Abstract
Data have been presented indicating that Staphylococcus aureus cell surface protein can be degraded by extracellular proteases produced by the same bacterium. We have found that in sarA mutant cells, which produce high amounts of four major extracellular proteases (staphylococcal serine protease [V8 protease] [SspA], cysteine protease [SspB], aureolysin [metalloprotease] [Aur], and staphopain [Scp]), the levels of cell-bound fibronectin-binding proteins (FnBPs) and protein A were very low compared to those of wild-type cells, in spite of unaltered or increased transcription of the corresponding genes. Cultivation of sarA mutant cells in the presence of the global protease inhibitor α2-macroglobulin resulted in a 16-fold increase in cell-bound FnBPs, indicating that extracellular proteases were responsible for the decreased amounts of FnBPs in sarA mutant cells. The protease inhibitor E64 had no effect on the level of FnBPs, indicating that cysteine proteases were not involved. Inactivation of either ssp or aur in the prototype S. aureus strain 8325-4 resulted in a threefold increase in the amount of cell-bound FnBPs. Inactivation of the same protease genes in a sarA mutant of 8325-4 resulted in a 10- to 20-fold increase in cell-bound protein A. As the serine protease requires aureolysin to be activated, it can thus be concluded that the serine protease is the most important protease in the release of cell-bound FnBPs and protein A.
Staphylococcus aureus produces several cell surface proteins which bind specifically to different host extracellular matrix proteins and plasma proteins (12, 13, 32). For many of the cell surface proteins a role in colonization and virulence has been demonstrated in animal models of infection (17, 23, 27, 33). Two highly homologous fibronectin-binding proteins (FnBPA and FnBPB), encoded by fnbA and fnbB, have been characterized (14, 21, 25, 41) and shown to be involved in adherence to damaged heart valves (23) and to promote internalization of S. aureus by epithelial cells (9). Although S. aureus is primarily considered to be an extracellular pathogen, the intracellular niche could promote long-term colonization and maintenance of chronic infections.
Protein A (Spa), which binds immunoglobulin G (IgG) by the Fc segment, is a major surface protein present in virtually all strains of S. aureus (10, 11). Strains of S. aureus with a high content of Spa are more resistant to phagocytosis by human neutrophils in vitro than strains with less Spa (34). Reduced virulence of a spa mutant compared to that of the corresponding wild type was demonstrated in a mouse intraperitoneal infection (31).
We have recently shown that transcription of the fnbA and fnbB genes is negatively regulated by agr and by an agr-independent mechanism that restricts fnb mRNA synthesis to the early exponential phase of growth (38). A similar temporal control of fnb transcription was also found in another strain of S. aureus (Newman) (43). However, only fnbA appeared to be regulated by agr in this strain. It was also found that fnbA, but not fnbB, was positively regulated by sarA. As for fnbA and fnbB, transcription of spa is negatively regulated by agr (20). However, unlike for fnbA, transcription of spa is negatively controlled by sarA (3, 42).
Data from recent studies indicate that both FnBPs and protein A may be degraded by extracellular proteases (3, 26, 42). Four major extracellular proteases are produced by S. aureus (1): staphylococcal serine protease (V8 protease) (SspA), a metalloprotease named aureolysin (Aur), a cysteine protease (Scp) named staphopain (18), and a second cysteine protease (SspB) encoded within the same operon as SspA (2, 36). All four proteases appear to be synthesized as preproenzymes, which are proteolytically cleaved to generate the mature enzymes. In the case of the serine protease the proform is enzymatically inactive and needs to be cleaved by aureolysin to become active (8). The proform of SspB that appeared to possess enzyme activity seems to be processed by SspA (36). Which enzymes are involved in the processing of aureolysin and staphopain remains to be determined.
The synthesis of extracellular proteases is positively regulated by agr and negatively regulated by sarA (2, 20) in such a way that protease production takes place mainly during the postexponential phase of growth, when synthesis of cell surface proteins has ceased. Because of the sensitivity of FnBPs and a limited number of unidentified cell surface proteins to degradation by staphylococcal serine protease, it has been suggested that this enzyme participates in the transition of S. aureus cells from an adhesive to an invasive phenotype (26). However, since there are four major proteases which are all regulated in the same way and which are involved in the maturation of each other, we decided to analyze which enzyme(s) is involved in the degradation of FnBPs and protein A in growing cultures of S. aureus. By studying the effects of different protease inhibitors and protease knockout mutants, we have come to the conclusion that staphylococcal serine protease (V8 protease) is the most important for degrading FnBPs and protein A.
