Background:
NOTCH1 pathogenic variants are implicated in multiple types of congenital heart defects including hypoplastic left heart syndrome, where the left ventricle is underdeveloped. It is unknown how NOTCH1 regulates human cardiac cell lineage determination and cardiomyocyte proliferation. In addition, mechanisms by which NOTCH1 pathogenic variants lead to ventricular hypoplasia in hypoplastic left heart syndrome remain elusive.
Methods:
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 genome editing was utilized to delete NOTCH1 in human induced pluripotent stem cells. Cardiac differentiation was carried out by sequential modulation of WNT signaling, and NOTCH1 knockout and wild-type differentiating cells were collected at day 0, 2, 5, 10, 14, and 30 for single-cell RNA-seq.
Results:
Human NOTCH1 knockout induced pluripotent stem cells are able to generate functional cardiomyocytes and endothelial cells, suggesting that NOTCH1 is not required for mesoderm differentiation and cardiovascular development in vitro. However, disruption of NOTCH1 blocks human ventricular-like cardiomyocyte differentiation but promotes atrial-like cardiomyocyte generation through shortening the action potential duration. NOTCH1 deficiency leads to defective proliferation of early human cardiomyocytes, and transcriptomic analysis indicates that pathways involved in cell cycle progression and mitosis are downregulated in NOTCH1 knockout cardiomyocytes. Single-cell transcriptomic analysis reveals abnormal cell lineage determination of cardiac mesoderm, which is manifested by the biased differentiation toward epicardial and second heart field progenitors at the expense of first heart field progenitors in NOTCH1 knockout cell populations.
Conclusions:
NOTCH1 is essential for human ventricular-like cardiomyocyte differentiation and proliferation through balancing cell fate determination of cardiac mesoderm and modulating cell cycle progression. Because first heart field progenitors primarily contribute to the left ventricle, we speculate that pathogenic NOTCH1 variants lead to biased differentiation of first heart field progenitors, blocked ventricular-like cardiomyocyte differentiation, and defective cardiomyocyte proliferation, which collaboratively contribute to left ventricular hypoplasia in hypoplastic left heart syndrome.
Keywords: cardiac lineage differentiation, cardiomyocyte proliferation, hypoplastic left heart syndrome, induced pluripotent stem cells, mesoderm, NOTCH1, single-cell RNA-seq
Novelty and Significance.
What Is Known?
During development, NOTCH signaling is essential for cardiac cell fate determination and cardiovascular morphogenesis.
Disruption of NOTCH signaling leads to impaired myocardial differentiation and proliferation.
Pathogenic variation in NOTCH1 is associated with hypoplastic left heart syndrome and aortic valve disease in humans.
What New Information Does This Article Contribute?
In human induced pluripotent stem cells (iPSCs), NOTCH1 disruption blocks human ventricular-like cardiomyocyte differentiation but promotes atrial-like cardiomyocyte generation.
NOTCH1 deficiency leads to impaired proliferation of early human cardiomyocytes, possibly through downregulation of pathways that are involved in cell cycle progression and mitosis.
Single-cell transcriptomic analysis reveals the abnormal cell lineage specification and imbalanced differentiation of the first heart field, second heart field and epicardial progenitors in NOTCH1 deficient iPSCs.
NOTCH signaling is an evolutionarily conserved pathway and involved in multiple aspects of cell fate determination and proliferation during development. Though NOTCH signaling is essential for heart development in mammals, it remains unclear how NOTCH1 modulates human cardiac cell lineage specification during development. By leveraging CRISPR/Cas9 genome editing technology, we generated multiple NOTCH1 knockout (N1KO) human iPSC lines. Human N1KO iPSCs can generate functional cardiomyocytes and endothelial cells, indicating that NOTCH1 is not required for mesoderm differentiation and cardiovascular development in vitro. However, NOTCH1 is required for human ventricular-like cardiomyocyte differentiation by limiting atrial-like cardiomyocyte generation. Single-cell transcriptomics analysis reveals NOTCH1 modulates cell fate determination of early cardiac mesoderm towards the first heart field, second heart field, and epicardial progenitors. Human N1KO iPSC-derived cardiomyocytes display cell proliferation defects, suggesting that NOTCH1 is required for cell cycle progression and mitosis during the early phases of cardiomyocyte proliferation in human heart development. As pathogenic NOTCH1 variants are associated with hypoplastic left heart syndrome in humans, this study provides novel insights on how NOTCH1 dysfunction may contribute to ventricular hypoplasia and structural anomalies in congenital heart defects.
Meet the First Author, see p 153
Editorial, see p 205
NOTCH signaling pathway is highly conserved across the animal kingdom and is widely utilized to regulate cell fate determination and cell proliferation in development.1,2 In mammals, the core components of the NOTCH signaling pathway include 4 receptors (NOTCH1-4) and 5 ligands (JAG1-2, DLL1, DLL3, and DLL4). Receptor-ligand interactions lead to proteolytic cleavage and release of the NOTCH intracellular domain from the membrane.3 The NOTCH intracellular domain then translocates to the nucleus and forms an activation complex with RBPJ (also known as CBF1) by evicting a histone deacetylase corepressor. The activation complex further recruits the histone acetyltransferase and MAML1, and triggers the transcriptional activation of NOTCH downstream target genes including the bHLH transcriptional repressors HEY1 and HEY2.4
NOTCH signaling pathway plays an essential role in cell fate determination and morphogenesis in the developing heart.5–7 During embryonic heart development, NOTCH receptors and ligands are primarily expressed in the developing endocardium and myocardium, respectively. NOTCH activation in the endocardium stimulates the transcription of EFNB2 (ephrin B2). EFNB2 then activates the secretory protein NRG1 (neuregulin-1), which promotes the differentiation of adjacent cells into trabecular cardiomyocytes.8,9 In parallel, NOTCH activation in the endocardium modulates BMP10 (bone morphogenetic protein 10) in the neighboring cardiomyocytes, promoting their proliferation.8 During mouse ventricular chamber development, NOTCH signaling first connects endocardium and myocardium to support trabeculation, then coordinates ventricular patterning and compaction with coronary artery development to form the mature chamber.10
NOTCH signaling disruption leads to impaired myocardial differentiation and proliferation, which are manifested by ventricular malformations such as hypoplastic left heart syndrome (HLHS)11,12 and left ventricular non-compaction cardiomyopathy in humans.13 HLHS is a severe type of congenital heart defect that occurs in approximately 1 out of 3841 live births.14 Typically, the left ventricle is under-developed in HLHS, accompanied by structural defects in the mitral valve, aortic valve, and ascending aorta.15 Although pathogenic NOTCH1 mutations have been linked to HLHS and aortic valve disease,11,16–18 molecular and cellular etiologies underlying the ventricular hypoplasia remain unknown.19 In this study, we leveraged CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 genome editing to generate NOTCH1 homozygous knockout (NOTCH1−/−, denoted as NOTCH1 knockout [N1KO] hereafter) human induced pluripotent stem cells (iPSCs). We differentiated N1KO-iPSC into cardiomyocytes (iPSC-CMs) and endothelial cells (iPSC-ECs), and profiled the transcriptional dynamics of differentiating cells at key developmental stages by single-cell RNA sequencing. We found that human cardiac lineage determination and cardiomyocyte proliferation were disrupted due to loss of function in NOTCH1. Our results provide mechanistic insights on how NOTCH1 mutations in humans lead to abnormal cardiomyocyte differentiation and proliferation, and contribute to the left ventricular hypoplasia in HLHS.
Methods
Data Availability
Detailed methods and Major Resource Table are provided in the Supplemental Material. Data will be made available upon reasonable request, by contacting the corresponding author. Raw and processed RNA-seq and single-cell RNA sequencing data are available in the NCBI GEO database (GSE195559 and GSE196632). The graphic abstract was generated using BioRender.com.
Results
NOTCH1 Is Not Required for Mesodermal and Vascular Endothelial Differentiation of Human iPSCs
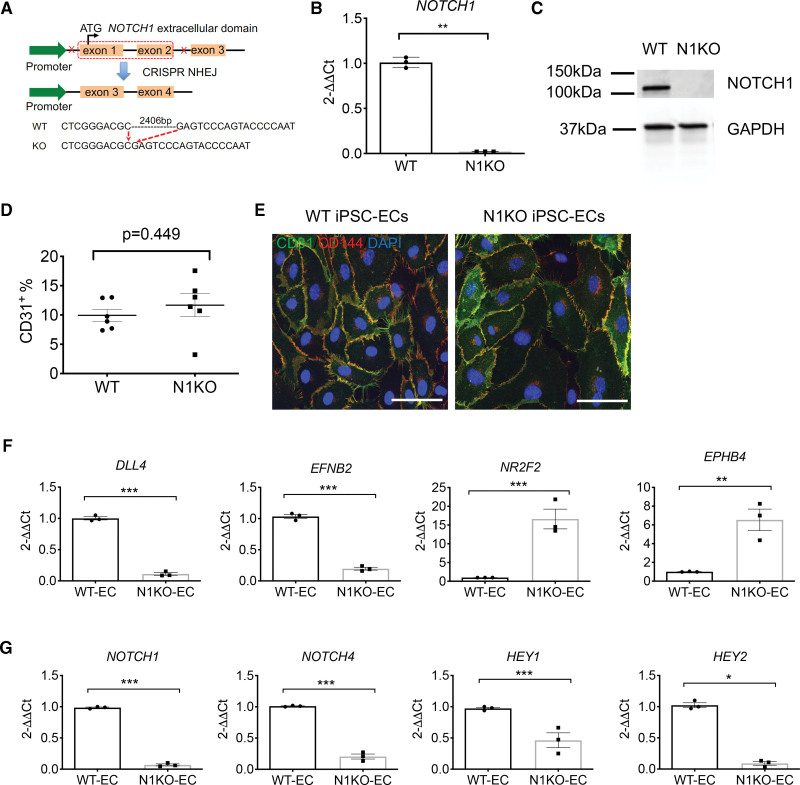
Notch signaling is essential for post-implantation embryonic development in mammals and homozygous Notch1 knockout mouse embryos die by the mid-gestation.20 Additional studies have shown that Notch1 knockout mouse embryos display severe defects in vascular morphogenesis.21 To illustrate the regulatory role of NOTCH1 in human cardiac development and congenital heart disease, we generated NOTCH1 homozygous knockout iPSC lines from 3 healthy subjects using CRISPR/Cas9 genome editing. We designed sgRNA pairs (Table S1) that targeted NOTCH1 exon 1 and exon 2 to completely disrupt the transcriptional initiation (Figure 1A). We utilized an EGFP-containing CRISPR vector PX45822 and enriched EGFP+ transfected cells by flow cytometry (Figure S1A). We designed pairs of primers within and flanking the deleted region to identify homozygous iPSC clones (Figure S1B). For homozygous knockouts, a smaller (409 bp) PCR product would be amplified using outside primers and no PCR product generated using inside primers (Table S2). Sanger sequencing confirmed deletion of a 2.4 kb NOTCH1 genomic segment in the N1KO iPSC clones (Figure S1C and S1D). Results from qPCR and Western blotting confirmed the absence of NOTCH1 at both mRNA and protein levels in the homozygous N1KO iPSCs (Figure 1B and 1C; Figure S1E). All 3 N1KO human iPSC lines showed normal karyotypes (Figure S1F).
