Abstract
Introduction
The transition to digital pathology has been carried out by several laboratories across the globe, with some cases described in Portugal. In this article, we describe the transition to digital pathology in a high-volume private laboratory, considering the main challenges and opportunities.
Material and methods
Our process started in 2020, with laboratory workflow adaptation and we are currently using a high-capacity scanner (Aperio GT450DX) to digitize slides at 20×. The visualization system, Aperio eSlide Manager WebViewer, is integrated into the Laboratory System. The validation process followed the Royal College of Pathologists Guidelines.
Results
Regarding validation, the first phase detected an error rate of 6.8%, mostly due to digitization errors. Phase optimization and collaboration with technical services led to improvements in this process. In the second validation phase, most of the slides had the desired quality for evaluation, with only an error rate of 0.6%, corrected with a new scan. The interpathologist correlation had a total agreement rate of 96.87% and 3.13% partial agreement.
Conclusion
The implementation and validation of digital pathology was a success, being ready for prime time. The total integration of all laboratory systems and the acquisition of new equipment will maximize their use, especially with the application of artificial intelligence algorithms.
Keywords: Digital pathology, Validation, Implementation
Introduction
The Germano de Sousa - Centro de Diagnóstico Histopatológico (CEDAP) is a private laboratory in Portugal, located in Coimbra. We currently have circa 150 000 exams per year (and growing), including cytology specimens. It provides diagnoses for private institutions and public departments, including biomarker identification and molecular analysis, and has a role in laboratory technicians training.
In the last years, digital pathology transition has been arising with success, in a similar manner to what happened to radiology in the past.1 The evolution of glass slides scanners was a very important component of this possibility, with faster equipment with better image acquisition.2
Several successful digital transformations with pathology workflow adaptation have been published,3, 4, 5, 6, 7 with their specific challenges and workflow. The implementation of this modality has recognized advantages, namely cost reduction and quicker workflows with consequent faster results, and facilitate multidisciplinary tumor boards.8 Despite these advantages, the number of Pathology Laboratories falls below the expectations, mainly due to high initial cost of implementation and the belief that it mainly for educational and research objectives.4 The majority of laboratories that have been successful in this task are major public laboratories resorting to external funding and private laboratories.
Digital pathology is more than acquiring a slide scanner and, contemplates leadership roles, involving the entire group (pathologists, technicians, administrative staff, etc.) and motivating people for a change in mindsets and workflow.
This article aims to report the CEDAP experience in the Digital Pathology transition, highlighting the process, emphasizing the problems and exploring the respective solutions and future perspectives.
Material & methods
Description of the laboratory
The CEDAP corresponds to a laboratory, based in Coimbra, a rather strategic location between Porto and Lisbon, being able to provide a diagnosis to all the samples collected by the Germano de Sousa group. It is a high-volume laboratory, with 103 358 cases in 2021, corresponding to a 57 276 histology cases, 40 947 cytology samples, and 5135 molecular tests. Due to the major widespread of the group, we receive samples from all around Portugal. Currently, we have 12 pathologists, full and part-time, and 2 dermatopathologists.
Due to the nature of our laboratory, this digital pathology system was implemented mainly for diagnostic purposes and supporting multidisciplinary meetings. As a secondary application, it can be used also for teaching, research, and algorithm validation.
Beginning of the process
The digital transition started in 2020, with the subsequent training/preparation of the laboratory staff, special reformulation of the laboratory, equipment acquisition, information technology (IT) infrastructure, and validation of the digital analysis by a pathologist. Due to the COVID-19 pandemic, some of these processes were delayed.
IT infrastructure and imaging technology
Our Laboratory Information System (LIS) and sample tracking software at the time of implementation was the XStore™ (Forum SI, Coimbra Portugal), but during this process, we performed the transition to SISTPAT™ (JSalgado, Porto, Portugal).
Regarding the scanner, we have 1 Aperio GT450DX Scanner by Leica Biosystems. These scanners each have a 450-slide capacity and enable brightfield applications and digitizing at 40× equivalent resolution (0.26 μm/pixel).
The scanning in our laboratory includes standard H&E and special histochemical and immunohistochemical stains. We scanned slides produced at our laboratory but also slides from external pathology departments that are sent to us—mostly from public institutions that have their Pathology Departments and can mount and stain their slides. Immunofluorescence slides were not scanned due to the need for a specific scanner and the low volume of these cases. Cytology was not included due to the need for Z-stack image acquisition which prompts additional time and size.9 We do not digitize immunofluorescence or cytology slides. Scanning of the frozen section was also not performed by option, since it will need further validation for routine purposes.
