Abstract
Arcanobacterium pyogenes is an opportunistic pathogen, associated with suppurative infections in domestic animals. In addition to pyolysin, a pore-forming, cholesterol-binding toxin, A. pyogenes expresses a number of putative virulence factors, including several proteases and neuraminidase activity. A 3,009-bp gene, nanH, was cloned and sequenced and conferred neuraminidase activity on an Escherichia coli host strain. The predicted 107-kDa NanH protein displayed similarity to a number of bacterial neuraminidases and contained the RIP/RLP motif and five copies of the Asp box motif found in all bacterial neuraminidases. Recombinant His-tagged NanH was found to have pH and temperature optima of 5.5 to 6.0 and 55°C, respectively. Insertional deletion of the nanH gene resulted in the reduction, but not absence, of neuraminidase activity, indicating the presence of a second neuraminidase gene in A. pyogenes. NanH was localized to the A. pyogenes cell wall. A. pyogenes adhered to HeLa, CHO, and MDBK cells in a washing-resistant manner. However, the nanH mutant was not defective for adherence to epithelial cells. The role of NanH in host epithelial cell adherence may be masked by the presence of a second neuraminidase in A. pyogenes.
Arcanobacterium pyogenes is a common inhabitant of the upper respiratory, urogenital (12, 54), and gastrointestinal tracts (35; B. H. Jost, K. W. Post, and S. J. Billington, unpublished data) of many domestic animal species. However, a physical or microbial insult to the host can lead to a variety of suppurative A. pyogenes infections, such as mastitis in dairy cows (27) and goats (2), liver abscesses in feedlot cattle (31, 34), and pneumonia in pigs (26) and various species of wildlife (17, 42, 57). A. pyogenes can also infect avian species (10) and humans (5, 16, 19), although infections in humans are rare.
A. pyogenes elaborates a number of extracellular proteins, including the hemolytic exotoxin pyolysin (PLO) (8), several proteases (48, 51), a DNase (30), and at least one neuraminidase (47). While all these proteins are putative virulence factors, only for PLO is there definitive evidence of involvement in the pathogenesis of infections by A. pyogenes (29).
Recently, there has been interest in the neuraminidases of bacterial pathogens and the potential role they play in pathogenesis. Neuraminidase (N-acetylneuraminyl hydrolase; EC 3.2.1.18) removes sialic acid from glycolipids, glycoproteins, and poly- and oligosaccharides. Bacterial neuraminidases have only 20 to 30% amino acid sequence identity, but they contain two conserved motifs, the RIP/RLP motif (Arg-Ile/Leu-Pro) and the Asp box motif (Ser-X-Asp-X-Gly-X-Thr-Trp), which occurs four or five times in the enzyme (14, 21, 43).
Neuraminidases are virulence factors, especially in bacteria that inhabit mucosal surfaces (20, 22, 52, 55), and they may play several roles in virulence. This enzyme can make sialic acid available as a carbon source to promote growth in a nutrient-limited environment (11, 23). The action of neuraminidase can decrease mucus viscosity (24), possibly enhancing colonization of the underlying tissues. Desialylation by neuraminidases can increase the susceptibility of mucosal IgA to bacterial proteases (18, 41). Neuraminidase can enhance bacterial adhesion and colonization (9, 13, 22, 56) and susceptibility of the host to the action of toxins (20), by exposing cryptic host cell receptor molecules.
This paper describes the cloning and characterization of a neuraminidase expressed by A. pyogenes. In addition, we show that A. pyogenes can adhere to epithelial cells in a washing-resistant manner, and we have investigated whether neuraminidase plays a role in this adhesion.
MATERIALS AND METHODS
Bacteria and growth conditions.
A. pyogenes strain BBR1 was isolated from a bovine abscess. Other A. pyogenes strains used in this study were from veterinary diagnostic laboratories or personal collections. A. pyogenes strains were grown on brain heart infusion (BHI; Difco) agar plates, supplemented with 5% bovine blood, at 37°C and 5% CO2 or in BHI broth supplemented with 5% bovine calf serum at 37°C with shaking. Escherichia coli DH5αMCR strains (Gibco-BRL) were grown at 37°C on Luria-Bertani (LB; Difco) agar or in LB broth with shaking. Antibiotics (Sigma) were added as appropriate; for A. pyogenes strains, erythromycin (EM) at 15 μg/ml and kanamycin (KM) at 30 μg/ml; for E. coli strains, ampicillin at 100 μg/ml, chloramphenicol at 30 μg/ml, EM at 200 μg/ml, and KM at 50 μg/ml.
Preparation of CSF, CWE, and protoplasts.
Culture supernatant fluid (CSF) was prepared from liquid cultures of A. pyogenes grown overnight to an optical density at 600 nm (OD600) of approximately 3.0 to 4.0. Cells were removed by centrifugation at 5,000 × g, and the CSF was filtered through a 0.22-μm-pore-size filter. A. pyogenes cell wall extract (CWE) and protoplasts were prepared as previously described for Streptococcus pneumoniae (36). Protoplasts were resuspended in distilled water and were lysed by several cycles of freezing and thawing. Total protein concentration was determined using the Bradford protein assay reagent (Bio-Rad).
DNA techniques.
Preparation of plasmid DNA and electroporation-mediated transformation of A. pyogenes strains were performed as previously described (28). Genomic DNA from A. pyogenes was isolated by the method of Pospiech and Neumann (40). A library of A. pyogenes BBR1 genomic DNA was constructed in λGEM-12, according to the manufacturer's instructions (Promega). Methods for growth and purification of bacteriophage were essentially as described by Ausubel et al. (4). DNA was prepared from bacteriophage as previously described (46) and was further purified using the Wizard DNA clean-up system (Promega). E. coli plasmid DNA extraction, transformation, DNA restriction, ligation, agarose gel electrophoresis, and Southern transfer of DNA to nitrocellulose membranes were performed essentially as described elsewhere (4). Preparation of DNA probes, DNA hybridization, and probe detection were performed using a digoxigenin DNA labeling and detection kit (Roche), as recommended by the manufacturer. PCR DNA amplification was performed using Taq DNA polymerase (Fisher Scientific) with the supplied reaction buffer for 35 cycles consisting of 1 min at 94°C, 1 min at 55°C, and 1 min/kb at 72°C, with a final extension step of 72°C for 5 min.