Figure 1.
Generation of NOTCH1 knockout (N1KO) human induced pluripotent stem cells (iPSCs) and endothelial differentiation. A, Schematic overview of deleting a 2.4-skb segment including exon 1 and exon 2 in the NOTCH1 gene using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 genome editing. B, Quantitative PCR (qPCR) analysis shows mRNA levels in N1KO iPSCs (t test, n=3). C, Western blot indicates the absence of NOTCH1 in the homozygous N1KO iPSCs at the protein level. GAPDH is used as an internal control. D, Endothelial differentiation efficiency measured by the percentage of CD31+ cells at D10 of differentiation (1-way ANOVA, n=6). E, Expression of endothelial markers CD31 and CD144 in wild-type (WT) and N1KO iPSC-ECs. F, qPCR analysis shows reduced expression of arterial endothelial genes (DLL4 and EFNB2) and enhanced expression of venous endothelial genes (NR2F2 and EPHB4) in homozygous N1KO iPSC-ECs. G, qPCR data indicate downregulation of NOTCH1, NOTCH4, HEY1, and HEY2 in N1KO compared with WT iPSC-ECs (t test, n=3). All bar graphs show mean±SEM. *P<0.05, **P<0.01, ***P<0.001. Scale bars = 50 μm. EC indicates endothelial cell.
To test whether NOTCH1 deletion disrupted mesodermal and endothelial cell differentiation in human iPSCs, we differentiated homozygous N1KO iPSCs into endothelial cells by stimulation of the Wnt signaling pathway using the small chemical CHIR99021. Homozygous NOTCH1 deletion did not block mesodermal differentiation, as TBXT+ and TBX6+ mesodermal cells were present in N1KO cells at D2 of differentiation (Figure S2). We observed no significant difference in endothelial differentiation efficiency between wild-type (WT) and N1KO as assessed by the percentage of CD31+ cells at D10 of differentiation (Figure 1D). Both WT and N1KO iPSC-ECs expressed endothelial cell-specific markers CD31 and CD144 (Figure 1E). Arterial endothelial-specific genes DLL4, EFNB, NOTCH1, NOTCH4, HEY1, and HEY2 were significantly downregulated in N1KO iPSC-ECs compared with WT iPSC-ECs (Figure 1F and 1G). In contrast, expression levels of venous endothelial cell marker genes such as NR2F2 and EPHB4 were dramatically increased in N1KO iPSC-ECs (Figure 1F). Collectively, these results indicate that NOTCH1 is not required for mesoderm differentiation, but is essential for arterial endothelial cell specification of human iPSCs.
Next, we carried out RNA-seq to profile transcriptomic changes in endothelial cells due to NOTCH1 disruption. We identified 895 differentially expressed genes (DEGs; false discovery rate [FDR]<0.05, fold change >2) between N1KO and WT iPSC-ECs (Figure S3A). Principal component analysis indicated whole transcriptomic differences between N1KO and WT iPSC-ECs (Figure S3B). There were 451 upregulated and 444 downregulated DEGs in N1KO iPSC-ECs. Expression levels of NOTCH pathway ligands DLL4 and receptor NOTCH1, and downstream targets (HEY2, HEYL, and HES4) were significantly decreased in N1KO iPSC-ECs (Figure S3C). In contrast, transcription factors NR2F2 and NKX2-5 were upregulated in N1KO iPSC-ECs (Figure S3C), indicating the promotion of venous endothelial cell specification due to NOTCH1 dysfunction.23,24 Gene ontology pathway enrichment analysis showed that the upregulated DEGs (in red) in N1KO iPSC-ECs were primarily associated with cell-cell adhesion, extracellular matrix organization, and epithelial tube morphogenesis, whereas downregulated DEGs (in blue) were enriched in the neural crest cell migration, pulmonary valve morphogenesis, aortic valve development, atrioventricular valve development, and cardiac chamber development (Figure S3D). This is consistent with the role of NOTCH1 variants in CHD, in which disruption of these biological pathways leads to bicuspid aortic valve, tetralogy of Fallot, and HLHS.11,16,18,25 QIAGEN Ingenuity Pathway Analysis (IPA) identified network interaction modulated by NOTCH downstream regulator RBPJ (CBF1; Figure S3E).
Ventricular-Like Cardiomyocyte Differentiation Is Impaired Whereas Atrial-Like Cardiomyocyte Determination Is Promoted in N1KO iPSCs
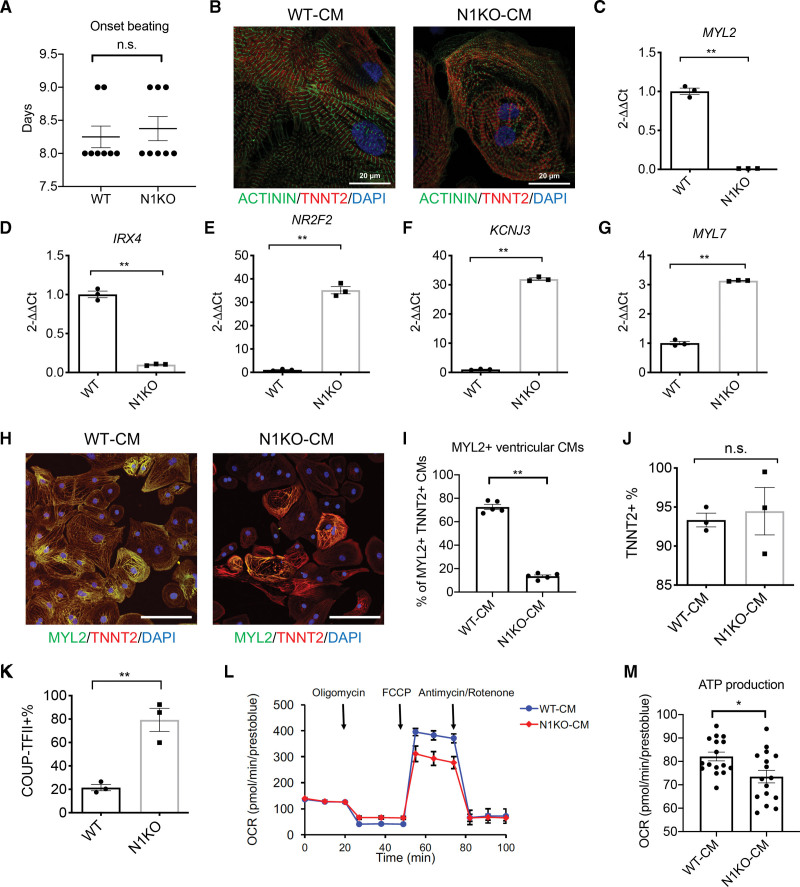
We examined how NOTCH1 deficiency affects human cardiac differentiation using a small molecule-mediated cardiac differentiation protocol.26 N1KO did not delay cardiac differentiation as illustrated by similar onset of beating (days 8 and 9) for N1KO and WT iPSCs (Figure 2A). N1KO and WT iPSC-CMs displayed typical sarcomere structures with intercalated distribution of cardiac troponin T (TNNT2) and α-actinin (Figure 2B). Because iPSC-CMs are a mixed population of ventricular-, atrial-, and nodal-like cells,27,28 we assessed whether NOTCH1 knockout influenced the specification of these cardiac subtypes. NOTCH1 deficiency resulted in significantly downregulated ventricular cardiomyocyte-specific genes myosin regulatory light chain 2, ventricular muscle isoform (MYL2) and Iroquois-class homeodomain protein IRX4 (Figure 2C and 2D). Interestingly, NOTCH1 disruption enhanced expression of genes that are enriched in human atria (eg, nuclear receptor NR2F2; potassium channel KCNJ3; and myosin regulatory light 2 chain 2, atrial isoform MYL7) in iPSC-CMs (Figure 2E through 2G). In addition, immunofluorescence staining using antibodies against TNNT2 and MYL2 showed significantly lower percentage of MYL2+TNNT2+ ventricular-like cardiomyocytes in N1KO (~10%) compared with WT iPSC-CMs (~70%) at D45 (Figure 2H and 2I). Notably, the percentages of cardiac troponin T (TNNT2+) cardiomyocytes were not significantly different between N1KO and WT iPSC-CMs as assayed by flow cytometry (Figure 2J), suggesting that NOTCH1 minimally affects cardiomyocyte differentiation efficiency in human iPSCs.
Figure 2.
NOTCH1 deficiency suppresses ventricular-like cardiomyocyte differentiation and promotes atrial-like cardiomyocyte differentiation of human induced pluripotent stem cells (iPSCs). A, Days until onset of cardiomyocyte beating in the differentiation of NOTCH1 knockout (N1KO) and wild-type (WT) iPSCs (t test, n=8). B, Sarcomere structure is visualized by co-staining with antibodies against cardiac troponin T (TNNT2, in red) and α-actinin (in green). Nuclei are labeled by DAPI (in blue). Scale bars = 20 μm. C–G, Quantitative PCR (qPCR) analysis indicates that ventricular cardiomyocyte-specific marker genes (MYL2 [myosin regulatory light chain 2], IRX4 [iroquois homeobox protein 4]) are downregulated whereas atrial cardiomyocyte-specific marker genes (NR2F2, KCNJ3, MYL7) are upregulated in N1KO versus WT iPSC-CMs (t test, n=3 for each group). H, Ventricular-like iPSC-derived cardiomyocytes (iPSC-CMs) are identified by immunocytochemistry staining with antibodies against ventricular cardiomyocyte-specific marker MYL2 (in green) and pan-cardiomyocyte marker TNNT2 (in red). Nuclei are labeled by DAPI (in blue). Scale bars = 50 μm. I, Quantitative analysis of MYL2+TNNT2+ ventricular-like cardiomyocytes in N1KO and WT iPSC-CMs (t test, n=5). J, Percentages of TNNT2+ cardiomyocytes in the population of N1KO and WT iPSC-CMs revealed by flow cytometry (t test, n=3). K, Flow cytometry analysis shows the percentage of atrial-like cardiomyocytes (COUP-TFII+) is elevated in N1KO versus WT iPSC-CMs (t test, n=3). L, Seahorse analysis indicates that maximal respiration capacity is decreased in N1KO versus WT D30 iPSC-CMs. M, ATP production is reduced in N1KO compared with WT D30 iPSC-CMs (t test, n=16). All bar graphs and dot plots show mean±SEM. *P<0.05, **P<0.01. n.s. indicates not significant.
In contrast, the proportion of atrial-like cardiomyocytes, which were labeled by COUP-TFII (encoded by NR2F2), was significantly increased in N1KO iPSC-CMs (Figure 2K). Because mitochondrial oxidative capacity is higher in ventricular myocardium in explanted human heart compared with atrial myocardium, we compared the mitochondrial respiration in iPSC-CMs using the oxygen consumption rate as an indicator. At maximal respiration level, the oxygen consumption rate was significantly lower in N1KO compared with WT iPSC-CMs (Figure 2L). ATP production was also significantly downregulated in N1KO iPSC-CMs (Figure 2M), possibly due to reduced mitochondrial respiration capacity, which is likely associated with fewer ventricular-like cardiomyocytes in the population. MitoTracker assay further showed that mitochondria mass was lower in N1KO than WT iPSC-CMs, indicating reduced number of mitochondria due to NOTCH1 disruption (Figure S4). Collectively, these results indicate that ventricular-like cardiomyocyte differentiation is compromised whereas atrial-like cardiomyocyte differentiation is promoted due to NOTCH1 disruption during human cardiac lineage commitment.
