Abstract
Rationale
Healthcare-associated transmission of nontuberculous mycobacteria (NTM) among people with cystic fibrosis (pwCF) has been investigated at CF centers worldwide, with conflicting conclusions. We investigated transmission at the Colorado Adult CF Program.
Objectives
To systematically investigate healthcare-associated transmission and/or acquisition of NTM to determine similarity among respiratory and environmental isolates, and to compare home residence watershed mapping among pwCF having genetically similar NTM isolates.
Methods
Whole-genome sequencing of NTM isolates from 80 pwCF was conducted to identify genetically similar isolate clusters (⩽30 SNP differences). Epidemiology, comparison of respiratory and environmental isolates, and home residence watershed mapping were analyzed.
Measurements and Main Results
Whole-genome sequencing analysis revealed 11 clusters of NTM [6 Mycobacterium abscessus subspecies (ssp.) abscessus, 1 M. abscessus ssp. massiliense, 2 Mycobacterium avium, and 2 Mycobacterium intracellulare] among pwCF. Epidemiologic investigation demonstrated opportunities for healthcare-associated transmission in two M. abscessus and two M. avium clusters. Respiratory and healthcare environmental isolate comparisons revealed no genetic similarity. Individuals comprising one M. abscessus cluster, with no plausible healthcare-associated transmission, resided in the same watershed.
Conclusions
This study suggests healthcare-associated transmission of M. abscessus is rare and includes a report of potential healthcare-associated transmission of M. avium among pwCF. One M. abscessus cluster possibly had common acquisition arising from residing in the same watershed. The presence of genetically similar isolates is insufficient to demonstrate healthcare-associated NTM transmission. Standardizing epidemiologic investigation, combined with environmental sampling and watershed analysis, will improve understanding of the frequency and nature of healthcare-associated NTM transmission among pwCF.
Keywords: cystic fibrosis, nontuberculous mycobacteria, whole-genome sequencing, transmission, epidemiology
At a Glance Commentary
Scientific Knowledge on the Subject
Healthcare-associated transmission of nontuberculous mycobacteria (NTM) among people with cystic fibrosis (CF) has been investigated at CF centers worldwide, with conflicting conclusions. The presence of genetically similar isolates is insufficient by itself to demonstrate healthcare-associated NTM transmission.
What This Study Adds to the Field
The HALT NTM study (Healthcare-associated Links in Transmission of Nontuberculous Mycobacteria in Cystic Fibrosis) provides a systematic, evidence-based approach to investigating potential episodes of healthcare-associated NTM transmission within a single CF care center. The HALT NTM study provides a framework to standardize epidemiologic investigation, coupled with healthcare environmental sampling and home of residence watershed analysis, and thereby improves understanding of the frequency and nature of healthcare-associated NTM transmission among people with CF as well as the potential to identify where acquisition might be at a higher probability.
Pulmonary disease caused by nontuberculous mycobacteria (NTM) is one of the most challenging infections among people with cystic fibrosis (pwCF) (1, 2). Healthcare-associated transmission of Mycobacterium abscessus among pwCF remains an ongoing concern (3–6). One study reported widespread global transmission of dominant clones of M. abscessus, potentially via person-to-person transmission of fomites and aerosols (7). Two multicenter prevalence studies of NTM found no nosocomial transmission among pwCF (8, 9). Other studies concluded no transmission of M. abscessus occurred within an individual center (10, 11). Improved surveillance combined with a standardized, systematic, evidence-based approach to epidemiologic investigation of potential episodes of healthcare-associated transmission is recognized as an unmet need to inform evidence-based infection prevention and control (IPC) measures for pwCF (12).
The sources, modes of transmission, and exposure risks for NTM among pwCF are poorly understood, but it is reported that NTM can be environmentally acquired from exposures to NTM-laden bioaerosols produced by soil and water (13–15). Importantly, NTM colonize municipal water systems and have been identified in healthcare facilities (16–18). NTM are found in water and biofilms on water supply pipes, both implicated in healthcare-associated outbreaks and pseudo-outbreaks (19). In addition to water sources, clinically relevant NTM have been cultured from environmental dust in homes of patients with NTM (20). Risk of NTM has also been associated with watershed-specific water-quality constituents (21).
The Colorado Research Development Program served as a biorepository for CF NTM isolates to study NTM genetics nationwide (22, 23). The Colorado Research Development Program has the largest collection of whole-genome sequencing (WGS) data from respiratory CF NTM isolates in the United States. WGS analysis of core genome similarities identified clusters of CF NTM isolates among pwCF who were cared for at the same center among multiple U.S. care centers (22, 23), heightening concern for potential healthcare-associated NTM acquisition originating from patient-to-patient transmission and/or common environmental source(s).
The HALTNTM study (Healthcare-associated Links in Transmission of Nontuberculous Mycobacteria in Cystic Fibrosis; www.clinicaltrials.gov identifier NCT04024423) provides a systematic, evidence-based approach to investigating potential episodes of healthcare-associated NTM transmission within a single care center (24). Here, we report a formal investigation of the Colorado Adult CF Program of 11 clusters of NTM among pwCF who received care at our center between January 1, 2012, and December 31, 2018. We identified six clusters of M. abscessus subspecies (ssp.) abscessus, one cluster of M. abscessus ssp. massiliense, two clusters of Mycobacterium avium, and two clusters of Mycobacterium intracellulare. Some of the results of these studies have been previously reported in the form of abstracts (25, 26).
Methods
Overview
This is a retrospective investigation of healthcare-associated transmission and/or acquisition of NTM among pwCF having genetically similar NTM isolates. A stepwise process was used (Figure 1A) to perform 1) WGS analysis of NTM isolates to determine relatedness at the core genome level; 2) epidemiologic investigation of pwCF, with genetically similar NTM isolates, in the healthcare system via electronic health record (EHR) review; 3) accessory genome analysis to determine relatedness at the pan genome level; 4) healthcare environmental sampling for NTM by microbiological culture and WGS comparison to respiratory isolates from pwCF; and 5) watershed mapping of home residences to assess the likelihood of shared acquisition from the home environment.
Figure 1.
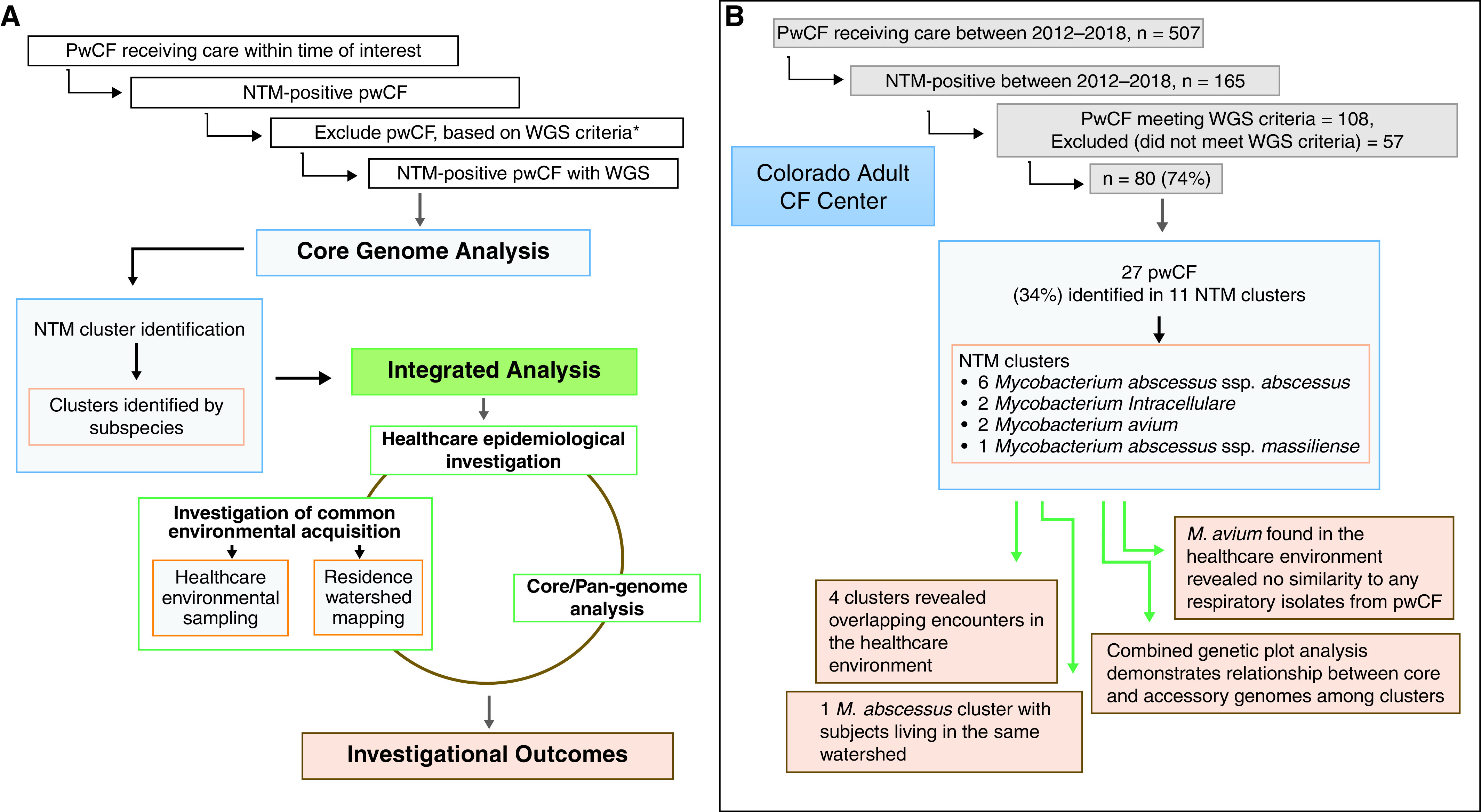
Healthcare-associated transmission of nontuberculous mycobacteria (NTM) investigation. The flow diagram (A) overview and (B) Colorado Adult CF Center investigation results. *WGS criteria defined as more than three positive NTM cultures in 7 years. CF = cystic fibrosis; pwCF = people with cystic fibrosis; WGS = whole-genome sequencing.
