Abstract
Background
In clinical genetics, establishing an accurate nosology requires analysis of variations in both etiology and the resulting phenotypes. At the phenotypic level, recognizing typical facial gestalts has long supported clinical and molecular diagnosis; however, the objective analysis of facial phenotypic variation remains underdeveloped. In this work we propose exploratory strategies for assessing facial phenotypic variation within and among clinical and molecular disease entities and deploy these techniques on cross-sectional samples of four RASopathies: Costello syndrome (CS), Noonan syndrome (NS), cardiofaciocutaneous syndrome (CFC), and neurofibromatosis type 1 (NF1).
Methods
From 3D dense surface scans, we model the typical phenotypes of the four RASopathies as average ‘facial signatures’ and assess individual variation in terms of direction (what parts of the face are affected and in what ways) and severity of the facial effects. We also derive a metric of phenotypic agreement between the syndromes and a metric of differences in severity along similar phenotypes.
Results
CFC shows a relatively consistent facial phenotype in terms of both direction and severity that is similar to CS and NS, consistent with the known difficulty in discriminating CFC from NS based on the face. CS shows a consistent directional phenotype that varies in severity. Although NF1 is highly variable, on average it shows a similar phenotype to CS.
Conclusions
We established an approach that can be used in the future to quantify variations in facial phenotypes between and within clinical and molecular diagnoses to objectively define and support clinical nosologies.
Keywords: Phenotype, methods, diagnosis
Introduction
Nosology concerns the definition and delineation of diseases. It requires a balance between ‘lumping’ similar diseases together and ‘splitting’ others into separate entities [1]. In clinical genetics, this process requires consideration of both etiology and the resulting phenotype [2]. At the etiological level, next generation sequencing has greatly informed our understanding of the genetics underlying many syndromes. This includes genotype-first approaches, which have revealed that many monogenic syndromes, previously thought to be well-defined, exhibit a much broader phenotypic spectrum than previously realized [3, 4]. These findings have forced the definitions of certain syndromes and syndrome families to be revised [e.g. 5-7].
At the phenotypic level, inter- and intra- syndrome variation can be difficult to define objectively. Facial phenotyping in clinical genetics usually relies on recognition of a facial gestalt that is considered typical for a given clinical or molecular diagnosis and the translation of observations into standardized terminology [8]. In recent years this has been augmented with computer software, such as ‘Face2Gene’ [9, 10], for automated recognition of facial gestalts typical of various syndromes. Nevertheless, recognizing facial gestalts does not yield straightforward measures of phenotypic variation and similarity. So-called ‘clinical face phenotype spaces’ [11, 12] are high-dimensional spaces derived from patient images, wherein distances correspond to facial similarities such that similar patients and phenotypically similar disorders are positioned together. Despite the potential to inform nosological discussions, it remains unclear how phenotypic variation and similarity should be assessed within such a space. Different molecular or clinical entities can exhibit similar facial changes to the same parts of the face, but with different severity [13]. Alternatively, different parts of the face may be affected, or the same parts may be affected in different ways. In this work we develop an exploratory strategy for quantitatively describing the variation within, and similarities and differences among, facial phenotypes using dense 3D surface scans. We deploy this approach to analyze four RASopathies. RASopathies are a family of well-studied disorders of the RAS/MAPK pathway, comprising Costello syndrome (CS), Noonan syndrome (NS), cardiofaciocutaneous syndrome (CFC) and neurofibromatosis type I (NF1), among others. Facial similarity and variation are well-understood clinically within this family of disorders, making them an interesting test case for the proposed methodology.
Materials and methods
Sample
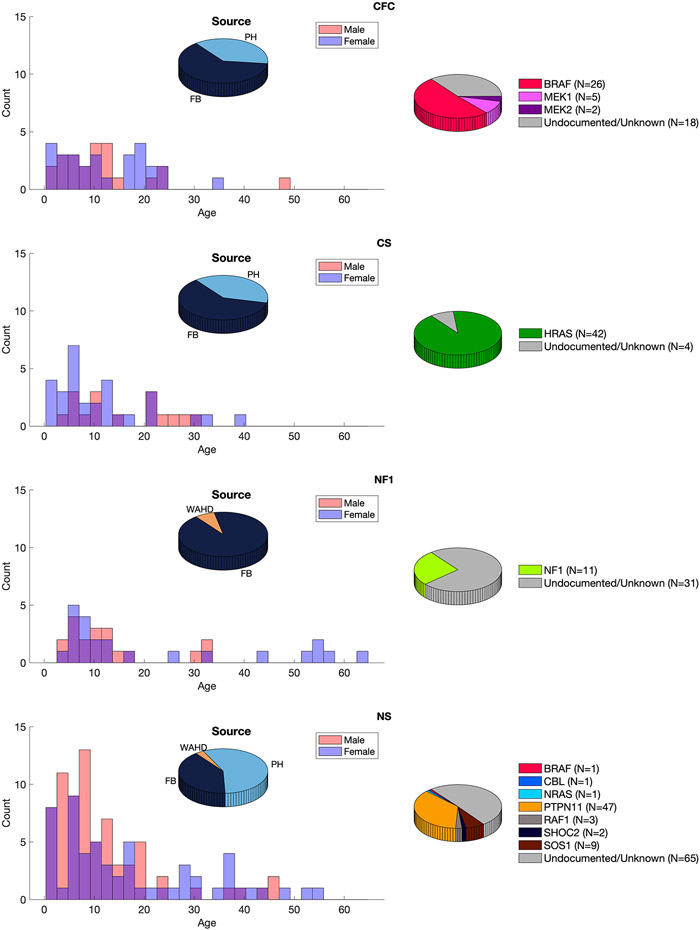
The RASopathy patient sample is derived from three established resources: 1) the FaceBase repository (www.facebase.org; FB00000861) [14]; 2) the database of the Western Australian Health Department; and 3) Peter Hammond’s legacy 3D dysmorphology dataset hosted at KU Leuven, Belgium. Data ascertainment is described in Supplementary Text 1. The study used all available data after excluding participants who 1) had no image of acceptable quality; 2) did not have necessary demographic data (age, sex and ancestry) reported; 3) failed image registration (see below) 4) were a second image of a patient already included in the analysis; or 5) were of non-European ancestry, This last criterion was applied because each patient is assessed relative to an openly available normative reference for 3D facial shape (see below), which is based only on participants of European ancestry. The final dataset comprised patients with a diagnosis of NS (N=129; 57 female), NF1 (N=42; 23 female), CFC (N=51, 28 female), or CS (N=46; 30 female). Supplementary Table 1 shows numbers of participants remaining after each exclusion criterion was applied. Figure 1 illustrates the final dataset broken down by age, sex, clinical diagnosis and molecular diagnosis at the level of the affected gene. Supplementary Table 2 reports further details of the molecular diagnosis per participant.
