Abstract
In attempts to effectively improve RNAi function, we herein report a new RNAi approach using X-shaped RDNA and Dgel (RNA interfering DNA hydrogel, Ri-Dgel). X-shaped RDNA is a 4 branched nanostructure which was composed of three dsDNA branches and one dsRNA branch, and the structure was made by annealing partially complementary ssDNAs and chimeric RNA–DNA oligonucleotides. Ri-Dgel was synthesized through the ligation of the X-shaped RDNAs using their palindromic sticky ends. In MDCK/GFP cells transfected with 1 μM of each format of siRNA, Ri-Dgel and X-RDNA, the intensity of GFP fluorescence was significantly reduced by 65% and 56%, respectively, while dsRNA which is a conventional siRNA format showed a relatively weak reduction intensity of 7% compared with a negative control. We also observed the decreased intensity of GFP fluorescence by approximately 59% in MDA-MB-231/GFP cells transfected with 5 nM Ri-Dgel. Furthermore, the Ri-Dgel showed persistent RNAi efficiency up to 6 days from the treatment. The use of Ri-Dgel to trigger RNAi resulted in enhanced efficacy and longer duration at lower concentration compared to traditional dsRNA implying the nanostructured DNA–RNA hybrid materials have great potential as a platform technology for RNAi-based therapy.
A new RNAi approach was developed using an X-RDNA and Ri-Dgel. The nanostructured materials of dsRNA–DNA hybrids showed higher efficient and longer lasting RNA interference effect compared with conventional dsRNA.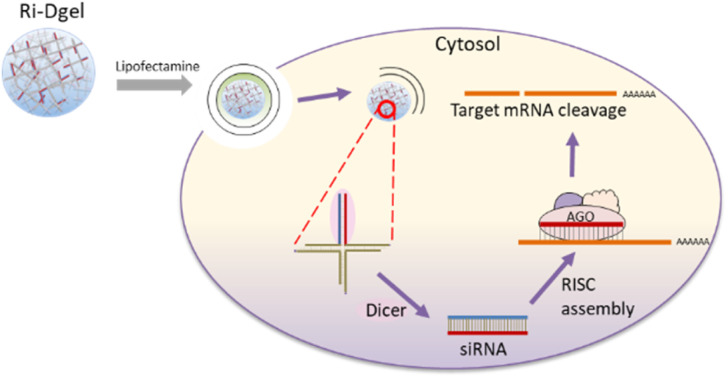
Introduction
To treat some disease, interfering with expression of the disease inducing proteins is essential. RNA interference (RNAi) is a prominent method by which the expression of a target gene is effectively silenced or knocked down by complementary (antisense) small interfering RNAs (siRNAs).1–6 RNase III class endoribonuclease, Dicer, plays a critical role in this process by cleaving delivered double-stranded RNAs (dsRNA) into 21–23 nucleotide (nt) siRNAs, which are assembled into RNA-induced silencing complexes (RISC) and lead to cleavage of the target mRNA in a sequence-specific manner resulting in silencing the target gene.7 The specific gene silencing effect of siRNA is promising in medical applications as therapeutic and diagnostic tools.8 In August 2018, the first siRNA-based drug Patisiran (Alnylam Pharmaceuticals Onpattro) was approved by the U.S. food and drug administration (FDA) for the treatment of hereditary variant transthyretin amyloidosis in adults.9 Furthermore, dozens of siRNA-based products are already in clinical trials at different stages.10 Accordingly, with the increasing importance of siRNA, the delivery methods, one of the key technologies required for efficient RNAi, were also extensively studied. Viruses as well as inorganic and organic nanoparticles were used to deliver siRNA into cells and some of them showed successful results. However, there are still obstacles to overcome for effective and selective siRNA delivery and furthermore stability to targeted cells in vivo.
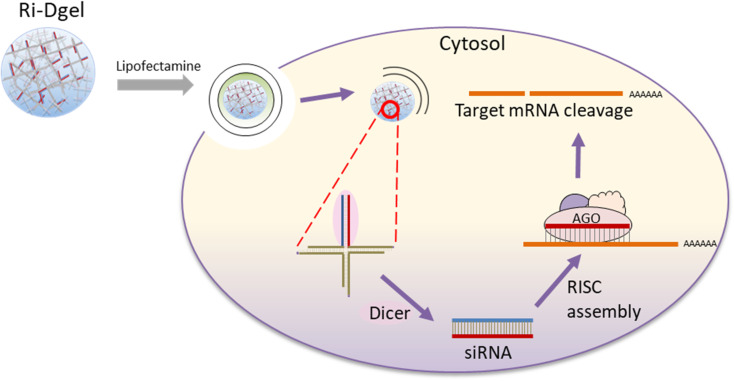
DNA which is well-known as a carrier of genetic information can also be used as a building block for self-assembly into molecular nanostructures because of its convenient programmability, adequate biocompatibility, biodegradability, and precise molecular recognition ability.11–15 Especially, DNA hydrogels (Dgel) made by ligating branched DNA nanostructures have been reported for many applications, including biosensing, drug delivery, immunotherapy, and tissue engineering.16,17 In attempts to effectively improve RNAi function, we here report a new approach of siRNA delivery using dsRNA introduced DNA hydrogel. dsRNA has been introduced at one of the four ends of X shaped DNA (X-RDNA) and hydrogel has been synthesized by ligating those X-RDNAs. The RNA embedded nanoscale DNA hydrogel was delivered into cells and siRNAs were generated by enzymatic cleavage with Dicer followed by interference on mRNA of a protein (Fig. 1). The dsRNA-conjugated DNA hydrogel (Ri-Dgel) showed longer lasting and more potent RNAi effects compared with conventional dsRNA.
