Abstract
Aims
The osteogenesis of human bone marrow mesenchymal stem cells (hBMSCs) plays a critical role in fracture healing. Osteogenic differentiation is regulated by a variety of post-translational modifications, but the function of protein palmitoylation in osteogenesis remains largely unknown.
Methods
Osteogenic differentiation induction of hBMSCs was used in this study. RT‒qPCR and immunoblotting assays (WB) were used to test marker genes of osteogenic induction. Alkaline phosphatase (ALP) activity, ALP staining and Alizarin red staining were performed to evaluate osteogenesis of hBMSCs. Signal finder pathway reporter array, co-immunoprecipitation and WB were applied to elucidate the molecular mechanism. A mouse fracture model was used to verify the in vivo function of the ZDHHC inhibitor.
Key findings
We revealed that palmitic acid inhibited Runx2 mRNA expression in hBMSCs and identified ZDHHC16 as a potential target palmitoyl acyltransferase. In addition, ZDHHC16 decreased during osteogenic induction. Next, we confirmed the inhibitory function of ZDHHC16 by its knockdown or overexpression during osteogenesis of hBMSCs. Moreover, we illustrated that ZDHHC16 inhibited the phosphorylation of CREB, thus inhibiting osteogenesis of hBMSCs by enhancing the palmitoylation of CREB. With a mouse femur fracture model, we found that 2-BP, a general inhibitor of ZDHHCs, promoted fracture healing in vivo. Thus, we clarified the inhibitory function of ZDHHC16 during osteogenic differentiation.
Significance
Collectively, these findings highlight the inhibitory function of ZDHHC16 in osteogenesis as a potential therapy method for fracture healing.
Keywords: Palmitoylation, BMSCs, Fracture, ZDHHC16, CREB
1. Introduction
Bone fractures are one of the most common traumatic damage [1], and approximately one-third of people will experience in their lifetime [2]. About 5% of fractures will suffer a failure of normal fracture healing and require extensive surgery for bone healing, which causes big clinical and economic problems [3]. Fracture repair is a complicated, regenerative process driven by a series of biological events in which bone marrow mesenchymal stem cells (BMSCs) make a major contribution to fracture healing [[4], [5], [6]].
Mesenchymal stem cells (MSCs) are a group of cells with self-renewal ability that can differentiate into cell types (osteoblasts and chondrocytes) related to bone formation during the fracture healing process [[7], [8], [9]]. Multiple signaling pathways are involved in the osteogenic differentiation of BMSCs and also participate in the process of bone regeneration and bone reconstruction, such as the Notch signaling pathway [10], Hedgehog signaling pathway [11], bone morphogenetic protein (BMP)/SMAD signaling pathway [12], WNT/β-CATENIN pathway [13], mitogen-activated protein kinase (MAPK) signaling pathway [14] and cAMP/PKA/CREB pathway [15], etc., which are intertwined to form a complex regulatory network in regulating osteogenic differentiation. In bone tissue, PTH (parathyroid hormone) binds PTH receptors and generates cyclic adenosine monophosphate (cAMP) from adenosine triphosphate (ATP). This leads to the activation of protein kinase A (PKA) and the phosphorylation of cAMP response element binding protein (CREB) [16]. Activated CREB binds to the cAMP response element (CRE) and triggers the cascaded expression of osteogenesis related genes [17]. However, other post-translational modifications of CREB are rarely reported. A better understanding of the mechanisms underlying osteogenesis of BMSCs is essential for their clinical application.
The differentiation of stem cells is modulated by a variety of post-translational modifications, as is the case for BMSCs [18,19]. Protein modifications have been reported to regulate the osteogenic differentiation of MSCs, such as trimethylated histone 3 lysine 27 (H3K27me3) [20,21], histone acetylation regulated by HDAC1 [22], and so on. As a newly emerging post-translational modification, cysteine palmitoylation mainly regulates protein targeting, trafficking, and stability [[23], [24], [25], [26]]. S-palmitoylation is catalyzed by 23 mammalian palmitoyl acyltransferases known as DHHCs due to their conserved Asp-His-His-Cys (DHHC) sequence motif [27]. 2-BP, one of the most commonly used palmitoylation inhibitors, directly and irreversibly inhibits the palmitoyltransferase activity of all the DHHC (Asp-His-His-Cys) proteins. Although numerous proteins are modified by palmitoylation [28], its physiological role remains largely unclear, and how DHHCs affect the osteogenic differentiation of BMSCs has not been reported.
In this study, we revealed that palmitic acid inhibited runt-related transcription factor 2 (Runx2) mRNA expression in osteogenesis of hBMSCs and found that knockdown of ZDHHC16 increased Runx2 mRNA expression. In addition, ZDHHC16 decreased during osteogenic induction. The inhibitory function of ZDHHC16 in osteogenesis of hBMSCs was further confirmed by knockdown and overexpression of ZDHHC16. We then found that ZDHHC16 inhibited CREB phosphorylation by promoting its palmitoylation. Using a mouse femur fracture model, we demonstrated that 2-BP, a general inhibitor of the ZDHHCs, promoted fracture healing in vivo. Our findings further demonstrate that the therapeutic inhibition of ZDHHC16 in BMSCs may lead to phosphorylation of CREB and suggest a role for ZDHHC16 in the pathophysiological process leading to reduced bone formation.
2. Materials and methods
2.1. Isolation, culture, and transfection of human BMSCs (hBMSCs)
This experiment was approved by the Institutional Ethics Committee of Xin Hua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine. Informed consent was obtained from all donors. Human BMSCs were isolated from healthy volunteers and amplified according to the previously reported methods [29]. The BMSCs used in the following experiment were obtained before the 4th generation. The osteogenic medium was α-MEM supplemented with 10% FBS, 50 mg/ml ascorbic acid, 10 mM glycerophosphate, 100 nM dexamethasone and antibiotics (Sigma; St. Louis, MO, USA) for osteogenic differentiation induction. The maintenance medium consisted of DMEM (Gibco, Invitrogen) containing 10% FBS and 100 U/mL penicillin/streptomycin (Invitrogen), and the hBMSCs were maintained at 37 °C and 5% CO2.
