Abstract
Mitochondria play a central role in oxidative phosphorylation (OXPHOS), bioenergetics linked with ATP production, fatty acids biosynthesis, calcium signaling, cell cycle regulation, apoptosis, and innate immune response. Severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infection manipulates the host cellular machinery for its survival and replication in the host cell. The infectiaon causes perturbed the cellular metabolism that favours viral replication leading to mitochondrial dysfunction and chronic inflammation. By localizing to the mitochondria, SARS CoV proteins increase reactive oxygen species (ROS) levels, perturbation of Ca2+ signaling, changes in mtDNA copy number, mitochondrial membrane potential (MMP), mitochondrial mass, and induction of mitophagy. These proteins also influence the fusion and fission kinetics, size, structure, and distribution of mitochondria in the infected host cells. This results in compromised bioenergetics, altered metabolism, and innate immune signaling, and hence can be a key player in determining the outcome of SARS-CoV infection. SARS-CoV infection contributes to stress and activates apoptotic pathways. This review summarizes how mitochondrial function and dynamics are affected by SARS-CoV and how the mitochondria-SARS-CoV interaction benefits viral survival and growth by evading innate host immunity. We also highlight how the SARS-CoV-mediated mitochondrial dysfunction contributes to post-COVID complications. Besides, a discussion on targeting virus-mitochondria interactions as a therapeutic strategy is presented.
Keywords: SARS, CoV, Mitochondria, Virus, ROS, Mitochondrial localization signal
1. Introduction
Mitochondria are versatile cellular organelles that play a central role in many biochemical pathways linked with ATP production, biosynthesis of fatty acids, calcium signaling, cell cycle regulation, apoptosis, and innate immune response (Friedman and Nunnari, 2014). Human mitochondrial DNA (mtDNA) is 16569 bp in size, encodes for 13 polypeptides, two ribosomal RNA, and 22 transfer RNA genes (Young and Copeland, 2016). The 13 polypeptides are essential for oxidative phosphorylation pathway function, and the nuclear genome encodes all the remaining proteins required for the structure and function of the mitochondria (Young and Copeland, 2016). Mitochondria are highly dynamic organelles that undergo a coordinated cycle of fusion and fission. These transient morphological adaptations of mitochondria are essential for several molecular processes such as control of cell cycle, immune function, mitochondrial quality, and apoptosis (Tilokani et al., 2018).
The mitochondria are highly vulnerable to both physiological and environmental stress including bacterial, fungal, and viral infections (Khan et al., 2015). Viral infections have been shown to adversely affect mitochondrial structure and functions, and impact the metabolism and immune signalling (Khan et al., 2015). Recent scientific investigations have suggested the critical role of mitochondria in eliciting parts of innate and adaptive immune responses as several immune signaling pathways converge inside the mitochondria (Banoth and Cassel, 2018). Mitochondria-mediated immune signalling has been associated with activation, transcription, differentiation as well as the survival of various types of immune cells (Angajala et al., 2018). It is now clear that viruses use various mechanisms to target host cell mitochondria for their growth and survival, further weakening the host cellular immune response and enhancing cell killing (Ganji and Reddy, 2020). Linking mitochondrial functions as a mechanism of the viral hijacking of host immune response is an emerging and promising field that can be exploited for clinical management of viral infection and its control.
RNA viruses are the virus-containing single-stranded or double-stranded RNA as genetic material protected by a capsid covered by glycoproteins (Carrasco-Hernandez et al., 2017). RNA viruses cause several human diseases including Severe Acute Respiratory Syndrome (SARS), polio, influenza, hepatitis, among others (Carrasco-Hernandez et al., 2017). Mitochondrial changes documented in response to RNA virus infection include mitochondrial depolarization, oxidative stress, changes to mitochondrial fusion and fission kinetics, and mitophagy (Elesela and Lukacs, 2021). Targeting mitochondrial dynamics by proteins encoded by RNA virus including SARS-CoV appears to be one of the mechanisms by which viruses escape from the anti-viral innate immune response (Burtscher et al., 2020, Ganji and Reddy, 2020, Nunn et al., 2020). For instance, Open reading frame-9b (orf9b) protein encoded by Severe acute respiratory syndrome coronavirus 1 (SARS-CoV1) degrades dynamin-related protein 1 (DRP1) leading to fusion of mitochondrial and host cell interferon (IFN) response to virus infection (Shi et al., 2014b). Orf9b severely affects the host cell interferon response by degrading MAVS, TRAF3, and TRAF6 (Shi et al., 2014b). A study by Jiang and the group showed the high-affinity interaction of orf9b and TOM70 in inhibiting type I interferon response (Jiang et al., 2020b). Additionally, the orf9b expression leads to the induction of autophagy in the host cell. RNA viruses are known regulators of mitochondrial-mediated apoptosis, which is an essential mechanism for the survival, propagation, and dissemination of virus inside the host cell. For example, PB1-F2 protein encoded by Influenza A virus localizes to mitochondria and disrupts the organization of mitochondria induction of cell death. HIV viral protein R targets Mfn2 disrupts MMP and induces cell death (Chanturiya et al., 2004).
2. SARS family of coronaviruses and mitochondria:
Coronaviruses are a class of positive-sense single-stranded RNA viruses belonging to the Coronavirinae subfamily with a genome size ranging from 26 to 32 kb consisting of at least six open reading frames (ORFs) and four structural proteins (Yang et al., 2020). The structural proteins include spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The major difference between the SARS-CoV and SARS-CoV-2 are in orf3b, spike S1, orf8 (Chan et al., 2020). SARS coronavirus, commonly known as SARS-CoV causes severe acute respiratory syndrome (SARS) in humans. The common human coronaviruses included alpha coronavirus (229E, NL63), and beta coronavirus (OC43, HKU1). MERS-CoV, SARS-CoV, and SARS-CoV-2 are other human coronaviruses discovered recently causing respiratory syndromes in humans.
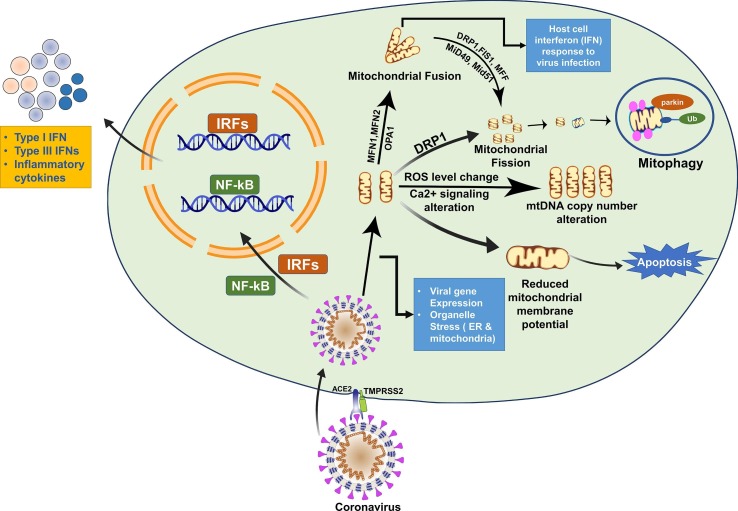
Mitochondria acts as a master regulator of apoptosis. SARS genome encoded proteins may target and modulate anterograde and retrograde signaling to control the mitochondrial function (Burtscher et al., 2020, Singh et al., 2020). Besides, SARS also modulates the function of mitochondria as a mechanism to escape from the host immune system (Burtscher et al., 2020). For instance, a study by Ying-Xim and co-workers showed that protein 7A encoded by SARS-CoV-1 targets Bcl-Xl, a pro-survival gene, to induce apoptosis (Tan et al., 2007). SARS-CoV-1 7a induces apoptosis in a variety of cell types such as the lung, kidney, and liver by activation of caspases (McBride and Fielding, 2012a). Also, protein 7a is found in the Golgi body (GB), endoplasmic reticulum (ER), and partially localized within the mitochondria (McBride and Fielding, 2012a). The 7a protein-mediated apoptosis is a p38-MAPK dependent process and has been reported to inhibit protein synthesis (Kopecky-Bromberg et al., 2006). The 7a protein of SARs-CoV-1 show 85.37% similarity with its counterpart of SARS-CoV-2 and hence may impart a similar effect on mitochondria. SARS-infected patients show CD4+, CD8+, and lymphocytes depletion (Diao et al., 2020). Thus, detection of the viruses in these types of cells and their depletion suggests that SARS might induce apoptosis in them. Based on these observations, it is proposed that induction of apoptosis might be an important mechanism to escape from the host immune system. This review summarizes the current knowledge on the effect of SARS-CoV on mitochondrial structure and function and associated signaling pathways as an escape mechanism from the host immune system. Besides, the review article also discusses the potential of targeting the virus-mitochondria network via mitochondrial pharmacological approach. The effect of SARS-CoV infection on mitochondrial structure and function is demonstrated (Fig. 1 ). Table 1 lists proteins of SARS-CoV which show deleterious effects on host cells in association with mitochondria. The preceding section describes the role of SARS-CoV proteins and their interaction with mitochondria.
Fig. 1.
Illustration demonstrating the effect of SARS-CoV infection on mitochondrial structure and function. Host cell interferon (IFN) response to virus infection: ORF-9b degrades DRP1 leading to mitochondrial fusion and host cell interferon (IFN) response to virus infection. Changes in mtDNA copy number: SARS-CoV proteins can influence ROS level, perturbation of Ca2 + signaling, changes in mtDNA copy number. Apoptosis: ORF9b degrades MAVS, TRAF3 and TRAF6 induce structural and functional change resulting in apoptosis.
Table 1.
