Abstract
Treponema denticola does not appear to produce siderophores, so it must acquire iron by other pathways. Indeed, T. denticola has been shown to have an iron-regulated 44-kDa outer membrane protein (HbpA) with hemin binding ability. To characterize the HbpA protein, its gene was cloned from genomic DNA libraries of T. denticola. Sequence analysis of the hbpA open reading frame indicated that it encoded a 42.8-kDa protein with a 23-amino-acid signal peptide. HbpA has no significant homology to any proteins in the databases. Southern blot analysis demonstrated that hbpA is present in several T. denticola ATCC strains and clinical isolates, but not in Treponema pectinovorum, Treponema socranskii, or Escherichia coli. HbpA, expressed as a recombinant protein in E. coli and purified by antibody affinity chromatography, has hemin binding activity as determined by lithium dodecyl sulfate-polyacrylamide gel electrophoresis with tetramethylbenzidine staining. Northern blot analysis showed that there were two hbpA-containing transcripts, of approximately 1.3 and 2.6 kb, and that the RNA levels were low-iron induced. Interestingly, the 2.6-kb mRNA also encoded a second protein with significant homology to hbpA. This downstream gene, called hbpB, was cloned and sequenced and its product was expressed as a fusion protein in E. coli. The hbpB gene product is 49% identical to HbpA and binds hemin. Thus, T. denticola has two novel hemin binding proteins which may be part of a previously unrecognized iron acquisition pathway.
There is mounting evidence for the roles of several oral anaerobic spirochetes in periodontal disease, with the best characterized species being Treponema denticola. This organism is frequently found in high numbers at periodontally diseased sites (2, 48), and its presence is strongly associated with the pathogenesis of human periodontal disease (32, 48). This organism is also considered a putative pathogen because it can produce a variety of virulence factors (9), including proteolytic enzymes (8, 33, 55), a hemolysin (5, 14), and immunosuppressive factors (46). These virulence activities are presumed to be important for establishing T. denticola in the periodontal pocket and possibly for causing some of the tissue destruction seen in periodontal disease.
Iron is an essential nutrient for the growth and metabolism of most bacteria (15, 31, 57). In order to survive in vivo, pathogenic bacteria have developed several mechanisms for acquiring iron in the host environment. Most bacteria secrete iron-chelating siderophores (37), which remove iron from host iron-binding proteins and then interact with bacterial membrane receptors to transport iron into the bacterium. In addition, some organisms have low-iron-inducible outer membrane proteins (OMPs) that bind host iron-binding proteins, such as transferrin or lactoferrin, and remove the iron (39, 43). Finally, some gram-negative human pathogens acquire iron from heme compounds. In these systems, a bacterial hemin-binding protein serves as a surface receptor to bind heme protein, and then the hemin is transported into the cells by a protein complex containing the inner membrane TonB protein (40, 58). The ability to utilize hemin and hemin-containing compounds for nutritional iron uptake has been documented for numerous pathogenic bacteria, including the periodontal pathogen Porphyromonas gingivalis (19, 38, 58).
T. denticola does not appear to produce siderophores (45), so it must have other mechanisms for iron acquisition. Indeed, T. denticola can bind lactoferrin (50) and hemin (7, 44). In addition, T. denticola produces a hemolysin (6), which could lyse erythrocytes in vivo and provide heme compounds for the bacterium. Previous studies have shown that T. denticola contains at least two hemin binding molecules: a 47-kDa constitutively produced hemin binding protein from strain ATCC 35405 (45) and a 44-kDa hemin binding protein (HbpA), which is up-regulated under iron-restricted growth, that is found in strains GM-1, MS-25, ATCC 33520, and ATCC 33404 (7). However, the role of these proteins in iron acquisition is unproven and the mechanism of binding and iron transport by these proteins has not been characterized. The 44-kDa HbpA has been purified and its amino-terminal sequence has been determined (7). The limited sequence had no homology with other proteins involved in iron uptake, suggesting that T. denticola may use a novel system to acquire iron. In order to understand the mechanism of this T. denticola iron uptake system and eventually determine its role in pathogenesis, the gene (hbpA) for the 44-kDa hemin binding protein from T. denticola ATCC 35404 was cloned, sequenced, and characterized, as was a downstream homologue called hbpB. The protein products of these genes are shown to bind hemin. Thus they may contribute to hemin utilization by T. denticola.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
T. denticola ATCC 35404 (TD-4) was used as the source of genomic DNA and in the RNA studies. T. denticola strains GM-1 and MS25 were originally isolated from human periodontal pockets (56). Strain SW-1 is a clinical isolate recovered by Steve Walker from a patient with periodontal disease. Treponema socranskii and Treponema pectinovorum are ATCC strains 35536 and 33768, respectively. The cells were grown anaerobically (5% CO2, 10% H2, 85% N2) at 37°C in GM-1 medium (56). To grow cells in iron-restrictive conditions, 2,2′-bipyridyl (BPD) was added to a final concentration of 200 μM.
Plasmids pUC18 and pBluescript II KS were used as vectors for cloning. Escherichia coli strains DH5α and DH10b were used as host strains for plasmids and were routinely grown in Luria-Bertani broth or on Luria-Bertani agar plates supplemented with ampicillin (50 μg/ml) when appropriate.
Chromosomal DNA isolation and Southern blot analysis.
T. denticola chromosomal DNA was isolated using a detergent-proteinase K lysis procedure that included treatment with cetyltrimethylammonium bromide to remove polysaccharides and cell wall debris (3). For Southern blot analyses, chromosomal DNAs were digested with the indicated restriction endonucleases, separated on 0.7% agarose gels, and transferred to nylon membranes. Hybridization of the membranes was carried out in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight at 56°C and the filters were washed three times with 6× SSC at 56°C. DNA fragments used as hybridization probes were isolated from plasmid or amplified by PCR and then isolated from agarose gels by a freeze-thaw-phenol extraction procedure (47). DNA probes were labeled with [α-32P]dATP using a random primer DNA labeling system (Life Technologies, Gaithersburg, Md.) or, if oligonucleotides were used, end labeled with [γ-32P]ATP by T4 polynucleotide kinase.
