Abstract
Cetaceans are a group of aquatic mammals with the largest body sizes among living animals, including giant representatives such as blue and fin whales. To understand the genetic bases of gigantism in cetaceans, we performed molecular evolutionary analyses on five genes (GHSR, IGF2, IGFBP2, IGFBP7, and EGF) from the growth hormone/insulin-like growth factor axis, and four genes (ZFAT, EGF, LCORL, and PLAG1) previously described as related to the size of species evolutionarily close to cetaceans, such as pigs, cows, and sheep. Our dataset comprised 19 species of cetaceans, seven of which are classified as giants because they exceed 10 m in length. Our results revealed signs of positive selection in genes from the growth hormone/insulin-like growth factor axis and also in those related to body increase in cetacean-related species. In addition, pseudogenization of the EGF gene was detected in the lineage of toothless cetaceans, Mysticeti. Our results suggest the action of positive selection on gigantism in genes that act both in body augmentation and in mitigating its consequences, such as cancer suppression when involved in processes such as division, migration, and cell development control.
Subject terms: Evolution, Molecular evolution
Introduction
Gigantism results from species evolving huge body sizes relative to their small-bodied ancestors. This phenomenon has been extensively studied because it affects critical life-history traits such as longevity, fecundity, and health1–4. However, it is still unclear how natural selection favors great body size from an evolutionary perspective. Gigantism may bring consequences such as an overall reduction in the genetic effective population size (Ne) due to lower population densities5, lower reproductive output6, and the need to develop suppression mechanisms for diseases such as cancer, given the large number of cells needed to constitute a giant organism7. Despite this, several lineages of terrestrial and aquatic animals became giants throughout the history of life. Examples of these lineages can be found across the entire tree of life, such as tortoises8, sloths9, dinosaurs10, and aquatic animals such as the extinct Jaekelopterus rhenaniae—the largest arthropod ever found11—and the reptiles Mosasaurus hoffmanni12 and Shonisaurus sikanniensis13.
Aquatic and terrestrial habitats impose different selective pressures on body size, with aquatic organisms typically reaching larger proportions than their terrestrial relatives14. Several reasons have been proposed to explain this difference, for example, thermoregulation, abundant high-quality food in the aquatic environment, and a wider space available to explore new niches and specializations15,16.
Cetaceans (whales, porpoises, and dolphins) are aquatic mammals that evolved from small terrestrial ancestors around 50 million years ago during the Eocene17. The recolonization of the aquatic environment was followed by many morphological and physiological modifications, such as streamlined bodies, loss of body hair to reduce friction during swimming, reduced olfactory and gustatory systems, and hindlimb loss18. Currently, there are approximately 86 species of cetaceans, and these animals are divided into two groups: odontocetes (toothed whales) and mysticetes (whales with baleen that allow the filtration of food)19. One notable characteristic of cetaceans is the large size of several species. For example, the blue whale (Balaenoptera musculus) is the largest animal known to have existed, measuring 30 m long, and weighing more than 150 tons20. Also, the sperm whale (Physeter catodon) can reach up to 20 m and is the largest toothed animal living today. Several other cetaceans are known for their large bodies, such as the 25 m long fin whale (Balaenoptera physalus)21, 19 m long humpback whale (Megaptera novaeangliae)22, 17 m long bowhead whale (Balaena mysticetus)23, and 15 m long gray whale (Eschrichtius robustus)24. One of the main hypotheses to explain the large size of cetaceans relates to how they obtain food. It is argued that toothed cetaceans (i.e., odontocetes) developed large bodies due to the ability to dive and exploit prey from the seabed using a powerful biosonar, while in baleen whales (i.e., mysticetes) the evolution of gigantism is associated with the highly efficient exploitation of small prey25,26.
From a molecular point of view, body length is a complex character associated with many genes27. In mammals, research is mainly focused on domesticated species related to meat and milk production, due to their economic importance. In this context, many genes involved in body size growth have been described, such as the transcription factor LCORL (Ligand Dependent Nuclear Receptor Corepressor Like), which is responsible for size differences in sheep; NCAPG (Non-SMC Condensin I Complex Subunit G), PLAG1 (Pleomorphic Adenoma Gene 1), which acts in prenatal growth and is associated to body size of cattle; and ZFAT (Zinc Finger And AT-Hook Domain Containing) related to embryonic development and growth in human populations and horses28–30. Also, the growth hormone/insulin-like growth factor (GH-IGF) axis has been associated with growth rates, such GHSR (Growth Hormone Secretagogue Receptor), IGFBP2 (Insulin-Like Growth Factor Binding Protein 2), IGFBP7 (Insulin-Like Growth Factor Binding Protein 7), IGF2 (Insulin-Like Growth Factor 2), and EGF (Epidermal Growth Factor) genes31. Furthermore, since the somatotropic axis plays a central role in regulating growth, any locus expressing hormones, factors, or peptides within this system may reasonably represent a potential gene of significant importance in enhancing growth.
Accordingly, we aimed to investigate the molecular evolution of genes related to body size in cetaceans in a phylogenetic framework. We focused on five genes from the growth hormone/insulin-like growth factor (GH-IGF) axis, and four genes previously associated with increased body size in other cetartiodactyl species. Our main goal is to expand our understanding of the genetic basis of the morphological phenotypic variability of cetaceans.
Results
Among the nine selected genes to perform the molecular evolution analyses, we found that in the EGF gene, stop codons resulted in the interruption of the reading frame only in the Mysticeti cetacean lineage. To our knowledge, this is the first time that EGF pseudogenization has been reported for mysticetes. As this work focuses on the coding regions, only the results for the other eight genes are described below.
Selection analyses
Branch model
Branch analyses were performed using species trees. First, we labeled the ancestral branch that led to the giant cetaceans, i.e., the stem mysticete lineage and the branch of the sperm whale (Physeter catodon), since this is the only giant species of odontocete group, exceeding 10 m in body size (Fig. 1). Then we compare the rates between giant and non-giant cetaceans, labeling all species with body size larger than 10 m as one group.
Figure 1.
The species tree used in this study illustrates the group of giant cetaceans and the other mammals included in our dataset. In red, selection of ancestral branches that give rise to gigantism. Mammalian phylogeny is based on Beck and Baillie86, while phylogenetic relationships among cetaceans are based on McGowen et al.87.