Viral and host function of coronavirus genes.
| Gene | Start | Stop | Size | Protein | Protein Similarity | Viral Function | Host Factor | References |
|---|---|---|---|---|---|---|---|---|
| S (SARS-CoV-1) | 21,492 | 25,259 | 3768 | Spike Protein | 75.96% | Facilitates viral entry and replication | Induces apoptosis | Chow et al., 2005, Jin and Zheng, 2009 |
| S (SARS-CoV-2) | 21,563 | 25,384 | 3821 | |||||
| SARS3a (SARS-CoV-1) | 25,268 | 26,092 | 825 | Hypothetical protein sars3a | 72.36% | Antiviral interferon (IFN) response | Induces Necrosis and apoptosis | Kopecky-Bromberg et al., 2007, Law et al., 2005 |
| ORF3a (SARS-CoV-2) | 25,393 | 26,220 | 827 | |||||
| SARS3b (SARS-CoV-1) | 25,689 | 26,153 | 465 | Hypothetical protein sars3b | – | Antiviral interferon (IFN) response | Induces both necrosis and apoptosis | Kopecky-Bromberg et al., 2007 |
| E (SARS-CoV-1) | 26,117 | 26,347 | 231 | Envelope protein | 94.74% | Controls viral replication, inhibition of anti-apoptotic protein | Stress response and apoptosis pathways | DeDiego et al., 2011, Yang et al., 2005 |
| E (SARS-CoV-2) | 26,245 | 26,472 | 227 | |||||
| M (SARS-CoV-1) | 26,398 | 27,063 | 666 | Matrix protein | 90.54% | Defines the shape of the viral envelope, Allow the budding of virions | Pro-apoptotic functions in host cells | Neuman et al., 2011, Tsoi et al., 2014 |
| M (SARS-CoV-2) | 26,523 | 27,191 | 668 | |||||
| SARS6 (SARS-CoV-1) | 26,913 | 27,265 | 353 | Hypothetical protein sars6 | 68.85% | Reduces production of INF | Induces host cell apoptosis | Neuman et al., 2011, Ye et al., 2008 |
| ORF6 (SARS-CoV-2) | 27,202 | 27,387 | 185 | |||||
| SARS7a (SARS-CoV-1) | 27,273 | 27,641 | 369 | Hypothetical protein sars7a | 85.25% | Activates antiviral signaling | Caspase-dependent apoptosis | Tan et al., 2007 |
| ORF7a (SARS-CoV-2) | 27,394 | 27,759 | 365 | |||||
| SARS7b (SARS-CoV-1) | 27,638 | 27,772 | 135 | Hypothetical protein sars7b | 85.37% | Pathogenesis of SARS-CoV infection | Induces apoptosis | Schaecher et al., 2007a |
| ORF7b (SARS-CoV-2) | 27,756 | 27,887 | 131 | |||||
| SARS8a (SARS-CoV-1) | 27,779 | 27,898 | 120 | Hypothetical protein sars8a | – | Viral replication | Induces apoptosis | Chen et al., 2007, Shi et al., 2019a |
| ORF8 (SARS-CoV-2) | 27,894 | 28,259 | 365 | ORF8 protein | – | |||
| SARS8b (SARS-CoV-1) | 27,864 | 28,118 | 255 | Hypothetical protein sars8b | – | Viral replication | Activation of autophagy | Chen et al., 2007, Shi et al., 2019a |
| N (SARS-CoV-1) | 28,120 | 29,388 | 1269 | Nucleocapsid protein | 90.52% | Actin reorganization | Antagonist of IFN signaling | Kopecky-Bromberg et al., 2007, Surjit et al., 2004 |
| N (SARS-CoV-2) | 28,274 | 29,533 | 1259 | |||||
| SARS9b (SARS-CoV-1) | 28,130 | 28,426 | 297 | Hypothetical protein sars9b | – | Suppress the innate immunity | Induces apoptosis | Shi et al., 2014b |
| ORF10 (SARS-CoV-2) | 29,558 | 29,674 | 116 | ORF10 protein | – | – | – | – |
2.1. Non-structural proteins (NSP)
SARS-CoV genome encodes 16 non-structural proteins designated as nsp1-nsp16 (Malone et al., 2022). Nsps plays a vital role in suppressing the host immune pathway. Various nsp proteins have been reported to target host cellular proteins that are required for maintaining the mitochondrial structure and function. For instance, it is reported that nsp2, by targeting mitochondrial biogenesis genes, affect the intracellular signaling pathways linked to apoptosis and anti-viral defense (Cornillez-Ty et al., 2009). Nsp2 interacts with prohibitin 1 (PHB1) and prohibitin 2 (PHB2). The PHB1 and PHB2 are coupled with cell cycle, migration, differentiation, and mitochondrial biogenesis (Cornillez-Ty et al.). Thus, the interaction between nsp2 with PHB1 and PHB2 suggests its effect on mitochondrial biogenesis, and host cell proliferation and differentiation, which might facilitate the growth and proliferation of the viruses as well as escape from the host defense (Cornillez-Ty et al., 2009). To the best of our knowledge, nsp3 does not directly target mitochondria. However, the murine cells infected with coronavirus (Alb ts6 icv) shows partial localization of nsp3 and nsp4 to mitochondria (Clementz et al., 2008). Besides, the infected cells show larger mitochondria with vacuoles. Bioinformatic analysis of SARS-CoV-2 interactome identified nsp4, nsp8, Orf9c to interact with the mitochondria. Lai et al. (2007) demonstrated that in HL-CZ cells, 36% of genes upregulated in response to SARS-CoV 3CLpro overexpression were linked with mitochondrial function and pro-apoptotic genes such as apoptosis-inducing factor (AIF), ATP synthase beta chain (Atp2), and cytochrome c oxidase (Lai et al., 2007). This suggested that CoV 3CLpro might play a key role in mitochondria-mediated apoptosis. SARS-CoV nsp6 protein is associated with autophagosomes formation from ER (Cottam et al., 2011). Further, studies have reported the lipids for membranes of autophagosomes can also come from mitochondria. In this connection, more detailed studies are required to understand the contribution of mitochondria in the formation of autophagosomes and its relation with mitophagy and host cell apoptosis. The computational analysis identified nsp8 of SARS-CoV-2 to interact with the proteins associated with mitochondrial function (Gordon et al., 2020b). Nsp10 is another non-structural protein encoded by SARS-CoV. A study by Li and co-workers showed that nsp10 interacts with both host genes (BTF3 and ATF5) and mitochondrial genes (NADH 4L subunit and cytochrome oxidase II) (Li et al., 2005). Nsp10 alters the NADH-cytochrome activity in lung fibroblast cells (Li et al., 2005). Besides, nsp10 also depolarizes the inner mitochondrial membrane to induce severe damage to the cells (Li et al., 2005). Nsp15 encoded from SARS–CoV by inhibiting mitochondrial anti-viral signaling adaptor protein induces apoptosis. By regulating pRB, nsp15 controls the cell cycle and is proposed to impact metabolic status and immune response (Bhardwaj et al., 2012). Those nsps which does not target mitochondria are reported to target other cell organelles (ER and GB) important for an anti-viral response. Thus, SARS-CoV uses multiple nsp to induce organelle stress, activate stress-responsive cell death pathways to induce a cytotoxic effect. Analysis to identify mitochondrial localization was performed on nsp using MitoFates tool (Fukasawa et al., 2015), potential MMP cleavage sites and the probability score along with the function of each nsp is shown in Table 2 .
Table 2.
Function of non-structural peptides.
| Mature Peptide | Nucleotide Start | Nucleotide End | #Base | aa- size | Protein Product |
MPP Cleavage site |
Function | References | |
|---|---|---|---|---|---|---|---|---|---|
| Position | Probability | ||||||||
| nsp1 | 266 | 805 | 540 | 180 | Leader protein | 30G | 0.044 | Antagonism against IFN signaling | Kamitani et al., 2009 |
| nsp2 | 806 | 2719 | 1914 | 638 | Counterpart of MHV p65 | 47G | 0.014 | Aid in growth and proliferation of the viruses | Cornillez-Ty et al., 2009 |
| nsp3 | 2720 | 8554 | 5834 | 1944 | Nsp3-pp1a/pp1ab | 31I | 0.000 | SG-mRNA synthesis | LaStarza et al., 1994 |
| nsp4 | 8555 | 10,054 | 1500 | 500 | Nsp4-pp1a/pp1ab | 56D | 0.041 | Causes disrupted mitochondrial morphology | Freundt et al., 2009 |
| nsp5 | 10,055 | 10,972 | 918 | 306 | 3C-like proteinase | 41H | 0.003 | Proteolytic processing of the replicase polyproteins | Stobart et al., 2013 |
| nsp6 | 10,973 | 11,842 | 870 | 290 | Nsp6-pp1a/pp1ab (TM3) | 6 T | 0.019 | Initiate cellular autophagy and a general ER stress response | Fung et al., 2016, Prentice et al., 2004 |
| nsp7 | 11,843 | 12,091 | 249 | 83 | Nsp7-pp1a/pp1ab | 22 V | 0.000 | Viral replication | Subissi et al., 2014 |
| nsp8 | 12,092 | 12,685 | 594 | 198 | Nsp8-pp1a/pp1ab | 58 K | 0.000 | Viral RNA synthesis | Subissi et al., 2014 |
| nsp9 | 12,686 | 13,024 | 339 | 113 | Nsp9-pp1a/pp1ab | 11Q | 0.065 | Involved in the replicative cycle | Slanina et al., 2021, Sutton et al., 2004 |
| nsp10 | 13,025 | 13,441 | 417 | 139 | Formerly known as growth-factor-like protein (GFL) | 79C | 0.000 | Regulates replication | Bouvet et al., 2014 |
| nsp11 | 13,442 | 13,480 | 39 | 13 | Nsp11-pp1a short peptide | 7F | 0.005 | Viral replication | Deming et al., 2007 |
| nsp12 | 13,442 | 16,236 | 2769 | 923 | RNA-dependent RNA polymerase | 34A | 0.035 | Viral replication | Subissi et al., 2014 |
| nsp13 | 16,237 | 18,039 | 1803 | 601 | Nsp13-pp1ab (ZD, NTPase/HEL; RNA 5′-triphosphatase) | 23P | 0.037 | Viral replication | Jang et al., 2020 |
| nsp14 | 18,040 | 19,620 | 1581 | 527 | 3′-to-5′ exonuclease | 54L | 0.035 | Proofreading exoribonuclease | Ma et al., 2015 |
| nsp15 | 19,621 | 20,658 | 1038 | 346 | EndoRNAse | 62 N | 0.000 | Limits apoptosis in macrophages | Deng et al., 2017 |
| Nsp16 | 20,659 | 21,552 | 893 | 297 | 2′-O-ribose methyltransferase | 20 M | 0.037 | Viral RNA synthesis and viral replication | Wu et al., 2020 |
2.2. Orf3a protein of SARS-CoV
It is the first accessory protein encoded by the SARS-CoV genome. Initial studies have indicated Orf3a protein is localized to the ER, plasma membrane (PM), and GB. In SARS infected lung section, ORF3a is predominantly found in the cytoplasm of pneumocytes (McBride and Fielding, 2012b). The Orf3a mediated pro-apoptotic function involves both caspase 8 and caspase 9, suggesting the involvement of both extrinsic and intrinsic pathways of apoptosis (Ren et al., 2020). Besides, Orf3a also targets Bax, p53, and p38MAPK for the induction of host cell death (Hemmat et al., 2021). More specifically, Orf3a induces apoptosis by activation of p38MAPK and subsequent leakage of cytochrome C (Hemmat et al., 2021). These data suggest that Orf3a can directly affect the mitochondrial function to induce apoptosis.
2.3. Structural protein
The four structural proteins encoded by the SARS-CoV genome are spike, envelope, nucleocapsid, and membrane (Hemmat et al., 2021). Herein, we discuss the role of various structural proteins and their role in altering mitochondrial function. The spike protein (S protein) is a highly glycosylated type I transmembrane protein that assembles as trimers on the surface of the virions to give the crown-like appearance. The interaction between the Spike proteins and the Angiotensin-converting enzyme 2 (ACE2) receptor facilitates the entry of the virus into the host cell. McBride and co-workers have demonstrated that spike protein contains an ER retrieval signal sequence for interaction with the membranes (McBride and Fielding, 2012a). In addition to facilitating the entry of the virus inside the cells, spike protein also induces apoptosis in Vero E6 cells (Wu et al., 2004). A recent study by Kalashnyk et al. (2021), demonstrated that the SARS-Cov-2 spike protein α7 nAChR-binding portion prevents mitochondrial-driven apoptosis (Kalashnyk et al., 2021). This study shows that when the virus is inside the cell and when uncoated, will facilitate the viral replication cycle and make the host cell vulnerable. Detailed studies are required to understand the precise interaction between mitochondria and spike proteins.
