Abstract
Background:
PAX-fusion negative rhabdomyosarcoma (FN RMS) is driven by alterations in the RAS/MAP kinase pathway and is partially responsive to MEK inhibition. Overexpression of IGF1R and its ligands is also observed in FN RMS. Preclinical and clinical studies have suggested that IGF1R is itself an important target in FN RMS. Our previous studies revealed preclinical efficacy of the MEK1/2 inhibitor, trametinib, and an IGF1R inhibitor, BMS-754807, but this combination was not pursued clinically due to intolerability in preclinical murine models. Here, we sought to identify a combination of an MEK1/2 inhibitor and IGF1R inhibitor that would be tolerated in murine models and effective in both cell line and patient derived xenograft models of RAS-mutant FN RMS.
Methods:
Using proliferation and apoptosis assays, we studied the factorial effects of trametinib and ganitumab (AMG 479), a monoclonal antibody with specificity for human and murine IGF1R, in a panel of RAS-mutant FN RMS cell lines. The molecular mechanism of the observed synergy was determined using conventional and capillary immunoassays. The efficacy and tolerability of trametinib/ganitumab was assessed using a panel of RAS-mutated cell-line and patient-derived RMS xenograft models.
Results:
Treatment with trametinib and ganitumab resulted in synergistic cellular growth inhibition in all cell lines tested and inhibition of tumor growth in four out of six models of RAS-mutant RMS. The combination had little effect on body weight and did not produce thrombocytopenia, neutropenia, or hyperinsulinemia in tumor-bearing SCID beige mice. Mechanistically, ganitumab treatment prevented the phosphorylation of AKT induced by MEK inhibition alone. Therapeutic response to the combination was observed in models without a mutation in the PI3K/PTEN axis.
Conclusions:
We demonstrate that combined trametinib and ganitumab is effective in a genomically diverse panel of RAS-mutated FN RMS preclinical models. Our data also show that the trametinib/ganitumab combination likely has a favorable tolerability profile. These data support testing this combination in a phase I/II clinical trial for pediatric patients with relapsed or refractory RAS-mutated FN RMS.
Keywords: RAS, MAP kinase, rhabdomyosarcoma, PTEN, IGF1R
Introduction
Rhabdomyosarcoma (RMS) accounts for 3% of all childhood cancers. This tumor is sub-divided into two major genetic and histologic subtypes: embryonal, which is usually PAX3-FOXO1 fusion negative (FN), and alveolar, which is usually PAX3-FOXO1 fusion positive (FP). The FN subtype carries a better prognosis than the FP subtype. However, the 5-year overall survival rate for patients with metastatic or relapsed RMS of either subtype is poor at 26% or 17%, respectively (1,2). Regimens of cytotoxic chemotherapeutic agents including vinorelbine and cyclophosphamide are sometimes active in the relapsed setting, but targeted agents are needed to improve overall survival in relapsed and metastatic FN RMS (3).
Comprehensive genomic analyses of human RMS tumors indicate that the most common somatic mutation in FN RMS is an oncogenic change in one of the three RAS isoforms, NRAS, HRAS, or KRAS (4,5). In a recent consortium study of 641 RMS tumors, 32% of evaluated FN RMS tumors demonstrated a mutation in one of the RAS isoforms (6). In addition, a subset of FN RMS tumors (17%) harbor mutations in the RAS GTPase-activating protein NF1 (6), which may lead to aberrant RAS signaling in these tumors. This mutational profile suggests that the RAS/RAF/MEK/ERK MAP kinase pathway plays a role in FN RMS tumorigenesis. Interestingly, although RAS mutation does not impact event-free or overall survival in patients with FN RMS (6,7), functional studies have verified that MAP kinase signaling is important for FN RMS cell proliferation and tumor growth (8). However, in preclinical models, FN RMS tumors rapidly acquire resistance to MEK inhibitors when given as a single agent, suggesting that a combination of a MEK inhibitor with another agent will be required to provide therapeutic benefit for patients with FN RMS (9–11).
The IGF1R signaling pathway is also recurrently altered in FN RMS. IGF1R is a receptor tyrosine kinase (RTK) that signals through the RAS/RAF/MEK/ERK and PI3 kinase/AKT pathways when stimulated by one of its high affinity ligands, IGF1 or IGF2. Both IGF2 and IGF1R are recurrently overexpressed in FN RMS. Overexpression of IGF2 due to loss of heterozygosity with uniparental disomy at 11p15.5, and is observed in the majority of FN RMS tumors, including 65% of RAS-mutant FN RMS tumors (5). In contrast, the overexpression of IGF1R that is observed in FN RMS is due to genomic amplification (12,13) and is associated with inferior survival. Inhibition of IGF1R signaling inhibits FN RMS cell proliferation and tumor growth, invasion, and metastasis (12–14). However, FN RMS tumors rapidly acquire resistance to inhibitors of IGF1R signaling when they are used as single agents (15–17).
Previously, we established that the combination of the MEK inhibitor, trametinib, and a small molecule inhibitor of IGF1R, BMS-754807, synergistically inhibits FN RMS cell proliferation and tumor growth in one of two orthotopic cell line xenograft models tested. Treatment with BMS-754807 prevented trametinib-induced activation of IGF1R in RAS-mutant RMS cell lines (11). However, trametinib/BMS-754807 caused excessive body weight loss in SCID beige mice, indicating intolerability in the murine models that likely predicts intolerability in humans. In the current study, we assessed the efficacy and tolerability of combining the FDA-approved trametinib with ganitumab (AMG 479), a fully human monoclonal antibody against IGF1R, in mouse models of RAS-mutant RMS. Ganitumab functions by blocking ligand binding to the extracellular domain of IGF1R, which induces IGF1R internalization and degradation (18), whereas BMS-754807 is a reversible, ATP-competitive antagonist of IGF1R and related proteins (19). Clinically, ganitumab is well tolerated. Adverse events due to ganitumab, when they occur, include lymphopenia, thrombocytopenia, and hyperglycemia (20). Here we demonstrate that combined trametinib and ganitumab treatment inhibits tumor growth in four of six heterogenous RAS-mutant RMS preclinical models, and that this combination is well tolerated in immunodeficient mice at clinically achievable doses of both agents (20,21). These data support testing this combination in a phase I/II clinical trial for pediatric patients with relapsed or refractory RAS-mutant RMS.
Materials and Methods
Cell lines, patient-derived xenografts, and reagents
The RMS cell lines RD (RRID:CVCL1649), SMS-CTR (RRID:CVCL_A770), and BIRCH (RRID:CVCL_M599), were obtained as previously reported (11) (Supplemental Table 1). The patient-derived xenograft (PDX) models SJRHB013758, SJRHB000026, and SJRHB010927 were obtained from the Childhood Solid Tumor Network at St. Jude (22). All cell lines were confirmed to be mycoplasma negative using the MycoAlert kit (Lonza) and their identity was confirmed by STR fingerprinting (Genetica/LapCorp) prior to experimental use. Published single nucleotide variants in the PDX models were confirmed by whole exome sequencing (WES, see Supplemental Data File 1). Copy number variants were determined by either SNP array or WES. BIRCH was grown in RPMI with 10% FBS. The other cell lines (including PDX models adapted to cell culture) were grown in DMEM with 10% FBS. Trametinib was obtained from the NIH Developmental Therapeutics Program. Ganitumab was obtained through the Cancer Therapy Evaluation Program for nonclinical use. Vincristine was obtained from the NIH Division of Veterinary Resources (DVR) veterinary pharmacy. Cycloheximide (01810) was obtained from Sigma.
Genetic modification of FN RMS cells
PLKO.1 lentiviral shRNA particles targeting PTEN (TRCN0000002746) were obtained from Sigma. PLKO.1 control vector targeting GFP was obtained from Addgene (30323). Myc-tagged PTEN pLenti lentivirus particles were obtained from Origene (cat # RC202627L3V). The empty pLenti vector was also obtained from Origene. SMS-CTR or RD cells were transduced with viral particles in the presence of 8 μg /mL polybrene, incubated for 48 hours, and then split into selection with 2 μg/mL puromycin. Cells were cultured in the presence of puromycin for one week prior to confirming PTEN knock-down or over-expression by immunoblot. Puromycin was removed one week prior to treatment studies.
Cell growth assay
Time-course viability curves were generated by quantifying percent cell confluence from phase contrast images using an Incucyte SX5 (Essen Bioscience) as previously described (23). Cells were plated to achieve 20% confluence at the time of drug dosing. Each condition was assayed in quadruplicate (i.e., four observational units per experimental unit (24)). EC50 values were calculated using the log inhibitor versus response – variable slope non-linear curve fit function in GraphPad Prism (8.4.3).
Matrix combination assays
Assay-ready plates (Aurora E8; white, solid-bottom, 1,536-well) were prepared by acoustic-droplet spotting of 25 nL DMSO-solvated trametinib (Echo 550) and 1 μL culture media-solvated ganitumab (Echo 525) to each well. Cells were harvested with trypsin (Gibco; 25300054) and dispensed (Combi MultiDrop) into assay-ready plates to yield 500 cells/well in 5 μL assay volume. Plates were covered with breathable metal lids (Kalypsys) and incubated at 37 °C, 5% CO2 for 72 hours. To determine cell viability, 2.5 μL CellTiter-Glo reagent (Promega) was dispensed (BioRaptr) into each well. Luminescence was read on a ViewLux instrument (Perkin-Elmer). Values were normalized to DMSO control and synergy calculations were completed as reported previously (25). To deconvolute synergistic synergy versus potency, data were further analyzed by Multi-dimensional Synergy of Combinations (MuSyC) Analysis (26,27) using an online tool (https://musyc.lolab.xyz) and are presented as mean +/− 95% confidence interval.
Annexin V assay
RD or SMS-CTR cells were treated with vehicle (DMSO), 10 nM trametinib, 100 or 1000 nM ganitumab, or trametinib/ganitumab for 48 hours prior to analysis as described (23). Six biological replicates (experimental units) per condition are shown. The six treatment conditions comprise a 2 X 3 factorial structure. The two levels were trametinib (0, 10 nM) and ganitumab (0, 100, 1000 nM). Preliminary analysis indicated that the residuals were heterogeneous, so an arcsine square root transformation was performed on the data prior to formal analysis. A factorial ANOVA was conducted on the transformed annexin positivity data to estimate the trametinib and ganitumab interaction effect and simple effects (Supplemental Data File 2).
Capillary immunoassays
Cells in culture were treated as indicated, pelleted, and flash frozen. Cell lysates were prepared in MPER (Thermo). Fresh frozen tumor samples were prepared in TPER (Thermo) using a TissueRuptor. Lysates were analyzed as described (11). Primary antibodies are listed in Supplemental Table 2. For the cell culture experiments, the data were made up of three outcome variables (pERK, pAKT, IGF1R) from two cell lines (SMS-CTR, RD). For each cell line, there were seven treatments of which six comprised a 2 × 3 factorial treatment structure. Trametinib was administered at 0 nM for 48 hrs, or at 10 nM for either 6 hr or 48 hr. Ganitumab was administered at three doses (nM): 0, 100, 1000. Each of the seven treatments were comprised of four experimental units. Preliminary analysis indicated that the residuals were heterogeneous, so a Box-Cox transformation was performed on the data to determine what type of transformation would homogenize the residuals. After data transformation, and by cell line, each variable was subjected to a one-way ANOVA. Graphical examination of the residuals confirmed that they were both approximately normally distributed and homogeneous. Contrasts were constructed to test for several effects detailed in Supplemental Data File 3. In a post hoc fashion, additional pairwise comparisons were made, and are also detailed in Supplemental Data File 3. The two-tailed F-test p-values from these pairwise comparisons were adjusted using Holm’s method.