Oligonucleotides and DNA amplification by PCR.
For amplification of the DNA fragment encoding the amino terminus of the hbpA gene, a degenerate, 29-base oligonucleotide (5′ AARACIATGCCIGCIGCIAARGAYAAYAA 3′, where I = inosine, R = A or G, and Y = C or T) was synthesized based upon the previously determined amino acid sequence of the N terminus of HbpA (7). T. denticola genomic DNA was digested by PstI and EcoRI, and the resulting fragments were ligated into the PstI/EcoRI sites of pUC18. The ligation mixture served as the template in a PCR synthesis. The hbpA 29-mer oligonucleotide and the M13 forward primer were used as primers in the PCR. An initial denaturation step (5 min, 94°C) was followed by 40 cycles of amplification (1 min at 94°C, 1 min at 48°C, and 3 min at 72°C) in a model PTC-100 thermocycler from MJ Research.
Construction and screening of partial libraries of T. denticola genomic DNA.
T. denticola genomic DNA (80 μg) was digested with the appropriate restriction endonucleases and electrophoresed on a 0.7% agarose gel. A region of the gel containing DNA fragments corresponding in size to the hybridizing DNA fragment, as determined by preliminary Southern blotting, was excised and the DNA was extracted using a Qiaex kit (Qiagen Inc., Chatsworth, Calif.). These size-separated DNA fragments were ligated into either pBluescript or pUC18 and transformed into E. coli DH5α. Colonies from these partial libraries were transferred onto nylon membranes and screened for the correct insert by colony blot hybridization. The hybridization and washing conditions were the same as those described above for Southern blot analysis.
DNA sequencing.
Segments of various clones were sequenced in the Center for Advanced DNA Technology at the University of Texas Health Science Center at San Antonio using an Applied Biosystems model 373A sequencing system. All sequences were determined independently from both strands. The deduced amino acid sequences were compared to the nonredundant, combined protein databases at the National Center for Biotechnology Information using the BLAST algorithm (1) and the PROSITE program.
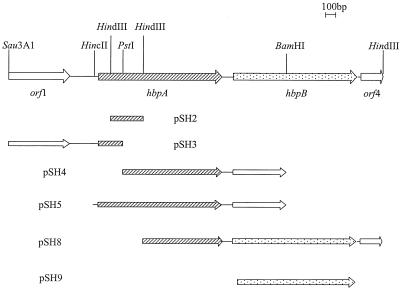
Expression and purification of rHbpA.
To express HbpA in E. coli, the 1.6-kb PstI-BamHI fragment from pSH4, which encodes the carboxy end of hbpA, was first ligated into pBluescript. The resulting plasmid was cleaved with HincII and PstI, and a 245-bp HincII-PstI fragment from pSH3, which encodes the amino portion of hbpA, was ligated in. This reconstructed the entire hbpA gene in one plasmid (pSH5). The plasmid was transformed into E. coli DH5α and the expression of recombinant HbpA (rHbpA) was assessed by Western blot analysis. To purify rHbpA, growth supernatant from E. coli/pSH5 cells was applied to an anti-HbpA affinity column. The affinity column was made from anti-HbpA rabbit serum (7), which had been absorbed against E. coli DH5α cells, purified by salt precipitation, and cross-linked to Affi-Gel 10 (Bio-Rad Laboratories, Richmond, Calif.). After washing of the column with pH 9.0 Tris buffer and pH 6.3 phosphate-buffered saline, the protein was eluted with pH 2.5 glycine-HCl buffer. The eluate was neutralized immediately with pH 8.0 Tris buffer. The combined eluate was put into dialysis tubing, concentrated by placement in polyethylene glycol until the volume was reduced to 0.5 to 1.0 ml, and then dialyzed against pH 8.0 Tris buffer.
Expression of rHbpB as a fusion protein.
The coding region, without the signal peptide, of the hbpB gene was amplified by PCR with T. denticola genomic DNA as the template. The primers (AACTGCAGATGTAAGTCGATGCCTAAC and CGGAATTCTCTCCTATAAAAAG) used in the PCR contained a PstI or an EcoRI site, one on either primer, that was not encoded by the hbpB gene. The PCR product was digested with PstI and EcoRI and ligated, in frame, into the expression vector pRsetA (Invitrogen, San Diego, Calif.). The recombinant plasmid was transformed into E. coli BL21(DE3). The expression of rHbpB was induced by the addition of isopropyl-β-d-thiogalactopyranoside to cells containing the expression plasmid.
SDS-PAGE and Western blot analysis.
The discontinuous gel system of Laemmli (25) was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resolving gel consisted of 12% acrylamide in 0.125 M Tris (pH 8.8), and the stacking gel contained 4% acrylamide in 0.125 M Tris (pH 6.8). Total E. coli proteins were prepared by pelleting 5 ml of stationary cells, suspending the pellet in 1 ml of 1× sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, 1 mM bromophenol blue), and heating to 100°C for 5 min. The OMPs of T. denticola were isolated by the freeze-thaw method (34). After electrophoresis, the gels were either stained with Coomassie brilliant blue R250 or transferred onto Immobilon-P membranes (Millipore Corp., Bedford, Mass.) for Western blotting (54). For Western blotting, the membrane was blocked with 3% bovine serum albumin in Tris-buffered saline (20 mM Tris, 0.5 M NaCl [pH 7.5]) and then incubated with anti-HbpA antibodies followed by alkaline phosphatase-conjugated secondary antibodies at the appropriate dilution. The rabbit anti-HbpA serum was a generous gift of L. Chu and was prepared from a rabbit immunized with purified HbpA from T. denticola (7). The membranes were developed with a 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium colorimetric system. Bio-Rad prestained molecular weight standards were used to estimate the molecular masses of the proteins.
LDS-PAGE and TMBZ staining.
