Abstract
Multidrug-resistant Salmonella enterica serovar Typhimurium phage type DT104 has become a widespread cause of human and other animal infection worldwide. The severity of clinical illness in S. enterica serovar Typhimurium DT104 outbreaks has led to the suggestion that this strain possesses enhanced virulence. In the present study, in vitro and in vivo virulence-associated phenotypes of several clinical isolates of S. enterica serovar Typhimurium DT104 were examined and compared to S. enterica serovar Typhimurium ATCC 14028s. The ability of these DT104 isolates to survive within murine peritoneal macrophages, invade cultured epithelial cells, resist antimicrobial actions of reactive oxygen and nitrogen compounds, and cause lethal infection in mice were assessed. Our results failed to demonstrate that S. enterica serovar Typhimurium DT104 isolates are more virulent than S. enterica serovar Typhimurium ATCC 14028s.
Food-borne bacterial infections due to Salmonella remain a serious threat to human health in both developing and industrialized countries. Between 1997 and 1998, 37,842 cases of human salmonellosis were reported to the Centers for Disease Control and Prevention. The estimated number of human Salmonella infections in the United States exceeds 1.4 million annually (21). In 1995, 24% of all reported Salmonella infections in the United States were caused by Salmonella enterica serovar Typhimurium, second only to serovar Enteritidis phage group 4. Of the S. enterica serovar Typhimurium isolates, 32% were found to be multidrug-resistant S. enterica serovar Typhimurium DT104 (17), characterized as resistant to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (1). S. enterica serovar Typhimurium DT104 was first isolated in England in 1984 but is now routinely isolated worldwide (22, 24). A British study conducted in 1994 reported a 3% mortality rate due to infection with S. enterica serovar Typhimurium DT104 (1), a rate substantially higher than in historical controls. However, in a separate study examining Salmonella bacteremia in England and Wales from 1994 to 1996, the percentage of fatal salmonellosis cases due to S. enterica serovar Typhimurium DT104 was no greater than those due to non-multidrug-resistant S. enterica serovar Typhimurium (23). Attention became focused on S. enterica serovar Typhimurium DT104 in the United States when members of a Vermont family became gravely ill after consuming contaminated unpasteurized milk from their own dairy herd (C. R. Friedman, R. C. Brady, M. J. Celotti, S. E. Schoenfeld, R. H. Johnson, P. D. Galbraith, J. K. Carney, K. Robbins, and L. Slutsker, presented at Int. Conf. Emerg. Infect. Dis., Atlanta, Ga., 8 to 11 March 1998).
Because of the reported severity of disease caused by this organism and the increased frequency of isolation, S. enterica serovar Typhimurium DT104 has been proposed to have enhanced virulence in domestic animals and humans (24). Previous work by Carlson et al. (7, 8) indicated that most multidrug-resistant S. enterica serovar Typhimurium isolates do not have enhanced ability to invade or adhere to human epithelial tissue culture cells. In this study, we have examined additional in vitro and in vivo phenotypes associated with Salmonella virulence.
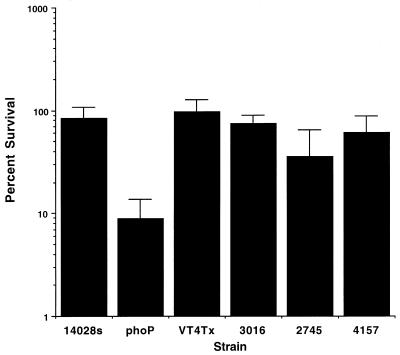
The ability of Salmonella to survive and replicate in host phagocytes is an essential component of Salmonella virulence. Mutants that are unable to survive in macrophages are attenuated for virulence in vivo (14). We compared several S. enterica serovar Typhimurium DT104 isolates from different geographic locales for their ability to survive within murine macrophages. The strain numbers and source of these strains are shown in Table 1. The intracellular survival of S. enterica serovar Typhimurium DT104 isolates and control strains in peritoneal macrophages from BALB/c mice was determined as previously described (5). Macrophages were infected with opsonized S. enterica serovar Typhimurium, and survival was measured 18 h postinfection. A phoP S. enterica serovar Typhimurium mutant was used as a macrophage-sensitive control (13). S. enterica serovar Typhimurium DT104 isolates were able to survive and replicate in activated murine macrophages at levels similar to those of S. enterica serovar Typhimurium 14028s (Fig. 1).
TABLE 1.
