Abstract
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H− have emerged as important causes of diarrheal diseases and the hemolytic-uremic syndrome in Germany. In this study, we characterized a 32-kb fragment of the plasmid of SF EHEC O157:H−, pSFO157, which differs markedly from plasmid pO157 of classical non-sorbitol-fermenting EHEC O157:H7. We found a cluster of six genes, termed sfpA, sfpH, sfpC, sfpD, sfpJ, and sfpG, which mediate mannose-resistant hemagglutination and the expression of fimbriae. sfp genes are similar to the pap genes, encoding P-fimbriae of uropathogenic E. coli, but the sfp cluster lacks homologues of genes encoding subunits of a tip fibrillum as well as regulatory genes. The major pilin, SfpA, despite its similarity to PapA, does not cluster together with known PapA alleles in a phylogenetic tree but is structurally related to the PmpA pilin of Proteus mirabilis. The putative adhesin gene sfpG, responsible for the hemagglutination phenotype, shows significant homology neither to papG nor to other known sequences. Sfp fimbriae are 3 to 5 nm in diameter, in contrast to P-fimbriae, which are 7 nm in diameter. PCR analyses showed that the sfp gene cluster is a characteristic of SF EHEC O157:H− strains and is not present in other EHEC isolates, diarrheagenic E. coli, or other Enterobacteriaceae. The sfp gene cluster is flanked by two blocks of insertion sequences and an origin of plasmid replication, indicating that horizontal gene transfer may have contributed to the presence of Sfp fimbriae in SF EHEC O157:H−.
Adherence is one of the prerequisites for the successful colonization of a eukaryotic host by a bacterium. One of the best understood mechanisms of adherence is that mediated by rod-shaped proteinaceous appendages of the bacterial surface called fimbriae or pili. Well-studied adhesion systems of pathogenic bacteria are the S-fimbria superfamily (30, 31) and the P (pyelonephritis-associated)-fimbriae of uropathogenic Escherichia coli. The latter are composed of a thin tip fibrillum (2 nm in diameter) carrying the adhesin at its distal end and joined at its proximal end to a more rigid, 7-nm-diameter pilus rod (10). The pap gene cluster consists of 11 genes encoding the main component of the pilus rod (PapA), several minor fimbrial subunits (PapHKEF), the adhesin (PapG), the assembly machinery (PapCDJ), and two regulatory proteins (PapIB) (for a review see reference 17). P-fimbriae are part of a family of adhesive organelles that are characterized by an assembly machinery consisting of a periplasmic chaperone (PapD) and a pore-forming outer membrane usher (PapC) (22).
Enterohemorrhagic E. coli (EHEC) O157:H7 are well known as causative agents of diarrhea, hemorrhagic colitis, and the hemolytic-uremic syndrome (HUS) (29, 45). Whereas the Shiga toxins, the most well-established virulence factor of EHEC, have been extensively studied, the mechanisms underlying the adherence of the bacteria to epithelial cells are only partly understood. One adherence system shared by E. coli O157:H7 and enteropathogenic E. coli (EPEC) is the attaching and effacing mechanism encoded by a pathogenicity island called the locus of enterocyte effacement (42). Recently, Tarr et al. (53) described an adherence-conferring protein of EHEC O157:H7 termed Iha, which is similar to the product of an iron-regulated gene of Vibrio cholerae. The presence of fimbriae on the surface of E. coli O157:H7 has been reported by several investigators (15, 18, 27, 58), but their genetic representation and their molecular structure are still unknown.
A biochemical characteristic of E. coli O157:H7 is that they do not ferment sorbitol, and such non-sorbitol-fermenting (NSF) strains have been isolated throughout the world. In Germany, however, nonmotile EHEC O157, which are able to ferment sorbitol (SF EHEC O157:H−), have also been implicated in outbreaks of HUS (1, 28). SF EHEC O157:H− strains are thought to represent a clonal lineage which has branched off at an early stage during the evolution from an EPEC-like E. coli O55:H7 ancestor to EHEC O157:H7 (16). Therefore, detailed comparison of the genomes of SF O157:H− and NSF O157:H7 strains could give further insight into the emergence of highly pathogenic EHEC. In a recent study striking differences were found in the gene compositions of plasmid pO157 of NSF EHEC O157:H7 and the plasmid of SF EHEC O157:H−, henceforth called pSFO157 (7). During our investigations to further characterize these differences, we discovered a gene cluster present only on pSFO157 and mediating mannose-resistant hemagglutination and the expression of a novel type of fimbriae.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli K-12 strains DH5α and HB101 were used as hosts for recombinant plasmids. The SF EHEC O157:H− strain 3072/96 was isolated from a patient suffering from HUS in Germany in 1996. This strain harbors the stx2 gene as well as the eae gene. The clinical E. coli isolates and the other Enterobacteriaceae used in this study were from our strain collection. Characteristics of the diarrheagenic E. coli strains were described earlier (3, 4, 7).
Bacteria were grown in Luria broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] sodium chloride, pH 7.5) or in CDMT [13 mM K2HPO4, 6 mM KH2PO4, 8 mM (NH4)2SO4, 2 mM sodium citrate, 0.4 mM MgSO4, 0.2% Casamino Acids, 0.2% glucose, 5 μM CaCl2, 0.01% tryptone, pH 7.4]. Solid media were prepared by the addition of 1.6% (wt/vol) Bacto agar.
General recombinant DNA techniques.
Plasmid DNA was purified with Qiagen tip-100 cartridges according to the instructions of the supplier. Purification of DNA from agarose gels was performed using a Prep-A-Gene kit (Bio-Rad). Restriction enzymes and T4 DNA ligase were purchased from Gibco-BRL and New England Biolabs. Plasmids pK18 (43) and pBluescript II KS(+) (Stratagene) were used as cloning vectors. Restriction enzyme analysis, ligation, and transformation were conducted according to standard procedures (49).
Sequence analysis.
Nucleotide sequencing was performed with an ABI Prism Big Dye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems) according to the instructions of the supplier. Sequencing reactions were run on an ABI Prism 377 automatic DNA sequencer (Perkin-Elmer). Both strands of the DNA were sequenced stepwise using customized oligonucleotide primers (ARK Scientific), and each base was determined three times on average.
Sequence analysis was conducted with the Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.) as well as with the Dnasis program (Hitachi Software). Promoter prediction was performed using the NNPP2.1 algorithm (promoter prediction by neural network; http://www.fruitfly.org /seq_tools/promoter.html). To illustrate phylogenetic relationships between SfpA and other major fimbrial subunits, an alignment of the predicted mature peptides was made using the CLUSTAL W program (21). This alignment was then used to calculate a distance matrix from which an unrooted phylogenetic tree was inferred according to the neighbor-joining method (48) using the Phylip 3.57c program package (http://evolution.genetics.washington.edu/phylip.html).
PCR.
PCR was performed using the GeneAmp 9600 PCR system (Perkin-Elmer). Table 1 shows the primers and conditions used for amplification of the sfpA gene (primers sfpA-U and sfpA-L), the sfpDG region (primers sfpDG-U and sfpDG-L), and the region connecting the cloned fragments pSFO157-E14 and pSFO157-E11 (primers wprom-3 and wprom-4). Amplification was carried out in a total volume of 50 μl containing 5 μl of bacterial cell suspension (about 103 bacterial cells suspended in 0.85% [wt/vol] sodium chloride), 30 pmol of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, 5 μl of 10× GeneAmp PCR buffer II, 3 μl of 25 mM MgCl2, and 2 U of AmpliTaq DNA polymerase (Perkin-Elmer). After an initial denaturation step of 5 min at 94°C, the samples were subjected to 30 cycles of denaturing, annealing, and extension (see Table 1). The reaction was completed with a final extension step of 5 min at 72°C. Each reaction was conducted at least twice in independent reactions.
TABLE 1.