For the first labeled scheme, codeML free-ratio test fitted our data significantly better than the one-ratio model for all genes. However, the two-ratio model, used to estimate whether giant cetaceans have a different omega value compared to the other species, fitted better for PLAG1. In the second scheme, no statistically significant differences were found using codeML.
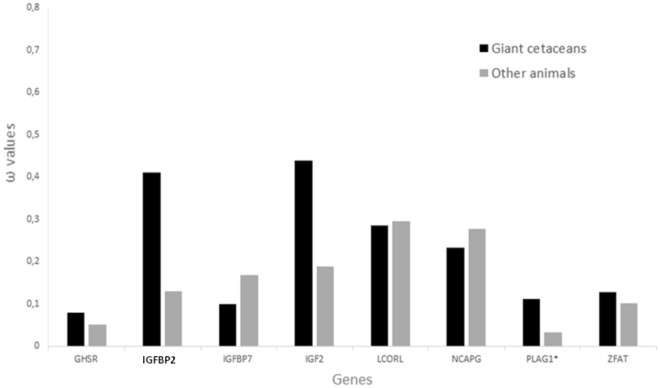
For RELAX analysis, the IGFBP2 gene was found to be under intensified selection (K > 1) using the first labeled scheme with the ancestral branch from mysticete lineage and sperm whale (Fig. 2), while no evidence of intensified selection was found using the second scheme, in which all giant cetaceans were labeled as a one group.
Figure 2.
Comparison of ω values calculated using the two-ratio model in codeML for genes related to body size in giant cetaceans and other mammals. The two-model was better suited to the data (p < 0.05) in the PLAG1 gene, marked with an asterisk. The IGFBP2 gene, in bold, represents the statistically significant intensified selection that the RELAX program detected (p < 0.05) in giant cetaceans.
Branch-site models
To estimate selective pressures acting in specific sites on the giant cetacean lineages, we performed branch-site models using BUSTED, aBSREL, and codeML. BUSTED indicated the occurrence of positive selection in at least one site in at least one branch for the GHSR gene (p-value ≤ 0.05), aBSREL resulted in episodic positive selection in Eschrichtius robustus lineage for 0.34% of sites on the same gene, and CodeML found significant positive selection for site 211 for GHSR. CodeML also found positive selection for sites 134 and 353 for the NCAPG gene and the site 278 for IGFBP7. No significant positive selection was detected for the IGF2, LCORL, PLAG1, and ZFAT genes.
Site models
To find sites under positive selection within giant cetaceans, we used SLAC, MEME, and FUBAR (Table 1). SLAC resulted in some codons with ω values greater than 1 but no statistically significant signatures of positive selection. MEME found episodic selection/diversifying selection for sites 243 and 249 for GHSR, and 241, 466, and 885 in the NCAPG gene. FUBAR detected thirteen sites under episodic selection/diversifying selection for NCAPG (348, 373, 464, 466, 630, 885, 906, 908, 925, 940, 953, 969, and 991), and the 211 codon was identified as being under selection for GHSR gene.
Table 1.
Codon positions under positive selection detected by the site model using FUBAR and MEME using genes trees.
| Gene | Site model | Branch-site model | ||
|---|---|---|---|---|
| FUBAR | MEME | codeML | aBSREL | |
| GHSR | 211 (0.93) | 243(0.03); 249(0.04) | 211 (0.984) | 0;34% |
| IGF2 | 0 | 0 | 0 | 0 |
| IGFBP2 | 0 | 0 | 0 | 0 |
| IGFBP7 | 0 | 0 | 278 (0.974) | |
| EGF | 0 | 0 | 0 | 0 |
| LCORL | 0 | 0 | 0 | 0 |
| PLAG1 | 0 | 0 | 0 | 0 |
| ZFAT | 0 | 0 | 0 | 0 |
| NCPAG | 348 (0.91), 373 (0.95); 464 (0.95), 466 (0.94), 630 (0.97); 885 (0.93); 906 (0.95); 908 (0.94); 925 (0.92); 940 (0.93); 953 (0.94); 969 (0.92); 991 (0.93) | 241 (0.03); 466 (0.03); 885 (0.04) | 134 (0.953); 353 (0.950) | 0 |
Significance was assessed by Posterior Probability (PP) > 90% in FUBAR and p value < 0.05 in MEME. Detection of the position of codons under positive selection was carried out through the branch-site model using codeML and aBSREL. Significance is obtained through BEB (Posterior Probability (PP) > 0.90) for codeML and p-value < 0.05 in aBSREL.
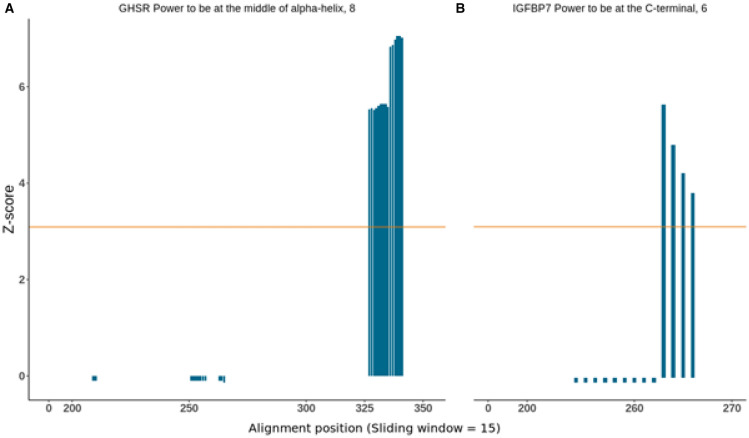
We found evidence for positive selection in the power to be at the middle of alpha-helix physicochemical property using TreeSAAP, with global z-scores > 3.09 (p < 0.001) for the GHSR gene, and in the power to be at the C-terminal physicochemical property for the IGFBP7 gene (Fig. 3).
Figure 3.
Detection of significant physicochemical amino acid changes using TreeSAAP and genes trees. This analysis was performed on the genes that presented higher ω values identified by codeML analysis in giant cetaceans. Only GHSR (A) and IGFBP7 (B) showed significant results. A highly significant z-score (z > 3.09, p < 0.01), represented here by the regions above the orange line, indicates more non-synonymous substitutions than assumed under the neutral model and therefore are interpreted as a result of positive selection. Respective property and category are shown above the graphs.
