Abstract
Nonalcoholic fatty liver disease (NAFLD) is a common liver disease with a wide spectrum of liver conditions ranging from hepatic steatosis to nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and hepatocellular carcinoma. The prevalence of NAFLD varies across populations, and different ethnicities have specific risks for the disease. NAFLD is a multi-factorial disease where the genetics, metabolic, and environmental factors interplay and modulate the disease’s development and progression. Several genetic polymorphisms have been identified and are associated with the disease risk. This mini-review discussed the NAFLD’s genetic polymorphisms and focusing on the differences in the findings between the populations (diversity), including of those reports that did not show any significant association. The challenges of genetic diversity are also summarized. Understanding the genetic contribution of NAFLD will allow for better diagnosis and management explicitly tailored for the various populations.
Keywords: NAFLD, genetics, diversity, ethnic difference, polymorphism, steatosis
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is a spectrum of liver disease that ranges from simple steatosis to nonalcoholic steatohepatitis (NASH), fibrosis, and ultimately cirrhosis and hepatocellular carcinoma (HCC). Previous studies suggest that NAFLD is strongly associated with other metabolic disorders, including metabolic syndrome, obesity, insulin resistance (IR), and type 2 diabetes (T2D) [1,2]. Currently, NAFLD is the most common liver disease affecting 30% of the global population, with a higher prevalence in Asia, Latin America, and Middle East-North Africa [3,4]. However, specific ethnicities are more protected against NAFLD than others [5,6], whereas some individuals having the same genetic polymorphisms would increase their predisposition to NAFLD [5,7].
Genetic and environmental factors modulate the risk of NAFLD disease development and its progression. Several genetic polymorphisms associated with NAFLD risk have been published before by genome-wide association studies (GWAS) and single or multiple-gene studies [8,9,10]. These genetic polymorphisms could implicate the pathways involved in NAFLD, such as IR, fatty acid metabolism, oxidative stress, and inflammation. Thus, this mini-review discussed the NAFLD’s genetic polymorphisms focusing on the differences between the populations (diversity), including no significant association. The challenges of genetic diversity are also addressed.
2. NAFLD Pathogenesis and Progression
Understanding the pathogenesis of NAFLD disease is vital in determining the genetics’ role in disease development and progression. NAFLD disease development is widely agreed to be a “multiple hits” theory, assuming the complex interplay of metabolic, genetic and epigenetics, and environmental factors [11,12,13].
One of the critical processes in NAFLD disease development is the dysregulation of the lipid metabolism that drives hepatic lipid accumulation or steatosis. The majority of the hepatic lipids or triglycerides (TG) content comes from the adipocytes-derived circulating free fatty acids (FFAs), which are mediated by the lipase enzymes [14,15]. One such enzyme is the adipose triglyceride lipase (ATGL), encoded by the Patatin-like phospholipase domain containing 2 (PNPLA2) gene. ATGL enzyme initiates the TG breakdown by hydrolyzing the ester bond into producing the diacylglycerol. This step leads to the downstream cascade of lipid breakdown by recruiting other lipases to produce the end products of glycerol and FFAs [16]. In the presence of IR, uncontrolled lipolysis could cause a significant rise in circulating FFAs [17]. Therefore, genetic modifications in these lipid metabolism regulators could contribute to NAFLD development.
Among the patients with steatosis, about 25% of them will have inflammation, hepatocyte ballooning, and cell death (NASH) [18]. Although various hypotheses are reported to explain this NASH progression, the actual molecular mechanism is partly understood. Excess lipids trigger oxidative stress, which, in turn, dramatically reduces the ability of FFAs removal by the mitochondrial β-oxidation and induces endoplasmic reticulum stress [19]. Reactive oxygen species (ROS) suppresses the expression of peroxisome proliferator-activated receptor-α (PPARA), an essential transcriptional factor of FFA oxidation [20,21], thus contributing to further lipid dysregulation. In another aspect, ROS also interact with unsaturated FFAs to enable lipid peroxidation and produce the Malondialdehyde, a marker of oxidative damage [22]. Therefore, ROS-mediated mitochondrial dysfunction and lipid dysregulation could compromise FFAs removal and oxidative phosphorylation, thus initiating a continuous cycle of mitochondrial dysfunction.
In addition to mitochondrial dysfunction, aberrant cytokine and inflammatory factors are evident in NASH and fibrosis progression. Hepatic resident-macrophage (Kupffer cells) play a vital role in liver inflammation. Activated Kupffer cells produce chemokines and cytokines such as Interleukin 6 (IL-6), Tumor necrosis factor-α (TNF-α), and C-C-motif chemokine ligand-2 (CCL2) that further magnify the effects of IR and inflammation thus triggering the hepatocyte injury and fibrosis [23]. The activation of M1 Kupffer cells (proinflammatory type) contributes to fibrosis pathogenesis, whereas the activation of M2 Kupffer cells (anti-inflammatory type) protects such progression [24,25]. Furthermore, the animal model of high-fat diet mice showed that the ratio of M1 to M2 Kupffer cells leads to different outcomes, as mice with a higher M2:M1 ratio are less likely to develop liver lesions, and vice versa for the mice with a high M1:M2 ratio [26]. Damaged hepatocytes and activated Kupffer cells release various proinflammatory cytokines and fibrotic inducers such as TNF-α, platelet-derived growth factor, CCL3, CCL5, and transforming growth factor-β, which, in turn, activates HSCs proliferation [23,27]. This HSCs activation increases expressions of α-smooth muscle actin, forming the stress fibers and depositing extracellular matrix (ECM) components [28]. Previous studies showed that genetic alterations or inhibition of the chemokines and their receptors had improved NASH in mice [29,30,31,32,33], indicating the possible role of genetic modulation in NASH and fibrosis progression.
3. Genetic Polymorphisms of NAFLD
The first reported GWAS study of NAFLD patients was in 2008 in a multi-cohort of Hispanics, African Americans, and European Americans [34]. In this study, the genetic variant of Patatin-like phospholipase domain-containing 3 (PNPLA3), rs738409, also known as I148M, was associated with greater lipid contents, even after adjustments for the ethnicity, body mass index (BMI), diabetes status, and alcohol intake [34]. After that, several studies also reported the effects of the I148M variant and other variants associated with NAFLD in different populations (Table 1).
Table 1.
Summary of the commonly reported single nucleotide polymorphism (SNP) associated with NAFLD.
| Gene | SNP | Population Study | SNP Effects | Reference(s) |
|---|---|---|---|---|
| PNPLA3 | rs738409 C > G (I148M) | Caucasians, African, Japanese, Latinos, Native Hawaiians, and European Americans, Amish, Family Heart Study, Framingham Heart Study, Finnish, Chinese, Taiwanese | Associated with NAFLD risk and steatosis | [34,35,36,37,38,39,40,41,42,43,44,45] |
| German, Japanese | Associated with NAFLD risk, steatosis, and fibrosis | [46,47,48] | ||
| Italian | Associated with NAFLD risk, steatosis, fibrosis, and cirrhosis | [49,50,51,52] | ||
| Belgians | Associated with NAFLD and NASH risk | [53] | ||
| Brazilian, Malaysians (Malays, Chinese, and Indians) | Associated with NAFLD and NASH risk, but not steatosis | [54,55] | ||
| Turkish | Associated with NAFLD and NASH risk, and fibrosis | [56,57] | ||
| Chinese | Associated with NAFLD risk but not steatosis | [58] | ||
| Chinese | Associated with NAFLD risk in lean individuals | [59] | ||
| Japanese | Associated with NAFLD risk and fibrosis, but not steatosis | [60,61] | ||
| Korean | Associated with NAFLD risk and fibrosis | [62,63] | ||
| Indian, Sri Lankans, Bangladeshi, Iranian, Singaporean (Chinese) | Associated with NAFLD risk | [64,65,66,67,68,69,70,71] | ||
| Indian | Associated with NAFLD and NASH risk, and steatosis | [72] | ||
| Filipinos | No association with NAFLD risk | [73] | ||
| rs2896019 T > G | Japanese | Associated with NAFLD risk | [74] | |
| rs6006460 G > T (S453I) | African American | Associated with lower hepatic fat content | [34] | |
| TM6SF2 | rs58542926 G > A (E167K) | Caucasian, African, Hispanic, and European Americans, Swedish, European, Finnish, Polish, and Italian | Associated with NAFLD risk and steatosis | [49,52,75,76,77,78,79,80,81,82] |
| German | Associated with NAFLD risk and steatosis but not fibrosis | [47] | ||
| Caucasians, African, and Hispanic Americans, Chinese | Associated with NAFLD risk, steatosis, and fibrosis | [43,83,84,85,86,87] | ||
| Italian, Finnish, and Swedish | Associated with NASH progression | [88] | ||
| Korean | Associated with NASH and fibrosis | [89] | ||
| Indian, Argentinian | Associated with NAFLD risk | [66,77,90] | ||
| Brazilian, Japanese, Singaporean (Chinese) | No association with NAFLD | [54,71,91] | ||
| GCKR | rs780094 C > T | Amish, Family Heart Study, and Framingham Heart Study, Caucasian, African, and Mexican Americans | Associated with NAFLD risk and steatosis | [39,92] |
| German | Associated with NAFLD and fibrosis severity | [46] | ||
| Italian | Associated with liver fibrosis severity | [93] | ||
| Taiwanese, Uygur | Associated with NAFLD risk | [94,95] | ||
| Malaysians (Malays, Chinese, and Indians) | Associated with NAFLD, NASH, and fibrosis | [96] | ||
| Iranian, Chinese, Singaporean (Chinese) | No association with NAFLD | [71,97,98] | ||
| rs1260326 C > T (P446L) | Italian, Japanese, Uygur, and Chinese | Associated with NAFLD risk | [49,74,95,99,100] | |
| Finnish | Associated with steatosis | [101] | ||
| Malaysians (Malays, Chinese, and Indians) | Associated with NAFLD, NASH, and fibrosis | [96] | ||
| Chinese | No association with NAFLD | [98,102] | ||
| MBOAT7 | rs641738 C > T | Dallas Heart Study and Europeans, Italian | Associated with NAFLD risk and steatosis | [49,52,103] |
| Finnish | Associated with NASH and fibrosis | [104] | ||
| German | Associated with fibrosis only | [47] | ||
| Eastern European | No association with fibrosis or cirrhosis | [105] | ||
| Americans, Italian, Argentinian, Chinese, Taiwanese, and Korean | No association with NAFLD risk | [37,89,102,106,107,108] | ||
| HSD17B13 | rs72613567 T > TA | Caucasian Americans | Associated with steatosis and lower risk of inflammation and NASH | [109,110] |
| European, Finnish | Associated with lower NASH and fibrosis risk | [111,112] | ||
| Argentinian, Japanese | Associated with lower NASH risk | [113,114] | ||
| Korean | Associated with lower NAFLD risk | [115] |
Abbreviation: GCKR: glucokinase regulatory protein; HSD17B13: hydroxysteroid 17β-dehydrogenase; MBOAT7: membrane-bound O-acyltransferase domain-containing 7; NAFLD: nonalcoholic fatty liver disease; NASH: nonalcoholic steatohepatitis; PNPLA3:patatin-like phospholipase domain-containing 3; TM6SF2: transmembrane 6 superfamily member 2.
3.1. PNPLA3 Loci
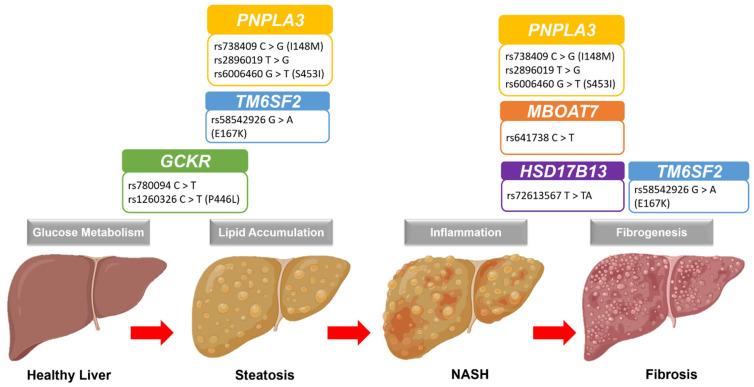
The gene PNPLA3 produces a triacylglycerol lipase (adiponutrin) responsible for triacylglycerol hydrolysis. The presence of the I148M variant reduces this lipase enzymatic activity and promotes hepatic steatosis (Figure 1) [116]. A recent meta-analysis of the I148M variant on NAFLD risk showed that individuals carrying the minor G-allele had a 19% higher risk of developing NAFLD, and the risk increased to 105% for individuals with both GG alleles [117]. Notably, this effect of the I148M variant was independent of IR and lipid levels [118], though diets may interact with the effects. A study of Hispanic children with the I148M variant showed high hepatic lipid contents when consuming carbohydrate-rich diets [119]. High levels of carbohydrates facilitate lipid metabolism via the activation of transcription factor sterol regulatory binding protein-1c that regulates the PNPLA3 expression [120,121]. Therefore, the effect of the I148M variant may be higher for those individuals that consume high carbohydrate-rich diets, suggesting the gene-environment interplay.
Figure 1.
Graphical representation of the NAFLD progression and the most common genetic polymorphisms. Abbreviation: GCKR: glucokinase regulatory protein; HSD17B13: hydroxysteroid 17β-dehydrogenase; MBOAT7: membrane-bound O-acyltransferase domain-containing 7; NAFLD: nonalcoholic fatty liver disease; NASH: nonalcoholic steatohepatitis; PNPLA3: patatin-like phospholipase domain-containing 3; TM6SF2: transmembrane 6 superfamily member 2.