The Image Managing System (IMS) we use is the Aperio eSlide Manager WebViewer viewing software (Leica Biosystems). Our DP server is a Dell 740 XD with 21TB of storage provided by Leica, running Aperio eSlide Manager. We have 1 Gigabit per second (Gbps) internal and external client networks and the connection between the server and our internal network is 10 Gbps.
Digital workflow
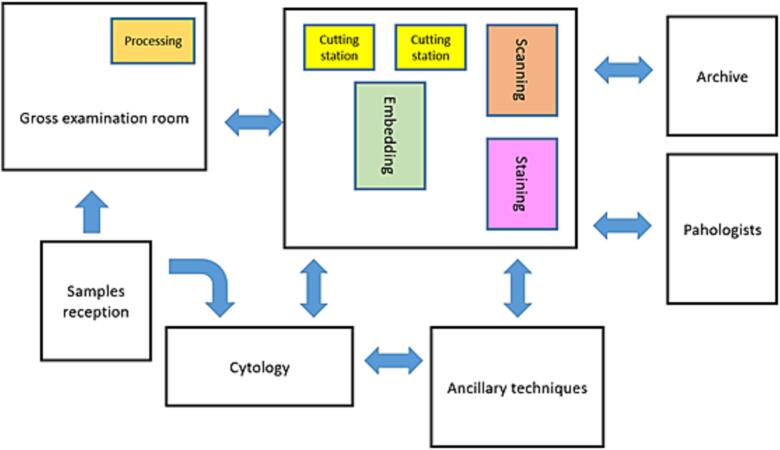
To have a continuous and functional flow in the laboratory, a LEAN methodology was put in place. The laboratory stations were restructured according to Fig. 1.
Fig. 1.
Schematic representation of the laboratory stations.
Summarily there was an investment in the reorganization of the laboratory equipment, without major infrastructure modification, allowing space for the slide scanner, next to the staining equipment, as it can be seen in Fig. 2.
Fig. 2.
The proximity of the scanner to the working stations is fundamental for a fluid workflow.
The restructuration was fundamental to allow a continuous workflow in the laboratory and achieve a complete slide creation and digitization of multiple slides in the same day (a maximum of little more than 300) and without perturbing the distribution to the pathologists.
The process did not involve an increase in the number of technicians, and after training, the scanning process was integrated into the routine work.
Regarding gross examination, our laboratory has implemented automatic access to the case by scanning the ID barcode. Gross photographs are regularly taken and stored in a shared folder. Samples are placed in printed cassettes with the correct QR code, for use in subsequent working stations.
In the embedding, an effort has been made to place the fragments together and in the center of the slide, to diminish the area to be scanned, and to not have fragments in the (possibly) non-scanned edges of the glass slide.
Regarding sectioning, the scanning of the QR code in the paraffin block allows the generation of one or more glass slides using a devoted printer and specific protocols, reducing work time and preventing errors.
Before scanning, all glass slides received a QR code on their label to assure the correct identification and case allocation by the Aperio Software. For the glass slides arriving from another institution, the solution was to print a new label, with our internal ID case number.
The scanning station was in the same room as the staining and coverslipping machine (HistoCore SPECTRA Workstation) ensuring an easy load of tracks directly into the Aperio 450. The scanning process was performed by proper-trained laboratory technicians, according to the manufacturer’s instructions using a 20× protocol.
After scanning, the glass slides were assembled in trays, as usual, and distributed to the pathologist, as described in section “validation”.
Pathologist workstation
The pathologists’ workstations at CEDAP are composed of a computer with an Inter Core i5-9500 CPU, with 3.00 GHz Clock Speed, and 16GB RAM with CPU integrated video card. The monitor used for WSI visualization is an ASUS 27 inch with a resolution of 2560 × 1440, LED type, and with a refresh rate of 75Hz. The equipment has all the necessary features and is in line with equipment used in other institutions.10
LIS and IMS integration
The majority of the instruments and systems in the laboratory were integrated through a Health Level 7 (HL7), which is the standard proceeding.10 Until the end of the present year, all the equipment is expected to be connected.
Validation
Regarding validation, we followed the guidelines/recommendations of the Royal College of Pathologists.11
Each pathologist had a short training session (circa 1h) with a trainer, to learn: (1) the basic digital pathology workflow and layout of the software; (2) how to use the system to open a case/slide and pan and zoom; (3) how to use the system to annotate a case and other advanced functions as necessary; (4) how to access the documentation for the system; and (5) how to identify gross scanning artifacts.
The pathologist reviews 1–2 cases on the digital system, with guidance and provides immediate feedback from the teacher about how to use the system.