MATERIALS AND METHODS
Bacterial strains and plasmids and cultivation conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus strains were precultured in tryptic soy broth for 16 to 18 h. Cells from 2 ml of preculture were inoculated into 100 ml of brain heart infusion (BHI) broth in a 1-liter baffled flask to an initial optical density at 600 nm (OD600) of 0.25 to 0.3 and were incubated on a rotary shaker (180 rpm) at 37°C. To test the effect of protease inhibitors, parallel overnight cultures of the sarA mutant strain 11D2 were transferred to 50 ml of BHI broth (OD600 of 0.1) with or without 0.4 U of the universal protease inhibitor α2-macroglobulin (Boehringer Mannheim) ml−1 or 10 μM cysteine protease-specific inhibitor E64 [l-trans-epoxysuccinyl-leucylamido-(4-guanidino) butane] (Sigma). Samples of cell lysates and supernatants were taken at indicated time points, and cell-associated FnBPs were analyzed by Western blotting.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Strain used for propagation of all plasmid constructs | Promega Corp. |
| S. aureus | ||
| 8325-4 | Prototype wild-type strain; rsbU mutant | 30 |
| RN4220 | Restriction-deficient mutant of 8325-4 | 22 |
| DB | Wild-type blood clinic isolate | 5 |
| 11D2 | DB; sar::Tn917 (Emr) | 6 |
| PC1839 | 8325-4; sarA::km (Kmr) | 2 |
| AK1 | 8325-4; aur::ermB (Emr) | This study |
| AK10 | RN4220; Δaur::pAK1 (Emr) | This study |
| AK2 | 8325-4; Δssp::ermB (Emr) | This study |
| AK20 | RN4220; ssp::pAK2 (Emr) | This study |
| AK3 | 8325-4; Δaur::ermB sarA::km (Emr Kmr) | This study |
| AK5 | 8325-4; Δssp::ermB sarA::km (Emr Kmr) | This study |
| Plasmids | ||
| pKT4 | pGEM-T Easy containing ermB | 42 |
| pAK1 | pKT4 containing upstream and downstream aur fragments inserted at either side of ermB | This study |
| pAK2 | pKT4 containing upstream and downstream ssp fragments inserted at either side of ermB | This study |
Kmr, resistance to kanamycin and neomycin; Emr, resistance to erythromycin and lincomycin.
Northern blot analysis.
Total S. aureus RNA was prepared by extraction of lysostaphin-treated cells with hot phenol as described previously (20). Northern blot analysis of fnb mRNA and RNAIII, using digoxigenin-labeled antisense RNA probes, was performed as described earlier (38). Serine protease (sspA), aureolysin (aur), and staphopain (scp) mRNAs were analyzed using 32P-labeled probes as described previously (28). DNA fragments of 889 bp (ssp), 1,197 bp (aur), and 407 bp (scp), encompassing the beginning of the structural genes, were generated by PCR using S. aureus 8325-4 chromosomal DNA as the template. PCR fragments were radiolabeled to a specific activity of 1 × 108 to 5 × 108 cpm μg−1 with [α-32P]dCTP using a random primer labeling kit (Boehringer Mannheim Biochemica). Northern blot images were processed and quantitated using the PhosphorImager 445SI and Image QuanT software (Molecular Dynamics).
Construction of plasmids and protease mutants by allelic replacement.
The plasmids were constructed in Escherichia coli DH5α, and molecular biology techniques and recombinant DNA manipulations were done as previously described (37). Plasmid DNA was extracted using the Qiagen plasmid mini-kit. E. coli transformants were selected on Luria-Bertani (Difco) plates containing 100 μg of ampicillin ml−1.
For construction of strain AK1 (aur allelic replacement mutant) two PCR fragments, of 1,064 and 953 bp, encompassing the first 197 and the last 61 codons of the 508-codon aureolysin gene were amplified using forward primers with added PstI and EcoRI restriction sites and a reverse primer with added SpeI and NcoI restriction sites. The PCR fragments were inserted in two steps at either side of the ermB cassette in pKT4 to generate plasmid pAK1. The plasmid was then transformed into S. aureus RN4220 by electroporation (39). Clones of bacteria were selected on erythromycin plates, and spontaneous chromosomal mutations were eliminated on lincomycin plates (2). The correct insertions were verified by restriction mapping and PCR analysis. Integration of pAK1 into the aur gene was verified in one transformant (AK10). Transduction of the aur mutation into S. aureus 8325-4 using phage Φ11 (29) was performed in order to obtain a double crossover and an allelic replacement of the metalloprotease gene. The aur mutation in AK1 was verified by PCR and by Southern blotting using an aur-specific probe. The lack of metalloprotease in AK1 was verified by Western blotting (see below).
