Abstract
Kinesins are motor proteins involved in microtubule (MT)-mediated intracellular transport. They contribute to key cellular processes, including intracellular trafficking, organelle dynamics and cell division. Pathogenic variants in kinesin-encoding genes underlie several human diseases characterized by an extremely variable clinical phenotype, ranging from isolated neurodevelopmental/neurodegenerative disorders to syndromic phenotypes belonging to a family of conditions collectively termed as ‘ciliopathies.’ Among kinesins, kinesin-1 is the most abundant MT motor for transport of cargoes towards the plus end of MTs. Three kinesin-1 heavy chain isoforms exist in mammals. Different from KIF5A and KIF5C, which are specifically expressed in neurons and established to cause neurological diseases when mutated, KIF5B is an ubiquitous protein. Three de novo missense KIF5B variants were recently described in four subjects with a syndromic skeletal disorder characterized by kyphomelic dysplasia, hypotonia and DD/ID. Here, we report three dominantly acting KIF5B variants (p.Asn255del, p.Leu498Pro and p.Leu537Pro) resulting in a clinically wide phenotypic spectrum, ranging from dilated cardiomyopathy with adult-onset ophthalmoplegia and progressive skeletal myopathy to a neurodevelopmental condition characterized by severe hypotonia with or without seizures. In vitro and in vivo analyses provide evidence that the identified disease-associated KIF5B variants disrupt lysosomal, autophagosome and mitochondrial organization, and impact cilium biogenesis. All variants, and one of the previously reported missense changes, were shown to affect multiple developmental processes in zebrafish. These findings document pleiotropic consequences of aberrant KIF5B function on development and cell homeostasis, and expand the phenotypic spectrum resulting from altered kinesin-mediated processes.
Introduction
Kinesins are motor proteins involved in microtubule (MT)-mediated intracellular transport. They contribute to key cellular processes, including intracellular trafficking, organelle dynamics and cell division (1–3). Function of kinesins is required for a number of biological processes, including central nervous system development and function, body axis determination and organ specification (4). Pathogenic variants in kinesin-encoding genes have been reported to underlie at least 30 human diseases that are characterized by an extremely variable clinical phenotype, ranging from isolated neurodevelopmental/neurodegenerative disorders (NDDs) to syndromic phenotypes belonging to a family of conditions collectively termed as ‘ciliopathies’ (5,6) defining a new family of disorders known as ‘kinesinopathies’ (7).
Among kinesins, kinesin-1 is the most abundant MT motor for transport of cargoes towards the plus end of MTs. This motor typically works as tetramer consisting of two identical heavy chains (110–120 kD) and two light chains (60–70 kD) (3). Kinesin-1 is essential for the transport of mitochondria, lysosomes, endoplasmic reticulum- and Golgi-derived vesicles, messenger RNAs, tubulin and intermediate filament subunits, and has a major role in anterograde axonal transport in neurons (8). Three kinesin-1 heavy chain isoforms exist in mammals, which are encoded by the KIF5A (MIM: 602821), KIF5B (MIM: 602809) and KIF5C (MIM: 604593) paralogs, originating from gene duplication events (9), and working as homo/heterodimers (10). While KIF5B is ubiquitously expressed, KIF5A and KIF5C are both neuronal specific in vertebrates (10,11).
In mice, loss-of-function (LoF) phenotypes have been reported for all three Kif5 genes. Kif5a null mutants normally develop but are neonatal lethal due to failure to inflate their lungs; on the other hand, postnatal Kif5a inactivation in neurons induced sensory neuron degeneration (12). Similarly, Kif5c knock-out (KO) mice show a mild reduction in brain size and motoneuron number but are otherwise normal and viable (10). In sharp contrast, the phenotype associated with Kif5b LoF is more severe as KO mice do not survive past 11.5 days of development (13). Such striking difference in the severity of phenotype is likely due to the wider expression pattern of Kif5b compared to the other paralogs, as well as the occurrence of functional redundancy in neurons partially compensating for either Kif5a or Kif5c loss.
Because of the severity and early embryonic lethality of Kif5b mutants, the function of this kinesin has poorly been investigated in vivo. Inactivation of Kif5b in neurons revealed its specific role in the development of excitatory synapses and the dendritic transport of mRNA (14). These defects were reported to cause defective synaptic plasticity and memory formation in mice (14). A specific role of this protein in cilia formation has been proposed, as its depletion results in abnormally elongated cilia (15). Moreover, a relevant role of this motor protein in lysosomal, autophagosome and mitochondrial transport has been demonstrated (13,16–18). Kif5b also plays a central role in skeletal morphogenesis promoting chondrocyte maintenance in zebrafish (18), and determining specific defects in cartilage formation with intracellular accumulation of collagen in chondrocytes in mice (19).
Dominantly acting missense variants in KIF5A cause autosomal-dominant spastic paraplegia 10 (SPG10, MIM: 604187) and Charcot-Marie-Tooth type 2 (CMT2), while KIF5A truncating and splice site variants underlie a neonatal form of intractable myoclonus (NEIMY, MIM: 617235) and are associated with an increased susceptibility to amyotrophic lateral sclerosis 25 (ALS25, MIM: 617921), respectively (20,21). Missense variants in KIF5C have been reported to cause a complex NDD characterized by cortical dysplasia associated with other brain malformations (MIM: 615282) (22). More recently, four subjects with de novo missense variants in KIF5B and affected with a syndromic skeletal condition characterized by kyphomelic dysplasia (KD), dysmorphisms and variable degrees of hypotonia with or without DD/ID were reported (23). The authors defined this new condition as KIF5B-related KD. Here we report two additional heterozygous missense variants and a single residue deletion in KIF5B resulting in a clinically wide phenotypic spectrum, ranging from cardiomyopathy with late-onset ophthalmoplegia and progressive skeletal myopathy to syndromic NDD with or without seizures. In vivo analyses provide evidence that the identified disease-associated KIF5B variants, including one of the previously reported variants associated with KD, have consistent pleiotropic consequences. In vitro analyses show that these variants disrupt proper lysosomal, autophagosome and mitochondrial organization as well as cilium biogenesis. Our findings and the critical review of the previous series identify a clinically heterogeneous family of disorders sharing hypotonia, facial dysmorphisms and DD/ID as recurrent features.
Results
Genetic and clinical findings
In the context of an intramural research program dedicated to subjects affected by undiagnosed diseases (Ospedale Pediatrico Bambino Gesù [OPBG], Rome, Italy), trio-based whole-exome sequencing (WES) was performed on an individual with a molecularly unexplained syndromic NDD (Fig. 1A, Table 1, Supplementary Material, Table S1, Supplemental Material, Note: clinical reports) after exclusion of any clinically relevant structural variant by high-resolution SNP array. Clinical data and DNA samples were collected, stored and used as approved by the OPBG Institutional Review Board (ref. 1702_OPBG_2018), after written informed consent. Target enrichment, parallel sequencing, and data processing, sequence alignment to GRCh37, variant filtering and prioritization by allele frequency, predicted functional impact, and inheritance models were performed as previously described (Supplemental Material, Note: methods) (24–27), according to the GATK’s Best Practices (28). WES statistics and data output are reported in Supplementary Material, Table S2. We did not identify functionally relevant variant(s) compatible with known Mendelian disorders based on the expected inheritance model and clinical presentation; however, a de novo missense change (c.1610 T > C, p.Leu537Pro) in KIF5B emerged as an excellent candidate. Additionally, four individuals from two unrelated families reported as heterozygous for different variants in KIF5B were identified via GeneMatcher (Table 1, Supplementary Material, Table S1) (29). The two probands had been investigated by WES using a family-based strategy. Target enrichment kits, sequencing platforms and WES statistics and data output are reported in Supplementary Material, Table S2. In these cases, variant validation and co-segregation analyses confirmed either the de novo origin of the variant (subject 3) or its co-segregation with the trait (family 2). None of the identified variants was present in the gnomAD database (http://exac.broadinstitute.org), and data from the ExAC database revealed that KIF5B is intolerant for LoF variants (pLI = 0.99) and partially intolerant for missense variants (Z-score + 3.06) (http://gnomad.broadinstitute.org). In all subjects, WES data analysis excluded the occurrence of functionally relevant variants affecting known disease genes.
Figure 1.

Clinical features of subject 1 and structural data. (A) The subject (8 years) showed hypotonic face, bitemporal narrowing, bushy and straight eyebrow, hypertelorism, long eyelashes, long palpebral fissures, large ears with thick lobe, broad nasal bridge, anteverted nares, short deep philtrum, everted upper lip and prominent upper and lower vermilion. Explicit permission was obtained to publish photographs of the subject. (B) Multiple sequence alignment of KIF5B orthologs and paralogs around the mutated residues. Residues that are invariant (at least down to Drosophila melanogaster) are grayed. (C) The 3D structure of the human kinesin-1 heavy chain motor domain complexed with tubulin and darpin (PDB 4HNA) showing Asn255 (balls and sticks with red meshes), its interactions (dotted lines) with nearby residues (sticks), the co-crystallized ADP (magenta sticks), and Mg2+ cation (blue). The residues mutated in KIF5B-related KD (Lys91, Thr195, and Gly234; balls and sticks with green meshes) are also shown. The pathogenic missense changes are predicted to perturb KIF5B interaction with ATP/ADP. (D) 3D structure of the coiled-coil region (dimeric) of human kinesin-1 heavy chain (PDB 6IGV) including the site affected by the p.Leu498Pro e p.Leu537Pro substitutions (displayed on both monomers). Leu498 in one monomer interacts with Val509 on the bound monomer, while Leu537 on one monomer interacts with Met470 on the bound monomer.
Table 1.