Patients and Respiratory Sample Collection
We collected respiratory isolates as part of routine CF management from 507 pwCF receiving care at the Colorado Adult CF Program from 2012 to 2018. A threshold of more than three positive NTM culture results during this time frame was imposed to select for participants with both relatively high healthcare use and evidence of persistent NTM burden. After excluding patients who had three or fewer positive NTM cultures in the 2012–2018 time frame, WGS was performed on NTM-positive respiratory isolates from 80 pwCF and environmental NTM isolates collected from the clinic and hospital healthcare settings.
WGS and Phylogenetic Analysis
DNA extraction, WGS, and SNP analysis were performed as previously described (22, 27). NTM respiratory isolates were considered highly related and falling into a cluster using a SNP threshold of 30 or less for M. abscessus ssp. and 20 or less for M. avium complex species. SNP thresholds were based on the maximum number of SNPs identified among within-patient longitudinal isolate pairs and corresponded to previously identified thresholds for potential transmission of NTM (4, 6–8, 11, 22, 28, 29). Our threshold estimations are based upon the isolate cohort included in the WGS analyses. We observed no more than 30 SNPs within a single patient’s longitudinal M. abscessus isolates, and we observed a maximum of 20 SNPs within a single patient’s longitudinal M. avium isolates. Phylogenetic trees were created using the first isolate sequenced from each person with CF, and cluster network analysis was performed as described (22). Combined genetic plots were created using GraphPad Prism 8.0. WGS data are available from the National Center for Biotechnology Information under BioProject PRJNA319839.
Pan-Genome Analysis
NTM isolate genomes were assembled with Unicycler (30), and pan-genome analysis was performed using Roary (31). Pairwise accessory genome comparisons were performed as previously described (32).
Epidemiologic Investigation
A Centers for Disease Control and Prevention standardized, validated healthcare-associated infection outbreak investigation form and toolkit (33, 34) were adapted to analyze date and location data to identify possible transmission events in the healthcare setting of subjects determined to fall within a cluster based on WGS and phylogenetic analysis. Demographic, patient location, and microbiological data were collected from the EHR, then assembled and managed using REDCap electronic data capture tools hosted at National Jewish Health.
Environmental Sample Collection
Environmental samples from the clinic and hospital were collected between September 2019 and March 2021. NTM were recovered from environmental samples using standard microbiological approaches as described elsewhere (35). Coffee machine water was filtered through 0.2-μm polyethersulfone membrane filters and plated on Middlebrook 7H10 agar plates with oleic acid/glycerol enrichment. Genomic DNA was extracted from bacterial pellets (27), and isolates were identified to the species or subspecies level through amplification and sequencing of the rpoB (RNA polymerase β-subunit) gene (35–37).
Watershed Data Collection
Patient addresses were extracted on the basis of the year of the first positive NTM culture. Addresses were converted to latitude and longitude coordinates using the R package ggmap (38). Mapping visualization was performed using ArcGIS 10.2 (Esri). The U.S. Geological Survey Hydrologic Unit Code level 8 watersheds (39) were used for watershed mapping. An analysis of the probability of three subjects falling within the same genetic cluster and watershed was performed. The rate of shared watersheds among independent pairs of subjects within clusters was determined to calculate probabilities of interest. For a cluster of three subjects, there are three possible pairs but two independent pairs. For example, the pairing of subjects a and b and of a and c creates two independent pairs; watershed commonality status for the pair b and c can then be determined from the first two.
Results
Participant Characteristics and NTM Cluster Characteristics
The baseline characteristics of the participants and clusters are shown in Table 1.
Table 1.
Subject Characteristics within Nontuberculous Mycobacteria Clusters
| General Demographics | |
|---|---|
| Total patients | 27 |
| Females, n (%) | 18 (66.6) |
| Average age, yr | 38.48 |
| Race | |
| White | 25 |
| Unknown/decline to answer | 2 |
| Ethnicity, n (%) | |
| Non-Hispanic | 24 (88.8%) |
| Hispanic | 1 (3.7%) |
| Unknown/decline to answer | 2 (7.4%) |
| Clinical descriptives | |
| Mutation, n (%) | |
| F508del:F508del | 11 (40.7%) |
| F508del:other | 13 (48.1%) |
| Other than F508del | 3 (11.2%) |
| Average of CF centers visited by subjects, n (range) | 1.4 (1–3) |
| Clinic visits per subject, n (range)* | 9.4 (3–18) |
| Admissions, n (range)* | 2.9 (0–18) |
| Subjects on NTM treatment, n (%) | 19 (70.3) |
| Subjects with ATS NTM criteria, n (%) | 15 (55.5) |
| Patients with known CF contact, n (%) | 2 (7.4) |
| Deceased by the end of study, n (%) | 4 (14.8) |
| Females, n (%) | 2 (50) |
| Average age, n (range) | 39.5 (25–79) |
| Cause of death | |
| Respiratory/cardiorespiratory | 2 (50) |
| Liver disease/liver failure | 1 (25) |
| Unknown | 1 (25) |
| Subjects with Mycobacterium abscessus | 3 (75) |
| Subjects with M. avium | 1 (25) |
| Number of clusters, n (%) | 11 |
| M. abscessus clusters | 6 (54.5) |
| Clusters in the dominant circulating clone | 3† |
| M. abscessus ssp. massiliense clusters | 1 (9.09) |
| M. avium | 2 (18.2) |
| M. intracellulare | 2 (18.2) |
| Number of patients in a cluster, average (range) | 2.45 (2–4) |
| Total overlap clinics | |
| Clinic | 6 |
| Admissions | 6 |
| Length of admission overlap in d, average (range) | 13.16 (5–17) |
Definition of abbreviations: ATS = American Thoracic Society; CF = cystic fibrosis; NTM = nontuberculous mycobacteria.
During the extraction range period, per cluster.
Fifty percent of the M. abscessus clusters.
Genomic Analysis
NTM were cultured at least once in 165 patients cared for at the Colorado Adult CF Program between 2012 and 2018. pwCF who had three or fewer NTM-positive cultures were excluded from analysis (n = 57) to select for individuals with relatively high healthcare use and evidence of a persistent NTM burden. Of the remaining 108 NTM-positive pwCF, 80 (74%) isolates, collected in the clinical setting and available for research investigation, underwent WGS. Core genome analysis revealed a total of 27 (34%) subjects identified among 11 clusters of NTM among pwCF, including 6 clusters of M. abscessus (A, E, F, G, J, and K), 1 cluster of M. massiliense (B), 2 clusters of M. avium (D and I), and 2 clusters of M. intracellulare (C and H) (Figure 2). M. abscessus clusters A, E, G, and J are members of the previously described dominant circulating clone (DCC1) (7, 22). Cluster network analysis with core genome SNP differences is shown (Figure 3). In all clusters, subject 1 was defined as the individual with the longest period with NTM-positive culture, subject 2 was defined as the individual with the second longest period with NTM-positive culture, and so forth.