2.2. Alkaline phosphatase (ALP) activity
We measured ALP activity using an ALP activity kit (Nanjing Jiancheng Biotech). In short, MSCs were lysed centrifuged at 12,000 rpm, 4 °C for 30 min. Then, 50 μl reaction buffer was added to 100 μl of supernatant and incubated at 37 °C for 15 min. Then, color development was stopped with 150 μl of stop solution, and the absorbance was measured at 405 nm. A protein determination kit was used to determine the total protein content (Pierce, Rockford, IL, USA). ALP activity was expressed as units per gram of protein per 15 min (U/gpro/15 min).
2.3. ALP staining
ALP staining was performed according to the manufacturer's instructions. In short, the cell layer was washed 3 times with PBS and then fixed in 4% paraformaldehyde for 10 min at room temperature. Cells were then incubated with a buffer containing 0.1% naphthol AS-Bi phosphate (Sigma‒Aldrich, St. Louis, MO, USA) and 2% Fast Violet B (Sigma‒Aldrich). After incubating for 1 h at 37 °C, the cell layer was washed with deionized water.
2.4. shRNA knockdown study
The plvx-lentiviral shRNA vector targeting ZDHHC16 and the non-silent plvx-shRNA1 control vector were synthesized by Open Biosystems (Thermo Fisher Scientific). The lentivirus was packaged by co-transfection HEK293T cells with shRNA-containing lentiviral vector plasmids, and the packaging plasmids psPAX2 and pMD2.G using the ViaFect™ transfection reagent (Promega) at a ratio of 3:2:1. 48 h later, the virus supernatant was collected and used to infect MSCs. ShZDHHC16 sequences:
F:gatccGCAGAGTATTTAGGAATCCTTCTCGAGAAGGATTCCTAAATACTCTGCTTTTTTg R:aattcAAAAAAGCAGAGTATTTAGGAATCCTTCTCGAGAAGGATTCCTAAATACTCTGCg
2.5. Alizarin red staining
Cells were fixed in 70% ice-cold ethanol for 1 h, washed with ddH2O, and then stained with 40 mM Alizarin Red S (pH 4.9, Sigma) under gentle agitation for 15 min. After staining, the cells were washed 5 times with ddH2O. To quantitatively assess the degree of mineralization, the red stain was eluted with 10% (w/v) cetylpyridinium chloride (Sigma‒Aldrich) for 1 h and then quantified by spectrophotometric absorbance at OD 570 nm.
2.6. Signal finder pathway reporter array
Transfect BMSC with ZDHHC16 overexpression or control plasmids, and then use an Amaxa Nucleofector 96-well shuttle system to electroporate each reporter gene test and the negative control from the BMSC osteogenic pathway reporter gene array plate (SABiosciences). After 48 h of electroporation, a dual Glo luciferase reporter gene test (Promega) was performed, and the activity was monitored by a Synergy H4 Hybrid Multi-Mode Microplate Reader.
2.7. RNA extraction and RT‒qPCR
We extracted total RNA using TRIzol reagent (Invitrogen), and 0.5 μg RNA was subjected to reverse transcription using an RT‒PCR kit (KAPA, Sigma) as instructed by the manufacturer. An ABI-7500 Real-Time PCR Detection System was used to perform all RT‒qPCR experiments. β-actin was used as a control to normalize the amplified transcript level of each indicated gene. The primers used were as follows.
Zdhhc16 F: 5′- ACTGGCTGGTAGACAACGTG -3′; Zdhhc16 R: 5′- TGGCGATATCATTCCTGCCC -3′;
β-actin F: 5′- GAAGGATTCCTATGTGGGCGA -3′;
β-actin R: 5′- GATAGCACAGCCTGGATAGCAA -3′; Runx2 F: 5′- GACTGTGGTTACCGTCATGGC -3′; Runx2 R: 5′- ACTTGGTTTTTCATAACAGCGGA -3′; Ocn F: 5′- GACAAGTCCCACACAGCAACT -3′; Ocn R: 5′- GGACATGAAGGCTTTGTCAGA -3′; Col1a1 F: 5′- GCTCCTCTTAGGGGCCACT -3′; Col1a1 R: 5′- ATTGGGGACCCTTAGGCCAT -3′;
2.8. Immunoblotting assay (WB)
We used loading buffer to lyse the cells. The Lowry protein assay was used to calculate the protein concentration. Then, we used SDS‒PAGE analysis to separate 10 μg of protein lysate. The gel was transferred to a PVDF membrane (Solarbio). The designated protein was detected by immunoblotting with specific antibodies in 5% bovine serum albumin. The antibodies used were as follows: GAPDH (sc-20357, Santa Cruz); RUNX2 (Affinity Biosciences); CREB (AY0330, Abways); Phosphorylation-CREB (AY3301, Abways); Phosphorylation-PKA (Cell Signaling, No. 9621); ZDHHC16 (AB105658, Abcam); PKA (Cell Signaling, No. 4782).
2.9. Acyl-biotin exchange (ABE) palmitoylation assay
Cells were lysed in lysis buffer (1% IGEPAL CA-630; 50 mM Tris-HCl, pH 7.5; 150 mM NaCl 10% glycerol; protease inhibitors) with 50 mM N-ethylmaleimide and then Flag-CREB was purified by specific antibodies or beads. The purified CREB protein was subjected to lysis treatment buffer plus 1 M hydroxylamine (Sigma‒Aldrich) and placed at room temperature for 1 h. Finally, the beads were gently washed with lysis buffer pH 6.2 and incubated with lysis buffer pH 6.2 plus 2 μM biotin-BMCC (Thermo Fisher Scientific Inc.) at 4 °C for 1 h. After washing 3 times, the protein was used for Western blot analysis.
2.10. Co-immunoprecipitation
Cells were washed twice with ice-cold PBS and lysed in lysis buffer (50 mM Tris pH 7.4; 1% Triton X-100; 0.5 mM EDTA; 0.5 mM EGTA; 150 mM sodium chloride; 10% glycerol; 1 mM phenylmethylsulfonyl fluoride; complete protease inhibitor cocktail) on ice for 30 min, centrifuged at 15,000 g for 15 min at 4 °C. The supernatant was collected, treated with DNase (15 U ml-1, Pierce), precleared with 20 μl Protein G Agarose Beads (Thermo Fisher Scientific Inc.) and then incubated with primary antibody overnight at 4 °C or Flag/HA beads (Sigma‒Aldrich) for 2 h. The samples were added 20 μl Protein G Sepharose beads and rotated at 4 °C for 1 h. After washing 3 times with 1 ml lysis buffer, the bound protein was released by boiling in 30 μl SDS loading buffer.