Immunoblot experiments
Antibodies used in this study are listed in Supplemental Table 2. Cells were treated as indicated and lysates were analyzed essentially as described (11). Membranes probed with phospho-specific antibodies were subsequently stripped with Restore Plus stripping buffer (Thermo) and re-probed with antibodies for the corresponding total protein.
Cell line xenograft experiments
Xenograft studies were approved by the Animal Care and Use Committee (ACUC) of the NCI-Bethesda or the Frederick National Laboratory for Cancer Research (FNLCR). FNLCR is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. SCID beige (CB17.Cg-PrkdcscidLystbg-J/Crl) mice were purchased from Charles River laboratories. All animals were female, and all were injected at 4–8 weeks of age. For the SMS-CTR and RD therapeutic enhancement studies, 2 million cells in a 1:1 mixture of reduced growth factor basement membrane extract (BME, BioTechne) and Hanks balanced salt solution (HBSS) were injected orthotopically into the gastrocnemius muscle in the left hind leg of 75 SCID beige mice. When tumors reached 100 to 200 mm3, 50 of the mice were randomized based on tumor volume and body weight using Studylog Animal Study Workflow Software into vehicle, trametinib, ganitumab, trametinib/ganitumab combination, and vincristine treatment groups with 10 mice per group. No formal power calculation was performed prior to the start of this exploratory study, but the sample size of 10 mice per group is the standard used in the animal facility. Please see Supplemental Data File 4 for sample size tables and Supplemental Data File 5 for summary statistics of baseline tumor volume and body weight for all mouse experiments.
Trametinib suspensions were prepared in vehicle (0.5% hydroxypropylmethylcellulose/0.2% Tween 80/5% sucrose) weekly and stored at 4 °C until use. Trametinib was dosed in the indicated groups in the morning 5 days a week at a dose of 3 mg/kg by oral gavage (OG) (28); the vehicle group received the same volume of vehicle by OG. Ganitumab was resuspended in normal saline at a concentration of 3 mg/mL and was administered to indicated groups at a flat dose of 300 μg (100 μL) by intraperitoneal injection (IP) in the afternoon twice a week (29). The vincristine group received 1 mg/kg weekly, diluted in PBS immediately prior to injection in the tail vein (30).
Assigned treatments were administered for a period of 28 (SMS-CTR) or 35 (RD) days, after which the mice were observed for tumor development and body weight changes. In the SMS-CTR group, on day 71 after initial randomization, mice that had received the trametinib/ganitumab combination were re-randomized to receive either tumor monitoring (n = 4; because one mouse was euthanized prior to day 71 due to a non-treatment related injury) or a second 28-day cycle of trametinib/ganitumab (n = 5, average tumor volume roughly equivalent per group). The remaining 25 mice per cell line were randomized and treatment began when tumors reached between 540 and 1000 mm3 (SMS-CTR) or between 250 and 900 mm3 (RD). Mice were treated as above. Two mice per group were euthanized 4 hours after the fifth dose of vehicle or trametinib for pharmacodynamic assessment and the remaining mice were observed for tumor response (n = 3 per group).
For studies to determine the long-term efficacy of trametinib/ganitumab, 2 million SMS-CTR or BIRCH cells were injected orthotopically into SCID beige mice as described above (20–24 mice per cell line). When tumors reached about 200 mm3 (see Supplemental Data File 5), animals were randomized to receive either vehicle or trametinib/ganitumab. Animals were treated for a period of 70 days and then observed for tumor progression or recurrence and tolerability (assessed by body weight). Two mice per group were euthanized 4 hours after the fifth dose of trametinib for pharmacodynamic studies, leaving 8–10 mice per group for long-term follow up.
In all xenograft experiments, the tumor dimensions were measured twice a week (every 3–4 days) with digital calipers to obtain two diameters of the tumor sphere, from which the tumor volume was determined using the equation ((L x W2) / 2 (where L = the maximum diameter and W = the minimum diameter) (31). The whole hindlimb was included in the measurement. Animals were euthanized when they reached tumor endpoint, defined as when the leg measured greater than 18 mm in any direction. Animals were also euthanized if the tumors showed signs of ulceration or caused significant discomfort to the animal, in accordance with the humane endpoints recommended by the Institutional Animal Care and Use Committee (IACUC).
Patient-derived xenograft experiments
PDX were provided from the St. Jude Childhood Solid Tumor Network as a single cell suspension at a concentration of 6 million cells/mL in 90% FBS/10% DMSO (22). PDX were propagated essentially as described (22). PDX passage #3 was used for drug treatment experiments. In these experiments, NSG mice were injected orthotopically into the gastrocnemius muscle in the left hind leg with SJRHB013758, SJRHB000026, or SJRHB010927 cells as single cell suspensions in 1:1 mixture of HBSS and BME at a concentration of 1 million cells per 100 μL. When tumors reached about 200 mm3 (see Supplemental Data File 5), mice were randomized to receive either vehicle or trametinib/ganitumab (n = 10–12 mice per group). Two mice per group per PDX were harvested 4 hours after the fifth dose of trametinib or vehicle for pharmacodynamic analyses. The remaining mice were treated for 70 days and then monitored for tumor progression or recurrence (n = 8–10 per group).
Trametinib pharmacokinetics assay
For studies evaluating the pharmacokinetics of trametinib in the presence or absence of ganitumab, 2 million SMS-CTR cells were injected orthotopically into SCID beige mice as described above. When tumors reached 114–167 mm3, the mice were randomized to receive either trametinib alone or trametinib/ganitumab (n = 6 per group). Mice received trametinib at a dose of 3 mg/kg by oral gavage daily for 6 continuous days. Ganitumab (3 mg/kg) was given IP twice a week for a total of three doses. Whole blood was collected in K2EDTA tubes (Thermo) via terminal bleed (cardiac puncture) immediately before the seventh dose of trametinib would have been administered (“Pre” group: three mice per group, six doses of trametinib given) and 4 hours after the seventh dose of trametinib was administered (“Post” group: three mice per group, seven doses of trametinib given).
Plasma was prepared by centrifugation at 3000 rpm for 10 minutes. Plasma samples were measured via LC-MS/MS run alongside an eight-point standard curve ranging from 0.25–500 ng/mL (duplicate observations units). Sufficient accuracy and precision (< 20%) as well as good correlation to expected values (r ≥ 0.99) were met. The plasma samples were extracted using a 96-well solid-phase procedure (Evolute Express ABN, 30mg, Biotage Corp) with the aid of deuterated olaparib ([2H]8-olaparib) as an internal standard. Trametinib and the internal standard were eluted off the plate using acetonitrile, which was evaporated under a stream of nitrogen gas, and reconstituted with a water/acetonitrile mixture. The extracted samples were chromatographically separated on a Waters XSelect HSS pentafluorophenyl column (2.1 × 50mm, 2.5um) followed by tandem mass spectrometric detection using a Waters Quattro Micro triple quadrupole mass spectrometer.
Tolerability studies
For studies evaluating the tolerability of trametinib/ganitumab, 2 million SMS-CTR cells were injected orthotopically into SCID beige mice. When tumors reached 100–215 mm3, the mice were randomized into four treatment groups (vehicle, trametinib, ganitumab and trametinib/ganitumab) with 15 mice per group. Whole blood was collected in K2EDTA tubes by terminal bleed (cardiac punction) in three mice per group prior to initiation of treatment, and then in three mice per group weekly. Twenty microliters of whole blood were used for automated complete blood count determination (Hema vet 950). Plasma was prepared from the remaining blood samples by centrifugation at 3000 rpm for 10 minutes. Plasma insulin concentration in the fed state was assayed using the mouse ultrasensitive insulin ELISA kit (Alpco, 80-INSMSU-E01) according to the manufacturer’s instructions. The assay plate was read on a Spectromax M5 (Molecular Devices). Normal serum insulin levels were defined as per (32). We assayed plasma growth hormone levels using an ELISA kit (Alpco, 22-GHOMS-E01) in a similar manner.
Electrochemilluminescence assay
Lysates from RMS cells and PDX adapted to cell culture were prepared according to the manufacturer’s instructions. Protein concentration was determined by BCA assay and lysates were diluted to a final total protein concentration of 2 mg/mL. Twenty-five μL of these lysates were used in the insulin signaling panel (total protein kit, Meso Scale Discovery, K15152C-1 and phospho protein kit, Meso Scale Discovery, K15151C-1). Lysates were assayed in duplicate (two observational units per experimental unit). The assay was read on a Meso Sector S600. Lysates from MCF-7 cells (Meso Scale Discovery, Insulin Signaling Panel Whole Cell Lysate Set, C1151–1), with or without growth factor stimulation, were used as a positive control.
Data Availability Statement
The data generated in this study are available within the manuscript and its supplemental data files. Gene expression data analyzed in this study were obtained from GEO (GSE108022 and GSE66533).
Statistical Analyses
Please see the supplemental material for the statistical analysis details.
Results
Drugs that specifically inhibit the MEK1/2 and the IGF2/IGF1R pathway synergistically inhibit proliferation of RAS-mutated rhabdomyosarcoma cells
The combination of the MEK inhibitor trametinib and the IGF1R inhibitor BMS-754807 was effective in murine models of RAS-mutated RMS but the potential clinical translation of trametinib/BMS-754807 was hindered by intolerability (11). We hypothesized that this intolerability is likely due to the poly-pharmacology of BMS-754807, which potently inhibits the activity of several receptor tyrosine kinases (insulin receptor, TrkB, Met) and nonreceptor kinases (Aurora A, Aurora B, Jak2) in cell free-assays (19). To determine if MEK inhibition would synergize with more specific inhibitors of IGF1R in RAS-mutant RMS cells, we performed a 10X10 matrix combination assay in which viability of SMS-CTR (HRAS Q61K) or RD (NRAS Q61H) cells were determined in the presence of varying concentrations of both the MEK inhibitor trametinib and the IGF1R monoclonal antibody ganitumab (Figure 1A). Using the Bliss Independence model, synergy was observed between trametinib and ganitumab across multiple concentrations of each drug in both cell lines tested. Since Bliss analysis conflates synergistic potency and efficacy into a single score, we further assessed these parameters independently by Multi-dimensional Synergy of Combinations (MuSyC) Analysis (26,27). We observed a >10-fold increase in the potency of ganitumab (EC50 shift) in the presence of trametinib in both cell lines (SMS-CTR: 78-fold [95% confidence interval 16–147], RD: 12-fold [2.4–77]). We also observed a 6.2-fold [5.1–7.3] increase in the potency of trametinib in the presence of ganitumab in SMS-CTR cells, while the potency of trametinib in RD was unaffected by the presence of ganitumab (1.0-fold [0.35–2.5]). The efficacy of the combination approached 100% (0% cell viability) in SMS-CTR cells at high doses of trametinib, thus combination therapy had little effect on the overall efficacy. In contrast, in RD cells, the trametinib/ganitumab combination showed a 42 [37–55] % increase in efficacy over either drug alone. These results suggest that trametinib and ganitumab are synergistically potent in RD and SMS-CTR, and synergistically efficacious in RD.