Next, we performed RNA-seq to profile gene expression changes in response to NOTCH1 knockout. Principal component analysis plot revealed that N1KO iPSC-CMs were separated from WT iPSC-CMs at a global transcriptional level (Figure S5A). We identified 860 DEGs between N1KO and WT iPSC-CMs (FDR<0.05 and fold change>2; Figure S5B), with 408 upregulated and 452 downregulated in N1KO versus WT cells. The top enriched pathways upregulated in N1KO iPSC-CMs were involved in cardiac muscle cell differentiation, heart contraction, and muscle system process (Figure S5C). In contrast, downregulated pathways enriched in N1KO iPSC-CMs were associated with ventricular cardiac muscle tissue morphogenesis, action potential, and epithelial tube morphogenesis (Figure S5C). Gene ontology cellular component enrichment showed the upregulation of voltage-gated sodium channel complex and downregulation of cell-cell junction, sarcomere, myofibril, contractile fiber, Z disc, and I band in N1KO iPSC-CMs (Figure S5D). In addition, top upstream regulators predicted by IPA analysis included HEY2, CTNNB1 (β-catenin), NKX2-5, BMP4, and WNT3A (Figure S5E). Together, these results suggest an essential role of NOTCH1 in transcriptional modulation of human cardiac differentiation.
Electrophysiological Characterization of N1KO Cardiomyocytes
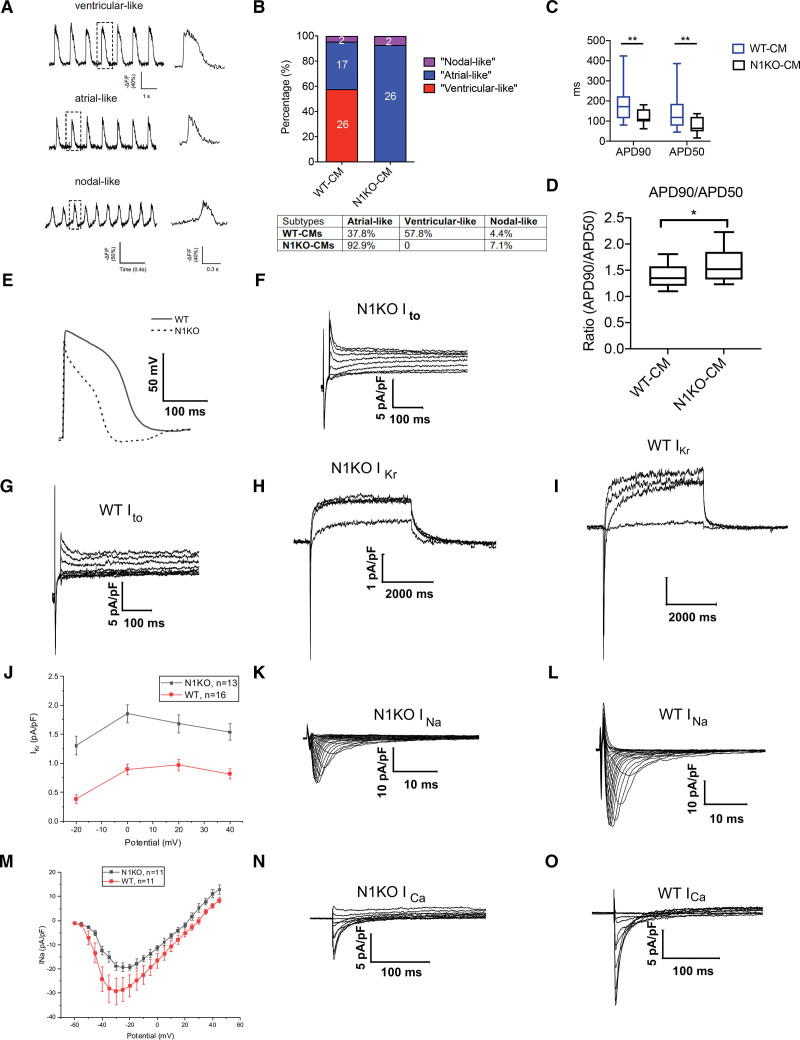
During mouse heart development, ectopic Notch activation in cardiomyocytes triggers Purkinje-like state and leads to prolongation of action potential duration through modulation of voltage-gated potassium (K+) currents.29,30 To elucidate how NOTCH1 disruption interferes with electrophysiological behaviors of human cardiomyocytes, we employed optical mapping to classify cardiac subtypes by measuring ASAP2 fluorescence dynamics.31 We grouped iPSC-CMs into 3 categories: ventricular-like, atrial-like, and nodal-like cardiomyocytes (Figure 3A) based on EP characterization. Under regular cardiac differentiation protocol using small molecules CHIR99021 and IWR-1, the majority (57.8%) of D45 WT iPSC-CMs showed ventricular-like electrophysiological behavior, while the remaining cardiomyocytes were classified as atrial-like (37.8%) or nodal-like (4.4%) cardiomyocytes (Figure 3B), consistent with previous reports.32,33 In contrast, most (92.9%) N1KO iPSC-CMs displayed atrial-like EP characteristics, with a small percentage of nodal-like cardiomyocytes (7.1%) and no detectable ventricular-like cardiomyocytes (Figure 3B). These results further demonstrate that homozygous NOTCH1 deletion blocks the differentiation of human ventricular-like cardiomyocytes and promotes atrial-like EP phenotypes in iPSC-CMs.
Figure 3.
NOTCH1 disruption alters electrophysiological behaviors of human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) by shortening action potential durations. A, Representative ventricular-, atrial-, and nodal-like electrophysiological characteristics in iPSC-CMs inferred by optical mapping. B, Percentages of ventricular-, atrial-, and nodal-like cardiomyocytes in wild-type (WT) and NOTCH1 knockout (N1KO) iPSC-CMs. C and D, APD90, APD50, and APD90/APD50 in wild-type (WT; n=20) and N1KO (n=20) iPSC-CMs measured by whole-cell patch clamp (t test, n=20). E, Patch clamp analysis shows shortened repolarization phase in N1KO versus WT iPSC-CMs. F and G, Transient outward K+ current is increased in N1KO (F) compared with WT (G) iPSC-CMs. H – J, Delayed rectifier K+ current (IKr tail current) is increased in N1KO (H) versus WT (I) iPSC-CMs. K – M, Inward Na+ currents are reduced in N1KO (K) compared with WT (L) iPSC-CMs across different member potentials. N and O, Amplitudes of inward Ca2+ currents are decreased in N1KO (N) versus WT (O) iPSC-CMs. Data are presented as mean±SEM. *P<0.05, **P<0.01.
To illustrate how NOTCH1 deletion affects ion currents in human cardiomyocytes, we conducted whole-cell patch clamp on single iPSC-CMs. We found that NOTCH1 deletion significantly shortened action potential durations at both action potential duration 90 (APD90) and APD50 in iPSC-CMs (Figure 3C). However, the ratio (APD90/APD50) was significantly increased in N1KO iPSC-CMs compared with that in WT controls (Figure 3D). Compared with WT cardiomyocytes, N1KO iPSC-CMs showed a shortened phase of repolarization that was similar to the EP characteristics of atrial-like cardiomyocytes (Figure 3E). As the initial repolarizing force, Ito (transient outward K+ current) was upregulated in the N1KO CMs and contributed to the shortened action potential duration (Figure 3F and 3G). Based on the current traces, N1KO iPSC-CMs exhibited stronger tail currents of IKr (delayed rectifier K+ current) than WT iPSC-CMs at different depolarizing membrane potentials (Figure 3H through 3J), which was likely to complete the repolarization phase and result in the shortened action potential duration. Inward Na+ currents were downregulated in N1KO-CMs compared with WT-CMs across the different membrane potentials (Figure 3K through 3M), which narrowed the depolarizing phase and thus produced the shortened action potential duration. The influx of Ca2+ through the voltage-gated Ca2+ channel can extend the repolarization stage. N1KO CMs displayed a decreased amplitude of inward Ca2+ currents (Figure 3N and 3O; Figure S6A), which further accelerated the repolarizing process and led to shortened action potential duration.
To explore whether NOTCH1 knockout affects genes involved in the regulation of action potential duration, we compared gene expression profiles between WT and N1KO iPSC-CMs. Our RNA-seq data revealed that N1KO-CMs showed a reduced expression level of SCN5A (Figure S6B) which encodes the α subunits of voltage-gated Na+ channel 1.5 (Nav1.5), a significant contributor to depolarization. Downregulation of inward Na+ currents in the N1KO-CMs was possibly caused by the reduced expression of Nav1.5. Furthermore, N1KO-CMs exhibited significant upregulation of KCNIP2 (Figure S6C), which encodes an accessory subunit of Kv4.2/Kv4.3, the primary mediators of Ito in the repolarizing phase. The expansion of Ito in N1KO-CMs could be attributed to the increased proportion of voltage-gated K+ channels on the plasma membrane through KCNIP2-mediated channel trafficking and gating.34–36 The expression level of KCNJ3, which encodes the α subunit of Kir3.1, was nearly 8 times higher in the N1KO-CMs than WT-CMs (Figure S6D). As KCNJ3 is predominantly expressed in the atrium,37 these results further demonstrate the prevalence of atrial-like cardiomyocytes in the population of N1KO iPSC-CMs.
To elucidate the potential downstream regulation in response to NOTCH1 disruption, we administered DAPT during cardiac differentiation to mimic the genetic deletion of NOTCH1 in control iPSC lines. DAPT blocks NOTCH pathway by inhibiting γ-secretase, which is responsible for the cleavage of NOTCH receptors. Compared with vehicle controls, DAPT treatment led to reduced percentage of ventricular-like cardiomyocytes in the population of iPSC-CMs, as shown by the immunofluorescence staining using an antibody against ventricular marker MYL2 (Figure S7A and S7B). In addition, whole-cell patch clamp recordings indicate that DAPT-treated iPSC-CMs displayed decreased APD90 and APD50, and shorted action potential duration (Figure S7C through S7E), which are similar to the electrophysiological characteristics of N1KO iPSC-CMs. Together, these results suggest that pharmacological blockage of NOTCH signaling pathway interferes with ventricular cardiomyocyte differentiation and highlight the essential role of NOTCH pathway in cardiac subtype specification.