Discussion
This study presents the molecular evolution of genes related to body size in mammals, focusing on giant cetaceans. We found molecular signs of selection in the GHSR, IGFBP7, PLAG1, and NCAPG genes from the nine genes included in our dataset, with results converging with different algorithms. Furthermore, in the EGF gene, the presence of stop codons resulted in the interruption of the reading frame in all Mysticeti cetacean species, which is unique to this group.
The presence of stop codons in the epidermal growth factor (EGF) gene is a pseudogenization indicator. The stop codons start at position 948 (exon 3) in the alignment, and remain present until the last exon in different and multiple sites for all species of the Mysticeti group. Some stop codons likely share the same locus between particular mysticete species, but because the reading frame is very corrupted, the alignment is unreliable only in the group of toothless cetaceans, and we cannot state the site of the first mutation in stem mysticete lineage that led to the EGF pseudogenization in this group. To our knowledge, the inactivation of EGF in mysticetes is reported for the first time in this paper. The inactivation of protein-coding genes has already been associated with several traits of cetaceans32, such as vision33, loss of taste receptors34, hair loss35, and teeth in Mysticeti36, among others. From an evolutionary perspective and considering the occupation of the aquatic environment, the loss of function of these genes can be understood as part of the adaptation process and not only because of the relaxation of selection37. EGF binds to the epidermal growth factor receptor (EGFR), which then dimerizes or forms ErbB-2, ErbB-3, or ErbB-4 homologs, increasing the intracellular activity of tyrosine kinase, activating effects such as cell proliferation, apoptosis and angiogenesis, embryonic growth, and tissue regeneration38. In addition, EGF has been associated with the development and eruption of teeth, being found within the dental follicle, in the alveolar bone related to ameloblasts (the cells that form tooth enamel), and during the pre-functional stage of tooth eruption in rats, animals in which EGF injections in neonates significantly stimulated the eruption of the incisor teeth39–41.
Teeth loss occurred in the common ancestor of all extant mysticete cetaceans, and ontogenetic evidence suggests that teeth develop rudimentarily in fetuses in this group; however, they are later aborted and reabsorbed before enamel formation42,43. Thus, evidence of pseudogenization of the EGF gene only in Mysticeti is likely related to the loss of teeth and the appearance of baleen. The baleen, for this group, was an evolutionary innovation that allowed whales to exploit a new foraging niche: filtration, which was previously identified as a probable trigger for gigantism26,44. Furthermore, the loss of EGF functionality and its role in important components of homeostasis, such as the kidneys, would be compensated for by the role of other genes in the EGF family and other pathways, an overlap of functions that has been reported in rats that did not have active EGF and yet had a normal and healthy phenotype44.
Cetacean body size seems to respond to intense selective pressures imposed by the aquatic environment. Factors such as thermoregulation, feeding ecology, and space availability shaped the gigantic body proportions of these animals14–16. Furthermore, migratory behavior, like the one performed by the blue whale that transits in polar waters, affects body size following Bergmann's rule, which states that animals living in colder climates are generally larger than those living in warmer regions45. The combination of these external factors can now be studied at the molecular level due to advances in genomic technologies. For example, a recent study reported signs of positive selection in body size genes in cetaceans: in small species, the genes under selection related to short size were ACAN, OBSL1, and GRB10; in the large cetaceans, the selection was identified in the CBS, EIF2AK3 and PLOD1 genes, all related to the large size46. Together, their results and the results from this study aid to our understanding of the evolutionary panorama of large body evolution, which is a complex feature that affects many genetic pathways.
Our results identified the GHSR with evidence for positive selection in the physicochemical property Power to be at the middle of alpha-helix, with global z-scores > 3.09 (p < 0.001) using TreeSAAP. BUSTED found evidence of positive selection for the GHSR gene (p-value ≤ 0.05) and aBSREL in the sperm whale for 0.32% of sites. Also, codons 243 and 259 were identified as being under positive selection by MEME, and 211 were identified by the codeML branch-site model (98%) and FUBAR (0.93 p.p). In this site, the sperm whale (Physeter catodon) was the only one to present glycine (G). This nonpolar amino acid that is compatible with hydrophilic and hydrophobic environments, while all other animals had threonine (T), which is polar and highly soluble in water. This modification is important because glycine is considered a "helix breaker" once it disrupts the regularity of the α helical backbone conformation since it lacks a β carbon, which is associated with more conformational freedom than other residues47,48. As mentioned before, the results reinforce that large phenotypes may evolve by different paths. It is worth noting that changes in specific species, such as the different site in the sperm whale—the only odontocete classified as a giant, may be related to characteristics of that species, involving the large body size or other characteristics affected by the gene. GHSR is an endogenous ligand that can stimulate, through its ghrelin ligand, the release of growth hormone through the pituitary gland and thus increase appetite, regulate body weight, energy metabolism, and fat accumulation49. In addition, it is associated with the secretion of gastric acid, control of cell proliferation, apoptosis, lactation, and cardiovascular pressure50–52. This gene has been linked to increased body size in cattle, sheep, and pigs30,53–55. In most cases, the increase in body size in these animals results from changes in a few sites, but these changes were not found in giant cetaceans. Nevertheless, this may indicate that minor changes in this gene can result in phenotypic modifications.
IGFBP7 is a 27 kD protein and a member of the IGFBP superfamily, responsible for the viability of insulin-like growth factors (IGFs)—molecules involved in promoting cell growth and division56. Evidence suggests that IGFBP7 acts as an oncosuppressor gene in prostate, breast, lung, and colorectal cancer due to its regulatory action related to cell proliferation, cell adhesion, cell senescence, and angiogenesis57,58. This repressor activity may arise with the interruption of the cell cycle in the G1 phase, induction of senescence, and an increase in the level of cell death through apoptotic cells59. In addition, it has already been observed that the higher the body mass index, the greater the expression of IGFBP7. This is possibly associated with the fact that obesity is an agent related to senescence, and IGFBP7 is secreted by senescent cells. This relationship that may indicate a compensation mechanism for organisms that reach high body mass60. In our analyses, we found evidence for positive selection in the physicochemical property power to be at the C-terminal with global z-scores > 3.09 (p < 0.001) using TreeSAAP. Furthermore, the branch-site implemented in codeML inferred that site 278 is under positive selection. In this site, the blue whale (Balaenoptera musculus) shows a loss of codons and no expressed amino acids. In contrast, the gray whale (Eschrichtius robustus) presents the nonpolar and hydrophobic methionine (M), while the other animals had glutamate (E), a polar amino acid. In summary, it seems that IGFBP7 is related to two main characteristics of giant cetaceans: increase in body size and suppression of cancer. Some cancer suppressor genes have already been reported to be under positive selection for cetaceans61, and IGFBP7 is likely to be one more.