Figure 1.
The RASopathy sample. Left shows the distribution of ages for each sex per group and the proportion of subjects coming from each of the three databases. Right shows numbers of molecularly confirmed (broken down by the affected gene) and unconfirmed cases. FB, the FaceBase repository; PH, Peter Hammond’s legacy three-dimensional dysmorphology collection; WAHD, the Western Australian Department of Health.
Image pre-processing
Image processing
To obtain standard facial representations, each 3D facial photograph was non-rigidly registered with a standard template using the ‘MeshMonk’ MATLAB toolbox [15, 16], resulting in a representation of each face as a standard set of 7160 points. Images were visually inspected and were excluded if the registration had failed.
Each standardized point configuration was then converted into a ‘facial signature’ [17] which codes the deviations of each point on each patient from an age and sex matched normative reference face as z-scores. This essentially removes variation due to normal growth and sex differences on the face. This was done using the open-source 3D Growth Curves and Facial Assessment Toolbox in MATLAB, a normative reference for 3D facial shape based on a sample with European ancestry [18]. Facial signatures were calculated along the x (lateral-medial), y (inferior-superior), and z (anterior-posterior) direction as well as the direction normal to the facial surface at each quasi-landmark. Facial signatures in the x, y and z directions were concatenated to define a single feature vector for each patient, which was used for all subsequent computation. These procedures essentially define a position for each patient in a high-dimensional space where each element of the feature vector is an axis and distance corresponds to facial similarity. The signatures along the surface normal were only used in visualizations. Furthermore, for visualization, an age- and sex-normalized facial shape of each patient was created by subtracting the coordinates of the age and sex-specific expected face from the coordinates of the patient and adding back on the coordinates of the overall average face of the 3D Growth Curve training data.
Assessing phenotypic consistency within the RASopathies
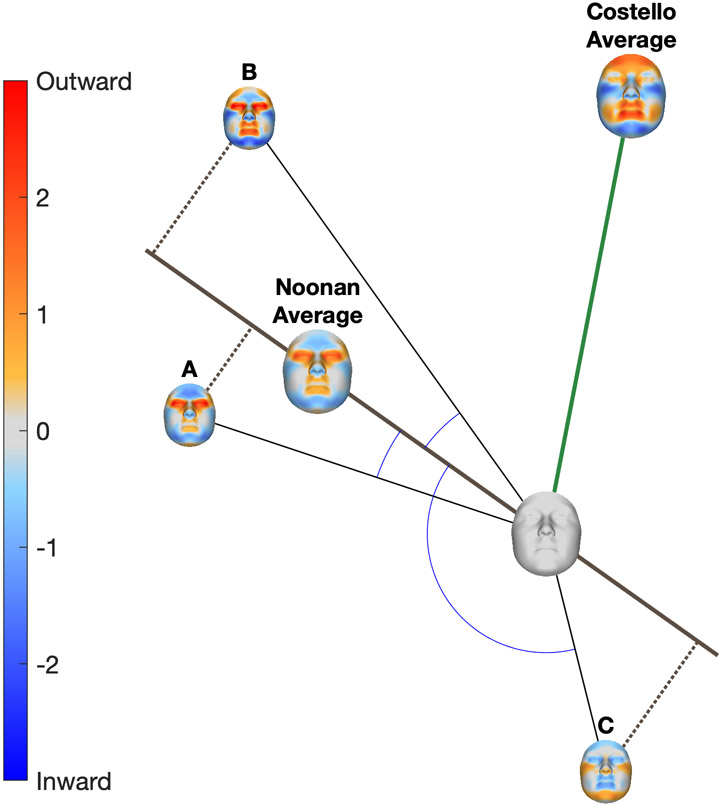
We assessed phenotypic consistency in terms of variation in direction (what parts of the face are affected and in what ways) and severity (to what degree is the face affected in a manner that is typical of the syndrome). The foregoing calculations can be interpreted geometrically as illustrated in Figure 2. The aggregate phenotype of each RASopathy was defined as the mean feature vector (mean signature) of all patients with the syndrome. Directional similarity of the ith individual to the jth syndrome mean signature was calculated as the cosine distance between the individual and group feature vectors:
where ai is the feature vector of the individual and μj is the average feature vector of the jth syndrome. This defined the typical phenotype of each syndrome as a transformation away from normal. Geometrically speaking, this was a direction or vector and similarity to it was measured as an angle (Figure 2). Anatomically, an example direction might, for instance, be loosely verbally described as the distance between the eyes widening in conjunction with a shrinking chin. Patients displaying this pattern (irrespective of severity) will have a small cosine distance to the mean. In the example, a patient with severe hypertelorism and micrognathia will have a low cosine distance, as will a patient with less severe hypertelorism and micrognathia, as well as a patient that is within normal range but has relatively widely spaced eyes and a small chin. A patient with the inverse difference (e.g., hypotelorism and macrognathia) will have a high cosine distance. The cosine distance is readily interpretable as it is on a normalized scale from 0-2. Values greater than 1 indicate the patient is better described displaying the inverse of the typical pattern than the typical pattern.
Figure 2.