The envelope (E) protein is another protein encoded by SARS-CoV required to complete the viral life cycle. The envelope protein of SARS-CoV is demonstrated to participate in stress response and apoptosis pathways. The E protein downregulates the inositol-requiring eanzyme 1 (IRE-1) of the unfolded protein response to reduce the apoptosis rate (DeDiego et al., 2011). This suggests that SARS-CoV controls the apoptosis of the host cell as per the requirement of the stages of viral life cycle. In vitro transfection experiments indicated that the E protein alone managed to reduce the mitochondrial stress by inhibiting the expression of hsp10 E1 (DeDiego et al., 2011). Yang et al. (2005) reported the role of E protein in SARS-CoV-induced lymphopenia. Transfection of E gene of SARS-CoV in Jurkat T-cells showed induction apoptosis by inhibition of anti-apoptotic protein Bcl-Xl( Yang et al., 2005 ). Although E protein does not localize to the mitochondria, it effectively regulates nuclear-encoded genes associated with mitochondrial functions to regulate stress response and apoptosis. This may be important for controlling viral replication.
The nucleocapsid (N) is another structural protein of SARS-CoV. In COS-1 cells, N protein-induced mitochondria-mediated apoptosis pathway (Zhang et al., 2007). Besides, the induction of apoptosis by N protein also involves actin reorganization. The membrane Glycoprotein M protein induces apoptosis in HEK293T cells via altering the expression of PDK-1 and Akt kinase and the release of cytochrome c (Chan et al., 2007). Another study by Tsoi and co-workers demonstrated that M−protein induces apoptosis by targeting PDK1-PKB/Akt signaling and induction of caspases 8 and 9 (Tsoi et al., 2014). Thus, approaches targeting the interaction between M−protein and PDK1 may be useful for treating SARS-CoV.
2.4. Accessory proteins
The accessory proteins are a group of proteins encoded by SARS-CoV that are not required for replication of the coronavirus. However, accessory proteins are very important to counteract the host immune system. The SARS coronavirus encodes eight accessory proteins designated as orf3a, orf3b, orf6, orf7a, orf7b, orf8a, orf8b, and orf9b (Astuti and Ysrafil, 2020a). Many of the accessory proteins target mitochondria and are described below.
2.5. Orf3b
The localization of orf3b inside mitochondria is controversial. A study showed that SARS-CoV orf3b localizes to mitochondria in infected Vero E6 cells (Chan et al., 2005). Further, the study also demonstrated that the amino acids from 80 to 138 were critical for orf3b mitochondrial translocation. Iitially, Orf3b accumulates within the nucleolus, then translocates to the OMM and take part in G0/G1 arrest as well as induction of apoptosis. Activation of type-I interferon (IFN-β) is required for activation of mitochondria-mediated anti-viral response. orf3b downregulates IFN-β, thereby inhibiting mitochondrial-mediated anti-viral response (Burtscher et al., 2020). Orf3b interacts with RUNX1b, activates AP-1 via JNK/ERK signaling, and induces both necrosis and apoptosis (Varshney and Lal, 2011). The upregulation of cytokines and chemokines has also been reported to interfere with both the structure and function of the mitochondria and the induction of apoptosis and necrosis.
2.6. Orf6
The orf6 of SARS-CoV 2 is approximately 61 amino acid protein in size (Dehipawala et al., 2021). The orf6 is found in the ER and GB of the infected cells and is likely to impact the viral replication and pathogenesis by antagonizing the function of STAT1. Orf6 along with orf3b and N proteins are reported as antagonists of IFN signalling (Lei et al., 2020). Further, orf6 is predominantly localized in ER and GB (Lee et al., 2021). Inhibition of interferon production by orf6 is coupled with induction of caspase 3 with concomitant ER stress and JNK dependent apoptosis. Mitochondria play a decisive role in caspases activation and caspase-mediated apoptosis. Although there is no report of orf6 localization inside the mitochondria, activation of caspase 3 suggests the involvement of mitochondria in SARS-CoV induced host cell apoptosis (Ye et al., 2008). By localizing to the membranes of ER and GB of the host cells, orf6 interferes with karyopherin alpha 2 and karyopherin beta 1 mediated nuclear import complex formation leading to loss of Signal Transducer and Activator of Transcription 1 (STAT1) nuclear transport (Frieman et al., 2007). STAT1 nuclear transport is linked with antiviral signaling. By preventing the nuclear translocation of the STAT1, orf3b down modulates the host antiviral defense system. Zhongde and co-workers showed that orf6 induces apoptosis in infected host cells by caspase-3- ER-stress-JNK axis (Ye et al., 2008). These data suggested that orf6 targets nuclear transport machinery to antagonize the host interferon production.
2.7. Orf7
The orf7 codes for two accessory proteins, namely orf7a and orf7b. Orf7a is associated with caspase-dependent apoptosis (Schaecher et al., 2007b). Orf7a interacts with Bcl-xL protein for induction of apoptosis in the infected host cell (Tan et al., 2007). Additionally, orf7a inhibits protein synthesis in the infected cell via activation of NF-kB and p38MAPK and inhibition of the cell cycle progression (Schaecher et al., 2007b). Additionally, orf7 protein interacts with transcription factor SGT and anti-apoptotic protein Bcl-Xl. Bone marrow stromal antigen 2 (BST2) acts as antiviral genes and target diverse viral families by tethering budding virions and restricting their release. Upon viral infection, BST2 activates LILRA4/ILT7 antiviral signaling in plasmacytoid dendritic cells. Through physical interaction, orf7a inhibits the glycosylation of BST2 (Taylor et al., 2015). Orf7a is found to be localized in GB, ER, ER-GB intermediate compartment, mitochondria, and cytoplasm of the infected cells. In contrast to this, ORF7b is predominantly localized to GB and is alone capable of inducing apoptosis. Cao and co-workers showed that SARS-CoV-2 manipulates the host ubiquitin system to enhance the ability of orf7a to antagonize the INF-I response (Cao et al., 2021).
2.8. Orf8
The orf8 encodes for two proteins, namely orf8a and orf8b. Orf8a is important for SARS-CoV replication. Orf8a induced apoptosis involves mitochondria-mediated caspase-3 activation (Fang et al., 2021). Besides, overexpression studies demonstrated that orf8a is localized inside the mitochondria and induced MMP, and ROS production. Recently, orf8b involvement in the activation of NLR family pyrin domain containing 3 (NLRP3) inflammasomes and stress pathways is reported (Shi et al., 2019b). Further, orf8b aggregates in the cytoplasm cause ER stress, mitochondrial dysfunction, damage to the lysosome, and activation of autophagy. In epithelial cells and macrophages, orf8b causes cell death and inflammasomes, respectively.
2.9. Orf9b
SARS-CoV uses orf9b to suppress innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome (Shi et al., 2014b). By localizing to the OMM, orf9b induces mitochondrial elongation by inhibiting the expression of DRP1 (Shi et al., 2014b). Another study by Gordon et al. revealed that orf9b forms a complex with mitochondrial import receptor TOM70 in SARS-CoV and SARS-CoV2 (Gordon et al., 2020a, Gordon et al., 2020b). Besides, orf9b limits the interferon response of the host cell by degrading MAVS, TRAF3, and TRAF6 (Jiang et al., 2020, Shi et al., 2014, Shi et al., 2014). Thus, orf9b induces both structural and functional changes to the mitochondria to escape from host defense and immune evasion.
3. SARS-CoV regulation of mitochondrial dynamics
SARS-CoV-mitochondrial interaction is one of the mechanisms by which the virus escapes from mitochondria mediates immunity. SARS-CoV infection leads to changes in the morphology of the infected cells and alters the expression of nuclear-encoded genes associated with mitochondrial functions. Therefore, SARS-CoV may have the ability to affect and compromise mitochondrial function. SARS-CoV can affect bioenergetics, innate immunity, apoptosis, and mitophagy by affecting retrograde and anterograde signaling. Herein, we discussed the role of SARS-CoV and its effect on mitochondrial structure and function (Table 3 ).
Table 3.
Coronavirus genes and mitochondrial targeting.
| Gene | Mitochondrial Function | References |
|---|---|---|
| ORF1ab | Cause disruption of mitochondrial morphology | Freundt et al., 2009 |
| S | Prevents mitochondria-driven apoptosis when the virus is uncoated inside the cell | Kalashnyk et al., 2021 |
| ORF3a | Affect mitochondrial function to induce apoptosis. ORF3a induces mitochondrial damage leading to activation of NLRP3 inflammasome | Law et al., 2005, Shi et al., 2014 |
| ORF3b | Activation of mitochondria-mediated anti-viral response. ORF3b at first accumulates within the nucleolus and then gets translocated to the OMM and demonstrated to take part in G0/G1 arrest and induction of apoptosis | McBride and Fielding, 2012a |
| E | Reduce mitochondrial stress. Regulates nuclear-encoded genes associated with mitochondrial functions to regulate stress response and apoptosis | DeDiego et al., 2011 |
| M | Induce mitochondria-mediated apoptosis, also induces apoptosis via altering the expression of PDK-1 and Akt kinase and the release of cytochrome c | Surjit et al., 2004, Tsoi et al., 2014 |
| ORF6 | Activation of the mitochondrial signaling pathway. Involves in activation of caspase 3 with involvement of mitochondria to induce host cell apoptosis | Sawicki et al., 2005 |
| ORF7a | Involved in Caspase-dependent mitochondria-mediated apoptosis | Tan et al., 2007 |
| ORF7b | Involved in Caspase-dependent mitochondria-mediated apoptosis | Tan et al., 2007 |
| ORF8a | ORF8a is localized inside the mitochondria and induced MMP and rROS production. Involved in Caspase-3 dependent mitochondria-mediated apoptosis | Chen et al., 2007 |
| ORF8b | Involve in mitochondrial dysfunction, damage to the lysosome, and activation of autophagy | Shi et al., 2019b |
| N | Reduces mitochondrial membrane potential, increased ROS production, and apoptosis induction by cytochrome release into the cytoplasm | Zhang et al., 2007 |
| ORF9b | Induces both structural and functional changes to the mitochondria also induces apoptosis | Shi et al., 2014 |
Transfection studies in HEK293 showed that orf9b is localized inside the mitochondria and induces the proteasomal degradation of Drp1 and affects mitochondrial fusion (Shi et al., 2014b). Further, reduced Drp1 levels also affect MAVS signaling. By promoting degradation of MAVS, TRAF3, and TRAF6, orf9b disrupts MAVS signaling and production of IFN-γ (Jiang et al., 2020, Jiang et al., 2020). Targeting of poly (rC) binding protein 2 (PCBP2) and the HECT domain E3 ligase (AIP4) by orf9b results in repression of the MAVS signaling pathway (Shi et al., 2014b). The localization of orf9b inside the mitochondria facilitates the interaction between PCBP2 and AIP4 ( Shi et al., 2014b ). The impact of coronavirus proteins on mitochondrial structure and functions is depicted in Table 4 . Analysis of mitochondrial DNA mutation in whole transcriptome data sets from control and SARS-CoV-2 infected patients lung tissue, blood, and infected cell line model showed increased mtDNA variation in SARS-CoV-2 infected lung tissue (Table 5, Table 6 ). Interestingly, we observed variation in nuclear mitochondrial genes which are associated with mitochondrial DNA replication (POLG, POLG2, PRIMPOL, TOP1MT, TOP3A), and repair (APEX1, DDX5, UNG) (Table 7 ). Gene expression of nuclear-encoded mitochondrial genes involved in mtDNA maintenance was downregulated in SARS-CoV-2 infected lung tissue (Table 8 ).