Stage 1 validation is designed to train the pathologist on the diagnostic appearances of cases using digital pathology. This step includes exposure to cases anticipated to be challenging and will encompass a variety of case types (simple and complex) and stains. Two pathologists reviewed this set, making notes on the digital diagnosis and immediately compare with the glass slide diagnosis. A comment on concordance is done, including pitfalls detected and identification of difficulties. A score on the confidence of the diagnosis is also attributed to both digital and glass evaluation, from 1 to 7, being not confident at all and 7 very confident.
A minimum of 20 cases is recommended but can be more if necessary.
This process ensures familiarity with the digital proceeding and after this, validation stage 2 was performed.
During step #2 of validation, the glass slides were physically delivered to the corresponding pathologist, to perform the comparison.
In this stage, the pathologist dual reports all of their cases using both digital and glass, to gain experience and confidence in using the technology. All cases in the domain are evaluated prospectively, thus introducing digital analysis into the routine.
The sample size and duration of the validation are variable but should be of sufficient length and detail so the pathologist develops proficiency and confidence in digital pathology. This procedure took 3–4 months. The sample was comprised of a mixture of small biopsies and large resection cases and was representative of the routine, including easy and difficult cases, non-neoplastic and oncological. The cases were not randomly selected because they were representative of the pathologist routine. Cases from outside of the laboratory had a new and dedicated label for scanner reading.
Throughout this validation process, cases identified as challenging were selected for step 3, so-called inter-pathologist concordance. This “library” of cases, up to 20–40 cases, was assembled and it was going to be evaluated by a different pathologist, with a final concordance assessment.
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS).
Results
During this process, 28 473 glass slides were scanned with an average of 67.5 s per slide (normalized 15 mm×15 mm slide scanned of 43.2 s). The average size of slide scanned was of 351.8 mm2 and average file size of 821MB.
This number do not reflect all the cases from our laboratory, since not all the cases were digitized for validation purposes.
In validation stage 1, 88 slides, corresponding to 35 cases, were scanned. The slides were representative of the following stains: hematoxylin and eosin (H&E) modified Giemsa, periodic acid Schiff (PAS), Masson trichrome, and diverse immunostains.
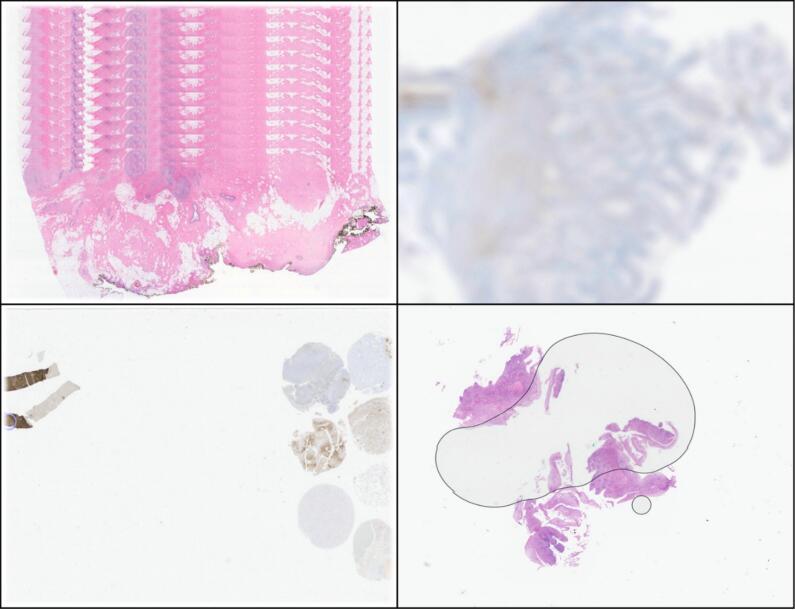
The comparison between digital and glass slides was satisfactory, and there were reported scanning errors in 6 slides (6.8%)—stripped slides, out of focus, and incomplete images, as demonstrated in Fig. 3.
Fig. 3.
Some common errors detected during Validation stage 1, namely stripped slides (upper left), out of focus (upper right), an incomplete images (lower left), and bubbles (lower right). The errors were easily corrected with a new scan (and bubble removal).