Fig. 1. Schematic illustration of dsRNA-DNA hybrid hydrogel for RNA interference (Ri-Dgel).
Experimental section
Materials
Chemicals including diethyl pyrocarbonate (DEPC), Magnesium chloride (MgCl2), Tris–HCl, tris/borate/EDTA (TBE), phosphate buffered saline (PBS), and adenine ribose triphosphate nucleotide (rATP) were obtained from Sigma Aldrich. Dulbecco's Modified Eagle's Medium/High glucose (DMEM), fetal bovine serum, and 1% antibiotic-antimycotic were purchased from HyClone (Utah, USA). MDCK/GFP and MDA-MB-231/GFP cells were purchased from Gentarget Inc (San Diego, CA, USA). All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA, USA). The oligonucleotides used in this study are listed in ESI Table S1.†
Methods
Synthesis of dsRNA-conjugated DNA hydrogel
X-DNA sequences in the previously reported literature were modified and used.18 Two RNA-conjugated DNA oligonucleotides (EGFP-X01 and EGFP-BamHI-X04) and two single DNA oligonucleotides (BamHI-X02 and BamHI-X03) were mixed with DEPC water including 10 mM MgCl2 to a final concentration of 100 μM. The mixture was annealed with the following condition: denature at 95 °C for 10 minutes and 60 °C for 5 minutes, 60 °C for 5 minutes, reduce 1 °C to maintain the temperature for 1 minute, 20 °C to maintain 1 minute, and reduce to 4 °C. For buffer exchange, the annealed X-RDNA solution was centrifuged for 10 min at 14 000 g and 4 °C with 3K Amicon filter (Merk Millipore, USA) for 4 times. Purified 8 μL of X-RDNA was ligated with T4 DNA ligase (TaKaRa, Japan) in a 30 μL reaction volume for 24 h at 16 °C.
ζ-potential measurements
A Malvern Nano Zetasizer (Spectris, United Kingdom) was used to measure the ζ-potential of dsRNA, dsRDNA, X-RDNA, and Ri-Dgel. The samples allowed to equilibrate for 10 min before being vortexed and inserted into the sample chamber. The ζ-potential was determined by taking the average of three sets of individual measurements.
In vitro dsRNA cleavage assay
1 μM sample of dsRNA, dsRDNA, X-RDNA, or Ri-Dgel was mixed with 2 μL of recombinant human DICER protein and incubated at 37 °C for a series of time lengths. All reactions contain 6.6 mM MgCl2 and 1 mM rATP in the 66 mM Tris–HCl (pH 7.6) reaction buffer. Cleavage reactions were analyzed by electrophoresis with 8% polyacrylamide gels in 1X TBE with 0.1% SDS.
Quantitative RT-PCR and stem-loop RT-PCR
Total and small RNA use in our experiments were isolated using TRIzol reagent (Invitrogen, USA) with the PureLink RNA (Invitrogen, USA) following the manufacturer's instructions. For reverses transcription, 4 μL of isolated RNA was reverse transcribed using RevertAid H Minus first strand cDNA synthesis Kit (Thermo Fisher Scientific, USA) with step-loop reverse transcription (RT) and oligo (dT)15 primers for siRNA and mRNA, respectively. The stem-loop RT primer is an oligonucleotide that forms a stem-loop structure including a universal reverse primer sequence on the loop and six-nucleotide extension at 3′ end that are complementary to siRNA of GFP and RNU6.19 Sequence data are presented in ESI Table S2.†
Quantitative real-time PCR was performed using 2 μL of the obtained cDNA and Solg 2X Real-Time PCR Smart mix (SolGent, Korea) following the manufacturer's instructions and run on StepOne Real-time PCR System (Applied biosystems, USA). For analysis of mRNA expression, each 1 μL of GFP and GAPDH specific primer (5 μM) sets was used. GFP-AS2 and RNU6 primers were used with universal primer for the study of siRNA detection. The PCR mixtures were heated to 95 °C for 15 min and then subjected to 40 cycles of amplification (20 s 95 °C, 60 s 60 °C, 60 s 72 °C).