Figure 2.
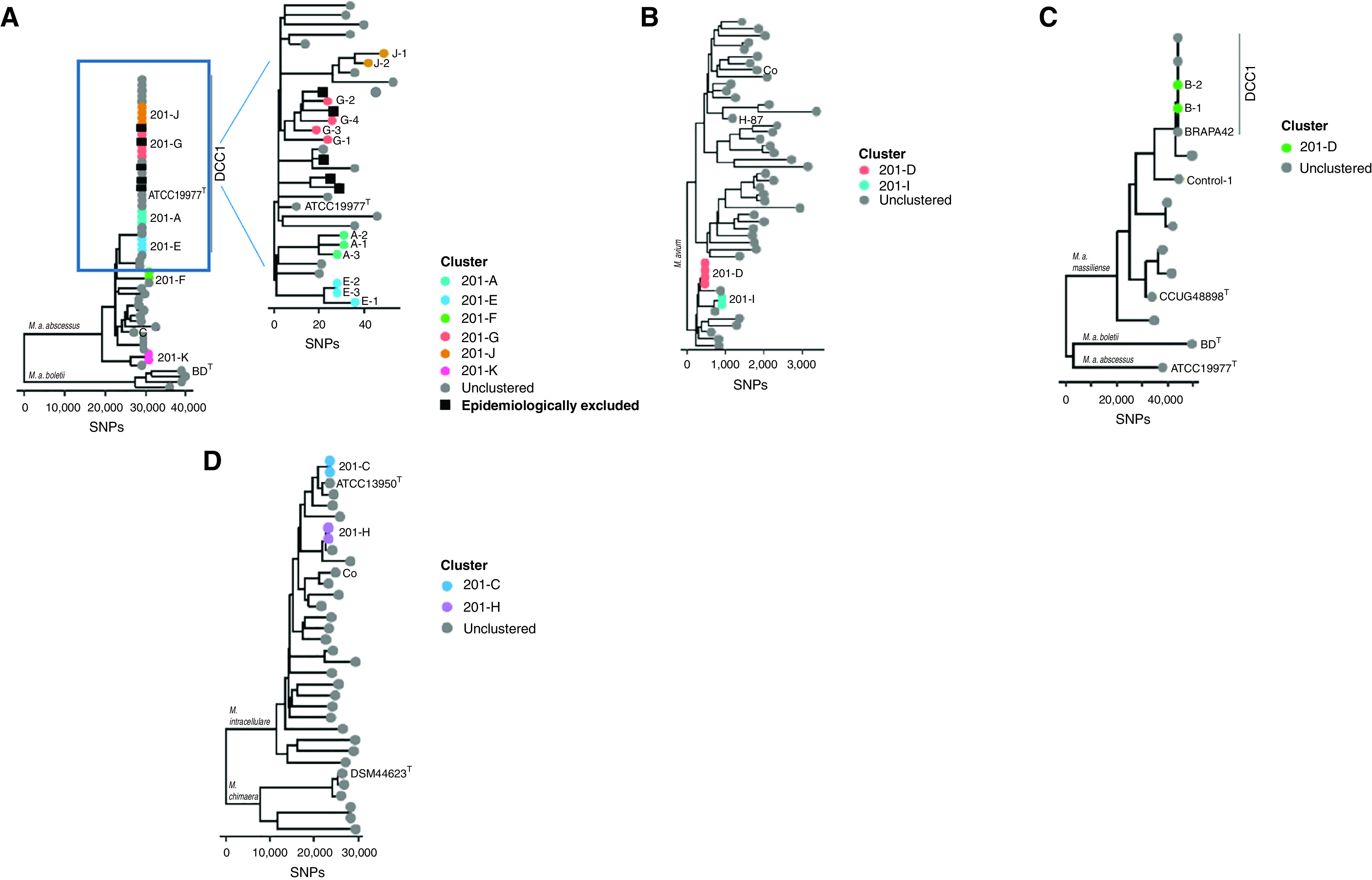
Phylogenetic trees comprising all locally identified clusters among people with cystic fibrosis at the Colorado Adult CF Program are shown. Reference isolates, including type strains and previously published isolate genomes, are shown as gray circles with labels. Epidemiologically excluded subjects are identified by black squares. The 11 sets of clustered nontuberculous mycobacteria isolates are represented as colored circles, with each letter representing a cluster (A–K). The number in the cluster identifies the subject order based on the first positive culture. The six Mycobacterium abscessus subspecies (ssp.) abscessus clusters are shown with four clusters in the dominant circulating clone 1 (DCC1), (A) enlarged for detail. (B) Two Mycobacterium avium clusters, (C) one Mycobacterium abscessus ssp. massiliense cluster, and (D) two Mycobacterium intracellulare clusters are shown. CF = cystic fibrosis.
Figure 3.
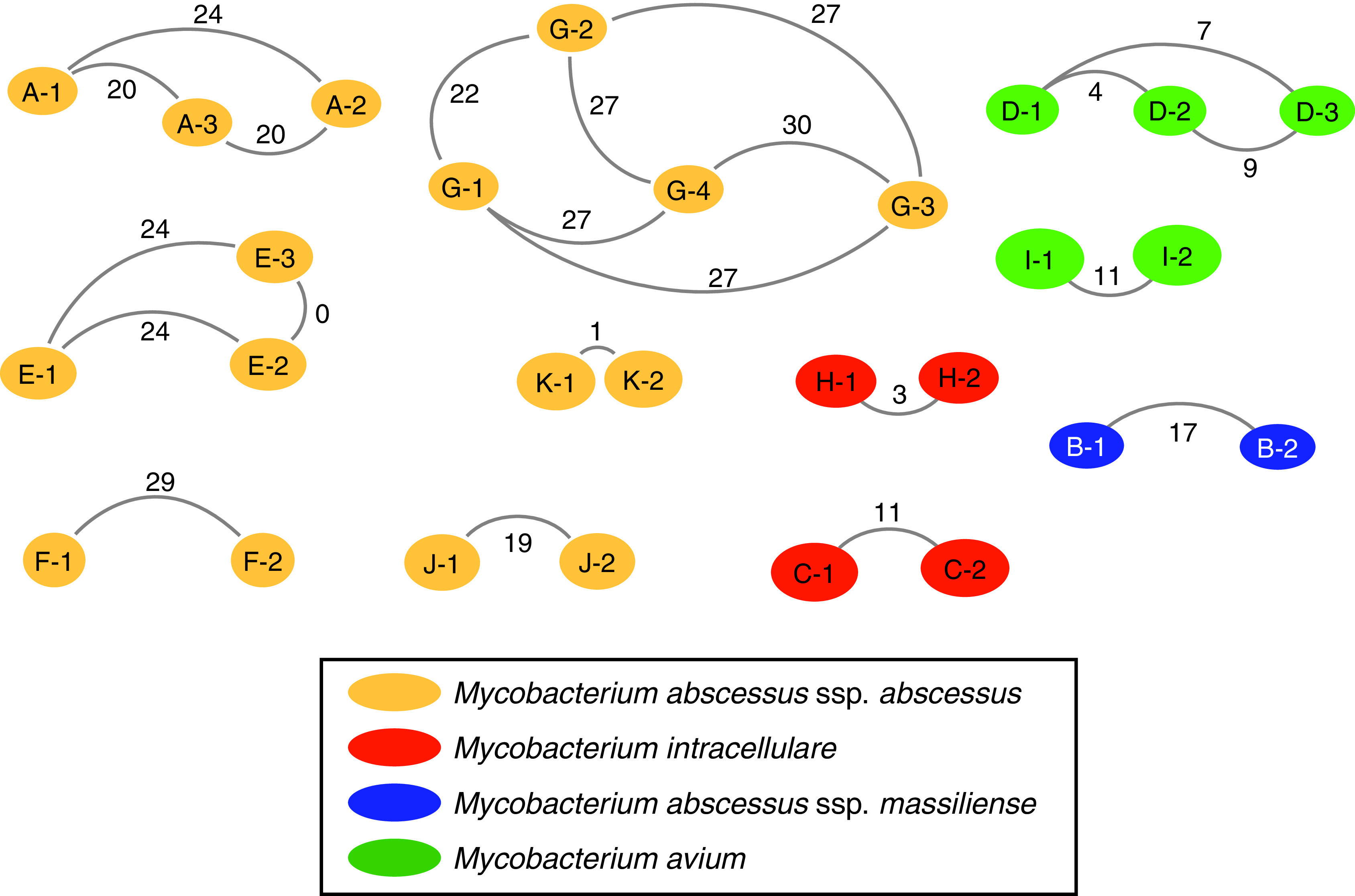
Cluster network analysis. Nontuberculous mycobacteria species are identified by color, and the letter identifies the cluster: Mycobacterium abscessus subspecies (ssp.) abscessus shown in yellow, Mycobacterium intracellulare shown in orange, Mycobacterium abscessus ssp. massiliense shown in blue, and Mycobacterium avium shown in green. Nodes represent each subject within a cluster. The number in the node identifies the subject order based on first positive culture. Core genome SNP distance is shown with the line connecting nodes.
