Abstract
Circulating cell-free tumor DNA (ctDNA) can serve as a real-time biomarker of tumor burden and provide unique insights into the evolving molecular landscape of cancers under the selective pressure of immunotherapy. Tracking the landscape of genomic alterations detected in ctDNA may reveal the clonal architecture of the metastatic cascade and thus improve our understanding of the molecular wiring of therapeutic responses. While liquid biopsies may provide a rapid and accurate evaluation of tumor burden dynamics during immunotherapy, the complexity of antitumor immune responses is not fully captured through single-feature ctDNA analyses. This underscores a need for integrative studies modeling the tumor and the immune compartment to understand the kinetics of tumor clearance in association with the quality of antitumor immune responses. Clinical applications of ctDNA testing in patients treated with immune checkpoint inhibitors have shown both predictive and prognostic value through the detection of genomic biomarkers, such as tumor mutational burden and microsatellite instability, as well as allowing for real-time monitoring of circulating tumor burden and the assessment of early on-therapy responses. These efforts highlight the emerging role of liquid biopsies in selecting patients for cancer immunotherapy, monitoring therapeutic efficacy, determining the optimal duration of treatment and ultimately guiding treatment selection and sequencing. The clinical translation of liquid biopsies is propelled by the increasing number of ctDNA-directed interventional clinical trials in the immuno-oncology space, signifying a critical step towards implementation of liquid biopsies in precision immuno-oncology.
Keywords: Tumor Biomarkers, Immunotherapy, Genetic Markers
Video Abstract.
Disclaimer: this video summarises a scientific article published by BMJ Publishing Group Limited (BMJ). The content of this video has not been peer-reviewed and does not constitute medical advice. Any opinions expressed are solely those of the contributors. Viewers should be aware that professionals in the field may have different opinions. BMJ does not endorse any opinions expressed or recommendations discussed. Viewers should not use the content of the video as the basis for any medical treatment. BMJ disclaims all liability and responsibility arising from any reliance placed on the content.
Introduction
Immune checkpoint inhibitors (ICIs) have revolutionized the treatment landscape for patients with advanced solid tumors,1–4 exemplified in the first tissue-agnostic drug approvals for patients whose tumors harbor microsatellite instability (MSI) or high tumor mutation burden (TMB).2 5–11 Similar promise has now been demonstrated in the early stage disease setting, given the FDA approval of ICIs for use in the adjuvant12–14 and neoadjuvant treatment paradigm.15 16 Despite the success of ICI therapy, clinical responses are often heterogeneous and only a subset of patients will attain long-term clinical outcomes, highlighting an urgent clinical need to precisely identify those most likely to benefit through improved patient selection and stratification biomarker strategies. This is particularly important in the context of an ever-increasing number of immunotherapy monotherapy or combination treatment strategies.17 18 Equally importantly, for patients who attain sustained responses to ICIs, the optimal duration of treatment remains unclear and conventional response assessments by radiographic imaging may fail to enable clinical decision making for treatment de-escalation.19 These observations highlight the pressing need for molecularly-informed strategies to enable early and accurate response assessment to immunotherapy and to facilitate long-term monitoring.
To this end, biomarker strategies to enable the tailored administration of immunotherapy according to tumor molecular features and dynamics represent a pivotal step forward for precision immuno-oncology. Snapshot biomarkers that are currently used to guide therapy selection, such as the measurement of PD-L1 expression, MSI, and TMB, often rely on tumor tissue samples obtained through invasive biopsies, which only partially capture the constellation of host-related, tumor-related, and tumor microenvironment (TME)-related factors that collectively modulate antitumor responses.20 Moreover, the complex interactions between tumor and host immune cell populations are continuous, dynamic and evolving under the selective pressure of ICI treatment, highlighting the need for longitudinal analyses to capture the nature and timing of clinical responses.21–23 Liquid biopsies are emerging as powerful approaches to monitor tumor burden during cancer evolution as cancer cells go through therapy-induced evolutionary bottlenecks and escape immune surveillance. In this Review article, we discuss the biology, evolution and clinical relevance of circulating cell-free tumor (ctDNA) focusing on the unique implications for cancer immunotherapy.
Approaches for ctDNA detection
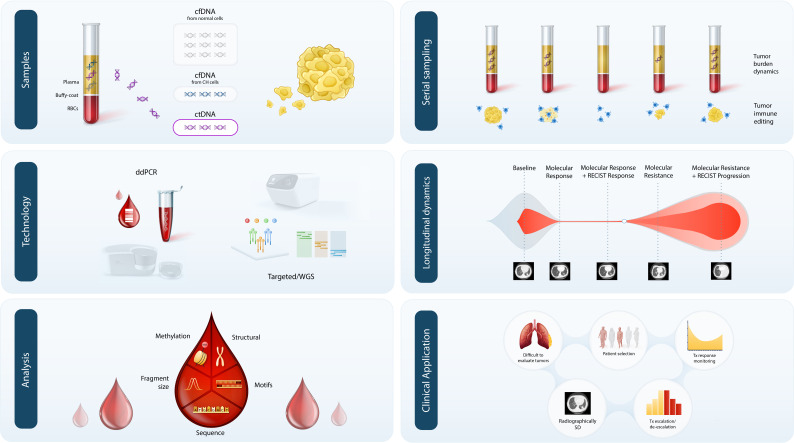
Cell-free DNA (cfDNA) is released predominantly from cells through apoptosis and necrosis24 25; a small fraction of cfDNA is tumor-derived, referred to as ctDNA, levels of which have been shown to correlate with tumor stage across a broad range of solid tumors26 (figure 1). However, significant tumor stage type-related and cancer type-related heterogeneity has been observed27 and the exact mechanisms responsible for ctDNA release have yet to be elucidated. Additionally, over the past two decades, a variety of ctDNA-based technologies have been developed for interrogating a broad range of alterations, including, sequence mutations,26 28 29 copy number changes,30 chromosomal rearrangements,31 methylation patterns,32 fragmentation lengths,33 nucleosomal occupation,34 and viral nucleic acid sequences35 36 (figure 1, reviewed in depth in the review articles by Chaudhuri and colleagues and Velculescu and colleagues, as part of this JITC liquid biopsy special review series). These approaches can be tumor informed or tumor agnostic and are being tested in a variety of different clinical diagnostic applications, including, early detection of cancer, high-risk patient identification after definitive treatment, genotype-based patient selection for targeted therapies, therapeutic response monitoring, and non-invasive tumor profiling at disease progression. In considering the clinical implementation of liquid biopsies, it is important to select the liquid biopsy approach based on its diagnostic application and intended use, to ensure that biologically relevant alteration types are assessed, and that the technical performance of the approach is sufficient in order to minimize the potential for technical limitations, such as assay sensitivity or biological limitations related to alterations associated with clonal hematopoiesis (CH).27 37
Figure 1.
Approaches for the detection and analysis of ctDNA in the context of immuno-oncology. cfDNA isolated from plasma contains a complex mixture of fragments of varying cellular origin, most of which are derived from normal or hematopoietic cells undergoing apoptosis. A proportion of cfDNA can also originate from clonally expanded hematopoietic cells and contains somatic mutations associated with clonal haematopoiesis (CH). In patients with cancer, a variable proportion of cfDNA fragments are tumor-derived, known as ctDNA, which can be detected through tumor-agnostic or tumor-informed approaches, using optimized PCR-based methods, such as droplet digital PCR, or next-generation sequencing, including targeted and whole genome sequencing. ctDNA can be analyzed using a variety of approaches to determine the landscape of sequence alterations, structural changes and methylation patterns, alongside fragment size profiles, end-motifs and other cfDNA fragment physical properties. In the context of immunotherapy, serial sampling and longitudinal ctDNA analyses can pinpoint the evolving tumor burden. As a real-time biomarker of tumor burden, longitudinal ctDNA assessments can also provide insights into emergence of potential therapeutic resistance mechanisms, which can be detected prior to radiographic responses derived from conventional imaging. Integration of ctDNA analyses into clinical use for immuno-oncology may provide added utility for the study of difficult to evaluate tumor-types (eg, mesothelioma), patient selection for immunotherapy and subsequent response monitoring, particularly in radiographically stable disease. Accurate response monitoring can inform decisions on the optimal duration of immunotherapy and appropriate strategies for treatment escalation/de-escalation based on ctDNA molecular responses. cfDNA, cell-free DNA; ctDNA, cell-free tumor DNA; ddPCR, droplet digital PCR, WGS; whole genome sequencing.
Tracking the coevolution of tumor and immune cells through non-invasive approaches
Liquid biopsies uniquely allow for evaluation of emerging genomic alterations and tumor evolution during therapy; nevertheless, coevolution of tumor and immune cells under the selective pressure of immunotherapy is a complex and dynamic process20 38 that is hard to capture by liquid biopsies alone. Analyses of early-stage NSCLC tumors have shown that longitudinal ctDNA assessments can provide a comprehensive insight into the evolving clonal architecture of tumors following surgery and adjuvant therapy.39 Subclonal tumor dynamics can be captured through ctDNA analyses, using bespoke approaches to longitudinally track patient-specific tumor-derived alterations, and used to derive a phylogenetic characterization of post-operative residual disease39 and recurrent tumors.40 41 These studies highlight the role of ctDNA analyses to assess the landscape of intratumoral heterogeneity and delineate molecular mechanisms of tumor resistance to systemic therapies that can be extended to cancer immunotherapy. Future studies to investigate the utility of ctDNA to differentiate between the selection and expansion of pre-existing tumor subclones, or the emergence of new subclonal populations driven by the de novo acquisition of genetic alterations following ICI will be important to improve our understanding of the mutational processes that govern shifts in the clonal stoichiometry of solid tumors during adaptive selection and immunoediting. As the loss of genomic material through chromosomal instability may affect antitumor immune responses,22 42 it will be important to consider both sequence and copy number alterations when evaluating the evolutionary trajectory of tumors through ctDNA analyses.
