Abstract
Background
PTPN2, a member of the non-receptor protein tyrosine phosphatases family, holds a crucial role in tumorigenesis and cancer immunotherapy. However, most studies on the role of PTPN2 in cancer are limited to specific cancer types. Therefore, this study aimed to investigate the prognostic significance of PTPN2 in human cancers and its function in the tumor microenvironment.
Methods
To shed light on this matter, we investigated the expression level, prognostic value, genomic alterations, molecular function, immune function, and immunotherapeutic predictive ability of PTPN2 in human cancers using the TCGA, GTEx, CGGA, GEO, cBioPortal, STRING, TISCH, TIMER2.0, ESTIMATE, and TIDE databases. Furthermore, the CCK-8 assay was utilized to detect the effect of PTPN2 on cell proliferation. Cell immunofluorescence analysis was performed to probe the cellular localization of PTPN2. Western blot was applied to examine the molecular targets downstream of PTPN2. Finally, a Nomogram model was constructed using the TCGA-LGG cohort and evaluated with calibration curves and time-dependent ROCs.
Results
PTPN2 was highly expressed in most cancers and was linked to poor prognosis in ACC, GBM, LGG, KICH, and PAAD, while the opposite was true in OV, SKCM, and THYM. PTPN2 knockdown promoted the proliferation of melanoma cells, while significantly inhibiting proliferation in colon cancer and glioblastoma cells. In addition, TC-PTP, encoded by the PTPN2 gene, was primarily localized in the nucleus and cytoplasm and could negatively regulate the JAK/STAT and MEK/ERK pathways. Strikingly, PTPN2 knockdown significantly enhanced the abundance of PD-L1. PTPN2 was abundantly expressed in Mono/Macro cells and positively correlated with multiple immune infiltrating cells, especially CD8+ T cells. Notably, DLBC, LAML, OV, and TGCT patients in the PTPN2-high group responded better to immunotherapy, while the opposite was true in ESCA, KIRC, KIRP, LIHC, and THCA. Finally, the construction of a Nomogram model on LGG exhibited a high prediction accuracy.
Conclusion
Immune checkpoint PTPN2 is a powerful biomarker for predicting prognosis and the efficacy of immunotherapy in cancers. Mechanistically, PTPN2 negatively regulates the JAK/STAT and MEK/ERK pathways and the abundance of PD-L1.
Keywords: PTPN2, Pan-cancer, Prognosis, Immunotherapy, Biomarker
1. Introduction
T-cell protein tyrosine phosphatase (TC-PTP), encoded by the protein tyrosine phosphatase non-receptor type 2 (PTPN2) gene, is a non-transmembrane protein consisting of a conserved catalytic structural domain and a non-catalytic c-terminal structural domain [1]. PTPN2, a member of the family of intracellular non-receptor PTPs (PTPNs), belongs to the largest family of class I cysteine PTPs and is essential for the regulation of various biological processes by dephosphorylating a variety of substrate proteins, including but not limited to epidermal growth factor receptor (EGFR), insulin receptor (IR), Janus kinase 1 (JAK1), JAK3, signal transducer and activator of transcription 1 (STAT1), STAT3, and Src family kinases [2]. Since the regulatory network of PTPN2 is extremely sophisticated and serves a unique function in various cancers, it can negatively regulate many signaling pathways and thus exert its pro- or anti-cancer function. For instance, PTPN2 acts as a tumor suppressor in breast and skin cancers. Specifically, loss of PTPN2 in breast cancer results in the activation of oncogenic signaling pathways, such as protein kinase B (AKT), Src family kinases (SFK), and STAT3 signaling pathways, and is also associated with resistance to tamoxifen [3,4]. In skin cancer, PTPN2 suppresses cancer cell proliferation and induces apoptosis by negatively regulating STAT1, STAT3, STAT5, phosphoinositide 3-kinase (PI3K)/AKT, and fetal liver kinase-1 (Flk-1)/c-Jun N-terminal kinase (JNK) signaling pathways [[5], [6], [7]]. In contrast, patients with pancreatic cancer, glioma, and glioblastoma with high expression of PTPN2 have a poor prognosis [8,9]. What's more, inflammatory responses or oxidative stress can induce up-regulation of PTPN2 expression, thereby promoting progression in patients with thyroid cancer, laryngeal cancer, and glioma [10–12]. To elucidate the role of PTPN2 in human cancers, a comprehensive pan-cancer analysis of PTPN2 is needed for further exploration.
There is accumulating evidence indicating that PTPN2 occupies a crucial position in immunomodulation and immunotherapy. PTPN2 is a crucial anti-inflammatory regulator that restricts the activation of T cells by dephosphorylating and inactivating Src family kinases, thereby reducing the inflammatory response [13,14]. Furthermore, PTPN2 holds a similar role in dendritic cells (DC) [15]. Of note, PTPN2 abolition in tumor cells enhances interferon-γ (IFN-γ)-mediated antigen presentation [16], while the depletion of PTPN2 in CD8+ T cells increases the proliferative capacity and cytotoxicity of Tim-3+ cells [17], both of which ultimately achieve antitumor effects. In brief, PTPN2 performs a “brake-like” function in the immune system, which is considered an emerging target for cancer immunotherapy.
However, most studies on the role of PTPN2 in cancer are limited to specific cancer types and there are no pan-cancer analyses of PTPN2. In this study, we explored the function of PTPN2 in pan-cancer using multiple databases, including the Cancer Genome Atlas (TCGA), the Genotype-Tissue Expression (GTEx), Chinese Glioma Genome Atlas (CGGA), the Gene Expression Omnibus (GEO), cBioPortal, STRING, the Tumor Immune Single-cell Hub (TISCH), TIMER2.0, ESTIMATE, and the Tumor Immune Dysfunction and Exclusion (TIDE) databases. First, we evaluated the expression level of PTPN2 and its relationship with prognosis and further explored the cellular localization and molecular function of PTPN2. Next, we elucidated in detail the role of PTPN2 in the tumor microenvironment (TME) and utilized the TIDE algorithm to predict the response to immunotherapy. Finally, a Nomogram model was constructed to predict the overall survival in patients with brain lower-grade glioma (LGG) based on the expression of PTPN2.