NOTCH1 Knockout Leads to Defective Proliferation of Human Early Cardiomyocytes
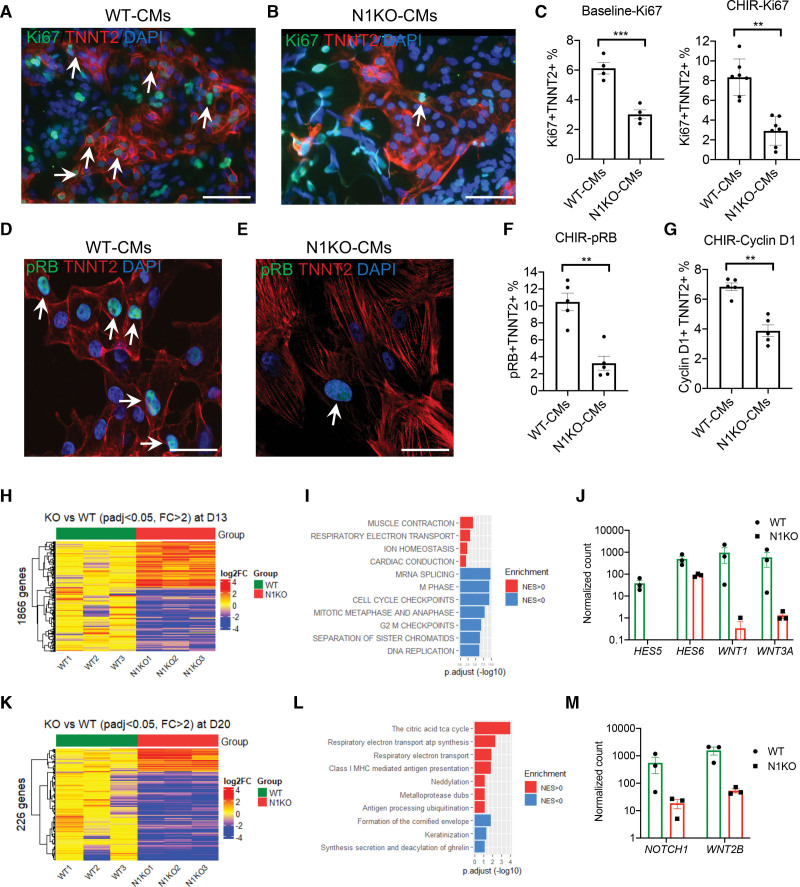
To understand how loss of NOTCH1 impacts human cardiomyocyte proliferation, we set up an in vitro assay to quantify the percentage of dividing cardiomyocytes (Figure S8A). Because human cardiomyocytes do not divide frequently,38 we utilized a small molecule, CHIR99021, to stimulate the proliferation of early iPSC-CMs.32,39 We used TNNT2 to identify cardiomyocytes and Ki67 to label dividing cells. TNNT2+ Ki67+ cells were considered proliferating cardiomyocytes. In the presence of CHIR99021, there was a significant increase of proliferating WT cardiomyocytes (Figure S8B), from ~1% without to ~9% with CHIR99021 treatment (Figure S8C). Although CHIR99021 promoted the robust proliferation of early iPSC-CMs in WT controls, it failed to stimulate the proliferation of N1KO iPSC-CMs. The percentage of dividing cardiomyocytes in N1KO iPSC-CMs was significantly lower than WT controls with and without CHIR99021 treatment when Ki67 was used to quantify dividing cardiomyocytes (Figure 4A through 4C). In addition, we used other cell cycle regulators, cyclin D1 and pRB (phosphorylated retinoblastoma protein), to label dividing cardiomyocytes, as both are required for cell cycle progression from G1 phase towards S phase. Similar to Ki67 labeling, we observed robust increase in the proliferation of WT iPSC-CMs in the presence of CHIR99021 using Cyclin D1 or pRB to identify dividing cardiomyocytes, respectively (Figure S9). However, the proliferation of early cardiomyocytes was significantly diminished in N1KO iPSC-CMs, resulting in a significantly lower percentage of dividing cardiomyocytes in N1KO cells (Figure 4D through 4G). These results indicate that NOTCH1 disruption leads to defective human cardiomyocyte proliferation, possibly through impairing cell cycle progression.
Figure 4.
Loss of function in NOTCH1 leads to defective proliferation of early cardiomyocytes. A and B, Immunofluorescence staining identifies dividing cardiomyocytes (Ki67+ TNNT2+) in wild-type (WT; A) and NOTCH1 knockout (N1KO; B) induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). Arrows indicate proliferating cardiomyocytes. TNNT2 is labeled in red whereas Ki67 is marked in green. Nuclei are stained with DAPI in blue. Scale bars = 100 μm. C, Quantitative analysis of ratios of Ki67+ TNNT2+ versus TNNT2+ cardiomyocytes with and without CHIR99021 in WT and N1KO iPSC-CMs (t test, n=4 for baseline, n=7 for CHIR). D and E, Detection of dividing cardiomyocytes using antibodies against pRB and TNNT2 in WT (D) and N1KO (E) iPSC-CMs. Scale bars = 50 μm. F, Quantitative analysis of ratios of pRB+ TNNT2+ versus TNNT2+ cardiomyocytes in the presence of CHIR99021 in WT and N1KO iPSC-CMs (t test, n=5). G, Quantitative analysis of ratios of Cyclin D1+ TNNT2+ versus TNNT2+ cardiomyocytes in the presence of CHIR99021 in WT and N1KO iPSC-CMs (t test, n=5). H, Heatmap of 1866 differentially expressed genes between N1KO and WT D13 iPSC-CMs (fold change >2, false discovery rate [FDR]<0.05). I, Gene Set Enrichment Analysis (GSEA) shows pathways that are associated with differentially expressed genes (DEGs) between N1KO and WT D13 iPSC-CMs. Upregulated pathways (NES>0) are marked in red whereas downregulated (NES<0) pathways are labeled in blue. J, Downregulation of NOTCH and WNT signaling components in N1KO versus WT D13 iPSC-CMs (q-value <0.01, n=3). K, Two hundred twenty-six differentially expressed genes are identified between N1KO and WT D20 iPSC-CMs (fold change >2, FDR<0.05). L, Relevant pathways associated with upregulated and downregulated genes in N1KO versus WT D20 iPSC-CMs inferred by GSEA. M, Downregulation of WNT2B in N1KO versus WT D20 iPSC-CMs (q-value <0.01, n=3). Normalized counts are log10 transformed, and median values of WT samples are used for baseline transformation for each gene. Data are presented as mean±SEM. **P<0.01, ***P<0.001.
Next, we carried out RNA-seq to profile transcriptomic difference between WT and N1KO cardiomyocytes at D13, D20, and D20 with CHIR99021 treatment (D20-C). Using FDR <0.05 and fold change >2, we identified 1866 DEGs in D13 iPSC-CMs between WT and N1KO samples (Figure 4H). Of these DEGs, 779 were upregulated and 1087 genes were downregulated in N1KO versus WT D13 iPSC-CMs. We then performed gene set enrichment analysis and found that the top enriched downregulated pathways were cell cycle checkpoint, mitotic metaphase and anaphase, G2-M phase checkpoints, DNA replication, separation of sister chromatids, and mRNA splicing (Figure 4I). NOTCH signaling downstream targets (HES5 and HES6) and WNT signaling components (WNT1 and WNT3A) were among those genes with significant fold changes (downregulated in N1KO; Figure 4J). These results suggest that genes and pathways related to cell cycle progression and cell proliferation are downregulated in N1KO iPSC-CMs (D13) at the transcriptional level.
We then examined the transcriptional profiles of D20 iPSC-CMs that were more differentiated and less proliferative compared with D13 cardiomyocytes. Interestingly, there were fewer DEGs (226, FDR <0.05 and fold change >2) between N1KO and WT iPSC-CMs at D20 than those at D13 (Figure 4K). The top enriched upregulated pathways were respiratory electron transport ATP synthesis and citric acid TCA cycle (Figure 4L). NOTCH signaling component (NOTCH1) and WNT pathway member (WNT2B) remained downregulated in D20 N1KO iPSC-CMs (Figure 4M). In contrast, cell cycle progression and cell proliferation pathways were not enriched at D20. Next, we explored whether CHIR99021 treatment could alter the transcriptional difference between N1KO and WT iPSC-CMs. CHIR99021 treatment greatly reduced the transcriptional disparity between N1KO and WT samples, to only 55 DEGs identified in the presence of CHIR99021 (Figure S10A and S10B). Principal component analysis plotting indicated that a transcriptional shift was induced by CHIR99021 treatment in both WT and N1KO iPSC-CMs (Figure S10C and S10D). Reactome analysis showed that the top enriched pathways were associated with respiratory electron transport, complex I biogenesis, and glycogen metabolism (Figure S10E). WNT pathway elements WNT2B, DKK2, and FRZB were downregulated in N1KO versus WT iPSC-CMs (Figure S10F). Across developmental stages (D13 versus D20 cardiomyocytes), NOTCH and WNT pathways were both downregulated in N1KO iPSC-CMs, regardless of CHIR99021 treatment (Figure 4J and 4M; Figure S10F). Together, these results reveal the developmental stage-dependent transcriptional dynamics of defective cardiac proliferation in N1KO iPSC-CMs, which may be attributed to the dysregulation of cell cycle regulators.
Single-Cell Transcriptomic Analysis Reveals That Loss of Function in NOTCH1 Alters Cardiac Cell Lineage Determination
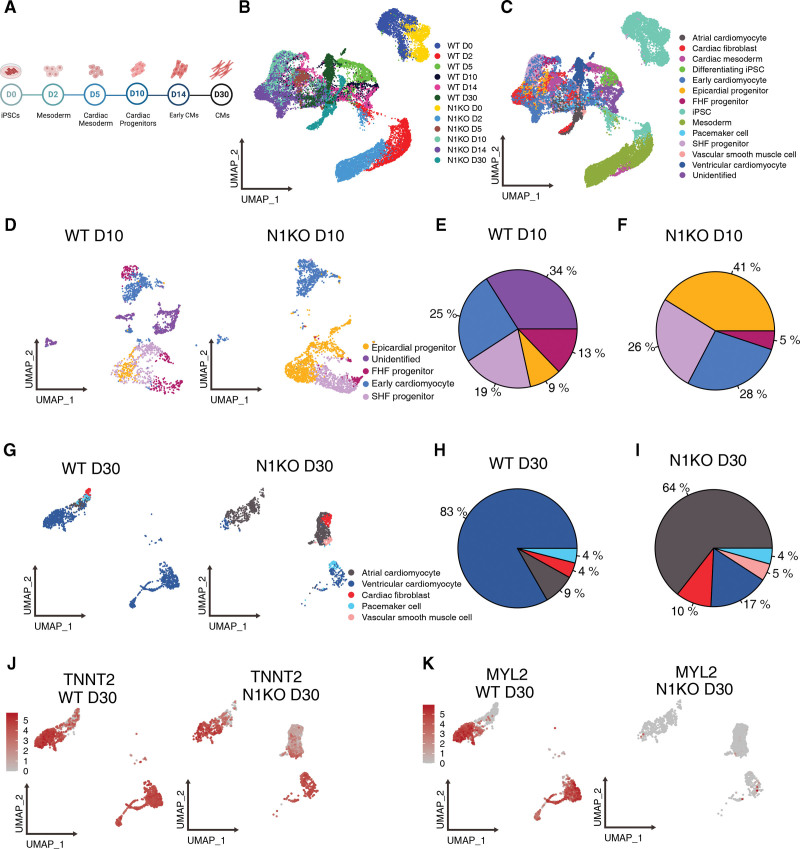
To elucidate how cardiac lineage determination is altered due to NOTCH1 disruption, we performed single-cell RNA sequencing at multiple developmental stages on differentiating cells collected at D0 (iPSC), D2 (mesoderm), D5 (cardiac mesoderm), D10 (cardiac progenitor), D14 (early cardiomyocyte), and D30 (fetal cardiomyocyte) during stepwise cardiac differentiation (Figure 5A). In total, we profiled the transcriptomes of 32 189 single cells including iPSCs, intermediate differentiating cells, and cardiomyocytes. Uniform manifold approximation and projection plot showed the topological structure of the 12 samples, with D0 and D2 cell populations distinguished from more differentiated cells (Figure 5B). Subsequently, we annotated 13 cell populations based on cell type-specific marker genes (Figure S11A): iPSC, differentiating iPSC, mesoderm, cardiac mesoderm, epicardial progenitor, first heart field (FHF) progenitor, second heart field (SHF) progenitor, early cardiomyocyte, cardiac fibroblast, atrial cardiomyocyte, ventricular cardiomyocyte, pacemaker cell, and vascular smooth muscle cell (Figure 5C). Cell-type specific markers that were used for identification of cell types are summarized in Table S4.