Nucleotide sequence determination.
The sequence of nanH was determined from plasmids pJGS274 and pJGS292 and appropriate subclones using automated DNA sequencing. Sequencing was performed on both strands, crossing all restriction sites, using KS, SK, or T7 sequencing primers or oligonucleotide primers based on the sequence of nanH. Oligonucleotide primers were synthesized by Sigma-Genosys. Sequencing reactions were performed by the DNA Sequencing Facility at The University of Arizona, using a 377 DNA sequencer (Applied Biosystems Inc.).
Computer sequence analysis.
Nucleotide sequence data were compiled using the Sequencher program (GeneCodes). Database searches were performed using the BlastX and BlastP algorithms (3). Sequence analysis was performed using the suite of programs available through the Genetics Computer Group, Inc. (University of Wisconsin). Signal sequence prediction was performed using SignalP (38). Multiple sequence alignments were performed using CLUSTAL W (53).
Cloning and purification of a recombinant, six-His-tagged NanH (His-NanH).
The nanH gene, lacking the coding region for the signal sequence, was amplified from A. pyogenes BBR1 genomic DNA by PCR with a 5′ primer containing an NheI site (5′-GGTTGCAGCTAGCGCCCCGAGCACAG-3′) and a 3′ primer containing a PstI site (5′-GCGTTATCGCGCTGCAGATTTAGCCC-3′) (restriction sites are underlined). These primers amplified a 2.9-kb product from bases 120 to 3009 of the plo gene. The PCR fragment was digested with NheI-PstI and cloned into NheI-PstI-digested pTrcHis B (Invitrogen) to generate pJGS306. pJGS306 encoded His-NanH, a 977-amino-acid protein comprising 963 amino acids of the mature NanH with an N-terminal extension of 14 amino acids encoded by pTrcHis B, including a six-His sequence.
Cultures for preparation of His-NanH were grown to an OD600 of 0.6, prior to induction with 2.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Gold Biotechnology) for 3 h. Cells were harvested by centrifugation at 5,000 × g, and the cell pellet was resuspended in 20 mM Tris-HCl–100 mM NaCl, pH 8.0. The cells were disrupted by two passages through a French pressure cell (Aminco) at 138 MPa, and the insoluble material was removed by centrifugation at 12,000 × g. His-NanH was purified from the soluble fraction using TALON metal affinity resin (Clontech), per the manufacturer's instructions. His-NanH was eluted from the resin with 50 mM imidazole–20 mM Tris-HCl–100 mM NaCl, pH 8.0 (TALON elution buffer). Total protein concentration was determined using the Bradford protein assay reagent (Bio-Rad).
Preparation of goat antiserum to His-NanH.
A female goat was immunized with 500 μg of His-NanH in a Ribi adjuvant system (Corexa) intramuscularly in the hind leg at two sites. A similar booster immunization of 500 μg of His-NanH in RAS was administered on days 29, 43, and 64. Blood was collected on day 75, and serum was harvested from the clotted blood by centrifugation at 400 × g. Preimmune serum was prepared in a similar manner, prior to immunization.
SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
E. coli whole cells or purified His-NanH was mixed 1:1 with sodium dodecyl sulfate (SDS) sample buffer (0.2 M Tris-HCl [pH 6.8], 2.5% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.013% bromophenol blue) and boiled for 10 min prior to electrophoresis in a 10% (wt/vol) SDS-polyacrylamide gel, essentially as described previously (4). Proteins were transferred to nitrocellulose (Schleicher and Schuell), and Western blots were immunostained as previously described (4) using goat anti-His-NanH and rabbit anti-goat immunoglobulin G (heavy plus light chains)-peroxidase conjugate (Kirkegaard & Perry Laboratories) as the primary and secondary antibodies, respectively. When required, sera were absorbed overnight at 4°C with an equal volume of bacterial cells which had been previously disrupted by two passages through a French pressure cell (Aminco) at 138 MPa. The sera were sterilized by passage through a 0.22-μm-pore-size filter prior to use.
Neuraminidase activity assay.
Neuraminidase activity was assayed using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUAN) (Sigma) (33), essentially as described by Winter et al. (60). One unit was defined as the release of 1 μmol of 4-methylumbelliferone from MUAN per min at 37°C.
Screening of large numbers of A. pyogenes or E. coli strains for neuraminidase activity was performed using a filter paper assay. Number 2 filter paper (Whatman) was wetted with 100 μM MUAN in 0.1 M sodium acetate, pH 4.5. Colonies were patched onto the filter paper and incubated at 37°C for 15 min, prior to excitation with a UV source (254 nm). The presence of a cyan fluorescence indicated a positive result.
Epithelial cell adhesion assay.
The epithelial cell lines used in the adherence experiments were Chinese hamster ovary (CHO) cells, human cervical epithelial cells (HeLa), and Madin-Darby bovine kidney (MDBK) cells. All cell lines were cultured in Iscove's modified Dulbecco's medium (Life Technologies) supplemented with 10% fetal bovine serum (Omega Scientific) (IMDM-10%) with 100 μg of gentamicin/ml (Sigma) in a humidified, 5% CO2 atmosphere at 37°C. For adherence assays, epithelial cells in IMDM-10% without gentamicin were seeded into 24-well plates at 2 × 105 cells per well in 1-ml volumes. The cells were incubated at 37°C and 5% CO2 for 18 h prior to addition of bacteria (freshly grown to an OD600 of 1.0) and His-NanH, where applicable. The plates were centrifuged for 10 min at 400 × g to increase the bacterium-host cell contact. Bacterial adhesion was assessed after 1 h of incubation at 37°C and 5% CO2. Cell monolayers were washed three times with 0.01 M phosphate-buffered saline, pH 7.2 (PBS), to remove nonadherent bacteria. Bacteria were recovered by treatment of the cell monolayers with 1 ml of 0.1% Triton X-100 for 10 min at 0°C, and viable bacteria were enumerated by dilution plating. All experiments were performed in triplicate on three separate occasions.
Nucleotide sequence accession number.
The nanH sequence data were submitted to the DDBJ/EMBL/GenBank databases under accession number AF298154.