2.11. Mouse fracture model
The procedures used in this study were as humane as possible. The animal study protocol was reviewed and approved by the institution's Animal Ethics Committee of the Shanghai Jiao Tong University School of Medicine (SJTUSM). This model used six-week-old male or female mice. The mice were anesthetized with chloral hydrate for the dislocation of the patella laterally to expose the femoral condyle. Then, the patella was repositioned. The fracture was performed at the midpoint of the femur using a dentist's micro drill. The muscle was re-equated, and a 6/0 nylon suture was used to close the skin. Radiography was used to track fracture repair with a weekly MX2 X-ray system (6 s at 932 kV).
2.12. Micro CT
The mouse femurs were wrapped with wet gauze and placed into a sample tube for micro-CT scanning (μCT 80; SCANCO Medical AG, Bathersdorf, Switzerland). In short, the scan range was set to 9.6 mm (600 slides), and the fracture line was set in the middle of this range. The resolution was 16 μm/voxel and 1024 × 1024 pixels. The region of interest (ROI) was selected for the mineralized tissue from two-dimensional (2D) images with standardized thresholds (>165). The three-dimensional (3D) reconstruction of the external callus was subjected to quantitative analysis of bone volume/tissue volume (BV/TV), including all 600 slides of 2D images.
2.13. Safranin O/fast green staining
After deparaffinization, the sections were stained with hematoxylin for 3 min and differentiated with 1% acid alcohol for 15 s. This step was followed by dyeing in 0.02% fast green aqueous solution for 3 min and then counterstaining with 0.1% safranin O for 3 min. Finally, the sections were dehydrated in ethanol serial dilutions, clarified in xylene, and fixed on glass slides using neutral gum. Each stained section was observed under an optical microscope, and pictures were taken using a microscope equipped with a CCD camera.
2.14. Statistical analysis
The data are expressed as the mean ± SEM. (n is the number of tissue preparations, cells, or experimental repetitions). In order to compare datasets, a two-tailed Student's t-test was used. A value of p < 0.05 is considered statistically significant.
3. Results
3.1. ZDHHC16 participated in osteogenic differentiation of hBMSCs
As one of the well-known osteogenic marker genes, RUNX2 is upregulated during the osteogenic differentiation of BMSCs. To validate the regulatory role of protein palmitoylation in osteogenesis, we measured Runx2 mRNA expression as a marker for osteogenic induction of BMSCs. After 7 days of osteoblastic differentiation in osteogenic medium, hBMSCs were serum starved for 1 day and cultured in medium with different fatty acids. We found that Runx2 mRNA expression was inhibited only by palmitic acid, not by other lipids (Fig. 1a). As reported, palmitic acid is essential for cysteine residue palmitoylation, and the results (Fig. 1a) indicated that protein palmitoyl modification might be involved in osteogenesis of hBMSCs. To further confirm this supposition, we administered 2-bromopalmitate (2-BP), a general palmitoylation inhibitor that inhibits the palmitoyltransferase activity of ZDHHCs, in an osteogenic induction experiment and found that 2-BP completely restored the decreased Runx2 mRNA expression inhibited by palmitic acid (Fig. 1b). At the same time, the expression levels of other osteogenic marker genes, Ocn and Col1a1, were also inhibited by palmitic acid, but the expression levels were restored to near normal levels after adding 2-BP (Fig. 1c and d). These data suggested that protein palmitoylation was important in osteogenesis of hBMSCs.
Figure 1.
ZDHHC16 is related to the osteogenic differentiation process of hBMSCs. (a) RT‒qPCR analysis of the expression of Runx2. hBMSCs were induced to osteogenesis and simultaneously treated with BSA conjugated fatty acids (300 μM) for 6 h. (b) RT‒qPCR analysis of the expression of Runx2. hBMSCs were exposed to palmitic acid with or without 2-BP (50 μM) for 48 h. (c) RT‒qPCR analysis of the expression of Ocn. hBMSCs were exposed to palmitic acid with or without 2-BP (50 μM) for 48 h. (d) RT‒qPCR analysis of the expression of Col1a1. hBMSCs were exposed to palmitic acid with or without 2-BP (50 μM) for 48 h. (e) RT‒qPCR analysis of the expression of Runx2 after knockdown of ZDHHCs in hBMSCs. (f) RT‒qPCR and Western blotting analysis of the expression of Zdhhc16 at different time points of hBMSCs osteogenic differentiation. All experiments were performed at least three times. Data are the mean ± SEM. (*p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student's t-test).
Protein palmitoylation is catalyzed by Asp-His-His-Cys (DHHC)-containing palmitoyl acyltransferases. To identify which palmitoyl acyltransferases were responsible for osteogenesis of hBMSCs, we knocked down each member of the ZDHHC family separately using specific short hairpin RNA (shRNA). During the induction of osteogenic differentiation of hBMSCs, significantly increased Runx2 mRNA expression was observed only in Zdhh16 knockdown cells (Fig. 1e), which indicated that ZDHHC16 participated in osteogenesis of hBMSCs. In addition, we evaluated the expression pattern of Zdhh16 on different days during osteogenic differentiation and observed that the expression of Zdhh16 decreased gradually in hBMSCs (Fig. 1f). This result suggested that ZDHHC16 played an inhibitory role during osteogenesis of hBMSCs.
Overall, these data indicated that palmitoylation mediated by ZDHHC16 might play an important role during the osteogenic differentiation of hBMSCs.
