Abstract
Simple Summary
Next–Generation Sequencing (NGS) has provided a deeper genetic understanding of acute myeloid leukemia (AML) that has been recently incorporated into AML classification and risk–stratification guidelines. Single molecular analysis has become inefficient and molecular testing based on NGS is emerging as an irreplaceable diagnostic tool in clinical settings. The PETHEMA cooperative group has constituted a nationwide NGS network with centralized analysis in seven high–skilled laboratories. The study of molecular profiles in the “real–life” PETHEMA cohort supports the increasing role of NGS on the clinical management of AML patients.
Abstract
Next–Generation Sequencing (NGS) implementation to perform accurate diagnosis in acute myeloid leukemia (AML) represents a major challenge for molecular laboratories in terms of specialization, standardization, costs and logistical support. In this context, the PETHEMA cooperative group has established the first nationwide diagnostic network of seven reference laboratories to provide standardized NGS studies for AML patients. Cross–validation (CV) rounds are regularly performed to ensure the quality of NGS studies and to keep updated clinically relevant genes recommended for NGS study. The molecular characterization of 2856 samples (1631 derived from the NGS–AML project; NCT03311815) with standardized NGS of consensus genes (ABL1, ASXL1, BRAF, CALR, CBL, CEBPA, CSF3R, DNMT3A, ETV6, EZH2, FLT3, GATA2, HRAS, IDH1, IDH2, JAK2, KIT, KRAS, MPL, NPM1, NRAS, PTPN11, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1 and WT1) showed 97% of patients having at least one mutation. The mutational profile was highly variable according to moment of disease, age and sex, and several co–occurring and exclusion relations were detected. Molecular testing based on NGS allowed accurate diagnosis and reliable prognosis stratification of 954 AML patients according to new genomic classification proposed by Tazi et al. Novel molecular subgroups, such as mutated WT1 and mutations in at least two myelodysplasia–related genes, have been associated with an adverse prognosis in our cohort. In this way, the PETHEMA cooperative group efficiently provides an extensive molecular characterization for AML diagnosis and risk stratification, ensuring technical quality and equity in access to NGS studies.
Keywords: acute myeloid leukemia, Next–Generation Sequencing, cross–validations, mutational profile, genomic classification, clinical validation
1. Introduction
Introduction of Next–Generation Sequencing (NGS) into routine molecular diagnosis has provided deep molecular knowledge of acute myeloid leukemia (AML). These findings have allowed for the refinement of classification and risk stratification systems based on recurrent genetic abnormalities.
In 2016, Papaemmanuil et al. proposed the first genomic classification of AML that identifies 11 molecular classes, each with distinct diagnostic features and clinical outcomes [1]. This classification has been recently revised and updated in Tazi et al., 2022, proposing 16 molecular classes based on cytogenetics and the mutational status of 32 genes [2]. The importance of genomic characterization has also been reflected in the recently revised World Health Organization (WHO) Classification [3], new International Consensus Classification (ICC) [4] and European LeukemiaNet (ELN) risk stratification [5], which prioritize genetic abnormalities to establish diagnosis and prognosis to evaluate measurable residual disease (MRD) and to select treatment.
In this situation, molecular analysis by single–gene techniques has become inefficient in order to provide a complete characterization of AML. In contrast, NGS represents a more sensitive tool to capture all the relevant molecular markers in one assay and is widely recommended to study the molecular landscape of this disease [6].
NGS implementation to perform accurate diagnosis in AML is currently demanded by physicians and patients. However, introduction of NGS into clinical routine faces novel challenges [7]. NGS requires large batches of samples in order to be cost–effective, workflows are time–consuming, and interpretation needs highly qualified specialists. Moreover, the diversity of NGS panels, platforms and quality control criteria might prevent the success of the approach [8]. Hence, to efficiently introduce NGS into routine molecular diagnostics, it is necessary to establish quality requirements and to standardize gene panels and variant reporting.
In order to provide comprehensive NGS studies to AML patients and to guarantee equity of access, the PETHEMA cooperative group (Programa Español de Tratamientos en Hematología) has established a nationwide network of central laboratories aimed to harmonize NGS results under consensual criteria in newly diagnosed and relapsed/refractory AML patients [9].
