Abstract
Pseudomonas aeruginosa is an important pathogen as it can cause hospital-acquired infections. Additionally, it can also colonize in patients and in other various environments. Hence, this study aimed to investigate the antimicrobial susceptibility, and to study the molecular features, of colonizing isolates of P. aeruginosa from Songklanagarind Hospital, Thailand. Genomic DNA extraction, whole-genome sequencing (WGS), and bioinformatics analysis were performed in all studied isolates. The findings demonstrated that the majority of isolates were non-susceptible to colistin and carbapenem. For in silico study, multilocus sequence typing (MLST) revealed one novel sequence type (ST) 3910 and multiple defined STs. The isolates carried several antimicrobial resistance genes (blaOXA-50, aph(3′)-IIb, etc.) and virulence-associated genes (fleN, waaA, etc.). CRISPR-Cas sequences with different spacers and integrated bacteriophage sequences were also identified in these isolates. Very high SNPs were found in the alignments of the novel ST-3910 isolate with other isolates. A comparative genomic analysis exhibited phylogenetic clustering of our colonizing isolates with clinical isolates from many countries. Interestingly, ST-3981, ST-3982, ST-3983, ST-3984, ST-3985, ST-3986, ST-3986, ST-3986, ST-3987, and ST-3988, the new STs from published genomes, were assigned in this study. In conclusion, this WGS data might be useful for tracking the spread of P. aeruginosa colonizing isolates.
Keywords: Pseudomonas aeruginosa, antimicrobial susceptibility, molecular feature, whole-genome sequencing, bioinformatics analysis, novel sequence type
1. Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative opportunistic bacterium that commonly infects and colonizes humans, animals, and plants, and it can be found in various environments, such as soil and water [1,2]. This bacterium is a member of the ESKAPEE pathogens, a group of antibiotic-resistant nosocomial pathogens, which includes Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli [3]. The Centers for Disease Control and Prevention (CDC) reported on the incidence of multidrug-resistant (MDR) P. aeruginosa in American hospitalized patients in 2017. This report outlined the causes of a variety of hospital-acquired infections, including respiratory tract infections (RTIs), bloodstream infections (BIs), urinary tract infections (UTIs), and surgical site infections (SSIs); all of which are considered to have a serious threat level [4]. In this report, there were an estimated 32,600 cases, with 2700 estimated deaths, and $767,000,000 estimated attributable healthcare costs in the United States of America (USA) [4]. Additionally, infection and colonization by P. aeruginosa, particularly MDR strains, are classified as the third group of Gram-negative pathogens in hospitals within Thailand [5].
Over the past decade, multiple studies have reported on the whole-genome sequencing (WGS) data of P. aeruginasa from several regions. In the study of Quick et al. (2014), WGS and bioinformatics analysis were conducted for 141 P. aeruginosa isolates from patients and contaminated sources (hospital water and the ward environment) [6]. The phylogenetic tree showed eight clades (A to H), which contained many clades of clinical isolates and a single clade of environmental isolates. The single nucleotide polymorphisms (SNPs) in the genes associated with intrinsic resistance mechanisms (mutations) to antimicrobial agents were identified in the studied isolates. Cottalorda et al. (2021) also sequenced 108 P. aeruginosa urinary isolates obtained from two to five samples of seven hospitalized patients in a French hospital [7]. Their results demonstrated that a single clone type of P. aeruginosa colonized in each patient, which was considered by <6000 SNPs between a pair of the isolates. Another study by Zhu et al. (2021) reported the WGS data of 151 carbapenem-resistant P. aeruginosa from China [8]. Eleven defined STs that included ST-463 as the most frequent and one undefined ST were found, and eight plasmid types with 13 plasmid patterns were observed. In the study in Thailand, Cazares et al. (2020) performed WGS in 23 of 48 MDR P. aeruginosa clinical isolates at a teaching hospital at Mahidol University, Bangkok, Central Thailand [9]. In this study, the majority of the isolates belonged to the dominant ST-253 (n = 3) in addition to many other STs (e.g., ST-233, ST-244, ST-270, ST-357, ST-491, ST-708, ST-1330, and so on); however, some isolates could not be assigned to the STs. Additionally, Soonthornsit et al. (2022) sequenced 10 antimicrobial-resistant P. aeruginosa isolates from a veterinary teaching hospital in Nakhon Pathom, Central Thailand [10]. They also found a high-risk clone ST-235 (n = 3) as well as other STs (ST-244, ST-485, ST-606, and ST-3405). Moreover, the multilocus sequence typing (MLST) and SNP analysis exhibited a correlation between the isolates from ward rooms to those from operating, wound care, and examination rooms. Even though these studies have reported genomic insights into P. aeruginosa, there is still a lack of data on whole-genome sequences of P. aeruginosa, especially the colonizing isolates from Southern Thailand.
Hence, the objectives of this study were to evaluate antimicrobial susceptibility and to study the molecular characteristics of P. aeruginosa isolates colonized in the patients at the Medicine Ward, Songklanagarind Hospital, Southern Thailand.
2. Results
2.1. Antimicrobial Susceptibility Profiles
Antimicrobial susceptibility profiles in 13 colonizing isolates of P. aeruginosa showed that all isolates were susceptible to almost all tested antimicrobial agents, including piperacillin-tazobactam, ceftolozane-tazobactam, amikacin, gentamicin, tobramycin, ciprofloxacin, and levofloxacin (Table 1). Meanwhile, a total of nine, three, two, and one isolates were resistant to colistin with minimum inhibitory concentrations (MICs) ranging from 4 to 8 µg/mL, imipenem with MICs ranging from 16 to 32 µg/mL, meropenem with MICs ranging from 8 to 16 µg/mL, and doripenem with MIC at 8 µg/mL, respectively. The results also exhibited that all (n = 13) and over a half (n = 9) of the isolates were non-susceptible (intermediate to resistant) to colistin and carbapenems, respectively.
Table 1.
Antimicrobial susceptibility in the colonizing isolates of P. aeruginosa.
| Isolate Code | Minimum Inhibitory Concentration (µg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactam Combination Agents |
Carbapenems | Lipopeptide | Aminoglycosides | Fluoroquinolones | |||||||
| TZP | C/T | IPM | MEM | DOR | CST | AMK | GEN | TOB | CIP | LVX | |
| PA01 | 16 (S) | 4 (S) | 16 (R) | 16 (R) | 8 (R) | 4 (R) | 2 (S) | 1 (S) | 2 (S) | 0.25 (S) | 1 (S) |
| PA02 | 4 (S) | 0.5 (S) | 64 (R) | 0.25 (S) | 0.25 (S) | 4 (R) | 8 (S) | 2 (S) | 4 (S) | 0.06 (S) | 0.25 (S) |
| PA03 | 8 (S) | 2 (S) | 4 (I) | 0.5 (S) | 0.5 (S) | 8 (R) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) | 0.5 (S) |
| PA04 | 8 (S) | 1 (S) | 4 (I) | 0.5 (S) | 0.5 (S) | 4 (R) | 4 (S) | 1 (S) | 2 (S) | 0.25 (S) | 0.5 (S) |
| PA05 | 8 (S) | 2 (S) | 4 (I) | 0.5 (S) | 0.5 (S) | 4 (R) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) | 0.25 (S) |
| PA06 | 16 (S) | 0.5 (S) | 4 (I) | 2 (S) | 1 (S) | 2 (I) | 2 (S) | 1 (S) | 2 (S) | 0.25 (S) | 0.5 (S) |
| PA07 | 8 (S) | 0.5 (S) | 4 (I) | 1 (S) | 0.5 (S) | 4 (R) | 2 (S) | 1 (S) | 0.5 (S) | 0.25 (S) | 0.5 (S) |
| PA08 | 8 (S) | 0.5 (S) | 2 (S) | 0.5 (S) | 0.5(S) | 2 (I) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) | 0.5 (S) |
| PA09 | 8 (S) | 1 (S) | 1 (S) | 0.5 (S) | 0.5 (S) | 2 (I) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) | 0.5 (S) |
| PA10 | 8 (S) | 2 (S) | 2 (S) | 0.5 (S) | 0.5 (S) | 4 (R) | 4 (S) | 1 (S) | 2 (S) | 0.25 (S) | 0.5 (S) |
| PA11 | 16 (S) | 0.5 (S) | 2 (S) | 0.5 (S) | 0.5 (S) | 2 (I) | 2 (S) | 1 (S) | 1 (S) | 0.25 (S) | 0.25 (S) |
| PA13 | 8 (S) | 0.5 (S) | 32 (R) | 8 (R) | 2 (S) | 4 (R) | 4 (S) | 1 (S) | 2 (S) | 0.25 (S) | 0.5 (S) |
| PA14 | 16 (S) | 1 (S) | 4 (I) | 0.5 (S) | 0.5 (S) | 8 (R) | 2 (S) | 1 (S) | 0.5 (S) | 0.25 (S) | 0.5 (S) |
S, susceptible; I, intermediate; R, resistant; TZP, piperacillin-tazobactam; C/T, ceftolozane-tazobactam; CST, colistin; IPM, imipenem; MEM, meropenem; DOR, doripenem; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin.
