Abstract

Ruthenium(II) polypyridyl complexes (RPCs) that emit from metal-to-ligand charge transfer (MLCT) states have been developed as DNA probes and are being examined as potential anticancer agents. Here, we report that MLCT-emissive RPCs that bind DNA undergo Förster resonance energy transfer (FRET) with Cy5.5-labeled DNA, forming mega-Stokes shift FRET pairs. Based on this discovery, we developed a simple and rapid FRET binding assay to examine DNA-binding interactions of RPCs with diverse photophysical properties, including non-“light switch” complexes [Ru(dppz)2(5,5′dmb)]2+ and [Ru(PIP)2(5,5′dmb)]2+ (dppz = dipyridophenazine, 5,5′dmb = 5,5′-dimethyl-2,2′-bipyridine, PIP = 2-phenyl-imidazo[4,5-f][1,10]phenanthroline). Binding affinities toward duplex, G-quadruplex, three-way junction, and mismatch DNA were determined, and derived FRET donor–acceptor proximities provide information on potential binding sites. Molecules characterized by this method demonstrate encouraging anticancer properties, including synergy with the PARP inhibitor Olaparib, and mechanistic studies indicate that [Ru(PIP)2(5,5′dmb)]2+ acts to block DNA replication fork progression.
Introduction
Ruthenium(II) polypyridyl complexes (RPCs) that bind DNA by noncovalent mechanisms have been proposed for a multitude of uses in biology, including cellular imaging agents,1 anticancer therapeutics,2 and photosensitizers for photodynamic therapy.3 Based on the discovery that [Ru(bpy)2(dppz)]2+ (bpy = 2,2′-bipyridine, dppz = dipyrido[3,2-a:2′,3′-c]phenazine) binds DNA with high affinity and an accompanying increase in metal-to-ligand charge transfer (3MLCT) luminescence—the DNA “light switch” effect4—RPCs utilizing intercalating ligands have been developed as high-affinity DNA binders.5 Judicious selection or chemical modification of ancillary or intercalating ligands can then generate RPCs that demonstrate site- or structure-selectivity6−9 along with tuneable biological activity.10 The increase in MLCT emission upon binding presents a convenient method to determine nonspecific binding site affinity via the generation of Scatchard plots that fit the McGhee and von Hippel equation.11 However, this equation is unsuitable for oligonucleotides of known sequences, which are vital in determining site- or structure-specific binding,8,12 and reliance on MLCT emission also inherently disfavors the study of RPC/DNA combinations that possess variable, weak, or complete absence of “light switch” properties. This latter category of RPCs includes molecules of considerable biological interest such as [Ru(N∧N)2(PIP)]2+ (PIP = 2-phenyl-imidazo[4,5-f][1,10]phenanthroline)13 and [Ru(dppz)2(N∧N)]2+ complexes.14 Although luminescence-independent techniques exist to examine and quantify DNA binding, including ultraviolet–visible (UV–vis) absorption15 and isothermal titration calorimetry (ITC),16 these methods are low throughput and comparisons of results obtained using different techniques between studies can be challenging. As such, a single assay suitable for a range of DNA structures and RPCs with diverse photophysical properties and compatible with high-throughput screening methods would be advantageous.
Förster resonance energy transfer (FRET) involves dipole–dipole coupling from the excited state of a donor luminophore (D) to the ground state of an acceptor luminophore (A)—a FRET pair.17 Due to its inverse-sixth-power distance dependence, FRET provides proximity detection at the nanometer scale.17 Biomolecules labeled with FRET pairs are utilized in bioassays to probe the biomacromolecular structure, protein–protein or protein–DNA interactions,18−20 including screening for small molecules that inhibit (or promote) these processes,21 while inherently fluorescent or fluorescently labeled compounds and target receptors facilitate the development of advanced binding assays.22,23 Advantages of FRET are its high sensitivity, broad dynamic range, and compatibility with high-throughput screening.17 MLCT-emissive phosphorescent RPCs hold significant potential as FRET donors, where the low energy and large Stokes shifts of MLCT phosphorescence (typically >150 nm) would be advantageous in addressing current limitations arising from the use of two organic fluorophores, which include poor signal-to-noise and spectral bleed through.24 Despite this, few studies have examined RPCs in FRET bioassays. Work to date has included a [Ru(bpy)3]2+-based enzyme-cleavable sensor25 and DNA sequences covalently labeled with RPCs to examine DNA–DNA assembly.26,27 Studies employing a DNA-binding RPC as a FRET donor are even rarer, yet the potential for this was explored by Lakowicz et al., who demonstrated successful RET between [Ru(bpy)2(dppz)]2+ and BO-PRO3 when both molecules were intercalated to DNA.28,29 A disadvantage of this was that two reversibly binding DNA molecules were employed, thereby introducing an additional variable that makes assay development problematic.
Here, we report the ability of 3MLCT emission from DNA-binding RPCs to undergo resonance energy transfer with a fluorophore covalently linked to DNA (Figure 1a). Using fluorophore-labeled oligonucleotides, we utilize this effect to characterize structure- and sequence-specific DNA binding of four RPCs with diverse photophysical properties, including non-“light switch” molecules, determining affinity—and proximity—of binding in a single, rapid assay. We show that molecules characterized by this method demonstrate encouraging anticancer properties, including impacting DNA replication and synergy with the PARP inhibitor Olaparib in a combination therapy approach.
Figure 1.
(a) Schematic diagram illustrating DNA-binding ruthenium(II) polypyridyl complexes as FRET donors for fluorophore-labeled DNA. (b) Chemical structures of ruthenium(II) polypyridyl complexes used in this study. Complexes were used as a mixture of stereoisomers.