This study summarized the NGS–AML project (NCT03311815), reporting quality control assays and the molecular profile of 2668 AML patients reported in the PETHEMA AML registry. We show co–occurring and mutual exclusion relationships among genes and distinct molecular profiles according to disease stage, age and sex and genomic classification in the “real–life PETHEMA cohort”.
2. Materials and Methods
2.1. Development of the Diagnostic Platform
Implementing NGS studies in the routine molecular diagnosis of AML patients requires specialization, budgetary stability and logistical support. The PETHEMA cooperative group established a nationwide network of NGS studies for fast and standardized molecular diagnosis of AML. This strategy aims to provide coverage of NGS studies to 38.5 million inhabitants distributed in geographical areas ranging from 2.2 to 8.9 million habitants. For this purpose, seven centers with logistical and technical capacity for the management of a high number of samples were designed as reference laboratories for the centralization of samples submitted by PETHEMA institutions in each area. In this way, a large territory and population was covered. The platform was supported by PETHEMA in logistical management, as well as, closer and well–established relationships between the sample referral institution and the assigned central laboratory. The designated reference centers for NGS analysis concentrate a large number of AML samples, allowing for the rapid completion of the sequencing runs and their management by highly specialized staff.
2.2. Study Design and Reference Laboratories
We show a prospective, multi–center, non–interventional and translational biomedical research, performed in seven Spanish PETHEMA reference laboratories: Hospital Universitario La Fe (HULF, Valencia, Spain), Hospital Universitario de Salamanca (HUS, Salamanca, Spain), Hospital Universitario 12 de Octubre (H12O, Madrid, Spain), Hospital Universitario Virgen del Rocío (HUVR, Sevilla, Spain), Hospital Universitario Reina Sofía (HURS, Córdoba, Spain), Hospital Universitario de Gran Canaria Dr. Negrín (HUDN, Las Palmas de Gran Canaria, Spain), CIMA LAB Diagnostics (UNAV, Pamplona, Spain).
2.3. Consensus Genes Establishment
The development of the diagnostic platform required several meetings to coordinate criteria on which genes should be analyzed based on their clinical relevance in AML. After extensive bibliographic revision of current molecular basis of AML, all reference laboratories should assess by NGS the mutational status of genes that define the diagnosis and prognosis as well as guide treatment options (ASXL1, BCOR, CEBPA, EZH2, FLT3, IDH1, IDH2, NPM1, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, ZRSR2 and TP53). Moreover, there was a recommendation for the study of other genes with proven evidence on their relevance in AML pathogenesis (ABL1, BRAF, CALR, CBL, CSF3R, DNMT3A, ETV6, GATA2, HRAS, JAK2, KIT, KRAS, MPL, NRAS, PTPN11, SETBP1, TET2 and WT1).
The establishment of consensus genes has enabled laboratories to work with the NGS platforms and panels according to their individual requirements, which have largely enabled the development and maintenance of the diagnostic platform. NGS panels and platforms used by each center are described in Supplementary Materials Tables S1 and S2.
2.4. NGS Standardization Procedures and Cross–Validation Rounds
The diagnostic platform established a quality control assay by exchanging control samples among reference laboratories every 9–12 months. To date, three cross validation (CV) rounds have been performed. As previously reported, in the first and second CV rounds, minimum quality parameters [uniformity (>85%) and mean read depth of 1000X] and consensus recommendations for variant report [All centers should report: (1) mutations in relevant genes for clinical guidelines, targeted therapy and risk stratification and (2) all pathogenic variants detected with VAF >5% excepting those described at hotspot regions which will be reported up to 1% VAF] were stablished. For both CV rounds, 10 samples harboring 54 variants were distributed. In first CV round, VAF for all variants was >5% while in the second CV round, 5 from 30 total variants had low VAF ranging from 1.8–4.9%. In the third CV round, to explore the accuracy and reliability of low VAF variants (<5%) detection, 4 samples harboring 32 variants (11 VAF < 5%) were analyzed, according to previously established criteria [9].
