Abstract
The complete nucleotide sequence and organization of the Yersinia enterocolitica serotype 0:8 low-calcium-response (LCR) plasmid, pYVe8081, were determined. The 67,720-bp plasmid encoded all the genes known to be part of the LCR stimulon except for ylpA. Eight of 13 intact open reading frames of unknown function identified in pYVe8081 had homologues in Yersinia pestis plasmid pCD1 or in Y. enterocolitica serotype 0:9 plasmid pYVe227. A region of approximately 17 kbp showed no DNA identity to pCD1 or pYVe227 and contained six potential new genes, a possible new replicon, and two intact insertion sequence (IS) elements. One intact IS element, ISYen1, was a new IS belonging to the IS256 family. Several vestigial IS elements appeared different from the IS distribution seen in the other LCR plasmids. The RepA proteins encoded by Y. enterocolitica serotype 0:8 pYVeWA and pYVe8081 were identical. The putative pYVe8081 replicon showed significant homology to the IncL/M replicon of pMU407.1 but was only distantly related to the replicons of pCD1 and pYVe227. In contrast, the putative partitioning genes of pYVe8081 showed 97% DNA identity to the spy/sopABC loci of pCD1 and pYVe227. Sequence analysis suggests that Yersinia LCR plasmids are from a common ancestor but that Y. enterocolitica serotype 0:8 plasmid replicons may have evolved independently via cointegrate formation following a transposition event. The change in replicon structure is predicted to change the incompatibility properties of Y. enterocolitica serotype 0:8 plasmids from those of Y. enterocolitica serotype 0:9 and Y. pestis LCR plasmids.
Pathogenic Yersinia enterocolitica is a well-established food-borne pathogen (29). Infection usually results in a self-limiting gastroenteritis, but in immunocompromised individuals septicemia and hepatic abscesses may occur. Postinfection complications include arthritis and erythema nodosum (8). Y. enterocolitica is a serologically diverse species that includes saprophytes as well as pathogens. Certain serotypes are consistently associated with human infection (30). Serotypes 0:3 and 0:9 are most frequently isolated in Europe, Japan, and Canada, while serotype 0:8 causes most infections in the United States. Serotype 0:8 is also associated with more severe invasive disease (3, 5). So far, only one phenotypic trait has been identified in Y. enterocolitica 0:8 to account for these observed differences in pathogenicity (37).
Essential virulence genes are carried on a ca.-70-kb plasmid in Y. enterocolitica, Yersinia pestis, and Yersinia pseudotuberculosis (42, 43). The virulence plasmid encodes virulence proteins called Yops (Yersinia outer proteins), a type III secretion system, the V antigen, and regulatory proteins. The virulence plasmid encodes the low-calcium response (LCR) (53), which refers to a complex response to in vitro growth conditions of temperature (37°C) and extracellular calcium concentration (less than 2.5 mM Ca2+). Under these conditions, pathogenic Yersinia shifts from vegetative growth to the production and secretion of virulence proteins. In vitro conditions probably mimic a signal in the mammalian host where the LCR results in paralysis of defenses at the site of infection and extreme suppression of cell-mediated immunity (6). Collectively, plasmid genes turned on by the LCR comprise the LCR stimulon (38, 52).
The LCR plasmids in Yersinia spp. are structurally, as well as functionally, related. Yop secretion involves 28 genes at four adjacent loci, virA, virB, virG, and virC. The virA locus consists of seven genes: yopN, tyeA, sycN, yscXY, lcrD/yscV, and lcrR. The virB operon is comprised of eight genes (yscN to yscU), and the virC operon contains 13 genes, yscA to yscM. virG (yscW in Y. pestis [39]) is a small monocistronic gene located between virB and the transcriptional regulator virF. Contiguous with virA is the lcrGVsycDyopBD operon, which is involved in translocation of Yops (21). The lcrV gene in this operon encodes the V antigen that is required for virulence. These genes form a contiguous cluster in all Yersinia LCR plasmids. Other effector genes (yopM, yopT, yopQ, yopP, yopO, yopE, and yopH) and their corresponding chaperones (sycT, sycE, and sycH) flank the main cluster.
Only virF, sycE, yopE, and yadA have been sequenced from Y. enterocolitica 8081 serotype 0:8. The predicted products of yopE, sycE, and virF are at least 95% identical to homologous proteins produced by the other pathogenic Yersinia strains. However, YadA encoded by Y. enterocolitica 8081 shows only 81% identity to YadA encoded by Y. enterocolitica serotype 0:9. Based on the results of DNA cross-hybridization studies, plasmids from Y. enterocolitica serogroups 0:9, 0:3, and 0:5 show 90% nucleotide identity with one another but share only 75% DNA identity with plasmids from Y. enterocolitica serogroup 0:8 (19). Plasmids from Y. enterocolitica serotype 0:8 show 55% nucleotide identity with the virulence plasmids from Y. pestis and Y. pseudotuberculosis (43). Taken together, these facts suggest that the LCR stimulon evolved as a cluster but that other parts of the LCR plasmid were able to evolve independently.
Recently, the LCR plasmids from Y. pestis (designated pCD1) and Y. enterocolitica serotype 0:9 (designated pYVe227) have been sequenced and analyzed. These comparative studies have provided useful clues to the evolution of these plasmids that are critical for virulence of members of the genus Yersinia and as well have aided in the identification of potential new virulence determinants (20, 21, 39). In this paper, we describe the nucleotide sequence of Y. enterocolitica serotype 0:8 LCR plasmid pYVe8081 and its relationship to the other known Yersinia LCR plasmids. Our analysis has revealed a potential new replicon and has increased our knowledge pertaining to the evolution of this important virulence determinant.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Y. enterocolitica 8081 (serotype 0:8) was used for the isolation of the pYVe8081 plasmid. Y. enterocolitica WA (serotype 0:8) was obtained from Peter Feng, Food and Drug Administration, Washington, D.C. Y. enterocolitica 8081 was obtained from two sources, Virginia Miller at Washington University, St. Louis, Mo., and Peter Feng.
Library construction, sequencing, and PCR.
Plasmid DNA was prepared from bacteria using a plasmid isolation kit (Qiagen, Santa Clarita, Calif.) according to the manufacturer's specifications. Y. enterocolitica was cultured in brain heart infusion broth (BD Biosciences, San Jose, Calif.) at 30°C. Plasmid DNA was extracted from Escherichia coli containing libraries of cloned pYVe8081 restriction fragments after the strains were grown overnight at 37°C in Luria broth (47). Separate libraries were prepared from ApoI-, BamHI-, HindIII-, and EcoRI-digested pYVe8081 DNA in vector pSK+ (Stratagene, La Jolla, Calif.) as described elsewhere (47). DNA templates were purified from random library clones and sequenced using Prism dye terminator (FS) labeled fluorescent cycle sequencing kits (Applied Biosystems, Foster City, Calif.) and an ABI 377XL automated sequencer (Applied Biosystems). Sequences were edited and assembled using Sequencher 3.0 software (Gene Codes Corp., Ann Arbor, Mich.). Gaps between contiguous sequences were amplified by PCR using the original plasmid DNA as template followed by sequencing of the PCR products. PCRs were performed with Hot Tub DNA polymerase (Amersham Corp., Arlington Heights, Ill.) following optimization of PCR conditions using the PCR Optimizer kit (Invitrogen, Carlsbad, Calif.). A ca.-10.5-kbp DNA segment located between yadA and yopO (see Fig. 1) was PCR amplified using the Expand Long Template PCR system (Roche Molecular Biochemicals, Indianapolis, Ind.) and sequenced by Fred Blattner's group (University of Wisconsin, Madison). Sequencing of the ca.-10.5-kbp PCR fragment was performed by random shearing and cloning as previously described (4, 26). In all cases, disagreements between our results and those of other laboratories were analyzed by PCR amplification and sequencing of the resultant products. Final assembly was confirmed by PCR amplification and restriction enzyme digestion of pYVe8081 to ensure accuracy of our assembly. The putative repA gene in Y. enterocolitica WA (serotype 0:8) was PCR amplified using forward primer CCGCCCAAAATGAGTGTG, located upstream from repA in pYVe8081, and reverse primer GGTTAGGAATACTTTGCGGC, located downstream from repA in pYVe8081. The PCR product was purified with QIAquick PCR purification columns (Qiagen) to remove the primers and then sequenced, as described above.