Table 4.
Coronavirus protein impact on mitochondrial structure and functions.
| Mitochondrial Impact | Coronavirus Protein | Virus Type | Targeting gene, enzyme, and pathway | Reference |
|---|---|---|---|---|
| Mitochondrial Dynamics | ORF9b | SARS-CoV | DRP1, IFN-g, PCBP2, AIP4, MAVS signalling, TRAF3, and TRAF6 | Gordon et al., 2020a |
| mtDNA Copy Number | – | SARS-CoV-2 | ROS, Ca2+ signaling | Valdes-Aguayo et al., 2021, Wiedmer et al., 2008 |
| Mitochondrial Membrane Potential | ORF8a, nsp10 | SARS-CoV | ROS, caspase-3 activation, | Keng et al., 2006 |
| Mitochondrial Structure, Size | ORF9b, ORF7a, ORF7b, ORF3b, ORF1ab | SARS-CoV | DRP1, MAVS, TRAF3 & TRAF6 | Freundt et al., 2009, Shi et al., 2014 |
| Oxidative Stress | ORF7a, nsp5 (3C-like protease), Nucleocapsid, ORF3a | SARS-CoV-2 SARS-CoV |
Ap4A-hydrolase, ROS, NF-kB signalling, IL-1β | Chernyak et al., 2020, Lin et al., 2006, Zhang et al., 2007 |
| Mitophagy | ORF9b, ORF10 | SARS-CoV-2 | DRP1, NIX | Li et al., 2022, Zhu et al., 2016 |
| Antiviral Immunity | ORF9a | SARS-COV | MAVS, NLRP3 inflammasome pathway, IL-1β and IL-18 | Zhou et al., 2012 |
| Mitochondrial Fusion | ORF9b | SARS-CoV-2 SARS-CoV |
DRP1, MFN1, MFN2, OPA1 | Alavi and Fuhrmann, 2013, Astuti and Ysrafil, , 2020, Shi et al., 2014 |
| Mitochondrial Fission | ORF9b | SARS-CoV-2 SARS-CoV |
DRP1, FIS1, MFF, MiD49, Mid51 | Astuti and Ysrafil, , 2020, Shi et al., 2014 |
Table 5.
Mitochondrial DNA variants identified in whole transcriptome data of control and SARS-CoV-2 infected patients’ tissue, blood, and infected cell line model.
| Subjects | SARS-CoV-2 status | Subject ID | Tissue type | No. of variants | Reference |
|---|---|---|---|---|---|
| Autopsy samples from patients deceased due to SARS-Cov2 infection | Positive | SRR12340086 SRR12340087 SRR12340088 SRR12340089 |
Lung-RLL Lung-RML Lung-RUL Lung-LUL |
79 68 73 77 |
Desai et al., 2020 |
| Transcriptional profile of leukocytes in PCR negative COVID-19 controls | Negative | SRR12313439_control SRR12313441_control SRR12313442_control SRR12313444_control SRR12313446_control SRR12313449_control SRR12313450_control |
leukocyte | 32 12 17 11 47 8 16 |
Gill et al., 2020 |
| Transcriptional profile of leukocytes in PCR positive COVID-19 patients | Positive | SRR12313440_covid SRR12313443_covid SRR12313445_covid SRR12313447_covid SRR12313448_covid SRR12313451_covid SRR12313452_covid |
leukocyte | 10 14 29 28 22 23 25 |
|
| Primary human lung epithelium (NHBE) were mock-treated | Negative | NHBE_1 NHBE_2 NHBE_3 |
Primary human lung epithelium (NHBE) | 15 15 15 |
Blanco-Melo et al., 2020 |
| Primary human lung epithelium (NHBE) were infected with SARS-CoV-2. | Positive | NHBE_4 NHBE_5 NHBE_6 |
Primary human lung epithelium (NHBE) | 15 15 16 |
|
| Uninfected human lung biopsies | Negative | Healthy_1 Healthy_2 |
Lung tissue | 22 35 |
|
| lung samples derived from COVID-19 deceased patient | Positive | Covid_1 Covid_2 |
Lung tissue | 50 55 |
# Lung-RLL, right lower lobe, Lung-RML, right middle lobe, Lung-RUL, right upper lobe,
Table 6.
Number of mitochondrial DNA variants in mitochondrial protein-coding genes identified in whole transcriptome data of control and SARS-CoV-2 infected patients tissue specimens.
|
Desai et al., (2020) |
Blanco-Melo et al., (2020) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SRR12340086 | SRR12340087 | SRR12340088 | SRR12340089 | COVID-1 | COVID-2 | Healthy-1 | Healthy-2 | NHBE-1 | NHBE-2 | NHBE-3 | NHBE-4 | NHBE-5 | NHBE-6 |
| ND1 | 4 | 4 | 4 | 4 | 3 | 5 | 1 | |||||||
| ND2 | 4 | 2 | 2 | 4 | 3 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| COX1 | 7 | 7 | 5 | 5 | 4 | 5 | 1 | 2 | ||||||
| COX2 | 1 | 1 | 1 | 1 | 4 | 2 | ||||||||
| ATP8 | 2 | 2 | 3 | 1 | ||||||||||
| ATP6 | 3 | 3 | 4 | 5 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| COX3 | 4 | 3 | 3 | 3 | 2 | 1 | ||||||||
| ND3 | 3 | 2 | 2 | 2 | 1 | 2 | 1 | |||||||
| ND4L | 2 | 2 | 2 | 2 | 1 | |||||||||
| ND4 | 3 | 3 | 2 | 3 | 4 | 4 | 3 | |||||||
| ND5 | 13 | 12 | 11 | 12 | 11 | 6 | 1 | 2 | ||||||
| ND6 | 2 | 2 | 2 | 2 | 1 | 1 | ||||||||
| CYTB | 4 | 3 | 4 | 4 | 9 | 6 | 2 | 5 | 2 | 2 | 2 | 2 | 2 | 2 |
| 52 | 46 | 45 | 48 | 42 | 37 | 8 | 19 | 4 | 4 | 4 | 4 | 4 | 4 | |
Table 7.
Deleterious protein-coding variants in nuclear mitochondrial genes identified in whole transcriptome data of SARS-CoV-2 infected patients lung tissue specimens.
| Gene symbol | Function | Mutation identified | Mutation type | SIFT | PolyPhen | Subject ID | Sample type | Reference |
|---|---|---|---|---|---|---|---|---|
| APEX1 APEX1 DDX5 |
mtDNA repair DEAD-box RNA helicase |
p.Pro311Ser p.Pro311Leu p.Ala441Val |
missense_variant missense_variant missense_variant |
Deleterious Deleterious Deleterious |
probably_damaging probably_damaging probably_damaging |
SRR12340086 |
Lung tissue | Desai et al., 2020 |
| UNG POLG |
mtDNA repair mtDNA replication |
p.Val272Ala p.Pro412Leu |
missense_variant missense_variant |
Deleterious Deleterious |
probably_damaging probably_damaging |
SRR12340087 | Lung tissue | |
| TOP3A POLG2 |
mtDNA decatenation and segregation mtDNA replication |
p.Ile70Asn p.Trp295Arg |
missense_variant missense_variant |
Deleterious Deleterious |
probably_damaging probably_damaging |
SRR12340088 |
Lung tissue | |
| PRIMPOL TOP1MT POLG TOP3A POLG2 |
mtDNA replication |
p.Tyr402Cys p.Leu552Arg p.Thr599Ile p.Asp484Tyr p.Val441Phe |
missense_variant missense_variant missense_variant missense_variant missense_variant |
Deleterious Deleterious Deleterious Deleterious Deleterious |
probably_damaging probably_damaging probably_damaging probably_damaging probably_damaging |
SRR12340089 | Lung tissue |
Table 8.
Downregulation of mitochondrial DNA maintenance genes identified in whole transcriptome data of SARS-CoV-2 infected patients lung tissue (from: Blanco-Melo et al., 2020).
| Gene symbol | logFC | Gene description | Function |
|---|---|---|---|
| RECQL4 | 3.301125548 | RecQ like helicase 4 | mtDNA maintenance |
| METTL4 | 0.960866022 | methyltransferase like 4 | mtDNA maintenance |
| POLB | 0.775953276 | DNA polymerase beta | mtDNA maintenance |
| APEX1 | 0.184002807 | apurinic/apyrimidinic endodeoxyribonuclease 1 | mtDNA maintenance |
| PIF1 | 0.064341063 | PIF1 5′-to-3′ DNA helicase | mtDNA maintenance |
| SSBP1 | −0.095574337 | single stranded DNA binding protein 1 | mtDNA maintenance |
| POLDIP2 | −0.215238656 | DNA polymerase delta interacting protein 2 | mtDNA maintenance |
| PPA2 | −0.222606157 | inorganic pyrophosphatase 2 | mtDNA maintenance |
| DNA2 | −0.290150169 | DNA replication helicase/nuclease 2 | mtDNA maintenance |
| EXOG | −0.314869688 | exo/endonuclease G | mtDNA maintenance |
| MTERF1 | −0.323074621 | mitochondrial transcription termination factor 1 | mtDNA maintenance |
| LIG3 | −0.363345113 | DNA ligase 3 | mtDNA maintenance |
| POLG2 | −0.423754726 | DNA polymerase gamma 2, accessory subunit | mtDNA maintenance |
| UNG | −0.431007241 | uracil DNA glycosylase | mtDNA maintenance |
| MGME1 | −0.936039411 | mitochondrial genome maintenance exonuclease 1 | mtDNA maintenance |
| OGG1 | −1.067414664 | 8-oxoguanine DNA glycosylase | mtDNA maintenance |
| MTERF2 | −1.133486683 | mitochondrial transcription termination factor 2 | mtDNA maintenance |
| POLRMT | −1.320205504 | RNA polymerase mitochondrial | mtDNA maintenance |
| TFAM | −1.324537142 | transcription factor A, mitochondrial | mtDNA maintenance |
| MUTYH | −1.34437367 | mutY DNA glycosylase | mtDNA maintenance |
| POLG | −1.536793488 | DNA polymerase gamma, catalytic subunit | mtDNA maintenance |
| ENDOG | −1.94800646 | endonuclease G | mtDNA maintenance |
| PRIMPOL | −2.032059322 | primase and DNA directed polymerase | mtDNA maintenance |
| TOP1MT | −1.906133073 | DNA topoisomerase I mitochondrial | mtDNA maintenance |
| TFB2M | −2.002097354 | transcription factor B2, mitochondrial | mtDNA maintenance |
| RNASEH1 | −2.236099433 | ribonuclease H1 | mtDNA maintenance |
| TOP3A | −3.016484079 | DNA topoisomerase III alpha | mtDNA maintenance |
| ATAD3A | −3.560998784 | ATPase family AAA domain containing 3A | mtDNA maintenance |
| TWNK | −3.597778413 | twinkle mtDNA helicase | mtDNA maintenance |
| ATAD3B | −4.024036831 | ATPase family AAA domain containing 3B | mtDNA maintenance |
4. SARS-CoV alter mtDNA copy number
Alteration in mtDNA copy number is known to affect both the structure and function of the mitochondria. Infection with viruses such as Herpes simplex virus 1 and Epstein Barr virus is known to alter the mtDNA content. For example, the UL12.5 protein of HSV1 induces mtDNA depletion during productive infection of mammalian cells (Saffran et al., 2007). Another study reported that the Zta protein of EBV depleted mtDNA in the host cells (Wiedmer et al., 2008). By interacting with mitochondrial single-stranded DNA binding protein, the Zta protein of EBV reduces mtDNA replication. HCV via induction of ROS and NO-mediated pathways induces mtDNA damage. Further, mtDNA depletion is also reported in HIV-HCV confected humans. A study by Collins et al. (2004) reported the inflammatory nature of the mtDNA (Collins et al., 2004). SARS-CoV impact on mtDNA copy number requires detailed investigation.