PCR primers and amplification conditions used in this study
| Primer | Sequence | Amplification conditions (temp [°C], time [s])
|
Product length (bp) | ||
|---|---|---|---|---|---|
| Denaturing | Annealing | Extension | |||
| sfpA-U | 5′-AGCCAAGGCCAAGGGATTATTA-3′ | 94, 30 | 59, 60 | 72, 60 | 440 |
| sfpA-L | 5′-TTAGCAACAGCAGTGAAGTCTC-3′ | ||||
| sfpDG-U | 5′-GATGCTGGCACCGAAGTCAA-3′ | 94, 30 | 57, 60 | 72, 60 | 1,682 |
| sfpDG-L | 5′-TCCTGCAGCATCCAAGTCAC-3′ | ||||
| wprom-3 | 5′-GCTGGACGCCGGTGCTTATT-3′ | 94, 30 | 61, 60 | 72, 150 | 1,900 |
| wprom-4 | 5′-CCAGATTGCCGCTCGCTTCAG-3′ | ||||
| sfpG-UApa | 5′-CCCGGGCCCGGCTGCGCTGCTGGTGAG-3′ | 94, 15 | 55, 30 | 68, 180a | 3,162 |
| sfpG-LApa | 5′-CCCGGGCCCTCATCTGCCGGTCCCTTAC-3′ | ||||
This PCR was performed using the Expand High Fidelity PCR system. From cycle 11 to 30 the extension time was prolonged 5 s with each cycle. The PCR was completed by a final elongation step of 7 min at 72°C.
Amplification of the sfpG gene (primers sfpG-UApa and sfpG-LApa) for complementation purposes was performed using the Expand High Fidelity PCR system (Roche Molecular Biochemicals) according to the recommendations of the supplier (Table 1).
Colony blot hybridization.
Probes specific for sfpA and sfpDG for colony blot hybridization were prepared by random labeling of amplicons derived from sfpA and sfpDG PCR using DNA of strain 3072/96 as the template. The PCR products were purified from agarose gels and labeled with a Digoxigenin Labeling and Detection kit (Roche Molecular Biochemicals) according to the instructions of the supplier. Colony blot hybridization was performed as described previously (7).
Preparation of fimbriae.
Strains were grown in CDMT for 20 h at 37°C with shaking. Bacteria were harvested by centrifugation (20 min, 1,700 × g, 4°C) and resuspended in 1/10 volume of a solution of 75 mM NaCl and 0.5 mM Tris-HCl, pH 7.4 (32). The cell suspension was incubated for 90 min at 60°C and the bacteria were removed by centrifugation. The thermoeluted fimbrial proteins were concentrated from the supernatant by precipitation with trichloroacetic acid and dissolved in Laemmli sample buffer.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoresis was performed on SDS–15% polyacrylamide gels in a Mini-PROTEAN II Dual Slab Cell (Bio-Rad) according to the method of Laemmli (34). Gels were either stained with Coomassie blue G or the proteins were blotted onto nitrocellulose membranes for immunologic detection.
Amino acid sequencing.
N-terminal sequencing of proteins was performed by automated Edman degradation. Heat-extracted fimbrial proteins were separated by SDS-PAGE and blotted onto a ProBlott membrane (Applied Biosystems). After Coomassie blue staining of the proteins, the 17-kDa band was excised and subjected to protein sequencing employing an Applied Biosystems 476A sequencer.
Preparation of anti-SfpA antiserum and immunoblot analysis.
For preparation of a polyclonal serum to SfpA, we purified SfpA from thermoeluted fimbrial extracts using SDS-PAGE followed by electroelution of the 17-kDa protein from gel slices as described previously (9). The protein eluate was emulsified with ABM-S (Linaris) as the adjuvant and administered subcutaneously to a New Zealand White rabbit. One booster injection was given 4 weeks later. A further 4 weeks later the rabbit was bled, and the resulting serum was investigated for reactivity against SfpA.
Immunoblot analysis was performed as described previously (9). In order to minimize nonspecific reactions, the sera were adsorbed against a vector control strain as follows. A crude extract of E. coli HB101/pK18 was diluted to a final concentration of 10 mg/ml in phosphate-buffered saline and mixed with 1 volume of the serum diluted 1:10. The mixture was incubated for 1 h at 4°C and centrifuged (15 min, 15,000 × g, 4°C), and the supernatant was used in immunoblot analysis. Rabbit normal serum was used at a final dilution of 1:100, and rabbit anti-SfpA-serum was used at a dilution of 1:20,000.
Hemagglutination assays.
Quantitative hemagglutination assays were performed in 96-well, round-bottom microtiter plates. Bacteria were grown in CDMT broth for 20 h at 37°C with shaking, harvested by centrifugation (15 min, 1,700 × g, 4°C), and adjusted in saline to 1010 cells ml−1. Twofold serial dilutions of the bacterial suspensions were prepared in saline. Human erythrocytes were diluted in saline to a 0.5% (vol/vol) suspension, and 100 μl of each bacterial dilution and 100 μl of the erythrocyte suspension were mixed in microtiter plates. After being shaken, the plates were incubated for 4 h at room temperature and then examined for hemagglutination. Wells containing a small pellet of erythrocytes at the bottom were considered hemagglutination negative, and those containing an even sheet of erythrocytes across the well were considered hemagglutination positive. The hemagglutination titer was expressed as the reciprocal of the greatest dilution of bacterial cells that resulted in positive hemagglutination.
For fast qualitative tests of hemagglutination activity, 15 μl of a bacterial culture or a colony suspended in saline was mixed with 15 μl of human erythrocytes (diluted 1:50 in saline) on glass slides. Agglutination of the erythrocytes was examined by eye after 1 min of rocking at room temperature. Hemagglutination assays were performed both in the presence and in the absence of 0.5% methyl α-d-mannopyranoside, a nonmetabolizable mannose analog.
Electron microscopy.
Bacteria grown on Luria broth agar plates were suspended in phosphate-buffered saline and allowed to adhere to a Pioloform-coated grid for 2 min. The grid was then negatively stained with 0.25% uranyl acetate for 60 min and examined with a Zeiss EM10 transmission electron microscope operated at 80 kV. Electron microscopy was performed at the Central Division of Electron Microscopy of the University of Würzburg.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to the DDBJ, EMBL, and GenBank databases under accession number AF228759.
RESULTS
Cloning of a novel fimbrial gene cluster.
In order to compare the properties of plasmid pSFO157 of SF EHEC O157:H− with those of pO157, we performed shotgun cloning of pSFO157 fragments derived from SF EHEC O157:H− strain 3072/96 in the vector pBluescript II KS. The ends of the cloned fragments were sequenced using M13/pUC forward and reverse primers. The sequences were compared to that of pO157 of NSF EHEC O157:H7 reference strain EDL933 (11). One 10.9-kb EcoRI fragment, termed pSFO157-E11, showed no homology to pO157. After subcloning, we obtained a sequence homologous to that of the papA gene, which encodes the major subunit of P-fimbriae of uropathogenic E. coli (UPEC). In addition, we found that a recombinant E. coli K-12 strain harboring pSFO157-E11, but not a vector control strain, was able to agglutinate human red blood cells. From this we hypothesized the presence of pap-like adhesin genes present on pSFO157-E11 and determined the complete sequence of this fragment.
Sequence analysis revealed nine open reading frames (ORFs), eight of which were transcribed in the same direction. The amino acid sequences deduced from six of these were markedly homologous to those of the F13 P-fimbrial proteins of UPEC strain J96, PapA, PapH, PapC, PapD, PapJ, and PapF (36). We therefore termed the ORFs in analogy to the pap fimbrial genes sfpA, sfpH, sfpC, sfpD, sfpJ, and sfpF, standing for “sorbitol-fermenting EHEC O157 fimbriae, plasmid-encoded” (Fig. 1). A further ORF 21 bp downstream of sfpF and without homology to known proteins was termed sfpG, because it shared some properties with the papG gene encoding the adhesin molecule of P-fimbriae (see below).
