Tertile 2 | Tertile 3 | Per double increase |
|---|---|---|---|---|
| eTibia lead, μg/g | <3.6 | 3.6–3.9 | >3.9 | |
| Model 1 | 1.00 (Reference) | 0.98 (0.84–1.13) | 0.99 (0.84–1.16) | 0.96 (0.80–1.15) |
| Model 2 | 1.00 (Reference) | 0.99 (0.85–1.15) | 1.04 (0.89–1.22) | 1.02 (0.85–1.23) |
| Model 3 | 1.00 (Reference) | 0.96 (0.83–1.12) | 1.03 (0.88–1.21) | 0.99 (0.83–1.19) |
| ePatella lead, μg/g | <4.3 | 4.3–4.5 | >4.5 | |
| Model 1 | 1.00 (Reference) | 0.92 (0.79–1.07) | 1.00 (0.85–1.17) | 1.02 (0.85–1.22) |
| Model 2 | 1.00 (Reference) | 0.91 (0.78–1.06) | 1.04 (0.88–1.22) | 1.08 (0.90–1.29) |
| Model 3 | 1.00 (Reference) | 0.90 (0.77–1.05) | 1.03 (0.88–1.21) | 1.07 (0.89–1.29) |
| eBlood lead, μg/dL | <1.4 | 1.4–1.7 | >1.7 | |
| Model 1 | 1.00 (Reference) | 1.04 (0.89–1.21) | 0.95 (0.81–1.12) | 1.02 (0.83–1.25) |
| Model 2 | 1.00 (Reference) | 1.08 (0.92–1.26) | 1.01 (0.86–1.18) | 1.07 (0.87–1.30) |
| Model 3 | 1.00 (Reference) | 1.07 (0.92–1.25) | 1.00 (0.85–1.17) | 1.06 (0.87–1.30) |
Model 1: adjusted for sex, smoking status (never, former, or current), body mass index (kg/m2), genetic principal components, and immune cell types (CD8+ cells, CD4+ cells, natural killer cells, B cells, and monocytes). Model 2: further adjusted for low‐density lipoprotein cholesterol (mg/dL), high‐density lipoprotein cholesterol (mg/dL), and diabetes status (yes/no). Model 3: further adjusted for systolic blood pressure (mm Hg), hypertension treatment (yes/no), and estimated glomerular filtration rate (mL/min per 1.73 m2). All models included center of recruitment as a strata term, and age was accounted for in the follow‐up times of all models. Tertiles were calculated on log2‐transformed lead epigenetic biomarker concentrations. CVD indicates cardiovascular disease; eBlood, DNA methylation–based biomarkers of lead exposure in blood; ePatella, DNA methylation–based biomarkers of lead exposure in patella; eTibia, DNA methylation–based biomarkers of lead exposure in tibia; and HR, hazard ratio.
The associations between lead epigenetic biomarkers and CVD mortality were modified by sex (Figure 1). The HR (95% CI) of CVD mortality for an interquartile range increase in tibia lead was 1.42 (1.17–1.72) for men versus 1.04 (0.89–1.23) for women (P value for interaction=0.014). The corresponding HRs (95% CIs) for eBlood and ePatella lead for men were 1.27 (1.09–1.49) and 1.12 (0.95–1.31), respectively (eBlood P value for interaction=0.231, and ePatella P value for interaction=0.976). Furthermore, 38% of probes (113 of 270) used in this analysis differed between men and women at a nominal P<0.05 after controlling for the false discovery rate (Table S2). Effect modification models for incident CVD were not significant for eTibia and ePatella lead by any participant characteristic evaluated, including sex, although the association for tibia lead was nonsignificantly stronger in men than women (Figure 2). For eBlood lead, the association with CVD incidence was nonsignificantly stronger for men versus women (HR, 1.13 [95% CI, 1.01–1.26] versus 1.04 [95% CI, 0.93–1.16]; P for interaction=0.281) and significantly modified by age, with oldest participants showing higher risk (P for interaction=0.007) and by study center (P=0.002), with participants from North Dakota and South Dakota showing an increased risk (HR, 1.27 [95% CI, 1.13–1.42]), whereas no association was found in Arizona (HR, 1.04 [95% CI, 0.78–1.38]) and Oklahoma (HR, 0.95 [95% CI, 0.84–1.07]).