Figure 5.
Single-cell transcriptomic analysis uncovers that NOTCH1 disruption alters cell lineage differentiation during human cardiac differentiation. A, Schematic summary showing sample collections during cardiac differentiation of human induced pluripotent stem cells (iPSCs) for scRNA-seq. B and C, Uniform manifold approximation and projection (UMAP) plotting shows the topological structures of D0, D2, D5, D10, D14, and D30 cell populations clustered by samples (B) and cell types (C). D, UMAP plotting of D10 wild-type (WT) and NOTCH1 knockout (N1KO) cells, which include epicardial progenitors, first heart field (FHF) progenitors, second heart field (SHF) progenitors, and committed cardiomyocytes. E and F, Percentages of various cell types in WT (E) and N1KO (F) differentiating cells at D10. G, UMAP plotting of D30 WT and N1KO cells which include ventricular-like cardiomyocytes, atrial-like cardiomyocytes, pacemaker-like cells, cardiac fibroblasts, and vascular smooth muscle cells. H and I, Percentages of respective cell types in WT (H) and N1KO (I) D30 cells. J and K, UMAP plotting shows the expression of pan-cardiomyocyte marker TNNT2 and ventricular cardiomyocyte marker MYL2 in WT (J) and N1KO (K) D30 cell populations.
Next, we compared single-cell transcriptomic profiles between N1KO and WT at each developmental stage. On D2 of cardiac differentiation, the majority of cells were at mesodermal stage, which were labeled by early mesoderm marker MIXL1 (Figure S11B and S11C). Most differentiating cells started to express mesoderm markers, while a small portion (9%) of cells developed into cardiac mesoderm in WT but not in N1KO samples (Figure S11D through S11F). On D5, the majority of differentiating cells were cardiac mesoderm precursors (MESP1+PDGFRA+), whereas a small percentage of SHF progenitors (ISL1+NKX2-5+) were detected in both WT and N1KO cells (Figure S11G). More cardiac mesoderm cells were present in N1KO cells compared with WT on D5 (Figure S11H and S11I). On D10, beating cardiomyocytes and early cardiac progenitors including FHF, SHF, and epicardial progenitors were present (Figure 5D). The proportion of early cardiomyocytes was similar between N1KO and WT cells (Figure 5E and 5F). However, the percentage of SHF progenitors (19% in WT versus 26% in N1KO) and epicardial progenitors (9% in WT versus 41% in N1KO) were higher in N1KO compared with WT cells. In contrast, the proportion of FHF progenitors was lower in N1KO (5%) than WT (13%) cells (Figure 5E and 5F). Immunofluorescence staining using antibodies against these progenitor cell lineage markers confirmed the elevated percentages of epicardial and SHF progenitors and reduced percentage of FHF progenitors in N1KO versus WT D10 cells (Figure S12). Together, these results indicated that cell lineage differentiation of cardiac mesoderm was biased towards epicardial and SHF progenitors at the expense of FHF progenitors.
Cell type composition at D14 was primarily early cardiomyocytes and cardiac fibroblasts in both N1KO and WT cells (Figures S11J through S11L), which were identified by TNNT2 (Figure S11M) and DDR2 (Figure S11N), respectively. At D30, cardiomyocytes were more committed and can be classified into atrial-like, ventricular-like, and nodal-like subtypes (Figure 5G). For WT, ventricular-like cardiomyocytes were the predominant cell type (83%), with the remainder atrial-like cardiomyocytes (9%), pacemaker cells (4%), and cardiac fibroblasts (4%; Figure 5H). In contrast, atrial-like cardiomyocytes (64%) were predominant whereas ventricular-like cardiomyocytes comprised only 17% of the population in N1KO-CMs at D30 (Figure 5I). Non-cardiomyocytes, including vascular smooth muscle cells (5%) and cardiac fibroblasts (10%) were more prevalent in D30 N1KO than WT cells (Figure 5I). The different proportions of ventricular-like cardiomyocytes were identified by the co-expression of ventricle-specific marker MYL2 and pan-cardiomyocyte marker TNNT2 (Figure 5J and 5K). These results suggest that ventricular-like cardiomyocyte differentiation is compromised in early cell fate determination due to NOTCH1 disruption, which may alternatively boost atrial-like cardiomyocyte differentiation. The increase in non-cardiomyocytes (ie, vascular smooth muscle cells and cardiac fibroblasts) may be caused by the enhanced differentiation of epicardial progenitors at an earlier developmental stage (D10) in N1KO cells.
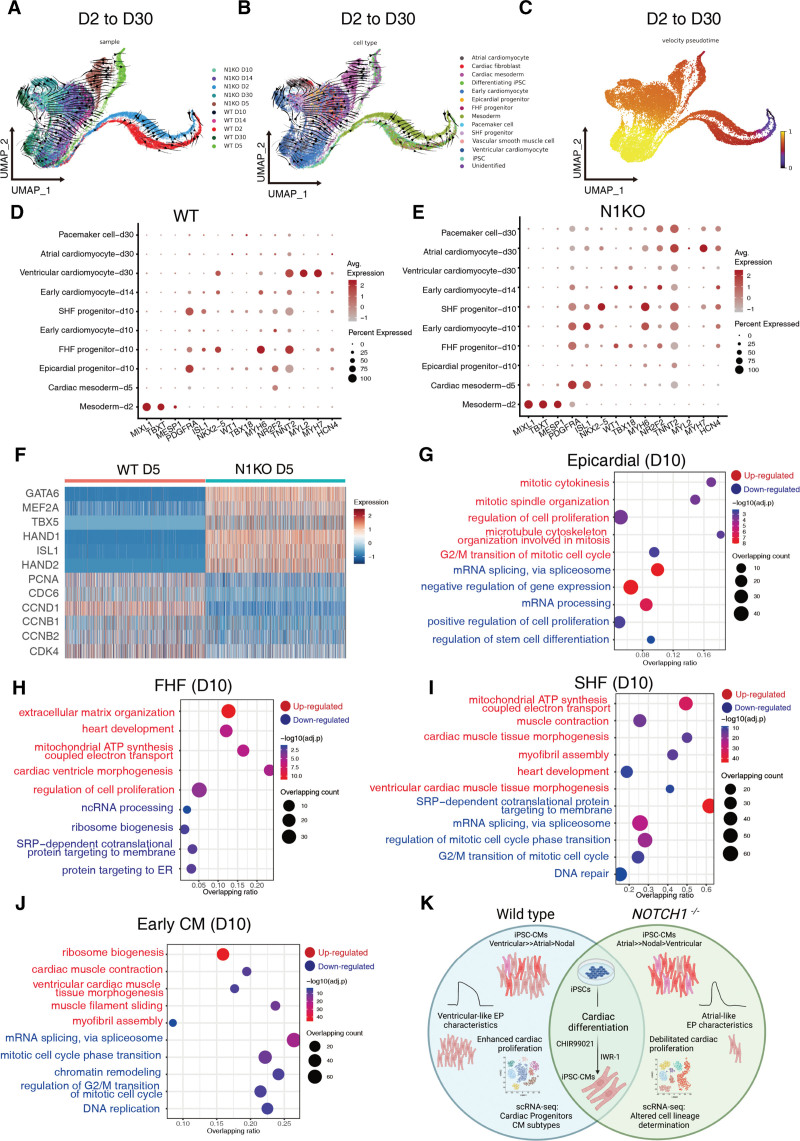
We explored how NOTCH1 disruption alters the developmental trajectory dynamics during cardiac differentiation using RNA velocity.40,41 Velocity pseudotime was inferred based on the rate of mRNA splicing during cardiac differentiation. Diffusion map prediction based on single-cell transcriptomic profiles42 in each cell type coincided with the cardiac differentiation trajectory: from undifferentiated iPSCs to mesoderm, cardiac progenitors, and terminally differentiated atrial-, ventricular-, and pacemaker-like cardiomyocytes (Figure 6A through 5C). Pseudotime order also reflected the cellular differentiation status across time points, with D30 (cardiomyocyte) and D2 (mesoderm) samples were plotted at distal ends of developmental trajectories (Figure 6C). Next, we assessed the divergent developmental trajectories between WT and N1KO cells at adjacent transition stages. Uniform manifold approximation and projection plotting showed that developmental pseudotime disparity in the first transition phase (D0 to D2; iPSCs to mesoderm) was quite distant (Figure S13A through S13C), with pluripotent stem cells and mesoderm cells located at distinct developmental states. In the second transition phase (D2 to D5), cellular differentiation paths between WT and N1KO cells converged at the end (Figure S13D through S13F). In the third transition phase (D5 to D10), developmental trajectories of WT and N1KO cells were both poised toward a more advanced stage (early cardiomyocyte; Figure S13G through S13I). In the fourth transition phase (D10 to D14), developmental trajectories converged between WT and N1KO cells and moved toward more differentiated cardiomyocytes (Figure S13J through S13L). Finally, in the fifth transition phase (D14 to D30), WT and N1KO cells followed divergent cellular differentiation pathways. D30 WT cells were at more developmentally advanced stages compared with D30 N1KO cells based on pseudotime prediction (Figure S13M through S13O). Collectively, these developmental trajectory prediction results indicate that N1KO and WT cellular differentiation routes differ in the early transition phase, converge in the middle phases, and diverge toward different mature cell types at the end of cardiac differentiation.
Figure 6.