Strain AK2 (ssp allelic replacement mutant) was constructed similarly to AK1. PCR fragments (244 bp [nucleotides 7 to 251] and 332 bp [nucleotides 2693 to 3025], GeneBank accession no. 309515) flanking the ssp operon were generated using forward and reverse primers with the same restriction sites as in the construction of pAK1. The PCR fragments were inserted at either side of the ermB casette in pKT4 to generate plasmid pAK2, which was then transferred into S. aureus RN4220 by electroporation. Recombinants were selected on erythromycin plates and lincomycin plates as described above. Restriction mapping and PCR analysis of one transformant (AK20) using primers internal to the ssp operon and flanking primers in the erythromycin cassette confirmed the integration of pAK2 in the ssp operon. The ssp mutation of AK20 was transduced in S. aureus 8325-4 using phage Φ11 (29) to obtain a double crossover and replacement of the ssp operon by the ermB cassette. Erythromycin-resistant clones were grown on casein agar plates, and colonies with small zones of proteolysis were checked for allelic replacement by PCR and Southern blotting. The loss of SspA in one clone, AK2, was verified by Western blotting (see below).
Strains AK3 (sarA aur double mutant) and AK5 (sarA ssp double mutant) were constructed by transfer of the sarA::km mutation from PC1839 into AK1 and AK2 using the transducing phage Φ11 as described before. The mutations were confirmed by PCR.
Western blotting of cell wall proteins and secreted proteins.
Cell-associated FnBPs and protein A were released by lysostaphin treatment of equal numbers of bacterial cells (1.2 OD600 units) as described previously (20). Released proteins were separated in sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gels (24) and transferred to polyvinylidene difluoride-based membranes (Immobilon-P; Millipore) using a Bio-Rad Mini Trans-Blot electrophoretic transfer cell as recommended by the supplier. Monoclonal antibodies (goat) against the fibronectin-binding D domains of FnBPA and FnBPB from S. aureus (a gift from L. K. Rantamäki, University of Helsinki, Finland) were used and were detected with horseradish peroxidase (HRP)-conjugated rabbit anti-goat antibodies (DAKO A/S, Glostrup, Denmark). Protein A was detected by Western ligand blotting using IgG2a from mouse (monoclonal antibody to human CD14; Nordic BioSite Inc.). IgG2a bound to protein A was detected using HRP-conjugated sheep anti-mouse antibodies (Amersham Life Science). For Western blot analysis of the metalloprotease and the serine protease, culture supernatants corresponding to a bacterial density of 0.12 OD600 units were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride-based membranes as described above. Polyclonal rabbit anti-metalloprotease and -serine protease antibodies (19) were used and detected with HRP-conjugated sheep anti-rabbit antibodies (Amersham Life Science).
Quantitation of FnBPs and protein A on Western blots was made by the Personal Densitometer SI and Image QuanT software (Molecular Dynamics).
Preliminary sequence data for the S. aureus strain COL were obtained from The Institute for Genomic Research website at http://www.tigr.org and for S. aureus strain 8325 were obtained from the University of Oklahoma Genome sequencing project website at http://www.genome.ou.edu/staph.html.
RESULTS
Analysis of fnb transcripts and cell-associated FnBPs in S. aureus DB (wild type) and the sarA mutant 11D2.
It has been reported that S. aureus sarA mutant cells bind less fibronectin than the corresponding parental cells (4, 7, 16, 43). This could be explained at least in part by the recent observation that transcription of fnbA, but not fnbB, was down-regulated in a sarA mutant (43). However, the decreased fibronectin binding in the sarA mutant could also be the result of increased production of extracellular proteases (2).