Figure 1. Hazard ratio (HR) (95% CI) of cardiovascular disease mortality by a doubling increase in epigenetic lead biomarker concentrations (log2 transformed) of tibia (A), patella (B), and blood (C) biomarkers, corresponding to the interquartile range (75th–25th percentile) within subgroups.
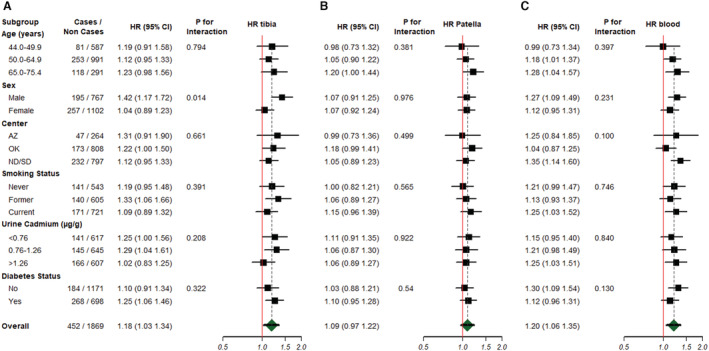
AZ indicates Arizona; ND, North Dakota; OK, Oklahoma; and SD, South Dakota.
Figure 2. Hazard ratio (HR) (95% CI) of cardiovascular disease incidence by a doubling increase in epigenetic lead biomarker concentrations (log2 transformed) of tibia (A), patella (B), and blood (C) biomarkers, corresponding to the interquartile range (75th–25th percentile) within subgroups.
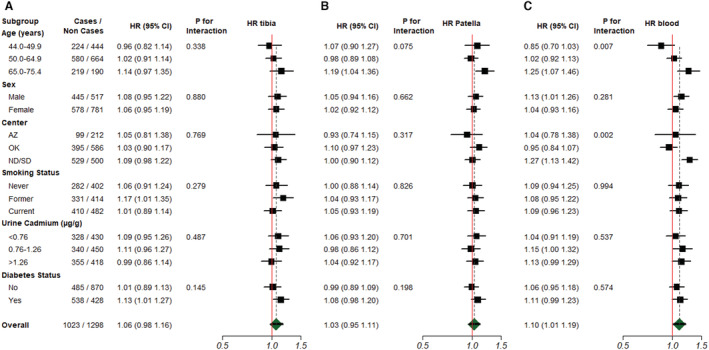
AZ indicates Arizona; ND, North Dakota; OK, Oklahoma; and SD, South Dakota.
DISCUSSION
eTibia and eBlood lead biomarkers were associated with increased risk of CVD mortality in the SHS. The association of eTibia and eBlood lead biomarkers with CVD mortality was modified by sex, with a positive association found in men and no association found in women. This finding is likely explained by the creation of these epigenetic biomarkers in the NAS, an all‐male population. The biomarkers have yet to be validated in women. However, findings remained significant for the whole population. eTibia and eBlood lead biomarkers were not associated with CVD incidence overall, but a positive association, which was significant for eBlood and borderline significant for eTibia, was found among men. No significant association was observed in models incorporating the ePatella lead biomarker with either CVD mortality or incidence.
This analysis builds on the development of bone and blood lead epigenetic biomarkers, 12 and is the first study to evaluate their relation with cardiovascular outcomes. Compared with the NAS subset that was used to generate these lead biomarkers, SHS participants were younger, had similar proportions of ever smokers, and included both men and women (Table S1). Lead levels (mean±SD) measured in tibia (21.1±12.9 μg/g), patella (27.4±17.7 μg/g), and blood (4.0±2.3 μg/dL) among the NAS subset were higher than epigenetic biomarker concentrations estimated in the SHS subset (eTibia: 14.1±3.5 μg/g; ePatella: 22.4±6.7 μg/g; and eBlood: 3.1±0.8 μg/dL), which is a reasonable finding as the SHS population is younger and resides in rural areas and small towns that have been less historically affected by traffic and leaded gasoline compared with the Boston, MA, area.