Divergent developmental trajectories and differential gene expression profiles in cardiac mesoderm, cardiac progenitors, and early cardiomyocytes during differentiation of wild-type (WT) and NOTCH1 knockout (N1KO) induced pluripotent stem cells (iPSCs). A – C, Uniform manifold approximation and projection (UMAP) plotting of overall developmental trajectories from induced pluripotent stem cells (iPSCs) to cardiomyocytes (D2 through D30) predicted by RNA velocity and clustered by samples (A), cell types (B), and pseudotime (C). D and E, Differential gene expression levels of cell type-specific markers and percentages of marker gene-positive cells in individual cell types in WT (D) and N1KO (E) samples. F, Heatmap of representative differentially expressed genes in D5 cardiac mesoderm between WT and N1KO. G through J, Upregulated and downregulated pathways that are enriched in D10 epicardial progenitors (G), first heart field (FHF) progenitors (H), second heart field (SHF) progenitors (I), and early cardiomyocytes (J) in N1KO versus WT samples. The x-axis shows the ratio of enriched pathways versus background. The y-axis represents the term of enriched pathways that are upregulated (in red) or downregulated (in blue). The sizes of the dots indicate the number of target genes in a given pathway whereas colors of the dots reflect log-transformed adjusted P. K, Graphic summary of this study. NOTCH1 disruption leads to skewed cardiac lineage differentiation and defective cardiomyocyte proliferation, possibly through balancing the cell fate determination of cardiac mesoderm towards epicardial, FHF, and SHF lineages, and modulating mitotic cell cycle progression in the early phase of cardiomyocyte expansion.
NOTCH1 Deficiency Leads to Differential Gene Expression Profiles in Human Cardiac Mesoderm, Progenitors, Cardiomyocytes, and Cardiac Fibroblasts Across Cardiac Differentiation
Using at least 2 markers for each cell type, we found differences in the expression levels and percentages of positive cells between N1KO and WT samples (Figure 6D and 6E). We selected the top 3 highest expressed genes in each cell type (Figure S14A and S14B). Transcriptional profiles were similar in the early phase of differentiation (ie, mesoderm), then differed gradually during differentiation. Next, we examined DEGs and their associated pathways in a given cell type during cardiac differentiation between N1KO and WT cells. In D5 cardiac mesoderm (MESP1+ PDGFRA+), several transcription factors associated with cardiac development including GATA6, MEF2A, TBX5, HAND1, ISL1, and HAND2 were upregulated; in contrast, cell cycle relevant genes PCNA1, CDC6, CCND1, CCNB1, CCNB2, and CDK4 were downregulated in N1KO versus WT cells (fold change >2, FDR <0.05, Figure 6F). Gene ontology analysis showed that the top enriched pathways for upregulated genes in N1KO cells were heart development, respiratory electron transport, circulatory system development, regulation of cell migration, and cardiac ventricle morphogenesis (FDR<0.05; Figure S14C). In contrast, the top enriched pathways for downregulated genes in N1KO versus WT were ribosome biogenesis, DNA replication/repair, G1/S transition of mitotic cell cycle, and mitotic cell phase transition (Figure S14C). These results suggest that cardiogenesis initiates earlier in N1KO cardiac mesoderm and NOTCH1 deficiency leads to reduced cell proliferation ability.
At D10 of cardiac differentiation, we assessed DEGs of epicardial, FHF and SHF progenitors, and early cardiomyocytes. Epicardial progenitors (WT1+ TBX18+) contribute to coronary smooth muscle, coronary endocardium, cardiac fibroblasts, and a small portion of cardiomyocytes.43,44 Upregulated DEGs in N1KO epicardial progenitors were involved in regulation of cell proliferation, mitotic cytokinesis, G2/M transition of mitotic cell cycle, and mitotic cytokinesis (Figure 6G), implying enhanced cell proliferation capacity in epicardial progenitors due to NOTCH1 deficiency. In contrast, mRNA splicing, gene expression, and translational activity were downregulated in N1KO epicardial progenitors (Figure 6G). FHF progenitors (NKX2-5+ ISL1-) primarily give rise to the left ventricle and part of the atria (atrial myocytes) whereas SHF progenitors (NKX2-5+ ISL1+) contribute to the right ventricle (RV) and conduction myocytes, part of the atria (atrial myocytes), and outflow tract.44 Upregulated DEGs in N1KO FHF cells were associated with heart development, extracellular matrix organization, respiratory electron transport chain, and cardiac ventricle morphogenesis whereas downregulated DEGs were involved in ribosome biogenesis, ncRNA processing, and protein targeting to ER (Figure 6H). Upregulated pathways in N1KO SHF progenitors (D10) were mitochondrial ATP synthesis coupled electron transport, muscle contraction, myofibril assembly, and ventricular cardiac muscle tissue morphogenesis, whereas downregulated DEGs were associated in mRNA splicing, mitotic cell cycle phase transition, and DNA repair (Figure 6I). Next, we ran gene regulatory network analysis and identified top ranked TFs in D10 SHF progenitors by regulon specificity scores using the IRIS3 Web server. Top 3 ranked TFs were TAL1, WT1, and PITX2 for WT D10 SHF progenitors, whereas top 3 ranked TFs were IRF3, PURA, and STAT3 for N1KO D10 SHF progenitors (Figure S15A and S15B). There were 2 common TFs for WT and N1KO SHF progenitors (Figure S15C): VEZF1 and PRDM6, which play essential roles in cardiac structure and function, cardiac development, and angiogenesis during heart development.45–49 Interestingly, VEZF1 and PRDM6 regulate a group of DEGs between N1KO and WT SHF cells, including upregulated genes associated with cardiac structure and physiological functions (MYBPC3, MYH6, ACTN2, and KCNIP2) and downregulated genes involved in cell cycle regulation (TOP2A, BIRC5, and CENPF). Taken together, these data suggest that NOTCH1 disruption triggers enhanced cell proliferation in epicardial progenitors and promotes cardiac morphogenesis and cellular differentiation in both FHF and SHF progenitors.
Next, we probed gene expression differences in committed cardiomyocytes (D10), early cardiomyocytes (D14), and fetal cardiomyocytes (D30). For committed cardiomyocytes (D10), upregulated DEGs in N1KO cells were related to ribosome biogenesis, cardiac muscle contraction, muscle filament sliding, and myofibril assembly; downregulated pathways in N1KO were involved in mRNA splicing via spliceosome, mitotic cell cycle phase transition, DNA replication, and regulation of G2/M transition of mitotic cell cycle (Figure 6J). For early cardiomyocytes (D14), top-enriched pathways in upregulated DEGs in N1KO cells were associated with extracellular matrix organization, regulation of cell migration, heart development, Wnt signaling pathway, and ventricular septum development, whereas downregulated DEGs in N1KO were involved in muscle contraction, muscle filament sliding, and myofibril assembly (Figure S14D). For D30 atrial-like cardiomyocytes (NR2F2+MYH6+TNNT2+), upregulated pathways were mitochondrial ATP synthesis coupled electron transport, Wnt signaling pathway, mitochondrial translational elongation, muscle contraction, and negative regulation of mitotic cell cycle phase transition, whereas downregulated pathways included mRNA splicing, chromatin remodeling, and mRNA processing in N1KO versus WT (Figure S14E). In N1KO ventricular-like cardiomyocytes (MYL2+MYH7+TNNT2+), increased pathway activity included extracellular matrix organization, cardiac right ventricle morphogenesis, and atrial septum morphogenesis, and decreased pathway activity in N1KO included ribosome biogenesis, respiratory electron transport chain, cardiac muscle contraction, and ventricular cardiac muscle tissue development (Figure S14F). N1KO D30 nodal-like cardiomyocytes (HCN4+TNNT2+) had upregulated pathways involved in mitochondrial ATP synthesis coupled electron transport, muscle contraction, mitochondrial translational elongation, and heart development, and downregulated pathways involved in mRNA splicing, mRNA processing, mRNA transport, and translation (Figures S14G). Taken together, these results suggest that NOTCH1 disruption leads to decreased cell cycle progression and cell proliferation in early cardiomyocytes, elevated mitochondrial ATP synthesis in atrial- and nodal-like cardiomyocytes, and enhanced cardiac right ventricle morphogenesis in ventricular-like cardiomyocytes, respectively.
Cardiac fibroblasts are essential for maintaining heart function and cardiac remodeling.50 Thus, we surveyed gene expression prolife changes in cardiac fibroblasts caused by NOTCH1 deficiency. Despite comprising a minor fraction of cardiac cell types identified on D14 and D30, cardiac fibroblasts might be required for cardiomyocyte proliferation and differentiation. Upregulated DEGs in N1KO D14 cardiac fibroblasts (DDR2+ TCF21+) were primarily associated with extracellular matrix organization, cardiac ventricle morphogenesis, ventricular septum development, and outflow tract septum morphogenesis, whereas downregulated DEGs were involved in mRNA splicing, mitotic sister chromatic segregation, mitotic cell cycle phase transition, mitotic cytokinesis, and centromere complex assembly (Figure S14H). Upregulated pathways in N1KO D30 cardiac fibroblasts included mitochondrial ATP synthesis coupled electron transport, ribosome biogenesis, extracellular matrix organization, and regulation of apoptotic process, whereas downregulated pathways were involved in mRNA splicing, mRNA processing, chromatin remodeling, cell cycle regulation, and DNA repair (Figure S14I). Collectively, these data indicate that cardiac fibroblasts share common pathways (enhanced outflow tract morphogenesis, mitochondrial ATP synthesis, and extracellular matrix organization) with cardiomyocytes and that both were impacted by defects in mitotic cell cycle progression and cell proliferation at their respective developmental stages due to NOTCH1 dysfunction.
Discussion
In this study, we demonstrate that NOTCH1 is dispensable for mesodermal and cardiovascular differentiation of human iPSCs. NOTCH1 disruption leads to enhanced atrial-like cardiomyocyte differentiation but compromised ventricular-like cardiomyocyte generation from human iPSCs. In addition, NOTCH1 deficiency causes defective cardiomyocyte proliferation and downregulated pathways associated with mitotic cell cycle progression and cellular proliferation in N1KO cardiomyocytes. Single-cell transcriptomic analysis indicates skewed differentiation of epicardial, FHF, and SHF progenitors from cardiac mesoderm due to NOTCH1 disruption. Developmental trajectory prediction suggests that cellular differentiation routes converge and diverge throughout cardiac differentiation between N1KO and WT cells. Our study provides novel insights into the mechanisms by which NOTCH1 mutations impair human ventricular-like cardiomyocyte differentiation and proliferation, suggesting a potential mechanism for the left ventricular hypoplasia in HLHS.