2. Materials and methods
2.1. PTPN2 expression levels and relationship with prognosis in cancers
The RNA-seq data of PTPN2 in each tumor and normal tissues were obtained from the TCGA database (https://portal.gdc.cancer.gov/) and GTEx database by UCSC XENA (https://xena.ucsc.edu/). Three types of prognosis data including overall survival (OS), disease-specific survival (DSS), and disease-free interval (DFI) were obtained from the TCGA database. Subsequently, CGGA (http://www.cgga.org.cn/index.jsp) (n = 420), GSE63885 (n = 70), and GSE54467 (n = 79) were employed to validate the prognostic value of PTPN2 in LGG, ovarian serous cystadenocarcinoma (OV), and skin cutaneous melanoma (SKCM), respectively. Among them, the continuous variable of PTPN2 expression data was used for univariate Cox regression analysis, and bivariate PTPN2 expression levels were performed for Kaplan-Meier curve analysis, whose cutoff was chosen by the “surv-cutpoint” function of the “survminer” R package. Data on the immune subtypes of LGG patients in TCGA were obtained from the paper published by Vesteinn Thorsson et al. [18].
2.2. Mutational analysis and pathway enrichment analysis of PTPN2
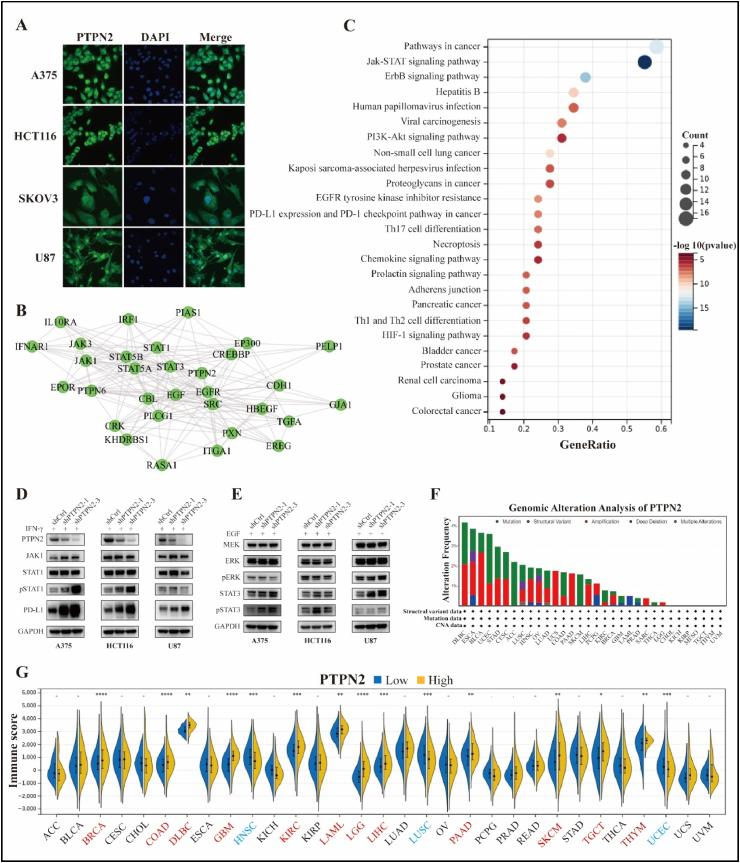
PanCancer Atlas Studies in the cBioPortal database (http://cbioportal.org) were performed for PTPN2 genomic mutation analysis, including 10,967 samples. A protein-protein interaction network (PPI) with an interaction score of 0.400 was constructed using the STRING (https://www.string-db.org/) online tool. Next, we performed Kyoto Gene and Genome Encyclopedia (KEGG) pathway enrichment analysis of PTPN2-related genes using the R package “clusterProfiler” and visualized them using the R package “ggplot2".
2.3. ESTIMATE score
ESTIMATE (https://bioinformatics.mdanderson.org/estimate/index.html) was used to download stromal and immune scores for each sample across all TCGA tumor types. The tumor purity is lower if the stromal and immune scores are higher. Here, the differential distribution of immune scores between the PTPN2-high and -low groups was assessed according to the median PTPN2 expression.
2.4. Single-cell analysis of PTPN2
Single-cell analysis of PTPN2 was performed using the TISCH online tool (http://tisch.comp-genomics.org/), which aims to characterize the tumor microenvironment at single-cell resolution. The detailed procedure for single-cell analysis of PTPN2 in all cancers was as follows: Gene (PTPN2), Cell-type annotation (Major-lineage), and Cancer-type (All cancers). PTPN2 expression in each cancer type was quantified and visualized in the form of heat and scatter plots.
2.5. Immune cell infiltration analysis in TIMER2
The TIMER2.0 database (http://timer.comp-genomics.org/) contains several algorithms for assessing the abundance of immune cell infiltration, including TIMER, CIBERSORT, quanTIseq, xCell, MCP-counter, and EPIC methods. We then downloaded data on immune cells from all cancer patients in TCGA and analyzed the correlation with PTPN2 expression levels. Among them, immune cells include CD4+ T cells, cancer-associated fibroblasts (CAF), progenitors, endothelial cells (Endo), eosinophils (Eos), hematopoietic stem cells (HSC), follicular helper T cells (Tfh), gamma delta T cells (Tgd), natural killer T cells (NKT), regulatory T cells (Treg), mast cells, B cells, neutrophils, monocytes (Mono), macrophages (Macro), dendritic cells, natural killer cells (NK), and CD8+ T cells.
2.6. Tumor heterogeneity and prediction of immunotherapy response
Firstly, we downloaded the unified standardized dataset TCGA Pan-Cancer (PANCAN, n = 10535) from the UCSC database and extracted the expression data of PTPN2 in each sample. The tumor mutation burden (TMB) of each tumor was calculated using the R software package “MAfTools”. Data on microsatellite instability (MSI) scores for each tumor were obtained from the paper published by Russell Bonneville et al. [19]. Data on tumor purity, tumor ploidy, homologous recombination deficiencies (HRD), and neoantigens were obtained from a paper published by Vesteinn Thorsson et al. [18]. We then integrated the above tumor heterogeneity data and analyzed the correlation with PTPN2 expression levels. Finally, we adopted the TIDE algorithm to predict the efficacy of immunotherapy by using the RNAseq data (level3) of all tumors in the TCGA database and the corresponding patient clinical information, and further analyzed the distribution of TIDE scores in the PTPN2-high and -low groups according to the median PTPN2 expression. Patients with high TIDE scores may not respond to immunotherapy, while patients with low TIDE scores may benefit from immunotherapy.
2.7. Construction and evaluation of the Nomogram model
Univariate and multivariate Cox regression analyses were conducted to screen independent risk factors for LGG patients. Then, PTPN expression and partial clinical data of LGG patients were jointly utilized to construct a Nomogram model. The construction of the Nomogram model and the plotting of the calibration curves were performed using the R package “rms” and the R package “survival”. Time-dependent receiver operating characteristic (ROC) curves were analyzed by the R package “timeROC” and visualized by the R package “ggplot2".
