Abstract
The association of Porphyromonas gingivalis to periodontal disease is not clearly understood. Similar proportions of P. gingivalis may be cultivated from both inactive and actively degrading periodontal pockets. Differences in virulence among strains of P. gingivalis exist, but the molecular reason for this remains unknown. We examined the population structure of P. gingivalis to obtain a framework in which to study pathogenicity in relation to evolution. Phylogenetic trees derived from the sequencing of fragments of four housekeeping genes, ahp, thy, rmlB, and infB, in 57 strains were completely different with no correlation between clustering of strains in the four dendrograms. Combining the various alleles of the four gene fragments sequenced resulted in 41 different sequence types. The index of association, IA, based on a single representative of each sequence type was 0.143 ± 0.202, indicating a population at linkage equilibrium. Inclusion of all isolates for the calculation of IA resulted in a value of 0.206 ± 0.171. This suggests an epidemic population structure supported by the finding of genetically identical strains in different parts of the world. We observed a random distribution of two virulence-associated mobile genetic elements, the ragB locus and the insertion sequence IS1598, among 132 strains tested. In conclusion, P. gingivalis has a nonclonal population structure characterized by frequent recombination. Our study suggests that particular genotypes, possibly with increased pathogenic potential, may spread successfully in the human population.
The close association of Porphyromonas gingivalis with human periodontal disease has been demonstrated in numerous studies (37). The almost complete absence by cultivation of this organism at healthy gingival sites has led some researchers to consider it a specific pathogen. Recently developed, more-sensitive techniques such as PCR, however, have demonstrated the presence of P. gingivalis at subcultivable levels at healthy gingival sites in adults, as well as in children (18, 22). In addition, P. gingivalis may be present at almost similar proportions in periodontal pockets undergoing destruction and pockets with unaltered pocket depth (2.5 and 1% of the cultivable flora, respectively) (7). Theoretically, this could be indicative of a heterogeneous species with subpopulations of low and high pathogenicity. Indeed, numerous studies have revealed variations among strains with regard to pathogenicity in experimental infections (8, 9, 12, 17, 21) and in the expression of potential virulence factors (16, 36, 38, 39, 40). In addition, specific genotypes of the fimA gene encoding fimbrillin and specific heteroduplex types of the ribosomal intergenic spacer region have been linked to periodontal disease in humans (3, 10). In order to evaluate whether differences in pathogenicity and virulence factors in P. gingivalis reflect the existence of particularly pathogenic evolutionary lineages, it is necessary to obtain information on the basic population structure of the species.
Bacterial species with a clonal population structure are characterized by a strong linkage disequilibrium caused by nonrandom association of alleles at different loci. In such populations the mutation rate is high compared to the rate of interstrain recombination, i.e., horizontal gene transfer does not occur frequently enough to break up clonal associations (24). Some studies of traditional, exogenous pathogens have revealed significant differences in the disease association of particular clones (5, 26, 27, 28, 31). Thus, for Haemophilus influenzae, Bordetella, and Staphylococcus aureus disease, even on a worldwide basis, is caused by a limited number of clones (26, 27, 28). The remaining part of the population may act as opportunistic pathogens or is seldom associated with disease. Provided there is a strict clonal population structure, any phenotypic trait associated with the virulent clones may identify them and thereby facilitate diagnosis. For a bacterial species exhibiting a nonclonal population structure evolutionary lineages are broken up due to frequent interstrain recombination relative to mutations. This may result in a random or near-random distribution of virulence-associated alleles of particular genes in the population. Consequently, the search for markers of particularly virulent strains must concentrate on individual genes or phenotypic traits responsible for virulence (15).
In two previous studies of P. gingivalis using multilocus enzyme electrophoresis (MLEE) and DNA fingerprinting, it was concluded that the population structure is clonal based on the finding of the same genotype of P. gingivalis in geographically widespread locations (21, 25). However, the significance of recovery of strains with an identical genotype is highly dependent on the differentiating power of the typing method used. In the case of MLEE, further analysis is necessary to verify whether the identical electophoretic type in two isolates represents a combination of frequent alleles rather than the same clone (23). Besides, in an epidemic species with a panmictic population structure such as that of Neisseria meningitidis, the repeated isolation of a particular genotype may be caused by its epidemic dissemination and does not reflect the long-term evolution of the species (1, 5). An indication of clonality is if the index of association (IA), which is a measure of linkage disequilibrium among alleles of different loci, is significantly different from zero, indicating that particular alleles co-occur (24). Another strong evidence for a clonal population structure is if similar or identical phylogenetic trees are derived from sequencing of different genes located at different parts of the chromosome (19, 23). This has been demonstrated in populations of Salmonella spp. (30).