Description of cosine distance and severity measures. Each patient (A, B and C) is represented by their facial signature. The facial signature is shown graphically as a colour-coded map where red indicates the point on the face of the patient is displaced outwardly relative to normal and blue indicates it is displaced inwardly. The average signatures of all patients with Noonan syndrome and all patients with Costello syndrome are shown by the larger colour maps. Geometrically, each signature can be interpreted as a vector. Anatomically each signature can be interpreted as a particular transformation of facial shape away from average (blank signature), for example, eyes widening (red) in combination with a shrinking (blue) chin. Similarity, in terms of what parts of the face are affected in what ways is measured by the angle between two vectors. For example, patients A and B have small angles (low cosine distance) to the Noonan phenotype, indicating they are affected in a way that is characteristic of Noonan syndrome.
For the ith individual, their severity on the jth mean signature was:
Geometrically, this was a projection onto the average signature and was a scalar measure of the magnitude of the facial effect in the direction of the average signature. In the example introduced above, a patient with more extreme micrognathia and hypertelorism will have a higher severity score than one with these features to a lesser degree. Variation in direction within a syndrome was measured as the root mean squared cosine distance from their average signature; we call this the ‘directional variation statistic’. Variation in severity was measured by the standard deviation of their severity scores; we call this the ‘severity variation statistic’.
Assessing phenotypic variation among the RASopathies
Two clinical or molecular entities may drive variation along similar or different directions. We assess this for each pair of disorders using the cosine of the angle between the mean signatures of the two disorders μI and μj:
We call this this the ‘phenotype agreement score’. This ranges from 1 indicating the two signatures are collinear and point in the same direction to −1, indicating the two signatures are collinear and point in opposite directions.
Clinical or molecular disease entities may produce similar directional phenotypes but differ in severity. Therefore, we suggest a complementary measure: the ‘severity difference score’. For each pair of syndromes, we compute the mean of the two average signatures and compute the severity scores of patients along this combined phenotype. We then compare the distributions of severity scores between the two syndromes using Cohen’s d statistic, which is the difference between the syndrome means divided by the average of the standard deviations of the two groups. Throughout we use a leave-one-out approach, where the patient being scored is excluded from the estimation of the average signature. To estimate confidence intervals of each statistic observations within each syndrome were randomly resampled 1000 times and the cosine distances, severity and derived statistics were recalculated.
Interpreting the magnitude of the statistics
To estimate what constitutes moderate, strong and very strong consistency, we computed the two variation statistics on a sample of 39 syndromes and craniofacial malformations. We defined values indicating 'moderate’, ‘strong’, and ‘very strong’ consistency as those lower than the 20th, 10th and 5th percentiles, respectively, of the distributions of these statistics. Similarly, to determine what is a ‘moderate’, ‘strong’, or ‘very strong’ phenotype agreement or severity difference we computed these statistics for all pairs of syndromes in the expanded dataset and used thresholds corresponding to the 80th, 90th, and 95th percentiles, respectively. These thresholds are arbitrary but quite conservative compared to common guidelines for interpreting other effect sizes. For comparison commonly used thresholds for Cohen’s d (small=0.3, mod=0.5, large=0.8) indicate the mean of the second sample is positioned at the 62nd 69th and 79th percentiles of the first sample, respectively. The data used are described in Supplementary Table 3, and the distributions of the statistics are reported in Supplementary Figure 1 and Supplementary Table 4.
Results
Average Signatures
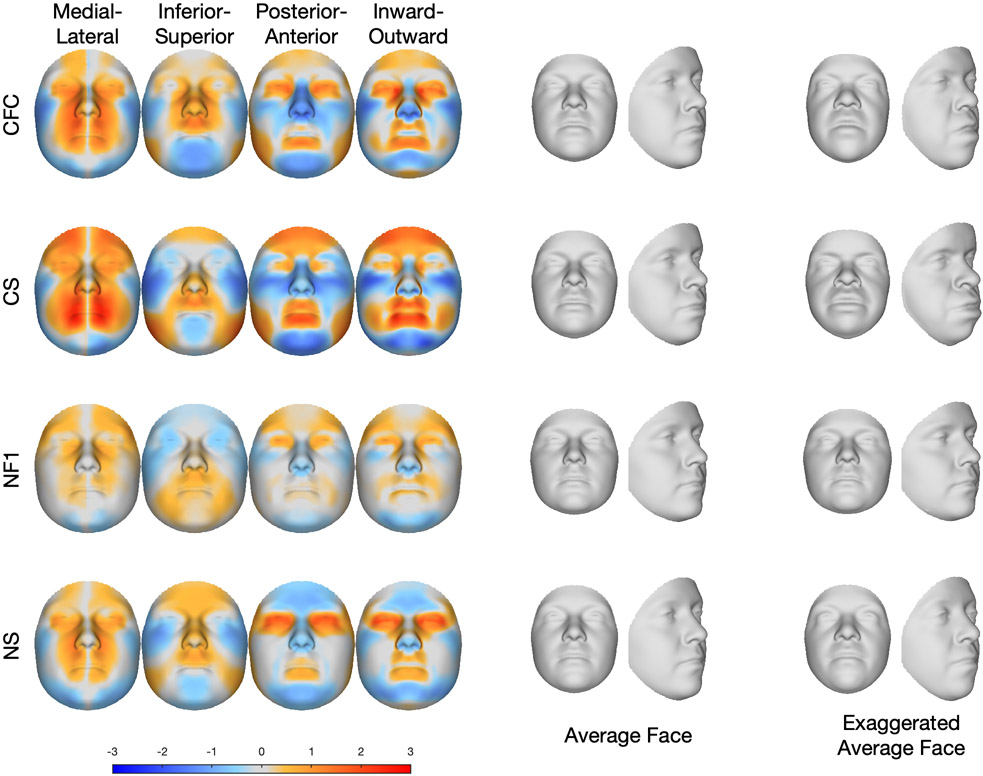
In this section we assess the typical facial phenotypes of each RASopathy. Figure 3 illustrates the average signature of each of the RASopathies. The signatures in the left panel describe the transformation visually in the horizontal (blue indicates lateral displacement, red indicates medial displacement), vertical (blue indicates inferior displacement, red indicates superior displacement), and depth (blue indicates posterior displacement, red indicates anterior displacement) directions as well as the directions locally perpendicular to the surface (blue indicates locally inward displacement, red indicates locally outward displacement). Many dysmorphic features typically used in clinical genetics can be inferred from particular color patterns in these signatures: anterior and locally outward displacement of the points on the lips is consistent with full lips; posterior and inward displacement of the zygomatic region is consistent with malar hypoplasia; superior and posterior displacement of the points of the nose is consistent with a short, depressed nose; and inward and posterior displacement of points on the chin, together with a medial displacement of the left and right sides of the chin, indicate a retruded chin and narrow jaw consistent with micrognathia. These four clinical features, while most pronounced in CS, are present to some degree in all four of the RASopathy syndromes. Anterior displacement of the forehead indicates forehead prominence, which occurs to differing degrees in CFC, CS and NF1. Complex changes in all three dimensions, although also coded in the facial signatures, are most easily visually appreciated by inspecting the average and exaggerated faces. For example, the eyes in CFC and NS are prominent and wide-spaced with down-slanting palpebral fissures.