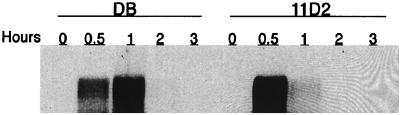
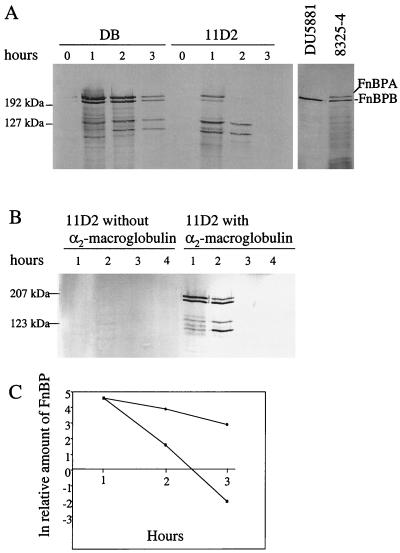
Inactivation of sarA in the clinical S. aureus strain DB had little effect on the maximum concentration of fnb mRNA, although the peak level was reached after 0.5 h (OD = 0.35) in the mutant compared to 1 h (OD = 0.7) in the wild-type strain (Fig. 1). There was no significant difference in growth rate between the strains. It should be pointed out that we used a probe that recognized both fnbA and fnbB mRNA. In accordance with the transcription analysis, synthesis of cell wall-associated FnBPs was restricted to the first hour of growth in both strains (Fig. 2A). However, in spite of roughly equal levels of fnb mRNAs, the amounts of FnBPs were 6- to 10-fold lower in sarA mutant cells than in wild-type cells (Fig. 2A). Full-length FnBPs appear on SDS-PAGE as bands with molecular masses of approximately 200 kDa (14, 15, 21). Control experiments using the fnbA knockout mutant DU5881 (Fig. 2A), together with previous experiments with the fnbA fnbB double mutant DU5883 (38), identified the largest band as FnBPA and the slightly smaller band as FnBPB. The amounts of FnBPA and FnBPB were equally reduced in the sarA mutant and the wild-type strain. The intensity of several smaller protein bands previously shown to represent degradation products (38) was also reduced in the sarA mutant. As the antiserum is specific for the fibronectin-binding domains of FnBPA and FnBPB, these results are consistent with the reduced binding of fibronectin to sarA mutant cells (6, 43).
FIG. 1.
Northern blot analysis of fnb transcripts in S. aureus DB (wild type) and the sarA mutant 11D2. Equal amounts (10 μg) of total cellular RNA isolated at the indicated time points during growth were analyzed using an RNA probe complementary to conserved regions of fnbA and fnbB.
FIG. 2.
(A) Cell wall-associated FnBPs in S. aureus DB (wild type) and sarA mutant 11D2 at different time points during growth. A control experiment with S. aureus 8325-4 (wild type) and the fnbA knockout mutant DU5881 is shown on the right. (B) sarA mutant (11D2) cells cultured with and without 0.4 U of α2-macroglobulin (protease inhibitor) ml−1. Cell surface proteins from equivalent numbers of cells were separated by SDS-PAGE and analyzed by Western immunoblotting using a monoclonal antibody against the conserved fibronectin-binding domains of FnBPA and FnBPB. Sizes of marker proteins (Kaleidoscope prestained standards) are indicated. (C) Rate of disappearance of FnBPs from cells of strain DB (circles) and 11D2 (squares) during growth. Relative densities of full-length FnBPs from panel A were plotted against time.
The slightly altered temporal expression of fnb mRNAs in the sarA mutant cannot alone explain the reduced level of FnBPs, suggesting either an impaired translation of the fnb transcripts in the sarA mutant or an increased release of FnBPs from the mutant cells. Densitometric analysis of Western blots revealed that after de novo synthesis of FnBPs seemed to have stopped (1 h of growth), the concentration of FnBPs per cell decreased about five times faster in the mutant (half-life = 10 min) than in the wild type (half-life = 46 min) (Fig. 2C). As the bacterial growth rate was the same for both strains, this suggests that FnBPs were actively released from mutant cells. However, no significant accumulation of FnBPs or larger degradation products in culture supernatants from sarA mutant cells could be demonstrated (data not shown), suggesting that the FnBPs were extensively degraded.