Although these epigenetic biomarkers have been estimated in 2 cohorts to predict Parkinson disease, 14 further research is needed to determine their transportability across different cohorts with diverse study populations and different health outcomes. This aforementioned study observed that increased concentrations of the tibia epigenetic biomarker was associated with Parkinson disease status in the Parkinson's Environment and Genes cohort and the System Genomics of Parkinson's Disease cohort, 14 whereas patella epigenetic biomarker concentrations were inversely associated with Parkinson disease status in the System Genomics of Parkinson's Disease cohort. 14 In both these cohorts, DNAm estimated tibia and patella lead concentrations were lower compared with the present analysis in the SHS, and these cohorts consisted of participants who were older and had a higher proportion of male participants than the present analysis. Notably, smoking information was missing in the System Genomics of Parkinson's Disease cohort, which is an important confounder that was included in the present study. The results presented in the current analysis are consistent in identifying that the tibia epigenetic biomarker was most strongly associated with disease.
The observed associations with the eTibia biomarker could stem from the longer half‐life of lead in tibia, a cortical bone, in comparison to patella, a trabecular bone. 4 Evidence suggests that lead in trabecular bone is more biologically active and that lead is exchanged into the bloodstream more readily than in cortical bones. 4 , 12 As the half‐life of tibia lead is longer than patella lead, tibia lead is believed to be more representative of cumulative lead exposure. The K‐shell X‐ray fluorescence spectroscopy measurement technique has also been cited as having greater measurement uncertainty for trabecular bones than cortical bones, attributable to a lower comparative mineral density in trabecular bones. 38 Thus, the greater precision of lead measurements in tibia could also explain the stronger associations reported between eTibia, rather than ePatella, biomarkers. Furthermore, the eBlood biomarker is a marker of short‐term lead exposure, which creates potential challenges in its use as a predictive marker for disease, although blood lead reflects both endogenous and exogenous sources of exposure. We found significant associations between the eBlood biomarker and increased risk of CVD mortality, as well as between eBlood lead and CVD incidence among men, suggesting that this epigenetic biomarker is relevant in capturing disease risk from lead exposure. The stronger association with mortality has also been observed among other risk factors in the SHS, such as urinary arsenic 49 and urinary cadmium concentrations. 19 This relationship may also be related to measurement error and differences in clinical care across sites for morbid events, where mortality represents a more definitive and robust end point. An alternative explanation is that lead exposure results in more severe disease, which would reflect a stronger relationship with CVD mortality than incidence.
The present study adds to the weight of evidence of DNAm‐based biomarkers to predict disease risk in the SHS. Prior research has identified that differential methylation at CpG sites and differentially methylated regions were associated with increased incidence of lymphatic‐hematopoietic, solid, and overall cancers, 50 smoking, 51 arsenic 52 and cadmium, 51 and coronary heart disease. 53 One challenge of using these biomarkers, however, is replicating their ability to capture risk across diverse populations. 53 Although the present study implements 3 epigenetic biomarkers of lead exposure, these results would be strengthened through replication in different populations, including in other American Indian cohorts and other racial and ethnic groups.
One limitation of this analysis is that the SHS does not have bone and blood lead measured concurrently with DNAm to compare the accuracy of the epigenetic biomarkers. As a consequence, we cannot confirm that these biomarkers directly reflect lead exposure in the SHS. Another alternative explanation is that the biomarkers reflect DNAm pathways affected by lead that could also be affected by other exposures, but that nevertheless are part of the mechanisms by which lead induces CVD. Furthermore, the epigenetic biomarkers were originally created in the NAS, an elderly, all‐male, and mainly White cohort, which could limit their generalizability to other cohorts like the SHS. In our analysis of sex‐dependent effects, the epigenetic biomarkers of tibia lead and blood lead remained associated with cardiovascular mortality and cardiovascular incidence primarily in men, whereas the association was practically null in women, suggesting that these epigenetic lead biomarkers are primarily relevant for men. Given the sensitivity of DNA methylation biomarkers to the availability of specific probes, we recommend that future analyses estimating epigenetic biomarkers of lead exposure consider the availability of the probes used herein. Given the potential of epigenetic biomarkers to act as noninvasive approximations of lead exposure, it is vital to perform further validation of these epigenetic biomarkers in cohorts where bone lead measures are available, and determine their relation to disease risk in both men and women.
CONCLUSIONS
In the SHS, recently developed tibia and blood lead epigenetic biomarkers were associated with increased risk of cardiovascular mortality, potentially reflecting the cardiovascular impact of cumulative and ongoing lead exposures. Future work must perform further validation of these lead epigenetic biomarkers in different populations, given their potential to capture disease risk.