The aim of the present study was to investigate to what extent recombination affects the population structure of P. gingivalis by comparing phylogenetic trees derived from sequencing fragments of four housekeeping genes. This was combined with an analysis of the prevalence and distribution of two genes with reported association to pathogenicity: the ragB locus contained within a newly discovered pathogenicity island of P. gingivalis (6, 11) and the insertion sequence IS1598 (35).
MATERIALS AND METHODS
Bacterial strains.
This study included a total of 132 different strains of P. gingivalis. Five strains (ATCC 33277T, ATCC 49417, 10-2-1, W50, and HG 184) were represented twice, one (W83) was represented three times, and one (381) was represented four times by cultures received from different laboratories. All strains were from human sources, the majority being from periodontally diseased subjects and four strains were from gingivally healthy sites. The strains had been isolated in 22 states or countries located on four continents. Each strain was originally identified as P. gingivalis by the investigator from whom it was obtained. Upon receipt, each strain was given a PGF number. They were revived on BGA agar plates (as described in reference 13, modified by the use of freeze-thawed blood and the omission of antibiotic according to J. Carlsson, Umeå, Sweden [personal communication]). All strains were subjected to Southern blot analysis and tested for presence of the ragB locus and the insertion sequence IS1598. Sequence analyses were performed on 57 strains and on 4 duplicate strains, the designations, isolation site, and geographic origin of which are given in Results.
Southern blot analysis.
Whole-cell DNA was prepared from a 14-ml liquid culture of plaque medium (14) as described previously (32), except that lysozyme treatment was omitted. The quality and concentration of DNA was determined by agarose gel electrophoresis. Approximately 2 μg of total cellular DNA was subjected to restriction endonuclease digestion with MspI (Boehringer Mannheim GmbH, Mannheim, Germany) according to the manufacturer's instructions. During the last 30 min of digestion 0.05 μg of DNase-free RNase (Boehringer Mannheim) was added. The DNA fragments were subjected to overnight electrophoresis in 1% agarose gels at 1.5 V/cm in TAE buffer and visualized by staining with ethidium bromide (34). The patterns obtained were used in the DNA fingerprinting analysis. The DNA in the gel was blotted onto Nytran nylon membranes (Schleicher & Schuell, Dassel, Germany) as described previously (34). Hybridization was carried out as described by Sambrook et al. (34), except that the filters were soaked in 1% (vol/vol) Triton X-100 prior to prehybridization and 0.02% sodium pyrophosphate was included in all solutions. The same filters were used for hybridization with each of the two probes, and the filters were stripped between hybridization experiments by immersing in 1 liter of boiling 0.1% sodium dodecyl sulfate (SDS) and then left to cool for 15 min. The probes used for hybridizations (see below) were labeled with [32P]dCTP using a random-primed DNA labeling kit (Boehringer Mannheim). After hybridization the filters were washed in 1× SET (0.15 M NaCl; 0.5 mM EDTA; 20 mM Tris-HCl, pH 7.0)–0.1% SDS–0.1% sodium pyrophosphate at 60°C. Probes for hybridization were prepared by PCR. The primer set for part of the ragB locus was 5′-GACTCTTGGCTCGTTTACTG-3′ combined with 5′-ACGCAAACGACTACCCTCA-3′, resulting in an amplicon of 436 bp when whole-cell DNA from strain W50 (PGF 111) was used as a template for the PCR (11). For the IS1598 locus the primer set was 5′-TATCCGCCTGTTGCCTCGTCC-3′ combined with 5′-GGCGGAG-GCGTAAACGGCTCA-3′, resulting in an amplicon of 1,100 bp when whole-cell DNA from W83 (PGF 7) was used as a template in the PCR (35). The identity of the amplicons was verified by partial DNA sequencing. The two PCR products were purified from agarose gels upon electrophoresis using the Qiaex II Gel Extraction Kit (Qiagen, Valencia, Calif.).
Nucleotide sequencing of gene fragments.
