Abstract
Doublecortin like kinase 1 (DCLK1) plays a crucial role in several cancers including colon and pancreatic adenocarcinomas. However, its role in squamous cell carcinoma (SCC) remains unknown. To this end, we examined DCLK1 expression in head and neck squamous cell carcinoma (HNSCC) and anal squamous cell carcinoma (ASCC). We found that DCLK1 is elevated in patient SCC tissue, which correlated with cancer progression and poorer overall survival. Furthermore, DCLK1 expression is significantly elevated in HPV negative HNSCC, which are typically aggressive with poor responses to therapy. To understand the role of DCLK1 in tumorigenesis, we used specific shRNA to suppress DCLK1 expression. This significantly reduced tumor growth, spheroid formation, and migration of HNSCC cancer cells. To further the translational relevance of our studies, we sought to identify a selective DCLK1 inhibitor. Current attempts to target DCLK1 using pharmacologic approaches have relied on non-specific suppression of DCLK1 kinase activity. Here, we demonstrate that DiFiD [3,5-bis (2,4-difluorobenzylidene)-4-piperidone] binds to DCLK1 with high selectivity. Moreover, DiFiD mediated suppression of DCLK1 led to G2/M arrest and apoptosis and significantly suppressed tumor growth of HNSCC xenografts and ASCC patient derived xenografts, supporting that DCLK1 is critical for SCC growth.
Keywords: DCLK1, HNSCC, Anal SCC, therapy, DiFiD, patient derived xenograft
Introduction
Squamous cell carcinoma (SCC) originates in the mucosal epithelium at several sites including the oral cavity, oropharynx, and anus. Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide [1]. Etiologic factors associated with HNSCC include tobacco and alcohol abuse, and human papilloma virus (HPV) infection. Despite therapeutic advances, HNSCC remains difficult to treat; due, in part, to late-stage tumor presentation, as well as high rates of local recurrence and distant metastases that contribute to poor five-year survival [2, 3]. Anal squamous cell carcinoma (ASCC) is a rare cancer, which like HNSCC is associated with HPV infection, and its incidence is increasing. Accordingly, the identification of novel targets and therapeutics is needed to improve SCC treatment outcomes. There remains a paucity of preclinical descriptions of ASCC biology or potential drug targets.
Recently, doublecortin like kinase 1 (DCLK1) was found to be highly expressed in salivary gland tumors and is associated with low overall and disease-free survival [4]. DCLK1, encodes a member of the doublecortin family and protein kinase superfamily [5, 6]. DCLK1 contains two N-terminal doublecortin domains and a C-terminal kinase domain homologous to Ca2+/calmodulin-dependent kinases (CamK). Doublecortin domains participate in microtubule polymerization, whereas the serine/threonine protein kinase domain and a serine/proline-rich domain, mediate multiple protein-protein interactions. The activity of each domain is independent of one another [7]. DCLK1 is expressed in various cell types including neurons, osteoblasts, and tuft cells in the colon. Initial reports by our group [5, 6, 8, 9] and that of multiple laboratories [10-12], have reported DCLK1 expression is observed in a small percentage of cells (<5%) in colon and pancreatic adenocarcinomas. Further, it has been established as a marker of tumor-initiating cells [13]. The role of DCLK1 in SCCs remains unknown.
Presently, we determine that DCLK1 expression is upregulated in SCC. Further, DCLK1 regulates HNSCC proliferation, invasion, and migration. We also report for the first time that DiFiD [3,5-bis (2,4-difluorobenzylidene)-4-piperidone], which has shown efficacy in preclinical models of pancreatic cancer, binds to and inhibits DCLK1 activity and demonstrated antitumor effects in SCC [14]. These studies demonstrate the potential therapeutic value of targeting DCLK1 in SCC.
Methods and Materials
Cell lines, patient tissues, and reagents
Well-characterized HNSCC cell lines UM-SCC-1 (from Dr. Tom Carey, University of Michigan, Ann Arbor, MI), OSC19 (from Theresa Whiteside, University of Pittsburgh, Pittsburgh, PA), HN5 (from Dr. Jeffrey Myers, The University of Texas MD Anderson Cancer Center, Houston, TX) and FaDu (ATCC) were used in this study [15]. Het-1A, an immortalized non-cancerous esophageal squamous epithelial cell line was obtained from ATCC. Established cell lines were authenticated by short tandem repeat profiling at Johns Hopkins in 2018 using the Promega GenePrint 10 kit and analyzed using GeneMapper v4.0 software. FaDu were maintained in EMEM (Corning) with 10% heat inactivated FBS. All other cell lines were maintained in DMEM (Corning) with 10% heat-inactivated FBS (Sigma-Aldrich) without antibiotics. Cells were incubated at 37°C in the presence of 5% CO2. Tissue Microarray (#HN483) was purchased from US Biomax, Inc. Sections from paraffin embedded blocks of seventeen deidentified anal squamous cell carcinoma (ASCC) patient samples were obtained from the Department of Pathology, University of Kansas Medical Center. After review, the institutional IRB determined that this study (STUDY00140502) was not research involving human subjects. DiFiD and EF24 were acquired as previously reported [14, 16, 17]. KN-62 was purchased from Tocris Bioscience (Minneapolis, MN).
Cloning of DCLK1 DCX Domain
The DCLK1 DCX domain was amplified by PCR. cDNA was run on 2% agarose gels and bands were purified using the GeneJET gel purification kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. DCLK1 DCX DNA was cloned into the N-Terminal pFLAG 3 vector (Addgene) by restriction digest using NotI and BamHI restriction enzymes. FaDu cells were transfected and selected using 400 μg/mL G418 sulfate salt. Primers used for DCX Domain:
DCLK1 NotI FP: 5’-GATCGCGGCCGCGATGTCCTTCGGCAGA-3’
DCLK1 BamHI DCX RP: 5’-GATCGGATCCCTAGCCATCGTTCTCATC-3’
DCLK1 Kinase assay
Purified recombinant DCLK1, CamKIIα, CamKIIβ and CamKIV (150 ng, Signalchem) were incubated in reaction buffer (Invitrogen) with 9 nM peptide substrate (Gs peptide), 10 μM ATP (Invitrogen), and either vehicle DMSO (dimethyl sulfoxide), EF24, DiFiD or KN-62 (a pan-CAMK inhibitor) for 30 min at 37°C. We used Gs peptide because it has been used as a substrate for CamK assays [18, 19]. Our assays were adapted based on these studies. Fold change was calculated relative to ATP counts in the control. Consequently, 100 μl of luciferin-luciferase mixture (ATP determination kit, Invitrogen) was added into each well and luminance was monitored at 560 nm by using Synergy™ NEO microplate reader.
Molecular docking
All docking calculations were carried out with AutoDock Vina software [20] (Molecular Graphics Lab, Scripps Research Institute, http://vina.scripps.edu/) to analyze DiFiD interactions with the 3D structure of kinase domains of DCLK1 (Protein Data Bank ID: 5JZN). Default parameters on Autodock tools were used to analyze the docking. Total Kollman and Gasteiger charges were added to the protein and the ligand prior to docking. We used Lamarckian GA to find the best conformations and chose approximately 10 conformations for further analyses. The most stable compound conformation was selected based on the scoring function and the lowest binding energy, and visualized using Pymol (https://pymol.org/2/) [21].
Surface Plasmon Resonance
SPR analysis of DiFiD-DCLK1 binding affinity was performed at Ametek Reichert Technologies, using Reichert4SPR instrumentation. A dextran sensor chip (Reichert part# 13206066) was activated for 240s with the mixing 400 mM EDC (1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride) and 100 mM NHS (N-hydroxysuccinimide) at a flow rate of 10 μL/min. Recombinant DCLK1 protein (Signalchem) was dissolved in running buffer (10mM HEPES, 138mM NaCl, 2.7 mM KCL, 0.05% Tween-20, 5% DMSO, ph7.4) and injected to the sample channel at a flow rate of 10 μL/min to reach immobilization. The chip was deactivated by 1M ethanolamine hydrochloride-NaOH at a flow rate of 25 μL/min for 240s. DiFiD was diluted in running buffer at a flow rate 25 μL/min with an association phase of 120s, followed by 120s dissociation. The KD value of DiFiD against DCLK1 protein were obtained by Biacore 8k evaluation software.
