Abstract
Response rates to immunotherapy in solid tumors remain low due in part to the elevated prevalence of terminally exhausted T cells, a hypofunctional differentiation state induced through persistent antigen and stress signaling. However, the mechanisms promoting progression to terminal exhaustion in the tumor remain undefined. Using the low-input chromatin immunoprecipitation sequencing method CUT&RUN, we profiled the histone modification landscape of tumor-infiltrating CD8+ T cells throughout differentiation. We found that terminally exhausted T cells had unexpected chromatin features that limit their transcriptional potential. Terminally exhausted T cells had a substantial fraction of active chromatin, including active enhancers enriched for bZIP/AP-1 transcription factor motifs that lacked correlated gene expression, which was restored by immunotherapeutic costimulatory signaling. Reduced transcriptional potential was also driven by an increase in histone bivalency, which we linked directly to hypoxia exposure. Enforced expression of the hypoxia-insensitive histone demethylase Kdm6b was sufficient to overcome hypoxia, increase function, and promote antitumor immunity. Our study reveals the specific epigenetic changes mediated by histone modifications during T cell differentiation that support exhaustion in cancer, highlighting that their altered function is driven by improper costimulatory signals and environmental factors. These data suggest that even terminally exhausted T cells may remain competent for transcription in settings of increased costimulatory signaling and reduced hypoxia.
INTRODUCTION
Immunotherapy such as blockade of “checkpoint” molecules like cytotoxic T lymphocyte associated protein 4 (CTLA4) and programmed cell death protein 1 (PD1) for cancer aims to reinvigorate or augment the antitumor immune response (1). In many patients, T cells fail to control cancer due to their differentiation into an exhausted state, leading to limited proliferative and killing capacity, metabolic dysfunction, and lower cytokine expression (2). Recent studies found that a less differentiated progenitor pool of exhausted CD8+ T cells have increases in proliferation and cytotoxic capacity upon PD1 blockade, whereas terminally differentiated exhausted T cells remain poorly responsive (3, 4). The prevalence of exhausted T cells in tumors correlates with resistance to PD1 blockade (5, 6). It remains critical to understand the underlying processes of how T cells progress to terminal exhaustion to design strategies that specifically target these cells and restore immune function.
The transcriptome and epigenome of exhausted T cells suggest a distinct differentiation trajectory from effector and memory T cells (5, 7, 8). This is driven by the convergence of various chronic signals, including the antigen receptor and metabolic stress responses. For both chronic infection and cancer, differentiation to exhaustion is progressive, defined by at least two transcriptionally and functionally distinct states: one progenitor or stem-like and another terminally differentiated. Progenitor cells have increased proliferative potential, effector cytokine production, and metabolic capacity and are identified by low expression of PD1, elevated expression of Slamf6/Ly108, and the transcription factor Tcf1. Elevated and sustained PD1 expression, concomitant with multiple inhibitory receptors (like Tim3, Tigit, and CD39), and the transcription factor Tox define terminally exhausted T cells (5, 9–12). These states also have distinct open chromatin landscapes as measured by assay for transposase-accessible chromatin using sequencing (ATAC-seq) (5).
The chromatin states controlling transcription are complex. Understanding how histone modifications contribute to gene regulation is an active area of research, but several well-described modifications provide a more nuanced view of epigenomic relationships that regulate the transcriptome of cellular states (13, 14). Advances in low cell input assays have overcome technical hurdles assessing the epigenetic landscape of tumor-infiltrating immune cells by histone modifications rather than transposase accessibility. Using cleavage under targets and release under nuclease (CUT&RUN) and cleavage under targets and tagmentation (CUT&TAG), we determined the epigenetic landscape of tumor-infiltrating T cells progressing to terminal exhaustion by interrogating four well-d escribed histone modifications. We also defined the relationship of Tox with the epigenetic landscape and transcriptome (15, 16). We identified unexpected epigenetic features in terminally exhausted CD8+ T cells: an active chromatin landscape where a substantial fraction of genes does not correlate with active transcription and an increased fraction of bivalent chromatin. In addition, we reveal the relationship of the epigenome with the transcriptome, finding that impaired costimulatory signaling and exposure to hypoxia in the tumor microenvironment contribute to altered gene regulation. Our study lays the groundwork for future studies of how the dysregulated epigenome of exhausted T cells may be targeted to repurpose these cells therapeutically.
RESULTS
Progenitor and terminally exhausted tumor-infiltrating T cells have distinct chromatin features
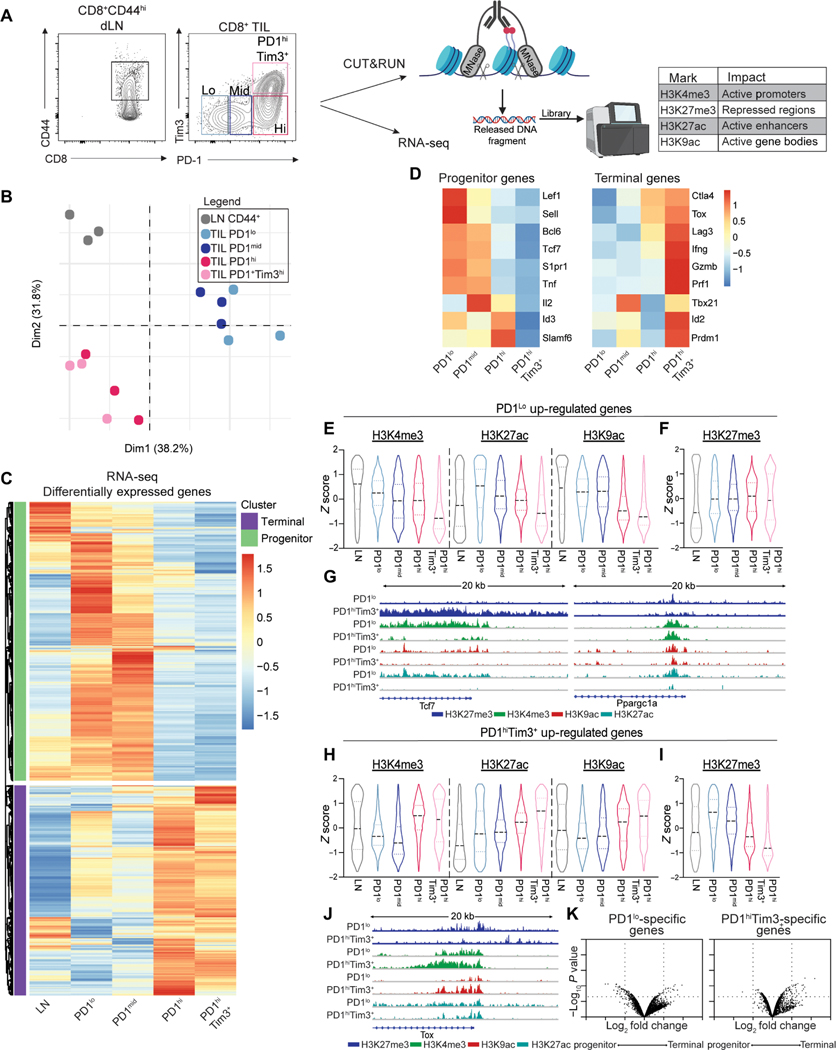
Single-cell RNA sequencing and ATAC-seq demonstrated that progenitor and terminally exhausted states have distinct transcriptomes and chromatin accessibility in both chronic viral infection and tumors, yet how histone modifications contribute to the transition from progenitor to terminal exhaustion has not been explored (5, 7, 8, 17, 18). We used B16F10 melanoma (B16), an aggressive murine tumor cell line that predictably promotes T cell exhaustion and is insensitive to PD1 blockade, to better understand the relationship between the epigenome and transcriptome of CD8+ tumor-infiltrating lymphocytes (TILs) as they differentiated from a progenitor state (PD1loTim3−) to a terminally exhausted state (PD1hiTim3+) (19). CD8+ T cells isolated from B16 tumors were sorted into four populations based on expression of PD1 and Tim3 (PD1lo, PD1mid, PD1hi, and PD1hiTim3+). Antigen-experienced CD8+ T cells (CD44hi) from paired draining lymph nodes (dLNs) were isolated as controls (Fig. 1A). Transcriptome analysis confirmed the distinction between TIL subsets and activated CD44hiCD8+ T cells in the dLN (Fig. 1B). PD1lo and PD1mid TILs were closely related, whereas PD1hi and PD1hiTim3+ cells were more similar, suggesting a transition between PD1mid to PD1hi cells discriminating progenitor and terminally exhausted states (Fig. 1B). Analysis of differentially expressed genes (DEGs) revealed similarity of gene expression among PD1lo and PD1mid T cells, and CD44hi cells from the dLNs and a separation from PD1hi and PD1hiTim3+ cells (Fig. 1C). DEG in PD1lo cells confirmed a signature similar to that described for progenitor exhausted cells (Slamf6, Tcf7, Bcl6, Id3, Lef1, and Tnf), whereas PD1hiTim3+ cells had a signature consistent with terminal exhaustion (Lag3, Tox, Ifng, Prdm1, and Id2) (Fig. 1D). Pearson correlation with published RNA-seq of progenitor and terminal cells isolated from tumors demonstrated that PD1lo cells were similar to Slamf6+ progenitor cells, whereas PD1hiTim3+ were similar to Slamf6−Tim3+ terminally exhausted cells (fig. S1A) (5). Thus, PD1 and Tim3 expression identified progenitor and terminal states of exhaustion that transition from the PD1mid to the PD1hi stage.
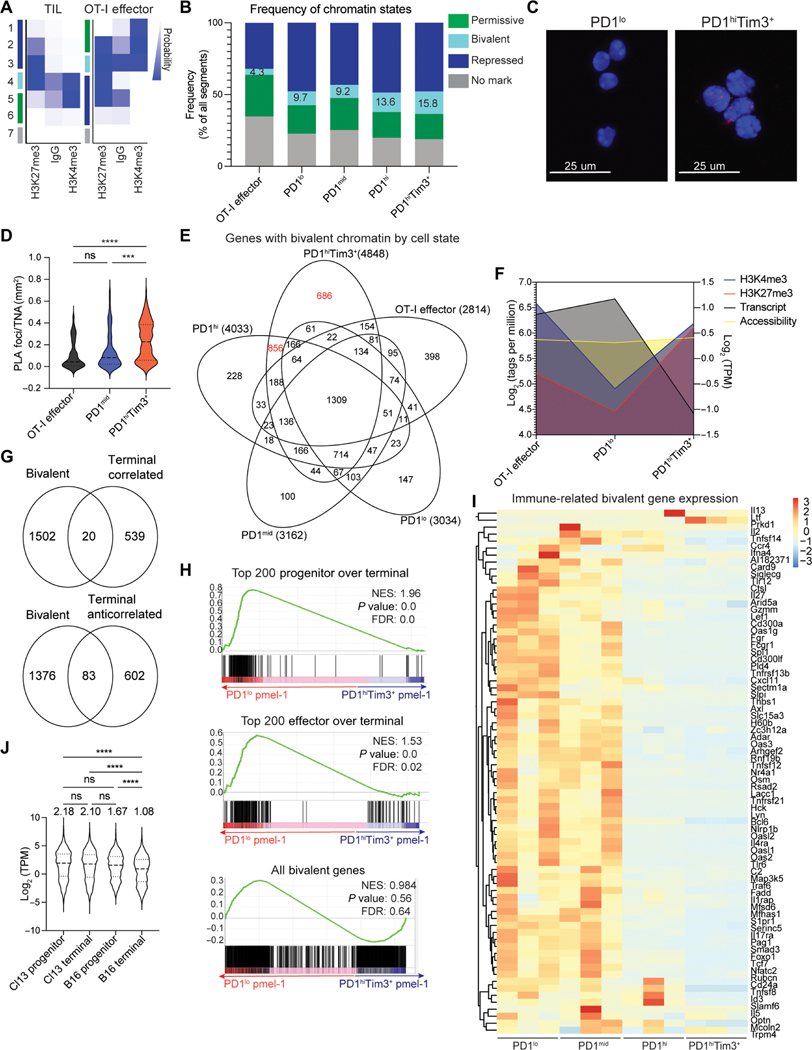
Fig. 1. Transcriptional signatures of exhausted T cells are set by histone landscapes.