RESULTS
A. pyogenes strains express neuraminidase activity.
A total of 53 strains of A. pyogenes isolated from a variety of animals, including bovine, porcine, and avian species were tested for neuraminidase expression by the filter paper method. All 53 strains were positive for neuraminidase activity, indicating that this enzyme is probably expressed by all A. pyogenes isolates.
In addition, neuraminidase activity was expressed throughout the cell cycle. A. pyogenes BBR1 cells, grown to an OD600 of 0.45, 1.0, or 5.2, were washed once with PBS, and an equal number of cells was applied to filter paper saturated with MUAN. All the samples fluoresced strongly, indicating that at least in vitro, neuraminidase activity was present throughout the A. pyogenes cell cycle.
Cloning and nucleotide sequence determination of nanH.
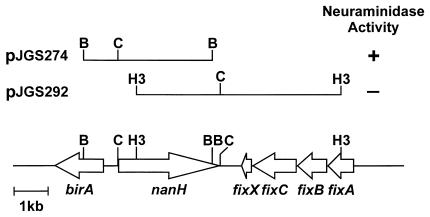
During a project to clone A. pyogenes promoter sequences in the promoter-probe vector pKK232-8 (Amersham Pharmacia Biotech), sequences homologous to the neuraminidase gene, nedA, of Micromonospora viridifaciens (45) were identified. In order to clone the entire gene, a probe was prepared from a region spanning bases 1639 to 1898 of the nanH gene and was used to probe a λGEM-12 library of BBR1 genomic DNA. Several plaques hybridized strongly with the probe and were selected for further analysis. One of these, λJGS6, contained an approximately 16-kb partial Sau3AI fragment. DNA purified from λJGS6 was digested with BamHI or HindIII and cloned into similarly digested pBC KS (Stratagene). Two overlapping clones encompassing the entire nanH gene region were obtained: pJGS274, containing a 3,773-bp BamHI fragment, and pJGS292, containing a 6,151-bp HindIII fragment (Fig. 1). pJGS274, but not pJGS292, conferred neuraminidase activity on the E. coli host, as determined by the MUAN filter paper assay.
FIG. 1.
Map of the nanH gene region. BamHI (B), ClaI (C), and HindIII (H3) sites used to clone portions of the nanH gene are shown.
The DNA sequence of the nanH gene region was deduced from these clones. There are several in-frame ATG codons at the 5′ end of the nanH gene. The first and second ATG appear to be equal candidates for start codons, as they both have consensus ribosome-binding sites. Assuming translation from the first ATG, the 3,009-bp nanH gene encoded a protein with a predicted molecular mass of 107.3 kDa. A gram-positive signal sequence, with a cleavage site between Ala 40 and Ala 41, was predicted by SignalP (38). A putative rho independent terminator (ΔG = −17.0 kcal/mol) was identified 26 bases downstream of the nanH stop codon. No E. coli ς70-like promoter sequences were apparent upstream of the nanH gene.
Upstream of nanH, an open reading frame (ORF), birA, was identified, and its protein product had similarity to biotin ligase from Deinococcus radiodurans (59). Downstream sequences contained ORFs whose protein products had similarity to the electron transport proteins, FixABCX, involved in nitrogen fixation in Bradyrhizobium japonicum (25, 58). birA and the putative fixABCX operon were also transcribed in the opposite direction to nanH (Fig. 1), suggesting that nanH is monocistronic.
Analysis of the primary structure of NanH.
Cleavage at the predicted signal peptide sequence of NanH would result in a mature protein with a predicted molecular mass of 103.2 kDa and a pI of 6.3. NanH showed similarity to a number of bacterial neuraminidases, including those from Actinomyces viscosus (31.2% identity, 61.8% similarity), M. viridifaciens (21.4% identity, 42.3% similarity), and Bacteroides fragilis (13.7% identity, 33.4% similarity). In addition, the NanH protein contained the conserved catalytic RIP/RLP motif, as well as five copies of the Asp box motif (Ser-X-Asp-X-Gly-X-Thr-Trp) associated with bacterial neuraminidases (14, 21, 43) (Fig. 2).
FIG. 2.
Amino acid alignment of the active site region of A. pyogenes NanH, A. viscosus NanH (61), M. viridifaciens NedA (45), and B. fragilis NanH (1). Amino acids identical to the NanH sequence are boxed in black. Conservative substitutions are boxed in gray. The RIP/RLP motif and the two additional catalytic arginine residues are indicated by the arrows. The Ser-X-Asp-X-Gly-X-Thr-Trp (Asp box) motifs are underlined. Amino acid numbers for each protein are indicated on the right.
At the C terminus of the NanH protein there was a sequence similar to the cell wall sorting signals found in surface-expressed proteins of gram-positive bacteria (49). The terminal 33 amino acids of NanH (LVHTGATVVGLSVAAAVLLLAGGAIAIIRRRQG) consisted of an LPXTG cleavage motif (bold), a hydrophobic domain (underlined), and a positively charged stop transfer sequence (italics). However, in the case of the A. pyogenes NanH protein, the cleavage motif was LVHTG, instead of LPXTG. In addition, a Pro-rich repetitive region, which is thought to facilitate spanning of the cell wall peptidoglycan (49), was found directly upstream of the LPXTG motif.
Cloning and expression of His-NanH.
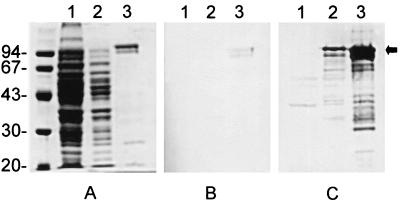
To facilitate the purification of recombinant NanH from E. coli, the NanH-coding sequence, lacking the sequence for the putative signal peptide was cloned into pTrcHis B. SDS-PAGE and Coomassie brilliant blue staining of IPTG-induced cultures of DH5αMCR(pJGS306) did not reliably indicate the presence of an overexpressed protein of approximately 104 kDa, compared to similarly induced cultures of DH5αMCR(pTrcHis B) (Fig. 3A). However, His-NanH was purified from DH5αMCR(pJGS306) to >90% homogeneity using TALON resin (Fig. 3A), and the size of this protein corresponded to that of the predicted molecular mass of His-NanH. His-NanH routinely purified as a doublet, which may indicate some processing of the protein in E. coli (Fig. 3A). Purified His-NanH retained neuraminidase activity as determined using the fluorometric assay with MUAN as a substrate.