NCAPG (Non-SMC Condensin I Complex Subunit G) is a gene strongly associated with increased body size and weight gain. It has been reported to be linked to birth weight, withers height, feeding efficiency, and pubertal growth in bovine species62–64. Besides cattle, this gene has been linked to growth in horses, donkeys (Equus asinus), pigs, humans, and chickens65–71. NCAPG also presented many sites evolving under positive selection, thirteen in the FUBAR program (348, 373, 464, 466, 630, 885, 906, 908, 925, 940, 953, 969, and 991), three in MEME (241, 466, and 885) and two in the codeML branch-site model (134 and 353) with some of them recovered by different methods, such as the 466, and 885 identified by FUBAR and MEME. Accordingly, this gene is probably the one that may be most directly involved in cetacean gigantism from our dataset, acting directly on two important characteristics—growth and weight gain.
In the same direction, PLAG1 (Pleomorphic Adenoma Gene 1) is a gene associated with growth in cattle72, pigs73, and sheep74, mainly in traits such as height, knuckle, biceps, and shank75. This gene encodes a zinc finger protein family, a nuclear protein transcription regulator76, playing an important role in the transcriptional regulation of growth factors such as IGF2, which is related to embryo growth and cell survival77. This is a candidate gene for future analysis with new parameters since it is related to growth in several animals, and mutations have been described as promoting changes in height78. In our analyses, PLAG1 was the only gene with evidence of positive selection by the codeML branch model.
Collectively, our results indicate four genes likely to be involved in increasing body size in giant cetaceans. Some of these genes, such as GHSR and IGFBP7, may also be responsible for mitigating the possible consequences of extreme size, as they control important aspects of the cell cycle. Hypothetically, being a giant has severe consequences, such as increased chances of developing cancer, in addition, cetaceans are long-lived animals, which is also related to this disease. Giant cetacean species (larger than 10 m) included in this study live longer than 30 years, with the humpback whale (Megaptera novaeangliae) reaching 50 years, and the blue whale (Balaenoptera musculus) and the fin whale (Balaenoptera physalus) reaching up to 90 years, while the bowhead whale (Balaena mysticetus) is the longest lived mammal known, with a lifespan of 200 years79. Despite these triggers, giant animals are less likely to develop cancer than small animals, a logical contradiction called Peto's Paradox that suggests the existence of a mitigation mechanism7. Thus, genes that act on body growth and control of the negative aspects mentioned above through cell control, cell division, and tumor suppression could be targets of natural selection, allowing these animals to become giants, live longer, and have great body mass.
It is worth remembering that body size is a complex characteristic that involves many factors and molecular pathways. Throughout cetacean evolutionary history, different lineages had different selective pressures on different genes that could result in the same large body phenotype. This could be the case for the sperm whale, an odontocete as large as a mysticete. Interestingly, genes previously reported as associated with large sizes in artiodactyls, such as LCORL and ZFAT, apparently do not show the same effect in cetaceans, highlighting how large sizes may arise from different pathways from different genes in different lineages.
Conclusion
In summary, here we investigated the molecular evolution of genes possibly related to increased body size in giant cetaceans. We found evidence for positive selection at the coding level for sites in the GHSR, IGFBP7, PLAG1, and NCAPG genes. Besides that, we found evidence of pseudogenization of the EGF gene in the Mysticeti lineage, an event likely related to teeth loss in these cetaceans, which could be connected with the emergence of the baleen plate filter system. In conclusion, our study provides new perspectives on the evolution of cetacean gigantism, reinforcing the selective pressures of the aquatic environment, the various possibilities of action of natural selection on different genes that have similar functions depending on specific characteristics for each species, and indicating that pseudogenization is also an adaptive process for this group.
Material and methods
Sample data
We focused on the genes GHSR, IGF2, IGFBP2, IGFBP7, and EGF from the growth hormone/insulin-like growth factor axis and the genes NCAPG, LCORL, PLAG1, and ZFAT that are associated with increased body size in artiodactyls. The cetacean group was composed of 12 odontocetes (Lagenorhynchus obliquidens, Neophocaena asiaeorientalis, Delphinapterus leucas, Tursiops truncatus, Orcinus orca, Monodon monoceros, Globicephala melas, Lipotes vexillifer, Physeter catodon, Phocoena sinus, Sotalia fluviatilis, and Sotalia guianensis) and seven mysticetes (Balaenoptera acutorostrata, Balaena mysticetus, Eschrichtius robustus, Eubalaena japonica, Megaptera novaeangliae, Balaenoptera physalus, and Balaenoptera musculus), totaling 19 species. The coding sequences for the species Balaena mysticetus came from the public platform The Bowhead Whale Genome Resource. In addition, Sotalia fluviatilis and Sotalia guianensis were sequenced by our laboratory (data not published). All other coding sequences were retrieved from GenBank and the accession numbers can be found in the Table 1. The sequences were retrieved according to their availability in the databases and quality. Thus, the dataset is not the same for all genes, but at least one giant cetacean is present for all of them. The sequences were aligned using the MUSCLE algorithm80 and we used Geneious software v. 7.1.381 to remove fragmented sequences that were larger or smaller than expected.
True gigantism in cetaceans is defined as body length above 10 m82. According to this criterion, the species classified as giants were Physeter catodon, Balaena mysticetus, Eschrichtius robustus, Megaptera novaeangliae, Eubalaena japonica, Balaenoptera physalus, and Balaenoptera musculus (Table 2).
Table 2.