2.2. Genome Assembly Quality
The genome assembly quality of all the studied isolates is demonstrated in Table S1 and Figure S1. In the QUAST results, the genome lengths of 6,202,244 to 6,974,963 bps were obtained from 13 P. aeruginosa isolates, and N50 ranged from 391,215 to 931,346 bps. A total of 62 to 266 contigs were generated from these isolates, and L50 ranged from three to seven contigs. In the BUSCO assessment results, even though one duplicated gene and a few fragmented genes were detected in our P. aeruginosa genomes, very high completeness without missing genes was found in all the genomes.
2.3. Sequence Types and Serotypes
Multilocus sequence typing (MLST) was performed in all colonizing isolates of P. aeruginosa to identify the sequence types (STs). The results revealed that two isolates belonged to ST-162, while 10 isolates belonged to different STs (ST-266, ST-270, ST-313, ST-500, ST-532, ST-647, ST-980, ST-1097, ST-1197, and ST-1240), as presented in Table 2. Notably, the PA02 isolate was not assigned to any ST due to its new allelic profile. Afterward, the assembled genome of the PA02 isolate was submitted with a new MLST profile into PubMLST (https://pubmlst.org/, accessed on 28 April 2022), and ST-3910 was then released for the PA02 isolate. In the prediction of P. aeruginosa serotypes, the results revealed that O11, O6, O3, O1, O5, and O10 were identified in four, four, two, one, one, and one isolates, respectively (Table 2).
Table 2.
Sequence types (STs) and serotypes of the colonizing isolates of P. aeruginosa.
| Isolate Code | Patient’s Sex | Specimen | Serotype | Multilocus Sequence Typing (MLST) | Accession No. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | ST | |||||
| PA01 | Female | Rectum | O6 | 17 | 5 | 6 | 3 | 4 | 15 | 10 | 647 | JANTQX000000000 |
| PA02 | Male | Rectum | O11 | 20 | 30 | 64 | 26 | 30 | 24 | 32 | 3910 * | JANTQW000000000 |
| PA03 | Female | Rectum | O3 | 16 | 5 | 11 | 3 | 4 | 12 | 3 | 980 | JANTQV000000000 |
| PA04 | Female | Throat | O6 | 6 | 5 | 1 | 30 | 4 | 6 | 27 | 1097 | JANTQU000000000 |
| PA05 | Female | Rectum | O3 | 28 | 5 | 11 | 7 | 1 | 6 | 61 | 1240 | JANTQT000000000 |
| PA06 | Male | Rectum | O10 | 5 | 8 | 119 | 6 | 12 | 6 | 3 | 1197 | JANTQS000000000 |
| PA07 | Female | Throat | O11 | 5 | 4 | 5 | 5 | 5 | 20 | 4 | 532 | JANTQR000000000 |
| PA08 | Female | Throat | O5 | 22 | 3 | 17 | 5 | 2 | 10 | 7 | 270 | JANTQQ000000000 |
| PA09 | Male | Rectum | O6 | 11 | 57 | 7 | 3 | 4 | 15 | 1 | 500 | JANTQP000000000 |
| PA10 | Male | Throat | O6 | 16 | 5 | 11 | 72 | 44 | 7 | 52 | 266 | JANTQO000000000 |
| PA11 | Male | Throat | O1 | 47 | 8 | 7 | 6 | 8 | 11 | 40 | 313 | JANTQN000000000 |
| PA13 | Male | Throat | O11 | 6 | 5 | 6 | 34 | 27 | 3 | 7 | 162 | JANTQM000000000 |
| PA14 | Male | Throat | O11 | 6 | 5 | 6 | 34 | 27 | 3 | 7 | 162 | JANTQL000000000 |
* Novel ST containing a new allelic profile.
2.4. Acquired Antimicrobial Resistance Genes
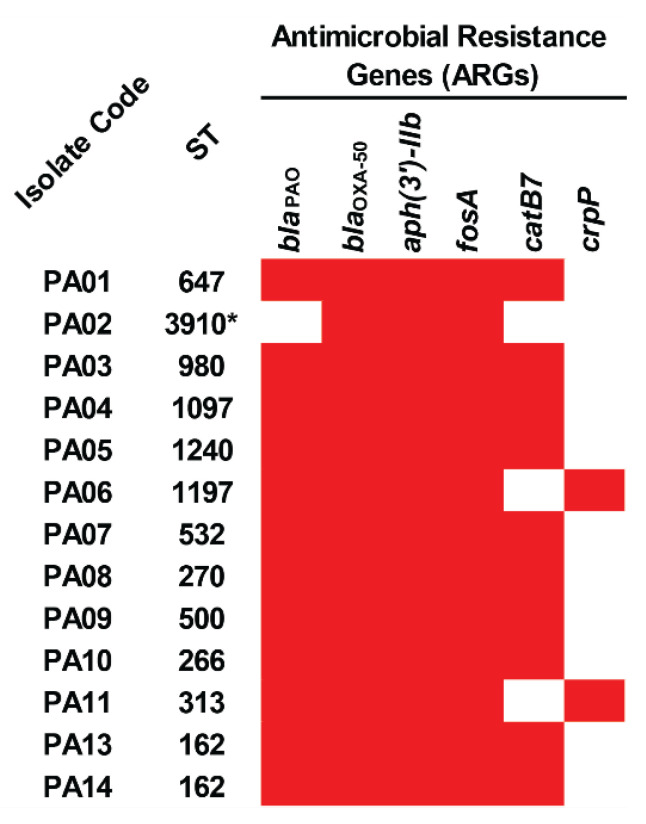
In the detection of acquired antimicrobial resistance genes (ARGs), it was found that all the colonizing isolates of P. aeruginosa carried blaOXA-50, aph(3′)-IIb, and fosA, while 12, 10, and two isolates harbored blaPAO, catB7, and crpP genes (Figure 1 and Table S2). Among the studied isolates, a novel ST-3910 (PA02) possessed only three ARGs, including blaOXA-50, aph(3′)-IIb, and fosA. These genes may provide resistance to β-lactam (blaOXA-50 and blaPAO), aminoglycoside (aph(3′)-IIb), fosfomycin (fosA), chloramphenicol (catB7), and ciprofloxacin (crpP).
Figure 1.
Distribution of antimicrobial resistance genes (ARGs) in the colonizing isolates of P. aeruginosa. The red box represents the gene presence. ST, sequence type. * Novel ST.
