Abstract
We have identified an 85-kDa outer membrane protein that is expressed by all tested strains of Haemophilus ducreyi. Studies of related proteins from other pathogenic bacteria, including Haemophilus influenzae, Pasteurella multocida, Neisseria gonorrhoeae, Neisseria meningitidis, and Shigella dysenteriae, suggested a role for these proteins in pathogenesis and immunity. In keeping with the first such described protein from Haemophilus influenzae type B, we termed the H. ducreyi protein D15. The gene encoding the H. ducreyi D15 protein was cloned and sequenced, and the deduced amino acid sequence was found to be most similar to sequences of the D15-related proteins from other Pasteurella spp. The arrangement of the flanking genes was similar to that of H. influenzae Rd and suggested that D15 was part of a multigene operon. Attempts to make a null mutation of the D15 gene were unsuccessful, paralleling results in other D15 gene studies. Overexpression of H. ducreyi D15 in Escherichia coli resulted in a source of recombinant D15 (rD15) from which it was readily purified. rD15 was immunogenic, and it was found that immunization of rabbits with an rD15 vaccine preparation conferred partial protection against a virulent challenge infection. Antisera to an N-terminal peptide recognized all tested strains of H. ducreyi.
Haemophilus ducreyi is the etiologic agent of chancroid, a sexually transmitted genital ulcerative disease that is prevalent in Africa, Asia, and certain other developing countries (1, 54). Recently, several studies have demonstrated a role for chancroid as an important independent risk factor for the heterosexual transmission of human immunodeficiency virus (23, 28, 54). Reinfection with H. ducreyi occurs after natural (54) or experimental (6) human infection, suggesting that infection does not confer immunity. No vaccine or practical field diagnostic test currently exists for chancroid.
H. ducreyi is an obligate human pathogen. The bacterium is difficult to grow in vitro, where it requires the essential nutrient heme, presumably supplied in vivo by host hemoglobin, heme, or catalase (19). The ability of H. ducreyi to resist the bactericidal nature of complement-intact normal human serum and its partial resistance to phagocytic killing in vitro may partially explain its ability to survive in chancroid lesions, which contain these host defenses mechanisms (18, 32, 35–37).
Because of the importance of chancroid as a risk factor for the acquisition of human immunodeficiency virus, recent studies from several laboratories have begun to reveal potential virulence factors and potential vaccine candidates of H. ducreyi. Several outer membrane proteins have been characterized at the molecular level, including a hemoglobin receptor (15, 17), a pilin protein (8), a lipoprotein (26; T. Hiltke, C. Lau, S. Gurunathan, A. Juarez, A. Campagnari, and S. Spinola, presented at the 94th General Meeting of the American Society for Microbiology, Las Vegas, Nev., 1994) two OmpA-like proteins (30), a P6-like protein (49), a heme receptor (51), and a protein required for expression of serum resistance, termed DsrA (18). Three secreted proteins likely to be involved in virulence include a hemolysin (2, 14, 38–40, 53), a cytolethal distending cytotoxin (9, 41), and two filamentous hemagglutinin-like proteins termed LspA1 and LspA2 (55). A cytoplasmic superoxide dismutase (45, 46), an enzyme involved in resistance to polymorphonuclear leukocyte killing, has been found to be required for full virulence expression in an animal model (47). For most of these putative virulence factors, separate isogenic mutants, each lacking one factor, have been constructed and tested in the human challenge model of H. ducreyi infection. To date only the hemoglobin receptor mutant (4), the P6 mutant (5), and the dsrA mutant (6a) have been found to be highly attenuated.
Flack et al. first reported an 85-kDa outer membrane protein which is highly conserved in typeable and nontypeable strains of H. influenzae (21). They termed it D15. Since then, several studies of animal models of H. influenzae infection demonstrated that D15 confers protection against homologous strains (33, 52, 56); in one study, D15 was protective against heterologous strains (33). Similarly, Oma 87, a highly related protein of Pasteurella multocida, has been shown to elicit protection in an animal model of infection (44). Since the D15 and D15-like proteins had previously been shown to elicit protective immunity in these other systems, we sought evidence for D15-induced immunity in the H. ducreyi system.
We report here the cloning and sequencing of the gene encoding the D15 protein of H. ducreyi. Recombinant D15 (rD15) was overexpressed and purified from Escherichia coli for the purpose of immunobiological studies. We show that rD15 is immunogenic in rabbits and that D15 shares certain attributes of successful vaccines.
MATERIALS AND METHODS
Strains and media.
We used the extensively characterized H. ducreyi strain 35000 for most assays. Other strains used were previously described by us (15) or others (3, 14, 35). The strains used in Western blots are listed in the legend to Fig. 5. An additional 26 strains from a characterized panel (14) were used in other Western blots but are not listed because there was no difference in reactivity or mobility (data not shown). For routine growth, H. ducreyi was maintained on chocolate agar plates prepared by the UNC Hospitals media lab. The basal medium for chocolate agar was Mueller-Hinton agar containing 1% IsoVitaleX and 1% autoclaved hemoglobin. The E. coli strains were DH5αMCR and BL21(DE3)pLysS. E. coli strains harboring plasmids were grown on Luria-Bertani agar or in Luria-Bertani broth containing appropriate antibiotics. The antibiotics used for E. coli included ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), kanamycin (30 μg/ml), and rifampin (200 μg/ml). Outer membranes were isolated as previously described (15).
FIG. 5.
Western blots of immune response to rD15. A preparative SDS-polyacrylamide gel containing 25 μg of H. ducreyi outer membrane proteins was electrophoresed and electroblotted to nitrocellulose. After blocking, the membrane was placed in a Miniprotean MultiScreen apparatus and probed with representative anti-rD15 rabbit antisera obtained after two immunizations with rD15-His, but before the challenge with strain 35000 (1:1,000 dilution). Protein A-alkaline phosphatase and BCIP-NBT were used for detection of bound immunoglobulins. Lane 1, pooled preimmune serum; lanes 2 to 7, postimmune serum from rabbits 7, 8, 9, 76, 77, 78, 79, respectively; lane 8, affinity-purified anti-peptide IgG; lane 9, no primary antibody. The preimmune sera were pooled in this experiment; in other experiments using individual preimmune sera, no reaction was observed (n = 3). No reactivity was noted in mock-immunized rabbits (data not shown).
N-terminal amino acid sequencing and antipeptide antibody production.