In mammals, the NOTCH signaling pathway plays essential roles in patterning embryonic endocardium into prospective regions for valve and cardiac chamber development in early embryonic stages, and modulating the late developmental processes involving cardiac valve morphogenesis, outflow tract formation, and trabeculae compaction in the ventricular chamber.51 Defined roles of NOTCH1 in human early heart development remain unknown. Here, using single-cell transcriptomic analysis, we have found that NOTCH1 disruption promotes epicardial and SHF cell fate determination, but inhibits the differentiation of FHF progenitors from cardiac mesoderm. A recent study shows that noncanonical NOTCH signaling pathway suppresses SHF formation in the precardiac mesoderm of mouse embryos.52 NOTCH1 inhibition within SHF leads to abnormal migration of cardiac neural crest cells and defective endothelial-mesenchymal transition within the outflow tract endocardial cushions.53 Epicardial NOTCH1 is required for coronary artery development and smooth muscle cell differentiation.54,55 In this study, NOTCH1 deficiency not only stimulates human epicardial lineage differentiation but also promotes the proliferation of epicardial cells, suggesting a cell type-specific response to NOTCH signaling during embryonic heart development. The enhanced epicardial progenitor differentiation and proliferation in N1KO cardiac mesoderm may underlie the abnormal coronary artery in HLHS.56,57
In this study, NOTCH1 disruption leads to enhanced atrial-like cardiomyocyte differentiation, compromised ventricular-like cardiomyocyte differentiation, and shortened action potential duration. During embryonic heart development, NOTCH signaling exhibits stage-specific effects on ventricular chamber development including trabeculation and compaction.10 Early studies indicated that NOTCH pathway is a crucial regulator of cardiac differentiation in human embryonic stem cells.58 Chemical inhibition of NOTCH signaling promotes cardiac mesoderm differentiation and cardiomyocyte generation of human and murine PSCs.59,60 NOTCH1 disruption in cardiac differentiation leads to upregulation of the transcription factor COUP-TFII that promotes vein identity in the vasculature development61 and determines the atrial identity in the heart.62 In vitro deletion of the NOTCH pathway downstream target gene HEY2 significantly increases the proportion of atrial-like cardiomyocytes during cardiac differentiation of human iPSCs.63 Our findings highlight that modulation of NOTCH pathway could be potentially used for enriching atrial- or ventricular-like cardiomyocytes from patient-specific iPSCs.28 Conditional NOTCH1 knockout and expression rescue experiments in human iPSCs would be the next step to illustrate how NOTCH1 impacts cardiac cell lineage determination and cardiomyocyte proliferation at different developmental stages.
Originally identified as a proto-oncogene in T-cell acute lymphoblastic leukemia, NOTCH1 can serve as both an oncogene and a tumor suppressor in cancer development.64 NOTCH signaling modulates G1/S phase progression in human T cells through regulating cyclin D3, CDK4, and CDK6.65 In this study, we uncover that NOTCH1 disruption leads to defective cell proliferation and mitotic cell cycle progression in cardiac mesoderm, cardiac progenitors, and early cardiomyocytes. During mammalian heart development, endocardial NOTCH intracellular domain activation induces BMP10 in the underlying myocardium, which promotes cardiomyocyte proliferation by suppressing the cell cycle inhibitor, p57. Conditional deletion of Notch1 in the endocardium of mouse embryos leads to defective cardiomyocyte proliferation and impairs cardiac differentiation in ventricular chambers.7,8 Ectopic Notch activation in neonatal cardiomyocytes triggers cell cycle reentry and proliferation through transcriptional regulation of cyclin D, whereas Notch inhibition prevents the proliferation of embryonic cardiomyocytes through apoptosis.66,67 Following studies reveal that non-canonical NOTCH signaling interacts with Wnt pathway to regulate the proliferation and differentiation of cardiac progenitors in mouse embryos.68,69 Notch1 deficiency results in enhanced proliferation Isl1+ cardiac progenitors and inhibits their cardiac differentiation, which can be rescued by stabilizing β-catenin.70 However, WNT pathway activation by CHIR99021 can’t stimulate the proliferation of N1KO iPSC-CMs (Figure 4). Therefore, we speculate that RBPJ/CBF1 dependent canonical NOTCH pathway may be involved in the cell cycle regulation in human early cardiomyocytes.
In HLHS, several structures on the left side of the heart are not full developed, including the left ventricle, mitral valve, aortic valve, and ascending portion of the aorta. Pathogenic NOTCH1 variants seem to be responsible for a spectrum of developmental defects in the left side of the heart in HLHS.11 Our study provides informative insights on how NOTCH1 dysfunction may contribute to HLHS. (1) We reveal that NOTCH1 disruption suppresses ventricular-like cardiomyocyte differentiation and inhibits cardiomyocyte proliferation, which could contribute to left ventricle hypoplasia in HLHS patients with NOTCH1 mutations. (2) NOTCH1 knockout disrupts the differentiation of cardiac mesoderm and partially blocks the generation of FHF progenitors which primarily contribute to left ventricle. This could help explain the developmental etiology of HLHS. (3) Genome-edited human iPSC lines could provide a versatile platform to investigate how endocardial-myocardial communication is dysregulated in HLHS due to NOTCH1 mutations. Future studies using HLHS patient-derived iPSCs with NOTCH1 mutations are needed to elucidate detailed cellular and developmental etiologies of HLHS.
In conclusion, NOTCH1 is required for human ventricular-like cardiomyocyte differentiation through balancing cell fate determination of early cardiac mesoderm toward epicardial, FHF, and SHF lineages and by limiting atrial-like cardiomyocyte generation (Figure 6K). In addition, NOTCH1 modulates cell cycle progression and proliferation of cardiac progenitors and early cardiomyocytes during human cardiac differentiation, possibly through a RBPJ dependent canonical NOTCH pathway. Given that NOTCH1 variants are linked to HLHS and aortic valve disease, our data will provide insights into the underlying genetic and developmental mechanisms by which NOTCH1 dysfunction contributes to ventricular hypoplasia and abnormal structures of outflow tract in congenital heart disease.
Limitations of Study
While this study using genome-edited N1KO iPSCs provides some mechanistic insights on how disruption of NOTCH1 interferes with human cardiac cell lineage determination and cardiomyocyte proliferation, future studies using HLHS patient-specific iPSCs that harbor pathogenic variants in NOTCH1 could provide more direct insights about the genetic contribution of pathogenic variation in NOTCH1 to the pathogenesis of HLHS. Furthermore, we acknowledge the limitations of using monolayers of iPSC-derived cardiomyocytes to study normal cardiac development and congenital heart disease. Future work using iPSC-derived 3D cardiac organoids is warranted to elucidate the 3D structural mechanisms of cardiac cell lineage commitment and intercellular communication under normal and diseased conditions.
Article Information
Acknowledgments
We would like to thank Dr Dennis Lewandowski in the Heart Center at Nationwide Children’s Hospital for critically reading and editing this article.
Sources of Funding
This work was partially supported by the American Heart Association (AHA) Career Development Award 18CDA34110293 (M-T. Zhao), NIH/NHLBI R01HL155282 (M-T. Zhao), R21HL165406 (M-T. Zhao), NIH Diversity Supplement R01HL155282-02S1 (M-T. Zhao and M. Alonzo), R01HL096962 (I. Deschenes), R01HL132520 (I. Deschenes), R01GM131399 (Q.M.), R01HL163680 (J.C. Wu), R01HL146690, R01HL126527 (J.C. Wu), and R01 HL130020 (J.C. Wu), Additional Ventures Innovation Fund (AVIF) and Single Ventricle Research Fund (SVRF; V. Garg and M-T. Zhao). This work was also supported by the Startup Fund from the Abigail Wexner Research Institute at Nationwide Children’s Hospital (M-T. Zhao).
Disclosures
None.
Supplemental Material
Supplemental Methods
Tables S1–S4
Figures S1–S15
Supplementary Material
Nonstandard Abbreviations and Acronyms
- APD
- action potential duration
- DEG
- differentially expressed genes
- FHF
- first heart field
- HLHS
- hypoplastic left heart syndrome
- iPSC
- induced pluripotent stem cell
- iPSC-CM
- iPSC-derived cardiomyocyte
- iPSC-EC
- iPSC-derived endothelial cell
- N1KO
- NOTCH1 knockout
- SHF
- second heart field
- WT
- wild type
S. Ye, C. Wang, and Z. Xu contributed equally.
For Sources of Funding and Disclosures, see page 202.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.122.321398.
References
- 1.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- 2.Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical Notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017 [DOI] [PubMed] [Google Scholar]
- 4.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol.2012;4:a011213. doi: 10.1101/cshperspect.a011213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279 [DOI] [PubMed] [Google Scholar]
- 6.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacGrogan D, Munch J, de la Pompa JL. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol. 2018;15:685–704. doi: 10.1038/s41569-018-0100-2 [DOI] [PubMed] [Google Scholar]
- 8.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell.2009;138:257–270. doi: 10.1016/j.cell.2009.04.060 [DOI] [PubMed] [Google Scholar]
- 10.D’Amato G, Luxan G, del Monte-Nieto G, Martinez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20 doi: 10.1038/ncb3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17:2886–2893. doi: 10.1093/hmg/ddn187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin MD, Cadar AG, Williams CH, Guo Y, Bichell DP, Su YR, Hong CC. Hypoplastic left heart syndrome sequencing reveals a novel NOTCH1 mutation in a family with single ventricle defects. Pediatr Cardiol. 2017;38:1232–1240. doi: 10.1007/s00246-017-1650-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luxan G, Casanova JC, Martinez-Poveda B, Prados B, D’Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046 [DOI] [PubMed] [Google Scholar]
- 14.Mai CT, Isenburg JL, Canfield MA, Meyer RE, Correa A, Alverson CJ, Lupo PJ, Riehle-Colarusso T, Cho SJ, Aggarwal D, et al. ; National Birth Defects Prevention NetworkNational Birth Defects Prevention Network. National population-based estimates for major birth defects, 2010-2014. Birth Defects Res.2019;111:1420–1435. doi: 10.1002/bdr2.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barron DJ, Kilby MD, Davies B, Wright JG, Jones TJ, Brawn WJ. Hypoplastic left heart syndrome. Lancet.2009;374:551–564. doi: 10.1016/S0140-6736(09)60563-8 [DOI] [PubMed] [Google Scholar]
- 16.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature.2005;437:270–274. doi: 10.1038/nature03940 [DOI] [PubMed] [Google Scholar]
- 17.Riley MF, McBride KL, Cole SE. NOTCH1 missense alleles associated with left ventricular outflow tract defects exhibit impaired receptor processing and defective EMT. Biochim Biophys Acta. 2011;1812:121–129. doi: 10.1016/j.bbadis.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theis JL, Hrstka SC, Evans JM, O’Byrne MM, de Andrade M, O’Leary PW, Nelson TJ, Olson TM. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum Genet. 2015;134:1003–1011. doi: 10.1007/s00439-015-1582-1 [DOI] [PubMed] [Google Scholar]
- 19.Grossfeld P, Nie S, Lin L, Wang L, Anderson RH. Hypoplastic left heart syndrome: a new paradigm for an old disease?. J Cardiovasc Dev Dis.2019;6:10. doi: 10.3390/jcdd6010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707 [DOI] [PubMed] [Google Scholar]
- 21.Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352 doi: 10.1101/gad.14.11.1343 [PMC free article] [PubMed] [Google Scholar]