2.5. Insertion Sequence Elements
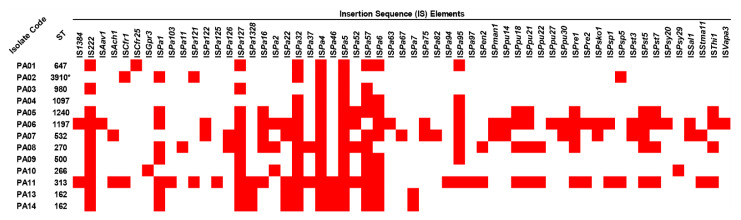
To look for mobile genetic elements (MGEs), insertion sequence (IS) elements and integrons in all colonizing isolates of P. aeruginosa were identified. The results showed that 57 IS elements were present in the isolates. Among them, ISPa4 and ISPa5 were detected in all isolates, while ISPa32, IS222, ISPa127, ISPa57, and ISPa6 were found in 12, 11, 11, 11, and 10, respectively (Figure 2 and Table S3). In contrast, ISAav1, ISCfr25, ISGpr3, ISPa103, ISPa11, ISPa125, ISPa63, ISPa67, ISPa82, ISPa94, ISPa97, ISPpu27, ISPsy20, ISPsy29, and ISVapa3 were only found in one isolate. Among these isolates, the ST-313 (PA11) isolate possessed the highest number of IS elements, followed by ST-1197 (PA06), ST-532 (PA07), ST270 (PA08), and so on. However, via in silico analysis, integrons were not found in the studied isolates.
Figure 2.
Distribution of insertion sequence (IS) elements in the colonizing isolates of P. aeruginosa. The red box represents the gene presence. ST, sequence type. * Novel ST.
2.6. Virulence-Associated Genes
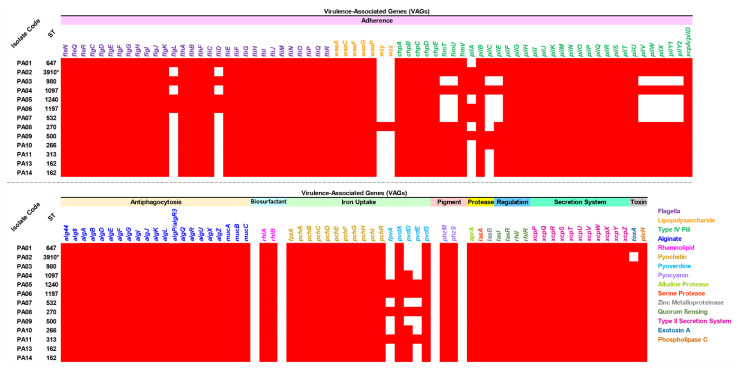
Virulence-associated genes (VAGs) were explored in all the colonizing isolates of P. aeruginosa, which possessed 115 to 127 genes encoding several virulence factors, as shown in Figure 3 and Table S4. All isolates carried all detected genes encoding antiphagocytosis (alginate), biosurfactant (rhamnolipid), iron uptake (pyochelin), pigment (pyocyanin), protease (alkaline protease, serine protease, and zinc metalloproteinase), regulation (quorum sensing), secretion system (type II secretion system), and toxin (phospholipase c). Furthermore, high numbers of genes encoding adherence (flagella, lipopolysaccharide, and type IV pili) were found in all isolates. A gene encoding exotoxin A (toxA) was also present in all isolates, except for the novel ST-3910 (PA02) isolate. On the other hand, among the genes encoding lipopolysaccharide, only the ST-270 (PA08) isolate carried wzy and wzz genes.
Figure 3.
Distribution of virulence-associated genes (VAGs) in the colonizing isolates of P. aeruginosa. The red box represents the gene presence. ST, sequence type. * Novel ST.
2.7. Bacteriocins
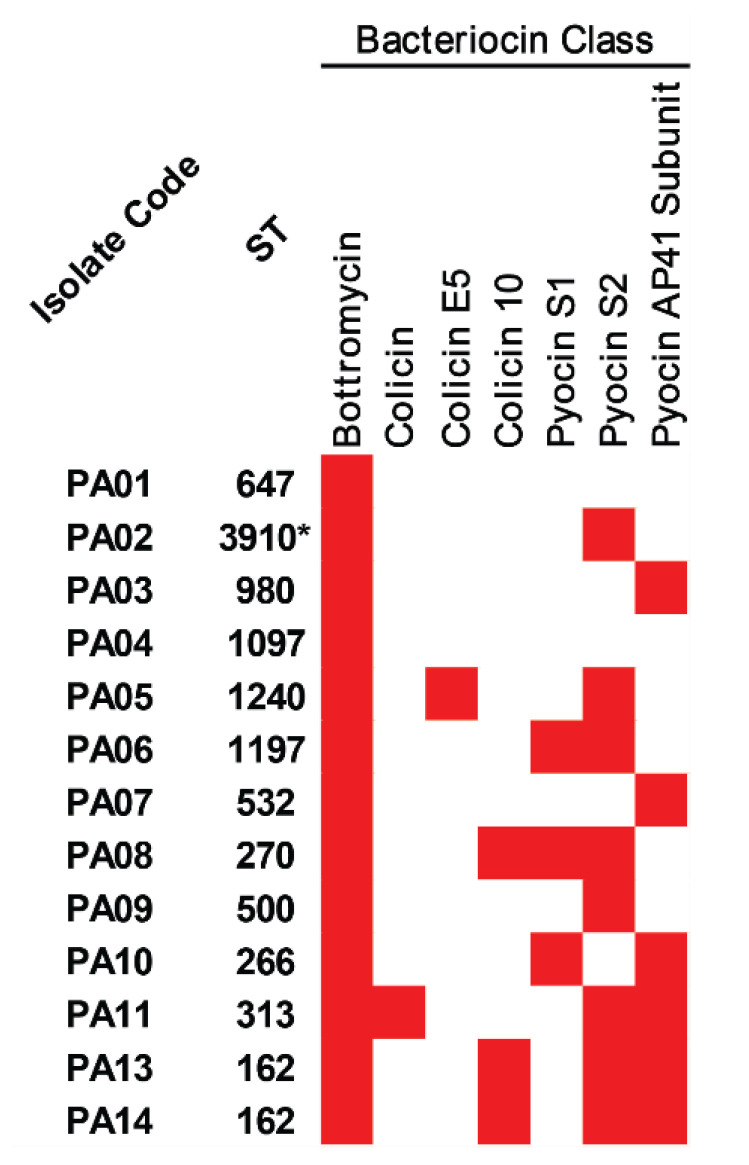
With regards to bacterial competition, bacteriocin-encoding genes were predicted in all colonizing isolates of P. aeruginosa, and the results are illustrated in Figure 4 and Table S5. Bottromycin and pyocin AP41 subunit were positive in all and eight isolates, respectively, while colicin 10 and pyocin S1 were found in three isolates. In addition, colicin and colicin E5 were only detected in the ST-313 (PA11) and ST-1240 (PA05) isolates, respectively.
Figure 4.
Bacteriocins in the colonizing isolates of P. aeruginosa. The red box represents the gene presence. * Novel ST.
2.8. CRISPR-Cas System
To evaluate the bacterial defense system, a clustered, regularly interspaced, short palindromic repeats and CRISPR-associated protein (CRISPR-Cas) system was explored in all colonizing isolates of P. aeruginosa. We found the CRISPR-Cas regions in seven isolates, as listed in Table 3 and Table S6. Even though the Cas region was not detected in the ST-266 (PA10) isolate, one CRISPR region carrying nine spacer sequences was detected in this isolate. Remarkably, nine CRISPR loci with four to twenty-seven different spacer sequences and two Cas types (IF and IE) were harbored by the novel ST-3910 (PA12) isolate. Two groups of the cas genes (cas1_cas3-cas2_csy1_csy2_csy3_cas6 and cas6_csy3_csy2_ csy1_cas3-cas2_cas1) were classified as Cas type IF, while the other two groups (cas2_cas1_cas6_cas5_cas7_cse2_cse1_cas3 and cas3_cse1_cse2_cas7_cas5_cas6_cas1_cas2) were predicted as Cas type IE.
Table 3.