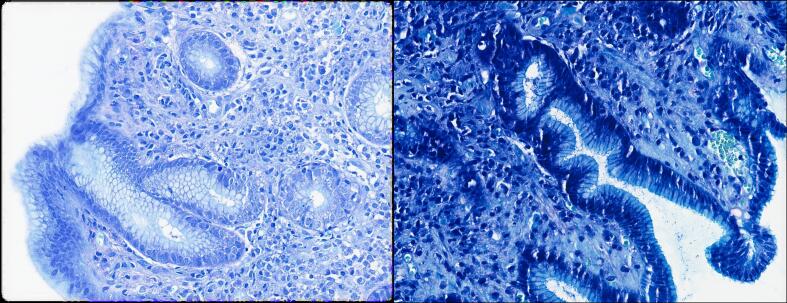
An interesting finding was that in some slides referred from external laboratories, the assessment of H. pylori infection with the modified Giemsa stain was a little more difficult due to the lack of some staining contrast. A minor modification in the protocols was enough to solve this issue, as demonstrated in Fig. 4.
Fig. 4.
Giemsa before (left) and after (right) protocol modifications ensuring a higher contrast and easily identifiable H. pylori.
In the first scanned immunohistochemical slides, there were minor interface issues with the BenchMark ULTRA, Roche Diagnostics® platform, but easily solved.
Regarding validation stage 2, a total of 2991 cases were evaluated, with an average of 2.8±1.8 glass slides per case (minimum of 1 and maximum of 24), corresponding to a total of 8474 scanned slides, with comparison of the quality between glass and scanned slides.
The majority of cases were satisfactory for evaluation on the digital slides (score 7 in the evaluation criteria). There were 51 slides (0.6%) in which a rescan was necessary. This procedure was able to solve the problem. The main reasons for a rescan were (from major to minor) misplaced labels with QR codes that prompted a non-allocation of the case, thick slides, excess glue/mounting medium, and air bubbles resulting in an out-of-focus slide. There was no record of slide breakages or scanner malfunction due to incorrect handling by the technicians.
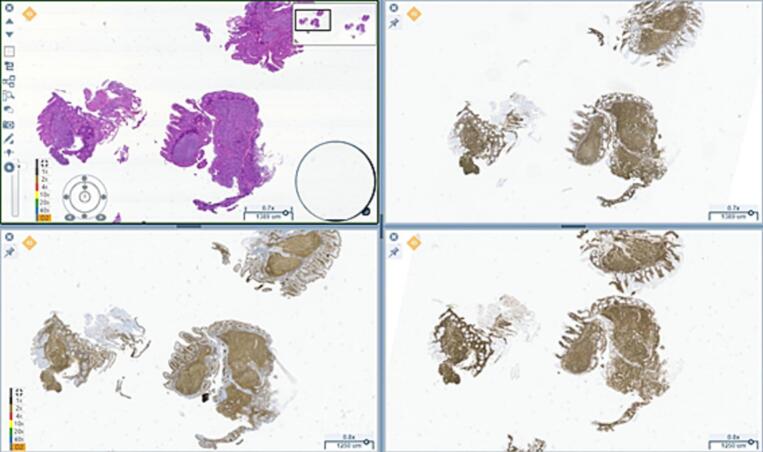
Picture acquisition was also feasible, with rapid results. One of the advantages reported was the possibility of viewing up to 4 slides simultaneously facilitating the H&E and immunohistochemistry interpretation, as it is shown in Fig. 5.
Fig. 5.
A case of a follicular duodenal lymphoma, where the simultaneous visualization of the H&E and the immunohistochemistry renders an easy diagnosis.
For the concordance between pathologists, 31 cases were compiled and scanned, comprising several organs and conditions and subjected to the evaluation of 6 pathologists.
After evaluation and statement of the diagnosis, there was a total agreement of the digital evaluation in 96.87% (31 out of 32), with partial agreement in 3.13% (1 case), with some stating mild chronic gastritis and others going for reactive gastritis.
Beside confidence in the diagnosis, pathologists were satisfied with the system ergonomics and velocity in loading slides from the server.
Discussion
Digital pathology has registered a remarkable evolution since the introduction of WSI.12 Microscopic examination of stained glass slides is still the gold-standard for rendering a diagnosis, but the improvement of WSI methodology (high image resolution, high capacity and velocity scanners, image management software, and algorithms for image analysis) and cost reduction (particularly of storage) have prompted a wider use and adoption of this modality.13
The now natural digital movement was promoted in 1992 by Kayser, and formalized in 2016 with the creation of the European Society for Digital Pathology (ESDIP)14 and is becoming a reality worldwide. The recent publication of the ESDIP recommendations15 will sure be valuable for digital pathology implementation.
According to the results, our implementation was a success with the rapid adoption of Digital Pathology for diagnostic purposes. The diagnoses were interchangeable between a light microscope and a computer monitor, especially after some training and adjustment to the methodology. The sharing of difficult cases was also easier with the methodology, allowing annotated comments.