5. SARS-CoV alters the mitochondrial membrane potential (MMP)
The infection with SARS-CoV causes an extensive cytopathic effect. MMP plays an important role in the segregation and sorting of the defective mitochondria for subsequent repair or elimination process in which inner and outer mitochondria membrane proteins play a crucial role. The SARS-CoV nucleocapsid protein reduced the MMP while inducing apoptosis in COS-1 cells (Zhang et al., 2007). Overexpression of orf3b did not significantly alter the MMP of the host cell (Kopecky-Bromberg et al., 2007). The perturbation of MMP by orf8a of SARS-CoV is also reported. Nsp10 interacts with the cellular oxidoreductase system and causes the cytopathic effect. Transfection of nsp10 gene to KMB-17 cells showed a decrease in MMP at 24hrs followed by a recovery at 48 hrs post-transfection (Li et al., 2005). Also, pull down and Western blot analysis showed that nsp10 interacts with cytochrome oxidase complex, thus affecting the mitochondrial oxidoreductase system. A study by Schneider et al. reported the loss of MMP in mitochondria upon viral infection (Schneider et al., 2019).
6. SARS-CoV and mitochondrial mass
Several studies have reported that viral infection alters mitochondrial mass. Viral genome encoded proteins induce mitophagy to reduce the mitochondrial mass (Gou et al., 2017). Total mitochondrial mass is an essential factor contributing to MMP. Transmissible gastroenteritis virus (TGEV) infected IPEC-J2 showed a reduced mitochondrial mass (Zhu et al., 2016). The reduction of mitochondrial mass could be due to the degradation of mitochondria by activation of mitochondrial degradation machinery.
7. SARS-CoV proteins influences size, structure, and distribution of mitochondria
SARS-CoV infection exerts its effect via targeting the structure, distribution, and function of mitochondria. The orf9b of SARS-CoV localizes inside the mitochondria and causes elongation of mitochondria in HEK293 cells (Barbier et al., 2017). Besides, orf9b activates ubiquitination and proteasomal degradation of DRP1, thereby inhibiting mitochondria fission. Besides, the elongated mitochondria also showed interaction with autophagosomes. The interaction between autophagosome and mitochondria may induce mitophagy. Thus, alteration in the mitochondrial structure may contribute to compromised antiviral response and signaling. Not all the SARS-CoV encoded proteins are translocated to mitochondria. For example, a study by Schaecher and colleagues (2007) showed in Vero cells that, SARS-CoV orf7a and orf7b are localized to GB but not to mitochondria. Orf4b is another protein that is not reported inside mitochondria (Schaecher et al., 2007b). A study by Matthews et al.(2014) showed that Middle East respiratory syndrome coronavirus (MERS-CoV) encoded orf4b localizes to the nucleus and not to mitochondria (Matthews et al., 2014). Another study by Freundt et al. (2009) showed that, inside the host cells, the protein encoded by the SARS-CoV are distributed between the mitochondria and nucleus (Freundt et al., 2009). Live imaging and confocal microscopy studies showed that orf3b is localized in both nucleus and mitochondria in Vero cells. Very interestingly, orf3b is also reported to affect the spatiotemporal distribution within the infected cells. For instance, orf3b was initially detected in the nucleus and gets translocated to the mitochondria over time with the help of a mitochondrial translocation signal (Freundt et al., 2009). Besides, orf3b has a nuclear export sequence rich in leucine to be transported from the nucleus. The interaction between leucine-rich nuclear export and CRM1 facilitates the nuclear transport of orf3b. Murine coronavirus replication protein nsp4 is partially localized to mitochondria and causes disrupted mitochondrial morphology (Freundt et al.). Also, the cells infected with Alb ts6 icv showed larger and extensively vacuolated mitochondria (Clementz et al., 2008). Changes in the mitochondrial structure are also linked with host cell response against viruses. For instance, orf9b by inducing degradation of DRP1 to elongate mitochondria, which is linked with host cell interferon response against SARS-CoV. SARS-CoV also targets the respiratory chain complex I proteins. A study by Pfefferle and co-workers have shown that the respiratory chain complex I proteins is one of the key targets of SARS-CoV (Pfefferle et al., 2011).
8. SARS-CoV induction of oxidative stress
It is now well established that viruses induce oxidative stress in the host cell for their survival. Studies on SARS-CoV also suggest a similar mechanism. A study by Lin and colleagues (2006) has demonstrated that SARS-CoV induces oxidative stress in the host cells. An increase in ROS and defective ROS scavenging/anti-oxidant systems contributes to oxidative stress, inflammation, and activation of immune responses. SARS-CoV 3CLpro significantly increases the ROS levels and activation of the NF-kB pathway in HL-CZ cells (Lin et al., 2006). Besides, the increase in ROS was linked with the induction of apoptosis in HL-CZ cells. While 3CLpro induced nuclear factor-kappa B pathway, it significantly inhibited AP1-dependent transcription. Oxidative stress-induced transcription of NF-kB signaling is reported as an inducer of apoptosis in several types of cells. Another study by Zhang et al. (2007) demonstrated that the apoptosis induction by the SARS-CoV nucleocapsid protein is a mitochondrial-dependent pathway. Transfection of nucleocapsid gene into COS-1 cells reduced MMP, increased ROS production, and apoptosis induction by cytochrome release into the cytoplasm, caspase-3 activation, and PARP cleavage (Zhang et al., 2007). Very interestingly, the SARS-CoV membrane gene (M) and spike gene (S) were incapable of inducing apoptosis in COS-1 cells.
Acute lung injury upon infection with SARS-CoV is one of the causes of respiratory failure and a high mortality rate. Activation of oxidative stress pathway together with pro-inflammatory host response is proposed to contribute to the onset of lung injury. Oxidative stress is reported to induce the expression of pro-inflammatory cytokine genes (Mirowsky et al., 2016). SARS-CoV infected aged macaques showed a stronger pro-inflammatory response as opposed to young macaques (Smits et al., 2010). Orf7a encoded by SARS-CoV is reported in various pathological condition, including inappropriate induction of apoptosis and inhibition of protein synthesis inside the host cell. Screening by yeast two-hybrid approach and co-immunoprecipitation analysis identified orf7a to interact with Ap4A-hydrolase. Further, the Ap4A-hydrolase level gets elevated upon oxidative stress.
SARS-CoV orf3a protein stimulates the secretion of IL-1β via efflux of K+ and ROS production. By disrupting intracellular ionic concentration, orf3a induces mitochondrial damage leading to activation of NLRP3 inflammasome. SARS-CoV papain-like protease (PLpro) upregulates the pro-fibrotic responses in lung cells. PLpro triggered TGF-β1 activation in an Egr-1 dependent manner by ROS/p38 MAPK/STAT3 pathway. By localizing inside the mitochondria and targeting the MAVS/TRAF3/TRAF6 axis orf9b suppresses innate immunity and targets the MAVS/TRAF3/TRAF6 axis to suppress innate immunity (Shi et al., 2014a). Further, another SARS-CoV protein, orf8a is reported within mitochondria. Orf8a protein plays a critical role in increasing MMP and ROS production, cellular oxygen consumption rate, inducing apoptosis by caspase-3 activation in Vero, HEK293, and Huh7 cells (Keng et al., 2006).
9. SARS-CoV proteins induce mitophagy
Virus-induced mitophagy is implicated in viral propagation. Studies have documented the induction of mitophagy upon infection by SARS-CoV. Zhu et al. (2016) showed the induction of mitophagy upon TGEV infection. Virus infection induces mitophagy to escape from host immunity and completion of the viral life cycle. Orf9b counteracts the stress, which fragments and aggregates mitochondria to promote host cell survival during viral replication (Zhu et al., 2016).
10. Molecular determinants of mitochondrial localization of SARS-CoV
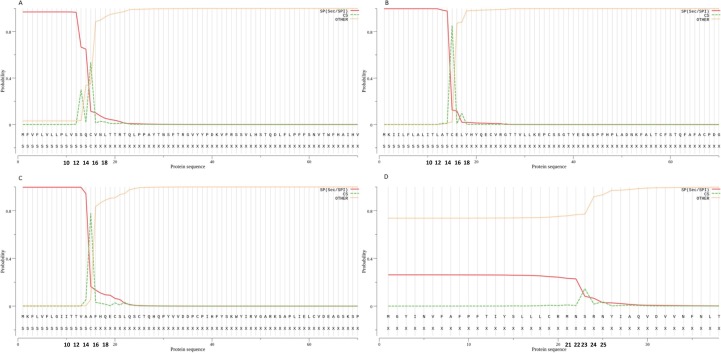
SARS-CoV encoded proteins have been shown to affect both structure and function of mitochondria. The mitochondrial dysfunction could be due to the localization of SARS-CoV proteins into mitochondria. Interestingly, both proteins and RNA of SARS-CoV possess a distinct mitochondrial translocation signal (Singh et al., 2020). The accessory protein orf3b encoded by the SARS-CoV2 genome is localized in the outer membranes of mitochondria. Besides mitochondria, orf3b is also localized to the nucleus. Orf3b possesses a nuclear export sequence-dependent on CRM1 and is predicted to process an amphipathic α-helix sequence containing two lysine residues, which may bind to the outer membranes of the mitochondria (Freundt et al., 2009). Another study reported the presence of mitochondrial translocation signals in the 5′- and 3′-untranslated regions of SARS-CoV2 (Singh et al., 2020, Wu et al., 2020). Further, we have predicted the mitochondrial signal peptides, signal peptide cleavage site, and mitochondrial processing peptidase cleavage site in SARS-CoV2 encoded proteins using SignalP v5.0 tool, MitoFates, and TargetP tools (Almagro Armenteros et al., 2019, Emanuelsson et al., 2000, Fukasawa et al., 2015). Our bioinformatic analysis of the SARS-CoV-2 genome identified Spike protein, orf7a, and orf8 to possess a mitochondrial signal peptide cleavage site (Fig. 2 ). However, these findings need to be experimentally confirmed. Experiments are ongoing in our laboratories to identify the function of these proteins in manipulating host mitochondria by SARS-CoV-2.