Clustered regularly interspaced short palindromic repeats and CRISPR-associated protein (CRISPR-Cas) system in the colonizing isolates of P. aeruginosa.
| Isolate Code | ST | CRISPR-Cas | ||||
|---|---|---|---|---|---|---|
| Region | Element | No. of Spacer | No. of cas Gene (Cas Type) | Direct Repeat (DR) Consensus/cas Gene | ||
| PA01 | 647 | 1 | CRISPR | 13 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |
| 2 | CRISPR | 11 | GTTCACTGCCGTATAGGCAGCTAAGAAA | |||
| 3 | Cas cluster | 6 (IF) | cas6, csy3, csy2, csy1, cas3-cas2, cas1 | |||
| 4 | CRISPR | 7 | TTTCTTAGCTGCCTACACGGCAGTGAAC | |||
| PA02 | 3910 * | 1 | CRISPR | 10 | GTTCACTGCCGTGTAGGCAGCTAAGAAA | |
| 2 | Cas cluster | 6 (IF) | cas1, cas3-cas2, csy1, csy2, csy3, cas6 | |||
| 3 | CRISPR | 4 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| 4 | CRISPR | 7 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| 5 | CRISPR | 27 | CGGTTCATCCCCACGCATGTGGGGAACAC | |||
| 6 | Cas cluster | 8 (IE) | cas2, cas1, cas6, cas5, cas7, cse2, cse1, cas3 | |||
| 7 | CRISPR | 4 | CGGTTCATCCCCACACCCGTGGGGAACAC | |||
| 8 | CRISPR | 4 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| 9 | CRISPR | 6 | TTTCTTAGCTGCCTACACGGCAGTGAAC | |||
| 10 | CRISPR | 10 | GTTCACTGCCGTGTAGGCAGCTAAGAAA | |||
| 11 | CRISPR | 5 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| PA03 | 980 | 1 | CRISPR | 22 | GTTCACTGCCGTATAGGCAGCTAAGAAA | |
| 2 | CRISPR | 11 | GTTCACTGCCGTGTAGGCAGCTAAGAAA | |||
| 3 | Cas cluster | 6 (IF) | cas1, cas3-cas2, csy1, csy2, csy3, cas6 | |||
| 4 | CRISPR | 13 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| PA04 | 1097 | 1 | CRISPR | 19 | GTTCACTGCCGTGTAGGCAGCTAAGAAA | |
| 2 | Cas cluster | 6 (IF) | cas1, cas3-cas2, csy1, csy2, csy3, cas6 | |||
| 3 | CRISPR | 22 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |||
| PA05 | 1240 | 1 | CRISPR | 7 | GTTCACTGCCGTATAGGCAGCTAAGAAA | |
| 2 | Cas cluster | 6 (IF) | cas6, csy3, csy2, csy1, cas3-cas2, cas1 | |||
| 3 | CRISPR | 14 | TTTCTTAGCTGCCTACACGGCAGTGAAC | |||
| PA06 | 1197 | ND | ND | ND | ND | |
| PA07 | 532 | 1 | CRISPR | 11 | GTGTTCCCCACGGGTGTGGGGATGAACCG | |
| 2 | Cas cluster | 8 (IE) | cas3, cse1, cse2, cas7, cas5, cas6, cas1, cas2 | |||
| 3 | CRISPR | 12 | GTGTTCCCCACATGCGTGGGGATGAACCG | |||
| PA08 | 270 | ND | ND | ND | ND | |
| PA09 | 500 | ND | ND | ND | ND | |
| PA10 | 266 | 1 | CRISPR | 9 | TTTCTTAGCTGCCTATACGGCAGTGAAC | |
| PA11 | 313 | ND | ND | ND | ND | |
| PA13 | 162 | ND | ND | ND | ND | |
| PA14 | 162 | ND | ND | ND | ND | |
ND, not detected. * Novel ST.
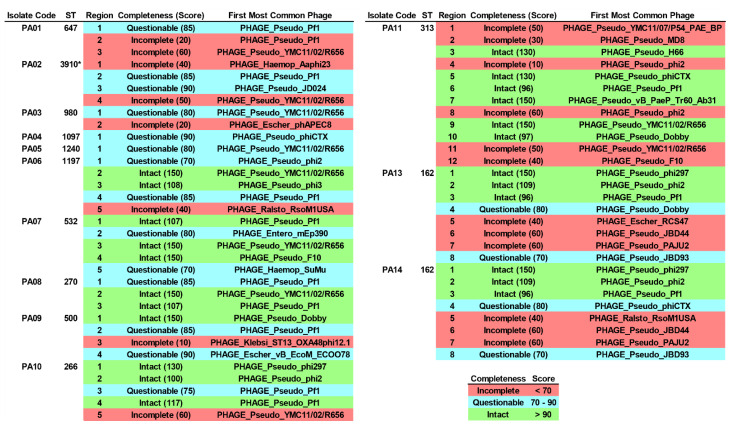
2.9. Integrated Bacteriophage Genomes
Lysogenic bacteriophage infection is one of the factors for driving genes (e.g., ARGs, VAGs, etc.) to other bacterial cells. Here, integrated bacteriophage genomes (IBGs) in all the colonizing isolates of P. aeruginosa were screened for. At least a part of the bacteriophage sequences had been integrated into the genomes of the studied isolates, as shown in Figure 5 and Table S7. A total of 23, 20, and 18 regions were identified as intact, incomplete, and questionable, respectively. A majority of those regions were classified as Pseudomonas phages (n = 52), while some regions were classified as Escherichia phages (n = 3), Haemophilus phages (n = 2), Ralstonia phages (n = 2), an Enterobacter phage (n = 1), and a Klebsiella phage (n = 1). The partial sequence of PHAGE_Pseudo_Pf1 (n = 13) was dominantly found in the isolates, followed by PHAGE_Pseudo_YMC11/02/R656 (n = 10), PHAGE_Pseudo_phi2 (n = 6), PHAGE_Pseudo_phiCTX (n = 3), PHAGE_Pseudo_Dobby (n = 3), PHAGE_Pseudo_phi297 (n = 3), PHAGE_Pseudo_phiCTX (n = 3), and so on. Most of the sequences were encoded for bacteriophage composition proteins (head/capsid, tail, and plate) as well as bacteriophage enzymes (integrase, terminase, recombinase, and transposase).
Figure 5.
Integrated bacteriophage genomes (IBGs) in the colonizing isolates of P. aeruginosa. ST, sequence type. * Novel ST.
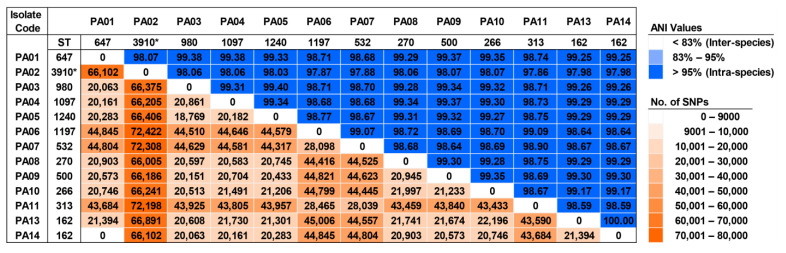
2.10. Genomic Diversity and Phylogenetic Relationship
To study the genomic diversity of the studied isolates, pairwise average nucleotide identity (ANI) and pairwise single nucleotide polymorphism (SNP) distance were analyzed, and these results are demonstrated in Figure 6. The ANI values ranged from 97.86–100%, showing that all colonizing isolates were intra-species of P. aeruginosa. An ANI value of 100% was only found in an alignment of the ST-162 genomes (PA13 and PA14). Although a total of 18,769 to 72,422 SNPs were found in the pairs of these sequences, none of the SNPs were identified in an alignment between the ST-647 (PA01) and ST-162 (PA14) genomes. On the contrary, 21,394 SNPs were detected in a pair of ST-162 sequences (PA13 and PA14). Over 66,000 SNPs were predominantly observed in the pairs of the novel ST-3910 (PA02) sequences with other sequences.
Figure 6.
The matrix of pairwise average nucleotide identity (ANI) values and pairwise single nucleotide polymorphism (SNP) distance among the colonizing isolates of P. aeruginosa. ST, sequence type. * Novel ST.