Lithium dodecyl sulfate (LDS)-PAGE (Sigma, St. Louis, Mo.) and tetramethylbenzidine (TMBZ) (Sigma) staining were employed to assess the hemin binding ability of proteins (53). The resolving gel consisted of 12% acrylamide in 0.375 M Tris buffer (pH 8.8) containing 0.1% LDS. The stacking gel contained 4% acrylamide and 0.1% LDS. Where indicated, the protein samples, consisting of 250 ng of purified recombinant protein or 10 μg of T. denticola OMP, were incubated with hemin (20 μg/ml) at 37°C for 30 min and diluted 4:1 with sample loading buffer before loading onto the gel. The proteins were electrophoresed at a constant 220 V in the dark at 4°C. Detection of hemin-protein complexes was accomplished by staining the gel with TMBZ, a chromogen that is turned blue by proteins with hemin-associated peroxidase activity. The gel was fixed in 0.25 M sodium acetate (pH 5.0) and stained in TMBZ solution, and the color was developed by adding 30 mM hydrogen peroxide to the staining solution. The gel then was washed in 0.25 M sodium acetate–isopropanol (8:2) and dried immediately.
Northern blot analysis.
Total RNA of T. denticola was isolated using the Ultraspec RNA isolation system (Biotecx Laboratories Inc., Houston, Tex.). RNA was extracted from 30 ml of T. denticola cells which had been growing for 2 days in GM-1 with or without BPD after being inoculated with 3 ml of a 3-day-old culture of T. denticola cells. Cells were harvested and suspended in 1 ml of Ultraspec RNA solution for cell lysis. After the addition of 0.2 ml of chloroform and centrifugation, the RNA in the aqueous phase was precipitated with an equal volume of isopropanol and washed twice with 75% ethanol. The RNA pellet was dissolved in 50 μl of diethyl pyrocarbonate-treated water. The yield of RNA was determined by absorbance spectrophotometry at 260 nm. The indicated amount of RNA were electrophoresed in 1% agarose formaldehyde gels and transferred to a nylon membrane by capillary transfer. Probes were prepared and hybridizations were done as described above for Southern blotting.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in GenBank with accession numbers AF196837 and AF332358.
RESULTS
Cloning of the hemin binding protein gene (hbpA).
Based on the previously determined amino-terminal amino acid sequence of HbpA (7), a degenerate 29-base oligonucleotide (29-mer) potentially encoding amino acids 2 to 11 of HbpA was synthesized. Samples of T. denticola genomic DNA were digested with various restriction enzymes and the resulting collections of fragments were ligated into appropriately digested pUC18. These ligation mixtures served as templates in PCR amplifications with the 29-mer oligonucleotide and either M13 forward or M13 reverse primers. The PstI-EcoRI ligation mixture gave a 167-bp PCR product with the 29-mer and M13 forward primers (data not shown). Sequence analysis of the PCR fragment indicated that a 107-bp region in the fragment could encode a 46-amino-acid-long polypeptide whose first 20 amino acids matched the previously determined amino-terminal sequence of the 44-kDa protein.
A 107-bp HindIII/PstI fragment from this PCR product corresponding to amino acids 20 to 46 of HbpA was used as a hybridization probe in colony blot hybridization against various T. denticola genomic DNA libraries. No stable positive colonies were found. Since it has been reported that overexpression of some T. denticola membrane proteins may be toxic to E. coli (10), we decided to clone the rest of the hbpA gene in several fragments. The 107-bp HindIII/PstI fragment was used as hybridization probe to clone a 333-bp HindIII fragment from a size-limited T. denticola genomic library (Fig. 1, pSH2). The insert from pSH2 was then used as a hybridization probe against another T. denticola genomic library to clone the 3′ half of the hbpA gene on a 1.6-kb PstI/BamHI fragment (Fig. 1, pSH4). Cloning of the upstream region of hbpA proved to be more difficult; E. coli cells with plasmids containing the beginning of the hbpA gene and some upstream DNA grew much more slowly, as if the plasmid or its product was toxic to E. coli. However, we were able to clone the 5′ end of the hbpA gene as a 1.1-kb Sau3AI/PstI DNA fragment (Fig. 1, pSH3) by screening a Sau3AI/PstI genomic DNA library, using the insert from pSH2 as a hybridization probe.
FIG. 1.
Molecular organization of hbpA region from T. denticola strain TD-4. Key restriction endonuclease sites are marked. Inserts pSH2 and pSH5 were cloned into pBluescript. The pSH3, pSH4, and pSH8 inserts were cloned into pUC18. The pSH9 PCR product was cloned into pRsetA.
The HbpA gene encodes a novel protein with a secretion signal.
Analysis of the 2,703-bp sequence contained in pSH2, pSH3, and pSH4 revealed one complete open reading frame (ORF) in the middle and two incomplete ORFs at either end (Fig. 1). All three ORFs were in the same orientation and they were 283 bp (orf1 to hbpA) and 90 bp (hbpA to hbpB) apart. The G+C content of the sequenced DNA was 38.8%, which is similar to the T. denticola overall G+C content of 37 to 38% (49), suggesting that hbpA is not part of a pathogenicity island. The promoter prediction program NNPP (41) indicates that there is a possible bacterial ς70-like promoter between nucleotides 811 and 841, just upstream of the HbpA start codon (Fig. 2A). However, these data must be interpreted cautiously since the consensus promoter sequence is based upon E. coli promoters, although several T. denticola promoters have been characterized (13, 16, 24, 30). A potential ribosome binding site (Shine-Dalgarno) is located 7 bp upstream of the HbpA start codon. Finally, an inverted repeat is found in the region between hbpA and hbpB (Fig. 2A). This could encode a rho-independent transcription termination site.
FIG. 2.