Cellular thermal shift assay (CETSA)
The ability of DiFiD to interact with and stabilize DCLK1 in cells was determined using CETSA [22]. Briefly, cells (8 × 106) were treated with media containing DMSO or DiFiD (5 μM) for 4 hours. After treatment, the cells were aliquoted into PCR tubes and exposed to a temperature gradient. Subsequently, cells were lysed using three repeated freeze-thaw cycles in liquid nitrogen followed by centrifugation. The resultant lysates were then utilized in downstream western blot analyses.
Drug affinity responsive target stability (DARTS)
The ability of DiFiD to interact with and stabilize DCLK1 in cells was studied by using DARTS [23]. FaDu cells were cultured and grown up to 70-80% confluency. Cells were washed three times with ice-cold PBS and. lysed using M-PER lysis buffer supplemented with protease inhibitor cocktail tablet (Roche) and collected by scraping off with a cell scraper. Cells were lysed on ice for 10 minutes, clarified by centrifugation, and protein estimated using BCA method. Cell lysates were divided into equal concentration aliquots and incubated with DMSO or DiFiD (5 μM) for 30 minutes with gentle shaking. Following incubation, lysates were treated with pronase (10 mg/ml stock) in 1:1/100, 1:1/200, 1:1/400, 1:1/800, 1:1/1600, 1:1/3200, 1:1/6400 protein:pronase ratio for 15 minutes. The protease digestion was stopped using 20X protease inhibitor cocktail (Roche) for 10 minutes on ice. The resultant protein samples were diluted with 4X Laemmli buffer, heated at 70°C for 10 minutes and loaded on to 10% SDS-PAGE gel, transferred to PVDF membrane and incubated with DCLK1 antibody at a concentration of 1:1000. Protein levels on western blot were pictured by Bio-Rad ChemiDoc-XRS+ instrument and analyzed by image lab software. Band intensity was calculated relative to the lowest dilution of pronase.
Animal studies
All protocols were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center. To assess DCLK1 expression at various stages of disease progression, 4-nitro-quinoline-oxide (4-NQO,100 ppm in sterile drinking water ad libitum) was administered for 16 weeks to C3H mice (n=20) using a previously reported protocol [24]. Mice were then given sterile drinking water for 3 weeks, at which time animals were sacrificed and tongues excised. Immunohistochemistry was then used to assess DCLK1 expression.
Five-week-old female Foxn1/nude mice (Charles River Laboratory) were injected with 1×106 HN5 or FaDu cells in the flank. One week following implantation, mice were randomized into two groups with 7 mice per group in the HN5 injected mice and 10 mice per group in the FaDu treated mice. Animals were treated with either vehicle control (2.5% DMSO in water) or DiFiD (2 mg/kg body weight), administered intraperitoneally daily for 15 days. Tumor growth was measured every 2-3 days by a blinded observer measuring tumor diameters using vernier calipers and volume was calculated (Tumor volume = longest dimension × shortest dimension2 x 0.52) as previously described [25]. At the end of treatment, animals were euthanized, and the tumors were collected, weighed, and processed for downstream analytical assays.
Additionally, we established a patient-derived xenograft from an anal SCC that metastasized to the liver. The tumor was passaged twice through NOD SCID gamma (NSG) mice. ASCC tumors from the third passage were implanted subcutaneously into flanks of 8-week-old mice. Mice were randomized based on the tumor volume into two groups with 10 mice (5 female and 5 male) in the control group and 14 mice (9 female and 5 male) in the DiFiD treatment group. Average tumor volumes across both groups were 50 ± 8.9 mm3. Mice were treated daily with either vehicle control (DMSO in water) or DiFiD (2 mg/kg body weight) intraperitoneally daily for 15 days. Tumor growth was measured, and the volume calculated as mentioned before. Fractional tumor volumes were calculated as per previously published protocols [26].
Statistical analysis
Data are reported as mean ± SEM. Parametric, one-tailed t-test with Welch correction was used to assess significance in all experiments unless stated. Outliers were detected using Graphpad software. Significant outliers were identified using the Grubb’s Test with Alpha=0.05. Significant outliers were removed from further statistical analysis. For in vivo studies, repeated measures ANOVA test was employed to assess the level of significance in tumor volumes between treatment arms. For The Cancer Genome Atlas (TCGA) survivorship comparison, log-rank (Mantel-Cox) test assessed differences between curves. To generate a best expression cut off, patients were stratified into two groups and association between survival and reads per kilobase (RPKM) was examined. The RPKM value that yields the maximum difference between survival of the two groups at the lowest log-rank P-value determined best expression cut off. From this, a high/low expression cut off (0.37) was applied. All statistical calculations were performed on GraphPad Prism software (version 6.03), with significance determined by p<0.05.
Results
DCLK1 is upregulated in Squamous Cell Cancers
DCLK1 is associated with pro-survival signaling in various cancers, including colorectal and pancreatic cancers [8, 18, 19]. Its expression is elevated in HNSCC tumor samples compared to normal oral epithelial tissue (Fig. 1A and B, p<0.01). TCGA analysis show that increased DCLK1 expression correlated with increased tumor histological grade (Supplementary Fig. S1A), and increase expression was higher in HPV-negative patients (Supplementary Fig. S1B).
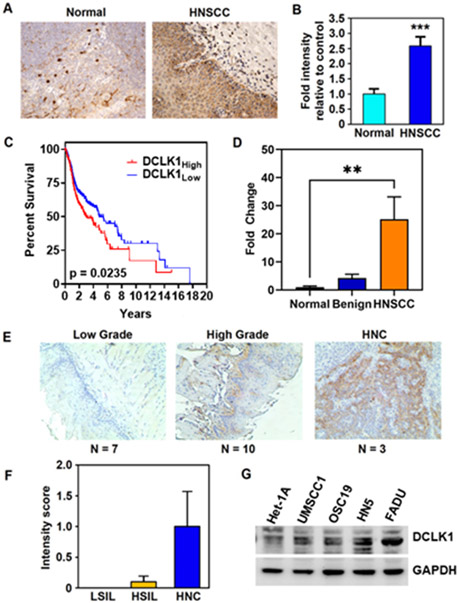
Figure 1. DCLK1 is overexpressed in HNSCC.
(A) IHC staining for DCLK1 (arrows) in normal tonsil and HNSCC tissue (B) Fold change in mean intensity of DCLK1 in tissue microarray from HNSCC (n=40) relative to normal oral mucosa (tonsillar) (n=8) samples determined by Leica Aperio ScanScope XT slide scanner (20 images scored per sample) (p<0.003 by two-tailed T-test). (C) TCGA head and neck squamous cancer (HNSC) data demonstrate that high DCLK1 levels trend towards a decrease in overall survival (High, n=192; low, n= 307 *p<0.05). (D) DCLK1 mRNA expression from a cDNA array of HNSCC tumor samples represented as fold change normalized to normal oral mucosa (Normal, n=4; benign, n=3; HNSCC, n=13). (E) Bright field images and (F) scoring of DCLK1 stained oral mucosa tissue, representing low (LSIL, n=7) and high grade (HSIL, n=10), and HNC (n=3) from a 4NQO-carcinogen induced mouse model. (G) Western blot for DCLK1 protein expression in HNSCC cell lines and immortalized non-cancerous squamous esophageal cell line, Het-1A. β-actin used as a loading control. *p<0.05, ***p<0.001.
Kaplan-Meier survival curves generated from TCGA HNSCC mRNA expression datasets demonstrated a trend towards lower overall survival in patients with DCLK1 high expressing tumors (n=192) compared to DCLK1 low expressing tumors (n=307) (Fig. 1C). Patients with DCLK1 low expressing tumors survived on average 4.827 years post diagnosis; patients with DCLK1 high expressing tumors had a shorter median survival of 2.995 years.