Internal fragments of the following housekeeping genes were used for sequencing: ahp encoding alkyl hydroperoxide reductase, thy encoding thymidylate synthetase, rmlB encoding dTDP–glucose-4,6-dehydratase, and infB encoding initiation factor 2. The four genes were selected from the unfinished genome sequence of P. gingivalis W83 (http://www.forsyth.org/pggp/) on the basis of a high degree of homology to the corresponding genes in Escherichia coli and, thus, they were assumed to be evolutionarily conserved among species. Primers for PCR amplification of fragments of the genes were designed from regions showing a high degree of homology with the corresponding genes from other bacteria as tested by searching the GenBank database. The gene fragments were amplified from whole-cell DNA by PCR using the following pairs of primers: ahp (5′-CCTTACCTTTGTATGCCCGACGGA-3′ and 5′-TCACGCCACTTGGCCGGACATAC-3′), resulting in an amplicon of 253 bp of the 564 bp gene; thy (5′-GAATGGGCTGATGA-GAACGGC-3′ and 5′-CGGATGCGGATCATACCCTTC-3′), resulting in a 359-bp amplicon of the 795-bp gene; rmlB (5′-GGCGG-TGCCGGTTTTATCGGA-3′ and 5′-AGCCTTGCTGGCCGAATAGGG-3′), resulting in amplicon of 331 bp of the 1,066-bp gene; and infB (5′-CGATCGTTACGGTCATGGGAC-3′ and 5′-GGCTTTCAGCTCTAAGATATC-3′), resulting in an amplicon of 372 bp of the 2,940-bp gene. For amplification we used approximately 1 ng of whole-cell DNA as template and Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Uppsala, Sweden). The temperature profile for the PCR was an initial denaturation at 94°C for 5 min, followed by 30 cycles at 94°C for 1 min, 60°C for 2 min, 72°C for 2 min, and then a final extension at 72°C for 8 min in a Perkin-Elmer type 480 DNA thermal cycler (Perkin-Elmer Applied Biosystems, Norwalk, Conn.). The PCR products were purified using Wizard Minicolumns (Promega, Madison, Wis.). For DNA sequencing we used the same primers as for the PCR, and sequencing of both strands of the amplified fragments was achieved by using a Thermo Sequenase dye terminator cycle sequencing kit (Amersham Life Science, Cleveland, Ohio); the resulting products were analyzed with an Applied Biosystems PRISM 377 automated sequencer (Perkin-Elmer Applied Biosystems).
Phylogenetic analyses.
The unweighted-pair-group method with arithmetic means (UPGMA) (a distance matrix method) provided in the PileUp program in the GCG package (Genetics Computer Group, Madison, Wis.) was used for multiple alignment of sequences and construction of dendrograms.
The genetic diversity (h) at an enzyme locus among strains, i.e., the probability that two randomly chosen strains have different alleles of the locus, was calculated from allele frequencies as follows: h = 1 − Σxi2 [n/(n − 1)], where xi is the frequency of the ith allele of the locus and n is the number of strains.
The index of association (IA) is a measure of linkage disequilibrium defined by the equation IA = VO/VE − 1, where VO is the observed variance of the mean number of loci at which two strains differ and VE is the expected variance of the mean number of loci at which two P. gingivalis strains differ assuming a random association of alleles (24). The statistical null hypothesis is that the alleles are in linkage equilibrium and an IA value that is significantly different from zero is indicative of a clonal population structure. IA was calculated using the ETLINK program of T. S. Whittam (Institute of Molecular Evolutionary Genetics, University of Pennsylvania).
RESULTS
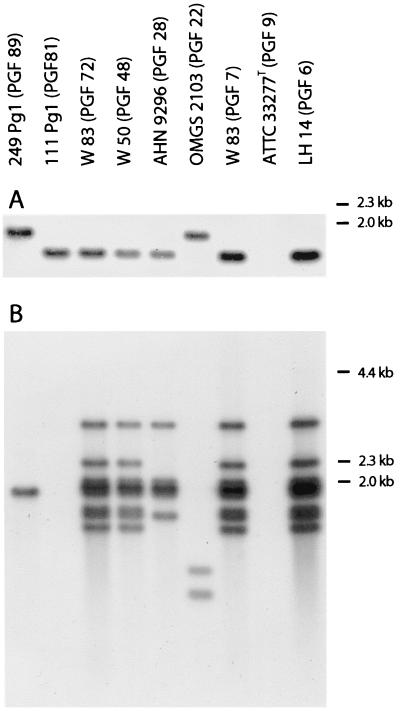
Whole-cell DNA digested with MspI of all 132 strains of P. gingivalis and the 10 duplicates from different laboratories was subjected to Southern blot analysis using a DNA probe of 436 bp recognizing the ragB locus (6, 11) (Fig. 1A) and a DNA probe of 1,100 bp recognizing the insertion sequence IS1598 (35) (Fig. 1B). The ragB locus was found in 17 of the 132 strains, and two patterns of hybridization were recorded each for 14 and 3 strains, respectively (Fig. 1A). The IS1598 probe revealed a very diverse hybridization pattern recognizing from one to multiple DNA fragments in 91 of the 132 strains, whereas 41 strains were devoid of this insertion sequence. No attempts were made to score the different patterns. All duplicate strains showed hybridization patterns identical to those of the parent strains with both probes.