Transfection experiments
Cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic in a well humidified atmosphere of 5% CO2 at 37 °C. Transfection of each siRNA format to MDCK/GFP and MDA-MB-231/GFP cells was performed with lipofectamine 3000 according to manufacturer's instructions. Briefly, the cells were trypsinized, counted, and plated at 7000 per well in 96-well plates. After incubation for 1 day at 37 °C with 5% CO2, each RNA with 0.35 μL of Lipofectamine 3000 was applied in the final volume of 100 μL complete DMEM respectively. To study whether the silencing effect was processed in a dose-dependent manner, we transfected MDCK/GFP cells with sample of 1, 10, 100, 500, 1000 nM at 48 h without replacing the medium until subsequent assays. The cells were incubated for 48 h until they were ready to assay without replacing the medium.
To study the silencing effect of those siRNA formats in a time-dependent manner, we transfected MDCK/GFP cells with 500 nM of each siRNA and evaluated RNAi effect at four preselected time points i.e., 3 days, 4 days, and 5 days in plated at 2000 per well in 96-well plates after the transfection. After 3 days of transfection, the complete medium was replaced for the subsequent assays.
Assessment of GFP expression
After replacing the media to PBS, the GFP expression was observed under fluorescent microscopy. Then, the fluorescence intensity of GFP was analyzed by synergy HTX multi-mode microplate reader (BioTek, USA) with excitation and emission settings of 488 and 530 nm, respectively. The number of viable cells was determined by the CellTiter-Glo Luminescent Cell Viability Assay (Promega, USA) according to the protocol. The fluorescence intensity was normalized to luminescence of the corresponding cell viability, then analyzed and compared with the measurements of untreated control or transfection of dsRNA.
Result and discussion
Structure and design of dsRNA-conjugated DNA hydrogel
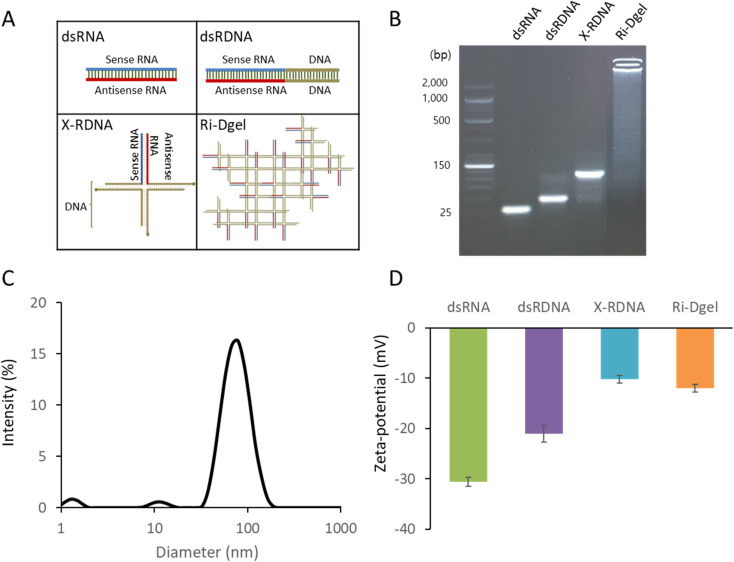
As an alternative format of siRNA to effectively enhance RNAi function, dsRNA-conjugated DNA hydrogel (Ri-Dgel) was proposed and designed. The Ri-Dgel was synthesized by ligating X-shaped DNAs using T4 DNA ligase. The X-shaped DNAs were composed of four oligonucleotides which were partially complementary each other and made by annealing those strands. The four strands were named as EGFP-X01, BamHI-X02, BamHI-X03, and EGFP-BamHI-X04, respectively (ESI Table S1†). Among those four oligonucleotides, two strands EGFP-X01 and EGFP-BamHI-X04, were designed to be chimeric sequences including 27 bases of EGFP gene sequence specific sense and antisense RNA sequences for RNAi function on each strand in addition to DNA sequences for making X-shaped nanostructure (X-RDNA). The length of siRNA on the X-RDNA was decided based on a previous study by Kim et al., (2005) showing that the duplex length of 27 bp has better inhibitory activity than the 21-mer siRNA.20 In mammalian cells, dsRNAs longer than 30 bp can trigger the interferon signaling pathway appearing nonspecific inhibition of gene expression, and also leading to apoptosis.21 As shown in Fig. 2A, four oligonucleotides (EGFP-X01, BamHI-X02, BamHI-X03, and EGFP-BamHI-X04) were annealed to form X-RDNAs followed by ligation resulting Ri-Dgel. Stepwise synthesis of X-RDNA and Ri-Dgel was verified by agarose gel electrophoresis (Fig. 2B). The Ri-Dgel was further characterized by measuring size and surface charge. As shown in Fig. 2C and D, the measured gel diameter was 78.8 ± 11.6 nm which is appropriate size for cell uptake and the surface was negatively charged as expected (−12.05 ± 0.8 mV). In addition to the X-RDNA and Ri-Dgel, 27 bp of dsRNA (EGFP-R01 and EGFP-R04) and 47 bp of chimeric RDNA (EGFP-X01 and EGFP-X01-rev) were also prepared as controls to evaluate the RNAi efficiency of newly developed siRNA formats for the cell experiments (Fig. 2A).