Strains used in this study
| Strain | Relevant information | Reference or Source |
|---|---|---|
| 14028s | ATCC | |
| 14028s phoP | 14 | |
| 14028s hilA | 2 | |
| 14028s invA | 15 | |
| VT4Tx | Vermont DT104 bovine isolate | USDA, Athens, Ga. |
| 3016 | Georgia DT104 poultry isolate | USDA, Athens, Ga. |
| 2745 | Washington DT104 human isolate | J. Gay, Washington State University |
| 4157 | Washington DT104 bovine isolate | J. Gay, Washington State University |
FIG. 1.
Macrophage survival. Survival of Salmonella strains was determined in BALB/c (itys) peritoneal macrophages. Peritoneal macrophages were harvested from mice 4 days after injection with 5 mM sodium periodate and plated at a density of 3 × 105 to 5 × 105/well. They were infected 24 h later at a multiplicity of infection of 5:1 with opsonized S. enterica serovar Typhimurium 14028s, S. enterica serovar Typhimurium DT104 isolates, or macrophage-sensitive mutant phoP S. enterica serovar Typhimurium. Extracellular bacteria were killed using amikacin (100 μg/ml) since the DT104 isolates are resistant to gentamicin. Results are expressed as percent survival and represent the average of two independent assays. Error bars indicate standard deviation.
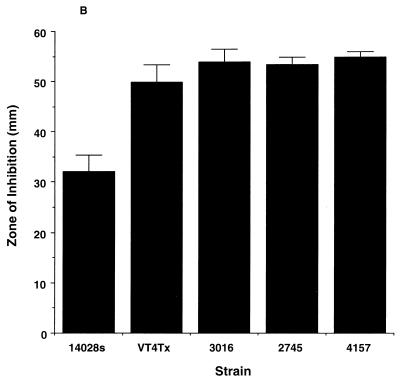
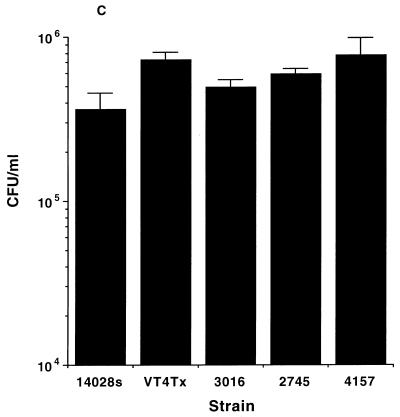
Another important characteristic of Salmonella is its ability to resist reactive oxygen and nitrogen species produced by host phagocytes (4, 6, 11, 18). The resistance of S. enterica serovar Typhimurium DT104 isolates to reactive oxygen species and reactive nitrogen species was assayed as previously described by DeGroote et al. (10–12) and Lu et al. (20). S. enterica serovar Typhimurium DT104 isolates did not demonstrate enhanced susceptibility to hydrogen peroxide compared with S. enterica serovar Typhimurium 14028s (Fig. 2A). Similar results were observed for paraquat susceptibility (Fig. 2B). S. enterica serovar Typhimurium DT104 isolates showed similar resistance to acidified nitrite after 3 h of incubation (Fig. 2C) (20). Furthermore, all DT104 isolates were able to grow to the same extent in acidified (pH5) Luria-Bertani (LB) broth lacking sodium nitrite (data not shown).
FIG. 2.
Susceptibility to reactive oxygen and nitrogen species. (A and B) Salmonella cultures were grown overnight in LB medium, diluted in saline, and plated (100 μl containing 106 CFU) on M9 minimal plates containing 0.2% glucose. Then 15 μl of 3% hydrogen peroxide (A) or paraquat (1 mM) (B) was spotted onto 6-mm-diameter paper disks, the disks were placed onto the bacterial lawn, and the plates were incubated overnight at 37°C. The zone of inhibition was measured on two axes, averaged, and plotted. (C) Susceptibility to acidified nitrite was determined by adding 20 μl of an overnight culture to 2 ml of acidified LB (pH 5) with or without 20 mM sodium nitrite. Bacteria were incubated at 37°C with aeration for 3 and 6 h, and viable cells were enumerated by plating dilutions onto LB agar.