Since the production of proteases is up-regulated in sarA mutants (2) and since FnBPs can be degraded by the staphylococcal serine protease SspA (V8) (26), it seems reasonable to believe that the decreased amount of FnBPs on 11D2 cells was the result of increased protease production. In addition to the V8 protease S. aureus produces at least three other extracellular proteases (Aur, Scp, and SspB), which are all up-regulated at the transcriptional level in strain 11D2 (data not shown) and could be responsible for the degradation of FnBPs. To test this, strain 11D2 was cultivated in the presence of α2-macroglobulin, a universal protease inhibitor that inhibits the activity of all major staphylococcal proteases (26, 35; unpublished results), or the cysteine protease inhibitor E64, which inhibits the staphylococcal cysteine proteases (18, 35, 36). At least 16 times more cell-associated FnBPs were found on cells grown in the presence of α2-macroglobulin than on cells grown in the absence of inhibitor (Fig. 2B). The addition of the inhibitor had no effect on bacterial growth rate (data not shown). No increase in the amount of FnBPs could be seen when the sarA mutant was cultivated in the presence of E64 (data not shown), indicating that cysteine proteases are not involved in the degradation of FnBPs. The amount of FnBPs on sarA mutant cells grown in the presence of α2-macroglobulin was roughly the same as that on wild-type cells grown without protease inhibitor (Fig. 2A), which is in agreement with the roughly equal levels of fnb mRNAs in these strains (Fig. 1).
Effect of ssp and aur knockout mutations on FnBP levels.
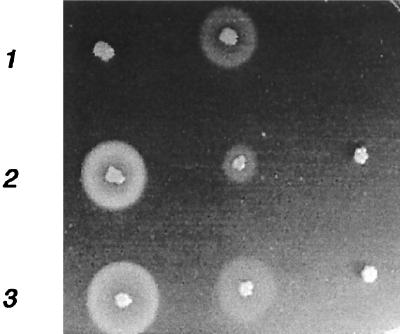
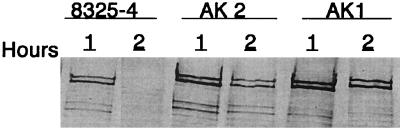
Our results indicate that serine protease and aureolysin are the major players in the degradation of FnBPs. Specific knockout mutants were therefore constructed. Because of difficulties in genetically manipulating strains DB and 11D2, the mutations were made in the prototype S. aureus strain 8325-4, which produces high amounts of proteases and possesses relatively low levels of cell wall-associated FnBPs. The serine protease gene (sspA) is part of an operon containing two additional open reading frames, one (sspB) coding for cysteine protease and the other (sspC) coding for a 12-kDa cytoplasmic protein with unknown function (2, 36). The aureolysin gene, on the other hand, seems to be mono-cistronic. The ssp operon and the metalloprotease gene, aur, were inactivated by allelic replacement as described in Materials and Methods. Mutants AK1 (aur) and AK2 (ssp) showed dramatically reduced zones of proteolysis on casein agar plates (Fig. 3). Inactivation of either protease gene resulted in the same (three- to fivefold, in three different experiments) increase in the amount of cell wall-associated FnBPs as for the wild-type strain and a slower disappearance of FnBPs from the cell surface (Fig. 4). Inactivation of ssp and aur in the sarA mutant PC1839 gave similar but less-pronounced results (data not shown). As the serine protease is produced as an inactive proenzyme which is activated through cleavage by the metalloprotease (8), inactivation of aur also leads to the loss of serine protease activity. As inhibition of cysteine proteases had no effect on FnBP levels and as the increase in FnBPs was roughly the same in AK1 and AK2, it can be concluded that the serine protease is the most important protease for the degradation of FnBPs.
FIG. 3.
Zones of proteolysis around S. aureus strains grown on a casein agar plate. Row 1, DB (wild type) and 11D2 (sarA mutant); row 2, 8325-4 (wild type), AK2 (ssp mutant), and AK1 (aur mutant); row 3, PC1839 (sarA mutant), AK5 (ssp sarA double mutant), and AK3 (aur sarA double mutant).
FIG. 4.
Cell wall-associated FnBPs in S. aureus 8325-4 (wild type), AK2 (ssp mutant), and AK1 (aur mutant) harvested after 1 and 2 h of growth in BHI broth. Cell surface proteins from equivalent numbers of cells were separated by SDS-PAGE and analyzed by Western immunoblotting using a monoclonal antibody against the conserved fibronectin-binding domains of FnBPA and FnBPB.
Effect of serine protease and metalloprotease on protein A.