An approximately 85-kDa D15 protein from H. ducreyi strain 35000 was subjected to Edman degradation using an outer membrane preparation electroblotted from a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel to a polyvinylidene difluoride membrane as previously described (15). The sequence APFVLKDIRIDGVQTETNA was obtained. This sequence was determined by the UNC/PMBB MicroProtein Chemistry Facility (Program in Molecular Biology and Biotechnology, University of North Carolina at Chapel Hill).
A predicted antigenic peptide derived from the N-terminal sequence was synthesized by standard fluoroenylmethoxycarbonyl chemistry by the UNC/PMBB MicroProtein Chemistry Facility. The peptide sequence was CGGKDIRIDGVQTETGNA. The three underlined residues are not part of the D15 protein sequence (see above). The initial C functioned for sulfhydryl coupling, and the following GG functioned as a spacer. After peptide purification by reverse-phase high-performance chromatography, the sequence was confirmed by fast atom bombardment-mass spectrophotometry. The peptide was conjugated to keyhole limpet hemocyanin, and rabbits were immunized a total of five times at 3-week intervals with 0.5 mg of peptide conjugate per immunization. Freund's complete adjuvant was used for the first immunization; Freund's incomplete adjuvant was used with succeeding doses. Immune serum was affinity purified using D15 peptide immobilized on a thiopropyl-agarose column as previously described (15).
Initial PCR and cloning of the gene encoding the D15 protein (pUNCH 1215).
Initial attempts to obtain genomic plasmid clones using the antipeptide immunoglobulin G (IgG) described above in an expression library were unsuccessful. Similarly, we were unable to obtain clones using degenerate oligonucleotide probes. We speculated that production of D15 with a leader sequence might be toxic to the host E. coli, as had been our experience with several other outer membrane proteins. Therefore, we used a PCR strategy to amplify a 2.4-kb product lacking the putative leader sequence and cloned it into pCRII. To initially confirm that the PCR was correct, inserts were subcloned from pCRII into an expression system (pET30). After induction with isopropyl-β-d-thiogalactopyranoside (IPTG), recombinants expressing D15 protein were identified in Western blots with the anti-synthetic peptide IgG described above. Subsequently, sequencing (see below) confirmed that we had cloned D15. The 5′ degenerate oligonucleotide (5′GGATCCGAATTCGCICCITTYGTIGTIAARGAYATHMG) was deduced from the N-terminal amino sequence of H. ducreyi and H. influenzae Rd DNA and amino acid sequences. The first H. ducreyi codon began at the first amino acid (alanine) of the mature protein. The 3′ degenerate amplimer (5′CGCCGGCGTTCGACCICCIATISWRAAYTGKAAYTGYTC) was derived from the sequence of the H. influenzae Rd D15 amino acid sequence and encoded the terminal phenylalanine and stop codon of D15. The underlined bases indicate restriction sites or other bases not part of the D15 sequence. The PCR conditions were 94°C for 1 min, 42°C 1 min, and 68°C for 4 min, each for 30 cycles. Ready To Go PCR tubes (Amersham Pharmacia Biotech Inc.) were used for PCR, and the final magnesium concentration was adjusted to 3.75 mM. The initial expression clone, designated pUNCH 1215, expressed full-length immunoreactive protein (data not shown).
Genomic cloning of D15 and its flanking DNA.
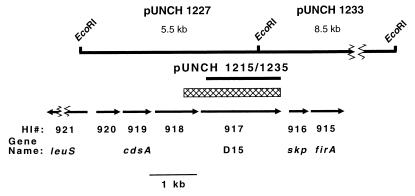
Since pUNCH 1215 was constructed with degenerate amplimers (including sequence derived from H. influenzae), it was imperfect. We obtained genomic clones in order to determine the correct H. ducreyi sequence, to understand the gene arrangement and organization of the D15 region, to identify promoter sequences, and to determine the structure of the leader sequence. An EcoRI site was identified in the middle of the H. ducreyi D15 gene from pUNCH 1215 (Fig. 1) and exploited in the construction of D15 genomic clones. Using both halves of pUNCH 1215 as probes in Southern blots, we identified upstream and downstream genomic EcoRI fragments of 5.5 and 8.0 kb, respectively. Appropriate size-selected DNA fragments were gel purified and ligated to alkaline phosphatase-treated pMCL210 vector (36). Colony hybridizations were performed using the appropriate pUNCH 1215 EcoRI probe fragment (Genius protocol; Boehringer Mannheim). The upstream genomic clone (pUNCH 1227 [Fig. 1]) was initially confirmed by internal PCR using primers derived from pUNCH 1215 sequence (data not shown). The downstream genomic clone (pUNCH 1233) was initially confirmed by Southern blotting (data not shown).
FIG. 1.
Restriction map of the H. ducreyi D15 region and the clones described in this study. The lower portion shows the corresponding H. influenzae region. A total of 14 kb of contiguous genomic DNA from H. ducreyi is contained within pUNCH 1227 and pUNCH 1233. The coding sequence of the mature form of rD15 found in pUNCH 1215 and 1235 is indicated (the hexahistidine leader present in pUNCH 1215 and 1235 is not shown). HI# indicates the H. influenzae ORF number and gene designation. The cross-hatched box indicates the H. ducreyi sequence presented in Fig. 2. The broken lines indicate additional flanking sequences not shown.
DNA sequencing.
Both strands of the D15 gene in pUNCH 1215 were sequenced. Comparison to D15-like genes from other bacteria suggested that we had indeed cloned the D15 gene of H. ducreyi. Initial sequencing of the genomic clones pUNCH 1227 and pUNCH 1233 was done on single strands only. Any discrepancies between pUNCH 1215 and the genomic clones were resolved by double-stranded sequencing the relevant regions of genomic clones.
Construction of an rD15 expression clone (pUNCH 1235) and purification of rD15 protein.
Some of the details of our expression and purification of pUNCH 1235 are described elsewhere (20) and are therefore briefly described here. Our strategy for construction of expression clone pUNCH 1235 was identical to that used for pUNCH 1215, but it used correct homologous H. ducreyi primers (after we had obtained genomic sequence). Primers were designed with unique restriction sites for in-frame fusion to the expression plasmid pET30a with an N-terminal hexahistidine leader sequence (20). The primers were 5′-GGATCCGAATTCGCACCATTTGTAGTAAAAGAT) and 3′-CGCCGGCGTTCGAATTAGAATGTGCTGCCAACTG. The first amino acid of pUNCH 1235 was the N terminus of the mature protein, and the final amino acid was the terminal phenylalanine. PCR products were ligated into plasmid pCRII and transformed into E. coli DH5αMCR. At least four white colonies containing the appropriate-size insert were selected. Inserts were removed following digestion with appropriate restriction endonucleases, pooled, and ligated into pET30a that had been cut with the same enzymes. After transformation into E. coli [(BL21(DE3)pLysS or Nova Blue (DE3)]) and induction, several transformants were analyzed in Western blots probed with affinity-purified anti-D15 peptide IgG to identify clones expressing full-length product (20).