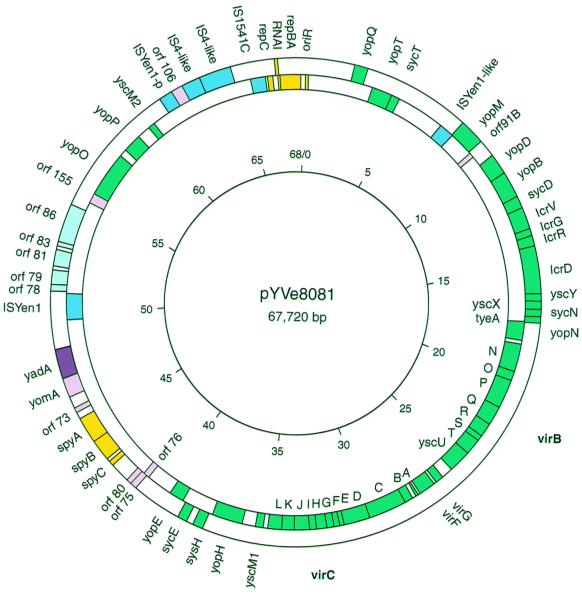
FIG. 1.
Map of pYVe8081 showing significant genes, IS elements, and replication and partition regions. The direction of transcription is clockwise for genes shown inside the circle and counterclockwise for genes shown outside the circle. Green boxes indicate genes comprising the LCR stimulon, and a purple box indicates yadA. Yellow boxes indicate genes with replication and partition functions. Pink boxes indicate previously identified genes of unknown function, and potential new genes are indicated by light blue boxes. IS elements are indicated by dark blue boxes. IS remnants shown in this figure are discussed in the text. IS-like indicates IS element remnants with less than 90% DNA identity to the GenBank match. ISYen-p is a truncated homologue of ISYen1. The positions of the virB operon (yscN to yscU) and the virC operon (yscA to yscM1) are noted in boldface on the outside of the circle. The inner circle shows the scale in kilobase pairs.
Sequence analysis.
Open reading frames (ORFs) capable of encoding peptides at least 50 amino acids long were identified using Sequencher 3.0, DNAStar (Lasergene, Madison, Wis.), and ORF Finder from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/gorf/gorf.html). In the absence of a significant homologue included in GenBank, the start codon giving the longest ORF was used. Putative ORFs were then selected by a combination of GenBank matches using BLASTX (1) and the presence of a potential ribosome-binding site (59). We searched for protein function identification using the ISREC Profile Scan Server (www.isrec.isb-sib.ch/software/PFSCAN_form.html) and eMOTIF Search (dna.Stanford.EDU/identify/).
Nucleotide sequence accession number.
The annotated sequence was deposited in GenBank under accession no. AF336309, AY026194, and AY026195.
RESULTS AND DISCUSSION
Molecular arrangement of pYVe8081.
The entire circular DNA sequence of pYVe8081 was 67,720 bp in length and was similar to the previously determined size of 66,000 bp (41). Significant ORFs, sites, and insertion sequence (IS) elements are shown in Fig. 1 and summarized in Table 1. All of the previously identified virulence-associated genes were present in pYVe8081 except for ylpA, which is expressed only in pYVe227 (20, 21, 39). The virABC loci and the lcrGVH-yopBD operon showed 98 to 99% nucleotide identity to these loci in pCD1 and pYVe227; virG and virF were 97 to 98% identical in all three plasmids.
TABLE 1.
Features identified in Y. enterocolitica pYVe8081
| Genea | Position (bp)b | Size (amino acids) | Homologous gene by BLASTc | Functiond |
|---|---|---|---|---|
| oriR | 1–165 | Putative replication site | ||
| yopQ | c2291–2839 | 182 | AF102990 | Yop translocation control |
| yopT | 3352–4320 | 322 | AF102990 | Yop effector |
| sycT | 4320–4712 | 130 | AF102990 | YopT chaperone |
| yopM | c7723–8826 | 367 | AF102990 | Yop effector |
| orf 91B | 8949–9167 | 72 | AF080156 | Unknown |
| yopD | c9653–10573 | 306 | AF102990 | Yop translocator |
| yopB | c10592–11797 | 401 | AF102990 | Yop translocator |
| sycD | c11775–12281 | 168 | AF074612 | YopB, YopD chaperone |
| lcrV | c12284–13258 | 324 | AF102990 | YopB, YopD secretion |
| lcrG | c13260–13547 | 95 | M26405 | Control of Yop release |
| lcrR | c13589–14029 | 146 | AF102990 | Unknown |
| lcrD (yscV) | c14026–16140 | 704 | AF102990 | Yop secretion |
| yscY | c16127–16471 | 114 | AF102990 | Yop secretion |
| yscX | c16468–16836 | 122 | AF074612 | Yop secretion |
| sycN | c16833–17204 | 123 | AF102990 | YopN chaperone |
| tyeA | c17191–17469 | 92 | AF102990 | Control of Yop release |
| yopN | c17450–18331 | 293 | AF074612 | Control of Yop release |
| yscN | 18529–19848 | 439 | AF074612 | Yop secretion |
| yscO | 19845–20309 | 154 | AF074612 | Yop secretion |
| yscP | 20309–21670 | 453 | AF074612 | Yop secretion |
| yscQ | 21667–22590 | 307 | AF102990 | Yop secretion |
| yscR | 22587–23240 | 217 | AF102990 | Yop secretion |
| yscS | 23242–23508 | 88 | AF074612 | Yop secretion |
| yscT | 23505–24290 | 261 | AF074612 | Yop secretion |
| yscU | 24290–25354 | 354 | AF102990 | Yop secretion |
| virG (yscW) | 25930–26325 | 131 | AF102990 | YscC localization |
| virF | 26449–27264 | 271 | X96798 | Yop regulation |
| yscA | 27344–27442 | 32 | AF102990 | Unknown |
| yscB | 27667–28080 | 137 | AF102990 | Unknown |
| yscC | 28086–29909 | 607 | AF102990 | Yop secretion |
| yscD | 29906–31162 | 418 | AF102990 | Yop secretion |
| yscE | 31159–31362 | 67 | AF102990 | Yop secretion |
| yscF | 31359–31622 | 87 | AF102990 | Yop secretion |
| yscG | 31624–31971 | 115 | AF102990 | Yop secretion |
| yscH | 31968–32465 | 165 | AF102990 | Unknown |
| yscI | 32466–32813 | 115 | AF102990 | Yop secretion |
| yscJ | 32820–33554 | 244 | AF102990 | Yop secretion |
| yscK | 33554–34183 | 209 | AF102990 | Yop secretion |
| yscL | 34129–34800 | 223 | AF102990 | Yop secretion |
| yscM1 | 35019–35366 | 115 | AF102990 | Yop regulation |
| yopH | 36048–37454 | 468 | AF102990 | Yop effector |
| sycH | c37676–38107 | 143 | AF102990 | YopH chaperone |
| sycE | c38461–38853 | 130 | M34278 | YopE chaperone |
| yopE | 39046–39705 | 219 | M34278 | Yop effector |
| orf76 | 40996–41292 | 98 | AF074612 | Unknown |
| orf75 | c41131–41433 | 100 | AF074612 | Unknown |
| orf80 | c41426–41668 | 80 | AF056093 | Unknown |
| spyC | c42557–42781 | AF074612 | pYV partition site | |
| spyB | c42930–43892 | 320 | AF102990 | pYV partition protein |
| spyA | c43892–45058 | 388 | AF102990 | pYV partition protein |
| orf73 | c45372–45587 | 71 | T00231 | Unknown |
| yomA | c46108–46983 | 291 | AF056092 | Unknown |
| yadA | c47053–48321 | 422 | X13881 | Adhesin |
| ISYen1 | 49547–50841 | AF074611 | Unknown | |
| orf78 | c50936–51244 | 102 | No database match | Unknown |
| orf79 | c51244–51873 | 209 | AF302424 | Unknown |
| orf81 | c52048–52782 | 244 | AP001918 | Putative recombinase |
| orf83 | c52919–53116 | 65 | No database match | Unknown |
| orf86 | c54868–55134 | 88 | AF264951 | Unknown |
| orf155 | 55296–55727 | 143 | AF074612 | Putative Yop chaperone |
| yopO | 55745–57934 | 729 | AF102990 | Yop effector |
| yopP | 58301–59167 | 288 | AF102990 | Yop effector |
| yscM2 | 59786–60136 | 116 | AF102990 | Yop regulation |
| orf106 | c61616–62167 | 183 | AF074611 | Unknown |
| IS1541C | 65189–65896 | AJ238015 | Unknown | |
| repC | 65992–66237 | 81 | U27345 | Putative replication protein |
| RNAI | c66399–66481 | U27345 | Putative ctRNA | |
| repB | 66499–66555 | 18 | U27345 | Putative replication protein |
| repA | 66548–67570 | 340 | U27345 | Putative replication protein |
Y. enterocolitica terminology is used for the LCR stimulon genes; alternate gene designations are in parentheses. Intact IS elements and putative replication-partition features are included.