Fig. 2.
Signal peptide cleavage sites in SARS-CoV2 peptides. A) spike protein with a probability score of 0.968. B) ORF7a protein with a probability score of 0.998. C) ORF8 protein with a probability score of 0.997. D) ORF10 protein with a probability score of 0.262.
11. SARS-CoV target mitochondria for anti-viral immunity
Mitochondria play a critical role in generating anti-viral immunity to protect the host cell. Mitochondria, through MAVS, plays a vital role in the production of IFN. Orf9a physically binds and promotes K48 mediated ubiquitination-dependent proteasomal degradation of MAVS to disrupt the production of IFN. Mechanistic studies have reported that both ROS or mtDNA from damaged mitochondria are reported to activate the NLRP3 inflammasome pathway. By associating with mitochondrial antiviral signaling (MAVS) or mitofusin 2, activated NLRP3 is translocated to the outer mitochondrial membrane leading to caspase-1 activation. Active caspase-1 triggers IL-1β and IL-18 secretion. Another study showed the upregulation of genes encoded by the mtDNA in the peripheral blood mononuclear cell (PBMC) of convalescent SARS patients (Zhou et al., 2012). Besides, gene-related to oxidative stress, heat shock proteins, and cytokines were also significantly elevated. The co-upregulation of mitochondrial genes and cytokines connects the possible cross-talk between mitochondria and antiviral immunity. However, comprehensive studies are required to investigate the molecular mechanism behind the role of compromised mitochondrial function with antiviral immunity.
12. Prospects of targeting the virus-mitochondria interactions
The proteins of SARS-CoV2 have been shown to trigger apoptosis and cytokine storm in the host cell. Patients with severe and acute respiratory syndrome show significantly higher levels of pro-inflammatory cytokines in their peripheral blood (Costela-Ruiz et al., 2020). The uncontrolled increase in pro-inflammatory cytokines or cytokine storm has been shown to manifest severe distress leading to organ dysfunction (Ye et al., 2020). Mitochondrial dysfunction is a key source of ROS and has been speculated to trigger inflammatory response and induction of cytokine storm. Both mitochondrial dysfunctions leading to increased ROS and inflammation have been linked with innate immune system activation and induction of NLRP3 inflammasome, and induction of cytokine storms (Kaivola et al., 2021). For instance, CoV Envelope protein, ORF3a, and ORF8b have been shown to activate the inflammasome. The CoV envelope protein-induced calcium influx stimulates mitochondria to generate ROS (Kaivola et al., 2021). ORF3a induced K + efflux have been shown to participate and promote the assembly of inflammasomes via the promotion of TRAF3-ORF3a interaction resulting in NF- κβ activation and transcription of pro-inflammatory cytokines notably pro–IL-1β and IL-18 (Siu et al., 2019, Yap et al., 2020). ORF8b has been shown to directly interact and stimulate NLRP3. Activation of NLRP3 has been shown to induce pores in mitochondria and plasma membranes and may stimulate the IL-1β/IL-18 secretion and induction of a series of cascades leading to cytosolic secretion of cytochrome-C and apoptosis (Shah, 2020). These data collectively suggest that the SARS-COV2 encoded proteins induced structural and functional alteration in mitochondria can induce mitophagy. Thus, SARS-CoV2 infection can initiate signaling events leading to apoptosis activation via mitochondrial damage, induction of ROS and inflammasome, and cytokine storm.
Given the important role of mitochondrial role in inflammation and cytokine storm during SARS-COV2 pathogenesis, protecting the mitochondria from inflammation-induced damage might be very attractive. In recent times, several molecules that can protect mitochondria have been developed. Since, mitochondrial damage appears to be an upstream event in the induction of inflammation and inflammasome assembly, targeting this crosstalk could be a promising approach for improved management of COVID-19 patients. We thus, propose that targeting the virus and mitochondria nexus may boost the host immune pathways and promote cell survival and prevent premature apoptosis of the cell. Towards this, mitochondrial pharmacology may be attempted to target the virus-mitochondria interaction. Besides, mitochondria-mediated ROS is one of the drivers of inflammation. The use of anti-oxidant agents may be an attractive and safe approach to inhibit ROS and inflammation. OXPHOS modulators, mitochondrial pyruvate carrier, pharmacological induction of mitochondrial biogenesis, mitochondrial redox state, mitochondrial dynamic needs detailed investigation as pharmacological targets for the management of COVID-19 patients. Moreover, blocking the interaction between SARS-CoV protein with mitochondria and induction of inflammasome by RNAi approach may represent a viable strategy to prevent the anti-immune signaling for SARS-CoV2 and prevent the upstream events amplifying the effects of ROS, inflammasome, and cytokine storm.
13. Altered mitochondrial function in individuals with post-COVID complications
The COVID-19 pandemic has ravaged most of the world, with Asian countries, including India, being most affected. Earlier studies have shown that viral infections are linked with impaired mitochondrial function causing long-term cognitive and metabolic perturbation in patients (Katz et al., 2010, Sweetman et al., 2020). Studies have shown that individuals with primary mitochondrial disorders are at high risk of developing severe complications post-SARS-CoV-2 exposure (Pizzamiglio et al., 2022, Singh et al., 2020). The mechanism of altered metabolic adaptation is due to the SARs virus regulating the host metabolism machinery and redirecting them to viral replication (Elesela and Lukacs, 2021). Research studies have also indicated that patients with post-COVID complications, including long COVID, have been associated with chronic fatigue, and neuropsychiatric and neurometabolic disorders (Kedor et al., 2022). These studies indicated that there is a possibility that post-SARS-CoV-2 infection, patients with long COVID symptoms involving CNS symptoms, myalgic encephalomyelitis, and cognitive dysfunction due to perturbed mitochondrial metabolic pathway (Booth et al., 2012, Paul et al., 2021). Post-COVID complications may arise from perpetual metabolic imbalance contributed by mitochondrial dysfunction and chronic inflammation leading to long COVID symptoms (de Boer et al., 2022, Nunn et al., 2022). An earlier study has shown that SARS-CoV-2 can regulate mitochondrial oxidative phosphorylation (OXPHOS), apoptosis, and ATP levels in the airway epithelial cells in the host, contributing to hypoxemia in patients (Archer et al., 2022). Yet another study has shown that patients with long COVID may have limited capacity to return to normal exercise routine due to impaired oxygenation in muscle tissue (Singh et al., 2022). Mitochondrial therapy, including antioxidant CoQ10, glycolysis inhibitors, Vitamin E, and minerals, along with regular exercise, may help patients with long COVID symptoms for faster recovery (Wood et al., 2021).
14. Conclusion
Despite significant progress, the role of mitochondria in the pathophysiology of COVID-19 remains poorly understood. Experimental evidence suggests the potential role of mitochondria in controlling immunity against SARS-CoV-2. Research has shown that coronavirus infection leads to alteration in both structure and function of the mitochondria. These mitochondrial alterations are likely to alter cross-talk between mitochondria and the nucleus during SARS-CoV infection. The relevance of these interactions during SARS pathogenesis requires detailed investigation. Mechanistic studies about mitochondrial dynamics may open new avenues to design and target novel therapeutic strategies against SARS-CoV. Since many SARS-CoV proteins localize to mitochondria, inhibiting their translocation can be useful as a treatment strategy against SARS infection. In conclusion, studies focusing on SARS-CoV-2 manipulation of key mitochondrial functions such as energetics, anterograde and retrograde signaling, anti-viral signaling, crosstalk between mitochondria, and crosstalk between mitochondria and other organellar should fill the existing gap to advance prevention or treatment of COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Dr. TMA Pai structured Ph.D. fellowship and ICMR-Senior Research fellowship, Government of India [BMI/11(107)/2020] to Mr. Pradyumna and Dr. TMA Pai Foundation Endowment award for Dr. K. Satyamoorthy from Manipal Academy of Higher Education. All the authors thank Manipal Academy of Higher Education, Manipal, Technology Information Forecasting and Assessment Council (TIFAC)-Core in Pharmacogenomics at MAHE, Manipal, Fund for Improvement of S&T Infrastructure (FIST) and Karnataka Fund for Infrastructure Strengthening in Science and Technology (K-FIST), Government of Karnataka.
Author contributions
S.P. Kabekkodu , S. Chakrabarty and K. Satyamoorthy designed study, P. Jayaram and S. Mallya analyzed data, SP. Kabekkodu, S. Chakrabarty and K. Satyamoorthy wrote the paper, K. Thangaraj, K.K. Singh and K. Satyamoorthy revised and updated the manuscript.
Data availability
The raw data is available from public database Gene Expression Omnibus with GEO IDs: GSE150316, GSE147507 and GSE154998. The analyzed result are provided in the respective table in the manuscript.