Sources of Funding
This work was funded by the National Institute of Environmental Health Sciences (T32 ES007322). This work was supported by grants by the National Heart, Lung, and Blood Institute (under contract numbers 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030) and previous grants (R01HL090863, R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319) and cooperative agreements (U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521), by the National Institute of Environmental Health Sciences (R01ES021367, R01ES025216, P42ES010349, and P30ES009089) and by the Spanish Funds for Research in Health Sciences, Carlos III Health Institute, cofunded by European Regional Development Fund (CP12/03080 and PI15/00071). A. Domingo‐Relloso was supported by a fellowship from “la Caixa” Foundation (identifier 100010434) (fellowship code “LCF/BQ/DR19/11740016”). Dr Gao was supported by the Peking University Start‐up Grant (BMU2021YJ044). During the preparation of this article, Dr Colicino was supported by R01 ES032242 and P30 ES023515. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Indian Health Service. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or decision to submit the manuscript for publication.
Disclosures
None.
Supporting information
Data S1
Table S1–S4
Figures S1–S7
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.122.026934
For Sources of Funding and Disclosures, see page 9.
REFERENCES
- 1. Bakulski KM, Rozek LS, Dolinoy DC, Paulson HL, Hu H. Alzheimer's disease and environmental exposure to lead: the epidemiologic evidence and potential role of epigenetics. Curr Alzheimer Res. 2012;9:563–573. doi: 10.2174/156720512800617991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weisskopf MG, Proctor SP, Wright RO, Schwartz J, Spiro A III, Sparrow D, Nie H, Hu H. Cumulative lead exposure and cognitive performance among elderly men. Epidemiology (Cambridge, Mass). 2007;18:59–66. [DOI] [PubMed] [Google Scholar]
- 3. Schober SE, Mirel LB, Graubard BI, Brody DJ, Flegal KM. Blood lead levels and death from all causes, cardiovascular disease, and cancer: results from the NHANES III mortality study. Environ Health Perspect. 2006;114:1538–1541. doi: 10.1289/ehp.9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Toxicological Profile for Lead . Agency for Toxic Substances and Disease Registry. 2020. Available at: https://www.atsdr.cdc.gov/toxprofiles/tp13.pdf [PubMed]
- 5. Landrigan PJ, Bellinger D. It's time to end Lead poisoning in the United States. JAMA Pediatr. 2021;175:1216–1217. doi: 10.1001/jamapediatrics.2021.3525 [DOI] [PubMed] [Google Scholar]
- 6. Lead . United States Department of Labor ‐ Occupational Safety and Health Administration. Available at: https://www.osha.gov/lead
- 7. Nigra AE, Navas‐Acien A. Racial inequalities in drinking water lead exposure: a wake‐up call to protect patients with end stage kidney disease. J Am Soc Nephrol. 2021;32:2419–2421. doi: 10.1681/ASN.2021060793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanphear BP, Weitzman M, Eberly S. Racial differences in urban children's environmental exposures to lead. Am J Public Health. 1996;86:1460–1463. doi: 10.2105/ajph.86.10.1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moody HA, Darden JT, Pigozzi BW. The relationship of neighborhood socioeconomic differences and racial residential segregation to childhood blood Lead levels in metropolitan Detroit. J Urban Health. 2016;93:820–839. doi: 10.1007/s11524-016-0071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakai T. Biomarkers of lead exposure. Ind Health. 2000;38:127–142. doi: 10.2486/indhealth.38.127 [DOI] [PubMed] [Google Scholar]
- 11. Somervaille LJ, Chettle DR, Scott MC, Tennant DR, McKiernan MJ, Skilbeck A, Trethowan WN. In vivo tibia lead measurements as an index of cumulative exposure in occupationally exposed subjects. Br J Ind Med. 1988;45:174–181. doi: 10.1136/oem.45.3.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colicino E, Just A, Kioumourtzoglou MA, Vokonas P, Cardenas A, Sparrow D, Weisskopf M, Nie LH, Hu H, Schwartz JD, et al. Blood DNA methylation biomarkers of cumulative lead exposure in adults. J Expo Sci Environ Epidemiol. 2021;31:108–116. doi: 10.1038/s41370-019-0183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paul KC, Horvath S, Del Rosario I, Bronstein JM, Ritz B. DNA methylation biomarker for cumulative lead exposure is associated with Parkinson's disease. Clin Epigenetics. 2021;13:59. doi: 10.1186/s13148-021-01051-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The strong heart study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757 [DOI] [PubMed] [Google Scholar]
- 16. Harris S, Harper BL. Lifestyles, diets, and native American exposure factors related to possible Lead exposures and toxicity. Environ Res. 2001;86:140–148. doi: 10.1006/enrs.2001.4250 [DOI] [PubMed] [Google Scholar]