To assess correlation of DCLK1 with disease progression, we evaluated the expression of DCLK1 in a cDNA panel (Origene), from normal oral mucosa (n=8), benign (n=10), and HNSCC (n=16) patient tissues. DCLK1 was significantly upregulated in cancerous tissue compared to normal (Fig. 1D, p<0.01). To evaluate DCLK1 expression through various stages of tumor progression, tongue tissue from 4NQO treated mice was collected (n=20). Mice developed various stages of disease with 7/20 mice developing low grade squamous intraepithelial lesions (LSIL), 10/20 developing high grade squamous intraepithelial lesions (HGSIL) and 3/20 developing invasive HNSCC. Levels of DCLK1 expression progressively increased in dysplastic and invasive lesions, with highest expression observed in HNSCC (Fig. 1E and F). We then evaluated DCLK1 expression in HNSCC cell lines; OSC19, HN5, UM-SCC-1, and FaDu and Het-1A, a non-cancerous immortalized esophageal epithelial line. DCLK1 was differentially expressed with the highest expression observed in HN5 and FaDu cells, and lowest expression in Het-1A cells (Fig. 1G). Immunofluorescence revealed DCLK1 to be expressed in HN5 spheroids (Supplementary Fig. S1C). Therefore, HN5 and FaDu were used for subsequent studies. These data suggest that DCLK1 is elevated in HNSCC compared to normal oral mucosa and is associated with tumor progression. Further, anal squamous cell carcinoma (ASCC) patient samples (n=17) were stained for DCLK1 expression. DCLK1 was more significantly expressed in ASCC tumor compared to normal anal mucosa (Supplementary Fig.S1D and E). We observed increased nuclear staining along the tumor invasive front (Supplementary Fig. S1D).
DCLK1 suppression inhibits HNSCC growth
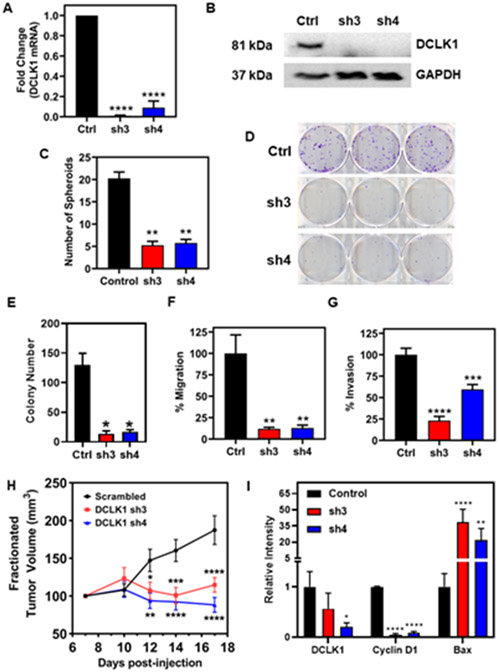
To evaluate the antitumor efficacy of targeting DCLK1 in HNSCC, we attenuated DCLK1 levels using two shRNA constructs, sh3 and sh4. DCLK1 mRNA and protein were significantly reduced (Fig. 2A and Fig. 2B) in FaDu cells stably transduced with sh3 or sh4 compared to scrambled controls. DCLK1 has been shown to play a critical role in anchorage independent growth in vitro. Hence, we assessed the effects of DCLK1 knockdown on spheroid formation. DCLK1 knockdown suppressed spheroid formation in FaDu cells compared to scrambled controls (p<0.01, Fig. 2C). DCLK1 knockdown also resulted in stunted colony formation (Fig. 2D). Functionally, DCLK1 shRNA knockdown reduced HNSCC migration and invasion in vitro (Fig. 2F and Fig. 2G, respectively). To demonstrate that DCLK1 is an important driver of cancer in vivo, FaDu cells stably transduced with shRNA targeting DCLK1 (sh3 and sh4), or a scrambled control shRNA were subcutaneously injected into NOD SCID gamma (NSG) mice. Suppression of DCLK1 resulted in slower growing tumors compared to scrambled controls (Fig. 2I). Tumor lysates were then analyzed by immunoblot for DCLK1, cyclin D1, and Bax (Fig. 2H and Supplementary Fig. 2). Expectedly, DCLK1 expression was diminished in both shRNA groups. Cell cycle regulator, cyclin D1 expression was significantly reduced, while pro-apoptotic, Bax expression was greatly increased in DCLK1 knockdown tumors. Collectively, these data indicate DCLK1 may help drive cancer progression, and DCLK1 suppression may lead to cell cycle arrest and apoptosis.
Figure 2. DCLK1 suppression inhibits tumor growth, invasion, and migration.
(A) Fold change in mRNA expression of DCLK1 in FaDu cells stably transduced with shRNA targeting DCLK1 (termed: sh3 and sh4) relative to cells transduced with a scrambled control shRNA. Data represent cumulative results from three independent experiments with ± SEM. (B) Western blot of DCLK1 protein in control and stably knocked down DCLK1 sh3 and sh4 FaDu clones. GAPDH served as a loading control. (C) Quantification of spheroid number from spheroid formation assay. Data represent cumulative results from two independent experiments with ± SEM (D) Representative images of colony formation assays for DCLK1 control, sh3 and sh4 FaDu clones. (E) Quantification of colony number. Data represents cumulative results from three independent experiments with ± SEM (F) Migration and (G) invasion assays are normalized for differences in proliferation rates over the duration of the assay. Data represents cumulative results from three independent experiments with ± SEM. (H) Tumor volumes from NSG mice that were inoculated subcutaneously with 1x106 FaDu cells stably transduced with shRNA targeting DCLK1 (sh3 and sh4) or scrambled control shRNA into their right flank (n=10 animals per group). Tumor measurements began 7 days post-injection of cells. Error bars represent ± SEM (I) Quantification of western blot analysis from lysates of a cohort of xenograft tumors. Error bars represent ± SEM (Western blots shown in Supplementary Figure 5A) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
DiFiD binds to DCLK1 and inhibits its activity
The small molecule, 3,5-bis (2,4-difluorobenzylidene)-4-piperidone (DiFiD) was previously shown to suppress the growth of pancreatic cancer cells in vitro and in vivo [14]. However, the precise target for DiFiD remained unknown. DCLK1 is implicated in pancreatic and colorectal cancer progression. Therefore, we first determined compound-protein interaction. Molecular docking predicted a DiFiD interacts with DCLK1 by forming hydrogen bonds with aspartic acid 533, with a binding energy of −7.9 kcal/mol, as shown in ribbon and surface views (Fig. 3C).
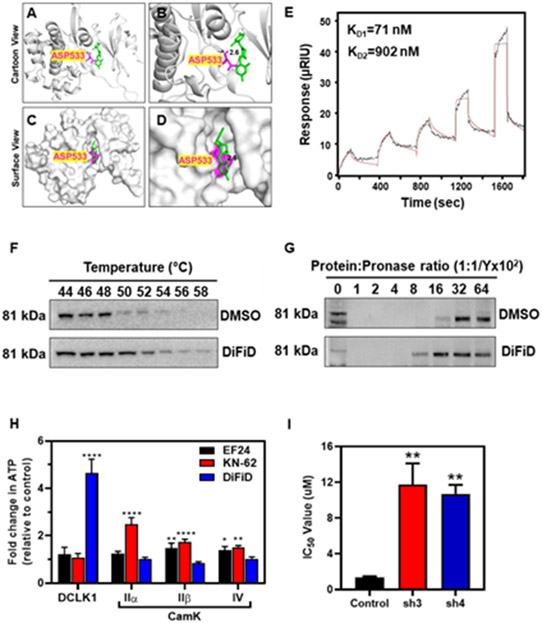
Figure 3. DiFiD binds to DCLK1.