“c” denotes a gene encoded on the minus strand.
The accession number of the GenBank match with the highest probability value was used. In cases where the match with the highest probability value was found in more than one Yersinia spp., the Y. enterocolitica pYVe227 sequence was used.
Functions of LCR stimulon genes are from a review by Cornelis et al. (6).
A comparison of the plasmid maps of pCD1, pYVe227, and pYVe8081 revealed a similar organization of shared genes (Fig. 2). The following differences between pYVe227 and pYVe8081 are notable. yscM2 is located between yadA and the partition region (spyABC) in pYVe227 but was located upstream from yopP in pYVe8081. A block of inserted DNA containing putative replication genes flanked by IS-like elements in pYVe8081 interrupted the sequence between yopP and yopQ encoded by pYVe227. This inserted DNA replaced the ylpA gene encoded by pYVe227. Also, the orientation of the yomA/yadA genes and the yopM gene was reversed in pYVe8081.
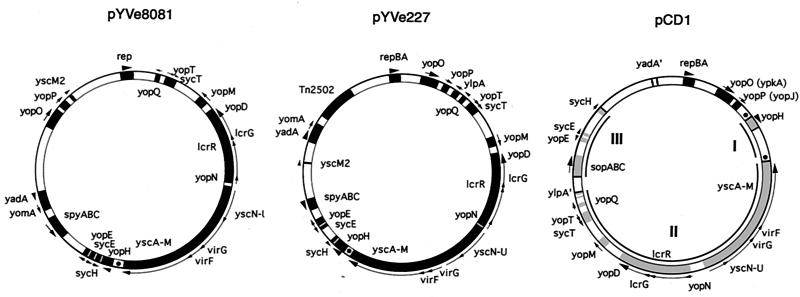
FIG. 2.
Comparison of LCR plasmid maps of pYVe8081, pYVe227, and pCD1. Only representative genes are shown, to facilitate orientation. The maps of pYVe227 (21) and pCD1 (39) are redrawn. Arrows outside the circles indicate the direction of transcription. YadA′ indicates the yadA pseudogene in pCD1, and Tn2502 is the arsenic resistance transposon in pYVe227. Regions in pCD1 involved in the inversions referred to in the text are shown in gray and numbered. The dots represent GTATT direct repeats in pYVe8081, pYVe227, and pCD1.
Gross differences in the organization of pCD1 and the other two plasmids result from the inversion of at least two regions (20, 21, 39). One region in pCD1 extends from the beginning of sopA to the end of sycH (shown as region III on the pCD1 map in Fig. 2). The other inverted region in pCD1 starts with the ylpA pseudogene and continues to the beginning of yopH (regions II and I in Fig. 2). The absence of mobile genetic elements near the yopH end of the ylpA-to-yopH region makes it difficult to explain how this segment may have inverted (21). Our comparison of LCR plasmid sequences suggests the possibility of a third inversion (region I in Fig. 2) that could explain the current orientation of the Yop gene cluster found on the Y. enterocolitica plasmids. Inversion of region I in pCD1 could have resulted from IS-promoted recombination near the yopH region. Specifically, Perry et al. (39) found a copy of IS100 near yopH on pCD1. As a consequence of this possible inversion, yopH and yscM would be next to each other and oriented in the same direction. If this event was followed by the inversion of the segment encompassing regions I and II, the orientation of the Yop gene cluster in pCD1 would be similar to that seen now in the Y. enterocolitica plasmids. As further support of mobile gene activity in this region, we found short direct repeats (GTATT) that may be vestiges of illegitimate recombination events in the ancestral LCR plasmid. In pCD1, two copies of this repeat are located just upstream of yopH and downstream of yscM (shown as dots flanking region I in Fig. 2). In contrast, Y. enterocolitica pYVe8081 and pYVe227 each have a single copy of the identical sequence between yopH and yscM. Although we cannot be certain of the molecular events that led to the current architecture seen in these three LCR plasmids, it is obvious from DNA sequence analysis that multiple mobile genetic events have participated in the construction of this virulence gene cluster.
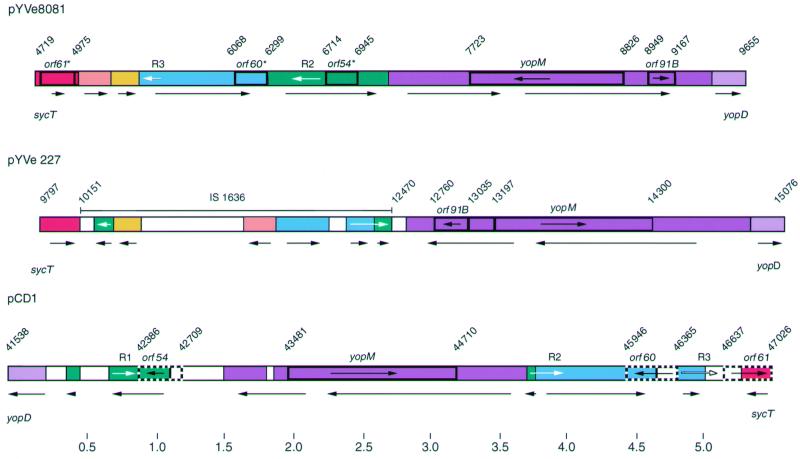
The molecular arrangement of pYVe8081 in the region between sycT and yopD was significantly different from that of both pYVe227 and pCD1 (Fig. 3). The sycT-to-yopD region was located on the opposite side of the Yop gene cluster in both Y. enterocolitica plasmids compared to the Y. pestis plasmid (Fig. 2) (21, 39). The yopM locus was transcribed in the same direction in pYVe8081, pCD1, and Y. pseudotuberculosis plasmid pIB1 but transcribed in the opposite direction in pYVe227. In addition, homologues of pCD1 (39) and pYVe227 (21) genes encoded by pYVe8081 were disrupted by these rearrangements. Specifically, pYVe8081 contained only remnants of the intact IS1636 located in the sycT-to-yopD region of pYVe227, as well as remnants of orf54, orf60, and orf61 identified in the same region on pCD1 (Fig. 3). These genetic rearrangements have resulted in size variation of the sycT-to-yopD region in all three plasmids. Iriarte and Cornelis noted rearrangements in this region between pCD1 and pYVe227 (21). They proposed that homologous exchange between long repeats could account for these differences. Despite these changes, the sycT-to-yopD region of pYVe8081 retained approximately 95% nucleotide identity with the homologous segments encoded by pCD1 and pYVe227.