Evolutionary studies leveraging ctDNA analyses may also provide unique insights into the neoantigen landscape and the dynamic acquisition or loss of neoantigen-encoding mutations during tumor clonal reshaping in the context of ICI. Changes in tumor neoantigen profiles during ICI have been linked to host immune cell repertoire reshaping and clonotypic expansion of neoantigen-specific T cells.22 43 However, tracking of mutation-associated neoantigens through ctDNA has not been studied widely to date,44 and comprehensive assessments of tumor-derived and ctDNA-derived neoantigen profiles are required in large prospectively sampled patient cohorts to delineate the role of ctDNA in tracking neoantigen evolution during ICI treatment.
Longitudinal ctDNA assessments can be combined with dynamic on-treatment assessment of the T cell receptor (TCR) repertoire in peripheral blood to understand the coevolution of the tumor and immune compartments during ICI. Pretreatment TCR repertoire diversity,45 the early turnover of peripheral T cells and overall T cell repertoire reshaping, attributed to clonotypic modulation during ICI, have been linked to antitumor immune responses.46–48 Peripheral blood immune profiling in patients with advanced melanoma has shown that the extent of reinvigoration of circulating exhausted CD8 T cells can provide a dynamic predictor of ICI response.49 50 Accordingly, the persistence of exhausted TCR clonotypes in peripheral blood has been shown to correlate with poor responses to immunotherapy.50 The proportion of shared TCR clonotypes in peripheral and tumor-infiltrating T cells (TIL) and dynamic on-treatment clonotypic expansions of peripheral T cell populations have all been shown to correlate with both pathological response and clinical outcomes to immunotherapy and will be important factors to analyze more closely in relation to evolving ctDNA dynamics.23 48 51 While capturing tumor and immune repertoire dynamics during immunotherapy provides critical insights in the biology of tumor immunoediting, ctDNA-focused liquid biopsies have been shown to more consistently capture clinical outcomes with cancer immunotherapy and as such are closer to clinical implementation.
ctDNA-informed patient selection for cancer immunotherapy: blood TMB
TMB has been linked with therapeutic responses to ICI52–56; as a high number of somatic mutations would encode for neoepitopes presented on the surface of tumor cells, triggering an antitumor immune response.43 However, measurement of tumor tissue-derived TMB (tTMB) can be limited by high clonal complexity, which may not be sufficiently captured through a single biopsy, and can be unreliable for tumor samples with low tumor purity.21 In contrast, the ctDNA pool is representative of mutations originating from distinct primary and metastatic tumor sites, as well as tumor subclones, and may offer greater accuracy for TMB determination compared with single tissue biopsies.39 41 Efforts to derive estimates of TMB using blood-based analyses—referred to as blood TMB (bTMB)—have overall shown concordance between bTMB and tTMB57–60 and bTMB has been proposed as an emerging biomarker of response to ICI.61 62
The predictive value of bTMB was initially explored in a retrospective manner across several ICI clinical trials (POPLAR,60 OAK,60 MYSTIC63; table 1). The POPLAR and OAK studies in patients with metastatic NSCLC that received atezolizumab compared with docetaxel showed an improvement in both progression-free survival (PFS; POPLAR: 4.2 months vs 2.9 months) and overall survival (OS; POPLAR: 13 months vs 7.4 months; OAK: 13.5 months vs 6.8 months) for patients with a high bTMB (defined by the presence of >16 SNVs across 394 genes interrogated by NGS).60 Similarly, in the MYSTIC study, bTMB>20 mutations/Mb predicted clinical benefit from durvalumab and tremelimumab versus chemotherapy in patients with advanced NSCLC.63
Table 1.
Summary of studies evaluating blood TMB as a predictive biomarker for response to immune checkpoint blockade
| Study | Analysis | Approach | NGS assay | Panel size | Cohort size | Tumor type | Disease stage | Trial ID | Treatment | TMB cut-off |
| Khagi et al, 2017145 | Retrospective | Targeted NGS | Guardant 360 (73 genes) | 200 Kb | n=69 | Pan-cancer | Not reported | N/A | Anti-PD1/PDL1/CTLA4 | VUS, >3 alterations |
| Gandara et al, 201860 | Retrospective | Targeted NGS | Custom NGS assay (bait set version T7, Integrated DNA Technology, >300 genes) | 1.1 Mb | n=259 | NSCLC | Advanced/metastatic | POPLAR (NCT01903993); OAK (NCT02008227) | Atezolizumab vs docetaxel | 16 Mutations/Mb |
| Wang et al, 2019146 | Retrospective | Targeted NGS | Custom assay (NCC-GP150, 150 genes) | Not reported | n=48 cohort 1; n=50 cohort 2 | NSCLC | Advanced/metastatic | N/A | Anti-PD-1/PD-L1 | 6 Mutations/Mb |
| Si et al, 202163 | Retrospective | Targeted NGS | Guardant OMNI (500 genes) | 2 Mb | n=1001 | NSCLC | Metastatic | MYSTIC (NCT02453282) | Durvalumab and tremelimumab vs chemotherapy | 20 Mutations/Mb |
| de Castro Jr et al, 202265 | Prospective | Targeted NGS | Guardant OMNI (500 genes) | 2 Mb | n=512 | NSCLC | Metastatic | NEPTUNE (NCT02542293) | Durvalumab and tremelimumab versus chemotherapy | 20 Mutations/Mb |
| Kim et al, 202266 | Prospective | Targeted NGS | Foundation Medicine (>300 genes) | 1.1 Mb | n=152 | NSCLC | Locally advanced/metastatic | B-F1RST (NCT02848651) | Atezolizumab | 16 Mutations/Mb |
| Peters et al, 202268 | Prospective | Targeted NGS | Foundation Medicine (>300 genes) | 1.1 Mb | n=472 | NSCLC | Advanced/metastatic | BFAST (NCT03178552) | Atezolizumab versus chemotherapy | 16 Mutations/Mb |
| He et al, 202269; Schenker et al, 202270 | Prospective | Targeted NGS | Foundation Medicine (>300 genes) | 1.1 Mb | n=212 | Pan-cancer | Advanced/metastatic | CheckMate 848 (NCT03668119) | Nivolumab+ipilimumab vs nivolumab monotherapy | 10 Mutations/Mb |
BFAST, Blood First Assay Screening Trial; bTMB, blood tumor mutation burden; N/A, not available; NGS, next-generation sequencing; NSCLC, Non-small cell lung cancer; VUS, variants of unknown significance.
Despite the promising findings in retrospective studies, prospective analyses using predefined bTMB thresholds, have not confirmed the predictive role of bTMB for ICI therapeutic response (table 1). The phase 3 NEPTUNE trial of durvalumab and tremelimumab did not meet its primary endpoint of OS in patients with metastatic NSCLC and a bTMB >20 Mutations/Mb.64 65 In the phase 2, B-F1RST trial of first-line atezolizumab monotherapy in patients with locally advanced or metastatic NSCLC, a high bTMB of >16 mutations/Mb was associated with an increased overall response rate (ORR) with further incremental dose-dependent increases in ORR observed with greater bTMB thresholds.66 However, these findings did not translate into a significant difference in PFS between high and low bTMB subgroups.66 Similarly, the phase 3 randomized Blood First Assay Screening Trial, that prospectively assessed bTMB as a predictive biomarker for ICI response, revealed no significant differences in investigator assessed PFS in patients with NSCLC and bTMB>16 mutations/Mb receiving first-line atezolizumab.67 68 In line with results from the B-F1RST trial, exploratory analyses revealed an increase in the predictive power of bTMB with the selection of higher thresholds,68 supporting the evolving role of bTMB in a dose-dependent manner. These findings were in-line with the CheckMate 848 trial that showed an ORR with nivolumab and ipilimumab in patients with tTMB-high tumors of 35.3% compared with 22.5% for patients with a bTMB-H result using a cut-off of 10 mutations/Mb for both assays.69 70 Notably, for patients with tTMB-H tumors with bTMB<10 mutations/Mb, ORR was maintained at 35.0%, however, for patients with bTMB-H and tTMB <10 mutations/Mb, ORR decreased to 9.7%.69 70 Further exploratory analyses in the CheckMate 848 study demonstrated an improvement in the correlation between tTMB and bTMB when the plasma variants considered had a maximum somatic allele frequency ≥1%.69 70 Collectively, these findings support the notion that modeling continuous and even non-linear predictive effects across bTMB ranges may improve the predictive value of bTMB for ICI.
Overall, in light of the recent results of the B-FIRST and B-FAST studies that did not meet their primary endpoints, we believe that bTMB in its current state requires additional standardization and harmonization prior to its clinical use as a predictor of response to immunotherapy. To this end, evidence across multiple studies demonstrates the need for further analyses to understand the biological and technical confounders in bTMB analyses. These may include adjusting for the depth of sequencing and region of interest of the panel used, improving the assay sensitivity of detection of low abundance ctDNA, accurate exclusion of alterations that may be associated with CH and importantly harmonizing threshold selection. Additionally, while insertions and deletions (indels) resulting from frameshift alterations have been shown to be more immunogenic than single nucleotide substitutions, these can be difficult to detect at low allelic fractions in ctDNA, presenting a key technical limitation that should be addressed in future studies.71 Refinement of bTMB to account for confounding biological factors, such as loss-of-heterozygosity of the human leukocyte antigen class I (HLA-I) locus, will also be critical as impaired neoantigen presentation because of somatic HLA-I LOH can attenuate the value of bTMB assessment.72 To this end, recent findings suggest that the utility of bTMB may be improved when combined with other relevant biomarkers (such as PD-L1 status) as part of multifeature models, which warrant further analysis.73
ctDNA-informed patient selection for cancer immunotherapy: blood MSI
Similar to TMB, the assessment of MSI resulting from underlying deficiencies in DNA mismatch repair through ctDNA analyses can also be used to predict ICI response.74–76 While the current gold standard for determining tumor MSI status is through PCR-based detection of targeted microsatellites,77 MSI detection can be achieved in ctDNA using droplet digital PCR approaches or panel NGS.74–76 78 Deep next-generation sequencing combined with molecular barcoding has been used in combination with novel bioinformatics algorithms to enable error correction of NGS artifacts by several orders of magnitude, to allow for direct detection of subtle changes in microsatellite lengths (≥0.10% sensitivity) in a high background of cfDNA.79 For a subset of metastatic cancer patients included in the KEYNOTE-016 study, MSI status determined by ctDNA analysis was associated with improved PFS.76 Similarly, blood-based assessment of MSI using NGS approaches was found to be highly specific and sensitive when compared with tissue-based evaluations and correlated with PFS in patients treated with PD-L1 blockade.74 75 In a study evaluating real world outcomes with ICI, patients with advanced GI cancer with ctDNA derived MSI-high status were observed to have response rates to immunotherapy similar to published data based on tissue MSI assessments.80 While the predictive value of cfDNA-based assessments of MSI has not been widely and prospectively explored in patients receiving ICI; these findings support the use of ctDNA-based assessment of MSI status, allowing for more rapid identification of patients who can benefit from immunotherapy and expanding its use in patients in whom tissue biopsies cannot be obtained.
