Abstract
Spectroscopic single-molecule localization microscopy (sSMLM) captures the full emission spectra of individual molecules while simultaneously localizing their spatial locations at a precision greatly exceeding the optical diffraction limit. To achieve this, sSMLM uses a dispersive optical component to separate the emitted photons into dedicated spatial and spectral imaging channels for simultaneous acquisition. While adding a cylindrical lens in the spatial imaging channel enabled three-dimensional (3D) imaging in sSMLM, the inherent astigmatism leads to technical hurdles in spectral calibration and nonuniform lateral resolution at different depths. We found that implementing the biplane method based on the already established spatial and spectral imaging channels offers a much more attractive solution for 3D sSMLM. It allows for more efficient use of the limited photon budget and provides homogeneous lateral resolution compared with the astigmatism-based method using a cylindrical lens. Here we report 3D biplane sSMLM and demonstrate its multi-color 3D imaging capability by imaging microtubules and mitochondria in fixed COS-7 cells immunostained with Alexa Fluor 647 and CF 660C dyes, respectively. We showed a lateral localization precision of 20 nm at an average photon count of 550, a spectral precision of 4 nm at an average photon count of 1250, and an axial localization resolution of 50 nm.
1. INTRODUCTION
Spectroscopic single-molecule localization microscopy (sSMLM) has recently emerged as a new tool for obtaining molecular insights into cellular dynamics by capturing the spatial information of single molecules with their inherent spectroscopic signatures [1–4]. sSMLM enables direct observation of the nanoscopic features of biological systems beyond the diffraction limit. Its ability to probe spectroscopic variations associated with interactions between single molecules and their local environment has allowed several functional studies to reveal subcellular hydrophobicity changes and photochemical transformation with single-molecule sensitivity [4–7]. In addition, by identifying individual molecules based on minute differences in their emission spectra, sSMLM has demonstrated its versatile imaging applications including multi-color imaging, fluorescent impurity rejection, chemical recognition, and high-throughput fluorescence spectroscopy at the single-molecule level [1–3,8–13].
Along with spectroscopic single-molecule imaging, extending sSMLM’s imaging capability from two-dimensional (2D) to three-dimensional (3D) in the spatial domain is essential for uncovering rich information about single molecules for fundamental understanding of subcellular structures. In conventional single-molecule localization microscopy (SMLM), several methods have been developed to unambiguously determine the axial position of single molecules together with their in-plane lateral coordinates [14–28]. The 3D SMLM imaging is commonly realized by manipulating the point-spread function (PSF) using artificially introduced astigmatism [14];however, this method is inherently sensitive to optical aberrations in thick samples [15]. In addition, the ellipticity of the PSF affects the localization precision and thus causes inhomogeneous lateral precision along the axial position [16,17]. Alternatively, the biplane method encodes the axial position of the same single molecules into distinctly defocused PSFs at two axially separated image planes [16–19]. In this approach, the lateral precision is not affected by the depth variation. However, this method divides the total number of photons emitted into two detection channels and reduces the photon budget in each channel, which consequently restricts the localization precision in either channel. The third method is based on a 4π interferometric configuration [20,21]. Despite its outstanding performance, its usage is restricted to a thin sample limited by its imaging depth. Additionally, the alignment procedure using multiple cameras is quite demanding.
With sSMLM, researchers have already reported 3D imaging capability [1,5,27]. We and other groups have implemented 3D imaging using the astigmatism-based approach [1,5]. Along with its drawbacks described above, this method suffered from the reduced signal-to-noise ratio (SNR) in sSMLM when the PSF was considerably enlarged. More importantly, it caused challenges in wavelength calibration, as the two beam paths for spatial and spectral images experienced different spherical aberrations, which is mainly caused by inserting a cylindrical lens only into the zeroth order (spatial imaging path). Reported by Smith et al. [29], a diffractive optic was used to obtain the 3D spatial information of single molecules and the emission color, but it offers relatively low spectral precision and thus limits the range of functional imaging applications in sSMLM.
Methods focused only on manipulating PSFs in spatial images using double-helix PSFs and spherical aberration were also reported for SMLM [23–28]. These adaptive optics-based 3D imaging methods, which use a spatial light modulator or a deformable mirror, not only increase the complexity of system alignment but also result in undesirable attenuation of the photons, especially in sSMLM.
In fact, sSMLM fundamentally suffers from a limited photon budget because the emitted photons from single molecules need to be divided into two detection channels to simultaneously capture the spatial and spectral images [1–7,9]. This fundamental constraint restricts the direct adoption of many different approaches for 3D imaging. Although the spectral image contains both the spatial and spectral information of single molecules, only the spectral information has been used, and the spatial information was often ignored in sSMLM [1–7,9].