A similar effect of the I148M variant is evident for more advanced stages of NAFLD. In a meta-analysis of 16 studies, the homozygous GG alleles confer a 3.5-fold greater risk of having NASH and a 3.2-fold higher risk of having fibrosis [122]. In a more recent meta-analysis of 13,817 individuals with NAFLD, this I148M variant confers a 2.54-fold greater risk of having NASH [123], with a significant dose-dependent of the G allele [117,123]. A similar association was also observed between the I148M variant and progression to cirrhosis, in which the presence of one G allele confers a 2-fold higher risk of cirrhosis, and homozygous GG alleles confer 3-fold higher risk compared to CC genotypes [51]. Thus, the PNPLA3 I148M variant has been incorporated into various predictive models to diagnose NAFLD disease severity and progression [115,124,125,126].
3.2. TM6SF2 Loci
Another reported SNP in NAFLD is the rs58542926 (E167K) variant from the Transmembrane 6 superfamily, member 2 (TM6SF2) gene. This E167K variant was associated with a higher risk of having NAFLD, hepatic steatosis, and advanced fibrosis but not inflammation (Figure 1) [127,128,129]. Moreover, this effect of the E167K variant is more significant in children than adults [128]. Although the effect of the E167K variant is lower compared to PNPLA3 I148M, the individuals carrying both I148M and E167K variants had a double or additive risk of having NAFLD [86], indicating the gene–gene interplay in the disease. There are also leaner NAFLD individuals carrying the E167K variant than obese or overweight individuals [130], thus highlighting its specific role in developing NAFLD. Notably, most previous studies of “lean” NAFLD are from Asia [131], and this is consistent with the high frequency of the E167K variant in East Asians. Across the populations, the minor T allele frequency is more common in East Asians (~34%) than in Europeans (~26%), Hispanics (~10%), and Africans (~6%) [132]. However, there is a lack of genetic diversity to conclude this relationship due to the underrepresentation of other populations.
Although it is debatable, there is evidence that NAFLD individuals with the E167K variant have a low risk of having coronary artery disease (CAD) [88,133,134]. TM6SF2 is an ER transmembrane protein predominantly found in the liver, kidney, and small intestine cells and responsible for regulating the secretion of lipoproteins [135]. The E167K variant causes a loss of this protein function and consequently reduces the secretion of very low-density lipoprotein (VLDL) [136]. This protective effect of the E167K variant was further replicated in a large meta-analysis GWAS study of 60,801 CAD individuals [137] and an exome study of more than 300,000 individuals [138]. This dual conflicting effect of the E167K variant on NAFLD and CAD risks adds more to the complex pathophysiology of NAFLD and its related cardiometabolic risk.
3.3. GCKR Loci
The GCKR gene encodes a glucokinase regulator by forming a complex with glucokinase and diverts its location to the nucleus, affecting hepatic glucose storage and metabolism (Figure 1) [139]. Inhibition of glucokinase activity is one of the mechanisms for controlling the rate of hepatic glucose metabolism and lipogenesis [140]. Thus, any variant that affects the functionality of GCKR protein may contribute to the NAFLD risk. One of the most reported GCKR variants is the rs780094, and the minor T-allele increases the risk of having NAFLD and hepatic steatosis [141,142]. Interestingly, other studies showed that the rs780094 variant was protective against T2D risk [143]. Consistent with the role of GCKR in glucose metabolism, this variant’s presence could increase the glucokinase rate due to a lack of inhibition from the GCRK protein. This lack of GCRK inhibition is further evident in the studies investigating the effect of another GCKR rs1260326 (P446L) variant. The loss of GCKR function due to the P446L variant was associated with lower fasting glucose and insulin levels but higher hepatic lipid content [50,88,144]. Uncontrolled glucokinase activity causes a high malonyl-CoA (product of glucose metabolism) level blocking the FFA oxidation via the inhibition of carnitine-palmytoyltransferase-1, and malonyl-CoA also promotes lipogenesis by becoming its substrate, thus increasing lipid accumulation [140]. Moreover, both GCKR-P446L and PNPLA3-I148M variants have synergistic effects on the NAFLD risk, particularly on hepatic steatosis. A study of children and adolescents with obesity showed that both variants were independently associated with hepatic lipid contents. The combined analysis of both variants’ effects explained the hepatic lipid content variability in different ethnicities (39% in African Americans, 32% in Caucasians, and 15% in Hispanics) [145], indicating the gene–gene interplay for lipid regulation in the youth.
3.4. MBOAT7 Loci
Another reported SNP in NAFLD is rs641738 from the membrane-bound O-acyltransferase domain-containing 7 (MBOAT7) gene. Individuals carrying the minor T-allele have reduced expression of MBOAT7 and hepatic inflammation and fibrosis (Figure 1) [146,147]. MBOAT7 gene encodes the lysophosphatidylinositol acyltransferase enzyme that adds the arachidonic acid (AA) into the membrane phospholipids, phosphatidylinositol (PI) [148], and a deficiency of MBOAT7 protein leads to hepatic inflammation-mediated fibrosis [149]. Consistent with this, T-allele in rs641738 was associated with liver fibrosis in NAFLD individuals [47,103,104].
3.5. HSD17B13 Loci
Among the SNPs associated with NAFLD, one splicing site SNP rs72613567 in hydroxysteroid 17β-dehydrogenase (HSD17B13) gene confers a protective effect on NAFLD risk, particularly lowering the inflammation, NASH, and fibrosis (Figure 1) [150]. The insertion of TA nucleotides causes an early termination and produces a truncated protein of hepatic lipid-droplet protein HSD17B13, resulting in a loss of this protein function [111]. Although the information about this protein is limited, individuals carrying the rs72613567 variant have a lower risk of NASH and fibrosis, though no effect was observed on lipid levels or hepatic steatosis [109,110,111,112,113,115]. Interestingly, this protective effect of the rs72613567 variant is evident even in individuals carrying the PNPLA3 I148M variant by lowering fibrosis risk [111,114]. However, a recent study reported that the protective effects of the rs72613567 variant could be limited to specific groups of individuals that are either women or individuals aged more than 45 years or having diabetes or obesity and individuals with the PNPLA3 I148M variant [110], though these findings need further clarifications.
3.6. Other Genetic Loci
There are also rare variants associated with NAFLD. Most of these SNPs have been explicitly identified for a single population or have only been reported by a few studies (Table 2).
3.6.1. Loci in Energy Metabolism
One NAFLD variant involved in energy metabolism is the rs4240624 variant near the protein phosphatase-1 regulatory subunit-3B (PPP1R3B) gene. This noncoding rs4240624 variant was associated with hepatic steatosis, higher cholesterol, and lower fasting glucose [39]. Further study also showed that the PPP1R3B protein promotes glycogen storage in the liver via activating glycogen synthase and inhibiting glycogen breakdown [151], suggesting the role of this gene in the development of NAFLD. Another SNP of PPP1R3B, rs61756425, was associated with NAFLD disease severity [49], though more validation is needed to confirm this relationship.
Another reported SNP is the rs8192678 (G482S) from the PPARG coactivator-1 alpha (PPARGC1A) gene. This G482S variant was associated with a higher risk of having NAFLD in Iranians, and Chinese Han populations, though no association was observed in the lipid or glucose profile [152,153]. A recent study in the Chinese Hans population showed that this G482S was also associated with NASH [154], and this finding was replicated in the Japanese population but with another PPARGC1A SNP of rs2290602. Although the effects of PPARGC1A SNPs are only reported in these three populations, the polymorphisms of PPARGC1A have been linked to many other metabolic diseases [155], thus highlighting its possible contribution to NAFLD disease.
Table 2.
Summary of the rarely reported single nucleotide polymorphism (SNP) associated with NAFLD.
| Gene | SNP | Population Study | SNP Effects | Reference(s) |
|---|---|---|---|---|
| PPP1R3B | rs4240624 G > A/C | Amish, Family Heart Study, and Framingham Heart Study | Associated with NAFLD risk | [39] |
| Caucasian, African, and Mexican Americans | Associated with steatosis | [92] | ||
| rs61756425 G > T | Italian | Associated with NAFLD severity | [49] | |
| PPARGC1A | rs8192678 G > A (G482S) | Iranian, Chinese | Associated with NAFLD and NASH risk | [152,153,154] |
| rs2290602 G > T | Japanese | Associated with NAFLD and NASH risk | [156] | |
| SAMM50 | rs3761472 A > G | Chinese, Korean, and Japanese | Associated with NAFLD risk | [48,157,158,159] |
| Indian | Associated with NASH | [72] | ||
| rs738491 C > T | Chinese, Japanese | Associated with NAFLD risk | [48,157] | |
| rs2143571 G > A | Chinese, Korean, Japanese | Associated with NAFLD risk | [48,157,159] | |
| APOC3 | rs2070666 T > A | Chinese | Associated with hepatic lipid content | [160] |
| rs2854116 (T-455C) | European | No association with NAFLD risk | [161] | |
| Indian | Associated with NAFLD and lipid content | [162,163] | ||
| rs2854117 (C-482T) | European | No association with NAFLD risk | [161] | |
| Indian | Associated with NAFLD and lipid content | [162,163] | ||
| ATGR1 | rs2276736 A > G/T rs3772630 T > A/C rs3772627 A > G |
Malaysians (Malays, Chinese, and Indians) | Associated with NAFLD risk in the presence of PNPLA3 I148M variant | [164] |
| Japanese | Associated with NAFLD risk and fibrosis | [165] | ||
| GATAD2A | rs4808199 G > A | Japanese | Associated with NAFLD | [74] |
| IL27 | rs4788084 C > T | Indian | Associated with hepatic lipid content | [72] |
| LPIN1 | rs13412852 C > T | Italian | Tended to associate with lower NASH | [166] |
| LYPLAL1 | rs12137855 C > T | Amish, Family Heart Study, Framingham Heart Study, Finnish | Associated with NAFLD risk and steatosis | [39,101] |
| PEMT | rs7946 C > T (V175M) | Chinese, Caucasian | Associated with NAFLD risk | [167,168,169] |
| Indian | Associated with NAFLD risk in lean individuals | [170] | ||
| Korean | No association with NAFLD risk | [171] | ||
| MTTP | -493C/G | Japanese | Associated with NASH | [172] |
| Italian, Brazilian | No association with NAFLD risk | [173,174,175] | ||
| PARVB | rs5764455 A > C/G | Japanese | Associated with NASH | [48] |
| rs2073080 C > A/T | Indian, Finnish | Associated with steatosis | [66,101] | |
| SOD | rs4880 A > G (C47T) | Italian and European | Associated with hepatic steatosis and fibrosis | [176] |
| Japanese | Associated with NASH | [172] | ||
| REPIN1 | 12 bp deletion | Germany | Associated with lower NAFLD severity | [177] |
| UCP3 | rs1800849 C > T (-55CT) | Spaniards | Associated with NASH | [178] |
Abbreviation: AGTR1: angiotensin II receptor type 1; APOC3: apolipoprotein C3; GATAD2A: GATA zinc finger domain containing 2A; IL27: interleukin 27; LPIN1: lipin-1; LYPLAL1: lysophospholipase-like 1; MTTP: microsomal triglyceride transfer protein; NAFLD: nonalcoholic fatty liver disease; NASH: nonalcoholic steatohepatitis; REPIN1: replication initiator 1; PARVB: parvin beta; PEMT: phosphatidylethanolamine N-methyltransferase; PPARGC1A: PPARG coactivator 1 alpha; PPP1R3B: protein phosphatase-1 regulatory subunit 3B; SAMM50: SAMM50 sorting and assembly machinery component; SOD2: superoxide dismutase 2; UCP3: uncoupling protein 3.
3.6.2. Loci in Mitochondrial Regulation
The SAMM50 sorting and assembly machinery component (SAMM50) gene polymorphisms also contribute to NAFLD risk. Three SNPs, rs3761472, rs738491, and rs2143571, were associated with a greater risk of having NAFLD in East Asians, including Chinese Han, Korean, and Japanese populations [48,157,158,159]. This finding was replicated again in the Indian population with a significant association with NASH, though it is for one SNP only (rs3761472) [72]. The exact mechanism of how polymorphism in the SAMM50 gene could lead to NAFLD is partly understood. Since SAMM50 is responsible for maintaining the mitochondrial structure and assembly of respiratory chain complexes, any modifications on the protein will lead to mitochondrial dysfunction and FFA oxidation [158], known processes in NAFLD disease development. In addition to SAMM50, other oxidative stress-associated SNPs, such as rs4880 (C47T) in superoxide dismutase 2 (SOD2) [172,176] and rs1800849 (-55CT) in uncoupling protein 3 (UCP3) [178] genes, also associated with NAFLD, particularly with NASH. Despite limited findings on the oxidative stress-related SNPs in NAFLD, the role of cellular oxidative stress is well explored in the development of NAFLD and NASH.