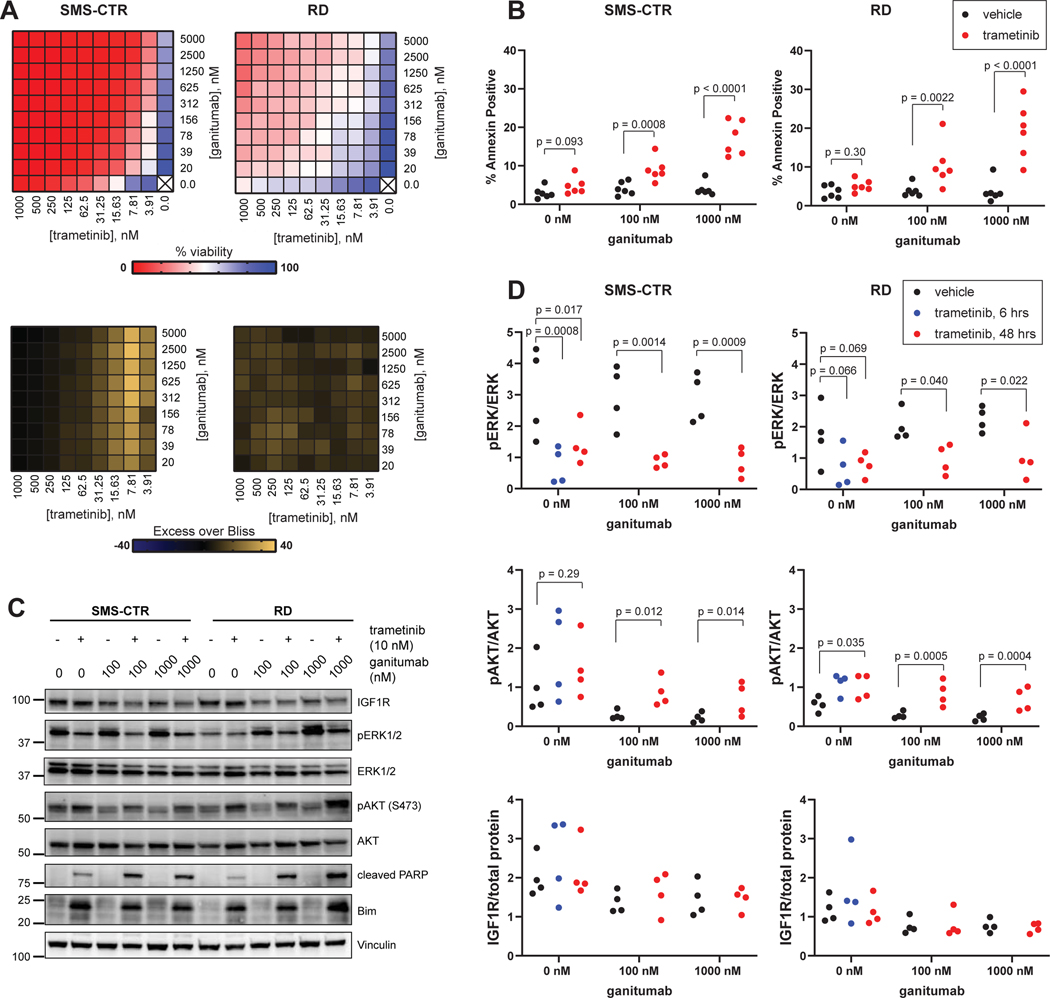
Figure 1: Trametinib and ganitumab synergistically inhibit RAS-mutant RMS viability.
(A) Matrix (10X10) plot for the combination of trametinib (0 to 1000 nM) and ganitumab (0 to 5000 nM) in both viability (CellTiter-Glo, top) and Excess over Bliss (bottom) format for SMS-CTR (left) and RD (right). (B) SMS-CTR (left) or RD (right) cells were treated with vehicle, 10 nM trametinib, 100 nM ganitumab, 1000 nM ganitumab, or trametinib/ganitumab in a factorial treatment structure for 48 hours, after which cells were stained with Annexin V-APC and Sytox Green then analyzed by flow cytometry. Percent Annexin positive cells were defined as the percentage of cells that were Annexin V positive and Sytox positive or negative. Individual biological replicates (experimental units) are shown. X-axis is not to scale. Exact p-values for pairwise comparisons are shown. ANOVA was performed on transformed data. (C) SMS-CTR (left) or RD (right) cells were treated with vehicle, 10 nM trametinib, 100 nM or 1000 nM ganitumab, or trametinib/ganitumab for 48 hours, after which cells were harvested and the resulting cells analyzed for IGF1R protein levels; phosphorylated and total forms of ERK and AKT; as well as the pro-apoptotic proteins cleaved PARP and Bim. (D) SMS-CTR (left) or RD (right) cells were treated with vehicle or 10 nM trametinib for 6 or 48 hours prior to treatment with either vehicle, 100 nM, or 1000 nM ganitumab for the final 6 hours (factorial treatment structure plus the 6 hour trametinib treatment condition). The treated cells were harvested and analyzed by capillary immunoassay to quantitate the ratio of phosphorylated ERK to total ERK (top), phosphorylated AKT to total AKT (middle), and total IGF1R to total protein (bottom). Individual biological replicates (experimental units) are shown. X-axis is not to scale. Exact p-values for pairwise comparisons are shown. ANOVA was performed on transformed data.
We also assessed the impact of the combination of trametinib and ganitumab on apoptosis in FN-RMS using both flow cytometric assessment of annexin positivity (Figure 1B) and immunoblotting for apoptotic markers (Figure 1C). A one-way factorial ANOVA was performed to analyze the effect of trametinib and ganitumab on annexin positivity (Supplemental Data File 2). Evidence strongly suggests that there was an interaction between the effects of trametinib and ganitumab in both SMS-CTR (numerator/denominator degrees of freedom [df] = 2/30; F-test p-value = 0.0002) and RD (df = 2/30; p = 0.0010). Further examination of the simple effects strongly indicates that ganitumab alone did not affect the percent of annexin positive cells in SMS-CTR (df = 2/30; p = 0.47) or RD (df = 2/30; p = 0.95). Conversely, simple effects analysis strongly suggests that trametinib with or without ganitumab increased the percent of annexin positive cells in SMS-CTR (df = 2/30; p < 0.0001) and RD (df = 2/30; p < 0.0001). Pairwise analysis (comparing trametinib 0 nM with trametinib 10 nM at the different ganitumab concentrations) strongly suggests that the percent of annexin positive cells is increased in the presence of trametinib in SMS-CTR and RD cells that were co-treated with either 100 nM or 1000 nM ganitumab, but not in the absence of ganitumab (Figure 1B). Increased PARP cleavage was also observed in SMS-CTR and RD cells treated with the trametinib/ganitumab combination as compared to cells treated with either single agent. Expression of the pro-apoptotic protein Bim, which mediates the apoptotic response to MEK inhibition (33), was increased in both cell lines treated with trametinib in the presence or absence of ganitumab (Figure 1C). These results suggest that trametinib/ganitumab induces apoptosis in RAS-mutated RMS cells in part through the upregulation of Bim.
We used quantitative capillary immunoassays to assess the mechanism of synergy between trametinib and ganitumab in FN-RMS. Our evidence suggests that the interaction effect between trametinib and ganitumab on ERK and AKT phosphorylation or total IGF1R expression was weak (Supplemental Data File 3). However, similar to what was observed with 100 nM trametinib (11), treatment for 6 or 48 hours with a clinically achievable concentration of trametinib (10 nM) decreased ERK phosphorylation in SMS-CTR to a greater extent than RD. Ganitumab alone did not affect ERK phosphorylation in either cell line, however, decreased ERK phosphorylation was observed when ganitumab (100 nM or 1000 nM) was added for the final 6 hours of trametinib treatment, an effect that is more pronounced in SMS-CTR (Figure 1D, top panel). Trametinib treatment alone increased AKT phosphorylation in RD cells to a greater extent than SMS-CTR cells, while ganitumab alone decreased AKT phosphorylation in both cell lines. The induction of AKT phosphorylation due to trametinib treatment appeared to be attenuated in SMS-CTR but not RD cells treated with both trametinib and ganitumab (Figure 1D, middle panel). Ganitumab alone and in combination with trametinib decreased expression of IGF1R in RD cells to a greater extent than SMS-CTR cells (Figure 1D, bottom panel), consistent with the antibody-mediated receptor internalization and degradation observed in other cellular contexts (18). Phosphorylated IGF1R was not reliably quantifiable in this assay due to the decrease in total IGF1R. These data suggest that the synergy observed between ganitumab and trametinib in RAS-mutated RMS cells is due in part to ganitumab attenuating trametinib-induced AKT phosphorylation.
Combined trametinib and ganitumab treatment is efficacious in murine orthotopic xenograft models of RAS-mutated rhabdomyosarcoma
We next sought to test the efficacy of trametinib/ganitumab in orthotopic RAS-mutated RMS xenograft models. SCID beige mice were implanted with SMS-CTR or RD xenografts. When tumors were palpable the animals were randomized to receive one of five treatments: vehicle, trametinib, ganitumab, trametinib/ganitumab, or vincristine, a chemotherapy agent that is part of the standard-of-care regimen for upfront and relapsed FN RMS.
Mice bearing SMS-CTR xenografts were treated for a 28-day cycle, at which point treatment was stopped and the animals were observed for tumor regrowth. We observed tumor shrinkage in the single agent trametinib, ganitumab, and vincristine arms, but tumors regrew in these conditions immediately upon cessation of treatment. The tumor shrinkage in the ganitumab arm was notable given the modest efficacy of this agent in vitro, suggesting ganitumab may play a role in modulating the tumor microenvironment in addition to inhibiting the growth of the tumor cells themselves. Combined trametinib and ganitumab treatment led to a 40-day off-treatment period during which no tumors recurred. After tumors were observed to regrow in these animals (between days 70 and 74), the mice in the initial trametinib/ganitumab group were re-randomized to either observation or to a second 28-day cycle of trametinib/ganitumab. The tumors again fully regressed with trametinib/ganitumab treatment; however, the period of remission was shorter after cessation of cycle 2 than after cycle 1 (Figure 2A, left and Supplemental Figure 1A, top, as well as Supplemental Data File 6).