Figure 3.
Average facial signatures in the horizontal direction (medial displacement=red; lateral displacement=blue); vertical direction (inferior displacement=blue, superior displacement=red); and depth (posterior displacement=blue, anterior displacement=red) and the direction locally perpendicular to the face (locally inward displacement=blue, locally outward displacement=red). The middle panel shows the average age and sex normalised face of each group, and the final panel shows an exaggerated version of theage and sex normalised average face.
Phenotypic consistency within the RASopathies
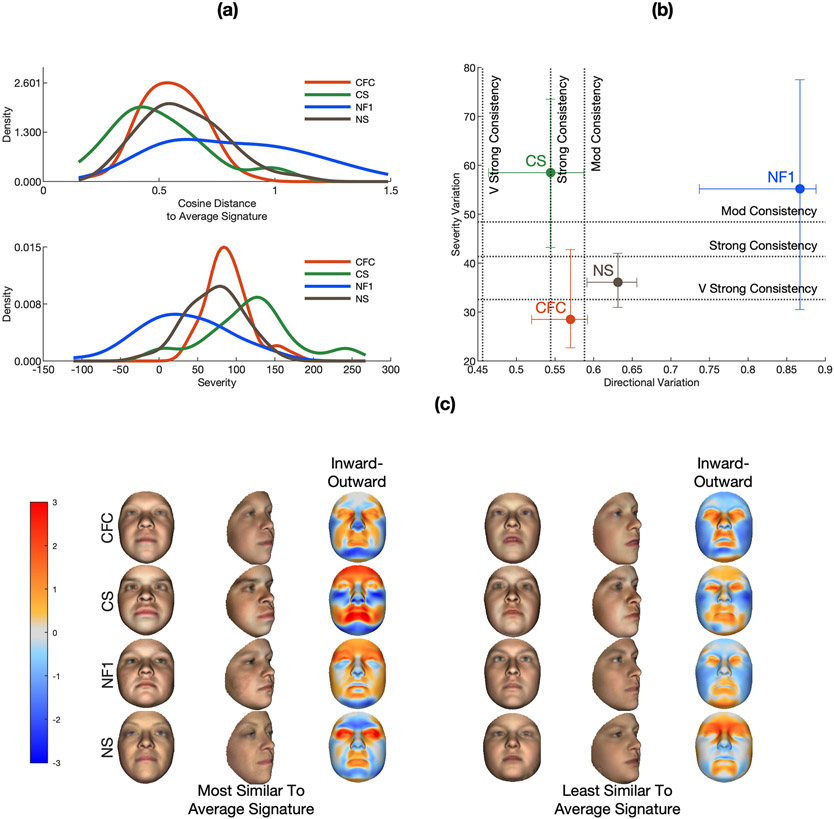
In this section we assess the consistency of the facial phenotypes displayed within the samples of each of the four RASopathies. As described in the methods, similarity to the average signature and severity was measured for each individual. Kernel densities, fitted to the distributions of these statistics for each syndrome, are shown in Figure 4a. Syndromes in which the face is typically affected in the same direction (the same facial features are transformed in the same way, though potentially to different degrees) will have generally low cosine distances. This is indicated by the central tendency of these distributions and is summarized for each syndrome in the directional variation statistic (Figure 4b). Individuals may also vary in the degree of this transformation (‘severity’), and syndromes may vary in how much individuals within the syndrome vary in severity. This is captured in the dispersion of the distributions of severity scores (Figure 4a) which is measured for each syndrome in the severity variation statistic (Figure 4b). The central tendency of the severity distributions also partially reflects the average magnitude of the effect on the face; however, only in the direction modelled by the average signature; for example, faces exhibiting an extreme phenotype that is dissimilar to the average signature could have a low severity score. To put the two variation statistics into context, the estimated cut-offs indicating moderate, strong and very strong consistency are plotted as vertical and horizontal lines on Figure 4b. To visually illustrate the breadth of the phenotypes observed within each syndrome Figure 4c shows average faces and signatures of the five patients that are most and the five patients that are least similar to the average signature (left). The phenotypes in CS and CFC show moderate to strong consistency in direction. NS shows slightly lower consistency and NF1 shows very low consistency. CS and NF1 shows low consistency in severity whereas CFC and NS are more consistent. This indicates that within CS and within CFC, and to a lesser extent within NS, the face is usually affected in similar directions (facial features are affected in similar ways). CS stands out because the direction of the effect is very consistent, though with variable severity (facial features may be affected to a greater or lesser degree). The phenotype of NF1 shows low consistency in terms of both direction and severity.
Figure 4.
Consistency of the phenotypes of the four RASopathies. Figure part A plots fitted kernel densities to distributions of severity scores and cosine distance for each of the four syndromes. Figure part B plots the directional and severity variation statistics along with the defined cut-offs for mod, strong and very strong consistency. Error bars indicate the 95% CIs of the statistics estimated by resampling (see Materials and methods). Figure part C shows the average age-normalised and sex-normalised faces and of the five patients that are most and the five patients that are least similar (in terms of cosine distance) to the average signatures for each of the four groups as well as their average facial signatures, computed along the surface normals (red indicates locally outward displacement; blue indicates locally inward displacement).