Molecular docking studies demonstrating (A) ribbon, (B) magnified ribbon, (C) surface, and (D) magnified surface views of the DCLK1 kinase domain binding to DiFiD with a binding energy (B.E.) of −7.8 Kcal/mol. (E) Surface Plasmon Resonance assay demonstrating the binding affinity of DiFiD analyte to DCLK1 ligand (Kd1=71 nm, Kd2 = 902nm). (F) Representative immunoblot of CETSA binding assay of HNSCC (FaDu) treated with 5 μM DiFiD or DMSO for 4 hours then subjected to thermal denaturation. (G) Representative immunoblot of DARTS binding assay of HNSCC (FaDu) treated with 5 μM DiFiD or DMSO, then subjected to enzymatic degradation at various target: enzyme ratios for 15 minutes. (H) In vitro kinase assay demonstrating that DiFiD has high selectivity for and greater inhibition of its target DCLK1 relative to other Cam kinase targets. Other inhibitors, EF24 (IKK inhibitor) and KN-62 (pan CamKII inhibitor) demonstrate no significant inhibition of DCLK1. Data represent cumulative results from three independent experiments with ± SEM. **p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (I) IC50 of DiFiD when applied to FaDu cells stably transduced with scrambled control shRNA (control), or shRNA directed against DCLK1-3 (sh3) or DCLK1-4 (sh4). Data represents cumulative data from 3 independent experiments with ± SEM. **p<0.01
Additionally, the binding kinetics of DiFiD and DCLK1 were studied via surface plasmon resonance (SPR). We observed dissociation constants of KD1 = 71 nM and KD2 =902 nM (Fig. 3E). Taken together this data shows that DiFiD binds DCLK1 with high affinity. To confirm that DCLK1 is a binding target of DiFiD in the cells, we performed cellular thermal shift assay (CETSA) to assess protein stability following thermal denaturation. FaDu cells were treated with 5 μM DiFiD at 37°C for 4 hours. Cells were aliquoted into equal volumes, subjected to a thermal gradient and DCLK1 expression was then evaluated by western blot. Thermal denaturation of DCLK1 occurred at 54°C in the DMSO treated control group, which increased to 58°C in the presence of DiFiD (Fig. 3F). To further validate DiFiD binding with DCLK1, we performed the drug affinity responsive target stability (DARTS) assay. Briefly, FaDu cell lysates were incubated with DMSO or DiFiD (5 μM) for 30 minutes. We then incubated cells with increasing protein:pronase ratio. We observed that DiFiD protected DCLK1 from protease-mediated degradation, as DCLK1 expression was extended to 1: 1/800 protein: pronase ratio compared to 1: 1/1600 in the DMSO control arm (Fig. 3G). Altogether, these data suggest that DiFiD binds to DCLK1.
To confirm DiFiD specifically interacts with the kinase domain of DCLK1, we performed CETSA analyses. For this, we stably expressed the N-terminal DCX domain (lacking the kinase domain) in HNSCC cells (Supplementary Fig. S3A and S3B). To detect DCX domain fragments in FaDu cells, we utilized DCLK1 antibody produced using residues near the N-terminus. This specifically recognizes DCLK1 protein isoforms containing 82 kDa DCX sequences. We found that cells expressing the predicted 51 kDa N-terminal DCX domain did not exhibit thermal stabilization when treated with DiFiD (Supplementary Fig. S3C). This suggests that DiFiD does not stabilize DCLK1 lacking the kinase domain. Therefore, we conclude that DiFiD preferentially binds to the kinase domain of DCLK1, further confirming molecular docking predictions.
DCLK1 belongs to the family of kinases with homology to calmodulin kinases, but it does not depend on Ca2+/calmodulin for its kinase activity [27]. Since DiFiD interacts with the kinase domain, we next assessed the effect of DiFiD on DCLK1 kinase activity by performing an in vitro kinase assay. Here, we incubated recombinant DCLK1 with GS peptide as substrate to assess the ability of DCLK1 to phosphorylate the peptide [18, 19]. Along with DiFiD, we also tested the ability of EF24 (IKK inhibitor) and KN62, a pan-CaMKII inhibitor. We observed greater inhibition of DCLK1-mediated ATP consumption following incubation with DiFiD, than with either EF24 or KN-62 (Fig. 3H). To determine the specificity of DiFiD to DCLK1, we also performed studies with CaMKIIα, CaMKIIβ, and CaMKIV. We observed that DiFiD did not affect the activities of these CaMK proteins. These data demonstrate a higher selectivity and specificity of DiFiD for DCLK1.
Since DiFiD has favorable binding to DCLK1, inhibits its kinase activity, and is relatively ineffective at reducing Het1A proliferation (Supplementary Fig. 4B), we hypothesized that cells expressing lower levels of DCLK1 would be resistant to DiFiD. Consequently, we performed hexosaminidase assays with cells where DCLK1 was knocked down using specific shRNA. There was an increase in their respective half maximal inhibitory concentration (IC50) values from 1.3 μM to greater than 11 μM compared to control cells (Fig. 3I). These data demonstrate DiFiD activity is dependent upon the presence of DCLK1, and DiFiD has poor activity against cells with low DCLK1 expression. These data further suggest that DCLK1 is a highly specific direct target of DiFiD.
DiFiD demonstrates potent cytotoxicity in HNSCC in vitro
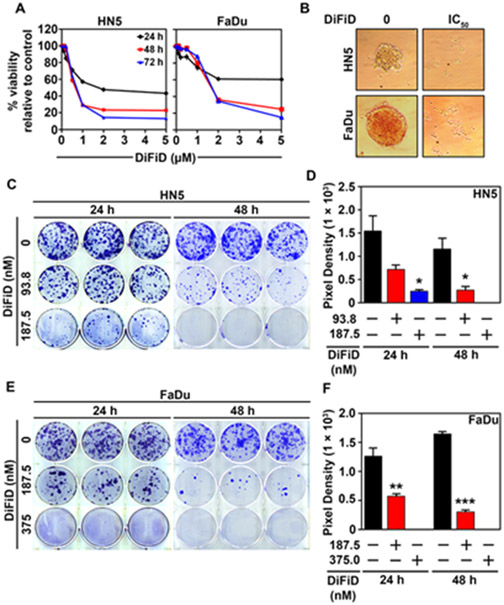
Since DiFiD inhibits DCLK1 activity, we sought to determine the effect of DiFiD on HNSCC cancer cell viability. We observed that DiFiD inhibits HNSCC viability in a dose and time-dependent manner (Fig. 4A and Supplementary Fig. S3A). We identified an effective dose to assess the mechanism of action. The IC50 was measured by hexosaminidase assay and observed within 48 hours at concentrations of 750 nM and 1.5 μM in HN5 and FaDu cell lines, respectively. In addition, we tested toxicity of DiFiD on an immortalized non-cancerous cell line, Het1A [28] (Fig. 1G). We observed that the IC50 for Het1A at 48 hours was 9 μM, a 6-12-fold increase compared to FaDu and HN5 cells, respectively (Supplementary Fig. S4B). In addition, we observed that DiFiD attenuates spheroid growth in both HN5 and FaDu cell lines when treated at IC50 concentrations, suggesting that it affects anchorage independent growth (Fig. 4B). We next determined the effect of DiFiD on clonogenicity of HNSCC cells. Initial studies with IC50 doses demonstrated complete suppression of colony formation. However, when treated at lower doses of 93.75 nM (1/8th IC50) and 187.5 nM (1/4th IC50) for 48 hours, we observed a dose dependent decrease in both the number and size of colonies for HN5 cells (Fig. 4C and D, p<0.01). Similarly, FaDu cells treated with a 1/4th (375 nM) and 1/8th (187.5 nM) IC50 dose of DiFiD exhibited a significant inhibition in colony number and size (Fig. 4E and F, p<0.01). These data suggest that DiFiD has potent cytotoxic effects on HNSCC that are long lasting.
Figure 4. DiFiD inhibits HNSCC proliferation in 2-D and 3-D culture models.