Results and Discussion
Synthesis and Photophysical Properties
[Ru(bpy)2(dppz)]2+ (1) and [Ru(5,5′dmb)2(dppz)]2+ (5,5′dmb = 5,5′dimethyl bpy) (2) were prepared by previously reported methods.4,81H NMR, mass spectroscopy, and elemental analysis were in agreement with the published data. The novel complexes [Ru(dppz)2(5,5′dmb)]2+ (3) and [Ru(PIP)2(5,5′dmb)]2+ (4) were prepared in a similar manner via the preparation of the intermediate compounds [Ru(dppz)2Cl2] and [Ru(PIP)2Cl2] and characterized by 1H NMR, 13C NMR, high-resolution mass spectrometry (HRMS), Fourier-transform infrared spectroscopy (FTIR), and elemental analysis. Full synthetic and characterization details are in the Supporting Information (Figures S1–S10). Excitation/emission spectra of the hexafluorophosphate salts of 3 and 4 in acetonitrile demonstrated characteristic 3MLCT emission upon 1MLCT excitation (Figure S11 and Table S1); however, in aqueous media as their chloride salts, the emission of 3 is almost completely quenched, while 4 retains a relatively high MLCT intensity (Figure S12a). The addition of calf-thymus DNA to each complex in aqueous media reveals a varied photophysical response: 2 acts as a conventional “light switch” molecule, albeit with reduced intensity compared to 1, 3 does not exhibit “light switch” behavior as only a 2.4-fold enhancement of MLCT emission is seen, and the aqueous MLCT emission of 4 is quenched by the addition of DNA (Table 1 and Figure S12b).
Table 1. Photophysical Properties of DNA-Bound 1–4 [Donor] and Cy5.5 [Acceptor] FRET Pairsa.
| complex | DNA kb (M–1) | n (base pairs) | λex (max) (nm) | λem (max) (nm) | I(DNA)/I(aq.) | Φ(DNA) | J (λ) (M–1 cm–1·nm4) | Ro (Å) |
|---|---|---|---|---|---|---|---|---|
| 1 | 2.513 × 106 | 1.26 | 450 | 620 | 117 | 0.008 | 1.50 ± 0.23 × 1016 | 38.9 ± 1.02 |
| 2 | 2.632 × 106 | 1.73 | 440 | 635 | 13 | 0.004 | 1.68 ± 0.24 × 1016 | 35.1 ± 0.85 |
| 3 | 1.248 × 106 | 1.14 | 440 | 630 | 2.4 | 0.0005 | 1.50 ± 0.08 × 1016 | 38.9 ± 0.34 |
| 4 | nd | nd | 470 | 605 | 0.56 | 0.042 | 1.34 ± 0.16 × 1016 | 50.2 ± 0.97 |
Buffer: 5 mM Tris, 200 mM NaCl, pH 7.5. J (λ) = spectral overlap integral. Ro = Förster radius. ND = not determined.
Binding affinities were determined by fitting the McGhee von Hippel binding model11 to luminescence titrations, showing that 2 and 3 each bind calf-thymus DNA with binding affinities comparable to 1 (equilibrium binding constants, Kb of 2.63 × 106 and 1.25 × 106 M–1 for 2 and 3, respectively, Table 1). The binding affinity of 4 could not be determined by either fluorescence or UV–visible absorption titrations; however, apparent binding constants, Kapp, from the ethidium bromide displacement assay indicates that 4 possesses a DNA-binding affinity comparable to 3 (Figure S13).
MLCT-Cy5.5 FRET Pair Characterization
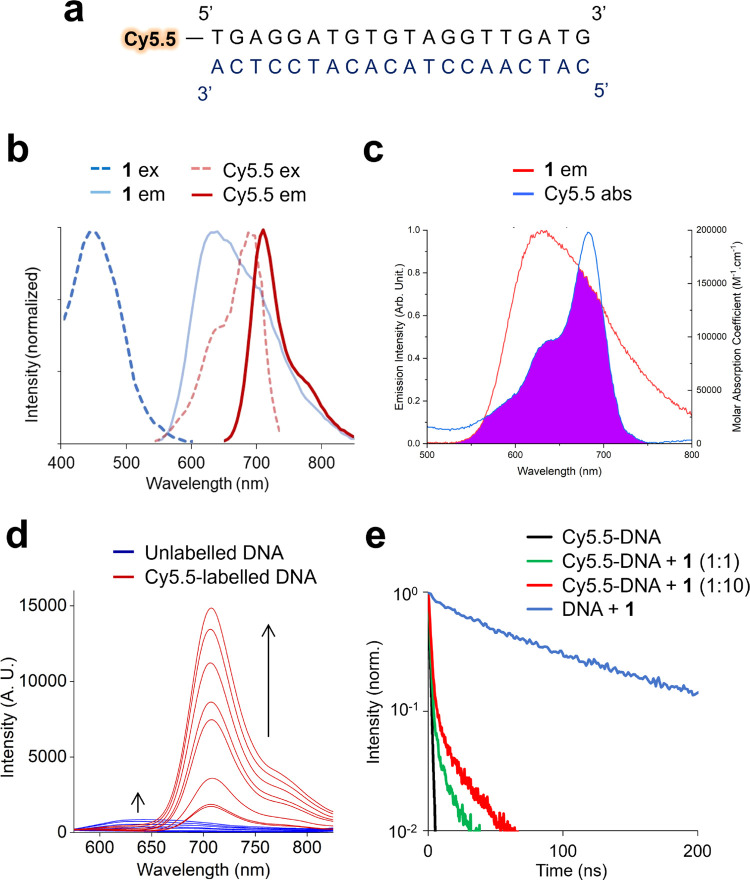
Based on the emission spectra of DNA-bound 1–4 (Figure S14 and Table 1), we hypothesized that each RPC would be a suitable donor for the cyanine dye Cy5.5 (λex[max] = 683 nm, λem[max] = 703 nm) to act as the acceptor. Using a Cy5.5-labeled 20-mer duplex (Figure 2a and Table 2), the sufficient spectral overlap of the MLCT emission of DNA-bound 1 and Cy5.5 was demonstrated (Figure 2b,c and Table 1) and the addition of 1 to the Cy5.5-labeled 20-mer followed by 450 nm excitation resulted in an intense emission peak at 710 nm (Figure 2d). These wavelengths correspond to 1MLCT excitation of 1 and Cy5.5 emission. Comparing these results to experiments conducted using the unlabeled 20-mer, negligible 3MLCT emission at 630 nm was observed for the Cy5.5-labeled 20-mer (Figure 2d), demonstrating successful 3MLCT emission quenching in the presence of Cy5.5, consistent with an RET process. In addition to this, a clear increase in the lifetime of the 710 nm Cy5.5 peak in the presence of the donor was observed (Figure 2e), resulting in an average lifetime of 2.4–3.6 ns for the 1-Cy5.5 FRET pair (average lifetime for Cy5.5-DNA = 1.2 ns, Table S2). Similar results for 2–4 were also achieved (Figures S14 and S15 and Table 1), confirming successful RET from the MLCT excited states of the DNA-binding 1–4 to Cy5.5-labeled DNA in each case. Förster radii (R0) were calculated17 and found to range from 3.5 nm (2-Cy5.5) to 5.0 nm (4-Cy5.5) (Table 1). Each new MLCT-Cy5.5 FRET pair has a Stokes shift value of ∼250 nm, some of the largest values described to date, signifying that they classify as “mega-Stokes shift” FRET pairs.30
Figure 2.