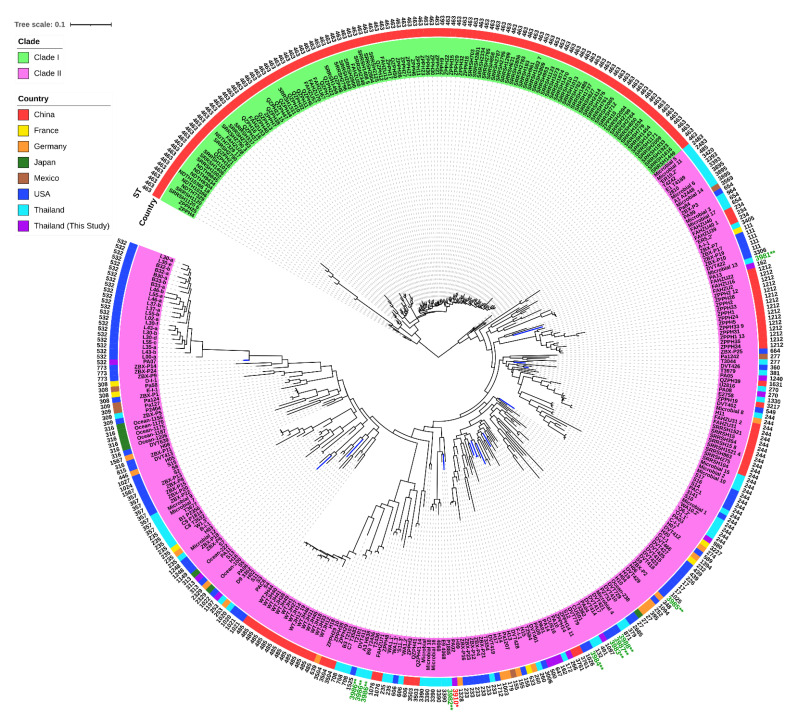
In addition, our genomes (n = 13) were compared with previously published genomes (n = 357) of P. aeruginosa clinical isolates, and the metadata of the included genomes is illustrated in Table S8. The pan-genome profiles of 370 genomes showed 35,430 pan genes, which included 4004 (11.30%) core genes and 31,426 (88.70%) accessory genes (Figure S2 and Table S9). Many genes encode for hypocritical proteins. Interestingly, the ST-463 isolates in the upper monophyletic group (clade I) harbored 38 unique genes. These genes encode for hypothetical proteins (n = 29) and other identified proteins (n = 9), including tyrosine recombinase XerC, HTH-type transcriptional repressor GlaR, putative NADH-specific resorcinol 4-hydroxylase, hexuronate transporter, ATP-dependent DNA helicase Rep, ATP-dependent RNA helicase RhlE, RNA polymerase-associated protein RapA, GTPase Era, and ADP-ribosylarginine hydrolase Tri1. More importantly, 127 genes-encoding unique proteins, including hypothetical proteins (n = 109) and other identified proteins (n = 18), had the highest frequency that exited in the novel ST-3910 (PA02) isolate, compared with the other 12 genomes of our colonizing isolates. The SNP-based phylogenetic tree of 370 genomes revealed two major clades (Figure 7), which is similar to the pan-genome matrix against the phylogenetic tree based on accessory genes (Figure S2). Clade I contained only the ST-463 isolates (n = 116) from China, while clade II included multiple isolates (n = 254) belonging to 99 different STs from various countries. Our colonizing isolates of P. aeruginosa were distributionally found in several subclades of clade II. Besides this, a total of 10 published genomes from other studies in Thailand (n = 5) and the USA (n = 5) could not be assigned with STs. These genomes were then submitted into PubMLST, and 10 novel STs (ST-3981, ST-3982, ST-3983, ST-3984, ST-3985, ST-3986, ST-3986, ST-3986, ST-3987, and ST-3988) were assigned accordingly. Notably, some published genomes from other areas of Thailand as well as our genomes were clustered in different subclades.
Figure 7.
A single nucleotide polymorphism (SNP) phylogenetic tree of our genomes (blue linage) and other available genomes of P. aeruginosa. ST, sequence type. * Novel ST from our P. aeruginosa genome (red text). ** Novel STs from previously published genomes of P. aeruginosa (green text).
3. Discussion
P. aeruginosa, especially antibiotic-resistant strains, are categorized as the third pathogen of Gram-negative bacteria causing nosocomial infections in Thailand [5]. Importantly, multidrug-resistant P. aeruginosa is considered as a serious threat level by the Centers for Disease Control and Prevention (CDC), 2019 [4]. In addition to the infections, colonization by this pathogen is raising concerns and challenges in controlling the spread of P. aeruginosa, both within the same ward or different wards, as well as intra- or inter-hospitals.
The results from this study revealed that 12 colonizing isolates of P. aeruginosa belonged to 11 defined STs with ST-162 being the most prevalent; however, the novel ST-3910 was assigned for the PA02 isolate. Among the defined STs, ST-270 was similarly found in a previous study from the central part of Thailand [9]. The antimicrobial susceptibility results demonstrated that the colonizing isolates were still susceptible to many selected antibiotics. Certain isolates were resistant to colistin and carbapenem (imipenem, meropenem, and/or doripenem). In WGS analysis, the detection of ARGs showed that the aminoglycoside resistance gene (aph(3′)-IIb) was present in all colonizing isolates of P. aeruginosa. In contrast, the isolates were phenotypically susceptible to all selected aminoglycosides (amikacin, gentamicin, and tobramycin). It was therefore hypothesized that the aph(3′)-IIb gene carried by these isolates might be truncated and cannot be expressed, leading to aminoglycoside susceptibility. Furthermore, three ARG patterns were detected in the isolates and the novel ST-3910 (PA02) harbored the lowest number of ARGs. All the ARGs detected in this study are commonly found in P. aeruginosa clinical isolates, especially the blaOXA-50 and blaPAO genes [11,12,13].
In the prediction of factors causing genetic movements, such as MGEs, insertion sequences ISPa4 and ISPa5 were identified in all studied isolates. ISPa4 and ISPa5 are unclassified IS elements that were first detected with lengths of 2564 and 965 bps, respectively, in the mucoid isolates of P. aeruginosa PAO-muc (accession number U16785) collected from cystic fibrosis patients [14]. A previous study showed that these IS elements were present together and might have originated from plasmids and/or insertion sequences [14]. Moreover, 500 bp of both IS elements contained 94% similarity with ISPa6; additionally, these three IS elements were located upstream of the exotoxin A-encoding gene (toxA) [14,15,16]. Nevertheless, we found that although all the studied isolates possessed ISPa4 and ISPa5, the toxA gene was harbored by almost all isolates, except for the novel ST-3910 (PA02) isolate. Thus, these findings are not consistent with the phenomenon from earlier studies [14,15,16].
Besides the toxA gene, many other VAGs were also observed in at least four studied isolates. This was with the exception of the wzy and wzz genes-encoding lipopolysaccharide, which generally plays a role in adherence, as this was only found in the ST-270 (PA08) isolate. The Wzy–Wzz interaction has been previously reported, and these two proteins were associated with B-band LPS synthesis in P. aeruginosa [17,18]. Our isolates contained nine major virulence factors, including adherence, antiphagocytosis, biosurfactant, iron uptake, pigment, protease, regulation, secretion system, and toxin. These findings indicate a high pathogenicity of the colonizing isolates of P. aeruginosa, which may contribute to acute and chronic infections [19,20]. In the prediction of bacterial competition, the studied isolates contained at least one bacteriocin-encoding gene being found. The bottromycin detected in all isolates has an activity to inhibit aminoacyl tRNA in the connection with the A site on the 50S ribosome in DNA replication. Prior studies reported that bottromycin has antibacterial activity combatting vancomycin-resistant Enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) [21,22].
In addition, CRISPR-Cas regions, the bacterial adaptive immune system, were identified in the studied isolates. These findings revealed type IF and/or IE CRISPR-Cas systems in seven isolates. Among them, the novel ST-3910 (PA12) isolate harbored the highest number of CRISPR loci, with many distinct spacers and two Cas types. This could confirm their evolution against invaded foreign genetic elements from bacteriophage infections and/or external plasmids [23]. Moreover, the evidence of bacteriophage infections was also assessed. The findings demonstrated various bacteriophage sequences encoding composition proteins as well as significant enzymes, and most of them were identified from Pseudomonas phages, especially P. aeruginosa filamentous 1 (Pf1) phage (PHAGE_Pseudo_Pf1). The Pf1 phage has been reported as one of the key factors that promote the pathogenicity of P. aeruginosa, including biofilm formation and antiphagocytosis [24]. It was therefore speculated that our colonizing isolates have been infected with lysogenic bacteriophages and these viruses might be the important factors driving genetic materials, the so-called horizontal gene transfers (HGTs), from infecting to colonizing isolates, resulting in the increased virulence in the colonizing isolates.