- 17. Malcoe LH, Lynch RA, Keger MC, Skaggs VJ. Lead sources, behaviors, and socioeconomic factors in relation to blood lead of native american and white children: a community‐based assessment of a former mining area. Environ Health Perspect. 2002;110(Suppl 2):221–231. doi: 10.1289/ehp.02110s2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller CJ, Noonan CJ, MacLehose RF, Stoner JA, Lee ET, Best LG, Calhoun D, Jolly SE, Devereux RB, Howard BV. Trends in cardiovascular disease morbidity and mortality in American Indians over 25 years: the strong heart study. J Am Heart Assoc. 2019;8:e012289. doi: 10.1161/jaha.119.012289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tellez‐Plaza M, Guallar E, Howard BV, Umans JG, Francesconi KA, Goessler W, Silbergeld EK, Devereux RB, Navas‐Acien A. Cadmium exposure and incident cardiovascular disease. Epidemiology (Cambridge, Mass). 2013;24:421–429. doi: 10.1097/EDE.0b013e31828b0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tellez‐Plaza M, Guallar E, Fabsitz RR, Howard BV, Umans JG, Francesconi KA, Goessler W, Devereux RB, Navas‐Acien A. Cadmium exposure and incident peripheral arterial disease. J Circulation. 2013;6:626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franceschini N, Fry RC, Balakrishnan P, Navas‐Acien A, Oliver‐Williams C, Howard AG, Cole SA, Haack K, Lange EM, Howard BV, et al. Cadmium body burden and increased blood pressure in middle‐aged American Indians: the strong heart study. J Hum Hypertens. 2017;31:225–230. doi: 10.1038/jhh.2016.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oliver‐Williams C, Howard AG, Navas‐Acien A, Howard BV, Tellez‐Plaza M, Franceschini N. Cadmium body burden, hypertension, and changes in blood pressure over time: results from a prospective cohort study in American Indians. J Am Soc Hypertens. 2018;12:426–437.e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lamas GA, Ujueta F, Navas‐Acien A. Lead and cadmium as cardiovascular risk factors: the burden of proof has been met. J Am Heart Assoc. 2021;10:e018692. doi: 10.1161/jaha.120.018692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajagopalan S, Landrigan PJ. Pollution and the heart. N Engl J Med. 2021;385:1881–1892. doi: 10.1056/NEJMra2030281 [DOI] [PubMed] [Google Scholar]
- 25. Jain NB, Potula V, Schwartz J, Vokonas PS, Sparrow D, Wright RO, Nie H, Hu H. Lead levels and ischemic heart disease in a prospective study of middle‐aged and elderly men: the VA normative aging study. Environ Health Perspect. 2007;115:871–875. doi: 10.1289/ehp.9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welty TK, Lee ET, Yeh J, Cowan LD, Go O, Fabsitz RR, Le NA, Oopik AJ, Robbins DC, Howard BV. Cardiovascular disease risk factors among American Indians. The strong heart study. Am J Epidemiol. 1995;142:269–287. doi: 10.1093/oxfordjournals.aje.a117633 [DOI] [PubMed] [Google Scholar]
- 27. Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Team RC . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 29. Aryee MJ, Jaffe AE, Corrada‐Bravo H, Ladd‐Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England). 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics (Oxford, England). 2017;33:558–560. doi: 10.1093/bioinformatics/btw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Triche TJ Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low‐level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina methylation BeadChip. Bioinformatics (Oxford, England). 2016;32:2659–2663. doi: 10.1093/bioinformatics/btw285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off‐target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. 2016;9:22–24. doi: 10.1016/j.gdata.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD, Zhang Y, Torres LC. sva: surrogate variable analysis. R Package version. 2019;3:882–883. [Google Scholar]
- 36. Illumina. Infinium MethylationEPIC Product Files. 2017. Available at: https://emea.support.illumina.com/downloads/infinium‐methylationepic‐v1‐0‐product‐files.html
- 37. Bell B, Rose CL, Damon AJA, Development H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Devel. 1972;3:5–17. [Google Scholar]
- 38. Hu H, Rabinowitz M, Smith D. Bone lead as a biological marker in epidemiologic studies of chronic toxicity: conceptual paradigms. Environ Health Perspect. 1998;106:1–8. doi: 10.1289/ehp.981061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aro AC, Todd AC, Amarasiriwardena C, Hu H. Improvements in the calibration of 109Cd K x‐ray fluorescence systems for measuring bone lead in vivo. Phys Med Biol. 1994;39:2263–2271. doi: 10.1088/0031-9155/39/12/009 [DOI] [PubMed] [Google Scholar]
- 40. Fan J. Sure independence screening for ultrahigh dimensional feature space. J Roy Stat Soc: Series B. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown SD. Zeeman effect‐based background correction in atomic absorption spectrometry. Anal Chem. 1977;49:1269A–1281A. [Google Scholar]
- 42. Hu H, Aro A, Payton M, Korrick S, Sparrow D, Weiss ST, Rotnitzky A. The relationship of bone and blood Lead to hypertension: the normative aging study. JAMA. 1996;275:1171–1176. doi: 10.1001/jama.1996.03530390037031 [DOI] [PubMed] [Google Scholar]
- 43. 2017 SHS . Strong Heart Study Phase VI Manual of Operations. 2017.