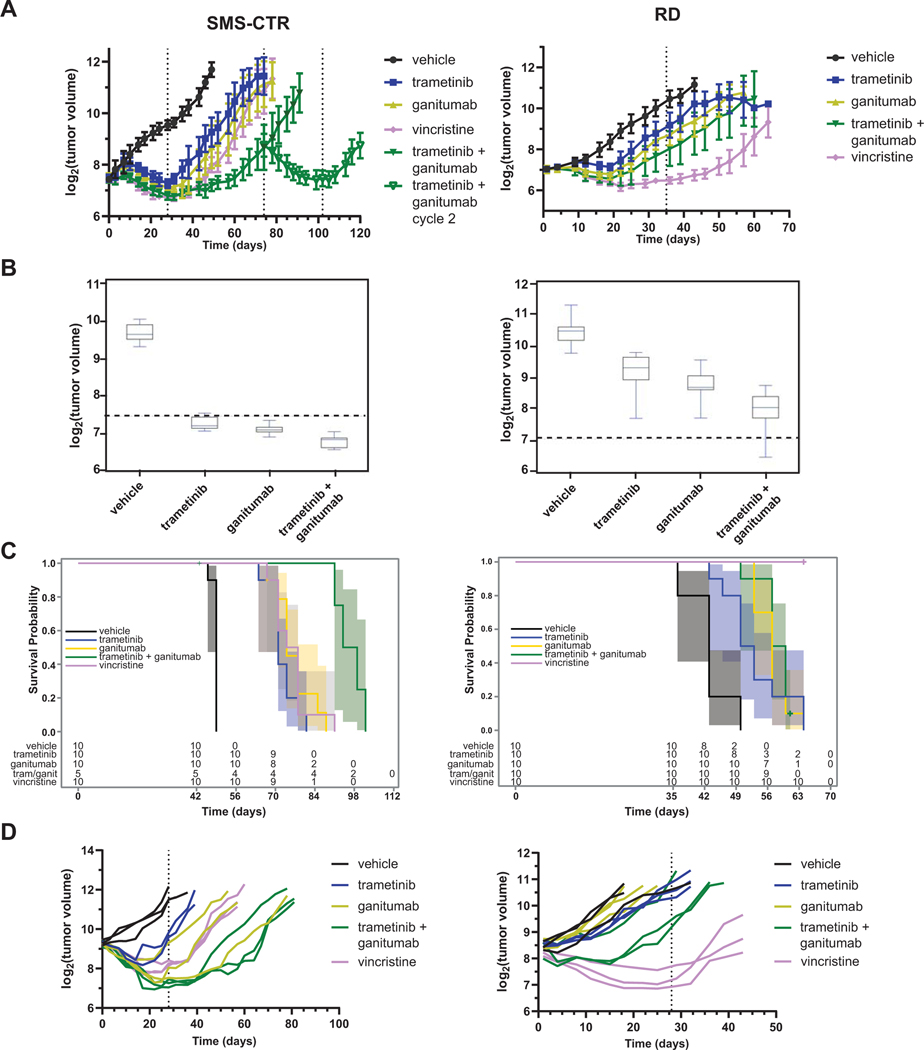
Figure 2: Combining ganitumab with trametinib inhibits tumor growth and prolongs survival in SMS-CTR but not RD xenografts.
(A) SCID beige mice bearing 100–200 mm3 SMS-CTR (left) or RD (right) xenografts were randomized to receive the indicated treatments (10 mice per group). SMS-CTR xenograft-bearing mice received treatment for 28 days and were then observed for tumor regrowth. When tumors became palpable in the mice previously treated with the combination of trametinib and ganitumab, these mice were re-randomized to receive either vehicle or trametinib/ganitumab (cycle 2). RD xenograft-bearing mice received treatment for 35 days. Mean tumor volume ± one standard deviation is plotted on a log2 scale. Means are displayed only if there are three or more tumor volume measurements at that time point. Dashed lines indicate the beginning or end of a treatment cycle. Line graphs for individual mice are shown in Supplemental Figure 1A. (B) Box plots of tumor volumes at the end of the treatment period for SMS-CTR (left, 28 days) and RD (right, 35 days) xenografts. The boxes represent quartiles, and the whiskers represent 1.5 X IQR. Dashed lines indicate the tumor volume at the start of treatment. See Supplemental Data Files 4 (SMS-CTR) and 5 (RD) for statistical analyses of these data. (C) Overall survival of mice bearing SMS-CTR (left, excluding mice treated with a second cycle of trametinib/ganitumab) or RD (right) xenografts given the indicated treatments. The 95% confidence intervals are represented by shading. The number of mice at risk for a given time point are indicated below the Kaplan-Meier plots. (D) Line graphs of tumor volume as a function of treatment duration for mice bearing SMS-CTR (left) or RD (right) xenografts receiving the indicated treatment. In this experiment, tumors were allowed to grow to a volume of 540–1100 mm3 (SMS-CTR) or 250–900 mm3 (RD) prior to the randomization of mice into treatment arms (n = 3 per group). Each line represents data from a single mouse. One mouse with an SMS-CTR xenograft that received trametinib treatment (Animal 140) was determined to have an outlier response and is omitted from this graph. See Supplemental Data Files 2 and 3 for sample sizes over time and baseline tumor volume/body weight summary statistics, respectively.
In RD xenograft models, vehicle-treated mice did not reach endpoint at 28 days due to slower tumor growth, therefore, the mice were treated for a period of 35 days. After initial tumor regression, on-treatment tumor regrowth was observed in each of the treatment arms except the vincristine arm. Tumors regrew in the vincristine arm immediately after the cessation of treatment (Figure 2A, right and Supplemental Figure 1A, bottom, as well as Supplemental Data File 7). These data suggest that the trametinib/ganitumab combination is more efficacious in SMS-CTR than RD xenografts, a conclusion that is consistent with the results of the trametinib/BMS-754807 study (11) and the in vitro trametinib/ganitumab experiments (Figure 1). We had previously observed greater mouse body weight loss from day 7 to day 21 of treatment with trametinib/BMS-754807 than with single agents (11). In the current study, we did not observe decreased body weight during treatment. We observed decreased body weight after treatment cessation in the trametinib, ganitumab, and vincristine groups in the mice bearing SMS-CTR xenografts but not RD xenografts (Supplemental Figure 1B), presumably due to difficulty accessing food with a large hindlimb tumor upon tumor growth. This weight loss post-treatment introduced a degree of nonlinearity in the data such that the rate of body weight change in the different treatment groups could not be directly compared using linear regression. However, the data suggest that the trametinib/ganitumab combination has little effect on weight in tumor-bearing SCID beige mice.
To further characterize the interaction between trametinib and ganitumab in vivo, we compared the mean tumor volumes at the end of the first treatment cycle in SMS-CTR and RD xenografts (Figure 2B). In both SMS-CTR and RD, the effect of trametinib on tumor volume was more pronounced in the absence of ganitumab, and the effect of ganitumab on tumor volume was more pronounced in the absence of trametinib (Supplemental Data Files 6 and 7, LS-means). Bliss analysis of tumor volumes at the end of the first treatment cycle reveals additivity between the treatments (Supplemental Data Files 6 and 7, Bliss), however; factorial ANOVA of the data suggests that the interaction effect may be antagonistic at the current dosing schedule. Despite the lack of evidence for synergy between trametinib and ganitumab in vivo, evidence does suggest tumor growth was inhibited best by the combination of agents.
In the SMS-CTR xenograft model, our data suggest that mice treated with one cycle of trametinib/ganitumab had a survival advantage compared to mice treated with vehicle (log-rank unadjusted p < 0.001), trametinib (p = 0.003), ganitumab (p = 0.062), or vincristine (p = 0.054) (Figure 2C, left, and Supplemental Data File 6). The effect of trametinib on the median survival was similar in the presence or absence of ganitumab, and the effect of ganitumab on the median survival was similar in the presence or absence of trametinib. Interestingly, however, one cycle of trametinib/ganitumab was associated with a more pronounced survival advantage than what would be expected due to effects of trametinib and ganitumab as single agents (p = 0.0003, Supplemental Data File 6). In contrast, for the RD xenograft model, our data suggest that only vincristine treatment conferred a survival advantage (Figure 2C, right, and Supplemental Data File 7). Overall, these results are again consistent with the trametinib/ganitumab combination being more effective in SMS-CTR than RD models.
To better characterize the response to trametinib/ganitumab in the SMS-CTR and RD models, we delayed randomization and treatment initiation until larger tumors had developed (n = 5 mice per treatment group). In this experiment, two mice per group were euthanized after 5 days of treatment for histopathologic and pharmacodynamic assessment. In mice bearing large SMS-CTR tumors, we observed complete tumor regression with trametinib, ganitumab, trametinib/ganitumab, and vincristine treatment (Figure 2D, left). Tumors regrew in each of the single agent treatment arms when the treatment was stopped; however, mice in the trametinib/ganitumab group experienced a 20-day remission off treatment. In mice bearing large RD tumors, trametinib and ganitumab had modest effects, and only vincristine induced tumor regression (Figure 2D, right). Increased fibrosis was observed in tumors treated with the combination of trametinib and ganitumab in SMS-CTR (Supplemental Figure 1C, H&E, left). In addition, decreased total IGF1R and decreased phosphorylated ERK were observed, indicating target engagement for both ganitumab and trametinib with this dosing schedule (Supplemental Figure 1C and 1D, left). Total IGF1R was also decreased in RD tumors treated with trametinib and ganitumab. However, ERK phosphorylation was largely unaffected by treatment with trametinib and ganitumab in the RD xenografts, suggesting the dose of trametinib used was insufficient to lead to a decrease in intratumoral ERK phosphorylation in this model (Supplemental Figure 1C and 1D, right).
Preclinical tolerability of the trametinib-ganitumab combination
Because ganitumab binds to murine IGF1R with high affinity (34), we were able to assess the impact of ganitumab on the metabolism and tolerability of trametinib in our RAS-mutant RMS xenograft models. To assess if ganitumab affects the plasma exposure of trametinib, we treated SMS-CTR tumor-bearing SCID beige mice with either trametinib alone or trametinib in combination with ganitumab for 7 days continuously. Trough (immediately before dose would be due) and peak (4 hours after dose delivered) plasma trametinib levels at steady-state (day 7) were determined by LC-MS/MS. Our data suggest that ganitumab did not affect trough or peak levels of trametinib (Figure 3A). Importantly, the 3 mg/kg dose of trametinib resulted in plasma trametinib levels close to the human Cmax (Novartis, trametinib package insert), indicating that clinically relevant dosing was achieved in these models.
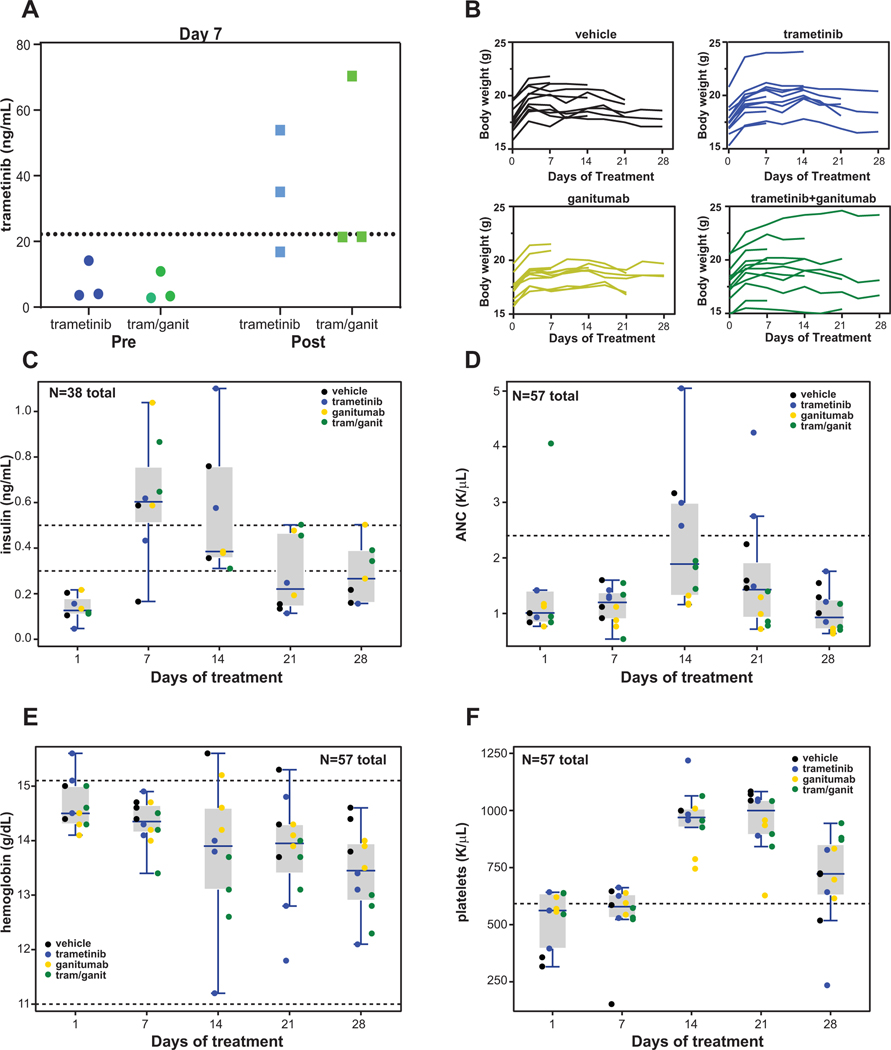
Figure 3: The combination of trametinib and ganitumab is well tolerated in SCID beige mice.