(A) Sort strategy of CD44hi CD8+ T cells from the dLNs and B16 melanoma tumors for RNA-seq and CUT&RUN analysis. TILs sorted by PD1 and Tim3. (B) Principal components analysis of transcriptomes of LN CD44hi CD8+ T cells and TIL subsets. (C) DESeq2 of transformed log2 (TPM) normalized transcript expression of DEG between LN CD44hi CD8+ T cells and TIL subsets. (D) Select DEGs associated with progenitor and terminal exhausted T cells. (E and F) H3K4me3, H3K27ac, H3K9ac (E), and H3K27me3 (F) coverage of genes up-regulated in PD1lo cells. Z score of tag counts per million (CPM) of 10 kb regions surrounding the TSS. (G) Genome browser views of Tcf7 and Ppargc1a in indicated subsets. (H and I) H3K4me3, H3K27ac, H3K9ac (H), and H3K27me3 (I) coverage of genes up-regulated in PD1hi Tim3+ cells. Z score of CPM of 10-kb regions surrounding the TSS. (J) Genome browser views of Tox. (K) Fold change and P value of changes in chromatin accessibility determined by ATAC-seq (GSE122713) in progenitor and terminally exhausted T cells from B16 melanoma at genes specific to PD1lo and PD1hiTim3+ CD8+ TILs (log2 fold change >1 and log10 P value <0.05). Coverage determined for 10-kb regions surrounding the TSS. RNA-seq generated from three individual mice treated as biological replicates. CUT&RUN for each mark was performed twice with 8 to 10 mice pooled per experiment.
Next, we used CUT&RUN to generate a foundational map of chromatin states to assess active chromatin (H3K4me3, H3K9ac, and H3K27ac) and repressed chromatin (H3K27me3) (Fig. 1A). To determine whether chromatin states that underlie the transcriptomes were consistent with gene expression, 20-kb regions of chromatin around the transcription start site (TSS) of DEG up-regulated in PD1lo progenitor cells or PD1hi Tim3+ terminal cells were assessed to include proximal and more distal elements regulating gene expression. Genes up-regulated in PD1lo cells displayed an increased amount of active chromatin (H3K4me3, H3K27ac, and H3K9ac) compared with terminally exhausted cells (PD1hiTim3+) and had less repression (H3K27me3) (Fig. 1, E and F). Terminally exhausted cells (PD1hiTim3+) exhibited a bimodal distribution of H3K27me3 at progenitor-specific genes (Fig. 1F). In terminally exhausted cells, some progenitor-specific genes had increased H3K27me3 (i.e.,Tcf7), indicative of chromatin repression as cells differentiated to a terminal state. Yet some genes remained low for H3K27me3 (i.e., Ppargc1a), suggesting that loss of active histone modifications may be sufficient to down-regulate gene expression (Fig. 1, F and G).
A similar pattern was observed for genes specific to terminal exhaustion with increased active chromatin (H3K4me3, H3K27ac, and H3K9ac) at genes up-regulated in PD1hiTim3+ (Fig. 1H). In contrast to bimodal distribution for progenitor-specific genes, H3K27me3 at terminal specific genes was considerably increased in PD1lo cells and reduced in PD1hiTim3+. These data suggest that progression to terminal exhaustion requires H3K27 demethylation for genes to become up-regulated (Fig. 1I). For example, at the Tox locus, H3K27me3 is reduced in PD1hiTim3+ cells, whereas active marks are increased (Fig. 1J). We next determined whether changes in chromatin accessibility measured by ATAC-seq in publicly available datasets had similar alterations when looking at DEGs (5). Unexpectedly, we found limited changes in accessibility, suggesting that changes in histone modifications provide a more nuanced view of epigenomic control of transcriptional programs for states of exhaustion (Fig. 1K). These data suggest that gene expression alterations during progression to exhaustion are due to epigenetic changes regulating transcriptional control of gene expression.
We next compared TIL subsets with “bona fide” effector CD8+ T cells generated using antigen-specific effector CD8+ T cells (OT-I T cells) responding to acute viral infection, Vaccinia virus expressing ovalbumin (Vaccinia-OVA) (fig. S1B). TILs segregated away from effector cells, whereas progenitor and terminally exhausted T cells separated from each other (fig. S1C). DEG identified transcriptional signatures specific to Vaccinia-OVA effector T cells, TIL progenitor, or TIL terminal exhaustion states. However, some genes highly expressed by effector cells had low to moderate expression in progenitor cells (fig. S1D). Chromatin states in effector cells tracked with gene expression (fig. S1E). Both H3K4me3 and H3K27me3 were increased in terminally exhausted cells at effector gene loci (fig. S1E). The epigenetic state of effector cells at progenitor- and terminal-specific genes also had a mixed profile. H3K4me3 was decreased at progenitor genes, with bimodal distribution at terminal specific genes, suggesting that some genes associated with terminal exhaustion may have active chromatin in effector cells (fig. S1, F and G). We compared our CUT&RUN datasets with previously published chromatin immunoprecipitation sequencing (ChIP-seq) analysis of naïve, effector, and memory CD8+ T cells (20). In line with our above analysis, H3K4me3 in effector CD8+ T cells had similarities to terminal exhaustion, whereas naïve, memory, and memory precursor cells had distinct profiles of active promoters (fig. S1H). In contrast, active enhancers marked by H3K27ac showed little correlation between TIL subsets and effector, memory, or naïve cells (fig. S1I). Repressed chromatin had similarities between memory and progenitor exhausted cells, suggesting a shared landscape of repressed genes (fig. S1J). Thus, states of exhaustion in TILs are distinct from naïve, effector, and memory CD8+ T cells responding to infection at both the transcriptome and epigenome.
Terminally exhausted T cells are characterized by regions of active chromatin with low correlative transcription
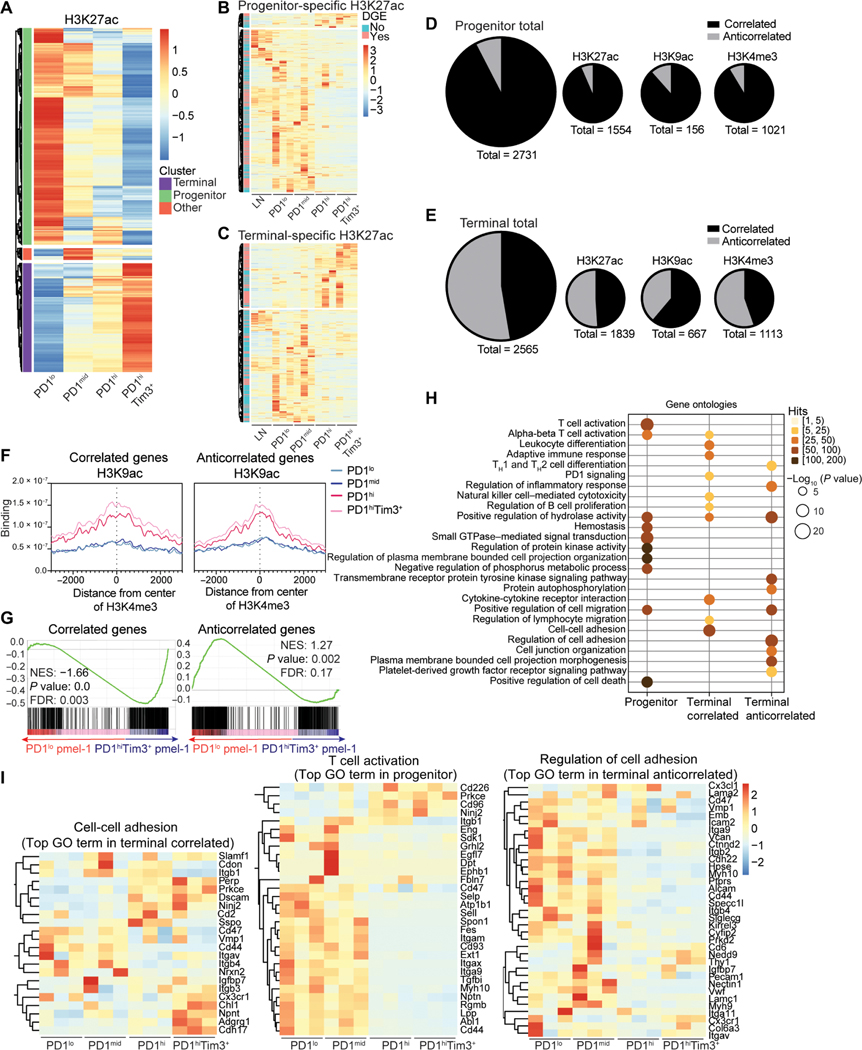
H3K4me3, H3K27ac, and H3K9ac mark active chromatin permissive for transcription (13). To identify active enhancers specific to progenitor or terminal states, we performed differentially enriched peak (DEP) analysis of H3K27ac across TIL subsets (Fig. 2A). Enhancers promote gene expression; thus, we interrogated transcription of genes found closest to enhancers specific for each population of cells. In progenitor cells, the majority of active enhancers (~85%) correlated with increased gene expression that decreased as cells differentiated to terminal exhaustion (Fig. 2, B and D). In contrast, in terminally exhausted T cells, only ~55% of active enhancers correlated with gene expression; an extensive fraction (~45%) had anticorrelative gene expression (Fig. 2, C and E). Although distal enhancers may account for a fraction of these, it is unlikely that enhancer distance would be vastly different in TIL populations (Fig. 2, D and E). Genes associated with effector T cells were among both correlated enhancers (Havcr2, Prdm1, and Ifng) and anticorrelated enhancers (Il10, Runx2, and Irf8) (fig. S2). Enhancer activity correlated with other marks of active chromatin (H3K9ac for active gene transcription and H3K4me3 for active promoters) (fig. S2). We therefore explored how H3K9ac and H3K4me3 related to gene expression in TIL populations (fig. S3, A and B). Similar to active enhancers, progenitor-specific H3K4me3 and H3K9ac primarily correlated with gene expression, whereas terminal-specific H3K4me3 and H3K9ac had a substantial fraction that was not correlated (Fig. 2, D and E, and fig. S3, C to F). In contrast, effector Vaccinia-OVA–elicited OT-I T cells primarily had correlative gene expression at active promoters, suggesting that the presence of active chromatin with anticorrelative gene expression was specific to terminal exhaustion (fig. S3, G to I). We next determined whether the combined presence of multiple active chromatin marks was key to gene expression. Unexpectedly, we found that similar amounts of H3K9ac and H3K27ac were associated with H3K4me3-specific peaks in both correlated genes and anticorrelated genes (Fig. 2F and fig. S3J). Thus, accumulation of active chromatin marks alone is insufficient to predict gene expression in terminally exhausted TILs.