The possibility of annotated images and instantaneous consultation is a major key player in pathology, improving the diagnosis, especially in second consultations.16 The digital annotations can also be useful for molecular purposes, by selecting the correct area for analysis.3
Routine scanning as 20x was sufficient since the viewer allows a 200% magnification and achieving a 40× digital view.
Measuring the time of analysis was not a main purpose of this article, and it was not proper measured, however there seems to be a decrease in turnaround time in the digital diagnosis, especially when measuring distances was fundamental (surgical margins; deep of tumor infiltration).
Even with the validation of digital slides for diagnostic purposes, there will be a support in hybrid format (digital + physical) for newcomers in the field, in order to allow the pathologist to be more familiar and confident with digital diagnosis, undergoing a learning curve. In order to facilitate adaption, any new recruited pathologist will undergo a 3–4-week hybrid format, or more if necessary, until it feels confident for digital only diagnostic.
The concordance between pathologists was high, with only minor discrepancies in a gastric biopsy, with some favoring reactive gastritis and others a mild chronic gastritis. This in accordance with the literature in which the diagnosis of gastritis in not always reproducible between pathologists and exhibits some variability.17
The LEAN approach was also fundamental for a fluid workflow, and in the near future with the fully implementation of the new LIS in the laboratory, complete traceability will be possible, allowing adequate monitoring and further improvement possibilities.
In order to keep up with the laboratory growth, we are in the process of acquiring and installing an extra scanner with high slide capacity will be installed, allowing for a higher scanning capacity, which is in line with the current ESDIP recommendations.15 The LEAN methodology adopted allows us a continuous loading of the scanner, with a daytime scanning routine, but if necessary overnight scanning will be performed.
The rapid growth and demand of faster reports was accompanied by an increased in the number of external pathologist working for CEDAP, meaning that we were forced to adapt procedures for sending glass slides and requisitions to pathologists that were not working in the CEDAP physical facilities, including pathologist that were working in different cities.
Digital allocation will also be pivotal for laboratory technicians and administrative staff, since with the direct allocation of cases to the pathologists, skipping the manual assembly and glass slide delivery to the pathologists, with the latter consuming up to 24 h in case of an external pathologist to the Laboratory.
The almost instantaneously attribution of cases will provide our laboratory with a precious gain in time, resulting in faster reports, with an expected average of 3–5 days since the sample arrives at CEDAP. In addition, eliminating the need of sending the physical slides will result in a lesser allocation of resources (human and financial resources) and the journey to a paper-free laboratory will be environment friendly, which is applauded by our staff.
As stated previously, gross photograph documentation is performed by routine, and the pictures are currently accessed by demand, and in the near future will be integrated into the LIS. We are currently testing voice recognition at this step, which will allow direct text conversion, saving time and preventing transcription errors.
Frozen section digitation will be an objective in a near future. We are at the moment gathering frozen section tissue slides, from intra-operatory request but also from fresh tissue submitted to regular gross examination in order to compare the evaluation, similarly to the described by Cima et al.18 This has the potential to be applied for cancer evaluation and transplant pathology, especially if it could be used with smaller devices such as the Aperio Scan Scope™, Nanozoomer™ or Grundium Ocus™, expanding the area of action.
A fully implementation of WSI images would allow to apply IVD validated tools in order to improve turnaround time and better diagnosis and IHC quantification,19 and has also the potential of creating test datasets for the development of new artificial intelligence algorithms.20 There are already FDA and EMA approved algorithms with wide diffusion, such as Paige Prostate™ and Visiopharm™, among others,21 with several being proposed as auxiliary methodology for lymph node metastases,22, 23, 24 histological classification of tumors,25 and even translating spatial biomarker dynamics.26 The evolution of scanning systems and emergence of vendor-neutral archives2,10 may be important is testing several solutions and minimize the impact of scanner variability.27 The implementation of this methodology will also allow cancer screening algorithms, case triage, and forecasting molecular changes.28 The amount of data in the millions of pixels of a WSI is enormous and only possible to analyze using AI computational methods, prompting a digital transition that will probably represent the next major revolution in pathology, sometimes designated as the Computer Vision for Pathology21 with an impact compared with immunohistochemistry and molecular pathology.29 The pathology laboratories that are not able to perform this transition will definitively become outdated.