The ability of Salmonella to invade the intestinal epithelium is a necessary step for the initial phase of Salmonella infection. To determine the relative invasive ability of S. enterica serovar Typhimurium DT104 isolates, standard epithelial cell invasion assays were performed with cultured HEp-2 cells, a human epithelial carcinoma cell line. As shown in Fig. 3, S. enterica serovar Typhimurium 14028s and DT104 isolates invaded HEp-2 cells to a similar extent, while the noninvasive S. enterica serovar Typhimurium mutants, hilA and invA, showed significantly reduced invasion, as previously described (16, 19). Thus, the invasive properties of the S. enterica serovar Typhimurium DT104 isolates tested are not significantly different from those of S. enterica serovar Typhimurium 14028s.
FIG. 3.
Invasion of HEp-2 cells by Salmonella strains. HEp-2 cells were grown to confluence and infected at a multiplicity of infection of 10 to 50 with S. enterica serovar Typhimurium 14028s, S. enterica serovar Typhimurium DT104 isolates, and hilA and invA mutant strains of S. enterica serovar Typhimurium grown overnight in LB without aeration at 37°C. Nonadherent bacteria were removed by washing three times with phosphate-buffered saline; then, RPMI plus 10% fetal calf serum containing amikacin at 100 μg/ml was added and the plates were incubated for an additional 1 h. The cells were washed three times with phosphate-buffered saline and lysed with 1% Triton X-100. Bacterial invasion was determined by plating serial dilutions onto LB agar. The results, expressed as percent invasion, are representative of three independent assays. Error bars indicate standard deviation.
The standard in vitro assays described above indicate that S. enterica serovar Typhimurium DT104 isolates do not demonstrate an increased ability to invade tissue culture cells, survive within murine macrophages, or withstand reactive oxygen or nitrogen species. However, in vivo virulence cannot always be predicted from in vitro phenotypic assays (22). Therefore, we tested whether DT104 isolates exhibit increased in vivo virulence relative to the well-characterized strain S. enterica serovar Typhimurium 14028s in the murine model of Salmonella infection, including a competitive infection assay. The virulence of S. enterica serovar Typhimurium DT104 isolates in susceptible mice was compared to that of S. enterica serovar Typhimurium 14028s. Salmonella-susceptible BALB/c mice were infected orally with different inocula of one of four geographically diverse S. enterica serovar Typhimurium DT104 isolates or S. enterica serovar Typhimurium 14028s. Following oral administration of ∼108 CFU, the ability of S. enterica serovar Typhimurium VT4Tx (a bovine strain isolated from the Vermont outbreak) to cause lethal infection in mice was essentially identical to that of S. enterica serovar Typhimurium 14028s. Similar observations were made following oral administration of clinical DT104 isolates 3016 (a chicken isolate from Georgia) and 4157 (a bovine isolate from Washington State) (Table 1). However, strain 2745, a human clinical DT104 isolate (from Washington State), did not cause lethal infection in BALB/c mice following inoculation of 108 CFU. Salmonella-resistant C3H/HeN mice that were infected with S. enterica serovar Typhimurium DT104 isolates did not succumb to infection or show clinical signs of salmonellosis when the bacterium was given at doses ranging up to 108 CFU (Table 2). PCR analysis of this strain using primers for four loci known to be required for Salmonella virulence (spvC, hilA, sodCI, and invF) (9, 11, 15, 19) indicated that strain 2745 carried these genes and suggested that its decreased virulence is not attributable to the absence of these loci. S. enterica serovar Typhimurium DT104 isolates had similar in vitro growth characteristics in LB broth, formed smooth colonies on LB agar plates, and grew well on M9 minimal medium supplemented with 0.2% glucose (data not shown).
TABLE 2.