2.8. Cell culture and lentiviral packaging and infection
All cell lines in this study were obtained from ATCC. HEK293T (ATCC, CRL-3216), melanoma cell line A375 (ATCC, CRL-1619), and glioblastoma cell line U87 (ATCC, HTB-14) were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS, Gibco, USA) and penicillin/streptomycin (Sigma, USA). Colon cancer cell line HCT116 (ATCC, CCL-247) and ovarian cancer cell line SKOV3 (ATCC, HTB-77) were cultured in RPMI 1640 medium (RPMI 1640, Gibco, USA) with 10% FBS and penicillin/streptomycin. All cell lines were cultured at 37 °C in a constant temperature incubator containing 5% CO2.
HEK293T cells were used for lentiviral production. Lentiviral expression vector pLKO was used to construct the PTPN2 knockdown vector (shPTPN2). During preparation, 500 ng of target gene plasmid was added to 100 μl opti-MEM together with 50 ng VSVG, 500 ng pR8.74, and 3 μl transfection reagent PEI, mixed sufficiently and left for 15 min, and then the mixture was co-transfected into approximately 80% confluent HEK293T cells in 12-well plates. The supernatant containing lentivirus was harvested 72 h after transfection, filtered through a 0.45 mM PES filter, and then stored at −80 °C for backup. Subsequently, A375, HCT116, and U87 cells were seeded in six-well plates, and 24 h later 50 μl of viral solution and polybrene (1:1000) were added. 24 h after infection, the cell culture medium containing viral solution was replaced with a fresh complete cell culture medium with puromycin added.
The target sequences of the shPTPN2 included:
shPTPN2-1: CCGGGATGACCAAGAGATGCTGTTTCTCGAGAAACAGCATCTCTTGGTCATCTTTTT
shPTPN2-2: CCGGTGCAAGATACAATGGAGGAGACTCGAGTCTCCTCCATTGTATCTTGCATTTTT
shPTPN2-3: CCGGGAAGATGTGAAGTCGTATTATCTCGAGATAATACGACTTCACATCTTCTTTTT
shPTPN2-4: CCGGGTGCAGTAGAATAGACATCAACTCGAGTTGATGTCTATTCTACTGCACTTTTT
shPTPN2-5: CCGGCTCACTTTCATTATACTACCTCTCGAGAGGTAGTATAATGAAAGTGAGTTTTT.
2.9. Western blot and reverse transcription and quantitative real-time PCR (RT-qPCR)
A375, HCT116, and U87 cells were collected separately in 1.5 ml EP tubes, RIPA lysis buffer containing protease inhibitors and phosphatase inhibitors was added, and cells were lysed sufficiently to obtain cellular proteins. Protein concentrations were determined using the BCA Protein Concentration Assay Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturer's instructions. Equal amounts of proteins were separated in 12% SDS-PAGE, then proteins were transferred to PVDF membranes, blocked with 5% skim milk powder for 1 h at room temperature, and then incubated with a 1:1000 dilution of protein primary antibody at 4 °C overnight. The membranes were washed three times with TBST, then incubated with a secondary antibody for 1 h at room temperature, and washed three more times with TBST.
Total RNA was extracted using RNA-easy Isolation Reagent (Vazyme, China) according to the manufacturer's protocol. RNA concentration was quantified using NanoDrop ND2000 (Thermo Fisher, Waltham, MA, USA). 1 μg of RNA per sample was reverse transcribed into cDNA using the Tiangen Reverse Transcription Kit, and the obtained cDNA products were diluted to a final concentration of 10 ng/μl. Real-time PCR was performed using 2 × SYBR Green Premix Ex Taq (Takara, Shiga, Japan) on an ABI 7500 PCR system (Applied Biosystems, CA, USA). Primer pairs are listed below. Analyses were performed using the comparative cycle threshold (CT) method, and all samples were normalized for GAPDH expression. The sequences of primers used were as follows:
GAPDH forward: GGAGCGAGATCCCTCCAAAAT
GAPDH reverse: GGCTGTTGTCATACTTCTCATGG
PTPN2 forward: GAAGAGTTGGATACTCAGCGTC
PTPN2 reverse: TGCAGTTTAACACGACTGTGAT
2.10. Cell proliferation assay
PTPN2 knockdown in A375, HCT116, and U87 cell lines to assay the proliferative capacity of the cells. The 96-well plates were seeded with 2 × 103 cells per well and incubated in an incubator at 37 °C with 5% CO2. Then, 10 μl of CCK-8 solution was added to each well, and the absorbance at 450 nm was measured using an enzyme marker after 0, 24, 48, 96, and 120 h, respectively.
2.11. Cell immunofluorescence
Cell lines A375, HCT116, U87, and SKOV3 were utilized for immunofluorescence analysis. Cells were fixed and incubated with primary antibodies at a dilution of 1:100, fluorescence dye-conjugated secondary antibodies, and DAPI, according to standard protocols. Cells were examined using a Liver SR super-resolution turntable microscope (Live SR CSU W1, Nikon, Japan) with a 60 × oil immersion objective.
2.12. Detection of downstream targets of PTPN2
The cell lines A375, HCT116, and U87 were employed to analyze the downstream pathways regulated by PTPN2. Before the assay, one group of cells was treated with IFN-γ (11725-HNAS, Sino Biological) at a concentration of 100 ng/ml for 36 h, and the other group was treated with EGF (PHG0311, Invitrogen) at a concentration of 100 ng/ml for 30 min. The immunoblot assay was identical to the workflow described above. Antibody information was as follows: Antibodies against PTPN2 (58935, CST), JAK1 (3344, CST), STAT1(19994, CST), Phospho-STAT1(Y701) (7649, CST), PD-L1 (13684, CST), STAT3 (4904, CST), Phospho-STAT3 (Y705) (9145, CST), p44/42 MAPK (Erk1/2) (4695, CST), Phospho-p44/42 MAPK (Erk1/2) (4370, CST), and GAPDH (60004-1-Ig, Proteintech) were used.
2.13. Statistical analysis
In this study, all statistical analyses were processed on R Studio software and P value < 0.05 was considered statistically significant. All analyses were preceded by a log 2 transformation of all RNA-seq data. Wilcoxon rank-sum test and Two-way repeated measures ANOVA were used for comparison between the two groups. Spearman's test was used for all correlation analyses. Univariate Cox regression analysis and log-rank test were employed to evaluate the prognostic value of PTPN2 in cancers.