Summary of the clinical and molecular data of the present and previously published subjects with pathogenic KIF5B variants
| Present report | Itai et al. (23) | |||||||
|---|---|---|---|---|---|---|---|---|
| Patient | Subject 1 | Subject 2 | Subject 3 | Individual 1 | Individual 2 | Individual 3 | Individual 4 | Total (%) |
| Sex | Male | Male | Female | Female | Female | Female | Male | 3 M; 4F |
| Age at last evaluation | 8y3m | 67y | 6y | 3y3m | 24y | 7y6m | 2y1m | 2y1m-67y |
|
KIF5B nucleotide change (amino acid change) |
c.1610 T > C (p.Leu537Pro) |
c.765_767delCAA (p.Asn255del) | c.1493 T > C (p.Leu498Pro) | c.584C > A (p.Thr195Lys) |
c.701G > T (p.Gly234Val) |
c.272A > G (p.Lys91Arg) |
c.272A > G (p.Lys91Arg) |
|
| Inheritance | De novo | Paternally inherited | De novo | De novo | De novo | De novo | De novo | |
| Anomalous neonatal coursea | Yes | Uneventful | Yes | Yes | Yes | Yes | Yes | 6/7 (86%) |
| Feeding/swallowing problemsa | Yes | No | Yes | Yes | Yes | Yes | Yes | 6/7 (86%) |
| Growth (at last evaluation) | 3y | 3y3m | 24y | 9y5m | 2y1m | |||
| Height | 116 cm (−2 SD) |
on average | 92 cm (−0.68 SD) |
89 cm (−1.5 SD) |
142.2 cm (<−2.5 SD) |
102 cm (−5.1 SD) |
71.2 cm (−4.9 SD) |
|
| Weight | 26 kg (+0.06 SD) |
78 kg (+0.70 SD) |
12.40 kg (−1.09 SD) |
11.6 kg (−1.1 SD) |
37.5 kg (<−2 SD) |
21.2 kg (−1.5 SD) |
9.2 kg (−2.2 SD) |
|
| Cognition and behavior | ||||||||
| Global developmental delay | Severe | No | No | No | No | Mild | NR | 2/6 (33%) |
| Intellectual disability | Severe | No | Mild | No | No | Yes | Yes | 4/7 (57%) |
| Neurological features | ||||||||
| Seizures | Yes | No | No | No | Yes | No | No | 2/7 (28%) |
| Hypotonia | Severe | No | Yes | NR | Velopharyngeal insufficiency | Glossoptosis | Tracheomalacia | 5/6 (83%) |
| CNS anomalies | Cortical and subcortical atrophy |
No | NR | Spinal arachnoid cyst, cord herniation | Diffuse low-density areas in white matter | NR | NR | 3/4 (75%) |
| Dysmorphic featuresa | Yes | No | Yes | Yes | Yes | Yes | Yes | 5/6 (83%) |
| Dilated cardiomyopathy | No | Yes (onset 50y) | No | No | No | No | No | 1/7 (14%) |
| Musculo/skeletal anomalies | ||||||||
| Joint contracture/clubfoot | No | Clubfoot | No | Clubfoot, joints contractures | Clubfoot | No | Clubfoot | 4/7 (57%) |
| Scoliosis/kyphosis | Yes | No | No | No | Yes | No | No | 2/7 (28%) |
| Skeletal dysplasia | No | No | No | Yes | Yes | Yes | Yes | 4/7 (57%) |
| Abductor weakness | No | Yes | No | NR | NR | NR | NR | 1/3 (33%) |
| Myopatic features | Yes | Yes | Yes | No | Yes | Yes | Yes | 6/7 (86%) |
Nucleotide and amino acid positions are according to NM_004521.3 and NP_004512.1, respectively. ClinVar ID: SCV002524131 (c.1610 T > C), SCV002524132 (c.765_767delCAA) and SCV002524133 (c.1493 T > C).
NR: not reported.
aSee Supplementary Material, Table S1 for details.
The clinical phenotype characterizing the collected cases was variable. Subject 1 (p.Leu537Pro), a 9-year-old male, showed a NDD characterized by severe DD/ID, early-onset refractory epilepsy, hypotonia, feeding difficulties requiring percutaneous gastrostomy, brain abnormalities (i.e. progressive cortical atrophy and thin corpus callosum) and dysmorphic features (i.e. hypotonic face, bitemporal narrowing, bushy and straight eyebrows, hypertelorism, ptosis, long eyelashes, long palpebral fissures, large ears with thick lobes, broad nasal bridge, anteverted nares, short deep philtrum, everted upper lip, prominent upper and lower vermilion, and mild gingival overgrowth with abnormal teeth eruption). His perinatal period was complicated by respiratory distress requiring ventilation, and recurrent respiratory infections and chronic respiratory insufficiency were reported. Severe scoliosis was reported at 9 years. Subject 2, a 67-year-old male, and his two affected sisters (p.Asn255del) belonged to a family segregating an autosomal dominant form of adult-onset dilated cardiomyopathy (DCM) requiring heart transplant in three members, which was associated with a skeletal myopathy deteriorating over time with ophthalmoplegia, elevated creatine phosphokinase (CPK) levels, contractures of joints and a peripheral neuropathy with progressive ‘stocking’ sensory loss (Supplementary Material, Fig. S1). Subject 3 (p.Leu498Pro) was a 6-year-old female with mild DD, severe hypotonia, swallowing and feeding difficulties since infancy requiring G-tube feeding for the first 7 months of life, and recurrent vomiting. Mild dysmorphic features (i.e. wide forehead with prominent glabella and nasal bridge, smaller chin and nose, and micrognathia) were also noted. A summary of the clinical features of each affected individual is reported in Table 1, while a detailed clinical description of each patient is reported in the supplementary section (Supplemental Note: clinical reports and Supplementary Material, Table S1).
Structural and biochemical characterization of the identified KIF5B variants
The identified variants affect two different regions of the protein. Specifically, Asn255 is a highly conserved residue located within the motor domain of the kinesin (Fig. 1B). Based on the crystal structure of the human kinesin-1 heavy chain motor domain complexed with tubulin and darpin (PDB 4HNA), the in-frame deletion was expected to cause a structural rearrangement of a key helix contributing to the motor domain conformation and its interaction with the tubulin alpha chain, suggesting a possible impact on motor domain functions (Fig. 1C). Of note, three recently reported missense variants implicated in KIF5B-related KD affected residues (Lys91, Thr195 and Gly234) also mapping at the motor domain, clustering in the pocket mediating ATP/ADP binding (Fig. 1C). The two substitutions at codons 498 and 537 involved two highly conserved leucine residues located in the coiled coil region of the protein (Fig. 1B), which has a key function in mediating KIF5B dimerization (30). The available crystal structure of the coiled coil region (dimeric) of the human kinesin-1 heavy chain (PDB 6IGV) was used to explore their predicted structural and functional impact. The two Leu-to-Pro changes affected sites directly contributing to the hydrophobic interactions stabilizing the intermolecular association of the coiled coils into a dimer (Fig. 1D). Remarkably, in both substitutions the replacing residue was a proline, which is a helix-breaker and is expected to affect the proper orientation and conformation of the coiled coil.
To investigate the impact of the identified variants on the stability of the protein, the level of individual mutants in 293 T cells was assessed by transiently transfected experiments, which documented a negligible impact of each variant on protein stability (Supplementary Material, Fig. S2A). A slightly reduced level of the endogenous protein was only documented in primary fibroblasts heterozygous for the KIF5BN255del protein (Supplementary Material, Fig. S2B).
In vitro functional characterization studies
A role of kinesin molecular motors in processes controlling cilia formation and function has been established (5). In particular, a number of kinesins are required in processes mediating cilia length and disassembly as well as in those governing motile cilia-specific functions (31). Recent studies demonstrated that KIF5B localizes at the cilium basal body and regulates ciliogenesis by interacting with CCDC28B, a protein implicated in Bardet-Biedl syndrome 1 (BBS, MIM: 209900) (15). In particular, KIF5B LoF has been reported to promote abnormally elongated cilia formation, indicating a role in cilia length regulation (15). Based on such evidence, we first explored the effect of altered KIF5B function on primary cilium biogenesis and morphology in primary fibroblasts from affected subjects carrying the heterozygous p.Asn255del, p.Leu498Pro and p.Leu537Pro variants. The analysis performed in starved fibroblasts revealed the presence of aberrant primary cilium formation in all patients’ cell lines (Fig. 2). Specifically, cilia with altered morphology (either short or characterized by a basal body in absence of any visible cilium [dot cilium]) were invariably observed in fibroblasts heterozygous for the p.Leu537Pro and p.Asn255del KIF5B mutants, whereas primary fibroblasts carrying the p.Leu498Pro variant showed a large proportion of cells having cilia altered in length and thickness (Fig. 2). These findings indicated a dominant impact of each of the identified variants on cilium biogenesis.
Figure 2.
KIF5B variants cause an aberrant primary cilium morphology. Confocal images showing altered primary cilium morphology in patients’ fibroblasts compared to control cells. Specifically, cilia with altered morphology (either short or characterized by a basal body in absence of any visible cilium [dot cilium], zoomed images) were invariably observed in fibroblasts heterozygous for the variants encoding KIF5BLeu537Pro and KIF5BAsn255del. Similarly, primary fibroblasts carrying the KIF5BLeu498Pro protein showed a large proportion of cells having cilia altered in length and thickness (zoomed images). Primary cilia are labeled with ARL13B (red), basal bodies and nuclei are labeled with pericentrin (green) and DAPI (blue), respectively. Scale bars are respectively 10 μm (left) and 2 μm (right). Cells were analyzed for each line over three independent experiments (50 cells/line each) for a total of 150 cells/line scored. P values were calculated by one-way ANOVA with Tukey’s correction for multiple testing. Graph bars show mean ± SEM.