Lethality and competitive infection of S. enterica serovar Typhimurium 14028s and DT104 isolates for BALB/c mice
| Strain | Dose (CFU)a | Mortality (no. dead/total no.)a | Dose (CFU)b | Spleen CIb | Liver CIb |
|---|---|---|---|---|---|
| 14028s | 3.6 × 106 | 0/3 | |||
| 3.6 × 107 | 1/3 | ||||
| 3.6 × 108 | 3/3 | ||||
| VT4Tx | 2.8 × 106 | 0/3 | 1.4 × 108 | 0.3620 ± 0.701c | 0.3627 ± 0.698c |
| 2.8 × 107 | 2/3 | ||||
| 2.8 × 108 | 3/3 | ||||
| 3016 | 3.1 × 106 | 0/3 | 1.3 × 108 | 0.0108 ± 0.022 | 0.0046 ± 0.008 |
| 3.1 × 107 | 1/3 | ||||
| 3.1 × 108 | 3/3 | ||||
| 2745 | 2.8 × 106 | 0/3 | 1.3 × 108 | 0.0039 ± 0.005 | 0.0506 ± 0.096 |
| 2.8 × 107 | 0/3 | ||||
| 2.8 × 108 | 0/3 | ||||
| 4157 | 3.0 × 106 | 0/3 | 1.3 × 108 | 0.0306 ± 0.051 | 0.0124 ± 0.015 |
| 3.0 × 107 | 3/3 | ||||
| 3.0 × 108 | 2/3 |
Groups of three 6-week-old BALB/c female mice (Charles River Laboratories) were infected orally with S. enterica serovar Typhimurium ATCC 14028s or DT104 isolates grown overnight at 37°C in LB broth. Prior to oral infection, food and water were withheld from the mice for 4 h. Bacteria were diluted in phosphate-buffered saline and administered to the mice by allowing them to drink 20 μl from the end of a pipette tip. Infected mice were observed daily for up to 3 weeks, and moribund mice were euthanized.
For the competitive infection assay, each S. enterica serovar Typhimurium DT104 isolate was mixed 1:1 with S. enterica serovar Typhimurium ATCC 14028s for coinfection of four BALB/c female mice per group. The mice were challenged using the same procedure as described in footnote a. Results are listed as the median CI for day 6 postinfection ± standard deviation.
Variation due to one mouse having a CI greater than 1.
A competitive-infection assay was performed to further compare the virulence of the S. enterica serovar Typhimurium DT104 isolates with that of S. enterica serovar Typhimurium 14028s. Groups of four BALB/c mice were orally infected with ∼108 CFU containing a 1:1 mixture of S. enterica serovar Typhimurium 14028s and each of the four S. enterica serovar Typhimurium DT104 isolates: VT4Tc, 3016, 2745, and 4157. On days 4 and 6 postinfection, mice were euthanized and tissues were collected for bacterial enumeration. The tissues were homogenized in 10 ml of sterile water, and 10-fold serial dilutions were plated on XLD (Difco) and XLD containing chloramphenicol at 20 μg/ml, to distinguish S. enterica serovar Typhimurium 14028s (chloramphenicol susceptible) from the multidrug-resistant DT104 isolates (chloramphenicol resistant). The number of CFU of S. enterica serovar Typhimurium 14028s per organ was calculated by subtracting the number of colonies on the XLD-chloramphenicol plates from the number of colonies on the corresponding XLD plates. The competitive index (CI) was calculated as the ratio of the CFU of each S. enterica serovar Typhimurium DT104 isolate to the CFU of S. enterica serovar Typhimurium 14028s recovered from the spleen and liver. None of the four S. enterica serovar Typhimurium DT104 isolates tested were able to colonize the spleen or liver of infected mice as well as S. enterica serovar Typhimurium 14028s did during mixed infections, as demonstrated by a CI ratio of less than 1 (Table 2). Nevertheless, three of the four DT104 strains were able to cause lethal infections in mice when administered singly.
In conclusion, we have utilized in vitro and in vivo virulence assays to compare four geographically diverse S. enterica serovar Typhimurium DT104 clinical isolates with a well-characterized virulent S. enterica serovar Typhimurium strain. S. enterica serovar Typhimurium DT104 isolates from Washington, Vermont, and Georgia did not demonstrate enhanced resistance to reactive oxygen or nitrogen species, nor were these isolates able to survive and replicate in activated murine macrophages or invade cultured epithelial cells to a greater extent than S. enterica serovar Typhimurium ATCC 14028s. When tested for virulence in susceptible mice, most DT104 isolates showed similar lethality, although one DT104 isolate (2745) was unable to cause lethal infection. In a mixed-infection assay, none of the S. enterica serotype Typhimurium DT104 isolates demonstrated an enhanced ability to compete with S. enterica serovar Typhimurium ATCC 14028s. The increasing frequency of S. enterica serovar Typhimurium DT104 isolation from both humans and domestic animals cannot be attributed to enhanced virulence-associated phenotypes detectable by conventional assays. Of course, the conditions that permit S. enterica serovar Typhimurium DT104 to disseminate efficiently under field conditions cannot be completely replicated in the laboratory.