Here we present 3D biplane sSMLM, which provides simultaneous acquisition of 3D spatial information and the spectroscopic signatures of single molecules by taking advantage of the two already existing spatial and spectral imaging channels. By recognizing that sSMLM already divides the photons into two independent channels, 3D imaging can be efficiently implemented using the biplane method without the need for excessive additional optical elements or tedious modification of the system only in the spatial channel. Therefore, the drawback of dividing photons into two imaging channels becomes an added benefit. In this work, we demonstrate spectroscopic 3D imaging of single molecules using 3D biplane sSMLM and show multiplexed 3D imaging of subcellular structures in fixed cells.
2. 3D BIPLANE SSMLM SYSTEM
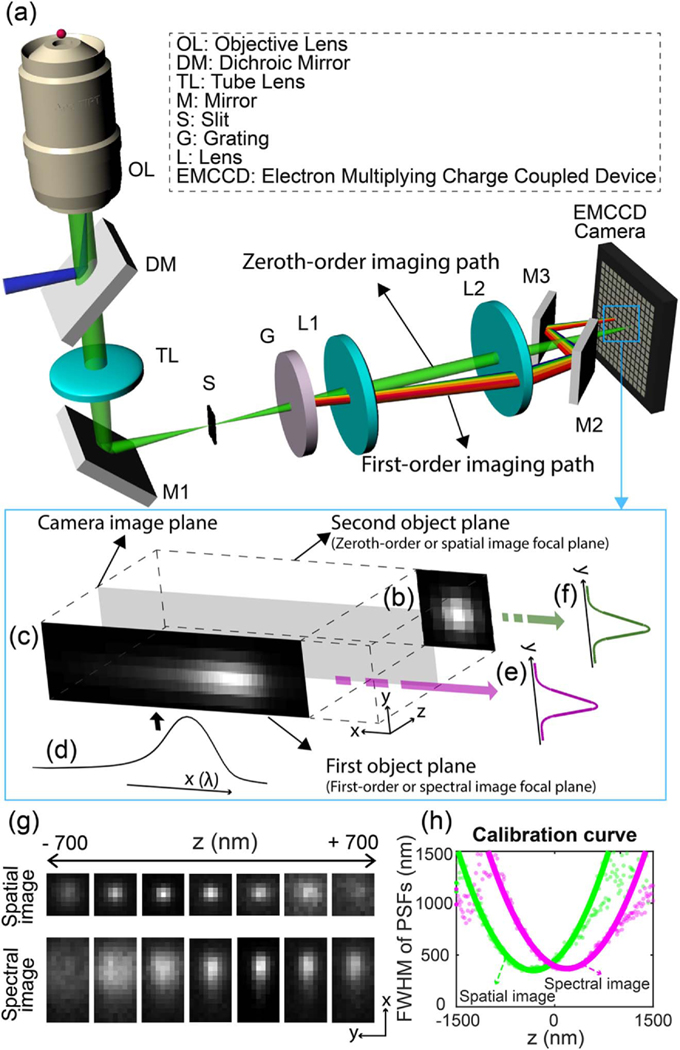
Figure 1(a) shows the schematic of the 3D biplane sSMLM. To divide the emitted photons into the zeroth- and first-order imaging paths for simultaneous spatial and spectral imaging, we added a diffraction grating to a conventional SMLM system. First, we focused a 640 nm CW laser to the back aperture of the objective lens (CFI Apochromat 100×, NA = 1.49, Nikon) by a dichroic beamsplitter (FF649-DI01–25X36, Semrock). We offset the excitation beam from the optical axis of the objective lens for highly inclined and laminated optical sheet (HILO) illumination. Then the emitted fluorescence photons were transmitted to the intermediate image plane by a mirror after passing through an emission filter (BLP01–647R-25, Semrock) and a tube lens inside an inverted microscope body (Eclipse Ti-U, Nikon). We inserted a slit in front of the spectrometer to confine the field of view (FOV) of the spatial distribution of the emitted fluorescence at the intermediate image plane. Subsequently, we inserted a transmission grating with a ratio of 1:3 between the zeroth order and the first order in the far-red spectral range (100 grooves/mm, Star Analyser 100, Paton Hawksley Education Ltd.), to separate the spatial and spectral images.
Fig. 1.
(a) Schematic and (b–f) working principle of 3D biplane sSMLM. (b) The detected spatial image and (c) the spectral image of a single-molecule emission. (c) The spectral image is the result of the convolution of the diffraction-limited PSF of individual stochastic fluorescent-emitting molecules in the spectral imaging plane and (d) the linearly spread spectroscopic signature. By integrating the spectral and spatial images along the x axis, (e), (f) 1D PSFys are retrieved from both images and used for biplane imaging. (g) The experimentally acquired spatial (top row) and spectral (bottom) images at the different axial positions from single emitters. (h) The experimentally obtained depth calibration curve.