Since expression clone pUNCH 1235 lacked its native signal peptide, induction of expression resulted in the accumulation of rD15 into inclusion bodies as described by Qi et al. (42). We followed the general method for purification of inclusion bodies proposed by Qi et al. (42). pUNCH 1235, which contained a vector hexahistidine leader fused to the N terminus, enabled us to further purify the protein by metal chelate chromatography under denaturing conditions (6 M urea) as described by the manufacturer and as modified by us previously (20). We attempted to renature purified recombinant protein slow reduction of urea concentration. The protein was first diluted to 1 mg/ml, and the urea concentration was reduced to 2 M. Zwittergent 3,14 was added to 5%, and the urea was gradually removed by dialysis against phosphate-buffered saline (PBS) containing decreasing amounts of urea over several days at 4°C until no urea was present.
Vaccine trials using rD15 in the TDRM of H. ducreyi infection.
We used the temperature-dependent rabbit model (TDRM) of H. ducreyi infection as first described by Hansen et al. (25) and as previously modified by our group (12). Eight-week-old New Zealand White rabbits (supplied by Charles River Inc.; weighing approximately 1.5 kg when first vaccinated) were immunized twice, 4 weeks apart, using rD15-His tag immunogen in Zwittergent 3,14 or in 350 μl of PBS or 100 μg of rFetA as a control. The first dose was in Freund's complete adjuvant, and the second was in Freund's incomplete adjuvant. In both immunizations, half of the dose was given intramuscularly and the other half was given subcutaneously. Rabbits were bled for serologic assays at weeks 0, 2, 4, 6, and 8 from the first immunization and after infection at 8 weeks. During the 2-week period between weeks 6 and 8, the rabbits were acclimatized to a temperature of 15°C. The day before infection, the backs of the rabbits were shaved. The following day, the rabbits were intradermally inoculated with 105 to 103 CFU of broth-grown mid-log-phase H. ducreyi strain 35000 in triplicate. Every second day for a period of 20 days, two lesions from each inoculum size were scored macroscopically on a scale of 0 to 4 (0, nil; 1, erythema; 2, induration; 3, suppuration; and 4, ulcer), while the third lesion was cultured by sidewise aspiration of the lesion for the presence of H. ducreyi. All measurements were conducted by an operator blinded to the immunization group or status of the individual animal.
Immunological studies.
Enzyme-linked immunosorbent assays (ELISAs) and Western blotting were performed as previously described (16). Protein A-alkaline phosphatase was used as the secondary conjugate. p-Nitrophenol phosphate and 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) substrates were used for development of ELISAs and Western blots, respectively.
Whole-cell binding assays.
Whole-cell dot blots were performed using chocolate agar-grown H. ducreyi strain 35000 for the antigen as previously described (16), with the following modifications. H. ducreyi (optical density = 0.2 at 600 nm) was suspended in PBS containing Ca and Mg supplements (the 100× stock contains, per liter, 2.1 g of MgCl2, 1.7 g of CaCl2, and 0.7 g of MgSO4); 100 μl of this bacterial suspension was briefly suctioned onto a nitrocellulose membrane (Nitroplus; Osmonics, Inc.). All subsequent incubations were performed under gentle agitation. The membrane was blocked in 2% bovine serum albumin for 15 min, and primary antibody dilutions were applied for 1 h. After three 5-min washes in PBS-Tween (0.05%), protein A-alkaline phosphatase conjugate (1:5,000; Sigma) was applied for 1 h. After washing again as described above, the substrate BCIP–NBT was added for development without shaking.
Bactericidal assays.
To measure the ability of anti-rD15 antibodies to elicit a bactericidal response, bactericidal assays were performed as previously described (16). Briefly, chocolate agar-grown bacteria were diluted in GCB broth to contain approximately 2,000 CFU/ml. Preimmune or postimmune sera from the rabbits represented in Fig. 5 were pooled for use in bactericidal assays. These sera were mixed with the bacteria in a total volume of 100 μl in a microtiter dish. After 15 min of incubation with heat-inactivated rabbit anti-rD15 antibody, 10 μl of complement-intact, pooled normal human serum (NHS; approximately 10%, final concentration) or normal rabbit serum (10%; Pel Freeze Biological, Rogers, Ark.) (27) was added, and incubation continued for 30 min. Controls included no rabbit antibody (to control for preimmune killing). Controls for complement activity in the NHS included testing the complement source (serum) against serum-susceptible organisms (H. ducreyi dsrA mutant FX517) (18). Controls using heat-inactivated NHS (complement source) were done to determine if a reduction in CFU was the result of agglutination by antibody.
Nucleotide sequence ascession number.
The DNA sequence of the H. ducreyi D15 gene has been assigned GenBank accession number AF329831.
RESULTS
Identification of a D15 protein from H. ducreyi.
During an unrelated study to search for TonB-dependent outer membrane receptor proteins, several proteins in the range of 70 to 100 kDa were subjected to N-terminal amino acid sequencing. The sequence obtained for a protein of approximately 85 kDa was similar to the sequence of protective antigens described for H. influenzae and P. multocida. Like H. ducreyi, these two related bacteria are members of the family Pasteurelleaceae. An antipeptide serum to the N-terminal amino acid sequence of the H. ducreyi protein was prepared, and all tested strains of H. ducreyi expressed an immunoreactive protein of mobility similar to that of the D15 protein of strain 35000 (34 strains [data not shown]). Therefore, based on these data, we chose to study the D15 protein from H. ducreyi at the molecular level and to determine its potential as a vaccine candidate.
Cloning and sequencing of the D15 gene.
Initially, we used degenerate and heterologous primers in PCR to amplify, clone, and sequence a 2.4-kb gene encoding the D15 protein from H. ducreyi strain 35000 (pUNCH 1215 [Table 1]). pUNCH 1215 was fused with an N-terminal leader sequence from the vector. After induction with IPTG, an immunoreactive protein was produced which migrated slightly more slowly (as expected due to the hexahistidine leader fusion) than did native D15 from H. ducreyi (data not shown). We further confirmed that we had cloned a D15 gene by sequencing pUNCH 1215 and comparing the sequence to those of other D15 genes. We then used pUNCH 1215 to obtain two genomic EcoRI clones spanning approximately 13 kb of DNA which included all of the D15 gene and flanking DNA (Fig. 1).