It should be noted that not all subjects with the M. abscessus DCC1 were included in clusters. Overall, 31 subjects had the M. abscessus DCC1 clone; however, SNP distances among all DCC1 isolates ranged from 0 to 99 SNPs (see Table E1 in the online supplement). After epidemiological investigation, six subjects with DCC1 were excluded on the basis of previously described exclusion criteria. Among the remaining 25 subjects with M. abscessus DCC1 isolates, 12 (48%) had 30 or fewer SNP differences from at least 1 other subject (e.g., clusters A, E, G, and J).
Epidemiologic Investigation
Healthcare-associated opportunities for NTM transmission between patients in identified clusters were investigated by plotting outpatient visits and inpatient hospitalizations along with individual NTM culture data. Timeline overlap analysis (Figure 4) of M. abscessus clusters A and J as well as M. avium clusters D and I demonstrated episodes of healthcare-associated opportunities for transmission in a total of 4 (36%) of 11 clusters. Two (33%) of the M. abscessus clusters (clusters A and J) had identified healthcare overlaps. In M. abscessus cluster A, two of three subjects (A1 and A2) shared four hospitalization overlaps totaling 59 days, including one hospitalization in which the first subject had a positive NTM smear result within 1 year of the second subject becoming NTM-positive. The third subject (A3) had no identifiable healthcare overlaps. In M. abscessus cluster J, both subjects (J1 and J2) had one clinic overlap within 24 hours and one 15-day hospitalization overlap during which subject 2 became NTM positive. M. abscessus cluster E revealed one clinic overlap in a 24-hour period 4 years before NTM conversion of the second subject (not shown). Both the M. avium clusters D and I demonstrated healthcare overlaps. M. avium cluster D revealed one clinic overlap within 12 hours between D1 and D2 (within 2 yr of D2 becoming NTM positive) as well as between D2 and D3 (both NTM positive during same clinic day overlap). Both subjects in M. avium cluster I had one clinic overlap within 12 hours, followed 8 days later by a 5-day hospital overlap during which the first subject was NTM smear positive and the second subject developed a first-time NTM-positive culture.
Figure 4.
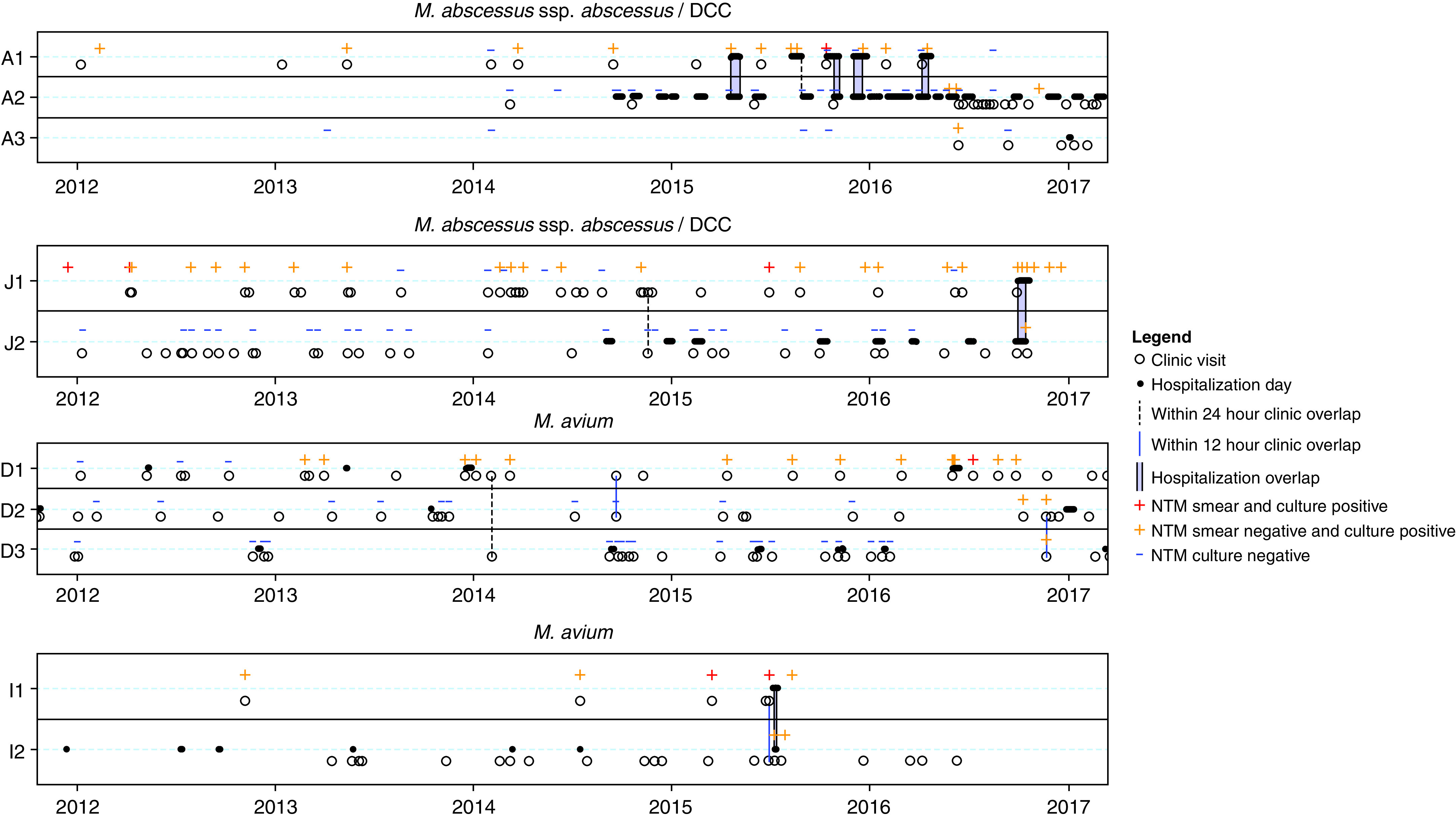
Timeline overlap analysis of people with cystic fibrosis identified in nontuberculous mycobacteria (NTM) clusters demonstrating clinic visits (open circles), hospitalization days (closed circles), within–24-hour clinic overlap (vertical dashed lines), within–12-hour clinic overlap (vertical solid lines), and hospitalization overlap (vertical purple bars). Subject NTM status is represented as smear and culture positive (red plus), smear negative and culture positive (yellow plus), and smear and culture negative (blue dash). The letter identifies the cluster, and the number identifies the subject order based on first positive culture date. Clusters that did not have overlaps or had overlaps occurring >2 years before the second subject becoming NTM positive are not shown. DCC = dominant circulating clone. M. = Mycobacterium.
Three (50%) M. abscessus clusters (F, K, and G), one (100%) M. massiliense cluster (B), and two (100%) M. intracellulare clusters (C and H) revealed no healthcare-associated opportunities for transmission (not shown). There were no known social links, defined as siblings, spouses, or housemates with CF, between subjects outside the healthcare system, as determined by EHR review.