In addition to blood-derived TMB and MSI assessment, overall pre-treatment levels of ctDNA, measured through the number of mutant copies per milliliter of plasma81 82 or the mutant allele fraction of tumor-specific alterations,83 84 may also be a useful surrogate for tumor burden. The presence or absence of ctDNA itself may also hold predictive value in determining ICI responses, as significant improvements in response rates to atezolizumab have been shown in NSCLC patients with undetectable ctDNA.85
MRD detection and ctDNA-driven stratification for adjuvant/neoadjuvant immunotherapy
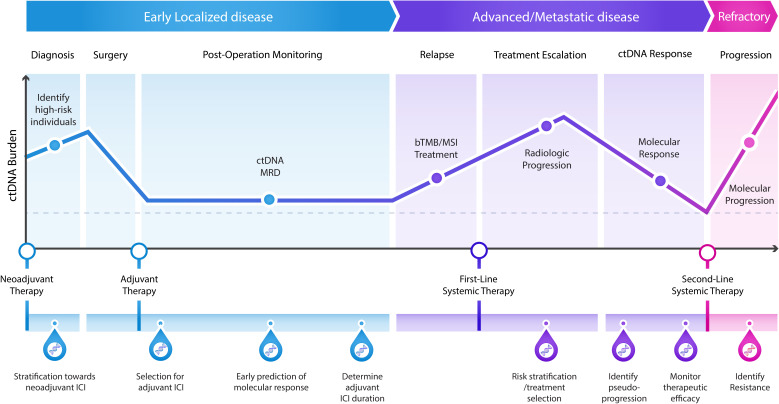
In addition to assessment of bTMB and MSI prior to ICI, longitudinal ctDNA tracking can provide added clinical value for determining tumor burden dynamics and improve the accuracy of minimal residual disease (MRD) (early-stage paradigm) and response monitoring (metastatic paradigm) (figure 2). One of the first applications of liquid biopsies in the MRD setting was that of tracking the BCR-ABL fusion transcript level in patients with chronic myeloid leukemia, which laid the path forward for the use of molecular response as a surrogate endpoint in registration studies.86 87 In solid malignancies, ctDNA analyses in the neoadjuvant setting are now being evaluated in assessing early on-therapy efficacy in patients with localized disease. The CheckMate-816 trial in patients with NSCLC who received neoadjuvant ICI highlighted the association of post-ICI treatment ctDNA elimination, as determined using bespoke tumor-informed ctDNA assays, with pathological complete response, presenting a new avenue for evaluating tumor regression in early-stage disease.15 Further prospective studies are required in the neoadjuvant setting to further explore the association of ctDNA-based approaches with pathological complete response and event-free or OS.
Figure 2.
Potential applications of ctDNA to guide clinical decision-making in patients receiving immunotherapy. In patients with early localized disease, ctDNA analyses at the time of clinical diagnosis can enable the stratification of high-risk individuals towards neoadjuvant treatment with ICI. Furthermore, the detection of ctDNA after treatment with curative intent, which is indicative of MRD, could aid in the identification of high-risk patients that could benefit from adjuvant immunotherapy. Longitudinal ctDNA assessment can also inform optimization of therapy duration and guide maintenance therapy. In patients with metastatic disease, blood-based assessments of predictive genomic biomarkers of ICI response, including bTMB and MSI, can improve the selection of patients for immunotherapy regimens. Longitudinal monitoring of ctDNA levels during immunotherapy can expedite clinical response assessments and allow for pseudoprogression to be accurately identified and distinguished from true disease progression. Finally, in refractory disease, ctDNA tracking can be used to detect the emergence of resistance mutations leading to molecular progression prior to radiologic progression. bTMB, blood TMB; ctDNA, cell-free tumor DNA; ICI, immune checkpoint inhibitors; MRD, minimal residual disease; MSI, microsatellite instability.
ICIs are incorporated into adjuvant therapeutic strategies,11 88 89 where the risk of serious immune-related adverse effects and financial toxicity must be balanced with potential therapeutic benefit, highlighting a need for improved high-risk patient assessment strategies. To this end, leveraging ctDNA to detect MRD associated with micrometastatic disease prior to development of overt metastases may provide a way forward to maximize the clinical efficacy of immunotherapy for patients with early-stage disease (figure 2). Postoperative ctDNA detection can be used to identify patients with MRD and an increased risk of disease relapse, and to enrich for the subset of patients most likely to benefit from adjuvant immunotherapy. ctDNA studies in patients with colorectal and urothelial carcinomas have shown that MRD can be detected as early as 3 days postoperatively using tumor-agnostic ctDNA profiling.90–92 This outlines an important clinically actionable window of opportunity to select patients for adjuvant immunotherapy to prevent disease relapse in selected patients, while spare treatment-related toxicity in others.90 For patients with operable urothelial carcinoma in the IMvigor010 trial (NCT02450331), the presence of ctDNA post-operatively was associated with improved disease-free and OS in the atezolizumab arm versus the observation arm, demonstrating the utility of ctDNA in identifying high-risk patients and informing therapeutic interventions that improved outcomes.92 Similarly, for patients with localized stage I-III lung cancer treated with curative intent, ctDNA-confirmed MRD identified 100% of cases who ultimately relapsed and preceded radiographic progression in 72% of patients, with a median lead time of 5.2 months, opening a window of therapeutic opportunity for ICI approaches.58 Nevertheless, the verdict as to whether detectable ctDNA at the MRD timepoint is predictive or prognostic is still out. In exploratory analyses of the IMpower010 study, adjuvant atezolizumab improved disease-free survival independent of ctDNA clearance for patients with NSCLC,93 highlighting the need for further exploration of the predictive nature of ctDNA MRD. ctDNA-based MRD detection has also demonstrated proof of concept for the selection of patients for consolidation ICI following chemoradiation therapy (CRT) in the locally advanced setting.94 In 65 patients with stage IIB-IIIB NSCLC, of whom 28 received consolidation ICI following CRT, the kinetics of ctDNA after CRT and during early ICI treatment were associated with clinical outcomes as assessed for both PFS and OS.94
Recently, the DYNAMIC clinical trial investigated whether a ctDNA-guided approach could reduce the use of adjuvant chemotherapy without compromising the risk of recurrence for patients with stage II colon cancer. A lower percentage of patients in the ctDNA-guided arm received adjuvant therapy (15% vs 28% in the control arm), while at the same ctDNA-guided management was noninferior to standard management with respect to recurrence-free survival (93.5% vs 92.4%). These findings indicate that ctDNA may be used to guided therapeutic decision and tailor adjuvant chemotherapy to patients at highest risk for recurrence without compromising recurrence-free survival.95 It is important to acknowledge that in the ctDNA-guided group of the DYNAMIC trial, recurrence or death occurred in 6% of ctDNA-negative patients; therefore, a fraction of MRD ctDNA negative patients still experience disease recurrence despite the use of a tumor-informed sensitive ctDNA detection approach. Importantly, the clinical sensitivity of MRD is higher when longitudinal MRD assessment is implemented compared with single MRD analysis, therefore, longitudinal monitoring may increase the probability of ctDNA detection and distinguish true MRD ctDNA positive cases.96 Overall, as ctDNA technologies continue to evolve we need to broaden our understanding of ctDNA negative MRD in the context of a given ctDNA assay, cancer lineage and therapeutic setting and design ctDNA interventional clinical trials based on landmark or longitudinal ctDNA MRD detection.97
ctDNA-driven early on-treatment ICI response evaluation in the metastatic setting
A key application of ctDNA-based liquid biopsies in tailoring immunotherapy is to enhance the interpretation of patterns of tumor response and progression during treatment in the metastatic setting (figure 2). Accurately assessing the efficacy of systemic immunotherapy has proven a significant challenge due to limitations with radiographic response assessments.98 Monitoring of early on-therapy ctDNA kinetics can enable real-time circulating tumor burden assessment and track tumor responsiveness to ICIs.23 99 100 There is an ever-increasing number of studies supporting the role of ctDNA molecular responses as an early endpoint for ICI response.83 100–105 Different measures have been used to monitor circulating tumor burden during ICI including the mean106 107 or maximum28 106 108 mutant allelic fraction (MAF) of tumor-derived alterations as well as MAFs of driver mutations.23 Tumor-agnostic, white blood cell (WBC) DNA-informed approaches, can improve the specificity for tracking circulating tumor load, compared with plasma-only approaches.109–111 Matched next-generation sequencing of WBC DNA enables the identification and effective removal of germline alterations and importantly CH variants detected in plasma. Such approaches are particularly suitable for cell-free tumor burden tracking in the metastatic setting, where tumor shedding and quantities are sufficient and can be captured by first-generation hybrid capture next generation sequencing assays without prior tumor profiling.