- 22.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc.2013;8:2281–2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aranguren XL, Beerens M, Coppiello G, Wiese C, Vandersmissen I, Lo Nigro A, Verfaillie CM, Gessler M, Luttun A. COUP-TFII orchestrates venous and lymphatic endothelial identity by homo- or hetero-dimerisation with PROX1. J Cell Sci. 2013;126:1164–1175. doi: 10.1242/jcs.116293 [DOI] [PubMed] [Google Scholar]
- 24.Sissaoui S, Yu J, Yan A, Li R, Yukselen O, Kucukural A, Zhu LJ, Lawson ND. Genomic characterization of endothelial enhancers reveals a multifunctional role for NR2F2 in regulation of arteriovenous gene expression. Circ Res. 2020;126:875–888. doi: 10.1161/CIRCRESAHA.119.316075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page DJ, Miossec MJ, Williams SG, Monaghan RM, Fotiou E, Cordell HJ, Sutcliffe L, Topf A, Bourgey M, Bourque G, et al. Whole exome sequencing reveals the major genetic contributors to nonsyndromic tetralogy of fallot. Circ Res. 2019;124:553–563. doi: 10.1161/CIRCRESAHA.118.313250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao MT, Chen H, Liu Q, Shao NY, Sayed N, Wo HT, Zhang JZ, Ong SG, Liu C, Kim Y, et al. Molecular and functional resemblance of differentiated cells derived from isogenic human iPSCs and SCNT-derived ESCs. Proc Natl Acad Sci USA. 2017;114:E11111–E11120. doi: 10.1073/pnas.1708991114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Protze SI, Lee JH, Keller GM. Human pluripotent stem cell-derived cardiovascular cells: from developmental biology to therapeutic applications. Cell Stem Cell.2019;25:311–327. doi: 10.1016/j.stem.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 28.Zhao MT, Shao NY, Garg V. Subtype-specific cardiomyocytes for precision medicine: where are we now?. Stem Cells. 2020;38:822–833. doi: 10.1002/stem.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rentschler S, Yen AH, Lu J, Petrenko NB, Lu MM, Manderfield LJ, Patel VV, Fishman GI, Epstein JA. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation.2012;126:1058–1066. doi: 10.1161/CIRCULATIONAHA.112.103390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khandekar A, Springer S, Wang W, Hicks S, Weinheimer C, Diaz-Trelles R, Nerbonne JM, Rentschler S. Notch-mediated epigenetic regulation of voltage-gated potassium currents. Circ Res. 2016;119:1324–1338. doi: 10.1161/CIRCRESAHA.116.309877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JZ, Termglinchan V, Shao NY, Itzhaki I, Liu C, Ma N, Tian L, Wang VY, Chang ACY, Guo H, et al. A human iPSC double-reporter system enables purification of cardiac lineage subpopulations with distinct function and drug response profiles. Cell Stem Cell.2019;24:802–811.e5. doi: 10.1016/j.stem.2019.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye S, Wan X, Su J, Patel A, Justis B, Deschenes I, Zhao MT. Generation and expansion of human cardiomyocytes from patient peripheral blood mononuclear cells. J Vis Exp.2021; 168:e62206. doi: 10.3791/62206 [DOI] [PubMed] [Google Scholar]
- 33.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim LA, Furst J, Butler MH, Xu S, Grigorieff N, Goldstein SA. Ito channels are octomeric complexes with four subunits of each Kv4.2 and K+ channel-interacting protein 2. J Biol Chem. 2004;279:5549–5554. doi: 10.1074/jbc.M311332200 [DOI] [PubMed] [Google Scholar]
- 35.Bahring R, Dannenberg J, Peters HC, Leicher T, Pongs O, Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J Biol Chem. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200 [DOI] [PubMed] [Google Scholar]
- 36.Foeger NC, Marionneau C, Nerbonne JM. Co-assembly of Kv4 {alpha} subunits with K+ channel-interacting protein 2 stabilizes protein expression and promotes surface retention of channel complexes. J Biol Chem. 2010;285:33413–33422. doi: 10.1074/jbc.M110.145185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleischmann BK, Duan Y, Fan Y, Schoneberg T, Ehlich A, Lenka N, Viatchenko-Karpinski S, Pott L, Hescheler J, Fakler B. Differential subunit composition of the G protein-activated inward-rectifier potassium channel during cardiac development. J Clin Invest. 2004;114:994–1001. doi: 10.1172/JCI15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, et al. Dynamics of cell generation and turnover in the human heart. Cell.2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 39.Buikema JW, Lee S, Goodyer WR, Maas RG, Chirikian O, Li G, Miao Y, Paige SL, Lee D, Wu H, et al. Wnt activation and reduced cell-cell contact synergistically induce massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell.2020;27:50–63.e5. doi: 10.1016/j.stem.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. RNA velocity of single cells. Nature.2018;560:494–498. doi: 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 42.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics.2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325 [DOI] [PubMed] [Google Scholar]
- 43.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature.2008;454:109–113. doi: 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. Circ Res. 2015;116:341–353. doi: 10.1161/CIRCRESAHA.116.302752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyashita H, Kanemura M, Yamazaki T, Abe M, Sato Y. Vascular endothelial zinc finger 1 is involved in the regulation of angiogenesis: possible contribution of stathmin/OP18 as a downstream target gene. Arterioscler Thromb Vasc Biol. 2004;24:878–884. doi: 10.1161/01.ATV.0000126373.52450.32 [DOI] [PubMed] [Google Scholar]
- 46.Gewies A, Castineiras-Vilarino M, Ferch U, Jahrling N, Heinrich K, Hoeckendorf U, Przemeck GK, Munding M, Gross O, Schroeder T, et al. Prdm6 is essential for cardiovascular development in vivo. PLoS One. 2013;8:e81833 doi: 10.1371/journal.pone.0081833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.AlAbdi L, He M, Yang Q, Norvil AB, Gowher H. The transcription factor Vezf1 represses the expression of the antiangiogenic factor Cited2 in endothelial cells. J Biol Chem. 2018;293:11109–11118. doi: 10.1074/jbc.RA118.002911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paavola J, Alakoski T, Ulvila J, Kilpio T, Siren J, Perttunen S, Narumanchi S, Wang H, Lin R, Porvari K, et al. Vezf1 regulates cardiac structure and contractile function. EBioMedicine.2020;51:102608. doi: 10.1016/j.ebiom.2019.102608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong L, Li N, Gasque V, Mehta S, Ye L, Wu Y, Li J, Gewies A, Ruland J, Hirschi KK, et al. Prdm6 controls heart development by regulating neural crest cell differentiation and migration. JCI Insight.2022;7:e156046. doi: 10.1172/jci.insight.156046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivey MJ, Tallquist MD. Defining the cardiac fibroblast. Circ J. 2016;80:2269–2276. doi: 10.1253/circj.CJ-16-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luxan G, D’Amato G, MacGrogan D, de la Pompa JL. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118:e1–e18. doi: 10.1161/CIRCRESAHA.115.305350 [DOI] [PubMed] [Google Scholar]
- 52.Miyamoto M, Andersen P, Sulistio E, Liu X, Murphy S, Kannan S, Nam L, Miyamoto W, Tampakakis E, Hibino N, et al. Noncanonical Notch signals have opposing roles during cardiac development. Biochem Biophys Res Commun. 2021;577:12–16. doi: 10.1016/j.bbrc.2021.08.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.del Monte G, Casanova JC, Guadix JA, MacGrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL. Differential Notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ Res. 2011;108:824–836. doi: 10.1161/CIRCRESAHA.110.229062 [DOI] [PubMed] [Google Scholar]
- 55.Grieskamp T, Rudat C, Ludtke TH, Norden J, Kispert A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ Res. 2011;108:813–823. doi: 10.1161/CIRCRESAHA.110.228809 [DOI] [PubMed] [Google Scholar]
- 56.Baffa JM, Chen SL, Guttenberg ME, Norwood WI, Weinberg PM. Coronary artery abnormalities and right ventricular histology in hypoplastic left heart syndrome. J Am Coll Cardiol. 1992;20:350–358. doi: 10.1016/0735-1097(92)90101-r [DOI] [PubMed] [Google Scholar]
- 57.Crucean A, Alqahtani A, Barron DJ, Brawn WJ, Richardson RV, O’Sullivan J, Anderson RH, Henderson DJ, Chaudhry B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J Rare Dis. 2017;12:138. doi: 10.1186/s13023-017-0683-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a [DOI] [PubMed] [Google Scholar]
- 59.Jang J, Ku SY, Kim JE, Choi K, Kim YY, Kim HS, Oh SK, Lee EJ, Cho HJ, Song YH, et al. Notch inhibition promotes human embryonic stem cell-derived cardiac mesoderm differentiation. Stem Cells. 2008;26:2782–2790. doi: 10.1634/stemcells.2007-1053 [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Li P, Liu K, He Q, Han S, Sun X, Li T, Shen L. Timely inhibition of Notch signaling by DAPT promotes cardiac differentiation of murine pluripotent stem cells. PLoS One. 2014;9:e109588. doi: 10.1371/journal.pone.0109588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature.2005;435:98–104. doi: 10.1038/nature03511 [DOI] [PubMed] [Google Scholar]
- 62.Wu SP, Cheng CM, Lanz RB, Wang T, Respress JL, Ather S, Chen W, Tsai SJ, Wehrens XH, Tsai MJ, et al. Atrial identity is determined by a COUP-TFII regulatory network. Dev Cell. 2013;25:417–426. doi: 10.1016/j.devcel.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen SY, Chen WY, Chetal K, et al. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat Commun. 2018;9:4906. doi: 10.1038/s41467-018-07333-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood.2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, Osborne BA. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood.2009;113:1689–1698. doi: 10.1182/blood-2008-03-147967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campa VM, Gutierrez-Lanza R, Cerignoli F, Diaz-Trelles R, Nelson B, Tsuji T, Barcova M, Jiang W, Mercola M. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J Cell Biol. 2008;183:129–141. doi: 10.1083/jcb.200806104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Collesi C, Zentilin L, Sinagra G, Giacca M. Notch1 signaling stimulates proliferation of immature cardiomyocytes. J Cell Biol. 2008;183:117–128. doi: 10.1083/jcb.200806091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22:257–265. doi: 10.1016/j.tcb.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alkan F, Wenzel A, Anthon C, Havgaard JH, Gorodkin J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol. 2018;19:177. doi: 10.1186/s13059-018-1534-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joachim A, Ye S, Zhao MT. Generation of cardiomyocytes and endothelial cells from human iPSCs by chemical modulation of Wnt signaling. Methods Mol Biol. 2022;2549:335–344. doi: 10.1007/7651_2021_427 [DOI] [PubMed] [Google Scholar]
- 73.Chamberland S, Yang HH, Pan MM, Evans SW, Guan S, Chavarha M, Yang Y, Salesse C, Wu H, Wu JC, et al. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife.2017;6:e25690. doi: 10.7554/eLife.25690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal.2011;17:10. doi: 10.14806/ej.17.1.200 [Google Scholar]
- 75.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics.2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics.2014;30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 78.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. Integrated analysis of multimodal single-cell data. Cell.2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97 doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics.2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 82.Ma A, Wang C, Chang Y, Brennan FH, McDermaid A, Liu B, Zhang C, Popovich PG, Ma Q. IRIS3: integrated cell-type-specific regulon inference server from single-cell RNA-Seq. Nucleic Acids Res. 2020;48:W275–W286. doi: 10.1093/nar/gkaa394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Detailed methods and Major Resource Table are provided in the Supplemental Material. Data will be made available upon reasonable request, by contacting the corresponding author. Raw and processed RNA-seq and single-cell RNA sequencing data are available in the NCBI GEO database (GSE195559 and GSE196632). The graphic abstract was generated using BioRender.com.