2.5. Patients and Inclusion Criteria
All adult patients (≥18 years) with newly diagnosed or relapsed/refractory AML (excluding acute promyelocytic leukemia) according to the World Health Organization criteria (2008 and 2016), regardless of the treatment received, were eligible for mutational profile study by NGS. The Institutional Ethics Committee for Clinical Research of each institution approved this study. Written informed consent in accordance with the recommendations of the Declaration of Human Rights, the Conference of Helsinki, and institutional regulations were obtained from all patients.
2.6. Clinical Validation
Clinical validation was performed based on the new genomic classification which proposes unified framework for disease classification and risk–stratification in AML based on cytogenetic analysis and an NGS–panel of 32 genes [2]. Molecular class defining genes were: NPM1, TP53, WT1, CEBPA, DNMT3A, IDH1, IDH2, ZRSR2, U2AF1, SRSF2, SF3B1, ASXL1, STAG2, BCOR, RUNX1, EZH2, MLL, PHF6, SF1, NF1, CUX1, SETBP1, FLT3 and TET2. * Bold genes represent PETHEMA consensus genes.
This new classification categorizes AML in 16 molecular classes with different prognostic values and encompass the established WHO entities: “WHO2016 set 1” [inv(16), t(8;21) and NPM1] and “WHO2016 set 2” [t(11;x), t(6;9), inv(3) and CEBPAbi]; and also novel categories: “TP53 and complex karyotype (CK)”, “sAML1” (Mutated SRSF2, SF3B1, U2AF1, ASXL1, EZH2, RUNX1 or SETBP1), “sAML2” (More than one mutations in sAML1 genes including DNMT3A and TET2), “WT1”, “Trisomies”, “DNMT3A + IDH1/2”, “Not class defining mutations (mNOS)” and “No events” category. Since our study excludes acute promyelocytic leukemia, category “t(15;17)” is not applicable.
This classification also proposes an integrated risk score based on the 16 molecular classes: “NPM1”, “inv(16)”, “t(8;21)”, “CEBPAbi” and “No events” define the favorable risk group; “sAML1”, “t(6;9)”, “WT1”, “mNOS”, “t(11;X)”, “DNMT3A–IDH1/2” and “trisomies” the intermediate and “TP53–CK”, “sAML2” and “inv(3)” the adverse risk group. Genomic groups and sub–classifications are summarized in Supplementary Table S3. Internal tandem duplications (ITD) in FLT3 are the only genetic alterations with independent prognosis value from class membership. These mutations were not considered as “class defining” alterations as they are represented in all classes but modulate risk groups classification as follows: In the favorable risk group, patients with mutated NPM1 who also harbored a FLT3–ITD mutation were reclassified to the intermediate risk group. Similarly, a FLT3–ITD mutation reclassifies intermediate risk patients to an adverse risk group. Moreover, in order to assess the prognostic impact of TP53 configurations, we classified patients according TP53 mono–allelic or multi–hit as described by Tazi et al.: Mono–allelic: One TP53 mutation with VAF ≤ 65%; and multi–hit: Two TP53 mutations or one TP53 mutation with VAF > 65% or one TP53 mutation + del(17).
2.7. Statistics
All statistics were performed using SPSS version 22 (IBM, Armonk, NY, USA) and GraphPad Prism 4 (GraphPad, La Jolla, CA, USA) software programs. Chi square test was used to assess associations between categorical variables. Survival analyses were performed using the Kaplan–Meier method and the log–rank test. The Cox proportional–hazards model was used to evaluate the risk of death among groups. p–value (p) < 0.05 was considered as a statistically significant test.