Fig. 2. Active chromatin landscapes in terminally exhausted T cells do not correlate with gene expression.
(A) Tag counts (normalized per million) of H3K27ac at differential peaks between TIL subsets (DESeq2). (B and C) Normalized expression of genes nearest to progenitor-specific (B) and terminal-specific (C) H3K27ac peaks. Genes that meet DEGs cutoffs (fold change > 2, P < 0.05) in pink to the left of heatmap. (D and E) Number of genes with and without corresponding expression in progenitor (D) and terminally exhausted T cells for (E) H3K27ac, H3K9ac, H3K4me3, and the total of all three. (F) H3K4me3 coverage at correlated and anticorrelated H3K9ac peaks. (G) Gene set enrichment analysis of tumor-specific pmel-1 progenitor and terminally exhausted T cell transcriptomes for correlated or anticorrelated genes defined in (E). (H) Enrichment of GO terms via Metascape in genes with active chromatin landscapes in indicated groups. (I) Log2-normalized expression of select genes enriched in GO pathways in (H). FDR, false discovery rate; NES, normalized enrichment score.
Although expression of exhaustion-associated molecules PD1 and Tim3 reliably identify tumor antigen–specific CD8+ T cells, sorted CD44hiPD1lo T cells may be contaminated by so-called “bystander” T cells that display an activated cell signature without T cell receptor (TCR) engagement in tumor (21). To determine whether correlated and anticorrelated gene expression were preserved in tumor-specific T cells, we performed RNA-seq on adoptively transferred TCR-Tg pmel-1 T cells specific for the melanoma antigen gp100. By day 12, most transferred pmel-1 T cells display high PD1 and Tim3 expression. Thus, we staggered transfers of pmel-1 T cells 12 and 7 days before sacrifice and used congenic Thy1.1 expression to identify differentiated PD1hiTim3+ pmel-1 T cells transferred early in tumor progression and newly infiltrating PD1lo pmel-1 T cells transferred later (fig. S3K). PD1hi Tim3+ pmel-1 T cells exhibit increased expression of correlated genes compared with PD1lo pmel-1 T cells, whereas genes defined as anticorrelated were unchanged and enriched in expression in PD1lo pmel-1 T cells (Fig. 2G and fig. S3L). Thus, the presence of anticorrelated genes is likely not due to a difference in responsiveness to tumor antigens and is a hallmark of terminal exhaustion.
Last, we explored transcriptional pathways associated with progenitor-specific and terminal-specific active chromatin regions, breaking terminal-specific active chromatin into correlative and anticorrelative genes. Gene Ontology (GO) pathway analysis demonstrated both overlapping and distinct pathways to each group of genes, including T cell activation and lymphocyte or leukocyte pathways, PD1 signaling, cell adhesion, and cell migration (Fig. 2H). Genes involved in T cell activation/function were among those specifically expressed in progenitor- and terminal-specific regions of active chromatin (Fig. 2I). Together, these data reveal genes critical for T cell activation and function that were not up-regulated in exhausted T cells despite the presence of multiple active chromatin marks. Thus, terminally exhausted TILs have a chromatin landscape characterized by the presence of active histone modifications but without corresponding active gene transcription.
Anticorrelated enhancers of terminally exhausted T cells are defined by AP-1 binding motifs
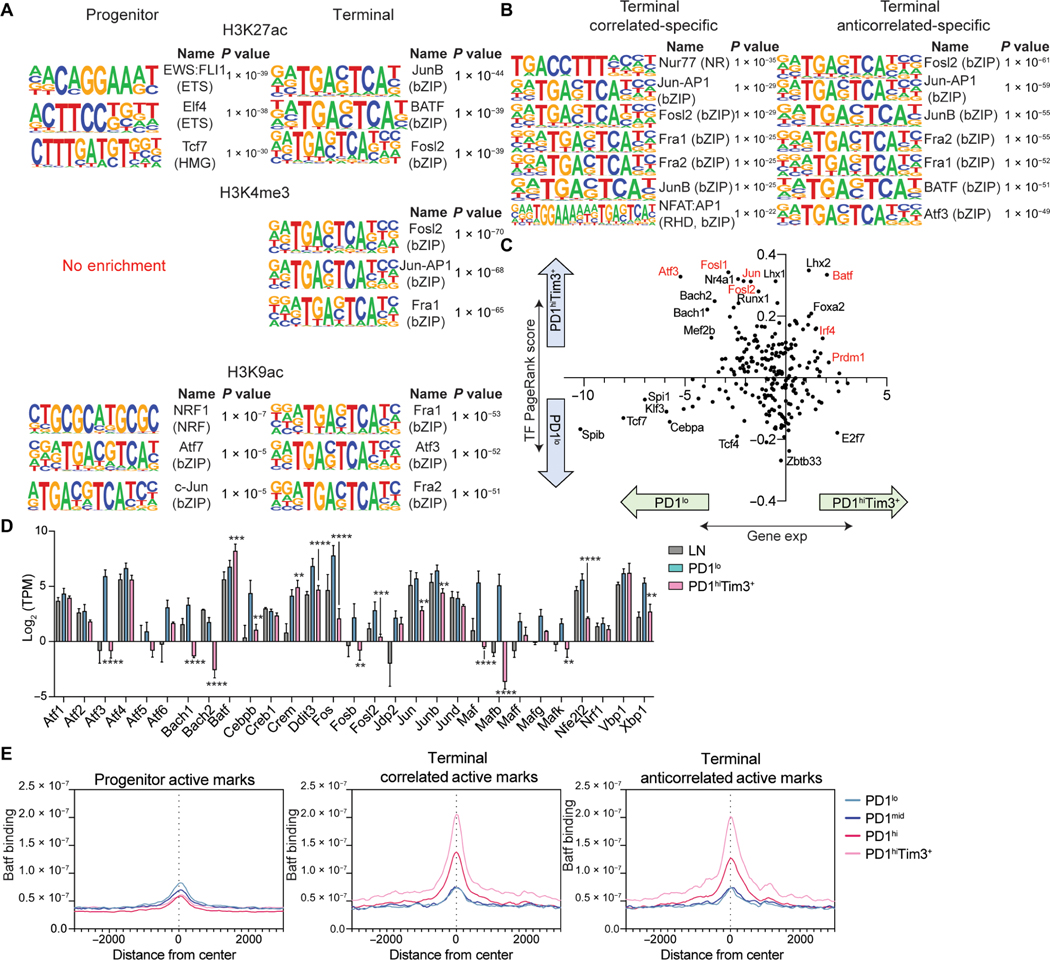
We next sought to understand why a significant fraction of genes in terminally exhausted cells maintain active chromatin but lack gene expression. Increased H3K27me3 might repress these genes; however, levels were similar at both correlated and anticorrelated genes (fig. S3M). Active chromatin is permissive, but transcription factors are required to recruit machinery to drive gene expression (13). Progenitor-specific enhancers were enriched for motifs from the erythroblast transformation specific (ETS) family and TCF1, whereas terminal-specific enhancers had enrichment for the basic leucine zipper domain (bZIP) consensus sequence bound by activator protein 1 (AP-1) family members (Fig. 3A). Terminal-specific H3K4me3 and H3K9ac peaks were also enriched for bZIP motifs, whereas progenitor-specific H3K4me3 and H3K9ac peaks had less specific enrichment of any motifs. bZIPs were enriched in enhancers of both correlated genes and anticorrelated genes; however, two notable motifs were enriched only in correlated genes: Nur77 and the composite nuclear factor of activated T cells (NFAT):AP-1 motif (Fig. 3B). Nur77 (encoded by Nr4a1) activity correlates with recent TCR stimulation, and NFAT:AP-1 complexes occur in response to TCR and costimulatory signaling, suggesting that, consistent with prior work, activating signals promote expression of terminal exhaustion–specific genes (18, 22).
Fig. 3. bZIP/AP-1 family motifs are enriched in active chromatin of terminally exhausted TILs.
(A) Top results from HOMER motif analysis of DESeq2-defined H3K27ac, H3K4me3, and H3K9ac differential peaks in progenitor or terminally exhausted T cells. Terminally exhausted–specific peaks used as background for progenitor exhausted–specific analysis and vice versa. (B) Top results from HOMER motif analysis of correlated and anticorrelated peaks compiled for each mark. Progenitor exhausted–specific peaks used as background for both correlated and anticorrelated analyses. (C) Fold change in PageRank score (y axis) and fold change in gene expression (x axis) for transcription factors. H3K4me3, H3K27ac, gene expression data, and HOMER motif list were provided to Taiji. (D) Expression of AP-1 family members in LN, PD1lo, and PD1hiTim3+ (mean and SD). P value (generated by a Wald test in DESeq2); **P < 0.01, ***P < 0.001, and ****P < 0.0001. (E) Batf coverage (n = 1) at progenitor, correlated, and anticorrelated active peaks defined by presence of H3K27ac, H3K9ac, or H3K4me3.
Exhaustion is driven by continuous antigen receptor stimulation; thus, altered TCR signaling may contribute to the loss of gene expression despite the presence of active chromatin. Chronic antigen stimulation–driven exhaustion also occurs in the context of chronic viral infection; therefore, we sought to determine whether bZIP motifs were enriched in terminally exhausted CD8+ T cells responding to chronic virus. Using previously published ATAC-seq datasets (5) of progenitor and terminal CD8+ T cells isolated from both chronic (clone 13) lymphocytic choriomeningitis virus (LCMV) and B16 tumor models, we identified five clusters that could be distinguished as progenitor specific (cluster 2), LCMV specific (cluster 1), tumor specific (cluster 3), LCMV terminal specific (cluster 5), and tumor terminal specific (cluster 4) (fig. S4A). Motif analysis identified Tcf7 and Lef1 consensus sequences specific to progenitor cells (cluster 2), whereas bZIP motifs were highly enriched in tumor-specific and tumor terminal–specific clusters (clusters 3 and 4) (fig. S4B). In contrast, ETS motifs were enriched in LCMV-specific and LCMV terminal–specific clusters (clusters 1 and 5) (fig. S4B). Thus, enrichment of bZIP/AP-1 motifs in terminal active chromatin regions was specific to tumor-mediated exhaustion.