(A) Sequences of the noncoding DNA segments around the hbpA and hbpB genes. The dotted lines around a gene name represent the coding region sequences for orf1, hbpA, hbpB, and orf4. The possible promoter region for hbpA is indicated by −10 and −35. The inverted repeats, which may serve as rho-independent terminators, are marked by pairs of arrows. SD, Shine-Dalgarno site. (B) Comparison of deduced amino acid sequences of HbpA and HbpB from T. denticola. The putative signal peptide sequences are underlined. The asterisks denote positions with identical amino acids in the two proteins. The dashes indicate gaps introduced to maximize the alignment.
The middle ORF encoded the HbpA protein, since its deduced sequence matched positions 2 to 23 of the previously determined amino-terminal amino acid sequence of the hemin binding protein (7). Not unexpectedly, the deduced sequence contained additional amino acids at the amino terminus, suggesting that the hbpA gene encodes a 23-amino-acid signal peptide (Fig. 2B) which is cleaved during translocation across the membrane. Indeed, the deduced sequence of the putative 23-amino-acid signal peptide is a typical signal sequence; it has positively charged residues at the N terminus, hydrophobic residues in the middle of the peptide, and a potential peptidase cleavage site at the end. A BLAST search of the protein databases with the deduced HbpA sequence revealed that there were no significant homologues among other proteins, including other hemin binding proteins. This indicates that HbpA is a novel hemin binding protein.
Interestingly, the deduced 178-amino-acid sequence from the incomplete ORF downstream of hbpA shared 50% identity with the HbpA protein; thus, the gene was named hbpB. The protein encoded by hbpB also has a putative signal peptide, indicating that it is a membrane or secreted protein. Since the partial sequence of HbpB shared so much identity with HbpA, the rest of the hbpB gene was cloned. Basically, a DNA fragment corresponding to the amino terminus of hbpB was amplified by PCR and used to probe a library of T. denticola genomic DNA digested with HindIII. The insert from the resulting clone, pSH8 (Fig. 1), was sequenced. The entire hbpB gene encodes a 403-amino-acid protein with a 23-amino-acid putative signal peptide; these are the exact sizes found for HbpA. However, the deduced molecular weight of the mature HbpB protein is 420 daltons smaller than that of HbpA. Overall, HbpB and HbpA are 49% identical and 60% similar in amino acid sequence (Fig. 2B). A BLAST search with the HbpB sequence indicated that no proteins in the National Center for Biotechnology Information nonredundant protein database were homologous to HbpB.
Finally, there are two partial ORFs in the sequenced DNAs. orf1, just upstream of hbpA, appears to encode 194 amino acids at the carboxy end of a protein. However, the possible role of this ORF in the cell is unknown, since it has no homologues in the protein databases. orf4, which is 36 bp downstream of hbpB, appears to encode the start of a protein. However, the possible function of Orf4 is unclear since a BLAST search with the limited sequence available did not reveal any homologue in the protein databases.
The hbpA gene is present in various T. denticola strains, but not in T. pectinovorum, T. socranskii, or E. coli.
Since Scott et al. (45) detected a 47-kDa hemin binding protein, but not a 44-kDa protein, in another strain of T. denticola, it was of interest to determine if the hbpA gene was present in various strains of T. denticola or in other oral spirochetes. Therefore, Southern blot analysis was performed on genomic DNA isolated from six different T. denticola strains, including ATCC 35405, which is the strain that was used by Scott et al. (45), and ATCC 33520, which is the strain being sequenced by The Institute for Genomic Research. In addition, DNAs from the oral treponemes T. pectinovorum and T. socranskii were examined. Using a probe from the first third of the hbpA gene, all of the T. denticola strains gave a 333-bp band when cut with HindIII (data not shown). This is the size expected from the sequence analysis of strain ATCC 35404. The T. pectinovorum, T. socranskii, and E. coli DNAs did not hybridize to the hbpA probe. To see if the gene organization around hbpA was also conserved among the T. denticola strains, the same filter was hybridized with orf1- and hbpB-specific probes. All of the T. denticola strains had a 2.9-kb band with the orf1 probe and a 2.3-kb band with the hbpB probe, while T. pectinovorum, T. socranskii, and E. coli gave no signal with either probe (data not shown). These results indicate that all of the strains of T. denticola tested, including the one used by Scott et al. in identifying a 47-kDa hemin binding protein, have the hbpA gene and that the gene organization around hbpA is identical in all T. denticola strains. This conclusion was reinforced by a search of the preliminary T. denticola genome sequence at The Institute for Genomic Research. One contig contained most of hbpA and all of hbpB, and the sequence from strain ATCC 33520 differed from our sequence at five bases in hbpA and four bases in hbpB. These base changes led to only two amino acid changes in HbpA and two in HbpB.
The hbpA gene has two transcripts which are low-iron induced.
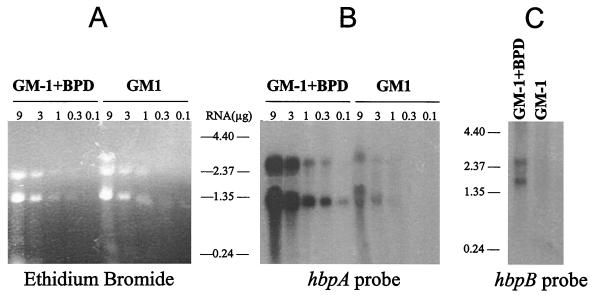
Bacterial genes are often organized in operons that are involved in the same metabolic or regulatory pathways. To see if hbpA is part of an operon, especially with hbpB, RNA from T. denticola was analyzed by Northern blotting. Total RNA was isolated from bacteria growing either in GM-1 medium or in GM-1 medium with 200 μM BPD to see if the hbpA RNA, like the protein, was induced under iron-restrictive growth conditions. Two hbpA transcripts were seen: a 1.3-kb band and a less intense 2.6-kb RNA species (Fig. 3B). Both transcripts must be from the hbpA gene, since Southern blot analyses with multiple restriction endonucleases and the same hybridization probe demonstrated that hbpA is a single-copy gene (data not shown). The 1.3-kb RNA was only 100 bases larger than the size of the hbpA ORF, thus indicating that the major hbpA transcript is monocistronic. However, the presence of the less intense 2.6-kb transcript indicated that hbpA was also cotranscribed with another gene(s). To determine which adjacent gene, orf1 or hbpB, was part of the hbpA operon, Northern blots were also hybridized with hbpB and orf1 probes. The orf1 probe did not hybridize with a 2.6-kb transcript (data not shown). However, the hbpB probe did hybridize with a 2.6-kb band (Fig. 3C), indicating that hbpA is cotranscribed with hbpB. Since bacterial RNA is relatively unstable, the Northern blotting results were confirmed by reverse transcription-PCR using oligonucleotide pairs that would produce PCR products if an RNA were transcribed across the orf1-hbpA, hbpA-hbpB, or hbpB-orf4 coding region junctions. Only the hbpA-hbpB oligonucleotide pair gave a product in the reverse transcription-PCRs (data not shown), demonstrating that only hbpA and hbpB are cotranscribed. The hbpB probe also hybridized with a 1.8-kb RNA species in Northern blot analyses (Fig. 3C), suggesting that it too can be transcribed monocistronically. Interestingly, all three hbpA and hbpB RNA species are induced at least 10-fold in RNA from cells grown under iron-restrictive conditions. This strongly suggests that the iron regulation of the 44-kDa HbpA protein and the HbpB protein is at the level of transcription or RNA stability.