3. Results
3.1. PTPN2 expression levels and the relationship with prognosis
The overview of the process used in our study was shown in Fig. 1. To investigate the expression of PTPN2 in human cancers, we integrated normal tissue data from the GTEx database with tumor tissue data from the TCGA database. PTPN2 was highly expressed in the majority of cancers, including bladder urothelial carcinoma (BLCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), LGG, pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS), while it was down-regulated in adrenocortical carcinoma (ACC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), OV, prostate adenocarcinoma (PRAD), SKCM, and thyroid carcinoma (THCA) (Fig. 2A). To gain further insight into the association between PTPN2 expression level and prognosis, we performed survival analyses for each cancer, including OS, DSS, and PFI. As shown in Fig. 2B, PTPN2 exerted completely distinct roles in various cancer types and was associated with poor patient prognosis in most cancers, but was a protective factor in a small number of cancers. Two indicators, DSS and PFI, were associated with cancer patients’ treatment outcomes, in high agreement with the results of OS analysis. Specifically, univariate Cox regression analysis and Kaplan–Meier survival curves collectively showed that up-regulation of PTPN2 expression was a perilous factor that seriously affected the overall survival of patients with ACC, GBM, LGG, kidney chromophobe (KICH), and PAAD, but the opposite results were found in OV, SKCM, and THYM (Fig. 2C–K). Therefore, we tentatively speculated that PTPN2 may exert carcinogenic effects in GBM, LGG, and PAAD while exhibiting tumor suppressive properties in OV and SKCM in combination with PTPN2 expression levels as well as prognosis.
Fig. 1.
Flow chart of this study.
Fig. 2.
The expression level of PTPN2 and the relationship with prognosis. (A) Comparison of PTPN2 mRNA expression between tumor and normal tissues in TCGA and GTEx databases. (B) The correlation between PTPN2 expression and overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) was summarized based on univariate Cox regression and Kaplan-Meier survival curves. Red indicates PTPN2 is a risk factor and green indicates a protective factor. (C) Forest plot demonstrating the prognostic value of PTPN2 in human cancers based on univariate Cox regression analysis. Kaplan-Meier overall survival curves of PTPN2 in ACC (D), GBM (E), LGG (F), KICH (F), PAAD (H), OV (I), SKCM (J), and THYM (K). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To further confirm this speculation, we conducted a validation using independent cohorts and CCK-8 assay. High expression of PTPN2 was indeed a danger factor for poor prognosis in LGG patients utilizing 420 cases from the CGGA database (Fig. 3A). As well, analysis of immune subtypes supported this conclusion, with immune subtype C4 carrying a significantly worse prognosis than C3 in LGG, and PTPN2 expression in C4 being superior to that in C3 (Fig. 3B). Furthermore, high PTPN2 expression was beneficial in prolonging the survival of OV and SKCM patients using 70 cases in GSE63885 and 79 cases in GSE54467, respectively (Fig. 3C and D). Next, we investigated the effect of PTPN2 knockdown on the proliferative capacity in different types of cancer cells. Western blot and RT-qPCR were utilized to verify the knockdown efficiency of PTPN2, and we selected shPTPN2-1 and shPTPN2-3 for follow-up experiments (Fig. 3E). PTPN2 knockdown promoted the proliferation of melanoma A375 cells, while significantly inhibiting the proliferation of colon cancer HCT116 cells and glioblastoma U87 cells (Fig. 3F–H). Collectively, these data demonstrated that the function of the PTPN2 was not entirely consistent, but depended heavily on the type of cancer.
Fig. 3.
Validation of the prognostic value and proliferative capacity of PTPN2. (A) Kaplan-Meier overall survival curves of PTPN2 in LGG based on CGGA database. (B) PTPN2 was differentially expressed among various immune subtypes in LGG. C3: inflammatory, C4: lymphocyte depleted, C5: immunologically quiet. (C) Kaplan-Meier overall survival curves of PTPN2 in OV based on GSE63885 cohort. (D) Kaplan-Meier overall survival curves of PTPN2 in SKCM based on GSE54467 cohort. (E) Western blot and RT-qPCR were utilized to verify the knockdown efficiency of PTPN2 in A375 cells. The original WB images were available in the supplementary file. (F–H) CCK-8 assay to detect the effect of PTPN2 knockdown on cell proliferation of A375, HCT116, and U87 cells, respectively. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.2. Cellular localization and molecular function of PTPN2
TC-PTP, encoded by the PTPN2 gene, can shuttle between the nucleus and cytoplasm in response to intracellular stimuli [20]. Immunofluorescence images also revealed that TC-PTP was mainly localized in the nucleus, followed by the cytoplasm in A375, HCT116, SKOV3, and U87 cell lines (Fig. 4A). To explore the unique molecular functions of PTPN2, we constructed a PPI network using the STRING database and performed a KEGG enrichment analysis. The results were that PTPN2-related genes were mainly involved in various oncogenic pathways, such as JAK/STAT, EGFR, and programmed cell death protein 1 (PD-1)/programmed cell death ligand 1 (PD-L1) signaling pathways, and were significantly related to the development of human cancer (Fig. 4B and C). To verify this finding, we constructed PTPN2 knockdown models of A375, HCT116, and U87 cells. To mimic the microenvironment of tumor cells in vivo to a greater extent in vitro, we used IFN-γ to induce and stimulate the cells. As a result, PTPN2 knockdown facilitated STAT1 phosphorylation and significantly activated the JAK1/STAT1 pathway in A375 and HCT116 cells, whereas it was not obviously altered in U87 cells. Strikingly, PTPN2 knockdown stimulated PD-L1 expression to varying degrees in all three cell lines, most notably in A375 cells (Fig. 4D). Next, to investigate the effect of PTPN2 on downstream targets of the EGFR pathway, we first used epidermal growth factor (EGF) to process the cells. The results were that PTPN2 knockdown promoted STAT3 phosphorylation and activation of the JAK/STAT3 pathway in A375 and HCT116 cells while boosting the extracellular signal-regulated kinase (ERK) phosphorylation and activation of the MEK/ERK pathway in U87 cells (Fig. 4E). Overall, PTPN2 could dephosphorylate the substrate proteins STAT1/3 and ERK and negatively regulate the expression of PD-L1. Multiple studies have shown that the mutational inactivation of PTPN2 can promote inflammation and carcinogenesis in part through activation of the JAK/STAT pathway [21,22]. However, PTPN2 mutation rates were generally low in cancers, with the highest mutation rate in DLBC at 4.17% (Fig. 4F). In addition, PTPN2-related genes were also engaged in regulating the differentiation of Th1, Th2, and Th17 cells (Fig. 4C), and Immune scores were higher in the PTPN2-high group in most cancers, including BRCA, COAD, DLBC, GBM, KIRC, LAML, LGG, LIHC, PAAD, SKCM, TGCT, and THYM (Fig. 4G), which collectively suggested a critical role of PTPN2 in the immune system.