(A) SCID beige mice bearing SMS-CTR xenografts in the left hindlimb were treated with trametinib alone or trametinib and ganitumab (combination) daily for a period of 7 days. Plasma was obtained from mice (n = 3 per group) prior to (trough) or 4 hours after (peak) the final (right) dose of trametinib and the trametinib concentration was determined by LC-MS/MS. Individual data points from biological replicates (3 experimental units per condition) are displayed. The Cmax of trametinib in adults given a single dose of 2 mg (22.2 ng/mL) (Novartis, trametinib package insert) is indicated with a dashed line. (B) Line graphs of body weight as a function of treatment duration for four treatment groups of mice used in the tolerability study; each line represents data from a single mouse. Days (n per group): 0 (15); 3, 7 (12); 10, 14 (9); 17, 21 (6); 24, 28 (3). Please see Supplemental Data File 2 for further sample size information. (C) Box plots of serum insulin determined by ELISA (Ultra Mouse Insulin ELISA, ALPCO) from SCID beige mice bearing SMS-CTR xenografts and treated as indicated. The normal range for serum insulin for BALB/c and C57BL/6 mice (0.3–0.5 ng/mL) is indicated by dashed lines (32). (D) Box plots of absolute neutrophil count (ANC) determined from SCID beige mice bearing SMS-CTR xenografts in the indicated treatment groups. The upper limit of normal for ANC (2.4 K/μL) for mice is indicated by a dashed line. (E) Box plots of hemoglobin from SCID beige mice bearing SMS-CTR xenografts treated as indicated as a function of duration of treatment. The normal range for hemoglobin for mice (11.0–15.1 g/dL) is indicated by dashed lines. (F) Box plots of platelet count determined from SCID beige mice bearing SMS-CTR xenografts treated as indicated as a function of treatment duration. The lower limit of normal for platelets (592 K/μL) for mice is indicated by a dashed line. In panels C through F, the boxes represent quartiles, and the whiskers indicate the maximum value within 1.5 X IQR; also, x-axis values are not to scale. Values from individual animals are shown and are colored according to the treatment group. In panels C through F, day 1 values are pre-treatment.
Clinically, both trametinib (Novartis, trametinib package insert) and ganitumab (20,21) have hematologic and metabolic side effects. To assess these effects when the drugs were given in combination, we monitored body weight, analyzed complete blood counts (CBC; 20 variables), and measured serum insulin levels in SCID beige mice bearing SMS-CTR tumors being treated with either vehicle, trametinib, ganitumab, or trametinib/ganitumab for a full 28-day cycle. In this tolerability study, we observed the expected tumor growth inhibition in all three treatment groups (Supplemental Figure 2A). Also as expected, we did not observe body weight loss on treatment during this tolerability study (Figure 3B); however, the sample sizes are small after day 14, so these results should be interpreted with caution.
The small sample sizes in this study (n = 3 per group per time point) precluded formal statistical analysis of how the different treatments interacted with the CBC parameters and insulin levels over time. However, three subset analyses are reported here (Supplementary Information, Statistical Analysis Details): 1) the factorial interaction effect (trametinib x ganitumab) pooled over days 7 to 21; 2) the treatment effect pooled over days 7 to 21; and 3) the time effect (days 1 to 28) pooled over treatment. For the third analysis, if the time effect p-value was < 0.0025 then pair-wise comparisons were made between day 1 (untreated controls) and the other days (Supplemental Data File 8).
Prior to initiation of treatment (Day 1), serum insulin levels were below the normal range, then increased to above the normal range on days 7 and 14, and mostly returned to below the normal range on days 21 and 28 (Figure 3C and Supplemental Figure 2B). Quantile regression results suggested that measured insulin levels were affected over time in this study (p = 0.0003, Supplemental Data File 8); however, our data suggest that the factorial interaction effect between trametinib and ganitumab on serum insulin was weak (p = 0.55). The data indicate a potential risk for transient hyperinsulinemia early in the treatment course with trametinib and ganitumab; however, since 37% (22/60) of the insulin data are missing due to sample hemolysis, the results for insulin should be viewed cautiously. Of note, we attempted to measure serum growth hormone (GH) levels from the samples obtained in this study because mouse GH is known to be affected by ganitumab treatment (34); however, GH levels were below the limit of detection in all samples.
The absolute neutrophil count (ANC) was within normal limits prior to treatment initiation and on day 7 of treatment, increased somewhat to above the normal range on day 14, and then decreased to within the normal range on day 21 and 28 (Figure 3D and Supplemental Figure 2C). Importantly, no neutropenia was observed in the study. Quantile regression results indicated both a time effect (p < 0.0001, days 1–28) and treatment effect (p = 0.0003, days 7–21) on ANC. There was evidence for a treatment effect for the related CBC variables, white blood count, percentage of neutrophils, and percentage of lymphocytes, as well (p < 0.0025, quantile regression, Supplemental Data File 8). However, our data suggest that the factorial interaction effect between trametinib and ganitumab was weak for ANC (p = 0.83). These effects on ANC in SCID beige mice are consistent with prior studies in athymic nude mice (34).
Hemoglobin levels did not decrease below the lower limit of normal (11.0 g/dL) during the tolerability study, but our data suggest that hemoglobin levels did decrease over the treatment course (p = 0.0003, Figure 3E and Supplemental Figure 2D). Moreover, evidence suggests that the factorial interaction between the effects of trametinib and ganitumab on hemoglobin was weak (p = 0.89, quantile regression). Similarly, the mean corpuscular volume (MCV) decreased steadily over the treatment course (p < 0.0001, quantile regression, Supplemental Figure 2E), but remained within the normal range for the course of the study. These data suggest that careful monitoring for microcytic anemia should be performed for longer treatment courses of either trametinib, ganitumab, or the combination.
The platelet count was at the lower limit of normal prior to treatment initiation, increased to within the normal range on day 14 and day 21, and decreased to near the lower limit of normal on day 28 (Figure 3F and Supplemental Figure 2F). The platelet count did not exceed the upper limit of normal over the course of the study (2972 K/μL), and our data suggest that the factorial interaction between the effects of trametinib and ganitumab on the platelet count was weak (p = 0.61, quantile regression). None of the remaining CBC variables demonstrated a strong time effect (p ≥ 0.0025, quantile regression). In summary, the CBC and serum insulin data suggest that the trametinib/ganitumab combination did not cause clinically significant hematological effects or sustained hyperinsulinemia in tumor-bearing SCID beige mice in this tolerability study (28 days).
Determinants of intrinsic resistance to trametinib/ganitumab in orthotopic RAS-mutant rhabdomyosarcoma models
The trametinib/ganitumab combination showed therapeutic efficacy in SMS-CTR but not RD (Figure 2) xenograft models. Previous studies have demonstrated a low degree of association between in vitro induction of apoptosis and in vivo response to ganitumab as a single agent (35). Therefore, to further assess the efficacy of trametinib/ganitumab, we employed multiple additional mouse models of RAS-mutated FN-RMS, including one additional cell line xenograft model, BIRCH, and three patient-derived xenograft (PDX) models (Supplemental Data File 9). In these experiments, we wanted to determine the efficacy of long-term exposures to trametinib/ganitumab, so mice were treated until tumors began to grow on treatment or for at least 45 days. To that end, the SMS-CTR experiment was repeated, and instead of treating mice in 28-day cycles, mice were treated for 70 days. Mice with SMS-CTR tumors had tumor regression to a size undetectable with digital calipers in the presence of trametinib/ganitumab and tumors did not regrow during the prolonged treatment period; in fact, the tumors did not recur until 40 days after the end of treatment (Figure 4A, top left). In the BIRCH cell line xenograft model, which like SMS-CTR harbors an HRAS Q61K mutation, mice treated with trametinib/ganitumab had initial tumor regression, but the tumors did begin to regrow at the end of the 70-day treatment period (Figure 4A, top right). The response of RD cell line xenografts, in which mice had tumor regression that was short-lived, is also presented in Figure 4A for comparison (top center). Our data suggest that long-term treatment with trametinib and ganitumab increased progression-free survival in each of the cell line xenograft models tested (Figure 4B, top).
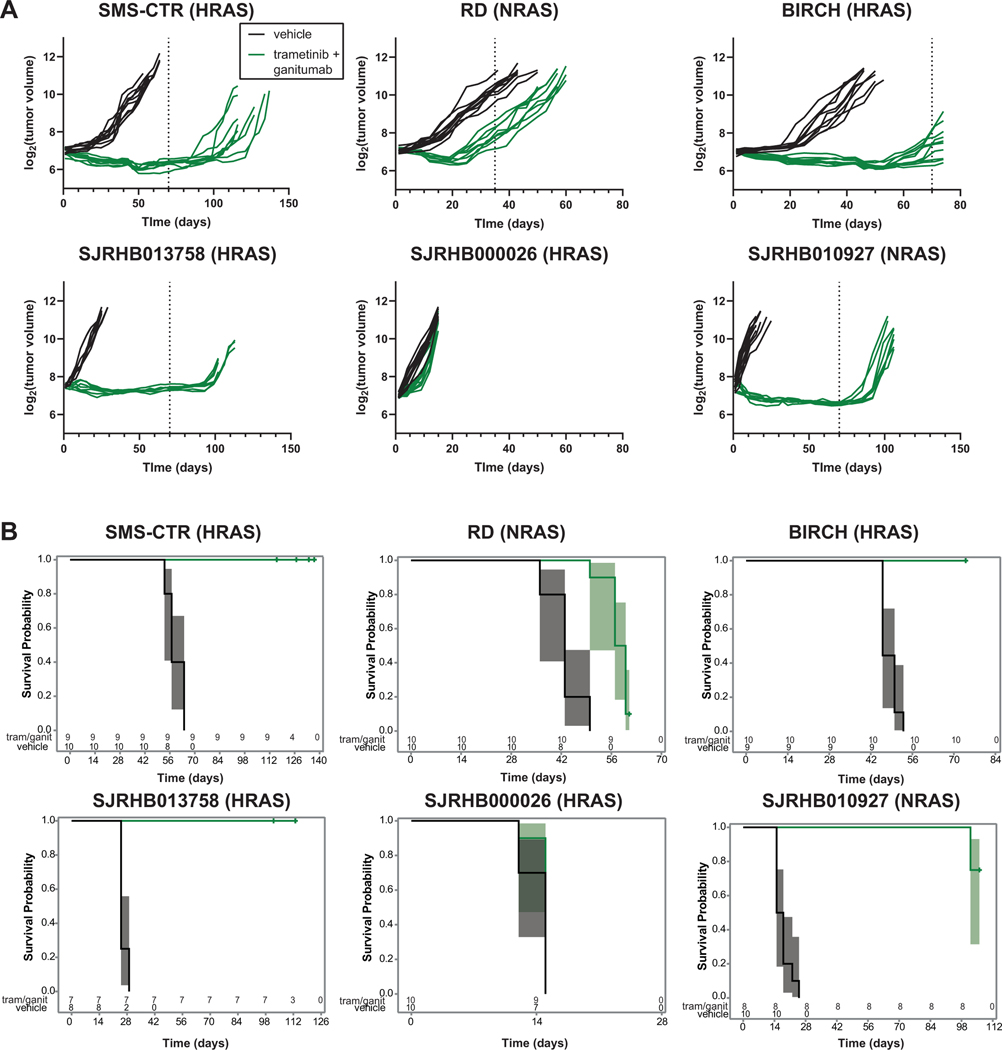
Figure 4: Prolonged treatment with trametinib/ganitumab inhibits tumor growth and prolongs survival in cell line and patient-derived xenograft models of RAS-mutant RMS.