FIG. 1.
MspI-digested chromosomal DNA from P. gingivalis strains hybridized to a probe recognizing the ragB locus (A) and a probe recognizing the insertion sequence IS1598 (B).
Internal fragments of four housekeeping genes, ahp, thy, rmlB, and infB, were amplified by PCR and sequenced. A search in the unfinished genome sequence of P. gingivalis W83 revealed that the four genes were separated by a minimum distance of 382 kb on the chromosome. The 57 strains included in the sequence analyses were chosen among the 132 strains according to the following criteria: (i) representation of strains from diverse geographical areas (22 countries or states on four continents), (ii) inclusion of all four strains from healthy gingival sites, and (iii) inclusion of all ragB-positive strains. Four duplicate strains received from different laboratories were also subjected to sequence analyses to test whether they did indeed represent the parent strains.
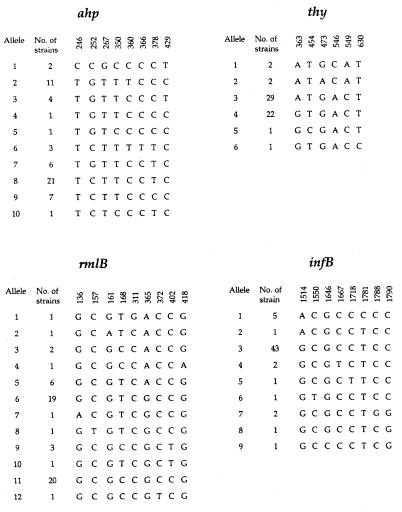
After alignment, none of the sequences contained deletions or insertions. The four duplicate strains had identical sequences in all four genes sequenced compared to the parent strains. Figure 2 shows the sites of variations in each gene fragment sequenced and the number of strains of each allelic type. The eight sites of variation in the ahp gene fragment sequenced were all silent site mutations, and they gave rise to 10 alleles, four of which were predominant, with each being found in six or more strains. The genetic diversity, h, of this locus was 0.77. A relatively high diversity was also found in the rmlB gene fragment, where the nine sites of variation resulted in 12 alleles, of which three were predominant (h = 0.73). The variation in four of the nine sites resulted in amino acid substitutions. The thy gene fragment had six sites of variation, of which two were nonsynonymous, leading to a change in the encoded amino acid. The six sites of variation gave rise to only six alleles, of which two predominated (h = 0.50). The lowest genetic diversity was found in the infB gene fragment, which had a genetic diversity of 0.36 due to one predominant allele despite the presence of eight polymorphic nucleotide positions and nine alleles. Only one of these polymorphic sites was nonsynonymous.
FIG. 2.
Sites of variation among sequenced fragments of four housekeeping genes in 57 P. gingivalis strains. The numbering is with the A in the ATG start codon as position 1.
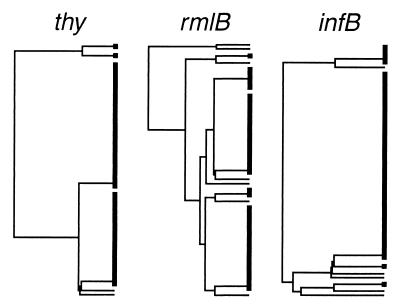
A phylogenetic tree with unscaled branches based on multiple alignment of the ahp sequences using the UPGMA method in the PileUp program is presented in Fig. 3. The alleles of the other gene fragments sequenced are given by numbers that refer to the alleles presented in Fig. 2. For comparison, the structures of the phylogenetic trees based on the other genes sequenced are shown in Fig. 4. No correlation between the clustering of strains in the different dendrograms was observed. In general, strains with identical gene sequences in one gene had quite different alleles in another gene and vice versa. For example, strains with the type 2 allele of the ahp sequence represented three alleles of the thy gene, namely, alleles 4, 3, and 1, and these three alleles of the thy gene were found in strains representing two, six, and seven different alleles of the ahp gene, respectively. More examples are evident from Fig. 3.
FIG. 3.