3.2. ZDHHC16 suppressed osteogenesis of hBMSCs
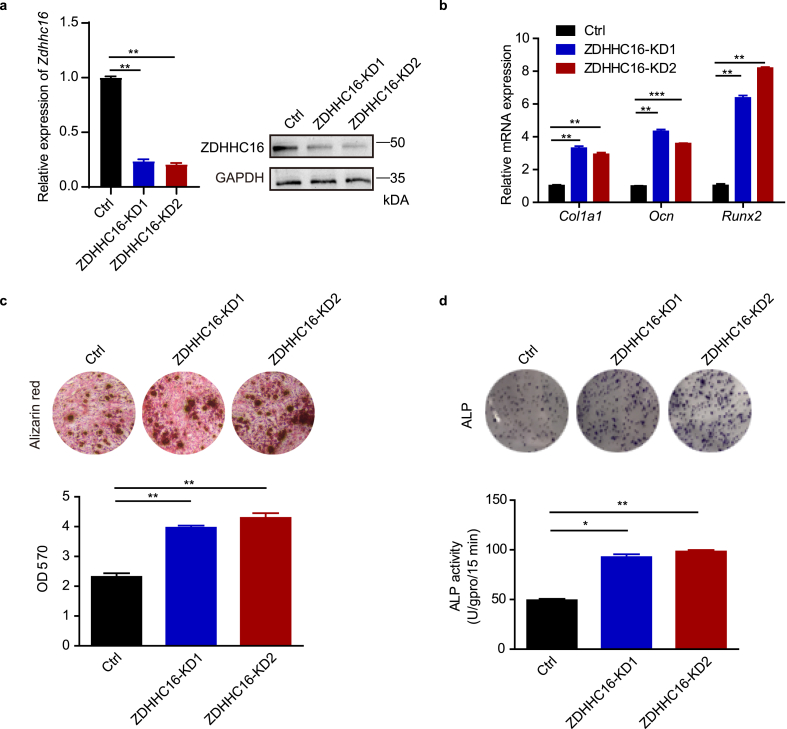
As the expression profile of Zdhh16 decreased during osteogenesis of hBMSCs (Fig. 1e), we knocked down ZDHH16 with two different shRNA sequences (ZDHHC16-KD1 and ZDHHC16-KD2) in normal hBMSCs to further explore the direct role of ZDHHC16 in osteogenic differentiation of hBMSCs. Both shRNAs resulted in efficient knockdown of ZDHHC16, which was confirmed by qPCR and Western blot experiments (Fig. 2a). Next, we evaluated the expression of osteogenic marker genes (Col1a1, Ocn, and Runx2) during osteogenesis of hBMSCs by RT‒qPCR. We found that knockdown of ZDHHC16 enhanced the mRNA expression of Col1a1, Ocn, and Runx2 (Fig. 2b). Consistent with the above mRNA results, knockdown of ZDHHC16 also significantly increased the mineralized deposits in the Alizarin red staining assay (Fig. 2c) and significantly increased the alkaline phosphatase (ALP) activity in the ALP assays (Fig. 2d). These results further supported the conclusion that ZDHHC16 inhibits osteogenesis of hBMSCs.
Figure 2.
ZDHHC16 knockdown promotes osteogenic differentiation of hBMSCs. (a) q-PCR and Western blotting analysis of the indicated proteins in control (Ctrl) or ZDHHC16 knockdown hBMSCs. (b) RT‒qPCR analysis of Col1a1, Ocn and Runx2 mRNA expression in hBMSCs infected with lentiviral-Ctrl or lentiviral-shZDHHC16 after 7 days of osteogenic induction. (c) Alizarin red S staining of hBMSCs infected with lentiviral-Ctrl or lentiviral-shZDHHC16 after 21 days of osteogenic induction was detected at OD 570 and quantified. (d) ALP staining (upper) and ALP activity (lower) analysis of hBMSCs infected with lentiviral-Ctrl or lentiviral-shZDHHC16 after 7 days of osteogenic induction. All experiments were performed at least three times. Data are the mean ± SEM. (*p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student's t-test)
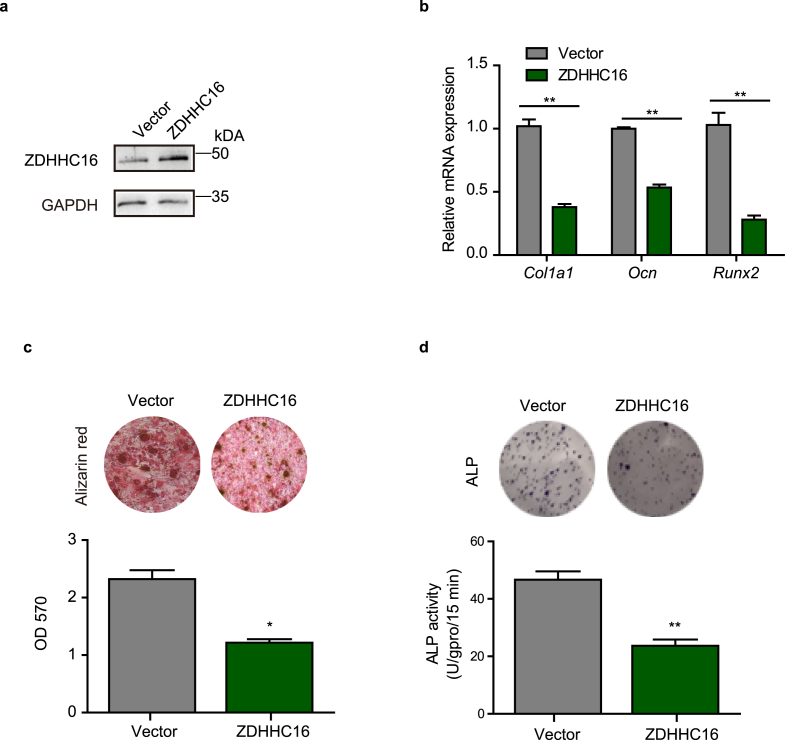
Furthermore, we overexpressed ZDHHC16 in hBMSCs to validate that ZDHHC16 inhibited osteogenesis of hBMSCs (Fig. 3a) and investigated that overexpression of ZDHHC16 significantly inhibited the mRNA expression of osteogenic marker genes (Col1a1, Ocn, and Runx2) (Fig. 3b). In addition, we conducted Alizarin red staining assays and ALP assays to evaluate the function of ZDHHC16 in osteogenesis and found that overexpressed ZDHHC16 dramatically decreased the proportion of mineralization (Fig. 3c) and significantly inhibited ALP activity (Fig. 3d). These data indicated that ZDHHC16 overexpression inhibited osteogenesis of hBMSCs.
Figure 3.
