Abstract
The epidermal growth factor receptor (EGFR) is commonly upregulated in multiple cancer types, including breast cancer. In the present study, evidence is provided in support of the premise that upregulation of the EGFR/MEK1/MAPK1/2 signaling axis during antiestrogen treatment facilitates the escape of breast cancer cells from BimEL-dependent apoptosis, conferring resistance to therapy. This conclusion is based on the findings that ectopic BimEL cDNA overexpression and confocal imaging studies confirm the pro-apoptotic role of BimEL in ERα expressing breast cancer cells and that upregulated EGFR/MEK1/MAPK1/2 signaling blocks BimEL pro-apoptotic action in an antiestrogen-resistant breast cancer cell model. In addition, the present study identified a pro-survival role for autophagy in antiestrogen resistance while EGFR inhibitor studies demonstrated that a significant percentage of antiestrogen-resistant breast cancer cells survive EGFR targeting by pro-survival autophagy. These pre-clinical studies establish the possibility that targeting both the MEK1/MAPK1/2 signaling axis and pro-survival autophagy may be required to eradicate breast cancer cell survival and prevent the development of antiestrogen resistance following hormone treatments. The present study uniquely identified EGFR upregulation as one of the mechanisms breast cancer cells utilize to evade the cytotoxic effects of antiestrogens mediated through BimEL-dependent apoptosis.
Keywords: MEK1/MAPK1/2 inhibitors, pro-survival autophagy, BimEL, apoptosis, antiestrogen, antiprogestin, EGFR, breast cancer, antiestrogen resistance, senescence
Introduction
Our long-term goal is to identify molecular targets to circumvent the development of endocrine resistance and breast cancer progression. Breast cancer is the most common cancer among women in the United States; ~70% of breast cancers express estrogen receptor alpha (ERα). ERα expressing tumors (ER+), which include the luminal A and luminal B subtypes, are typically treated with a selective estrogen receptor modulator (SERM), a selective estrogen receptor down-regulator (SERD), or an aromatase inhibitor (AI). These antiestrogen therapies are administered prior to and/or following surgery, localized radiation, and/or chemotherapy depending on stage and tumor subtype (1). Tamoxifen (TAM) has been the most commonly used SERM (2) and still remains the standard adjuvant treatment for pre-menopausal women with ER+ breast cancer. However, AIs show increased efficacy compared with TAM therapy (3) for post-menopausal women with all stages of ER+ breast cancer and are becoming the preferred treatment.
Regardless of the hormone used as a first-line endocrine treatment of breast cancer, intrinsic or acquired resistance remains a significant clinical challenge (4). Once resistance to a specific antiestrogen modality is identified, clinical benefit is often achieved if the patient is placed on a different antiestrogen therapy. However, the continued development/expression of antiestrogen resistance often persists and ER+ breast cancers that are refractory to hormone therapy are the most common cause of breast cancer death (5). Multiple mechanisms of endocrine resistance have been identified (6), primarily from pre-clinical studies utilizing appropriate breast cancer cell models that have been recently reviewed (7). CDK4/6 inhibitors are now commonly combined with antiestrogen treatment to improve outcomes and reduce the development of antiestrogen resistance for advanced breast cancer (8). Yet, resistance to CDK4/6 inhibitors also develops leading to relapse (9) with minimal information available on the mechanisms of resistance (10).
With the goal of improving the efficacy of antiestrogen treatment and reducing the emergence of antiestrogen-resistant breast cancer cells, our early studies established that the combined treatment of an antiestrogen plus an antiprogestin, such as mifepristone (MIF), compared with antiestrogen treatment alone, more effectively induced apoptosis and growth arrest of ER+ breast cancer cells (11-14). These earlier studies were motivated by small-scale clinical trials that showed efficacy for the antiprogestin action of MIF as a single agent for breast cancer treatment (15). Although the efficacy of this combined treatment was superior to that of antiestrogen single-agent treatment, a robust apoptotic response required the additional targeting of MEK1, the mitogen activated protein (MAP) kinase (16). In the aforementioned previous study, it was established by the authors that targeting MEK1 blocked the activation of the mitogen activated kinases MAPK1/2, also referred to as extracellular signal-regulated protein kinases ERK1 and ERK2. Targeting MAPK1/2 resulted in elevated cellular levels of dephosphorylated Bim extra-long (dBimEL), a pro-apoptotic member of the BH3 family. MEK1 targeting reduced active, phosphorylated MAPK1/2 (pMAPK1/2), established downstream effectors of MEK1, and the reduction of pMAPK1/2 activity increased the levels of dBimEL in breast cancer cells (16). Further, siRNA targeting studies identified dBimEL to be required for the cytotoxic effects of 4-hydroxytamoxifen (4-OHT) and/or MIF treatments, particularly if these treatments were conducted under conditions of MEK1/MAPK1/2 blockade (16).
BimEL is an established pro-apoptotic member of the BH3 family, and a major isoform encoded by the Bim gene, designated Bcl-2-like protein 11 (BCL2L11). A key role for Bim deletion polymorphisms may be involved in the response of patients to kinase-targeted therapies (17). Pre-clinical studies also identified a key role for BimEL in mediating sensitivity to histone deacetylase inhibitors (18) and oncogene-targeted therapies, which have previously been reviewed (19). Additionally, a previous study identified Bim deletion polymorphisms as predictive of breast cancer progression in young Asian women (20). To our knowledge, a role for BimEL in the hormonal sensitivity of breast cancer in the clinic has not been explored, but both the Raf/MEK/MAPK1/2 and PI3K/AKT pro-survival signaling cascades that are implicated in endocrine resistance stringently downregulate the levels of Bim in breast cancer cells (21). In the present study, it was hypothesized that BimEL expression and/or function would be downregulated to facilitate the development of antiestrogen resistance. To test this hypothesis, the antiestrogen-sensitive ER+ MCF-7 cells and the antiestrogen-resistant breast cancer cell model TR5 were utilized, established by a step wise 4-OHT selection of MCF-7 cells (22). TR5 cells have been used in our previous studies that uniquely identified a pro-survival role for autophagy in response to antiestrogen treatment (22,23).
Autophagy is a highly conserved cellular catabolic process that requires the expression of at least 28 autophagy (ATG) genes. Autophagy can be induced above basal levels in eukaryotic cells by various stressors, including reactive oxygen species (ROS) and nutrient deprivation to protect normal and cancer cells from death (24). The functional unit of autophagy is the autophagosome, a double-membrane organelle that must fuse with the lysosome (generating the autolysosome) to allow degradation of sequestered contents into basic macromolecules that can be released to the cell for survival purposes. Expression of LC3-II, encoded by the ATG8 gene, is commonly used as a marker of autophagy. LC3 was originally identified as a microtubule associated protein and named 'microtubule-associated-protein light-chain-3'. LC3-II is a 16 kDa protein generated from a covalent conjugation of phosphatidylethanolamine to LC3-I. LC3-I, a cytosolic 18-kDa protein, is generated by cleavage of the LC3 and is not involved in autophagosome membrane formation or function. LC3-II, however, increases and becomes peripherally membrane-associated during autophagy and is degraded with the turnover of the autolysosomal membrane. During the initial 4-OHT selection, TR5 cells showed high level LC3-II expression localized to autophagosomes, and siRNA ATG6 targeting in 4-OHT-selected TR5 cells resulted in a robust apoptotic response (22). In the present study, further analysis of the antiestrogen-resistant TR5 cells establishes a central role for the EGFR/MEK1/MAPK1/2 signaling axis as a mechanism to evade hormonally induced, dBimEL-mediated cell death and identifies pro-survival autophagy as the overriding response to EGFR and MEK1/MAPK1/2-targeted therapy.
Materials and methods
Cell culture
The antiestrogen-sensitive, ERα expressing MCF-7 breast adenocarcinoma cells were purchased from the American Type Culture Collection (ATCC). The TR5 antiestrogen-resistant cell line was established in our laboratory as previously described (22). MCF-7 and TR5 cells are routinely checked for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (cat. no. LT07-418; Lonza Bioscience). During the course of the present study and prior to manuscript submission, the antiestrogen-resistant TR5 cells and MCF-7 parent cells were subjected to STR profiling by the ATCC and confirmed to be a match to HTB-22 ATCC human MCF-7 cell line. Cells were routinely cultured in DMEM complete medium supplemented with 10% fetal bovine serum (FBS), 2% antibiotics-antimycotics (cat. no. 15240-062; Invitrogen; Thermo Fisher Scientific, Inc.), 1% sodium pyruvate (cat. no. SH3023901; Thermo Fisher Scientific, Inc.), and 10 µg/ml insulin (cat. no. I9278; MilliporeSigma). Prior to hormonal treatments, cells were placed in DMEM-F12 medium (cat. no. 11039-021; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% dextran-coated charcoal stripped FBS, designated DCC FBS, (cat. no. SFBU32-0500; Equitech-Bio Inc.) plus insulin (10.0 µg/ml). The serum concentration was stepped down to 5% DCC FBS as previously described (11,22,23). Treatments were conducted in cells seeded in the absence of insulin. Cells were allowed to adhere to the culture vessel, which required ~16-24 h. For all experiments, cells were plated at a density where overcrowding did not result due to seeding densities, minimizing effects of contact inhibition. Typically, cells were harvested at early time points (i.e., 24 or 48 h) for analysis of autophagy levels, and at later time points (72-144 h) for evaluation of cell number and/or apoptosis. To be able to reproducibly detect E2-stimulation of breast cancer cells by MTT and cell counts, a minimum of 72-96 h was required. Treatments were: 10 nM estradiol (E2; MilliporeSigma), 10 nM E2 plus 1 µM or 5 µM 4-OHT, 10 nM E2 plus 10 µM MIF in the presence and absence of 4-OHT (Sigma-Aldrich; Merck KGaA), U0126 at 5 µM or 10 µM (MilliporeSigma), chloroquine (CQ) at 10 µM (Sigma-Aldrich; Merck KGaA), Z-VAD-FMK at 10 µM (R&D systems, Inc.), MG132 at 1 µM (Sigma-Aldrich; Merck KGaA), erlotinib (ERL) at 5 or 10 µM, spautin-1 at 10 µM, and compound 19 vps34 inhibitor at 1-5 µM (Selleck Chemicals).
Cell counts
Cells were seeded in triplicate at a density to attain 50-70% confluence within 24 h. Adherent cells were treated with drugs and/or hormones for the indicated times as described in the figure legends. For cell counts, the adherent, monolayer cells were released from the culture dish by trypsinization, collected, and pooled with any detached cells collected from the culture medium. A single cell suspension was obtained by syringing three times with a 25 gauge 7/8″ needle. For some experiments, prior to cell counts, trypan blue (TB; 0.08%; Sigma-Aldrich; Merck KGaA) was added to the cell suspension for 5 min to identify non-viable cells with compromised membrane permeability. Cell counts were conducted with a hemocytometer or a Coulter Counter following dilution in Isoton II.