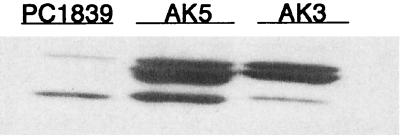
Previous studies in our laboratory have shown that the increased transcription of spa (protein A) in the sarA mutant PC1839 was not accompanied by a corresponding increase in protein A production (42). To determine whether this was due to degradation of protein A by extracellular proteases, the aur and ssp knockout mutations were transferred by phage transduction to the sarA mutant PC1839 to form strain AK3 (aur sarA mutant) and AK5 (ssp sarA mutant), respectively. Mutations were verified by PCR using primers specific for the inserted ermB gene and the flanking DNA regions. Compared to the parental strain PC1839, AK5 had a slightly smaller zone of proteolysis that was less dense (Fig. 3). Strain AK3, on the other hand, showed no zone of proteolysis. Both mutants showed a 10- to 20-fold increase in cell wall-associated protein A compared to the level in the parental strain PC1839 (Fig. 5). All immunoglobulin-binding bands seen in Fig. 5 represent protein A as they were absent in a spa knockout mutant (data not shown). Inactivation of ssp or aur in strain 8325-4 resulted in comparable increases in protein A (data not shown). Since inactivation of the ssp operon (sspA and sspB) resulted in roughly the same increase in protein A as inactivation of aur did, it can be concluded that SspA is the most important enzyme in the degradation of protein A.
FIG. 5.
Cell wall-associated protein A in postexponential-phase (4 h) cells of S. aureus PC1839 (sarA mutant), AK5 (ssp sarA double mutant), and AK3 (aur sarA double mutant). Cell surface proteins from equivalent numbers of cells were separated by SDS-PAGE and analyzed by Western ligand blotting using mouse IgG.
Transcription of ssp and hld (RNAIII) in DB (wild type) and 11D2 (sarA).
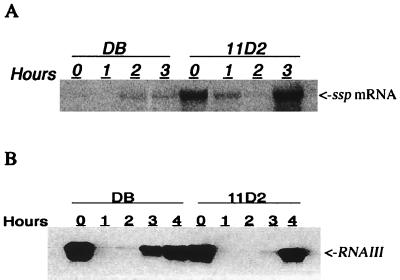
As the serine protease appears to be responsible for the degradation of FnBPs, transcription of sspA in S. aureus strains DB and 11D2 was analyzed. Northern blot analysis confirmed that transcription of the ssp operon was up-regulated in the sarA mutant strain 11D2 compared to that in wild-type strain DB (Fig. 6A). In mutant cells the level of ssp transcript increased dramatically during the postexponential phase of growth (2 to 3 h). However, significant amounts of ssp transcript were seen at time zero, suggesting that SspA was produced during the early exponential phase of growth at the same time that FnBPs were produced (Fig. 1). Increased protease production by the sarA mutant 11D2 was also indicated by large zones of precipitation around bacteria grown on casein agar plates (Fig. 3).
FIG. 6.
Northern blot analysis of ssp mRNA (A) and RNAIII (B) in S. aureus DB (wild type) and the sarA mutant 11D2 at different time points during growth.
As transcription of ssp is stimulated by agr (42), levels of RNAIII were also analyzed. Transcription of ssp during the early exponential phase of growth of 11D2 was consistent with the presence of RNAIII in cells taken at time zero (Fig. 6B). Although the initial levels of RNAIII were essentially the same in both strains, transcription of ssp was severely repressed in the wild-type strain, probably because of high SarA levels. As expected, levels of RNAIII were very low during the mid-exponential phase of growth (1 and 2 h) and increased dramatically during the late exponential and postexponential phases. Notably, the increase in RNAIII appeared later (OD600 = 12.0) in the sarA mutant than in the wild-type strain (OD600 = 8.4), consistent with the expression of RNAIII being positively controlled by sarA (16).
DISCUSSION
In the present study we have demonstrated that the decreased expression of FnBPs in 11D2 (sar mutant) cells compared to that of the wild-type cells (DB) was due to increased proteolytic degradation of FnBPs rather than to decreased transcription of the fnb genes. We also found that the decreased amount of protein A in sarA mutant cells was the result of increased protease production. The major protease responsible for the degradation of FnBPs and protein A appeared to be the staphylococcal serine protease (V8).