To study the genomic diversity, a comparative genomic analysis was performed. All pairwise ANI values exhibited intra-species of P. aeruginosa. Meanwhile, pairwise SNP distances showed different numbers in almost all pairs of the alignments, and high SNPs were found in the pairs of the novel ST-3910 (PA02) with other isolates. Concerning the PA13 and PA14 isolates, although they belonged to the same ST (ST-162) and their genomic features were similar with an ANI value of 100%, a total of 21,394 SNPs were identified and these were clustered in the distinct subclades within the clade II (Figure 6 and Figure 7). Thus, it was speculated that many recombination events might have occurred in other parts within the core genomes of these isolates, not in the seven housekeeping genes used in MLST, leading to a high SNP distance [25]. On the other hand, although the PA01 and PA14 isolates belonged to different STs (ST-647 and ST-162, respectively) and their genomic features were different with the ANI value of 99.25%, none of the core genome SNP distances were observed and they were grouped in the same subclade within clade II (Figure 6 and Figure 7). It was hypothesized that this may be the opposite of a previously mentioned case (PA13 and PA14, the ST-162 isolates). The core genome of the PA01 isolate was probably identical to that of the PA14 isolate, whereas some SNPs should be found in the MLST alleles that resulted in defining distinct STs. Ambiguous nucleotides and dashes might be found in the short-read WGS data of these isolates, which is not included in the running of the SNP-dists program, leading to an SNP distance of zero.
In comparison with the published genomes of the clinical isolates from several countries, these findings revealed a high level of accessory genes, indicating an open pan-genome and extensive genomic diversity of P. aeruginosa. Nevertheless, genes encoding unique and specific proteins in our genomes of colonizing isolates were not observed, compared with previously published genomes of infecting isolates. Additionally, the SNP-based phylogenetic tree demonstrated that our genomes and other published genomes were similarly clustered within the phylogenetic clade II. This phenomenon indicated a close genomic relatedness within the colonizing and infecting isolates.
In comparison with previous studies, some ARG, VAGs, IS elements, and IBGs patterns of the genomes of P. aeruginosa, carbapenem-resistant Acinetobacter baumanii (CRAB), and carbapenem-resistant Enterobacterales (CRE) were different [26,27,28,29,30]. For example, in the detection of β-lactam resistance genes, the blaOXA-23, blaNDM, and blaPAO genes were only found in CRAB, CRE, and P. aeruginosa, respectively. These findings confirmed that the acquisition of specific genes is dependent on bacterial species. Furthermore, the studies of IBGs in different bacterial species could confirm the narrow/specific host range of bacteriophages.
Finally, even though the genomic insights into the draft genomes of P. aeruginosa colonizing isolates were analyzed, further experiments may be required in the future. For example, one limitation of this study was that the complete plasmids from short-read WGS data could not be identified. Therefore, long-read WGS data would be beneficial to identify both classified and unclassified plasmids as well as integrons. Identifying ARGs and VAGs on the plasmids and/or integrons can confirm HGTs, which can track the spread of these particular genes among P. aeruginosa and other Gram-negative bacteria. Importantly, complete genomes by long-read sequencing would be used as the reference genomes of P. aeruginosa colonizing isolates.
4. Materials and Methods
4.1. Colonizing Isolates of Pseudomonas aeruginosa
In 2017, a total of 13 P. aeruginosa isolates were collected from patients, admitted to the Medicine Ward of Songklanagarind Hospital, Songkhla, Thailand, who were suffering from various underlying diseases, except for P. aeruginosa infection. The species of P. aeruginosa was identified by biochemical testing and confirmed by 16S rRNA amplicon sequencing. Among 13 isolates, 6 and 7 isolates were obtained from the rectum and the throats of patients, respectively.
4.2. Antimicrobial Susceptibility Testing
The susceptibility of P. aeruginosa to 11 antimicrobial agents (piperacillin-tazobactam, ceftolozane-tazobactam, doripenem, imipenem, meropenem, colistin, gentamicin, tobramycin, amikacin, ciprofloxacin, and levofloxacin) was evaluated by the broth microdilution method, following Clinical and Laboratory Standard Institute (CLSI) 2018 guidelines [31]. Briefly, the 2-fold serial concentration of antimicrobial agent was prepared in a 96-well culture plate. Afterward, the P. aeruginosa culture was adjusted to a 0.5 MacFarland standard and further diluted to 1:100. The desired concentration of P. aeruginosa was added to a 96-well culture plate containing serial dilution of antimicrobial agents and they were incubated at 37 °C for 18 h. The results were measured using resazurin dye and interpreted according to the CLSI breakpoint. P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as quality controls.
4.3. DNA Extraction and Whole-Genome Sequencing
Genomic DNA (gDNA) of 13 P. aeruginosa isolates was extracted using GF-1 Bacterial DNA Extraction Kit, following the manufacturer’s instructions. The concentration and purity of extracted gDNA were checked using Thermo Scientific NanoDrop 2000/2000c Spectrophotometers and agarose gel electrophoresis. The qualified DNA sample was sent to perform short-read whole-genome sequencing (WGS) with the MGISEQ-2000 (MGI, Shenzhen, China).
4.4. Genome Assembly and Annotation
The quality of sequence reads was initially assessed by MGI Tech Co. Ltd. The qualified sequence reads were de novo assembled using Unicycler v0.4.7 [32]. The quality and completeness of assembled genomes were investigated using QUAST v4.0 [33] and BUSCO v5.2.2 [34], respectively. The contaminant sequences were evaluated and removed using Kraken2 v2.0.7 [35] and Geneious R10.26 [36]. Then, the assembled genome was annotated using Prokka v1.12 [37].
4.5. Sequence Analysis
The sequence types (STs) and acquired antimicrobial resistance genes (ARGs) were identified using staramr v0.7.2 (https://github.com/phac-nml/staramr, accessed on 1 October 2022) [38,39,40]. Serotypes of P. aeruginosa were predicted using Pseudomonas aeruginosa serotyper (PAst) 1.0 in the center for genomic epidemiology (CGE) [39,41]. Virulence-associated genes (VAGs) were investigated using BLASTN v2.12.0 with 80% identity and 1e-30 E-value cut-offs against the major VAGs in Pseudomonas spp. in virulence factor database (VFDB) of (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi, accessed on 1 October 2022). To look for mobile genetic elements (MGEs), insertion sequence (IS) elements were searched using BLASTN v2.12.0 with 80% identity and 1e-30 E-value cut-offs against ISfinder database (https://isfinder.biotoul.fr/, accessed on 1 October 2022) [42], while integrons were predicted using integron_finder v2.0 (https://github.com/gem-pasteur/Integron_Finder, accessed on 1 October 2022) [43]. Furthermore, CRISPR-Cas regions were investigated using CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index, accessed on 1 October 2022) [44], while bacteriophage sequences that integrated into our P. aeruginosa genomes were detected using the phage search tool enhanced release (PHASTER) (http://phaster.ca, accessed on 1 October 2022) [45,46]. Gene-encoding bacteriocins were explored using the bacteriocin genome mining tool (BAGEL4) (http://bagel4.molgenrug.nl/databases.php, accessed on 1 October 2022) [47].
4.6. Genomic Diversity and Phylogenetic Analyses
Pairwise average nucleotide identity (ANI) and pairwise single nucleotide polymorphism (SNP) distances were evaluated using FastANI v1.32 [48] and SNP-dists v0.8.2 (https://github.com/tseemann/snp-dists, accessed on 1 October 2022), respectively. Additionally, the pan-genome profiles of the studied 13 genomes were compared with 357 previously published genomes of P. aeruginosa from the National Center for Biotechnology Information (NCBI) database, using Roary v3.13.0 [49]. The metadata of the published genomes from several countries, including China, the USA, Germany, Japan, France, and Mexico [7,8,50,51,52,53], as well as other areas of Thailand [9,10] are exhibited in Table S8. A phylogenetic tree against a pan-genome matrix was generated using roary_plots (https://github.com/sanger-pathogens/Roary/tree/master/contrib/roary_plots, accessed on 1 October 2022). SNPs were called from core gene alignment using SNP-sites v2.4.1 [54]. Then, a core genome SNP-based phylogenetic tree was constructed using a geneious tree builder in Geneious R10.26 [36], with a selection of the neighbor-joining method and 500 bootstrap replicates. The circular tree was created using an online tool, the interactive tree of life (iTOL) (https://itol.embl.de/, accessed on 1 October 2022) [55].