Archiving is also influenced by digital pathology implementation. Despite our intention of becoming a paper-free laboratory, we are still demanded to keep paper copies of our reports. This archiving has already IT support, with computer-recorded positions for faster retrieval, if necessary. With the LIS transition, our block and glass slide archive will follow the same methodology, prompting faster archiving and less time consumption. In addition, due to the private nature of our laboratory we are constantly demanded to send blocks and/or slides due to patient referral to oncology centers, the faster identification and withdraw of samples will be of major value. If the block is absent due to additional ancillary techniques (p.e.), the system will display a message stating this condition and identifying the responsible. The advantages of this fully automated and computer-guided archive are well described.10
As seen in our results, only a minority of slides exhibited scanning issues, and this issue was solved with a rescan of the glass slide. This prompted the introduction of a rescan order option through the LIS in a similar way as an ancillary technique. Some authors promote a routine check of all WSI by an assigned technician,3 however in a high-volume laboratory, this is a highly time-consuming task for just 0.6%. The upper mentioned solution seems to us the right way to address this situation, a view also shared by Fraggetta et al.10
Despite not being the primary objective of the laboratory, the transition to digital pathology may also be an important opportunity for education with teaching or sharing interesting cases,30 which would be especially valuable in pandemic outbreaks31 using online platforms or even apps.32 A construction of a digital repository would also be of benefit for research purposes and for algorithm validation, especially when supported by AI tools, and this can be performed on a national basis, including both public and private institutions, similar to what is being developed in other countries.33
Digital archiving is still not totally consensual, but resorting to some modifications for image size diminishing, such as smaller fragments, concentration of the fragments in one place, images compression, etc.3,10 may be an option. Nevertheless, since glass slides and paraffin blocks have still to be archived for several years, it is always possible to rescan them.
In conclusion, our transition to digital pathology has started and has been as success regarding diagnosis and workflow modifications. There was no affection of the quality of the diagnosis. The transition to a different LIS and the integration of all the systems and equipment in the laboratory, expected by the end of the year, will allow us to have a full benefit of a digital environment, resulting in less expense of resources, faster turnaround time in reports, and AI applications.
Funding
This work was no object of funding.
Work contribution
Study design and conception: IF, CSM, RCO. Digital pathology validation: CSM, SM, SC, ACA, CA, RCO. Laboratory modifications and scanner application: IF, DC, MP, JMR. Work Supervision: HG and RCO. RCO drafted the manuscript. All authors have approved the manuscript final version.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Reiner B.I., Siegel E.L., Siddiqui K. Evolution of the digital revolution: a radiologist perspective. J Digital Imag [Internet]. 2003;16(4):324. doi: 10.1007/s10278-003-1743-y. [cited 2022 Sep 29]. Available from: /pmc/articles/PMC3044070/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantanowitz L., Sharma A., Carter A.B., Kurc T., Sussman A., Saltz J. Twenty years of digital pathology: an overview of the road travelled, what is on the horizon, and the emergence of vendor-neutral archives. J Pathol Inform [Internet]. 2018 Jan 1;9(1) doi: 10.4103/jpi.jpi_69_18. https://pubmed.ncbi.nlm.nih.gov/30607307/ [cited 2022 Sep 29]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eloy C., Vale J., Curado M., et al. Digital pathology workflow implementation at IPATIMUP. Diagnostics (Basel, Switzerland) [Internet] 2021 Nov 1;11(11) doi: 10.3390/diagnostics11112111. https://pubmed.ncbi.nlm.nih.gov/34829458/ [cited 2022 Sep 29]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraggetta F., Garozzo S., Zannoni G., Pantanowitz L., Rossi E. Routine digital pathology workflow: the catania experience. J Pathol Inform [Internet]. 2017;8(1) doi: 10.4103/jpi.jpi_58_17. https://pubmed.ncbi.nlm.nih.gov/29416914/ [cited 2022 Sep 29]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Retamero J.A., Aneiros-Fernandez J., del Moral R.G. Complete digital pathology for routine histopathology diagnosis in a multicenter hospital network. Arch Pathol Lab Med [Internet]. 2020;144(2):221–228. doi: 10.5858/arpa.2018-0541-OA. https://pubmed.ncbi.nlm.nih.gov/31295015/ [cited 2022 Sep 29]. Available from. [DOI] [PubMed] [Google Scholar]