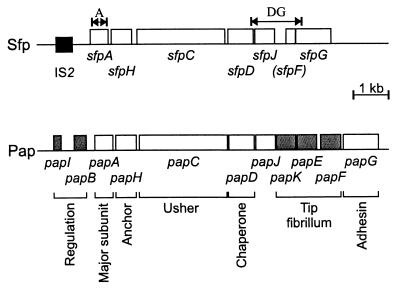
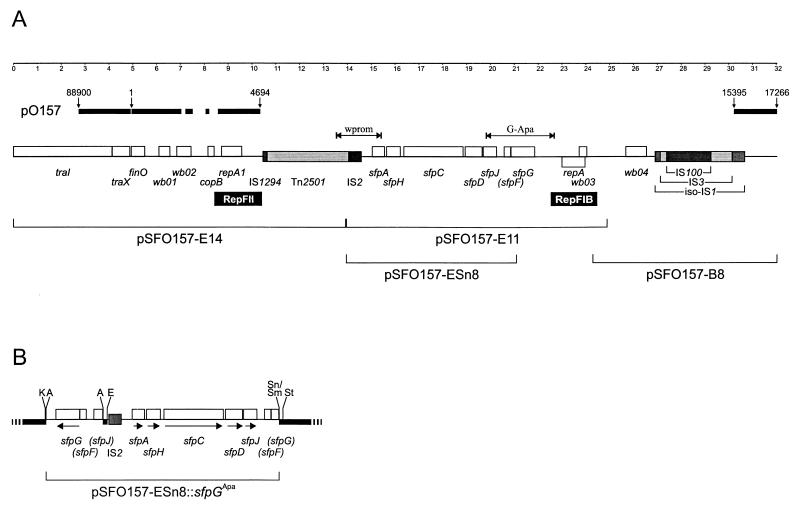
FIG. 1.
Genetic organization of the sfp fimbrial gene cluster and comparison with the pap cluster. Functions of Pap proteins (22) are shown below the map. Shaded boxes indicate components only present in the pap cluster. The black box depicts a DNA region with high homology to IS2. The double arrows indicate regions amplified by sfpA PCR (A) and sfpDG PCR (DG).
The properties of the proteins deduced from sfp genes are summarized in Table 2 and are compared to those of corresponding Pap proteins. As shown by the method of von Heijne (60), all Sfp proteins, with the exception of SfpF, possess putative N-terminal signal peptides like the Pap pilus subunits and assembly proteins (Table 2).
TABLE 2.
Properties of putative sfp gene products and comparison to the corresponding Pap proteinsa
| Protein | Molecular mass overall (kDa) | Signal peptide (amino acids)c | Molecular mass of mature protein (kDa) | pI of mature protein | G+C content of respective gene (%) |
|---|---|---|---|---|---|
| SfpA | 17.9 [18.7] | 20 | 15.9 [16.6] | 4.93 [4.72] | 41.9 [44.1] |
| SfpH | 21.3 [21.8] | 19 | 19.1 [19.3] | 7.17 [5.36] | 53.2 [48.8] |
| SfpC | 91.9 [91.1] | 27 | 89.1 [88.3] | 6.08 [5.92] | 56.1 [52.5] |
| SfpD | 26.9 [26.8] | 26 | 24.2 [24.6] | 9.72 [9.01] | 47.7 [45.6] |
| SfpJ | 21.1 [20.7] | 20 | 19.1 [17.8] | 4.80 [7.11] | 40.2 [55.5] |
| SfpFb | 9.3 [17.6] | None | 34.5 [42.9] | ||
| SfpG | 36.2 [38.3] | 22 | 33.5 [35.9] | 5.52 [5.59] | 36.0 [38.3] |
Numbers in brackets indicate values for the corresponding pap protein.
Translated starting from the first ATG codon of the putative nonfunctional sfpF gene; see the text for further details.
Predicted using the method of von Heijne (60).
Major pilus subunit SfpA.
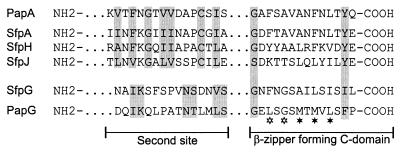
The predicted mature SfpA protein had 62% amino acid similarity to the major pilin of F13 P-fimbriae, PapAF13 (36), 41% similarity to the E. coli type I pilin, FimA (41), and 38% similarity to the major pilin of E. coli S-fimbriae, SfaA (51). SfpA showed structural properties of the so-called class I pilins (12): a pair of cysteine residues (positions 19 and 58 of the predicted mature protein) known to establish a disulfide bridge in pilins of type I, P-fimbriae, and S-fimbriae; a variable inner region; and a conserved C-terminal β-zipper-forming domain (residues 161 to 174) responsible for the interaction of pilins with the periplasmic chaperone during the assembly of the pilus (Fig. 2). This domain contains invariant glycine and aromatic residues separated by a series of alternating hydrophobic amino acids (33). Another site of interaction between the chaperone and the pilin, the so-called second site near the amino terminus (17), was also conserved in SfpA (Fig. 2).
FIG. 2.
Conserved structural motifs within Sfp subunits. Putative amino acid sequences of the C-terminal β-zipper-forming motif as well as the second site of interaction between pilins and the periplasmic chaperone are compared to the respective motifs of PapA and PapG (17). The most conserved residues are indicated by shaded boxes, and the asterisks mark the alternating hydrophobic amino acids in the C domain (open asterisks stand for residues conserved in all pilins with the exception of SfpJ).
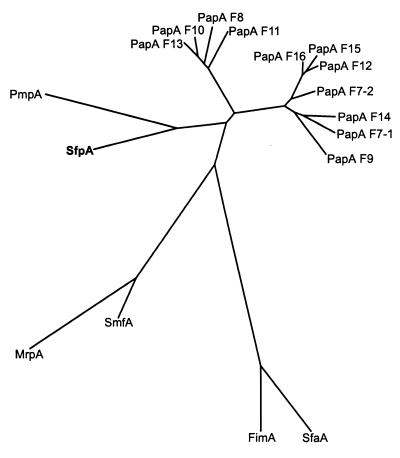
Recently, Girardeau et al. (19) classified class I pilins into seven subfamilies and described intrasubfamily signature motifs. Based on these characteristics, the SfpA pilin is a member of the Ic subfamily, together with E. coli PapA, Serratia marcescens SmfA, and Proteus mirabilis MrpA and PmpA. However, the SfpA characteristics differed from the subfamily's signature: the S1 motif, GxG[KT]V[TS]FxG[TS]V[VI]DAP (strongly conserved residues are shown in boldface), is present as GQGIINFKGIIINAP in SfpA (differences from the conserved residues are underlined). In contrast to all other members of the Ic subfamily, SfpA carries asparagine in place of aspartate at position 13 of the motif. The valine residues at positions 5 and 11 are replaced by isoleucine in contrast to PapA but similar to the P. mirabilis pilins PmpA and MrpA as well as SmfA of S. marcescens. Furthermore, SfpA did not possess an insertion of 8 to 11 residues in loop L4, which is a further characteristic of the Ic subfamily and especially of the Pap pilins. Again, the S. marcescens and P. mirabilis pilins SmfA and MrpA are similar to SfpA in lacking this insertion. Obviously, SfpA is not a close member of the Pap pilin family, despite having the highest sequence similarity to PapAF13. Phylogenetic analyses using CLUSTAL and Phylip programs supported this conclusion. As shown in Fig. 3, SfpA did not cluster together with the most homologous PapAF13 or other PapA alleles (24) but had a common branch with PmpA from canine uropathogenic P. mirabilis (5), a protein showing 58% amino acid similarity to the mature SfpA protein.
FIG. 3.