Fig. 2. Various dsRNA formats and characterization. (A) Structure of each dsRNA format. (B) Agarose gel electrophoresis image confirmation of each dsRNA, (C) DLS size analysis of synthesized Ri-Dgel. (D) Zeta potential of each dsRNA format. Error bars refer to standard deviations from three replicates.
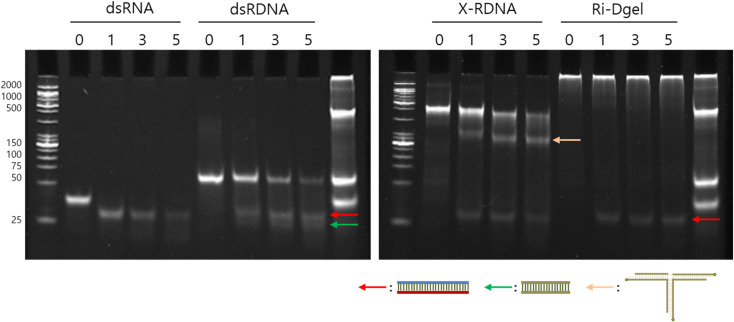
The stability of Ri-Dgel was evaluated by treating with Dicer and monitoring the enzymatic degradation with varied time intervals. Dicer is a kind of RNase which cleaved dsRNAs producing siRNAs in cells. Here the RNase was employed to see a resistance of various formats of RNAs against enzymatic degradation. To compare the stability, conventional double stranded 27 bp RNA, linear RNA–DNA chimeric strands (dsRDNA), and X-RDNA were treated with Dicer as controls for Ri-Dgel. According to the agarose gel electrophoresis of the samples treated with Dicer for 1, 3, and 5 hours, Ri-Dgel showed highest resistance against the RNase attack compared with other formats (Fig. 3). The intensity of the bands on the gel was quantified by imageJ program. As shown in gel image, dsRNAs were almost completely cleaved by Dicer after 1 hour of incubation. After 5 hours of incubation, of initial amount of dsRDNA and X-RDNA were enzymatically digested by about 83% and 64%, respectively. On the contrary, just a small part of RNAs were cleaved from Ri-Dgel after the 5 hours of digestion reaction. Because dsDNAs of Ri-Dgel were ligated by T4 DNA ligase, only one band, dsRNA was observed after DICER treatment, and there was no dsDNA size in the gel. These results meant that the Ri-Dgel was an effective format of protecting RNA from RNase attack and this will play a beneficial role during delivery process.
Fig. 3. Stability assay of each dsRNA format against digestion with RNase. Each of dsRNA sample was incubated with recombinant human Dicer enzyme for a series of time periods (0, 1, 3, and 5 hours) and analyzed by SDS-PAGE. Each arrow indicates fragments cleaved from RNA–DNA hybrid nanostructures. Red indicates siRNA fragments, green indicates dsDNA fragments, and yellow indicates tri-branch DNA structures which left after the cleavage of siRNA from X-RDNA.
In vitro evaluation of RNA interference ability of Ri-Dgel
RNAi efficiency of the Ri-Dgel was studied by treating GFP expressing cells with various types of siRNA. GFP was chosen as a target protein with its ease of monitoring and quantification of expression. As GFP expressing cells, Madin–Darby Canine Kidney (MDCK)/GFP and the human breast cancer cell line MDA-MB-231/GFP cell were employed for representing normal and cancer cells, respectively.