Fig. 4.
Cellular localization and molecular function of PTPN2. (A) Immunofluorescence analysis of PTPN2 protein, nucleus, and merge images of A375, HCT116, SKOV3, and U87 cell lines. (B) The PPI network of PTPN2-related genes. (C) KEGG enrichment analysis for PTPN2-related genes. (D, E) Western blot to detect the downstream pathway of PTPN2 using A375, HCT116, and U87 cell lines after IFN-γ and EGF treatment, respectively. The original WB images were available in the supplementary file. (F) Genomic mutations of PTPN2 in cancers. (G) Differences in Immune score between the PTPN2-high and –low groups in cancers. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
3.3. Single-cell analysis and immune cell infiltration analysis of PTPN2
It is well known that PD-L1 was primarily expressed on the surface of cancer cells to restrict the activation of T cells through binding to PD-1. Thus, the abundance of PD-L1 was related to the immunosuppressive state in the tumor microenvironment. As previously mentioned, PTPN2 could negatively regulate PD-L1 expression, but the degree of regulation varies in different cancer types, which also implies different immune cell infiltration. Hence, a comprehensive understanding of the correlation between PTPN2 expression and immune cell infiltration in all cancers is beneficial for the precision immunotherapy of cancer.
To explore the expression of PTPN2 in immune cells, stromal cells, and malignant cells in the tumor microenvironment, we performed single-cell analyses using 79 datasets from the TISCH database. As shown in Fig. 5, PTPN2 was mainly expressed in Mono/Macro in all cancers, followed by malignant cells. Additionally, we observed that PTPN2 was highly expressed in glioma and colorectal cancer (CRC). Specifically, the GSE102130 dataset contained 3321 cells from 6 glioma patients, and PTPN2 was highly expressed in Mono/Macro, malignant cells, and oligodendrocytes. In the GSE146771 dataset, which contains 10,468 cells from 10 CRC patients, PTPN2 was widely expressed in a variety of cells, such as proliferating T cells, regulatory T cells, plasma cells, monocytes/macrophages, etc. These data suggested that PTPN2 was expressed on a variety of immune cells and may be involved in the regulation of their differentiation and development.
Fig. 5.
Summary of PTPN2 expression of 33 cancer types in 79 single-cell datasets.
As shown in Fig. 6, PTPN2 expression was significantly positively correlated with a variety of immune cells, especially CD8+ T cells, neutrophils, and a subset of CD4+ T cells, dendritic cells, B cells, and Mono/Macro cells. In contrast, PTPN2 expression was negatively related to HSC and NKT cells. Furthermore, we also observed significant differences in the correlation of PTPN2 with immune cells, depending on the type of cancer. Especially in GBM, LGG, breast invasive carcinoma (BRCA), and THYM, PTPN2 was closely associated with the infiltration of immune cells. The role of immune cells in cancer therapy should not be ignored, and our results noted that PTPN2 may influence cancer development, prognosis, and treatment by regulating immune cells.
Fig. 6.
Correlation between PTPN2 and immune cells in TME, including CD4+T cells, CAF, progenitor, Endo, Eos, HSC, Tfh, Tgd, NKT, Treg, mast cells, B cells, neutrophils, monocytes, macrophages, dendritic cells, NK cells, and CD8+T cells. *P < 0.05, and **P < 0.01.
3.4. Relationship between PTPN2 and tumor heterogeneity and its prediction for immunotherapy
PTPN2 has been proven to negatively regulate the expression of PD-L1 and holds a critical role in the immune system. Next, we further investigated the role of PTPN2 in the prediction of immunotherapy efficacy. First, we evaluated PTPN2 expression with hallmarks of tumor heterogeneity, including TMB, MSI, tumor purity, neoantigens, polyploidy, and HRD. Among them, TMB was highly correlated with the efficacy of PD-1/PD-L1 inhibitors, and this indicator can reasonably predict the efficacy of immunotherapy [23]. We revealed that PTPN2 was positively associated with TMB in most cancers, including GBM/LGG, LGG, COAD, STAD, COAD/READ, HNSC, LUAD, stomach and esophageal cancer (STES), UCS, and BLCA, but negatively in THYM, THCA, CHOL, and liver hepatocellular carcinoma (LIHC) (Fig. 7A). Furthermore, MSI was an important tumor biomarker caused by defective DNA mismatch repair function in tumor tissues [24]. Positive correlations with MSI were identified in LUAD, STAD, COAD, LUSC, STES, COAD/READ, HNSC, and BRCA, and negative correlations were discovered in DLBC, GBM/LGG, and KIPAN (KICH + KIRC + KIRP) (Fig. 7B). Tumor purity was significantly correlated with clinical characteristics, genomic expression, and biological characteristics of patients. PTPN2 was positively relevant to tumor purity in HNSC, LUSC, STAD, and CESC, and negatively relevant to tumor purity in THYM, GBM, KIPAN, TGCT, LIHC, sarcoma (SARC), KIRC, COAD, COAD/READ, BLCA, and BRCA (Fig. 7C). Neoantigens were very attractive targets for tumor immunotherapy, and vaccines against neoantigens have been developed in clinical trials for various solid tumors. PTPN2 was positively correlated with neoantigens only in COAD, COAD/READ, and BRCA (Fig. 7D). Similarly, polyploidy was strongly associated with chromosomal instability involved in cancer development. Positive correlations with polyploidy were found in TGCT, SARC, STAD, LGG, STES, and OV, and negative correlations were discovered in UCS, COAD, COAD/READ, and THCA (Fig. 7E). Clinical studies have demonstrated that HRD status was strongly linked to the sensitivity of platinum-based chemotherapeutic agents and PARP inhibitors, which was a critical indicator of treatment choice and prognosis for various tumors [25]. Of note, PTPN2 was positively associated with HRD in several cancers, including KICH, UCEC, LGG, PAAD, LIHC, BRCA, LUAD, GBM/LGG, HNSC, STES, LUSC, KIPAN, BLCA, STAD, and KIRC. However, the correlation was negative in THYM, LAML, GBM, COAD, and COAD/READ (Fig. 7F).