MTT assay
Cells were seeded at a density of 7,500 cells per well (48-72 h harvest) or 5,000 cells per well (120-144 h harvest) in 96-well clear bottom microplates (cat. no. 3603; Corning, Inc.), allowed to adhere for 16-24 h, and then treated in DMEM-F12 with 5% DCC FBS as indicated in the figure legends. At the end of the treatment period, the colorimetric MTT assay was conducted according to the manufacturer's directions (MilliporeSigma). Following solubilization of the purple formazan product that correlates to the number of viable cells per well, plates were read on a TECAN Spectrafluor Plus with a test wavelength of 570 nm and a reference wavelength of 630 nm.
Proteolysis of long-lived proteins
Analysis of long-lived protein turnover was conducted as previously described (23). In brief, cells were seeded in DMEM-F12 with 5% DCC FBS plus 10 µg/ml insulin for 24 h. The adherent cells were then incubated for 24 h at 37°C with 0.2 µCi/ml [14C (U)] L-valine (Moravek Biochemicals, Inc.). Excess radioisotope was removed with phosphate-buffered saline, 1× PBS (pH 7.4), prior to a 2 h incubation in DMEM/F12 media with 5% DCC FBS, 10 mM unlabeled valine (cat. no. V0500; MilliporeSigma), and the appropriate drug combination as described in the figure legends. A 2 h incubation at 37°C allowed for short-lived protein turnover, after which the medium was aspirated and cells were placed in the same treatment medium and incubated at 37°C 24 and 48 h, allowing for long-lived protein turnover. trichloroacetic acid precipitable and soluble radioactivity was collected from cells and medium, respectively, quantified by liquid scintillation counting (LS6500; Beckman Coulter, Inc.), and expressed as % long-lived protein degradation which is the ratio of TCA-soluble radioactivity to radioactivity in the precipitated proteins. In certain experiments, proteolysis was measured in the absence and presence of CQ, a lysosomotropic agent that enters the lysosome as a protonated compound and increases intracellular pH, blocking autolysosomal turnover along with any cellular constituents contained within the autolysosome.
Protein harvest, immunoblotting, and densitometry
Cell lysates were harvested for total protein as previously described (11,12,22,23). For all studies, with the exception of the experiments performed for Fig. 1D, cell lysates were derived from total cell populations (adherent and detached cells) that were collected and combined, prior to cellular lysis. For the studies presented in Fig. 1D, cell lysates were harvested from adherent and detached (apoptotic) cell populations separately. Immunoblotting was conducted according to the manufacturer's protocol using the following primary antibodies: BimEL (cat. no. CS2933; 1:500 dilution), GAPDH (cat. no. CS5174; 1:1,000 dilution), LC3 (cat. no. CS12741; 1:500 dilution), cleaved-Lamin A (cat. no. CS2035; 1:1,000 dilution), phosphorylated MAPK (cat. no. CS9101; 1:200 dilution), p62 (cat. no. CS5114; 1:1,000 dilution), cleaved-PARP (cat. no. CS9541; 1:500 dilution), phosphorylated EGFR (cat. no. CS3777; 1:250 dilution) and EGFR (cat. no. CS4267; 1:500 dilution), all from Cell Signaling Technology, Inc.; ERK1 (cat. no. SC-94; 1:200 dilution), ERK2 (cat. no. SC-154; 1:200 dilution) and ERα (cat. no. SC-8002; 1:1,000 dilution), all from Santa Cruz Biotechnology, Inc.; β-actin (cat. no. A5441; 1:2,000 dilution; Sigma-Aldrich; Merck KGaA) and GFP (cat. no. 2273995; 1:500 dilution; MilliporeSigma). Secondary antibodies included anti-mouse IgG (cat. no. 715-035-150; 1:10,000) and anti-rabbit IgG (cat. no. 711-035-152; 1:10,000) both from Jackson ImmunoResearch Laboratories, Inc. Immunodetection was performed using the ECL detection system (cat. no. 34080; Pierce; Thermo Fisher Scientific, Inc.) and HyBlot CL autoradiography film (cat. no. E3012; Denville Scientific, Inc.). Densitometry was performed in triplicate, employing three differing levels of exposure each analyzed using the Adobe Photoshop CS5 histogram function to determine the intensity of signal. For each lane of the gel, the signal intensity of the specific protein analyzed was divided by the signal intensity of the loading control to normalize protein loading variation per lane. For each cell population undergoing the specified treatment, the normalized signal intensity was divided by the normalized signal intensity of the control cells (in lane 1 of each gel/western blot) to determine the relative fold change of protein expression. The signal intensity in control cells was always assigned an arbitrary value of 1 to allow direct comparisons of signal intensity (calculated fold changes) to be readily identified. Loading controls in the present study utilized β-actin, GAPDH, total MAPK1/2 levels (25,26), and/or Ponceau S (cat. no. P7170; Sigma-Aldrich; Merck KGaA) staining for total protein normalization (27). In previous studies by the authors, it was determined that the levels of total MAPK1/2 do not change in response to hormonal treatments or MEK1 blockade; only the level of phosphorylated MAPK1/2 changes with treatments (16). In some analyses, duplicate blots were required for MAPK detection due to the interference of residual pMAPK signal after stripping. When protein loading was similar and signal intensity differences clearly identifiable between the experimental groups relative to the control group, densitometry was not performed, e.g. Fig. 1C and D.
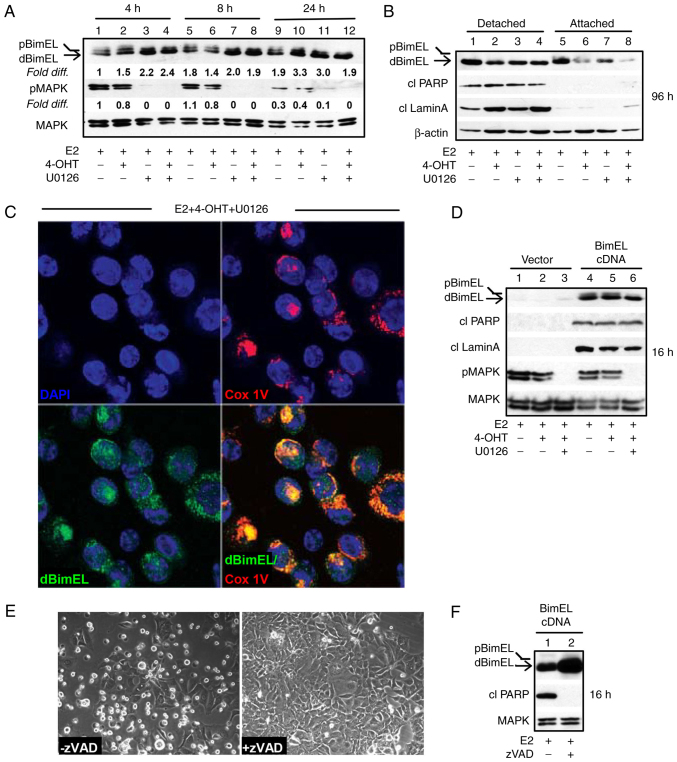
Figure 1.
BimEL is a key effector of apoptosis as determined by selective localization of dBimEL to the mitochondrial membrane of non-adherent, apoptotic MCF-7 breast cancer cells and overexpression studies. (A) A time course (4-24 h) of MEK1 blockade analyzed by immunoblotting studies showed that within 4 h of MEK1 blockade with U0126, a >2-fold increase in dBimEL was detected relative to BimEL levels in the E2-treated control breast cancer cells. The signal intensity of BimEL in the control cells (E2-treated) was arbitrarily assigned a value of 1 after corrections for loading were calculated as described in materials and methods. (B) Increased levels of dBimEL are selectively present along with cleaved PARP and cleaved Lamin A in the apoptotic cells detached from the adherent monolayer within 96 h. Total protein was isolated from the detached vs. adherent cell populations for this analysis; β-actin signal serves as the loading control showing minor variations in protein loading per lane. (C) dBimEL is localized to the mitochondrial membranes of detached (apoptotic) cells collected from cell populations undergoing treatment with E2 + 4-OHT + U0126 for 96 h as determined by confocal studies of antibody staining to BimEL and/or to the mitochondrial specific protein COx IV. (D) Immunoblotting revealed ectopic overexpression of BimEL cDNA in cells treated with E2, E2 + 4-OHT, and E2 + 4-OHT + U0126 with dephosphorylated BimEL being the dominant species expressed and induction of apoptosis by 16 h, as evidenced by increased levels of cleaved PARP and cleaved lamin A (lanes 4-6 compared with lanes 1-3). (E and F) BimEL-induced apoptosis is caspase-dependent based on the ability of the pan-caspase inhibitor zVAD-fmk to reduce both the detachment of cells from (E) the monolayer and the (F) cytosolic levels of cleaved PARP within 16 h. In panels A & D corrections for loading variations per lane were determined using total MAPK1/2 intensity to allow determination of efficacy of U026-mediated blockade of MEK (reduced pMAPK1/2 levels). 4-OHT, 4-hydroxytamoxifen.
Assessment of autophagic flux by LC3-II turnover
Per independent experiment, cells were seeded in duplicate at a density of 2×105 cells per 60-mm culture dish for each treatment such that in one dish the treatment was conducted in the presence of 10 µM CQ. The CQ-treated cell population allowed the steady state level of LC3-II to be determined for each treatment which was compared with the LC3-II level in cells undergoing the same treatment under conditions of active LC3-II flux. LC3-II signal intensities, after corrections for protein loading variations per lane, can be compared between cell populations undergoing the same treatment in the absence vs. presence of CQ; CQ allows the steady state LC3-II levels to be determined. These comparative levels of LC3-II are used to calculate autophagic flux as previously described (28). The following formula was utilized to approximate the flux in each treated cell population: [(+CQ signal intensity) minus (-CQ signal intensity)]/(+CQ signal intensity) as follows: the signal intensity of LC3-II in the protein lysate from cells treated in the absence of CQ was subtracted from the signal intensity of LC3-II (steady state LC3-II levels) in duplicate cell populations undergoing the same treatment in the presence of CQ. This value was then divided by the signal intensity of the steady state level of LC3-II to approximate the percent of LC3-II actually fluxed in the cell population. To graph the relative flux indexes, the percent of LC3-II flux in the control group, E2-treated cells were arbitrarily set equal to 1.