(a) Cy5.5-labeled 20-mer duplex DNA. (b) Excitation (ex, dashed) and emission (em, solid) spectra of DNA-bound RPC [Ru(bpy)2(dppz)]2+ (blue) and Cy5.5 (red). (c) Spectral overlap of the MLCT emission of DNA-bound 1 and Cy5.5 absorption. (d) Red: the emission spectra (λex = 450 nm) of Cy5.5-labeled 20-mer (1 μM) with increasing concentration of 1 (0.1–20 μM). Blue: the addition of 1 to unlabeled DNA showing MLCT emission (λex = 450 nm). (e) Lifetime of 710 nm emission in the absence and presence of 1. Lifetime of unlabeled 20-mer with 1 at 630 nm included for comparison. Buffer: 5 mM Tris, 200 mM NaCl, pH 7.5.
Table 2. DNA Sequences Used in this Work.
| structure | sequences (all written 5′ to 3′) |
|---|---|
| 20-mer duplex | S1: [cyanine55]TGAGGATGTGTAGGTTGATG |
| S2: CATCAACCTACACATCCTCA | |
| 3WJ | S1: [cyanine55]TGAGGATGTGTAGGTTGATG |
| S2: CATCAACCTAAGAATGAGAC | |
| S3: GTCTCATTCTCACATCCTCA | |
| G-quadruplex | [cyanine55]AGGGTTAGGGTTAGGGTTAGGG |
| matched duplex | S1: [cyanine55]GACCAGCTTATCACCCCTAGATACCAT |
| S2: ATGGTATCTAGGGGTGATAAGCTGGTC | |
| mismatch 1 duplex | S1: [cyanine55]GACCAGCTTATCACCCCTAGATACCAT |
| S2: ATGGTATCTAGGGGTGATAAGCTCGTC | |
| mismatch 2 duplex | S1: [cyanine55]GACCAGCTTATCACCCCTAGATACCAT |
| S2: ATGGTATCTAGGGCTGATAAGCTGGTC | |
| mismatch 3 duplex | S1: [cyanine55]GACCAGCTTATCACCCCTAGATACCAT |
| S2: ATGCTATCTAGGGGTGATAAGCTGGTC |
Structure-Specific DNA Binding
Identifying small molecules that bind and stabilize the nonduplex G-quadruplex (G4) and three-way junction (3WJ) structures is a topic of current interest within medicinal chemistry.31−33 RPCs are well-established to interact with G-quadruplexes;34−36 however, their binding to 3WJs has not been investigated. To examine whether MLCT-Cy5.5 FRET could determine binding affinities of 1–4 toward different DNA structures, Cy5.5-labeled oligonucleotides that form a duplex, G-quadruplex (G4),37 or a three-way junction (3WJ)38 (Figure 3a and Table 2) were treated with a concentration gradient of each complex and the resultant FRET intensity was determined. A 96-well plate and plate reader format was employed to improve throughput, allowing all 12 RPC/DNA combinations to be examined in a single experiment (Figure 3b). Resultant FRET binding curves were fit to a dose–response model to allow derivation of equilibrium dissociation constants, Kd (Figures 3c and S16 and Table 3), an established means of quantifying small molecule–nucleic acid binding in high-throughput screening.39
Figure 3.
(a) Schematic diagrams of a Cy5.5-labeled duplex, G-quadruplex (G4), and three-way junction (3WJ). (b) Left: an example of a typical experimental setup. Right: the sample emission spectra of Cy5.5-labeled DNA with increasing concentrations of 1–4 and successful FRET for each DNA structure/compound combination (λex = 450 nm). See Table 2 for precise DNA sequences used to form each structure. (c) FRET intensity (left) and the dose–response curve (right) generated from the addition of 2 or 4 (0.1–20 μM) to Cy5.5-labeled duplex, G4, or 3WJ DNA (1 μM). λex = 450 nm, λem = 710 nm. FRET intensity was background corrected. 3WJ and duplex buffer: 5 mM Tris, 200 mM NaCl, pH 7.5. G4 buffer: 100 mM KCl, 10 mM Tris.HCl, pH 7.4.
Table 3. Dissociation Constants (Kd) for Binding of 1–4 to 20-mer Duplex, G-Quadruplex (G4), and Three-Way Junction (3WJ) DNA Measured Using Cy5.5-Labeled Oligonucleotides and FRETa.