Intracellular transport and proper MT dynamics is fundamental for cellular function, survival and morphogenesis (1,2). Kinesin superfamily proteins are important molecular motors that directionally transport various cargos, including membranous organelles, protein complexes and mRNAs, via their interaction with MTs and the modulation of their dynamic organization (2,32). Based on these considerations, we investigated the distribution and morphology of the major intracellular compartments and organelles (i.e. lysosomes, autophagosomes, Golgi apparatus and mitochondria) in patients’ fibroblasts. Confocal microscope analysis performed on cells endogenously expressing each of the three KIF5B mutated alleles showed a marked increase in the size and number of lysosomes, which also display an anomalous distribution that was particularly enriched at the cell periphery (Fig. 3; Supplementary Material, Figs S3 and S4). No grossly altered distribution and morphology of the mitochondrial network was observed. We also noticed an anomalous localization of autophagosomes in all primary cell lines. This compartment, which was characterized by enlarged vesicles, mainly accumulated in the perinuclear region (Fig. 4A), and did not co-localize with lysosomes following autophagic flux induction or bafilomycin treatment (Fig. 4B). Given the involvement of kinesins in Golgi apparatus maintenance (33), we also evaluated the Golgi structure by immunofluorescence analysis, demonstrating a variably scattered pattern of cisternae suggestive of a disarranged Golgi organization, which was particularly evident in cells endogenously expressing the p.Asn255del and p.Leu537Pro KIF5B proteins (Fig. 5).
Figure 3.

Fibroblasts heterozygous for KIF5B variants show an aberrant morphology of lysosomes and their anomalous pericellular aggregation. Confocal imaging shows morphology and distribution of lysosomes, which are enlarged and more numerous in patients’ cells compared to what is observed in control cells. Their distribution is principally in cell periphery (see also Supplementary Material, Fig. S4). In the same panels staining for mitochondria show no grossly altered distribution and morphology of the mitochondrial network. Scale bar is 5 μm. In these experiments for immunofluorescence analyses cells were stained using antibodies against Lamp1 (lysosome marker, green), TOM20 (mitochondria marker, red) and DAPI (DNA marker, blue).
Figure 4.
Disease-causing KIF5B mutations affect autophagosomes morphology and subcellular localization. (A) Confocal microscopy analyses show a peculiar subcellular localization of autophagosomes in patients’ cells compared to control cells. In particular, these organelles localize near the nucleus and are enlarged compared to that observed in control cells, and a few number colocalized with lysosomes residing in the peripheral region of the cell. Scale bars are respectively 10 μm (left) and 2 μm (right). (B) In these panels, this behavior is exacerbated by treatment with bafilomycin. Scale bars are respectively 10 μm (left) and 5 μm (right). The cells were stained with antibodies against Lamp1 (lysosomes marker, red), LC3I/II (autophagosomes marker, green) and DAPI (DNA marker, blue).
Figure 5.

Disease-causing KIF5B mutations affect Golgi network distribution and morphology. Representative confocal images reporting the altered morphology, distribution of Golgi cisternae. Immunofluorescence analysis, performed with anti GM130 (Golgi marker, green) and DAPI (blue), shows Golgi stacks clustered in the perinuclear region in control cells, whereas a scattered pattern of cisternae suggestive of Golgi fragmentation were observed in patients’ cells, with particular evidence in fibroblasts endogenously expressing the KIF5BAsn255del and KIF5Leu537Pro variants. Scale bar is 5 μm.
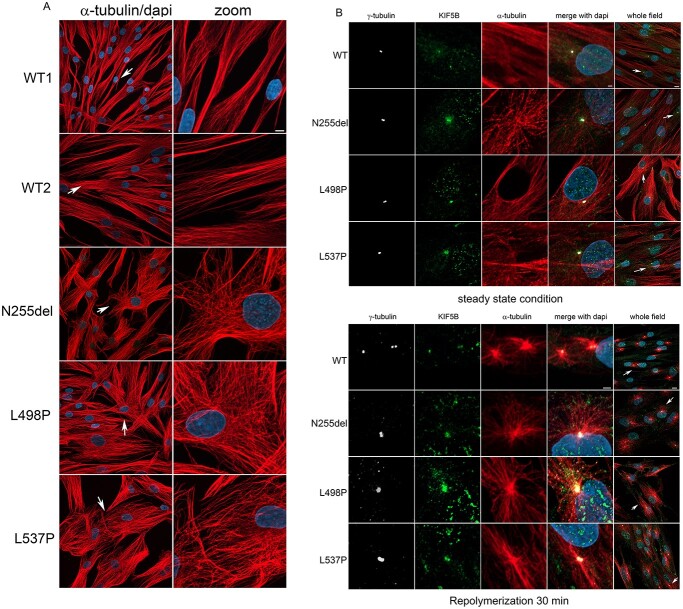
Kinesin-1 is involved in the control of MT dynamics by stimulating MT elongation and rescue (34,35). Since defects in MT dynamics can affect Golgi structures, organelles subcellular localization and ciliogenesis (31,35,36), we hypothesized an altered function of the MT network as the unifying event underlying the observed endophenotype of cells expressing each of the KIF5B variants. To test this hypothesis, MT depolymerization in cells was induced by nocodazole treatment, and Golgi reorganization was assessed. As expected, a scattered Golgi distribution was observed both in patients’ and control cells after treatment (Fig. 6). However, while induction of MT repolymerization rescued proper compact perinuclear organization of the Golgi elements in control cells, patients’ cells failed in properly reorganizing the Golgi network, supporting the involvement of defective MT function in the observed aberrant Golgi organization. To confirm this hypothesis, we analyzed the MT network organization in patients’ cells, which demonstrated a misoriented MT growth (Fig. 7A). Of note, a constitutive localization of the protein at the MTs organizing center (MTOC) was observed in patients’ cells, as shown by γ-tubulin staining (Fig. 7B). Overall, these findings identified an altered MT dynamics and organization as a unifying event in primary cells from the subjects with KIF5B variants causing aberrant vesicular distribution.
Figure 6.
Dependency of Golgi distribution from MTs dynamics. Confocal microscopy analyses show distribution of Golgi cisternae. Images show the perinuclear localization of Golgi in control cell in steady state condition compared to patients’ cell that display a more relaxed Golgi cisternae distribution. Treatment with nocodazole (10 μM) for 35 min promote a substantial MT depolymerization associated with Golgi apparatus fragmentation and dispersion, effect more evident in control cells. After nocodazole washed-out MT were allowed to re-polymerize in a nocodazole free medium for 30 min. Panels show the control MT network normally organized associated to complete re-clustering of Golgi elements in a perinuclear region, unlike the patients’ cells showing a more relaxed Golgi apparatus and a disorganized MT network. This experiment indicates that MTs play a role in maintaining the integrity and location of the Golgi apparatus in cells in interphase. After detergent-extraction in PEM buffer, cells were fixed with ice methanol and then stained with anti-α-tubulin antibody (red), anti Golgin 97 (Golgi marker, green) and DAPI (blue). Scale bar is 5 μm.
Figure 7.
Defective KIF5B function impacts MT orientation and cell shape and localize to MTOC during MT repolymerization as well as in interphase. (A) Confocal microscopy analyses show an evident aberrant orientation of MT in patients’ cells in interphase compared to control cells. Scale bar is 5 μm (B) Panel show a colocalization of KIF5B with the MTOC. In patients’ cells this colocalization is evident during MT repolymerization but persist during cell interphase. In WT cells, it is more evident during MT repolymerization and less in cells in steady state condition. This experiment suggests a role of KIF5B on microtubular dynamics. Scale bar is 2 μm (zoomed images) and 10 μm (whole field) respectively. Cells were stained with anti-α-tubulin antibody (MT marker, red), anti-γ-tubulin (MTOC marker, white), KIF5B (green) and DAPI (blue).
In vivo functional characterization studies
To investigate the developmental impact of each variant in vivo, wild-type (WT) and KIF5B cDNAs containing the presently identified variants and the c.584C > A missense substitution (p.Thr195Lys), the latter as representative of the pathogenic variants identified in individuals presenting with KIF5B-related KD (23), were transcribed in vitro and mRNAs were first injected into 1-cell stage zebrafish transgenic embryos where a cardiac-specific promoter driving green fluorescent protein (GFP) expression allowed to visualize their effect on heart development (Fig. 8A and C). The injected concentration was set based on the amount tolerated by the larvae to not induce any gross phenotypic defect when injected with the WT allele. Injected embryos were imaged at 2 days post-fertilization (dpf) when the major components of the heart have formed and composed of two distinct chambers separated by a contracting ring (37). Overexpression of KIF5BThr195Lys and KIF5BAsn255del resulted in a wide range of morphological defects in heart size and anatomy, which were scored as mild or severe, and embryos expressing these variants presented heart oedemas at 2 dpf (Fig. 8A–C). By contrast, no significant effect on heart morphology was observed following the overexpression of the other mutants or the WT KIF5B mRNA. Using the same strategy, we next evaluated the effect of KIF5B variants on the formation of craniofacial structures by performing Alcian blue staining at 3 dpf to specifically focus on cartilage formation (Fig. 8D and E). Although all the major cartilaginous elements were present, the ceratohyal cartilage angle was less acute in all of the embryos injected with mutant KIF5B variants compared to those expressing the WT protein or non-injected embryos. These results are in line with the observed phenotypes in the zebrafish null mutant (18), and support the hypothesis that these variants behave as dominant alleles in vivo. Together, these findings indicated that altered KIF5B function can lead to cardiac and craniofacial malformations during embryonic development.
Figure 8.