- 44. 2001 SHS . Strong Heart Study Operations Manual Phase IV. Volume II: Morbidity and mortality surveillance procedures. 2001.
- 45. Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The Strong Heart Study A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757 [DOI] [PubMed] [Google Scholar]
- 46. Li S, Wang J, Zhang B, Liu Y, Lu T, Shi Y, Shan G, Dong L. Urinary lead concentration is an independent predictor of cancer mortality in the US general population. Front Oncol. 2018;8:242. doi: 10.3389/fonc.2018.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scheer J, Findenig S, Goessler W, Francesconi KA, Howard B, Umans JG, Pollak J, Tellez‐Plaza M, Silbergeld EK, Guallar E, et al. Arsenic species and selected metals in human urine: validation of HPLC/ICPMS and ICPMS procedures for a long‐term population‐based epidemiological study. Anal Methods. 2012;4:406–413. doi: 10.1039/c2ay05638k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, Klengel T, Mehta D, Binder EB, Epstein MP, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–241. doi: 10.1002/gepi.21789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moon KA, Guallar E, Umans JG, Devereux RB, Best LG, Francesconi KA, Goessler W, Pollak J, Silbergeld EK, Howard BV, et al. Association between exposure to low to moderate arsenic levels and incident cardiovascular disease. A prospective cohort study. Ann Intern Med. 2013;159:649–659. doi: 10.7326/0003-4819-159-10-201311190-00719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Domingo‐Relloso A, Huan T, Haack K, Riffo‐Campos AL, Levy D, Fallin MD, Terry MB, Zhang Y, Rhoades DA, Herreros‐Martinez M, et al. DNA methylation and cancer incidence: lymphatic‐hematopoietic versus solid cancers in the strong heart study. Clin Epigenetics. 2021;13:43. doi: 10.1186/s13148-021-01030-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Domingo‐Relloso A, Riffo‐Campos AL, Haack K, Rentero‐Garrido P, Ladd‐Acosta C, Fallin DM, Tang WY, Herreros‐Martinez M, Gonzalez JR, Bozack AK. Cadmium, smoking, and human blood DNA methylation profiles in adults from the strong heart study. Environ Health Perspect. 2020;128:067005. doi: 10.1289/EHP6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bozack AK, Domingo‐Relloso A, Haack K, Gamble MV, Tellez‐Plaza M, Umans JG, Best LG, Yracheta J, Gribble MO, Cardenas A. Locus‐specific differential DNA methylation and urinary arsenic: an epigenome‐wide association study in blood among adults with low‐to‐moderate arsenic exposure. Environ Health Perspect. 2020;128:067015. doi: 10.1289/EHP6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Navas‐Acien A, Domingo‐Relloso A, Subedi P, Riffo‐Campos AL, Xia R, Gomez L, Haack K, Goldsmith J, Howard BV, Best LG, et al. Blood DNA methylation and incident coronary heart disease: evidence from the strong heart study. JAMA Cardiol. 2021;6:1237–1246. doi: 10.1001/jamacardio.2021.2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1
Table S1–S4
Figures S1–S7