The relative positions between the grating and the intermediate image plane determined a pixel-wavelength relationship, referred to as the spectral dispersion [nanometer per pixel (nm/pixel)] in the spectral image (see definition and principle in Method A) [30]. We used a spectral dispersion of 8 nm/pixel. In addition, after a relay optics (focal length = 150 mm) we added a pair of mirrors in the first-order (spectral) imaging path in front of the electron-multiplying charge-coupled device (EMCCD) camera (iXon 897, Andor) to introduce a delay line. This delay created an axial separation of 500 nm between the spatial and spectral imaging object planes for biplane imaging. We placed the EMCCD camera in the middle of the two axially separated object planes, which enabled the simultaneous capturing of the spatial and spectral images with different PSF sizes corresponding to the relative axial position to the optical focal plane. Except for the pair of mirrors, no additional optical elements were necessary
Figures 1(b)–1(f) show the working principle of the 3D biplane sSMLM. Figures 1(b) and 1(c) are illustrations of the detected spatial and spectral images of a single-molecule emission. Note that the camera image plane (the gray plane) is in the middle of the zeroth-order (or spatial imaging) and first-order (or spectral imaging) focal planes. The spectral image shown in Fig. 1(c) is the convolution of the PSF of individual stochastic fluorescent-emitting molecules in the spectral imaging plane and the linearly spread spectral profile shown in Fig. 1(d). Therefore, we can retrieve the emission spectrum [Fig. 1(d)] and the one-dimensional (1D) PSF [defined as PSFy, Fig. 1(e)] by integrating the spectral image along the y axis and x axis, respectively. Meanwhile, we can calculate a similar 1D PSFy from the spatial image as shown in Fig. 1(f). These two 1D PSFys collectively provide the z-axis information of the fluorescent emitter with respect to the camera image plane. Meanwhile, the emission spectrum, shown in Fig. 1(d), is still used for spectroscopic analysis. Experimentally acquired spatial (top row) and spectral (bottom) images at different axial positions (−700 to 700 nm) from single molecules are shown in Fig. 1(g).
For every single-molecule emission, we integrated the simultaneously acquired spectral and spatial images along the x axis to extract the 1D PSFys from the two focal planes as shown in Figs. 1(e) and 1(f). We then took the ratio of the full-widthat-half-maximum (FWHM) values of the two 1D PSFys and compared it with an experimentally obtained depth calibration curve [Fig. 1(h)] to determine the axial position. By combining with x–y localization from the spatial image, we can achieve 3D localization. Details of a full reconstruction procedure, including the spectral and 3D calibration procedures, are provided in Methods B and C.
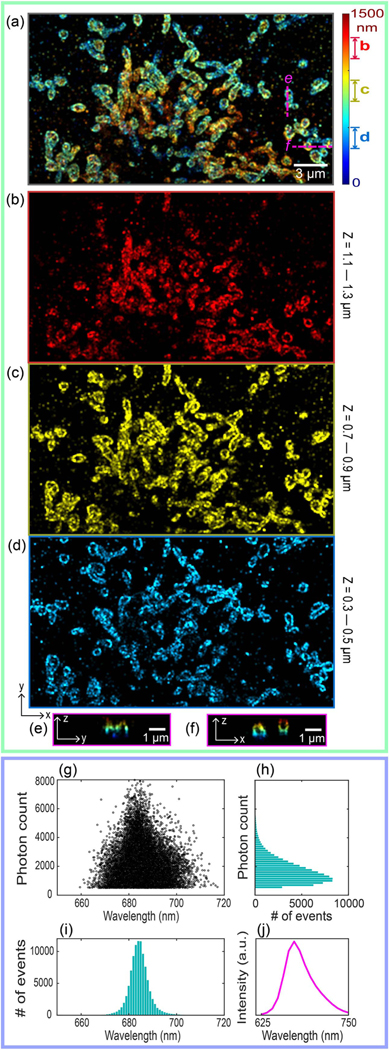
To demonstrate the 3D imaging capability of this new biplane design, we imaged COS-7 cells with Alexa Fluor 647 (AF647)-labeled mitochondria (see sample preparation in Method D). The illumination power was ~15 kW∕cm2, the EMCCD exposure time was 20 ms, and the axial separation between the spatial and spectral images was 500 nm. The FOV of the acquired images was ~15 × 30 μm2 (see details in Supplement 1). The average density of the fluorophores was <15 emitters per raw image (camera frame). The number of raw images was 25,000, and the total acquisition time was ~9 min for one reconstructed super-resolution image. Figure 2(a) is the overall projection image of the reconstructed 3D sSMLM with pseudocolors corresponding to the z-axis positions. We acquired three 3D biplane sSMLM datasets with 450 nm axial translation at each acquisition and combined them to form the overall 3D dataset, covering a depth range of 1.5 μm. Figures 2(b), 2(c), and 2(d) are the projection images from three 200 nm thick volumes, as highlighted in Fig. 2(a). Figures 2(e) and 2(f) are cross-sectional images of mitochondria along the y–z and x–z planes, respectively, as highlighted in Fig. 2(a). As we can see, the nanostructures of the mitochondria are clearly distinguished along all three-dimensional directions.