Healthcare Environmental Sampling
Healthcare water biofilms and dust were sampled (n = 161), including 44 clinic water biofilm sites (sink faucet, water fountain, water bottle fillers, water bath, and buffer container), 40 clinic dust samples (ceiling vents), 74 hospital biofilm sites (sink faucet, showerhead face, showerhead screen, shower hose, water dispensers, ice dispensers, coffee machines, and coffee machine water), and 3 hospital dust samples (ceiling vents). Of 161 healthcare environmental samples, 11 (6.8%) were positive for NTM. On the basis of rpoB sequencing, Mycobacterium lentiflavum (one clinic sink), Mycobacterium mucogenicum (one clinic water fountain), Mycobacterium setense (one clinic filtered water station), Mycobacterium gordonae (two clinic sinks, one hospital sink, and one hospital shower hose), Mycobacterium chelonae (two hospital coffee machines), Mycobacterium avium (two hospital coffee machines and a hospital coffee machine water supply line), and Mycobacterium peregrinum (one clinic ceiling vent) were recovered from the 11 NTM-positive healthcare environmental samples. Two of the healthcare environmental samples cultured both M. avium and M. chelonae, which were isolated from the two common-space coffee machines in the CF hospital ward. M. avium was also isolated from the single-source water supply line to one of the coffee machines, but a 10-ounce water aliquot from the coffee machines failed to culture NTM. Of 43 healthcare dust samples collected, one (2.3%) clinic ceiling vent was positive for M. peregrinum. The diversity of environmental healthcare NTM-positive samples is shown in Figure 5A. Clinically relevant environmental healthcare samples were analyzed further by WGS and revealed no genetic similarity between the environmental isolates and any CF NTM respiratory isolates at our center (data not shown).
Figure 5.
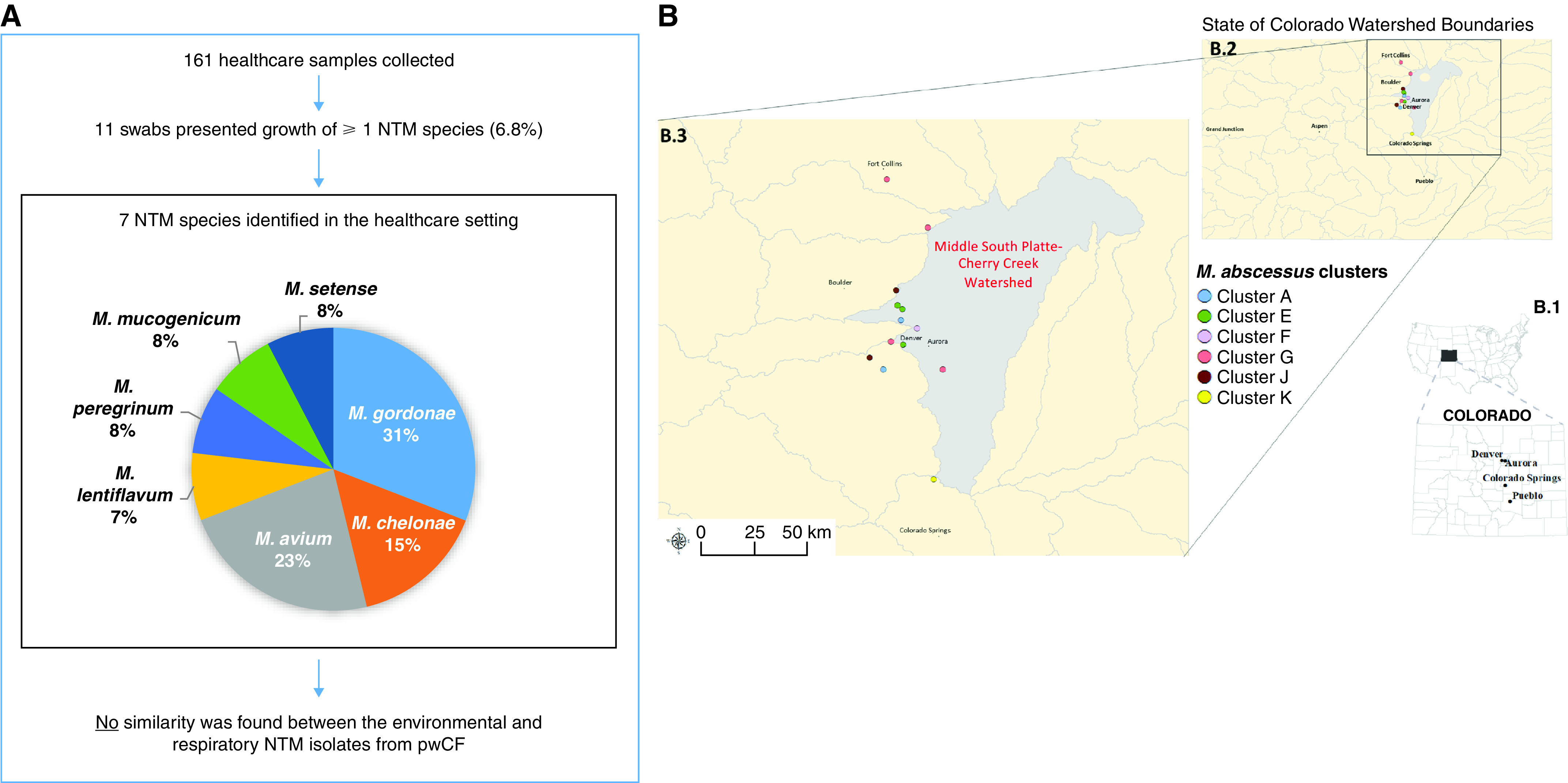
(A) Healthcare environmental sample collection of the clinic and hospital settings demonstrating nontuberculous mycobacteria (NTM) diversity and comparison with respiratory isolates from people with cystic fibrosis (pwCF) is shown. (B) Within the state of Colorado, (B.1) cities and county lines are shown. (B.2) Watershed boundaries are shown, and (B.3) home of residence mapping of pwCF in Mycobacterium abscessus subspecies (ssp.) abscessus clusters demonstrates M. abscessus cluster E shared the same home residence watershed in the absence of any evidence of healthcare-associated overlaps. Mycobacterium intracellulare, Mycobacterium abscessus ssp. massiliense, and Mycobacterium avium clusters (not shown) did not demonstrate shared home of residence watershed.
Watershed Analysis
We analyzed addresses on the basis of watershed distribution for 27 NTM-positive patients with CF in the previously described 11 NTM clusters. Five patients had addresses outside of Colorado. No group of pwCF in the M. avium or M. intracellulare clusters resided in the same Colorado watershed, with the exception of M. avium cluster D, in which two of three patients lived within the same watershed (Middle South Platte–Cherry Creek [MSP-CC]). For the M. massiliense cluster, only one patient had a home of residence in Colorado. Among the M. abscessus clusters, three (F, J, and K) had only one patient per cluster with a home of residence in Colorado. All three subjects in M. abscessus cluster E resided within the same watershed: the MSP-CC watershed (Figure 5B). We used a binomial method to determine how likely it would be for this result to occur by chance. Given that 3 of 16 (19%) independent pairs of subjects within the 11 clusters lived in the same watershed, the probability that 2 independent pairs (i.e., a cluster with three subjects) are in the same cluster by chance is 0.19 × 0.19 = 0.04.
Pan-Genome Analysis
Accessory genome analysis was performed to assess pan-genome comparisons of all isolates within NTM clusters. Integrated pan-genome analysis was performed by plotting core genome SNP differences of isolate pairs within clusters versus the percentage of shared accessory genome of the same isolate pairs (Figure 6). M. abscessus clusters E and K together with M. avium cluster D pan-genome analyses fell within cutoff points of ⩽10 SNPs and ⩾95% shared accessory genome, representing the most stringent level of isolate similarity. A summary of investigational outcomes is shown in Figure 1B.
Figure 6.