5. Conclusions
This study exhibited antimicrobial resistance profiles and WGS data of the colonizing isolates of P. aeruginosa isolated from non-infected patients in a teaching hospital in Southern Thailand. Our findings revealed that all isolates were susceptible to many antimicrobial agents, except for colistin and carbapenems. Interestingly, a novel ST-3910 from the present study and 10 novel STs from other prior studies were assigned. The key genetic patterns, which might be used to predict their genetic evolution as well as their adaptation mechanism, were reported. Importantly, these data could probably be necessary as one of the important data points for tracking and managing the colonization and infection caused by this organism in the future.
Acknowledgments
In this study, the English language was edited by Andrew Jonathan Tait, International Affair Office, Prince of Songkla University, Thailand.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12010165/s1, Figure S1: BUSCO assessment results; Figure S2: Pan-genome matrix against phylogenetic tree based on accessory genes; Table S1: QUAST results of the colonizing isolates of P. aeruginosa; Table S2: Acquired antimicrobial resistance genes (ARGs) in the colonizing isolates of P. aeruginosa; Table S3: Insertion sequence (IS) elements in the colonizing isolates of P. aeruginosa; Table S4: Virulence-associated genes (VAGs) in the colonizing isolates of P. aeruginosa; Table S5: Bacteriocins in the colonizing isolates of P. aeruginosa; Table S6: CRISPR-Cas system in the colonizing isolates of P. aeruginosa; Table S7: Integrated bacteriophage genomes (IBGs) in the colonizing isolates of P. aeruginosa; Table S8: Metadata of previously published genomes from the NCBI database; Table S9: Pan-genome profiles of our genomes (n = 13) and previously published genomes (n = 357).
Author Contributions
Conceptualization, A.C. and K.S. (Komwit Surachat); methodology, A.C., K.S. (Komwit Surachat), K.J., R.P., S.C. (Sarunyou Chusri), K.S. (Kamonnut Singkhamanan), S.C. (Sanicha Chumtong), C.S., K.S. (Kuwanhusna Saroeng) and M.W.; software A.C., K.S. (Komwit Surachat) and K.J.; validation, A.C., K.S. (Komwit Surachat), K.J. and S.C. (Sanicha Chumtong); formal analysis, A.C., K.S. (Komwit Surachat), R.P., S.C. (Sarunyou Chusri) and K.S. (Kamonnut Singkhamanan); investigation, A.C., K.S. (Komwit Surachat), K.J., S.C. (Sanicha Chumtong), C.S. and K.S. (Kuwanhusna Saroeng) resources, A.C., K.S. (Komwit Surachat), S.C. (Sarunyou Chusri), R.P., K.S. (Kamonnut Singkhamanan) and K.J.; data curation, A.C. and K.S. (Komwit Surachat); writing—original draft preparation, A.C. and K.S. (Komwit Surachat); writing—review and editing, A.C., K.S. (Komwit Surachat), R.P., S.C. (Sarunyou Chusri), K.S. (Kamonnut Singkhamanan), P.S. and M.W.; visualization, A.C., K.S. (Komwit Surachat), K.J. and S.C. (Sanicha Chumtong); supervision, A.C., K.S. (Komwit Surachat), R.P., S.C. (Sarunyou Chusri) and K.S. (Kamonnut Singkhamanan); project administration, K.S. (Komwit Surachat), S.C. (Sarunyou Chusri), R.P. and A.C.; funding acquisition, K.S. (Komwit Surachat) and S.C. (Sarunyou Chusri). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Human Research Ethics Committee (HREC) of Prince of Songkla University (protocol code: 64-284-14-1, date of approval: 9 June 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The assembled genomes of all P. aeruginosa isolates in this study have been deposited in the NCBI GenBank under BioProject number PRJNA871949, with BioSample numbers SAMN30433208 to SAMN30433220.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Science, Research and Innovation Fund (NSRF) and Prince of Songkla University, Thailand, grant number MED6505096c. In addition, this research was also supported by the Postdoctoral Fellowship from Prince of Songkla University, Thailand.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Yamada K., Aoki K., Nagasawa T., Sasaki M., Murakami H., Ishii T., Ujiie S., Morita T., Ishii Y., Tateda K. Complete whole-genome sequence of the novel Pseudomonas species strain TUM18999, isolated from a patient with a burn wound in Japan. J. Glob. Antimicrob. Resist. 2021;24:395–397. doi: 10.1016/j.jgar.2021.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Cabot G., López-Causapé C., Ocampo-Sosa A.A., Sommer L.M., Domínguez M.Á., Zamorano L., Juan C., Tubau F., Rodríguez C., Moyà B. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob. Agents Chemother. 2016;60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cain A.K., Nolan L.M., Sullivan G.J., Whitchurch C.B., Filloux A., Parkhill J. Complete genome sequence of Pseudomonas aeruginosa reference strain PAK. Microbiol. Resour. Announc. 2019;8:e00865-19. doi: 10.1128/MRA.00865-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC . Antibiotic Resistance Threats in the United States, 2019 (2019 AR Threats Report) Centers for Disease Control and Prevention (CDC); Atlanta, GA, USA: 2019. [(accessed on 4 January 2020)]. Available online: https://www.cdc.gov/drugresistance/Biggest-Threats.html. [Google Scholar]
- 5.Lob S.H., Kazmierczak K.M., Chen W.-T., Siddiqui F., DeRyke C.A., Young K., Motyl M.R., Sahm D.F. In vitro activity of ceftolozane/tazobactam against Gram-negative isolates collected from ICU patients with lower respiratory tract infections in seven Asian countries—SMART 2017–2019. J. Glob. Antimicrob. Resist. 2022;29:527–533. doi: 10.1016/j.jgar.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Quick J., Cumley N., Wearn C.M., Niebel M., Constantinidou C., Thomas C.M., Pallen M.J., Moiemen N.S., Bamford A., Oppenheim B. Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: An observational study using whole-genome sequencing. BMJ Open. 2014;4:e006278. doi: 10.1136/bmjopen-2014-006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottalorda A., Leoz M., Dahyot S., Gravey F., Grand M., Froidure T., Aujoulat F., Le Hello S., Jumas-Bilak E., Pestel-Caron M. Within-host microevolution of Pseudomonas aeruginosa urinary isolates: A seven-patient longitudinal genomic and phenotypic study. Front. Microbiol. 2021;11:611246. doi: 10.3389/fmicb.2020.611246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y., Chen J., Shen H., Chen Z., Yang Q.-w., Zhu J., Li X., Yang Q., Zhao F., Ji J. Emergence of ceftazidime-and avibactam-resistant Klebsiella pneumoniae carbapenemase-producing Pseudomonas aeruginosa in China. Msystems. 2021;6:e00787-21. doi: 10.1128/mSystems.00787-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cazares A., Moore M.P., Hall J.P., Wright L.L., Grimes M., Emond-Rhéault J.-G., Pongchaikul P., Santanirand P., Levesque R.C., Fothergill J.L. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 2020;11:1370. doi: 10.1038/s41467-020-15081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soonthornsit J., Pimwaraluck K., Kongmuang N., Pratya P., Phumthanakorn N. Molecular epidemiology of antimicrobial-resistant Pseudomonas aeruginosa in a veterinary teaching hospital environment. Vet. Res. Commun. 2022;1:1–14. doi: 10.1007/s11259-022-09929-0. [DOI] [PubMed] [Google Scholar]
- 11.Irum S., Naz K., Ullah N., Mustafa Z., Ali A., Arslan M., Khalid K., Andleeb S. Antimicrobial resistance and genomic characterization of six new sequence types in multidrug-resistant Pseudomonas aeruginosa clinical isolates from Pakistan. Antibiotics. 2021;10:1386. doi: 10.3390/antibiotics10111386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed O.B. Detection of antibiotic resistance genes in Pseudomonas aeruginosa by whole genome sequencing. Infect. Drug Resist. 2022;3:6703–6709. doi: 10.2147/IDR.S389959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsson A., Wistrand-Yuen P., Nielsen E.I., Friberg L.E., Sandegren L., Lagerbäck P., Tängdén T. Efficacy of antibiotic combinations against multidrug-resistant Pseudomonas aeruginosa in automated time-lapse microscopy and static time-kill experiments. Antimicrob. Agents Chemother. 2020;64:e02111-19. doi: 10.1128/AAC.02111-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol P.A., Luan M.-Z., Storey D.G., Thirukkumaran P. Genetic rearrangement associated with in vivo mucoid conversion of Pseudomonas aeruginosa PAO is due to insertion elements. J. Bacteriol. 1994;176:553–562. doi: 10.1128/jb.176.3.553-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovlev E. The mechanism of formation of Pseudomonas aeruginosa biofilm, a type of structured population. Microbiology. 2002;71:249–254. doi: 10.1023/A:1015866123848. [DOI] [PubMed] [Google Scholar]