TABLE 1.
Results for the immunization protocols tested
| Immunization group | Inoculum titer | No. of rabbits | Mean inoculum size (CFU) ± SD | Mean no. of days culture positive ± SD | Mean cumulative lesion size (mm) ± SD | Mean peak lesion score ± SD | No. (%) of rabbits that developed ulcers | Proportion (%) of lesions that developed into ulcers | Mean duration of ulcers (days) ± SD |
|---|---|---|---|---|---|---|---|---|---|
| Sham | 105 | 14 | 5 ±1.9 | 10.1 ± 4.2 | 51.4 ± 6.3 | 4 ± 0 | 13 (93) | 27/28 (96) | 12 ± 3.2 |
| 104 | 14 | 4.2 ± 2.1 | 10.7 ± 2.8 | 47.2 ± 8.8 | 4 ± 0 | 14 (100) | 28/28 (100) | 11.5 ± 3.3 | |
| 103 | 14 | 3.7 ± 2 | 9.1 ± 2.6 | 30.4 ± 6.9 | 3.5 ± 0.5 | 6 (43) | 17/28 (61) | 4.7 ± 3.4 | |
| rD15 | 105 | 13 | 7.6 ± 2.8a | 6.8 ± 2.7a | 43.9 ± 13 | 3.8 ± 0.3 | 9 (69) | 21/26 (81) | 7.9 ± 4.5b |
| 104 | 13 | 6.1 ± 2.5a | 8.5 ± 3.2 | 40.4 ± 13 | 3.8 ± 0.6 | 10 (77) | 22/26a (85) | 8.5 ± 4 | |
| 103 | 13 | 5.8 ± 2.4a | 7.4 ± 3.9 | 24 ± 14.2 | 3.3 ± 0.7 | 4 (31) | 10/26 (38) | 3.3 ± 4.1 | |
| rFetA | 105 | 8 | 7.1 ± 4 | 9 ± 4.1 | 52.6 ± 14 | 4 ± 0 | 8 (100) | 16/16 (100) | 11.8 ± 3.7 |
| 104 | 8 | 5.7 ± 2 | 6.8 ± 3.2a | 46 ± 10 | 4 ± 0 | 8 (100) | 16/16 (100) | 11.4 ± 3.2 | |
| 103 | 8 | 6 ± 2.7a | 7.8 ± 4.1 | 29.5 ± 12 | 3.5 ± 0.7 | 4 (50) | 10/16 (63) | 5.9 ± 4.7 |
P < 0.05; one-way repeated measures analysis of variance.
The DNA sequence for the H. ducreyi D15 gene contained an open reading frame (ORF) of 2,382 bp and predicted a protein of 794 amino acids. A putative ribosome-binding site (AGGA) was separated by 8 nucleotides from the D15 ATG initiation codon. The deduced amino acid sequence revealed a 17-amino-acid N-terminal leader sequence. The VXA sequence resembled a signal peptidase I cleavage site, and our Edman degradation data confirmed cleavage. Cleavage at this site would yield a mature protein of 86,915 Da, which is in agreement with the migration of native D15 from outer membranes subjected to SDS-polyacrylamide gel electrophoresis. Concordant with outer membrane localization, D15 contained a carboxyl-terminal motif ending with a phenylalanine, which is found in the majority of integral outer membrane proteins (50). A search of databases with the D15 sequence identified several similar proteins from other pathogenic bacteria, including P. multocida, H. influenzae, Neisseria gonorrhoeae, N. meningitidis, E. coli, Shigella dysenteriae, and Brucella abortus. The H. ducreyi D15 protein is most similar to the other members of the Pasteurellaceae for which there is sequence available. Compared to the H. ducreyi D15 protein, the P. multocida strain PM70 protein (http://www.ncbi.nlm.nih.gov/Micro_blast/unfinishedgenome.html) was 66% similar and 56% identical, whereas the H. influenzae type B strain CA protein (22) was 63% similar and 50% identical (Fig. 2).
FIG. 2.
Comparison of the H. ducreyi D15 amino acid sequence with D15 sequences from other Pasteurellaceae. The mature coding region of the H. ducreyi D15 protein was analyzed using the program Pretty Box. Identical sequences are shaded and boxed. H. ducr, H. ducreyi strain 35000; H. influ, H influenzae type B strain CA; P. mult, P. multocida strain PM70. Three variable regions for the H. ducreyi D15 protein are underlined.
A short intergenic region (49 nucleotides) separated the D15 ORF from the upstream ORF and contained no obvious promoter. The upstream ORF (Fig. 1) was very similar to a protein (ORF HI 918) of unknown function. A 102-bp intergenic region between D15 and the downstream ORF contained a 14-base inverted repeat with 12 exact matches, which may represent a transcriptional terminator. Downstream of D15 were two ORFs similar to skp and firA described for P. multocida (11).
D15 expression in E. coli.
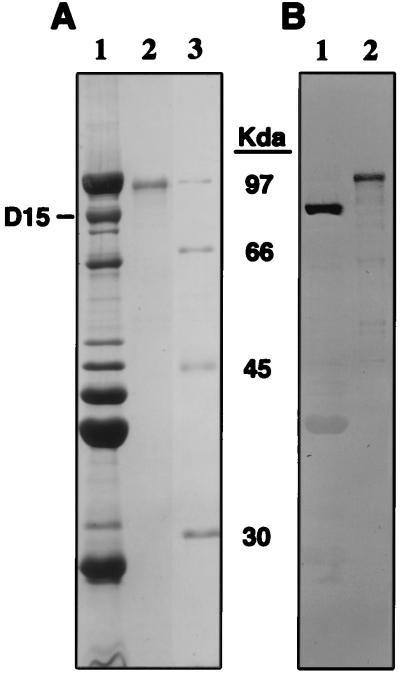
Although we had already constructed expression clone pUNCH 1215, it had been made with degenerate, heterologous primers. Therefore, we reconstructed a plasmid equivalent to pUNCH 1215, but using the appropriate primers to form pUNCH 1235. After induction of rD15 protein and its purification from E. coli on a nickel column, the rD15-His protein was putatively refolded. The final purified rD15-His protein used for rabbit challenge experiments was analyzed for purity by SDS-PAGE (Fig. 3A, lane 2). It should be noted that the larger size of the recombinant protein in lane 2 is consistent with the presence of a hexahistidine leader in this form of rD15. The antigenicity of the rD15-His protein was analyzed by Western blotting using the independently derived affinity-purified anti-D15 synthetic peptide IgG as the primary antibody (Fig. 3B, lane 2).