To further identify transcription factors that play a role in gene regulation, we performed Taiji PageRank analysis, which uses an integrative multiomics framework to identify regulatory networks and candidate driver transcription factors (20, 23). Highly ranked transcription factors in progenitor cells correlated with increased expression of those factors (Fig. 3C). In contrast, many of the transcription factors highly ranked in terminally exhausted cells were decreased in expression, with notable exceptions (Batf, Prdm1, and Irf4) (Fig. 3C). Many of these down-regulated but highly ranked factors were AP-1 family members (Atf3, Jun, and Fosl2). Most bZIP transcription factors were down-regulated in terminal cells, with few exceptions including Batf (Fig. 3D). CUT&RUN revealed that Batf was not bound at sites of active chromatin (H3K27ac, etc.) in progenitor cells but was highly enriched in terminally exhausted cells (Fig. 3E). No differences were noted between correlated genes and anticorrelated genes. These data suggested that the AP-1 family member Batf was bound specifically in terminal cells at places where active chromatin is present.
Tox associates with transcription factors to promote exhaustion-specific transcriptional programs
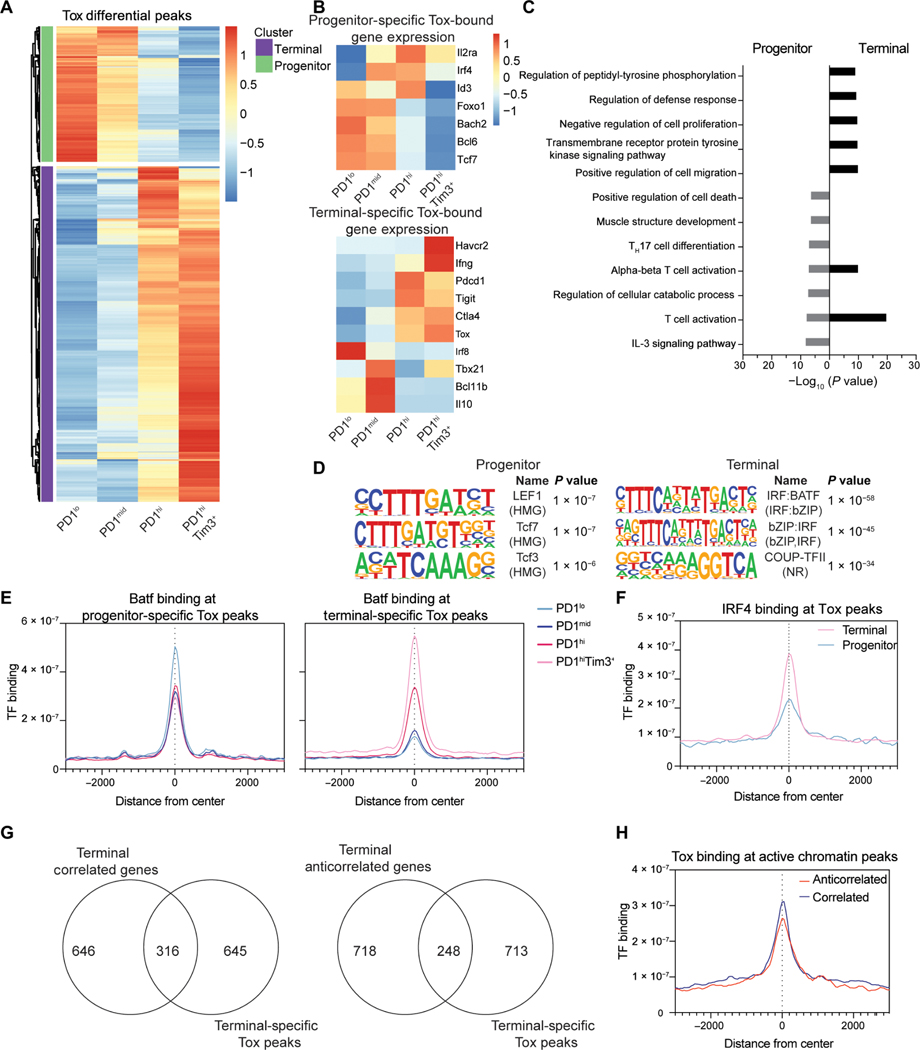
Tox, a transcription factor associated with terminal exhaustion, would not be identified by motif analysis because Tox does not bind a consensus motif but rather to secondary structure (24). To determine the relationship of Tox binding to the epigenetic landscape in TILs, we performed CUT&RUN for Tox. Although Tox increases in expression as cells progress toward terminal exhaustion, Tox is expressed at low levels in progenitor cells (10, 25–28). Therefore, we identified DEP of Tox in both progenitor and terminally exhausted CD8+ T cells (Fig. 4A). In progenitor cells, Tox bound several genes associated with the progenitor program, including Tcf7, Bcl6, Bach2, Foxo1, and Id3 that were mostly associated with increased gene expression, although some were decreased, including Il2ra and Irf4 (Fig. 4B and fig. S4C). Similarly, in terminal cells, Tox bound more than 300 genes, many associated with the transcriptional program of terminal cells, including Havcr2 (encoding Tim3), Tbx21, Ifng, Ctla4, Pdcd1, Tigit, and Tox itself. Again, Tox binding was associated with both expression and repression (Irf8, Bcl11b, and Il10) (Fig. 4B and fig. S4C). Pathway analysis revealed programs in both populations associated with T cell activation, although enrichment was more significant in terminal cells than progenitor cells (Fig. 4C). Motif analysis revealed cell state–specific motifs; in progenitor cells, Tox was associated with Lef1 and Tcf7 motifs, whereas in terminal cells, interferon regulatory factor (IRF):Batf motifs were strongly enriched (Fig. 4D). IRF4, together with Batf, has been associated with promoting T cell exhaustion. We found that Batf binding significantly overlapped Tox in terminal cells (Fig. 4E). Furthermore, IRF4 was more enriched at regions bound by Tox in terminally exhausted cells than in progenitor cells, confirming enrichment of IRF4:Batf motifs (Fig. 4F). Next, we determined the relationship of Tox to active chromatin (H3K4me3, H3K27ac, and H3K9ac) in terminal cells. Tox overlapped with correlated and anticorrelated active marks, although there was slightly more overlap in the correlated group (Fig. 4G). Tox was enriched at sites of active chromatin, and binding was increased at sites correlated with expression compared with anticorrelated genes (Fig. 4H and fig. S4D). These data suggest that Tox binds to active chromatin enriched in IRF:Batf composite motifs and works together with IRF4 and Batf to promote expression at key genes associated with terminal exhaustion. In contrast, in progenitor cells, Tox binds Tcf7 motifs to promote expression of the progenitor transcriptome.
Fig. 4. Tox associates with state-specific transcription factors TCF1 and Batf/IRF4.
(A) Normalized CPM of Tox at differential peaks identified between TIL subsets (DESeq2). (B) Normalized TPM at selected genes associated with progenitor-specific and terminal-specific Tox peaks. (C) −Log10 (P value) values for GO term pathways identified by Metascape enriched for genes with progenitor and terminally exhausted–specific Tox binding. (D) Top results from HOMER motif analysis of DESeq2-defined Tox differential peaks in progenitor or terminally exhausted T cells. Terminally exhausted–specific peaks used as background for progenitor exhausted–specific analysis and vice versa. (E) Batf coverage at progenitor and terminal-specific DESeq2-defined differential Tox peaks. n = 1. (F) IRF4 binding (GSE54191) at progenitor and terminal-specific DESeq2-defined differential Tox peaks. IRF4 ChIP-seq is in vitro–activated T cells (61). (G) Terminal specific peaks of active chromatin (H3K4me3, H3K27Ac, and H3K9Ac) grouped by those with correlated or anticorrelated gene expression associated with terminal-specific Tox peaks. (H) Tox coverage in terminally exhausted cells at active chromatin defined as having anticorrelated and correlated gene expression as in Fig. 2. CUT&RUN for Tox (n = 2) and Batf (n = 1) was performed with 8 to 10 mice pooled per experiment before sorting.
Immunotherapies that alter costimulatory signaling drive expression of anticorrelated genes
Our results suggest that exhausted T cells harbor enhancers defined by AP-1 binding sites not actively promoting transcription. AP-1 is driven by costimulation, and NFAT in the absence of AP-1 drives programs like anergy and exhaustion (22). Exhausted T cells express high levels of both inhibitory and costimulatory receptors (2), most notably 4–1BB (CD137, encoded by Tnfrsf9) (29). Agonistic antibodies to 4–1BB are under intense clinical investigation, and we and others have shown that 4–1BB agonists restore function in exhausted T cells (30). 4–1BB signaling promotes activation of AP-1 family members (31).
Thus, we asked whether checkpoint blockade or costimulatory agonist therapy could restore transcription at the defined anticorrelated loci. B16-bearing mice were treated with either anti-PD1 or anti–4-1BB immunotherapy, and progenitor or terminally exhausted subsets were sorted for RNA-seq (Fig. 5A). Only anti–4-1BB treatment led to increased expression of AP-1 family members, including Atf3, Batf3, and Mafb (Fig. 5B), yet both immunotherapies increased inflammatory response genes (fig. S5, A and B). As expected, progenitor-exhausted cells had no significant enrichment of anticorrelated genes between controls and treated animals, reflecting the limited expression of these genes and the low expression of 4-1BB by progenitor T cells (fig. S5, C and D). Conversely, terminally exhausted cells isolated from anti-PD1–treated mice were enriched in both correlated and anticorrelated genes, indicating increased expression of genes with active chromatin states (Fig. 5C and fig. S5E). 4–1BB–treated exhausted T cells only increased expression of anticorrelated genes, with limited enrichment of correlated genes, suggesting that agonism of 4–1BB increases nuclear AP-1 and restores the expression of genes that retain active enhancers but lack gene expression at the steady state (Fig. 5D and fig. S5F).
Fig. 5. Immunotherapies that alter costimulatory signaling drive expression of anticorrelated genes.
(A) Experimental design of immunotherapy treatments. (B) Log2-normalized expression (mean and SD) of AP-1 family members in terminally exhausted TILs in IgG−, anti–4-1BB−, or anti-PD1–treated mice. P value (generated by a Wald test in DESeq2); ***P < 0.001. (C and D) Gene set enrichment analysis of transcriptomes comparing terminally exhausted (PD1hiTim3+) TILs isolated from tumors in control (IgG) versus PD1-treated (C) and 4-1BB–treated (D) mice. Gene lists of anticorrelated enhancers (left) and correlated enhancers (right) defined as in Fig. 2 (table S1). (E) Terminal anticorrelated genes that change in expression upon treatment. (F) Enrichment of GO terms defined by Metascape in anticorrelated genes modified by treatment with anti-PD1 or anti–4-1BB. RNA-seq data were generated from three individual mice per treatment group sorted into progenitor (PD1lo) or terminal (PD1hiTim3+) CD8+ TILs. GTPase, guanosine triphosphatase; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase.