(A) Hexosaminidase viability assay of HNSCC (HN5 and FaDu) treated for 24, 48, and 72 hours at increasing concentrations of DiFiD (0, 0.125, 0.25, 0.5, 1, 2, 5 μM). Figure is representative of three experimental replicates with ± SEM. (B) DiFiD abrogated spheroid growth in HNSCC (HN5 and FaDu) at IC50 concentrations. (C-F) Colony formation assay was performed to assess long term efficacy of 24 h or 48 h DiFiD treatment. Colony formation assay and quantification of pixel density representing the colony size using ImageJ of HN5 (C, D) and FaDu (E, F), 14 days post DiFiD treatment. Graphs represents cumulative results from three independent experiments with ± SEM. *p<0.05, **p<0.01, ***p<0.001.
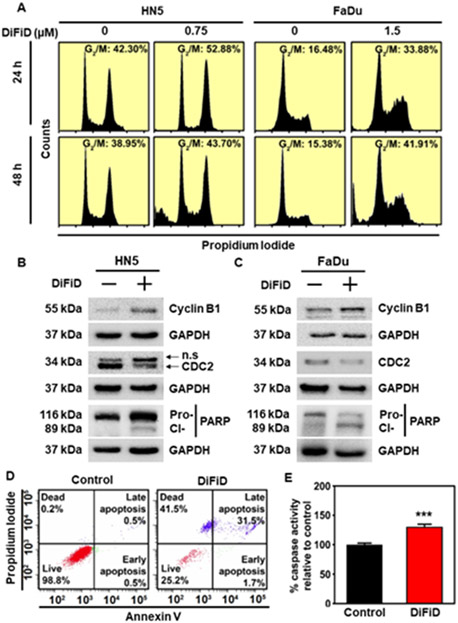
DiFiD induces G2/M arrest and apoptosis
To further characterize the effects of DiFiD treatment on HNSCC proliferation, we used flow cytometry to evaluate cell cycle progression. HN5 and FaDu cell lines were treated with DiFiD. Within 24 hours, DiFiD treatment increased the percentage of cells arrested at the G2/M phase (Fig. 5A and Supplementary Fig. S5). At 48 hours, there was an increase in the sub G0, or fragmented DNA stage following treatment, suggesting increased cell death. To confirm G2/M arrest, western blot analysis was performed for G2/M associated proteins cyclin B1 and cell division cycle protein 2 (CDC2). HN5 and FaDu cancer cells were treated with IC50 doses of DiFiD for 24 hours, in which time dependent increases in cyclin B1 and simultaneous decreases in CDC2 expression were observed in the treatment arm (Fig. 5B and 5C). We also performed western blot analysis for pro- and cleaved-PARP protein. DiFiD treatment induced PARP cleavage in both HN5 and FaDu cell lines (Fig. 5B and 5C). This suggests that DiFiD further induces apoptosis in HNSCC cancer cell lines associated with a G2/M arrest. This was confirmed by Annexin V/PI staining that demonstrated an increased percentage of DiFiD treated cells were in the apoptotic and dead fractions compared to vehicle control (Fig. 5D). To assess the role of caspases in the induction of apoptosis by DiFiD, we performed caspase 3/7 assay to assess effector caspase activity. DiFiD induced a significant upregulation in the activity of the effector caspases (Fig. 5E, p<0.001). Taken together, these data confirmed that DiFiD mediated suppression of DCLK1 activity contributes to mitotic catastrophe of HNSCC.
Figure 5. DiFiD induces G2/M arrest and apoptosis.
(A) Cell cycle analysis was performed in HN5 and FaDu cell lines after 24 or 48 h treatment with IC50 concentrations of DiFiD, (HN5=750 nM; FaDu=1.5 μM). Figure is representative of three independent studies. (B) Representative immunoblot of HN5 cells treated with 750 nM DiFiD for 24 hours demonstrates expression of cyclin B1, CDC2, Pro- and cleaved-PARP protein with GAPDH used as a loading control. (C) Representative immunoblot of FaDu cells treated with 1.50 μM DiFiD for 24 hours demonstrates expression of cyclin B1, CDC2, Pro- and cleaved-PARP protein with GAPDH used as a loading control. (D) Annexin V assay of HN5 cells following treatment with DiFiD at the IC50 dose. HN5 cells were treated with DiFiD at the IC50 dose for 48 h, stained with Annexin V (FITC) and PI, and analyzed by flow cytometry. (E) Caspase 3/7 assay demonstrates caspase cleavage in HN5 cells 48 h post treatment with DiFiD. Data represents cumulative results from three independent experiments with ± SEM ***p<0.001.
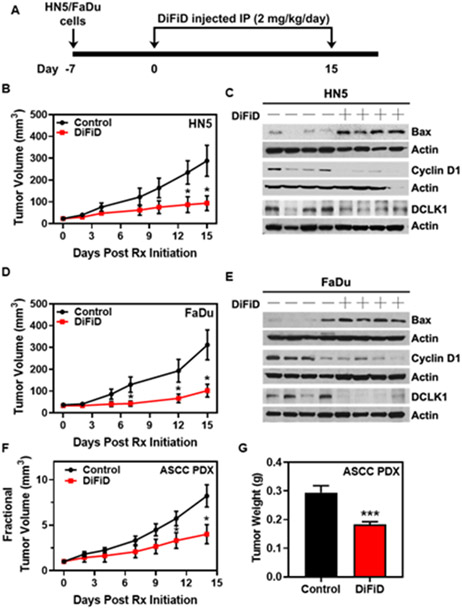
DiFiD has antitumor effects in vivo
To determine the in vivo antitumor activity of DiFiD, we treated FaDu and HN5 subcutaneous tumors in Foxn1nu/nu mice. Briefly, HN5 or FaDu cells were injected subcutaneously into the flanks of nude mice and were subsequently treated with DiFiD at 2 mg/kg/day for 15 days (Fig. 6A). DiFiD treatment significantly inhibited tumor growth in HN5 xenografts compared to vehicle control (DMSO) treated tumors (n=7, p<0.05, Fig. 6B and Supplementary Fig. 6A). Similarly, with FaDu xenograft subcutaneous tumors, DiFiD treatment significantly reduced tumor growth compared to the vehicle control treatment arm (n=10, p<0.05, Fig, 6C and Supplementary Fig. 6E). Lastly, the antitumor effect of DiFiD was tested in an ASCC patient-derived xenograft model. DiFiD treatment significantly reduced PDX tumor growth as evidenced by tumor volume and weight of ASCC PDX tumors when compared to the control group (p<0.05, Fig. 6D and p<0.001, Fig. 6E). These preclinical data indicate that DiFiD is a promising, well-tolerated, therapeutic agent for the management of SCC.
Figure 6. DiFiD inhibits SCC growth in vivo.
(A) DiFiD treatment protocol for xenograft studies. HN5/FaDu (1 x 106) cells were inoculated subcutaneously into the right flank of athymic Foxn1nu/nu mice. DiFiD (2 mg/kg) or vehicle control (DMSO) was administered by intraperitoneal injection once daily for 15 days. (B) HN5 tumor volumes (n=7 mice/group) are depicted. (C) HN5 xenograft tumors were subjected to biomarker analyses. Representative immunoblots from a small cohort of HN5 xenograft tumors were probed for Bax, cyclin D1 and DCLK1. (D) FaDu tumor volumes (n=10 mice/group) are depicted. (E) Representative immunoblots from a small cohort of FaDu xenograft tumors were probed as in (C). ASCC PDX was transplanted into the flanks of NSG mice. DiFiD (2 mg/kg) or vehicle control (DMSO) was administered by intraperitoneal injection once daily for 15 days. (F) Fractional tumor volume and weight (G) are depicted (n=10 mice/group for control and n=14 mice/group for DiFiD treatment). Error bars represent ± SEM. *p<0.05, ***p<0.001.