FIG. 3.
Overexpression and purification of His-NanH. Whole-cell lysates of IPTG-induced cultures of DH5αMCR(pTrcHis B) (lanes 1) and DH5αMCR(pJGS306) (lanes 2) and 100 ng of purified His-NanH (lanes 3) were subjected to SDS–10% PAGE. Separated proteins were stained with Coomassie brilliant blue (A) or were transferred to nitrocellulose by Western blotting and immunostained with 1/100 E. coli-adsorbed preimmune serum (B) or 1/100 E. coli-adsorbed His-NanH antiserum (C). The positions of the molecular size standards are shown (in kilodaltons) on the left. The arrow indicates the position of His-NanH.
Reaction of His-NanH with specific antibodies.
In Western blots, antiserum prepared against His-NanH detected a protein of approximately 104 kDa in IPTG-induced cultures of DH5αMCR(pJGS306) and preparations of purified His-NanH but not IPTG-induced cultures of DH5αMCR(pTrcHis B) (Fig. 3C). Preimmune serum also weakly reacted with His-NanH (Fig. 3B). This is not surprising, as A. pyogenes is a common inhabitant of the bacterial flora in domestic animals, and sera from normal goats contain antibodies to NanH and other A. pyogenes proteins, such as PLO (8; B. H. Jost and S. J. Billington, unpublished data).
Determination of optimal conditions for enzyme activity.
In order to determine the pH optimum for this enzyme, the activity of 25 ng of His-NanH was measured by the fluorometric assay using MUAN as a substrate in 100 mM citrate-phosphate buffer with a pH ranging from 3.0 to 8.5. The pH optimum for His-NanH was 5.5 to 6.0 (data not shown). Similarly, the temperature optimum for His-NanH was determined by incubation with MUAN in 100 mM citrate-phosphate buffer, pH 6.0, at temperatures ranging from 23 to 75°C. The temperature optimum for His-NanH was 55°C (data not shown). The enzyme was quite robust, with approximately 45% of the maximal activity after 1 h at 75°C. Because His-NanH had >90% of maximal activity at 37°C and 37°C is a physiologically relevant temperature, 37°C was chosen as the standard incubation temperature. Purified His-NanH had a specific activity of 1.3 U/mg at pH 6.0 and 37°C.
Previous reports have suggested that the large bacterial neuraminidases require Ca2+ ions for activity (14). The requirement for divalent cations was tested by incubation of 25 ng of His-NanH in 100 mM citrate-phosphate buffer, pH 6.0, for 1 h at 37°C with final concentrations of 1 mM CaCl2, 1 mM MgCl2, or 10 mM EDTA. Addition or removal of divalent cations did not significantly affect the activity of His-NanH (data not shown).
Determination of the prevalence of the nanH gene by DNA dot blotting.
In order to determine whether nanH was present in all A. pyogenes strains, genomic DNA was prepared from 53 A. pyogenes strains and was subjected to hybridization at high stringency with a nanH-specific probe that spanned bases 1639 to 1898 of the nanH ORF. The DNA from all 53 strains hybridized strongly to the probe (data not shown), indicating that the nanH gene is present in all A. pyogenes strains.
Construction and characterization of a nanH mutant.
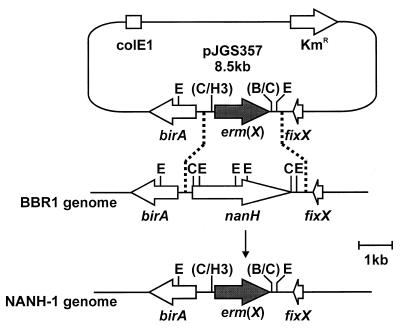
Construction of the nanH mutant used an allelic exchange plasmid in which the nanH gene was completely replaced by an erm(X) cassette (Fig. 4). This plasmid was constructed by deletion of the 2.6-kb ClaI fragment containing the nanH gene sequences from pJGS292 (Fig. 1), resulting in the recombinant plasmid pJGS326. The 0.73-kb ClaI fragment of pJGS293, containing sequences upstream of nanH, was cloned into similarly digested pJGS326 to form pJGS354. The entire 4.3-kb HindIII insert of pJGS342 was cloned into similarly digested pHSS20 (37) to form pJGS356. The KM resistance gene in pHSS20 is functional in A. pyogenes and was used to identify the presence of recombinants which arose by a single crossover event. A 1.65-kb HindIII-BamHI fragment containing the erm(X) gene from pNG2 (50) was treated with T4 DNA polymerase (Promega). This fragment was cloned into the similarly treated unique ClaI site in pJGS356 to generate the recombinant plasmid pJGS357 (Fig. 4). As pJGS357 was based on a ColE1 replicon, it acted as a suicide plasmid in A. pyogenes (29). pJGS357 plasmid DNA was introduced into A. pyogenes BBR1 cells by electroporation, and recombinants were selected on BHI-blood agar containing EM. Emr Kms colonies were chosen for further analysis.
FIG. 4.
Scheme for insertional inactivation of the A. pyogenes nanH gene. A ClaI fragment containing nanH was replaced with the erm(X) gene of pNG2, to construct pJGS357. This plasmid was introduced into A. pyogenes strain BBR1 by electroporation. Reciprocal recombination, indicated by the dashed lines, resulted in replacement of the endogenous nanH gene in the BBR1 chromosome with the erm(X) gene to construct NANH-1. B, BamHI; C, ClaI; E, EcoRI; H3, HindIII. Only the insert portion of pJGS357 is to scale.
Southern blotting of A. pyogenes genomic DNA digested with EcoRI revealed hybridizing bands of 1.9, 1.0, 0.9, and 0.3-kb in BBR1, when probed with a nanH-specific probe (spanning bases 120 to 3009 of the nanH ORF). No hybridizing bands were observed in a nanH mutant (NANH-1) that was probed similarly, indicating the complete loss of the nanH gene in this strain. A 2.6-kb band was apparent in EcoRI-digested NANH-1 but not BBR1 genomic DNA when an erm(X)-specific probe was used. Neither BBR1 nor NANH-1 genomic DNA hybridized with a pHSS20-specific (vector) probe (data not shown). These data confirm deletion of the nanH gene in NANH-1 by a double-crossover event.