Contrary to the present results, it was recently shown that transcription of fnbA, but not fnbB, was down-regulated in a sarA mutant (43). In strain Newman, used in a previous study (43), FnBPA appeared to be the dominating FnBP, while in strains DB and 8325-4, which we used, roughly equal amounts of FnBPA and FnBPB were produced (Fig. 2A). In strain 8325-4, FnBPA and FnBPB also seemed to contribute equally to the adherence of S. aureus cells to fibronectin-coated surfaces (15). The different relative levels of FnBPA and FnBPB and the different transcription patterns of fnbA and fnbB with respect to their regulation by sarA may be explained by differences between the fnb promoter sequences (16, 43). Strain-dependent differences may, however, also be due to variations in the levels of expression of sarA and other regulators (e.g., agr). Expression of both fnbA and fnbB was negatively controlled by agr in strain 8325-4 (38), while only fnbA appeared to be regulated by agr in strain Newman (43).
The observation that cultivation of S. aureus in the presence of E64 did not significantly affect the amount of cell wall-associated FnBPs strongly suggests that cysteine proteases (SspB and Scp) were not involved in the degradation of FnBPs. The increase in FnBPs seen in the ssp knockout mutant was therefore most likely due to the loss of SspA activity. This was supported by the observation that deletion of the metalloprotease that is required for the activation of SspA resulted in the same increase in FnBPs as the ssp deletion and by the finding that SspA alone can degrade cell wall-associated FnBPs (26). However, a nonpolar sspA mutation in strain SP6391 (derived from 8325-4) did not enhance fibronectin binding, suggesting that the degradation of FnBPs might be the result of the combined activity of two or more proteases (36). However, this discrepancy could also be explained by the observation that binding of soluble fibronectin is not proportional to the amount of cell wall-associated FnBPs. A 16-fold increase in FnBPs resulted in only a twofold increase in fibronectin binding (38). A similar discrepancy between the level of cell-bound FnBPs and binding of soluble fibronectin can also be deduced from data reported previously (43).
FnBPs, protein A, and other cell surface proteins are covalently linked to the cell wall peptidoglycan (40). The release of FnBPs and protein A from the cell wall could therefore also be due to increased autolysin activity. In sarA mutants both proteases and autolysin activity are up-regulated, suggesting that the decreased levels of FnBPs and protein A might be the combined result of increased proteolysis and cell wall turnover. However, the increased levels of protein A in the ssp sarA and aur sarA double mutants compared to that in the sarA mutant (Fig. 5) strongly indicate that proteolytic degradation is the most important factor.
Previous reports clearly indicated that the production of FnBPs is inhibited by agr and stimulated by sarA (38, 43), while the production of proteases is inhibited by sarA and stimulated by agr (2, 42). This means that the production of serine protease would generally be down-regulated when fnbA and fnbB are expressed. Under conditions where no extracellular proteases are produced, cell wall-associated FnBPs appeared to be very stable and disappeared from the bacterial surface at a rate that was proportional to the cell doubling time, meaning that complete down-regulation of the fibronectin-binding phenotype would take several generations. It therefore seems reasonable that FnBPs can be actively released from the bacterial surface soon after de novo synthesis has been turned off. Depending on the regulation of sarA and agr expression and the relative levels of sarA and RNAIII, the amount of FnBPs on the bacterial surface can be modulated to meet the specific requirements during the course of infection.
In the case of protein A the situation seems more complicated, as sarA represses transcription of both spa and ssp. Thus, increased protein A production, as a result of down-regulation of sarA, will be counteracted by the concomitant increase in protease production. However, as agr is also positively regulated by sarA (7), down-regulation of sarA might result in a decreased level of RNAIII, which in turn results in decreased protease production and therefore less degradation of protein A. This is consistent with the observation that an agr sarA double mutant produced higher amounts of protein A than a sarA mutant (42).
In our study we demonstrated that cell wall-associated FnBPs and protein A can be released through the activity of the staphylococcal serine protease. The relevance of this in adaptation to various environmental conditions during the infection process remains to be determined.
ACKNOWLEDGMENTS
We thank Agneta Wahlquist and Lena Norénius for skillful technical assistance. We are also grateful to A. Cheung for providing us with strains DB and 11D2.
This work was supported by grant 4513 from the Swedish Medical Research Council.