Phylogenetic relationship between SfpA and different PapA alleles. Included are 11 PapA variants of E. coli (F7–1, F7–2, F8, F9, F10, F11, F12, F13, F14, F15, and F16), the Pap-related SmfA, MrpA, and PmpA pilins of S. marcescens and P. mirabilis, and the E. coli type I (FimA) and S-fimbriae (SfaA) major subunits. The unrooted tree was based on (predicted) mature peptides and was constructed using the neighbor-joining method. The presumed evolutionary distance between any two members of the tree equals the sum of the lengths of the branches connecting them.
Minor subunits and assembly genes.
A second ORF of the gene cluster, termed sfpH, is located 79 bp downstream of sfpA (Fig. 1). The predicted mature SfpH protein showed 71% amino acid sequence similarity to PapHF13 (36). However, the pI of the mature SfpH protein is markedly different from that of PapH (Table 2). Like SfpA, SfpH possessed a C-terminal chaperone binding motif as well as a conserved second site (Fig. 2). In view of the homology to PapH, we suggested that SfpH is a minor component involved in the anchoring of the pilus to the outer membrane and modulating its length (2).
We detected two genes downstream of sfpH, sfpC, and sfpD, highly homologous to genes encoding components involved in the assembly of P-fimbriae. The predicted mature SfpC protein showed 76% similarity to the PapC outer membrane usher protein and also had a similar molecular weight and pI (Table 2). PapC has been shown to form oligomeric channels in the outer membrane which facilitate the transport of the nascent pilus out of the cell (56).
The mature SfpD protein showed 83% sequence similarity to the periplasmic chaperone PapD. This protein forms stable complexes with pilus subunits in the periplasm and presents these subunits to the outer membrane usher (17). PapD is a member of a large group of periplasmic chaperones of fimbrial systems of gram-negative bacteria. All these chaperones are characterized by invariant amino acid residues Arg-8 and Lys-112 necessary for the binding of subunits during pilus assembly (33, 52). In SfpD, Arg-8 and Lys-112 are conserved, indicating that this protein is the chaperone necessary for the assembly of Sfp fimbriae.
The genes in the 3′ part of the sfp cluster differed more from their corresponding pap genes than did sfpA, sfpH, sfpC, and sfpD. The sequence of SfpJ showed only 38% overall sequence similarity to PapJ, and the mature SfpJ had a pI of 4.8, whereas that of PapJ was 7.11. However, the molecular masses of the two proteins, about 21 kDa, are similar. Interestingly, the sequence homology was not equally distributed over the whole protein. Whereas the N-terminal 76 residues of the mature protein (amino acids 21 to 96) showed 53% sequence similarity, the C-terminal part (residues 97 to 187) had only 25% similarity. SfpJ possessed structural motifs for the interaction with the periplasmic chaperone at the N and C termini (Fig. 2). The C-terminal motif, however, seemed to be conserved to a lesser extent than that of SfpA, SfpH, and SfpG. Differences similar to these are described in a comparison of PapJ to Pap pilus subunits (55). On the other hand, some characteristics which distinguish PapJ from typical pilins were not detected in SfpJ. The mature SfpJ proteins possess only two cysteine residues, whereas PapJ has five. Furthermore, a hydrophobic domain and a sequence suggested to act as an ATP binding site in PapJ were not detected in SfpJ. We assume, therefore, that SfpJ is a more typical pilin than PapJ and might be a component of the pilus structure. PapJ, however, has been shown not to be part of the Pap pilus but to be necessary for its integrity and is thought to work as a cochaperone (55).
The region downstream of sfpJ showed 53% nucleotide sequence identity over 516 bp with the sequence of the P-pilus minor subunit gene papF. However, the ATG start codon seemed to be changed to a TTG codon, and the protein deduced from the ORF, starting at the first ATG codon, represents only the C-terminal half of a protein homologous to PapF. In addition, the deduced amino acid sequence did not show the properties of a signal peptide necessary for transport through the cytoplasmic membrane. We suggested, therefore, that sfpF is a nonfunctional ORF.
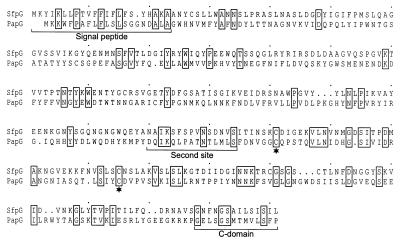
The gene located 21 bp downstream of sfpF and termed sfpG showed significant homology to neither a pap gene nor any other known sequence when its product was compared to amino acid or nucleotide sequence databases using Fasta or Blast algorithms. However, in addition to its position immediately downstream of sfpF (Fig. 1), it shared properties with the papG gene, encoding the adhesin of P-fimbriae. The protein deduced from sfpG showed a molecular mass and pI similar to those of PapG (Table 2). Pairwise comparison of SfpG and PapGF13 sequences revealed local similarities, especially in the C-terminal half (Fig. 4). This C-terminal domain is known as the assembly domain of PapG and is necessary for the interaction with other pilus subunits and the assembly machinery, whereas the N-terminal half of PapG is responsible for adhesin specificity (23). As for SfpA, SfpH, and SfpJ, a C-terminal chaperone recognition motif was conserved in SfpG with the exception of the penultimate aromatic residue, which is changed to an isoleucine residue (Fig. 2). Nevertheless, the P-fimbrial adhesin allele PrsG similarly possesses a penultimate leucine in place of an aromatic residue (36). Some residues of the second site of interaction between PapG and the periplasmic chaperone (61) were also conserved in SfpG (Fig. 2). Furthermore, a pair of cysteine residues (positions 195 and 229 of the mature protein) with a 33-residue spacing in the C-terminal half of the protein is similar to the cysteine pair with a 31-residue spacing establishing a conserved disulfide bridge in PapG (22) (Fig. 4).
FIG. 4.
Sequence comparison between SfpG and PapGF13 (36). The alignment was produced using CLUSTAL W. Identical amino acid residues are depicted by boxes. The predicted signal peptide as well as the sites responsible for the interaction between PapG and the periplasmic chaperone (second site and C domain) are indicated by the brackets. Conserved pairs of cysteine residues in the C-terminal protein domain are marked by asterisks.
Structure of the sfp gene cluster and comparison with the pap cluster.
Sequence comparison of the whole sfp gene cluster with the pap cluster showed that several pap genes had no equivalent in the sfp system (Fig. 1). DNA regions, homologous to papK and papE, which encode subunits of the tip fibrillum of P-fimbriae, could not be detected. In addition, the homologue of papF, the third gene encoding a component of the tip fibrillum, appeared to be nonfunctional. From these data we suggest that the structure of the Sfp pilus is different from that of the Pap pilus, especially with regard to the tip fibrillum. We also found no genes corresponding to the regulatory genes papI and papB and also no homology to an ORF encoding a 17-kDa protein which is typically present downstream of the papG gene (36). Moreover, the respective intergenic regions between the sfp and pap genes were much more different from one another, both in length and sequence, than the genes themselves. The region between sfpH and sfpC in particular was three times longer than that between papH and papC. A search using the program NNPP revealed putative promoter sequences starting 111 bp upstream of sfpA, 91 bp upstream of sfpC, and 117 bp upstream of sfpG.
The G+C content of the sfp genes varies over the gene cluster (Table 2). Whereas the sfpH, sfpC, and sfpD genes have a G+C content close to the typical G+C content of E. coli genes, the sfpA, sfpJ, and sfpG genes contain only 36 to 42% G+C. Such variations in the base composition have also been reported for the pap genes (36). The sfpJ gene, however, showed a G+C content of 40.2%, whereas papJ has 55.5%. As shown for the homology between sfpJ and papJ (see above), the G+C content also was not equally distributed over the sfpJ gene: the papJ homologous N-terminal part had a G+C content of 48% compared to only 32% G+C in the C-terminal half of the gene.
DNA sequences adjacent to the sfp gene cluster.