Fig. 7.
Relationship between PTPN2 and tumor heterogeneity and its prediction for immunotherapy. Correlation analysis of PTPN2 expression with (A) TMB, (B) MSI, (C) tumor purity, (D) neoantigens, (E) ploidy, and (F) HRD. (G) PTPN2 expression predicts immunotherapy response in cancer patients based on the TIDE algorithm. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The TIDE algorithm provided a powerful tool for predicting response to tumor immunotherapy. Tumor patients with high TIDE scores were likely to be non-responders to immunotherapy [26]. The results were that TIDE scores were higher in the PTPN2-high group in ESCA, KIRC, KIRP, LIHC, and THCA, suggesting that high expression of PTPN2 in these cancers was detrimental to the immunotherapy of patients. However, the opposite was true for DLBC, LAML, OV, and TGCT (Fig. 7G). In conclusion, the combination of immune-related gene expression, immune cell infiltration, tumor heterogeneity analysis, and TIDE scores was a valid approach in cancer research.
3.5. Construction and evaluation of the Nomogram model
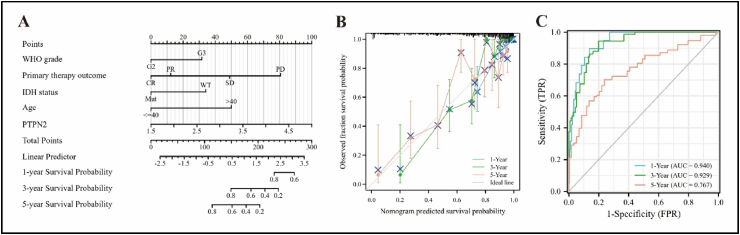
Given the prognostic value of PTPN2 in LGG and its specific role in TME, we proceed to construct a clinical prognostic model to predict the probability of overall survival in LGG patients. As shown in Table 1, univariate and multivariate Cox regression analysis indicated that PTPN2 expression, WHO grade, primary therapy outcome, isocitrate dehydrogenase (IDH) status, and age were risk factors for the survival of LGG patients. Therefore, we constructed a Nomogram and plotted a calibration curve with a C-index value of 0.854 (95% CI: 0.835–0.872) (Fig. 8A and B). Additionally, time-dependent ROC curves showed that the accuracy of the model in predicting 1-, 3-, and 5-year overall survival in LGG patients was 0.940, 0.929, and 0.767, respectively (Fig. 8C).
Table 1.
Univariate and multivariate Cox regression analysis of clinical data of LGG patients.
| Characteristics | Total (N) | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| PTPN2 | 527 | 2.651 (1.902–3.693) | <0.001 | 1.891 (1.185–3.018) | 0.008 |
| WHO grade | 466 | ||||
| G2 | 223 | Reference | |||
| G3 | 243 | 3.059 (2.046–4.573) | <0.001 | 2.025 (1.275–3.215) | 0.003 |
| Primary therapy outcome | 457 | ||||
| PD | 110 | Reference | |||
| SD | 146 | 0.439 (0.292–0.661) | <0.001 | 0.495 (0.304–0.806) | 0.005 |
| PR | 64 | 0.175 (0.076–0.402) | <0.001 | 0.218 (0.078–0.608) | 0.004 |
| CR | 137 | 0.122 (0.056–0.266) | <0.001 | 0.166 (0.075–0.370) | <0.001 |
| IDH status | 524 | ||||
| WT | 97 | Reference | |||
| Mut | 427 | 0.186 (0.130–0.265) | <0.001 | 0.468 (0.284–0.773) | 0.003 |
| Age | 527 | ||||
| ≤40 | 264 | Reference | |||
| >40 | 263 | 2.889 (2.009–4.155) | <0.001 | 3.047 (1.936–4.796) | <0.001 |
CR, complete remission/response; PD, progressive disease; PR, partial remission/response; SD, stable disease.
Fig. 8.
Nomogram model was constructed based on the expression of PTPN2 in LGG. (A) Nomogram was constructed combining clinicopathological characteristics and PTPN2 expression in LGG. (B) Calibration curves for Nomogram model in LGG. (C) The capability of time-dependent ROC curves to predict 1-, 3-, and 5-year overall survival in patients with LGG.
4. Discussion
PTPN2, as a member of the intracellular non-receptor protein tyrosine phosphatase family, is antagonistic to protein tyrosine kinases (PTKs), as are other protein tyrosine phosphatases. Physiologically, PTPN2 is implicated in hematopoietic, inflammatory responses, immune system, and cell proliferation activities. In addition, a large body of evidence suggests that PTPN2 is coupled with tumor proliferation and chemoresistance, indicating that PTPN2 may be an emerging therapeutic target. PTPN2 was overexpressed in most cancers and high levels of PTPN2 predicted a poor prognosis for patients with thyroid cancer, laryngeal cancer, glioma, and KRAS-mutated pancreatic cancer [[9], [11],27]. However, there is limited information on the prognostic value of PTPN2 in other solid cancer types. Here, we also revealed that PTPN2 was a risk factor in ACC and KICH while a protective factor in THYM, SKCM, and OV, suggesting that PTPN2 was a double-edged sword with different functions in various cancers. Controversially, PTPN2 suppressed the tumorigenicity of Delta EGFR (the most common mutation site) -expressing glioblastoma cells in vivo by inhibiting Delta EGFR-mediated activation of ERK2 [28]. However, we found that PTPN2 knockdown remarkably inhibited the proliferation of glioblastoma U87 cells in our study. We speculated that there are several reasons, firstly, there may be some differences in various experimental settings and tumor cell lines. Secondly, there are also differences in the molecular mutations in tumor cell lines. If the regulators of PTPN2 are mutated, this may lead to alterations in the function of PTPN2, resulting in opposite functions in the same cancer. Third, PTPN2 possessed structural domains of phosphatase activity that could regulate the dephosphorylation of many substrate proteins, including tumor promoters or repressors. Therefore, focusing on only one molecular pathway may bias the overall role of PTPN2 in tumors. To address this controversial issue, combining in vitro cell lines and in vivo mouse genetic studies by knocking out specific regulators and effectors will contribute to a better understanding of the specific signaling pathways regulated by PTPN2.