To elucidate the molecular mechanism whereby DiFiD exerts its antitumor effects on HNSCC, we analyzed xenograft tumors using western blotting. Tumor samples were subjected to electrophoresis, and subsequently, expression of Bax, Cyclin D1, and DCLK1 were determined. DiFiD significantly induced the expression of Bax in HN5 (Fig. 6F and Supplementary Fig. 6B) and FaDu (Fig. 6G and Supplementary Fig. 6F) tumor samples relative to vehicle control. Additionally, DiFiD significantly reduced expression of Cyclin D1 (Fig. 6F and Supplementary Fig. 6C) and DCLK1 (Fig. 6F and Supplementary Fig. 6D) in HN5 tumors, suggesting inhibition of proliferation and cell cycle entrance. FaDu tumor xenografts also demonstrated decreased expression of cyclin D1 (Fig. 6G and Supplementary Fig. 6G) and DCLK1 (Fig. 6G and Supplementary Fig. 6H), further substantiating our observations in vivo. Taken together, these studies demonstrate that DiFiD inhibits tumor growth by targeting DCLK1 and inducing cellular apoptosis.
Discussion
SCCs of the head and neck and anus are aggressive malignancies with high rates of local recurrence, distant metastasis, and poor clinical outcomes, including reduced survival. Current standard of care include surgery followed by chemoradiotherapy. However, despite aggressive treatment, the survival rate remains low highlighting a significant unmet medical need in SCC patients. In this article, we demonstrate that DCLK1 is a clinically relevant target for HNSCC and ASCC. DCLK1 is well characterized as a reserve, stress induced stem cell marker in pancreatic and colorectal cancers [5, 6]. Using multiple platforms, we demonstrate DCLK1 expression is significantly upregulated in HNSCC compared to normal oral mucosa of patient tissues. Furthermore, high expression of DCLK1 correlates with poor clinical outcomes. These findings agree with a recent report associating high DCLK1 levels with poor HNSCC patient survival [4].
Genetic knockdown of DCLK1 has demonstrated promising findings in neuroblastoma, colorectal, and pancreatic tumors [29, 30]. DCLK1 knockdown triggers apoptosis and inhibits proliferation, mitochondrial function, and ATP synthesis in neuroblastoma cells [30]. TCGA analysis has shown DCLK1 to be positively correlated with NOTCH signaling in HNSCC [31]. Additionally, in vitro inhibition of DCLK1 via siRNA or inhibitor (LRRK2-IN-1) in HNSCC cell lines showed decreased levels of downstream NOTCH effectors, HEY1, HES1, and HES5 [31]. Furthermore, DCLK1 siRNA nanoparticle delivery to colorectal and pancreatic tumor xenografts resulted in the significant inhibition of tumor growth with seemingly high tolerance [29]. Our data presented herein supports these previous findings, as we observed decreased spheroid growth, colony formation, migration, and invasion following knockdown of DCLK1. As such, due to the high expression of DCLK1 in HNSCC tumor tissues and cell lines, we postulated that DCLK1 may be a potential therapeutic target for HNSCC.
Currently there is a DCLK1 inhibitor, DCLK1-IN-1, which has been reported to inhibit DCLK1 activity with high specificity [32]. Nevertheless, DCLK1-IN-1 has not been tested in vivo. This compound was derived from the 5, 11-dihydro-6H-benzo[e]pyrimido[5,4-b][1,4]diazepin-6-one scaffold, which includes the LRKK2 and LRRK2-IN-1 inhibitors. Weygant et al., in targeting leucine-rich repeat kinase 2 (LRRK2), with the small molecule inhibitor LRRK2-IN-1, reported inhibition of DCLK1, and subsequent attenuation of HCT116 (colon) and AsPC-1 (pancreatic) growth, and invasiveness [33]. However, direct binding of DCLK1 by LRRK-IN-1 was not examined in this study and the effects are most likely due to the indirect effects of LRRK inhibition on DCLK1 activity [34]. Furthermore, several kinase inhibitors with anti-tumor activity, such as XMD8-92 (MAPK7 inhibitor), BI-2536 (PLK1 inhibitor), and TAE-684 (ALK inhibitor), demonstrate non-specific activity, with a comparable affinity towards DCLK1 as much as their target kinases [35]. Therefore, while inhibition of DCLK1 may play a role in the therapeutic activity of these compounds, it is highly valuable to identify a specific DCLK1 inhibitor for future clinical applications that has potent antitumor activity.
Previously, we reported that DiFiD shows antitumor activity towards pancreatic cancer cells [11]. However, the direct target for DiFiD was not elucidated. We used an in vitro kinase assay to demonstrate that DiFiD effectively inhibits DCLK1 kinase activity, but not related CaMK family members, suggesting that the compound is a specific competitive inhibitor of DCLK1. We further confirmed a direct interaction through in vitro binding assays and SPR analysis. The CETSA and DARTs binding assays involved the uptake of the compound by cells before thermal or enzymatic denaturation, respectively [22]. We showed that DCLK1 was robustly stabilized by DiFiD. Additionally, cells stably transfected with expression plasmids containing only the DCX domain did not exhibit thermal stabilization as was observed with full length DCLK1 containing the kinase domain. Interestingly, we found that DCX domain required higher temperatures to denature compared to full length protein. This is likely due to enhanced microtubule association and stabilization of the domain, as this domain in DCLK1 is critical for its microtubule binding and polymerization activity [7, 36]. SPR analysis, which is the gold standard for target interactions, identified strong binding of DiFiD to DCLK1 with low nM concentration equilibrium dissociation constants (KD). Taken together, these data demonstrate that DiFiD has high selectivity for DCLK1 with binding sites located in the C-terminal kinase domain.
We demonstrated that DiFiD induces cell-cycle arrest and apoptosis, attenuating HNSCC proliferation and colony formation in vitro, and tumor growth in vivo. Het1A, non-cancerous cells expressing low amounts of DCLK1 in comparison to the HNSCC cell lines, HN5 and FaDu, demonstrated lower DiFiD mediated toxicity. When we knocked-down DCLK1 in FaDu cells using shRNA, we observed a 10-fold decrease in DiFiD toxicity, further demonstrating the specificity of DiFiD for DCLK1. DiFiD increased cell cycle arrest in the G2/M phase resulting in mitotic catastrophe, data consistent with previously published studies [14, 33]. This is further supported by our observation of a significant increase in the sub-G0 population following treatment with DiFiD. Moreover, we observed an upregulation of the G2/M associated protein, cyclin B1 and a decrease in CDC2, proteins that are abnormally regulated when cells undergo mitotic catastrophe [37, 38]. These data corroborate our previous reported results demonstrating the induction of p21 and a reduction in cyclins A1 and D1 following DiFiD treatment in pancreatic cancer [14].
ASCC has an incidence rate of 0.2-4.4 per 100,000 people per year which has risen worldwide over the last three decades, especially in homosexual men (35 per 100,000 per year) and those with HIV (75-135 per 100,000 per year) [39, 40]. Despite these rising numbers, the standard of care treatment for this cancer comprised of fluorouracil and mitomycin C has remained essentially unchanged since its inception [41]. The addition of intensity-modulated radiation therapy and cisplatin has shown similar efficacy to fluorouracil and mitomycin C in a large, randomized trial [42, 43]. The relative rarity of ASCC presents challenges in conducting pivotal clinical trials. In addition, validated preclinical models of ASCC that accurately replicate clinical observations are limited. Existing preclinical models of ASCC include a cell line derived from a lymph node metastasis, two transgenic mouse models and a xenograft from a single patient [44-47]. Here we present a patient derived xenograft model and provide preliminary evidence of a promising, new therapeutic target for the management of ASCC.
In our studies, we observed significant antitumor effects in SCC preclinical models following treatment with DiFiD. DCLK1 is expressed in various normal cell types, including neurons, osteoblasts, and colon stem cells [48], and is involved in physiological processes, including retrograde transport, neuronal migration, and neurogenesis. The data presented in this article suggest that DiFiD is well tolerated at doses that demonstrated antitumor activity, as mice maintained normal weight gain and ambulation. DiFiD tolerance was observed in an earlier study [14], further supporting the notion that it is well tolerated at the doses used to inhibit cancer growth.