Dendrogram constructed from sequencing a fragment of the ahp gene from 57 P. gingivalis strains. The columns on the right show the strain designation, including our own reference number in parentheses, the allele types of the four genes sequenced, the presence or absence and pattern of hybridization to the probe identifying the ragB locus, the presence (+) or absence (−) of hybridization to the probe identifying IS1598, the isolation site and clinical diagnosis as presented by the contributor of the strain, and the geographic origin of the strain. Sequence types represented by multiple strains are contained in with boxes designated by capital letters. CCUG = Culture Collection University of Göteborg, Göteborg, Sweden; ATCC = American Type Culture Collection, Rockville, Md; Strain OMZ 488 = B262 (PGF56) showed positive catalase reaction; UK, United Kingdom.
FIG. 4.
Dendrograms based on partial sequences of thy, rmlB, and infB genes in 57 P. gingivalis strains.
The topologically different trees derived from sequencing genes in different areas of the bacterial genome (Fig. 3 and 4) and the lack of correlation between the clustering of strains in the trees indicated either no or a weak genetic linkage of the loci sequenced. In order to statistically analyze the extent of linkage, the IA was calculated based on the combined results of the four gene fragments sequenced. A total of 41 different combinations, termed sequence types, of the alleles in the four genes were revealed among the 57 strains. The value of IA based on a single representative of each of the 41 sequence types was 0.143 ± 0.202, which is not significantly different from zero, thus indicating that the population is at linkage equilibrium due to frequent recombination. When including all isolates, the IA was 0.206 ± 0.171.
The presence of the ragB locus and the insertion sequence IS1598 relative to the four dendrograms in Fig. 3 and 4 revealed that neither of these mobile genetic elements was associated with any particular allele of the four loci ahp, thy, rmlB, and infB. The ragB locus was found in strains of 10 different sequence types, and the insertion sequence IS1598 was found in strains of 25 different sequence types. The four isolates from healthy gingival sites (DCR 2006, 13 JC, HG 1319, and HG 1338) represented three different sequence types, and they were also heterogenic with regard to presence of ragB and insertion sequence IS1598.
The eight sequence types represented by multiple isolates are marked with boxes and capital letters in the ahp-based dendrogram (Fig. 3). Sequence type C included three strains from the same study of an Indonesian population, sequence type H included two strains from the same study of a Romanian population, while sequence type B included two strains from the same collection and with no source information. All strains in each of these sequence types were identical with regard to the presence or absence of ragB and insertion sequence IS1598. To further test for identity, DNA fingerprinting of whole-cell DNA digested with MspI was performed, which revealed identical patterns within each of the three sequence types described above. It cannot be ruled out that the strains representing each of these three sequence types originated from the same person since data on this were not available. However, this possibility can be excluded for the other five sequence types represented by multiple strains (A, D, E, F, and G) since for each of these types the strains originated from different countries and for three of the sequence types even different continents. Some heterogeneity was found in sequence types D, E, and G when the information on the presence of ragB and IS1598 was included in the analysis (Fig. 3). DNA fingerprinting of whole-cell DNA digested with MspI confirmed this heterogeneity and revealed heterogeneity also in sequence type F. Still, some strains in three of the five sequence types were identical in all parameters tested. These were as follows: the two strains of sequence type A; strains OMGS 2101 and P03/318 of sequence type D; and strains 32.2.4, 13 JC, W83, LH 14, and OMZ 308 (W50) of sequence type E.
Sequence type E, represented by seven strains, may represent a clone because limited diversity was observed for other parameters tested. Six of the seven strains had both the insertion sequence IS1598 and the same pattern of the ragB locus, while one strain (19A4) was devoid of these genetic elements. Whole-cell DNA of five of the six strains revealed an identical pattern when subjected to digestion with MspI and, likewise, hybridization with the probe detecting insertion sequence IS1598 gave rise to an identical pattern for these five strains. The remaining strain (AHN 9296) had a pattern of shared but fewer bands for both the MspI digest and the IS1598 hybridization, indicating that it was genetically very closely related but not identical to the five strains.