FIG. 3.
Northern blot analyses of the hbpA and hbpB mRNAs. (A) Total RNA was isolated from T. denticola ATCC 35404 grown in GM-1 medium plus 200 μM BPD or in GM-1 medium. The indicated amounts of RNA were separated on a 1% agarose gel and stained with ethidium bromide. (B and C) The RNA was then transferred to a membrane and hybridized with an hbpA probe (a 333-bp HindIII fragment internal to the hbpA gene) (B) or an hbpB probe (a PCR product internal to the hbpB gene) (C). Each sample in panel C contained 3 μg of total RNA.
Purified rHbpA shows hemin binding ability in LDS-PAGE with TMBZ staining.
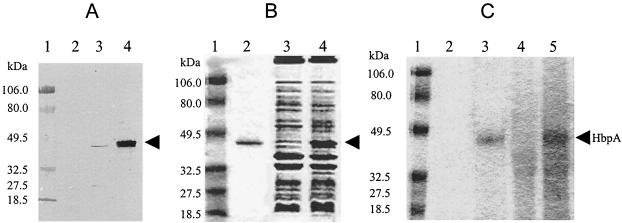
To facilitate the purification and characterization of the HbpA protein, the entire hbpA gene was reconstructed in one plasmid, pSH5 (Fig. 1). E. coli transformants carrying pSH5 grew more slowly than E. coli with vector alone, suggesting that production of the HbpA protein was somewhat toxic to E. coli. Consistent with this finding, HbpA did not appear to be overproduced in E. coli; when proteins from E. coli cells carrying pSH5 were compared to proteins from E. coli with vector alone on stained gels, rHbpA was not detected (data not shown). However, the HbpA protein is made in E. coli, since Western blot analysis of total protein from pSH5-containing cells had a protein band with the same size as native HbpA (Fig. 4A, lane 3). Mature rHbpA was also found in culture supernatants of pSH5-containing E. coli cells (data not shown). Finally, there was also a faint band at 27 kDa in the pSH5 sample. This could be a breakdown product of the HbpA in E. coli or it could arise from mistranslation from an internal ATG, of which there are several. Our results cannot distinguish between these possibilities.
FIG. 4.
Characterization of HbpA protein expressed in E. coli. (A) Total protein samples were prepared from the cells indicated and separated by SDS-PAGE before being transferred to a nylon membrane. The primary antibody used in the subsequent Western blotting was anti-HbpA antiserum prepared previously against HbpA purified from T. denticola. Lanes: 1, molecular size standards; 2, protein from E. coli DH5α with pBluescript vector alone; 3, protein from E. coli DH5α with pSH5; 4, OMP from T. denticola grown in GM-1 plus 200 μM BPD. (B) Various protein samples were subjected to LDS-PAGE and stained with Coomassie brilliant blue. Lanes: 1, prestained molecular size standards; 2, 250 ng of purified rHbpA from E. coli; 3, 10 μg of OMP from T. denticola grown in GM-1 medium; 4, 10 μg of OMP from T. denticola grown in GM-1 plus 200 μM BPD. (C) The indicated protein samples were subjected to LDS-PAGE and reacted with TMBZ. Lanes: 1, prestained molecular size standards; 2, 250 ng of purified rHbpA not treated with hemin; 3, 250 ng of purified rHbpA preincubated with hemin; 4, 10 μg of OMP from T. denticola grown in GM-1 medium and then preincubated with hemin; 5, 10 μg of OMP from T. denticola grown in GM-1 plus 200 μM BPD and then preincubated with hemin. The position of the 44-kDa HbpA protein is marked by an arrow.
Immunoaffinity chromatography was employed to purify rHbpA protein from the E. coli culture supernatant. rHbpA which eluted from an anti-HbpA column appeared as a single band on SDS-PAGE (Fig. 4B, lane 2). To test the hemin binding ability of the recombinant protein, its heme-dependent peroxidase activity was assessed. Purified rHbpA was incubated with or without hemin for 30 min at 37°C, electrophoresed on an LDS-polyacrylamide gel, and then reacted with TMBZ, which turns blue in the presence of a peroxidase. The rHbpA incubated with hemin gave a strong reaction (Fig. 4C, lane 3), while HbpA without hemin did not react with TMBZ (Fig. 4C, lane 2), indicating that the rHbpA can bind hemin. Thus, despite the fact that it has no homology with other hemin binding proteins, the T. denticola HbpA has hemin binding activity.
Fusion HbpB protein is expressed in E. coli and shows hemin binding ability.