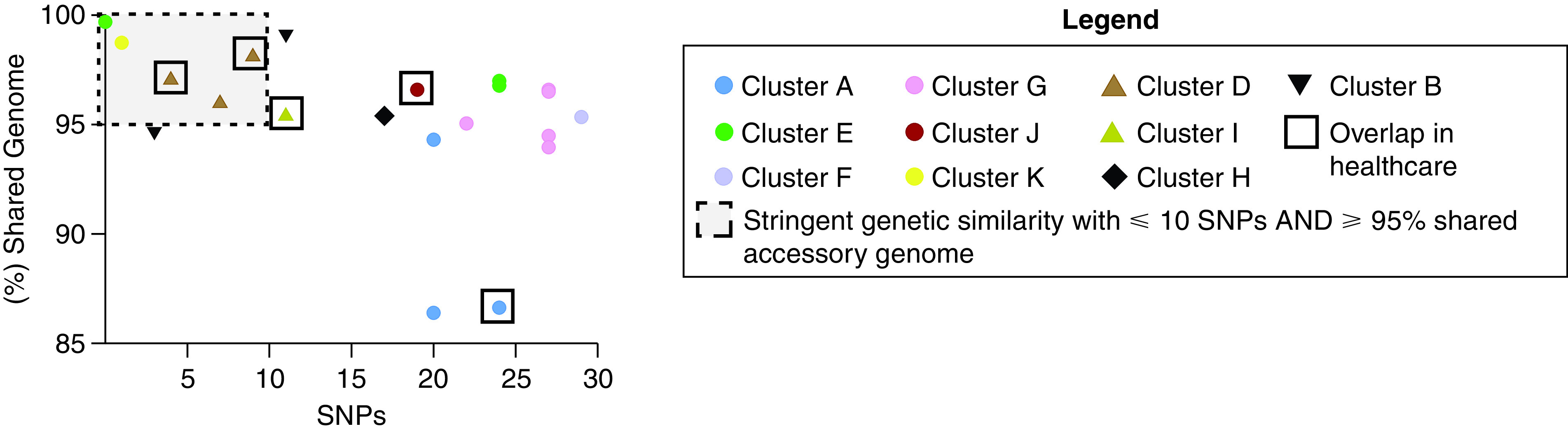
Integrated pan-genome analysis comparing SNP differences between pairs of nontuberculous mycobacteria isolates within clusters (x-axis) versus percentage shared accessory genome (y-axis). Mycobacterium abscessus (ssp.) abscessus clusters are represented by circles, Mycobacterium avium clusters are represented by triangles, the Mycobacterium intracellulare cluster is represented by a diamond, and the Mycobacterium abscessus ssp. massiliense cluster is represented by an upside-down triangle. Clusters A (blue circles) and J (scarlet circles), as well as M. avium clusters D (caramel triangles) and I (chartreuse triangles), demonstrated episodes of healthcare-associated opportunities for transmission. Cutoff points of ⩽10 SNPs and ⩾95% shared accessory genome are noted in the dashed line box, representing the most stringent degree of clustered isolate similarity. M. avium cluster D, with sequential healthcare-associated opportunity for transmission from D1 to D2 and D2 to D3, M. abscessus cluster K with no evidence of healthcare-associated opportunity for transmission, and M. abscessus cluster E, with no evidence of healthcare-associated opportunity for transmission but residing in the same watershed, fall within the stringent genetic similarity cutoffs.
Discussion
We report our investigation of healthcare-associated transmission of NTM among pwCF receiving care at the largest adult CF program in the United States. Between 2012 and 2018, 165 pwCF receiving care at the program had at least one positive NTM culture. Core genome analysis identified 11 clusters of NTM among pwCF, suggesting recent common acquisition or shared transmission. Of the 11 clusters, 4 clusters (2 M. abscessus and 2 M. avium) revealed opportunity for healthcare-associated transmission with overlapping encounters in the healthcare environment. One M. abscessus cluster, without evidence of overlapping encounters, shared home of residence in the same watershed. Respiratory and healthcare environmental isolates revealed no genetic similarity. Pan-genome analysis supported stringent isolate similarity in one M. avium cluster (with healthcare overlap) and one M. abscessus cluster (residing in the same watershed). Using integrated analysis of epidemiologic investigations, healthcare environmental sampling, mapping home of residence watershed, and determination of strength of cluster relatedness with pan-genome analysis, this study suggests healthcare-associated transmission of NTM is uncommon in our center.
Intense focus has been placed on understanding human-transmissible M. abscessus and M. massiliense (40). Findings of healthcare-associated opportunities for transmission of isolates with ⩽30 SNP differences in the core genome in M. abscessus and M. massiliense clusters have previously been attributed to possible transmission events (4, 7, 41). In our investigation, M. massiliense cluster A had no evidence of common environmental acquisition but revealed a total of 59 days of hospital overlap among two subjects, with subject 1 being smear positive during one overlapping period. There was no evidence of healthcare overlap with the third subject in the cluster. Similarly, M. abscessus cluster J had no evidence of common environmental acquisition but shared a single episode of 15 days of hospitalization overlap between both subjects in the setting of one subject consistently having NTM-positive cultures. However, pan-genome analysis revealed that neither of these clusters met the stringent criteria for genetic relatedness of ⩽10 SNP differences and ⩾95% shared accessory genome (Figure 6), suggesting decreased likelihood of healthcare-associated transmission. The remaining M. abscessus clusters that did not have overlaps or had overlaps occurring >2 years before the second subject becoming NTM positive are not biologically plausible sources to support healthcare-associated epidemiological linkage. Notably, members of M. abscessus cluster E, with no evidence of healthcare-associated opportunity for transmission, were found to all reside in the same watershed, and two of the subjects had no SNP differences, and >98% shared an accessory genome, suggesting common acquisition of M. abscessus from the home environment in two of the three patients in the cluster. The finding of highly related NTM isolates among pwCF living in a common watershed may warrant future investigation.
Similarly to other reports (6, 22), we found the majority of our M. abscessus clusters and our only M. massiliense cluster were within previously described dominant circulating clones (7). At our center, 48% of subjects with M. abscessus DCC1 isolates had ⩽30 SNP differences from at least one other subject, suggesting that this SNP threshold is applicable for dominant clones and other M. abscessus genotypes. Moreover, the additional layer of accessory genome analysis provides further discrimination of isolates that are closely related at the core genome level (42).
We describe the possibility of healthcare-associated transmission of M. avium among pwCF, a finding that, to our knowledge, has not been reported previously. Both M. avium clusters had no evidence of common environmental acquisition, but they had shared opportunities for transmission in the healthcare environment. M. avium cluster D shared sequential single-day healthcare-associated opportunity for transmission from D1 to D2 and from D2 to D3 in the setting of a biologically plausible time frame for acquisition, and it met the stringent criteria for relatedness (⩽10 SNP differences and ⩾95% shared accessory genome). M. avium cluster I had a single-day healthcare opportunity, followed by a 5-day hospital overlap among both subjects, with subject 1 being smear positive during the overlapping period and having a SNP difference of 11 with >96% accessory genome relatedness, suggestive of a transmission event.
Environmental sampling revealed 6.8% of samples were NTM-positive in our healthcare setting, with the majority of organisms not being clinically significant. The only clinically relevant environmental NTM were M. chelonae and M. avium. There were no clusters of M. chelonae observed in our center. Notably, none of the environmental M. avium identified in the healthcare setting was genetically related to any of the clinical M. avium respiratory samples occurring either in clusters or in individual pwCF at our center. Since starting inpatient CF care at our hospital in 2015, contact precautions have been required for the care of all pwCF as recommended in the 2013 CF guidelines (43). During the study time frame, pwCF were allowed direct access in a CF inpatient common area to independently prepare coffee from a pod-style single-serve coffee machine, which cultured M. avium from the machine and source water supply. Simulated coffee preparation, without a coffee pod, followed by hot water collection from the coffee machines did not culture NTM. Environmental coffee machine–associated M. avium isolates were genetically distinct from all M. avium respiratory isolates collected from pwCF during the study time frame, suggesting that direct acquisition of M. avium from a commonly used hospital device is a low risk for pwCF. In an effort to strengthen infection control standards, hospitalized patients are no longer allowed direct access to prepare coffee.
It is noteworthy that the geographic region studied revealed relatively low endemic healthcare-setting NTM colonization. Importantly, no NTM were cultured from healthcare showerheads, a source known to have high rates of clinically relevant NTM in other studies (44). However, this finding is limited by the low sampling of healthcare showerheads and requires further investigation. Other studies have linked M. abscessus healthcare-associated outbreaks to hospital tap water (45), but our study did not demonstrate growth of M. abscessus within sites tested in our healthcare system. This result suggests that geographic region may be important when considering modification of CF NTM IPC guidelines.