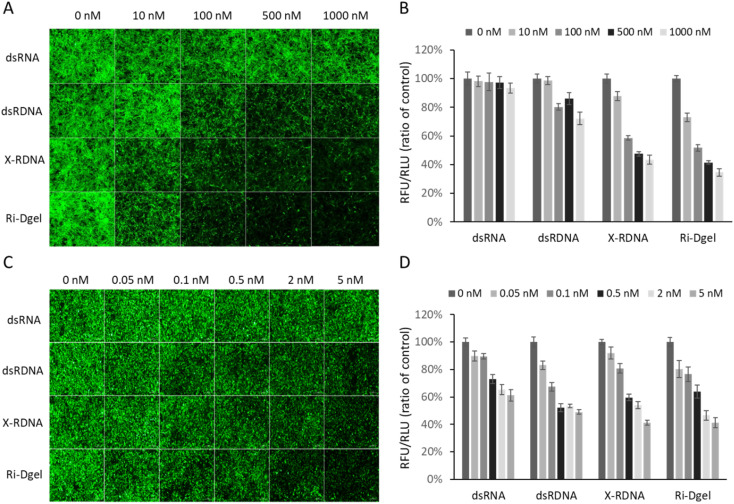
To see if the RNAs properly reduce GFP expression, firstly, the MDCK/GFP cells were treated with 10, 100, 500, and 1000 nM of each RNA sample (dsRNA, dsRDNA, X-RDNA, or Ri-Dgel) and the expressed GFPs were monitored after 48 hours by imaging with fluorescence microscope. As shown in Fig. 4A, no significant fluorescence reduction was observed in the cells when they were treated with conventional dsRNA, which means the dsRNA could not effectively interfere protein expression. On the other hand, when treated with dsRDNA, which is chimeric double stranded oligonucleotides composed of fused RNA and DNA sequences, reduction of fluorescence intensity from GFPs in the cells were clearly observed at 100 to 500 nM samples. It is an interesting result that simple fusion of RNA and DNA could improve RNAi efficiency compared with dsRNA. The improved efficiency was rationally expected with the enhanced stability of the dsRDNAs generating siRNA as already shown in Fig. 3. According to the fluorescence microscope images, further improved RNAi efficiency were observed when the cells were treated with X-RDNA or Ri-Dgel. The reduced GFP expression were clearly observed from 10 nM samples of both X-RDNA and Ri-Dgel. The remarkable increase of RNAi effect of those nanostructured and crosslinked RDNAs also seemed to be attributed to the stability improvement. To further investigate the RNAi efficiency improvement depending on a different dsRNA formats, the GFP expressions were quantified by measuring the total fluorescence from the cell culturing wells using a microplate reader. Fig. 4B is showing fluorescence intensity normalized with the number of live cells from each RNA treated MDCK/GFP cell culturing well. Compared with a negative control which was not treated with siRNA, GFP expression was reduced by only 7% in treated cells even with 1000 nM of dsRNA. The expression interference efficiencies of other samples were increased correspondingly with the stabilities of each dsRNA format as expected. When 10 nM of X-RDNA and Ri-Dgel was treated to the cells, the fluorescence intensity of X-RDNA decreased by 12%, whereas that of Ri-Dgel decreased by 27%. Our result shows that Ri-Dgel has the significantly stronger activity at a lower concentrations compared to X-RDNA. When 1000 nM of each RNA sample was treated to the wells, 28, 56, and 65% of fluorescence intensity was reduced for dsRDNA, X-RDNA, and Ri-Dgel, respectively.
Fig. 4. RNA interference effects of nanostructured RNA–DNA hybrid materials. In MDCK/GFP and MDA-MB-231/GFP transfected with Ri-Dgel in a dose manner for a 48 h exposure. (A) Fluorescence microscope images of MDCK/GFP cells treated with each dsRNA format of various concentrations. Images were taken after 48 hours of incubation. (B) Quantified fluorescence intensity from each sample measured using microplate reader. The values were normalized with negative control sample. (C) Fluorescence microscope images of MDA-MB-231/GFP cells. (D) Quantified fluorescence intensity from each sample measured using microplate reader. Each green fluorescence was normalized with luminescence intensity measured in relative light units (RLU) with CellTiter-Glo Luminescent Cell Viability Assay. Error bars refer to standard deviations from three replicates.
In terms of cell viability of MDCK/GFP cells treated with increasing concentrations of dsRNA, dsRDNA, X-RDNA, and Ri-Dgel, there were no significant cytotoxicity was observed in all samples with RNA amounts used in the experiments (Fig. S1†).
A cancer cell, MDA-MB-231/GFP cell, was also transfected with 0.05, 0.1, 0.5, 2, 5 nM of each RNA sample (dsRNA, dsRDNA, X-RDNA, or Ri-Dgel) with Lipofectamine 3000. The RNAi efficiencies in the cancer cells were measured using a fluorescence microscope and plate reader as same as MDCK/GFP cells. As shown in Fig. 4C and D, the trend of RNAi efficiency of each format of RNA on the cell was correspondingly similar to that on the MDCK/GFP cells. In other words, the order of RNAi efficiency was Ri-Dgel, X-RDNA, dsRDNA, and dsRNA.