Transfections with BimEL cDNA
The BimEL cDNA expression vector (EX-O0071-M029) and control vector (EX-Neg-M02) were purchased from GeneCopoeia, Inc. The vectors/plasmids were isolated using a commercially available midi-prep kit (cat. no. 12143; Qiagen GmbH) following the manufacturer's protocol. For transfections, cells were seeded in DMEM-F12 medium with 5% DCC FBS to yield ~50% confluence. Adherent cells were transfected with plasmids (4.0 µg) by using either x-fect reagent (cat. no. 631317; Takara Bio USA, Inc.) or lipofectamine LTX (11668-019; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following a 16-24 h transfection at 37°C, cells were treated for various times at 37°C and harvested for analysis as described in figure legends.
Immunocytochemistry/confocal microscopy
Detached cells were collected via centrifugation (300 × g for 5 min at 4°C) of treatment media removed from culture dishes. Cell pellets were resuspended in DMEM-F12 plus 5% DCC FBS (SFBU32-0500; Equitech-Bio, Inc.) media and aliquoted into cytospin funnels (cat. no. 5991040; Global Medical Instrumentation, Inc.) and centrifuged (300 × g for 5 min at 4°C) using a Shandon Elliot Cytospin for collection on slides. Cells were fixed in 4% paraformaldehyde (cat. no. 50980487; Thermo Fisher Scientific, Inc.) at room temperature (RT) for 15 min, washed 3 times by gentle agitation in 1X PBS (cat. no. 46-013-CM; Mediatech/Cellgro; Thermo Fisher Scientific, Inc.) for 5 min at RT, and permeabilized in ice-cold methanol at 20°C for 10 min. Cells were blocked in 15% FBS, in 1× PBS, for 1 h at RT. Primary antibody was incubated with cells overnight (ON) at 4°C; secondary antibody incubations were for 1-2 h at 37°C. Antibodies diluted in Normal Antibody Diluent (NAD) (cat. no. ABB125; ScyTek Laboratories, Inc.) included: BimEL (1:400; CS2819; Cell Signaling Technology, Inc.) and COx IV (1:200; cat. no. 4D11B3E8; Invitrogen; Thermo Fisher Scientific, Inc.); AlexaFluor αRabbit (1:1,000) and Cy3 αMouse (1:800) (cat. nos. 711-545-152 and 715-165-150, respectively; both from Jackson ImmunoResearch Laboratories, Inc.). Coverslips were mounted with Vectashield DAPI Hard Mount (cat. no. XH-1500; Jackson ImmunoResearch Laboratories, Inc.). Images of the slides were captured using a Zeiss LSM 780 upright confocal microscope (Carl Zeiss AG) equipped with 63× 1.4 NA Plan-Apochromat objective, a digital AxioCam camera, and a motorized z-axis stage accessory. MCF-7 cells with BimEL and COx IV antibodies were excited using an Ar-laser: 488 nm 10 and 561 nm 8%, respectively. Confocal Z-axis stacks were separated by 0.37 µm with volume rendering of Z-stacks compiled using Zeiss LSM Image Browsing software.
Electron microscopy
For EM analysis, adherent cells were collected, washed in 2× PBS, and fixed for 1 h in ice-cold 3% glutaraldehyde/0.1 M cacodylate buffer, pH 7.4, rinsed overnight at 4°C in 0.1 M sucrose/0.1 M cacodylate buffer; post-fixed for 1 h at 4°C in 1% OsO4/0.1 M cacodylate buffer, and embedded in Epon. Sections (0.1 µm) were stained with uranylacetate/lead citrate (Fluka) and examined with a JEOL 1010 transmission electron microscope in the Electron Microscopy & Histology Core Facility (Dept. of Cellular Biology and Anatomy, Augusta University).
Statistical analysis
All data are presented as the mean ± SD values (n≥3). Sigma Plot 11.0 for Windows (Inpixon) was utilized to perform statistical analyses. Statistical differences between two groups were determined by paired Student's t-test, a one-way ANOVA followed by a Dunnett's post hoc, or a Holm-Šídák test. For comparisons of greater than three groups, a one-way ANOVA was performed, followed by Tukey's multiple comparison post hoc test to determine significance. *P<0.05 was considered to indicate a statistically significant difference.
Results
BimEL localizes to the mitochondrial membrane and induces apoptosis in ER+ breast cancer cells
In a previous study by the authors, siRNA knockdown was utilized to establish that dephosphorylated BimEL (designated dBimEL throughout) mediated ROS-dependent apoptosis in ER+ breast cancer cells induced by antiestrogen and antiprogestin treatments and the direct targeting of MEK1 (16). In the present study, the pro-apoptotic action of BimEL was further characterized. Focus was addressed on ER+ MCF-7 breast cancer cells undergoing treatments with E2 and E2 + 4-OHT in the absence vs. presence of the small molecule, reversible MEK1 inhibitor U0126. Within 4 h, treatment of MCF-7 cells with U0126 resulted in a greater than 2-fold upregulation of dBimEL; whereas dBimEL upregulation in response to 4-OHT treatment required a minimum of 24 h. A representative western blot showing dBimEL upregulation by MEK1 blockade (Fig. 1A) revealed the increased signal intensity for dBimEL in U0126-treated cells (lanes 3 and 4) relative to the E2-treated control cells (lane 1). Cell detachment due to dBimEL-dependent apoptosis, however, is not typically detectable until ~36-48 h of treatment, with optimal detection of the cleaved forms of PARP and lamin A detectable at 72-96 h (16). Thus, to address a potentially selective role of dBimEL in apoptosis, the detached, dying cells were collected separately from the adherent, viable monolayer of MCF-7 cells after 96 h of treatment with E2, E2 + 4-OHT, and E2 + 4-OHT + U0126 for western blotting. These studies determined that dBimEL was present at high levels in the detached, apoptotic cells, while the adherent cells expressed predominantly pBimEL (i.e. the non-apoptotic form of the protein). The representative western blot in Fig. 1B shows this selective localization of dBimEL, cleaved PARP, and cleaved lamin A in detached cells (lanes 1-4) as compared with barely detectable levels in adherent cells (lanes 5-8). The β-actin is present at similar levels due to equal loading of protein for the detached and adherent cells and does not reflect the number of detached apoptotic cells in the populations undergoing the various treatments. For example, to accomplish equal protein loading, detached cells were collected from five 60 mM dishes of E2-treated MCF-7 cells for total lysate preparation. This low-level of cell detachment and apoptosis occurs in the E2-treated cell population as a consequence of the depletion of E2 between 72 & 96 h after E2 supplementation. By contrast, cell detachment and apoptosis are readily induced by treatment with 4-OHT +/- U0126, thus detached cells collected from one 60 mM dish was sufficient. Overall, this analysis showed cleaved PARP, cleaved lamin A, and dBimEL selectively localized in the detached cell population undergoing BimEL-mediated apoptosis. Furthermore, the detached, non-viable cells showed co-localization of dBimEL with COx IV, a mitochondrial outer membrane protein. Representative images of this co-localization are provided from analyses of cells treated with 4-OHT + U0126 (Fig. 1C). This co-localization was not detected in the viable adherent cells (data not shown) and is consistent with our previous study identifying mitochondrial membrane permeabilization and ROS production to be dependent on BimEL expression (16).
The lag between BimEL dephosphorylation and induction of apoptosis indicated that dBimEL is sequestered by pro-survival members of the Bcl-2 protein family (19,29) that require downregulation or inactivation for dBimEL to induce apoptosis. Thus, BimEL cDNA was overexpressed to increase the ratio of dBimEL to the intrinsically expressed anti-apoptotic BH3 family members. Ectopic overexpression of BimEL cDNA resulted in high level expression of dBimEL and a concomitant rapid apoptotic response, with cleavage of PARP and lamin A detectable in E2-treated cells within 16-24 h of transfection (Fig. 1D, lanes 4-6 compared with 1-3). The apoptosis induced by dBimEL overexpression was caspase-mediated as the pan-caspase inhibitor zVAD attenuated cell detachment from the monolayer (Fig. 1E) and reduced the levels of cleaved PARP in the cell populations at 16 h (Fig. 1F), in a manner similar to zVAD blockade of 4-OHT and MIF-induced apoptosis previously described (11) and verified for the present study (data not shown). Altogether, these studies confirmed that dBimEL is a key effector of caspase-dependent apoptosis in ER+ antiestrogen-sensitive breast cancer cells.
Constitutive activation of MEK1/MAPK1/2 in antiestrogen-selected TR5 cells circumvents BimEL-dependent apoptosis
MEK1/MAPK1/2 signaling has been implicated in antiestrogen resistance (30); though, it is not known whether MAPK1/2 regulation of BimEL levels is involved. Thus, it was investigated if the MEK1/MAPK1/2/BimEL signaling axis was altered in an antiestrogen-resistant cell line, designated TR5, established in our laboratory (22). When initially selected, MCF-7 cells were subjected to a stepwise drug selection that allowed cells to adapt to increasing concentrations of 4-OHT in medium supplemented with E2. The final step of 4-OHT selection was adaptation to 5.0 µM 4-OHT, a concentration detected in the tissues of patients undergoing tamoxifen therapy (31). As TR5 cells were undergoing 4-OHT selection, parent MCF-7 cells were similarly passaged in E2. For the present study, TR5 and MCF-7 parent cells were thawed from cryostorage. Once stably adapted to 5.0 µM 4-OHT (~2 weeks), TR5 cells showed similar characteristics as described in our original study, including lack of growth inhibition and induction of apoptosis in response to 1.0 µM 4-OHT (whereas the sensitive cells show a 30% decline in viable cell number), a reduction in growth in 4-OHT concentrations of >4.0 µM (Fig. 2A), and detectable autophagosomes/autolysosomes in the cytosol (Fig. 2B, top panel). The MCF-7 parent cells showed a 4-OHT sensitivity profile similar to that of early passage MCF-7 cells obtained from the ATCC (Fig. 2A) and did not show elevated levels of cytoplasmic autophagosomes (Fig. 2B, bottom panel). Immunocytochemistry studies confirmed the presence of LC3-II puncta and showed detectable co-localization of p62 in the autophagosomes and autolysosomes of 4-OHT selected TR5 cells. For these studies, the lysosomotropic agent CQ, was added 4 h prior to harvest to block autolysosomal turnover (Fig. 2C), which allows the identification of autophagosomes with LC3-II and p62 localization (32). Both LC3-II and p62, encoded by the SQSTM1 gene, are involved in autophagosome formation and function and are commonly used as markers of functional autophagy (33).