A previous study used watersheds, an area of land in which all the water that enters it drains into a common water body and can be understood as a construct with defined, natural boundaries and environmental relevance (46), to explore the role of water exposure on NTM infection risk in Colorado (47). Consequently, two individuals living far from one another within the same watershed may have more similar exposures than two individuals living near one another in different watersheds. When examining NTM species, watersheds were used as a proxy for general water exposure, and the MSP-CC watershed had a relative risk of 4.1 compared with the mean risk of all 92 watersheds in Colorado (47). All M. abscessus cluster E subjects resided within the MSP-CC watershed, cultured almost genetically identical NTM isolates, and had no hospital overlap. These observations support the possibility that infections may be acquired environmentally, potentially through common water exposure. In contrast, an M. abscessus outbreak investigation among pwCF in Hawaii demonstrated that no outbreak subjects resided within a shared aquifer (drinking water arising from water-containing porous rock saturated with groundwater) (5). Knowing whether a particular region has higher endemic exposure risk is valuable information for susceptible populations. However, in this study, we were unable to sample the subject’s residences, thereby limiting our conclusions.
An additional limitation of the study is the inability to perform WGS on all clinical NTM isolates. Of 165 NTM-positive pwCF, 57 were excluded because of infrequent healthcare use, inadequate long-term culture data, or lack of clinical follow-up. Of 108 eligible subjects, 80 underwent WGS because not all clinical isolates were available for research investigation. In this study, environmental sampling did not culture all conceivable sources of NTM in the environment. Culture and WGS evaluation of specific healthcare-associated sources included surface dust and plumbing biofilms but excluded WGS evaluation of other sources such as soil. Clinically relevant NTM have been cultured from environmental dust in homes of patients with NTM (20). This is the first study, to our knowledge, that compares healthcare-associated environmental dust with clinical NTM isolates. The decision to sample water biofilms rather than free water was based on the fact that hydrophobic NTM cells prefer surface adherence over remaining suspended in water, resulting in more concentrated NTM numbers in biofilms (1,000–15,000 cfu/cm2) compared with water (10–100 cfu/ml) (48). In addition, clinically relevant NTM readily form biofilms on common plumbing materials (49). There is also good evidence that individual NTM clones are stable in biofilms over long periods of time, including in hospital sources. One study reported that NTM from water biofilms have been repeatedly isolated over a 41-month period from a recirculating hospital hot water system (50). We excluded evaluation of home and other outside environmental sources of NTM acquisition among patients with highly similar NTM strains within our program. Although additional healthcare environmental sampling and home environmental sampling are both interesting and important, such sampling was beyond the scope of this study.
Conclusions
This standardized epidemiologic investigation, combined with environmental sampling, watershed analysis, and integrated pan-genome analysis, was performed to evaluate the potential for healthcare-associated NTM acquisition and/or transmission among pwCF. We report potential healthcare-associated transmission of M. avium among pwCF. Both M. avium clusters had biologically plausible episodes of healthcare-associated opportunities for transmission, and one M. avium cluster met the stringent threshold of ⩽10 SNPs and ⩾95% shared accessory genome, and the other cluster differed in criteria only by 11 SNPs. We found that healthcare-associated transmission of M. abscessus is uncommon and unlikely in our program. On the basis of our center data, there is not substantial evidence of healthcare-associated transmission to warrant additional IPC measures beyond those already recommended (43). However, this may not be generalizable to other centers where IPC measures and design of the healthcare setting may differ. Two M. abscessus clusters demonstrated episodes of healthcare-associated opportunities for transmission but did not meet the stringent threshold of isolate similarity to support healthcare-associated transmission. One M. abscessus cluster, in which patients resided in the same watershed, revealed no episodes of healthcare-associated opportunities for transmission but high genetic similarity at the core and accessory genome, supporting environmental acquisition. Finally, our results suggest that the presence of genetically similar isolates is insufficient by itself to demonstrate healthcare-associated NTM transmission. The HALT NTM study provides a framework to standardize epidemiologic investigation, coupled with healthcare environmental sampling and home of residence watershed analysis, and thereby improves understanding of the frequency and nature of healthcare-associated NTM transmission among pwCF as well as the potential to identify where acquisition might be at a higher probability. Concurrent parallel investigations are underway at several CF centers in the United States using the same algorithm.
Footnotes
Supported by the Departments of Pediatrics and Medicine and the Center for Genes, Environment, and Health at National Jewish Health as well as by the Cystic Fibrosis Foundation (GROSS19Q0 [J.E.G.], NICK20Y2SVC [J.A.N.], NICK20Y2OUT [J.A.N.], and NICK15RO [J.A.N.]).
Author Contributions: Conceptualization: J.E.G. and J.A.N. Study design: J.E.G., C.L.D., and J.A.N. Data collection: J.E.G., S.C., K.P., N.A.H., R.M.D., L.E.E., E.L., C.V., V.C.N.d.M., and J.R.H. Data analysis: J.E.G., S.C., K.P., N.A.H., F.J., R.M.D., L.E.E., E.L., C.V., J.R.H., M. Strand, M. Strong, and J.A.N. Funding acquisition: J.E.G. and J.A.N. All authors contributed to the manuscript and approved the final version submitted for publication.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202108-1911OC on January 27, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Qvist T, Pressler T, Høiby N, Katzenstein TL. Shifting paradigms of nontuberculous mycobacteria in cystic fibrosis. Respir Res . 2014;15:41. doi: 10.1186/1465-9921-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, et al. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax . 2016;71:i1–i22. doi: 10.1136/thoraxjnl-2015-207360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, Tonelli MR, et al. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med . 2012;185:231–232. doi: 10.1164/ajrccm.185.2.231. [DOI] [PubMed] [Google Scholar]
- 4. Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet . 2013;381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston DI, Chisty Z, Gross JE, Park SY. Investigation of Mycobacterium abscessus outbreak among cystic fibrosis patients, Hawaii 2012. J Hosp Infect . 2016;94:198–200. doi: 10.1016/j.jhin.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 6. Yan J, Kevat A, Martinez E, Teese N, Johnson K, Ranganathan S, et al. Investigating transmission of Mycobacterium abscessus amongst children in an Australian cystic fibrosis centre. J Cyst Fibros . 2020;19:219–224. doi: 10.1016/j.jcf.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 7. Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, et al. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous Mycobacterium. Science . 2016;354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doyle RM, Rubio M, Dixon G, Hartley J, Klein N, Coll P, et al. Cross-transmission is not the source of new Mycobacterium abscessus infections in a multicenter cohort of cystic fibrosis patients. Clin Infect Dis . 2020;70:1855–1864. doi: 10.1093/cid/ciz526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olivier KN, Weber DJ, Wallace RJ, Jr, Faiz AR, Lee JH, Zhang Y, et al. Nontuberculous Mycobacteria in Cystic Fibrosis Study Group Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med . 2003;167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 10. Harris KA, Underwood A, Kenna DT, Brooks A, Kavaliunaite E, Kapatai G, et al. Whole-genome sequencing and epidemiological analysis do not provide evidence for cross-transmission of Mycobacterium abscessus in a cohort of pediatric cystic fibrosis patients. Clin Infect Dis . 2015;60:1007–1016. doi: 10.1093/cid/ciu967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tortoli E, Kohl TA, Trovato A, Baldan R, Campana S, Cariani L, et al. Mycobacterium abscessus in patients with cystic fibrosis: low impact of inter-human transmission in Italy. Eur Respir J . 2017;50:1602525. doi: 10.1183/13993003.02525-2016. [DOI] [PubMed] [Google Scholar]