Persistent or increasing ctDNA levels following immunotherapy treatment have been widely associated with progressive disease.23 100–105 In patients with advanced solid tumors receiving pembrolizumab, rising ctDNA levels from baseline to 6–7 weeks on-therapy were shown to be predictive of therapeutic resistance, with a positive predictive value of 97.5%.83 Similar findings have been reported in patients in metastatic melanoma and gastric adenocarcinoma where rising ctDNA levels were shown to correlate with progressive disease assessed by radiographic imaging112 and even precede radiologic progression with >3 months lead time.100 104
While ctDNA persistence or rise in ctDNA levels, uniformly signifies molecular progression, the definition of ctDNA molecular response is less clear and likely dependent on the NGS assay employed. Any ctDNA reduction,83 >50% reduction in ctDNA levels102 106 or complete elimination,15 23 have been proposed to determine ctDNA molecular response and have been variably associated with ICI therapeutic response. Importantly, ctDNA complete elimination has also been associated with favorable long-term clinical outcomes in patients, suggesting that this metric may be the most accurate longitudinal predictor of durable clinical responses.23 92 94 101 113
Employing ctDNA dynamics as an early endpoint of ICI response can be particularly informative in situations where radiographic imaging fails to capture tumor regression, exemplified in pseudoprogression as well as in characterizing the heterogenous nature of radiographically stable disease. ctDNA molecular response has been shown to distinguish patients with radiographically stable disease that attain long PFS, such that patients with stable disease that cleared ctDNA had significantly longer PFS compared with those that did not clear.23 These findings suggest that radiographic imaging failed to detect the magnitude of therapeutic response for some patients with stable disease as determined by imaging and that ctDNA response may be of particular value in assessing therapeutic response in this setting. Monitoring such changes in ctDNA kinetics may also help to differentiate pseudoprogression from true disease progression.114 115 In patients with metastatic melanoma and radiographically progressive disease at first restaging on ICI, a greater than tenfold reduction in ctDNA levels within 12 weeks of treatment initiation was strongly predictive of pseudoprogression, with 90% sensitivity and 100% specificity.114
ctDNA dynamics have also been shown to predict the therapeutic efficacy of other immunotherapies, including CD19-targeted chimeric antigen receptor (CAR)-T cell therapy in relapsed or refractory diffuse large B-cell lymphoma.116–119 Approaches using shallow whole genome sequencing for the analysis of genome-wide tumor-derived structural alterations have shown reductions in ctDNA levels in patients responding to CAR-T cell therapy and increasing ctDNA levels in patients who did not attain responses.117 ctDNA dynamics may correlate and could be combined with the abundance of CAR-T cell construct-derived DNA in peripheral blood.117 Higher levels of CAR-T-derived DNA were observed in patients responding to axi-cel treatment, indicating a relative expansion of the CAR-T cell population.120 Similarly, tracking of lymphoma-specific variable, diversity and joining gene segments has shown utility for predicting patient outcomes following CAR T-cell therapy.119 In this setting, higher pretreatment ctDNA concentrations and the persistence of detectable ctDNA following treatment have been associated with progression after axi-cel infusion.119 Collectively, these findings demonstrate the expanding applications of liquid biopsies for therapeutic response across the spectrum of cancer immunotherapy.
In addition to the clinical value of longitudinal ctDNA assessment as a potential endpoint of ICI response, ctDNA molecular response can be used in early drug development to evaluate the therapeutic effect of novel treatments faster and more accurately compared with radiographic imaging, ultimately expediting drug development. As a field, we have come to agree that ctDNA persistence or rise in ctDNA levels signifies molecular and clinical progression; therefore, ctDNA molecular progression can be used as a ‘quick fail’ approach to triage drugs in development unlikely to result in circulating tumor load clearance, which in turn would indicate unfavorable clinical outcomes. Evidentiary steps needed to explore the value of ctDNA in drug development and regulatory decision-making include further prospective analyses to evaluate the concordance between ctDNA molecular response and radiographic responses and the association of ctDNA response with long-term clinical outcomes. Harmonized definitions of ctDNA molecular response as well as pinpointing the optimal timing for assessment of ctDNA responses, are required to enable integration of liquid biopsies in precision immuno-oncology.
ctDNA-driven interventional clinical trials in immuno-oncology
Given the evidence supporting ctDNA dynamics as an early endpoint for therapeutic response, an increasing number of clinical trials use liquid biopsies to guide adaptive immunotherapy-based interventions in solid cancers. These trials include ctDNA-guided intervention with ICIs in various clinical stages including early or locally advanced, resected or residual, and advanced or metastatic solid cancers (table 2).
Table 2.
ctDNA-guided interventional ICI clinical trials
| Trial (NCT ID) | Target enrollment | Indication | Treatment | ctDNA platform | Primary endpoint | Setting and references |
| NCT04966663 | 66 | Resected localized NSCLC | Adjuvant chemotherapy+nivolumab | Inivata RaDaR | DFS | T1-2N0M0 or T3/T4 multifocal tumors that are MRD+at 3–6 weeks post complete surgical resection are eligible |
| NCT04385368 (MERMAID-1) | 332 | Resected stage II-III NSCLC | Adjuvant chemotherapy+durvalumab vs SOC chemotherapy | ArcherDX | DFS in MRD+pts | Stratified by MRD status at 3–4 weeks postsurgery(147) |
| NCT04642469 (MERMAID-2) | 284 | Resected stage II-III NSCLC | Adjuvant chemotherapy as per SOC then durvalumab vs placebo | ArcherDX | DFS in PD-L1+pts | Pts who are MRD+within 96 weeks surveillance period after curative-intent therapy are randomized(148) |
| NCT04849364 (PERSEVERE) | 197 | Residual TNBC after pre-operative therapy | Atezolizumab+chemotherapy/PARP/PI3K combinations | Not reported | 2 year DFS | ctDNA-directed umbrella study for pts with detectable ctDNA postoperative |
|
NCT04138628 (TOMBOLA) |
282 | Resected urothelial carcinoma (radical cystectomy) | Atezolizumab | Not reported | % of patients with molecular and radiographic CR | Pts with detectable ctDNA postoperative are eligible |
| NCT04093167 | 50 | Treatment-naïve, advanced PD-L1+NSCLC | Pembrolizumab | PGDx | Concordance between molecular response and radiologic response | Molecular response after 1–3 cycles determines whether chemotherapy is added. |
| NCT04166487 | 40 | Treatment-naïve, advanced NSCLC | Pembrolizumab | Inivata InVision | 6 month PFS | Chemotherapy added based on ctDNA response after two cycles. |
| NCT03178552 (BFAST) | 440 | Treatment-naïve, advanced NSCLC | Atezolizumab versus SOC chemotherapy | Foundation ACT | PFS | ctDNA-directed umbrella study with arm for blood TMB+pts68 149 |
| NCT04585490 | 48 | Unresectable stage III NSCLC | Consolidative durvalumab+chemotherapy | AVENIO | Change in ctDNA level following consolidative chemotherapy | Pts with detectable ctDNA after chemoRT have chemotherapy added to SOC durvalumab |
| NCT03808441 (CAcTUS) | 40 | Advanced BRAF V600E/K/R melanoma | Nivolumab+ipilimumab | Not reported | % of patients meeting 80% ctDNA threshold | Pts are switched from BRAF targeted therapy to immunotherapy when BRAF ctDNA level is ≤80% from baseline(150) |
|
NCT04966676 (ATLAS) |
50 | Metastatic NSCLC | Nivolumab+ipilimumab | Guardant | 6 month PFS | Patients with increasing or stable ctDNA given platinum-doublet chemotherapy |
BFAST, Blood First Assay Screening Trial; chemoRT, chemoradiation; CR, complete response; ctDNA, cell-free tumor DNA; DFS, disease-free survival; ICI, immune checkpoint inhibitor; MRD, minimal residual disease; PFS, progression-free survival; SOC, standard of care; TMB, tumor mutational burden.
In the early stage setting, several randomized clinical trials are using ctDNA to select patients for adjuvant therapy or additional consolidative therapies for both resected and advanced/metastatic NSCLC. The Phase III MERMAID-1 clinical trial is using postoperative ctDNA-based MRD to identify MRD-positive (MRD+) patients for randomization to durvalumab or placebo with chemotherapy (NCT04385368). The MERMAID-2 trial randomizes patients who become MRD+to durvalumab versus placebo during longitudinal surveillance after surgical resection (NCT04642469). For patients who are MRD+after consolidation chemotherapy and immunotherapy for unresectable stage III NSCLC, the non-randomized NCT04585490 clinical trial is assessing ctDNA-driven treatment escalation. Similar MRD-based studies are conducted for patients with residual triple-negative breast cancer after neoadjuvant therapy (PERSEVERE; NCT04849364) resected urothelial carcinoma after radical cystectomy (TOMBOLA; NCT04138628).
A burgeoning area of clinical research focuses on adaptive on-treatment clinical trials, where ctDNA molecular response is used to guide therapeutic changes for patients with advanced/metastatic cancer.121 In BRAF V600E-mutated melanoma, the CAcTUS clinical trial is assessing longitudinal ctDNA level changes to inform when to switch from BRAF-targeted dabrafenib/trametinib therapy to dual ICI with ipilimumab and nivolumab (NCT03808441). In NSCLC, ongoing clinical trials are utilizing early ctDNA dynamics on single-agent ICI to assess concordance of ctDNA molecular response with radiographic response, as well as consideration of escalation with the addition of platinum-doublet chemotherapy (NCT04093167, NCT04166487). In the phase 2 BR36 clinical trial of treatment-naïve patients with ALK and EGFR mutation negative metastatic NSCLC, serial ctDNA analysis is being used to determine the optimal timing for molecular response assessment, assess concordance with radiographic responses and evaluate the correlation between depth of molecular responses and clinical outcomes including OS and PFS.122 In stage 2 of the study, patients will be randomized to evaluate the clinical benefit of treatment intensification based on ctDNA molecular responses (NCT04093167). Similarly, a molecular response-adaptive approach is investigated after 3 cycles of pembrolizumab monotherapy in a non-randomized fashion (NCT04166487). The ATLAS trial is evaluating the utility of ctDNA clearance to guide adaptive treatment of patients with metastatic NSCLC receiving dual nivolumab and ipilimumab treatment (NCT04966676); in this trial patients with increasing or stable ctDNA levels are considered for escalation with platinum doublet chemotherapy.