3.6.3. Loci in Cholesterol Polymorphism
The cholesterol gene polymorphism also contributes to NAFLD risk. The SNPs in apolipoprotein C3 (APOC3) rs2070666, rs2854116, and rs2854117 were associated with NAFLD risk [160,162,163]. This APOC3 gene encodes a component of VLDL (apolipoprotein C3) that inhibits lipoprotein and hepatic lipase, and loss of APOC3 protein function was associated with high triglycerides [179]. Another cholesterol related-gene is microsomal triglyceride transfer protein (MTTP), and its -493C/G intronic variant was reported to increase NASH risk in the Japanese population [172]. The heterodimeric MTTP protein is responsible for transporting lipid molecules, including triglycerides and cholesterol ester, and apolipoprotein B assembly. Therefore, any modification of the MTTP gene will lead to the absence of VLDL, known as abetalipoproteinemia [180] and, hence, more significant hepatic lipid accumulation. Previous studies also identified other NAFLD variants such as the Lipin-1 (LPIN1) rs13412852 [166], interleukin 27 (IL27) rs4788084 [72], GATA zinc finger domain containing 2A (GATAD2A) rs4808199 [74], lysophospholipase-like 1 (LYPLAL1) rs12137855 [39,101], angiotensin II receptor type 1 (ATGR1) rs2276736, rs3772630, and rs3772627, phosphatidylethanolamine N-methyltransferase (PEMT) rs7946 V175M [167,168,169,170,171], replication initiator 1 (REPIN1) 12-bp deletion [177], and parvin beta (PARVB) rs5764455 [48] and rs2073080 [66,101].
3.6.4. Copy Number of Variants (CNV)
Another genetic change associated with NAFLD is the copy number variants (CNV). Currently, five published papers reported CNV association with NAFLD risk. The first study used the GWAS analysis of CNV in 10 Malaysian individuals with simple steatosis and 39 individuals with NASH [181]. In this study, the most common CNV is the 14q11.2 region, which is present in 53.8% of NASH individuals, and within this region, there are a group of olfactory receptor (OR) family genes. Two novel CNVs were also identified, the 13q12.11 and 12q13.2 regions, consisting of the exportin 4 (XPO4) and phosphodiesterase 1B (PDE1B) genes, respectively, and these CNVs were associated with NASH progression [181]. Following that, a larger study of Malaysian NAFLD individuals confirmed that the duplication of 13q12.11 (XPO4 gene) was associated with NASH risk. The gain of an additional copy of 13q12.11 was associated with higher serum triglycerides and ALT enzyme levels [182]. The involvement of XPO4 CNV in NAFLD was replicated in another study of Caucasians with metabolic-NAFLD (MAFLD), and XPO4 CNV was associated with fibrosis [183]. Therefore, these findings confirm the possible involvement of XPO4 CNV in modulating the risk for NAFLD and fibrosis.
Other studies also reported different CNVs associated with NAFLD risk. A more recent NAFLD study of the Chinese population showed that 338 autosomal CNVs were identified for NAFLD, and the deletion of NLR Family Pyrin Domain Containing 4 (NLRP4) CNV was associated with NAFLD risk [184]. Another study of Chinese NAFLD individuals reported that the deletion of carboxylesterase 1 (CES1) CNV was associated with NAFLD risk (OR: 2.75) [185]. Although the evidence for CNV on NAFLD risk is limited, the replication of XPO4 CNV association in two different populations may indicate that the CNVs could modulate the NAFLD risk. Thus, more future study is needed to determine this relationship.
From these findings, most of the NAFLD genetic polymorphisms are within the genes involved in regulating hepatic lipid metabolism, and the genetic contribution to NAFLD risk is considered significant. Therefore, these genetic variants are potential diagnostic tools to screen susceptible individuals for NAFLD. However, some differences in these genetic variants between the populations and ethnicities need consideration.
3.7. Polygenic Risk Scores
Due to the limitation of a single genetic variant to explain the NAFLD risk, developing a polygenic risk score (PRS) is favorable. In a study to determine the causal relationship between steatosis and the development of liver damage, a PRS model was developed based on the risk alleles of four genes (PNPLA3, TM6SF2, GCKR, and MBOAT7) that were associated with hepatic steatosis [186]. Each variant association with liver damage was proportional to its association with steatosis. Notably, the PRS model (steatosis risk alleles) association with liver damage was stronger than the conventional risk association via histological steatosis assessments [186]. These findings indicate that genetic risk alleles may reflect the actual long-term effects of steatosis in contributing to liver damage, and the histological assessment of steatosis may undermine the effects of steatosis on liver damage. Other studies also used similar PRS model and improved the diagnosis of steatosis in adults [187] and children [188] and HCC risk [189]. A PRS model based on three common variants of PNPLA3, TM6SF2, and MBOAT7 genes was also associated with a higher risk of liver damage [47] and HCC risk [190].
In the Asian populations, a study of Korean NAFLD individuals showed that the PRS model (PNPLA3, TM6SF2, IR, liver enzymes, C-reactive protein, and diabetes status) predicted NASH risk with the area under the curve (AUROC) of 0.835 and 0.809 in NAFLD individuals with and without diabetes, respectively [115]. For Japanese individuals, a PRS model based on the (PNPLA3, GCKR, and GATAD2A) was associated with the NAFLD risk, and the risk was higher with the accumulation of the risk alleles [74]. Two Chinese studies showed that both PNPLA3 and TM6SF2 risk alleles confer a higher risk of having NAFLD in an additive manner [86,191].
Other studies reported the PRS model with the addition of the protective variant HSD17B13 gene and NAFLD risk. One study investigated the potential of a PRS model based on three variants (PNPLA3, TM6SF2, and HSD17B13) to predict cirrhosis. The PRS model was able to predict the cirrhosis risk by 12-fold compared to the general population [192]. A recent multiethnic study of Americans (Caucasian, African, and Hispanic Americans) investigated the potential of multiple candidate genes to predict HCC risk among NAFLD individuals [193]. In this study, two PRS models were developed, namely the PRS-HFC (a model calculated by the sum of the effects of PNPLA3-TM6SF2-MBOAT7-GCKR risk alleles) and the PRS-5 (a PRS-HFC model that also incorporated the protective allele of HSD17B13). Both PRS models predicted the HCC risk better than a single gene risk score in NAFLD individuals (three-fold higher). Moreover, the prediction of HCC risk was independent of severe fibrosis, particularly in younger individuals and individuals with T2D [193]. This finding is essential, as many individuals are often not screened for HCC risk if no evidence of fibrosis exists. Since PRSs models are easily measured based on a single blood test and often adjusted to possible environmental factors, predicting the future development of NAFLD complications in clinical settings is possible.
4. Genetic Diversity in NAFLD Risk
Since NAFLD is a multi-factorial disease, there is a considerable contribution of genetics in modulating the individual or ethnic variations for the disease severity and risk of mortality associated with NAFLD.
4.1. Genetic Diversity of PNPLA3 I148M
Among those above genetic variants, the PNPLA3 I148M is the most replicated variant in various populations (Table 1). Most studies showed similar predisposition effects on NAFLD, in which the individuals carrying the G allele have a greater risk of having steatosis and NAFLD. However, the prevalence of NAFLD and PNPLA3 I148M variant frequency differs among populations.
The frequency of the I148M variant was highest in the Hispanic (0.49) compared to European (0.23) and African (0.17) Americans. Consistently, the observed levels of hepatic lipid contents in those NAFLD individuals also have a similar trend, with Hispanics having the highest hepatic lipid content, whereas African Americans have the lowest [34]. Thus, this variant I148M may partly explain the hepatic steatosis variability between the different ethnicities. Moreover, another PNPLA3 variant, rs6006460 (S453I), was reported to associate with protective effects on NAFLD risk in African Americans. This protective effect was consistent with the higher frequency of this S453I variant in Africans (~10%) compared to other ethnicities (<1%) [34], confirming the genetic contribution to a lower risk of NAFLD among African individuals. Interestingly, the I148M variant is also associated with a “lean” NAFLD phenotype from Asia populations [5,63], suggesting a more specific relationship between the genetic variant and the subtypes of NAFLD geographically. However, a multiethnic Malaysian study showed that the I148M variant was associated with NAFLD risk, although the association was similar among the Malays, Chinese, and Indian ethnicities [55]. This discrepancy may be due to the small sample size for each ethnicity, which could undermine the statistical analysis. Furthermore, all three Malaysian ethnicities still belong to the Asian, and thus would have been different from the comparisons of Hispanics, Americans, Europeans, and Africans in other studies.
Another perspective is that the I148M variant could have sexual dimorphism on NAFLD risk or susceptibility. A meta-analysis of 16 studies reported a negative relationship between the male gender and the I148M variant on hepatic lipid content [122], and the effect of this variant on NAFLD risk was higher in women [39,122]. In contrast, the overall prevalence of NAFLD is higher in men globally [194], with countries such as the USA, Spain, and China having more men develop NAFLD [195]. However, other studies in Sri Lanka and Thailand showed an increase in NAFLD prevalence in women [195], and the overall median age of women who develop NAFLD is greater than that in men [196]. Consistent with this, the overall NAFLD prevalence in men becomes minimal in individuals over 50 years of age [194], implying the menopausal effects on the risk for NAFLD in women. Since estrogen is an essential hormone in lipid regulation and metabolism [197], the female gender could play a role in modulating the effects of I148M on NAFLD risk. However, this gender-specific effect requires further validation as most previous studies of the I148M variant did not perform gender-specific analysis.
4.2. Genetic Diversity of Other NAFLD Variants
The ethnic differences in other NAFLD variants are also evident. One such variant is the rs641738 in the MBOAT7, which was associated with NAFLD risk in Italians and Europeans, but not Asians (Table 1) [198]. In contrast, the association between the APOC3 rs2854116 [T-455C] variant with NAFLD risk and hepatic lipid content was observed in Indian populations [162,163,199] but not in Western populations [40,53]. Similarly, some of the common NAFLD variants associations identified in Western populations did not replicate in some Asian populations. For example, Iranian studies showed that the PPARGC1A G482S [152], not the GCKR rs780094 variant [97], is associated with the NAFLD risk. On the other hand, the TM6SF2 E167K variant did not associate with NAFLD risk in the Japanese [91] and Brazilian [54] populations. Interestingly, the PNPLA3 I148M variant did not associate with the NAFLD risk in Filipino, despite this variant being higher in frequency in cases [73]. However, the sample size is small for an accurate conclusion.
In Malaysia’s multiethnic studies, three SNPs (rs3772627, rs2276736, and rs3772630) in the AGTR1 gene have no association with the NAFLD risk in all patients. Though, a sub-analysis of the ethnicity showed that in the Indian ethnic group, these ATGR1 SNPs have a significant protective effect on NAFLD risk, and no such association was observed in the Malays and Chinese Malaysians [164]. A similar subethnic analysis for the association with NAFLD and hepatic lipid content was done for the Indian population. The PNPLA3 I149M variant was significantly associated with NAFLD risk in North Indians, whereas the TM6SF2 E167K variant was associated with NAFLD in South Indians [90]. These differences may be due to the recent findings of the genomic profile of the North Indians are similar to Europeans and the South Indians to Asians [200]; thus, they may have a different genetic susceptibility.
4.3. Discrepancies between the Same Populations or Ethnicities
There are also discrepancies in the genetic findings of NAFLD within the same population or ethnicities. For example, a Chinese study of 112 adult NAFLD patients and 120-matched controls showed that the PNPLA3 I148M variant increases the susceptibility of having NAFLD though no association was observed in steatosis grade [58]. However, another study of Chinese NAFLD individuals (203 adult patients with 202 matched controls) reported that the I148M variant was associated with the steatosis grade and other parameters such as BMI and plasma liver enzyme level [41]. Both of these studies have similar numbers of individuals; however, the method to confirm the steatosis was different. The latter study used an ultrasonography tool instead of a liver biopsy to confirm the steatosis [41]. Using ultrasound to diagnose steatosis is less accurate for individuals with lower-grade steatosis than higher-grade steatosis [201]. Thus, the diagnosis to exclude the presence of lower-grade steatosis may be compromised; therefore, the usage of ultrasound may contribute to the discrepancies in the findings.
Other discrepancies were also observed in studies of Japanese NAFLD individuals. The first Japanese study of 253 adult NAFLD individuals and 578 controls showed that the I148M variant increases the susceptibility of having NAFLD and fibrosis. However, no association was observed in steatosis grade [60]. Another study of 392 Japanese NAFLD individuals and 934 controls showed an association between the I148M variant with steatosis grade and fibrosis [48]. Both studies were done by the same group of researchers and used the same methods to diagnose and classify the clinical assessments, except that the latter study performed GWAS analysis, whereas the previous study used a candidate SNP approach. Significant evidence showed that the candidate-gene approach has shortcomings, including selection and publication bias and poor replication [202]. Therefore, this may explain the differences seen between these two Japanese studies.
Another NAFLD variant, the GCKR P446L variant, was reported with different findings among the Chinese populations. Two studies showed no association between the P446L variant and the susceptibility of NAFLD [98,102], whereas two studies reported that the P446L variant increases the susceptibility of having NAFLD [99,100]. The differences between these studies are the number of individuals included and the presence of other metabolic syndromes or diseases in the patients. Two studies that reported no association between the P446L variant relationship and NAFLD susceptibility also investigated the effects of metabolic syndrome [102] and coronary artery disease [98]. Thus, it may undermine the statistical analyses performed to determine the relationship between the P446L variant and NAFLD risk.
4.4. Challenges in NAFLD Genetic Diversity Research
Most of the NAFLD genetic polymorphism reported consistent effects between different populations. However, some discrepancies may be attributed to the ethnic-specific effect or the unknown genetic factors that have not been discovered yet. Furthermore, most previous findings are from the Western and East Asian populations and thus may undermine the role of genetics in NAFLD, especially in minorities. The Asia region has the second highest overall prevalence of NAFLD at 25% [203], and focusing on South East Asia, the overall prevalence was 42% [204]. Among the South East Asia countries, Indonesia ranks at the top with a NAFLD prevalence of 51%, followed by Singapore at 40% and Malaysia at 39% [204]. However, despite the high NAFLD prevalence, the genetic information from these South East Asians and the underrepresented Malay ethnicity was limited. Similarly, genetic polymorphism data is scarce for the Middle East region. More publications and reports from these two regions are needed to understand the effects of genetics in conferring the NAFLD risk. Moreover, genetics and environmental (geographic and socioeconomic) factors interact with each other to confer the NAFLD risk and resulting different subethnic effects. For example, the NAFLD prevalence among American Hispanics differs based on the ancestry origin of the individuals. NAFLD prevalence was highest for Hispanic individuals of Mexican origin (33%) compared to Hispanics of Puerto Rican origin (18%) and Dominican origin (16%) [205]. This finding is important as it shows that sub-ethnic diversity exists; thus, the traditional grouping of race or ethnicity may undermine the genetic contribution to NAFLD risk.