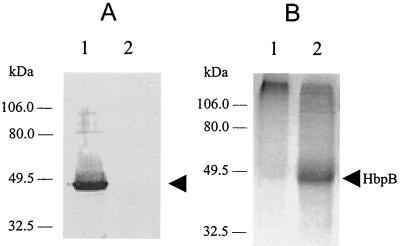
In order to determine if HbpB could bind hemin, it was expressed as a fusion protein in E. coli. The portion of the hbpB gene encoding the mature HbpB protein was ligated into the expression vector pRsetA to generate plasmid pSH9 (Fig. 1) This construct should express a protein with a 36-amino-acid polyhistidine-containing peptide fused to the amino terminus of the HbpB protein. Indeed, pSH9 expressed a protein of the expected size as determined by Western blot analysis with anti-His tag antibody (data not shown). The HbpB protein also showed cross-reactivity with anti-HbpA antibody in Western blotting because of the similarity of the two proteins (Fig. 5A, lane 1). To assess the hemin binding ability of HbpB, the cytosolic proteins from E. coli cells containing pSH9 and pRsetA were analyzed by LDS-PAGE and TMBZ staining. The result shows that recombinant fusion HbpB can bind hemin (Fig. 5B, lane 2).
FIG. 5.
Characterization of recombinant fusion HbpB protein. (A) Total protein samples were prepared from the cells indicated and then used in Western blotting as described in the legend for Fig. 4A. Lanes: 1, protein from E. coli BL21(DE3) with pSH9; 2, protein from E. coli BL21(DE3) with pRsetA vector alone. (B) The indicated protein samples were subjected to LDS-PAGE and then reacted with TMBZ. Lanes: 1, 10 μg of soluble protein from E. coli BL21(DE3) with pRsetA vector alone; 2, 10 μg of soluble protein from E. coli BL21(DE3) with pSH9. Both samples were preincubated with hemin before electrophoresis. The 46-kDa HbpB fusion protein is marked by an arrow.
DISCUSSION
The 44-kDa hemin binding protein (HbpA) from T. denticola has some properties in common with hemin binding receptors from other gram-negative bacteria. It is located in the outer membrane of the bacterium, it appears to have a signal peptide, and its expression is increased under iron-restricted growth conditions. However, the T. denticola HbpA is not homologous to any other hemin binding protein, nor does it share stretches of significant sequence identity with iron acquisition proteins from other bacteria. The inability to find a “hemin binding motif” in HbpA is not surprising, since no such motif has been identified in hemin binding proteins from other bacteria, despite the fact that most of the hemin binding proteins from other bacteria do show homology with each other. Thus, the 44-kDa OMP is a novel hemin binding protein, which is likely to serve as a hemin receptor for T. denticola.
Previous studies have shown that the hemin binding receptors in Yersinia enterocolitica, Haemophilus influenzae, and Shigella dysenteriae have a TonB box sequence at the amino terminus of the hemin receptor protein (21, 35, 36, 51). This protein motif is critical for the interaction of certain outer membrane receptors with the inner membrane protein TonB that provides the energy needed to carry out the uptake of specific substrates into the periplasmic space. Interestingly, there was no TonB sequence motif in either HbpA or HbpB from T. denticola. Thus, the mechanism used by HbpA and HbpB to get iron and hemin into the cell may be different. One possibility is that HbpA, HbpB, or both serve as the initial hemin receptor, which then passes off the molecule to another receptor which is TonB dependent. This second hemin receptor could be the previously identified 47-kDa OMP from T. denticola. Such a mechanism has been found in neisseria, where HpuA and HpuB are both hemoglobin receptors which act in a complex to internalize hemoglobin. In this case, only HpuB has a TonB box motif (28). Another possibility is that the hemin uptake system in T. denticola is unique and uses a non-TonB mechanism to provide the energy needed to internalize hemin and iron.
In bacteria, functionally related genes are often part of an operon. Sequence analysis of the region around the T. denticola hbpA gene revealed three adjacent ORFs, orf1, hbpB, and orf4. By Northern blot analysis, hbpB was shown to be part of an operon with hbpA, thus leading us to hypothesize that the HbpB protein has a role in hemin uptake by T. denticola. Indeed, HbpB binds hemin. Some other bacterial hemin uptake systems are encoded in operons. For example, in Y. enterocolitica, the hemin uptake operon contains four proteins: HemP, HemR, HemS, and HemT. HemR is the hemin receptor and its expression is regulated by iron. It has been proposed that HemT is involved in the transport of hemin into the cytoplasm, where it is degraded by HemS (52). However, hemin binding protein genes can also be transcribed monocistronically. For example, P. gingivalis hemR transcription is monocistronic (23). Finally, the cotranscribed hpuA and hpuB genes of Neisseria meningitidis are required as a coreceptor in the acquisition of iron from hemoglobin (27, 28). Mutant analysis is necessary to see if the T. denticola HbpA and HbpB proteins function in the same or parallel pathways in iron acquisition from hemin.
Some bacterial OMPs, such as the major OMP of T. denticola (10, 11), are toxic when overexpressed in E. coli and therefore their genes can be difficult to clone in certain vectors. For membrane proteins such as the major OMP from T. denticola, the toxic portion is usually the 5′ region of gene, which encodes the signal peptide and N-terminal region of the protein; the toxicity may be due to the blockage of general protein transport. Apparently, the amino terminus of HbpA is also somewhat toxic when expressed in E. coli. The two clones, pSH3 and pSH5, that contained the amino terminus of HbpA only formed small colonies. The toxicity appears to be due to the amino terminus of HbpA, since it is the only common element in pSH3 and pSH5.