Average size in meters of all cetacean species included in this study.
| Infraorder | Species | Size (m) |
|---|---|---|
| Odontocetes | Phocoena sinus | 1.4 |
| Sotalia fluviatilis | 1.5 | |
| Lipotes vexillifer | 2.5 | |
| Neophocaena asiaeorientalis | 2.0 | |
| Sotalia guianensis | 2.2 | |
| Lagenorhynchus obliquidens | 2.2 | |
| Tursiops truncatus | 3.8 | |
| Delphinapterus leucas | 4.2 | |
| Monodon monoceros | 5.0 | |
| Globicephala melas | 5.7 | |
| Orcinus orca | 8.0 | |
| Physeter catodon | 20.0 | |
| Mysticetes | Balaenoptera acutorostrata scammoni | 8.5 |
| Eschrichtius robustus | 15.0 | |
| Eubalaena japonica | 18.0 | |
| Balaena mysticetus | 17.0 | |
| Megaptera novaeangliae | 19.0 | |
| Balaenoptera physalus | 25.0 | |
| Balaenoptera musculus | 30.0 |
Significant values are in [bold].
True gigantism in cetaceans was defined as body length above 10 m. Physeter catodon, Balaena mysticetus, Eschrichtius robustus, Megaptera novaeangliae, Eubalaena japonica, Balaenoptera physalus, and Balaenoptera musculus are classified as giants. Information from Encyclopedia of Marine Mammals.
Molecular evolutionary analyses
To estimate the role of natural selection in our focus genes we estimated the value of ω (dN/dS), which is the ratio of the rate of non-synonymous substitutions (dN) to the rate of synonymous substitutions (dS), where ω < 1 indicates purifying selection, ω = 1 suggests neutral evolution, and ω > 1 indicates positive selection83,84. Different approaches were applied: the branch model that identifies how ω varies through the branches of the phylogeny, site-models that detect variations of ω in distinct sites, and the branch-site model that integrates both approaches83–85. The branch model and branch-site models were performed for a dataset with the species of interest and other mammals labeling the ancestral branches that resulted in these giant animals. In contrast, the site-models were performed only with the cetacean species classified as giants (Supplementary Figs. S1 and S2). Such tests were done on species trees, with relationships based on Beck and Baillie86 for mammals, while phylogenetic relationships among cetaceans are based on McGowen et al.87.
Branch models
To check whether the value of ω for giant cetaceans was different compared to other animals of the phylogenetic tree we used a branch model available at codeML within the PAML package85 that allows the variation of ω in the branches of the phylogeny. First, we used the one-ratio model that estimates a single value of ω for all branches. Then, the free-ratio model was applied, calculating ω for each branch. Finally, we used the two-ratio model, where we inferred a value of ω for giant cetaceans and another for the rest of the phylogeny. In this case, the interest group was identified as a foreground branch, while the algorithm treated the other unmarked ones as background branches88. We tested two scenarios, first labeling the ancestral branches that led to the stem mysticete lineage and the branch of the sperm whale (Physeter catodon), and second labeling all cetaceans' species with body size larger than 10 m classified as giants as one group to compare within non-giants cetaceans. The same configuration was used in RELAX, a method to test whether the selection was relaxed (K < 1) or intensified (K > 1) on a portion of branches specified a priori in the phylogeny89.
Site models
For site model analyses, the dataset comprised only the sequences of giant cetaceans. FUBAR software (Fast Unconstrained Bayesian AppRoximation) was used to identify sites that may have experienced generalized diversification or purifying selection by estimating the ratios between dN and dS substitution rates for each site where posterior probability (PP) of ω > 1 is greater than 95%90. SLAC (Single-Likelihood Ancestor Counting) was also used. This algorithm combines maximum-likelihood (ML) and counting approaches to calculate the ratios between dN and dS rates by site given a codon alignment91. Finally, MEME (Mixed-Effects Model of Evolution) looked for evidence of episodic or diversifying selection at individual sites allowing ω to change from site to site and branch to branch92.
TreeSAAP v.3.293 relies on the MM01 model implemented in baseML from the PAML package85 using phylogeny to reconstruct the most likely ancestral states for the gene sequences, detecting selection at the amino acid level. The software assigns weight values to the non-synonymous codon changes, for which overall physicochemical effects are assessed using a model with 31 physicochemical amino acid properties, with these changes ranging from 1 (conservative) to 8 (radical change). Positive selection is checked through a z-score to calculate deviation from neutral evolution. A highly significant z-score (z > 3.09, p < 0.01) indicates more non-synonymous substitutions than assumed under the neutral model, and only amino acid changes with a score between 6 and 8 and with a positive z-score < 0.001 were considered94.
Branch-site models
The branch-site model was used to identify whether some sites were subjected to the action of positive selection in the group of giant cetaceans. For this analysis in codeML, the interest group (i.e., all giant cetaceans) was labeled in the phylogeny as foreground branches, where sites with ω > 1 are allowed, and the rest of the tree was labeled as background branches, where sites with ω > 1 are not allowed. Model A was then used against the null model.
Two other branch-site tests were performed, from the HyPhy package on the DataMonkey portal91,95. BUSTED (Branch-Site Unrestricted Statistical Test for Episodic Diversification), which identifies a gene that experienced positive selection in at least one site in at least one branch96, and aBSREL (Adaptive Branch-Site Random Effects Likelihood), which allows positive selection in unspecified branches of the tree. To avoid excessive parameterization, aBSREL uses the Akaike Information Criterion correction (AICc) to estimate the ideal number of categories per branch instead of defining that each branch must be equipped with three classes. In addition, the Bonferroni-Holm approach was used to control false-positive rates84,97.
Supplementary Information
Acknowledgements
This study was funded by Coordination for the Improvement of Higher Education Personnel—Brasil (CAPES)—Finance Code 001 and FAPESP (2015/18269-1). LF was funded by FAPESP postdoctoral scholarship (2017/25058-2) and EKSR by FAPESP doctoral scholarship (2018/01236-1). We are grateful to the scientists that made available the gene sequences used in this study.
Author contributions
M.F.N. conceived the research hypothesis; F.A.S., E.M.S.S., E.K.S.R. and L.F. analyzed the data and drafted the manuscript; all authors contributed equally to the final version and reviewed the manuscript.