DISCUSSION
Our study demonstrates that recombination dominates over mutations in P. gingivalis because sequencing of fragments of four housekeeping genes located in different parts of the genome revealed different phylogenetic trees for each gene and lack of correlation between clustering of strains in the individual dendrograms (Fig. 3 and 4). This is considered to be strong evidence for a nonclonal population structure (19, 23). This conclusion is supported by a value of IA (0.143 ± 0.202) that is not significantly different from zero when including one representative of each of the 41 sequence types observed. The equation defining IA indicates an expected value of zero for a population at linkage equilibrium (24). Inclusion of all isolates gave a value of IA just significantly different from zero (0.206 ± 0.171). The combination of an IA > 0 when including all isolates and an IA that is not significantly different from zero when including only one representative of each sequence type suggests an epidemic population structure as reported for meningococci and pneumococci (1, 5, 20, 24). An epidemic population structure is characterized by being effectively sexual in the long term, but this may be obscured by temporary bursts (epidemics) of particular genotypes. Sampling of isolates from a certain limited time period may result in temporarily predominant genotypes being represented many times in the analysis, which may mask the actual panmictic structure of the population. Consequently, the linkage equilibrium may only be disclosed in the calculation of IA if a single representative of each genotype is included (24). In the present study DNA fingerprinting of MspI-digested whole-cell DNA from strains of the same sequence type confirmed the genetic identity of several strains in spite of their widely different geographic origin, thus supporting an epidemic population structure. Our conclusion contrasts with that of two previous reports on a clonal population structure of P. gingivalis based on isolation of the same genotype from geographically widespread locations (21, 25). As discussed in the introduction and as illustrated by our results, this is not necessarily an indication of clonality (23). The random distribution of serotype, invasive potential in experimental infections, and fimbrial type observed in the two studies supports our conclusion. Experimental evidence of specific mechanisms for genetic exchange has not been provided for P. gingivalis. However, the nonclonal population structure found in here provides evidence that horizontal gene transfer between strains followed by homologous recombination in housekeeping genes does occur in vivo. The mechanism is as yet unknown.
Information on the clinical status of the periodontal sites sampled did not lead to disclosure of potentially highly virulent sequence types of P. gingivalis; neither did the random distribution of the ragB locus and the insertion sequence IS1598. The lack of association between the presence of ragB locus and the insertion sequence IS1598 and any of the sequence-based phylogenetic trees supports the concept of a nonclonal population structure, but it is not evidence in itself because both are mobile genetic elements (11, 35).
The high diversity of P. gingivalis found in the studies of Loos et al. (21) and Ménard and Mouton (25) was confirmed in our study in which the combined analysis of only four gene fragments gave rise to 41 genotypes among 57 strains. Inclusion of more characters (the presence of ragB and insertion sequence IS1598 and pattern of MspI digests of whole-cell DNA) increased the diversity further. These results indicate that the considerably lower diversity reported by Ali et al. (2) based on ribotyping with two restriction enzymes is an underestimation of the true genetic polymorphism in P. gingivalis. It is also conceivable that these estimates are influenced by repeated inclusion of identical isolates from each individual.
In our study, six of the strains in sequence type E (Fig. 3) may constitute a clone associated with pathogenicity. Inclusion of more characters, i.e., patterns of whole-cell DNA digested with MspI and hybridization patterns with probes recognizing ragB and the insertion sequence IS1598, confirmed their close genetic relatedness. The six strains included W83 and W50, with known strong pathogenicity, and all had the ragB locus and the insertion sequence IS1598, both of which have been reported to be associated with pathogenicity. The association of human periodontal disease with strains having characteristics in common with W83 was recently reported by Griffen et al. (10), who used heteroduplex analysis of the ribosomal intergenic spacer region for typing the strains, and by Amano et al. (3, 4), who reported a strong association of fimA genotype IV, represented in W50 and W83, with periodontal disease. Rumpf et al. (33) even suggested an association between pathogenicity and the phylogeny of P. gingivalis based on sequencing of the ribosomal intergenic spacer region because strains W83 and ATCC 49417, whose heteroduplex types were strongly associated with periodontitis, also showed close proximity in the dendrogram. However, in our study these strains represented two different sequence types with identity in only two of the four genes sequenced. This illustrates the problems of inferring phylogenic conclusions based on a single gene, particularly in species with a nonclonal population structure.
In addition to the population genetic analysis based on selected housekeeping genes, we also investigated the distribution of mobile genetic elements. The ragB locus is a potential virulence determinant because it forms part of a pathogenicity island most likely acquired by horizontal gene transfer and is present in a proportion of P. gingivalis strains that includes highly virulent strains (11). The detection of this locus in a single isolate from a healthy gingival sulcus (Fig. 2) does not rule out this possibility. In this study the ragB locus was found in 13% of the 132 P. gingivalis strains studied in accordance with the findings of Hanley et al. (11). They reported that the ragB locus was detected by PCR in 36% of subgingival plaque samples from deep periodontal pockets. This does not necessarily suggest a prevalence of 36% among P. gingivalis isolates because it cannot be excluded that the ragB locus is present in other members of the oral flora since the locus is present on a mobile genetic element. However, the concomitant presence of the ragB locus and P. gingivalis had a statistically significantly stronger association to deep periodontal pockets than the presence of P. gingivalis alone (11).