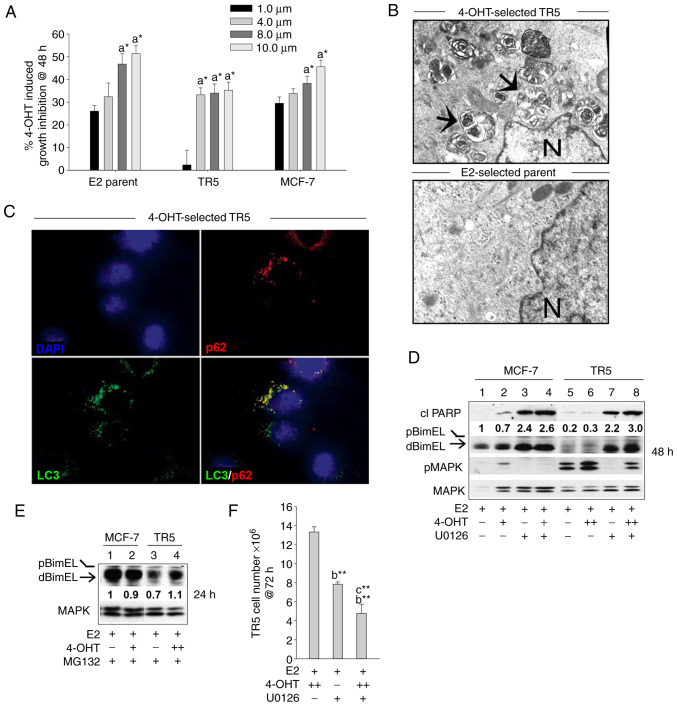
Figure 2.
BimEL-dependent apoptosis is suppressed in 4-OHT-selected, antiestrogen resistant TR5 breast cancer cells due to upregulation of MEK1/MAPK1/2 signaling. (A) Graphical representation of dose-dependent, 4-OHT-induced growth inhibition of parent MCF-7 and 4-OHT resistant TR5 cells at 48 h compared with MCF-7 ATCC early passage cells. Treatments with 4-OHT were conducted in the presence of 10 nM E2. Percentage of growth inhibition was determined by MTT assay. (B) High level autophagosome production in TR5 cells continuously passaged in 5.0 µM 4-OHT as identified by electron microscopy analyses; arrows designate autophagosomes and N, nucleus. (C) Immunofluorescence staining shows expression of LC3 and p62 in puncta in the cytosol of 4-OHT-selected TR5 cells; CQ was added 4 h prior to harvest to impede autolysosome turnover to allow the identification of LC3-II and p62 localization to autophagosomes. (D) Targeting MEK1/MAPK1/2 with U0126 restored detectable levels of activated dBimEL, designated by the arrow, and induced apoptosis as evidenced by cleavage of PARP at 24 h (compare signal intensity of dBimEL in lanes 7-8 to lanes 5-6). Total MAPK1/2 intensity was used as loading control to allow determination of efficacy of U026-mediated blockade of MEK (reduced pMAPK1/2 levels). (E) Proteasome inhibition with MG132 (1 µM) restored detectable levels of BimEL to TR5 cells undergoing 4-OHT selection; levels of BimEL in the parent MCF-7 cells during proteasomal blockade are shown for comparison and are similar to those identified in our previous study (16). (F) TR5 cells show a significant reduction in cell number following 72 h treatment with U0126 as a single agent or in combination with 5.0 µM 4-OHT. TR5 cells were seeded for 24 h in the absence of drug selection, which is E2 plus 5.0 µM 4-OHT. The adherent TR5 cells were then treated with E2 + 4-OHT, E2 + U0126, and E2 + 4-OHT + U0126. Cell counts were determined with a hemocytometer. Results are expressed as the mean ± SD values. Comparisons that were statistically significant include TR5 growth inhibition mediated by athe designated treatment compared with the E2 (10 nM) control cells; bthe designated treatment compared with growth in 5.0 µM 4-OHT and cthe designated treatment compared with growth in U0126. +, designates treatment with 1 µM 4-OHT; ++, 5 µM 4-OHT. *P<0.05 and **P<0.05. CQ, chloroquine; 4-OHT, 4-hydroxytamoxifen; p-, phosphorylated.
MEK1 activity levels (i.e., phosphorylated MAPK1/2) were next compared by western blot analyses of total protein isolated from TR5 and MCF-7 cells treated with E2, E2 + 4-OHT, and E2 + 4-OHT + U0126. MEK1 activity was assayed by immunoblotting studies that compared MAPK1/2 phosphorylation levels. Elevated levels of pMAPK1/2 and barely detectable levels of dBimEL (pro-apoptotic form) were identified in 4-OHT-selected TR5 cells as compared with the parent MCF-7 cells (Fig. 2D). Importantly, the targeting of MEK1 with U0126 in 4-OHT-selected TR5 cells reduced pMAPK1/2 levels and restored detectable BimEL expression, concomitant with induction of apoptotic cell death at 24 h as evidenced by cleavage of PARP (Fig. 2D; lanes 7 and 8 compared with 5 and 6). The fact that MEK1/MAPK1/2 blockade restored detectable dBimEL levels, indicated that BimEL was expressed primarily as pBimEL in TR5 cells and that pBimEL was being degraded via the proteasome, a mode of pBimEL degradation previously identified for MCF-7 cells (16). To investigate this, TR5 and MCF-7 cells were cultured in E2 or E2 plus 4-OHT in the presence of the proteasome inhibitor MG132. Treatment with MG132 resulted in an accumulation of pBimEL in TR5 cells, approaching levels observed in parent MCF-7 cells (Fig. 2E). Thus, the elevated pMAPK1/2 in TR5 cells mediated phosphorylation of BimEL and its subsequent degradation in the proteasome. Cell counts further showed that treatment of TR5 cells with U0126 as a single agent or in combination with 4-OHT significantly reduced TR5 cell number by 58.7±1.9 and 63.4±4.2%, respectively (Fig. 2F). Thus, MEK1/MAPK1/2 blockade generated pro-apoptotic dBimEL to levels that induced apoptosis and a significant reduction in TR5 cell number.
Targeting MEK1/MAPK1/2 in antiestrogen-resistant breast cancer cells does not effectively block pro-survival autophagy
Because a large subpopulation (36.6±4.2%) of the antiestrogen-resistant, 4-OHT-treated TR5 cells survived U0126-mediated MEK1/MAPK1/2 blockade (Fig. 2F), it was hypothesized that the surviving cells could be utilizing autophagy as a mode of survival. To examine this hypothesis, LC3-II and p62 turnover (flux) were analyzed as surrogate measures of functional autophagy. Due to its association with the autophagosome membrane, LC3-II turnover occurs concomitantly with autolysosomal flux (33,34). Moreover, when functioning as an autophagy receptor protein that shuttles misfolded proteins and damaged organelles to the autophagosome, p62 is degraded during autolysosome turnover (35). To analyze LC3-II and p62 levels and turnover (flux), hormonal treatments were conducted in duplicate; one culture dish for each treatment contained CQ to block autolysosomal turnover and allowed LC3-II and p62 steady state levels to be quantified. Following treatment, cells were harvested for total protein and western blot analyses were performed. As revealed in Fig. 3A, the relative steady state level of LC3-II in cell populations treated in the presence of CQ (lanes 5-8) was compared with LC3-II levels in the matched cell population undergoing the respective treatment in the absence of CQ (lanes 1-4), with all signal intensities calculated relative to the LC3-II signal intensity in the E2-treated control cells (lane 1) assigned an arbitrary value of 1 after corrections for loading variation per lane as described in materials and methods. Results from multiple independent experiments are graphically shown in Fig. 3B. These data identified: i) ~30% of LC3-II undergoes active flux in the 4-OHT-selected TR5 cells; and ii) the blockade of MEK1/MAPK1/2 does not attenuate the 4-OHT-induced LC3-II flux. A similar pattern of p62 flux can be discerned in Fig. 3A, comparing the steady state levels of p62 (lanes 5-8) to the p62 levels in cell populations undergoing the respective treatment in the absence of CQ (lanes 1-4). U0126 treatment effectively blocked pMAPK1/2 and induced the accumulation of pro-apoptotic dBimEL in the cell populations (Fig. 3A), consistent with experiments described in Fig. 2D. Immunocytochemistry further showed co-localization of LC3-II and p62 in cytosolic puncta in TR5 cells undergoing 4-OHT + U0126 treatment (Fig. 3C).
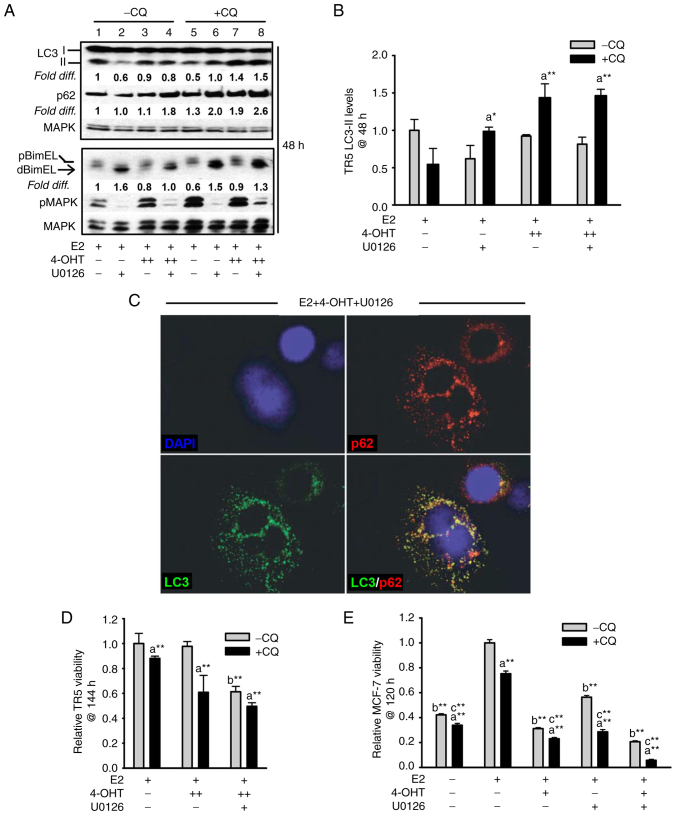
Figure 3.