As both anti-PD1 and anti–4-1BB therapies increased expression of anticorrelated genes, we next asked whether the genes restored contained a core signature. Of the 249 restored genes, only 90 genes were commonly restored by both therapies, whereas 137 or 22 genes were specifically changed by anti-PD1 or anti–4-1BB alone, respectively (Fig. 5E). Pathway analysis of these 90 genes revealed significant enrichment in inflammatory response and leukocyte differentiation pathways, whereas genes affected by anti-PD1 alone were in endocytosis or guanosine triphosphatase pathways, and anti–4-1BB alone regulated responses to glucocorticoids (Fig. 5F). Together, anti–4-1BB or anti-PD1 therapy can restore expression of genes primed by an active chromatin landscape in terminally exhausted T cells, but only anti–4-1BB does so by increasing AP-1 family member expression, suggesting that immunotherapies may be capable of “reinvigorating” some effector function of terminally exhausted cells by restoring gene expression of key inflammatory genes.
Bivalent chromatin is increased in terminally exhausted TILs
Our data were reminiscent of a transcriptionally repressed state known as bivalency. Bivalent chromatin is defined by both active H3K4me3 and repressive H3K27me3 with an absence of gene expression. In T cells, bivalency plays a role in effector CD8+ T cell differentiation, exhibiting bivalent chromatin at effector genes in naïve T cells (32). Bivalency has traditionally been associated with stem cell–like cells, poising genes for rapid expression or repression upon encounter with appropriate environmental stimuli for further differentiation (33, 34). However, recent studies have suggested that bivalent chromatin is also prevalent in terminally differentiated cells and may protect chromatin from excess DNA methylation (35, 36).
We asked whether there were intrinsic differences in bivalent chromatin in progenitor and terminal TILs using ChromHMM, a hidden Markov model developed to identify patterns of chromatin marks (37, 38). To identify chromatin “states,” replicates of TIL samples and OT-I CD8+ effector T cells were merged, and states were identified on the basis of the probability of antibody enrichment of chromatin regions (Fig. 6A). Immunoglobulin G (IgG) was included to eliminate any states enriched as background. An increased probability of IgG binding was typically found in permissive or bivalent chromatin states, in line with nonspecific IgG binding to open chromatin regions. Three states were identified as repressed on the basis of the high probability of binding only H3K27me3. Two permissive states had a high probability of H3K4me3 alone, with state 5 identified as the predominant permissive state. The probabilities of H3K4me3 and H3K27me3 binding were roughly equal in state 4, suggesting bivalency. State 7 had low probabilities for all marks. Seven chromatin states were also identified in OT-I effector cells (Fig. 6A). To determine whether transcription corresponded to the defined states, we assessed gene expression in terminally exhausted and OT-I effector cells using RNA-seq data. Repressive states had reduced gene expression compared with permissive states, and bivalent chromatin had reduced expression compared with permissive chromatin (fig. S6A) (33).
Fig. 6. Terminally exhausted T cells have increased bivalent genes.
(A) ChromHMM emission plots depicting defined states in TILs and Vaccinia-OVA effector cells. (B) Frequency of chromatin states identified in (A) in TIL subsets and Vaccinia-OVA effector cells. (C) Representative images from PLA for H3K4me3 and H3K27me3 in PD1lo and PD1hiTim3+ TILs showing PLA foci (red) and nuclear area (DAPI). (D) Quantification of PLA foci normalized by total nuclear area shown by DAPI. One-way ANOVA, ***P < 0.001 and ****P < 0.0001. (E) Genes identified in each subset as bivalent defined by both H3K4me3 and H3K27me3 in regions ± 1 kb of all TSSs and filtered for expression less than 2 TPM. (F) Summary plot showing changes in H3K4me3, H3K27me3, chromatin accessibility (ATAC-seq, GSE122713), and gene expression of exhaustion-specific bivalent genes identified in (D). Listeria-OVA infection data: GSE95237. (G) Exhaustion-specific bivalent genes compared with correlated and anticorrelated genes defined in Fig. 2. (H) Gene set enrichment analysis of tumor-specific pmel-1 progenitor and terminal exhaustion transcriptomes for top 200 genes that decrease from effector to terminal (top), from progenitor to terminal (middle), or all bivalent genes identified in (E). (I) Normalized expression of select immune-related genes identified as exhaustion-specific bivalent genes. (J) Log2-normalized expression of bivalent genes identified in (E) from progenitor and terminally exhausted cells from chronic LCMV or B16 melanoma tumors (GSE122713). Numbers above indicate mean TPM value. One-way ANOVA, ****P < 0.0001. ns, not significant.
We next compared the frequency of chromatin states among the TIL populations and in effector cells. Whereas all TILs had more bivalent chromatin than effector cells, terminally exhausted cells displayed the highest percentage of bivalent chromatin (Fig. 6B). As CUT&RUN data are from bulk populations of cells, we sought to confirm the presence of both histone marks within close proximity in single cells, indicative of true bivalency and not heterogeneity from cells that were either active or repressed at those loci. Fluorometric proximity ligation assay (PLA) is a highly sensitive and specific single-cell method for identification of two targets in close proximity, such as H3K4me3 or H3K27me3 (39, 40). Consistent with true bivalency, we found a significant increase in PLA foci within PD1hiTim3+ terminal T cells as compared with progenitor and OT-I effector CD8+ T cells (Fig. 6, C and D, and fig. S6, B to D). Thus, terminally exhausted T cells had an increase in bivalent chromatin, which may represent another mechanism by which transcription is controlled as cells become exhausted.
We next explored gene programs associated with bivalent chromatin in each TIL subset. We restricted our analysis to bivalent chromatin encompassing a 1-kb region around the TSS and further removed genes that had high expression, as this would not meet the criteria of bivalency. Terminally exhausted cells had an increased number of genes with bivalent promoter regions [4848; PD1hi Tim3+ (686), PD1hi(856)], compared with effector (398) or progenitor exhausted cells [PD1lo (147), PD1mid (100)] (Fig. 6E and table S1). Key genes such as Klf2, Smad3, and Bach2 showed both H3K4me3 and H3K27me3 peaks in terminal cells, whereas LN and progenitor cells had only H3K4me3 (fig. S6, E to G). Because gene repression can be a function of chromatin accessibility, we used available ATAC-seq data to explore the relationship between open and closed chromatin and bivalency. We found no changes in chromatin accessibility at terminal-specific bivalent regions in OT-I effector, progenitor, or terminal cells despite changes in H3K27me3 and transcript expression (Fig. 6F and fig. S7, A and B). Genes identified as bivalent have little overlap with genes described in terminally exhausted cells as having active chromatin but anticorrelated gene expression, in line with comparable H3K27me3 at these regions (Fig. 6G and fig. S3J). Furthermore, expression of bivalent genes was also decreased in tumor-specific pmel-1 T cells, suggesting that bivalency is not a function of comparing bystander PD1lo progenitor cells with tumor-reactive terminally exhausted cells (Fig. 6H and fig. S7C).
Terminal exhaustion bivalent genes were enriched for pathways involved in inflammatory response and leukocyte differentiation (Fig. 6I and fig. S7D). To determine whether bivalent genes associated with exhaustion were also present in chronic viral infection, we explored their expression in previously published datasets of progenitor and terminally exhausted states isolated from chronic viral infection (LCMV clone 13) or B16 tumors (5). Consistent with our results, expression was significantly decreased in tumor terminal exhaustion. In contrast, no change was observed in chronic viral infection, suggesting that the tumor microenvironment enforces a distinct terminal state in T cells and the increase in bivalent chromatin is specific to tumor-mediated terminal exhaustion (Fig. 6J). Thus, only tumor-mediated terminally exhausted cells have an increase in bivalent chromatin at promoters leading to decreased gene expression.
Tumor hypoxia supports increased bivalency in terminally exhausted cells
Terminally exhausted cells carry severe metabolic defects, demonstrating that exhausted T cell differentiation is the experience of oxidative stress and elevated reactive oxygen species (41). As oxygen is required for the dioxygenase reactions associated with histone and DNA demethylation, and terminally exhausted cells experience elevated hypoxia to progenitors, we asked whether environmental stressors like hypoxia can drive bivalency through alteration of demethylase activity (42, 43). To determine whether hypoxia could promote bivalent chromatin, we explored transcriptome data from our previously published in vitro system that produces exhaustion-like cells via continuous TCR stimulation and culture in hypoxia (41). Culture with continuous stimulation and hypoxia decreased expression of the top 200 bivalent genes in terminally exhausted TILs compared with cells stimulated acutely or continuously in normoxia, suggesting that continuous stimulation was not the main driver of bivalency (fig. S8, A to C). Similar decreases were found when looking at all bivalent genes, several of which are immune related (fig. S8, D and E). These data suggested that hypoxia may induce bivalency in terminally exhausted cells.
We reasoned that mitigating hypoxia in the tumor could reduce bivalent chromatin and restore gene expression. To test this, we used a genetically modified B16 tumor cell line with a deletion of a subunit of mitochondrial complex I (Ndufs4), resulting in a tumor that consumes less oxygen and produces less hypoxia but maintains baseline proliferation (41). Bivalent gene expression was partially recovered in terminally exhausted cells isolated from Ndufs4-deficient B16 (Fig. 7, A and B, and fig. S8F). To determine whether this recovery was due to changes in bivalency, we analyzed the presence of H3K4me3 and H3K27me3 in progenitor and terminally exhausted T cells isolated from Ndufs4-deficient and control B16 tumors. Although no significant changes occurred to H3K4me3, we observed a significant decrease in H3K27me3 at bivalent gene promoters in terminal cells isolated from tumors with less hypoxia (Fig. 7C). Furthermore, we found fewer PLA foci formed by proximity of H3K4me3 and H3K27me3 in cells isolated from tumors with less hypoxia, confirming that decreasing hypoxia was sufficient to decrease bivalent chromatin (Fig. 7, D and E, fig. S8, G to I). Thus, hypoxia promotes bivalent chromatin in tumor-infiltrating terminally exhausted T cells, representing a key microenvironmental signal that alters immune differentiation.
Fig. 7. Hypoxia is sufficient to promote terminal-specific bivalent chromatin and reduce gene expression.
(A) Log2-normalized expression of bivalent genes identified Fig. 6E. P value (generated by a Wald test in DESeq2); **P < 0.01 and ****P < 0.0001. (B) Gene set enrichment analysis of terminally exhausted TIL transcriptomes isolated from Ndufs4-deficient or wild-type (WT) B16 melanoma tumors. Top 200 bivalent genes defined in Fig. 6E that decrease from effector to terminally exhausted cells. (C) Log2-normalized coverage of bivalent genes from CUT&Tag data for H3K4me3 and H3K27me3. (D) Representative images from PLA for H3K4me3 and H3K27me3 in PD1lo and PD1hiTim3+ TILs showing PLA foci (red) and nuclear area (DAPI). (E) Quantification of PLA foci normalized by total nuclear area (TNA) shown by DAPI. One-way ANOVA, **P < 0.01. KO, knockout.