| duplex |
G4 |
3WJ |
||||
|---|---|---|---|---|---|---|
| complex | Kd (μM) | r (Å) | Kd (μM) | r (Å) | Kd (μM) | r (Å) |
| 1 | 2.8 ± 0.9 | 33.5 | 3.9 ± 1.3 | 26.8 | 4.3 ± 0.9 | 37.4 |
| 2 | 2.3 ± 0.5 | 33.1 | 3.8 ± 1.7 | 30.0 | 4.4 ± 0.7 | 35.0 |
| 3 | 6.3 ± 0.3 | 23.8 | nd | nd | nd | nd |
| 4 | 2.6 ± 1.0 | 55.9 | nd | nd | 6.4 ± 0.3 | 49.6 |
Low micromolar Kd values for 1–4 toward the 20-mer duplex compare favorably with known small molecule DNA aptamer affinities40 and provide a binding affinity order of 2 > 4 > 1 > 3 (Table 3). In addition to duplex DNA, both 1 and 2 showed high affinity toward G4 (Table 3) and the comparable binding affinities for 1 toward duplex and quadruplex DNA are in agreement with other works.35 In contrast to 1 and 2, however, neither 3 nor 4 reached binding saturation with G4 in the concentration range tested, indicating reduced affinity toward G4 for these two complexes. This may be explained by the greater steric bulk of each molecule as 3 and 4 each possess two intercalating ligands coordinated to an octahedral Ru(II) center and is consistent with the observation that the majority of quadruplex-binding small molecules are planar, typically interacting with G4 by G-tetrad stacking.32 In addition to duplex and G4 DNA, the FRET binding assay provided a clear indication of the binding of each complex with 3WJ with a binding order of 1 > 2 > 4 > 3 (Figures 3c and S16 and Table 3). All complexes displayed a reduced affinity for 3WJ compared to the duplex; however, this may be rationalized by the greater number of duplex binding sites of the 3WJ structure.
As FRET can also determine donor–acceptor proximity at the nanometer scale,17 mean D–A (r) distances were calculated. In our FRET assay, these values correspond to the average distance between the DNA-bound RPC (D) and Cy5.5 label (A). As the location of the Cy5.5 label in the DNA structure is known, this can provide an indication of potential binding sites of the RPC. D–A distances calculated in this manner were found to range from 2.4 to 5.6 nm (Table 3), values consistent with the size of DNA structures employed.41,42
Development of a Mismatch Detection Assay
In addition to structure selectivity, RPCs can exhibit sequence-specific binding, such as toward duplexes that contain non-Watson Crick mismatched base pairs (mismatches).7−9,43 Accordingly, the interaction of 1–4 with Cy5.5-labeled 27-mer duplexes containing a CC mismatch was examined by the FRET binding assay (Figure 4a). Duplexes were designed so that the mismatch was located at differing proximities from the Cy5.5 label: 4, 14, or 24 base pairs. FRET intensity and Kd values for each duplex/complex combination were determined in a similar manner as described previously and preferential mismatch binding was assessed by comparison to the well-matched control sequence. Employing this methodology, it was apparent that 3 displays the greatest FRET intensity with the addition of the “Mismatch 1” duplex that contains the CC mismatch at the 4 bp position (Figure 4b,c). Furthermore, when the CC mismatch is “moved” further from the Cy5.5 label in the case of the “Mismatch 2” and “Mismatch 3” duplexes, FRET intensity decreases. As the MLCT emission intensity of 3 remains constant with the addition of all four unlabeled 27-mers (Figures 4b and S17b), this confirms that the observed decrease in FRET intensity for the labeled sequences is the result of the CC mismatch being moved further away from the Cy5.5 label. Kd values show 3 has a higher binding affinity for the Mismatch 1 duplex compared to all other duplexes tested (Table 4 and Figure S18) and, perhaps most remarkably, D–A distances reveal that 3 is bound in closer proximity to Cy5.5 for this sequence, which contains the CC mismatch closest to the fluorescent label (Table 4). Similar behavior was also observed for 2, a molecule that has been previously reported to interact with CC mismatches,8 and relative FRET intensity was again found to be independent of MLCT emission intensity (Figures 4c and S17). Taken together, these results are consistent with 2 and 3 targeting with the CC mismatch and illustrate how MLCT-Cy5.5 FRET can be employed to identify mismatch interactive RPCs, including the non-“light switch” complex 3. It is also noteworthy that all complexes displayed reduced binding affinities toward the matched 27-mer compared to the 20-mer duplex above. This may be rationalized by the increased number of binding sites presented by the larger duplex requiring a greater concentration of each complex for binding saturation and this observation illustrates that internal controls of comparable DNA sizes are required for the interpretation of binding constant data.
Figure 4.
(a) Matched and mismatch-containing Cy5.5-labeled 27-mer DNA duplexes. (b) Emission spectra of 3 (5 μM) and unlabeled (left) or Cy5.5-labeled (right) 27-mers. 1 μM DNA was employed in each case. λex = 450 nm, all emission spectra were collected using the same optical parameters. (c) FRET intensity of 1–4 (3 μM) with the addition of Cy5.5-labeled 27-mer duplexes (1 μM). λex = 450 nm and λem = 710 nm. FRET intensity was normalized to matched DNA. Data are average of two independent repeats +/– SD. Buffer: 5 mM Tris, 200 mM NaCl, pH 7.5.
Table 4. Dissociation Constants (Kd) and Mean Donor–Acceptor Distances (r) for the Binding of 1–4 to Cy5.5-Labeled 27-mers Measured Using FRETa.
| matched |
mismatch 1 |
mismatch 2 |
mismatch 3 |
|||||
|---|---|---|---|---|---|---|---|---|
| Complex | Kd (μM) | r (Å) | Kd (μM) | r (Å) | Kd (μM) | r (Å) | Kd (μM) | r (Å) |
| 1 | 5.1 ± 0.4 | 47.6 | 5.0 ± 0.7 | 42.2 | 5.3 ± 0.6 | 45.4 | 6.5 ± 1.8 | 49.7 |
| 2 | 4.7 ± 0.1 | 47.7 | 4.6 ± 0.6 | 38.5 | 4.3 ± 0.6 | 42.9 | 5.1 ± 0.3 | 45.4 |
| 3 | 8.1 ± 1.7 | 54.7 | 6.4 ± 1.7 | 44.1 | 8.3 ± 3.0 | 56.3 | 8.1 ± 3.7 | 49.6 |
| 4 | 7.9 ± 2.5 | 49.9 | 7.3 ± 2.5 | 42.4 | nd | nd | 7.6 ± 1.9 | 52.5 |
ND = not determined (i.e., Kd > 20 μM). Mean +/– SD of two or three independent experiments.