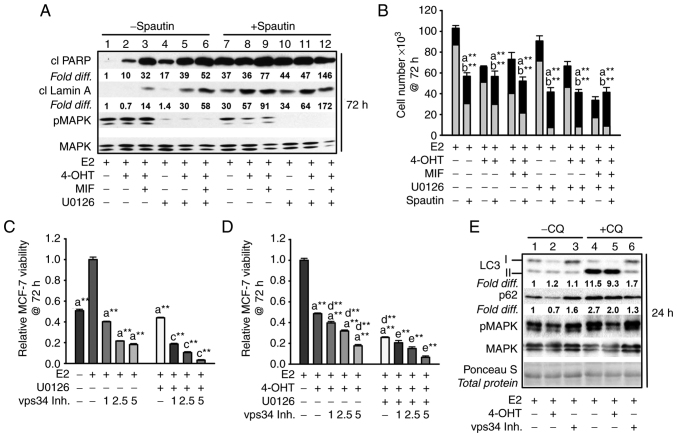
Targeting the MEK1/MAPK1/2 signaling axis in antiestrogen resistant TR5 cells induces pro-survival autophagy. (A) Immunoblot, representative of three independent experiments, determined LC3-II levels in TR5 cells treated with 4-OHT (5 µM) and/or U0126 (10 µM) as compared with levels in E2-treated cells. 4-OHT treatment, in the presence or absence of U0126, increased steady-state levels of LC3-II and p62 in TR5 cells. Relative signal intensity of LC3-II, p62, and BimEL is shown relative to their signal intensity in TR5 control cells (passaged for 24 h in E2-supplemented DMEM/F12 medium) which was given an arbitrary value of 1. Signal intensity of total MAPK1/2 (designated MAPK) was used to correct for loading variations per lane and determine the efficacy of U026-mediated blockade of MEK (reduced pMAPK1/2 levels). (B) A graphical representation of the data shows higher steady-state LC3-II levels in the cells undergoing treatments in the presence vs. absence of CQ, indicative of active autolysosomal flux under conditions of MEK1/MAPK1/2 blockade. (C) MEK1/MAPK1/2 blockade by U0126 for 48 h in 4-OHT-treated TR5 cells shows functional autophagy with co-localization of LC3-II and p62 as determined by immunocytochemistry and confocal microscopy as described in materials and methods. CQ was added to the treatment 4 h prior to harvest to block autolysosomal flux. (D and E) Relative viability of (D) antiestrogen resistant TR5 cells and (E) antiestrogen sensitive MCF-7 cells as determined by the MTT assay is shown following treatments with E2, and or E2 + 4-OHT in the absence or presence of U0126, plus and minus CQ (n=3) at 144 h for TR5 cells and 120 h for MCF-7 cells. CQ treatment reduced the viability of TR5 and MCF-7 cells treated with 4-OHT or 4-OHT + U0126, indicating that autophagic flux was cytoprotective under conditions of MEK1/MAPK1/2 blockade. Results are expressed as the mean ± SD values Comparisons that were statistically significant identify growth inhibition mediated by aCQ relative to the respective treatment conducted in the absence of CQ; bE2 + 4-OHT + U0126 relative to E2 + 4-OHT; and cthe designated treatment relative to E2. *P<0.05 and **P<0.05. CQ, chloroquine; 4-OHT, 4-hydroxytamoxifen; p-, phosphorylated.
The autophagy expressed in 4-OHT-selected TR5 cells was not cytotoxic and, therefore, not required for BimEL-dependent apoptosis induced by MEK1/MAPK1/2 blockade. This was determined by analyzing the effects of CQ on TR5 cell viability. It was reasoned that if autophagic catabolism was a prerequisite for BimEL-dependent cell death, CQ should attenuate the cytotoxic outcome of U0126 treatment of TR5 cells. However, CQ treatment combined with 5.0 µM 4-OHT or 5.0 µM 4-OHT + U0126 reduced TR5 cell viability (Fig. 3D), by 40±15% and significantly enhanced the cytotoxic effects of U0126 treatment, respectively (Fig. 3D). For comparison, antiestrogen-sensitive MCF-7 cells were similarly analyzed. The combined treatment of 4-OHT plus U0126 was highly cytotoxic by 120 h, with a greater than 4-fold reduction in MCF-7 cell viability as compared with a 3.2- and 1.7-fold reduction by 4-OHT and U0126 single agent treatments, respectively (Fig. 3E). Importantly, CQ significantly reduced cell viability in all the MCF-7 populations undergoing treatments. These data supported the conclusion that autophagy is protective against BimEL-dependent apoptosis in 4-OHT-selected TR5 breast cancer cells, but caution that the inhibition of autophagy during MEK1/MAPK1/2 blockade is not an effective strategy to eliminate the antiestrogen-resistant breast cancer cells.
Autophagy and dBimEL-dependent apoptosis are simultaneously detected in antiestrogen-sensitive breast cancer cells undergoing hormone treatments in the presence and absence of MEK1/MAPK1/2
It was next sought to characterize the relationship between autophagy and apoptosis in antiestrogen-sensitive cells undergoing hormone treatments in the presence and absence of MEK1 blockade. In addition to treatments with E2 and 4-OHT, the antiprogestin MIF was also used for these studies because the combined treatment of MIF plus 4-OHT induces a robust dBimEL-dependent apoptosis in MCF-7 cells independent of MEK1/MAPK1/2 co-targeting (16). It was first established that MCF-7 cells treated with 4-OHT, MIF, or MIF + 4-OHT expressed higher levels of autophagy than the E2-treated control cells. These studies, provided in Fig. S1, utilized EM to quantify autophagosomes per cell, long-lived protein turnover studies (23) as an independent read-out of autophagic catabolism, and cell counts to identify growth inhibition by 4-OHT ± MIF treatments relative to autophagy levels. When hormonal treatments (E2, E2 + 4-OHT, E2 + MIF, and E2 + 4-OHT + MIF) were conducted for 48 h in the absence and presence of U0126, apoptosis induced by MEK1/MAPK1/2 blockade was reproducibly detected. Furthermore, apoptosis was most pronounced in cells subjected to 4-OHT + MIF treatments ± U0126. This can be observed in the representative western blot shown in Fig. 4A (top panel), showing the highest levels of cleaved PARP and cleaved lamin A in lanes 3 and 6. Notably, LC3-II flux was also detected in the hormonally-treated cell populations even under conditions of MEK1/MAPK1/2 blockade at levels comparable to LC3-II flux in the E2-treated control cells. As revealed in Fig. 4A (bottom panel), the relative signal intensity of LC3-II in the cell populations undergoing the various treatments in the presence of CQ (lanes 7-12) compared with LC3-II levels not actively fluxed in cell populations undergoing the respective treatment in the absence of CQ (lanes 1-6). A graphical representation of LC3-II levels for each cell population undergoing the designated treatment in the presence vs. absence of CQ is provided in Fig. 4B. These data demonstrated an inability of MEK1/MAPK1/2 blockade by U0126 to effectively block autophagy (LC3-II flux). In independent studies, long-lived protein turnover (23) was analyzed. These studies utilized CQ to confirm lysosome-dependent long-lived protein turnover in the cell populations undergoing each treatment regimen. Collectively, these studies identified active autophagic catabolism along with detectable apoptosis in breast cancer cells undergoing hormonal treatments in the absence and presence of MEK1/MAPK1/2 blockade (Fig. 4C).
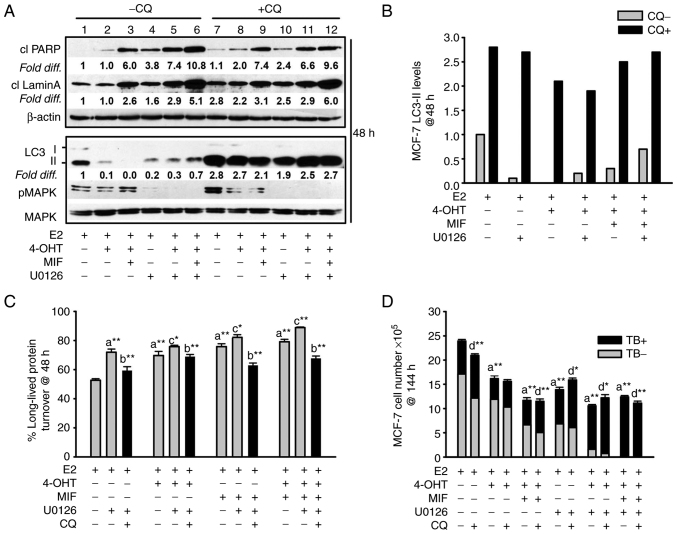
Figure 4.
Apoptosis and non-cytotoxic autophagy detected in antiestrogen sensitive breast cancer cell populations undergoing MEK1/MAPK1/2 blockade. (A, top panel) Increased levels of the cleaved forms of lamin A and PARP, two markers of apoptosis, were consistently detected by 48 h of hormonal treatments conducted in the presence of U0126 (top panel; signal intensities in lanes 4-6 were compared with lanes 1-3, with signal intensity for E2-treated control cells in lane 1 arbitrarily set to a value of 1 after corrections were calculated per lane using β actin as the loading control. (A, bottom panel) Increased steady-state LC3-II levels in cells undergoing the designated treatments for 48 h in the presence of CQ compared with the respective treatment conducted in the absence of CQ (signal intensity in lanes 7-12 was compared with lanes 1-6). Corrections for loading per lane were made using total MAPK as the loading control, also allowing the efficacy U0126-mediated blockade of MEK1/MAPK1/2 to be evaluated. (B) Graphical representation of the relative fold difference in the cells undergoing treatments in the presence vs. absence of CQ, shows higher steady-state LC3-II levels in the cells undergoing treatments in the presence of CQ, indicative of active autolysosomal flux under conditions of MEK1/MAPK1/2 blockade. (C) Long-lived protein catabolism (autophagic flux) was functional in hormonally-treated breast cancer cells in the absence and presence of MEK1/MAPK1/2 blockade by U0126 treatment, with catabolism in 4-OHT and/or MIF-treated cell populations showing increased catabolism relative to E2-treated control cells. CQ was added to duplicate cell populations to confirm the involvement of the lysosome in the long-lived protein turnover measured. (D) CQ-mediated blockade of autolysosomal flux did not increase the number of viable (trypan blue negative, designated TB-) cells, and modestly increased the number of non-viable cells (designated TB+) in cell populations undergoing hormonal treatments in the absence and presence of U0126. Results are expressed as the mean ± SD values. Comparisons that were statistically significant include: athe designated treatment compared with E2-treated control cells; bthe designated treatment conducted in the presence of CQ relative to the respective treatment conducted in the absence of CQ; cthe designated treatment conducted in the presence of U0126 relative to the respective treatment conducted in the absence of U0126; dCQ mediated inhibition of cell number or increases in trypan blue positive cells (TB+). *P<0.05 and **P<0.001. CQ, chloroquine; 4-OHT, 4-hydroxytamoxifen; TB+, trypan blue; MIF, mifepristone.