The current review is limited due to the lack of data from underrepresented countries and populations. The comparisons for genetic diversity might be biased and limit the generalizability of these genetic variants’ roles in the development of NAFLD disease. Another factor is that the small sample sizes and low statistical powers of the Asian studies [206] may undermine the actual NAFLD risk associations; therefore, their risk may not differ from the Western populations. Notably, these underpowered studies are often included in most meta-analysis studies despite there is a risk of publication bias. Underpowered studies tend to show extreme effects compared to large studies; though, this effect varied across meta-analyses [207]. In some meta-analyses with two or more large studies dominating the analysis, the underpowered studies have a minimal effect. Removing the underpowered studies will affect the precision if all studies have a similar sample size [207], thus performing the meta-analysis with a small sample size studies needs great cautions. Since sample size remains a significant issue in Asian studies, additional genetic information is vital to discover the unknown common and rare variants that could explain the individual risks and enhance understanding of NAFLD biology among Asians. Future studies could employ more in-depth genome sequencing (whole-exome sequencing or whole-genome sequencing) per individual to discover the low-frequency variants and CNVs [208]. This additional information from sequencing combined with GWAS is proven to work, as seen in the discovery of the new T2D variants that were otherwise missing from the GWAS studies alone [209]. Moreover, the extensive or deep phenotyping of the individuals could improve the analysis to detect new associations and heritability between the traits [210]. Participation in a large-scale international collaborative project will also allow for resources and knowledge transfer between the countries or populations and, thus, could improve the cohort sample size and diversity.
5. Conclusions
NAFLD prevalence and risk differ across populations. Genetic polymorphisms significantly influence the risk of having NAFLD and disease progression. However, most of the reported genetic studies are from Western populations and some from East Asians. There is a significant gap in the genetic information from other parts of Asia and minority ethnicities. The missing information is needed to address the ethnic-specific effects that could be used to tailor specific diagnostic tools and management programs. Future research is needed to address the genetic diversity of NAFLD and improve the understanding of the NAFLD disease development and progression.
Acknowledgments
A part of the figures was created using the BioRender.com via a licensed account.
Author Contributions
Conceptualization, S.A.S.; writing—original draft preparation, S.A.S.; writing—review and editing, S.A.S., M.N.H.A. and V.D.; and project administration and funding acquisition, S.A.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This manuscript is funded by the Fundamental Research Grant Scheme (FRGS) from the Ministry of Higher Education (MOHE), Malaysia (Grant code: FRGS/1/2019/SKK08/UKM/03/8).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Cotter T.G., Rinella M. Nonalcoholic fatty liver disease 2020: The state of the disease. Gastroenterology. 2020;158:1851–1864. doi: 10.1053/j.gastro.2020.01.052. [DOI] [PubMed] [Google Scholar]
- 2.Mohamed J., Nazratun Nafizah A.H., Zariyantey A.H., Budin S.B. Mechanisms of diabetes-induced liver damage: The role of oxidative stress and inflammation. Sultan Qaboos Univ. Med. J. 2016;16:e132–e141. doi: 10.18295/squmj.2016.16.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry L., Paik J., Younossi Z.M. Review article: The epidemiologic burden of non-alcoholic fatty liver disease across the world. Aliment. Pharmacol. Ther. 2022;56:942–956. doi: 10.1111/apt.17158. [DOI] [PubMed] [Google Scholar]
- 4.Younossi Z.M., Yilmaz Y., Yu M.L., Wai-Sun Wong V., Fernandez M.C., Isakov V.A., Duseja A.K., Mendez-Sanchez N., Eguchi Y., Bugianesi E., et al. Clinical and patient-reported outcomes from patients with nonalcoholic fatty liver disease across the world: Data from the global non-alcoholic steatohepatitis (nash)/non-alcoholic fatty liver disease (nafld) registry. Clin. Gastroenterol. Hepatol. 2022;20:2296–2306.e6. doi: 10.1016/j.cgh.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Han M.A.T., Yu Q., Tafesh Z., Pyrsopoulos N. Diversity in nafld: A review of manifestations of nonalcoholic fatty liver disease in different ethnicities globally. J. Clin. Transl. Hepatol. 2021;9:71–80. doi: 10.14218/JCTH.2020.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riazi K., Swain M.G., Congly S.E., Kaplan G.G., Shaheen A.A. Race and ethnicity in non-alcoholic fatty liver disease (nafld): A narrative review. Nutrients. 2022;14:4556. doi: 10.3390/nu14214556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yip T.C.-F., Vilar-Gomez E., Petta S., Yilmaz Y., Wong G.L.-H., Adams L.A., De Lédinghen V., Sookoian S., Wong V.W.-S. Geographical similarity and differences in the burden and genetic predisposition of nafld. Hepatology. 2022 doi: 10.1002/hep.32774. online ahead of print . [DOI] [PubMed] [Google Scholar]
- 8.Jonas W., Schürmann A. Genetic and epigenetic factors determining nafld risk. Mol. Metab. 2021;50:101111. doi: 10.1016/j.molmet.2020.101111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Campo J.A., Gallego-Durán R., Gallego P., Grande L. Genetic and epigenetic regulation in nonalcoholic fatty liver disease (nafld) Int. J. Mol. Sci. 2018;19:911. doi: 10.3390/ijms19030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu X., Xia M., Gao X. Update on genetics and epigenetics in metabolic associated fatty liver disease. Ther. Adv. Endocrinol. Metab. 2022;13:20420188221132138. doi: 10.1177/20420188221132138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wajsbrot N.B., Leite N.C., Salles G.F., Villela-Nogueira C.A. Non-alcoholic fatty liver disease and the impact of genetic, epigenetic and environmental factors in the offspring. World J. Gastroenterol. 2022;28:2890–2899. doi: 10.3748/wjg.v28.i25.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulaiman S.A., Muhsin N.I.A., Jamal R. Regulatory non-coding rnas network in non-alcoholic fatty liver disease. Front. Physiol. 2019;10:279. doi: 10.3389/fphys.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorairaj V., Sulaiman S.A., Abu N., Abdul Murad N.A. Extracellular vesicles in the development of the non-alcoholic fatty liver disease: An update. Biomolecules. 2020;10:1494. doi: 10.3390/biom10111494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pafili K., Roden M. Nonalcoholic fatty liver disease (nafld) from pathogenesis to treatment concepts in humans. Mol. Metab. 2021;50:101122. doi: 10.1016/j.molmet.2020.101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorairaj V., Sulaiman S.A., Abu N., Abdul Murad N.A. Nonalcoholic fatty liver disease (nafld): Pathogenesis and noninvasive diagnosis. Biomedicines. 2021;10:15. doi: 10.3390/biomedicines10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Wu Y., Rong X., Zheng C., Guo J. Anti-lipolysis induced by insulin in diverse pathophysiologic conditions of adipose tissue. Diabetes Metab. Syndr. Obes. 2020;13:1575–1585. doi: 10.2147/DMSO.S250699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bence K.K., Birnbaum M.J. Metabolic drivers of non-alcoholic fatty liver disease. Mol. Metab. 2021;50:101143. doi: 10.1016/j.molmet.2020.101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pais R., Maurel T. Natural history of nafld. J. Clin. Med. 2021;10:1161. doi: 10.3390/jcm10061161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., Herder C., Carstensen M., Krausch M., Knoefel W.T., et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Ma X., Cheng F., Yuan K., Jiang K., Zhu T. Lipid storage droplet protein 5 reduces sodium palmitate-induced lipotoxicity in human normal liver cells by regulating lipid metabolism-related factors. Mol. Med. Rep. 2019;20:879–886. doi: 10.3892/mmr.2019.10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J.L., Wang Q.Y., Luan H.Y., Kang Z.C., Wang C.B. Effects of l-carnitine against oxidative stress in human hepatocytes: Involvement of peroxisome proliferator-activated receptor alpha. J. Biomed. Sci. 2012;19:32. doi: 10.1186/1423-0127-19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engin A. Non-alcoholic fatty liver disease. Adv. Exp. Med. Biol. 2017;960:443–467. doi: 10.1007/978-3-319-48382-5_19. [DOI] [PubMed] [Google Scholar]
- 23.Zhu B., Chan S.L., Li J., Li K., Wu H., Cui K., Chen H. Non-alcoholic steatohepatitis pathogenesis, diagnosis, and treatment. Front. Cardiovasc. Med. 2021;8:1059. doi: 10.3389/fcvm.2021.742382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locati M., Curtale G., Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu. Rev. Pathol. 2020;15:123–147. doi: 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazankov K., Jørgensen S.M.D., Thomsen K.L., Møller H.J., Vilstrup H., George J., Schuppan D., Grønbæk H. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019;16:145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 26.Smith K. Liver disease: Kupffer cells regulate the progression of ald and nafld. Nat. Rev. Gastroenterol. Hepatol. 2013;10:503. doi: 10.1038/nrgastro.2013.140. [DOI] [PubMed] [Google Scholar]
- 27.Sulaiman S.A., Dorairaj V., Abdul Ghafar K.N., Abdul Murad N.A. Noncoding rnas interactions in hepatic stellate cells during hepatic fibrosis. Livers. 2021;1:263–285. doi: 10.3390/livers1040021. [DOI] [Google Scholar]
- 28.Yan Y., Zeng J., Xing L., Li C. Extra- and intra-cellular mechanisms of hepatic stellate cell activation. Biomedicines. 2021;9:1014. doi: 10.3390/biomedicines9081014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S.J., IglayReger H.B., Kadouh H.C., Bodary P.F. Inhibition of the chemokine (c-c motif) ligand 2/chemokine (c-c motif) receptor 2 pathway attenuates hyperglycaemia and inflammation in a mouse model of hepatic steatosis and lipoatrophy. Diabetologia. 2009;52:972–981. doi: 10.1007/s00125-009-1309-8. [DOI] [PubMed] [Google Scholar]
- 30.Baeck C., Wehr A., Karlmark K.R., Heymann F., Vucur M., Gassler N., Huss S., Klussmann S., Eulberg D., Luedde T., et al. Pharmacological inhibition of the chemokine ccl2 (mcp-1) diminishes liver macrophage infiltration and steatohepatitis in chronic hepatic injury. Gut. 2012;61:416–426. doi: 10.1136/gutjnl-2011-300304. [DOI] [PubMed] [Google Scholar]
- 31.Nio Y., Yamauchi T., Iwabu M., Okada-Iwabu M., Funata M., Yamaguchi M., Ueki K., Kadowaki T. Monocyte chemoattractant protein-1 (mcp-1) deficiency enhances alternatively activated m2 macrophages and ameliorates insulin resistance and fatty liver in lipoatrophic diabetic a-zip transgenic mice. Diabetologia. 2012;55:3350–3358. doi: 10.1007/s00125-012-2710-2. [DOI] [PubMed] [Google Scholar]
- 32.Herrero-Cervera A., Vinué Á., Burks D.J., González-Navarro H. Genetic inactivation of the light (tnfsf14) cytokine in mice restores glucose homeostasis and diminishes hepatic steatosis. Diabetologia. 2019;62:2143–2157. doi: 10.1007/s00125-019-4962-6. [DOI] [PubMed] [Google Scholar]
- 33.Ambade A., Lowe P., Kodys K., Catalano D., Gyongyosi B., Cho Y., Iracheta-Vellve A., Adejumo A., Saha B., Calenda C., et al. Pharmacological inhibition of ccr2/5 signaling prevents and reverses alcohol-induced liver damage, steatosis, and inflammation in mice. Hepatology. 2019;69:1105–1121. doi: 10.1002/hep.30249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A., Boerwinkle E., Cohen J.C., Hobbs H.H. Genetic variation in pnpla3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotman Y., Koh C., Zmuda J.M., Kleiner D.E., Liang T.J. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (pnpla3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorden A., Yang R., Yerges-Armstrong L.M., Ryan K.A., Speliotes E., Borecki I.B., Harris T.B., Chu X., Wood G.C., Still C.D., et al. Genetic variation at ncan locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum. Hered. 2013;75:34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansoor S., Maheshwari A., Di Guglielmo M., Furuya K., Wang M., Crowgey E., Molle-Rios Z., He Z. The pnpla3 rs738409 variant but not mboat7 rs641738 is a risk factor for nonalcoholic fatty liver disease in obese U.S. Children of hispanic ethnicity. Pediatr. Gastroenterol. Hepatol. Nutr. 2021;24:455–469. doi: 10.5223/pghn.2021.24.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park S.L., Li Y., Sheng X., Hom V., Xia L., Zhao K., Pooler L., Setiawan V.W., Lim U., Monroe K.R., et al. Genome-wide association study of liver fat: The multiethnic cohort adiposity phenotype study. Hepatol. Commun. 2020;4:1112–1123. doi: 10.1002/hep4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyysalo J., Stojkovic I., Kotronen A., Hakkarainen A., Sevastianova K., Makkonen J., Lundbom N., Rissanen A., Krauss R.M., Melander O., et al. Genetic variation in pnpla3 but not apoc3 influences liver fat in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2012;27:951–956. doi: 10.1111/j.1440-1746.2011.07045.x. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Xing C., Tian Z., Ku H.C. Genetic variant i148m in pnpla3 is associated with the ultrasonography-determined steatosis degree in a chinese population. BMC. Med. Genet. 2012;13:113. doi: 10.1186/1471-2350-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang X.-R., Song J.-Y., Liu F.-H., Ma J., Wang H.-J. Gwas-identified common variants with nonalcoholic fatty liver disease in chinese children. J. Pediatr. Gastroenterol. Nutr. 2015;60:669–674. doi: 10.1097/MPG.0000000000000662. [DOI] [PubMed] [Google Scholar]