At present it is not clear whether the increased prevalence of S. enterica serovar Typhimurium DT104 in many parts of the world is more likely to be a result of its resistance to multiple antimicrobial agents per se (3) or to greater competitive fitness related to other, unknown factors. Nevertheless, the continued widespread use of antimicrobial agents in the production of food animals is likely to provide a potent selection pressure for the emergence and persistence of multidrug-resistant strains such as S. enterica serovar Typhimurium DT104.
Acknowledgments
We thank T. Halsey, L. Hatcher, A. Treece, and B. J. Welker for their technical assistance and Rebecca Wilson and Dale Hancock for critical reading of the manuscript.
This work was supported by USDA grant 95-372042659 and NIH grants AI34397-06 (S.J.L.), AI10181 (A.V.T.), and AI39557 (F.C.F.).
REFERENCES
- 1.Akkina J E, Hougue A T, Angulo R J, Johnson R, Petersen K E, Saini P K, Fedorka-Cray P J, Schlosser W E. Epidemiologic aspects, control, and importance of multiple-drug resistant Salmonella typhimurium DT104 in the United States. J Am Vet Med Assoc. 1999;214:790–798. [PubMed] [Google Scholar]
- 2.Bajaj V, Lucas R L, Hwang C, Lee C A. Coordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 3.Bjorkman J, Huge D, Andersson D I. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier N A, Bossie S, Chen C-Y, Fang F C, Guiney D G, Libby S J. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect Immun. 1997;65:3725–3730. doi: 10.1128/iai.65.9.3725-3730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchmeier N A, Libby S J, Xu Y, Lowewen P, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Investig. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson S A, Browning M, Ferris K E, Jones B D. Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog. 2000;28:37–44. doi: 10.1006/mpat.1999.0322. [DOI] [PubMed] [Google Scholar]
- 8.Carlson S A, Willson R M, Crane A J, Ferris K E. Evaluation of invasion-conferring genotypes and antibiotic-induced hyperinvasive phenotypes in multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog. 2000;28:373–378. doi: 10.1006/mpat.2000.0355. [DOI] [PubMed] [Google Scholar]
- 9.Chikami G K, Fierer J, Guiney D G. Plasmid-mediated virulence in Salmonella dublin demonstrated by use of a Tn5-oriT construct. Infect Immun. 1985;50:420–424. doi: 10.1128/iai.50.2.420-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Groote M A, Fang F C. NO inhibitions: antimicrobial properties of nitric oxide. Clin Infect Dis. 1995;21(Suppl. 2):S162–S165. doi: 10.1093/clinids/21.supplement_2.s162. [DOI] [PubMed] [Google Scholar]
- 11.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazques-Torres A, Xu Y, Fang F C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide sythanse. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote M A, Testerman T, Xu Y, Stauffer G, Fang F C. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science. 1996;272:414–417. doi: 10.1126/science.272.5260.414. [DOI] [PubMed] [Google Scholar]
- 13.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 14.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galán J E, Curtiss R I. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosek G, Leschinsky D, Irons S, Safrank T J. Multidrug-resistant Salmonella serotype Typhimurium—U.S., 1996. Morb Mortal Wkly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 18.Humphreys S, Stevenson A, Bacon A, Weinhardt A B, Roberts M. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C A, Jones B D, Falkow S. Indentification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S, Manges A R, Xu Y, Fang F C, Riley L W. Analysis of virulence of clinical isolates of Salmonella enteritidis in vivo and in vitro. Infect Immun. 1999;67:5651–5657. doi: 10.1128/iai.67.11.5651-5657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mead P S, Slutsker L, Dietz V, McCaig L R, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Threlfall E J, Frost J A, Ward L R, Rowe B. Increasing spectrum of resistance in multiresistant Salmonella typhimurium. Lancet. 1996;347:1053–1054. doi: 10.1016/s0140-6736(96)90199-3. [DOI] [PubMed] [Google Scholar]
- 23.Threlfall E J, Ward L R, Rowe B. Multiresistant Salmonella typhimurium DT104 and salmonella bacteraemia. Lancet. 1998;352:287–288. doi: 10.1016/s0140-6736(05)60261-9. [DOI] [PubMed] [Google Scholar]
- 24.Villar R G, Macek M D, Simons S, Hayes P S, Goldoft M J, Lewis J H, Rowan L L, Hursh D, Patnode M, Mead P S. Investigation of multidrug-resistant Salmonella serotype DT104 infections linked to raw-milk cheese in Washington state. JAMA. 1999;281:1811–1816. doi: 10.1001/jama.281.19.1811. [DOI] [PubMed] [Google Scholar]