Enforced expression of the H3K27 histone demethylase Kdm6b is sufficient to recover T cell function and antitumor response
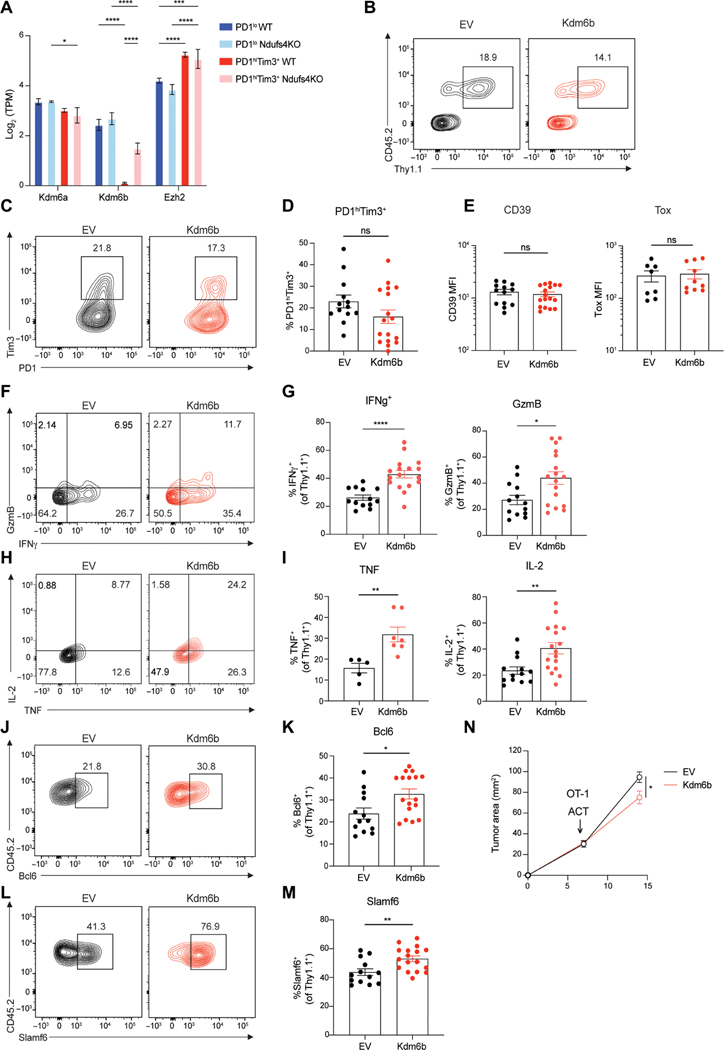
UTX and Jmjd3, encoded by Kdm6a and Kdm6b, are histone lysine demethylases (KDMs) that act on H3K27, whereas Ezh2 methylates H3K27 (44). Kdm6a is a direct oxygen sensor, and hypoxic conditions inhibit KDM activity, either directly or through hypoxia-inducible factor–mediated mechanisms (45). Kdm6b (Jmjd3), in contrast, is thought to be less sensitive to oxygen tension and known to affect effector CD8 T cell function and formation of memory (46). Thus, we hypothesized that altered function or expression of H3K27 histone modifiers may be critical to driving bivalency. We found that Kdm6b, the less oxygen-sensitive KDM, is significantly decreased in a hypoxia-dependent manner in terminally exhausted cells (Fig. 8A). In contrast, Ezh2 was increased in terminally exhausted cells and was not altered by hypoxia, suggesting that increased bivalency may be due to the dysregulation of chromatin mediators of H3K27 methylation (47).
Fig. 8. Kdm6b overexpression in tumor-specific T cells maintains their effector potential and augments the antitumor response.
(A) Log2-normalized gene expression (mean and SD) of histone modifiers that regulate H3K27me3 from subsets of exhausted TILs from WT and Ndufs4-deficient B16 melanoma. P value (generated by a Wald test in DESeq2); *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (B) Representative flow plots of CD45.2 OT-I T cells transduced with retrovirus for empty vector (EV) or Kdm6b. Transduced cells identified by Thy1.1. (C) PD1 and Tim3 expression in EV or Kdm6b expressing OT-I T cells isolated from B16-OVA. (D) Frequency of PD1hiTim3+ cells. (E) Mean fluorescent intensity (MFI) of exhaustion markers CD39 and Tox. (F) Interferon-γ (IFN-γ) and granzyme B (GzmB) staining gated on transduced OT-I T cells restimulated ex vivo with peptide (SIINFEKL). (G) Frequency of IFN-γ+ and GzmB+ cells. (H) Tumor necrosis factor (TNF) and IL-2 staining as in (F) and (G). (I) Frequency of TNF+ and IL-2+ cells. (J and L) for Bcl6 (J) and Slamf6 (L) staining gated on transduced OT-I T cells. (K and M) Frequency of Bcl6+ (K) and Slamf6+ (M) cells. (N) Tumor growth between adoptive transfer of OT-I–transduced T cells at day 7 and termination of experiment at day 14. Flow cytometry plots are representative of ≥3 experiments. ACT, adoptive cell therapy. Statistics are Mann-Whitney (D, E, G, I, K, and M) and two-way ANOVA (N) with *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Because Kdm6a is directly inhibited in settings of low oxygen, we hypothesized that restoring histone demethylase activity via enforced expression of Kdm6b, its less oxygen-sensitive counterpart, may limit bivalency and restore function to terminally exhausted cells. We overexpressed Kdm6b in CD45.2 TCR transgenic (OT-I) CD8+ T cells via retroviral transduction (Kdm6b-T2A-Thy1.1) (Fig. 8B and fig. S8J) then adoptively transferred these cells into CD45.1 recipients bearing palpable B16 tumors expressing ovalbumin (B16-OVA) (fig. S8K). Kdm6b overexpressing T cells expressed similar levels of the canonical exhaustion markers PD1, Tim3, CD39, and Tox compared with controls, suggesting that the core differentiation program to exhaustion, likely driven by antigen, was unchanged (Fig. 8, C to E). However, Kdm6b-overexpressing T cells displayed markedly increased cytokine production when restimulated with peptide, indicating that although they resembled exhausted T cells, they carried greater effector potential (Fig. 8, F to I). Kdm6b overexpression led to increased production of interleukin-2 (IL-2), a bivalent gene critical for T cell longevity and function (Fig. 8, H and I), as well as other bivalent genes, including Bcl6 and Slamf6, associated with stemness and memory (Fig. 8, J to M). Tumor growth was limited in mice receiving Kdm6b-overexpressing OT-I T cells, suggesting that these cells carried greater antitumor activity in vivo (Fig. 8N and fig. S8, L and M). Thus, maintenance of demethylase activity via Kdm6b overexpression limits the acquisition of a dysfunctional phenotype as T cells differentiate in response to persistent antigen, although these cells express canonical markers of exhaustion.
Our data support a model in which the hypoxic environment of solid tumors dysregulates the balance of H3K27 methylation, leading to bivalency at genes necessary for a robust effector response, whereas inhibitory receptor signaling reduces transcription by down-regulation of AP-1 family members needed for active chromatin at effector genes. Collectively, our data reveal the environmental burden of the tumor microenvironment upon infiltrating T cells and highlight how immunomodulatory therapy affecting transcription factor activity may synergize with hypoxia-mitigating regiments to support effective antitumor immune responses for cancer therapy.
DISCUSSION
Immunotherapies are limited by inadequate understanding of the transition from progenitor T cells to terminal exhaustion. How tumor microenvironments affect this progression at the epigenetic level is not known, and whether terminally exhausted T cells have therapeutic potential to gain effector capacity is unclear. By profiling histone modifications and transcriptomes of tumor-infiltrating CD8+ T cells as they progressed to exhaustion, we uncovered chromatin states with transcriptional potential that were limited by signals specific to the tumor microenvironment. When exploring regions of active chromatin that were unique to terminally exhausted cells, we found a large percentage of genes with active chromatin yet reduced gene expression. Although distal enhancers could account for this observation, it would be unexpected that terminally exhausted cells use more distal enhancers than progenitors, suggesting a more likely hypothesis that a loss of enhancer-promoter contacts occurs as cells progress to exhaustion. Identifying specific enhancer-promoter relationships requires chromatin conformation assays, technologies that are now limited by cell number or by the resolution of the chromatin space. Active chromatin in terminally exhausted T cells was highly enriched for bZIP/AP-1 family motifs, yet very few AP-1 family members were expressed. The AP-1 family selects cell type–specific enhancers in macrophages and fibroblasts (48, 49). Thus, loss of AP-1 may be sufficient to limit enhancer-promoter contacts. NFAT rendered incapable of binding AP-1 increases exhaustion; thus, the balance of NFAT downstream of TCR and AP-1 downstream of costimulation plays a key part in the progression to exhaustion (22, 50). We hypothesize that sustained costimulation and AP-1 expression drive productive enhancer-looping to promote gene expression; however, more studies are needed to formally demonstrate this link.
Immunotherapies that directly agonize costimulatory molecules alter AP-1. We reasoned that this could restore gene expression with active chromatin landscapes. 4–1BB agonism specifically altered anticorrelated genes, whereas PD1 blockade increased all genes in terminal cells. One challenge in comparing these two treatments is the difference in cellular targets. PD1 primarily targets progenitor cells and allows progression to a more terminal state while effector function is maintained, whereas 4–1BB agonist acts on terminally exhausted T cells that exhibit elevated 4–1BB expression (30). We sorted progenitor and terminally exhausted cells separately to account for changes to the cellular phenotype but were unable to parse these differences in a refined manner. Although both treatments affected anticorrelated genes, the precise genes within that group were not identical, suggesting that PD1 and 4–1BB therapies have different signaling pathways, leading to increased effector cell programs.
Our analysis suggests that Tox has roles in both progenitor and terminally exhausted states (10, 25, 26). Tox is an HMG protein that binds DNA in a sequence-independent manner and regulates chromatin architecture, suggesting that Tox may play a role in regulating enhancer-promoter looping (24). We found Tox pairs with distinct binding partners in each subset, TCF-1 in progenitor and Batf:IRF [AP-1–interferon regulatory factor (IRF) composite element (AICE)] in terminally exhausted cells (51). TCF-1 can act as a pioneer factor, binding to regions of heterochromatin to promote chromatin accessibility (52). However, intrinsic histone deacetylase activity and repressive function has also been associated with TCF-1, suggesting that either the binding partners or the chromatin landscape affects the precise role of TCF-1 (53). Both Batf and IRF4 are key transcription factors regulating exhaustion (12, 54, 55). Batf:IRF4 complexes bind to AICE to drive differentiation of TH17 cells, B cells, and dendritic cells (51). We found that terminal exhaustion ranked both Batf and Irf4 as important and positively correlated with expression, unlike AP-1 family members. Our data implicate a complex of Batf, Irf4, and Tox that is responsible for regulating gene expression in terminally exhausted cells.