Fig. 2.
(a) Overall projection image of the reconstructed 3D sSMLM with pseudocolors corresponding to the z-axis positions of individual molecules. (b–d) The projection images from three 200 nm thick sections as highlighted in (a). The cross-sectional images in (e) the y–z plane and (f) x–z plane corresponding to the three magenta-dashed lines as highlighted in (a). (g–j) Spectral analyses of same single AF647 molecules. (g) The scatterplot of the photo count versus the spectral centroids. (h) and (i) respectively show the statistics of the emission photon count versus the number of emission events and centroid wavelengths versus the number of emission events. (j) The averaged spectrum of AF647 of all emission events in (g).
To characterize the performance of the 3D biplane sSMLM, we experimentally quantified the spatial localization precisions and spectral precision using fluorescent nanospheres and compared them with the theoretical values. As shown in Fig. S1, we confirmed that the experimentally obtained values agree well with the theoretical expectations. We also compared the localization precision and spectral precision of the 3D biplane sSMLM with that of the astigmatism-based 3D sSMLM. As shown in Fig. S2, both methods showed comparable spatial precisions and spectral precision (see Supplement 1). We further measured the depth variation of the ratio between the localization precisions along the x and y directions (defined as lateral precision isotropy) and compared the depth variations of the precision isotropy in both the biplane- and astigmatism-based 3D imaging methods. As shown in Fig. S3, the biplane-based method had a nearly uniform lateral precision isotropy within the entire 800 nm depth range. In contrast, the lateral precision isotropy varied significantly along the depth in the astigmatism-based method due to nonuniform photon densities along the x and y axes induced by astigmatism [16,17].
Using the simultaneously captured spectral images, we characterized the spectroscopic signatures of the individual AF647 molecules based on the spectral centroid method [1–3,5–7,30]. The scatterplot of the photo count versus the spectral centroid is shown in Fig. 2(g). We extracted the spectral centroid values from 99,211 molecules in one 3D sSMLM dataset. We rejected emissions with a detected intensity below 500 photons. Figures 2(h) and 2(i) respectively show the statistics of the emission photon count versus the number of emission events and centroid wavelengths versus the number of emission events. We found that the average photon count was 1250 in the spectral images and 550 in the spatial images. As shown in Fig. 2(i), we observed a narrow wavelength centroid distribution, which corresponds to a spectral precision of 4.21 nm. Figure 2(j) shows the averaged spectrum of AF647 for all the detected emission events in Fig. 2(g).
3. SIMULTANEOUS MULTI-COLOR 3D IMAGING
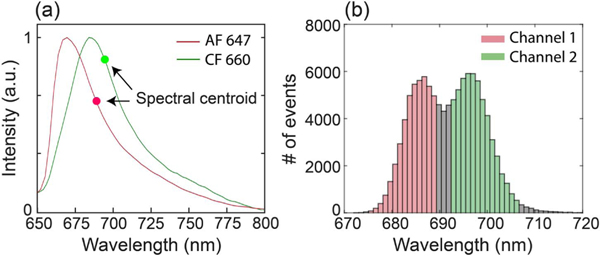
We further demonstrated simultaneous dual-color 3D imaging of two far-red molecular dyes with highly overlapping emission spectra using 3D biplane sSMLM. Several far-red dyes, which have similar fluorescent emission spectra (with typically 2.5 to 4.5 nm variation in the emission wavelength centroid) and can be excited by a single laser, have recently been reported for sSMLM [1]. We selected two dyes (AF647 and CF660C) with similar emission spectra to label the microtubule and mitochondria, respectively. Figure 3(a) shows the normalized emission spectra of AF647 (red) and CF660C (green) with their respective centroids (686 nm for AF647 and 695 nm for CF660C) highlighted. Figure 3(b) shows the histogram of the spectral centroid of the multi-color 3D sSMLM image of tubulin and TOM20, labeled with AF647 and CF660C, respectively. Most of the detected spectral centroids for the microtubules (labeled with AF647) are around 686 nm, and most of the detected spectral centroids for the mitochondria (labeled with CF660C) are around 695 nm. We separated them by defining different color channels in the spectral image: the first channel from 674 to 689 nm for AF647 (red) and the second channel from 692to 707 nm for CF660C (green). We confirmed the estimated misidentification between these two color channels was relatively low (<7%) (see Supplement 1, Fig. S4).
Fig. 3.