KIF5B is important for proper heart formation and contributes to craniofacial development in zebrafish. (A) Representative images of KIF5B variant-injected embryos for the different morphological classes of heart defects (from top to bottom, no defects, mild and severe defects). (B) Confocal imaging of Tg(cmlc2:eGFP) zebrafish transgenic hearts at 2 days post-fertilization (dpf). At this stage the zebrafish heart is formed by two chambers, the ventricule (Ven) and the atrium (Atr) separated by a contractile ring (yellow arrowheads). Injected transgenic embryos with the different KIF5B variants at 1-cell stage were imaged at 2 dpf, revealing heart formation defects specifically in embryos expressing KIF5BAsn255del variants. Scale bars: 150 μm. (C) Heart defects quantification of 2 dpf larvae injected with KIF5B alleles. With respect to non-injected controls and wild type KIF5B-injected embryos, larvae injected with the KIF5BAsn255del and the KIF5Bp.Thr195Lys variants show a significant increase in the number of embryos with mild and severe heart defects. (****) P-value < 0.0001. Ven: ventricle, Atr: atrium. (D) Alcian blue staining at 3 dpf of zebrafish larvae injected at 1-cell stage with (top. right) wild-type KIF5B (bottom, left) KIF5BLeu495Pro (bottom, right) KIF5BAsn255del, compared to the non-injected larvae (top, left). White dashed lines label the ceratohyal cartilages. (E) The ceratohyal angle (Ch-angle) for each condition was measured, revealing a wider angle at the ceratohyal cartilage intersection for all injected variants compared to non-injected control larvae or wild-type KIF5B. ch: ceratohyal; m: Meckel’s; pq: palatoquadrate. (**) P-value = 0.0081 and (****) < 0.0001. Scale bars: 100 μm.
As kinesin-1 is the major anterograde motor complex in axons, we asked whether the identified KIF5B variants could impair axonal transport of organelles, such as mitochondria. To test this hypothesis, we co-expressed each KIF5B variant with a mitochondrial fused protein (phb-GFP, labeling the inner mitochondrial membrane) into single mxn:kalT4 -expressing motoneuron cells labeled at the membrane by a lyn-tagRFP marker (Fig. 9A). The mitochondrial coverage along the axon of sparsely labeled KIF5B variant-expressing CaP motoneurons as well as the distance between the most distal mitochondria and their cell body were measured at 2 dpf when these cells are functional (38). For all injected variants, a similar trend could be observed in particular for p.Thr195Lys and p.Leu498Pro alleles where we measured a significant reduction in the mitochondrial coverage along the axon. In addition, the overexpression of the different KIF5B variants in these cells also significantly resulted in mitochondrial transport defects along their axon (Fig. 9A). Interestingly, this phenotype seemed to be not fully penetrant in each analyzed neuron with some cells showing a normal axonal distribution while other had almost no mitochondria in their axon. This is probably due to the variable extent to which each cell expressed the mutant protein, underling the possibility of a threshold level required to achieve a dominant negative effect to block the transport of mitochondria by endogenous kinesin-1. Overall, these findings suggested that KIF5B variants impair mitochondrial transport in neurons and can act as dominant negative alleles in vivo. Finally, using this same model system, we next assessed the lysosome morphology in vivo. To this goal, we labeled lysosomes in live zebrafish embryos expressing KIF5BWT, KIF5BAsn255del or KIF5BLeu537Pro mRNA. In line with the findings collected in patients’ fibroblasts, embryos injected with the mutant KIF5B mRNAs were characterized by a significant increase in lysosome size and with larger aggregates compared to KIF5BWT expressing siblings (Fig. 9B), as observed in the zebrafish KO mutant (18), supporting a role of KIF5B in lysosome trafficking.
Figure 9.

KIF5B variants affect lysosomes organization in zebrafish, and motoneuron mitochondrial transport in zebrafish and iPSC-derived motoneurons. (A) DNA constructs containing the different KIF5B variants cloned under the inducible UAS cassettes, the mitochondria labeling protein phb-GFP and membrane labeling tag lynRFP (UAS:KIF5B-variant-E2A-PHB-GFP;UAS:lyntagRFP) were injected in the transgenic line Tg(mnx1-hsp70:kalT4;cmlc2:eGFP) at 1-cell stage. In this line, the mnx1 promoter triggers the expression of the injected construct specifically in motoneurons. The motoneuron membrane is labeled by lyntagRFP (magenta), and mitochondria are stained by phb-GFP (green). Scale bar: 50 μm (left panels). Quantification of proportion of motoneuron axon covered by mitochondrial signal (GFP/lyntagRFP signal). Both KIF5BThr195Lys and KIF5BLeu498Pro variants display significant decrease in mitochondria coverage defects compared to KIF5BWT, KIF5BAsn255del and KIF5BLeu537Pro injected variants. (*) P-value = 0.0423 for KIF5BLeu498Pro variant and (**) P-value = 0.0038 for KIF5Bp.Thr195Lys variant (right upper panel). The distance between the cell body and the most distal detected mitochondria along the motoneuron axon was measured and normalized to the total axon length. All injected KIF5B variants display mitochondria displacement defects compared to the KIF5B WT injected variant. (****) P-value < 0.0001 for KIF5Bp.Thr195Lys variant, (**) P-value = 0.0037 for KIF5BLeu498Pro variant, (**) P-value = 0.0058 for KIF5BLeu537Pro variant and (*) P-value = 0.0146 for KIF5BAsn255del variant (right bottom panel). (B) Representative images showing confocal maximum intensity Z projections of zebrafish embryos at around 30 h post-fertilization (hpf) injected at 1-cell stage with KIF5BWT, KIF5BAsn255del and KIF5BLeu537Pro mRNAs and stained with LysoTracker Red DND-99. Dotted lines and circles indicate the head region and the eye, respectively. Asterisks indicate autofluorescence in the yolk tissue. Scale bar: 100 μm. The scatter plot shows the area of all the LysoTracker positive ROIs together analyzed in the head region of different animals in the different experimental groups (N = 8 for KIF5BWT and KIF5BLeu537Pro, N = 9 for KIF5BAsn255del), median with interquartile range of the pooled data are shown for each experimental grouped. Compared to KIF5BWT injected embryos, larvae injected with the variants KIF5BAsn255del and KIF5BLeu537Pro show a significant increase in lysosomes’ size, (*) P-value < 0.05. (C) Immunofluorescence microscopy analysis (left panels) showing staining for anti-TUJ1 antibody (green) and anti-mitochondria (red) in motoneurons differentiated from iPSC clones obtained from primary fibroblasts from Subject 1 (p.Leu537Pro) and an age- and sex-matched healthy control (WT). Scale bar = 5 μm. The bar graph (right panel) shows the quantification of the mitochondria-specific signal (anti-mitochondria antibody) normalized over the neurite’s area analyzed. Data are presented as mean ± SEM, n = 3. *P-value<0.05, according to Mann-Whitney test.
iPSC-derived neurons heterozygous for the p.Leu537Pro substitution show altered mitochondrial distribution along neurites
Assessment of mitochondrial distribution in motoneurons of zebrafish embryos expressing each of the KIF5B mutants documented a defective distal distribution of mitochondria along the axon, which however was not observed in primary fibroblasts of patients. To explore further this aspect in vitro, considering an equivalent cellular context, primary fibroblasts obtained from subject 1 (p.Leu537Pro) were reprogrammed using non-integrating episomal technology into induced pluripotent stem cells (iPSCs). Following pluripotency validation of the generated iPSC lines (data not shown), these clones and three iPSC clones derived from an age- and sex-matched healthy individual were differentiated into motoneurons, and the mitochondrial coverage along neurites was assessed by confocal laser microscopy experiments. For each analyzed motoneuron, neurite length was digitally divided into the proximal (closer to the cell soma) and distal (closer to the neurite ending) halves, and mitochondrial distribution was determined by direct counting. A statistically significant reduction in mitochondrial coverage in the distal part of neurites in motoneurons expressing KIF5BLeu537Pro was observed (Fig. 9C). A reduction in the number of mitochondria was also observed in the proximal part of neurites of these cells, though differences did not reach statistical significance. Overall, these findings provide evidence of the occurrence of an altered distribution of mitochondria along the neurites of motoneurons endogenously expressing the KIF5BLeu537Pro mutant, which likely results from a defective mitochondrial transport in neurons, confirming the in vivo data collected using zebrafish as model system.
Discussion
Kinesin superfamily members are highly conserved motor proteins that bind to MTs in an ATP-dependent and independent manner and can be involved in various functions, including transport of cargo (organelles, RNAs, protein complexes and lipid vesicles) along MTs, regulating the dynamics of cytoplasmic and spindle MTs (1,7,31). Due to its capability to bind numerous different cargoes, KIF5B is known to be involved in a vast number of cellular processes, including vesicle transport between the endoplasmic reticulum and plasma membrane as well as lysosomes and mitochondria positioning. Recently, several studies have revealed a relevant role of kinesin-1 in the control of MT re-growth and sliding (35,39). In particular, the MT sliding mechanism promoted by KIF5B in interphase generates a force that reorganizes the cytoskeleton and drives shape changes and polarization of cells. Despite multiple functions of KIF5B, only recently a syndromic skeletal dysplasia named KIF5B-related KD, characterized by hypotonia and craniofacial dysmorphisms with or without DD/ID, was reported to be mutated in KIF5B residues clustering at the 5′ portion of the gene (23). Here, we describe three additional heterozygous variants in KIF5B located at the more 3′ end of the gene in the terminal portion of the kinesin motor domain and in the coiled coil domain resulting in a clinically wide phenotypic spectrum, ranging from syndromic NDD, with or without seizures, to adult-onset cardiomyopathy with ophtalmoplegia and progressive skeletal myopathy.