3. Results
3.1. Third Cross Validation Round
In the third CV round, the error rate (ER) for variants with VAF > 5% decreased from previous rounds: 1st: 39%, 2nd: 14.4% and 3rd: 4.76%. However, the ER in variants with VAF < 5% increased from 28.6% (five variants with mean VAF 3.3%) to 59.6% (11 variants with mean VAF 1.2%). ER, mean VAF and standard deviation (SD) for the last CV round are summarized in Supplementary Table S4 and Figure S1. Therefore, the diagnostic platform maintained: (1) the cut–off of VAF > 5% to report clinically relevant variants and (2) the criteria to report only low VAF variants (<5%) in hotspot regions with strong clinical evidence, suggesting variant confirmation in an additional sample.
3.2. Baseline Demographics and Molecular Profile in NGS–AML Protocol Cohort
From October 2017 to October 2019, NGS analyses were performed in 1631 samples from 1471 AML patients enrolled in the NGS–AML protocol (NCT03311815), with available clinical date (i.e., treatment approach). Disease status at samples collection was: 1268 diagnosis (DX), 204 relapse (REL) and 159 refractoriness (RES) (Table 1).
Table 1.
Diagnosis cohort (N = 1268). Demographic and baseline characteristics.
| Characteristic | Mean | Median | Range | N | (%) |
|---|---|---|---|---|---|
| Age, years | 64.9 | 67.7 | 18–98 | 1268 | 100 |
| <65 | 540 | 42.6 | |||
| ≥65 | 728 | 57.4 | |||
| Sex | 1268 | 100 | |||
| Male | 712 | 56.2 | |||
| Female | 556 | 43.8 | |||
| ECOG | 1075 | 100 | |||
| 0 | 420 | 39.1 | |||
| 1 | 452 | 42.0 | |||
| 2 | 135 | 12.6 | |||
| 3 | 53 | 4.9 | |||
| 4 | 15 | 1.4 | |||
| Not available | 193 | ||||
| WBC (×109/L) | 32.8 | 8.8 | 0.24–407 | 1118 | |
| BM blast cells, % | 53.4 | 52.0 | 0–100 | 1026 | |
| Creatinine, mg/dL | 1.1 | 0.90 | 0.28–10.3 | 1071 | |
| MRC cytogenetic profile | 1011 | 100 | |||
| Favorable | 57 | 5.6 | |||
| Intermediate | 178 | 17.6 | |||
| Unfavorable | 269 | 26.6 | |||
| Normal karyotype | 507 | 50.1 | |||
| Not available | 257 | ||||
| AML FAB subtype | 715 | 100 | |||
| M0 | 88 | 12.3 | |||
| M1 | 144 | 20.1 | |||
| M2 | 126 | 17.6 | |||
| M4 | 173 | 24.2 | |||
| M5 | 144 | 20.1 | |||
| M6 | 31 | 4.3 | |||
| M7 | 9 | 1.3 | |||
| Not available | 553 | ||||
| Therapeutic approach | 1268 | 100 | |||
| Intensive | 695 | 54.8 | |||
| Non–intensive | 513 | 40.5 | |||
| Supportive care only | 60 | 4.7 | |||
| Type of AML | 1268 | 100 | |||
| De novo | 920 | 72.6 | |||
| Secondary | 348 | 27.4 |
An additional cohort of 1225 samples analyzed by NGS between November 2019–May 2021 has been included only for molecular characterization of AML: 1166 (DX), 47 (REL) and 12 (RES). Only those samples that met NGS quality requirements established by the diagnostic platform were included in the study. Overall, 2856 samples were analyzed (2434 DX, 251 REL, and 171 RES).