(A) Line graphs of tumor volume as a function of treatment duration (note different time scales for each model) for mice bearing cell line xenografts (top; SMS-CTR, RD, BIRCH) or patient-derived xenografts (bottom; SJRHB013758, SJRHB000026, SJRHB010927) receiving vehicle or trametinib/ganitumab. Each line represents data from a single mouse (n = 8–10 per group). The RD data are reproduced from Figure 2 for comparison. Dashed lines indicate the end of a treatment cycle. Data are plotted on a log2 scale. Tumor volumes from animals that were euthanized prematurely (and censored from survival analysis) or were determined to be outliers are not displayed but the raw data are available in Supplemental Data File 7. (B) Overall survival of mice bearing cell line xenografts (top), or patient-derived xenografts (bottom) treated with either vehicle or trametinib/ganitumab. The 95% confidence intervals are represented by shading. The number of mice at risk for a given time point are indicated below the Kaplan-Meier plots.
In the PDX models, trametinib/ganitumab induced tumor regression in both SJRHB013758 (HRAS G13R, Figure 4A, bottom left) and SJRHB010927 (NRAS G13D, Figure 4A, bottom right). The SJRHB000026 model (HRAS G13R and PIK3CA H1047R), however, was resistant to trametinib/ganitumab (Figure 4A, bottom center). Evidence strongly suggests that long term treatment with trametinib and ganitumab increased progression-free survival in SJRHB013758 and SJRHB010927 but not SJRHB000026 (Figure 4B, bottom).
Pharmacodynamic markers of response to trametinib/ganitumab, pERK, pAKT and total IGF1R, were assessed in each of these models using quantitative capillary immunoassays. ERK phosphorylation decreased as a result of trametinib/ganitumab treatment in each of the cell line xenograft models. In the PDX models, a decrease in ERK phosphorylation was observed only in lysates of SJRHB000026 tumors (Supplemental Figure 3A, left). AKT phosphorylation decreased in the models that responded to trametinib/ganitumab (SMS-CTR, BIRCH, SJRHB013758, and SJRHB010927; Supplemental Figure 3A, center). IGF1R expression decreased in just SMS-CTR, SJRHB013758 and SJRHB010927 (Supplemental Figure 3A, right). Immunohistochemical staining of SJRHB013758 tumors using both H&E, with which fibrotic regions are eosinophilic, and Masson’s trichrome, which stains fibrotic regions blue, (Supplemental Figure 3B) revealed that the trametinib/ganitumab combination increased tumor fibrosis, similar to what was observed in SMS-CTR tumors (Supplemental Figure 1C). The SJRHB013758 tumor stroma was positive for phosphorylated ERK, which complicates the tissue analysis and may explain why no change in ERK phosphorylation was observed by capillary immunoassay in this model (Supplemental Figure 3A).
To confirm on target activity of trametinib and ganitumab in the additional models of RAS-mutated RMS, we attempted to adapt the PDX models to cell culture. Both SJRHB013758 and SJRHB000026 were adapted to culture, but we recovered only tumor-associated fibroblasts in a culture of SJRHB010927. Importantly, both the SJRHB013758 and SJRHB000026 cells in culture express the myogenic master transcription factor MYOD1 (Supplemental Figure 4A), credentialling these cell lines as rhabdomyosarcoma lines. The SJRHB013758 cells are also able to undergo myogenic differentiation in the presence of trametinib (Supplemental Figure 4A). Trametinib treatment inhibited ERK phosphorylation in BIRCH, SJRHB013758, and SJRHB000026 cells (Supplemental Figure 4B), similar to the inhibition observed in RD and SMS-CTR cells (Figure 1D). Trametinib treatment increased AKT phosphorylation in each of these cell lines, but ganitumab did not impact AKT phosphorylation in the presence or absence of trametinib in these cell lines. Despite the efficacy of the trametinib/ganitumab combination in BIRCH and SJRHB013758 xenograft models, these agents did not induce PARP cleavage in these cells in vitro. Bim expression was induced by trametinib in SJRHB013758, but not BIRCH, and surprisingly, Bim expression was induced in SJRHB000026, which did not respond to trametinib and ganitumab (Supplemental Figure 4B), suggesting that Bim induction is not a reliable marker of response to trametinib/ganitumab.
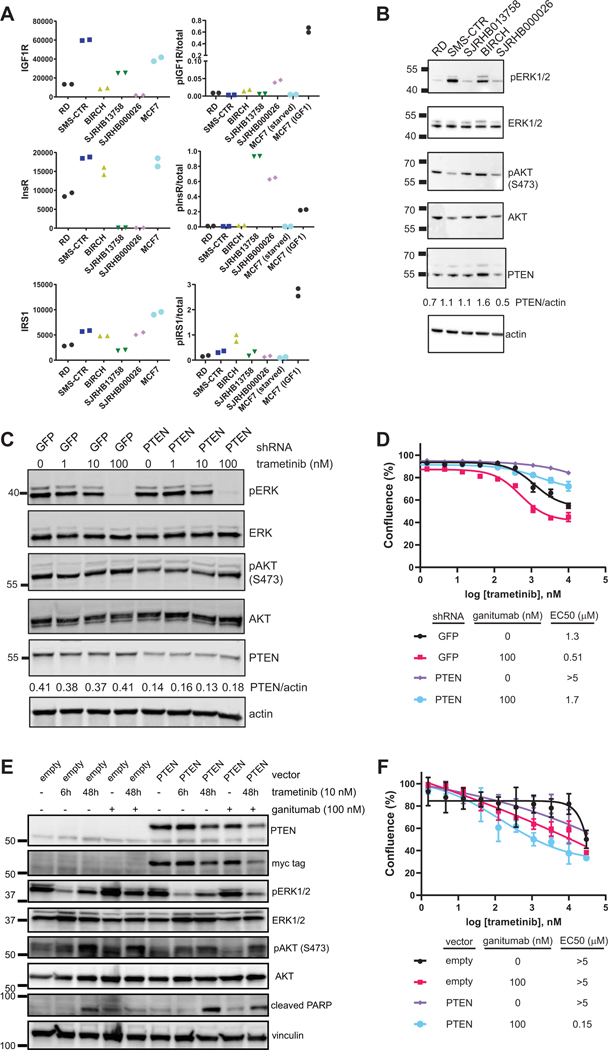
In RMS cells, the in vitro response to IGF1R monoclonal antibodies is associated with IGF1R expression (35). To determine if the in vivo response to trametinib/ganitumab is also associated with IGF1R expression, we performed a quantitative sandwich immunoassay on lysates from RMS cell lines and PDX cell lines. Lysates from serum starved or IGF1-stimulated MCF7 cells were used as positive controls for the assay. In this experiment, we confirmed that SMS-CTR has higher IGF1R expression than RD or BIRCH (Figure 5A). Despite lower IGF1R expression, however, BIRCH tumors regressed in the presence of trametinib/ganitumab. Notably, BIRCH cells express high levels of the insulin receptor (InsR), therefore, agents that target both IGF1R and InsR such as BMS-754807 or monoclonal antibodies specific for IGF1/2, may be more effective than ganitumab in this model. SJRHB013758 cells, however, had low IGF1R and InsR expression but SJRHB013758 tumors regressed in the presence trametinib/ganitumab. SJRHB000026, which had a poor response to trametinib/ganitumab, also had very low IGF1R and InsR expression. These results suggest that in vivo response to trametinib/ganitumab is weakly associated with IGF1R and InsR expression. In addition, response to trametinib/ganitumab is weakly associated with expression of the IGF1R/InsR adaptor IRS1, and the phosphorylation of IGF1R, InsR, and IRS1 (Figure 5A). Interestingly, SMS-CTR and SJRHB000026 have copy number gain of IGF1R, while both SMS-CTR and SJRHB013758 have copy number gain of IGF2 (Supplemental Data File 1). Both models had a robust response to trametinib/ganitumab, suggesting that IGF2 copy number gain might be a biomarker of sensitivity to trametinib/ganitumab.
Figure 5: Response to trametinib/ganitumab corresponds to PTEN expression but not IGF1R expression.
(A) Expression of IGF1R, insulin receptor (InsR), and IRS1 (left) as well as the ratio of phosphorylated to total levels of IGF1R, InsR, and IRS1 (right) in RMS cell lines and PDX cells adapted to cell culture were determined by an ECL-based sandwich immunoassay (MSD). MCF7 lysates in the absence (starved) and presence of IGF1 were included as positive controls. Technical duplicates (i.e., two observational units per experimental unit) are displayed. (B) ERK and AKT phosphorylation in 10% FBS, as well as AKT, ERK1/2, and PTEN expression were determined by immunoblot in RMS cell lines and PDXs adapted to cell culture. (C) ERK and AKT phosphorylation, as well as AKT, ERK1/2, and PTEN expression were determined by immunoblot from lysates of SMS-CTR cells engineered to express shRNA for either GFP (control) or PTEN that were treated with the indicated concentrations of trametinib for 6 hours. (D) Efficacy of trametinib and the trametinib/ganitumab combination was determined in SMS-CTR cells engineered to express shRNA for GFP or PTEN using percent confluence 72 hours after treatment as a marker of viability. Cells were treated with varying concentrations of trametinib in the presence or absence of ganitumab (100 nM). Percent confluence was determined by live cell imaging (Incucyte SX5). Means of quadruplicate samples (observational units) are presented, error bars indicate the standard deviation. (E) ERK and AKT phosphorylation, PARP cleavage, and AKT, ERK1/2, PTEN, and Myc-tag expression were determined by immunoblot from lysates of RD cells engineered to express empty pLenti vector or Myc-PTEN. Cells were treated with were treated with 10 nM trametinib for 6 hours, or vehicle, 10 nM trametinib, 100 nM ganitumab, or trametinib/ganitumab for 48 hours. (F) Efficacy of trametinib and the trametinib/ganitumab combination was determined in RD cells engineered to express either empty vector or Myc-PTEN. Means of quadruplicate samples (observational units) are presented, error bars indicate the standard deviation.
RMS tumors frequently display hypermethylation at the promoter of PTEN, a negative regulator of the PI3 kinase pathway. Interestingly, however, promoter hypermethylation is only weakly correlated with decreased mRNA expression of PTEN (Supplemental Figure 5A–C). RD cells have PTEN promoter hypermethylation and display decreased protein expression of PTEN, compared to other RMS cells (36). This decrease in steady-state protein expression is observed in RD cells despite maintained RNA expression (Supplemental Figure 5D). To test if the other models have alterations in PTEN expression, we evaluated PTEN protein expression by immunoblot (Figure 5B). In this experiment, we confirmed that PTEN expression in RD is decreased compared to the SMS-CTR and BIRCH cell lines. PTEN expression was also decreased in SJRHB000026 cells, which also harbors a PIK3CA mutation. Interestingly, AKT phosphorylation at S473 was not increased at baseline in the SJRHB000026 cells (Figure 5B). Baseline AKT or ERK phosphorylation is weakly associated with response to trametinib/ganitumab. However, the two cell lines with the lowest PTEN expression are also the cell lines that did show tumor regression with trametinib/ganitumab, indicating that PTEN expression could be a biomarker of response to this combination.