(a) Normalized emission spectra of AF647 (red) and CF660C (green) with their centroids (686 nm for AF647 and 695 nm for CF660C), respectively. (b) The histogram of spectral centroid of multi-color 3D sSMLM image of tubulin and TOM20, labeled with AF647 and CF660C, respectively. To separate the two dyes, we defined two different color channels: (1) 674 to 689 nm for AF647 and (2) 692 nm to 707 nm for CF660C.
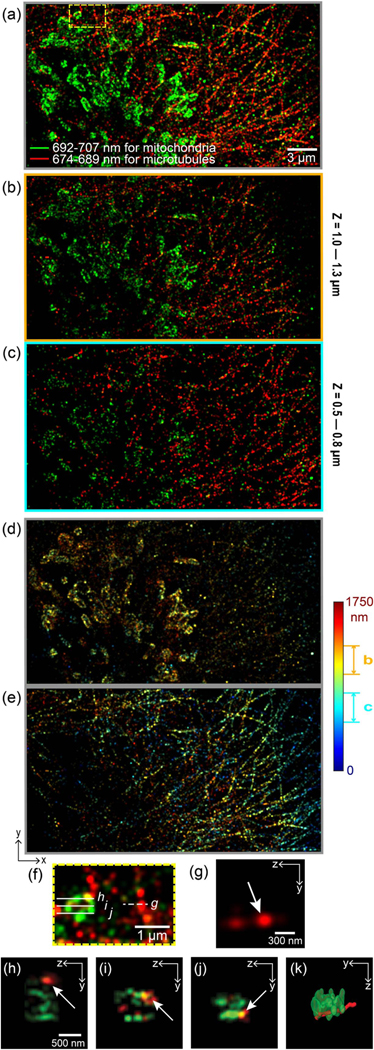
Figure 4 shows multi-color 3D sSMLM images of tubulin and TOM20 in COS-7 cells labeled with AF647 and CF660C, respectively. Similar to the data acquisition shown in Fig. 2, we combined three datasets translated along the z axis at a step size of 450 nm. Figure 4(a) shows the 2D projection of the whole depth range of 1.75 μm. The red and green colors in Fig. 4(a) represent the microtubules and mitochondria, respectively. We observed well-distinguished mitochondria and microtubules with negligible misidentification in their morphologies. In addition, we show the projection images with depth ranges from 1.0 to 1.3 μm and from 0.5to 0.8 μm in Figs. 4(b) and 4(c), respectively. We clearly observed different morphologies for both distinct subcellular structures at the different axial ranges over all the images. Figures 4(d) and 4(e) are separated 3D sSMLM images of mitochondria and microtubules, respectively. Figure 4(f) is a magnified view of the region, highlighted by the yellow-dashed box in Fig. 4(a), showing spatially overlapping mitochondria and microtubule structures. The cross-sectional image of a microtubule along the y–z plane is shown in Fig. 4(g) from the position highlighted by the dashed line in Fig. 4(f). From the axial line profiles of microtubules, we measured the axial resolution to be ~50 nm (see Supplement 1, Fig. S6). Figures 4(h)–4(j) show cross-sectional images of a mitochondrion and a microtubule and their spatial co-localization from positions highlighted by three white solid lines (h, i, and j) in Fig. 4(f). We provide a 3D surface-rendering result in Fig. 4(k), which shows that the microtubule is close to the bottom of the mitochondria. These results demonstrate that 3D biplane sSMLM has the potential to provide insight into nanoscopic intracellular structures.
Fig. 4.
(a) Overall 2D projection view over a whole depth range of 1.75 μm. The red and green colors represent AF647-labeled microtubules and CF660C-labeled mitochondria, respectively. (a) The projection images for different axial ranges from (b) 1.0 to 1.3 μm and from (c) 0.5 to 0.8 μm [color-coded along the spectral centroid in (a–c)]. The separated 3D sSMLM images for different color channels, (d) 692 to 707 nm for mitochondria and (e) 674–689 nm for microtubules [color-coded along the axial axis in (d–e)]. (f) The magnified view of the region indicated by the yellow-dashed box in (a). (g) The cross-section image corresponding to the white-dashed lines in(f).(h–j)Thecross-section images corresponding to the three white-solid lines in (f) [color-coded along the spectral centroid in (f–j)]. (k) The volumetric rendering of the region covering (h–j). The rendering was visualized with the interpolation for a microtubule.
4. CONCLUSION AND DISCUSSION
By taking advantage of the axial position information encoded in already existing spatial and spectral imaging channels, we developed 3D biplane sSMLM. This method allows for simultaneous 3D spectroscopic imaging of multiple dyes without the need for sophisticated optics modification for PSF manipulation. Compared with reported 3D sSMLM work, we only added a pair of mirrors to introduce an optical delay in the spectral imaging channel and maximized the benefits of the two already existing imaging channels. We showed a lateral localization precision of 20 nm at an average photon count of 550 with an axial localization precision of 50 nm in the multi-color 3D imaging. We also confirmed the spectral precision of 4 nm at an average photon count of 1250 in the single-color 3D imaging (see detailed analyses in Supplement 1), and we confirmed a nearly uniform lateral localization precision within an imaging depth range of 800 nm using fluorescent nanospheres.