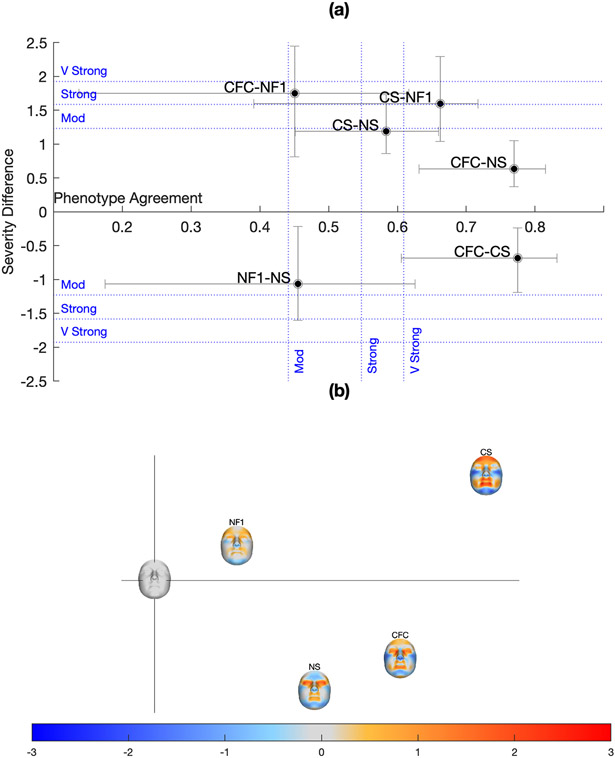
Phenotype agreement and severity differences among the RASopathies
In this section we assess similarities and differences, in terms of direction and severity, among the RASopathy phenotypes. The relationship among the four syndrome phenotypes is summarized graphically in Figure 5. Part (a) plots the phenotype agreement and severity difference statistics for each pair of syndromes. In terms of phenotype agreement, CFC is very strongly similar to NS and to CS, while CS is less similar (in the strongly-very strongly range) to NS. This indicates that CFC phenotype is intermediate between NS and CS. NF1 is very strongly similar to CS.
Figure 5.
Phenotype agreement and severity difference among the RASopathies. Figure part A plots the phenotype agreement and severity difference statistics for each pair of RASopathies. The sign of the severity difference indicates whether the first syndrome was more severe than the second (positive) or the opposite (negative). Error bars indicate the 95% CIs of the statistics. Figure part B plots the average signatures on the first two PCs of an uncentred principal components analysis of the average signatures and a signature that is all zeros (the blank face at the origin). The facial signatures shown here are computed along the surface normals (red indicates locally outward displacement; blue indicates locally inward displacement).
To assess differences in severity between two syndromes along a single direction, the average signatures of both syndromes were averaged and the two groups were ordinated and compared along this combined signature. This assessment is less meaningful when there is less agreement between the average signatures of both groups. NF1 shows a strong to very strong severity difference to CS and CFC, with NF1 being less severe, and is very strongly similar to CS. CFC paired with NS show less than moderate severity difference, as does CFC paired with CS, CS paired with NS, and NF1 paired with NS.
The overall relationship among the average signatures is also approximated graphically on the first two axes of an uncentered PCA of the average signatures and a signature that is all zeros (Figure 5b). Here, the similarity between NF1 and CS is shown by their position along a similar vector from the origin, but at different distances, reflecting the difference in severity.
Discussion
Over the past two decades, discovery of the molecular bases of many disorders has revealed unsuspected biological relationships among disorders that were previously thought to be unrelated [4]. At the same time, it has become clear that, for many Mendelian disorders, the range of associated phenotypes is considerably broader than was realized initially [3, 4]. In many cases this has resulted in substantial revision of syndrome nosology [e.g. 5-7]. The recognition of typical facial gestalts has long supported clinical delineation of craniofacial syndromes. However, as the range of associated phenotypes for a given syndrome has broadened, the facial phenotypic gestalt often has become unclear.
Clinical face phenotype spaces [11, 12] position individuals within a continuous high-dimensional space, where informative image-based features, derived for all patients, are axes. Facially similar individuals and groups of individuals are positioned closely together in the space. Despite their potential for informing syndrome nosology no established framework exists for establishing how syndromes are interrelated within such a space and how syndromes vary internally. In this work we establish a CFPS from dense 3D surface scans, modelling variation in four RASopathies and develop a framework for exploring phenotypic variation within and among them.
An intuitively appealing approach to appreciate variation in a clinical face phenotype space is to visualize the high-dimensional space as projections onto a low-dimensional space, via a dimension reduction technique such as principal components analysis [19, 20], t-SNE [12, 21] or signature graph analysis [17]. Any low-dimensional projection of a high-dimensional space will lose some aspects of the high-dimensional variation and a weakness of the aforementioned approaches is that the axes of the low-dimensional space are not necessarily biologically informative. Here, we began by modelling each syndrome group as a vector from average to the average signature of the syndrome. Projections onto this vector correspond to the degree to which an individual displays this combination of features. This differs from and complements other metrics of overall severity such as distance from the origin or the ‘signature weight’ [22] as it measures only variation that is most typical of the syndrome and has previously been shown to represent a biologically meaningful axis of ‘severity’. For example position along similarly-estimated axis was shown to correlate with the size of the causal deletion in Wolf-Hirschhorn syndrome [13]. The natural complement to this is angular variation with respect to the average signature vector, which corresponds to deviation from the typical facial transformation. To assess consistency of the facial phenotype, we use two univariate variation statistics based on angular similarity to and variation in the projection onto the average signature. These differ from other metrics of overall variation such as the trace of the within-class covariance matrix [23] in that they partition the variation into two separate components with different meanings. To investigate similarities and differences between facial phenotypes we measure directional similarity between each pair of average signatures and differences in severity between each pair of syndromes along a joint average signature. To interpret the magnitude of these statistics and to ascertain what are biologically meaningful values, we defined cut-offs based on distributions of similarly computed statistics from a larger sample of 39 genetic syndromes and craniofacial malformations. This constitutes a stronger approach than statistical hypthesis testing, which would only demostrate that the values of the statistics are non-zero in the population. However, this is limited by the particular constitution of this larger sample of syndromes, which cannot be assumed to be wholly representative of the population of all relevant conditions. Our particular choice of thresholds is conservative relative to most guidelines for interpreting effect sizes, although remains essentially arbitrary. More work is needed to understand fully how the magnitude of these statistics should be interpreted. Particularly how the variation statistics present in genetically homogeneous, as opposed to heterogeneous conditions can be investigated. Furthermore, it should be established how the statistics relate to measures of biological relatedness between syndromes, such as DNA methylation epi-signature similarity [24].