Bivalent chromatin is defined as the presence of both positive (H3K4me3) and negative (H3K27me3) histone modifications at the same nucleosome within promoter regions. Primarily found in pluripotent cells and associated with genes that regulate cell fate decisions, bivalent genes are poised for rapid induction or repression upon encounter with appropriate differentiation signals (33, 56). Recent studies have challenged this view, identifying bivalent chromatin in terminally differentiated cells and demonstrating protection from DNA methylation (35, 36). We identified an increase in bivalent chromatin in terminally exhausted cells using two methods: CUT&RUN with ChromHMM and at the single-cell level using PLA. We hypothesize that these are not cell fate genes ready for rapid expression but rather aberrant bivalency due to a loss of demethylase activity. Hypoxia is associated with increased bivalency, and terminally exhausted cells experience the most hypoxia (41, 45, 57). Changing hypoxia both in vitro and in vivo could alter expression of bivalent genes, and decreasing hypoxia supported H3K27me3. In support of this, overexpression of the hypoxia-insensitive Kdm6b drove up-regulation of bivalent genes, including IL-2, Bcl6, and Slamf6, and was sufficient to increase the function of terminally exhausted TILs, leading to improved antitumor immunity. However, we did not observe decreased expression of inhibitory receptors or Tox, suggesting that improved effector functionality may be further bolstered with additional checkpoint blockade. Studies exploring DNA methylation via Dnmt3a found that exhausted cells had increased de novo DNA methylation at effector genes, which could be relieved by loss of Dnmt3a or DNA-demethylating agents in combination with anti-PD1 therapy (58). As there is interest in combining therapies targeting the epigenome, including Dnmt3a and Ezh2, in combination with checkpoint blockade, our study underscores the need for further exploration of the molecular players driving alterations such as bivalency to understand how these therapeutic strategies may alter tumor immunity (58, 59).
Although CUT&TAG provided a technical advantage for analysis of histone modifications in small cell numbers, it is a bulk method, and thus, we are unable to resolve the level of heterogeneity at the single-cell level. Although single-cell methods may resolve some questions, there are limits of detection at the individual histone that are challenging to overcome with current technologies. An additional caveat of our study is that we limited analysis to B16 melanoma for practical reasons; however, although the precise chromatin states may vary somewhat between tumor lines, it is likely that our findings would be consistent for those that are highly infiltrated by immune cells and exhibit hypoxic niches.
Our data highlight the convergence of both altered immunologic signals and pathologic environmental signals that redirect differentiation to exhaustion and suggest that exhausted T cells remain in their dysfunctional state due to the “memory” of these previous stressors. However, our data collectively reveal a primed chromatin state in terminally exhausted T cells that may be available for reinvigoration given the right therapeutic inputs, and our study may be of use for developing combination therapies that take full advantage of all subsets of tumor-infiltrating T cells to eradicate cancer cells.
MATERIALS AND METHODS
Study design
Our research objective was to investigate the epigenetic and transcriptomic landscape of tumor-derived CD8+ T cells. Study design included controlled and observational laboratory experiments. Sample sizes for sequencing studies pooled animals to collect sufficient cell numbers and were repeated two to three times to generate biological replicates. All data were included, and outliers were only removed if there was sufficient evidence of contamination or technical flaws in the experimental process. All animal studies, including tumor endpoints, were approved under Institutional Animal Care and Use Committee protocol 20077737. Analysis of initial datasets were used to form hypotheses on the relationship of hypoxia and costimulatory signaling.
Tumor experiments
Mice were injected intradermally with 2.5 × 105 B16-F10 cells [B16; American Type Culture Collection (ATCC)], B16-OVA (MO5; from P. Basse and L. Falo at the University of Pittsburgh), or B16-Ndufs4KO (41). TILs and dLNs were harvested on day 14, when tumors were typically 8 to 10 mm in diameter. In immunotherapy experiments, B16 was injected intradermally into mice. On day 10 (5- to 6-mm average diameter), mice were treated with 200 μg of anti- PD1 (J43; BioXCell), 200 μg of anti–4-1BB (3H3; BioXCell), or equivalent isotype control and then a second dose after 48 hours. Tumors were isolated for TIL isolation 24 hours after the second dose.
Proximity ligation assay
Sorted T cells were attached to slides using an Epredia Cytospin Centrifuge for 10 min at 2000 rpm, 4°C, followed by immediate 4% paraformaldehyde fixation and serial permeabilization washes with 0.1% Triton X-100 in 1× phosphate-buffered saline. After blocking with Duolink buffer (Sigma-Aldrich), primary antibodies were directed against H3K4me3 (C36B11; Cell Signaling Technology) and H3K27me3 (ab6002; Abcam) and were added to Duolink antibody diluent and incubated on slides overnight at 4°C. Downstream staining was accomplished in accordance with the manufacturer’s protocol with Duolink In Situ Orange (Sigma-Aldrich). Slides were imaged with a Nikon 100× 1.40 numerical aperture objective on a Nikon Ti inverted microscope and point scanning confocal scan head. Stacks of 500-nm intervals were captured for 4′,6-diamidino-2-phenylindole (DAPI) and Cy3 channels and refined on NIS-Elements to capture the three-dimensional structure and accurately count PLA foci within the cell nucleus.
Kdm6b overexpression studies
Retrovirus for T cell transduction was generated with the Platinum-E (Plat-E) cell line (ATCC) and used Xfect Transfection Reagent (Takara) per the manufacturer’s protocol. Naive OT-I T cells were activated with plate-bound anti-CD3 (5 μg/ml; BioLegend), soluble anti-CD28 (1 μg/ml; BioLegend), and IL-2 (100 U; National Institutes of Health) for 24 hours. Retroviral supernatants were harvested from Plat-E cultures, filtered, and supplemented with polybrene (6 μg/ml) and IL-2 (100 U). Prepared retroviral supernatants were spun onto activated T cells (2200 rpm, 2 hours, 37°C) and then rested at 37°C for an additional 2 hours. T cells were washed thoroughly and cultured in fresh complete RPMI 1640 with IL-2 (50 U) for 3 days to allow for expansion and expression of vector cassette. After 3 days, 5 million T cells (per mouse) were retro-orbitally injected into CD45.1 mice carrying day 7 B16-OVA tumors (~6 × 6 mm2). Transferred T cells were identified by CD45.2 and vector expression by Thy1.1.
CUT&RUN and CUT&TAG assays
CUT&RUN and CUT&TAG assays were performed as previously described (15, 16). For CUT&RUN, live sorted cells were incubated overnight with concanavalin A beads (Bangs Laboratories Inc.) and antibodies recognizing H3K4me3 (Abcam), H3K27me3 (Cell Signaling Technology), H3K27ac (Abcam), H3K9ac (Abcam), Tox (Abcam), Batf (Brookwood Biomedical), or IgG (Cell Signaling Technology). Protein A–micrococcal nuclease (pA-MNase) was then added, followed by CaCl2 to cleave the antibody-bound chromatin. Phenol-chloroform extraction was then performed to isolate enriched DNA. Two replicates were performed for each antibody (except for Batf). Libraries were prepared using NEBNext Multiplex Oligos for Illumina (New England Biolabs) and either the sparQ DNA Library Prep Kit (Quantabio) or the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs). For CUT&TAG, nuclei were extracted from live sorted cells and incubated overnight with concanavalin A beads (Bangs Laboratories Inc.) and antibodies recognizing H3K4me3 (Abcam) and H3K27me3 (Cell Signaling Technology). A goat anti-r abbit secondary antibody (EpiCypher) and pAG-Tn5 (Epicypher) were then added, followed by MgCl2 to tagment the antibody-bound chromatin with Illumina-compatible adapters. After amplification of libraries, DNA was purified using AMPure beads (Beckman Coulter). Libraries were quantified by quantitative polymerase chain reaction (qPCR) using either the sparQ Library Quant Kit (Quantabio) or the NEBNext Library Quant Kit (New England Biolabs) for Illumina. Appropriate library size was confirmed by running amplified qPCR products on an agarose gel. Cluster generation and 75–base pair paired-end, dual-indexed sequencing was performed on an Illumina NextSeq500.
Analysis of chromatin data
Motif enrichment analysis was performed using HOMER findMotifsGenome (60). Differential peak files were used for these analyses to define the specific motifs that were changing. The background file was specified using progenitor as background for terminal analyses and vice versa, unless otherwise specified. All motifs shown are from the known motifs output of findMotifsGenome.
HOMER makeTagDirectory and AnnotatePeaks functions were used to make histograms. Alignment files were used in makeTagDirectories to make tag directories for each replicate. AnnotatePeaks was used with the peak file of interest and the tag directories to generate magnitude data for each replicate at a given point from the center of the peak. These values were normalized using the read count for each replicate and graphed using GraphPad Prism.
PageRank analysis was performed using Taiji (23). The analysis was performed using H3K4me3, transcriptome, and H3K27ac data to rank the transcription factors. The motifs used were from the Homer database. Fold change between progenitor and terminally exhausted PageRank values was then calculated to determine factors important in each context.
Bivalent chromatin was defined using ChromHMM. Bam files with duplicated and blacklisted regions removed were used in the BinarizeBam function with a bin size of 1000. The LearnModel function was then used with seven states defined and a bin size of 1000. Bivalent states were defined using the presence of both H3K27me3 and H3K4me3 and low gene expression.
Statistical analysis
Statistical significance of genomics data was determined using P values given by DESeq2. Any other methods of determining statistical significance are described in the figure legends.
Supplementary Material
Acknowledgments:
We thank S. Henikoff (Fred Hutch Cancer Center) for pA-MNase reagent and S. Hainer (U. Pittsburgh) for pA-MNase plasmid and associated CUT&RUN technical advice. We thank the UPMC Hillman Cancer Center Cytometry Flow core and the UPMC Children’s Hospital of Pittsburgh Flow Core for help with cell sorting and analysis. We thank the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh for Next Generation Sequencing Services and the University of Pittsburgh Center for Research Computing for computing cluster access and support.
Funding:
B.R.F. was supported by T32 CA082084; N.E.S. was supported by the NCI Predoctoral to Postdoctoral Fellow Transition Award (F99/K00) (no. F99CA222711). P.D.A.V. was supported by the National Cancer Institute of the National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award 1F30CA247034-01 and T32CA082084 and the National Institute of General Medical Sciences of the NIH (T32GM008208). R.P. was supported by T32AI089443. A.C.P. was supported by R01AI153104 and R21AI135027. G.M.D. was supported by an NIH New Innovator Award (DP2AI136598) and R21AI135367, the UPMC Hillman Cancer Center Melanoma/Skin Cancer (P50CA121973), Head and Neck Cancer SPORE (P50CA097190), the Alliance for Cancer Gene Therapy, the Mark Foundation for Cancer Research Emerging Leader Award, the Cancer Research Institute Lloyd J. Old STAR Award, and the Sy Holzer Endowed Cancer Immunotherapy Fund. This work used flow cytometry and animal facilities at UPMC Hillman Cancer Center, supported by P30CA047904. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
G.M.D. declares competing financial interests and has submitted patents covering the use of metabolic reprogramming in cell therapies that are licensed or pending and is entitled to a share in net income generated from licensing of these patent rights for commercial development. G.M.D. consults for and/or is on the scientific advisory board of BlueSphere Bio, Century Therapeutics, Nanna Therapeutics, Novasenta, Pieris Pharmaceuticals, and Western Oncolytics/Kalivir; has grants from bluebird bio, Nanna Therapeutics, Novasenta, Pfizer, Pieris Pharmaceuticals, TCR2, and Western Oncolytics/Kalivir; and owns stock in Novasenta.