In order to investigate the DNA regions neighboring the sfp gene cluster and to look for possible regulatory genes of the system, we cloned and sequenced restriction fragments of pSFO157 contiguous to pSFO157-E11 (Fig. 5A). A 7.7-kb BamHI fragment, termed pSFO157-B8 and located downstream of the sfp gene cluster, was identified by Southern blot hybridization of restricted pSFO157 DNA using pSFO157-E11 as the probe. Based on continuous sequence homologies to Tn2501 (see below), the 13.9-kb EcoRI fragment pSFO157-E14 was hypothesized to be located upstream of sfpA. In order to examine this hypothesis and to sequence the connection between the two fragments, we generated PCR primers wprom-3 and wprom-4 (Table 1 and Fig. 5A). A PCR using this primer pair and DNA of strain 3072/96 as the template revealed the expected product as having a length of 1,900 bp. The nucleotide sequence of the PCR product was identical to the ends of pSFO157-E14 and pSFO157-E11, and there was no additional DNA present between the two fragments.
FIG. 5.
(A) Genetic organization of the sfp gene cluster within plasmid pSFO157. White boxes indicate open reading frames, and shaded boxes depict regions with homology to IS elements. Putative origins of replication (RepFII and RepFIB) are indicated below the map. The brackets in the lower row show the localization of plasmid fragments cloned in this study. The double arrow labeled “wprom” indicates the PCR amplicon used for the sequencing of the connection between fragment pSFO157-E14 and pSFO157-E11. The arrow labeled “G-Apa” depicts the location of the PCR product used for the cloning of sfpG. Above the gene map, nearly identical sequences of plasmid pO157 of NSF EHEC O157:H7 strain EDL933 are depicted by the bold line together with their sequence positions (GenBank AF074613). The ruler depicts map positions of pSFO157 in kilobase pairs. (B) Map of the recombinant plasmid pSFO157-ESn8::sfpGApa. The G-Apa PCR amplicon was inserted into the ApaI restriction site of the sfpG mutant plasmid pSFO157-ESn8. Plasmid parts derived from the vector pBluescript II KS are depicted as a bold black line together with relevant restriction sites (A, ApaI; E, EcoRI; K, KpnI; Sm, SmaI; Sn, SnaBI; St, SstI). Arrows indicate the transcription direction of the ORFs. Nonfunctional ORFs are given in parentheses.
Nucleotide sequencing of pSFO157-E14 and pSFO157-B8 revealed no further sequences similar to that of fimbrial subunit or regulatory genes. We found a DNA region of 2,054 bp which was 97% identical to a RepFIB origin of replication 686 bp downstream of the sfp cluster. This sequence enclosed the complete minimal replication region of IncFI plasmids, including an ORF for the replication protein RepA (50) (Fig. 5A). The sfp gene cluster, together with this RepFIB origin, is flanked by two blocks composed of different insertion sequences (IS). Upstream of sfpA, a region of 3,415 bp has 76% similarity to the transposase and resolvase genes of Tn2501 (38, 39). Adjacent to the Tn2501-like sequences we detected homologies to IS2 (46) and IS1294 (54) (Fig. 5A). We found another IS-related region downstream of the RepFIB origin composed of an IS100 (99.6% identity but with an internal deletion of 95 bp) (37) inserted into an IS3 element (80% similarity) (57), which is itself inserted into an iso-IS1-like sequence (68% similarity) (40). None of the IS elements flanking the sfp gene cluster seemed to be functional, because in all cases the transposase gene was not fully conserved due to partial deletion, insertion, or frameshift mutations. Nevertheless, the terminal inverted repeats of Tn2501, iso-IS1, IS3, and IS100 are conserved. In addition, the IS100 and IS3 elements are flanked by direct repeats, indicating that these IS elements have been inserted into the plasmid by regular transposition events.
Upstream of the IS1294-like sequence we found a region of 2,017 bp homologous to the origin of replication of IncFII plasmid R100 (47), including an ORF encoding the replication protein RepA1 (Fig. 5A). At the distal end of pSFO157-E14 we detected further homologies to parts of plasmid R100, comprising traI, traX, and finO (Fig. 5A).
A DNA region similar to that of plasmid R100 is also present in plasmid pO157 of NSF EHEC O157:H7. We therefore compared the sequences adjacent to the sfp cluster to the complete sequence of pO157 (11) and found a large region upstream of the sfp cluster which was nearly identical in the two sequences and included traI, traX, finO, and the RepFII origin of replication (Fig. 5A). These identical sequences, however, are interrupted by segments which show differences. Downstream of the sfp gene cluster we also detected DNA identical to the sequence of pO157, beginning from the right part of iso-IS1 (Fig. 5A). In pO157, the region between the RepFII origin and the iso-IS1-like sequences is covered by the katP (8) and espP (9) genes together with the incomplete insertion elements IS91, IS600, and IS1203. We suggested, therefore, that the sfp cluster, together with an RepFIB origin of replication and several IS elements, is inserted into pSFO157 in the region where espP and katP reside in pO157.
Expression of Sfp fimbriae by recombinant E. coli strains.
In order to investigate the expression of Sfp fimbriae, we transformed pSFO157-E11 into E. coli K-12 strain HB101. This strain was used because it is known to lack type I fimbriae (6). The Fim− phenotype of our laboratory stock of E. coli HB101 was confirmed by its inability to agglutinate yeast cells.
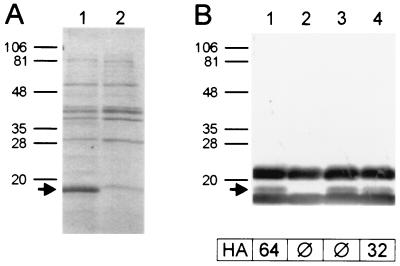
Analysis of thermoeluted extracts of strain HB101/pSFO157-E11 by SDS-PAGE revealed a protein with an apparent molecular mass of about 17 kDa which was not expressed by the vector control strain HB101/pBluescript II KS (Fig. 6A). N-terminal amino acid sequencing of this protein revealed the sequence ASQGQGIINF, corresponding to residues 21 to 30 of the deduced amino acid sequence of SfpA. The starting point corresponds to the predicted cleavage site of the signal peptide (Table 2).
FIG. 6.
Coomassie blue-stained SDS-PAGE (A) and immunoblot analysis using anti-SfpA antiserum (B) of thermoeluted fimbrial extracts taken from recombinant E. coli strains HB101/pSFO157-E11 (lane 1), HB101/pBluescript II KS (vector control) (lane 2), HB101/pSFO157-ESn8 (sfpG mutant) (lane 3), and pSFO157-ESn8::sfpGApa (lane 4). Positions and sizes of marker proteins (in kilodaltons) are given on the left. The arrows indicate the SfpA protein as identified by amino acid sequencing. Titers of mannose-resistant hemagglutination (HA) of the strains are given below the immunoblot. The bands strongly reacting with anti-SfpA antiserum above and below the SfpA protein and not visible in Coomassie blue-stained SDS-PAGE are suspected to represent nonproteinaceous components of the bacterial cell envelope present in the SfpA preparation used for immunization. These bands were not detected using rabbit normal serum, nor were they visible in silver-stained gels.