In the light of what was previously reported, we excluded occurrence of skeletal signs suggestive of KD in our patients (e.g. bent long bones, narrow thorax, and scoliosis) with the exception of scoliosis for subject 1. We next reviewed clinical features from the previously four published subjects to find common signs with the present series. On the whole, all the subjects showed variable degrees of hypotonia ranging from mild to severe (Table 1, Supplementary Material, Table S1). In particular, the four subjects affected with KD invariably presented with neonatal distress and respiratory failure requiring medical intervention in most cases. Tracheomalacia, glossoptosis and velopharyngeal insufficiency were also present in single individuals. In these subjects, the hypotonia seemed to partially resolve with age. The individuals of the present series were affected by variable degrees of hypotonia, as well, which was particularly severe in subject 1. Subject 3 (p.Leu498Pro) was affected by a multisystemic hypotonic condition starting since the prenatal period with decreased fetal movements. Neonatal period and childhood were characterized by feeding and swallowing difficulties, requiring g-tube feeding. Symptoms seemed to partially ameliorate with age, as well. She also presented with mild DD/ID. Finally, progressive skeletal myopathy, ophthalmoplegia and joint contractures were observed in the affected individuals from family 2, who also showed persistently elevated CPK levels.
We compared the clinical pictures from previously published patients and subject 1 in the present series. Although the small number of individuals and the bias of comparing them at different ages (3 newborns and 2 children aged 8 and 9 years, respectively), we managed to find recurrent facial signs also previously identified in the published series (i.e. bitemporal narrowing, hypertelorism, ptosis, midface hypoplasia with depressed nasal bridge, anteverted nares, a deep short and prominent philtrum, tented upper lip, and micrognathia) were common to almost all individuals (Supplementary Material, Table S1).
KDs are a heterogeneous group of rare skeletal disorders characterized by bent long bones, osteoporosis, narrow thorax and variable vertebral defects (40). They comprise Schwartz-Jampel syndrome type 1 (MIM 255800), Stuve-Wiedeman syndrome (MIM 601559), cartilage hair hypoplasia/methaphyseal dysplasia without hypotrichosis (MIM 250250 and MIM 250460), and osteogenesis imperfecta XVIII (MIM 617952), which are all characterized by severe skeletal involvement with DD/ID never or rarely reported. Moreover, they are all inherited as recessive traits. Three subjects affected by KIF5B-related KD and two individuals in our series presented with variable degrees of DD/ID. So we anticipate DD/ID, hypotonia, and a suggestive facial gestalt as recurrent features of this new class of disorders regardless any other associated signs, which may specifically be related to the individual affected domains. Notably, joint contractures were observed only in subjects carrying pathogenic variants within the kinesin motor domain (p.Asn255del, family 2 of the present series; p.Lys91Arg, p.Thr195Lys and p.Gly234Val, from Itai’s series) (23). These findings suggest that a close correlation between an altered function of the motor domain and joint contractures/club-foot may exist. Based on the wide spectrum of presentation of this heterogeneous group of phenotypes, we propose to group them as KIF5B-related diseases.
Cardiomyopathy is a feature rarely observed in kinesinopahies. To the best of our knowledge, one family with recurring restrictive cardiomyopathy associated with biallelic inactivating KIF20A variants has been reported (41). Among the seven unrelated affected individuals with pathogenic KIF5B variants, cardiomyopathy was observed as a segregating trait only in family 2 of the present series. This might be due to different reasons. First, all the affected members from family 2 (1 father and 3 sibs) developed DCM in their sixth decade of life without any other relevant signs before. Due to the young age of all the other affected subjects, with the exception of individual 2 in Itai’s series (24 years), we cannot exclude possible late-onset DCM in them. Cardiac surveillance is highly recommended in all these subjects. On the other hand, the in frame p.Asn255del could be exclusively related to a ‘cardiac phenotype’ and this is in line with the zebrafish model in which a wide range of cardiac defects has been evidenced. Of note, KIF5B overexpression was found in rats with pathological cardiac hypertrophy caused by polyephrine stimulation (42). In cultured neonatal rat cardiomyocytes, an abnormal and more peripheral localization of mitochondria was found. Silencing of Kif5b reverted this peripheral mitochondrial localization. This finding mirrors our results, which show a significant reduction in the mitochondrial distribution along the axon of motoneuron cells in zebrafish embryos overexpressing individual KIF5B variants and iPSC-derived motoneurons from subject 1. These data suggest that defects in mitochondria trafficking become evident in cells characterized by long cellular processes and intense metabolic activity that makes the transport of these organelles at the same time challenging and crucial.
Overall, our data document a pleiotropic impact of dysregulated KIF5B function on a multitude of cellular processes. Since the altered cilium biogenesis/morphology and aberrant organelle localization can be traced back to altered microtubular dynamics (35,36,43,44), we hypothesize that an altered function of the MT network could be the unifying event underlying the endophenotype of cells expressing each of the KIF5B variants. The aberrant positioning of mitochondria and lysosomes is expected to result in an altered interaction between these organelles, modifying and/or preventing lysosome-mitochondria contacts, a mechanism that is recognized to be important on mediate multiple cellular functions, including organelles network dynamics and metabolite transfer (e.g. calcium, iron, cholesterol and other lipids) (45). The alteration of this network, which has been documented to be involved in multiple NDDs, could contribute to disease pathogenesis, and requires dedicated effort to be investigated. Finally, the clinical heterogeneity characterizing the KIF5B-related diseases appears to mirror the distribution of individual variants. A larger series of affected individuals is anticipated to more accurately profile the genotype–phenotype correlations characterizing this phenotypic spectrum.
Materials and Methods
WES analysis
Subject 1. DNA of the affected subject and his parents was extracted from circulating leukocytes and sequenced by means of Illumina paired end technology coupled with the SureSelect AllExon V.5 (Aligent) enrichment kit. WES raw data were processed and analyzed using an in-house implemented pipeline previously described (25,46), according to the GATK’s Best Practices (28). The UCSC GRCh37/hg19 version of genome assembly was used as a reference for reads alignment by means of Burrows-Wheeler Aligner-Maximal Exact Match (BWA-MEM) tool and the subsequent variant calling with HaplotypeCaller (GATK v3.7) (28,47). Variants functional annotation was made by SnpEff v.4.3 (48) and dbNSFP v.4.0 tools (49). Most relevant in silico impact prediction were also evaluated, such as combined annotation dependent depletion (CADD) v.1.4 (50), Mendelian Clinically Applicable Pathogenicity (M-CAP) v.1.3 (51) and Intervar v.2.0.1 (52). By filtering against our population-matched database (~2000 WES) and public databases (dbSNP150 and gnomAD V.2.1), the analysis was focused on high-quality rare variants which affect coding sequences and splice site regions.
Subject 2. DNA was extracted from peripheral blood leukocytes using Puregene reagents (Gentra Systems Inc.). Genomic DNA was prepared with a customized reagent kit from Kapa Biosystems and captured using the NimbleGen SeqCap VCRome 2 exome capture reagent or xGen lockdown probes. Samples were sequenced on the Illumina HiSeq 2500 platform with v4 chemistry, generating 75 bp or 76 bp pair-end reads. We achieved coverage of ≥15x in ≥90% of targeted regions for all WES samples. Shared heterozygous variants among three affected family members were assessed given the family history suggestive of dominant inheritance. Segregation among remaining unaffected family members was assessed by Sanger sequencing of the KIF5B variant using standard methods.
Subject 3. Using genomic DNA from the proband and parents, the exonic regions and flanking splice junctions of the genome were captured using the IDT xGen Exome Research Panel v1.0 (Integrated DNA Technologies). Massively parallel (NextGen) sequencing was done on an Illumina system with 100 bp or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19, and analyzed for sequence variants using a custom-developed analysis tool. Reported variants were confirmed, if necessary, by an appropriate orthogonal method in the proband and, if submitted, in selected relatives. Additional sequencing technology and variant interpretation protocol has been previously described (53). The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page (http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/).
The pathogenic variants identified in this work have been submitted to ClinVar (SCV002524131 [c.1610 T > C], SCV002524132 [c.765_767delCAA] and SCV002524133 [c.1493 T > C]).
In vitro studies
Constructs
Single nucleotide changes resulting in the Asn255 in-frame deletion, and Leu498Pro and Leu537Pro substitutions were introduced by site-directed mutagenesis in a C-terminal Myc-tagged human KIF5B cDNA cloned in pcDNA3.1 vector (Invitrogen).
Cell cultures
293 T cells and primary fibroblasts were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin–streptomycin, at 37°C with 5% CO2. 293 T cells were seeded in six-well plates the day before transfection. Monolayer were transfected at 70 to 80% confluency with wild-type or mutants Myc-tagged KIF5B expression plasmids, using Fugene 6 transfection reagent (Promega). Forty-eight hours after transfection, cells were lysed and the level of KIF5B was assessed by immunoblotting with an anti-Myc (Cell Signaling) antibody. KIF5B endogenous level was evaluated on fibroblasts lysates using a rabbit polyclonal anti-KIF5B antibody (Abcam). Membranes were probed with an anti-GAPDH antibody (Santa Cruz) or mouse monoclonal β-actin antibody (Sigma) for protein normalization.
Confocal analysis
Confocal analysis was performed on a Zeiss LSM 980 with Airyscan2, using the 63x oil objective and excitation spectral laser lines at 405, 488, 546, 594 and 633 nm.
Primary cilium staining
Cells were plated onto cover slips, maintained 24 h in low serum medium to promote emission of cilia and then fixed in 4% paraformaldehyde (PFA). Primary cilium was stained with a rabbit polyclonal anti-ARL13B antibody (Abcam) followed by goat anti-rabbit Alexa Fluor 594 (red), while basal bodies were stained with mouse monoclonal anti-pericentrin (Abcam) followed by goat anti-mouse Alex Fluor 488 (green) and nuclei with DAPI (blue).
Staining of lysosomes, mitochondria, Golgi apparatus and autophagosomes
For immunofluorescence, patients’ and controls fibroblasts were seeded at the density of 20 x 103 in 24-well cluster plates onto 12-μm cover glasses. After 24 h of culture in complete medium, cells were fixed with 3% PFA (30 min at 4°C) or absolute chilled methanol for 10 min at −20°C. After permeabilization with 0.5% TritonX-100 (10 min at room temperature), fibroblasts were stained with mouse monoclonal anti-Lamp1 antibody (Cell signaling), and/or rabbit polyclonal TOM20 antibody (Santa Cruz) and rabbit polyclonal GM130 antibody (Abcam), followed by the appropriate secondary antibody (Life Technologies) and DAPI.