Conclusion
DCLK1 is critical for HNSCC and anal SCC growth. For the first time we demonstrated that DiFiD binds to DCLK1 and inhibits SCC growth in vitro and in vivo. Thus, our results strongly establish DCLK1 as a clinically relevant therapeutic target for SCC.
Supplementary Material
Supplementary Figure 1. (A) DCLK1 mRNA expression by histological tumor grade represented as fragments per kilobase of transcript per million (FPKM) mapped reads, obtained through TCGA analysis (G1, n=64; G2, n=312; G3, n=125) (B) DCLK1 mRNA expression in HPV negative (n=74) and HPV positive tumors (n=42) represented as fragments per kilobase of transcript per million (FPKM) mapped reads, obtained through TCGA analysis (C) HN5 spheroids stained with H&E, or DCLK1 (green) and DAPI (blue). (D) Representative images of DCLK1 staining in non- cancerous anal mucosa (n=3) and ASCC (n=14). (E) Cumulative results of composite score (intensity x % positivity) of DCLK1 staining in anal mucosa samples. *p<0.05, **p<0.01, ***p<0.001
Supplementary Figure 2. (A) Western blot analysis of xenograft tumor lysates demonstrate response DCLK1 regulation in vivo. FaDu (1x106) cells stably transfected with shRNA targeting DCLK1-3 (sh3), DCLK1-4 (sh4) or scrambled control shRNA were injected subcutaneously into NSG mice. Tumors were profiled for biomarker modulation.
Supplementary Figure 3. (A) Diagram of DCLK1 full length and DCX domain fragments. (B) Restriction digested plasmid constructs expressing either full length (FL) or DCX domain (DCX) fragments. (C) Western blot analysis of CETSA samples from DCX domain expressing FaDu cells treated with DMSO or DiFiD.
Supplementary Figure 4. (A) Bright field images of HN5 and FaDu cells at 100X magnification treated with increasing doses of DiFiD for 48 h. (B) Hexosaminidase assay for Het1A cells treated with DiFiD for up to 72 hours.
Supplementary Figure 5. Quantification of cell cycle data from three independent experiments with HN5 and FaDu cells treated with DiFiD for 24 h or 48 h.
Supplemental Figure 6. HN5 or FaDu (1 x 106) cells were inoculated subcutaneously into the right flank of athymic Foxn1nu/nu mice. DiFiD (2 mg/kg) or vehicle control (DMSO) was administered by intraperitoneal injection once daily for 15 days. HN5 Tumor weight (A) (n=7 mice/group), are depicted. HN5 xenograft tumors were subjected to biomarker analyses. Densitometric analysis of HN5 xenograft tumors immunoblot signals for (B) Bax (C) Cyclin D1 (D) DCLK1. (E) FaDu tumor weight (n=10 mice/group). Densitometric analyses of FaDu tumor signals for (F) Bax (G) Cyclin D1 (H) DCLK1 are depicted graphically, including ± SEM, *p<0.05.
Acknowledgements
We would like to thank The University of Kansas Cancer Center Biospecimen Repository Core Facility for their services in providing high quality tissue samples.
Funding
This study was supported in part by the University of Kansas Cancer Center under CCSG P30CA168524; an NIH Clinical and Translational Science Award grant (UL1TR000001, formerly UL1RR033179) awarded to the University of Kansas Medical Center, an internal Lied Basic Science Grant Program of the KUMC Research Institute, the Thomas P. O’Sullivan IV, and Marina O’Sullivan Family Fund.
List of abbreviations
- ASCC
Anal Squamous Cell Carcinoma
- CETSA
Cellular Thermal Shift Assay
- DARTS
Drug affinity responsive target stability assay
- DCLK1
Doublecortin-Like Kinase 1
- DiFiD
3,5-bis (2,4-difluorobenzylidene)-4-piperidone
- DMSO
Dimethyl Sulfoxide
- HGSIL
High grade squamous intraepithelial lesions
- HNSCC
Head and Neck Squamous Cell Carcinoma
- HPV
Human Papilloma Virus
- LRRK2
leucine-rich repeat kinase 2
- LSIL
Low grade squamous intraepithelial lesions
- NSG
NOD SCID gamma mice
- PDX
Patient derived xenograft
- RPKM
Reads Per Kilobase of transcript, per Million mapped reads
- SCC
Squamous Cell Carcinoma
- TCGA
The Cancer Genome Atlas
References
- 1.Ferlay J, et al. , Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer, 2015. 136(5): p. E359–86. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, et al. , Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin, 2011. 61(4): p. 212–36. [DOI] [PubMed] [Google Scholar]
- 3.Sahu N and Grandis JR, New advances in molecular approaches to head and neck squamous cell carcinoma. Anticancer Drugs, 2011. 22(7): p. 656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadletz L, et al. , Role of cancer stem-cell marker doublecortin-like kinase 1 in head and neck squamous cell carcinoma. Oral Oncol, 2017. 67: p. 109–118. [DOI] [PubMed] [Google Scholar]
- 5.Rangarajan P, et al. , Crocetinic acid inhibits hedgehog signaling to inhibit pancreatic cancer stem cells. Oncotarget, 2015. 6(29): p. 27661–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwatra D, et al. , Methanolic extracts of bitter melon inhibit colon cancer stem cells by affecting energy homeostasis and autophagy. Evid Based Complement Alternat Med, 2013. 2013: p. 702869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel O, et al. , Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1. Structure, 2016. 24(9): p. 1550–61. [DOI] [PubMed] [Google Scholar]
- 8.Sureban SM, et al. , Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology, 2009. 137(2): p. 649–59, 659 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponnurangam S, et al. , Quinomycin A targets Notch signaling pathway in pancreatic cancer stem cells. Oncotarget, 2016. 7(3): p. 3217–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge Y, et al. , Alternative splice variants of DCLK1 mark cancer stem cells, promote self-renewal and drug-resistance, and can be targeted to inhibit tumorigenesis in kidney cancer. Int J Cancer, 2018. 143(5): p. 1162–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandrakesan P, et al. , Dclk1, a tumor stem cell marker, regulates pro-survival signaling and self-renewal of intestinal tumor cells. Mol Cancer, 2017. 16(1): p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi Y, et al. , Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet, 2013. 45(1): p. 98–103. [DOI] [PubMed] [Google Scholar]
- 13.Westphalen CB, et al. , Dclk1 Defines Quiescent Pancreatic Progenitors that Promote Injury-Induced Regeneration and Tumorigenesis. Cell Stem Cell, 2016. 18(4): p. 441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramaniam D, et al. , 3,5-bis(2,4-difluorobenzylidene)-4-piperidone, a novel compound that affects pancreatic cancer growth and angiogenesis. Mol Cancer Ther, 2011. 10(11): p. 2146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CJ, et al. , Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck, 2007. 29(2): p. 163–88. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam D, et al. , Diphenyl difluoroketone: a curcumin derivative with potent in vivo anticancer activity. Cancer Res, 2008. 68(6): p. 1962–9. [DOI] [PubMed] [Google Scholar]
- 17.Subramaniam D, et al. , Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS One, 2012. 7(2): p. e30590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacchetto R, et al. , Glycogen synthase binds to sarcoplasmic reticulum and is phosphorylated by CaMKII in fast-twitch skeletal muscle. Arch Biochem Biophys, 2007. 459(1): p. 115–21. [DOI] [PubMed] [Google Scholar]
- 19.Anderson KA, et al. , Regulation and function of the calcium/calmodulin-dependent protein kinase IV/protein serine/threonine phosphatase 2A signaling complex. J Biol Chem, 2004. 279(30): p. 31708–16. [DOI] [PubMed] [Google Scholar]
- 20.Trott O and Olson AJ, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010. 31(2): p. 455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander N, Woetzel N, and Meiler J, bcl::Cluster : A method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int Conf Comput Adv Bio Med Sci, 2011. 2011: p. 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jafari R, et al. , The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc, 2014. 9(9): p. 2100–22. [DOI] [PubMed] [Google Scholar]