While acknowledging the above limitation these statistics can be interpreted in combination to more fully assess phenotypic variation among patient groupings, both within and between syndromes. Here we highlight some key findings in the context of clinical knowledge about the four RASopathies analysed in the study. CFC shows a relatively consistent phenotype in terms of both direction and severity. This is unexpected from a clinical point of view, as CFC is usually difficult to discriminate from CS and NS based on the face. This finding is more understandable when one also considers the phenotypic similarity among CS, CFC and NS. CFC is very strongly similar to CS and to NS. The average signatures illustrate that the phenotype of CFC entails a strong component of orbital hypertelorism as in NS while also displaying a more prominent forehead and full lips and cheeks, similar to CS. The face of CS is usually easy to recognize clinically, consistent with strong directional consistency of the phenotype, which comprises full-lips, malar hypoplasia, depressed nose, protruding forehead, and retrognathia. More surprising is the low consistency (high variation) in severity for CS. As CS is genetically relatively homogeneous and is, with very few exceptions, caused by variants in the HRAS gene, the biological underpinnings of this variation are an important avenue for future research. NF1 shows a generally mild but highly variable phenotype in terms of both direction and severity, consistent with clinical lore that NF1 does not have a distinctive facial phenotype. Nevertheless, the average signature of NF1 is very strongly similar to that of CS, albeit with a strong severity difference between the two syndromes, indicating that the phenotype of NF1 is to some extent a milder version of that of CS. While some cases of NF1 have previously been found to be similar to NS [25], the similarity between NF1 and NS is weaker than between NF1 and CS. Age-related changes to the statistics are investigated in Supplementary Text 1. Directional variation declines with age for both NS and CS. CFC and CS increase in severity with increasing age but variation in severity remains constant. Phenotype agreement between all pairs of syndromes, except NF1 paired with CS, declines with age. Severity difference increases with age for syndrome pairs CFC-NF1, CS-NF1 and NS-NF1.
The four RASopathies considered in this study are clinically defined entities which we treated as unified groups. It is possible that this may inflate variation and obscure interesting subtypes as these disorders are, to different degrees, genetically heterogeneous. CFC and NS can each be caused by variants in several genes including some genes (e.g., BRAF) that can cause both. NF1 is caused exclusively by variants in the NF1 gene, however patients with microdeletions show relatively severe NF1 phenotypes [26]. CS is almost exclusively caused by a small number of variants affecting various codons in the HRAS gene. Analysis of molecularly defined categories is not feasible given the small sample sizes of patients with less-common pathogenic variants available to us but is an important avenue for future work. Another possible effect of our reliance on clinical categorization may be to reduce variation. In the absence of molecular confirmation, or in cases where the pathogenic variant does not uniquely specify the clinical diagnosis, the facial phenotype may have already been factored into the diagnosis. This could have reinforced the facial differences between the groups. This may be especially the case for NS and CS where the face is commonly used as part of diagnosis. This is likely to have had little influence on NF1 where the presence of particular facial features is not a diagnostic criterion [27, 28]. Another possible impact of reliance on clinical diagnosis is the possible presence of misdiagnoses in the dataset, especially for patients where the diagnosis is not confirmed molecularly.
The application of next-generation sequencing in clinical genetics has rapidly expanded our understanding of the etiology of many disorders; however, the complementary development of deep phenotyping technology has lagged far behind. Here, we have developed an approach to quantitatively measure both within and between-syndrome phenotypic variation, consistency, and severity, and we apply these techniques to characterize both similarity and differences of both phenotype and severity. We applied these techniques to analyze a clinically well-studied group of related disorders characterized by variants in genes encoding protein components of the RAS/MAPK pathway, and we show how this approach can highlight phenotypic relationships among these related clinical entities. In the future, we anticipate that these techniques can contribute to developing a more objective nosology in clinical genetics.
Supplementary Material
Key messages:
What is already known on this topic?
Nosology in clinical genetics requires consideration of both etiology and the resulting phenotype. While next generation sequencing has advanced our understanding of the etiology of many genetic conditions, methods for analyzing facial phenotypic variation are underdeveloped.
What this study adds?
This study describes a method for analyzing variation in the facial phenotype within and among related conditions and applies the methods to samples of four RASopathies: Costello syndrome, Noonan syndrome, cardiofaciocutaneous syndrome and neurofibromatosis type 1.
How this study might affect research, practice or policy?
This study facilitates the objective study of variation in facial phenotypes with a view to the development of more objective nosologies in clinical genetics.
Acknowledgements
We acknowledge Professor Judith Allanson from the University of Ottawa and Professor Raoul Hennekam from the University of Amsterdam for their contributions to recruitment and data collection.
Funding
This work was supported by the research fund KU Leuven (BOF-C1, C14/15/081 & C14/20/081; PC); NIH-NIDCR (U01DE024440; RS, OK and BH); the Research Program of the Research Foundation Flanders (Belgium) (FWO, G078518N; PC). HP is a senior clinical investigator of the FWO (18B1521N).
Footnotes
Ethics approval statement
The study was approved by the ethical review board of KU Leuven and University Hospitals Gasthuisberg, Leuven (S56392). Patient recruitment and data collection at various centers was approved by: University College London Hospital (JREC00/E042); University of Calgary (REB14-0340); University of Colorado (14-0419); Stanford University (IRB-35709); University of Southern California (CCI-08-00150) University of California, San Francisco (14-12955); and the King Edward’s Memorial Hospital (RGS0000002376; RGS0000002682). All analyses were carried out in accordance with the relevant guidelines and regulations.