Data availability
The datasets analysed during the current study are available in the Supplementary Files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-24529-3.
References
- 1.Calder WAI. Size, Function, and Life History. Harvard University Press; 1984. pp. 139–161. [Google Scholar]
- 2.Pan H, et al. The genome of the largest bony fish, ocean sunfish (Mola mola), provides insights into its fast growth rate. GigaSci. 2016;5:36. doi: 10.1186/s13742-016-0144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Álvarez SH, Karlsson E, Ryder OA, Toh K, Crawford AJ. How to make a rodent giant: Genomic basis and tradeoffs of gigantism in the capybara, the world’s largest rodent. Mol. Biol. Evol. 2021;38:1715–1730. doi: 10.1093/molbev/msaa285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quesada V, Rodríguez FS, Miller J, Otín CL. Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evol. 2019;3:87–95. doi: 10.1038/s41559-018-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damuth J. Population density and body size in mammals. Nature. 1981;821:699–700. doi: 10.1038/290699a0. [DOI] [Google Scholar]
- 6.Leffler EM, et al. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy JD, Victor EM, Jenese HC. Why don't all whales have cancer? A novel hypothesis resolving Peto's paradox. ICB. 2007;47:317–328. doi: 10.1093/icb/icm062. [DOI] [PubMed] [Google Scholar]
- 8.Jaffe AL, Slater GJ, Alfaro ME. The evolution of island gigantism and body size variation in tortoises and turtles. Biol. Lett. 2011;7:558–561. doi: 10.1098/rsbl.2010.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomassini RL, et al. Gregariousness in the giant sloth Lestodon (Xenarthra): Multi-proxy approach of a bonebed from the Last Maximum Glacial of Argentine Pampas. Sci. Rep. 2020;10:10955. doi: 10.1038/s41598-020-67863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander PM, et al. Biology of the sauropod dinosaurs: The evolution of gigantism. Biol. Rev. 2011;86:117–155. doi: 10.1111/j.1469-185X.2010.00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braddy SJ, Poschmann M, Tetlie OE. Giant claw reveals the largest ever arthropod. Biol. Lett. 2008;4:106–109. doi: 10.1098/rsbl.2007.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soliar LT. Anatomy and functional morphology of the largest marine reptile known, Mosasaurus hoffmanni (Mosasauridae, Reptilia) from the Upper Cretaceous, Upper Maastrichtian of The Netherlands. Phil. Trans. R. Soc. Lond. B. 1995;347:155–180. doi: 10.1098/rstb.1995.0019. [DOI] [Google Scholar]
- 13.Nicholls EL, Manabe M. Giant ichthyosaurs of the Triassic—a new species of Shonisaurus from the Pardonet Formation (Norian: Late Triassic) of British Columbia. J. Vertebr. Paleontol. 2004;24:838–849. doi: 10.1671/0272-4634(2004)024[0838:GIOTTN]2.0.CO;2. [DOI] [Google Scholar]
- 14.Smith FA, Lyons SK. How big should a mammal be? A macroecological look at mammalian body size over space and time. Phil. Trans. R. Soc. B. 2011;366:2364–2378. doi: 10.1098/rstb.2011.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downhower JF, Bulmer LS. Calculating just how small a whale can be. Nature. 1988;335:675. doi: 10.1038/335675b0. [DOI] [Google Scholar]
- 16.Smith FA, et al. Similarity of mammalian body size across the taxonomic hierarchy and across space and time. Am. Nat. 2004;163:672–691. doi: 10.1086/382898. [DOI] [PubMed] [Google Scholar]
- 17.Thewissen J, Cooper L, Clementz M, Bajpai S, Tiwari BN. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature. 2007;450:1190–1194. doi: 10.1038/nature06343. [DOI] [PubMed] [Google Scholar]
- 18.Berta A, Sumich JL, Kovacs KM. Marine Mammals: Evolutionary Biology. Academic Press; 2005. pp. 178–194. [Google Scholar]
- 19.Mead, J. G. & Brownell R. L. Order Cetacea. In (eds. Wilson, D. E. & Reeder, D. M.) Mammal Species of the World: A Taxonomic and Geographic Reference 723–743 (University Press, 2005).
- 20.Sears R, Perrin WF. Blue whale: Balaenoptera musculus. Encyclopedia of Marine Mammals 2nd edition. Academic Press; 2009. pp. 120–124. [Google Scholar]
- 21.Aguilar A, García-Vernet R. Fin whale: Balaenoptera physalus. Encyclopedia of Marine Mammals. 3. Cambridge: Academic Press; 2018. pp. 368–371. [Google Scholar]
- 22.Clapham PJ. Humpback whale: Megaptera novaeangliae. Encyclopedia of Marine Mammals. 3. Academic Press; 2018. pp. 489–492. [Google Scholar]
- 23.Rugh DJ, Shelden EW. Bowhead whale: Balaena mysticetus. Encyclopedia of Marine Mammals. 2. Cambridge: Academic Press; 2009. pp. 131–133. [Google Scholar]
- 24.Jones ML, Swartz L. Gray whale: Eschrichtius robustus. Encyclopedia of Marine Mammals. 2. Academic Press; 2009. pp. 503–511. [Google Scholar]
- 25.Goldbogen JA, et al. Why whales are big but not bigger: Physiological drivers and ecological limits in the age of ocean giants. Science. 2019;366:1367–1372. doi: 10.1126/science.aax9044. [DOI] [PubMed] [Google Scholar]
- 26.Goldbogen JA, Madsen PT. The evolution of foraging capacity and gigantism in cetaceans. Exp. Biol. 2018;221:jrb166033. doi: 10.1242/jeb.166033. [DOI] [PubMed] [Google Scholar]
- 27.Kemper KE, Visscher PM, Goddard ME. Genetic architecture of body size in mammals. Genome Biol. 2012;13:244. doi: 10.1186/gb-2012-13-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, et al. Molecular characterization and expression of SPP1, LAP3 and LCORL and their association with growth traits in sheep. Genes. 2019;10:616. doi: 10.3390/genes10080616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016;87:159–167. doi: 10.1111/asj.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makvandi-Nejad S, et al. Four loci explain 83% of size variation in the horse. PLoS ONE. 2012;7:e39929. doi: 10.1371/journal.pone.0039929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worley KC, Warren WC, Rogers J, Locke D, Muzny DM. The common marmoset genome provides insight into primate biology and evolution. Nat. Genet. 2014;46:850–857. doi: 10.1038/ng.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huelsmann M, et al. Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Sci. Adv. 2019;5:eaaw671. doi: 10.1126/sciadv.aaw6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGowen MR, Tsagkogeorga G, Williamson J, Morin PA, Rossiter SJ. Positive selection and inactivation in the vision and hearing genes of cetaceans. Mol. Biol. Evol. 2020;37:2069–2083. doi: 10.1093/molbev/msaa070. [DOI] [PubMed] [Google Scholar]