Reference
- Alavi M.V., Fuhrmann N. Dominant optic atrophy, OPA1, and mitochondrial quality control: Understanding mitochondrial network dynamics. BioMed Central. 2013 doi: 10.1186/1750-1326-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Angajala A., Lim S., Phillips J.B., Kim J.H., Yates C., You Z., Tan M. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front. Immunol. 2018;9:1605. doi: 10.3389/fimmu.2018.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S.L., Dasgupta A., Chen K.H., Wu D., Baid K., Mamatis J.E., Gonzalez V., Read A., Bentley R.E., Martin A.Y., Mewburn J.D., Dunham-Snary K.J., Evans G.A., Levy G., Jones O., Al-Qazazi R., Ring B., Alizadeh E., Hindmarch C.C., Rossi J., Lima P.D., Falzarano D., Banerjee A., Colpitts C.C. SARS-CoV-2 mitochondriopathy in COVID-19 pneumonia exacerbates hypoxemia. Redox Biol. 2022;58 doi: 10.1016/j.redox.2022.102508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti, I., Ysrafil, 2020a. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab Syndr 14, 407-412. [DOI] [PMC free article] [PubMed]
- Banoth B., Cassel S.L. Mitochondria in innate immune signaling. Transl. Res. 2018;202:52–68. doi: 10.1016/j.trsl.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier V., Lang D., Valois S., Rothman A.L., Medin C.L. Dengue virus induces mitochondrial elongation through impairment of Drp1-triggered mitochondrial fission. Virology. 2017;500:149–160. doi: 10.1016/j.virol.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Liu P., Leibowitz J.L., Kao C.C. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012;86:4294–4304. doi: 10.1128/JVI.07012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo, D., Nilsson-Payant, B.E., Liu, W.-C., Uhl, S., Hoagland, D., Møller, R., Jordan, T.X., Oishi, K., Panis, M., Sachs, D., Wang, T.T., Schwartz, R.E., Lim, J.K., Albrecht, R.A., TenOever, B.R., 2020. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036-1045.e1039. [DOI] [PMC free article] [PubMed]
- Booth N.E., Myhill S., McLaren-Howard J. Mitochondrial dysfunction and the pathophysiology of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) Int. J. Clin. Exp. Med. 2012;5:208–220. [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtscher, J., Cappellano, G., Omori, A., Koshiba, T., Millet, G.P., 2020. Mitochondria: In the Cross Fire of SARS-CoV-2 and Immunity. iScience 23, 101631. [DOI] [PMC free article] [PubMed]
- Cao Z., Xia H., Rajsbaum R., Xia X., Wang H., Shi P.-Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 2021;18:746–748. doi: 10.1038/s41423-020-00603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Hernandez R., Jacome R., Lopez Vidal Y., Ponce de Leon S. Are RNA viruses candidate agents for the next global pandemic? A review. ILAR J. 2017;58:343–358. doi: 10.1093/ilar/ilx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Ma C.W., Chan W.Y., Chan H.Y. The SARS-Coronavirus Membrane protein induces apoptosis through modulating the Akt survival pathway. Arch. Biochem. Biophys. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanturiya A.N., Basanez G., Schubert U., Henklein P., Yewdell J.W., Zimmerberg J. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J. Virol. 2004;78:6304–6312. doi: 10.1128/JVI.78.12.6304-6312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-Y.C.Y., Ping, Y.-H.Y.H., Lee, H.-C.H.C., Chen, K.H.K.-H., Lee, Y.-M.Y.M., Chan, Y.J.Y.-J., Lien, T.-C.T.C., Jap, T.-S.T.S., Lin, C.-H.C.H., Kao, L.-S.L.S., Chen, Y.-M.A.Y.Ming A., 2007. Open Reading Frame 8a of the Human Severe Acute Respiratory Syndrome Coronavirus Not Only Promotes Viral Replication but Also Induces Apoptosis. The Journal of Infectious Diseases 196, 405-415. [DOI] [PMC free article] [PubMed]
- Chernyak B.V., Popova E.N., Prikhodko A.S., Grebenchikov O.A., Zinovkina L.A., Zinovkin R.A. COVID-19 and Oxidative Stress. Biochemistry (Mosc.) 2020;85:1543–1553. doi: 10.1134/S0006297920120068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K.Y.C., Yeung Y.S., Hon C.C., Zeng F., Law K.M., Leung F.C.C. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 2005;579:6699–6704. doi: 10.1016/j.febslet.2005.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz M.A., Kanjanahaluethai A., O'Brien T.E., Baker S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins L.V., Hajizadeh S., Holme E., Jonsson I.-M., Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 2004;75:995–1000. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., Kuhn P., Buchmeier M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 2 Interacts with a Host Protein Complex Involved in Mitochondrial Biogenesis and Intracellular Signaling. J. Virol. 2009;83:10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodriguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth F R. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E., Petrache I., Goldstein N.M., Olin J.T., Keith R.C., Modena B., Mohning M.P., Yunt Z.X., San-Millan I., Swigris J.J. Decreased Fatty Acid Oxidation and Altered Lactate Production during Exercise in Patients with Post-acute COVID-19 Syndrome. Am. J. Respir. Crit. Care Med. 2022;205:126–129. doi: 10.1164/rccm.202108-1903LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jiménez-Guardeño J.M., Regla-Nava J.A., Álvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011;7:e1002315–e. doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehipawala, S., Cheung, E., Tremberger, G., Cheung, T., 2021. Entropy and Fractal Dimension Study of the TDP-43 Protein Low Complexity Domain Sequence in ALS Disease Severity and SARS-CoV-2 Gene Sequences in Virulence Variability. Entropy (Basel) 23. [DOI] [PMC free article] [PubMed]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X., Hackbart, M., Mettelman, R.C., O'Brien, A., Mielech, A.M., Yi, G., Kao, C.C., Baker, S.C., 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. P Natl Acad Sci USA 114, E4251-E4260. [DOI] [PMC free article] [PubMed]
- Desai N., Neyaz A., Szabolcs A., Shih A.R., Chen J.H., Thapar V., Nieman L.T., Solovyov A., Mehta A., Lieb D.J., Kulkarni A.S., Jaicks C., Xu K.H., Raabe M.J., Pinto C.J., Juric D., Chebib I., Colvin R.B., Kim A.Y., Monroe R., Warren S.E., Danaher P., Reeves J.W., Gong J., Rueckert E.H., Greenbaum B.D., Hacohen N., Lagana S.M., Rivera M.N., Sholl L.M., Stone J.R., Ting D.T., Deshpande V. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elesela, S., Lukacs, N.W., 2021. Role of Mitochondria in Viral Infections. Life (Basel) 11. [DOI] [PMC free article] [PubMed]
- Emanuelsson O., Nielsen H., Brunak S., Von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fang P., Fang L., Zhang H., Xia S., Xiao S. Functions of coronavirus accessory proteins: overview of the state of the art. Viruses. 2021;13 doi: 10.3390/v13061139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Park E., Lenardo M.J., Xu X.-N. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J. Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y., Tsuji J., Fu S.C., Tomii K., Horton P., Imai K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, T.S., Liao, Y., Liu, D.X., 2016. Regulation of stress responses and translational control by coronavirus. MDPI AG. [DOI] [PMC free article] [PubMed]
- Ganji R., Reddy P.H. Impact of COVID-19 on Mitochondrial-Based Immunity in Aging and Age-Related Diseases. Front. Aging Neurosci. 2020;12 doi: 10.3389/fnagi.2020.614650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.E., dos Santos C.C., O’Gorman D.B., Carter D.E., Patterson E.K., Slessarev M., Martin C., Daley M., Miller M.R., Cepinskas G., Fraser D.D. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med. Exp. 2020;8 doi: 10.1186/s40635-020-00361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E., Jang, G.M., Bouhaddou, M., Xu, J., Obernier, K., Meara, M.J., Guo, J.Z., Swaney, D.L., Tummino, T.A., Huttenhain, R., Kaake, R.M., Richards, A.L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., Kim, M., Haas, P., Polacco, B.J., Braberg, H., Fabius, J.M., Eckhardt, M., Soucheray, M., Bennett, M.J., Cakir, M., McGregor, M.J., Li, Q., Naing, Z.Z.C., Zhou, Y., Peng, S., Kirby, I.T., Melnyk, J.E., Chorba, J.S., Lou, K., Dai, S.A., Shen, W., Shi, Y., Zhang, Z., Barrio-Hernandez, I., Memon, D., Hernandez-Armenta, C., Mathy, C.J.P., Perica, T., Pilla, K.B., Ganesan, S.J., Saltzberg, D.J., Ramachandran, R., Liu, X., Rosenthal, S.B., Calviello, L., Venkataramanan, S., Liboy-Lugo, J., Lin, Y., Wankowicz, S.A., Bohn, M., Sharp, P.P., Trenker, R., Young, J.M., Cavero, D.A., Hiatt, J., Roth, T., Rathore, U., Subramanian, A., Noack, J., Hubert, M., Roesch, F., Vallet, T., Meyer, B., White, K.M., Miorin, L., Rosenberg, O.S., Verba, K.A., Agard, D., Ott, M., Emerman, M., Ruggero, D., Garc, amp, amp, iacute-Sastre, A., Jura, N., von Zastrow, M., Taunton, J., Ashworth, A., Schwartz, O., Vignuzzi, M., Enfert, C., Mukherjee, S., Jacobson, M., Malik, H.S., Fujimori, D.G., Ideker, T., Craik, C.S., Floor, S., Fraser, J.S., Gross, J., Sali, A., Kortemme, T., Beltrao, P., Shokat, K., Shoichet, B.K., Krogan, N.J., 2020a. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. bioRxiv, 2020.2003.2022.002386-002020.002303.002322.002386.