The insertion sequence IS1598 was present in 69% of the 132 strains tested. One discrepancy between our results and those of Sawada et al. (35) was observed. They found that strain 381 had the IS1598, whereas it was absent in all four duplicates of strain 381 from different sources included in our study. The reported association of IS1598 with pathogenicity is based on its presence in three strains, of which two had a known high pathogenicity, and its absence in four strains (35). Further studies focusing on the various locations of this gene locus are necessary to understand the pathogenic significance of the IS1598 locus.
We conclude that P. gingivalis has an epidemic population structure characterized by frequent recombination. Such genetic events explain the random distribution of the potential virulence markers, the ragB locus and the IS1598 locus, in the P. gingivalis population observed in this study and the previously reported random distribution of serotype, invasive potential in experimental infections, and fimbrial type (21, 25). Our study suggests that particular genotypes, possibly with increased pathogenic potential, may spread successfully in the human population. Virulence in P. gingivalis is thus not confined to a distinct evolutionary lineage that may be easily defined. The search for virulence-associated clones must, therefore, focus on individual genes or groups of genes.
ACKNOWLEDGMENTS
We gratefully acknowledge the following scientists for providing many of the strains for the study: G. Dahlén, R. D. Gmür, E. Könönen, N. Skaug, R. Teanpaisan, and A. J. van Winkelhoff. We thank J. Aduse-Opoku for assistance with the sequence annotation from the P. gingivalis genome. The technical assistance of Lene Friis and Lise Hald is also much appreciated.
Financial support was obtained from The Foundations of the Danish Dental Association for Scientific and Practical Investigations in the Art in Dentistry and for Promotion of Scientific and Practical Dentistry, the Plasmid Foundation, the Velux Foundation, the Research Foundation of the University of Aarhus (grants no. F-1998-SUN-1-103 and E-1999-SUN-1-166), and the Danish Medical Research Council (grant no. 9702265).
REFERENCES
- 1.Achtman M. Clonal spread of serogroup A meningococci: a paradigm for the analysis of microevolution in bacteria. Mol Microbiol. 1994;11:15–22. doi: 10.1111/j.1365-2958.1994.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 2.Ali R W, Martin L, Haffajee A D, Socransky S S. Detection of identical ribotypes of Porphyromonas gingivalis in patients residing in the United States, Sudan, Romania and Norway. Oral Microbiol Immunol. 1997;12:106–111. doi: 10.1111/j.1399-302x.1997.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 3.Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 4.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–1430. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant D A, Mocca L F, Frasch C E, Frøholm L O, Zollinger W D, Selander R K. Genetic structure of Neisseria meningitidis populations in relation to serogroups, serotype, and outer membrane protein pattern. J Bacteriol. 1987;169:2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis M A, Hanley S A, Aduse-Opoku J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J Periodont Res. 1999;34:1–6. doi: 10.1111/j.1600-0765.1999.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 7.Dzink J L, Socransky S S, Haffajee A D. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 8.Genco C A, Cutler C W, Kapscynski D R, Maloney K, Arnold R R. A novel mouse model to study the virulence of and host response to Porp hyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–1263. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenier D, Mayrand D. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J Clin Microbiol. 1987;25:738–740. doi: 10.1128/jcm.25.4.738-740.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffen A L, Lyons S R, Becker M R, Moeschberger M L, Leys E J. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 1999;37:4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanley S A, Aduse-Opoku J, Curtis M A. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt S C, Ebersole J, Felton J, Brunsvold M, Kornman K S. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- 13.Hunt D E, Jones J V, Dowell V R., Jr Selective medium for the isolation of Bacteroides gingivalis. J Clin Microbiol. 1986;23:441–445. doi: 10.1128/jcm.23.3.441-445.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen S B, Löe H, Schiött C R, Theilade E. Experimental gingivitis in man. IV. Vancomycin induced changes in bacterial plaque composition as related to development of gingival inflammation. J Periodont Res. 1968;3:284–293. doi: 10.1111/j.1600-0765.1968.tb01934.x. [DOI] [PubMed] [Google Scholar]
- 15.Kilian M. Clonal basis of bacterial virulence. In: Guggenheim B, Shapiro S, editors. Oral biology at the turn of the century. S. Basel, Switzerland: Karger AG; 1998. pp. 131–142. [Google Scholar]