FIG. 3.
SDS-PAGE (A) and Western blot analysis (B) of H. ducreyi outer membrane proteins and purified recombinant proteins. (A) H. ducreyi outer membrane proteins prepared from strain 35000 or recombinant His-tagged proteins purified from E. coli as described in the text were solubilized at 100°C for 5 min under reducing conditions. After electrophoresis, the gel was stained with Coomassie blue. Lanes: 1, H. ducreyi outer membrane proteins (30 μg, heme limiting); 2, purified rD15-His (2 μg) from pUNCH 1235; 3, molecular weight standards. (B) Western blot of H. ducreyi outer membrane proteins and D15 proteins. Five micrograms of H. ducreyi outer membrane protein or 500 ng of recombinant protein was loaded in each lane of an SDS–10% polyacrylamide gel. Lane numbers correspond to those in panel A. After electrotransfer, D15 proteins were detected with affinity-purified anti-D15 peptide IgG.
Immunization with rD15-His modifies the course of a homologous challenge infection in the TDRM of H. ducreyi infection.
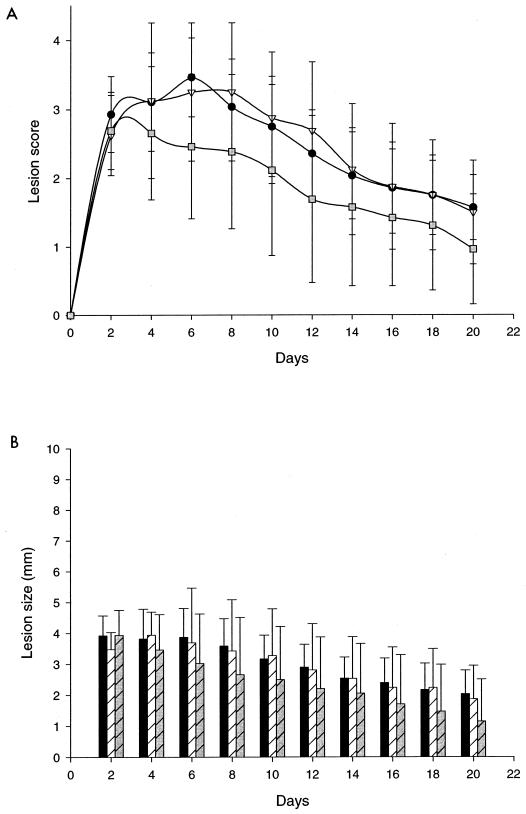
We used the TDRM to test the vaccine potential of rD15-His, in four groups of nine rabbits, including parallel controls. By chance, challenge inoculum titers were slightly but significantly higher in test-versus sham-immunized control animals (Table 1). Nevertheless, for inocula from 103 to 105 CFU (in 100 μl), the lesion score and size (Fig. 4), cumulative size, and proportion of rabbits that developed ulcers (Table 1) were slightly lower for the rD15-immunized rabbits than the sham-immunized rabbits. Duration of culture positivity (Table 1) was reduced by 2 to 3 days in rD15-immunized rabbits (P < 0.05 at 105 CFU). Similarly, the duration of ulcers (Table 1) was reduced by up to 4 days (P < 0.05 at 104 and 105 CFU). A negative control immunogen, rFetA-His, was used to seek a nonspecific effect of immunization with an irrelevant protein, expressed and purified from the same system as rD15-His. No pattern of differential virulence was observed between this group and sham-immunized rabbits (Fig. 4 and Table 1).
FIG. 4.
Immunization with rD15 modifies the course of a homologous challenge infection in the temperature-dependent rabbit model of H. ducreyi infection. Rabbits were vaccinated twice with 100 μg of rD15-His (n = 13) rFetA-His (n = 8) or sham were vaccinated with 350 μl of PBS (n = 14) as described in the text. Four weeks after the second vaccination, rabbits were challenged intraepithelially with 103 CFU of H. ducreyi strain 35000 in 100 μl of PBS. Lesion score (A) and lesion size (B) of rabbits vaccinated with rD15-His (gray boxes and gray bars) and rFetA-His (inverted white triangles and white hatched bars) are compared to those of controls (black circles and black bars) (scored as 0 [nil], 1 [erythema], 2 [papule], 3 [pustule], and 4 [ulceration]). Data from rD15- and rFetA-immunized rabbits were compared to data from sham-immunized controls by an analysis of variance.
Immune response to rD15 from the rabbit challenge experiments.
The immune response to the rD15-His from the rabbit challenge experiments was analyzed for specificity and assessed semiquantitatively. Rabbit antiserum, obtained after two immunizations but before the challenge infection (8 weeks), was specific for the SDS-denatured form of the D15 protein in Western blots of H. ducreyi outer membranes (Fig. 5).
ELISAs were performed using two different antigens. The first ELISA used purified rD15-His as the coating antigen, and the optical densities of the postimmune sera (1:1,000) from these rabbits varied between 0.640 and 2.145. In contrast, the second ELISA used outer membrane protein purified from strain 35000 as the coating antigen, and the postimmune optical densities were all less than 0.1 at the same dilution. Optical densities for preimmune sera were all less than 0.05. When these sera were tested for binding to whole cells in dot blots, we found that binding was weak or negative (data not shown). Significantly, some PBS (sham)-immunized rabbits appeared to have a similar weak rise in titer in dot blots, suggesting that the increase in binding was not due to D15 immunization.
Bactericidal assays.
The ability of antisera to direct complement-mediated killing of H. ducreyi was tested in bactericidal assays. As sources of complement, we used either NHS or normal rabbit serum, previously tested for complement activity. We observed no killing of H. ducreyi by serum at the tested dilutions (between 1:10 and 1:500 [data not shown]) in either the pre- or postimmune specimens.
Conservation of D15 in H. ducreyi.
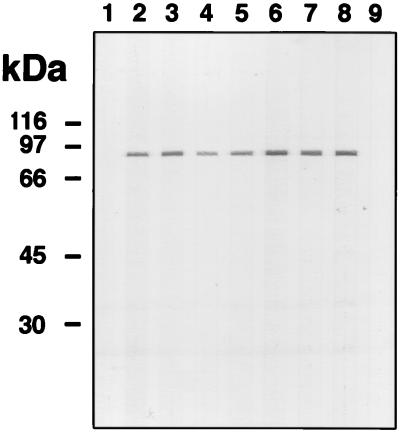
Western blot of analyses of geographically diverse strains were undertaken to examine the conservation of D15 expression and conservation of D15 antigenicity. We used the anti-peptide D15 IgG (34 strains [Fig. 6 and data not shown]), and all tested strains made an immunoreactive protein. The migration patterns of the D15 bands were very similar or indistinguishable, suggesting a high degree of amino acid conservation in the D15 proteins from unrelated strains.