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou H., Zhao M., Xu H., Yuan J., He W., Zhu M., Ding H., Yi L., Chen J. CSFV induced mitochondrial fission and mitophagy to inhibit apoptosis. Oncotarget. 2017;8:39382–39400. doi: 10.18632/oncotarget.17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmat N., Asadzadeh Z., Ahangar N.K., Alemohammad H., Najafzadeh B., Derakhshani A., Baghbanzadeh A., Baghi H.B., Javadrashid D., Najafi S., Ar Gouilh M., Baradaran B. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021;166:675–696. doi: 10.1007/s00705-021-04958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.-W., Zhang H.-N., Meng Q.-F., Xie J., Li Y., Chen H., Zheng Y.-X., Wang X.-N., Qi H., Zhang J., Wang P.-H., Han Z.-G., Tao S.-C. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.Y., Zheng B.J. Roles of spike protein in the pathogenesis of SARS coronavirus. Hong Kong Med. J. 2009;15:37–40. [PubMed] [Google Scholar]
- Kaivola, J., Nyman, T.A., Matikainen, S., 2021. Inflammasomes and SARS-CoV-2 Infection. Viruses-Basel 13. [DOI] [PMC free article] [PubMed]
- Kalashnyk O., Lykhmus O., Izmailov M., Koval L., Komisarenko S., Skok M. SARS-Cov-2 spike protein fragment 674–685 protects mitochondria from releasing cytochrome c in response to apoptogenic influence. Biochem. Biophys. Res. Commun. 2021;561:14–18. doi: 10.1016/j.bbrc.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, B.Z., Boas, S., Shiraishi, Y., Mears, C.J., Taylor, R., 2010. Exercise tolerance testing in a prospective cohort of adolescents with chronic fatigue syndrome and recovered controls following infectious mononucleosis. J Pediatr 157, 468-472, 472 e461. [DOI] [PMC free article] [PubMed]
- Kedor C., Freitag H., Meyer-Arndt L., Wittke K., Hanitsch L.G., Zoller T., Steinbeis F., Haffke M., Rudolf G., Heidecker B., Bobbert T., Spranger J., Volk H.D., Skurk C., Konietschke F., Paul F., Behrends U., Bellmann-Strobl J., Scheibenbogen C. A prospective observational study of post-COVID-19 chronic fatigue syndrome following the first pandemic wave in Germany and biomarkers associated with symptom severity. Nat. Commun. 2022;13:5104. doi: 10.1038/s41467-022-32507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng C.T., Choi Y.W., Welkers M.R.A., Chan D.Z.L., Shen S., Gee Lim S., Hong W., Tan Y.J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology. 2006;354:132–142. doi: 10.1016/j.virol.2006.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Syed G.H., Kim S.J., Siddiqui A. Mitochondrial dynamics and viral infections: A close nexus. BBA. 2015;1853:2822–2833. doi: 10.1016/j.bbamcr.2014.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Palese P. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 2006;80:785–793. doi: 10.1128/JVI.80.2.785-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Jou M.J., Huang S.Y., Li S.W., Wan L., Tsai F.J., Lin C.W. Proteomic analysis of up-regulated proteins in human promonocyte cells expressing severe acute respiratory syndrome coronavirus 3C-like protease. Proteomics. 2007;7:1446–1460. doi: 10.1002/pmic.200600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaStarza M.W., Lemm J.A., Rice C.M. Genetic analysis of the nsP3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J. Virol. 1994;68:5781–5791. doi: 10.1128/jvi.68.9.5781-5791.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan E., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M.D., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lee, J.-G., Huang, W., Lee, H., van de Leemput, J., Kane, M.A., Han, Z., 2021. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell & Bioscience 2021 11:1 11, 1-12. [DOI] [PMC free article] [PubMed]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang L., Dong C., Che Y., Jiang L., Liu L., Zhao H., Liao Y., Sheng Y., Dong S., Ma S. The interaction of the SARS coronavirus non-structural protein 10 with the cellular oxido-reductase system causes an extensive cytopathic effect. J. Clin. Virol. 2005;34:133–139. doi: 10.1016/j.jcv.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hou P., Ma W., Wang X., Wang H., Yu Z., Chang H., Wang T., Jin S., Wang X., Wang W., Zhao Y., Zhao Y., Xu C., Ma X., Gao Y., He H. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell. Mol. Immunol. 2022;19:67–78. doi: 10.1038/s41423-021-00807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Lin K.H., Hsieh T.H., Shiu S.Y., Li J.Y. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 2006;46:375–380. doi: 10.1111/j.1574-695X.2006.00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. P Natl Acad Sci USA. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.L., Coleman C.M., van der Meer Y., Snijder E.J., Frieman M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Multidisciplinary Digital Publishing Institute (MDPI) 2012:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirowsky J.E., Dailey L.A., Devlin R.B. Differential expression of pro-inflammatory and oxidative stress mediators induced by nitrogen dioxide and ozone in primary human bronchial epithelial cells. Inhal. Toxicol. 2016;28:374–382. doi: 10.1080/08958378.2016.1185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn, A.V.W., Guy, G.W., Brysch, W., Bell, J.D., 2022. Understanding Long COVID; Mitochondrial Health and Adaptation-Old Pathways, New Problems. Biomedicines 10. [DOI] [PMC free article] [PubMed]
- Nunn A.V.W., Guy G.W., Brysch W., Botchway S.W., Frasch W., Calabrese E.J., Bell J.D. SARS-CoV-2 and mitochondrial health: implications of lifestyle and ageing. Immun. Ageing. 2020;17:33. doi: 10.1186/s12979-020-00204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B.D., Lemle M.D., Komaroff A.L., Snyder S.H. Redox imbalance links COVID-19 and myalgic encephalomyelitis/chronic fatigue syndrome. PNAS. 2021;118 doi: 10.1073/pnas.2024358118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle, S., Schöpf, J., Kögl, M., Friedel, C.C., Müller, M.A., Carbajo-Lozoya, J., Stellberger, T., von Dall’Armi, E., Herzog, P., Kallies, S., Niemeyer, D., Ditt, V., Kuri, T., Züst, R., Pumpor, K., Hilgenfeld, R., Schwarz, F., Zimmer, R., Steffen, I., Weber, F., Thiel, V., Herrler, G., Thiel, H.-J., Schwegmann-Weßels, C., Pöhlmann, S., Haas, J., Drosten, C., von Brunn, A., 2011. The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors. PLoS Pathogens 7, e1002331-e1002331. [DOI] [PMC free article] [PubMed]
- Pizzamiglio C., Machado P.M., Thomas R.H., Gorman G.S., McFarland R., Hanna M.G., Pitceathly R.D.S., Mito C.-S.-G. COVID-19-related outcomes in primary mitochondrial diseases: an international study. Neurology. 2022;98:576–582. doi: 10.1212/WNL.0000000000200240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffran H.A., Pare J.M., Corcoran J.A., Weller S.K., Smiley J.R. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39–e. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 2007;81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Kurz S., Manske K., Janas M., Heikenwälder M., Misgeld T., Aichler M., Weissmann S.F., Zischka H., Knolle P., Wohlleber D. Single organelle analysis to characterize mitochondrial function and crosstalk during viral infection. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-44922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S., Qi H.-Y., Boularan C., Huang N.-N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discovery. 2019;5 doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I., Joseph P., Heerdt P.M., Cullinan M., Lutchmansingh D.D., Gulati M., Possick J.D., Systrom D.M., Waxman A.B. Persistent exertional intolerance after COVID-19: insights from invasive cardiopulmonary exercise testing. Chest. 2022;161:54–63. doi: 10.1016/j.chest.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K.K., Chaubey, G., Chen, J.Y., Suravajhala, P., 2020. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. American Journal of Physiology-Cell Physiology 319, C258-C267. [DOI] [PMC free article] [PubMed]
- Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanina, H., Madhugiri, R., Bylapudi, G., Schultheiß, K., Karl, N., Gulyaeva, A., Gorbalenya, A.E., Linne, U., Ziebuhr, J., 2021. Coronavirus replication–transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed]
- Smits, S.L., De Lang, A., Van Den Brand, J.M.A., Leijten, L.M., Van Ijcken, W.F., Eijkemans, M.J.C., Van Amerongen, G., Kuiken, T., Andeweg, A.C., Osterhaus, A.D.M.E., Haagmans, B.L., 2010. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathogens 6. [DOI] [PMC free article] [PubMed]
- Stobart C.C., Sexton N.R., Munjal H., Lu X., Molland K.L., Tomar S., Mesecar A.D., Denison M.R. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 2013;87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi, L., Posthuma, C.C., Collet, A., Zevenhoven-Dobbe, J.C., Gorbalenya, A.E., Decroly, E., Snijder, E.J., Canard, B., Imbert, I., 2014. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. P Natl Acad Sci USA 111, E3900-E3909. [DOI] [PMC free article] [PubMed]
- Surjit M., Liu B., Jameel S., Chow V.T.K., Lal S.K. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13–18. doi: 10.1042/BJ20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L.M., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 Replicase Protein of SARS-Coronavirus, Structure and Functional Insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman E., Kleffmann T., Edgar C., de Lange M., Vallings R., Tate W. A SWATH-MS analysis of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome peripheral blood mononuclear cell proteomes reveals mitochondrial dysfunction. J. Transl. Med. 2020;18:365. doi: 10.1186/s12967-020-02533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-J.-Y.-X., Tan T.H.P., Lee M.J.R., Tham P.-Y., Gunalan V., Druce J., Birch C., Catton M., Fu N.Y., Yu V.C., Tan Y.-J.-Y.-X. Induction of Apoptosis by the Severe Acute Respiratory Syndrome Coronavirus 7a Protein Is Dependent on Its Interaction with the Bcl-XL Protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T., Sundberg E.J., Frieman M.B. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89:11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilokani L., Nagashima S., Paupe V., Prudent J. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem. 2018;62:341–360. doi: 10.1042/EBC20170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi H., Li L., Chen Z.S., Lau K.F., Tsui S.K.W., Chan H.Y.E. The SARS-coronavirus membrane protein induces apoptosis via interfering with PDK1PKB/Akt signalling. Biochem. J. 2014;464:439–447. doi: 10.1042/BJ20131461. [DOI] [PubMed] [Google Scholar]
- Valdes-Aguayo, J.J., Garza-Veloz, I., Badillo-Almaraz, J.I., Bernal-Silva, S., Martinez-Vazquez, M.C., Juarez-Alcala, V., Vargas-Rodriguez, J.R., Gaeta-Velasco, M.L., Gonzalez-Fuentes, C., Avila-Carrasco, L., Martinez-Fierro, M.L., 2021. Mitochondria and Mitochondrial DNA: Key Elements in the Pathogenesis and Exacerbation of the Inflammatory State Caused by COVID-19. Medicina (Kaunas) 57. [DOI] [PMC free article] [PubMed]
- Varshney B., Lal S.K. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry. 2011;50:5419–5425. doi: 10.1021/bi200303r. [DOI] [PubMed] [Google Scholar]
- Wiedmer A., Wang P., Zhou J., Rennekamp A.J., Tiranti V., Zeviani M., Lieberman P.M. Epstein-barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 2008;82:4647–4655. doi: 10.1128/JVI.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E., Hall K.H., Tate W. Role of mitochondria, oxidative stress and the response to antioxidants in myalgic encephalomyelitis/chronic fatigue syndrome: a possible approach to SARS-CoV-2 'long-haulers'? Chronic Dis Transl Med. 2021;7:14–26. doi: 10.1016/j.cdtm.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.E., Fazal F.M., Parker K.R., Zou J., Chang H.Y. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst. 2020;11:102–108.e103. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.D., Shang B., Yang R.F., Yu H., Ma Z.H., Shen X., Ji Y.Y., Lin Y., Wu Y.D., Lin G.M., Tian L., Gan X.Q., Yang S., Jiang W.H., Dai E.H., Wang X.Y., Jiang H.L., Xie Y.H., Zhu X.L., Pei G., Li L., Wu J.R., Sun B. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiao Z., Ye K., He X., Sun B., Qin Z., Yu J., Yao J., Wu Q., Bao Z., Zhao W. SARS-CoV-2: characteristics and current advances in research. Virol. J. 2020;17:117. doi: 10.1186/s12985-020-01369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Xiong Z., Zhang S., Yan Y., Nguyen J., Ng B., Lu H., Brendese J., Yang F., Wang H., Yang X.F. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap J.K.Y., Moriyama M., Iwasaki A. Inflammasomes and Pyroptosis as Therapeutic Targets for COVID-19. J. Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wang B.L., Mao J.H. The pathogenesis and treatment of the 'Cytokine Storm' in COVID-19. J Infection. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Wong C.K., Li P., Xie Y. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Biophys. Acta Gen. Subj. 2008;1780:1383–1387. doi: 10.1016/j.bbagen.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.J., Copeland W.C. Human mitochondrial DNA replication machinery and disease. Curr. Opin. Genet. Dev. 2016;38:52–62. doi: 10.1016/j.gde.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wei L., Jiang D., Wang J., Cong X., Fei R. SARS-CoV nucleocapsid protein induced apoptosis of COS-1 mediated by the mitochondrial pathway. Artificial Cells, Blood Substitutes, and Biotechnology. 2007;35:237–253. doi: 10.1080/10731190601188422. [DOI] [PubMed] [Google Scholar]
- Zhou P., Li H., Wang H., Wang L.F., Shi Z. Bat severe acute respiratory syndrome-like coronavirus ORF3b homologues display different interferon antagonist activities. J. Gen. Virol. 2012;93:275–281. doi: 10.1099/vir.0.033589-0. [DOI] [PubMed] [Google Scholar]
- Zhu L., Mou C., Yang X., Lin J., Yang Q. Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget. 2016;7:27122–27141. doi: 10.18632/oncotarget.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data is available from public database Gene Expression Omnibus with GEO IDs: GSE150316, GSE147507 and GSE154998. The analyzed result are provided in the respective table in the manuscript.