Competing interests
The authors declare no competing interests.
References
- 1.McKusick VA. On Lumpers and Splitters, or the Nosology of Genetic Disease. Perspectives in Biology and Medicine 1969;12:298–312. [DOI] [PubMed] [Google Scholar]
- 2.Biesecker LG, Adam MP, Alkuraya FS, Amemiya AR, Bamshad MJ, Beck AE, Bennett JT, Bird LM, Carey JC, Chung B, Clark RD, Cox TC, Curry C, Dinulos MBP, Dobyns WB, Giampietro PF, Girisha KM, Glass IA, Graham JM, Gripp KW, Haldeman-Englert CR, Hall BD, Innes AM, Kalish JM, Keppler-Noreuil KM, Kosaki K, Kozel BA, Mirzaa GM, Mulvihill JJ, Nowaczyk MJM, Pagon RA, Retterer K, Rope AF, Sanchez-Lara PA, Seaver LH, Shieh JT, Slavotinek AM, Sobering AK, Stevens CA, Stevenson DA, Tan TY, Tan W-H, Tsai AC, Weaver DD, Williams MS, Zackai E, Zarate YA. A dyadic approach to the delineation of diagnostic entities in clinical genomics. The American Journal of Human Genetics 2021;108:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bögershausen N, Wollnik B. Mutational Landscapes and Phenotypic Spectrum of SWI/SNF-Related Intellectual Disability Disorders. Front Mol Neurosci 2018;11:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kline AD, Moss JF, Selicorni A, Bisgaard A-M, Deardorff MA, Gillett PM, Ishman SL, Kerr LM, Levin AV, Mulder PA, Ramos FJ, Wierzba J, Ajmone PF, Axtell D, Blagowidow N, Cereda A, Costantino A, Cormier-Daire V, FitzPatrick D, Grados M, Groves L, Guthrie W, Huisman S, Kaiser FJ, Koekkoek G, Levis M, Mariani M, McCleery JP, Menke LA, Metrena A, O’Connor J, Oliver C, Pie J, Piening S, Potter CJ, Quaglio AL, Redeker E, Richman D, Rigamonti C, Shi A, Tümer Z, Van Balkom IDC, Hennekam RC. Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet 2018;19:649–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang LX, Lemire G, Gonzaga-Jauregui C, Molidperee S, Galaz-Montoya C, Liu DS, Verloes A, Shillington AG, Izumi K, Ritter AL, Keena B, Zackai E, Li D, Bhoj E, Tarpinian JM, Bedoukian E, Kukolich MK, Innes AM, Ediae GU, Sawyer SL, Nair KM, Soumya PC, Subbaraman KR, Probst FJ, Bassetti JA, Sutton RV, Gibbs RA, Brown C, Boone PM, Holm IA, Tartaglia M, Ferrero GB, Niceta M, Dentici ML, Radio FC, Keren B, Wells CF, Coubes C, Laquerrière A, Aziza J, Dubucs C, Nampoothiri S, Mowat D, Patel MS, Bracho A, Cammarata-Scalisi F, Gezdirici A, Fernandez-Jaen A, Hauser N, Zarate YA, Bosanko KA, Dieterich K, Carey JC, Chong JX, Nickerson DA, Bamshad MJ, Lee BH, Yang X-J, Lupski JR, Campeau PM. Further delineation of the clinical spectrum of KAT6B disorders and allelic series of pathogenic variants. Genet Med 2020;22:1338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhoj EJ, Haye D, Toutain A, Bonneau D, Nielsen IK, Lund IB, Bogaard P, Leenskjold S, Karaer K, Wild KT, Grand KL, Astiazaran MC, Gonzalez-Nieto LA, Carvalho A, Lehalle D, Amudhavalli SM, Repnikova E, Saunders C, Thiffault I, Saadi I, Li D, Hakonarson H, Vial Y, Zackai E, Callier P, Drunat S, Verloes A. Phenotypic spectrum associated with SPECC1L pathogenic variants: new families and critical review of the nosology of Teebi, Opitz GBBB, and Baraitser-Winter syndromes. European Journal of Medical Genetics 2019;62:103588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortier GR, Cohn DH, Cormier-Daire V, Hall C, Krakow D, Mundlos S, Nishimura G, Robertson S, Sangiorgi L, Savarirayan R, Sillence D, Superti-Furga A, Unger S, Warman ML. Nosology and classification of genetic skeletal disorders: 2019 revision. Am J Med Genet A 2019;179:2393–419. [DOI] [PubMed] [Google Scholar]
- 8.Carey JC. Special Issue: Elements of morphology: Standard Terminology. American Journal of Medical Genetics Part A 2009;194A:1–127. [DOI] [PubMed] [Google Scholar]
- 9.Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz PM, Kamphausen SB, Zenker M, Bird LM, Gripp KW. Identifying facial phenotypes of genetic disorders using deep learning. Nature Medicine 2019;25:60–4. [DOI] [PubMed] [Google Scholar]
- 10.Latorre-Pellicer A, Ascaso Á, Trujillano L, Gil-Salvador M, Arnedo M, Lucia-Campos C, Antoñanzas-Pérez R, Marcos-Alcalde I, Parenti I, Bueno-Lozano G, Musio A, Puisac B, Kaiser FJ, Ramos FJ, Gómez-Puertas P, Pié J. Evaluating Face2Gene as a Tool to Identify Cornelia de Lange Syndrome by Facial Phenotypes. International Journal of Molecular Sciences 2020;21:1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferry Q, Steinberg J, Webber C, FitzPatrick DR, Ponting CP, Zisserman A, Nellåker C. Diagnostically relevant facial gestalt information from ordinary photos. eLife 2014;3:e02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh T-C, Bar-Haim A, Moosa S, Ehmke N, Gripp KW, Pantel JT, Danyel M, Mensah MA, Horn D, Rosnev S, Fleischer N, Bonini G, Hustinx A, Schmid A, Knaus A, Javanmardi B, Klinkhammer H, Lesmann H, Sivalingam S, Kamphans T, Meiswinkel W, Ebstein F, Krüger E, Küry S, Bézieau S, Schmidt A, Peters S, Engels H, Mangold E, Kreiß M, Cremer K, Perne C, Betz RC, Bender T, Grundmann-Hauser K, Haack TB, Wagner M, Brunet T, Bentzen HB, Averdunk L, Coetzer KC, Lyon GJ, Spielmann M, Schaaf C, Mundlos S, Nöthen MM, Krawitz P. GestaltMatcher: Overcoming the limits of rare disease matching using facial phenotypic descriptors. medRxiv 2021;:2020.12.28.20248193. [Google Scholar]