ZDHHC16 overexpression impaired osteogenic differentiation of hBMSCs. (a) Western blotting analysis of the indicated proteins in Ctrl or ZDHHC16-overexpressing hBMSCs. (b) RT‒qPCR analysis of Col1a1, Ocn and Runx2 mRNA expression in hBMSCs infected with lentiviral vector or lentiviral ZDHHC16 after 7 days of osteogenic induction.(c) Alizarin red S staining of hBMSCs infected with lentiviral vector or lentiviral ZDHHC16 after 21 days of osteogenic induction was detected at OD 570 and quantified. (d) ALP staining (upper) and ALP activity (lower) analysis of hBMSCs infected with lentiviral-Ctrl or lentiviral-shZDHHC16 after 7 days of osteogenic induction. All experiments were performed at least three times. Data are the mean ± SEM. (*p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student's t-test)
In general, these results demonstrated that ZDHHC16 plays a critical role in inhibiting osteogenic differentiation of hBMSCs.
3.3. ZDHHC16-mediated palmitoylation inhibited the phosphorylation of CREB during osteogenesis of hBMSCs
Multiple signaling pathways are involved in the osteogenic differentiation of hBMSCs. To determine which signaling pathway is regulated by ZDHHC16 during osteogenic induction of hBMSCs, we constructed a series of luciferase reporter assays. Interestingly, we found that only cAMP luciferase activity dramatically decreased in the ZDHHC16 overexpression group compared to the control group (Fig. 4a). This result indicated that the cAMP pathway might be regulated by ZDHHC16 during osteogenesis. Correspondingly, we also observed that the activity of the cAMP pathway was enhanced in ZDHHC16 knockdown cells (Fig. 4b). To further ascertain that the cAMP pathway was regulated by palmitoylation, we found that the activity of the cAMP pathway was also downregulated in the group treated with palmitic acid, which is essential for cysteine residue palmitoylation (Fig. 4c). We also found that the inhibitory effect was restored by a palmitoylation inhibitor (Fig. 4c). These results indicated that the cAMP pathway might be regulated by ZDHHC16. Mediated palmitoylation during osteogenesis of hBMSCs.
Figure 4.
ZDHHC16 inhibits the cAMP signaling pathway by inhibiting the phosphorylation of CREB. (a) Identification of signaling pathways modulated by ZDHHC16 overexpression in hBMSCs. x and y axes, normalized ratio of firefly/Renilla luciferase activities. (b) Luciferase activity (mean ± SD) in BMSCs treated with different lentiviruses for 48 h(c) cAMP luciferase activity (mean ± SD) in BMSCs exposed to palmitic acid with or without 2-BP (50 μM) for 48 h. (d) Western blotting analysis of protein expression in the cAMP signaling pathway after ZDHHC16 overexpression. OM stands for osteogenic medium. (e) IP analysis of the binding between ZDHHC16 and CREB protein by IP Myc in hBMSCs overexpressing Myc-ZDHHC16 after osteogenic induction. (f) ABE analysis of the CREB palmitoylation degree in different treatment groups. SS stands for serum starved. IP Flag in hBMSCs overexpressed Flag-CREB after osteogenic induction. hBMSCs were after 7 days of osteoblastic differentiation in osteogenic medium, serum starved for 1 day and simultaneously treated with BSA conjugated palmitic acids (300 mM) for 6 h. (g) IP analysis using anti-Flag antibody, followed by WB analysis with the indicated antibodies. Lysates were prepared from Ctrl, ZDHHC16-knockdown, or 2-BP-treated hBMSCs overexpressing Flag-CREB after osteogenic induction.
To elucidate how ZDHHC16 regulates the cAMP/PKA/CREB pathway, we measured the protein expression and modification of the cAMP/PKA/CREB pathway and found that ZDHHC16 had no effect on the protein expression of PKA and CREB and did not influence the phosphorylation of PKA but did modulate the phosphorylation of CREB (Fig. 4d), which is critical for CREB entering the nucleus, exerting its transcriptional function. These data indicated that ZDHHC16 modulated the cAMP/PKA/CREB pathway by inhibiting the phosphorylation of CREB during osteogenic induction of hBMSCs. As the phosphorylation of CREB decreased in the ZDHHC16 overexpression group, we wondered whether ZDHHC16 modulates the phosphorylation of CREB directly. We performed a co-immunoprecipitation (co-IP) assay to illustrate how ZDHHC16 regulates the phosphorylation of CREB and found that overexpressed Myc-tagged ZDHHC16 bound to endogenous CREB directly (Fig. 4e). Next, we conducted a palmitic acid treatment assay to determine whether CREB has palmitoylation modification and found that during osteogenesis, CREB did undergo palmitoylation with palmitic acid treatment and was inhibited by 2-BP (Fig. 4f).
To elucidate whether ZDHH16 modulates the phosphorylation of CREB in a manner dependent on its palmitoyltransferase activity, we overexpressed Flag-tagged CREB in ZDHHC16 knockdown hBMSCs, followed by IP using an antibody against the FLAG tag. We found that the palmitoylation of CREB decreased as ZDHHC16 was knocked down; simultaneously, the phosphorylation of CREB increased (Fig. 4g), which demonstrated that ZDHHC16-mediated palmitoylation of CREB modulated the phosphorylation of CREB. To further ascertain whether the decreasing phosphorylation of CREB was due to the palmitoylation activity of ZDHHC16, we used 2-BP in the IP assay. The IP results showed that 2-BP inhibited the palmitoylation of CREB without influencing the expression of ZDHHC16 and the expression of exogenous CREB but still promoted the phosphorylation of CREB as ZDHHC16 was knocked down (Fig. 4g). These results demonstrated that ZDHHC16 mediated the palmitoylation of CREB and that its palmitoyl acyltransferase activity inhibited the phosphorylation of CREB.
Overall, these data clarified that ZDHHC16 inhibited the cAMP/PKA/CREB pathway by directly inhibiting the phosphorylation of CREB through its palmitoyl acyltransferase activity during osteogenic differentiation of hBMSCs.