3.3. Summary Mutation Profile
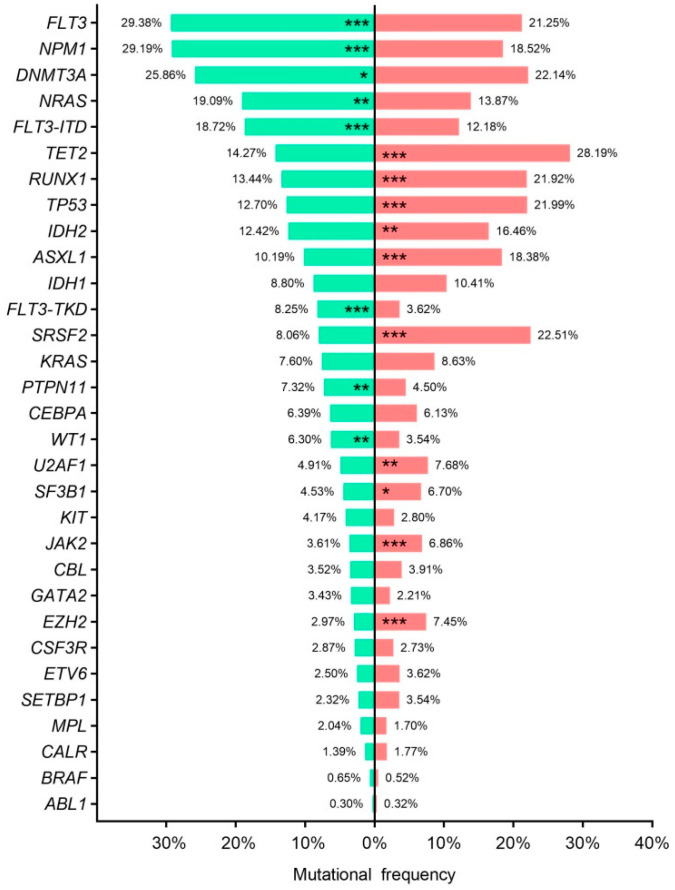
In the global cohort (N = 2856 samples), 7768 variants were detected. A total of 96.5% of samples showed at least 1 mutation, mean 2.7 mutations/sample (range 0–9). Most patients had three variants (21.1%), followed by patients with two (20.9%) and four (17.8%). FLT3 (24.6%), DNMT3A (24.3%), NPM1 (22.4%) and TET2 (21.6%) were the most frequently mutated genes (Supplementary Figure S2). According to ELN–2022 [5], 85.3% of patients at diagnosis showed at least one mutation in clinically relevant genes to establish the diagnosis, prognosis or to select treatment.
3.4. Co–Mutations and Exclusivity Patterns
NPM1, FLT3 and DNMT3A were significantly co–mutated for all combinations (p < 0.001). PTPN11 and NPM1 showed a strong association (p < 0.001) as well as PTPN11–DNMT3A (p = 0.0017) and PTPN11–FLT3 (p = 0.024). Mutations in NPM1 were highly associated with mutated IDH1 (p < 0.001) but were exclusive with R172–IDH2 (p < 0.001). In contrast, DNMT3A mutations were highly associated with both mutated IDH1 and IDH2 (p < 0.001). CEBPA was frequently co–mutated with GATA2 (p < 0.001); and ASXL1, RUNX1 and SRSF2 were strongly associated with each other (p < 0.001).
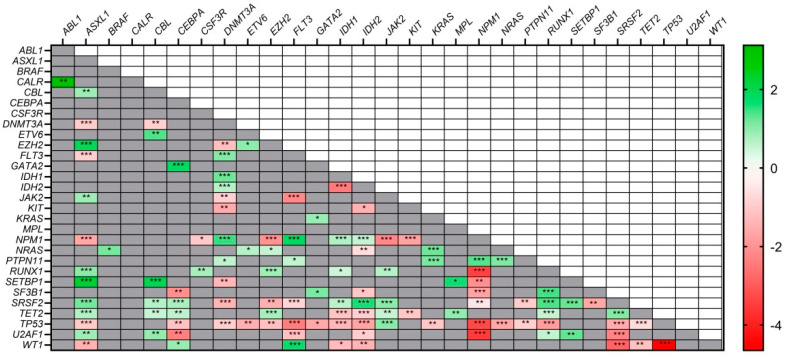
On the other hand, mutations in TP53 were the most exclusive with all the analyzed genes (p < 0.05). Mutations in IDH1 and IDH2 were also mutually exclusive of each other (p < 0.001). Mutated NPM1 was highly exclusive with mutations in: RUNX1 (p < 0.001), SRSF2 (p < 0.001) and ASXL1 (p < 0.001). The main association and exclusivity patterns for all genes are shown in Figure 1.
Figure 1.