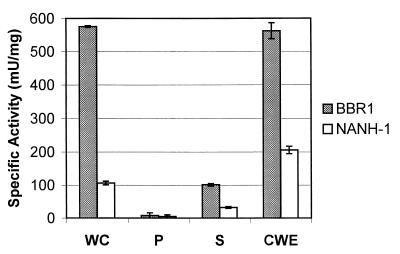
The neuraminidase activity of NANH-1 was compared to that of the wild-type strain BBR1. BBR1 and NANH-1 were grown overnight to an OD600 of 3.0 to 4.0, the cells were washed twice in PBS and resuspended at an OD600 of 3.0, and the neuraminidase activity was assessed by the fluorometric assay using MUAN as a substrate. NANH-1 had fivefold less neuraminidase activity than the wild-type (Fig. 5). Therefore, insertional deletion of nanH did not result in complete loss of expression of neuraminidase activity from NANH-1, indicating that NanH is not the only neuraminidase expressed by A. pyogenes.
FIG. 5.
Cell localization of neuraminidase activity. A total of 4 × 107 whole cells (WC), 4 × 107 lysed protoplasts (P), 20 μl of culture supernatant (S), or 200 μg of purified CWE from BBR1 or NANH-1 was assayed for neuraminidase activity using the fluorometric assay with MUAN as a substrate. Error bars indicate 1 standard deviation from the mean calculated from the averages of at least three independent experiments.
Localization of NanH.
Whole cells, CSF, and CWE were prepared from BBR1 and NANH-1 which had been grown overnight to an OD600 of 3.0 to 4.0. These samples were tested for neuraminidase activity with the fluorometric assay using MUAN as a substrate. The majority of neuraminidase activity was detected in whole cells and CWE from either BBR1 or NANH-1. However, BBR1 whole cells and CWE had significantly higher neuraminidase activity than whole cells and CWE from NANH-1 (Fig. 5). Some neuraminidase activity was detected in the CSF of both BBR1 and NANH-1 (Fig. 5), and this activity may have resulted from fragments of cell wall material present in the CSF. Negligible neuraminidase activity was detected in lysed protoplasts of BBR1 and NANH-1 (Fig. 5). These data indicate that the majority of NanH-specific neuraminidase activity was associated with the cell wall. In addition, the activity of the putative second neuraminidase also appeared to be cell wall associated.
Adherence of A. pyogenes to epithelial cells.
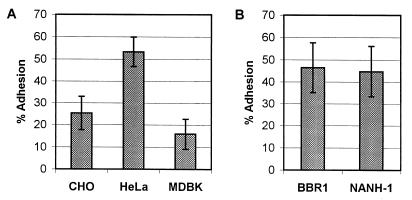
It was previously demonstrated that A. pyogenes adhered to HeLa cells (15). We also tested the ability of BBR1 to adhere to other epithelial cell lines. BBR1 adhered to CHO, HeLa, and MDBK cells in a washing-resistant manner (Fig. 6A). However, bacterial adherence was greatest using HeLa cells (Fig. 6A), and this cell line was used in all subsequent studies.
FIG. 6.
Adhesion of A. pyogenes strains to epithelial cell lines. A. pyogenes strains were added to cell monolayers and allowed to adhere for 1 h at 37°C prior to washing and recovery of cell-associated bacteria. (A) Adhesion of A. pyogenes BBR1 to CHO, HeLa, and MDBK cells. (B) Adhesion of BBR1 and NANH-1 to HeLa cells. Adhesion is shown as a percentage of the number of bacteria originally added to the cells. Error bars indicate 1 standard deviation from the mean calculated from the averages of at least three independent experiments.
The ability of BBR1 and NANH-1 to adhere to HeLa cells was tested. The wild-type BBR1 and nanH mutant strains adhered equally, with average adherences of 46.4 and 44.7%, respectively (Fig. 6B). In addition, the adherence of NANH-1 to CHO and MDBK cells was evaluated and was not significantly different from that of BBR1 (data not shown). These findings suggest that lack of NanH does not impair adhesion of NANH-1 to these epithelial cell lines.
In order to determine whether addition of exogenous NanH could enhance adhesion, 1 to 100 μg of purified His-NanH or an equal volume of TALON elution buffer (as a control) was added, either 1 h prior to or simultaneously with the addition of BBR1 or NANH-1 bacteria. No significant difference in the adhesion of the two strains was observed under any experimental condition (data not shown). These data indicate that addition of exogenous NanH did not increase the ability of BBR1 or NANH-1 to adhere to HeLa cells.
DISCUSSION
This is the first report of the cloning and sequencing of a neuraminidase gene, nanH, from A. pyogenes. In addition, this work provides indirect evidence that A. pyogenes expresses a second neuraminidase. NanH was localized to the cell wall, and the second neuraminidase also appeared to be cell wall associated. Furthermore, we demonstrated that neuraminidase activity is expressed by all A. pyogenes strains tested (n = 53).
The nanH gene of A. pyogenes was cloned and sequenced and appeared to exist in a monocistronic operon, surrounded by the housekeeping genes birA and fixABCX. nanH expressed a 103.2-kDa protein with neuraminidase activity, and the NanH protein was most closely related to the neuraminidase of A. viscosus (61). The NanH protein contained sequences consistent with its activity as a neuraminidase, including the RIP/RLP motif and five copies of the Asp box (14, 21, 43). In addition, the finding that NanH is localized to the A. pyogenes cell wall is consistent with the presence of an N-terminal signal peptide and C-terminal cell sorting signals, including an LPXTG-like cell anchor (49). However, in NanH, the cell wall anchor motif is LVHTG, which is slightly at variance with the consensus sequence observed in almost all cell wall-associated proteins identified to date. One exception is a sucrase expressed by Actinomyces naeslundii, which has the motif LARTG (39). In addition, two other cell wall-associated proteins have been identified in A. pyogenes, both of which have cell sorting signals where the Pro of the LPXTG motif has been replaced (B. H. Jost and S. J. Billington, unpublished data). Therefore, it appears that the A. pyogenes cell anchor sequence may be divergent from those seen in other bacteria.