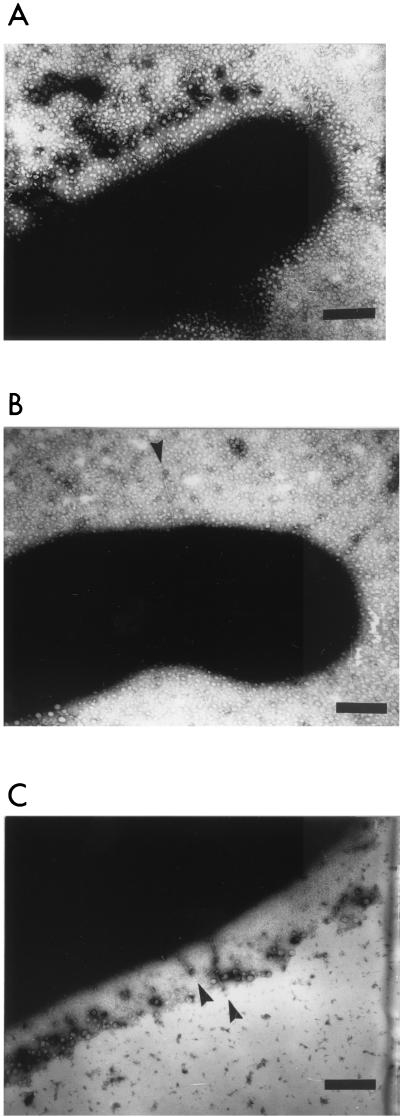
Recombinant E. coli HB101/pSFO157-E11 and the vector control strain HB101/pBluescript II KS were also analyzed by transmission electron microscopy for the presence of fimbriae. Whereas the vector control strain was not fimbriated (data not shown), we detected fimbriae on cells of strain HB101/pSFO157-E11 (Fig. 7A). The fimbriae were up to 0.4 μm in length and had a diameter of 3 to 5 nm.
FIG. 7.
Expression of fimbriae in recombinant E. coli K-12 strains harboring the sfp gene cluster shown by electron microscopy. (A) Strain HB101/pSFO157-E11; (B) sfpG mutant strain HB101/pSFO157-ESn8; (C) complemented mutant HB101/pSFO157-ESn8::sfpGApa. The bar represents 200 nm.
Taken together, our data showed that the sfp gene cluster in plasmid pSFO157-E11 is sufficient to mediate the expression of fimbriae in E. coli K-12 and that SfpA is the major subunit of Sfp fimbriae.
Construction and analysis of an sfpG deletion mutant.
In order to test our hypothesis that sfpG is part of the sfp cluster, even though it lacks homology to known fimbrial proteins, and that it encodes the adhesin subunit of the sfp fimbriae, we constructed a mutant lacking a functional sfpG gene. For this purpose we restricted plasmid pSFO157-E11 with the enzymes EcoRI and SnaBI and ligated the resulting 7,259-bp fragment into the vector pBluescript II KS, which had been restricted with EcoRI and SmaI. The resulting plasmid, pSFO157-ESn8, was transformed into E. coli HB101, and the deletion of sfpG was confirmed by nucleotide sequencing. Compared to plasmid pSFO157-E11, pSFO157-ESn8 lacked the RepFIB origin of replication and 683 bp of the 3′ part of sfpG but still retained sfpA, sfpH, sfpC, sfpD, and sfpJ (Fig. 5A).
The mutant strain HB101/pSFO157-ESn8 as well as strain HB101/pSFO157-E11 and the vector control strain HB101/pBluescript II KS were investigated for hemagglutination by a quantitative assay. Whereas the strain harboring the whole sfp cluster showed a mannose-resistant hemagglutination titer of 64, the mutant as well as the vector control strain was unable to agglutinate human red blood cells. On the other hand, immunoblot analysis of thermoeluted fimbrial proteins of the strains revealed that the mutant strain still possessed SfpA on its surface (Fig. 6B). Furthermore, we could detect fimbriae on cells of the sfpG mutant strain by electron microscopy (no more than a few per cell were detected, however) (Fig. 7B). The morphology of these fimbriae showed no obvious differences to those observed on strain HB101/pSFO157-E11 (Fig. 7A).
Attempts to clone sfpG in order to trans-complement the gene on a second plasmid failed, possibly due to a toxic side effect of SfpG when expressed without the chaperone and usher system (25). We therefore performed cis-complementation of sfpG. For this purpose we amplified sfpG by high-fidelity PCR using primers SfpG-UApa and SfpG-LApa, which contain ApaI restriction sites (Table 1 and Fig. 5A). The 3,162-bp amplicon was restricted with ApaI, ligated into ApaI-restricted plasmid pSFO157-ESn8, and transformed into E. coli HB101. The resulting plasmid, pSFO157-ESn8::sfpGApa, was verified using restriction enzyme analysis and nucleotide sequencing. In this construct the sfpG gene is inserted upstream and in the opposite direction of the other sfp genes (Fig. 5B).
As shown by immunoblot analysis, strain HB101/pSFO157-ESn8::sfpGApa produced the SfpA protein (Fig. 6B). The strain also exhibited a hemagglutination titer of 32, one step lower than that of strain HB101/pSFO157-E11. Electron microscopy revealed fimbriated cells; however, the number of fimbriae seemed to be reduced compared to that of strain HB101/pSFO157-E11 (Fig. 7C). It is possible that the coordinated expression of all fimbrial subunits is impaired in the artificial complementation construct, leading to problems in the assembly of the fimbriae.
Prevalence of the sfp gene cluster in E. coli and other Enterobacteriaceae
In order to determine the prevalence of the novel fimbrial gene cluster in pathogenic E. coli, we constructed PCR primer pairs specific for two different regions of the sfp cluster, namely, the sfpA gene and the region from sfpD to sfpG (Fig. 1 and Table 1). Using these primer pairs we examined 107 clinical isolates of E. coli, including 74 EHEC strains of different serotypes, 23 strains of other diarrheagenic E. coli (EPEC, enteroaggregative E. coli, enterotoxigenic E. coli, and enteroinvasive E. coli), and 10 UPEC strains. In addition, 15 isolates of other enterobacterial species, such as Enterobacter spp., Klebsiella pneumoniae, P. mirabilis, Salmonella enterica, S. marcescens, Yersinia enterocolitica, and Yersinia pseudotuberculosis were included in the study. Whereas all 14 isolates of SF EHEC O157:H−, including one strain isolated from a cow in the Czech Republic (4), possessed both the sfpA and sfpDG region, all the other 108 strains tested were negative in both PCRs (Table 3). The E. coli K-12 strains DH5α and HB101 used as hosts for cloning and expression of the sfp cluster were also negative by PCR. In order to exclude the possibility that the PCR had failed to detect the sfp genes due to minor sequence variations within the primer binding sites, we performed colony blot hybridization using probes specific for sfpA and sfpDG. The results of these hybridization experiments confirmed the PCR results in all cases. Therefore, we conclude that the sfp gene cluster is specific for the SF EHEC O157:H− group among human pathogenic E. coli.
TABLE 3.
Prevalence of the sfp gene cluster in human pathogenic E. coli and other Enterobacteriaceae examined by PCR and colony blot hybridization
| Pathogena or serotype | Total no. | No. sfpA positive | No. sfpDG positive |
|---|---|---|---|
| SF EHEC O157:H− | 14 | 14 | 14 |
| NSF EHEC O157:H7/H− | 7 | 0 | 0 |
| EHEC O26:H11/H− | 16 | 0 | 0 |
| EHEC O111:H2/H− | 7 | 0 | 0 |
| EHEC O103:H2 | 6 | 0 | 0 |
| Other EHECs | 24 | 0 | 0 |
| EPEC | 9 | 0 | 0 |
| EAEC | 6 | 0 | 0 |
| ETEC | 4 | 0 | 0 |
| EIEC | 4 | 0 | 0 |
| UPEC | 10 | 0 | 0 |
| Other Enterobacteriaceae | 15 | 0 | 0 |
EAEC, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EIEC, enteroinvasive E. coli.