Barriers to adoption of ctDNA analyses in clinical cancer care
Despite the recent US Food & Drug Administration (FDA) approvals of rapid non-invasive comprehensive genomic profiling and companion diagnostic platforms, such as Guardant360 CDx123 and FoundationOne Liquid CDx,124 the adoption of liquid biopsies has been limited by several factors ranging from limitations in assay sensitivity, access to these technologies, and financial toxicity concerns across a broader range of clinical applications. Significant variability within and across tumor types can be observed with regards to the amount of ctDNA that is shed, along with the burden and distribution of disease, which can also modify ctDNA detection.125 Somatic alterations derived from clonal haematopoiesis or other non-neoplastic tissue compartments, as well as germline alterations can complicate interpretation of liquid biopsy results.126
Practical operational challenges further limit the immediate adoption of liquid biopsy in the clinic. Lack of validated decentralized testing approaches and clinical care pathways that facilitate ordering and reporting of liquid biopsies represent barriers to broader use in already complex electronic health records systems.127 Even when ordering systems are integrated, the costs and insurance coverage for patients undergoing liquid biopsy is still emerging for Medicare and commercial payors, can often be unclear, and are less established than tissue-based assessments. Lastly, even when liquid biopsy test results are readily obtained, the reports can be challenging to interpret for actionability and may require additional consultation with molecular pathologists and molecular tumor boards. These expert resources are often limited in availability and primarily located within academic medical canters, super-imposing the challenge of precision oncology disparities.
While liquid biopsy-based analyses for genotype-matched therapy selection are the most well established, emerging ctDNA applications such as MRD detection in early-stage disease and molecular response monitoring in metastatic disease are less mature from a regulatory and routine cancer care perspective. The analytical and clinical validation framework for diagnostics approaches in these settings is currently being established by the US FDA128 and will require prospective, interventional clinical trials to understand the clinical utility of quantitative assessments of ctDNA from a prognostic and predictive perspective to drive broader adoption in clinical management (reviewed in depth in the review articles by Beaver and colleagues, as part of this JITC liquid biopsy special review series).
Future directions for liquid biopsies in the context of immunotherapy
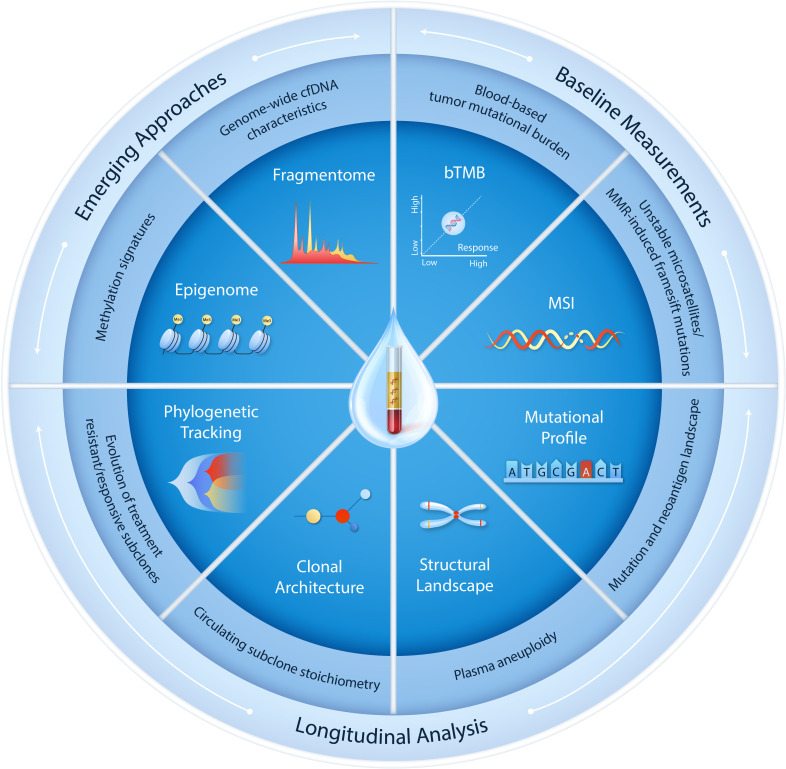
The potential of ctDNA analyses may be enhanced through the characterization of additional hallmark cfDNA features using integrative approaches. To this end, the assessment of structural and fragmentation features of cfDNA, as well as tumor-derived epigenetic signatures and nucleosomal footprints associated with the landscape of transcriptionally active genes, may provide additional features to inform the optimized use of liquid biopsies for patient selection, risk stratification, and treatment response monitoring for ICIs (figure 3). Multiparameter liquid biopsy testing has shown promising potential for cancer early detection, where the combined assessment of circulating proteins129 or clinical risk factors130 alongside mutations or fragmentation features in cfDNA has improved the sensitivity for detection of early disease. Similar approaches incorporating multiple non-invasive parameters (including baseline ctDNA measurements and on-therapy dynamic changes alongside pretherapy/on-therapy peripheral blood CD8+T cell abundance) can leverage both tumor and immune response components to derive a unified definition of molecular response to ICI therapy (online supplemental table S1).23 73 Higher peripheral neutrophil-to-lymphocyte ratios have also been shown to correlate with longitudinal ctDNA trends and have been associated with poorer clinical outcomes following immunotherapy treatment (online supplemental table S1).109 131 ctDNA features may also be combined with circulating tumor cells or circulating extracellular particles (the latter reviewed in depth in the review article by Panabieres and Pantel, as part of this JITC liquid biopsy special review series).
Figure 3.
Leveraging the hallmarks of cfDNA to expand current and future uses in immuno-oncology. Multiple hallmark features of cfDNA can be harnessed through either baseline measurements or longitudinal analyses. Blood-based tumor mutational burden and MSI, the latter detected through changes in microsatellite length or MMR-induced frameshift alterations giving rise to a MSI-high (MSI-H) tumor genotype, have shown significant promise for non-invasive assessment in cases where the availability of tissue biopsies is limited. In tandem, longitudinal ctDNA tracking using sequence alterations, structural changes and tumor-specific phylogenetic profiling can enable quantitative assessment of changes in circulating tumor load and provide a key insight into tumor immunoediting during immunotherapy. Several emerging approaches have gained traction and may enhance the future potential of ctDNA testing in immuno-oncology. In this context, analyses of cfDNA epigenetic features, genome-wide chromatin accessibility and fragmentation profiles may be implemented in integrative multi-feature ctDNA methodologies to detect and track circulating tumor burden during immunotherapy. bTMB, microsatellite instability; cfDNA, cell-free DNA; ctDNA, cell-free tumor DNA; MMR, mismatch repair; MSI, microsatellite instability.
jitc-2022-005924supp001.pdf (175.2KB, pdf)
To this end, the assessment of structural28 30 132 133 and fragmentation features of cfDNA,130 134 as well as tumor-derived epigenetic signatures32 and nucleosomal footprints associated with the landscape of transcriptionally active genes,34 135 136 may provide additional features to inform the optimized use of liquid biopsies for patient selection, risk stratification, and treatment response monitoring for ICIs. The ability to detect fragments of circulating microbiome DNA in peripheral blood has also been demonstrated in patients with melanoma, prostate and lung cancer137 and unveils new potential for liquid biopsies to characterize changes in the bacterial microbiome in the context of immunotherapy that may be reflective of clinical outcomes.
Conclusions
The value of landmark and longitudinal ctDNA profiling in immuno-oncology is supported by a growing number of studies in this rapidly evolving field. Liquid biopsies can be used to select and triage patients to therapeutic strategies based on the accurate and rapid evaluation of tumor molecular profiling as well as tumor burden dynamics in response to treatment. Liquid biopsies provide important insights in tumor clonal stoichiometry and immunoediting as cancer cells go through evolutionary bottlenecks in the context of immunotherapy. Are we ready to integrate liquid biopsies in clinical immuno-oncology? We are close, but certainly there are critical questions that have to be answered first. Next-generation assay harmonization and calibration for technical and biological noise in liquid biopsies remains an important next step. In our opinion, the most promising application of liquid biopsies is through assessment of longitudinal circulating tumor burden dynamics during therapy and we envision that several tumor-agnostic liquid biopsy assays, each one potentially coming with different thresholds and definitions of ctDNA molecular response would be used to guide immunotherapy escalation or de-escalation. Prospective testing in the clinical trial setting remains key to fully demonstrate the clinical utility of liquid biopsies for cancer immunotherapy and identify the clinical settings where ctDNA molecular responses are the most informative. To this end, we would like to highlight several national and international initiatives led by the Friends of Cancer Research,106 BloodPAC,138 the Foundation for the National Institutes of Health,139 European Society for Medical Oncology (ESMO),140 American Society of Clinical Oncology (ASCO),141 the International Association of Liquid Biopsy,142 International Association for the Study of Lung Cancer (IASLC),143 the International Quality Network for Pathology, the European Cancer Patient Coalition and the European federation of Pharmaceutical Industries and Associations144 that aim to tackle clinical, analytical, and technical challenges and through standardization and harmonization efforts and consensus guidelines enable the integration of liquid biopsies in precision immuno-oncology. We strongly believe that the future practice of precision immuno-oncology includes liquid biopsy approaches to enable making the earliest best decisions with precision for the increasing number of individuals receiving cancer immunotherapy.
Acknowledgments
This work was supported in part by the US National Institutes of Health grants CA121113 (VA), the Department of Defense Congressionally Directed Medical Research Programs grant CA190755 (VA), the Emerson Collective (VA) and the International Lung Cancer Foundation (LS).