The transcription start sites have been determined for four genes in T. denticola (13, 16, 24, 30), two of which appear to use a ς70-like promoter and two of which seem to use a ς28-like promoter. In the hbpA gene, promoter prediction by the NNPP program indicates that there is a possible ς70-like promoter between nucleotides 811 and 841, about 18 nucleotides upstream of the HbpA translation start codon. Sequence examination reveals a good −10 match in that region but a poor match to the E. coli −35 consensus sequence. This is very similar to the results for the T. denticola dmcA gene promoter (24), although other T. denticola promoter regions do have sequences that are very similar to both components of the E. coli ς70-like promoter consensus (10, 12). At the other end of the hbpA gene, there is a stem-loop structure located 9 nucleotides downstream of the hbpA translation stop codon. This structure fits the criteria for a rho-independent transcription termination site (42). However, either the hbpA transcription termination site is an inefficient site or there is an antitermination mechanism in T. denticola, since the major hbpA transcript also contains the downstream hbpB gene. Finally, the hbpA and hbpB RNAs are induced in cells growing under iron restriction. The low-iron induction of transcription of hemin binding protein genes has been described in several other systems (26, 35, 59, 60). In some cases, transcriptional regulation by iron is due to Fur or DtxR proteins, which act as iron-responsive DNA-binding repressor proteins (4, 17, 18, 27). Although the transcription of hbpA is regulated by iron, we did not find any matches to the Fur or DtxR consensus binding sequences in the hbpA promoter region. Thus, the mechanism for iron regulation of hbpA transcription needs further study.
Different strains of T. denticola have different OMP profiles. The msp gene, which encodes the major OMP of T. denticola, differs between strains, although there are regions of strong identity among the msp genes from the different strains (11); by Southern blot analysis, there are at least two restriction fragment length polymorphism patterns around the msp gene among strains. The restriction fragment length polymorphism pattern around the hemolysin gene from T. denticola ATCC 35404 is different from that of the hemolysin gene from a clinical isolate, GM-1 (22). Similarly, Southern blot analyses revealed that the flanking regions of the aspartate carbamoyltransferase gene and cystalysin gene differ between strains (20) (L. Chu, unpublished data). All of these results indicate that the genomes of different strains of T. denticola often vary. Interestingly, the gene organization around the hbpA gene appears to be conserved among the T. denticola strains tested: all six strains tested hybridized to the exact same size of HindIII fragments containing orf1, hbpA, and hbpB. All of these results indicate that this cluster of genes is conserved in T. denticola and thus may have an important role in the cell. The functions of each of these three genes can now be examined by the gene disruption methodology developed by Li and Kuramitsu (29).
ACKNOWLEDGMENTS
We thank A. Burgum for technical assistance and J. Ebersole and L. Chu for helpful discussions.
This work was supported by grants DE11368 and DE11771 from the National Institutes of Health.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Armitage G C, Dickinson W R, Jenderseck R S, Levine S M, Chambers D W. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J Periodontal. 1982;53:550–556. doi: 10.1902/jop.1982.53.9.550. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1994. [Google Scholar]
- 4.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu L, Burgum A, Kolodrubetz D, Holt S C. The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases. Infect Immun. 1995;63:4448–4455. doi: 10.1128/iai.63.11.4448-4455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu L, Holt S C. Purification and characterization of a 45 kDa hemolysin from Treponema denticola ATCC 35404. Microb Pathog. 1994;16:197–212. doi: 10.1006/mpat.1994.1020. [DOI] [PubMed] [Google Scholar]
- 7.Chu L, Song M, Holt S C. Effect of iron regulation on expression and hemin-binding function of outer-sheath proteins from Treponema denticola. Microb Pathog. 1994;16:321–335. doi: 10.1006/mpat.1994.1033. [DOI] [PubMed] [Google Scholar]
- 8.Fenno J C, Hannam P M, Leung W K, Tamura M, Uitto V J, McBride B C. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66:1869–1877. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenno J C, McBride B C. Virulence factors of oral treponemes. Anaerobe. 1998;4:1–17. doi: 10.1006/anae.1997.0131. [DOI] [PubMed] [Google Scholar]
- 10.Fenno J C, Muller K H, McBride B C. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–2497. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenno J C, Wong G W, Hannam P M, Muller K H, Leung W K, McBride B C. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol. 1997;179:1082–1089. doi: 10.1128/jb.179.4.1082-1089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene S R, Stamm L V. Identification, sequence, and expression of Treponema denticola mcpA, a putative chemoreceptor gene. FEMS Microbiol Lett. 1997;157:245–249. doi: 10.1111/j.1574-6968.1997.tb12780.x. [DOI] [PubMed] [Google Scholar]
- 13.Greene S R, Stamm L V. Molecular characterization of a chemotaxis operon in the oral spirochete, Treponema denticola. Gene. 1999;232:59–68. doi: 10.1016/s0378-1119(99)00115-8. [DOI] [PubMed] [Google Scholar]
- 14.Grenier D. Characteristics of hemolytic and hemagglutinating activities of Treponema denticola. Oral Microbiol Immunol. 1991;6:246–249. doi: 10.1111/j.1399-302x.1991.tb00485.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths E. Iron and bacterial virulence—a brief overview. Biol Methods. 1991;4:7–13. doi: 10.1007/BF01135551. [DOI] [PubMed] [Google Scholar]
- 16.Heinzerling H F, Olivares M, Burne R A. Genetic and transcriptional analysis of flgB flagellar operon constituents in the oral spirochete Treponema denticola and their heterologous expression in enteric bacteria. Infect Immun. 1997;65:2041–2051. doi: 10.1128/iai.65.6.2041-2051.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennecke H. Regulation of bacterial gene expression by metal-protein complexes. Mol Microbiol. 1990;4:1621–1628. doi: 10.1111/j.1365-2958.1990.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 18.Holmes R K. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis. 2000;181(Suppl. 1):S156–S167. doi: 10.1086/315554. [DOI] [PubMed] [Google Scholar]