- 43.Wang X., Liu Z., Wang K., Wang Z., Sun X., Zhong L., Deng G., Song G., Sun B., Peng Z., et al. Additive effects of the risk alleles of pnpla3 and tm6sf2 on non-alcoholic fatty liver disease (nafld) in a chinese population. Front. Genet. 2016;7:140. doi: 10.3389/fgene.2016.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia M.F., Ling Y., Bian H., Lin H.D., Yan H.M., Chang X.X., Li X.M., Ma H., Wang D., Zhang L.S., et al. I148m variant of pnpla3 increases the susceptibility to non-alcoholic fatty liver disease caused by obesity and metabolic disorders. Aliment. Pharmacol. Ther. 2016;43:631–642. doi: 10.1111/apt.13521. [DOI] [PubMed] [Google Scholar]
- 45.Wang C.W., Lin H.Y., Shin S.J., Yu M.L., Lin Z.Y., Dai C.Y., Huang J.F., Chen S.C., Li S.S., Chuang W.L. The pnpla3 i148m polymorphism is associated with insulin resistance and nonalcoholic fatty liver disease in a normoglycaemic population. Liver Int. 2011;31:1326–1331. doi: 10.1111/j.1478-3231.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- 46.Hudert C.A., Selinski S., Rudolph B., Bläker H., Loddenkemper C., Thielhorn R., Berndt N., Golka K., Cadenas C., Reinders J., et al. Genetic determinants of steatosis and fibrosis progression in paediatric non-alcoholic fatty liver disease. Liver Int. 2019;39:540–556. doi: 10.1111/liv.14006. [DOI] [PubMed] [Google Scholar]
- 47.Krawczyk M., Rau M., Schattenberg J.M., Bantel H., Pathil A., Demir M., Kluwe J., Boettler T., Lammert F., Geier A. Combined effects of the pnpla3 rs738409, tm6sf2 rs58542926, and mboat7 rs641738 variants on nafld severity: A multicenter biopsy-based study. J. Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitamoto T., Kitamoto A., Yoneda M., Hyogo H., Ochi H., Nakamura T., Teranishi H., Mizusawa S., Ueno T., Chayama K., et al. Genome-wide scan revealed that polymorphisms in the pnpla3, samm50, and parvb genes are associated with development and progression of nonalcoholic fatty liver disease in japan. Hum. Genet. 2013;132:783–792. doi: 10.1007/s00439-013-1294-3. [DOI] [PubMed] [Google Scholar]
- 49.Di Costanzo A., Belardinilli F., Bailetti D., Sponziello M., D’Erasmo L., Polimeni L., Baratta F., Pastori D., Ceci F., Montali A., et al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (nafld) by a candidate genes resequencing strategy. Sci. Rep. 2018;8:3702. doi: 10.1038/s41598-018-21939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenti L., Al-Serri A., Daly A.K., Galmozzi E., Rametta R., Dongiovanni P., Nobili V., Mozzi E., Roviaro G., Vanni E., et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin i148m polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 51.Vespasiani-Gentilucci U., Gallo P., Porcari A., Carotti S., Galati G., Piccioni L., De Vincentis A., Dell’Unto C., Vorini F., Morini S., et al. The pnpla3 rs738409 c > g polymorphism is associated with the risk of progression to cirrhosis in nafld patients. Scand. J. Gastroenterol. 2016;51:967–973. doi: 10.3109/00365521.2016.1161066. [DOI] [PubMed] [Google Scholar]
- 52.Longo M., Meroni M., Paolini E., Erconi V., Carli F., Fortunato F., Ronchi D., Piciotti R., Sabatini S., Macchi C., et al. Tm6sf2/pnpla3/mboat7 loss-of-function genetic variants impact on nafld development and progression both in patients and in in vitro models. Cell Mol. Gastroenterol. Hepatol. 2022;13:759–788. doi: 10.1016/j.jcmgh.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verrijken A., Beckers S., Francque S., Hilden H., Caron S., Zegers D., Ruppert M., Hubens G., Van Marck E., Michielsen P., et al. A gene variant of pnpla3, but not of apoc3, is associated with histological parameters of nafld in an obese population. Obesity. 2013;21:2138–2145. doi: 10.1002/oby.20366. [DOI] [PubMed] [Google Scholar]
- 54.Lisboa Q.C., Nardelli M.J., Pereira P.A., Miranda D.M., Ribeiro S.N., Costa R.S.N., Versiani C.A., Vidigal P.V.T., Ferrari T.C.A., Couto C.A. Pnpla3 and tm6sf2 polymorphisms in brazilian patients with nonalcoholic fatty liver disease. World J. Hepatol. 2020;12:792–806. doi: 10.4254/wjh.v12.i10.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zain S.M., Mohamed R., Mahadeva S., Cheah P.L., Rampal S., Basu R.C., Mohamed Z. A multi-ethnic study of a pnpla3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum. Genet. 2012;131:1145–1152. doi: 10.1007/s00439-012-1141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uygun A., Ozturk K., Demirci H., Oztuna A., Eren F., Kozan S., Yilmaz Y., Kurt O., Turker T., Vatansever S., et al. The association of nonalcoholic fatty liver disease with genetic polymorphisms: A multicenter study. Eur. J. Gastroenterol. Hepatol. 2017;29:441–447. doi: 10.1097/MEG.0000000000000813. [DOI] [PubMed] [Google Scholar]
- 57.Idilman R., Karatayli S.C., Kabacam G., Savas B., Elhan A.H., Bozdayi A.M. The role of pnpla3 (rs738409) c>g variant on histological progression of non-alcoholic fatty liver disease. Hepatol. Forum. 2020;1:82–87. doi: 10.14744/hf.2020.2020.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X., Zhao Q., Wu K., Fan D. I148m variant of pnpla3 confer increased risk for nonalcoholic fatty liver disease not only in european population, but also in chinese population. Hepatology. 2011;54:2276. doi: 10.1002/hep.24567. [DOI] [PubMed] [Google Scholar]
- 59.Lin H., Wong G.L., Whatling C., Chan A.W., Leung H.H., Tse C.H., Shu S.S., Chim A.M., Lai J.C., Yip T.C., et al. Association of genetic variations with nafld in lean individuals. Liver Int. 2022;42:149–160. doi: 10.1111/liv.15078. [DOI] [PubMed] [Google Scholar]
- 60.Hotta K., Yoneda M., Hyogo H., Ochi H., Mizusawa S., Ueno T., Chayama K., Nakajima A., Nakao K., Sekine A. Association of the rs738409 polymorphism in pnpla3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med. Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawaguchi T., Sumida Y., Umemura A., Matsuo K., Takahashi M., Takamura T., Yasui K., Saibara T., Hashimoto E., Kawanaka M., et al. Genetic polymorphisms of the human pnpla3 gene are strongly associated with severity of non-alcoholic fatty liver disease in japanese. PLoS ONE. 2012;7:e38322. doi: 10.1371/journal.pone.0038322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S.S., Byoun Y.S., Jeong S.H., Woo B.H., Jang E.S., Kim J.W., Kim H.Y. Role of the pnpla3 i148m polymorphism in nonalcoholic fatty liver disease and fibrosis in korea. Dig. Dis. Sci. 2014;59:2967–2974. doi: 10.1007/s10620-014-3279-z. [DOI] [PubMed] [Google Scholar]
- 63.Park J.H., Cho B., Kwon H., Prilutsky D., Yun J.M., Choi H.C., Hwang K.B., Lee I.H., Kim J.I., Kong S.W. I148m variant in pnpla3 reduces central adiposity and metabolic disease risks while increasing nonalcoholic fatty liver disease. Liver Int. 2015;35:2537–2546. doi: 10.1111/liv.12909. [DOI] [PubMed] [Google Scholar]
- 64.Bhatt S.P., Nigam P., Misra A., Guleria R., Pandey R.M., Pasha M.A. Genetic variation in the patatin-like phospholipase domain-containing protein-3 (pnpla-3) gene in asian indians with nonalcoholic fatty liver disease. Metab. Syndr. Relat. Disord. 2013;11:329–335. doi: 10.1089/met.2012.0064. [DOI] [PubMed] [Google Scholar]
- 65.Karoli R., Fatima J., Singh P.S., Siddiqi Z., Varshney S., Beg M.S., Khan M.A. Association of genetic non-alcoholic fatty liver disease with insulin resistance-are we different? J. Assoc. Physicians India. 2019;67:34–38. [PubMed] [Google Scholar]
- 66.Kanth V.V., Sasikala M., Rao P.N., Steffie Avanthi U., Rao K.R., Nageshwar Reddy D. Pooled genetic analysis in ultrasound measured non-alcoholic fatty liver disease in indian subjects: A pilot study. World J. Hepatol. 2014;6:435–442. doi: 10.4254/wjh.v6.i6.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narayanasamy K., Karthick R., Panneerselvam P., Mohan N., Ramachandran A., Prakash R., Rajaram M. Association of metabolic syndrome and patatin-like phospholipase 3–rs738409 gene variant in non-alcoholic fatty liver disease among a chennai-based south indian population. J. Gene Med. 2020;22:e3160. doi: 10.1002/jgm.3160. [DOI] [PubMed] [Google Scholar]
- 68.Niriella M.A., Pathmeswaran A., De Silva S.T., Kasturiratna A., Perera R., Subasinghe C.E., Kodisinghe K., Piyaratna C., Rishikesawan V., Dassanayaka A.S., et al. Incidence and risk factors for non-alcoholic fatty liver disease: A 7-year follow-up study among urban, adult sri lankans. Liver Int. 2017;37:1715–1722. doi: 10.1111/liv.13478. [DOI] [PubMed] [Google Scholar]
- 69.Alam S., Islam M.S., Islam S., Mustafa G., Saleh A.A., Ahmad N. Association of single nucleotide polymorphism at pnpla3 with fatty liver, steatohepatitis, and cirrhosis of liver. Indian J. Gastroenterol. 2017;36:366–372. doi: 10.1007/s12664-017-0784-y. [DOI] [PubMed] [Google Scholar]
- 70.Choobini N., Azarpira N., Mashayekhi M.R. Association of pnpla3 gene polymorphism (rs738409) and nonalcoholic fatty liver disease in southern iranian population. JABS. 2016;6:60–68. [Google Scholar]
- 71.Lee G.H., Phyo W.W., Loo W.M., Kwok R., Ahmed T., Shabbir A., So J., Koh C.J., Hartono J.L., Muthiah M., et al. Validation of genetic variants associated with metabolic dysfunction-associated fatty liver disease in an ethnic chinese population. World J. Hepatol. 2020;12:1228–1238. doi: 10.4254/wjh.v12.i12.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chatterjee A., Basu A., Das K., Chowdhury A., Basu P. Exome-wide scan identifies significant association of rs4788084 in il27 promoter with increase in hepatic fat content among indians. Gene. 2021;775:145431. doi: 10.1016/j.gene.2021.145431. [DOI] [PubMed] [Google Scholar]
- 73.Baclig M.O., Lozano-Kühne J.P., Mapua C.A., Gopez-Cervantes J., Natividad F.F. Genetic variation i148m in patatin-like phospholipase 3 gene and risk of non-alcoholic fatty liver disease among filipinos. Int. J. Clin. Exp. Med. 2014;7:2129–2136. [PMC free article] [PubMed] [Google Scholar]
- 74.Kawaguchi T., Shima T., Mizuno M., Mitsumoto Y., Umemura A., Kanbara Y., Tanaka S., Sumida Y., Yasui K., Takahashi M., et al. Risk estimation model for nonalcoholic fatty liver disease in the japanese using multiple genetic markers. PLoS ONE. 2018;13:e0185490. doi: 10.1371/journal.pone.0185490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjærg-Hansen A., Vogt T.F., Hobbs H.H., Cohen J.C. Exome-wide association study identifies a tm6sf2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eslam M., Mangia A., Berg T., Chan H.L., Irving W.L., Dore G.J., Abate M.L., Bugianesi E., Adams L.A., Najim M.A., et al. Diverse impacts of the rs58542926 e167k variant in tm6sf2 on viral and metabolic liver disease phenotypes. Hepatology. 2016;64:34–46. doi: 10.1002/hep.28475. [DOI] [PubMed] [Google Scholar]
- 77.Sookoian S., Castaño G.O., Scian R., Mallardi P., Fernández Gianotti T., Burgueño A.L., San Martino J., Pirola C.J. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61:515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Y., Llauradó G., Orešič M., Hyötyläinen T., Orho-Melander M., Yki-Järvinen H. Circulating triacylglycerol signatures and insulin sensitivity in nafld associated with the e167k variant in tm6sf2. J. Hepatol. 2015;62:657–663. doi: 10.1016/j.jhep.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 79.Krawczyk M., Stachowska E., Milkiewicz P., Lammert F., Milkiewicz M. Reduction of caloric intake might override the prosteatotic effects of the pnpla3 p.I148m and tm6sf2 p.E167k variants in patients with fatty liver: Ultrasound-based prospective study. Digestion. 2016;93:139–148. doi: 10.1159/000441185. [DOI] [PubMed] [Google Scholar]
- 80.Grandone A., Cozzolino D., Marzuillo P., Cirillo G., Di Sessa A., Ruggiero L., Di Palma M.R., Perrone L., Miraglia Del Giudice E. Tm6sf2 glu167lys polymorphism is associated with low levels of ldl-cholesterol and increased liver injury in obese children. Pediatr. Obes. 2016;11:115–119. doi: 10.1111/ijpo.12032. [DOI] [PubMed] [Google Scholar]