The autophagy/autophagic catabolism in antiestrogen-sensitive MCF-7 breast cancer cells undergoing the various treatments did not appear to be required for apoptosis induction by MEK1/MAPK1/2 blockade, previously identified as BimEL-dependent (16). The following data supported this conclusion: first, CQ treatment did not attenuate the levels of cleaved PARP or cleaved lamin A in the cell populations undergoing the various treatments (Fig. 4A, top panel, lanes 7-12 compared with lanes 1-6 for signal intensity of cleaved PARP and cleaved lamin A). Secondly, cell counts conducted with cell populations undergoing the various treatments for 144 h established that CQ-mediated blockade of autophagic flux was not cytotoxic; CQ did not increase total cell number, nor reduce the TB positive cell number (Fig. 4D). CQ treatment, however, showed only modest increased MCF-7 apoptotic cell death in response to the treatments. Thus, two other small molecule inhibitors that target early-stage autophagy were utilized: spautin-1, a small molecule inhibitor that targets Beclin (36) and compound 19, a selective vps34 inhibitor (37). These inhibitors reduced cell viability in MCF-7 cell populations undergoing MEK1/MAPK1/2 blockade in the presence and absence of antiestrogen treatment. Spautin-1 induced a robust death response by 72 h as evidenced by a measurable elevation in the levels of cleaved PARP (Fig. 5A), a reduction in cell number, and an increase in TB positive cells (Fig. 5B). In a similar manner, 72 h MTT assays showed pronounced effects of vps34 inhibition by compound 19 on hormonally-treated MCF-7 cells during MEK1/MAPK1/2 blockade (Fig. 5C and D). Vps34 inhibition utilizing 1.0 µM compound 19 resulted in blockade of p62 flux and a measurable reduction in LC3-II in cells undergoing MEK1/MAPK1/2 (Fig. 5E), consistent with effective vps34 inhibition (37). Altogether, these studies supported the conclusion that autophagy, when present, is protective against apoptosis and that early-stage autophagy inhibitors are more potent than CQ in blocking pro-survival autophagy in breast cancer cells undergoing MEK1/MAPK1/2 blockade.
Figure 5.
Inhibiting early-stage autophagy induces apoptosis in antiestrogen sensitive breast cancer cells undergoing MEK1/MAPK1/2 blockade. (A) Spautin-1, an inhibitor of early autophagy, induced apoptosis by 72 h of treatment as evidenced by increased levels of cleaved PARP and cleaved lamin A in MCF-7 cell populations treated with hormones in the presence or absence of U0126; signal intensities in lanes 7-12 compared with lanes 1-6. The signal intensity was arbitrarily set to a value of 1.0 in the E2-treated control cells (lane 1), after correction for loading variation per lane utilizing the signal intensity of total MAPK1/2 (designated MAPK), which also allowed the level of pMAPK1/2 activation (phosphorylation) to be determined following U0126 treatment. (B) Spautin-1 decreased cell number and increased the number of trypan blue positive (dead) cells when combined with hormone treatments in the absence and presence of U0126. (C and D) Vps34 inhibition by compound 19 (1.0-5.0 µM) reduced MCF-7 cell viability reproducibly detectable by 72 h in cell populations treated with E2 (C), or E2 + 4-OHT (D) in the presence and absence of U0126 at 10.0 and 5.0 µM for treatments in panel C and D, respectively. (E) Within 24 h, compound 19 at 1.0 µM blocked p62 turnover based on similar p62 levels in cells treated in the absence vs. presence of CQ (signal intensity compared in lanes 3 and 6, with signal intensity for E2-treated control cells given an arbitrary value of 1.0); decreased LC3-II lipidation, with higher levels of LC3-I as compared with LC3-II detected in cells; and did not show off target blockade of pMAPK. Total protein on the western blot was stained with Ponceau S to verify ~ even loading per lane. (Panel E). Comparisons that were statistically significant include: athe designated treatment vs. E2-treated; bthe designated treatment conducted in the presence vs. absence of spautin-1; cthe designated treatment conducted in the presence vs. absence of U0126; dthe designated treatment conducted in the presence vs. absence of 4-OHT; and ethe designated treatment conducted in the presence vs. absence of 4-OHT and U0126. *P<0.05 and **P<0.05. CQ, chloroquine; 4-OHT, 4-hydroxytamoxifen; MIF, mifepristone; p-, phosphorylated.
EGFR upregulation in antiestrogen-resistant breast cancer cells regulates MEK1/MAPK1/2 signaling in autophagy
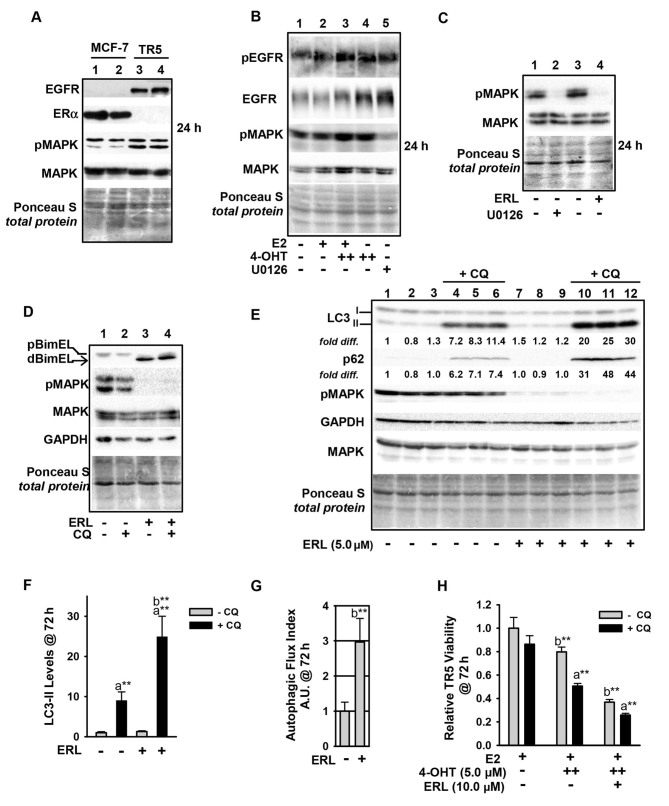
Due to the key role of MEK1/MAPK1/2 signaling in regulating BimEL and autophagy levels in the antiestrogen-resistant TR5 cells, it was next sought to determine the mechanism of MEK1/MAPK1/2 upregulation in 4-OHT-selected TR5 cells. Immunoblot analyses determined that EGFR expression was highly upregulated, while ERα expression was potently downregulated (Fig. 6A). The EGFR in 4-OHT-selected TR5 cells was phosphorylated at tyrosine (Tyr) 1068, and this phosphorylation (designated pEGFR) occurred independent of MEK1/MAPK1/2 kinase activity as treatment with U0126 did not reduce pEGFR in TR5 cells (Fig. 6B). By stark contrast, erlotinib (ERL), a reversible but highly selective EGFR inhibitor (38), effectively blocked MAPK1/2 phosphorylation in TR5 cells (Fig. 6B, lane 5; Fig. 6C, lane 4) and ERL-treated TR5 cells showed 4- to 5-fold increases in the levels of dBimEL (Fig. 6D). It was further determined that ERL treatment induced autophagy (Fig. 6, panels D and E), with more than a 2-fold increase in LC3-II and p62 turnover in ERL-treated TR5 cells (Fig. 6F). This induction can be observed by comparing LC3-II and p62 signal intensity in cells undergoing treatments in the presence vs. absence of CQ in which steady state levels can be directly compared between lanes 10-12 and lanes 4-6. A graphical representation of autophagic (LC3-II) flux is shown in Fig. 6G and was determined as described in materials and methods. Importantly, ERL treatment combined with 4-OHT significantly reduced TR5 cell viability, and CQ co-treatment enhanced this reduction and also reduced cell viability when TR5 cells were seeded in 4-OHT (Fig. 6H). Thus, autophagy induced by ERL served a cytoprotective role. Based on these data, it was hypothesized that the upregulation of EGFR/MEK1/MAPK1/2 facilitated TR5 cell escape from BimEL-dependent apoptosis during the initial stepwise 4-OHT selection and also provided proliferative capability to TR5 cells experiencing a high-level autophagy induction. A role of EGFR/MEK/MAPK1/2 signaling in the escape of breast cancer cells from the action of antiestrogens was proposed in Fig. 7.
Figure 6.
Upregulation of the EGFR/MEK1/MAPK1/2 signaling axis in antiestrogen resistant TR5 cells. (A) TR5 cells express EGFR, concomitant with loss of ERα expression as shown by immunoblotting. (B) EGFR is phosphorylated at Tyr1068 in a MEK1/MAPK1/2 independent manner and under conditions of 4-OHT selection at 1 and 5 µM (designated as + and ++, respectively). (C and D) ERL, a selective tyrosine kinase inhibitor of EGFR utilized at 5 µM, blocks pMAPK1/2 phosphorylation at 24 h as effectively as U0126-mediated blockade of MEK1/MAPK1/2 (signal intensity of pMAPK compared in lanes 2 and 4) (C), and increases dBimEL levels in TR5 cells (D). (E) Immunoblot analyses (n=3) shows increased autophagy in 4-OHT-selected TR5 cells treated with 5 µM ERL for 72 h; LC3-II and p62 are elevated in treatments conducted in the presence of CQ indicating active flux in the ERL-treated cell populations. (F and G) A graphical representation of LC3-II steady-state levels, (F) and flux (G) are provided for LC3-II. Autophagic flux was calculated as described in materials and methods. (H) MTT assay shows a decrease in the relative viability of TR5 cells undergoing ERL treatment at 10 µM plus and minus CQ for 72 h. In panels A-E, signal intensity of total MAPK provides the loading control, was used to correct for loading variations to establish relative signal intensities (D and E), and to determine efficacy of U0126-mediated blockade of MEK (reduced pMAPK1/2 levels). Comparisons that show statistically significant differences includea CQ relative to the respective treatment conducted in the absence of CQ; bthe designated treatment compared with no treatment or growth in E2. **P<0.05. CQ, chloroquine; 4-OHT, 4-hydroxytamoxifen; p-, phosphorylated.
Figure 7.
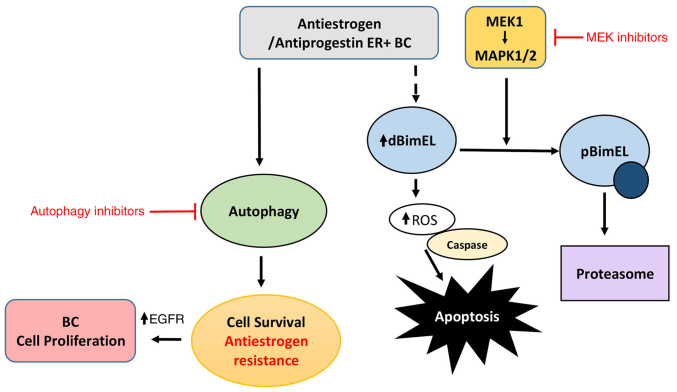
EGFR/MEK1/MAPK1/2 signaling axis is predicted to be a major regulator of apoptosis and autophagy in hormonally-treated breast cancer cells. A schematic showing that antiestrogen and antiprogestin treatment induces BimEL-dependent apoptosis more effectively if MEK1/MAPK1/2 is co-targeted as previously described (16), along with pro-survival autophagy as detailed in the present study. Based on data generated in the present study, this model predicts that breast cancer cells can upregulate EGFR/MEK1/MAPK1/2 to evade BimEL dependent apoptosis, facilitate the development of antiestrogen resistance, and proliferation (cancer cell growth).