- 6.Montezuma D., Monteiro A., Fraga J., et al. Digital pathology implementation in private practice: specific challenges and opportunities. Diagnostics (Basel, Switzerland) [Internet] 2022 Feb 1;12(2) doi: 10.3390/diagnostics12020529. https://pubmed.ncbi.nlm.nih.gov/35204617/ [cited 2022 Sep 29]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Temprana-Salvador J., López-García P., Vives J.C., et al. DigiPatICS: digital pathology transformation of the catalan health institute network of 8 hospitals—planification, implementation, and preliminary results. Diagnostics. 2022;12(4):852. doi: 10.3390/diagnostics12040852. https://www.mdpi.com/2075-4418/12/4/852/htm [Internet]. 2022 Mar 30 [cited 2022 Sep 29]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baidoshvili A., Bucur A., van Leeuwen J., van der Laak J., Kluin P., van Diest P.J. Evaluating the benefits of digital pathology implementation: time savings in laboratory logistics. Histopathology [Internet]. 2018 Nov 1;73(5):784–794. doi: 10.1111/his.13691. [cited 2022 Sep 29]. Available from. [DOI] [PubMed] [Google Scholar]
- 9.MG H, SE M, J C, J X, I A, L P Comparison of glass slides and various digital-slide modalities for cytopathology screening and interpretation. Cancer cytopathology [Internet]. 2017;125(9) doi: 10.1002/cncy.21880. https://pubmed.ncbi.nlm.nih.gov/28558124/ [cited 2022 Sep 29]. Available from. [DOI] [PubMed] [Google Scholar]
- 10.Fraggetta F., Caputo A., Guglielmino R., Pellegrino M.G., Runza G., L’imperio V. A survival guide for the rapid transition to a fully digital workflow: the “caltagirone example.”. Diagnostics (Basel, Switzerland) [Internet] 2021 Oct 1;11(10) doi: 10.3390/diagnostics11101916. https://pubmed.ncbi.nlm.nih.gov/34679614/ [cited 2022 Oct 1]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross S., Furness P., Igali L., Snead D., Treanor D. 2018. Best practice recommendations for implementing digital pathology Unique document number G162 Document name Best practice recommendations for implementing digital pathology. [Google Scholar]
- 12.Wright A.M., Smith D., Dhurandhar B., et al. Digital slide imaging in cervicovaginal cytology: a pilot study. Arch Pathol Lab Med [Internet]. 2013 May;137(5):618–624. doi: 10.5858/arpa.2012-0430-OA. https://pubmed.ncbi.nlm.nih.gov/22970841/ [cited 2022 Sep 29]. Available from. [DOI] [PubMed] [Google Scholar]
- 13.Jahn S.W., Plass M., Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med [Internet]. 2020 Nov 1;9(11):1–17. doi: 10.3390/jcm9113697. [cited 2022 Sep 29]. Available from: /pmc/articles/PMC7698715/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eloy C., Zerbe N., Fraggetta F. Europe unites for the digital transformation of pathology: the role of the new ESDIP. J Pathol Inform. 2021 Jan 1;12(1):10. doi: 10.4103/jpi.jpi_80_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraggetta F., L’imperio V., Ameisen D., et al. Best practice recommendations for the implementation of a digital pathology workflow in the anatomic pathology laboratory by the European Society of Digital and Integrative Pathology (ESDIP) Diagnostics (Basel, Switzerland) [Internet] 2021 Nov 1;11(11) doi: 10.3390/diagnostics11112167. https://pubmed.ncbi.nlm.nih.gov/34829514/ [cited 2022 Oct 5]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara S., Bychkov A., Munkhdelger J., et al. Substantial improvement of histopathological diagnosis by whole-slide image-based remote consultation. Virchows Archiv Int J Pathol [Internet]. 2022 Aug 1;481(2):295–305. doi: 10.1007/s00428-022-03327-2. https://pubmed.ncbi.nlm.nih.gov/35672584/ [cited 2022 Sep 30]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennelli G., Grillo F., Galuppini F., et al. Gastritis: update on etiological features and histological practical approach. Pathologica [Internet]. 2020 Sep 1;112(3):153. doi: 10.32074/1591-951X-163. [cited 2022 Oct 11]. Available from: /pmc/articles/PMC7931571/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cima L., Brunelli M., Parwani A., et al. Validation of remote digital frozen sections for cancer and transplant intraoperative services. J Pathol Inform [Internet]. 