Cytotoxicity and Impact on DNA Replication
RPCs that bind DNA by intercalation can stall DNA replication forks, inhibiting cell proliferation,44,45 and mismatch binding complexes have shown increased cytotoxicity toward mismatch-mediated repair (MMR)-deficient cancer cell lines.8,43 As our FRET binding studies show that 2–4 bind DNA with moderate affinity and also indicate that 2 and 3 interact with mismatches, their cytotoxic properties were examined in a small panel of cancer cell lines, including MMR-deficient HCT116 colorectal carcinoma cells. The low bioactivity of 1 has been reported elsewhere46 and so was not included in these studies. Assessing the impact on cell viability by the MTT assay, 3 and 4 each display moderate cytotoxicity toward the four cancer cell lines, particularly evident in HCC38 human triple-negative breast cancer (TNBC) cells (derived half inhibitory IC50 values of 3.8 and 16.5 μM, respectively, Figure S19 and Table 5). No clear evidence of enhanced cytotoxicity toward MMR-deficient HCT116 cells was observed for 2 or 3 and, considering that 3 also demonstrates a comparable cytotoxicity profile to the nonmismatch interactive 4, these results suggest cytotoxicity occurs through a mismatch-independent pathway. In comparison to their anticancer activity, both 3 and 4 showed significantly reduced effects toward normal MCF10A human breast epithelial cells (IC50s >100 and 63.1 μM for 3 and 4, respectively), indicating that both complexes demonstrate greater potency and enhanced cancer selectivity than cisplatin in MDA-MB-231 TNBC cells (Figure S19 and Tables 5 and S3).
Table 5. Half Inhibitory (IC50) Values of 2, 3, or 4 in MDA-MB-231 or HCC38 Breast Cancer, HCT116 Colorectal Carcinoma, T24 Bladder Cancer Cell Lines, or the MCF10A Non-Tumorigenic Epithelial Cell Line (72 h Treatment)a.
| complex | MDA-MB-231 | HCC38 | HCT116 | T24 | MCF10A |
|---|---|---|---|---|---|
| cisplatin | 32.5 ± 11.7 | 4.5 ± 0.3 | 5.0 ± 1.4 | 1.5 ± 0.5 | 26.5 ± 8.2 |
| 2 | >100 | >100 | 53.7 ± 9.9 | 34.4 ± 3.3 | >100 |
| 3 | 21.8 ± 2.9 | 3.8 ± 2.5 | 16.4 ± 1.4 | 28.0 ± 4.4 | >100 |
| 4 | 20.5 ± 9.6 | 16.5 ± 4.2 | 9.9 ± 1.8 | 29.9 ± 2.3 | 63.1 ± 21.0 |
Cisplatin treatment is included for comparison. Data are mean ± SD of three independent experiments.
To examine the impact of 3 and 4 on DNA replication, a DNA fiber assay on treated cells followed by sequential CldU and IdU labeling was performed.47 In this manner, treating MDA-MB-231 cells with 4 for 1 h results in a marked decrease in both CldU and IdU tract lengths compared to control cells (Figure 5a), thereby providing direct evidence that 4 acts to decrease DNA replication fork speed and interferes with DNA replication. Furthermore, treatment with 4 resulted in substantial disruption to cell-cycle progression, with an increase in S-phase cells and an accompanying decrease in the G2/M phase compared to the control (Figure 5b). Examining cell fate, an increase in cells with sub-G1 DNA content and annexin V positive population indicate the induction of apoptosis by 4 (Figure 5b,c). These results are consistent with 4 inhibiting DNA replication, inducing S-phase arrest, and ultimately triggering cell death by apoptosis. Interestingly, and despite possessing comparable DNA-binding affinities and IC50 concentrations to 4, 3 did not impact DNA replication nor cell-cycle progression in a similar manner, a finding that implies the PIP ligand is required for the DNA replication inhibition shown by 4.
Figure 5.
(a) Left: a DNA fiber assay scheme. MDA-MB-231 cells were treated with the DMSO control, 3 or 4 (20 μM, 1 h), then pulse-labeled with CldU followed by IdU for 30 mins each. Examples of fiber images after immunofluorescence are shown. Right: the quantification of CldU and IdU tract lengths from at least 150 fibers per experimental condition, two independent repeats. (b) Left: the representative cell-cycle plots of MDA-MB-231 cells treated with the stated single-agent (10 μM) alone or in combination with Olaparib (10 μM) for 72 h, as determined by flow cytometry. Right: the quantification of cell-cycle phase distributions. Data were expressed as mean ± SD of two independent experiments (n = 2). (c) Left: the annexin V-FITC assay and representative dot plots of MDA-MB-231 cells treated with the stated single-agent (10 μM) alone or in combination with Olaparib (10 μM) for 72 h. Right: the quantification of apoptotic (annexin V positive) cells. Data were expressed as mean ± SD of two independent experiments (n = 2).
Synergy with PARP Inhibitor Olaparib
Finally, RPCs that target DNA can achieve synergistic cancer cell killing with the PARP inhibitor Olaparib, a strategy that can enhance both cytotoxicity and cancer selectivity.48 Cotreatment of the panel of cancer cell lines with 2, 3, or 4 alongside a low dose (10 μM) of the PARP inhibitor Olaparib resulted in enhanced cytotoxicity of each complex compared to single-agent conditions (Figures S19 and S20). Derived IC50 concentrations and combination indices (CIs) provided evidence of a synergistic relationship between the two RPCs and Olaparib in MDA-MB-231 cells (Table S4 and Figure S21). Briefly examining the mechanism of synergy, MDA-MB-231 cells treated with 3 or 4 alongside Olaparib showed an increase in sub-G1 content and annexin V positive cells compared to single-agent conditions (Figure 5b,c). These results indicate elevated apoptosis as a result of each combination in a similar manner to previous work utilizing the structurally-related PARP hypersensitizer [Ru(dppz)2(PIP)]2+.49 That 3 and 4 demonstrate evidence of PARPi synergy is consistent with both molecules inducing DNA damage; however, when combined with the results above, this implies distinct mechanistic differences between the two complexes: 4 most likely achieves PARPi synergy via stalling DNA replication forks while 3 operates by an as yet unknown mechanism of DNA damage generation. Future work will explore this in more detail.