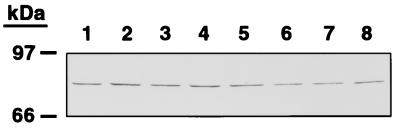
FIG. 6.
D15 protein is expressed and immunologically conserved in H. ducreyi. Protein from each indicated H. ducreyi strain (approximately 5 × 107 CFU or 15 μg per lane) was subjected SDS-PAGE and electroblotting using the affinity-purified anti-peptide D15 IgG. Lanes: 1, 35000; 2, CIP542; 3, 4V; 4, 26V; 5, C110; 6, 2753; 8, 90–244.
DISCUSSION
In this study, we identified an 85-kDa outer membrane protein in H. ducreyi which shows significant similarity to a protein found in several other pathogenic bacteria. This protein was of interest because it has been shown to be protective in animal models of infection in two other bacterial systems.
Overall the structure of the H. ducreyi, H. influenzae, and P. multocida D15 or D15-like proteins were conserved; however, it should be noted that small interspecies variable regions were present in the three proteins (Fig. 2). Compared to the other two species, three H. ducreyi variable regions resided in the C-terminal one-third of the protein (Fig. 2). The first H. ducreyi variable region was 16 amino acids long and contained an insertion of 6 or 7 amino acids compared to the P. multocida or H. influenzae protein, respectively. Interestingly, the next two H. ducreyi interspecies variable regions are in the identical positions of the interstrain H. influenzae D15 variable regions previously described by Loosmore et al. (33). In variable regions 2 and 3 of the H. ducreyi sequence, there were deletions of five and six amino acids, respectively, compared to H. influenzae and P. multocida. Other studies of integral outer membrane proteins have suggested that variation in surface-exposed portions (loops) is driven by immunologic pressure. However, in Western blots of 34 geographically diverse strains of H. ducreyi, a D15 protein of approximately the same molecular mass was observed, suggesting a high degree of interstrain conservation. Nevertheless, understanding the variability of H. ducreyi D15 will require additional sequencing of the D15 genes from other strains.
The region surrounding the D15 gene in H. ducreyi was very similar to the D15 region of H. influenzae Rd (22). The H. influenzae Rd D15 ORF is assigned number HI 917. We identified an ORF upstream of the H. ducreyi D15 (similar to Rd HI 918) and two downstream (similar to Rd HI 916 and 915). HI 916 and 915 show similarity to Skp and FirA from P. multocida (12). Skp has been implicated as a molecular chaperone (10, 48), whereas FirA has been shown to be an acyltransferase required for lipid A biosynthesis (29, 43). Furthermore, Skp is a protective antigen in H. influenzae (31). The short intergenic sequence between the upstream gene (49 bp) and the start codon for D15, along with the lack of obvious promoter sequence, suggests the possibility that D15 is part of a single transcriptional unit driven from a yet to be identified promoter.
Attempts to construct isogenic D15 mutants were unsuccessful. The first technique tried was allelic replacement (23), which had been successful in our laboratory for three H. ducreyi genes previously (17, 19, 51). The second mutagenesis procedure, which we have also used previously (18), was described by Bozue et al. (7). D15 mutants have not been reported from any of the organisms studied to date, suggesting that D15 may be necessary for the viability of gram-negative bacteria, perhaps by affecting expression or function of other vital genes. Construction of nonpolar and/or conditional D15 mutants will be critical to studies of the function and significance of D15.
Some data suggest that H. ducreyi D15 might be a vaccine candidate, whereas other data contradict this notion. D15 and related proteins have been shown to be protective antigens (21, 33, 44, 56). Our studies using the rabbit model of infection suggested that we were able to modify the course of an infection by immunizing with rD15-His. Although we were not able to completely prevent disease or infection, our results are not substantially different from those of three previous studies using outer membranes (25), pilin (12), or hemolysin (14) from H. ducreyi as immunogens. These rabbit experiments in inducible immunity differ from others reported in the blinding of the observer to the interventions, which may reduce biased observation, but at the expense of the apparent effect. Results of immunological studies indicate that H. ducreyi D15 is well conserved. A synthetic peptide based on D15 and rD15 proteins were immunogenic in rabbits. rD15 is readily produced in E. coli, where it can be induced to 50% of the total cellular protein. rD15 was readily purified, and it demonstrated partial protection from a homologous challenge. In humans, the D15 protein is apparently expressed in vivo, since an earlier study detected the presence of antibodies in humans (20).
However, neither the antibodies which we produced during this study nor those present after natural human infection are bactericidal. Indeed, the resistance of H. ducreyi to NHS or hyperimmune serum is well known (13, 27). No strongly bactericidal antibody against H. ducreyi has yet been reported (27). Likewise, we were unable to demonstrate bactericidal activity for anti-rD15 antibodies. Repeated infection with H. ducreyi is fairly common in areas where chancroid is endemic. D15 antibodies are produced after naturally occurring chancroid infections (20) but apparently do not prevent subsequent infections. It is possible that the antibodies produced after experimental immunization of animals or natural chancroid disease do not contain the proper characteristics that would lead to protection. Perhaps the antibodies are of low affinity or to the wrong D15 epitopes, they do not fix complement properly, or they do not lead to opsonophagocytic killing, none of which were measured in this study. Whether antibodies or a cellular response mediate the partial protection shown here in rabbits is not known. However, the lack of binding of antibodies to whole cells from rabbits immunized with rD15 suggest that a cellular immune response was responsible for protection, as has been suggested by another study using a different antigen (13). Alternatively, it might be that additional immunizations would generate antibodies which recognize D15 on whole cells. Additional rabbit challenge studies are required to clarify these issues. It should be noted that in the H. influenzae system, rD15 or passively transferred anti-rD15 antibodies protected in animal models of infection; however, these sera demonstrated only variable, inconsistent bactericidal activity (33). Thus, further studies with H. ducreyi D15 may clarify these vaccine issues and determine the function of this interesting protein.
ACKNOWLEDGMENTS
We thank Ralph Judd for helpful discussions, members of the Fred Sparling Laboratory for helpful discussions and critiquing the manuscript, and Annice Roundtree for expert technical assistance. We thank Pat Totten for the generous gift of H. ducreyi strains. Isabelle Leduc performed the experiments using the rabbit model of H. ducreyi infection in Bill Cameron's Laboratory.