2018 Jan 1;9(1) doi: 10.4103/jpi.jpi_52_18. [cited 2022 Sep 29]. Available from: /pmc/articles/PMC6187937/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancellotti C., Cancian P., Savevski V., et al. Artificial intelligence & tissue biomarkers: advantages, risks and perspectives for pathology. Cells [Internet] 2021 Apr 1;10(4) doi: 10.3390/cells10040787. [cited 2022 Sep 30]. Available from: /pmc/articles/PMC8066881/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homeyer A., Geißler C., Schwen L.O., et al. Recommendations on compiling test datasets for evaluating artificial intelligence solutions in pathology. Mod Pathol. 2022:1–11. doi: 10.1038/s41379-022-01147-y. https://www.nature.com/articles/s41379-022-01147-y [Internet]. 2022 Sep 10 [cited 2022 Sep 30]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel A.U., Shaker N., Mohanty S., et al. Cultivating clinical clarity through computer vision: a current perspective on whole slide imaging and artificial intelligence. Diagnostics (Basel, Switzerland) [Internet] 2022 Jul 22;12(8):1778. doi: 10.3390/diagnostics12081778. https://pubmed.ncbi.nlm.nih.gov/35892487/ [cited 2022 Oct 1]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang W.Y., Chen C.C., Yu W.H., et al. Identification of nodal micrometastasis in colorectal cancer using deep learning on annotation-free whole-slide images. Mod Pathol. 2021;34(10):1901–1911. doi: 10.1038/s41379-021-00838-2. https://www.nature.com/articles/s41379-021-00838-2 [Internet]. 2021 Jun 8 [cited 2022 Sep 30]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu P.H., Zhao L.L., Wu H.L., Zhao D.B., Chen Y.T. Artificial intelligence in gastric cancer: application and future perspectives. World J Gastroenterol [Internet] 2020 Sep 9;26(36):5408. doi: 10.3748/wjg.v26.i36.5408. [cited 2022 Oct 16]. Available from: /pmc/articles/PMC7520602/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Kohlberger T., Norouzi M., et al. Artificial intelligence-based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Me [Internet]. 2019;143(7):859–868. doi: 10.5858/arpa.2018-0147-OA. https://pubmed.ncbi.nlm.nih.gov/30295070/ [cited 2022 Oct 16]. Available from. [DOI] [PubMed] [Google Scholar]
- 25.Chen C.L., Chen C.C., Yu W.H., et al. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat Commun. 2021;12(1):1–13. doi: 10.1038/s41467-021-21467-y. https://www.nature.com/articles/s41467-021-21467-y [Internet]. 2021 Feb 19 [cited 2022 Sep 30]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchinson L.G., Grimm O. Integrating digital pathology and mathematical modelling to predict spatial biomarker dynamics in cancer immunotherapy. npj Digit Med. 2022;5(1):1–13. doi: 10.1038/s41746-022-00636-3. https://www.nature.com/articles/s41746-022-00636-3 [Internet]. 2022 Jul 12 [cited 2022 Sep 30]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan A., Janowczyk A., Müller F., et al. Impact of scanner variability on lymph node segmentation in computational pathology. J Pathol Inform. 2022 Jan 1;13:100127. doi: 10.1016/j.jpi.2022.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraggetta F. Clinical-grade computational pathology: Alea Iacta Est. J Pathol Inform [Internet]. 2019 Jan 1;10(1) doi: 10.4103/jpi.jpi_54_19. [cited 2022 Oct 1]. Available from: /pmc/articles/PMC6939341/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eloy C., Bychkov A., Pantanowitz L., et al. DPA-ESDIP-JSDP task force for worldwide adoption of digital pathology. J Pathol Inform [Internet]. 2021 Jan 1;12(1) doi: 10.4103/jpi.jpi_65_21. https://pubmed.ncbi.nlm.nih.gov/35070480/ [cited 2022 Oct 1]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassell L.A., Absar S.F., Chauhan C., et al. Pathology education powered by virtual and digital transformation. Archiv Pathol Lab Med [Internet]. 2022 Jul 25 doi: 10.5858/arpa.2021-0473-RA. https://pubmed.ncbi.nlm.nih.gov/35878400/ [cited 2022 Sep 30]; Available from. [DOI] [PubMed] [Google Scholar]
- 31.Ishak A., Alrawashdeh M.M., Meletiou-Mavrotheris M., Nikas I.P. Virtual pathology education in medical schools worldwide during the COVID-19 pandemic: advantages, challenges faced, and perspectives. Diagnostics [Internet]. 2022 Jul 1;12(7) doi: 10.3390/diagnostics12071578. [cited 2022 Sep 30]. Available from: /pmc/articles/PMC9321717/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajaram A., Olory C., Leduc V., et al. An integrated virtual pathology education platform developed using Microsoft Power Apps and Microsoft Teams. J Pathol Inform. 2022 Jan 1;13:100117. doi: 10.1016/j.jpi.2022.100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janowczyk A., Baumhoer D., Dirnhofer S., et al. Towards a national strategy for digital pathology in Switzerland. Virchows Archiv Int J Pathol [Internet]. 2022 Oct 1;481(4) doi: 10.1007/s00428-022-03345-0. https://pubmed.ncbi.nlm.nih.gov/35622144/ [cited 2022 Oct 16]. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]