Recombinant His-NanH protein was found to have optimal activity at pH 5.5 to 6.0 and 55°C, with no requirement for Ca2+ or Mg2+. These conditions correspond to those previously reported for a purified, native A. pyogenes neuraminidase (47). However, there were differences in the apparent size and cellular location of NanH (103.2 kDa, cell wall) and the purified, native A. pyogenes neuraminidase (50 kDa, CSF) (47). Size variation, probably as a result of specific breakdown or proteolytic cleavage, has been observed in other bacterial neuraminidases, with the truncated proteins retaining enzymatic activity. The 107-kDa S. pneumoniae NanA is observed as an enzymatically active 86-kDa species following purification (32). The size of the M. viridifaciens NedA protein varies between 41 and 68 kDa depending on the culture conditions (45). Indeed, upon prolonged storage at 4°C, His-NanH converted to a species of 62 kDa, with no loss of specific activity (data not shown). However, it is still uncertain whether NanH is the 50-kDa neuraminidase purified from A. pyogenes CSF (47). NanH was localized to the cell wall of A. pyogenes, but NanH was also present in CSF, as NANH-1 had significantly less neuraminidase activity in the CSF than BBR1 (Fig. 5). Schaufuss and Lämmler could detect significant neuraminidase activity in the CSF of only 2 out of 42 of A. pyogenes strains tested, and they did not report testing whole cells or CWE for the presence of neuraminidase activity (47).
The entire nanH ORF was deleted during the construction of NANH-1, as confirmed by Southern blotting (data not shown). Significant neuraminidase activity in NANH-1 indicated the presence of a second enzyme in A. pyogenes. In addition, the location of the second neuraminidase also appeared to be cell wall associated, as evidenced by the retention of some neuraminidase activity in the CWE of NANH-1.
The initial event in infection by many bacteria is their attachment to mammalian cells via specific recognition structures, leading to bacterial colonization of the host. Adherence of A. naeslundii to both a human epithelial cell line (9) and polymorphonuclear leukocytes (44) was enhanced by pretreatment with neuraminidase. Like A. pyogenes, S. pneumoniae expresses two distinct neuraminidases, NanA (7), which is cell associated, and NanB (6), which is thought to be secreted from the cell. Neuraminidase treatment of tracheal organ cultures increased the adherence of S. pneumoniae (56). Furthermore, S. pneumoniae mutants deficient in neuraminidase activity had reduced abilities to colonize and persist in the nasopharynx (55).
It is clear that neuraminidase activity plays a role in mediating host cell adhesion of some pathogens to mucosal surfaces. In order to determine whether this was the case for A. pyogenes, experiments assessing the adhesion of NANH-1, the neuraminidase mutant, were conducted. As previously reported, A. pyogenes can adhere to HeLa cells (15), and we demonstrated adherence to other epithelial cell lines, although A. pyogenes adhered best to HeLa cells. NANH-1 displayed no defect in adherence to HeLa cells compared with the wild-type, BBR1. If the neuraminidase activity does play a role in mediating adherence of A. pyogenes, it is possible that, at least under these conditions, the second neuraminidase was sufficient for maximal adherence. If this is the case, addition of exogenous His-NanH would not significantly affect adhesion to the host cell. Alternately, neuraminidase may play no role in the adhesion of A. pyogenes to cultured epithelial cells. The effects of neuraminidase activity may be more evident in vivo, where it may act to reduce mucus viscosity (24), assisting the adherence of A. pyogenes by other colonization factors, such as putative collagen- or fibronectin-binding proteins. The validity of these hypotheses is being tested in our laboratory by construction of a knockout mutant of the second neuraminidase in NANH-1 and assessment of the ability of this double mutant to adhere to host epithelial cells, both in vitro and in vivo.
ACKNOWLEDGMENTS
We thank Stefani Gilbert for construction of the λGEM-12 library and Hien Trinh and Dawn Bueschel for their excellent technical assistance.
Partial support for this work was provided by USDA/NRICGP awards (97-35204-4750 and 99-35204-7818).
REFERENCES
- 1.Akimoto S, Ono T, Tsutsui H, Kinouchi T, Kataoka K, Ohnishi Y. Complete sequence of the Bacteroides fragilis YCH46 neuraminidase-encoding gene. Biochem Biophys Res Commun. 1994;203:914–921. doi: 10.1006/bbrc.1994.2269. [DOI] [PubMed] [Google Scholar]
- 2.Al-Graibawi M A A, Sharma V K, Al-Shammari A J. Microbial pathogens from goat mastitis and phage-typing of Staphylococcus aureus isolates. Comp Immunol Microbiol Infect Dis. 1986;9:23–28. doi: 10.1016/0147-9571(86)90071-8. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 5.Barnham M. Actinomyces pyogenes bacteraemia in a patient with carcinoma of the colon. J Infect. 1988;27:231–234. doi: 10.1016/s0163-4453(88)96522-x. [DOI] [PubMed] [Google Scholar]
- 6.Berry A M, Lock R A, Paton J C. Cloning and characterization of nanB, a second Streptococcus pneumoniae neuraminidase gene, and purification of the NanB enzyme from recombinant Escherichia coli. J Bacteriol. 1996;178:4854–4860. doi: 10.1128/jb.178.16.4854-4860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry A M, Paton J C, Glare E M, Hansman D, Catcheside D E A. Cloning and expression of the pneumococcal neuraminidase gene in Escherichia coli. Gene. 1988;71:299–305. doi: 10.1016/0378-1119(88)90046-7. [DOI] [PubMed] [Google Scholar]