FIG. 3.
Comparison of the sycT-yopD region in pYVe8081 to corresponding regions in pYVe227 and pCD1. The different colors represent different DNA segments. White segments are not present in pYVe8081. Arrows under the plasmid maps show the orientation of DNA segments. Numbers indicate nucleotide positions in base pairs for each plasmid. Sequences of pYVe227 and pCD1 are from the GenBank database (accession no. AF102990 and AF074612, respectively). Remnants orf61*, orf60*, and orf54* are truncated ORFs in pYVe8081 that show homology to intact genes (orf61, orf60, and orf54) in pCD1. Genes encoding YopM and ORF91B are also labeled. Arrows within genes show the direction of transcription. White arrows indicate the positions of the long repeated sequences R1, R2, and R3, referred to in the text. Only intact IS elements are shown.
In contrast, two regions of pYVe8081 showed no DNA homology to either pCD1 or pYVe227. One region of approximately 7 kbp extended upstream from yadA to orf155, and the other region of approximately 10 kbp extended downstream from yscM2 to yopQ. These two areas contained six potential new genes, a possible new replicon, two intact IS elements, and a number of vestigial IS elements (Fig. 1).
The translated products of most virulence-associated genes in pYVe8081 showed at least 92% identity to corresponding proteins encoded by pCD1, pIB1, and pYVe227. However, the pYVe8081-encoded yscP gene product was only 78% identical to its homologue from pYVe227. YscP encoded by pYVe8081 was 60 amino acids shorter (51). The absent amino acids form a tandem duplication of amino acid residues 261 to 320 in the Y. enterocolitica 0:9 YscP protein. The duplicated amino acids are also missing from the predicted YscP proteins produced by Y. pseudotuberculosis and Y. pestis (36). Thus, YscP encoded by pYVe8081 was more closely related to those of Y. pestis and Y. pseudotuberculosis than to that of Y. enterocolitica 0:9. The only other identity less than 92% that we noted between the predicted protein sequences of any of the LCR stimulon-encoded proteins was within YopM. The difference in YopM was due in part to a difference in the number of internal leucine-rich repeats (LRR). YopM from both pYVe8081 and pYVe227 is 367 amino acids long and contains 13 LRRs compared to the 15 LRRs encoded by the pCD1 allele. As a result, the Y. pestis protein is 409 amino acids long and shows 81% identity with pYVe8081 YopM.
Putative ORFs of unknown function in pYVe8081.
Consistent with the strong relatedness shown by the Yop stimulon genes, putative ORFs of unknown function located between yopQ and yadA in pYVe8081 had homologues in pCD1 or pYVe227 (Fig. 1; Table 2). Outside this region, orf155 (bp 55745 to 57934) was homologous to previously identified potential syc genes located upstream from yopO in pIB1, pCD1, and pYVe227 (15, 21, 39). Located between IS remnants downstream from yscM2, orf106 encoded a putative inner membrane protein which belongs to the phospholipase D superfamily (40). ORF106 was 43% identical (63% similar) over a 123-amino-acid span with a 162-amino-acid-long putative endonuclease in Y. pestis pMT1 (24) and 41% identical over an 82-amino-acid span with a 180-amino-acid-long endonuclease in pYVe227 (21). ORF181, ORF91A, ORF156, and PprA, previously identified in pYVe227 (21), were not encoded by pYVe8081. ORF5, ORF84, and ORF85, previously identified in pCD1 (39), were not encoded by pYVe8081.
TABLE 2.
Putative genes of unknown function in pYVe8081
| Gene | Position (bp)a | Shine-Dalgarno sequence and start codon | Homologous protein by BLAST (target) | % Homologyb | Accession no. (reference) |
|---|---|---|---|---|---|
| orf91B | 8949–9167 | AGAAGN4ATG | Unknown protein from pYVe227 | 91/75 | AF080156 (21) |
| orf76 | 40996–41292 | AGGACGN4ATG | Unknown protein from pCD1 | 93/100 | AF074612 (39) |
| orf75 | c41131–41433 | GGTGN10GTG | Unknown protein from pCD1 | 94/100 | AF074612 (39) |
| orf80 | c41426–41668 | GGAGGN4ATG | Unknown protein from pYVe227 | 100/100 | AF056093 (21) |
| orf73 | c45372–45587 | AGGAGN6ATG | No database match | ||
| yomA | c46108–46983 | GGTGN10ATG | Unknown protein (YomA) from pYVe227 | 97/100 | AF056092 (21) |
| orf78 | c50936–51244 | GGAGGN3ATG | No database match | ||
| orf79 | c51244–51873 | AGGTGN5ATG | Partition protein from Actinobacillus actinomycetemcomitans | 26/99 | AF302424 |
| orf81 | c52048–52782 | GCGGAN7ATG | Possible site-specific recombinase on the F plasmid | 50/99 | AP001918 |
| orf83 | c52919–53116 | GGAGCN10ATG | No database match | ||
| orf86 | c54868–55134 | AGGAGGN5ATG | Unknown protein from Erwinia amylovora | 76/55 | AF264951 (28) |
| orf155 | 55296–55727 | AGAGN7GTG | Possible chaperone from pCD1 | 95/100 | AF074612 (39) |
| orf106 | c61616–62167 | GGCAN2ATG | Possible endonuclease from Y. pestis pMT1 | 43/76 | AF074611 (24) |
“c” denotes a gene encoded on the minus strand.
Percentage of identical amino acids compared with the percentage of the total target protein sequence.
The region between yadA and yopO contained five potential genes not found in pCD1 or pYVe227 (Fig. 1; Table 2). The translated product of the largest gene, orf81 (bp 52048 to 52782), was 244 amino acids long and was 50% identical over 99% of a probable site-specific recombinase encoded by the F plasmid (accession no. AP001918). A search of the pFam database revealed that ORF81 belongs to the phage integrase family of recombinases. Members include bacteriophage enzymes responsible for the integration of linear DNA into a host genome (18, 23). orf79 could encode a 211-amino-acid-long partition protein, since it was 26% identical over the entire length of an Actinobacillus actinomycetemcomitans partition protein. The Actinobacillus protein is a Walker-type ATPase from a type 1B partition locus. Type 1B partition loci are found in gram-positive and gram-negative bacteria (16). The finding of a remnant of a broad-host-range plasmid, specifically a protein associated with partitioning, suggests that pYVe8081 may have been generated by cointegration of distinct plasmids during its evolution.
IS elements.
Whole or partial IS elements, which represented seven IS element families (25), occupied 16% of the pYVe8081 plasmid (Table 3). In pCD1 and pYVe227, intact IS elements and most remnants belong to the IS3 family (21, 39). We considered IS-like sequences to be identical to previously identified IS elements if they showed greater than 90% nucleic acid identity or greater than 95% transposase (Tpase) amino acid identity according to the classification scheme of Mahillon and Chandler (25). The remaining remnants were named after the Tpase giving the highest GenBank match at the amino acid level.