Discussion
In a previous study, a key role was established for MAPK1/2 in the phosphorylation of BimEL which led to its proteasomal degradation in ER+ breast cancer cells and resistance to hormonally-induced ROS-dependent apoptosis (16). In the present study, the role of BimEL in antiestrogen sensitive and resistant breast cancer cells was further analyzed and it was demonstrated that: i) ectopic BimEL cDNA overexpression induced a robust and rapid apoptotic response; ii) intrinsically expressed dBimEL co-localized with COxIV to the outer mitochondrial membrane of apoptotic, but not viable adherent MCF-7 cells; and iii) upregulation of the EGFR/MEK1/MAPK1/2 signaling axis blocked dBimEL-dependent apoptosis in antiestrogen-selected, antiestrogen-resistant breast cancer cells. The antiestrogen-resistant breast cancer cell model used in the present study, designated TR5, was established by subjecting MCF-7 cells to a stepwise 4-OHT selection in the absence of clonal selection (22). Altogether, these data are consistent with a previous study by the authors identifying dBimEL as a key effector of ROS production induced by hormone treatments and MEK1 blockade in antiestrogen sensitive breast cancer cells (16). Of clinical relevance, MEK1/MAPK1/2 mediated signaling in breast cancer is associated with increased metastasis risk (19), as well as anti-estrogen resistance via cross-talk that occurs between the Raf/MEK/MAPK pathway and ERα (39). Consistent with a role for MEK1/MAPK1/2 in breast cancer metastasis, a pre-clinical study has identified the suppression of breast cancer metastasis by pro-apoptotic Bim (40). Thus, the targeting of MEK1/MAPK1/2 to activate BimEL-induced apoptosis of hormonally treated breast cancer cells has the potential to lead to improved clinical outcomes.
To date, the targeting of MEK1/MAPK1/2 signaling in clinical trials (41,42) has not recapitulated numerous of the promising pre-clinical in vitro and in vivo studies that established a role for MEK1/MAPK1/2 signaling in promoting breast cancer cell survival and progression (19,43-45). Even the use of a MEK inhibitor with AI has not proven effective for the treatment of advanced-stage breast cancer (46). Understanding the survival modes of breast cancer cells undergoing MEK1/MAPK1/2 blockade should help identify additional molecular targets to improve the use of selective MEK1 inhibitors in the clinic (19,41,46). Toward this goal, the present study identified functional autophagy in the anti-estrogen resistant TR5 cells and the antiestrogen sensitive parent MCF-7 cells undergoing MEK1/MAPK1/2 blockade. The autophagy was determined not to be a pre-requisite of BimEL-induced apoptosis but provided a cytoprotective role. Thus, it was indicated that pro-survival autophagy contributes to the lack of efficacy of MEK1/MAPK1/2 inhibitors in the clinic and should be considered a molecular target to improve the outcome of MEK1 targeting when utilized for the treatment of ER+ breast cancer. It will be important to identify the underlying mechanism(s) of autophagy in breast cancer cells that survive MEK1/MAPK1/2 targeting considering that the Ras/MEK/MAPK pathway is known to be upregulated in numerous breast cancers, particularly triple negative breast cancer (TNBC) (47). Moreover, it will be important to determine if targeting MEK1/MAPK1/2 in TNBC typically induces a pro-survival autophagy as it was identified in TR5 cells. Such studies are currently ongoing in our laboratories and have the potential to identify new molecular targets for the treatment of TNBC.
The targeting of EGFR in the antiestrogen resistant TR5 cells with the small molecule inhibitor ERL also induced a pro-survival autophagy, concomitant with reducing cell viability. Thus, the co-targeting of the EGFR/MEK1/MAPK1/2 signaling axis and autophagy along with hormonal therapy may circumvent the development of hormonal resistance in some ER+ breast cancers if used as a first line regimen. The proposed role for BimEL in the cytotoxic action of antiestrogens and antiprogestins is summarized in the schematic of Fig. 7, as previously demonstrated (16) and further elaborated in the present study; it demonstrates potential mechanisms to suppress the cytotoxic action of BimEL that may be critical for the development of antiestrogen resistance, i.e. upregulation of EGFR/MEK1/MAPK1/2 signaling axis and induction of autophagy. Clinically, the loss of ERα expression is known to occur in greater than 20% of breast cancers that relapse following endocrine therapy (48). Although there is a well-established inverse correlation between ERα expression and elevated EGFR, the role of EGFR in hormone status conversion in clinical breast cancer is not well defined (48). Importantly, EGFR has been recently identified as a therapeutic target to increase tamoxifen sensitivity in hormone receptor positive breast cancer (49). The aforementioned study showed EGFR upregulation, along with ERα downregulation, in breast cancer samples from patients with more advanced stage and higher-grade tumors.
During the present study, an increase in senescent-like cells in TR5 cell populations treated with inhibitors of EGFR, and to a lesser extent MEK1 (data not shown), was observed. These cells exhibited a flattened morphology, size enlargement, accumulation of vacuoles, and expression of β-galactosidase (β-gal), a marker of senescence (50). Although it was beyond the scope of the present study to determine if the autophagy induced by ERL and MEK inhibition facilitated the survival of the senescent-like cells, it is interesting to hypothesize that autophagy in the absence of MEK1/MAPK1/2 signaling is a prerequisite for a senescence-like state (or dormancy) and that adaptive, genetic, or epigenetic changes, such as EGFR upregulation regulate autophagy levels and allow reversal of the senescent state in breast cancer cells. This premise is not entirely inconsistent with the key role for EGFR recently reported by Chen et al (51) in regulating the switch between cell survival and cell death via autophagy regulation in cancer cells under hypoxic condition. To further understand the role of EGFR, our laboratories are currently investigating the role of EGFR/MEK1/MAPK1/2 signaling in regulating senescence, apoptosis, and autophagy in antiestrogen-resistant and sensitive ER+ breast cancer cells.
In conclusion, the present study demonstrated that the co-targeting of MEK1/MAPK1/2 and autophagy optimize BimEL-mediated cytotoxic effects of hormonal treatments of ER+ breast cancer cells. This combined treatment is predicted to be most effective as an initial systemic treatment approach for ER+ breast cancer because the development of antiestrogen resistance can involve adaptive, genetic, and/or epigenetic changes that make cells more resistant to the cytotoxic effects of autophagy blockade. For example, it was demonstrated that the upregulation of the EGFR/MEK1/MAPK1/2 signaling axis in an antiestrogen-resistant MCF-7 derived model of breast cancer that attenuates both the cytotoxic effects of BimEL and the cytostatic effects of autophagy. Knowledge of the mechanism(s) of autophagy induction by EGFR and/or MEK1/MAPK1/2 blockade is needed to identify new molecular targets to improve the use of MEK1/MAPK1/2 inhibitors in the clinic.
Supplementary Data
Acknowledgments
The authors would like to thank the Imaging Core Facility at Augusta University, directed by Dr. Graydon Gonsalvez. All imaging experiments were performed at the Augusta University Imaging Core Facility.