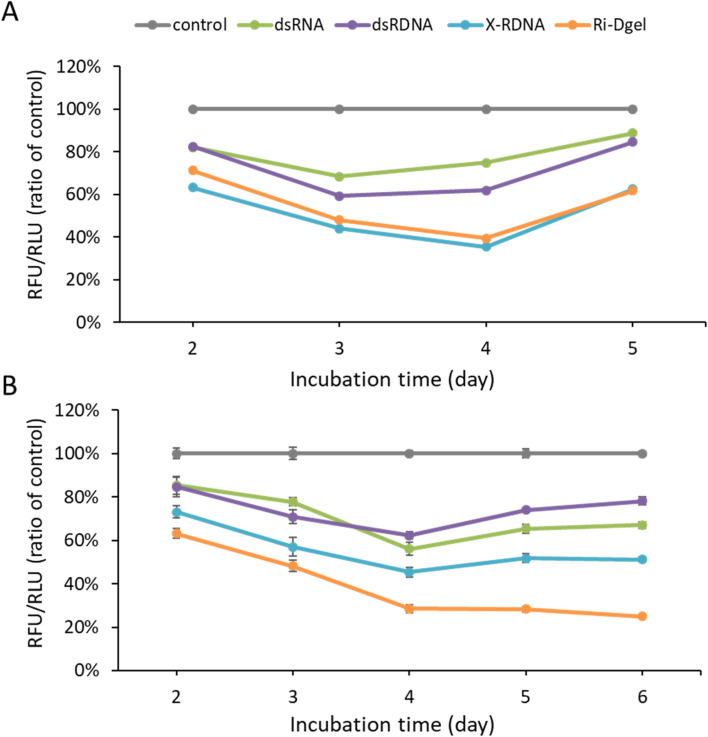
In addition to the efficiency evaluation, the durability of each RNA was investigated by time dependent monitoring of the GFP expression in cells. In detail, to see how long each RNA format sustain its RNAi effect, MDCK/GFP cells were treated with each format of RNAs and the GFP fluorescence intensity was measured in 2, 3, 4, and 5 days using microplate reader. According to the results in Fig. 5A, the X-RDNA and Ri-Dgel showed longer-lasting RNAi effect as well as higher efficiency inhibiting GFP expression. In case of dsRNA and dsRDNA, the GFP expressions were decreased to 68% and 59% by day 3, respectively, and gradually bounced up from day 4 to day 5, recovering to 85–89%. These results meant that the RNAi effect of dsRNA and dsRDNA lasted for 3 days and lost the effect from 4 days after treatment. In case of X-RDNA and Ri-Dgel, GFP fluorescence intensities were continuously decreased down to 36–39% by day 4, then recovered to 62% at day 5. The durability investigation experiment was reproduced using MDA-MB-231/GFP cells (Fig. 5B). Although the results were similar to those of MDCK/GFP cells, the Ri-Dgel showed even longer RNAi effect sustained for 6 days. Those results showed that the X-RDNA and Ri-Dgel provided longer-lasting and more effective RNAi efficiency inhibiting protein expression.
Fig. 5. Enhanced RNAi duration of nanostructured RNA–DNA hybrid materials. (A) Monitoring of GFP expression profiles in MDCK/GFP cells using fluorescence measurement after 2 to 5 days following 500 nM of dsRNAs treatment. (B) Monitoring of GFP expression profiles in MDA-MB-231/GFP cells using fluorescence measurement after 2 to 6 days following 0.1 nM of dsRNAs treatment. Error bars refer to standard deviations from three replicates.
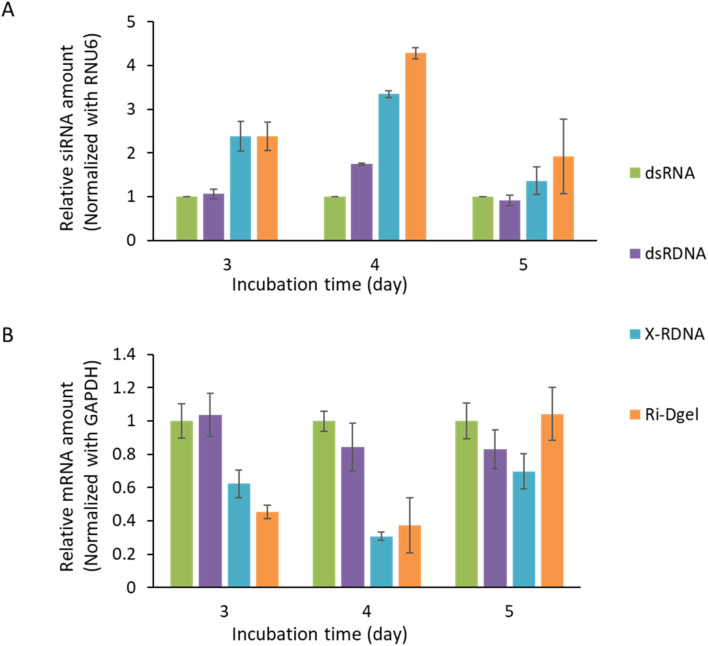
To ensure that effective knockdown of the target mRNA by each RNA format, the GFP mRNA and siRNA in treated MDCK/GFP cells were respectively quantified by total RNA extraction, reverse-transcription, and real-time qPCR. In case of siRNA amounts in cell lysates, siRNAs generated from plain dsRNAs remained less than any other formats of dsRNAs. As shown in Fig. 6A, in three days after the transfection, in the samples of linear dsRNA and dsRDNA left similar amounts of siRNAs which were less than half of those from X-RDNA and Ri-Dgel. On day 4, difference in the amount of left siRNAs was enlarged and the siRNAs from dsRNAs were less than 1/4 of Ri-Dgel. Although the amounts were generally reduced as days passed, these trends were consistent on day 5. These results showed that the dsRNA moieties were effectively protected from RNase attack in formats of nanostructured X-RDNA or Ri-Dgel and the more amounts of siRNAs generated from those dsRNA formats remained showing long-lasting RNAi effects in cells.