TABLE 3.
IS elements in pYVe8081
| IS element homologya | Accession no./organism | Position (bp) | Gene(s) present | Terminal repeats |
|---|---|---|---|---|
| IS1541C | AJ238015/Y. enterocolitica | 65189–65896 | Tpase | 2 target junctions |
| ISYen1 | AF074611/Y. pestis pMT1 | 49547–50841 | Tpase | 2 |
| ISYen1-partial | AF074611/Y. pestis pMT1 | 61626–61012 | Tpase-partial | 1 |
| ISYen1-like | AF074611/Y. pestis pMT1 | 7042–7717 | Tpase-partial | 2 |
| IS4-like | J01733/E. coli | 64438–63045 | Tpase-partial | 1 |
| IS4F-like | J01735/E. coli | 63044–62167 | Tpase-partial | 2 |
| IS1400 | AJ132945/Y. enterocolitica HPIb | 1207–1631 | orfA, orfB-partial | 1 |
| IS1400-like | AJ132945/Y. enterocolitica HPI | 48833–49484 | orfA-partial, orfB-partial | 1-partial |
| IS1400-like | AJ132945/Y. enterocolitica HPI | 53508–54155 | orfB-partial | None found |
| IS1353-like | AP000342/plasmid R100 | 587–1210 | orfA-partial | 1 |
| IS1353-like | AP000342/plasmid R100 | 1653–2138 | orfB-partial | 1 |
| IS1328-like | AJ132945/Y. enterocolitica HPI | 54574–54178 | Tpase-partial | 1 |
| IS1328-like | AJ132945/Y. enterocolitica HPI | 64446–65180 | Tpase-partial | 1 |
| IS1327-like | X87144/Erwinia herbicola | 42188–41782 | Tpase-partial | 1 |
| Tn1-like | L10085/plasmid RP4 | 60315–61006 | Tpase-partial | None found |
| ISpG5-like | AF224744/Porphyromonas gingivalis | 40159–39760 | Tpase-partial | None found |
| IS5377-like | X67862/Bacillus stearothermophius | 45731–45611 | Tpase-partial | None found |
| IS1477a-like | U62552/Xanthomonas campestris | 5174–5039 | orfB-partial | None found |
IS-like denotes an IS remnant with less than 90% DNA identity to the GenBank match. IS-partial denotes a truncated isoform.
HPI, high pathogenicity island.
Two IS elements in pYVe8081 were intact and therefore likely to be functional. We located an intact copy of IS1541C adjacent to the replication region (bp 65189 to 65896) in pYVe8081. The IS1541C element on the pYVe8081 plasmid was identical in size and sequence to IS1541C recently described but not mapped in Y. enterocolitica 8081 (12). IS1541C shows 94% DNA sequence identity to IS1541 located on the Y. pestis chromosome and on plasmid pMT1 and to isoforms IS1541A and IS1541B from Y. pseudotuberculosis, but only 85% DNA sequence identity to IS1541D from Y. enterocolitica 8081. We predict that IS1541D is located on the Y. enterocolitica chromosome, since no copies of it were found on Y. enterocolitica plasmids sequenced so far.
ISYen1 was a new 1,295-bp IS element located between yadA and yopO at bp 49547 to 50841. We designated this new element ISYen1 according to the nomenclature proposed by Mahillon and Chandler (25). We found 32-bp-long imperfect inverted repeats at both ends of ISYen1. The element was also flanked by 9-bp direct repeats that could have resulted from duplication of a target sequence at the site of insertion during transposition. We found a potential Shine-Dalgarno sequence and start codon, AGGAN7ATG, and a single ORF capable of encoding a 400-amino-acid long Tpase. The putative Tpase is a member of the transpo-mutator family of proteins, which includes the mutator element from maize (13). Tpases of the transpo-mutator protein family belong to the IS256 IS family (25). The ISYen1 Tpase showed 48% amino acid sequence identity over 92% of a Tpase from Y. pestis pMT1 (24) and 45% identity over 98% of ISRm3 from Rhizobium meliloti (61). The family shows an overall amino acid similarity of 22 to 44% across dissimilar genera, suggesting a common ancestor (25). However, members have also adapted to the G+C contents of their host species with a corresponding loss of nucleotide homology among family members. The group is of interest from an evolutionary standpoint because some of its members are highly mobile and therefore likely contributors to genome structure and evolution. Support for a common ancestor of these elements was also found in the sequence of the terminal inverted repeats in ISYen1, which were 53 to 63% identical to terminal repeats from IS285 (46), ISRm3, IS1132 (accession no. P35879), and IS1356 (57).
Evidence for the mobile nature of ISYen1 was obtained through sequence analysis. We found two disrupted elements homologous to ISYen1 on the plasmid. One remnant, located downstream from yscM2 (bp 61626 to 61012), was identical in nucleotide sequence to ISYen1 beginning with 8 bp of the direct repeat and including the left inverted repeat and the first 558 bp of the Tpase. The other remnant (bp 7042 to 7717) was 676 bp long and showed 82% nucleotide identity to ISYen1. Other IS elements in pYVe8081 are not likely to function, since their DNA sequences were either truncated at one or both ends or interrupted by the insertion of other IS elements or by deletions. The presence of many IS elements in a state of evolutionary decay in pYVe8081 suggests a history of gene transfer between this plasmid and plasmids from other strains and species as well as the chromosome. For example, a tandem duplication of IS4-like elements occupied 2.2 kbp upstream from IS1541C and showed 83% nucleotide identity over the entire length of IS4 (22). The presence of IS4-like elements is unique to pYVe8081 among LCR plasmids. Although the mobile genetic events that led to the accumulation of these remnants of IS elements cannot be determined, it is interesting that similar regions have been identified on other virulence plasmids (24).
We found a reverse transcriptase-like gene (bp 40239 to 40799) with 93% nucleotide identity over 37% of the enteropathogenic E. coli intA gene (56) and 92% nucleotide identity over 24% of the Shigella flexneri sfiA gene (55). Both intA and sfiA are group II intron-like sequences that encode reverse transcriptase-like proteins found within the introns of plants, fungi, and bacteria (45, 56). The enteropathogenic E. coli intA is found on the adherence factor plasmid (pB171), and sfiA is found in the she pathogenicity island in S. flexneri. The Y. pestis plasmid pMT1 also carries a homologous gene remnant (24). Retron-like elements are predicted to be involved in gene transfer.
Plasmid replication and partitioning.