3.4. 2-BP promoted osteogenesis in vivo
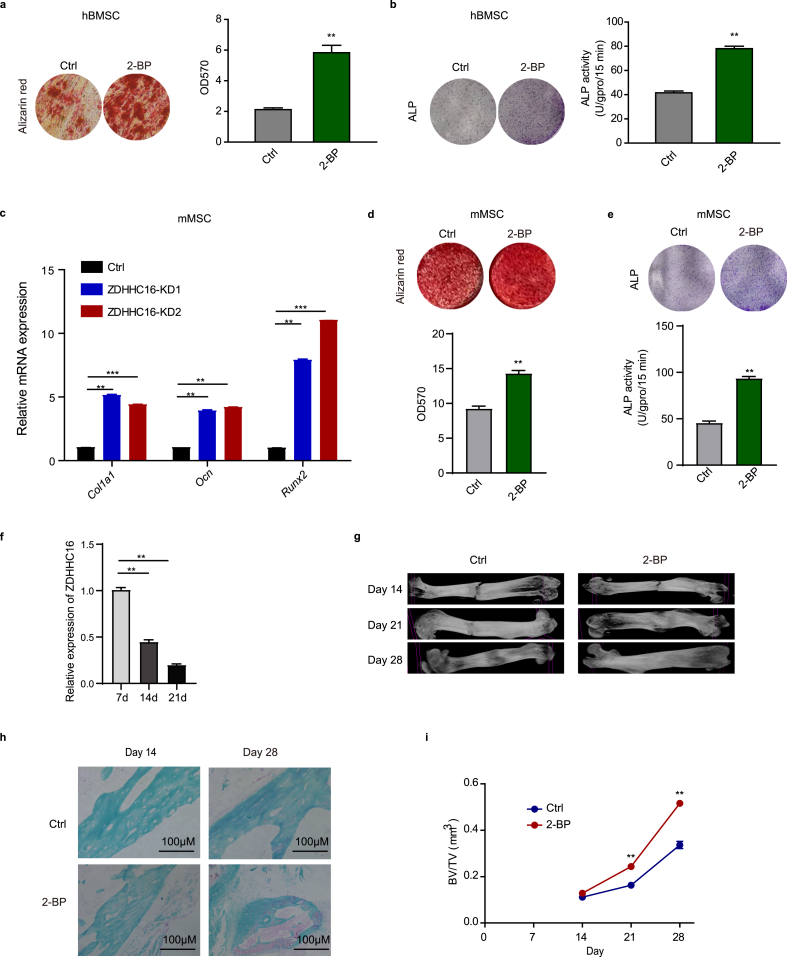
We explored the inhibitory function of ZDHHC16 in osteogenesis of hBMSCs in vitro, and then we used 2-BP, an inhibitor that inhibits ZDHHC enzyme activity, to confirm the inhibitory function of ZDHHC16 in vivo in a mouse femur fracture model. We first examined the function of 2-BP in osteogenesis of hBMSCs and found that 2-BP treatment dramatically increased the proportion of mineralization and ALP activity (Fig. 5a and b), which suggested that 2-BP promoted osteogenesis of hBMSCs. As the in vivo model is a mouse femur fracture model, we examined the function of ZDHHC16 and 2-BP during osteogenesis induction of mouse MSCs (mMSCs). We also found that ZDHHC16 knockdown increased the mRNA expression of Col1a1, Ocn, and Runx2 (Fig. 5c), and 2-BP treatment increased the proportion of mineralization and ALP activity (Fig. 5d and e). These results demonstrated that 2-BP also promoted osteogenesis of mMSCs.
Figure 5.
2-BP promotes fracture healing in mice. (a) Alizarin red S staining of hBMSCs treated with/without 2-BP (50 μM) after 21 days of osteogenic induction was detected at OD 570 and quantified. (b) ALP staining (upper) and ALP activity (lower) analysis of hBMSCs treated with/without 2-BP (50 μM) after 7 days of osteogenic induction. (c) (b) RT‒qPCR analysis of Col1a1, Ocn and Ruxn2 mRNA expression in mouse MSCs infected with lentiviral-Ctrl or lentiviral-shZDHHC16 after 7 days of osteogenic induction. (d) Alizarin red S staining of mouse MSCs treated with/without 2-BP (50 μM) after 21 days of osteogenic induction was detected at OD 570 and quantified. (e) ALP staining (upper) and ALP activity (lower) analysis of mouse MSCs treated with/without 2-BP (50 μM) after 7 days of osteogenic induction. (f) RT‒qPCR analysis of Zdhhc16 expression in the bone tissue of fracture sections at different time points after fracture. (g) Representative X-rays illustrating fracture callus development in the control vehicle group and 2-BP-treated group at 2, 3, and 4 weeks postosteotomy. (h) Callus sections were stained with safranin O/fast green staining after femoral fracture. Cartilage tissue combined with safranin appeared red, and bone tissue combined with fast green appeared green. (i) External fracture callus and related histomorphometric analysis by micro-CT. All experiments were performed at least three times. Data are the mean ± SEM. (*p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student's t-test)
In the mouse femur fracture model, we first tested Zdhhc16 mRNA expression in bone tissue of fracture sections and found that Zdhhc16 mRNA expression decreased during fracture healing (Fig. 5f), which indicated that ZDHHC16 played an inhibitory role during osteogenesis in vivo, similar to the result in Fig. 1f. This result further indicated that 2-BP might promote fracture healing in vivo. Thus, we applied 2-BP treatment in the mouse femur fracture model and observed hard callus with a fracture gap in both the control group and the 2-BP-treated group at Day 14 and Day 21 in the radiographs. However, the callus volume of the 2-BP-treated group was larger than that of the control group, and the fracture gap of the 2-BP-treated group was reduced compared with that of the control group (Fig. 5g). Especially at Day 28, the bone fractures were almost healed after treatment with 2-BP compared with the control group, with worse fracture healing (Fig. 5g). Consistent with the radiographic results, safranin O staining also showed enhanced bone fracture healing in the 2-BP-treated group (Fig. 5h). We also used microcomputed tomography (CT) to measure the bone volume fraction (BV/TV) and found that 2-BP treatment significantly augmented the bone volume fraction (Fig. 5i). These data demonstrated that 2-BP facilitated osteogenic differentiation in vivo.