Fig. 6. Quantitative analysis of RNA amount in dsRNA treated cells. (A) Amount of siRNA which remained in MDCK/GFP since the dsRNA treatment. siRNA levels were normalized to RNU6 expression. (B) Amount of GFP mRNA which remained in MDCK/GFP since the dsRNA treatment. Transcription levels were normalized to GAPDH expression. Error bars refer to standard deviations from three replicates.
In terms of mRNA amounts relative to the dsRNA treated sample, it is decreased consistently with the GFP expression level from dsRNA treated sample to Ri-Dgel, at day 3 after RNA transfection (Fig. 6B). More quantitatively, just half of mRNA levels were measured in the cells treated with X-RDNA and Ri-Dgel compared with those from dsRNA and dsRDNA transfected cells. By day 4, the mRNA levels were kept small in X-RDNA and Ri-Dgel showing about 1/3 of dsRNA and dsRDNA samples. As the incubation time went on, on day 5, mRNA amounts of all the samples bounced back showing similar levels. But mRNA amount of the sample treated with Ri-Dgel on day 5 day was 1.2–1.4 times higher than that of dsRDNA and X-RDNA. The increase in RNA amounts could be due to an increase in transcription or a decrease in siRNA, but the siRNA amount increased more than 1.4 to 2 times in Ri-Dgel treated sample compared to dsRDNA and X-RDNA. Consistently, it was observed that the fluorescence intensity of sample treated with Ri-Dgel were lower than those of dsRDNA and X-RDNA (Fig. 5).
As a result, the siRNA and mRNA amounts in cells transfected each dsRNA format consistently showed that RNA interference effect of Ri-Dgel was the highest among the siRNA generating dsRNA formats tested in this study.
Conclusion
Here, several types of dsRNAs which generate siRNAs by enzymatic cleavage in cells were developed and their persistence of effect as well as RNAi efficiency were studied. The RNAi efficiency were evaluated by treating GFP expressing cells with developed dsRNAs respectively. According to the results, hybrid form of DNA and RNA (dsRDNA) showed higher RNAi efficiency compared with dsRNA which is used as a conventional RNAi reagent. As the hybrids of RNA and DNA form more networked structures, the RNAi efficiency was improved further. In a specific condition, X-RDNA and Ri-Dgel showed 8 times and 9.3 times higher RNAi efficiencies than dsRNA, respectively. This improved efficiency was attributed to the branched structures of nucleic acids which protected the RNAs from RNase attack. The improved protection of RNA moieties in the nanostructures enhanced the persistence of the siRNAs and the RNAi effect lasted longer than simple dsRNAs. Based on the results, the nanostructured DNA–RNA hybrid materials are expected to be good candidates for RNA therapeutics with high and long lasting efficiencies and will expand the applicability of small dsRNAs for biomedicines. Furthermore, the hybrid nanostructures would provide an improved and safe strategies for long RNAs such as mRNAs which are currently spotlighted as new vaccines.
Author contribution
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the Samsung Research Funding Center of Samsung Electronics under Project Number SRFC-MA1802-08. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government(MSIT) (No. 2022R1A2C10090071113582110600101).