Incompatibility, defined as the inability of two plasmids to coexist in the same cell in the absence of external selection (33), is an indicator of relatedness that has been used to classify plasmids (7). Thus, plasmids in the same incompatibility (Inc) group share one or more elements of their replication or partition systems. Replicons from a number of different incompatibility groups have been sequenced, and their mechanisms of replication and replication control have been explained. Based on DNA and protein homologies as well as the shared use of countertranscript RNA (ctRNA) to control replication, the replicons of IncL/M, IncB, IncIα, and IncK plasmids have been grouped together to form the I complex (44). The current classification scheme is based on the translated products of their essential replication initiator genes, termed RepA (10, 11, 48).
orf125 (bp 66548 to 67570) was the only gene found on plasmid pYVe8081 that could encode a protein with homology to replication initiation proteins. The predicted 340-amino-acid-long protein was 54% identical over 96% of its length to RepA of pMU407.1, a naturally occurring conjugative plasmid belonging to the IncL/M group and the I-complex replicon group (9). ORF125 was 46% identical over 89% of its length to the replication initiator proteins from Col1b-P9 (IncIα) and pMu720 (IncB), which also belong to the I-complex replicon group. This putative protein had a molecular mass of 40,011 Da and an overall net positive charge, which are characteristic of DNA binding proteins (2). These findings support our designation of ORF125 as the pYVe8081 RepA homologue. Surprisingly, we found no homology in the GenBank database to the RepA proteins of pCD1, pIB1, or pYVe227. To confirm and extend our finding that pYVe8081 encoded a replication apparatus significantly different from that of other previously sequenced Yersinia LCR plasmids, we used PCR to amplify homologous sequences from a second plasmid pYVe8081 isolate (obtained from Virginia Miller) and from another serotype 0:8 Y. enterocolitica plasmid, pYVeWA. The DNA sequence of the amplified repA gene from the second pYVe8081 isolate was identical to our original sequence of plasmid pYVe8081 obtained from Peter Feng. The homologous DNA segment amplified from pYVeWA encoded repA with two silent nucleotide changes from our pYVe8081 sequence. Accordingly, the sequence of pYVe8081 repA that we obtained was not isolate specific and appears to be representative of Y. enterocolitica serotype 0:8 plasmids.
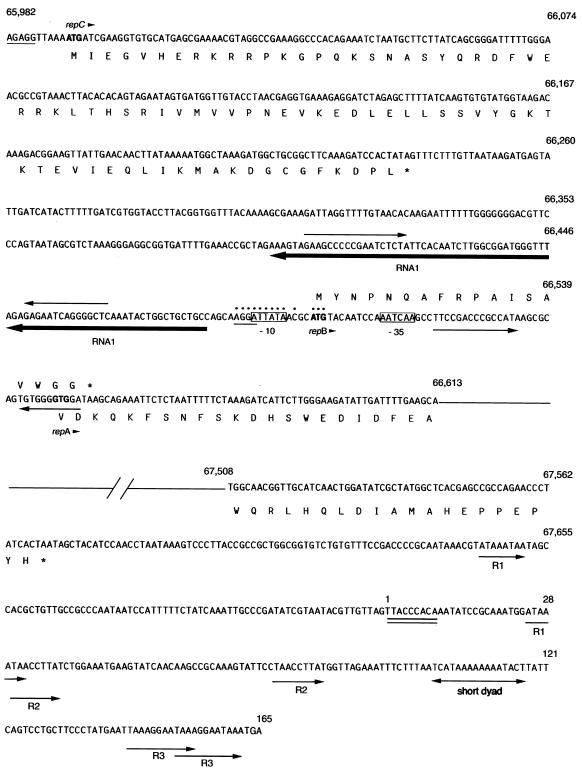
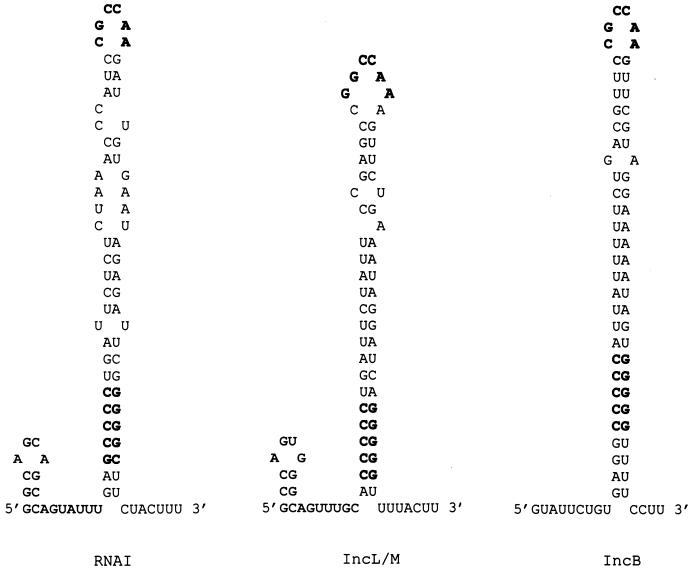
Upstream from repA in pYVe8081, we identified other essential elements found in I-complex replicons (Fig. 4). Homologues of RNAI in I-complex replicons encode ctRNAs that are the major incompatibility determinants and negative regulators of repA expression (2). Our pYVe8081 RNAI was 76% identical over 92% of its length to the antisense RNAI of IncL/M (2). RNAI from pYVe8081 was predicted to fold into a secondary structure (27, 62) composed of a minor stem-loop at its 5′ end, a major stem-loop containing a GC-rich region, and a short 3′ tail (Fig. 5). These features are also found in the stem-loop structures of the antisense molecules of IncL/M (Fig. 5). In addition, the loop sequence, CGCCAA, found in our RNAI transcript is conserved in the ctRNAs of previously sequenced I-complex replicons (34).
FIG. 4.
Nucleotide sequence of the putative replication region in pYVe8081. The numbers correspond to base pair positions in pYVe8081. The sequence shown is single stranded, with the deduced amino acid sequences given in single-letter amino acid code. The deduced amino acid sequence of RepB is shown above its nucleotide sequence. Possible start codons are shown in boldface, and an arrowhead following the gene name indicates the direction of transcription. An asterisk in the amino acid sequence depicts a nonsense codon. Possible Shine-Dalgarno sequences have a single line underneath them. Dots depict nucleotides that are identical in pYVe8081 and the pMU407 replicon. The possible −10 and −35 sequences for the antisense RNA (RNAI) are boxed. RNAI is underlined in boldface with an arrow that indicates the direction of transcription. Facing arrows indicate possible stem-loop structures in RNAI and repB. The double line under bp 1 to 9 denotes a possible DnaA protein-binding site. Arrows labeled R1, R2, and R3 indicate direct repeats within the AT-rich region.
FIG. 5.
Comparison of the sequences of the stem-loop structures predicted for RNAI molecules of pYVe8081, IncL/M (2), and IncB (49) plasmids based on the computer program of Zuker (27, 62). The conserved loop sequence (CGCCAA) and the GC-rich region in the major stem-loop are shown in boldface.
A second characteristic of I-complex replicons is the presence of repB homologues that encode small leader peptides involved in the regulation of repA expression. We found a putative repB upstream of repA (Fig. 4). In pYVe8081, repB was 55% identical over 94% of its length to repB from IncL/M plasmid pMU407.1. The repB nonsense codon (TAA) overlapped the repA start codon, suggesting translational coupling of these two genes, as has been observed previously for IncL/M plasmids (2).
Upstream from RNAI, we found an ORF whose product showed homology to the product of the repC gene, which is located in the corresponding position in the pMU407.1 replicon. However, ORF123 was only 36% identical over 78% of its length to RepC encoded by pMU407. ORF123 was also 36% identical over 74% of its length to RepB from both pCD1 and pYVe227. Both RepB and RepC appear to be involved in the control of repA expression in their respective replication systems (2, 39, 60). We designated the gene encoding ORF123 repC because of its location in the putative replicon of pYVe8081 and because its translated product was homologous to RepC from pMU407.1.
Members of the extended RepFIIA replicon family possess theta-type replicons with characteristic origins of replication (11). The theta-type origin of replication contains specific sequences that interact with the replication initiator protein. Additional features found at the origins of theta-replicating plasmids are an adjacent AT-rich region containing repeated sequences and a dnaA box, where the host DnaA protein binds. A potential dnaA box (TTACCCACA) almost identical to the dnaA box (TTATCCACA) of E. coli (14) was discovered approximately 150 bp downstream from the end of repA in pYVe8081 (Fig. 4). Since we could not find a sequence that matched the RepA binding site (17) in pYVe8081, we designated the first base of our dnaA box as the start of the possible oriR. The presence of repeated sequences (Fig. 4) and a 70% adenosine-plus-thymidine content in this region of pYVe8081 further suggest that this is the replication origin.