- 81.Mancina R.M., Sentinelli F., Incani M., Bertoccini L., Russo C., Romeo S., Baroni M.G. Transmembrane-6 superfamily member 2 (tm6sf2) e167k variant increases susceptibility to hepatic steatosis in obese children. Dig. Liver Dis. 2016;48:100–101. doi: 10.1016/j.dld.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Musso G., Cipolla U., Cassader M., Pinach S., Saba F., De Michieli F., Paschetta E., Bongiovanni D., Framarin L., Leone N., et al. Tm6sf2 rs58542926 variant affects postprandial lipoprotein metabolism and glucose homeostasis in nafld. J. Lipid Res. 2017;58:1221–1229. doi: 10.1194/jlr.M075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goffredo M., Caprio S., Feldstein A.E., D’Adamo E., Shaw M.M., Pierpont B., Savoye M., Zhao H., Bale A.E., Santoro N. Role of tm6sf2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: A multiethnic study. Hepatology. 2016;63:117–125. doi: 10.1002/hep.28283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y.L., Reeves H.L., Burt A.D., Tiniakos D., McPherson S., Leathart J.B., Allison M.E., Alexander G.J., Piguet A.C., Anty R., et al. Tm6sf2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L.Z., Luan G.P., Qun L.I.U., Hui G.A.O., Xin Y.N., Xuan S.Y. Association between tm6sf2 rs58542926 polymorphism and non-alcoholic fatty liver disease in qingdao han population and molecular mechanism. PLAMJ. 2019;44:127–131. [Google Scholar]
- 86.Xu M., Li Y., Zhang S., Wang X., Shen J., Zhang S. Interaction of tm6sf2 e167k and pnpla3 i148m variants in nafld in northeast china. Ann. Hepatol. 2019;18:456–460. doi: 10.1016/j.aohep.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 87.Wang X., Liu Z., Peng Z., Liu W. The tm6sf2 rs58542926 t allele is significantly associated with non-alcoholic fatty liver disease in chinese. J. Hepatol. 2015;62:1438–1439. doi: 10.1016/j.jhep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 88.Dongiovanni P., Petta S., Maglio C., Fracanzani A.L., Pipitone R., Mozzi E., Motta B.M., Kaminska D., Rametta R., Grimaudo S., et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 89.Koo B.K., Joo S.K., Kim D., Bae J.M., Park J.H., Kim J.H., Kim W. Additive effects of pnpla3 and tm6sf2 on the histological severity of non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2018;33:1277–1285. doi: 10.1111/jgh.14056. [DOI] [PubMed] [Google Scholar]
- 90.Bale G., Steffie A.U., Ravi Kanth V.V., Rao P.N., Sharma M., Sasikala M., Reddy D.N. Regional differences in genetic susceptibility to non-alcoholic liver disease in two distinct indian ethnicities. World J. Hepatol. 2017;9:1101–1107. doi: 10.4254/wjh.v9.i26.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akuta N., Kawamura Y., Arase Y., Suzuki F., Sezaki H., Hosaka T., Kobayashi M., Kobayashi M., Saitoh S., Suzuki Y., et al. Relationships between genetic variations of pnpla3, tm6sf2 and histological features of nonalcoholic fatty liver disease in japan. Gut Liver. 2016;10:437–445. doi: 10.5009/gnl15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hernaez R., McLean J., Lazo M., Brancati F.L., Hirschhorn J.N., Borecki I.B., Harris T.B., Nguyen T., Kamel I.R., Bonekamp S., et al. Association between variants in or near pnpla3, gckr, and ppp1r3b with ultrasound-defined steatosis based on data from the third national health and nutrition examination survey. Clin. Gastroenterol. Hepatol. 2013;11:1183–1190.e2. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petta S., Miele L., Bugianesi E., Cammà C., Rosso C., Boccia S., Cabibi D., Di Marco V., Grimaudo S., Grieco A., et al. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS ONE. 2014;9:e87523. doi: 10.1371/journal.pone.0087523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin Y.C., Chang P.F., Chang M.H., Ni Y.H. Genetic variants in gckr and pnpla3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am. J. Clin. Nutr. 2014;99:869–874. doi: 10.3945/ajcn.113.079749. [DOI] [PubMed] [Google Scholar]
- 95.Cai W., Weng D.H., Yan P., Lin Y.T., Dong Z.H., Mailamuguli, Yao H. Genetic polymorphisms associated with nonalcoholic fatty liver disease in uyghur population: A case-control study and meta-analysis. Lipids Health Dis. 2019;18:14. doi: 10.1186/s12944-018-0877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tan H.L., Zain S.M., Mohamed R., Rampal S., Chin K.F., Basu R.C., Cheah P.L., Mahadeva S., Mohamed Z. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: An interaction study with adiponutrin gene. J. Gastroenterol. 2014;49:1056–1064. doi: 10.1007/s00535-013-0850-x. [DOI] [PubMed] [Google Scholar]
- 97.Mohammadi S., Farajnia S., Shadmand M., Mohseni F., Baghban R. Association of rs780094 polymorphism of glucokinase regulatory protein with non-alcoholic fatty liver disease. BMC Res. Notes. 2020;13:26. doi: 10.1186/s13104-020-4891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao H., Liu S., Zhao Z., Yu X., Liu Q., Xin Y., Xuan S. Association of gckr gene polymorphisms with the risk of nonalcoholic fatty liver disease and coronary artery disease in a chinese northern han population. J. Clin. Transl. Hepatol. 2019;7:297–303. doi: 10.14218/JCTH.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yuan F., Gu Z., Bi Y., Yuan R., Niu W., Ren D., Zhang L., He G., Liu B.C. The association between rs1260326 with the risk of nafld and the mediation effect of triglyceride on nafld in the elderly chinese han population. Aging. 2022;14:2736–2747. doi: 10.18632/aging.203970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang Z., Wen J., Tao X., Lu B., Du Y., Wang M., Wang X., Zhang W., Gong W., Ling C., et al. Genetic variation in the gckr gene is associated with non-alcoholic fatty liver disease in chinese people. Mol. Biol. Rep. 2011;38:1145–1150. doi: 10.1007/s11033-010-0212-1. [DOI] [PubMed] [Google Scholar]
- 101.Sliz E., Sebert S., Würtz P., Kangas A.J., Soininen P., Lehtimäki T., Kähönen M., Viikari J., Männikkö M., Ala-Korpela M., et al. Nafld risk alleles in pnpla3, tm6sf2, gckr and lyplal1 show divergent metabolic effects. Hum. Mol. Genet. 2018;27:2214–2223. doi: 10.1093/hmg/ddy124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao S., An K., Liu Z., He H., An Z., Su Q., Li S. Genetic variants associated with metabolic dysfunction-associated fatty liver disease in western china. J. Clin. Lab. Anal. 2022;36:e24626. doi: 10.1002/jcla.24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R., Borén J., Montalcini T., Pujia A., Wiklund O., et al. The mboat7-tmc4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of european descent. Gastroenterology. 2016;150:1219–1230.e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luukkonen P.K., Zhou Y., Hyötyläinen T., Leivonen M., Arola J., Orho-Melander M., Orešič M., Yki-Järvinen H. The mboat7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J. Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 105.Basyte-Bacevice V., Skieceviciene J., Valantiene I., Sumskiene J., Petrenkiene V., Kondrackiene J., Petrauskas D., Lammert F., Kupcinskas J. Tm6sf2 and mboat7 gene variants in liver fibrosis and cirrhosis. Int. J. Mol. Sci. 2019;20:1277. doi: 10.3390/ijms20061277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zusi C., Morandi A., Maguolo A., Corradi M., Costantini S., Mosca A., Crudele A., Mantovani A., Alisi A., Miraglia Del Giudice E., et al. Association between mboat7 rs641738 polymorphism and non-alcoholic fatty liver in overweight or obese children. Nutr. Metab. Cardiovasc. Dis. 2021;31:1548–1555. doi: 10.1016/j.numecd.2021.01.020. [DOI] [PubMed] [Google Scholar]
- 107.Sookoian S., Flichman D., Garaycoechea M.E., Gazzi C., Martino J.S., Castaño G.O., Pirola C.J. Lack of evidence supporting a role of tmc4-rs641738 missense variant-mboat7- intergenic downstream variant-in the susceptibility to nonalcoholic fatty liver disease. Sci. Rep. 2018;8:5097. doi: 10.1038/s41598-018-23453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin Y.C., Chang P.F., Chang M.H., Ni Y.H. Genetic determinants of hepatic steatosis and serum cytokeratin-18 fragment levels in taiwanese children. Liver Int. 2018;38:1300–1307. doi: 10.1111/liv.13689. [DOI] [PubMed] [Google Scholar]
- 109.Ma Y., Belyaeva O.V., Brown P.M., Fujita K., Valles K., Karki S., De Boer Y.S., Koh C., Chen Y., Du X., et al. 17-beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69:1504–1519. doi: 10.1002/hep.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vilar-Gomez E., Pirola C.J., Sookoian S., Wilson L.A., Liang T., Chalasani N. The protection conferred by hsd17b13 rs72613567 polymorphism on risk of steatohepatitis and fibrosis may be limited to selected subgroups of patients with nafld. Clin. Transl. Gastroenterol. 2021;12:e00400. doi: 10.14309/ctg.0000000000000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P., Liu Y., Kozlitina J., Stender S., Wood G.C., et al. A protein-truncating hsd17b13 variant and protection from chronic liver disease. N. Engl. J. Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luukkonen P.K., Tukiainen T., Juuti A., Sammalkorpi H., Haridas P.A.N., Niemelä O., Arola J., Orho-Melander M., Hakkarainen A., Kovanen P.T., et al. Hydroxysteroid 17-β dehydrogenase 13 variant increases phospholipids and protects against fibrosis in nonalcoholic fatty liver disease. JCI Insight. 2020;5:e132158. doi: 10.1172/jci.insight.132158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pirola C.J., Garaycoechea M., Flichman D., Arrese M., San Martino J., Gazzi C., Castaño G.O., Sookoian S. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J. Lipid Res. 2019;60:176–185. doi: 10.1194/jlr.P089953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Seko Y., Yamaguchi K., Tochiki N., Yano K., Takahashi A., Okishio S., Kataoka S., Okuda K., Umemura A., Moriguchi M., et al. Attenuated effect of pnpla3 on hepatic fibrosis by hsd17b13 in japanese patients with non-alcoholic fatty liver disease. Liver Int. 2020;40:1686–1692. doi: 10.1111/liv.14495. [DOI] [PubMed] [Google Scholar]
- 115.Koo B.K., Joo S.K., Kim D., Lee S., Bae J.M., Park J.H., Kim J.H., Chang M.S., Kim W. Development and validation of a scoring system, based on genetic and clinical factors, to determine risk of steatohepatitis in asian patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2020;18:2592–2599.e10. doi: 10.1016/j.cgh.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 116.Pirazzi C., Adiels M., Burza M.A., Mancina R.M., Levin M., Ståhlman M., Taskinen M.R., Orho-Melander M., Perman J., Pujia A., et al. Patatin-like phospholipase domain-containing 3 (pnpla3) i148m (rs738409) affects hepatic vldl secretion in humans and in vitro. J. Hepatol. 2012;57:1276–1282. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 117.Salari N., Darvishi N., Mansouri K., Ghasemi H., Hosseinian-Far M., Darvishi F., Mohammadi M. Association between pnpla3 rs738409 polymorphism and nonalcoholic fatty liver disease: A systematic review and meta-analysis. BMC Endocr. Disord. 2021;21:125. doi: 10.1186/s12902-021-00789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kantartzis K., Peter A., Machicao F., Machann J., Wagner S., Königsrainer I., Königsrainer A., Schick F., Fritsche A., Häring H.U., et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 2009;58:2616–2623. doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis J.N., Lê K.A., Walker R.W., Vikman S., Spruijt-Metz D., Weigensberg M.J., Allayee H., Goran M.I. Increased hepatic fat in overweight hispanic youth influenced by interaction between genetic variation in pnpla3 and high dietary carbohydrate and sugar consumption. Am. J. Clin. Nutr. 2010;92:1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dubuquoy C., Robichon C., Lasnier F., Langlois C., Dugail I., Foufelle F., Girard J., Burnol A.F., Postic C., Moldes M. Distinct regulation of adiponutrin/pnpla3 gene expression by the transcription factors chrebp and srebp1c in mouse and human hepatocytes. J. Hepatol. 2011;55:145–153. doi: 10.1016/j.jhep.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 121.Perttilä J., Huaman-Samanez C., Caron S., Tanhuanpää K., Staels B., Yki-Järvinen H., Olkkonen V.M. Pnpla3 is regulated by glucose in human hepatocytes, and its i148m mutant slows down triglyceride hydrolysis. Am. J. Physiol. Endocrinol. Metab. 2012;302:E1063–E1069. doi: 10.1152/ajpendo.00125.2011. [DOI] [PubMed] [Google Scholar]