- 16.Laine M L, Appelmelk B J, van Winkelhoff A J. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J Periodont Res. 1996;31:278–284. doi: 10.1111/j.1600-0765.1996.tb00494.x. [DOI] [PubMed] [Google Scholar]
- 17.Laine M L, van Winkelhoff A J. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322–325. doi: 10.1111/j.1399-302x.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 18.Lamell C W, Griffin A L, McClellan D L, Leys E J. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in children. J Clin Microbiol. 2000;38:1196–1199. doi: 10.1128/jcm.38.3.1196-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenski R E. Assessing the genetic structure of microbial populations. Proc Natl Acad Sci USA. 1993;90:4334–4336. doi: 10.1073/pnas.90.10.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomholt H. Evidence of recombination and an antigenically diverse immunoglobulin A1 protease among strains of Streptococcus pneumoniae. Infect Immun. 1995;63:4238–4243. doi: 10.1128/iai.63.11.4238-4243.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loos B G, Dyer D W, Whittam T S, Selander R K. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–212. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons S R, Griffin A L, Leys E J. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–2365. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maynard-Smith J. Do bacteria have population genetics. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 1–12. [Google Scholar]
- 24.Maynard-Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ménard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musser J M, Hewlett E L, Peppler M S, Selander R K. Genetic diversity and relationships in populations of Bordetella spp. J Bacteriol. 1986;166:230–237. doi: 10.1128/jb.166.1.230-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musser J M, Kroll J S, Granoff D M, Moxon E R, Brodeur B R, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G, Hølby E A, Jonsdottir K E, Kabeer M, Kallings I, Khan W N, Kilian M, Knowles K, Koornhof H J, Li K I, Montgomery J, Pattison P E, Piffaretti J-C, Takala A K, Thong M L, Wall R A, Ward J I, Selander R K. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990A;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 28.Musser J M, Schlievert P M, Chow A W, Ewan P, Kreiswirth B N, Rosdahl V T, Naidu A S, Witte W, Selander R K. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc Natl Acad Sci USA. 1990B;87:225–229. doi: 10.1073/pnas.87.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodont Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 30.Nelson K, Whittam T S, Selander R K. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olyhoek T, Crowe B A, Achtman M. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev Infect Dis. 1987;8:665–691. doi: 10.1093/clinids/9.4.665. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen K, Hjorth P, Kilian M. Limited diversity of the immunoglo bulin A1 protease gene (iga) among Haemophilus influenzae serotype b strains. Infect Immun. 1988;56:987–992. doi: 10.1128/iai.56.4.987-992.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumpf R W, Griffen A L, Leys E J. Phylogeny of Porphyromonas gingivalis by ribosomal intergenic spacer region analysis. J Clin Microbiol. 2000;38:1807–1810. doi: 10.1128/jcm.38.5.1807-1810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sawada K, Kokeguchi S, Hongyo H, Sawada S, Miyamoto M, Maeda H, Nishimura F, Takashiba S, Murayama Y. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect Immun. 1999;67:5621–5625. doi: 10.1128/iai.67.11.5621-5625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapira L, Champagne C, Van Dyke T E, Amar S. Strain-dependent activation of monocytes and inflammatory macrophages by lipopolysaccharide of Porphyromonas gingivalis. Infect Immun. 1998;66:2736–2742. doi: 10.1128/iai.66.6.2736-2742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slots J, Ting M. Actnobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurence and treatment. Periodontology 2000. 1999;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 38.Sundqvist G, Figdor D, Hänström L, Sörlin S, Sandström G. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand J Dent Res. 1991;99:117–129. doi: 10.1111/j.1600-0722.1991.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 39.van Steenbergen T J M, Delemarre F G A, Namavar F, de Graaff J. Differences in virulence within the species Bacteroides gingivalis. Antonie Leeuwenhoek. 1987;53:233–244. doi: 10.1007/BF00393930. [DOI] [PubMed] [Google Scholar]
- 40.van Steenbergen T J M, Namavar F, de Graaff J. Chemiluminescence of human leukocytes by black-pigmented Bacteroides strains from dental plaque and other sites. J Periodont Res. 1985;20:58–71. doi: 10.1111/j.1600-0765.1985.tb00411.x. [DOI] [PubMed] [Google Scholar]