Footnotes
Twitter: @ValsamoA
Contributors: LS: Data curation, writing—original draft, writing—review and editing. JCM: Data curation, writing—original draft. JVC: Data curation, writing—original draft. BL: Data curation. JJ: Writing—review and editing. SS: Writing—review and editing. VL: Data curation, writing—original draft. BPL: Writing—review and editing. MS: Data curation, writing—review and editing. VA: Conceptualization, data curation, writing—original draft, writing—review and editing.
Competing interests: VA receives research funding to Johns Hopkins University from Astra Zeneca, Personal Genome Diagnostics and Delfi Diagnostics and has received research funding to Johns Hopkins University from Bristol-Myers Squibb in the past 5 years. VA is an inventor on patent applications (63/276,525, 17/779,936, 16/312,152, 16/341,862, 17/047,006 and 17/598,690) submitted by Johns Hopkins University related to cancer genomic analyses, ctDNA therapeutic response monitoring and immunogenomic features of response to immunotherapy that have been licensed to one or more entities. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. JVC has served on an advisory board for Illumina. JCM has served in a consulting role to MJH Life Sciences, Johnson & Johnson, and Doximity and has received research funding to his institution from Merck via the Conquer Cancer Young Investigators Award. BPL has served in a consultant/advisory role for Janssen, Daiichi Sankyo, AstraZeneca, Eli Lilly, Genentech, Mirati, Amgen, Pfizer, BMS, Guardant 360, and Foundation Medicine. SS has served in a consultant/advisory role for Genentech and MJH Life Sciences. VL has served in a consultant/advisory role for Takeda, Seattle Genetics, Bristol-Myers Squibb, AstraZeneca and Guardant Health and has received research funding from GlaxoSmithKline, BMS, Merck and Seattle Genetics. MS and JJ are employees of Personal Genome Diagnostics (Labcorp).
Provenance and peer review: Commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Bellmunt J, de Wit R, Vaughn DJ, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med 2017;376:1015–26. 10.1056/NEJMoa1613683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced Nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 2015;373:1803–13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015;372:2521–32. 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- 5.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcus L, Lemery SJ, Keegan P, et al. Fda approval summary: pembrolizumab for the treatment of microsatellite Instability-High solid tumors. Clin Cancer Res 2019;25:3753–8. 10.1158/1078-0432.CCR-18-4070 [DOI] [PubMed] [Google Scholar]
- 7.Felip E, Altorki N, Zhou C, et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet 2021;398:1344–57. 10.1016/S0140-6736(21)02098-5 [DOI] [PubMed] [Google Scholar]
- 8.Kelly RJ, Ajani JA, Kuzdzal J, et al. Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med 2021;384:1191–203. 10.1056/NEJMoa2032125 [DOI] [PubMed] [Google Scholar]
- 9.Luke JJ, Rutkowski P, Queirolo P, et al. Pembrolizumab versus placebo as adjuvant therapy in completely resected stage IIb or IIC melanoma (KEYNOTE-716): a randomised, double-blind, phase 3 trial. The Lancet 2022;399:1718–29. 10.1016/S0140-6736(22)00562-1 [DOI] [PubMed] [Google Scholar]
- 10.Choueiri TK, Tomczak P, Park SH, et al. Adjuvant pembrolizumab after nephrectomy in renal-cell carcinoma. N Engl J Med 2021;385:683–94. 10.1056/NEJMoa2106391 [DOI] [PubMed] [Google Scholar]
- 11.Bajorin DF, Witjes JA, Gschwend JE, et al. Adjuvant nivolumab versus placebo in muscle-invasive urothelial carcinoma. N Engl J Med 2021;384:2102–14. 10.1056/NEJMoa2034442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FDA . FDA approves nivolumab for adjuvant treatment of urothelial carcinoma, 2021. Available: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-nivolumab-adjuvant-treatment-urothelial-carcinoma
- 13.FDA . FDA approves pembrolizumab for adjuvant treatment of melanoma., 2021. Available: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adjuvant-treatment-melanoma
- 14.FDA . Fda approves pembrolizumab for adjuvant treatment of renal cell carcinoma., 2021. Available: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-pembrolizumab-adjuvant-treatment-renal-cell-carcinoma
- 15.Forde PM, Spicer J, Lu S, et al. Neoadjuvant nivolumab plus chemotherapy in resectable lung cancer. N Engl J Med 2022;386:1973–85. 10.1056/NEJMoa2202170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid P, Cortes J, Dent R, et al. Event-Free survival with pembrolizumab in early triple-negative breast cancer. N Engl J Med 2022;386:556–67. 10.1056/NEJMoa2112651 [DOI] [PubMed] [Google Scholar]
- 17.Meric-Bernstam F, Larkin J, Tabernero J, et al. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet 2021;397:1010–22. 10.1016/S0140-6736(20)32598-8 [DOI] [PubMed] [Google Scholar]
- 18.Ott PA, Hodi FS, Kaufman HL, et al. Combination immunotherapy: a road map. J Immunother Cancer 2017;5:16. 10.1186/s40425-017-0218-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marron TU, Ryan AE, Reddy SM, et al. Considerations for treatment duration in responders to immune checkpoint inhibitors. J Immunother Cancer 2021;9:e001901. 10.1136/jitc-2020-001901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anagnostou V, Landon BV, Medina JE, et al. Translating the evolving molecular landscape of tumors to biomarkers of response for cancer immunotherapy. Sci Transl Med 2022;14:eabo3958. 10.1126/scitranslmed.abo3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anagnostou V, Niknafs N, Marrone K, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat Cancer 2020;1:99–111. 10.1038/s43018-019-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 2017;7:264–76. 10.1158/2159-8290.CD-16-0828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anagnostou V, Forde PM, White JR, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res 2019;79:1214–25. 10.1158/0008-5472.CAN-18-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347–76. 10.1007/s10555-016-9629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139–42. 10.1016/s0009-8981(01)00665-9 [DOI] [PubMed] [Google Scholar]
- 26.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. 10.1126/scitranslmed.aan2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phallen J, Leal A, Woodward BD, et al. Early noninvasive detection of response to targeted therapy in non-small cell lung cancer. Cancer Res 2019;79:1204–13. 10.1158/0008-5472.CAN-18-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–55. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154. 10.1126/scitranslmed.3004742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015;7:1034–47. 10.15252/emmm.201404913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563:579–83. 10.1038/s41586-018-0703-0 [DOI] [PubMed] [Google Scholar]
- 33.Cristiano S, Leal A, Phallen J, et al. Genome-Wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385–9. 10.1038/s41586-019-1272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder MW, Kircher M, Hill AJ, et al. Cell-Free DNA comprises an in vivo nucleosome footprint that informs its Tissues-Of-Origin. Cell 2016;164:57–68. 10.1016/j.cell.2015.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung TH, Yim SF, Yu MY, et al. Liquid biopsy of HPV DNA in cervical cancer. J Clin Virol 2019;114:32–6. 10.1016/j.jcv.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 36.Cocuzza CE, Martinelli M, Sina F, et al. Human papillomavirus DNA detection in plasma and cervical samples of women with a recent history of low grade or precancerous cervical dysplasia. PLoS One 2017;12:e0188592. 10.1371/journal.pone.0188592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–90. 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gremel G, Lee RJ, Girotti MR, et al. Distinct subclonal tumour responses to therapy revealed by circulating cell-free DNA. Ann Oncol 2016;27:1959–65. 10.1093/annonc/mdw278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh AR, Leshchiner I, Elagina L, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 2019;25:1415–21. 10.1038/s41591-019-0561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forde PM, Anagnostou V, Sun Z, et al. Durvalumab with platinum-pemetrexed for unresectable pleural mesothelioma: survival, genomic and immunologic analyses from the phase 2 PrE0505 trial. Nat Med 2021;27:1910–20. 10.1038/s41591-021-01541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. 10.1126/science.aaf1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia Q, Chiu L, Wu S, et al. Tracking neoantigens by personalized circulating tumor DNA sequencing during checkpoint blockade immunotherapy in non-small cell lung cancer. Adv Sci 2020;7:1903410. 10.1002/advs.201903410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogan SA, Courtier A, Cheng PF, et al. Peripheral blood TCR repertoire profiling may facilitate patient stratification for immunotherapy against melanoma. Cancer Immunol Res 2019;7:77–85. 10.1158/2326-6066.CIR-18-0136 [DOI] [PubMed] [Google Scholar]
- 46.Valpione S, Galvani E, Tweedy J, et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer 2020;1:210–21. 10.1038/s43018-019-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poran A, Scherer J, Bushway ME, et al. Combined TCR repertoire profiles and blood cell phenotypes predict melanoma patient response to personalized neoantigen therapy plus anti-PD-1. Cell Rep Med 2020;1:100141. 10.1016/j.xcrm.2020.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Muzaffar J, Kirtane K, et al. T cell repertoire in peripheral blood as a potential biomarker for predicting response to concurrent cetuximab and nivolumab in head and neck squamous cell carcinoma. J Immunother Cancer 2022;10:e004512. 10.1136/jitc-2022-004512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang AC, Postow MA, Orlowski RJ, et al. T-Cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–5. 10.1038/nature22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira G, Stromhaug K, Klaeger S, et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 2021;596:119–25. 10.1038/s41586-021-03704-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274–8. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- 52.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cristescu R, Aurora-Garg D, Albright A, et al. Tumor mutational burden predicts the efficacy of pembrolizumab monotherapy: a pan-tumor retrospective analysis of participants with advanced solid tumors. J Immunother Cancer 2022;10:e003091. 10.1136/jitc-2021-003091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. 10.1016/S1470-2045(20)30445-9 [DOI] [PubMed] [Google Scholar]