- 34.Zhu K, Zhou X, Xu S. The loss of taste genes in cetaceans. BMC Evol. Biol. 2014;14:218. doi: 10.1186/s12862-014-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nery MF, Arroyo JI, Opazo JC. Increased rate of hair keratin gene loss in the cetacean lineage. BMC Genom. 2014;15:869. doi: 10.1186/1471-2164-15-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meredith RW, John G, Joyce C, Mark SS. Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of extant baleen whales. Proc. R. Soc. 2010;278:993–1002. doi: 10.1098/rspb.2010.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albalat R, Cañestro C. Evolution by gene loss. Nat. Rev. Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 38.Zeng F, Harris RC. Epidermal growth factor, from gene organization to bedside. Semin. Cell. Dev. Biol. 2014;28:2–11. doi: 10.1016/j.semcdb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norman J, Tsau YK, Bacay A, Fine LG. Epidermal growth factor accelerates functional recovery from ischaemic acute tubular necrosis in the rat: Role of the epidermal growth factor receptor. Clin. Sci. Lond. 1990;78:445–450. doi: 10.1042/cs0780445. [DOI] [PubMed] [Google Scholar]
- 40.Wise GE, Lin F, Fan W. Localization of epidermal growth factor and its receptor in mandibular molars of the rat prior to and during prefunctional tooth eruption. Dev. Dyn. 1992;195:121–126. doi: 10.1002/aja.1001950206. [DOI] [PubMed] [Google Scholar]
- 41.Cielinski MJ, Jolie M, Wise GE, Marks JSC. The contrasting effects of colonystimulating factor-1 and epidermal growth factor on tooth eruption in rat. Con. Tissue. Res. 1995;1:165–169. doi: 10.3109/03008209509013720. [DOI] [PubMed] [Google Scholar]
- 42.Deméré TA, McGowen MR, Berta A, Gatesy J. Morphological and molecular evidence for a stepwise evolutionary transition from teeth to baleen in mysticete whales. Syst. Biol. 2008;57:15–37. doi: 10.1080/10635150701884632. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa H, Amasaki H, Dohguchi H, Furuya A, Suzuki K. Immunohistological distributions of fibronectin, tenascin, type I, III and IV collagens, and laminin during tooth development and degeneration in fetuses of minke whale, Balaenoptera acutorostrata. J. Vet. Med. Sci. 1999;61:227–232. doi: 10.1292/jvms.61.227. [DOI] [PubMed] [Google Scholar]
- 44.Luetteke NC, et al. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 45.Blackburn TM, Gaston KJ, Loder N. Geographic gradients in body size: A clarification of Bergmann’s rule. Divers. Distrib. 1999;5:165–174. doi: 10.1046/j.1472-4642.1999.00046.x. [DOI] [Google Scholar]
- 46.Sun Y, et al. Insights into body size variation in cetaceans from the evolution of body-size related genes. BMC Evol. Biol. 2019;19:157. doi: 10.1186/s12862-019-1461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai K, Mitaku S. Mechanisms of secondary structure breakers in soluble proteins. Biophysics. 2005;1:55–65. doi: 10.2142/biophysics.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacob J, Duclohier H, Cafiso DS. The role of proline and glycine in determining the backbone flexibility of a channel-forming peptide. Biophys J. 1999;76:1367–1376. doi: 10.1016/S0006-3495(99)77298-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai JY, et al. Analysis of polymorphism of growth hormone secretagogue receptor in sheep. Pak. J. Zool. 2020;52:1161–1164. doi: 10.17582/journal.pjz/20190320070305. [DOI] [Google Scholar]
- 50.Korbonits M, Kojima M, Kangawa K, Grossman AB. Presence of ghrelin in normal and adenomatous human pituitary. Endocrine. 2001;14:101–104. doi: 10.1385/ENDO:14:1:101. [DOI] [PubMed] [Google Scholar]
- 51.Kojima M, Kangawa K. Ghrelin: Structure and function. Physiol. Rev. 2005;85:495–522. doi: 10.1152/physrev.00012.2004. [DOI] [PubMed] [Google Scholar]
- 52.Moreira A, Soares JB. Ghrelin and ghrelin receptor inhibitors: Agents in the treatment of obesity. Expert. Opin. Ther. Targets. 2008;12:1177–1189. doi: 10.1517/14728222.12.9.1177. [DOI] [PubMed] [Google Scholar]
- 53.Colinet FG, et al. Genomic location of the bovine Growth Hormone Secretagogue Receptor (GHSR) Gene and investigation of genetic polymorphism. Anim. Biotechnol. 2009;20:28–33. doi: 10.1080/10495390802602926. [DOI] [PubMed] [Google Scholar]
- 54.Zhang B, et al. Associations of polymorphism within the GHSR gene with growth traits in Nanyang cattle. Mol. Biol. Rep. 2009;36:2259–2263. doi: 10.1007/s11033-008-9442-x. [DOI] [PubMed] [Google Scholar]
- 55.Bahrami A, et al. Genetic polymorphisms and protein structures in growth hormone, growth hormone receptor, ghrelin, insulin-like growth factor 1 and leptin in Mehraban sheep. Gene. 2013;527:397–404. doi: 10.1016/j.gene.2013.05.066. [DOI] [PubMed] [Google Scholar]
- 56.Burger AM, Leyland-Jones K, Banerjee DD, Spyropoulos AK. Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur. J. Cancer. 2005;41:1515–1527. doi: 10.1016/j.ejca.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 57.Akaogi K, et al. Specific accumulation of tumor-derived adhesion factor in tumor blood vessels and in capillary tube-like structures of cultured vascular endothelial cells. Proc. Natl. Acad. Sci. 1996;93:8384–8389. doi: 10.1073/pnas.93.16.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sprenger CC, Damon SE, Hwa V, Rosenfeld RG, Plymate SR. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) is a potential tumor suppressor protein for prostate cancer. Cancer Res. 1999;59:2370–2375. [PubMed] [Google Scholar]
- 59.Wilson HM, Birnbaum RS, Poot M, Quinn LS, Swisshelm K. Insulin-like growth factor binding protein-related protein 1 inhibits proliferation of MCF-7 breast cancer cells via a senescence-like mechanism. Cell. Growth. Differ. 2002;13:205–213. [PubMed] [Google Scholar]