TC-PTP was localized in the nucleus and cytoplasm and can shuttle between the two in response to intracellular stimuli. It has been reported that PTPN2 is the first reported PTP gene to be knocked down in mice and modulate JAK/STAT signaling [29,30]. Similarly, our study also revealed that PTPN2 can negatively regulate the JAK/STAT and MEK/ERK pathways. However, it is worth noting that the pathways regulated by PTPN2 are not necessarily the same in various cancers. For instance, PTPN2 predominantly negatively modulated JAK/STAT3 signaling pathway in melanoma and colon cancer, but MEK/ERK signaling pathway in glioblastoma. These data indicated that the regulatory mechanism of PTPN2 was quite complicated, and a combination of in vivo and in vitro experiments was required to collectively identify exactly the role of PTPN2 as a cancer suppressor or promoter in specific cancer. Interestingly, PTPN2 and PTPN1 share analogous properties [31] that may be more beneficial to investigate them in combination to elucidate their roles.
The lines of evidence accumulated to the date indicated that PTPN2 is critical in the tumor microenvironment, for instance, inhibition of PTPN2 enhanced the efficacy of anti-PD1 and CAR-T in solid tumors [32,33]. PD-L1 is a transmembrane protein considered to be a co-suppressor of immune responses that binds to PD-1 and reduces the proliferation of PD-1+CD8+ T cells, thereby inhibiting their cytokine secretion and inducing apoptosis [34]. Strikingly, we revealed that PTPN2 can negatively regulate the abundance of PD-L1 and speculated that high expression of PTPN2 in the tumor microenvironment may increase the number of immune-infiltrating lymphocytes, especially CD8+ T cells. Notably, immune infiltration analysis showed a significant positive correlation between PTPN2 expression and CD8+ T cells, further confirming this conjecture. Furthermore, we found that PTPN2 negatively regulated PD-L1 to a greater extent in melanoma A375 and colon cancer HCT116 cells than in glioblastoma U87 cells, suggesting that the extent to which PTPN2 regulates PD-L1 expression depends on the type of cancer. Coincidentally, PTPN2 was significantly positively correlated with CD8+ T cells in COAD and SKCM, but not in GBM. Therefore, we can reasonably speculate that PTPN2 negatively regulates PD-L1 in THYM, DLBC, BRCA, and UVM to a higher extent according to the relationship between PTPN2, PD-L1, and infiltrating CD8+ T cells. However, high PTPN2 expression may increase the number of infiltrating CD8+ T cells, but not necessarily activate CD8+ T cells, as it has been shown that the lack of PTPN2 in T cells enhances the killing of tumor cells by T cells [17]. Collectively, these data indicated that PTPN2 holds a crucial role and performs an extremely sophisticated function in both immune cells and tumor cells. Once the imbalance of PTPN2 expression, thereby affecting tumor progression and patient prognosis by remodeling the tumor microenvironment.
Previous studies have demonstrated that low PTPN2 expression was associated with a higher relapse rate in patients with Luminal A and HER2-positive breast cancer, but not triple-negative tumors [35], indicating that the action of PTPN2 was associated with cancer subtypes. In the present study, we found that the expression of PTPN2 in LGG differed significantly among immune subtypes, and further studies should be conducted to assess how PTPN2 affects the prognosis and treatment response of each subtype. As we all know, antigen release by tumor cells is the first step in the cancer immune cycle. Effective antigen release will facilitate antigen presentation, thereby activating T cells to recognize and kill tumor cells. Several studies have shown a clear correlation between TMB and immune checkpoint inhibitors (ICI), and TMB reflects the number of cancer mutations, indicating a higher availability of neoantigens [36]. Of note, higher TMB is clinically associated with better immune checkpoint blockade (ICB) therapy in melanoma and non-small cell lung cancer [37,38]. But to escape, tumor cells exploit immunosuppressive checkpoints to limit the activation of T cells [37]. Strikingly, high-frequency MSI, especially in colorectal cancer, is an independent predictor of clinical features and prognosis [39]. Furthermore, tumor ploidy, purity, and HRD are hallmarks of cancer and key indicators of treatment options and prognosis in many cancers. From our results, PTPN2 expression was not only correlated with these tumor biomarkers in different types of cancer but also directly related to the efficacy of immunotherapy in cancer patients according to the TIDE algorithm. Combined with the results of DSS and PFI, these data collectively suggested that PTPN2 was appropriate for predicting treatment outcomes in cancer patients.
Currently, PTPN2 inhibitors are extensively studied and developed due to the critical role of PTPN2 in tumorigenesis and cancer immunity. As mentioned previously, several studies have shown that PTPN2 deficiency facilitates enhanced antitumor immunotherapy, such as anti-PD-1 and CAR-T therapies. However, our study showed that PTPN2 exerts distinct functions in various cancers, so the question is how to rationally utilize PTPN2 inhibitors. For example, using PTPN2 inhibitors may suppress the proliferation of colon cancer and glioblastoma cells while enhancing anti-tumor immunity, both of which play a synergistic anti-cancer role. Obviously, the use of inhibitors of PTPN2 in melanoma would be questionable, although perhaps enhancing anti-tumor immunity. In ovarian cancer, our results show that patients with high PTPN2 expression have a better prognosis and immunotherapeutic efficacy, indicating that PTPN2 inhibitors are not indicated.
In conclusion, PTPN2 was heterogeneously expressed in a variety of cancers and related to patient prognosis and efficacy of immunotherapy. PTPN2 knockdown promoted the proliferation of melanoma cells while significantly inhibiting the proliferation of colon cancer and glioblastoma cells. Mechanistically, PTPN2 negatively regulates the JAK/STAT and MEK/ERK pathways and PD-L1 abundance. In addition, PTPN2 positively correlated with a variety of immune infiltrating cells, particularly CD8+ T cells. What's more, the LGG clinical prognostic model constructed based on PTPN2 expression exhibited excellent predictive accuracy and facilitated clinical application. Overall, PTPN2 is a robust biomarker for predicting cancer prognosis and the efficacy of immunotherapy.
Author contribution statement
Xiaolong Tang: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Xue Sui: Analyzed and interpreted the data. Yongshuo Liu: Conceived and designed the experiments; Performed the experiments.