Funding Statement
The present study was supported by intramural finding by Augusta University as an Extramural Success Award and an Undergraduate Summer Student Training Research Award and by an extramural grant to PVS from the National Cancer Institute (grant no. NIHNCI1R01 CA121438-O1A1). Research in Dr Gewirtz's laboratory is supported by grants nos. CA268819 and CA239706 from the National Cancer Institute/National Institutes of Health and grant no. W81XWH 19-1-0490 from the Department of Defense Congressionally Directed Breast Cancer Research Program.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PVS, MT, DB and DG conceived and designed the present study. PVS, DG and MLH wrote the manuscript. AL, CJ, MM, SM, MLH, HC and AB conducted experiments. KL, MLH, MEM-L, DG and MEM-L performed data analysis. CJ, MLH and DG created the figures and diagram. JTB, WDH and PVS supervised the study. PVS and MLH confirm the authenticity of all of the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S. Primary breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v8–v30. doi: 10.1093/annonc/mdv298. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser-Weippl K, Badovinac-Crnjevic T, Fan L, Goss PE. Extended adjuvant endocrine therapy in hormone-receptor positive breast cancer. Breast. 2013;22(Suppl 2):S171–S175. doi: 10.1016/j.breast.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 4.Nardone A, De Angelis C, Trivedi MV, Osborne CK, Schiff R. The changing role of ER in endocrine resistance. Breast. 2015;24(Suppl 2):S60–S66. doi: 10.1016/j.breast.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodwell D, Wardley A, Johnston S. Postmenopausal advanced breast cancer: Options for therapy after tamoxifen and aromatase inhibitors. Breast. 2006;15:584–594. doi: 10.1016/j.breast.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Hartkopf AD, Grischke EM, Brucker SY. Endocrine-resistant breast cancer: Mechanisms and treatment. Breast Care (Basel) 2020;15:347–354. doi: 10.1159/000508675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke R, Jones BC, Sevigny CM, Hilakivi-Clarke LA, Sengupta S. Experimental models of endocrine responsive breast cancer: Strengths, limitations, and use. Cancer Drug Resist. 2021;4:762–783. doi: 10.20517/cdr.2021.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah M, Nunes MR, Stearns V. CDK4/6 inhibitors: Game changers in the management of hormone receptor-positive advanced breast cancer? Oncology (Williston Park) 2018;32:216–222. [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Zou W, Zhang J, Zhang Y, Xu Q, Li S, Chen C. Mechanisms of CDK4/6 inhibitor resistance in luminal breast cancer. Front Pharmacol. 2020;11:580251. doi: 10.3389/fphar.2020.580251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharifi MN, Anandan A, Grogan P, O'Regan RM. Therapy after cyclin-dependent kinase inhibition in metastatic hormone receptor-positive breast cancer: Resistance mechanisms and novel treatment strategies. Cancer. 2020;126:3400–3416. doi: 10.1002/cncr.32931. [DOI] [PubMed] [Google Scholar]
- 11.Gaddy VT, Barrett JT, Delk JN, Kallab AM, Porter AG, Schoenlein PV. Mifepristone induces growth arrest, caspase activation, and apoptosis of estrogen receptor-expressing, antiestrogen-resistant breast cancer cells. Clin Cancer Res. 2004;10:5215–5225. doi: 10.1158/1078-0432.CCR-03-0637. [DOI] [PubMed] [Google Scholar]
- 12.Schoenlein PV, Hou M, Samaddar JS, Gaddy VT, Thangaraju M, Lewis J, Johnson M, Ganapathy V, Kallab A, Barrett JT. Downregulation of retinoblastoma protein is involved in the enhanced cytotoxicity of 4-hydroxytamoxifen plus mifepristone combination therapy versus antiestrogen monotherapy of human breast cancer. Int J Oncol. 2007;31:643–655. [PubMed] [Google Scholar]
- 13.El Etreby MF, Liang Y, Wrenn RW, Schoenlein PV. Additive effect of mifepristone and tamoxifen on apoptotic pathways in MCF-7 human breast cancer cells. Breast Cancer Res Treat. 1998;51:149–168. doi: 10.1023/A:1006078032287. [DOI] [PubMed] [Google Scholar]
- 14.El Etreby MF, Liang Y. Effect of antiprogestins and tamoxifen on growth inhibition of MCF-7 human breast cancer cells in nude mice. Breast Cancer Res Treat. 1998;49:109–117. doi: 10.1023/A:1006098910000. [DOI] [PubMed] [Google Scholar]
- 15.Klijn JG, Setyono-Han B, Foekens JA. Progesterone antagonists and progesterone receptor modulators in the treatment of breast cancer. Steroids. 2000;65:825–830. doi: 10.1016/S0039-128X(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 16.Periyasamy-Thandavan S, Takhar S, Singer A, Dohn MR, Jackson WH, Welborn AE, LeRoith D, Marrero M, Thangaraju M, Huang S, Schoenlein PV. Insulin-like growth factor 1 attenuates antiestrogen- and antiprogestin-induced apoptosis in ER+ breast cancer cells by MEK1 regulation of the BH3-only pro-apoptotic protein Bim. Breast Cancer Res. 2012;14:R52. doi: 10.1186/bcr3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying HQ, Chen J, He BS, Pan YQ, Wang F, Deng QW, Sun HL, Liu X, Wang SK. The effect of BIM deletion polymorphism on intrinsic resistance and clinical outcome of cancer patient with kinase inhibitor therapy. Sci Rep. 2015;5:11348. doi: 10.1038/srep11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakraborty AR, Robey RW, Luchenko VL, Zhan Z, Piekarz RL, Gillet JP, Kossenkov AV, Wilkerson J, Showe LC, Gottesman MM, et al. MAPK pathway activation leads to Bim loss and histone deacetylase inhibitor resistance: Rationale to combine romidepsin with an MEK inhibitor. Blood. 2013;121:4115–4125. doi: 10.1182/blood-2012-08-449140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adeyinka A, Nui Y, Cherlet T, Snell L, Watson PH, Murphy LC. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin Cancer Res. 2002;8:1747–1753. [PubMed] [Google Scholar]
- 20.Lin CH, Shen CY, Lee JH, Huang CS, Yang CH, Kuo WH, Chang DY, Hsiung CN, Kuo KT, Chen WW, et al. High prevalence of the BIM deletion polymorphism in young female breast cancer in an East Asian country. PLoS One. 2015;10:e0124908. doi: 10.1371/journal.pone.0124908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sionov RV, Vlahopoulos SA, Granot Z. Regulation of Bim in health and disease. Oncotarget. 2015;6:23058–23134. doi: 10.18632/oncotarget.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT, Schoenlein PV. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 23.Periyasamy-Thandavan S, Jackson WH, Samaddar JS, Erickson B, Barrett JR, Raney L, Gopal E, Ganapathy V, Hill WD, Bhalla KN, Schoenlein PV. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy. 2010;6:19–35. doi: 10.4161/auto.6.1.10323. [DOI] [PubMed] [Google Scholar]
- 24.Towers CG, Wodetzki D, Thorburn A. Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. J Cell Biol. 2020;219:e201909033. doi: 10.1083/jcb.201909033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetti S, Gimond C, Chambard JC, Touboul T, Roux D, Pouysségur J, Pagès G. Extracellular signal-regulated kinases phosphorylate mitogen-activated protein kinase phosphatase 3/DUSP6 at serines 159 and 197, two sites critical for its proteasomal degradation. Mol Cell Biol. 2005;25:854–864. doi: 10.1128/MCB.25.2.854-864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong JE, Park JH, Kim CS, Lee SL, Chung HL, Kim WT, Lee EJ. Neuroprotective effects of erythropoietin against hypoxic injury via modulation of the mitogen-activated protein kinase pathway and apoptosis. Korean J Pediatr. 2017;60:181–188. doi: 10.3345/kjp.2017.60.6.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander H, Wallace S, Plouse R, Tiwari S, Gomes AV. Ponceau S waste: Ponceau S staining for total protein normalization. Anal Biochem. 2019;575:44–53. doi: 10.1016/j.ab.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palumbo C, De Luca A, Rosato N, Forgione M, Rotili D, Caccuri AM. c-Jun N-terminal kinase activation by nitrobenzoxadiazoles leads to late-stage autophagy inhibition. J Transl Med. 2016;14:37. doi: 10.1186/s12967-016-0796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ewings KE, Wiggins CM, Cook SJ. Bim and the pro-survival Bcl-2 proteins: Opposites attract, ERK repels. Cell Cycle. 2007;6:2236–2240. doi: 10.4161/cc.6.18.4728. [DOI] [PubMed] [Google Scholar]
- 30.Rani A, Stebbing J, Giamas G, Murphy J. Endocrine resistance in hormone receptor positive breast cancer-from mechanism to therapy. Front Endocrinol (Lausanne) 2019;10:245. doi: 10.3389/fendo.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kisanga ER, Gjerde J, Guerrieri-Gonzaga A, Pigatto F, Pesci-Feltri A, Robertson C, Serrano D, Pelosi G, Decensi A, Lien EA. Tamoxifen and metabolite concentrations in serum and breast cancer tissue during three dose regimens in a randomized preoperative trial. Clin Cancer Res. 2004;10:2336–2343. doi: 10.1158/1078-0432.CCR-03-0538. [DOI] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 35.Tang J, Li Y, Xia S, Li J, Yang Q, Ding K, Zhang H. Sequestosome 1/p62: A multitasker in the regulation of malignant tumor aggression (Review) Int J Oncol. 2021;59:77. doi: 10.3892/ijo.2021.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao S, Li S, Qin Y, Wang X, Yang Y, Bai H, Zhou L, Zhao C, Wang C. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int J Oncol. 2014;44:1661–1668. doi: 10.3892/ijo.2014.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda A, Harrington E, Cornella-Taracido I, Furet P, Knapp MS, Glick M, Triantafellow E, Dowdle WE, Wiedershain D, Maniara W, et al. Potent, selective, and orally bioavailable inhibitors of VPS34 provide chemical tools to modulate autophagy in vivo. ACS Med Chem Lett. 2015;7:72–76. doi: 10.1021/acsmedchemlett.5b00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Normanno N, Maiello MR, De Luca A. Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs): Simple drugs with a complex mechanism of action? J Cell Physiol. 2003;194:13–19. doi: 10.1002/jcp.10194. [DOI] [PubMed] [Google Scholar]
- 39.Thomas RS, Sarwar N, Phoenix F, Coombes RC, Ali S. Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-alpha activity. J Mol Endocrinol. 2008;40:173–184. doi: 10.1677/JME-07-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merino D, Best SA, Asselin-Labat ML, Vaillant F, Pal B, Dickins RA, Anderson RL, Strasser A, Bouillet P, Lindeman GJ, Visvader JE. Pro-apoptotic Bim suppresses breast tumor cell metastasis and is a target gene of SNAI2. Oncogene. 2015;34:3926–3934. doi: 10.1038/onc.2014.313. [DOI] [PubMed] [Google Scholar]
- 41.Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, Hamid O, Varterasian M, Asbury P, Kaldjian EP, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 42.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frogne T, Benjaminsen RV, Sonne-Hansen K, Sorensen BS, Nexo E, Laenkholm AV, Rasmussen LM, Riese DJ, II, de Cremoux P, Stenvang J, Lykkesfeldt AE. Activation of ErbB3, EGFR and Erk is essential for growth of human breast cancer cell lines with acquired resistance to fulvestrant. Breast Cancer Res Treat. 2009;114:263–275. doi: 10.1007/s10549-008-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiff R, Massarweh SA, Shou J, Bharwani L, Mohsin SK, Osborne CK. Cross-talk between estrogen receptor and growth factor pathways as a molecular target for overcoming endocrine resistance. Clin Cancer Res. 2004;10:331S–336S. doi: 10.1158/1078-0432.CCR-031212. [DOI] [PubMed] [Google Scholar]
- 45.Gee JM, Robertson JF, Ellis IO, Nicholson RI. Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int J Cancer. 2001;95:247–254. doi: 10.1002/1097-0215(20010720)95:4<247::AID-IJC1042>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 46.Zaman K, Winterhalder R, Mamot C, Hasler-Strub U, Rochlitz C, Mueller A, Berset C, Wiliders H, Perey L, Rudolf CB, et al. Fulvestrant with or without selumetinib, a MEK 1/2 inhibitor, in breast cancer progressing after aromatase inhibitor therapy: A multicentre randomised placebo-controlled double-blind phase II trial, SAKK 21/08. Eur J Cancer. 2015;51:1212–1220. doi: 10.1016/j.ejca.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Bartholomeusz C, Xie X, Pitner MK, Kondo K, Dadbin A, Lee J, Saso H, Smith PD, Dalby KN, Ueno NT. MEK inhibitor selumetinib (AZD6244; ARRY-142886) prevents lung metastasis in a triple-negative breast cancer xenograft model. Mol Cancer Ther. 2015;14:2773–2781. doi: 10.1158/1535-7163.MCT-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zattarin E, Leporati R, Ligorio F, Lobefaro R, Vingiani A, Pruneri G, Vernieri C. Hormone receptor loss in breast cancer: Molecular mechanisms, clinical settings, and therapeutic implications. Cells. 2020;9:2644. doi: 10.3390/cells9122644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong Y, Bae SY, You D, Jung SP, Choi HJ, Kim I, Lee SK, Yu J, Kim SW, Lee JE, et al. EGFR is a therapeutic target in hormone receptor-positive breast cancer. Cell Physiol Biochem. 2019;53:805–819. doi: 10.33594/000000174. [DOI] [PubMed] [Google Scholar]
- 50.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Henson ES, Xiao W, Huang D, McMillan-Ward EM, Israels SJ, Gibson SB. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12:1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.