4. Discussion
In the current study, we demonstrated that ZDHHC16 restrained osteogenesis of hBMSCs by inhibiting the cAMP/PKA/CREB signaling pathway. In detail, we first observed that palmitic acid inhibited Runx2 mRNA expression during osteogenic induction, which indicated that palmitoylation might play roles during osteogenesis. This supposition was further supported by the finding that 2-BP reversed the inhibition of Runx2 mRNA expression caused by palmitic acid. We then determined that ZDHHC16 was a candidate palmitoyl acyltransferase that played major roles during osteogenic differentiation of hBMSCs. Next, we overexpressed or knocked down ZDHHC16 in hBMSCs to demonstrate the inhibitory role of ZDHHC16 in osteogenesis of hBMSCs. ZDHHC16 regulated osteogenic differentiation via the cAMP/PKA/CREB pathway. Specifically, ZDHHC16 interacted with CREB and promoted its palmitoylation, thereby inhibiting the phosphorylation of CREB. In vivo, we were surprised to find that 2-BP, a general inhibitor of palmitoylation, promoted fracture healing. In addition, we observed increased ZDHHC16 expression in nonunion skeletal fracture patients. These data suggest that ZDHHC16 may be a promising clinical treatment target for fracture healing.
Protein S-palmitoylation exists in all eukaryotic cells and is a conserved post-translational modification of proteins that plays important roles in modulating protein stability, protein localization, trafficking, enzymatic activity and other cellular processes [24,25,[30], [31], [32], [33]]. Thus, numerous human diseases are related to an imbalance of protein S-palmitoylation, especially in cancers and neurological disorders [25,[33], [34], [35], [36]]. It has been reported that over 10% of human proteins are susceptible to palmitoylation, and researchers have identified and characterized at least 600 substrates [[37], [38], [39], [40], [41]]. Nevertheless, the role of palmitoylation in osteogenesis remains unclear. As a palmitoyl acetyltransferase, ZDHHC13 has been reported to be a key modulator of bone homeostasis in a Zdhhc13 KO mouse model [42]. In this mouse model, ZDHHC13 facilitates bone mass acquisition and postnatal skeletal development. In another study using cell culture models, palmitic acid was found to be toxic to osteoblasts [43], and palmitic treatment inhibited Alp, Ocn, and Runx2 expression and inhibited osteogenic differentiation. Moreover, Zdhhc1, Zdhhc 2, and Zdhhc 12 were significantly decreased by palmitic treatment, which suggested that palmitoylation inhibited the osteogenesis of osteoblasts. In our study, we found that palmitoylation mediated by ZDHHC16 inhibited osteogenesis using an hBMSC culture model. We also found that knockdown of other DHHC-containing palmitoyl acyltransferases (including ZDHHC1, 2, and 12) increased Runx2 mRNA expression (Fig. 1c). Thus, whether other DHHC-containing palmitoyl acyltransferases alone regulate osteogenic differentiation and whether there is synergy among them in regulating osteogenic differentiation need to be further studied.
In addition, we observed an opposite effect of palmitoylation in regulating osteogenic differentiation in vivo in our study compared to the research in Zdhhc13 KO mice. Although osteogenic differentiation is involved in both postnatal skeletal development and fracture healing, it is regulated by different signaling pathways under different physiological conditions. To elucidate the function of Zdhhc16 in vivo, Zdhhc16 KO mice are needed for a mouse femur fracture model, and we have planned to construct Zdhhc16 KO mice and even conditional KO mice in BMSCs. We are also interested in the effect of 2-BP treatment and Zdhhc16 on postnatal skeletal development.
Regarding the mechanism, we demonstrated that ZDHHC16 interacted with CREB and promoted the palmitoylation of CREB, inhibiting the phosphorylation of CREB. CREB is a 43 kDa transcription factor made up of different domains. A basic leucine zipper (bZIP) domain is essential for DNA binding and dimerization, and the kinase inducible domain (KID) is important for phosphorylation and activation [44]. We searched the protein sequence and found that there were three cysteines at the C-terminus, which could be palmitoylated, while the serine phosphorylated by PKA was in the middle of the protein. We wondered how palmitoylation inhibited CREB phosphorylation despite the relatively distant location of serine and cysteines. We speculate that palmitoylation occupies the binding position of PKA in the active CREB protein conformation, resulting in decreased phosphorylation. Another supposition is that the interaction between CREB and ZDHHC16 interferes with PKA binding to CREB, resulting in decreased phosphorylation. The hypothesis of the underlying mechanism is worthy of further exploration.
Taken together, we revealed that ZDHHC16 modulated osteogenesis of hBMSCs via inhibiting phosphorylation of CREB in vitro, and 2-BP promoted fracture healing in vivo. The results of this study provide a promising clinical treatment target for fracture healing.
5. Conclusion
Collectively, these findings highlight the inhibitory function of ZDHHC16 in osteogenesis as a potential therapy method for fracture healing.
Data availability
Please contact the corresponding author for reasonable data requests.
Funding
The authors received no specific funding for this work.
Author contributions
C.Z., S.L. made substantial contributions to conceived and designed the experiments. Z.L., Y.C., X.J., X.W., and F.W. performed the experiments and wrote the paper. Z.L. and S.L. analyzed and interpreted the data. All authors read and approved the final manuscript.
Consent for publication
All authors have provided their consent for publication.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We appreciate the valuable efforts and advice of our colleagues on this study.
Contributor Information
Shenghe Liu, Email: liush82@163.com.
Chao Zhang, Email: drzhangchao@sina.com.