It is worth mentioning that we found a wavelength calibration challenge in the astigmatism-based 3D sSMLM imaging (see detailed discussion in Supplement 1). When a cylindrical lens is inserted only into the zeroth-order (spatial imaging) path, the beam paths for the spatial and spectral imaging experience different spherical aberrations, and this error may cause a spectral bias. We observed a wavelength calibration error up to 5 pixels (corresponding to 40 nm spectral bias) at the 8-nm spectral dispersion [Supplement 1, Fig. S7]. Such spectral bias can be worse in sSMLM systems with lower spectral dispersion and larger FOVs. Therefore, the astigmatism-based 3D method can be applicable to sSMLM only after a careful spherical aberration compensation. In contrast, the biplane-based 3D method does not introduce an aberration difference between the two imaging paths, inducing no additional wavelength calibration error.
The active area of the EMCCD camera is relatively limited in sSMLM, as we simultaneously acquired both the spatial and spectral images. This is one of the key limitations to restrict the FOV and the density of fluorophores. Furthermore, spectroscopic signatures in the spectral image are dispersed into several pixels along the spectral axis (typically ~30 pixels under our experimental conditions), whereas the PSF along the y axis can be covered by 7 pixels (>99%), given the sigma value of the PSF is approximately 1 pixel under our experimental conditions. Therefore, the density of fluorophores is rather limited, and the overlapping issue between adjacent molecules in the spectral image is less tolerable in sSMLM.
The 3D biplane sSMLM offers several potential benefits to molecular biology investigations. First, it allows not only direct 3D observation of co-localized nanoscopic structures but also comprehensive analyses of multi-molecular functions and interactions. Second, 3D biplane sSMLM can reliably identify multiple molecular dyes, which can have highly overlapped emission spectra. Third, the system’s spectral dispersion can be tuned, and thus it can afford relatively high spectral dispersion for more in-depth single-molecule spectroscopic analysis. In addition, it further helps to effectively suppress undesirable background, including the autofluorescence of impurities, by separating them from target molecules based on their different spectroscopic signatures. Finally, 3D biplane sSMLM is compatible with and can be readily implemented in practically all existing photo-activated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM) systems. Therefore, biplane sSMLM provides an opportunity to investigate nanoscopic biological 3D structures at spectroscopic precision levels previously far out of reach.
5. METHODS
A. Definition of Spectral Dispersion
Spectral dispersion (nm/pixel) is defined as the wavelength range per individual pixel. Assuming the spectral dispersion is linear in a given sSMLM system, it can be expressed by
| (1) |
where W p is the camera pixel size [micrometers (μm)]; Rd is the reciprocal linear dispersion; d is the groove spacing; θ is the angle of diffraction at the mth order; m is the order of the maxima (typically 1); and f is the effective focal length of the relay optics in the spectrometer, which is defined as Df 2∕f 1, where D is the distance from the grating position to the intermediate image plane; f 1 is the focal length of the collimating lens (L1); and f 2 is the focal length of the focusing lens (L2). Accordingly, it shows the separation of wavelength [nanometers (nm)] per interval distance (μm). In this work, the spectral dispersion was adjusted to 8 nm/pixel.
B. Spectral Calibration Procedure
Inorderto obtain the emission spectra of single molecules from the recorded spectral images, a spectral calibration procedure is required and is specific to individual sSMLM systems. Using a calibration light source (Neon lamp, 6032, Newport), we first captured a reference image, which includes a straight line confined by a narrow slit and multiple spectral lines. These multiple spectral lines in the spectral image correspond tothe emissions centered at a specific wavelength of the calibration light source, whereas the straight line in the spatial image corresponds to the slit position. We used two main emission peaks at 640.23 and 703.24 nm. Then, the pixel positions and wavelengths of these spectra lines were fitted by a polynomial function. We used the fitted function to establish the spatial-spectral mapping relationship between the spatial image and spectral image. We point out that a grating offers a linear spectral dispersion within the spectral range.
Next, the obtained spatial-spectral mapping relationship was applied to individual stochastic emission events. We used the spatial images of the emission events as the reference locations for the spectral analysis. Accordingly, we arranged individual spectroscopic signatures in the spectral window with the same wavelength range. Finally, by integrating the spectral images along the y axis, we obtained the emission spectra of individual emission events.