To promote the autophagosomes formation fibroblasts were treated with EBSS medium (Earle’s Balanced Salt Solution) with or without bafilomycin for 4 hours, fixed with absolute chilled methanol for 10 min at −20°C. Cells were stained with a rabbit polyclonal anti-LC3I/II antibody (Cell Signaling) followed by the appropriate secondary antibodies (Life Technologies) and DAPI.
Microtubule dynamics analysis
Patients’ and controls fibroblasts were seeded at the density of 20 x 103 in 24-well cluster plates onto 12-μm cover glasses. After 24 h of culture in complete medium, cells were treated with nocodazole (10 μM) for 35 min (R/T) to depolymerize the MTs network. After nocodazole washout with PBS 1X, fibroblasts were incubated with pre-warmed nocodazole free-medium for 30 min to promote MT repolymerization. After detergent-extraction in PIPES, EGTA, magnesium (PEM) buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.9) and PEM Buffer supplemented with 0.1% Triton X-100 and taxol (20 μM), cells were fixed with absolute chilled methanol for 10 min at −20°C. Cells were stained with mouse monoclonal anti Golgin-97 (green) (Life Technologies) and rat monoclonal anti α-tubulin (red) (Abcam), followed by the appropriate secondary antibodies (Life Technologies) and DAPI.
KIF5B subcellular localization
Fibroblasts were seeded at the density of 20 x 103 in 24-well cluster plates onto 12-mm cover glasses and cultured for 24 h in complete medium. KIF5B subcellular localization was evaluated in steady state condition and during MT repolymerization after nocodazole treatment. After detergent-extraction in PEM buffer and PEM Buffer supplemented with 0.1% Triton X-100 and taxol (20 μM), cells were fixed with absolute chilled methanol for 10 min at −20°C. Staining was performed using mouse monoclonal ɣ-tubulin (white) (Abcam), rabbit polyclonal anti KIF5B (green) (Abcam) and rat monoclonal anti a-tubulin (red), followed by the appropriate secondary antibodies (Life Technologies) and DAPI.
Derivation, characterization and differentiation of patient-derived iPSC lines
iPSCs were derived from primary fibroblasts of subject 1 (p.Leu537Pro) reprogrammed by using the non-integrating episomal technology (Epi5 Episomal iPSC Reprogramming Kit, Thermofisher Scientific), following manufacturer’s instructions. Control iPSCs were derived from fibroblasts of an age- and sex-matched healthy subject using the same experimental approach. Pluripotency of the obtained iPSCs was verified by assessing positivity for alkaline phosphatase activity (86R, Leukocyte Alkaline Phosphatase Kit, Sigma Aldrich) and SOX2 (14-9811-82, Invitrogen, 1:100), POU5F1/OCT4 (MA5-14845, Thermofisher, 1:400), SSEA4 (sc-21 704, Santa Cruz, 1:250), PODXL/TRA-1-60 (sc-21 705, Santa Cruz, 1:100) expression by immunofluorescence microscopy analysis. iPSC lines were grown in feeder-free condition using Matrigel (BD Biosciences) in mTeSR1 Plus (Stemcell Technologies) at 37°C, 5% CO2. Differentiation of iPSC lines into motoneurons was performed as previously reported (54).
Assessment of mitochondrial distribution in iPSC-derived motoneurons
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature, washed with PBS, and blocked with 10% bovine serum and 0.1% Triton X-100 (Sigma Aldrich). Anti-TUJ1 (1:500, T2200, Sigma Aldrich) and anti-mitochondria (1:200, NB600-556, Novus Biologicals) were used as primary antibodies, and appropriate secondary antibodies conjugated with AlexaFluor 488 or AlexaFluor 555 (1:500, Life Technologies) were used. Coverslips were mounted using PBS/Glycerol (1:1). General morphological features of cells were assessed by using an inverted microscope Evos (Life Technologies), equipped with a dedicated capturing software. Slides processed for immunofluorescence were observed using a Leica Stellaris laser-scanning confocal microscope (Leica Microsystems). Sequential images were acquired using the LAS X software (Leica Microsystems). For mitochondrial distribution analyses, 16 and 26 samples for the control- and patient-derived cells were considered, respectively, by measuring the fluorescence intensity of anti-mitochondria signals normalized by the area covered by neurites (Image J software). Neurite length was divided in two regions (proximal and distal), and counts were performed to appreciate the density of mitochondria in the proximal and distal portions of neurites. Statistical analysis was performed using Prism software (GraphPad Software). Data were presented as means ± standard error of the mean (SEM) of n ≥ 3 independent experiments performed.
In vivo studies
Zebrafish embryo maintenance
Zebrafish (Danio rerio) were maintained at 28°C on a 14 h light/10 h dark cycle. The collected embryos were cultured in fish water containing 0.003% 1-phenyl-2-thiouera to prevent pigmentation and 0.01% methylene blue to prevent fungal growth in the dish. All fish were housed in the fish facility of our laboratory, which was built according to the local animal welfare standards. All animal procedures were performed in accordance with French and European Union animal welfare guidelines. Larval zebrafish were studied before the onset of sexual differentiation.
Transgenic lines
The mnx1hsp70:kalT4;cmlc2:eGFP vector has been generated by combining four plasmids using the MultiSite Gateway Technology (Invitrogen): p5E-mnx1hsp70 (38,55), pME-KalT4 (56), p3E-polyA and pDEST-cmcl2:eGFP containing Tol2 sites (57). Stable transgenic line Tg(mxn1hsp70:kalT4;cmlc2:eGFP) was generated by injection of the previously described plasmid at 20 ng/μl with tol2 transposase mRNA at 20 ng/μl in one-cell stage wild-type embryos. Tg(UAS:lynGFP;cmlc2:GFP) transgenic line was generated by injecting UAS:lynGFP;cmlc2:GFP (58) plasmid with at 10 ng/μl with tol2 transposase mRNA at 20 ng/μl in one-cell stage wild-type embryos.
Molecular cloning
The human cDNA variants for KIF5B (NM_004521.2, NCBI) and two other DNA cassettes: -E2A-phb-GFP-pA (59) and 10UAS:lyn-tagRFP-pA, were cloned into the pUAS:ubc-pA plasmid (60) with the Gibson Assembly Cloning Kit (E5510S, New England Biolabs).
For the generation of the pCS2:KIF5B_variant-E2A-lynGFP-pA constructs, the human cDNA variants for KIF5B and the E2A-lynGFP cassette were sub-cloned into a pCS2+ vector using the Gibson Assembly Cloning Kit (E5510S, New England Biolabs). KIF5B variants mRNA were produced from pCS2:KIF5B-E2A-lynGFP-pA DNA constructs linearized by restriction digest using Not I enzyme (R3189S, New England Biolabs) and mRNA synthesis was performed using the Sp6 minimal promoter present in the pCS2+ vector and the mMESSAGE mMACHINE SP6 Transcription Kit (AM1340, Invitrogen).
Microinjections
Injection of KIF5B variants mRNA at a concentration of 100 ng/μl at 1 cell stage was performed in Tg(mnx1hsp70:kalT4;cmlc2:eGFP) and Tg(UAS:lynGFP;cmlc2:eGFP) embryos to assess the effect of the different KIF5B variants on heart formation (through the cmlc2:eGFP expression) at 2 dpf and craniofacial development (by Alcian blue staining) at 3 dpf (58).
Consequences on mitochondrial transport was assessed by injecting each 14UAS:KIF5B_variant-E2A-phb-GFP-pA;10UAS:lyn-tagRFP-pA DNA constructs into Tg(mnx1hsp70: kalT4;cmlc2:eGFP) embryos at 1 to 4 cell stage, at a concentration of 25 ng/μl for single cell or sparse labeling.
Alcian blue staining
Alcian blue staining was performed using an acid-free protocol (61). Embryos were fixed in 100% ethanol overnight at 4°. Samples were the rehydrated in 50% ethanol in diluted in PBS for 10 min at room temperature. Embryos were then transferred in staining solution containing 0.02% Alcian blue solution (A5268, Sigma Aldrich), 40 mM MgCl2 in 70% ethanol solution. Cartilage was stained overnight at room temperature. The staining solution was then removed, embryos were washed once with H2O and bleached using 3% H2O2 (H1009, Sigma Aldrich) and 2% KOH (26668.296, VWR Chemicals) for 20 min at room temperature and no cover. Clearing was performed using sequential glycerol/KOH solutions: first 20% glycerol/0.25% KOH for 3 h, 50% glycerol/0.25% KOH for 3 h and finally stored in 50% glycerol/0.1% KOH at 4° until imaging.
Lysosome labeling
Embryos injected with KIF5B variants mRNA at 1-cell stage were live-stained at 24hpf with 70 μM LysoTracker Red DND-99 (#L7528, Invitrogen) in E3 medium at room temperature for 1 h and 30 min and were washed several times with E3 medium.
Imaging and statistical analyses
Zeiss LSM 780 and 880 confocal microscopes (Zeiss) were used for confocal microscopy, employing a 40x water immersion objective. Z-volumes were acquired with a 1 um resolution and images processed using ImageJ, Adobe Photoshop, Adobe Illustrator and Volocity software (v5.3). Axonal length and mitochondria coverage and distance to the cell body was performed using Fiji the plugin function NeuronJ in Fiji (62). For lysosome imaging, labeled embryos at 30 hpf were embedded in 1.5% low melting agarose (A9414, Sigma Aldrich) in E3 medium. Z-volumes were acquired at the Stellaris 5 confocal microscope (Leica) under 10x and 20x objective with 2.5 or 3 μm step size and analyzed with Fiji. Per each fish in the different experimental groups (N = 8 for KIF5B WT, N = 9 for p.Asn255del, p.Leu537Pro) the area of n = 10 clearly visible and randomly selected LysoTracker positive lysosomal vesicles in the head region was calculated using the ‘ROI manager’ plugin, by two independent researchers.