- 16.Rumbaugh K.P., Hamood A.N., Griswold J.A. Analysis of Pseudomonas aeruginosa Clinical isolates for possible variations within the virulence genes exotoxin a and exoenzymes. J. Surg. Res. 1999;82:95–105. doi: 10.1006/jsre.1998.5523. [DOI] [PubMed] [Google Scholar]
- 17.Rocchetta H.L., Lam J.S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huszczynski S.M., Coumoundouros C., Pham P., Lam J.S., Khursigara C.M. Unique regions of the polysaccharide copolymerase Wzz2 from Pseudomonas aeruginosa are essential for O-specific antigen chain length control. J. Bacteriol. 2019;201:e00165-19. doi: 10.1128/JB.00165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellatly S.L., Hancock R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 20.Oliver A., Mulet X., López-Causapé C., Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 2015;21:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y., Ichioka M., Hirose T., Nagai K., Matsumoto A., Matsui H., Hanaki H., Masuma R., Takahashi Y., Ōmura S. Bottromycin derivatives: Efficient chemical modifications of the ester moiety and evaluation of anti-MRSA and anti-VRE activities. Bioorganic Med. Chem. Lett. 2010;20:6116–6120. doi: 10.1016/j.bmcl.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 22.Brown D.G., Lister T., May-Dracka T.L. New natural products as new leads for antibacterial drug discovery. Bioorgan. Med. Chem. Lett. 2014;24:413–418. doi: 10.1016/j.bmcl.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 23.Hille F., Charpentier E. CRISPR-Cas: Biology, mechanisms and relevance. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:496. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiedoruk K., Zakrzewska M., Daniluk T., Piktel E., Chmielewska S., Bucki R. Two lineages of Pseudomonas aeruginosa filamentous phages: Structural uniformity over integration preferences. Genome Biol. Evol. 2020;12:1765–1781. doi: 10.1093/gbe/evaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Korne-Elenbaas J., Bruisten S.M., de Vries H.J., van Dam A.P. Within-host genetic variation in Neisseria gonorrhoeae over the course of infection. Microbiol. Spectr. 2022;9:e00313-22. doi: 10.1128/spectrum.00313-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chukamnerd A., Pomwised R., Phoo M.T.P., Terbtothakun P., Hortiwakul T., Charoenmak B., Chusri S. In vitro synergistic activity of fosfomycin in combination with other antimicrobial agents against carbapenem-resistant Klebsiella pneumoniae isolated from patients in a hospital in Thailand. J. Infect. Chemother. 2021;27:507–514. doi: 10.1016/j.jiac.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Chukamnerd A., Singkhamanan K., Chongsuvivatwong V., Palittapongarnpim P., Doi Y., Pomwised R., Sakunrang C., Jeenkeawpiam K., Yingkajorn M., Chusri S. Whole-genome analysis of carbapenem-resistant Acinetobacter baumannii from clinical isolates in Southern Thailand. Comput. Struct. Biotechnol. J. 2022;20:545–558. doi: 10.1016/j.csbj.2021.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaewnirat K., Chuaychob S., Chukamnerd A., Pomwised R., Surachat K., Phoo M.T.P., Phaothong C., Sakunrang C., Jeenkeawpiam K., Hortiwakul T. In vitro synergistic activities of fosfomycin in combination with other antimicrobial agents against carbapenem-resistant Escherichia coli harboring blaNDM-1 on the IncN2 plasmid and a study of the genomic characteristics of these pathogens. Infect. Drug Resist. 2022;15:1777. doi: 10.2147/IDR.S357965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phoo M.T.P., Ngasaman R., Indoung S., Naknaen A., Chukamnerd A., Pomwised R. Occurrence of NDM-5 and antibiotic resistance genes among Escherichia coli and Klebsiella pneumoniae in companion animals in Thailand. Southeast Asian J. Trop. Med. Public Health. 2020;51:391–405. [Google Scholar]
- 30.Chukamnerd A., Pomwised R., Jeenkeawpiam K., Sakunrang C., Chusri S., Surachat K. Genomic insights into blaNDM-carrying carbapenem-resistant Klebsiella pneumoniae clinical isolates from a university hospital in Thailand. Microbiol. Res. 2022;263:127136. doi: 10.1016/j.micres.2022.127136. [DOI] [PubMed] [Google Scholar]
- 31.CLSI . Performance Standards for Antimicrobial Susceptibility Testing, 28th ed. Volume 38 Clinical Laboratory Standard Institute (CLSI); Wayne, PA, USA: 2018. Approved Standard M100. [Google Scholar]
- 32.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comp. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seppey M., Manni M., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 35.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thrane S.W., Taylor V.L., Lund O., Lam J.S., Jelsbak L. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 2016;54:1782–1788. doi: 10.1128/JCM.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siguier P., Pérochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cury J., Jové T., Touchon M., Néron B., Rocha E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016;44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Néron B., Rocha E.P., Vergnaud G., Gautheret D., Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Jong A., van Hijum S.A., Bijlsma J.J., Kok J., Kuipers O.P. BAGEL: A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006;34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain C., Rodriguez L.M., Phillippy A.M., Konstantinidis K.T., Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nordstrom H.R., Evans D.R., Finney A.G., Westbrook K.J., Zamora P.F., Hofstaedter C.E., Yassin M.H., Pradhan A., Iovleva A., Ernst R.K. Genomic characterization of lytic bacteriophages targeting genetically diverse Pseudomonas aeruginosa clinical isolates. iScience. 2022;1:104372. doi: 10.1016/j.isci.2022.104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Botelho J., Tüffers L., Fuss J., Buchholz F., Utpatel C., Klockgether J., Niemann S., Tümmler B., Schulenburg H. Phylogroup-specific variation shapes pangenome dynamics in Pseudomonas aeruginosa. bioRxiv. 2022 doi: 10.1101/2022.04.24.489302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinosa-Camacho L.F., Delgado G., Miranda-Novales G., Soberón-Chávez G., Alcaraz L.D., Morales-Espinosa R. Complete genome sequences of two Pseudomonas aeruginosa strains isolated from children with bacteremia. Genome Announc. 2017;5:e00927-17. doi: 10.1128/genomeA.00927-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Espinosa-Camacho L.F., Delgado G., Soberón-Chávez G., Alcaraz L.D., Castañon J., Morales-Espinosa R. Complete genome sequences of four extensively drug-resistant Pseudomonas aeruginosa strains, isolated from adults with ventilator-associated pneumonia at a tertiary referral hospital in Mexico city. Genome Announc. 2017;5:e00925-17. doi: 10.1128/genomeA.00925-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Page A.J., Taylor B., Delaney A.J., Soares J., Seemann T., Keane J.A., Harris S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2:2314. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled genomes of all P. aeruginosa isolates in this study have been deposited in the NCBI GenBank under BioProject number PRJNA871949, with BioSample numbers SAMN30433208 to SAMN30433220.