- 55.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koeppel F, Blanchard S, Jovelet C, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One 2017;12:e0188174. 10.1371/journal.pone.0188174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017;7:1394–403. 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang N, Li Y, Liu Z, et al. The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer 2018;18:319. 10.1186/s12885-018-4199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gandara DR, Paul SM, Kowanetz M, et al. Blood-Based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441–8. 10.1038/s41591-018-0134-3 [DOI] [PubMed] [Google Scholar]
- 61.Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab with or without tremelimumab vs standard chemotherapy in first-line treatment of metastatic non-small cell lung cancer: the Mystic phase 3 randomized clinical trial. JAMA Oncol 2020;6:661–74. 10.1001/jamaoncol.2020.0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Velcheti V, Kim ES, Mekhail T, et al. Prospective clinical evaluation of blood-based tumor mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC): interim B-F1RST results. Journal of Clinical Oncology 2018;36:12001. 10.1200/JCO.2018.36.15_suppl.12001 [DOI] [Google Scholar]
- 63.Si H, Kuziora M, Quinn KJ, et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: results from the Mystic study. Clin Cancer Res 2021;27:1631–40. 10.1158/1078-0432.CCR-20-3771 [DOI] [PubMed] [Google Scholar]
- 64.PLC A. Update on the phase III NEPTUNE trial of Imfinzi plus tremelimumab in stage IV non-small cell lung cancer, 2019. [Google Scholar]
- 65.de Castro G, Rizvi NA, Schmid P, et al. NEPTUNE: phase 3 study of first-line Durvalumab plus tremelimumab in patients with metastatic NSCLC. J Thorac Oncol 2022. doi: 10.1016/j.jtho.2022.09.223. [Epub ahead of print: 12 Oct 2022]. [DOI] [PubMed] [Google Scholar]
- 66.Kim ES, Velcheti V, Mekhail T, et al. Blood-Based tumor mutational burden as a biomarker for atezolizumab in non-small cell lung cancer: the phase 2 B-F1RST trial. Nat Med 2022;28:939–45. 10.1038/s41591-022-01754-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dziadziuszko R, Peters S, Gadgeel SM, et al. 1281O Atezolizumab (atezo) vs platinum-based chemo in blood-based tumour mutational burden-positive (bTMB+) patients (pts) with first-line (1L) advanced/metastatic (m)NSCLC: Results of the Blood First Assay Screening Trial (BFAST) phase III cohort C. Annals of Oncology 2021;32:S950–1. 10.1016/j.annonc.2021.08.1883 [DOI] [Google Scholar]
- 68.Peters S, Dziadziuszko R, Morabito A, et al. Atezolizumab versus chemotherapy in advanced or metastatic NSCLC with high blood-based tumor mutational burden: primary analysis of BFAST cohort C randomized phase 3 trial. Nat Med 2022;28:1831–9. 10.1038/s41591-022-01933-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He J, Kalinava N, Doshi P, et al. Abstract 2139: evaluation of tissue- and plasma-derived tumor mutational burden and genomic alterations of interest from the CheckMate 848 clinical trial. Cancer Res 2022;82:2139. 10.1158/1538-7445.AM2022-2139 [DOI] [Google Scholar]
- 70.Schenker M, Burotto M, Richardet M, et al. Abstract CT022: CheckMate 848: a randomized, open-label, phase 2 study of nivolumab in combination with ipilimumab or nivolumab monotherapy in patients with advanced or metastatic solid tumors of high tumor mutational burden. Cancer Res 2022;82:CT022. 10.1158/1538-7445.AM2022-CT022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18:1009–21. 10.1016/S1470-2045(17)30516-8 [DOI] [PubMed] [Google Scholar]
- 72.Montesion M, Murugesan K, Jin DX, et al. Somatic HLA class I loss is a widespread mechanism of immune evasion which refines the use of tumor mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov 2021;11:282–92. 10.1158/2159-8290.CD-20-0672 [DOI] [PubMed] [Google Scholar]
- 73.Nabet BY, Esfahani MS, Moding EJ, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 2020;183:363–76. 10.1016/j.cell.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Georgiadis A, Durham JN, Keefer LA, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin Cancer Res 2019;25:7024–34. 10.1158/1078-0432.CCR-19-1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willis J, Lefterova MI, Artyomenko A, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res 2019;25:7035–45. 10.1158/1078-0432.CCR-19-1324 [DOI] [PubMed] [Google Scholar]
- 76.Le DT, Uram JN, Wang H, et al. Pd-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buza N, Ziai J, Hui P. Mismatch repair deficiency testing in clinical practice. Expert Rev Mol Diagn 2016;16:591–604. 10.1586/14737159.2016.1156533 [DOI] [PubMed] [Google Scholar]
- 78.Klouch KZ, Stern M-H, Trabelsi-Grati O, et al. Microsatellite instability detection in breast cancer using drop-off droplet digital PCR. Oncogene 2022;41:5289–97. 10.1038/s41388-022-02504-6 [DOI] [PubMed] [Google Scholar]
- 79.Ladas I, Yu F, Leong KW, et al. Enhanced detection of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic Acids Res 2018;46:e74. 10.1093/nar/gky251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kasi PM, Klempner SJ, Starr JS, et al. Clinical utility of microsatellite instability (MSI-H) identified on liquid biopsy in advanced gastrointestinal cancers (aGI). Journal of Clinical Oncology 2022;40:56. 10.1200/JCO.2022.40.4_suppl.056 [DOI] [Google Scholar]
- 81.Gray ES, Rizos H, Reid AL, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015;6:42008–18. 10.18632/oncotarget.5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JH, Long GV, Boyd S, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol 2017;28:1130–6. 10.1093/annonc/mdx026 [DOI] [PubMed] [Google Scholar]
- 83.Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 2020;1:873–81. 10.1038/s43018-020-0096-5 [DOI] [PubMed] [Google Scholar]
- 84.Cabel L, Riva F, Servois V, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017;28:1996–2001. 10.1093/annonc/mdx212 [DOI] [PubMed] [Google Scholar]
- 85.Socinski MA, Paul SM, Yun C, et al. Abstract CT194: exploratory subgroup analysis of atezolizumab (atezo) clinical characteristics in patients (PTS) with low circulating tumor DNA (ctDNA) in B-F1RST—a phase II trial evaluating blood-based tumor mutational burden (bTMB) in NSCLC. Cancer Res 2019;79:CT194. 10.1158/1538-7445.AM2019-CT194 [DOI] [Google Scholar]
- 86.Kantarjian H, Shah NP, Hochhaus A, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 2010;362:2260–70. 10.1056/NEJMoa1002315 [DOI] [PubMed] [Google Scholar]
- 87.Saglio G, Kim D-W, Issaragrisil S, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med 2010;362:2251–9. 10.1056/NEJMoa0912614 [DOI] [PubMed] [Google Scholar]
- 88.Eggermont AMM, Chiarion-Sileni V, Grob J-J, et al. Prolonged survival in stage III melanoma with ipilimumab adjuvant therapy. N Engl J Med 2016;375:1845–55. 10.1056/NEJMoa1611299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bellmunt J, Hussain M, Gschwend JE, et al. Adjuvant atezolizumab versus observation in muscle-invasive urothelial carcinoma (IMvigor010): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2021;22:525–37. 10.1016/S1470-2045(21)00004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (dynamic). Clin Cancer Res 2019;25:7058–67. 10.1158/1078-0432.CCR-19-1213 [DOI] [PubMed] [Google Scholar]
- 91.Zviran A, Schulman RC, Shah M, et al. Genome-Wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat Med 2020;26:1114–24. 10.1038/s41591-020-0915-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 2021;595:432–7. 10.1038/s41586-021-03642-9 [DOI] [PubMed] [Google Scholar]
- 93.Felip E. IMpower010: ctDNA status in patients (PTS) with resected NSCLC who received adjuvant chemotherapy (chemo) followed by atezolizumab (atezo) or best supportive care (BSC). ESMO Immuno-Oncology Congress 2022, Abstract 1O 2022 2022. [Google Scholar]
- 94.Moding EJ, Liu Y, Nabet BY, et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small cell lung cancer. Nat Cancer 2020;1:176–83. 10.1038/s43018-019-0011-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tie J, Cohen JD, Lahouel K, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med 2022;386:2261–72. 10.1056/NEJMoa2200075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moding EJ, Nabet BY, Alizadeh AA, et al. Detecting liquid remnants of solid tumors: circulating tumor DNA minimal residual disease. Cancer Discov 2021;11:2968–86. 10.1158/2159-8290.CD-21-0634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pellini B, Chaudhuri AA. Circulating tumor DNA minimal residual disease detection of non-small-cell lung cancer treated with curative intent. J Clin Oncol 2022;40:567–75. 10.1200/JCO.21.01929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Anagnostou V, Yarchoan M, Hansen AR, et al. Immuno-oncology trial endpoints: capturing clinically meaningful activity. Clin Cancer Res 2017;23:4959–69. 10.1158/1078-0432.CCR-16-3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Herbreteau G, Langlais A, Greillier L, et al. Circulating tumor DNA as a prognostic determinant in small cell lung cancer patients receiving Atezolizumab. J Clin Med 2020;9. doi: 10.3390/jcm9123861. [Epub ahead of print: 27 11 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Váraljai R, Wistuba-Hamprecht K, Seremet T, et al. Application of circulating cell-free tumor DNA profiles for therapeutic monitoring and outcome prediction in genetically heterogeneous metastatic melanoma. JCO Precis Oncol 2020;3:1–10. 10.1200/PO.18.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seremet T, Jansen Y, Planken S, et al. Undetectable circulating tumor DNA (ctDNA) levels correlate with favorable outcome in metastatic melanoma patients treated with anti-PD1 therapy. J Transl Med 2019;17:303. 10.1186/s12967-019-2051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goldberg SB, Narayan A, Kole AJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 2018;24:1872–80. 10.1158/1078-0432.CCR-17-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guibert N, Jones G, Beeler JF, et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer 2019;137:1–6. 10.1016/j.lungcan.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 104.Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 105.Keller L, Guibert N, Casanova A, et al. Early circulating tumour DNA variations predict tumour response in melanoma patients treated with immunotherapy. Acta Derm Venereol 2019;99:206–10. 10.2340/00015555-3080 [DOI] [PubMed] [Google Scholar]