The work presented was supported by grants R-29-AI40263 and AI31496 to C.E.E. This study was also supported by a Holderness Fellowship, the Research Educational Support Program of the National Institute for General Medical Science, and a Bristol Meyers Squibb Fellowship for Research in Academic Medicine to K.L.T.
REFERENCES
- 1.Albritton W L. Biology of Haemophilus ducreyi. Microbiol Rev. 1989;53:377–389. doi: 10.1128/mr.53.4.377-389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa M J, DeGagne P, Totten P A. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts. Infect Immun. 1996;64:2349–2352. doi: 10.1128/iai.64.6.2349-2352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa M J, Stevens M K, DeGagne P, Klesney-Tait J, Radolf J D, Hansen E J. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect Immun. 1995;63:1754–1761. doi: 10.1128/iai.63.5.1754-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Tawfiq J A, Fortney K R, Katz B P, Hood A F, Elkins C, Spinola S M. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J Infect Dis. 2000;181:1049–1054. doi: 10.1086/315309. [DOI] [PubMed] [Google Scholar]
- 5.Al-Tawfiq J A, Harezlak J, Katz B P, Spinola S M. Cumulative experience with Haemophilus ducreyi 35000 in the human model of experimental infection. Sex Transm Dis. 2000;27:111–114. doi: 10.1097/00007435-200002000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Al-Tawfiq J A, Palmer K L, Chen C Y, Haley J C, Katz B P, Hood A F, Spinola S M. Experimental infection of human volunteers with Haemophilus ducreyi does not confer protection against subsequent challenge. J Infect Dis. 1999;179:1283–1287. doi: 10.1086/314732. [DOI] [PubMed] [Google Scholar]
- 6a.Bong C T, Throm R E, Fortney K R, Katz B P, Hood A F, Elkins C, Spinola S M. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect Immun. 2001;69:1488–1491. doi: 10.1128/IAI.69.3.1488-1491.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozue J A, Tarantino L, Munson R S., Jr Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol Lett. 1998;164:269–273. doi: 10.1111/j.1574-6968.1998.tb13097.x. [DOI] [PubMed] [Google Scholar]
- 8.Brentjens R, Ketterer M, Apicella M, Spinola S. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J Bacteriol. 1996;178:808–816. doi: 10.1128/jb.178.3.808-816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Cock H, Schafer U, Potgeter M, Demel R, Muller M, Tommassen J. Affinity of the periplasmic chaperone Skp of Escherichia coli for phospholipids, lipopolysaccharides and non-native outer membrane proteins. Role of Skp in the biogenesis of outer membrane protein. Eur J Biochem. 1999;259:96–103. doi: 10.1046/j.1432-1327.1999.00010.x. [DOI] [PubMed] [Google Scholar]
- 11.Delamarche C, Manoha F, Behar G, Houlgatte R, Hellman U, Wroblewski H. Characterization of the Pasteurella multocida skp and firA genes. Gene. 1995;161:39–43. doi: 10.1016/0378-1119(95)00254-4. [DOI] [PubMed] [Google Scholar]
- 12.Desjardins M, Filion L G, Robertson S, Cameron D W. Inducible immunity with a pilus preparation booster vaccination in an animal model of Haemophilus ducreyi infection and disease. Infect Immun. 1995;63:2012–2020. doi: 10.1128/iai.63.5.2012-2020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins M, Filion L G, Robertson S, Kobylinski L, Cameron D W. Evaluation of humoral and cell-mediated inducible immunity to Haemophilus ducreyi in an animal model of chancroid. Infect Immun. 1996;64:1778–1788. doi: 10.1128/iai.64.5.1778-1788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutro S M, Wood G, Totten P. Prevalence of, antibody response to, and immunity induced by Haemophilus ducreyi hemolysin. Infect Immun. 1999;67:3317–3328. doi: 10.1128/iai.67.7.3317-3328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkins C. Identification and purification of a conserved heme regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect Immun. 1995;63:1241–1245. doi: 10.1128/iai.63.4.1241-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkins C, Barkley K B, Carbonetti N H, Coimbre A J, Sparling P F. Immunobiology of purified recombinant outer membrane porin protein I of Neisseria gonorrhoeae. Mol Microbiol. 1994;14:1059–1075. doi: 10.1111/j.1365-2958.1994.tb01339.x. [DOI] [PubMed] [Google Scholar]
- 17.Elkins C, Chen C J, Thomas C E. Characterization of the hgbA locus of Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkins C, Morrow K J, Jr, Olsen B. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect Immun. 2000;68:1608–1619. doi: 10.1128/iai.68.3.1608-1619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins C, Totten P A, Olsen B, Thomas C E. Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin. Infect Immun. 1998;66:151–160. doi: 10.1128/iai.66.1.151-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins C, Yi K, Olsen B, Thomas C, Thomas K, Morse S. Development of a serological test for Haemophilus ducreyi for seroprevalence studies. J Clin Microbiol. 2000;38:1520–1526. doi: 10.1128/jcm.38.4.1520-1526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flack F S, Loosmore S, Chong P, Thomas W R. The sequencing of the 80-kDa D15 protective surface antigen of Haemophilus influenzae. Gene. 1995;156:97–99. doi: 10.1016/0378-1119(95)00049-c. [DOI] [PubMed] [Google Scholar]
- 22.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J-F, Dougherty B, Merrick J, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J, Weidman J, Phillips C, Spriggs T, Hedblom E, Cotton M, Utterback T, Hanna M, Nguyen D, Saudek D, Brandon R, Fine L, Fritchman J, Fuhrmann J, Geoghagen N, Gnehm C, McDonald L, Small K, Fraser C, Smith H, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 23.Greenblatt R M, Lukehart S A, Plummer F A, Quinn T C, Critchlow C W, Ashley R L, DCosta L J, Ndinya A J O, Corey L, Ronald A R, et al. Genital ulceration as a risk factor for human immunodeficiency virus infection. AIDS. 1988;2:47–50. doi: 10.1097/00002030-198802000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Hansen E J, Latimer J L, Thomas S E, Helminen M, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen E J, Lumbley S R, Richardson J A, Purcell B K, Stevens M K, Cope L D, Datte J, Radolf J D. Induction of protective immunity to Haemophilus ducreyi in the temperature-dependent rabbit model of experimental chancroid. J Immunol. 1994;152:184–192. [PubMed] [Google Scholar]