- 122.Sookoian S., Pirola C.J. Meta-analysis of the influence of i148m variant of patatin-like phospholipase domain containing 3 gene (pnpla3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 123.Salameh H., Hanayneh M.A., Masadeh M., Naseemuddin M., Matin T., Erwin A., Singal A.K. Pnpla3 as a genetic determinant of risk for and severity of non-alcoholic fatty liver disease spectrum. J. Clin. Transl. Hepatol. 2016;4:175–191. doi: 10.14218/JCTH.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Y., Orešič M., Leivonen M., Gopalacharyulu P., Hyysalo J., Arola J., Verrijken A., Francque S., Van Gaal L., Hyötyläinen T., et al. Noninvasive detection of nonalcoholic steatohepatitis using clinical markers and circulating levels of lipids and metabolites. Clin. Gastroenterol. Hepatol. 2016;14:1463–1472.e6. doi: 10.1016/j.cgh.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 125.Hyysalo J., Männistö V.T., Zhou Y., Arola J., Kärjä V., Leivonen M., Juuti A., Jaser N., Lallukka S., Käkelä P., et al. A population-based study on the prevalence of nash using scores validated against liver histology. J. Hepatol. 2014;60:839–846. doi: 10.1016/j.jhep.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 126.Segura-Azuara N., Varela-Chinchilla C.D., Trinidad-Calderón P.A. Mafld/nafld biopsy-free scoring systems for hepatic steatosis, nash, and fibrosis diagnosis. Front. Med. 2021;8:774079. doi: 10.3389/fmed.2021.774079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Z., Que S., Zhou L., Zheng S., Romeo S., Mardinoglu A., Valenti L. The effect of the tm6sf2 e167k variant on liver steatosis and fibrosis in patients with chronic hepatitis c: A meta-analysis. Sci. Rep. 2017;7:9273. doi: 10.1038/s41598-017-09548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li X.Y., Liu Z., Li L., Wang H.J., Wang H. Tm6sf2 rs58542926 is related to hepatic steatosis, fibrosis and serum lipids both in adults and children: A meta-analysis. Front. Endocrinol. 2022;13:1026901. doi: 10.3389/fendo.2022.1026901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sookoian S., Pirola C.J. Meta-analysis of the influence of tm6sf2 e167k variant on plasma concentration of aminotransferases across different populations and diverse liver phenotypes. Sci. Rep. 2016;6:27718. doi: 10.1038/srep27718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen F., Esmaili S., Rogers G.B., Bugianesi E., Petta S., Marchesini G., Bayoumi A., Metwally M., Azardaryany M.K., Coulter S., et al. Lean nafld: A distinct entity shaped by differential metabolic adaptation. Hepatology. 2020;71:1213–1227. doi: 10.1002/hep.30908. [DOI] [PubMed] [Google Scholar]
- 131.Young S., Tariq R., Provenza J., Satapathy S.K., Faisal K., Choudhry A., Friedman S.L., Singal A.K. Prevalence and profile of nonalcoholic fatty liver disease in lean adults: Systematic review and meta-analysis. Hepatol. Commun. 2020;4:953–972. doi: 10.1002/hep4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kozlitina J. Genetic risk factors and disease modifiers of nonalcoholic steatohepatitis. Gastroenterol. Clin. N. Am. 2020;49:25–44. doi: 10.1016/j.gtc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 133.Holmen O.L., Zhang H., Fan Y., Hovelson D.H., Schmidt E.M., Zhou W., Guo Y., Zhang J., Langhammer A., Løchen M.L., et al. Systematic evaluation of coding variation identifies a candidate causal variant in tm6sf2 influencing total cholesterol and myocardial infarction risk. Nat. Genet. 2014;46:345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pirola C.J., Sookoian S. The dual and opposite role of the tm6sf2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology. 2015;62:1742–1756. doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 135.Luo F., Oldoni F., Das A. Tm6sf2: A novel genetic player in nonalcoholic fatty liver and cardiovascular disease. Hepatol. Commun. 2022;6:448–460. doi: 10.1002/hep4.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smagris E., Gilyard S., BasuRay S., Cohen J.C., Hobbs H.H. Inactivation of tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J. Biol. Chem. 2016;291:10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Simons N., Isaacs A., Koek G.H., Kuč S., Schaper N.C., Brouwers M. Pnpla3, tm6sf2, and mboat7 genotypes and coronary artery disease. Gastroenterology. 2017;152:912–913. doi: 10.1053/j.gastro.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 138.Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C., et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat. Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fang Y.L., Chen H., Wang C.L., Liang L. Pathogenesis of non-alcoholic fatty liver disease in children and adolescence: From “two hit theory” to “multiple hit model”. World J. Gastroenterol. 2018;24:2974–2983. doi: 10.3748/wjg.v24.i27.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eslam M., Valenti L., Romeo S. Genetics and epigenetics of nafld and nash: Clinical impact. J. Hepatol. 2018;68:268–279. doi: 10.1016/j.jhep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 141.Zain S.M., Mohamed Z., Mohamed R. Common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: A meta-analysis. J. Gastroenterol. Hepatol. 2015;30:21–27. doi: 10.1111/jgh.12714. [DOI] [PubMed] [Google Scholar]
- 142.Li J., Zhao Y., Zhang H., Hua W., Jiao W., Du X., Rui J., Li S., Teng H., Shi B., et al. Contribution of rs780094 and rs1260326 polymorphisms in gckr gene to non-alcoholic fatty liver disease: A meta-analysis involving 26,552 participants. Endocr. Metab. Immune Disord. Drug Targets. 2021;21:1696–1708. doi: 10.2174/1871530320999201126202706. [DOI] [PubMed] [Google Scholar]
- 143.Sparsø T., Andersen G., Nielsen T., Burgdorf K.S., Gjesing A.P., Nielsen A.L., Albrechtsen A., Rasmussen S.S., Jørgensen T., Borch-Johnsen K., et al. The gckr rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and ogtt-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 144.Beer N.L., Tribble N.D., McCulloch L.J., Roos C., Johnson P.R., Orho-Melander M., Gloyn A.L. The p446l variant in gckr associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Santoro N., Zhang C.K., Zhao H., Pakstis A.J., Kim G., Kursawe R., Dykas D.J., Bale A.E., Giannini C., Pierpont B., et al. Variant in the glucokinase regulatory protein (gckr) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55:781–789. doi: 10.1002/hep.24806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thabet K., Chan H.L.Y., Petta S., Mangia A., Berg T., Boonstra A., Brouwer W.P., Abate M.L., Wong V.W., Nazmy M., et al. The membrane-bound o-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis b. Hepatology. 2017;65:1840–1850. doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 147.Viitasalo A., Eloranta A.M., Atalay M., Romeo S., Pihlajamäki J., Lakka T.A. Association of mboat7 gene variant with plasma alt levels in children: The panic study. Pediatr. Res. 2016;80:651–655. doi: 10.1038/pr.2016.139. [DOI] [PubMed] [Google Scholar]
- 148.Lee H.C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. Caenorhabditis elegans mboa-7, a member of the mboat family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 2008;19:1174–1184. doi: 10.1091/mbc.e07-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thangapandi V.R., Knittelfelder O., Brosch M., Patsenker E., Vvedenskaya O., Buch S., Hinz S., Hendricks A., Nati M., Herrmann A., et al. Loss of hepatic mboat7 leads to liver fibrosis. Gut. 2021;70:940–950. doi: 10.1136/gutjnl-2020-320853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang P., Wu C.X., Li Y., Shen N. Hsd17b13 rs72613567 protects against liver diseases and histological progression of nonalcoholic fatty liver disease: A systematic review and meta-analysis. Eur. Rev. Med. Pharmacol. Sci. 2020;24:8997–9007. doi: 10.26355/eurrev_202009_22842. [DOI] [PubMed] [Google Scholar]
- 151.Stender S., Smagris E., Lauridsen B.K., Kofoed K.F., Nordestgaard B.G., Tybjaerg-Hansen A., Pennacchio L.A., Dickel D.E., Cohen J.C., Hobbs H.H. Relationship between genetic variation at ppp1r3b and levels of liver glycogen and triglyceride. Hepatology. 2018;67:2182–2195. doi: 10.1002/hep.29751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saremi L., Lotfıpanah S., Mohammadi M., Hosseinzadeh H., Hosseini-Khah Z., Johari B., Saltanatpour Z. Association between ppargc1a single nucleotide polymorphisms and increased risk of nonalcoholic fatty liver disease among iranian patients with type 2 diabetes mellitus. Turk. J. Med. Sci. 2019;49:1089–1094. doi: 10.3906/sag-1808-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin Y.C., Chang P.F., Chang M.H., Ni Y.H. A common variant in the peroxisome proliferator-activated receptor-γ coactivator-1α gene is associated with nonalcoholic fatty liver disease in obese children. Am. J. Clin. Nutr. 2013;97:326–331. doi: 10.3945/ajcn.112.046417. [DOI] [PubMed] [Google Scholar]
- 154.Zhang R.N., Shen F., Pan Q., Cao H.X., Chen G.Y., Fan J.G. Ppargc1a rs8192678 g>a polymorphism affects the severity of hepatic histological features and nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease. World J. Gastroenterol. 2021;27:3863–3876. doi: 10.3748/wjg.v27.i25.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bhatta P., Bermano G., Williams H.C., Knott R.M. Meta-analysis demonstrates gly482ser variant of ppargc1a is associated with components of metabolic syndrome within asian populations. Genomics. 2020;112:1795–1803. doi: 10.1016/j.ygeno.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 156.Yoneda M., Hotta K., Nozaki Y., Endo H., Uchiyama T., Mawatari H., Iida H., Kato S., Hosono K., Fujita K., et al. Association between ppargc1a polymorphisms and the occurrence of nonalcoholic fatty liver disease (nafld) BMC Gastroenterol. 2008;8:27. doi: 10.1186/1471-230X-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen L., Lin Z., Jiang M., Lu L., Zhang H., Xin Y., Jiang X., Xuan S. Genetic variants in the samm50 gene create susceptibility to nonalcoholic fatty liver disease in a chinese han population. Hepat. Mon. 2015;15:e31076. doi: 10.5812/hepatmon.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li Z., Shen W., Wu G., Qin C., Zhang Y., Wang Y., Song G., Xiao C., Zhang X., Deng G., et al. The role of samm50 in non-alcoholic fatty liver disease: From genetics to mechanisms. FEBS Open Bio. 2021;11:1893–1906. doi: 10.1002/2211-5463.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chung G.E., Lee Y., Yim J.Y., Choe E.K., Kwak M.S., Yang J.I., Park B., Lee J.E., Kim J.A., Kim J.S. Genetic polymorphisms of pnpla3 and samm50 are associated with nonalcoholic fatty liver disease in a korean population. Gut Liver. 2018;12:316–323. doi: 10.5009/gnl17306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang R.N., Zheng R.D., Mi Y.Q., Zhou D., Shen F., Chen G.Y., Zhu C.Y., Pan Q., Fan J.G. Apoc3 rs2070666 is associated with the hepatic steatosis independently of pnpla3 rs738409 in chinese han patients with nonalcoholic fatty liver diseases. Dig. Dis. Sci. 2016;61:2284–2293. doi: 10.1007/s10620-016-4120-7. [DOI] [PubMed] [Google Scholar]
- 161.Valenti L., Nobili V., Al-Serri A., Rametta R., Leathart J.B., Zappa M.A., Dongiovanni P., Fracanzani A.L., Alterio A., Roviaro G., et al. The apoc3 t-455c and c-482t promoter region polymorphisms are not associated with the severity of liver damage independently of pnpla3 i148m genotype in patients with nonalcoholic fatty liver. J. Hepatol. 2011;55:1409–1414. doi: 10.1016/j.jhep.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 162.Petersen K.F., Dufour S., Hariri A., Nelson-Williams C., Foo J.N., Zhang X.M., Dziura J., Lifton R.P., Shulman G.I. Apolipoprotein c3 gene variants in nonalcoholic fatty liver disease. N. Engl. J. Med. 2010;362:1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jain V., Kumar A., Ahmad N., Jana M., Kalaivani M., Kumar B., Shastri S., Jain O., Kabra M. Genetic polymorphisms associated with obesity and non-alcoholic fatty liver disease in asian indian adolescents. J. Pediatr. Endocrinol. Metab. 2019;32:749–758. doi: 10.1515/jpem-2018-0543. [DOI] [PubMed] [Google Scholar]
- 164.Zain S.M., Mohamed Z., Mahadeva S., Rampal S., Basu R.C., Cheah P.L., Salim A., Mohamed R. Susceptibility and gene interaction study of the angiotensin ii type 1 receptor (agtr1) gene polymorphisms with non-alcoholic fatty liver disease in a multi-ethnic population. PLoS ONE. 2013;8:e58538. doi: 10.1371/journal.pone.0058538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yoneda M., Hotta K., Nozaki Y., Endo H., Uchiyama T., Mawatari H., Iida H., Kato S., Fujita K., Takahashi H., et al. Association between angiotensin ii type 1 receptor polymorphisms and the occurrence of nonalcoholic fatty liver disease. Liver Int. 2009;29:1078–1085. doi: 10.1111/j.1478-3231.2009.01988.x. [DOI] [PubMed] [Google Scholar]
- 166.Valenti L., Motta B.M., Alisi A., Sartorelli R., Buonaiuto G., Dongiovanni P., Rametta R., Pelusi S., Fargion S., Nobili V. Lpin1 rs13412852 polymorphism in pediatric nonalcoholic fatty liver disease. J. Pediatr. Gastroenterol. Nutr. 2012;54:588–593. doi: 10.1097/MPG.0b013e3182442a55. [DOI] [PubMed] [Google Scholar]