Electronic supplementary information (ESI) available: Supplementary information includes nucleotide sequences for dsRNA, dsRDNA, X-RDNA, and Ri-Dgel. In addition, primers sequences for RT-qPCR are included. Cell viability test is also included. See DOI: https://doi.org/10.1039/d2ra06249f
References
- Elbashir S. M. Lendeckel W. Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe E. P. siRNAs and shRNAs: Tools for protein knockdown by gene silencing. Mater. Methods. 2013;3:197. [Google Scholar]
- Ambike S. Cheng C. Feuerherd M. Velkov S. Baldassi D. Afridi S. Q. Porras-Gonzalez D. Wei X. Hagen P. Kneidinger N. et al., Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 2022;50(1):333. doi: 10.1093/nar/gkab1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porosk L. Arukuusk P. Põhako K. Kurrikoff K. Kiisholts K. Padari K. Poogaa M. Langel Ü. Enhancement of siRNA transfection by the optimization of fatty acid length and histidine content in the CPP. Biomater. Sci. 2019;7:4363. doi: 10.1039/c9bm00688e. [DOI] [PubMed] [Google Scholar]
- Shen H. Sun T. Ferrari M. Nanovector Delivery of siRNA for Cancer Therapy. Cancer Gene Ther. 2012;19:367. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele E. Spinelli G. P. Miele E. Di Fabrizio E. Ferretti E. Tomao S. Gulino A. Nanoparticle-Based Delivery of Small Interfering RNA: Challenges for Cancer Therapy. Int. J. Nanomed. 2012;2012:3637. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M. Plasterk R. H. Dicers at RISC: the mechanism of RNAi. Cell. 2004;117(1):1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- Kim B. Park J. H. Sailor M. J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019;31(49):e1903637. doi: 10.1002/adma.201903637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urits I. Swanson D. Swett M. C. Patel A. Berardino K. Amgalan A. Berger A. A. Kassem H. Kaye A. Viswanath O. A review of patisiran (ONPATTRO®) for the treatment of polyneuropathy in people with hereditary transthyretin amyloidosis. Neurol Ther. 2020:1–15. doi: 10.1007/s40120-020-00208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. Zhong L. Weng Y. et al., Therapeutic siRNA: state of the art. Signal Transduction Targeted Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Xu X. Fang X. Cai S. Wang M. Xing C. Lu C. Yang H. Self-Assembled mRNA-Responsive DNA Nanosphere for Bioimaging and Cancer Therapy in Drug-Resistant Cells. Anal. Chem. 2020;92(17):11779. doi: 10.1021/acs.analchem.0c01895. [DOI] [PubMed] [Google Scholar]
- Ding F. Huang X. Gao X. Xie M. Pan G. Li Q. Song J. Zhu X. Zhang C. A Non-Cationic Nucleic Acid Nanogel for the Delivery of the CRISPR/Cas9 Gene Editing Tool. Nanoscale. 2019;11:17211. doi: 10.1039/c9nr05233j. [DOI] [PubMed] [Google Scholar]
- Pan G. Mou Q. Ma Y. Ding F. Zhang J. Guo Y. Huang X. Li Q. Zhu X. Zhang C. pH-Responsive and Gemcitabine-Containing DNA Nanogel To Facilitate the Chemodrug Delivery. ACS Appl. Mater. Interfaces. 2019;11(44):41082. doi: 10.1021/acsami.9b14892. [DOI] [PubMed] [Google Scholar]
- Li J. Zheng C. Cansiz S. Wu C. Xu J. Cui C. Liu Y. Hou W. Wang Y. Zhang L. et al., Self-assembly of DNA Nanohydrogels with Controllable Size and Stimuli-Responsive Property for Targeted Gene Regulation Therapy. J. Am. Chem. Soc. 2015;137(4):1412. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou L. Y. T. Zagorovsky K. Chan W. C. W. DNA assembly of nanoparticle superstructures for controlled biological delivery and elimination. Nat. Nanotechnol. 2014;9(2):148. doi: 10.1038/nnano.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gačanin J. Synatschke C. V. Weil T. Biomedical applications of DNA-based hydrogels. Adv. Funct. Mater. 2020;30(4):1906253. [Google Scholar]
- Lattuada E. Leo M. Caprara D. Salvatori L. Stoppacciaro A. Sciortino F. Filetici P. DNA-gel, novel nanomaterial for biomedical applications and delivery of bioactive molecules. Front. Pharmacol. 2020;11:1345. doi: 10.3389/fphar.2020.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Lee M. Kim T. Na J. Jung Y. Jung G. Y. Kim S. Park N. A RNA producing DNA hydrogel as a platform for a high performance RNA interference system. Nat. Commun. 2018;9(1):1–11. doi: 10.1038/s41467-018-06864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E. Hellens R. P. Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol. Biol. 2011;744:145–157. doi: 10.1007/978-1-61779-123-9_10. [DOI] [PubMed] [Google Scholar]
- Kim B. Park J. H. Sailor M. J. Rekindling RNAi therapy: materials design requirements for in vivo siRNA delivery. Adv. Mater. 2019;31(49):1903637. doi: 10.1002/adma.201903637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Carmichael G. G. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68(3):432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.