- 13.Hammond P, Hannes F, Suttie M, Devriendt K, Vermeesch JR, Faravelli F, Forzano F, Parekh S, Williams S, McMullan D. Fine-grained facial phenotype–genotype analysis in Wolf–Hirschhorn syndrome. European Journal of Human Genetics 2012;20:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [dataset] 14.Hallgrimsson B, Klein O, Spritz R. Developing 3D craniofacial morphometry data and tools to transform dysmorphology. FaceBase. September 22 2017. 10.25550/WWC [DOI] [Google Scholar]
- 15.White JD, Ortega-Castrillón A, Matthews H, Zaidi AA, Ekrami O, Snyders J, Fan Y, Penington T, Dongen SV, Shriver MD, Claes P. MeshMonk: Open-source large-scale intensive 3D phenotyping. Scientific Reports 2019;9:6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews HS, Burge JA, Verhelst P-JR, Politis C, Claes PD, Penington AJ. Pitfalls and promise of 3-dimensional image comparison for craniofacial surgical assessment. Plastic and Reconstructive Surgery – Global Open 2020;8:e2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond P, Suttie M, Hennekam RC, Allanson J, Shore EM, Kaplan FS. The face signature of fibrodysplasia ossificans progressiva. American Journal of Medical Genetics Part A 2012;158:1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews HS, Palmer RL, Baynam GS, Quarrell OW, Klein OD, Spritz RA, Hennekam RC, Walsh S, Shriver M, Weinberg SM, Hallgrimsson B, Hammond P, Penington AJ, Peeters H, Claes PD. Large-scale open-source three-dimensional growth curves for clinical facial assessment and objective description of facial dysmorphism. Sci Rep 2021;11:12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2016;374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mutsvangwa T, Douglas TS. Morphometric analysis of facial landmark data to characterize the facial phenotype associated with fetal alcohol syndrome. Journal of Anatomy 2007;210:209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Maaten L, Hinton G. Visualizing data using tSNE. Journal of Machine Learning 2008;9:2579–605. [Google Scholar]
- 22.Hammond P, Suttie M. Large-scale objective phenotyping of 3D facial morphology. Human mutation 2012;33:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallgrímsson B, Aponte JD, Katz DC, Bannister JJ, Riccardi SL, Mahasuwan N, McInnes BL, Ferrara TM, Lipman DM, Neves AB, Spitzmacher JAJ, Larson JR, Bellus GA, Pham AM, Aboujaoude E, Benke TA, Chatfield KC, Davis SM, Elias ER, Enzenauer RW, French BM, Pickler LL, Shieh JTC, Slavotinek A, Harrop AR, Innes AM, McCandless SE, McCourt EA, Meeks NJL, Tartaglia NR, Tsai AC-H, Wyse JPH, Bernstein JA, Sanchez-Lara PA, Forkert ND, Bernier FP, Spritz RA, Klein OD. Automated syndrome diagnosis by three-dimensional facial imaging. Genetics in Medicine 2020;22:1682–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aref-Eshghi E, Kerkhof J, Pedro VP, Barat-Houari M, Ruiz-Pallares N, Andrau J-C, Lacombe D, Van-Gils J, Fergelot P, Dubourg C, Cormier-Daire V, Rondeau S, Lecoquierre F, Saugier-Veber P, Nicolas G, Lesca G, Chatron N, Sanlaville D, Vitobello A, Faivre L, Thauvin-Robinet C, Laumonnier F, Raynaud M, Alders M, Mannens M, Henneman P, Hennekam RC, Velasco G, Francastel C, Ulveling D, Ciolfi A, Pizzi S, Tartaglia M, Heide S, Héron D, Mignot C, Keren B, Whalen S, Afenjar A, Bienvenu T, Campeau PM, Rousseau J, Levy MA, Brick L, Kozenko M, Balci TB, Siu VM, Stuart A, Kadour M, Masters J, Takano K, Kleefstra T, de Leeuw N, Field M, Shaw M, Gecz J, Ainsworth PJ, Lin H, Rodenhiser DI, Friez MJ, Tedder M, Lee JA, DuPont BR, Stevenson RE, Skinner SA, Schwartz CE, Genevieve D, Sadikovic B. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. The American Journal of Human Genetics 2020;106:356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denayer E, Legius E. Neurofibromatosis Type 1-Noonan Syndrome: What’s the Link? Noonan Syndrome and Related Disorders - A Matter of Deregulated Ras Signaling 2009;17:128–37. [Google Scholar]
- 26.Serra G, Antona V, Corsello G, Zara F, Piro E, Falsaperla R. NF1 microdeletion syndrome: case report of two new patients. Italian Journal of Pediatrics 2019;45:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neurofibromatosis: Conference Statement. Archives of Neurology 1988;45:575–8. [PubMed] [Google Scholar]
- 28.Legius E, Messiaen L, Wolkenstein P, Pancza P, Avery RA, Berman Y, Blakeley J, Babovic-Vuksanovic D, Cunha KS, Ferner R, Fisher MJ, Friedman JM, Gutmann DH, Kehrer-Sawatzki H, Korf BR, Mautner V-F, Peltonen S, Rauen KA, Riccardi V, Schorry E, Stemmer-Rachamimov A, Stevenson DA, Tadini G, Ullrich NJ, Viskochil D, Wimmer K, Yohay K, Huson SM, Evans DG, Plotkin SR. Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet Med 2021;23:1506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.