Conclusions
In conclusion, we have developed a FRET-based method that can be employed to examine the ability of RPCs to bind Cy5.5-labeled DNA. We show that MLCT-Cy5.5 FRET is compatible with DNA-binding RPCs with a range of MLCT-emissive properties and numerous DNA structures and sequences, where it may be used to quantify structure- and sequence-specific binding together in a single rapid assay. The advantages of MLCT-Cy5.5 FRET identified include a high signal-to-noise ratio, wavelength shifting and intensity enhancement of “light switch” emission, and the generation of a DNA-dependent FRET peak for non-“light switch” complexes. Furthermore, as the binding molecule is the FRET donor and the target DNA is the FRET acceptor, the derived proximity of the binding FRET pair provides an indication of the binding site. Employing this method, we identify a new mismatch-interactive RPC and show that molecules characterized by this method possess encouraging anticancer activity, including evidence of DNA replication inhibition and synergy with the PARP inhibitor Olaparib.
Acknowledgments
This work was supported by the Welsh Government and a Sêr Cymru Strategic Partner Acceleration Award 80761-SU-242 as well as the Royal Society of Chemistry (RSC) Research Fund and Research Enablement grants R20-8717 and E21-9540096197.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c11111.
Experimental section; characterization data for 3 and 4; photophysical properties of 3 and 4 as PF6- salts in acetonitrile; ethidium bromide displacement assay; emission spectra for compounds with DNA; additional dose-response curves; cell viability data; selectivity indices; IC50 values of 2, 3, or 4 in combination with Olaparib. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Gill M. R.; Thomas J. A. Ruthenium(II) Polypyridyl Complexes and DNA - from Structural Probes to Cellular Imaging and Therapeutics. Chem. Soc. Rev. 2012, 41, 3179–3192. 10.1039/c2cs15299a. [DOI] [PubMed] [Google Scholar]
- Poynton F. E.; Bright S. A.; Blasco S.; Williams D. C.; Kelly J. M.; Gunnlaugsson T. The Development of Ruthenium(II) Polypyridyl Complexes and Conjugates for in Vitro Cellular and in Vivo Applications. Chem. Soc. Rev. 2017, 46, 7706–7756. 10.1039/C7CS00680B. [DOI] [PubMed] [Google Scholar]
- Mari C.; Pierroz V.; Ferrari S.; Gasser G. Combination of Ru(II) Complexes and Light: New Frontiers in Cancer Therapy. Chem. Sci. 2015, 6, 2660–2686. 10.1039/C4SC03759F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. E.; Chambron J. C.; Sauvage J. P.; Turro N. J.; Barton J. K. A Molecular Light Switch for DNA: Ru(Bpy)2(Dppz)2+. J. Am. Chem. Soc. 1990, 112, 4960–4962. 10.1021/ja00168a052. [DOI] [Google Scholar]
- Zeglis B. M.; Pierre V. C.; Barton J. K. Metallo-Intercalators and Metallo-Insertors. Chem. Commun. 2007, 44, 4565–4579. 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüba E.; Hart J. R.; Barton J. K. [Ru(Bpy)2(L)]Cl2: Luminescent Metal Complexes That Bind DNA Base Mismatches. Inorg. Chem. 2004, 43, 4570–4578. 10.1021/ic0499291. [DOI] [PubMed] [Google Scholar]
- Song H.; Kaiser J. T.; Barton J. K. Crystal Structure of Δ-[Ru(Bpy)2dppz]2+ Bound to Mismatched DNA Reveals Side-by-Side Metalloinsertion and Intercalation. Nat. Chem. 2012, 4, 615–620. 10.1038/nchem.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton A. N.; Marcélis L.; Barton J. K. [Ru(Me4phen)2dppz]2+, a Light Switch for DNA Mismatches. J. Am. Chem. Soc. 2016, 138, 5020–5023. 10.1021/jacs.6b02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton A. N.; Marcélis L.; McConnell A. J.; Barton J. K. A Ruthenium(II) Complex as a Luminescent Probe for DNA Mismatches and Abasic Sites. Inorg. Chem. 2017, 56, 8381–8389. 10.1021/acs.inorgchem.7b01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett C. A.; Barton J. K. Methods to Explore Cellular Uptake of Ruthenium Complexes. J. Am. Chem. Soc. 2007, 129, 46–47. 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D.; von Hippel P. H. Theoretical Aspects of DNA-Protein Interactions: Co-Operative and Non-Co-Operative Binding of Large Ligands to a One-Dimensional Homogeneous Lattice. J. Mol. Biol. 1974, 86, 469–489. 10.1016/0022-2836(74)90031-X. [DOI] [PubMed] [Google Scholar]
- Rajput C.; Rutkaite R.; Swanson L.; Haq I.; Thomas J. A. Dinuclear Monointercalating Ru(II) Complexes That Display High Affinity Binding to Duplex and Quadruplex DNA. Chem. - Eur. J. 2006, 12, 4611–4619. 10.1002/chem.200501349. [DOI] [PubMed] [Google Scholar]
- Zeng L.; Chen Y.; Liu J.; Huang H.; Guan R.; Ji L.; Chao H. Ruthenium(II) Complexes with 2-Phenylimidazo[4,5-f][1,10]Phenanthroline Derivatives That Strongly Combat Cisplatin-Resistant Tumor Cells. Sci. Rep. 2016, 6, 19449 10.1038/srep19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierroz V.; Joshi T.; Leonidova A.; Mari C.; Schur J.; Ott I.; Spiccia L.; Ferrari S.; Gasser G. Molecular and Cellular Characterization of the Biological Effects of Ruthenium(II) Complexes Incorporating 2-Pyridyl-2-Pyrimidine-4-Carboxylic Acid. J. Am. Chem. Soc. 2012, 134, 20376–20387. 10.1021/ja307288s. [DOI] [PubMed] [Google Scholar]