In contrast to the putative replication region, the partitioning loci encoded by pYVe8081 appear to be alleles of the analogous regions on Y. pestis pCD1 (39) and Y. enterocolitica pYVe227 (21). A possible partitioning (par) locus at bp 42557 to 45058 (Fig. 1; Table 1) showed 98% nucleotide identity to the sopABC locus in pCD1 and approximately 97% nucleotide identity to the spyABC locus in pYVe227. The locus consisted of two trans-acting proteins and a cis-acting site, which had a genetic organization identical to that of the sop/par loci of low-copy-number plasmids F and P1 (16). The high DNA homology of the LCR plasmid partition systems is further evidence that they have a common evolutionary origin and suggests that these systems have not diverged appreciably since they were acquired.
Evolutionary aspects.
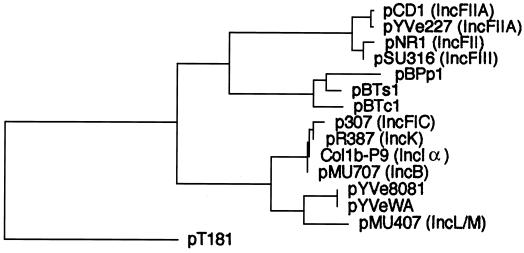
An unrooted phylogenetic tree based on the deduced protein sequences encoded by repA genes showed that the putative RepA from pYVe8081 and pYVeWA belonged to an extended replicon family which consisted of four distinct subgroups (Fig. 6). RepA from pYVe8081 and pYVeWA clustered with IncL/M RepA, while the other I-complex replicons (RepFIC, IncB, IncK, and IncIα) formed a separate subgroup. This is in agreement with the rooted tree constructed by Osborn et al., who proposed extending the RepFIIA family to include the I-complex replicons (34). Only 16% of pYVe8081 RepA residues matched with those in pCD1 and pYVe227, while the RepA proteins of pCD1 and pYVe227 differ from each other by only a single amino acid residue. The finding that pYVe8081 and pYVeWA replicons were only distantly related to the replicons of pCD1 and pYVe227 suggests that Y. enterocolitica serotype 0:8 plasmid replicons originated independently during the evolution of these plasmids. A model for replicon evolution involving cointegrate formation by two compatible plasmids has recently been described (54).
FIG. 6.
Unrooted phylogenetic tree of the translated products of repA genes from the extended RepFIIA family. Alignments were generated using the CLUSTAL program (DNAStar). Incompatibility groups are shown in parentheses. The replication initiator protein from rolling-circle replicating plasmid pT181 (31, 32) is included as an unrelated protein for a control. Buchnera aphidicola plasmids (50, 58) and their RepA (GenBank) accession numbers are as follows: pBPp1, CAA07300; pBTs1, CAA72701; and pBTc1, CAA72709. Other plasmids and their RepA (GenBank) accession numbers are as follows: pCD1, AAC69762; pYVe227, AF102990; pNR1, CAA26168; pSU316, AAA98204; pCol1b-P9, AAA23191; pMU707, AAA98176; pMU407.1, AAA87028; pR387, AAA98310; pYVe8081, AY026194; pYVeWA, AY026195; and p307, AAB17112.
The putative pYVe8081 oriR was similar to replication origins in the other RepFIIA family members since it lacked iterons and consisted instead of a dnaA box and an adjacent AT-rich region. The putative replication region in pYVe8081 also shared copy number control elements found in RepFIIA replicons: a small ctRNA and a small translated peptide encoded in the leader region of the repA mRNA. The presence of these elements suggests that pYVe8081 uses a mechanism of copy number control similar to that found in other RepFIIA family members. Based on recent analysis of RepFIIA replicons, it has been proposed elsewhere that the entire family consists of mosaic replicons that arose as a result of recombination events (35, 44). The likely site of recombination is at the junction of the sequence encoding the leader peptide and the repA gene.
In contrast to the divergence shown by the LCR plasmid replicons, the phenotypic traits exhibited by these plasmids are highly conserved. The segment that includes the virABC loci, virG, virF, and the GVH-yopBD operon in pYVe8081 showed at least 97% nucleotide identity with the corresponding region in pCD1 and pYVe227. This segment was part of a contiguous region, from yopQ to yadA, covering 68% of the pYVe8081 plasmid that has DNA homology to pCD1 and pYVe227 (Fig. 2). In addition, the region encoding ORF155, YopO, and YopP in pYVe8081 was 97% identical to the corresponding region in pCD1 and pYVe227 (Fig. 1). The conservation of virulence-related genes demonstrates the strong selective pressure that these genes are under due to the advantage that they confer on the host Yersinia. Although large regions of pYVe8081, pCD1, and pYVe227 are related, each plasmid has undergone considerable change in structure since it diverged, because deletions, insertions, and rearrangements of DNA sequences are required to explain differences in gene order and plasmid content.
The Yersinia LCR plasmids are nonconjugative, but remnants of transfer (tra) genes homologous to the tra locus of the F plasmid are found in pYVe227 and pCD1 (21, 39). We found no evidence of an ancestral tra locus in pYVe8081, but we did find the gene for a putative endonuclease (orf106) that was homologous to the Nuc protein at the end of the tra gene cluster in pYVe227. The presence of a vestigial tra locus on other LCR plasmids suggests that the ancestral Yersinia LCR plasmid had the ability to colonize other bacterial species.
Constructing evolutionary relationships among plasmids is challenging due to their mosaic nature, which results from the acquisition of genes by horizontal transfer. The sequence of pYVe8081 reveals a backbone with replication features that are clearly divergent from those previously reported for pCD1 and pYVe227 while the maintenance features in all three plasmids are nearly identical. Although analysis of our data in relation to those of others (21, 39) is not definitive, we suggest that the LCR plasmids arose from a common ancestor but that the replicons on Y. enterocolitica serotype 0:8 plasmids evolved independently through nonselective divergence following cointegrate formation. Regardless of the molecular events that led to the present structure of pYVe8081, our data strongly suggest that it should belong to a new incompatibility group. Confirmation of this possibility will await further testing.
ACKNOWLEDGMENTS
We thank Peter Feng and Virginia Miller for providing bacterial strains. We also thank Fred Blattner's group (University of Wisconsin, Madison), Karla Atkins, Emily Clements, Stuart Cohen, R. Lee Collins, and Nazma Jahan for technical support.