- 60.Bermejo AL, et al. Insulin resistance is associated with increased serum concentration of IGF-binding protein-related protein 1 (IGFBP-rP1/MAC25) Diabetes. 2006;55:2333–2339. doi: 10.2337/db05-1627. [DOI] [PubMed] [Google Scholar]
- 61.Tollis M, et al. Return to the sea, get huge, beat cancer: An analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae) Mol. Biol. Evol. 2019;36:1746–1763. doi: 10.1093/molbev/msz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eberlein A, et al. Dissection of genetic factors modulating fetal growth in cattle indicates a substantial role of the Non-SMC Condensin I Complex, Subunit G (NCAPG) Gene. Genetics. 2009;183:951–964. doi: 10.1534/genetics.109.106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weikard R, et al. Metabolomic profiles indicate distinct physiological pathways affected by two loci with major divergent effect on Bos taurus growth and lipid deposition. Physiol. Genom. 2010;42:79–88. doi: 10.1152/physiolgenomics.00120.2010. [DOI] [PubMed] [Google Scholar]
- 64.Setoguchi K, et al. The SNP c.1326T>G in the non-SMC condensin I complex, subunit G (NCAPG) gene encoding a p.Ile442Met variant is associated with an increase in body frame size at puberty in cattle. Anim. Genet. 2011;42:650–655. doi: 10.1111/j.1365-2052.2011.02196.x. [DOI] [PubMed] [Google Scholar]
- 65.Tetens J, Widmann P, Kuhn C, Thaller G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 2013;44:467–471. doi: 10.1111/age.12031. [DOI] [PubMed] [Google Scholar]
- 66.Shen J, et al. Genomic analyses reveal distinct genetic architectures and selective pressures in Chinese donkeys. JGG. 2020 doi: 10.21203/rs.3.rs-111083/v1. [DOI] [PubMed] [Google Scholar]
- 67.Rubin CJ, et al. Strong signatures of selection in the domestic pig genome. PNAS. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human heigh. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 69.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sasaki S, et al. Genetic mapping of quantitative trait loci affecting body weight, egg character and egg production in F2intercross chickens. Anim. Genet. 2004;35:188–194. doi: 10.1111/j.1365-2052.2004.01133.x. [DOI] [PubMed] [Google Scholar]
- 72.Fortes MRS, et al. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian Beef cattle. Anim Genet. 2013;44:636–647. doi: 10.1111/age.12075. [DOI] [PubMed] [Google Scholar]
- 73.Xu P, et al. Genome-wide association study for growth and fatness traits in Chinese Sujiang pigs. Anim Genet. 2020;51:314–318. doi: 10.1111/age.12899. [DOI] [PubMed] [Google Scholar]
- 74.Pan Y, et al. Indel mutations of sheep PLAG1 gene and their associations with growth traits. Anim. Biotechnol. 2021 doi: 10.1080/10495398.2021.1906265. [DOI] [PubMed] [Google Scholar]
- 75.Karim L, et al. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 2011;43:405–413. doi: 10.1038/ng.814. [DOI] [PubMed] [Google Scholar]
- 76.Juma AR, et al. Emerging role of PLAG1 as a regulator of growth and reproduction. J. Endocrinol. 2016;228:45–56. doi: 10.1530/JOE-15-0449. [DOI] [PubMed] [Google Scholar]
- 77.Erdenee S, et al. Sheep zinc finger proteins 395 (ZNF395): Insertion/deletion variations, associations with growth traits, and mRNA expression. Anim. Biotechnol. 2020;31:237–244. doi: 10.1080/10495398.2019.1585865. [DOI] [PubMed] [Google Scholar]
- 78.Fink T, et al. Functional confirmation of PLAG1 as the candidate causative gene underlying major pleiotropic effects on body weight and milk characteristics. Sci. Rep. 2017;7:44793. doi: 10.1038/srep44793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keane M, et al. Insights into the evolution of longevity from the bowhead whale genome. Cell. Rep. 2015;10:112–122. doi: 10.1016/j.celrep.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high-throughput. Nucleic. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lambert O, et al. The giant bite of a new raptorial sperm whale from the Miocene epoch of Peru. Nature. 2010;466:105–108. doi: 10.1038/nature09067. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 84.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 86.Beck RMD, Baillie C. Improvements in the fossil record may largely resolve current conflicts between morphological and molecular estimates of mammal phylogeny. Proc. R. Soc. B. 2018;285:20181632. doi: 10.1098/rspb.2018.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGowen MR, et al. Phylogenomic resolution of the cetacean Tree of Life using target sequence capture. Syst. Biol. 2020;69:479–501. doi: 10.1093/sysbio/syz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith MD, et al. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015;32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wertheim JO, et al. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015;2:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murrell B, et al. FUBAR: A fast, unconstrained Bayesian approximation for inferring selection. Mol. Biol. Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pond SL, Frost SD. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 92.Murrell B, et al. Detecting individual sites subject to episodic diversifying selection. PLoS. Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. TreeSAAP: Selection on amino acid properties using phylogenetic trees. Bioinformatics. 2003;19:671–672. doi: 10.1093/bioinformatics/btg043. [DOI] [PubMed] [Google Scholar]
- 94.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 95.Weaver S, et al. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murrell B, et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015;32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spielman SJ, et al. Evolution of viral genomes: Interplay between selection, recombination, and other forces. Methods. Mol. Biol. 2019;1910:427–468. doi: 10.1007/978-1-4939-9074-0_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analysed during the current study are available in the Supplementary Files.