- 8.Billington S J, Jost B H, Cuevas W A, Bright K R, Songer J G. The Arcanobacterium (Actinomyces) pyogenes hemolysin, pyolysin, is a novel member of the thiol-activated cytolysin family. J Bacteriol. 1997;179:6100–6106. doi: 10.1128/jb.179.19.6100-6106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan M J, Cisar J O, Vatter A E, Sandberg A L. Lectin-dependent attachment of Actinomyces naeslundii to receptors on epithelial cells. Infect Immun. 1984;46:459–464. doi: 10.1128/iai.46.2.459-464.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinton M K, Schellberg L C, Johnson J B, Frank R K, Halvorson D A, Newman J A. Description of osteomyelitis lesions associated with Actinomyces pyogenes infection in the proximal tibia of adult male turkeys. Avian Dis. 1993;37:259–262. [PubMed] [Google Scholar]
- 11.Byers H L, Homer K A, Beighton D. Sialic acid utilisation by viridans streptococci. Adv Exp Med Biol. 1997;418:713–716. doi: 10.1007/978-1-4899-1825-3_167. [DOI] [PubMed] [Google Scholar]
- 12.Carter G R, Chengappa M M. Essentials of veterinary bacteriology and mycology. 4th ed. Philadelphia, Pa: Lea and Febiger; 1991. [Google Scholar]
- 13.Childs W C, Gibbons R J. Selective modulation of bacterial attachment to oral epithelial cells by enzyme activities associated with poor oral hygiene. J Periodontal Res. 1990;25:172–178. doi: 10.1111/j.1600-0765.1990.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 14.Crennell S J, Garman E F, Philippon C, Vasella A, Laver W G, Vimr E R, Taylor G L. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J Mol Biol. 1996;259:264–280. doi: 10.1006/jmbi.1996.0318. [DOI] [PubMed] [Google Scholar]
- 15.Ding H, Lämmler C, Selim R S. Adherence of Actinomyces pyogenes to HeLa cells mediated by hydrophobic surface proteins. Zentralbl Bakteriol. 1993;279:299–306. doi: 10.1016/s0934-8840(11)80362-2. [DOI] [PubMed] [Google Scholar]
- 16.Drancourt M, Oulès O, Bouche V, Peloux Y. Two cases of Actinomyces pyogenes infection in humans. Eur J Clin Microbiol Infect Dis. 1993;12:55–57. doi: 10.1007/BF01997060. [DOI] [PubMed] [Google Scholar]
- 17.Foreyt W J, Jessup D A. Fatal pneumonia of bighorn sheep following association with domestic sheep. J Wildl Dis. 1982;18:163–168. doi: 10.7589/0090-3558-18.2.163. [DOI] [PubMed] [Google Scholar]
- 18.Frandsen E V G. Carbohydrate depletion of immunoglobulin A1 by oral species of gram-positive rods. Oral Microbiol Immunol. 1994;9:352–358. doi: 10.1111/j.1399-302x.1994.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 19.Gahrn-Hansen B, Frederiksen W. Human infections with Actinomyces pyogenes (Corynebacterium pyogenes) Diagn Microbiol Infect Dis. 1992;15:349–354. doi: 10.1016/0732-8893(92)90022-l. [DOI] [PubMed] [Google Scholar]
- 20.Galen J E, Ketley J M, Fasano A, Richardson S H, Wasserman S S, Kaper J B. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaskell A, Crennell S, Taylor G. The three domains of a bacterial sialidase: a β-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–1205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 22.Giebink G S. Otitis media: the chinchilla model. Microb Drug Resist. 1999;5:57–72. doi: 10.1089/mdr.1999.5.57. [DOI] [PubMed] [Google Scholar]
- 23.Godoy V G, Miller Dallas M, Russo T A, Malamy M H. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect Immun. 1993;61:4415–4426. doi: 10.1128/iai.61.10.4415-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottschalk A. Correlation between composition, structure, shape, and function of a salivary mucoprotein. Nature. 1960;186:949–951. doi: 10.1038/186949a0. [DOI] [PubMed] [Google Scholar]
- 25.Gubler M, Zurcher T, Hennecke H. The Bradyrhizobium japonicum fixBCX operon: identification of fixX and of a 5′ mRNA region affecting the level of the fixBCX transcript. Mol Microbiol. 1989;3:141–148. doi: 10.1111/j.1365-2958.1989.tb01803.x. [DOI] [PubMed] [Google Scholar]
- 26.Høie S, Falk K, Lium B M. An abattoir survey of pneumonia and pleuritis in slaughter weight swine from 9 selected herds. IV. Bacteriological findings in chronic pneumonic lesions. Acta Vet Scand. 1991;32:395–402. doi: 10.1186/BF03546970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson P, Olsson S-E, Olofson A-S, Fälth C, Holmberg O, Funke H. Bacteriological investigations of clinical mastitis in heifers in Sweden. J Dairy Res. 1991;58:179–185. doi: 10.1017/s0022029900029721. [DOI] [PubMed] [Google Scholar]
- 28.Jost B H, Billington S J, Songer J G. Electroporation-mediated transformation of Arcanobacterium (Actinomyces) pyogenes. Plasmid. 1997;38:135–140. doi: 10.1006/plas.1997.1299. [DOI] [PubMed] [Google Scholar]
- 29.Jost B H, Songer J G, Billington S J. An Arcanobacterium (Actinomyces) pyogenes mutant deficient in production of the pore-forming cytolysin pyolysin has reduced virulence. Infect Immun. 1999;67:1723–1728. doi: 10.1128/iai.67.4.1723-1728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lämmler C. Untersuchungen zu möglichen Pathogenitätsfaktoren von Actinomyces pyogenesÜbersichtsreferat. Berl Muench Tieraerztl Wochenschr. 1990;103:121–125. [PubMed] [Google Scholar]
- 31.Lechtenberg K F, Nagaraja T G, Leipold H W, Chengappa M M. Bacteriologic and histologic studies of hepatic abscesses in cattle. Am J Vet Res. 1988;49:58–62. [PubMed] [Google Scholar]
- 32.Lock R A, Paton J C, Hansman D. Purification and immunological characterization of neuraminidase produced by Streptococcus pneumoniae. Microb Pathog. 1988;4:33–43. doi: 10.1016/0882-4010(88)90046-0. [DOI] [PubMed] [Google Scholar]
- 33.Myers R W, Lee R T, Lee Y C, Thomas G H, Reynolds L W, Uchida Y. The synthesis of 4-methylumbelliferyl α-ketoside of N-acetylneuraminic acid and its use in a fluorometric assay for neuraminidase. Anal Biochem. 1980;101:166–174. doi: 10.1016/0003-2697(80)90056-1. [DOI] [PubMed] [Google Scholar]
- 34.Nagaraja T G, Laudert S B, Parrott J C. Liver abscesses in feedlot cattle. Part I. Causes, pathogenesis, pathology, and diagnosis. Comp Cont Educ Pract Vet. 1996;18:S230–S241. , S256. [Google Scholar]