- Ramu V.; Gill M. R.; Jarman P. J.; Turton D.; Thomas J. A.; Das A.; Smythe C. A Cytostatic Ruthenium(II)–Platinum(II) Bis(Terpyridyl) Anticancer Complex That Blocks Entry into S Phase by up-Regulating P27KIP1. Chem. - Eur. J. 2015, 21, 9185–9197. 10.1002/chem.201500561. [DOI] [PubMed] [Google Scholar]
- Mårtensson A. K. F.; Lincoln P. Competitive DNA Binding of Ru(Bpy)2dppz2+ Enantiomers Studied with Isothermal Titration Calorimetry (ITC) Using a Direct and General Binding Isotherm Algorithm. Phys. Chem. Chem. Phys. 2018, 20, 7920–7930. 10.1039/C7CP03184J. [DOI] [PubMed] [Google Scholar]
- Algar W. R.; Hildebrandt N.; Vogel S. S.; Medintz I. L. FRET as a Biomolecular Research Tool — Understanding Its Potential While Avoiding Pitfalls. Nat. Methods 2019, 16, 815–829. 10.1038/s41592-019-0530-8. [DOI] [PubMed] [Google Scholar]
- Krusiński T.; Ożyhar A.; Dobryszycki P. Dual FRET Assay for Detecting Receptor Protein Interaction with DNA. Nucleic Acids Res. 2010, 38, e108. 10.1093/nar/gkq049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J. W.; Warnasooriya C.; Kondo Y.; Nagai K.; Rueda D. Assembly and Dynamics of the U4/U6 Di-SnRNP by Single-Molecule FRET. Nucleic Acids Res. 2015, 43, 10963–10974. 10.1093/nar/gkv1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieb A. R.; D’Arcy S.; Kramer M. A.; White A. E.; Luger K. Fluorescence Strategies for High-Throughput Quantification of Protein Interactions. Nucleic Acids Res. 2012, 40, e33. 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renčiuk D.; Zhou J.; Beaurepaire L.; Guédin A.; Bourdoncle A.; Mergny J.-L. A FRET-Based Screening Assay for Nucleic Acid Ligands. Methods 2012, 57, 122–128. 10.1016/j.ymeth.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Karpenko I. A.; Margathe J.-F.; Rodriguez T.; Pflimlin E.; Dupuis E.; Hibert M.; Durroux T.; Bonnet D. Selective Nonpeptidic Fluorescent Ligands for Oxytocin Receptor: Design, Synthesis, and Application to Time-Resolved FRET Binding Assay. J. Med. Chem. 2015, 58, 2547–2552. 10.1021/jm501395b. [DOI] [PubMed] [Google Scholar]
- Hounsou C.; Margathe J.-F.; Oueslati N.; Belhocine A.; Dupuis E.; Thomas C.; Mann A.; Ilien B.; Rognan D.; Trinquet E.; Hibert M.; Pin J.-P.; Bonnet D.; Durroux T. Time-Resolved FRET Binding Assay to Investigate Hetero-Oligomer Binding Properties: Proof of Concept with Dopamine D1/D3 Heterodimer. ACS Chem. Biol. 2015, 10, 466–474. 10.1021/cb5007568. [DOI] [PubMed] [Google Scholar]
- Barnoin G.; Shaya J.; Richert L.; Le H.-N.; Vincent S.; Guérineau V.; Mély Y.; Michel B. Y.; Burger A. Intermolecular Dark Resonance Energy Transfer (DRET): Upgrading Fluorogenic DNA Sensing. Nucleic Acids Res. 2021, 49, e72 10.1093/nar/gkab237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-J.; Wong K. M.-C.; Yi C.; Yam V. W.-W. New Ruthenium(II) Complexes Functionalized with Coumarin Derivatives: Synthesis, Energy-Transfer-Based Sensing of Esterase, Cytotoxicity, and Imaging Studies. Chem. - Eur. J. 2012, 18, 8724–8730. 10.1002/chem.201103025. [DOI] [PubMed] [Google Scholar]
- Bichenkova E. V.; Yu X.; Bhadra P.; Heissigerova H.; Pope S. J. A.; Coe B. J.; Faulkner S.; Douglas K. T. DNA Mismatch Detection by Resonance Energy Transfer between Ruthenium(II) and Osmium(II) Tris(2,2′-Bipyridyl) Chromophores. Inorg. Chem. 2005, 44, 4112–4114. 10.1021/ic050586w. [DOI] [PubMed] [Google Scholar]
- Martí A. A.; Puckett C. A.; Dyer J.; Stevens N.; Jockusch S.; Ju J.; Barton J. K.; Turro N. J. Inorganic–Organic Hybrid Luminescent Binary Probe for DNA Detection Based on Spin-Forbidden Resonance Energy Transfer. J. Am. Chem. Soc. 2007, 129, 8680–8681. 10.1021/ja0717257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R.; Piszczek G.; Kang J. S. On the Possibility of Long-Wavelength Long-Lifetime High-Quantum-Yield Luminophores. Anal. Biochem. 2001, 288, 62–75. 10.1006/abio.2000.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. S.; Piszczek G.; Lakowicz J. R. Enhanced Emission Induced by FRET from a Long-Lifetime, Low Quantum Yield Donor to a Long-Wavelength, High Quantum Yield Acceptor. J. Fluoresc. 2002, 12, 97–103. 10.1023/A:1015375622992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan N. N. M. Y.; Idris A.; Abidin Z. H. Z.; Tajuddin H. A.; Abdullah Z. White Light Employing Luminescent Engineered Large (Mega) Stokes Shift Molecules: A Review. RSC Adv. 2021, 11, 13409–13445. 10.1039/D1RA00129A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duskova K.; Lamarche J.; Amor S.; Caron C.; Queyriaux N.; Gaschard M.; Penouilh M.-J.; de Robillard G.; Delmas D.; Devillers C. H.; Granzhan A.; Teulade-Fichou M.-P.; Chavarot-Kerlidou M.; Therrien B.; Britton S.; Monchaud D. Identification of Three-Way DNA Junction Ligands through Screening of Chemical Libraries and Validation by Complementary in Vitro Assays. J. Med. Chem. 2019, 62, 4456–4466. 10.1021/acs.jmedchem.8b01978. [DOI] [PubMed] [Google Scholar]
- Mitteaux J.; Lejault P.; Wojciechowski F.; Joubert A.; Boudon J.; Desbois N.; Gros C. P.; Hudson R. H. E.; Boulé J.-B.; Granzhan A.; Monchaud D. Identifying G-Quadruplex-DNA-Disrupting Small Molecules. J. Am. Chem. Soc. 2021, 143, 12567–12577. 10.1021/jacs.1c04426. [DOI] [PubMed] [Google Scholar]