- 106.Vega DM, Nishimura KK, Zariffa N, et al. Changes in circulating tumor DNA reflect clinical benefit across multiple studies of patients with non-small-cell lung cancer treated with immune checkpoint inhibitors. JCO Precis Oncol 2022;6:e2100372. 10.1200/PO.21.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burgener JM, Zou J, Zhao Z, et al. Tumor-Naïve multimodal profiling of circulating tumor DNA in head and neck squamous cell carcinoma. Clin Cancer Res 2021;27:4230–44. 10.1158/1078-0432.CCR-21-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jacob S, Davis AA, Gerratana L, et al. The use of serial circulating tumor DNA to detect resistance alterations in progressive metastatic breast cancer. Clin Cancer Res 2021;27:1361–70. 10.1158/1078-0432.CCR-20-1566 [DOI] [PubMed] [Google Scholar]
- 109.Hwang M, Canzoniero JV, Rosner S, et al. Peripheral blood immune cell dynamics reflect antitumor immune responses and predict clinical response to immunotherapy. J Immunother Cancer 2022;10:e004688. 10.1136/jitc-2022-004688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leal A, van Grieken NCT, Palsgrove DN, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 2020;11:525. 10.1038/s41467-020-14310-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hellmann MD, Nabet BY, Rizvi H, et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin Cancer Res 2020;26:2849–58. 10.1158/1078-0432.CCR-19-3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lipson EJ, Velculescu VE, Pritchard TS, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2014;2:42. 10.1186/s40425-014-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Q, Luo J, Wu S, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020;10:1842–53. 10.1158/2159-8290.CD-20-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee JH, Long GV, Menzies AM, et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with Anti-Programmed cell death 1 antibodies. JAMA Oncol 2018;4:717–21. 10.1001/jamaoncol.2017.5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ricciuti B, Jones G, Severgnini M, et al. Early plasma circulating tumor DNA (ctDNA) changes predict response to first-line pembrolizumab-based therapy in non-small cell lung cancer (NSCLC). J Immunother Cancer 2021;9:e001504. 10.1136/jitc-2020-001504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maluquer C, Bellosillo B, Mussetti A, et al. Liquid biopsy for disease monitoring after anti-CD19 chimeric antigen receptor T cell in diffuse large B-cell lymphoma. EJHaem 2021;2:109–11. 10.1002/jha2.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goodman AM, Holden KA, Jeong A-R, et al. Assessing CAR T-cell therapy response using genome-wide sequencing of cell-free DNA in patients with B-cell lymphomas. Transplant Cell Ther 2022;28:30.e1–30.e7. 10.1016/j.jtct.2021.10.007 [DOI] [PubMed] [Google Scholar]
- 118.Cherng H-JJ, Sun R, Sugg B, et al. Risk assessment with low-pass whole-genome sequencing of cell-free DNA before CD19 CAR T-cell therapy for large B-cell lymphoma. Blood 2022;140:504–15. 10.1182/blood.2022015601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frank MJ, Hossain NM, Bukhari A, et al. Monitoring of circulating tumor DNA improves early relapse detection after Axicabtagene Ciloleucel infusion in large B-cell lymphoma: results of a prospective multi-institutional trial. J Clin Oncol 2021;39:3034–43. 10.1200/JCO.21.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mika T, Thomson J, Nilius-Eliliwi V, et al. Quantification of cell-free DNAfor the analysis of CD19-CAR-T cells during lymphoma treatment. Mol Ther Methods Clin Dev 2021;23:539–50. 10.1016/j.omtm.2021.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Murray JC, Anagnostou V. Translating noninvasive molecular responses into clinical reality for cancer immunotherapy. Nat Rev Clin Oncol 2021;18:65–6. 10.1038/s41571-020-00450-4 [DOI] [PubMed] [Google Scholar]
- 122.Anagnostou V, Ho C, Wheatley-Price P, et al. FP05.02 a Biomarker-Directed, multi-center phase II study of molecular response adaptive Immuno-Chemotherapy in lung cancer. Journal of Thoracic Oncology 2021;16:S952. 10.1016/j.jtho.2021.08.219 [DOI] [Google Scholar]
- 123.Administration USFaD . U.S. food and drug administration: Guardant360 CDx—P200010/S002.
- 124.Administration USFaD . U.S. food and drug administration, FoundationOne® liquid Cdx, 2020. [Google Scholar]
- 125.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579–86. 10.1200/JCO.2012.45.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bauml J, Levy B. Clonal hematopoiesis: a new layer in the liquid biopsy story in lung cancer. Clin Cancer Res 2018;24:4352–4. 10.1158/1078-0432.CCR-18-0969 [DOI] [PubMed] [Google Scholar]
- 127.Warner JL, Jain SK, Levy MA. Integrating cancer genomic data into electronic health records. Genome Med 2016;8:113. 10.1186/s13073-016-0371-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.FDA . Use of Circulating Tumor DNA for Early Stage Solid Tumor Drug Development Guidance for Industry. In: Fda, 2022. https://www.fda.gov/media/158072/download [Google Scholar]
- 129.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–30. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mathios D, Johansen JS, Cristiano S, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun 2021;12:5060. 10.1038/s41467-021-24994-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jin J, Yang L, Liu D, et al. Association of the neutrophil to lymphocyte ratio and clinical outcomes in patients with lung cancer receiving immunotherapy: a meta-analysis. BMJ Open 2020;10:e035031. 10.1136/bmjopen-2019-035031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Douville C, Springer S, Kinde I, et al. Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (lines). Proc Natl Acad Sci U S A 2018;115:1871–6. 10.1073/pnas.1717846115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kirkizlar E, Zimmermann B, Constantin T, et al. Detection of clonal and Subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl Oncol 2015;8:407–16. 10.1016/j.tranon.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Medina JE, Roussos Torres ET, Leal A, et al. 1669P monitoring immune checkpoint inhibition in advanced solid tumors using genome-wide cfDNA fragmentomes. Annals of Oncology 2022;33:S1306–7. 10.1016/j.annonc.2022.07.1749 [DOI] [Google Scholar]
- 135.Ivanov M, Baranova A, Butler T, et al. Non-Random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics 2015;16 Suppl 13:S1. 10.1186/1471-2164-16-S13-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ulz P, Thallinger GG, Auer M, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet 2016;48:1273–8. 10.1038/ng.3648 [DOI] [PubMed] [Google Scholar]
- 137.Poore GD, Kopylova E, Zhu Q, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020;579:567–74. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Godsey JH, Silvestro A, Barrett JC, et al. Generic protocols for the analytical validation of next-generation sequencing-based ctDNA assays: a joint consensus recommendation of the BloodPAC's analytical variables Working group. Clin Chem 2020;66:1156–66. 10.1093/clinchem/hvaa164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Williams PM, Forbes T, P Lund S, et al. Validation of ctDNA quality control materials through a Precompetitive collaboration of the foundation for the National Institutes of health. JCO Precis Oncol 2021;5:910–20. 10.1200/PO.20.00528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pascual J, Attard G, Bidard F-C, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO precision medicine Working group. Ann Oncol 2022;33:750–68. 10.1016/j.annonc.2022.05.520 [DOI] [PubMed] [Google Scholar]
- 141.Chakravarty D, Johnson A, Sklar J, et al. Somatic genomic testing in patients with metastatic or advanced cancer: ASCO provisional clinical opinion. J Clin Oncol 2022;40:1231–58. 10.1200/JCO.21.02767 [DOI] [PubMed] [Google Scholar]
- 142.Krebs MG, Malapelle U, André F, et al. Practical considerations for the use of circulating tumor DNA in the treatment of patients with cancer: a narrative review. JAMA Oncol 2022;8:1830. 10.1001/jamaoncol.2022.4457 [DOI] [PubMed] [Google Scholar]
- 143.Rolfo C, Mack P, Scagliotti GV, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the International association for the study of lung cancer. J Thorac Oncol 2021;16:1647–62. 10.1016/j.jtho.2021.06.017 [DOI] [PubMed] [Google Scholar]
- 144.International Quality Network for Pathology (IQN Path) ECPCE, European federation of Pharmaceutical Industries and Associations (EFPIA) . Unlocking the potential of precision medicine in Europe – improving cancer care through broader access to quality biomarker testing, 2021. Available: https://www.efpia.eu/media/589727/unlocking-the-potential-of-precision-medicine-in-europe.pdf
- 145.Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res 2017;23:5729–36. 10.1158/1078-0432.CCR-17-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019;5:696–702. 10.1001/jamaoncol.2018.7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peters S, Spigel D, Ahn M, et al. P03.03 MERMAID-1: a phase III study of adjuvant Durvalumab plus chemotherapy in resected NSCLC patients with MRD+ post-surgery. Journal of Thoracic Oncology 2021;16:S258–9. 10.1016/j.jtho.2021.01.376 [DOI] [Google Scholar]
- 148.Spigel DR, Peters S, Ahn M-J, et al. 93TiP MERMAID-2: phase III study of durvalumab in patients with resected, stage II-III NSCLC who become MRD+ after curative-intent therapy. Journal of Thoracic Oncology 2021;16:S745–6. 10.1016/S1556-0864(21)01935-3 [DOI] [Google Scholar]
- 149.TSK M, Gadgeel S, Kim ES. Blood first line ready screening trial (B-F1RST) and blood first assay screening trial (BFAST) enable clinical development of novel blood-based biomarker assays for tumor mutational burden (TMB) and somatic mutations in 1L advanced or metastatic NSCLC. Annals of Oncology 2017;28. [Google Scholar]
- 150.Lee R, Rothwell DG, Chow S, et al. Cactus: a parallel arm, biomarker driven, phase II feasibility trial to determine the role of circulating tumor DNA in guiding a switch between targeted therapy and immune therapy in patients with advanced cutaneous melanoma. JCO 2021;39:TPS9587. 10.1200/JCO.2021.39.15_suppl.TPS9587 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005924supp001.pdf (175.2KB, pdf)