Statistical analyses were performed using the GraphPadPrism7.0a software. Multiple comparison analyses (ANOVA) using the Kruskal-Wallis test were performed to determine significance of KIF5B variants on CaP motoneuron axonal mitochondrial coverage and transport (in at least n = 13 CaP motoneurons for the different KIF5B alleles over n = 23 KIF5B WT expressing CaP cells) and craniofacial Ch-angle feature (in at least n = 6 embryos for the KIF5B variants compared to n = 8 WT KIF5B injected larvae). Effects on heart formation were statistically assessed by performing a Welch’s test (in n = 235 larvae at least for the tested KIF5B variants compared to 362 WT KIF5B injected embryos from three independent experiments). To assess statistical significance of the increased lysosomes size observed between the experimental groups, log transformed data were analyzed with hierarchical ANOVA to account for nested values, using two-stage procedure of Benjamini, Krieger and Yekutieli to correct for multiple comparison.
Supplementary Material
Acknowledgements
The authors thank the subjects and their families for participating in this study, and Serenella Venanzi (Istituto Superiore di Sanità) for her skillful technical assistance.
Conflict of Interest statement. K.McW. is an employee of GeneDx, LLC. The remaining authors declare no competing interests.
Lead contact: Marco Tartaglia
Contributor Information
Elisabetta Flex, Department of Oncology and Molecular Medicine, Istituto Superiore di Sanità, 00161 Rome, Italy.
Shahad Albadri, Sorbonne Université, INSERM, CNRS, Institut de la Vision, 17 Rue Moreau, F-75012 Paris, France.
Francesca Clementina Radio, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Serena Cecchetti, Core Facilities, Istituto Superiore di Sanità, 00161 Rome, Italy.
Antonella Lauri, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Manuela Priolo, UOSD Genetica Medica, Grande Ospedale Metropolitano "Bianchi Melacrino Morelli", 89124 Reggio Calabria, Italy.
Marta Kissopoulos, Department of Oncology and Molecular Medicine, Istituto Superiore di Sanità, 00161 Rome, Italy.
Giovanna Carpentieri, Department of Oncology and Molecular Medicine, Istituto Superiore di Sanità, 00161 Rome, Italy; Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Giulia Fasano, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Martina Venditti, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Valentina Magliocca, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Emanuele Bellacchio, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Carrie L Welch, Department of Pediatrics, Columbia University Irving Medical Center, NY, New York 10032, USA.
Paolo C Colombo, Department of Medicine, Columbia University Irving Medical Center, NY, New York 10032, USA.
Stephanie M Kochav, Department of Medicine, Columbia University Irving Medical Center, NY, New York 10032, USA.
Richard Chang, Division of Metabolic Disorders, Children's Hospital of Orange County (CHOC), CA, Orange 92868, USA.
Rebekah Barrick, Division of Metabolic Disorders, Children's Hospital of Orange County (CHOC), CA, Orange 92868, USA.
Marina Trivisano, Department of Neuroscience, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Alessia Micalizzi, Translational Cytogenomics Research Unit, Bambino Gesù Children's Hospital, IRCCS, 00146 Rome, Italy.
Rossella Borghi, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Elena Messina, Department of Oncology and Molecular Medicine, Istituto Superiore di Sanità, 00161 Rome, Italy; Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Cecilia Mancini, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Simone Pizzi, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Flavia De Santis, Institut Curie, PSL Research University, INSERM U934, CNRS UMR3215 Paris, France.
Marion Rosello, Sorbonne Université, INSERM, CNRS, Institut de la Vision, 17 Rue Moreau, F-75012 Paris, France.
Nicola Specchio, Department of Neuroscience, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Claudia Compagnucci, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Kirsty McWalter, GeneDx, Gaithersburg, MD 20877, USA.
Wendy K Chung, Department of Pediatrics, Columbia University Irving Medical Center, NY, New York 10032, USA; Department of Medicine, Columbia University Irving Medical Center, NY, New York 10032, USA.
Filippo Del Bene, Sorbonne Université, INSERM, CNRS, Institut de la Vision, 17 Rue Moreau, F-75012 Paris, France.
Marco Tartaglia, Genetics and Rare Diseases Research Division, Ospedale Pediatrico Bambino Gesù, IRCCS, 00146 Rome, Italy.
Funding
Ministero della Salute (Ricerca 5x1000, CCR-2017-23669081, RCR-2020-23670068_001 and RCR-2021-23671215 to M.T.; GR-2019-12368907 and Ricerca Corrente 2022, to C.C.; GR-2019-12368907 to A.L.; RF-2018-12366931, to F.C.R.); Ministero della Ricerca (FOE 2019, to M.T.); Horizon 2020; Marie Skłodowska-Curie IF (844636 to A.L.); Fondazione Umberto Veronesi (individual research fellowship to A.L.); Programme Investissements d’Avenir IHU FOReSIGHT (ANR-18-IAHU-01 to F.D.B.); Agence Nationale de la Recherche (ANR-18-CE16-0017-01 iReelAx, ANR-20-CE13-0011-02 CodeAX to F.D.B.); UNADEV/AVIESAN (UNADEV-19UU51-DELBENE, to F.D.B.) and Fondation pour la Recherche Médicale (MND202003011460, MND202003011485, to F.D.B.) M.R. was supported by a fellowship by Fondation pour la Recherche Médicale (ECO20170637481).
References
- 1. Hirokawa, N., Noda, Y., Tanaka, Y. and Niwa, S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol., 10, 682–696. [DOI] [PubMed] [Google Scholar]
- 2. Hirokawa, N. and Noda, Y. (2008) Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol., 88, 1089–1118. [DOI] [PubMed] [Google Scholar]
- 3. Verhey, K.J., Kaul, N. and Soppina, V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys., 40, 267–288. [DOI] [PubMed] [Google Scholar]
- 4. Konjikusic, M.J., Gray, R.S. and Wallingford, J.B. (2021) The developmental biology of kinesins. Dev. Biol., 469, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reiter, J.F. and Leroux, M.R. (2017) Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol., 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hildebrandt, F., Benzing, T. and Katsanis, N. (2011) Ciliopathies. N. Engl. J. Med., 364, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalantari, S. and Filges, I. (2020) 'Kinesinopathies': emerging role of the kinesin family member genes in birth defects. J. Med. Genet., 57, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirokawa, N., Niwa, S. and Tanaka, S. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron, 68, 610–638. [DOI] [PubMed] [Google Scholar]
- 9. Miki, H., Setou, M., Kaneshiro, K. and Hirokawa, N. (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. U. S. A., 98, 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanai, K., Okada, Y., Tanaka, Y., Harada, A., Terada, S. and Hirokawa, N. (2000) KIF5C, a novel neuronal kinesin enriched in motor neurons. J. Neurosci., 20, 6374–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Campbell, P.D. and Marlow, F.L. (2013) Temporal and tissue specific gene expression patterns of the zebrafish kinesin-1 heavy chain family, kif5s, during development. Gene Expr. Patterns, 13, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia, C.H., Roberts, E.A., Her, L.S., Liu, X., Williams, D.S., Cleveland, D.W. and Goldstein, L.S.B. (2003) Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J. Cell Biol., 161, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka, Y., Kanai, Y., Okada, Y., Nonaka, S., Takeda, S., Harada, A. and Hirokawa, N. (1998) Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell, 93, 1147–1158. [DOI] [PubMed] [Google Scholar]
- 14. Zhao, J., Fok, A.H.K., Fan, R., Kwan, P.Y., Chan, H.L., Hoi-Ying Lo, L., Chan, Y.S., Yung, W.H., Huang, J., Wan Lai, C.S. et al. (2020) Specific depletion of the motor protein KIF5B leads to deficits in dendritic transport, synaptic plasticity and memory. elife, 21, e53456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Novas, R., Cardenas-Rodriguez, M., Lepanto, P., Fabregat, M., Rodao, M., Fariello, M.I., Ramos, M., Davison, C., Casanova, G., Alfaya, L. et al. (2018) Kinesin 1 regulates cilia length through an interaction with the Bardet-Biedl syndrome related protein CCDC28B. Sci. Rep., 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakata, T. and Hirokawa, N. (1995) Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol., 131, 1039–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence, E.J., Boucher, E. and Mandato, C.A. (2016) Mitochondria-cytoskeleton associations in mammalian cytokinesis. Cell Div, 11, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cardoso, C.M.P., Groth-Pedersen, L., Høyer-Hansen, M., Kirkegaard, T., Corcelle, E., Andersen, J.S., Jäättelä, M. and Nylandsted, J. (2009) Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS One, 4, e4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santos-Ledo, A., Garcia-Macia, M., Campbell, P.D., Gronska, M. and Marlow, M.F. (2017) Kinesin-1 promotes chondrocyte maintenance during skeletal morphogenesis. PLoS Genet., 13, e1006918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He, S., Xue, W., Duan, Z., Sun, Q., Li, X., Gan, H., Huang, J. and Qu, J.Y. (2017) Biomed. Opt. Express, 8, 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicolas, A., Kenna, K.P., Renton, A.E., Ticozzi, N., Faghri, F., Chia, R., Dominov, J.A., Kenna, B.J., Nalls, M.A., Keagle, P. et al. (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron, 97, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. David Brenner, D., Yilmaz, R., Müller, K., Grehl, T., Petri, S., Meyer, T., Grosskreutz, J., Weydt, P., Ruf, W., Neuwirth, C. et al. (2018) Hot-spot KIF5A mutations cause familial ALS. Brain, 141, 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Itai, T., Wang, Z., Nishimura, G., Ohashi, H., Guo, L., Wakano, Y., Sugiura, T., Hayakawa, H., Okada, M., Saisu, T. et al. (2022) De novo heterozygous variants in KIF5B cause kyphomelic dysplasia. Clin. Genet., 102, 3–11. [DOI] [PubMed] [Google Scholar]