References
- 1.Einhorn T.A., Gerstenfeld L.C. Fracture healing: mechanisms and interventions. Nat. Rev. Rheumatol. 2015;11:45–54. doi: 10.1038/nrrheum.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Staa T.P., Dennison E.M., Leufkens H.G., Cooper C. Epidemiology of fractures in england and wales. Bone. 2001;29:517–522. doi: 10.1016/s8756-3282(01)00614-7. [DOI] [PubMed] [Google Scholar]
- 3.Garrison K.R., et al. Bone morphogenetic protein (BMP) for fracture healing in adults. Cochrane Database Syst. Rev. 2010:CD006950. doi: 10.1002/14651858.CD006950.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alwattar B.J., Schwarzkopf R., Kirsch T. Stem cells in orthopaedics and fracture healing. Bull. Hosp. Joint Dis. 2011;69:6–10. [PubMed] [Google Scholar]
- 5.Dong C.H., et al. The interplay of transcriptional and post-transcriptional regulation of migration of mesenchymal stem cells during early stages of bone fracture healing. Eur. Rev. Med. Pharmacol. Sci. 2017;21:5542–5547. doi: 10.26355/eurrev_201712_13990. [DOI] [PubMed] [Google Scholar]
- 6.Han D., Han N., Zhang P., Jiang B. Local transplantation of osteogenic pre-differentiated autologous adipose-derived mesenchymal stem cells may accelerate non-union fracture healing with limited pro-metastatic potency. Int. J. Clin. Exp. Med. 2015;8:1406–1410. [PMC free article] [PubMed] [Google Scholar]
- 7.Alm J.J., et al. Circulating plastic adherent mesenchymal stem cells in aged hip fracture patients. J. Orthop. Res. 2010;28:1634–1642. doi: 10.1002/jor.21167. [DOI] [PubMed] [Google Scholar]
- 8.Cheung W.H., Chin W.C., Wei F.Y., Li G., Leung K.S. Applications of exogenous mesenchymal stem cells and low intensity pulsed ultrasound enhance fracture healing in rat model. Ultrasound Med. Biol. 2013;39:117–125. doi: 10.1016/j.ultrasmedbio.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Granero-Molto F., et al. Mesenchymal stem cells expressing insulin-like growth factor-I (MSCIGF) promote fracture healing and restore new bone formation in Irs1 knockout mice: analyses of MSCIGF autocrine and paracrine regenerative effects. Stem Cell. 2011;29:1537–1548. doi: 10.1002/stem.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiddes I.T., et al. Human-specific NOTCH2NL genes affect Notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369. doi: 10.1016/j.cell.2018.03.051. e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carballo G.B., Honorato J.R., de Lopes G.P.F., Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. : CCS. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushioka J., et al. A novel negative regulatory mechanism of Smurf2 in BMP/Smad signaling in bone. Bone research. 2020;8:41. doi: 10.1038/s41413-020-00115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashiro T., et al. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452–462. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 14.Sasaki K., Hitora T., Nakamura O., Kono R., Yamamoto T. The role of MAPK pathway in bone and soft tissue tumors. Anticancer Res. 2011;31:549–553. [PubMed] [Google Scholar]
- 15.Park K.H., et al. Zinc promotes osteoblast differentiation in human mesenchymal stem cells via activation of the cAMP-PKA-CREB signaling pathway. Stem Cell. Dev. 2018;27:1125–1135. doi: 10.1089/scd.2018.0023. [DOI] [PubMed] [Google Scholar]
- 16.Qin L., Partridge N.C. Stimulation of amphiregulin expression in osteoblastic cells by parathyroid hormone requires the protein kinase A and cAMP response element-binding protein signaling pathway. J. Cell. Biochem. 2005;96:632–640. doi: 10.1002/jcb.20550. [DOI] [PubMed] [Google Scholar]
- 17.Sato K., et al. Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat. Med. 2006;12:1410–1416. doi: 10.1038/nm1515. [DOI] [PubMed] [Google Scholar]
- 18.Ezhkova E., et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lian J.B., et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 2006;7:1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 20.Jing H., et al. Suppression of EZH2 prevents the shift of osteoporotic MSC fate to adipocyte and enhances bone formation during osteoporosis. Mol. Ther. 2016;24:217–229. doi: 10.1038/mt.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemming S., et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cell. 2014;32:802–815. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 22.Lee H.W., et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 23.Fukata Y., Murakami T., Yokoi N., Fukata M. Local palmitoylation cycles and specialized membrane domain organization. Curr. Top. Membr. 2016;77:97–141. doi: 10.1016/bs.ctm.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Greaves J., Chamberlain L.H. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Linder M.E., Deschenes R.J. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 26.Peng T., Thinon E., Hang H.C. Proteomic analysis of fatty-acylated proteins. Curr. Opin. Chem. Biol. 2016;30:77–86. doi: 10.1016/j.cbpa.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linder M.E., Jennings B.C. Mechanism and function of DHHC S-acyltransferases. Biochem. Soc. Trans. 2013;41:29–34. doi: 10.1042/BST20120328. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H., et al. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118:919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauney J.R., et al. In vitro and in vivo evaluation of differentially demineralized cancellous bone scaffolds combined with human bone marrow stromal cells for tissue engineering. Biomaterials. 2005;26:3173–3185. doi: 10.1016/j.biomaterials.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Chamberlain L.H., Shipston M.J. The physiology of protein S-acylation. Physiol. Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hancock J.F., Magee A.I., Childs J.E., Marshall C.J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 32.Salaun C., Greaves J., Chamberlain L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeste-Velasco M., Linder M.E., Lu Y.J. Protein S-palmitoylation and cancer. Biochim. Biophys. Acta. 2015:107–120. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Chavda B., Arnott J.A., Planey S.L. Targeting protein palmitoylation: selective inhibitors and implications in disease. Expet Opin. Drug Discov. 2014;9:1005–1019. doi: 10.1517/17460441.2014.933802. [DOI] [PubMed] [Google Scholar]
- 35.Fukata Y., Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 36.Greaves J., Chamberlain L.H. New links between S-acylation and cancer. J. Pathol. 2014;233:4–6. doi: 10.1002/path.4339. [DOI] [PubMed] [Google Scholar]
- 37.Blanc M., David F.P.A., van der Goot F.G. SwissPalm 2: protein S-palmitoylation database. Methods Mol. Biol. 2019:203–214. doi: 10.1007/978-1-4939-9532-5_16. [DOI] [PubMed] [Google Scholar]
- 38.Blanc M., et al. SwissPalm: protein palmitoylation database. F1000Research. 2015;4:261. doi: 10.12688/f1000research.6464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowal L., Yang W., Freeman M.R., Steen H., Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood. 2011;118:e62–e73. doi: 10.1182/blood-2011-05-353078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang R., et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin B.R., Wang C., Adibekian A., Tully S.E., Cravatt B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods. 2011;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song I.W., et al. Palmitoyl acyltransferase, Zdhhc13, facilitates bone mass acquisition by regulating postnatal epiphyseal development and endochondral ossification: a mouse model. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al Saedi A., Myers D.E., Stupka N., Duque G. 1,25(OH)2D3 ameliorates palmitate-induced lipotoxicity in human primary osteoblasts leading to improved viability and function. Bone. 2020;141 doi: 10.1016/j.bone.2020.115672. [DOI] [PubMed] [Google Scholar]
- 44.Steven A., et al. What turns CREB on? And off? And why does it matter? Cell. Mol. Life Sci. : CM. 2020;77:4049–4067. doi: 10.1007/s00018-020-03525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact the corresponding author for reasonable data requests.