- Vuong S.; Stefan L.; Lejault P.; Rousselin Y.; Denat F.; Monchaud D. Identifying Three-Way DNA Junction-Specific Small-Molecules. Biochimie 2012, 94, 442–450. 10.1016/j.biochi.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Wilson T.; Costa P. J.; Félix V.; Williamson M. P.; Thomas J. A. Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence. J. Med. Chem. 2013, 56, 8674–8683. 10.1021/jm401119b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.; Geng X.; Zhao J.; Yao T.; Wang C.; Yang D.; Zheng L.; Ji L. Interaction of [Ru(Bpy)2(Dppz)]2+ with Human Telomeric DNA: Preferential Binding to G-Quadruplexes over i-Motif. Biochimie 2010, 92, 370–377. 10.1016/j.biochi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- McQuaid K.; Abell H.; Gurung S. P.; Allan D. R.; Winter G.; Sorensen T.; Cardin D. J.; Brazier J. A.; Cardin C. J.; Hall J. P. Structural Studies Reveal Enantiospecific Recognition of a DNA G-Quadruplex by a Ruthenium Polypyridyl Complex. Angew. Chem., Int. Ed. 2019, 58, 9881–9885. 10.1002/anie.201814502. [DOI] [PubMed] [Google Scholar]
- Waller Z. A. E.; Sewitz S. A.; Hsu S.-T. D.; Balasubramanian S. A Small Molecule That Disrupts G-Quadruplex DNA Structure and Enhances Gene Expression. J. Am. Chem. Soc. 2009, 131, 12628–12633. 10.1021/ja901892u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Haynes C. J. E.; Kieffer M.; Greenfield J. L.; Greenhalgh R. D.; Nitschke J. R.; Keyser U. F. FeII4L4 Tetrahedron Binds to Nonpaired DNA Bases. J. Am. Chem. Soc. 2019, 141, 11358–11362. 10.1021/jacs.9b03566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Luo Y.; Tran T.; Haniff H. S.; Nakai Y.; Fallahi M.; Martinez G. J.; Childs-Disney J. L.; Disney M. D. Defining RNA–Small Molecule Affinity Landscapes Enables Design of a Small Molecule Inhibitor of an Oncogenic Noncoding RNA. ACS Cent. Sci. 2017, 3, 205–216. 10.1021/acscentsci.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. L.; McKeague M.; Liang J. C.; Smolke C. D. Kinetic and Equilibrium Binding Characterization of Aptamers to Small Molecules Using a Label-Free, Sensitive, and Scalable Platform. Anal. Chem. 2014, 86, 3273–3278. 10.1021/ac5001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew-Fenn R. S.; Das R.; Harbury P. A. B. Remeasuring the Double Helix. Science 2008, 322, 446–449. 10.1126/science.1158881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A.; Jones N.; Arat N. Ö.; Fishel R.; Griffith J. D. Long Repeating (TTAGGG) (n) Single-Stranded DNA Self-Condenses into Compact Beaded Filaments Stabilized by G-Quadruplex Formation. J. Biol. Chem. 2018, 293, 9473–9485. 10.1074/jbc.RA118.002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. R.; Walker M. G.; Able S.; Tietz O.; Lakshminarayanan A.; Anderson R.; Chalk R.; El-Sagheer A. H.; Brown T.; Thomas J. A.; Vallis K. A. An 111In-Labelled Bis-Ruthenium(II) Dipyridophenazine Theranostic Complex: Mismatch DNA Binding and Selective Radiotoxicity towards MMR-Deficient Cancer Cells. Chem. Sci. 2020, 11, 8936–8944. 10.1039/D0SC02825H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. R.; Harun S. N.; Halder S.; Boghozian R. A.; Ramadan K.; Ahmad H.; Vallis K. A. A Ruthenium Polypyridyl Intercalator Stalls DNA Replication Forks, Radiosensitizes Human Cancer Cells and Is Enhanced by Chk1 Inhibition. Sci. Rep. 2016, 6, 31973 10.1038/srep31973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. R.; Jarman P. J.; Halder S.; Walker M. G.; Saeed H. K.; Thomas J. A.; Smythe C.; Ramadan K.; Vallis K. A. A Three-in-One-Bullet for Oesophageal Cancer: Replication Fork Collapse, Spindle Attachment Failure and Enhanced Radiosensitivity Generated by a Ruthenium(Ii) Metallo-Intercalator. Chem. Sci. 2018, 9, 841–849. 10.1039/C7SC03712K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzschneider U.; Niesel J.; Ott I.; Gust R.; Alborzinia H.; Wölfl S. Cellular Uptake, Cytotoxicity, and Metabolic Profiling of Human Cancer Cells Treated with Ruthenium(II) Polypyridyl Complexes [Ru(Bpy)2(N-N)]Cl2 with N-N = bpy, Phen, Dpq, Dppz, and Dppn. ChemMedChem 2008, 3, 1104–1109. 10.1002/cmdc.200800039. [DOI] [PubMed] [Google Scholar]
- Bennett L. G.; Wilkie A. M.; Antonopoulou E.; Ceppi I.; Sanchez A.; Vernon E. G.; Gamble A.; Myers K. N.; Collis S. J.; Cejka P.; Staples C. J. MRNIP Is a Replication Fork Protection Factor. Sci. Adv. 2022, 6, eaba5974 10.1126/sciadv.aba5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusoh N. A.; Ahmad H.; Gill M. R. Combining PARP Inhibition with Platinum, Ruthenium or Gold Complexes for Cancer Therapy. ChemMedChem 2020, 15, 2121–2135. 10.1002/cmdc.202000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusoh N. A.; Leong S. W.; Chia S. L.; Harun S. N.; Rahman M. B. A.; Vallis K. A.; Gill M. R.; Ahmad H. Metallointercalator [Ru(dppz)2(PIP)]2+ Renders BRCA Wild-Type Triple-Negative Breast Cancer Cells Hypersensitive to PARP Inhibition. ACS Chem. Biol. 2020, 15, 378–387. 10.1021/acschembio.9b00843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.