DISCUSSION
In this paper we describe a novel fimbrial gene cluster harbored by the large plasmid of SF EHEC O157:H− strains. Sfp fimbriae are highly homologous to P-fimbriae of UPEC regarding their assembly machinery. However, in phylogenetic analyses the major pilus subunit, SfpA, does not cluster together with typical PapA serotypes and also shares structural similarity with the major subunit of P. mirabilis Pmp fimbriae. Sfp fimbriae are also distinct from P-fimbriae with regard to the organization of their gene clusters. Compared to pap, the sfp cluster lacks genes homologous to the regulatory elements papI and papB and in particular lacks all genes coding for parts of the tip fibrillum (papK, papE, and papF). The absence of functional homologues of papKEF is not unique to strain 3072/96, from which the sfp cluster has been cloned. As we have shown by PCR, all SF O157:H− EHEC isolates possessing sfpA also gave an sfpDG PCR product having the same length as that obtained with strain 3072/96. Furthermore, nucleotide sequencing of the sfpDG PCR product of strain 493/89 isolated from a patient suffering from HUS in 1989 (26) revealed no differences compared to that of strain 3072/96, and this included the lack of an ATG start codon in the papF homologous region. The absence of tip subunit genes led us suppose either that Sfp fimbriae lack a tip fibrillum completely or that a similar structure is composed of other subunits, possibly SfpJ and/or SfpG.
The sfp gene differing most from pap is the ORF we have named sfpG. Using a mutated sfpG we showed that this gene actually contributes to the Sfp fimbriae in recombinant E. coli K-12 strains. Although strains lacking sfpG are able to express sfpA on their surface and to assemble fimbriae, the cells do not show mannose-resistant hemagglutination. Therefore, it seems likely that SfpG is the adhesin necessary for binding to erythrocytes and possibly to other eukaryotic cells, and this might be a prerequisite for the colonization of a metazoic host by SF EHEC O157:H−. Nevertheless, the observation that sfpG mutant bacteria express a reduced number of fimbriae even though the major pilus subunit, SfpA, is present on the surface led us to suppose that SfpG also has structural functions as a starter for the correct assembly of the fimbriae, as in the case of PapG and P-fimbriae (13).
The structure of the sfp cluster, which is not dissimilar to that of a reduced variant of a pap system, resembles the F17 fimbrial gene cluster. This cluster includes only four genes, f17-D and f17-C, encoding the chaperone and usher proteins, f17-A for the major pilus subunit, and the f17-G adhesin gene. A cluster of five genes is required for the expression of the F17-related fimbriae of Haemophilus influenzae, Hif and Haf. In addition to four proteins with functions similar to those of F17, the hif cluster encodes a minor subunit, HifD, probably serving as a terminator of pilus polymerization (35, 59). Another fimbrial system with only four structural genes (major subunit, adhesin, chaperon, and usher) and an additional regulator is involved in the expression of the SEF14 fimbriae of Salmonella enteritidis (14). In addition to the genetic structure, the morphology of Sfp fimbriae resembled that of F17 fimbriae and differed from that of P-fimbriae. Whereas the diameters of Sfp fimbriae and F17 fimbriae are 3 to 5 nm and 3 to 4 nm, respectively, P-pili have a diameter of 7 nm (10, 35). On the other hand, the sequence similarity of F17 and Sfp pilus subunits does not exceed 35%.
The role of plasmid-encoded Sfp fimbriae in the biology of SF EHEC O157:H− strains needs further investigation. It was not possible to detect Sfp fimbriae on wild-type EHEC O157:H− isolate 3072/96 under standard laboratory conditions, and variations in growth parameters such as type of media, temperature, aeration, osmolarity, and pH had no effect on their expression. The strain showed no mannose-resistant hemagglutination, and we were unable to detect the SfpA protein by immunoblot analysis. In order to exclude the possibility that the lack of fimbrial expression is a characteristic only of strain 3072/96, from which the sfp cluster had been cloned, we investigated 13 strains of SF EHEC O157:H− previously shown to harbor the fimbrial gene cluster. As in the case of strain 3072/96, we were also unable to detect Sfp fimbriae on these strains. We therefore concluded that, in contrast to the E. coli K-12 background of recombinant strains, the expression of Sfp fimbriae is strictly repressed in wild-type EHEC O157:H− strains. These findings resemble those of Elliott et al. (15), who examined the expression of fimbriae in EHEC O157:H7 and EPEC strains. The authors mutated a global regulator, termed Ler (locus of enterocyte effacement-encoded regulator), and found enhanced fimbrial expression and expression of novel types of fimbriae in the mutant strains. It is therefore possible that a similar repression of sfp genes by a strong repressor also takes place in SF EHEC O157:H− strains. If this is the case, then Sfp fimbriae may be expressed only under environmental conditions not easily mimicked in the laboratory and which occur only during an infection stage in humans, in an animal reservoir, or during a free phase in the environment.
Strains of SF EHEC O157:H− have been implicated in several outbreaks, as well as in sporadic cases, of HUS and diarrhea in Germany and the Czech Republic (1, 3, 20, 28). Although SF EHEC O157:H− strains represent a distinct clone within E. coli O157 (26), multilocus enzyme electrophoresis, sequence data of the β-glucuronidase gene, and multilocus sequence typing indicate that they are closely related to NSF EHEC O157:H7 strains (16, 44). It was hypothesized that both groups have evolved from a common EPEC-like O55:H7 ancestor and that the as-yet-unknown most recent common ancestor of SF EHEC O157:H− and NSF EHEC O157:H7 strains already possessed the large EHEC plasmid pO157. However, the sequence data provided in this study represent a segment of plasmid pSFO157 of SF EHEC O157:H− which differs markedly from pO157. The sfp fimbrial gene cluster, as we have shown, is unique to pSFO157 and seems to be inserted into a region corresponding to that where katP and espP reside in pO157. This is in accord with earlier observations showing that plasmids of SF EHEC O157:H− strains are distinct from those of NSF EHEC O157:H7/H− strains with regard to the katP and espP genes, which are present in NSF but not in SF strains (7). The remnants of different IS flanking the sfp cluster in pSFO157, as well as the katP and espP genes in pO157, lead us suppose that transposition processes took part in the creation of these differences. On the other hand, the existence of the RepFIB-like origin of replication downstream of the sfp cluster indicates that a second plasmid is the source of the fimbrial gene cluster and that this plasmid underwent replicon fusion with a pO157-like precursor of pSFO157. However, the gene pool from which SF EHEC O157:H− acquired the sfp fimbrial genes is not known, and the question of why this gene cluster, despite its putative mobility, is so unique among our collection of Enterobacteriaceae cannot be answered at this time.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant Ka 717/2–4).
We thank Stefanie Amersbach for excellent technical assistance.