The mechanism by which steady-state PTEN expression is decreased in RD is incompletely defined. To assess if PTEN protein stability is impacted in RD, we compared PTEN levels after cycloheximide (CHX) treatment in RD and SMS-CTR. CHX treatment decreased PTEN levels to a greater extent in RD than SMS-CTR, suggesting that PTEN protein stability is decreased in RD (Supplemental Figure 5E). Notably, RD is less sensitive to trametinib as a single agent as compared to SMS-CTR.
To further investigate if PTEN loss contributes to resistance to trametinib, we used shRNA to decrease PTEN expression in SMS-CTR. Approximately 50% knock-down in PTEN expression was achieved, as determined by immunoblot (Figure 5C). In cell viability assays, SMS-CTR cells expressing a control shRNA (GFP) remained sensitive to trametinib, and synergy with ganitumab was observed in these cells as evidenced by a shift in the IC50 to the left in the presence of 100 nM ganitumab (Figure 5D). In contrast, SMS-CTR cells expressing PTEN shRNA were resistant to trametinib, although ganitumab did sensitize these cells to trametinib (Figure 5D). Trametinib treatment (100 nM) decreased ERK phosphorylation in SMS-CTR cells expressing PTEN shRNA (Figure 5C), suggesting that these cells are less functionally dependent on the MAPK pathway (Supplemental Figure 6).
To confirm the role of PTEN in mediating RAS-mutated RMS sensitivity to trametinib, we overexpressed Myc-tagged PTEN in RD (Figure 5E). RD expressing Myc-PTEN remained resistant to trametinib, but the presence of ganitumab markedly sensitized these cells to trametinib (Figure 5F). PTEN overexpression potentiated the decrease in ERK and AKT phosphorylation and increased the PARP cleavage induced by trametinib/ganitumab treatment in RD cells (Figure 5E). These results are consistent with PTEN mediating sensitivity to trametinib/ganitumab in RAS-mutated RMS.
Discussion
In this study, we show that the MEK inhibitor trametinib and the IGF1R inhibitor ganitumab synergistically inhibit RAS-mutated RMS cell proliferation and induce apoptosis in cell culture models. Mechanistically, ganitumab relieves the hyperactivation of AKT that is induced by trametinib monotherapy. The trametinib/ganitumab combination induced tumor shrinkage to below the limits of detection by digital calipers in five of the six xenograft models tested (three cell line xenografts and two PDX). This tumor regression was durable off treatment in two models (SMS-CTR and SJRHB013758). Tumor regression was noted in large SMS-CTR tumors, suggesting that trametinib/ganitumab could have efficacy in patients with bulk disease. The response to trametinib/ganitumab is weakly associated with the specific RAS isoform mutation and the expression level of IGF1R. Interestingly, we observed that the cell line and PDX models that did not have a response to trametinib/ganitumab had alterations in the PI3K/PTEN/AKT axis. This observation could be clinically important given that 8% of FN-RMS tumors harbor a mutation in one of the RAS isoforms as well as PI3K/PTEN/AKT. Our current study is limited by small sample size (in some cases n = 1), and further preclinical and clinical validation is needed; however, our results suggest that hotspot mutations in PIK3CA and alterations in PTEN expression could function as biomarkers predictive of resistance to trametinib/ganitumab in RAS-mutant RMS.
The FN RMS models described in the current work are unique in that they have a functional dependency upon both the RAS/MAPK pathway and a specific RTK, IGF1R, such that co-targeting MEK and IGF1R specifically is efficacious. However, we cannot rule out the possibility that combined MEK and IGF1R inhibition in FN RMS cells might induce activation of an additional RTK as a bypass mechanism. Wild type RAS and some RAS mutants, such as KRASG12C, are responsive to upstream signaling from RTKs in RAS-mutated cancer cells (37). Several RTKs, including EGFR, FGFR1, AXL, and IGF1R have been implicated in the activation of wild type and mutant RAS in RAS-mutated cells, but the precise RTK required is often cell-type or cell-state specific (38). Activation of RAS downstream of RTK signaling requires the action of the RAS-selective guanine nucleotide exchange factors, SOS1 and SOS2, as well as the molecular scaffold SHP2, creating a functional dependency on these proteins in RAS-mutated cells (39–43). In addition, RAF/MEK/ERK MAPK pathway inhibition in cells expressing either wild type RAS or mutant RAS leads to relief of feedback inhibition resulting in the activation of several RTKs, although the specific RTK activated by MAPK inhibition is also context specific. For example, MEK inhibition induces upregulation and activation of the RTKs IGF1R and ERBB3 in KRAS-mutated non-small cell lung carcinoma and colorectal carcinoma cells (42,44,45). In KRAS-mutated pancreatic carcinoma cells, both upregulation and activation of EGFR, HER2, PDGFRα and AXL are observed (46). This phenomenon is also observed in cells expressing wild type RAS, including triple negative breast carcinoma cells, in which MEK inhibition upregulates and activates PDGFβ, AXL, VEGFR2 and RET (47); and BRAF-mutant melanoma cells, in which BRAF or MEK inhibition increases PDGFRβ and IGF1R phosphorylation (48,49). Both the ability of RTK signaling to activate mutant RAS and the upregulation of RTKs in response to MAPK inhibition underly the synergy between the MAPK inhibitors and either RTK or SHP2 inhibitors in cells expressing mutant RAS.
This study highlights the importance of a wild-type PI3K/PTEN/AKT axis for response to trametinib/ganitumab treatment. Activating mutations in PI3K might confer resistance to IGF1R inhibitors (50), such as ganitumab. Interestingly, we show that decreased PTEN expression confers intrinsic resistance to MEK inhibition in FN RMS. PTEN loss is also associated with intrinsic and acquired resistance to MAPK pathway inhibitors in AML (51), non-small cell lung cancer (52), and melanoma (53). One possible mechanism by which activating mutations in PI3K or PTEN loss could contribute to MEK inhibitor resistance is by decreasing dependency on the RAS/MAPK pathway (Supplemental Figure 6). PTEN expression and activity is controlled at multiple levels in cancer cells, including the epigenetic level, through promoter methylation; the transcriptional level, through activation of transcription factors downstream of the MAPK or NOTCH pathways; the post-transcriptional level, through the activity of multiple miRNAs; and the post-translational level, through modifications that affect PTEN stability, localization, or protein: protein interactions (54). Notably, a small percentage of RMS tumors have PTEN mutations or copy number loss (55). Pten deletion in a mouse model of FN RMS results in less differentiated and more aggressive tumors, underscoring the importance of this pathway in rhabdomyosarcoma tumor development (56). Our future studies will focus on identifying the molecular mechanism by which PTEN impacts sensitivity to MEK and IGF1R inhibitors in FN-RMS.
Although the plasma concentrations of trametinib achieved in this study were within range of clinically achievable levels, the differences in murine and human metabolism of trametinib limits our ability to fully confirm this. We used the maximum tolerated dose of trametinib in mice (3 mg/kg daily) (28). Trametinib has a longer half-life in humans (5.3 days) (57) than it does in mice (33 hours) (28), which complicates efforts to use a clinically relevant dose of trametinib in mouse models. Recent work has suggested that a Cmin of 10 ng/mL is achieved by the dose of trametinib that is FDA approved for adults (2 mg by mouth daily). This Cmin is achieved in nude mice (58) using a lower dose of trametinib than was used in this study, which employed SCID beige and NSG mice. We formulated trametinib differently than the nude mice study, and the bioavailablity of our trametinib formulation could be different. No pharmacokinetics studies of trametinib in pediatric populations have been published to date, however, a recent study in adults showed that the Cmax achieved by use of the oral solution of trametinib was higher than that achieved by the tablet form (59). Importantly, the dose of ganitumab used in this study (300 μg IP twice a week) achieved a Cmax (285 μg/mL) in mice (18) that is roughly equivalent to the Cmax observed in humans treated at the recommended phase II dose of ganitumab (341 μg/mL) (60), therefore, the ganitumab dose used in this study is clinically achievable. Future work may explore the efficacy of lower doses of trametinib in this combination.
In summary, combined trametinib and ganitumab treatment is efficacious in a panel of RAS-mutated FN RMS preclinical models at clinically relevant doses. The trametinib/ganitumab combination also had an improved tolerability profile compared to other IGF1R/MEK inhibitor combinations. The data presented here support the development of a phase I/II clinical trial evaluating the safety and efficacy of the combination of trametinib and ganitumab in pediatric patients with relapsed/refractory RAS-mutated RMS.
Supplementary Material
Statement of Translational Relevance:
The RAS/MAP kinase and IGF1R pathways are both recurrently altered in PAX fusion negative rhabdomyosarcoma. In this study, we show that co-targeting these pathways with trametinib, a MEK inhibitor, and ganitumab, a monoclonal antibody against IGF1R, is effective in RAS-mutant RMS preclinical models, and that this combination is well tolerated in immunodeficient mice at clinically achievable doses of both agents. The results support testing this combination in a phase I/II clinical trial for pediatric patients with relapsed or refractory RAS-mutant RMS.
Acknowledgements
The authors are grateful to John Glod, Christine Heske, Deborah Morrison, Rosa Nguyen, Paul Randazzo, Patricia Steeg, Naomi Taylor, Carol Thiele, and Brigitte Widemann for helpful discussions and to Jyoti Shetty, Bao Tran, and Yongmei Zhao for technical support. This research was supported by the Intramural Research Program of the NIH: Matthew Hall is supported by the intramural research program of the National Center for Advancing Translational Sciences and Marielle Yohe and Javed Khan are supported by the intramural research program of the National Cancer Institute (ZIA BC011804). Marielle Yohe is also supported by a Young Investigator Award from the Alex’s Lemonade Stand Foundation. This work was partially funded by the NCI’s Office of Translational Resources. The authors thank Yolanda L. Jones, NIH Library, for editing assistance.
This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Financial Support:
This research was supported by the Intramural Research Program of the NIH. Marielle E. Yohe is also supported by a Young Investigator Award from the Alex’s Lemonade Stand Foundation. This work was partially funded by the NCI’s Office of Translational Resources.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have declared that no competing interests exist.