C. Multi-Color 3D sSMLM Image Reconstruction
To acquire a 3D calibration curve, we first captured a few PSFs in the spatial and spectral images at different axial positions using far-red nanospheres (200 nm diameter; F8807, Invitrogen). We translated the sample from −1.5 to 1.5 μm along the z axis with a step size of 25 nm as shown in Fig. 1(h). Then we extracted the 1D PSFys in both the spatial and spectral images as shown in Figs. 1(e) and 1(f) by integrating the intensities along the x axis. After measuring the FWHM values of the two 1D PSFys, we calculated the ratio between them along the z axis to use as the depth calibration curve [Supplement 1, Fig. S8].
We reconstructed 2D images using ThunderSTORM [31] and 3D images using customized MATLAB codes. For each individual molecule, we first localized its x–y positions in the spatial image and then calculated the PSFy ratio between the spatial and spectral images to obtain its z position based on the depth calibration curve. Together with the spectral centroid calculated from the spectral image, we can obtain a 4D array for all individual molecules. During the spectral analysis, we rejected emissions with a detected intensity below 750 photons. We numerically corrected the slight magnification difference between the spatial and spectral imaging channels caused by their different path lengths before image reconstruction.
D. Sample Preparation
COS-7 cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco/Life Technologies) supplemented with 2 mM L-glutamine (Gibco/Life Technologies), 10% fetal bovine serum (Gibco/Life Technologies), and 1% penicillin/streptomycin (100 U mL−1, Gibco/LifeTechnologies) at 37°C with 5% CO2. The cells were plated on a No. 1 borosilicate-bottom eightwell Lab-Tek Chambered Coverglass with ~30% confluency. After 48 h, we fixed the cells in pre-warmed 3% paraformaldehyde and 0.1% glutaraldehyde in phosphate buffered saline (PBS) for 10 min, washed with PBS twice, quenched with freshly prepared 0.1% sodium borohydride in PBS for 7 min, and rinsed twice with PBS at 25°C. The fixed samples were permeabilized with a blocking buffer (3% bovine serum albumin (BSA), 0.5% Triton X-100 in PBS) for 20 min and then incubated with the primary antibodies (Mouse anti-TOM20, 2.5 μg mL−1, Santa Cruz, sc-17764; sheep anti-tubulin, 2.5 μg mL−1, Cytoskeleton, ATN02) in blocking buffer for 1 h. The samples were rinsed in a washing buffer (0.2% BSA, 0.1% Triton X-100 in PBS) three times for 5 min and further incubated with the corresponding secondary antibodies-dye conjugates (donkey anti-mouse Alexa Fluor 647 and/or anti-sheep CF660C, 2.5 μg mL−1, degree of labeling = 1–2 dyes per antibody, prepared according to [32]) for 40 min. Finally, the cells were washed thoroughly with PBS three times for 5 min and stored at 4°C. Prior to imaging acquisition, an imaging buffer (pH = ~8.0) containing 50 mM Tris, 10 mM NaCl, 0.5 mg mL−1 glucose oxidase (Sigma, G2133), 2000 U/mL catalase (Sigma, C30), 10% (w/v) D-glucose, and 100 mM cysteamine replaced PBS in the sample chamber and was imaged without further preparation.
Supplementary Material
Funding.
National Science Foundation (NSF) (CBET-1706642, EEC-1530734, EFMA-1830969); National Institutes of Health (NIH) (R01EY026078, R01EY029121).
Footnotes
Disclosures. C. Sun and H. F. Zhang have financial interests in Opticent Inc., which did not support this work. The other authors declare no conflicts of interest related to this work.
See Supplement 1 for supporting content.