REFERENCES
- 1.Ammon A, Petersen L R, Karch H. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H−. J Infect Dis. 1999;179:1274–1277. doi: 10.1086/314715. [DOI] [PubMed] [Google Scholar]
- 2.Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Schmidt H, Karmali M A, Khakhria R, Janda J, Bláhová K, Karch H. Isolation and characterization of sorbitol-fermenting Shiga toxin (Verocytotoxin)-producing Escherichia coli O157:H− strains in the Czech Republic. J Clin Microbiol. 1998;36:2135–2137. doi: 10.1128/jcm.36.7.2135-2137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielaszewska M, Schmidt H, Liesegang A, Prager R, Rabsch W, Tschäpe H, Cízek A, Janda J, Bláhová K, Karch H. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J Clin Microbiol. 2000;38:3470–3473. doi: 10.1128/jcm.38.9.3470-3473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bijlsma I G, Van Dijk L, Kusters J G, Gaastra W. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology. 1995;141:1349–1357. doi: 10.1099/13500872-141-6-1349. [DOI] [PubMed] [Google Scholar]
- 6.Blomfield I C, McClain M S, Eisenstein B I. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol Microbiol. 1991;5:1439–1445. doi: 10.1111/j.1365-2958.1991.tb00790.x. [DOI] [PubMed] [Google Scholar]
- 7.Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145:1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- 8.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 9.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 10.Bullitt E, Makowski L. Structural polymorphism of bacterial adhesion pili. Nature. 1995;373:164–167. doi: 10.1038/373164a0. [DOI] [PubMed] [Google Scholar]
- 11.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denich K, Blyn L B, Craiu A, Braaten B A, Hardy J, Low D A, O'Hanley P D. DNA sequences of three papA genes from uropathogenic Escherichia coli strains: evidence of structural and serological conservation. Infect Immun. 1991;59:3849–3858. doi: 10.1128/iai.59.11.3849-3858.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards R A, Schifferli D M, Maloy S R. A role for Salmonella fimbriae in intraperitoneal infections. Proc Natl Acad Sci USA. 2000;97:1258–1262. doi: 10.1073/pnas.97.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott S J, Sperandio V, Girón J A, Shin S, Mellies J L, Wainwright L, Hutcheson S W, McDaniel T K, Kaper J B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng P, Lampel K A, Karch H, Whittam T S. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 17.Fernández L A, Berenguer J. Secretion and assembly of regular surface structures in gram-negative bacteria. FEMS Microbiol Rev. 2000;24:21–44. doi: 10.1111/j.1574-6976.2000.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 18.Fratamico P M, Bhaduri S, Buchanan R L. Studies on Escherichia coli serotype O157:H7 strains containing a 60-MDa plasmid and on 60-MDa plasmid-cured derivatives. J Med Microbiol. 1993;39:371–381. doi: 10.1099/00222615-39-5-371. [DOI] [PubMed] [Google Scholar]
- 19.Girardeau J P, Bertin Y, Callebaut I. Conserved structural features in class I major fimbrial subunits (Pilin) in gram-negative bacteria. Molecular basis of classification in seven subfamilies and identification of intrasubfamily sequence signature motifs which might be implicated in quaternary structure. J Mol Evol. 2000;50:424–442. doi: 10.1007/s002390010045. [DOI] [PubMed] [Google Scholar]
- 20.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 22.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2730–2756. [Google Scholar]
- 23.Hultgren S J, Lindberg F, Magnusson G, Kihlberg J, Tennent J M, Normark S. The PapG adhesin of uropathogenic Escherichia coli contains separate regions for receptor binding and for the incorporation into the pilus. Proc Natl Acad Sci USA. 1989;86:4357–4361. doi: 10.1073/pnas.86.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson J R, Stell A L, Scheutz F, O'Bryan T T, Russo T A, Carlino U B, Fasching C, Kavle J, Van Dijk L, Gaastra W. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun. 2000;68:1587–1599. doi: 10.1128/iai.68.3.1587-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karch H, Heesemann J, Laufs R, O'Brien A D, Tacket C O, Levine M M. A plasmid of enterohemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen and for adhesion to epithelial cells. Infect Immun. 1987;55:455–461. doi: 10.1128/iai.55.2.455-461.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch H, Wiβ R, Gloning H, Emmrich P, Aleksic S, Bockemühl J. Hämolytisch-urämisches Syndrom bei Kleinkindern durch Verotoxin-produzierende Escherichia coli. Dtsch Med Wochenschr. 1990;115:489–495. doi: 10.1055/s-2008-1065036. [DOI] [PubMed] [Google Scholar]
- 29.Karmali M A, Steele B T, Petric M, Lim C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;i:619–620. doi: 10.1016/s0140-6736(83)91795-6. [DOI] [PubMed] [Google Scholar]
- 30.Khan A S, Kniep B, Oelschlaeger T A, van Die I, Korhonen T, Hacker J. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect Immun. 2000;68:3541–3547. doi: 10.1128/iai.68.6.3541-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan A S, Mühldorfer I, Demuth V, Wallner U, Korhonen T K, Hacker J. Functional analysis of the minor subunits of S fimbrial adhesion (SfaI) in pathogenic Escherichia coli. Mol Gen Genet. 2000;263:96–105. doi: 10.1007/pl00008680. [DOI] [PubMed] [Google Scholar]
- 32.Khan A S, Schifferli D M. A minor 987P protein different from the structural fimbrial subunit is the adhesin. Infect Immun. 1994;62:4233–4243. doi: 10.1128/iai.62.10.4233-4243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuehn M J, Ogg D J, Kihlberg J, Slonim L N, Flemmer K, Bergfors T, Hultgren S J. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Le Bouguenec C, Bertin Y. AFA and F17 adhesins produced by pathogenic Escherichia coli strains in domestic animals. Vet Res. 1999;30:317–342. [PubMed] [Google Scholar]
- 36.Marklund B I, Tennent J M, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 37.McDonough K A, Hare J M. Homology with a repeated Yersinia pestis DNA sequence IS100 correlates with pesticin sensitivity in Yersinia pseudotuberculosis. J Bacteriol. 1997;179:2081–2085. doi: 10.1128/jb.179.6.2081-2085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michiels T, Cornelis G. Detection and characterization of Tn2501, a transposon included within the lactose transposon Tn951. J Bacteriol. 1984;158:866–871. doi: 10.1128/jb.158.3.866-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michiels T, Cornelis G, Ellis K, Grinsted J. Tn2501, a component of the lactose transposon Tn951, is an example of a new category of class II transposable elements. J Bacteriol. 1987;169:624–631. doi: 10.1128/jb.169.2.624-631.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsubo H, Nyman K, Doroszkiewicz W, Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981;292:640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- 41.Orndorff P E, Falkow S. Nucleotide sequence of pilA, the gene encoding the structural component of type 1 pili in Escherichia coli. J Bacteriol. 1985;162:454–457. doi: 10.1128/jb.162.1.454-457.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perna N T, Mayhew G F, Pósfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 44.Reid S D, Herbelin C J, Bumbaugh A C, Selander R K, Whittam T S. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 45.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 46.Ronecker H J, Rak B. Genetic organization of insertion element IS2 based on a revised nucleotide sequence. Gene. 1987;59:291–296. doi: 10.1016/0378-1119(87)90337-4. [DOI] [PubMed] [Google Scholar]
- 47.Rosen J, Ryder T, Inokuchi H, Ohtsubo H, Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179:527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- 48.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Saul D, Spiers A J, McAnulty J, Gibbs M G, Bergquist P L, Hill D F. Nucleotide sequence and replication characteristics of RepFIB, a basic replicon of IncF plasmids. J Bacteriol. 1989;171:2697–2707. doi: 10.1128/jb.171.5.2697-2707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmoll T, Hacker J, Goebel W. Nucleotide sequence of the sfaA gene coding for the S-fimbrial protein subunit of Escherichia coli. FEMS Microbiol Lett. 1987;41:229–235. [Google Scholar]
- 52.Slonim L N, Pinkner J S, Brändén C I, Hultgren S J. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 1992;11:4747–4756. doi: 10.1002/j.1460-2075.1992.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarr P I, Bilge S S, Vary J, Jelacic S, Habeeb R L, Ward T R, Baylor M R, Besser T E. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavakoli N, Comanducci A, Dodd H M, Lett M C, Albiger B, Bennett P. IS1294, a DNA element that transposes by RC transposition. Plasmid. 2000;44:66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 55.Tennent J M, Lindberg F, Normark S. Integrity of Escherichia coli P pili during biogenesis: properties and role of PapJ. Mol Microbiol. 1990;4:747–758. doi: 10.1111/j.1365-2958.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 56.Thanassi D G, Saulino E T, Lombardo M J, Roth R, Heuser J, Hultgren S J. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmerman K P, Tu C P. Complete sequence of IS3. Nucleic Acids Res. 1985;13:2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toth I, Cohen M L, Rumschlag H S, Riley L W, White E H, Carr J H, Bond W W, Wachsmuth I K. Influence of the 60-megadalton plasmid on adherence of Escherichia coli O157:H7 and genetic derivatives. Infect Immun. 1990;58:1223–1231. doi: 10.1128/iai.58.5.1223-1231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Ham S M, van Alphen L, Mooi F R, van Putten J P M. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 1994;13:673–684. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 60.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Z, Jones C H, Haslam D, Pinkner J S, Dodson K, Kihlberg J, Hultgren S J. Molecular dissection of PapD interaction with PapG reveals two chaperone-binding sites. Mol Microbiol. 1995;16:1011–1020. doi: 10.1111/j.1365-2958.1995.tb02326.x. [DOI] [PubMed] [Google Scholar]