References
- 1.Oberlin O, Rey A, Lyden E, Bisogno G, Stevens MC, Meyer WH, et al. Prognostic factors in metastatic rhabdomyosarcomas: results of a pooled analysis from United States and European cooperative groups. J Clin Oncol 2008;26(14):2384–9 doi 10.1200/JCO.2007.14.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, et al. Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study Group. J Clin Oncol 1999;17(11):3487–93. [DOI] [PubMed] [Google Scholar]
- 3.Heske CM, Mascarenhas L. Relapsed Rhabdomyosarcoma. Journal of Clinical Medicine 2021;10(4):804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 2013;24(6):710–24 doi 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4(2):216–31 doi 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shern JF, Selfe J, Izquierdo E, Patidar R, Chou HC, Song YK, et al. Genomic Classification and Clinical Outcome in Rhabdomyosarcoma: A Report From an International Consortium. J Clin Oncol 2021;39(26):2859–71 doi 10.1200/jco.20.03060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agaram NP, Huang SC, Tap WD, Wexler LH, Antonescu CR. Clinicopathologic and survival correlates of embryonal rhabdomyosarcoma driven by RAS/RAF mutations. Genes Chromosomes Cancer 2022;61(3):131–7 doi 10.1002/gcc.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaseva AV, Yohe ME. Targeting RAS in pediatric cancer: is it becoming a reality? Curr Opin Pediatr 2020;32(1):48–56 doi 10.1097/MOP.0000000000000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia N, Del Pozo V, Yohe ME, Goodwin CM, Shackleford TJ, Wang L, et al. Vertical Inhibition of the RAF-MEK-ERK Cascade Induces Myogenic Differentiation, Apoptosis, and Tumor Regression in H/NRAS(Q61X) Mutant Rhabdomyosarcoma. Mol Cancer Ther 2022;21(1):170–83 doi 10.1158/1535-7163.Mct-21-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renshaw J, Taylor KR, Bishop R, Valenti M, De Haven Brandon A, Gowan S, et al. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res 2013;19(21):5940–51 doi 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yohe ME, Gryder BE, Shern JF, Song YK, Chou HC, Sindiri S, et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 2018;10(448) doi 10.1126/scitranslmed.aan4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crose LE, Linardic CM. Receptor tyrosine kinases as therapeutic targets in rhabdomyosarcoma. Sarcoma 2011;2011:756982 doi 10.1155/2011/756982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SY, Toretsky JA, Scher D, Helman LJ. The role of IGF-1R in pediatric malignancies. Oncologist 2009;14(1):83–91 doi 10.1634/theoncologist.2008-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins AS, Olmos D, Missiaglia E, Shipley J. Targeting the insulin-like growth factor pathway in rhabdomyosarcomas: rationale and future perspectives. Sarcoma 2011;2011:209736 doi 10.1155/2011/209736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham J, Prajapati SI, Nishijo K, Schaffer BS, Taniguchi E, Kilcoyne A, et al. Evasion mechanisms to Igf1r inhibition in rhabdomyosarcoma. Mol Cancer Ther 2011;10(4):697–707 doi 10.1158/1535-7163.Mct-10-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heske CM, Yeung C, Mendoza A, Baumgart JT, Edessa LD, Wan X, et al. The Role of PDGFR-beta Activation in Acquired Resistance to IGF-1R Blockade in Preclinical Models of Rhabdomyosarcoma. Transl Oncol 2016;9(6):540–7 doi 10.1016/j.tranon.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan X, Yeung C, Heske C, Mendoza A, Helman LJ. IGF-1R Inhibition Activates a YES/SFK Bypass Resistance Pathway: Rational Basis for Co-Targeting IGF-1R and Yes/SFK Kinase in Rhabdomyosarcoma. Neoplasia 2015;17(4):358–66 doi 10.1016/j.neo.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calzone FJ, Cajulis E, Chung YA, Tsai MM, Mitchell P, Lu J, et al. Epitope-specific mechanisms of IGF1R inhibition by ganitumab. PLoS One 2013;8(2):e55135 doi 10.1371/journal.pone.0055135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carboni JM, Wittman M, Yang Z, Lee F, Greer A, Hurlburt W, et al. BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol Cancer Ther 2009;8(12):3341–9 doi 10.1158/1535-7163.MCT-09-0499. [DOI] [PubMed] [Google Scholar]
- 20.Murakami H, Doi T, Yamamoto N, Watanabe J, Boku N, Fuse N, et al. Phase 1 study of ganitumab (AMG 479), a fully human monoclonal antibody against the insulin-like growth factor receptor type I (IGF1R), in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol 2012;70(3):407–14 doi 10.1007/s00280-012-1924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okusaka T, Ikeda M, Fukutomi A, Kobayashi Y, Shibayama K, Takubo T, et al. Safety, tolerability, pharmacokinetics and antitumor activity of ganitumab, an investigational fully human monoclonal antibody to insulin-like growth factor type 1 receptor, combined with gemcitabine as first-line therapy in patients with metastatic pancreatic cancer: a phase 1b study. Jpn J Clin Oncol 2014;44(5):442–7 doi 10.1093/jjco/hyu034. [DOI] [PubMed] [Google Scholar]
- 22.Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017;549(7670):96–100 doi 10.1038/nature23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalczyk JT, Wan X, Hernandez ER, Luo R, Lyons GC, Wilson KM, et al. Rigosertib induces mitotic arrest and apoptosis in RAS-driven rhabdomyosarcoma and neuroblastoma. Mol Cancer Ther 2020. doi 10.1158/1535-7163.MCT-20-0525. [DOI] [PMC free article] [PubMed]
- 24.Lazic SE, Clarke-Williams CJ, Munafo MR. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol 2018;16(4):e2005282 doi 10.1371/journal.pbio.2005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin GL, Wilson KM, Ceribelli M, Stanton BZ, Woo PJ, Kreimer S, et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci Transl Med 2019;11(519) doi 10.1126/scitranslmed.aaw0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer CT, Wooten DJ, Paudel BB, Bauer J, Hardeman KN, Westover D, et al. Quantifying Drug Combination Synergy along Potency and Efficacy Axes. Cell Syst 2019;8(2):97–108 e16 doi 10.1016/j.cels.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wooten DJ, Meyer CT, Lubbock ALR, Quaranta V, Lopez CF. MuSyC is a consensus framework that unifies multi-drug synergy metrics for combinatorial drug discovery. Nature Communications 2021;12(1):4607 doi 10.1038/s41467-021-24789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 2011;17(5):989–1000 doi 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 29.Beltran PJ, Chung YA, Moody G, Mitchell P, Cajulis E, Vonderfecht S, et al. Efficacy of ganitumab (AMG 479), alone and in combination with rapamycin, in Ewing’s and osteogenic sarcoma models. J Pharmacol Exp Ther 2011;337(3):644–54 doi 10.1124/jpet.110.178400. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J, George EO, Poquette CA, Cheshire PJ, Richmond LB, de Graaf SS, et al. Synergy of topotecan in combination with vincristine for treatment of pediatric solid tumor xenografts. Clin Cancer Res 1999;5(11):3617–31. [PubMed] [Google Scholar]
- 31.Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clinical & experimental metastasis 2000;18(3):261–71. [DOI] [PubMed] [Google Scholar]
- 32.Schreyer SA, Wilson DL, LeBoeuf RC. C57BL/6 mice fed high fat diets as models for diabetes-accelerated atherosclerosis. Atherosclerosis 1998;136(1):17–24 doi 10.1016/s0021-9150(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 33.Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest 2008;118(11):3651–9 doi 10.1172/JCI35437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moody G, Beltran PJ, Mitchell P, Cajulis E, Chung YA, Hwang D, et al. IGF1R blockade with ganitumab results in systemic effects on the GH-IGF axis in mice. J Endocrinol 2014;221(1):145–55 doi 10.1530/JOE-13-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao L, Yu Y, Darko I, Currier D, Mayeenuddin LH, Wan X, et al. Addiction to elevated insulin-like growth factor I receptor and initial modulation of the AKT pathway define the responsiveness of rhabdomyosarcoma to the targeting antibody. Cancer research 2008;68(19):8039–48 doi 10.1158/0008-5472.CAN-08-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan X, Helman LJ. Levels of PTEN protein modulate Akt phosphorylation on serine 473, but not on threonine 308, in IGF-II-overexpressing rhabdomyosarcomas cells. Oncogene 2003;22(50):8205–11 doi 10.1038/sj.onc.1206878. [DOI] [PubMed] [Google Scholar]
- 37.Haigis KM. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017;3(10):686–97 doi 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, et al. KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal 2019;12(583) doi 10.1126/scisignal.aaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, et al. BI-3406, a potent and selective SOS1::KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov 2020. doi 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed]
- 40.Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med 2018;24(7):961–7 doi 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 41.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 2018;20(9):1064–73 doi 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24(7):954–60 doi 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 43.Sheffels E, Sealover NE, Wang C, Kim DH, Vazirani IA, Lee E, et al. Oncogenic RAS isoforms show a hierarchical requirement for the guanine nucleotide exchange factor SOS2 to mediate cell transformation. Sci Signal 2018;11(546) doi 10.1126/scisignal.aar8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov 2016;6(7):754–69 doi 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov 2013;3(5):548–63 doi 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettazzoni P, Viale A, Shah P, Carugo A, Ying H, Wang H, et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer research 2015;75(6):1091–101 doi 10.1158/0008-5472.CAN-14-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 2012;149(2):307–21 doi 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nazarian RM, Prieto VG, Elder DE, Duncan LM. Melanoma biomarker expression in melanocytic tumor progression: a tissue microarray study. J Cutan Pathol 2010;37 Suppl 1:41–7 doi 10.1111/j.1600-0560.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 49.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 2010;18(6):683–95 doi 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guha M. Anticancer IGF1R classes take more knocks. Nat Rev Drug Discov 2013;12(4):250 doi 10.1038/nrd3992. [DOI] [PubMed] [Google Scholar]
- 51.Smith AM, Zhang CRC, Cristino AS, Grady JP, Fink JL, Moore AS. PTEN deletion drives acute myeloid leukemia resistance to MEK inhibitors. Oncotarget 2019;10(56):5755–67 doi 10.18632/oncotarget.27206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metzenmacher M, Goetz M, Herold T, Stuschke M, Aigner C, Darwiche K, et al. Acquired Resistance to BRAF/MEK Inhibitor Therapy in BRAF-V(600)-mutated Squamous Cell Lung Cancer: Concurrent Evolvement of PTEN and MEK1 Mutations. Clin Lung Cancer 2021;22(5):e668–e72 doi 10.1016/j.cllc.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Catalanotti F, Cheng DT, Shoushtari AN, Johnson DB, Panageas KS, Momtaz P, et al. PTEN Loss-of-Function Alterations Are Associated With Intrinsic Resistance to BRAF Inhibitors in Metastatic Melanoma. JCO Precis Oncol 2017;1 doi 10.1200/PO.16.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bermudez Brito M, Goulielmaki E, Papakonstanti EA. Focus on PTEN Regulation. Front Oncol 2015;5:166 doi 10.3389/fonc.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 2015;6:7557 doi 10.1038/ncomms8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langdon CG, Gadek KE, Garcia MR, Evans MK, Reed KB, Bush M, et al. Synthetic essentiality between PTEN and core dependency factor PAX7 dictates rhabdomyosarcoma identity. Nat Commun 2021;12(1):5520 doi 10.1038/s41467-021-25829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouellet D, Kassir N, Chiu J, Mouksassi MS, Leonowens C, Cox D, et al. Population pharmacokinetics and exposure-response of trametinib, a MEK inhibitor, in patients with BRAF V600 mutation-positive melanoma. Cancer Chemother Pharmacol 2016;77(4):807–17 doi 10.1007/s00280-016-2993-y. [DOI] [PubMed] [Google Scholar]
- 58.CDER. Pharmacology/Toxicology NDA Review and Evaluation Application 204114Orig1s000. 2013.
- 59.Cox DS, Allred A, Zhou Y, Infante JR, Gordon MS, Bendell J, et al. Relative bioavailability of pediatric oral solution and tablet formulations of trametinib in adult patients with solid tumors. Clin Pharmacol Drug Dev 2015;4(4):287–94 doi 10.1002/cpdd.152. [DOI] [PubMed] [Google Scholar]
- 60.Tolcher AW, Sarantopoulos J, Patnaik A, Papadopoulos K, Lin CC, Rodon J, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulin-like growth factor receptor 1. J Clin Oncol 2009;27(34):5800–7 doi 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the manuscript and its supplemental data files. Gene expression data analyzed in this study were obtained from GEO (GSE108022 and GSE66533).