- 12. Gross JE, Martiniano SL, Nick JA. Prevention of transmission of Mycobacterium abscessus among patients with cystic fibrosis. Curr Opin Pulm Med . 2019;25:646–653. doi: 10.1097/MCP.0000000000000621. [DOI] [PubMed] [Google Scholar]
- 13. Primm TP, Lucero CA, Falkinham JO., III Health impacts of environmental mycobacteria. Clin Microbiol Rev . 2004;17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falkinham JO., III Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerg Infect Dis . 2011;17:419–424. doi: 10.3201/eid1703.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Honda JR, Virdi R, Chan ED. Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front Microbiol . 2018;9:2029. doi: 10.3389/fmicb.2018.02029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. du Moulin GC, Stottmeier KD, Pelletier PA, Tsang AY, Hedley-Whyte J. Concentration of Mycobacterium avium by hospital hot water systems. JAMA . 1988;260:1599–1601. doi: 10.1001/jama.260.11.1599. [DOI] [PubMed] [Google Scholar]
- 17. Shin JH, Lee EJ, Lee HR, Ryu SM, Kim HR, Chang CL, et al. Prevalence of non-tuberculous mycobacteria in a hospital environment. J Hosp Infect . 2007;65:143–148. doi: 10.1016/j.jhin.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18. Williams MM, Armbruster CR, Arduino MJ. Plumbing of hospital premises is a reservoir for opportunistically pathogenic microorganisms: a review. Biofouling . 2013;29:147–162. doi: 10.1080/08927014.2012.757308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallace RJ, Jr, Brown BA, Griffith DE. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu Rev Microbiol . 1998;52:453–490. doi: 10.1146/annurev.micro.52.1.453. [DOI] [PubMed] [Google Scholar]
- 20. Kaevska M, Slana I, Kralik P, Reischl U, Orosova J, Holcikova A, et al. “Mycobacterium avium subsp. hominissuis” in neck lymph nodes of children and their environment examined by culture and triplex quantitative real-time PCR. J Clin Microbiol . 2011;49:167–172. doi: 10.1128/JCM.00802-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipner EM, French J, Bern CR, Walton-Day K, Knox D, Strong M, et al. Nontuberculous mycobacterial disease and molybdenum in Colorado watersheds. Int J Environ Res Public Health . 2020;17:3854. doi: 10.3390/ijerph17113854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davidson RM, Hasan NA, Epperson LE, Benoit JB, Kammlade SM, Levin AR, et al. Population genomics of Mycobacterium abscessus from U.S. cystic fibrosis care centers. Ann Am Thorac Soc . 2021;18:1960–1969. doi: 10.1513/AnnalsATS.202009-1214OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hasan NA, Davidson RM, Epperson LE, Kammlade SM, Beagle S, Levin AR, et al. Population genomics and inference of Mycobacterium avium complex clusters in cystic fibrosis care centers, United States. Emerg Infect Dis . 2021;27:2836–2846. doi: 10.3201/eid2711.210124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gross JE, Caceres S, Poch K, Hasan NA, Davidson RM, Epperson LE, et al. Healthcare-associated links in transmission of nontuberculous mycobacteria among people with cystic fibrosis (HALT NTM) study: rationale and study design. PLoS One . 2021;16:e0261628. doi: 10.1371/journal.pone.0261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross JE, Caceres S, Poch KR, Hasan NA, Davisson R, Honda JR, et al. North American Cystic Fibrosis Conference.
- 26.Gross JE, Caceres S, Poch KR, Hasan NA, Davisson R, Honda JR, et al. Healthcare-associated links in transmission of nontuberculous mycobacteria in patients with cystic fibrosis (HALT NTM) [abstract] Am J Respir Crit Care Med 2021203A1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Epperson LE, Strong M. A scalable, efficient, and safe method to prepare high quality DNA from mycobacteria and other challenging cells. J Clin Tuberc Other Mycobact Dis . 2020;19:100150. doi: 10.1016/j.jctube.2020.100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaw LP, Doyle RM, Kavaliunaite E, Spencer H, Balloux F, Dixon G, et al. Children with cystic fibrosis are infected with multiple subpopulations of Mycobacterium abscessus with different antimicrobial resistance profiles. Clin Infect Dis . 2019;69:1678–1686. doi: 10.1093/cid/ciz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon JK, Kim TS, Kim JI, Yim JJ. Whole genome sequencing of nontuberculous Mycobacterium (NTM) isolates from sputum specimens of co-habiting patients with NTM pulmonary disease and NTM isolates from their environment. BMC Genomics . 2020;21:322. doi: 10.1186/s12864-020-6738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol . 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics . 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davidson RM, Benoit JB, Kammlade SM, Hasan NA, Epperson LE, Smith T, et al. Genomic characterization of sporadic isolates of the dominant clone of Mycobacterium abscessus subspecies massiliense. Sci Rep . 2021;11:15336. doi: 10.1038/s41598-021-94789-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention. 2013. https://www.cdc.gov/hai/outbreaks/outbreaktoolkit.html
- 34.Centers for Disease Control and Prevention. 2013. www.cdc.gov/hai/pdfs/outbreaks/Response_Toolkit_Abstraction_Form-508.pdf
- 35. Virdi R, Lowe ME, Norton GJ, Dawrs SN, Hasan NA, Epperson LE, et al. Lower recovery of nontuberculous mycobacteria from outdoor Hawai’i environmental water biofilms compared to indoor samples. Microorganisms . 2021;9:224. doi: 10.3390/microorganisms9020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adékambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol . 2003;41:5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honda JR, Hasan NA, Davidson RM, Williams MD, Epperson LE, Reynolds PR, et al. Environmental nontuberculous mycobacteria in the Hawaiian islands. PLoS Negl Trop Dis . 2016;10:e0005068. doi: 10.1371/journal.pntd.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahle D, Wickham H. ggmap: spatial visualization with ggplot2. R J . 2013;5:144–161. [Google Scholar]
- 39.U.S. Department of Agriculture, Natural Resources Conservation Service (USDA-NRCS) Washington, DC: USDA-NRCS. Available from: http://datagateway.nrcs.usda.gov. [Google Scholar]
- 40. Bryant JM, Brown KP, Burbaud S, Everall I, Belardinelli JM, Rodriguez-Rincon D, et al. Stepwise pathogenic evolution of Mycobacterium abscessus. Science . 2021;372:eabb8699. doi: 10.1126/science.abb8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tettelin H, Davidson RM, Agrawal S, Aitken ML, Shallom S, Hasan NA, et al. High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks. Emerg Infect Dis . 2014;20:364–371. doi: 10.3201/eid2003.131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davidson RM, Nick SE, Kammlade SM, Vasireddy S, Weakly N, Hasan NA, et al. Genomic analysis of a hospital-associated outbreak of Mycobacterium abscessus: implications on transmission. J Clin Microbiol . 2022;60:e0154721. doi: 10.1128/JCM.01547-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saiman L, Siegel JD, LiPuma JJ, Brown RF, Bryson EA, Chambers MJ, et al. Cystic Fibrous Foundation Society for Healthcare Epidemiology of America. Infection prevention and control guideline for cystic fibrosis: 2013 update. Infect Control Hosp Epidemiol . 2014;35:S1–S67. doi: 10.1086/676882. [DOI] [PubMed] [Google Scholar]
- 44. Gebert MJ, Delgado-Baquerizo M, Oliverio AM, Webster TM, Nichols LM, Honda JR, et al. Ecological analyses of mycobacteria in showerhead biofilms and their relevance to human health. MBio . 2018;9:e01614-18. doi: 10.1128/mBio.01614-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baker AW, Lewis SS, Alexander BD, Chen LF, Wallace RJ, Jr, Brown-Elliott BA, et al. Two-phase hospital-associated outbreak of Mycobacterium abscessus: investigation and mitigation. Clin Infect Dis . 2017;64:902–911. doi: 10.1093/cid/ciw877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kolok AS, Beseler CL, Chen XH, Shea PJ. The watershed as a conceptual framework for the study of environmental and human health. Environ Health Insights . 2009;3:1–10. doi: 10.4137/EHI.S1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lipner EM, Knox D, French J, Rudman J, Strong M, Crooks JL. A geospatial epidemiologic analysis of nontuberculous mycobacterial infection: an ecological study in Colorado. Ann Am Thorac Soc . 2017;14:1523–1532. doi: 10.1513/AnnalsATS.201701-081OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Falkinham JO, III, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol . 2001;67:1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mullis SN, Falkinham JO., III Adherence and biofilm formation of Mycobacterium avium, Mycobacterium intracellulare and Mycobacterium abscessus to household plumbing materials. J Appl Microbiol . 2013;115:908–914. doi: 10.1111/jam.12272. [DOI] [PubMed] [Google Scholar]
- 50. von Reyn CF, Maslow JN, Barber TW, Falkinham JO, III, Arbeit RD. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet . 1994;343:1137–1141. doi: 10.1016/s0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]