REFERENCES
- 1.Arvidson S. Extracellular enzymes. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 379–385. [Google Scholar]
- 2.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar−lagr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung A L, Fischetti V A. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infect Immun. 1988;56:1061–1065. doi: 10.1128/iai.56.5.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Ying P. Regulation of alpha- and beta-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drapeau G R. Role of metalloprotease in activation of the precursor of staphylococcal protease. J Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziewanowska K, Patti J M, Deobald C F, Bayles K W, Trumble W R, Bohach G A. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect Immun. 1999;67:4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsgren A, Ghetie V, Lindmark R, Sjoquist J. Protein A and its exploitation. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. Vol. 2. London, United Kingdom: Academic Press Inc. Ltd.; 1983. pp. 429–480. [Google Scholar]
- 11.Forsgren A, Sjoquist J. Protein A from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol. 1966;97:822–827. [PubMed] [Google Scholar]
- 12.Foster T J, Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 13.Foster T J, McDevitt D. Surface-associated proteins of Staphylococcus aureus: their possible roles in virulence. FEMS Microbiol Lett. 1994;118:199–205. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]
- 14.Froman G, Switalski L M, Speziale P, Hook M. Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J Biol Chem. 1987;262:6564–6571. [PubMed] [Google Scholar]
- 15.Greene C, McDevitt D, Francois P, Vaudaux P E, Lew D P, Foster T J. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol Microbiol. 1995;17:1143–1152. doi: 10.1111/j.1365-2958.1995.mmi_17061143.x. [DOI] [PubMed] [Google Scholar]
- 16.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz S A, Schenning T, Heimahl A, Flock J-I. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis. 1996;174:83–88. doi: 10.1093/infdis/174.1.83. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann B, Schomburg D, Hecht H J. Crystal structure of a thiol proteinase from Staphylococcus aureus V-8 in the E-64 inhibitor complex. Acta Crystallogr. 1993;49(Suppl.):102. [Google Scholar]
- 19.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janzon L, Löfdahl S, Arvidson S. Evidence for a coordinate transcriptional control of alpha-toxin and protein A in Staphylococcus aureus. FEMS Microbiol Lett. 1986;33:193–198. [Google Scholar]
- 21.Jonsson K, Signas C, Muller H P, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 22.Kreiswirth B, Löfdahl S, Betly M J, O'Reilly M, Schleivert M P, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 23.Kuypers J M, Proctor R A. Reduced adherence to traumatized rat heart valves by low-fibronectin-binding mutant of Staphylococcus aureus. Infect Immun. 1989;57:2306–2312. doi: 10.1128/iai.57.8.2306-2312.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.McGavin M J, Raucci G, Gurusiddappa S, Hook M. Fibronectin binding determinants of the Staphylococcus aureus fibronectin receptor. J Biol Chem. 1991;266:8343–8347. [PubMed] [Google Scholar]
- 26.McGavin M J, Zahradka C, Kelly R, Scott J E. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, Francois P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morfeldt E, Janzon L, Arvidson S, Lofdahl S. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol Gen Genet. 1988;211:435–440. doi: 10.1007/BF00425697. [DOI] [PubMed] [Google Scholar]
- 29.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 30.Novick R P. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 31.Patel A H, Nowlan P, Weavers E D, Foster T. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect Immun. 1987;55:3103–3110. doi: 10.1128/iai.55.12.3103-3110.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 33.Patti J M, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Hook M. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson P K, Verhoef J, Sabath L D, Quie P G. Effect of protein A on staphylococcal opsonization. Infect Immun. 1977;55:3103–3110. doi: 10.1128/iai.15.3.760-764.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potempa J, Dubin A, Korzus G, Travis J. Degradation of elastin by a cysteine proteinase from Staphylococcus aureus. J Biol Chem. 1988;263:2664–2667. [PubMed] [Google Scholar]
- 36.Rice K, Peralta R, Bast D, de Azavedo J, McGavin M J. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect Immun. 2001;69:159–169. doi: 10.1128/IAI.69.1.159-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Saravia-Otten P, Muller H P, Arvidson S. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol. 1997;179:5259–5263. doi: 10.1128/jb.179.17.5259-5263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenk S, Laddaga R A. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 40.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 41.Signas C, Raucci G, Jonsson K, Lindgren P E, Anantharamaiah G M, Hook M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tegmark K, Karlsson A, Arvidson S. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2000;37:398–409. doi: 10.1046/j.1365-2958.2000.02003.x. [DOI] [PubMed] [Google Scholar]
- 43.Wolz C, Pöhlmann-Dietze P, Steinhuber A, Chien Y-T, Manna A W, van Wamell W, Cheung A L. agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol. 2000;36:230–243. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]