Footnotes
Competing interests: The other authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The datasets generated during this study are available at NCBI GEO repository GSE175443. Additional datasets not generated by this study are available at NCBI GEO repository under accession numbers GSE155192, GSE123235, GSE123236, GSE95237, and GSE54191. All materials used in this study are available upon request addressed to G.M.D. or A.C.P.
REFERENCES AND NOTES
- 1.Wei SC, Duffy CR, Allison JP, Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Wherry EJ, Kurachi M, Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, Curtis C, Bucktrout SL, Davis MM, Chang ALS, Chang HY, Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui I, Schaeuble K, Chennupati V, Marraco SAF, Calderon-Copete S, Ferreira DP, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, Luther SA, Speiser DE, Held W, Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, Yates KB, Lako A, Felt K, Naik GS, Manos M, Gjini E, Kuchroo JR, Ishizuka JJ, Collier JL, Griffin GK, Maleri S, Comstock DE, Weiss SA, Brown FD, Panda A, Zimmer MD, Manguso RT, Hodi FS, Rodig SJ, Sharpe AH, Haining WN, Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani AC, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane JP, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer-Rachamimov A, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G, Hacohen N, Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 176, 404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, Tonnerre P, Chung RT, Tully DC, Allen TM, Frahm N, Lauer GM, Wherry EJ, Yosef N, Haining WN, The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, Barnitz RA, Bartman C, Bengsch B, Huang AC, Schenkel JM, Vahedi G, Haining WN, Berger SL, Wherry EJ, Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, De Macedo MP, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba II, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA, Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, Utzschneider DT, von Hoesslin M, Cullen JG, Fan Y, Eisenberg V, Wohlleber D, Steiger K, Merkler D, Delorenzi M, Knolle PA, Cohen CJ, Thimme R, Youngblood B, Zehn D, TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin JX, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ, Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. U.S.A 116, 14113–14118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Utzschneider DT, Gabriel SS, Chisanga D, Gloury R, Gubser PM, Vasanthakumar A, Shi W, Kallies A, Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol 21, 1256–1266 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Natoli G, Maintaining cell identity through global control of genomic organization. Immunity 33, 12–24 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Talbert PB, Henikoff S, The Yin and Yang of histone marks in transcription. Annu. Rev. Genomics Hum. Genet 22, 147–170 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Skene PJ, Henikoff JG, Henikoff S, Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kaya-Okur HS, Janssens DH, Henikoff JG, Ahmad K, Henikoff S, Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc 15, 3264–3283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott-Browne JP, Lopez-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM, Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mognol GP, Spreafico R, Wong V, Scott-Browne JP, Togher S, Hoffmann A, Hogan PG, Rao A, Trifari S, Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. U.S.A 114, E2776–E2785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Lee LF, Fisher TS, Jessen B, Elliott M, Evering W, Logronio K, Tu GH, Tsaparikos K, Li X, Wang H, Ying C, Xiong M, VanArsdale T, Lin JC, Combination of 4–1BB agonist and PD-1 antagonist promotes antitumor effector/memory CD8 T cells in a poorly immunogenic tumor model. Cancer Immunol. Res 3, 149–160 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, Pereira RM, Crotty S, Chang JT, Pipkin ME, Wang W, Goldrath AW, Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, Duan K, Ang N, Poidinger M, Lee YY, Larbi A, Khng AJ, Tan E, Fu C, Mathew R, Teo M, Lim WT, Toh CK, Ong BH, Koh T, Hillmer AM, Takano A, Lim TKH, Tan EH, Zhai W, Tan DSW, Tan IB, Newell EW, Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, Togher S, Heissmeyer V, Zhang YC, Crotty S, Lamperti ED, Ansel KM, Mempel TR, Lahdesmaki H, Hogan PG, Rao A, The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity 42, 265–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Wang M, Zhao Y, Wang W, Taiji: System-level identification of key transcription factors reveals transcriptional waves in mouse embryonic development. Sci. Adv 5, eaav3262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Flaherty E, Kaye J, TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics 4, 13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, Werner MT, Huang C, Alexander KA, Wu JE, Attanasio J, Yan P, George SM, Bengsch B, Staupe RP, Donahue G, Xu W, Amaravadi RK, Xu X, Karakousis GC, Mitchell TC, Schuchter LM, Kaye J, Berger SL, Wherry EJ, TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott AC, Dundar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, Trivedi P, Menocal L, Appleby H, Camara S, Zamarin D, Walther T, Snyder A, Femia MR, Comen EA, Wen HY, Hellmann MD, Anandasabapathy N, Liu Y, Altorki NK, Lauer P, Levy O, Glickman MS, Kaye J, Betel D, Philip M, Schietinger A, TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo H, Chen J, Gonzalez-Avalos E, Samaniego-Castruita D, Das A, Wang YH, Lopez-Moyado IF, Georges RO, Zhang W, Onodera A, Wu CJ, Lu LF, Hogan PG, Bhandoola A, Rao A, TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. U.S.A 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurice NJ, Berner J, Taber AK, Zehn D, Prlic M, Inflammatory signals are sufficient to elicit TOX expression in mouse and human CD8+ T cells. JCI Insight 6, e150744 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams JB, Horton BL, Zheng Y, Duan Y, Powell JD, Gajewski TF, The EGR2 targets LAG-3 and 4–1BB describe and regulate dysfunctional antigen-specific CD8+ T cells in the tumor microenvironment. J. Exp. Med 214, 381–400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menk AV, Scharping NE, Rivadeneira DB, Calderon MJ, Watson MJ, Dunstane D, Watkins SC, Delgoffe GM, 4–1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J. Exp. Med 215, 1091–1100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharpe AH, Pauken KE, The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol 18, 153–167 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, Stock AT, Croom HA, Cullen JG, Nguyen ML, Rowe S, Olson MR, Finkelstein DB, Kelso A, Thomas PG, Speed TP, Rao S, Turner SJ, Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8+ T cell differentiation. Immunity 41, 853–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES, A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Blanco E, Gonzalez-Ramirez M, Alcaine-Colet A, Aranda S, Di Croce L, The bivalent genome: Characterization, structure, and regulation. Trends Genet. 36, 118–131 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Kumar D, Cinghu S, Oldfield AJ, Yang P, Jothi R, Decoding the function of bivalent chromatin in development and cancer. Genome Res. 31, 2170–2184 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah RN, Grzybowski AT, Elias J, Chen Z, Hattori T, Lechner CC, Lewis PW, Koide S, Fierz B, Ruthenburg AJ, Re-evaluating the role of nucleosomal bivalency in early development. bioRxiv 2021.2009.2009.458948 (2021).
- 37.Ernst J, Kellis M, ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M, Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hattori N, Niwa T, Kimura K, Helin K, Ushijima T, Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res. 41, 7231–7239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez D, Shankman LS, Nguyen AT, Owens GK, Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods 10, 171–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharping NE, Rivadeneira DB, Menk AV, Vignali PDA, Ford BR, Rittenhouse NL, Peralta R, Wang Y, Wang Y, DePeaux K, Poholek AC, Delgoffe GM, Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol 22, 205–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, Meng YJ, Meier SR, Jennings RB, Creech AL, Herbert ZT, McBrayer SK, Olenchock BA, Jaffe JD, Haigis MC, Beroukhim R, Signoretti S, Koivunen P, Kaelin WG, Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science (New York, N.Y.) 363, 1217–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batie M, Frost J, Frost M, Wilson JW, Schofield P, Rocha S, Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Manna S, Kim JK, Bauge C, Cam M, Zhao Y, Shetty J, Vacchio MS, Castro E, Tran B, Tessarollo L, Bosselut R, Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat. Commun 6, 8152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Batie M, Del Peso L, Rocha S, Hypoxia and chromatin: A focus on transcriptional repression mechanisms. Biomedicine 6, 47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Hardy K, Olshansky M, Barugahare A, Gearing LJ, Prier JE, Sng XYX, Nguyen MLT, Piovesan D, Russ BE, La Gruta NL, Hertzog PJ, Rao S, Turner SJ, KDM6B-dependent chromatin remodeling underpins effective virus-specific CD8+ T cell differentiation. Cell Rep. 34, 108839 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Nutt SL, Keenan C, Chopin M, Allan RS, EZH2 function in immune cell development. Biol. Chem 401, 933–943 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME, AP-1 Transcription factors and the BAF complex mediate signaldependent enhancer selection. Mol. Cell 68, 1067–1082.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phanstiel DH, Van Bortle K, Spacek D, Hess GT, Shamim MS, Machol I, Love MI, Aiden EL, Bassik MC, Snyder MP, Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol. Cell 67, 1037–1048.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yukawa M, Jagannathan S, Vallabh S, Kartashov AV, Chen X, Weirauch MT, Barski A, AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med 217, e20182009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, Hackney J, Nurieva R, Escalante CR, Ouyang W, Littman DR, Murphy KM, Singh H, A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson JL, Georgakilas G, Petrovic J, Kurachi M, Cai S, Harly C, Pear WS, Bhandoola A, Wherry EJ, Vahedi G, Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of t cells. Immunity 48, 243–257.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, Li Y, Phillips FC, Maina PK, Qi HH, Liu C, Zhu J, Pope RM, Musselman CA, Zeng C, Peng W, Xue HH, Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nat. Immunol 17, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Man K, Gabriel SS, Liao Y, Gloury R, Preston S, Henstridge DC, Pellegrini M, Zehn D, Berberich-Siebelt F, Febbraio MA, Shi W, Kallies A, Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Immunity 47, 1129–1141.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN, Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med 16, 1147–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE, Single-molecule decoding of combinatorially modified nucleosomes. Science (New York, N.Y.) 352, 717–721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prickaerts P, Adriaens ME, Beucken TVD, Koch E, Dubois L, Dahlmans VEH, Gits C, Evelo CTA, Chan-Seng-Yue M, Wouters BG, Voncken JW, Hypoxia increases genome-wide bivalent epigenetic marking by specific gain of H3K27me3. Epigenetics Chromatin 9, 46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghoneim HE, Fan Y, Moustaki A, Abdelsamed HA, Dash P, Dogra P, Carter R, Awad W, Neale G, Thomas PG, Youngblood B, De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim KH, Roberts CW, Targeting EZH2 in cancer. Nat. Med 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, Wherry EJ, Haining WN, The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol 15, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The datasets generated during this study are available at NCBI GEO repository GSE175443. Additional datasets not generated by this study are available at NCBI GEO repository under accession numbers GSE155192, GSE123235, GSE123236, GSE95237, and GSE54191. All materials used in this study are available upon request addressed to G.M.D. or A.C.P.