REFERENCES
- 1.Zhang Z, Kenny SJ, Hauser M, Li W, and Xu K, “Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy,” Nat. Methods 12, 935–938 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Dong B, Almassalha L, Urban BE, Nguyen TQ, Khuon S, Chew TL, Backman V, Sun C, and Zhang HF, “Super-resolution spectroscopic microscopy via photon localization,” Nat. Commun. 7, 12290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mlodzianoski MJ, Curthoys NM, Gunewardene MS, Carter S, and Hess ST, “Super-resolution imaging of molecular emission spectra and single molecule spectral fluctuations,” PLoS One 11, e0147506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongiovanni MN, Godet J, Horrocks MH, Tosatto L, Carr AR, Wirthensohn DC, Ranasinghe RT, Lee JE, Ponjavic A, Fritz JV, Dobson CM, Klenerman D, and Lee SF, “Multi-dimensional super-resolution imaging enables surface hydrophobicity mapping,” Nat. Commun. 7, 13544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong B, Soetikno BT, Chen X, Backman V, Sun C, and Zhang HF,“Parallel three-dimensional tracking of quantum rods using polarization-sensitive spectroscopic photon localization microscopy,” ACS Photon. 4,1747–1752 (2017). [Google Scholar]
- 6.Kim D, Zhang Z, and Xu K, “Spectrally resolved super-resolution microscopy unveils multipath reaction pathways of single spiropyran molecules,” J. Am. Chem. Soc. 139, 9447–9450 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Moon S, Yan R, Kenny SJ, Shyu Y, Xiang L, Li W, and Xu K,“Spectrally resolved, functional super-resolution microscopy reveals nanoscale compositional heterogeneity in live-cell membranes,” J. Am. Chem. Soc. 139, 10944–10947 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Broeken J, Rieger B, and Stallinga S, “Simultaneous measurement of position and color of single fluorescent emitters using diffractive optics,” Opt. Lett. 39, 3352–3355 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Davis JL, Dong B, Sun C, and Zhang HF, “Method to identify and minimize artifacts induced by fluorescent impurities in single-molecule localization microscopy,” J. Biomed. Opt. 23, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang L, Wojcik M, Kenny SJ, Yan R, Moon S, Li W, and Xu K,“Optical characterization of surface adlayers and their compositional demixing at the nanoscale,” Nat. Commun. 9, 1435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang T, Phelps C, Wang J, Lin LJ, Bittel A, Scott Z, Jacques S, Gibbs SL, Gray JW, and Nan X, “Simultaneous multicolor single-molecule tracking with single-laser excitation via spectral imaging,” Biophys. J. 114, 301–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Song K-H, Tang S, Ravelo L, Cusido J, Sun C, Zhang HF, and Raymo FM, “Far-red photoactivatable BODIPYs for the super-resolution imaging of live cells,” J. Am. Chem. Soc. 140, 12741–12745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Song K-H, Dong B, Davis JL, Shao G, Sun C, and Zhang HF, “Multicolor super-resolution imaging using spectroscopic single-molecule localization microscopy with optimal spectral dispersion,” Appl. Opt. 58, 2248–2255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang B, Wang W, Bates M, and Zhuang X, “Three-dimensional Super-resolution Imaging by Stochastic Optical Reconstruction Microscopy,” Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang B, Jones SA, Brandenburg B, and Zhuang X, “Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution,” Nat. Methods 5, 1047–1052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, and Bewersdorf J, “Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples,” Nat. Methods 5, 527–529 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Mlodzianoski MJ, Juette MF, Beane GL, and Bewersdorf J,“Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy,” Opt. Express 17, 8264–8277 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Ram S, Prabhat P, Chao J, Ward ES, and Ober RJ, “High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells,” Biophys. J 95, 6025–6043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winterflood CM, Platonova E, Albrecht D, and Ewers H, “Dual-color 3D superresolution microscopy by combined spectral-demixing and biplane imaging,” Biophys. J. 109, 3–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, and Hess HF, “Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure,” Proc.Natl. Acad. Sci. USA 106, 3125–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aquino D, Schonle A, Geisler C, Middendorff CV, Wurm CA, Okamura Y, Lang T, Hell SW, and Egner A, “Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores,” Nat. Methods 8, 353–359 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Xu K, Babcock HP, and Zhuang X, “Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton,” Nat. Methods 9, 185–188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, and Moerner WE, “Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function,” Proc. Natl. Acad. Sci. USA 106, 2995–2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grover G, DeLuca K, Quirin S, DeLuca J, and Piestun R, “Super-resolution photon-efficient imaging by nanometric double-helix point spread function localization of emitters (SPINDLE),” Opt. Express 20, 26681–26695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quirin S, Pavani SR, and Piestun R, “Optimal 3D single-molecule localization for superresolution microscopy with aberrations and engineered point spread functions,” Proc. Natl. Acad. Sci. USA 109, 675–679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia S, Vaughan JC, and Zhuang X, “Isotropic 3D super-resolution imaging with a self-bending point spread function,” Nat. Photonics 8, 302–306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aristov A, Lelandais B, Rensen E, and Zimmer C, “ZOLA-3D allows flexible 3D localization microscopy over an adjustable axial range,” Nat. Commun. 9, 2409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mlodzianoski MJ, Cheng-Hathaway PJ, Bemiller SM, McCray TJ,Liu S, Miller DA, Lamb BT, Landreth GE, and Huang F, “Active PSF shaping and adaptive optics enable volumetric localization microscopy through brain sections,” Nat. Methods 15, 583–586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith C, Huisman M, Siemons M, Grünwald D, and Stallinga S,“Simultaneous measurement of emission color and 3D position of single molecules,” Opt. Express 24, 4996–5013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song K, Dong B, Sun C, and Zhang HF, “Theoretical analysis of spectral precision in spectroscopic single-molecule localization microscopy,” Rev. Sci. Instrum. 89, 123703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovesny M, Krizek P, Borkovec J, Svindrych Z, and Hagen GM,“ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging,” Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dempsey GT, Vaughan JC, Chen KH, Bates M, and Zhuang X,“Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging,” Nat. Methods 8, 1027–1036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.