- 26.Hiltke T, Campagnari A, Spinola S. Characterization of a novel lipoprotein expressed by Haemophilus ducreyi. Infect Immun. 1996;64:5047–5052. doi: 10.1128/iai.64.12.5047-5052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiltke T J, Bauer M E, Klesney-Tait J, Hansen E J, Munson R S, Jr, Spinola S M. Effect of normal and immune sera on Haemophilus ducreyi 35000HP and its isogenic MOMP and LOS mutants. Microb Pathog. 1999;26:93–102. doi: 10.1006/mpat.1998.0250. [DOI] [PubMed] [Google Scholar]
- 28.Jessamine P G, Plummer F A, Ndinya A J O, Wainberg M A, Wamola I, DCosta L J, Cameron D W, Simonsen J N, Plourde P, Ronald A R. Human immunodeficiency virus, genital ulcers and the male foreskin: synergism in HIV-1 transmission. Scand J Infect Dis Suppl. 1990;69:181–186. [PubMed] [Google Scholar]
- 29.Kelly T M, Stachula S A, Raetz C R, Anderson M S. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 30.Klesney-Talt J, Hiltke T J, Maciver I, Spinola S M, Radolf J D, Hansen E J. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyd J M, Cripps A W. Potential of a novel protein, OMP26, from nontypeable Haemophilus influenzae to enhance pulmonary clearance in a rat model. Infect Immun. 1998;66:2272–2278. doi: 10.1128/iai.66.5.2272-2278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagergard T, Frisk A, Purven M, Nilsson L A. Serum bactericidal activity and phagocytosis in host defence against Haemophilus ducreyi. Microb Pathog. 1995;18:37–51. [PubMed] [Google Scholar]
- 33.Loosmore S M, Yang Y P, Coleman D C, Shortreed J M, England D M, Klein M H. Outer membrane protein D15 is conserved among Haemophilus influenzae species and may represent a universal protective antigen against invasive disease. Infect Immun. 1997;65:3701–3707. doi: 10.1128/iai.65.9.3701-3707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano Y, Yoshida Y, Yamshita Y, Doga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 35.Odumeru J A, Wiseman G M, Ronald A R. Relationship between lipopolysaccharide composition and virulence of Haemophilus ducreyi. J Med Microbiol. 1987;23:155–162. doi: 10.1099/00222615-23-2-155. [DOI] [PubMed] [Google Scholar]
- 36.Odumeru J A, Wiseman G M, Ronald A R. Role of lipopolysaccharide and complement in susccptibility of Haemophilus ducreyi to human serum. Infect Immun. 1985;50:495–499. doi: 10.1128/iai.50.2.495-499.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odumeru J A, Wiseman G M, Ronald A R. Virulence factors of Haemophilus ducreyi. Infect Immun. 1984;43:607–611. doi: 10.1128/iai.43.2.607-611.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer K L, Grass S, Munson R S. Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect Immun. 1994;62:3041–3043. doi: 10.1128/iai.62.7.3041-3043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer K L, Munson R S. Cloning and characterization of the genes encoding haemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 40.Palmer K L, Thornton A C, Fortney K R, Hood A F, Munson J R S, Spinola S M. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis. 1998;178:191–199. doi: 10.1086/515617. [DOI] [PubMed] [Google Scholar]
- 41.Purven M, Lagergard T. Haemophilus ducreyi, a cytotoxin-producing bacterium. Infect Immun. 1992;60:1156–1162. doi: 10.1128/iai.60.3.1156-1162.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi H L, Tai J Y, Blake M S. Expression of large amounts of neisserial porin proteins in Escherichia coli and refolding of the proteins into native trimers. Infect Immun. 1994;62:2432–2439. doi: 10.1128/iai.62.6.2432-2439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy A M, Coleman J. Mutations in firA, encoding the second acyltransferase in lipopolysaccharide biosynthesis, affect multiple steps in lipopolysaccharide biosynthesis. J Bacteriol. 1994;176:1639–1646. doi: 10.1128/jb.176.6.1639-1646.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruffolo C G, Adler B. Cloning, sequencing, expression, and protective capacity of the oma 87 gene encoding the Pasteurella multocida 87-kilodalton outer membrane protein. Infect Immun. 1996;64:3161–3167. doi: 10.1128/iai.64.8.3161-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Mateo L R, Hobbs M M, Kawula T H. Periplasmic copper-zinc superoxide dismutase protects Haemophilus ducreyi from exogenous superoxide. Mol Microbiol. 1998;27:391–404. doi: 10.1046/j.1365-2958.1998.00687.x. [DOI] [PubMed] [Google Scholar]
- 46.San Mateo L R, Toffer K L, Kawula T H. The sodA gene of Haemophilus ducreyi encodes a hydrogen peroxide-inhibitable superoxide dismutase. Gene. 1998;207:251–257. doi: 10.1016/s0378-1119(97)00642-2. [DOI] [PubMed] [Google Scholar]
- 47.San Mateo L R, Toffer K L, Orndorff P E, Kawula T H. Neutropenia restores virulence to an attenuated Cu, Zn superoxide dismutase-deficient Haemophilus ducreyi strain in the swine model of chancroid. Infect Immun. 1999;67:5345–5351. doi: 10.1128/iai.67.10.5345-5351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schafer U, Beck K, Muller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 49.Spinola S, Hiltke T, Fortney K, Shanks K. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect Immun. 1996;64:1950–1955. doi: 10.1128/iai.64.6.1950-1955.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struyve M, Moons M, Tommassen J. Carboxyl-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 51.Thomas C E, Olsen B, Elkins C. Cloning and characterization of tdhA, a locus encoding a TonB-dependent heme receptor from Haemophilus ducreyi. Infect Immun. 1998;66:4254–4262. doi: 10.1128/iai.66.9.4254-4262.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas W R, Callow M G, Dilworth R J, Audesho A A. Expression in Escherchia coli of a high-molecular weight protective surface antigen found in nontypeable and type b Haemophilus influenzae. Infect Immun. 1990;58:1909–1913. doi: 10.1128/iai.58.6.1909-1913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Totten P A, Norn D V, Stamm W E. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect Immun. 1995;63:4409–4416. doi: 10.1128/iai.63.11.4409-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward C K, Lumbley S R, Latimer J L, Cope L D, Hansen E J. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol. 1998;180:6013–6022. doi: 10.1128/jb.180.22.6013-6022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y, Thomas W R, Chong P, Loosmore S M, Klein M H. A 20-kilodalton N-terminal fragment of the D15 protein contains a protective epitope(s) against Haemophilus influenzae type a and type b. Infect Immun. 1998;66:3349–3354. doi: 10.1128/iai.66.7.3349-3354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]