- 167.Zhou Y.-J., Li Y.-Y., Nie Y.-Q., Yang H., Zhan Q., Huang J., Shi S.-L., Lai X.-B., Huang H.-L. Influence of polygenetic polymorphisms on the susceptibility to non-alcoholic fatty liver disease of chinese people. J. Gastroenterol. Hepatol. 2010;25:772–777. doi: 10.1111/j.1440-1746.2009.06144.x. [DOI] [PubMed] [Google Scholar]
- 168.Dong H., Wang J., Li C., Hirose A., Nozaki Y., Takahashi M., Ono M., Akisawa N., Iwasaki S., Saibara T., et al. The phosphatidylethanolamine n-methyltransferase gene v175m single nucleotide polymorphism confers the susceptibility to nash in japanese population. J. Hepatol. 2007;46:915–920. doi: 10.1016/j.jhep.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 169.Song J., Da Costa K.A., Fischer L.M., Kohlmeier M., Kwock L., Wang S., Zeisel S.H. Polymorphism of the pemt gene and susceptibility to nonalcoholic fatty liver disease (nafld) FASEB J. 2005;19:1266–1271. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bale G., Vishnubhotla R.V., Mitnala S., Sharma M., Padaki R.N., Pawar S.C., Duvvur R.N. Whole-exome sequencing identifies a variant in phosphatidylethanolamine n-methyltransferase gene to be associated with lean-nonalcoholic fatty liver disease. J. Clin. Exp. Hepatol. 2019;9:561–568. doi: 10.1016/j.jceh.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jun D.W., Han J.H., Jang E.C., Kim S.H., Kim S.H., Jo Y.J., Park Y.S., Chae J.D. Polymorphisms of microsomal triglyceride transfer protein gene and phosphatidylethanolamine n-methyltransferase gene in alcoholic and nonalcoholic fatty liver disease in koreans. Eur. J. Gastroenterol. Hepatol. 2009;21:667–672. doi: 10.1097/MEG.0b013e3283196adc. [DOI] [PubMed] [Google Scholar]
- 172.Namikawa C., Shu-Ping Z., Vyselaar J.R., Nozaki Y., Nemoto Y., Ono M., Akisawa N., Saibara T., Hiroi M., Enzan H., et al. Polymorphisms of microsomal triglyceride transfer protein gene and manganese superoxide dismutase gene in non-alcoholic steatohepatitis. J. Hepatol. 2004;40:781–786. doi: 10.1016/j.jhep.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 173.Gambino R., Cassader M., Pagano G., Durazzo M., Musso G. Polymorphism in microsomal triglyceride transfer protein: A link between liver disease and atherogenic postprandial lipid profile in nash? Hepatology. 2007;45:1097–1107. doi: 10.1002/hep.21631. [DOI] [PubMed] [Google Scholar]
- 174.Carulli L., Canedi I., Rondinella S., Lombardini S., Ganazzi D., Fargion S., De Palma M., Lonardo A., Ricchi M., Bertolotti M., et al. Genetic polymorphisms in non-alcoholic fatty liver disease: Interleukin-6-174g/c polymorphism is associated with non-alcoholic steatohepatitis. Dig. Liver Dis. 2009;41:823–828. doi: 10.1016/j.dld.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 175.Oliveira C.P., Stefano J.T., Cavaleiro A.M., Zanella Fortes M.A., Vieira S.M., Rodrigues Lima V.M., Santos T.E., Santos V.N., De Azevedo Salgado A.L., Parise E.R., et al. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2010;25:357–361. doi: 10.1111/j.1440-1746.2009.06001.x. [DOI] [PubMed] [Google Scholar]
- 176.Al-Serri A., Anstee Q.M., Valenti L., Nobili V., Leathart J.B., Dongiovanni P., Patch J., Fracanzani A., Fargion S., Day C.P., et al. The sod2 c47t polymorphism influences nafld fibrosis severity: Evidence from case-control and intra-familial allele association studies. J. Hepatol. 2012;56:448–454. doi: 10.1016/j.jhep.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 177.Abshagen K., Berger C., Dietrich A., Schütz T., Wittekind C., Stumvoll M., Blüher M., Klöting N. A human repin1 gene variant: Genetic risk factor for the development of nonalcoholic fatty liver disease. Clin. Transl. Gastroenterol. 2020;11:e00114. doi: 10.14309/ctg.0000000000000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Aller R., De Luis D.A., Izaola O., González Sagrado M., Conde R., Alvarez T., Pacheco D., Velasco M.C. Role of -55ct polymorphism of ucp3 gene on non alcoholic fatty liver disease and insulin resistance in patients with obesity. Nutr. Hosp. 2010;25:572–576. [PubMed] [Google Scholar]
- 179.Borén J., Packard C.J., Taskinen M.-R. The roles of apoc-iii on the metabolism of triglyceride-rich lipoproteins in humans. Front. Endocrinol. 2020;11:474. doi: 10.3389/fendo.2020.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hooper A.J., Burnett J.R., Watts G.F. Contemporary aspects of the biology and therapeutic regulation of the microsomal triglyceride transfer protein. Circ. Res. 2015;116:193–205. doi: 10.1161/CIRCRESAHA.116.304637. [DOI] [PubMed] [Google Scholar]
- 181.Zain S.M., Mohamed R., Cooper D.N., Razali R., Rampal S., Mahadeva S., Chan W.K., Anwar A., Rosli N.S., Mahfudz A.S., et al. Genome-wide analysis of copy number variation identifies candidate gene loci associated with the progression of non-alcoholic fatty liver disease. PLoS ONE. 2014;9:e95604. doi: 10.1371/journal.pone.0095604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zain S.M., Mohamed Z., Pirmohamed M., Tan H.L., Alshawsh M.A., Mahadeva S., Chan W.K., Mustapha N.R., Mohamed R. Copy number variation in exportin-4 (xpo4) gene and its association with histological severity of non-alcoholic fatty liver disease. Sci. Rep. 2015;5:13306. doi: 10.1038/srep13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Metwally M., Bayoumi A., Khan A., Adams L.A., Aller R., García-Monzón C., Arias-Loste M.T., Bugianesi E., Miele L., Alisi A., et al. Copy number variation and expression of exportin-4 associates with severity of fibrosis in metabolic associated fatty liver disease. eBioMedicine. 2021;70:103521. doi: 10.1016/j.ebiom.2021.103521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y.F., Zheng J., Peng H.W., Cai X.L., Pan X.T., Li H.Q., Hong Q.Z., Hu Z.J., Wu Y.L., Peng X.-E. Identifying potential biomarkers of nonalcoholic fatty liver disease via genome-wide analysis of copy number variation. BMC Gastroenterol. 2021;21:171. doi: 10.1186/s12876-021-01750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen B.B., Yan J.H., Zheng J., Peng H.W., Cai X.L., Pan X.T., Li H.Q., Hong Q.Z., Peng X.-E. Copy number variation in the ces1 gene and the risk of non-alcoholic fatty liver in a chinese han population. Sci. Rep. 2021;11:13984. doi: 10.1038/s41598-021-93549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S., Pelusi S., Pingitore P., Badiali S., Maggioni M., et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J. Intern. Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.De Vincentis A., Tavaglione F., Jamialahmadi O., Picardi A., Antonelli Incalzi R., Valenti L., Romeo S., Vespasiani-Gentilucci U. A polygenic risk score to refine risk stratification and prediction for severe liver disease by clinical fibrosis scores. Clin. Gastroenterol. Hepatol. 2022;20:658–673. doi: 10.1016/j.cgh.2021.05.056. [DOI] [PubMed] [Google Scholar]
- 188.Di Costanzo A., Pacifico L., Chiesa C., Perla F.M., Ceci F., Angeloni A., D’Erasmo L., Di Martino M., Arca M. Genetic and metabolic predictors of hepatic fat content in a cohort of italian children with obesity. Pediatr. Res. 2019;85:671–677. doi: 10.1038/s41390-019-0303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thomas C.E., Diergaarde B., Kuipers A.L., Adibi J.J., Luu H.N., Chang X., Dorajoo R., Heng C.K., Khor C.C., Wang R., et al. Nafld polygenic risk score and risk of hepatocellular carcinoma in an east asian population. Hepatol. Commun. 2022;6:2310–2321. doi: 10.1002/hep4.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Donati B., Dongiovanni P., Romeo S., Meroni M., McCain M., Miele L., Petta S., Maier S., Rosso C., De Luca L., et al. Mboat7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017;7:4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang Y., Kory N., BasuRay S., Cohen J.C., Hobbs H.H. Pnpla3, cgi-58, and inhibition of hepatic triglyceride hydrolysis in mice. Hepatology. 2019;69:2427–2441. doi: 10.1002/hep.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gellert-Kristensen H., Richardson T.G., Davey Smith G., Nordestgaard B.G., Tybjaerg-Hansen A., Stender S. Combined effect of pnpla3, tm6sf2, and hsd17b13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology. 2020;72:845–856. doi: 10.1002/hep.31238. [DOI] [PubMed] [Google Scholar]
- 193.Bianco C., Jamialahmadi O., Pelusi S., Baselli G., Dongiovanni P., Zanoni I., Santoro L., Maier S., Liguori A., Meroni M., et al. Non-invasive stratification of hepatocellular carcinoma risk in non-alcoholic fatty liver using polygenic risk scores. J. Hepatol. 2021;74:775–782. doi: 10.1016/j.jhep.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen X.Y., Wang C., Huang Y.Z., Zhang L.L. Nonalcoholic fatty liver disease shows significant sex dimorphism. World J. Clin. Cases. 2022;10:1457–1472. doi: 10.12998/wjcc.v10.i5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Younossi Z.M. Non-alcoholic fatty liver disease—A global public health perspective. J. Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 196.Torres D.M., Williams C.D., Harrison S.A. Features, diagnosis, and treatment of nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2012;10:837–858. doi: 10.1016/j.cgh.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 197.Bjune J.I., Strømland P.P., Jersin R., Mellgren G., Dankel S.N. Metabolic and epigenetic regulation by estrogen in adipocytes. Front. Endocrinol. 2022;13:828780. doi: 10.3389/fendo.2022.828780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ismaiel A., Dumitrascu D.L. Genetic predisposition in metabolic-dysfunction-associated fatty liver disease and cardiovascular outcomes-systematic review. Eur. J. Clin. Investig. 2020;50:e13331. doi: 10.1111/eci.13331. [DOI] [PubMed] [Google Scholar]
- 199.Wang J., Ye C., Fei S. Association between apoc3 polymorphisms and non-alcoholic fatty liver disease risk: A meta-analysis. Afr. Health Sci. 2020;20:1800–1808. doi: 10.4314/ahs.v20i4.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bamshad M., Kivisild T., Watkins W.S., Dixon M.E., Ricker C.E., Rao B.B., Naidu J.M., Prasad B.V., Reddy P.G., Rasanayagam A., et al. Genetic evidence on the origins of indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hernaez R., Lazo M., Bonekamp S., Kamel I., Brancati F.L., Guallar E., Clark J.M. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: A meta-analysis. Hepatology. 2011;54:1082–1090. doi: 10.1002/hep.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Menon D.K., Rosand J. Finding a place for candidate gene studies in a genome-wide association study world. JAMA Netw. Open. 2021;4:e2118594. doi: 10.1001/jamanetworkopen.2021.18594. [DOI] [PubMed] [Google Scholar]
- 203.Wong V.W., Chan W.K., Chitturi S., Chawla Y., Dan Y.Y., Duseja A., Fan J., Goh K.L., Hamaguchi M., Hashimoto E., et al. Asia-pacific working party on non-alcoholic fatty liver disease guidelines 2017-part 1: Definition, risk factors and assessment. J. Gastroenterol. Hepatol. 2018;33:70–85. doi: 10.1111/jgh.13857. [DOI] [PubMed] [Google Scholar]
- 204.Li J., Zou B., Yeo Y.H., Feng Y., Xie X., Lee D.H., Fujii H., Wu Y., Kam L.Y., Ji F., et al. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in asia, 1999–2019: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2019;4:389–398. doi: 10.1016/S2468-1253(19)30039-1. [DOI] [PubMed] [Google Scholar]
- 205.Fleischman M.W., Budoff M., Zeb I., Li D., Foster T. Nafld prevalence differs among hispanic subgroups: The multi-ethnic study of atherosclerosis. World J. Gastroenterol. 2014;20:4987–4993. doi: 10.3748/wjg.v20.i17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kumar A., Walia G.K., Gupta V., Sachdeva M.P. Genetics of nonalcoholic fatty liver disease in asian populations. J. Genet. 2019;98:29. doi: 10.1007/s12041-019-1071-8. [DOI] [PubMed] [Google Scholar]
- 207.Turner R.M., Bird S.M., Higgins J.P. The impact of study size on meta-analyses: Examination of underpowered studies in cochrane reviews. PLoS ONE. 2013;8:e59202. doi: 10.1371/journal.pone.0059202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Robinson M.R., Wray N.R., Visscher P.M. Explaining additional genetic variation in complex traits. Trends Genet. 2014;30:124–132. doi: 10.1016/j.tig.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Flannick J., Fuchsberger C., Mahajan A., Teslovich T.M., Agarwala V., Gaulton K.J., Caulkins L., Koesterer R., Ma C., Moutsianas L., et al. Sequence data and association statistics from 12,940 type 2 diabetes cases and controls. Sci. Data. 2017;4:170179. doi: 10.1038/sdata.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gershon E.S., Pearlson G., Keshavan M.S., Tamminga C., Clementz B., Buckley P.F., Alliey-Rodriguez N., Liu C., Sweeney J.A., Keedy S., et al. Genetic analysis of deep phenotyping projects in common disorders. Schizophr. Res. 2018;195:51–57. doi: 10.1016/j.schres.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.