- 23.Pai MY, et al. , Drug affinity responsive target stability (DARTS) for small-molecule target identification. Methods Mol Biol, 2015. 1263: p. 287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang XH, et al. , Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res, 2004. 10(1 Pt 1): p. 301–13. [DOI] [PubMed] [Google Scholar]
- 25.Wheeler SE, et al. , Enhancement of head and neck squamous cell carcinoma proliferation, invasion, and metastasis by tumor-associated fibroblasts in preclinical models. Head Neck, 2014. 36(3): p. 385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vishwakarma V, et al. , Potent Antitumor Effects of a Combination of Three Nutraceutical Compounds. Sci Rep, 2018. 8(1): p. 12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohmae S, et al. , Molecular identification and characterization of a family of kinases with homology to Ca2+/calmodulin-dependent protein kinases I/IV. J Biol Chem, 2006. 281(29): p. 20427–39. [DOI] [PubMed] [Google Scholar]
- 28.Liu T, et al. , Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis, 2007. 28(2): p. 488–96. [DOI] [PubMed] [Google Scholar]
- 29.Sureban SM, et al. , Nanoparticle-based delivery of siDCAMKL-1 increases microRNA-144 and inhibits colorectal cancer tumor growth via a Notch-1 dependent mechanism. J Nanobiotechnology, 2011. 9: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verissimo CS, et al. , Silencing of the microtubule-associated proteins doublecortin-like and doublecortin-like kinase-long induces apoptosis in neuroblastoma cells. Endocr Relat Cancer, 2010. 17(2): p. 399–414. [DOI] [PubMed] [Google Scholar]
- 31.Broner EC, et al. , Doublecortin-Like Kinase 1 (DCLK1) Is a Novel NOTCH Pathway Signaling Regulator in Head and Neck Squamous Cell Carcinoma. Front Oncol, 2021. 11: p. 677051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel O, et al. , Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1. Commun Biol, 2021. 4(1): p. 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weygant N, et al. , Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Mol Cancer, 2014. 13: p. 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luerman GC, et al. , Phosphoproteomic evaluation of pharmacological inhibition of leucine-rich repeat kinase 2 reveals significant off-target effects of LRRK-2-IN-1. J Neurochem, 2014. 128(4): p. 561–76. [DOI] [PubMed] [Google Scholar]
- 35.Davis MI, et al. , Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 2011. 29(11): p. 1046–51. [DOI] [PubMed] [Google Scholar]
- 36.Lipka J, et al. , Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J, 2016. 35(3): p. 302–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scaife RM, G2 cell cycle arrest, down-regulation of cyclin B, and induction of mitotic catastrophe by the flavoprotein inhibitor diphenyleneiodonium. Mol Cancer Ther, 2004. 3(10): p. 1229–37. [PubMed] [Google Scholar]
- 38.Choi HJ, Fukui M, and Zhu BT, Role of cyclin B1/Cdc2 up-regulation in the development of mitotic prometaphase arrest in human breast cancer cells treated with nocodazole. PLoS One, 2011. 6(8): p. e24312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernardi MP, et al. , Molecular biology of anal squamous cell carcinoma: implications for future research and clinical intervention. Lancet Oncol, 2015. 16(16): p. e611–21. [DOI] [PubMed] [Google Scholar]
- 40.Silverberg MJ, et al. , Risk of anal cancer in HIV-infected and HIV-uninfected individuals in North America. Clin Infect Dis, 2012. 54(7): p. 1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buroker TR, et al. , Combined therapy for cancer of the anal canal: a follow-up report. Dis Colon Rectum, 1977. 20(8): p. 677–8. [DOI] [PubMed] [Google Scholar]
- 42.Gunderson LL, et al. , Long-term update of US GI intergroup RTOG 98-11 phase III trial for anal carcinoma: survival, relapse, and colostomy failure with concurrent chemoradiation involving fluorouracil/mitomycin versus fluorouracil/cisplatin. J Clin Oncol, 2012. 30(35): p. 4344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng M, et al. , Australasian Gastrointestinal Trials Group (AGITG) contouring atlas and planning guidelines for intensity-modulated radiotherapy in anal cancer. Int J Radiat Oncol Biol Phys, 2012. 83(5): p. 1455–62. [DOI] [PubMed] [Google Scholar]
- 44.Stelzer MK, et al. , Rapamycin inhibits anal carcinogenesis in two preclinical animal models. Cancer Prev Res (Phila), 2010. 3(12): p. 1542–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stelzer MK, et al. , A mouse model for human anal cancer. Cancer Prev Res (Phila), 2010. 3(12): p. 1534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun ZJ, et al. , Inhibition of mTOR reduces anal carcinogenesis in transgenic mouse model. PLoS One, 2013. 8(10): p. e74888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeda A, et al. , Establishment and characterization of the human SaTM-1 anal canal squamous cell carcinoma cell line derived from lymph node metastasis. Int J Mol Med, 2009. 24(4): p. 465–72. [DOI] [PubMed] [Google Scholar]
- 48.Zou W, et al. , The microtubule-associated protein DCAMKL1 regulates osteoblast function via repression of Runx2. J Exp Med, 2013. 210(9): p. 1793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (A) DCLK1 mRNA expression by histological tumor grade represented as fragments per kilobase of transcript per million (FPKM) mapped reads, obtained through TCGA analysis (G1, n=64; G2, n=312; G3, n=125) (B) DCLK1 mRNA expression in HPV negative (n=74) and HPV positive tumors (n=42) represented as fragments per kilobase of transcript per million (FPKM) mapped reads, obtained through TCGA analysis (C) HN5 spheroids stained with H&E, or DCLK1 (green) and DAPI (blue). (D) Representative images of DCLK1 staining in non- cancerous anal mucosa (n=3) and ASCC (n=14). (E) Cumulative results of composite score (intensity x % positivity) of DCLK1 staining in anal mucosa samples. *p<0.05, **p<0.01, ***p<0.001
Supplementary Figure 2. (A) Western blot analysis of xenograft tumor lysates demonstrate response DCLK1 regulation in vivo. FaDu (1x106) cells stably transfected with shRNA targeting DCLK1-3 (sh3), DCLK1-4 (sh4) or scrambled control shRNA were injected subcutaneously into NSG mice. Tumors were profiled for biomarker modulation.
Supplementary Figure 3. (A) Diagram of DCLK1 full length and DCX domain fragments. (B) Restriction digested plasmid constructs expressing either full length (FL) or DCX domain (DCX) fragments. (C) Western blot analysis of CETSA samples from DCX domain expressing FaDu cells treated with DMSO or DiFiD.
Supplementary Figure 4. (A) Bright field images of HN5 and FaDu cells at 100X magnification treated with increasing doses of DiFiD for 48 h. (B) Hexosaminidase assay for Het1A cells treated with DiFiD for up to 72 hours.
Supplementary Figure 5. Quantification of cell cycle data from three independent experiments with HN5 and FaDu cells treated with DiFiD for 24 h or 48 h.
Supplemental Figure 6. HN5 or FaDu (1 x 106) cells were inoculated subcutaneously into the right flank of athymic Foxn1nu/nu mice. DiFiD (2 mg/kg) or vehicle control (DMSO) was administered by intraperitoneal injection once daily for 15 days. HN5 Tumor weight (A) (n=7 mice/group), are depicted. HN5 xenograft tumors were subjected to biomarker analyses. Densitometric analysis of HN5 xenograft tumors immunoblot signals for (B) Bax (C) Cyclin D1 (D) DCLK1. (E) FaDu tumor weight (n=10 mice/group). Densitometric analyses of FaDu tumor signals for (F) Bax (G) Cyclin D1 (H) DCLK1 are depicted graphically, including ± SEM, *p<0.05.