This research was supported by Research Area Director IV, which is part of the Medical Research and Materiel Command of the United States Army.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Athanasopoulos V, Praszkier J, Pittard A J. The replication of an IncL/M plasmid is subject to antisense control. J Bacteriol. 1995;177:4730–4741. doi: 10.1128/jb.177.16.4730-4741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brubaker R R. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burland V, Daniels D L, Plunkett G D, Blattner F R. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 1993;21:3385–3390. doi: 10.1093/nar/21.15.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelis G, Laroche Y, Balligand G, Sory M P, Wauters G. Yersinia enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couturier M, Bex F, Bergquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T L, Aber R C. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 9.Davey R B, Bird P I, Nikoletti S M, Praszkier J, Pittard J. The use of mini-Gal plasmids for rapid incompatibility grouping of conjugative R plasmids. Plasmid. 1984;11:234–242. doi: 10.1016/0147-619x(84)90029-5. [DOI] [PubMed] [Google Scholar]
- 10.del Solar G, Espinosa M. Plasmid copy number control: an ever-growing story. Mol Microbiol. 2000;37:492–500. doi: 10.1046/j.1365-2958.2000.02005.x. [DOI] [PubMed] [Google Scholar]
- 11.del Solar G, Giraldo R, Ruiz-Echevarria M J, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devalckenaere A, Odaert M, Trieu-Cuot P, Simonet M. Characterization of IS1541-like elements in Yersinia enterocolitica and Yersinia pseudotuberculosis. FEMS Microbiol Lett. 1999;176:229–233. doi: 10.1111/j.1574-6968.1999.tb13666.x. [DOI] [PubMed] [Google Scholar]
- 13.Eisen J A, Benito M I, Walbot V. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller R S, Kornberg A. Purified DnaA protein in initiation of replication at the Escherichia colichromosomal origin of replication. Proc Natl Acad Sci USA. 1983;80:5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galyov E E, Hakansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 17.Giraldo R, Diaz R. Differential binding of wild-type and a mutant RepA protein to oriRsequence suggests a model for the initiation of plasmid R1 replication. J Mol Biol. 1992;228:787–802. doi: 10.1016/0022-2836(92)90864-g. [DOI] [PubMed] [Google Scholar]
- 18.Guo F, Gopaul D N, van Duyne G D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 19.Heesemann J, Keller C, Morawa R, Schmidt N, Siemens H J, Laufs R. Plasmids of human strains of Yersinia enterocolitica: molecular relatedness and possible importance for pathogenesis. J Infect Dis. 1983;147:107–115. doi: 10.1093/infdis/147.1.107. [DOI] [PubMed] [Google Scholar]
- 20.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iriarte M, Cornelis G R. The 70-kilobase virulence plasmid of Yersiniae. In: Kaper J B, Hacker J, editors. Pathogenicity islands and other mobile elements. Washington, D.C.: American Society for Microbiology; 1999. pp. 91–127. [Google Scholar]
- 22.Klaer R, Kuhn S, Tillmann E, Fritz H J, Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181:169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- 23.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestisKIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahillon J, Kirkpatrick H A, Kijenski H L, Bloch C A, Rode C K, Mayhew G F, Rose D J, Plunkett G, 3rd, Burland V, Blattner F R. Subdivision of the Escherichia coliK-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene. 1998;223:47–54. doi: 10.1016/s0378-1119(98)00365-5. [DOI] [PubMed] [Google Scholar]
- 27.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 28.McGhee G C, Jones A L. Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovorastrain Ea88: gene organization and intraspecies variation. Appl Environ Microbiol. 2000;66:4897–4907. doi: 10.1128/aem.66.11.4897-4907.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilehn B. Studies on Yersinia enterocoliticawith special reference to bacterial diagnosis and occurrence in human acute enteric disease. Acta Pathol Microbiol Immunol Scand. 1969;206(Suppl.):1–48. [PubMed] [Google Scholar]
- 31.Novick R P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. . (Erratum, 44:10, 1990.) [DOI] [PubMed] [Google Scholar]
- 32.Novick R P, Adler G K, Majumder S, Khan S A, Carleton S, Rosenblum W D, Iordanescu S. Coding sequence for the pT181 repCproduct: a plasmid-coded protein uniquely required for replication. Proc Natl Acad Sci USA. 1982;79:4108–4112. doi: 10.1073/pnas.79.13.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick R P, Clowes R C, Cohen S N, Curtiss R D, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976;40:168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborn A M, da Silva Tatley F M, Steyn L M, Pickup R W, Saunders J R. Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146:2267–2275. doi: 10.1099/00221287-146-9-2267. [DOI] [PubMed] [Google Scholar]
- 35.Osborn M, Bron S, Firth N, Holsappel S, Huddleston A, Kiewiet R, Meijer W, Seegers J, Skurray R, Terpstra P, Thomas C M, Thorsted P, Tietze E, Turner S L. The evolution of bacterial plasmids. In: Thomas C M, editor. The horizontal gene pool. Amsterdam, The Netherlands: Harwood Academic Publishers; 2000. pp. 301–350. [Google Scholar]
- 36.Payne P L, Straley S C. YscP of Yersinia pestisis a secreted component of the Yop secretion system. J Bacteriol. 1999;181:2852–2862. doi: 10.1128/jb.181.9.2852-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry R D. Acquisition and storage of inorganic iron and hemin by the Yersiniae. Trends Microbiol. 1993;1:142–147. doi: 10.1016/0966-842x(93)90129-f. [DOI] [PubMed] [Google Scholar]
- 38.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestisKIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting C P, Kerr I D. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portnoy D A, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocoliticapathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Portnoy D A, Wolf-Watz H, Bolin I, Beeder A B, Falkow S. Characterization of common virulence plasmids in Yersiniaspecies and their role in the expression of outer membrane proteins. Infect Immun. 1984;43:108–114. doi: 10.1128/iai.43.1.108-114.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Praszkier J, Wei T, Siemering K, Pittard J. Comparative analysis of the replication regions of IncB, IncK, and IncZ plasmids. J Bacteriol. 1991;173:2393–2397. doi: 10.1128/jb.173.7.2393-2397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri shepathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rimpilainen M, Forsberg A, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosisshows extensive homology to YopH. J Bacteriol. 1992;174:3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Sesma A, Sundin G W, Murillo J. Phylogeny of the replication regions of pPT23A-like plasmids from Pseudomonas syringae. Microbiology. 2000;146:2375–2384. doi: 10.1099/00221287-146-10-2375. [DOI] [PubMed] [Google Scholar]
- 49.Siemering K R, Praszkier J, Pittard A J. Interaction between the antisense and target RNAs involved in the regulation of IncB plasmid replication. J Bacteriol. 1993;175:2895–2906. doi: 10.1128/jb.175.10.2895-2906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva F J, van Ham R C, Sabater B, Latorre A. Structure and evolution of the leucine plasmids carried by the endosymbiont (Buchnera aphidicola) from aphids of the family Aphididae. FEMS Microbiol Lett. 1998;168:43–49. doi: 10.1111/j.1574-6968.1998.tb13253.x. [DOI] [PubMed] [Google Scholar]
- 51.Stainier I, Bleves S, Josenhans C, Karmani L, Kerbourch C, Lambermont I, Totemeyer S, Boyd A, Cornelis G R. YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+ Mol Microbiol. 2000;37:1005–1018. doi: 10.1046/j.1365-2958.2000.02026.x. [DOI] [PubMed] [Google Scholar]
- 52.Straley S C, Bowmer W S. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestisinclude structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 54.Sykora P. Macroevolution of plasmids: a model for plasmid speciation. J Theor Biol. 1992;159:53–65. doi: 10.1016/s0022-5193(05)80767-2. [DOI] [PubMed] [Google Scholar]
- 55.Temin H M. Reverse transcriptases. Retrons in bacteria. Nature. 1989;339:254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- 56.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coliadherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyler S D, Rozee K R, Johnson W M. Identification of IS1356, a new insertion sequence, and its association with IS402 in epidemic strains of Burkholderia cepaciainfecting cystic fibrosis patients. J Clin Microbiol. 1996;34:1610–1616. doi: 10.1128/jcm.34.7.1610-1616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Ham R C, Moya A, Latorre A. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola(endosymbiont of aphids) J Bacteriol. 1997;179:4768–4777. doi: 10.1128/jb.179.15.4768-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Knippenberg P H. Aspects of translation initiation in Escherichia coli, In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 265–274. [Google Scholar]
- 60.Vanooteghem J C, Cornelis G R. Structural and functional similarities between the replication region of the Yersiniavirulence plasmid and the RepFIIA replicons. J Bacteriol. 1990;172:3152–3162. doi: 10.1128/jb.172.7.3600-3608.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics of RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]