- 35.Narayanan S, Nagaraja T G, Wallace N, Staats J, Chengappa M M, Oberst R D. Biochemical and ribotypic comparison of Actinomyces pyogenes and A. pyogenes-like organisms from liver abscesses, ruminal wall, and ruminal contents of cattle. Am J Vet Res. 1998;59:271–276. [PubMed] [Google Scholar]
- 36.Neeleman C, Geelen S P M, Aerts P C, Daha M R, Mollnes T E, Roord J J, Posthuma G, van Dijk H, Fleer A. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by the binding of complement regulatory protein factor H. Infect Immun. 1999;67:4517–4524. doi: 10.1128/iai.67.9.4517-4524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickoloff J A, Reynolds R J. Subcloning with new ampicillin- and kanamycin-resistant analogs of pUC19. BioTechniques. 1991;10:469–472. [PubMed] [Google Scholar]
- 38.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Norman J M, Bunny K L, Giffard P M. Characterization of levJ, a sucrase/fructanase-encoding gene from Actinomyces naeslundii T14V, and comparison of its product with other sucrose-cleaving enzymes. Gene. 1995;152:93–98. doi: 10.1016/0378-1119(94)00695-o. [DOI] [PubMed] [Google Scholar]
- 40.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 41.Reinholdt J, Tomana M, Mortensen S B, Kilian M. Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect Immun. 1990;58:1186–1194. doi: 10.1128/iai.58.5.1186-1194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhyan J C, Aune K, Ewalt D R, Marquardt J, Mertins J W, Payeur J B, Saari D A, Schladweiler P, Sheehan E J, Worley D. Survey of free-ranging elk from Wyoming and Montana for selected pathogens. J Wildlife Dis. 1997;33:290–298. doi: 10.7589/0090-3558-33.2.290. [DOI] [PubMed] [Google Scholar]
- 43.Roggentin P, Rothe B, Kaper J B, Galen J, Lawrisuk L, Vimr E C, Schauer R. Conserved sequences in bacterial and viral sialidases. Glycoconjugate J. 1989;6:349–353. doi: 10.1007/BF01047853. [DOI] [PubMed] [Google Scholar]
- 44.Ruhl S, Cisar J O, Sandberg A L. Identification of polymorphonuclear leukocyte and HL-60 cell receptors for adhesins of Streptococcus gordonii and Actinomyces naeslundii. Infect Immun. 2000;68:6346–6354. doi: 10.1128/iai.68.11.6346-6354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakurada K, Ohta T, Hasegawa M. Cloning, expression and characterization of the Micromonospora viridifaciens neuraminidase gene in Streptomyces lividans. J Bacteriol. 1992;174:6896–6903. doi: 10.1128/jb.174.21.6896-6903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos M A. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 1991;19:5442. doi: 10.1093/nar/19.19.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaufuss P, Lämmler C. Characterization of the extracellular neuraminidase produced by Actinomyces pyogenes. Zentralbl Bakteriol. 1989;271:28–35. doi: 10.1016/s0934-8840(89)80050-7. [DOI] [PubMed] [Google Scholar]
- 48.Schaufuss P, Sting R, Lämmler C. Isolation and characterization of an extracellular protease of Actinomyces pyogenes. Zentralbl Bakteriol. 1989;271:452–459. doi: 10.1016/s0934-8840(89)80104-5. [DOI] [PubMed] [Google Scholar]
- 49.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4083–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serwold-Davis T M, Groman N B. Mapping and cloning of Corynebacterium diphtheriae plasmid pNG2 and characterization of its relatedness to plasmids from skin coryneforms. Antimicrob Agents Chemother. 1986;30:69–72. doi: 10.1128/aac.30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi S, Kaidoh T, Azuma R. Assay of proteases from Actinomyces pyogenes isolated from pigs and cows by zymography. J Vet Med Sci. 1995;57:977–979. doi: 10.1292/jvms.57.977. [DOI] [PubMed] [Google Scholar]
- 52.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 53.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timoney J F, Gillespie J H, Scott F W, Barlough J E. Hagan and Bruner's microbiology and infectious diseases of domestic animals. 8th ed. Ithaca, N.Y: Cornell University Press; 1988. [Google Scholar]
- 55.Tong H H, Blue L E, James M A, DeMaria T F. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–924. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong H H, McIver M A, Fisher L M, DeMaria T F. Effect of lacto-N-neotetraose, asialoganglioside-GM1 and neuraminidase on adherence of otitis media-associated serotypes of Streptococcus pneumoniae to chinchilla tracheal epithelium. Microb Pathog. 1999;26:111–119. doi: 10.1006/mpat.1998.0257. [DOI] [PubMed] [Google Scholar]
- 57.Turnquist S E, Fales W H. Disseminated Actinomyces pyogenes infection in a free-ranging white-tailed deer. J Vet Diagn Investig. 1998;10:86–89. doi: 10.1177/104063879801000117. [DOI] [PubMed] [Google Scholar]
- 58.Weidenhaupt M, Rossi P, Beck C, Fischer H M, Hennecke H. Bradyrhizobium japonicum possesses two discrete sets of electron transfer flavoprotein genes: fixA, fixB and etfS, etfL. Arch Microbiol. 1996;165:169–178. doi: 10.1007/BF01692858. [DOI] [PubMed] [Google Scholar]
- 59.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Minton K W, Fleischmann R D, Ketchum K A, Nelson K E, Salzberg S, Smith H O, Venter J C, Fraser C M. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter A J, Comis S D, Osborne M P, Tarlow M J, Stephen J, Andrew P W, Hill J, Mitchell T J. A role for pneumolysin but not neuraminidase in the hearing loss and cochlear damage induced by experimental pneumococcal meningitis in guinea pigs. Infect Immun. 1997;65:4411–4418. doi: 10.1128/iai.65.11.4411-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeung M. Complete nucleotide sequencing of the Actinomyces viscosus T14V sialidase gene: presence of a conserved repeating sequence among strains of Actinomyces spp. Infect Immun. 1993;61:109–116. doi: 10.1128/iai.61.1.109-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]