- 19.Holt S C, Kesavalu L, Walker S, Genco C A. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 20.Ishihara K, Ishihara M, Takazoe I, Okuda K. Cloning and expression of the aspartate carbamoyltransferase gene from Treponema denticola. Appl Environ Microbiol. 1992;58:3399–3403. doi: 10.1128/aem.58.10.3399-3403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarosik G P, Sanders J D, Cope L D, Muller-Eberhard U, Hansen E J. A functional tonB gene is required for both utilization of heme and virulence expression by Haemophilus influenzae type b. Infect Immun. 1994;62:2470–2477. doi: 10.1128/iai.62.6.2470-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunakaran T, Holt S C. Cloning and expression of hemolysin genes from Treponema denticola strains ATCC 35404 (TD-4) and human clinical isolate GM-1 in Escherichia coli. Microb Pathog. 1994;16:337–348. doi: 10.1006/mpat.1994.1034. [DOI] [PubMed] [Google Scholar]
- 23.Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kataoka M, Li H, Arakawa S, Kuramitsu H. Characterization of a methyl-accepting chemotaxis protein gene, dmcA, from the oral spirochete Treponema denticola. Infect Immun. 1997;65:4011–4016. doi: 10.1128/iai.65.10.4011-4016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee B C. Isolation of an outer membrane hemin-binding protein of Haemophilus influenzae type b. Infect Immun. 1992;60:810–816. doi: 10.1128/iai.60.3.810-816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis L A, Gipson M, Hartman K, Ownbey T, Vaughn J, Dyer D W. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol Microbiol. 1999;32:977–989. doi: 10.1046/j.1365-2958.1999.01409.x. [DOI] [PubMed] [Google Scholar]
- 28.Lewis L A, Gray E, Wang Y P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Kuramitsu H K. Development of a gene transfer system in Treponema denticola by electroporation. Oral Microbiol Immunol. 1996;11:161–165. doi: 10.1111/j.1399-302x.1996.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 30.Limberger R J, Slivienski L L, Izard J, Samsonoff W A. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loesche W J. The role of spirochetes in periodontal disease. Adv Dent Res. 1988;2:275–283. doi: 10.1177/08959374880020021201. [DOI] [PubMed] [Google Scholar]
- 33.Makinen K K, Chen C Y, Makinen P L. Proline iminopeptidase from the outer cell envelope of the human oral spirochete Treponema denticola ATCC 35405. Infect Immun. 1996;64:702–708. doi: 10.1128/iai.64.3.702-708.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda K, Kawata T. Isolation, properties, and reassembly of outer sheath carrying a polygonal array from an oral treponeme. J Bacteriol. 1982;150:1405–1413. doi: 10.1128/jb.150.3.1405-1413.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills M, Payne S M. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 38.Otto B R, Verweij-van Vught A M, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 39.Pettersson A, Poolman J T, van der L P, Tommassen J. Response of Neisseria meningitidis to iron limitation. Antonie Leeuwenhoek. 1997;71:129–136. doi: 10.1023/a:1000179301748. [DOI] [PubMed] [Google Scholar]
- 40.Pidcock K A, Wooten J A, Daley B A, Stull T L. Iron acquisition by Haemophilus influenzae. Infect Immun. 1988;56:721–725. doi: 10.1128/iai.56.4.721-725.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reese M G, Harris N L, Eechman F H. Large scale sequencing specific neural networks for promoter and splice site recognition. In: Hunter L, Klein T, editors. Proceedings of the Pacific Symposium on Biocomputing, 1996. Kona, Hawaii: Klein; 1996. [Google Scholar]
- 42.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 822–843. [Google Scholar]
- 43.Schryvers A B, Stojiljkovic I. Iron acquisition systems in the pathogenic Neisseria. Mol Microbiol. 1999;32:1117–1123. doi: 10.1046/j.1365-2958.1999.01411.x. [DOI] [PubMed] [Google Scholar]
- 44.Scott D, Chan E C, Siboo R. Iron acquisition by oral hemolytic spirochetes: isolation of a hemin-binding protein and identification of iron reductase activity. Can J Microbiol. 1996;42:1072–1079. doi: 10.1139/m96-137. [DOI] [PubMed] [Google Scholar]
- 45.Scott D, Siboo I R, Chan E C, Klitorinos A, Siboo R. Binding of hemin and Congo red by oral hemolytic spirochetes. Oral Microbiol Immunol. 1993;8:245–250. doi: 10.1111/j.1399-302x.1993.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 46.Shenker B J, Listgarten M A, Taichman N S. Suppression of human lymphocyte responses by oral spirochetes: a monocyte-dependent phenomenon. J Immunol. 1984;132:2039–2045. [PubMed] [Google Scholar]
- 47.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 48.Simonson L G, Goodman C H, Bial J J, Morton H E. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56:726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smibert R M. Order I. Spirochaetales. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 49–57. [Google Scholar]
- 50.Staggs T M, Greer M K, Baseman J B, Holt S C, Tryon V V. Identification of lactoferrin-binding proteins from Treponema pallidum subspecies pallidum and Treponema denticola. Mol Microbiol. 1994;12:613–619. doi: 10.1111/j.1365-2958.1994.tb01048.x. [DOI] [PubMed] [Google Scholar]
- 51.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 53.Stugard C E, Daskaleros P A, Payne S M. A 101-kilodalton heme-binding protein associated with Congo red binding and virulence of Shigella flexneri and enteroinvasive Escherichia coli strains. Infect Immun. 1989;57:3534–3539. doi: 10.1128/iai.57.11.3534-3539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uitto V J, Pan Y M, Leung W K, Larjava H, Ellen R P, Finlay B B, McBride B C. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect Immun. 1995;63:3401–3410. doi: 10.1128/iai.63.9.3401-3410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinberg A, Holt S C. Interaction of Treponema denticola TD-4, GM-1, and MS25 with human gingival fibroblasts. Infect Immun. 1990;58:1720–1729. doi: 10.1128/iai.58.6.1720-1729.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 59.Worst D J, Otto B R, de Graaff J. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect Immun. 1995;63:4161–4165. doi: 10.1128/iai.63.10.4161-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, Hara Y, Tomochika K, Shinoda S. Utilization of hemin and hemoglobin as iron sources by Vibrio parahaemolyticus and identification of an iron-repressible hemin-binding protein. FEMS Microbiol Lett. 1995;128:195–200. doi: 10.1111/j.1574-6968.1995.tb07522.x. [DOI] [PubMed] [Google Scholar]