- 24. Michels, S., Foss, K., Park, K., Golden-Grant, K., Saneto, R., Lopez, J. and Mirzaa, G.M. (2017) Mutations of KIF5C cause a neurodevelopmental disorder of infantile-onset epilepsy, absent language, and distinctive malformations of cortical development. Am. J. Med. Genet. A, 173, 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flex, E., Martinelli, S., Van Dijck, A., Ciolfi, A., Cecchetti, S., Coluzzi, E., Pannone, L., Andreoli, C., Radio, F.C., Pizzi, S. et al. (2019) Aberrant function of the C-terminal tail of HIST1H1E accelerates cellular senescence and causes premature aging. Am. J. Hum. Genet., 105, 493–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motta, M., Pannone, L., Pantaleoni, F., Bocchinfuso, G., Radio, F.C., Cecchetti, S., Ciolfi, A., Di Rocco, M., Elting, M.W., Brilstra, E.H. et al. (2020) Enhanced MAPK1 function causes a neurodevelopmental disorder within the RASopathy clinical spectrum. Am. J. Hum. Genet., 107, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Radio, F.C., Pang, K., Ciolfi, A., Levy, M.A., Hernández-García, A., Pedace, L., Pantaleoni, F., Liu, Z., de Boer, E., Jackson, A. et al. (2021) SPEN haploinsufficiency causes a neurodevelopmental disorder overlapping proximal 1p36 deletion syndrome with an episignature of X chromosomes in females. Am. J. Hum. Genet., 108, 502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van der Auwera, G.A., Carneiro, M., Hartl, C., Poplin, R., del Angel, G., Levy-Moonshine, A., Jordan, T., Shakir, K., Roazen, D., Thibault, J. et al. (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics, 43, 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sobreira, N., Schiettecatte, F., Valle, D. and Hamosh, A. (2015) GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat., 36, 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeBoer, S.R., You, Y., Szodorai, A., Kaminska, A., Pigino, G., Nwabuisi, E., Wang, B., Estrada-Hernandez, T., Kins, S., Brady, S.T. et al. (2008) Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry, 47, 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reilly, M.L. and Benmerah, A. (2019) Ciliary kinesins beyond IFT: cilium length, disassembly, cargo transport and signalling. Biol. Cell., 111, 79–94. [DOI] [PubMed] [Google Scholar]
- 32. Hirokawa, N. (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 279, 519–526. [DOI] [PubMed] [Google Scholar]
- 33. Dafinger, C., Liebau, M.C., Elsayed, S.M., Hellenbroich, Y., Boltshauser, E., Korenke, G.C., Fabretti, F., Janecke, A.R., Ebermann, I., Nürnberg, G. et al. (2011) Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J. Clin. Invest., 121, 2662–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daire, V., Giustiniani, J., Leroy-Gori, I., Quesnoit, M., Drevensek, S., Dimitrov, A., Perez, F. and Poüs, C. (2009) Kinesin-1 regulates microtubule dynamics via a c-Jun N-terminal kinase-dependent mechanism. J. Biol. Chem., 284, 31992–32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marceiller, J., Drechou, A., Durand, G., Perez, F. and Poüs, C. (2005) Kinesin is involved in protecting nascent microtubules from disassembly after recovery from nocodazole treatment. Exp. Cell Res., 304, 483–492. [DOI] [PubMed] [Google Scholar]
- 36. Groth-Pedersen, L., Stampe Ostenfeld, L.M., Høyer-Hansen, M., Nylandsted, J. and Jäättelä, M. (2007) Vincristine induces dramatic lysosomal changes and sensitizes cancer cells to lysosome-destabilizing siramesine. Cancer Res., 67, 2217–2225. [DOI] [PubMed] [Google Scholar]
- 37. Brown, D.R., Samsa, L.A., Qian, L. and Liu, J. (2016) Advances in the study of heart development and disease using zebrafish. J. Cardiovasc. Dev. Dis., 3, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bercier, V., Hubbard, J.M., Fidelin, K., Duroure, K., Auer, T.O., Revenu, C., Wyart, C. and Del Bene, F. (2019) Dynactin1 depletion leads to neuromuscular synapse instability and functional abnormalities. Mol. Neurodegener., 14, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu, W. and Gelfand, V.I. (2017) Moonlighting motors: kinesin, dynein, and cell polarity. Trends Cell Biol., 27, 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cormier-Daire, V., Geneviève, D., Munnich, A. and Le Merrer, M. (2004) New insights in congenital bowing of the femora. Clin. Genet., 66, 169–176. [DOI] [PubMed] [Google Scholar]
- 41. Louw, J.J., Nunes Bastos, R., Chen, X., Verdood, C., Corveleyn, A., Jia, Y., Breckpot, J., Gewillig, M., Peeters, H., Santoro, M.M. et al. (2018) Compound heterozygous loss-of-function mutations in KIF20A are associated with a novel lethal congenital cardiomyopathy in two siblings. PLoS Genet., 14, e1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tigchelaar, W., de Jong, A.M., Bloks, V.W., van Gilst, W.H., de Boer, R.A. and Silljé, H.H. (2016) Hypertrophy induced KIF5B controls mitochondrial localization and function in neonatal rat cardiomyocytes. J. Mol. Cell. Cardiol., 97, 70–81. [DOI] [PubMed] [Google Scholar]
- 43. Conkar, D. and Firat-Karalar, E.N. (2021) Microtubule-associated proteins and emerging links to primary cilium structure, assembly, maintenance, and disassembly. FEBS J., 288, 786–798. [DOI] [PubMed] [Google Scholar]
- 44. Wehland, J., Henkart, M., Klausner, R. and Sandoval, I.V. (1983) Role of microtubules in the distribution of the golgi apparatus: effect of taxol and microinjected anti-alpha-tubulin antibodies. Proc. Natl. Acad. Sci. U. S. A., 80, 4286–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cisneros, J., Belton, T.B., Shum, G.C., Molakal, C.G. and Wong, Y.C. (2022) Mitochondria-lysosome contact site dynamics and misregulation in neurodegenerative diseases. Trends Neurosci., 45, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin, Y.C., Niceta, M., Muto, V., Vona, B., Pagnamenta, A.T., Maroofian, R., Beetz, C., van Duyvenvoorde, H., Dentici, M.L., Lauffer, P. et al. (2021) SCUBE3 loss-of-function causes a recognizable recessive developmental disorder due to defective bone morphogenetic protein signaling. Am. J. Hum. Genet., 108, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li, H. and Durbin, R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., Land, S.J., Lu, X. and Ruden, D.M. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu, X., Jian, X. and Boerwinkle, E. (2013) dbNSFP v2.0: a database of human nonsynonymous SNVs and their functional predictions and annotations. Hum. Mutat., 34, E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kircher, M., Witten, D.M., Jain, P., O'Roak, B.J., Cooper, G.M. and Shendure, J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jagadeesh, K., Wenger, A., Berger, M., Guturu, H., Stenson, P., Cooper, D., Bernstein, J. and Bejerano, G. (2016) M-CAP eliminates a majority of variants with uncertain significance in clinical exomes at high sensitivity. Nat. Genet., 48, 1581–1586. [DOI] [PubMed] [Google Scholar]
- 52. Li, Q. and Wang, K. (2017) InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet., 100, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Retterer, K., Juusola, J., Cho, M.T., Vitazka, P., Millan, F., Gibellini, F., Vertino-Bell, A., Smaoui, N., Neidich, J., Monaghan, K.G. et al. (2016) Clinical application of whole-exome sequencing across clinical indications. Genet. Med., 18, 696–704. [DOI] [PubMed] [Google Scholar]
- 54. Corti, S., Nizzardo, M., Simone, C., Falcone, M., Nardini, M., Ronchi, D., Donadoni, C., Salani, S., Riboldi, G., Magri, F. et al. (2012) Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci. Transl. Med., 4, 165ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dalgin, G., Ward, A.B., Hao, L.T., Beattie, C.E., Nechiporuk, A. and Prince, V.E. (2011) Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development, 138, 4597–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Distel, M., Wullimann, M.F. and Köster, R.W. (2009) Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. U. S. A., 106, 13365–13370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kwan, K.M., Fujimoto, E., Grabher, C., Mangum, B.D., Hardy, M.E., Campbell, D.S., Parant, J.M., Yost, H.J., Kanki, J.P. and Chien, C.B. (2007) The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn., 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- 58. Di Donato, V., De Santis, F., Albadri, S., Auer, T.O., Duroure, K., Charpentier, M., Concordet, J.P., Gebhardt, C. and Del Bene, F. (2018) An attractive reelin gradient establishes synaptic lamination in the vertebrate visual system. Neuron, 97, 1049–1062. [DOI] [PubMed] [Google Scholar]
- 59. Auer, T.O., Xiao, T., Bercier, V., Gebhardt, C., Duroure, K., Concordet, J.P., Wyart, C., Suster, M., Kawakami, K. and Wittbrodt, J. (2015) Deletion of a kinesin I motor unmasks a mechanism of homeostatic branching control by neurotrophin-3. elife, 4, e05061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Horstick, E.J., Jordan, D.C., Bergeron, S.A., Tabor, K.M., Serpe, M., Feldman, B. and Burgess, H.A. (2015) Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic Acids Res., 43, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walker, M.B. and Kimmel, C.B. (2007) A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem., 82, 23–28. [DOI] [PubMed] [Google Scholar]
- 62. Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, P., Rueden, C., Saalfeld, S., Schmid, B. et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.