Funding statement
Yongshuo Liu was supported by National Natural Science Foundations of China [81802400], China Postdoctoral Science Foundation [2020M670053].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest’s statement
The authors declare no competing interests.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e12873.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Morales L.D., et al. The role of T-cell protein tyrosine phosphatase in epithelial carcinogenesis. Mol. Carcinog. 2019;58(9):1640–1647. doi: 10.1002/mc.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang X., et al. Critical roles of PTPN family members regulated by non-coding RNAs in tumorigenesis and immunotherapy. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.972906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlsson E., et al. Loss of protein tyrosine phosphatase, non-receptor type 2 is associated with activation of AKT and tamoxifen resistance in breast cancer. Breast Cancer Res. Treat. 2015;153(1):31–40. doi: 10.1007/s10549-015-3516-y. [DOI] [PubMed] [Google Scholar]
- 4.Shields B.J., et al. TCPTP regulates SFK and STAT3 signaling and is lost in triple-negative breast cancers. Mol. Cell Biol. 2013;33(3):557–570. doi: 10.1128/MCB.01016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M., et al. Overexpression of TC-PTP in murine epidermis attenuates skin tumor formation. Oncogene. 2020;39(21):4241–4256. doi: 10.1038/s41388-020-1282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek M., et al. Epidermal-specific deletion of TC-PTP promotes UVB-induced epidermal cell survival through the regulation of Flk-1/JNK signaling. Cell Death Dis. 2018;9(7):730. doi: 10.1038/s41419-018-0781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H., et al. Targeted disruption of TC-PTP in the proliferative compartment augments STAT3 and AKT signaling and skin tumor development. Sci. Rep. 2017;7 doi: 10.1038/srep45077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuang W., et al. PTPN2, A key predictor of prognosis for pancreatic adenocarcinoma, significantly regulates cell cycles, apoptosis, and metastasis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.805311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P.F., et al. Molecular and clinical characterization of PTPN2 expression from RNA-seq data of 996 brain gliomas. J. Neuroinflammation. 2018;15(1):145. doi: 10.1186/s12974-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., et al. Upregulated PTPN2 induced by inflammatory response or oxidative stress stimulates the progression of thyroid cancer. Biochem. Biophys. Res. Commun. 2020;522(1):21–25. doi: 10.1016/j.bbrc.2019.11.047. [DOI] [PubMed] [Google Scholar]
- 11.Yang H.J., et al. Inflammatory response or oxidative stress induces upregulation of PTPN2 and thus promotes the progression of laryngocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020;24(8):4314–4319. doi: 10.26355/eurrev_202004_21012. [DOI] [PubMed] [Google Scholar]
- 12.Wu L., et al. PTPN2 induced by inflammatory response and oxidative stress contributed to glioma progression. J. Cell. Biochem. 2019;120(11):19044–19051. doi: 10.1002/jcb.29227. 10.1002/jcb.29227. [DOI] [PubMed] [Google Scholar]
- 13.Spalinger M.R., et al. PTPN2 controls differentiation of CD4(+) T cells and limits intestinal inflammation and intestinal dysbiosis. Mucosal Immunol. 2015;8(4):918–929. doi: 10.1038/mi.2014.122. [DOI] [PubMed] [Google Scholar]
- 14.Wiede F., et al. T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice. J. Clin. Invest. 2011;121(12):4758–4774. doi: 10.1172/JCI59492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hering L., et al. Protein tyrosine phosphatase non-receptor type 2 function in dendritic cells is crucial to maintain tissue tolerance. Front. Immunol. 2020;11:1856. doi: 10.3389/fimmu.2020.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manguso R.T., et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaFleur M.W., et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol. 2019;20(10):1335–1347. doi: 10.1038/s41590-019-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsson V., et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonneville R., et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol, 2017. 2017 doi: 10.1200/PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simoncic P.D., McGlade C.J., Tremblay M.L. PTP1B and TC-PTP: novel roles in immune-cell signaling. Can. J. Physiol. Pharmacol. 2006;84(7):667–675. doi: 10.1139/y06-012. [DOI] [PubMed] [Google Scholar]
- 21.Kleppe M., et al. Mutation analysis of the tyrosine phosphatase PTPN2 in Hodgkin's lymphoma and T-cell non-Hodgkin's lymphoma. Haematologica. 2011;96(11):1723–1727. doi: 10.3324/haematol.2011.041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parlato M., et al. Loss-of-Function mutation in PTPN2 causes aberrant activation of JAK signaling via STAT and very early onset intestinal inflammation. Gastroenterology. 2020;159(5):1968–1971. doi: 10.1053/j.gastro.2020.07.040. e4. [DOI] [PubMed] [Google Scholar]
- 23.Choucair K., et al. TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther. 2020;27(12):841–853. doi: 10.1038/s41417-020-0174-y. [DOI] [PubMed] [Google Scholar]
- 24.Sahin I.H., et al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer. 2019;121(10):809–818. doi: 10.1038/s41416-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frey M.K., Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol. Oncol. Res. Pract. 2017;4:4. doi: 10.1186/s40661-017-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang P., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z., et al. PTPN2 regulates the activation of KRAS and plays a critical role in proliferation and survival of KRAS-driven cancer cells. J. Biol. Chem. 2020;295(52):18343–18354. doi: 10.1074/jbc.RA119.011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klingler-Hoffmann M., et al. The protein tyrosine phosphatase TCPTP suppresses the tumorigenicity of glioblastoma cells expressing a mutant epidermal growth factor receptor. J. Biol. Chem. 2001;276(49):46313–46318. doi: 10.1074/jbc.M106571200. [DOI] [PubMed] [Google Scholar]
- 29.You-Ten K.E., et al. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J. Exp. Med. 1997;186(5):683–693. doi: 10.1084/jem.186.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simoncic P.D., et al. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol. 2002;12(6):446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 31.Heinonen K.M., et al. Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-gamma signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106(23):9368–9372. doi: 10.1073/pnas.0812109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh P.K., et al. PTPN2 elicits cell autonomous and non-cell autonomous effects on antitumor immunity in triple-negative breast cancer. Sci. Adv. 2022;8(8):eabk3338. doi: 10.1126/sciadv.abk3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiede F., et al. PTPN2 phosphatase deletion in T cells promotes anti-tumour immunity and CAR T-cell efficacy in solid tumours. EMBO J. 2020;39(2):e103637. doi: 10.15252/embj.2019103637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alsaab H.O., et al. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front. Pharmacol. 2017;8:561. doi: 10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veenstra C., et al. The effects of PTPN2 loss on cell signalling and clinical outcome in relation to breast cancer subtype. J. Cancer Res. Clin. Oncol. 2019;145(7):1845–1856. doi: 10.1007/s00432-019-02918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klempner S.J., et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncol. 2020;25(1):e147–e159. doi: 10.1634/theoncologist.2019-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jardim D.L., et al. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39(2):154–173. doi: 10.1016/j.ccell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodman A.M., et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Therapeut. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilar E., Gruber S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010;7(3):153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.