Co–occurrence and mutual exclusivity patterns among genes. Red: exclusive relationship; Green: co–occurring relationship. Higher color intensity indicates stronger association: * p < 0.05, ** p < 0.01, *** p < 0.001.
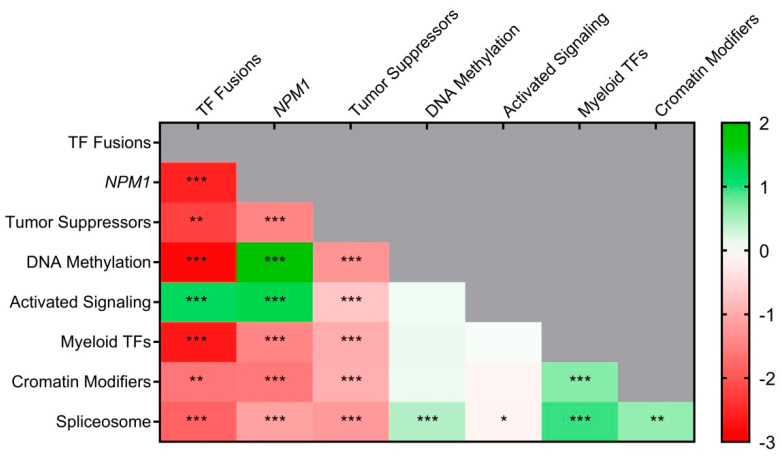
We also identified co–occurring and exclusivity patterns according to functional categories of AML [10]. Genes were grouped into nine categories based on their biological function (Supplementary Table S5). We identified commonly co–occurring events between “transcription–factor (TF) fusions” and “activating signaling genes” (p < 0.001). Associations were also found between “NPM1” with “DNA–methylation genes” (p < 0.001) and “activating signaling genes” (p < 0.001)” as well as co–occurring events between “Spliceosome–genes” with “myeloid TFs” (p < 0.001), “chromatin modifiers” (p < 0.01) and “DNA–methylation genes” (p < 0.001). On the other hand, several mutually exclusive relationships were observed: “NPM1” mutations were highly exclusive with “myeloid TFs” (p < 0.001), “Spliceosome–genes” (p < 0.001), and “chromatin–modifying” (p < 0.001); mutations in “Spliceosome–genes” were highly exclusive with “TF fusions” (p < 0.001) and remarkably, the “Tumor suppressor–genes” category was highly exclusive with all other functional categories (p < 0.001) (Figure 2).
Figure 2.
Heatmap of association and exclusivity patterns among functional categories. Red: exclusivity relationship; Green: co–occurring relationship. Higher color intensity indicates stronger association. * p < 0.05, ** p < 0.01, *** p < 0.001.
3.5. Disease Stage Mutational Profile
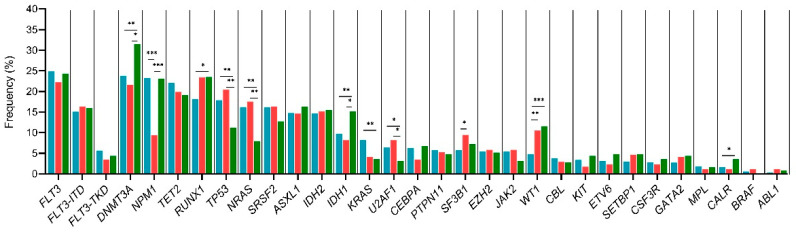
Subgroup analyses based on disease stage (DX or REL or RES) showed statistically significant results: NPM1 (p < 0.001) and signaling pathway genes such as KRAS (p = 0.007) and NRAS (p < 0.001) were more frequently mutated at diagnosis. In refractory AML, WT1 (p < 0.001) was more frequent, meanwhile relapse AML exhibited more mutations in RUNX1 (p = 0.037), DNMT3A (p = 0.018), IDH1 (p = 0.017) and WT1 (p < 0.001). Mutational frequency and p–values for each correlation are described in Supplementary Table S6A, Figure 3.
Figure 3.
Mutational frequency according to disease stage. Blue bars: Diagnosis, green bars: Relapse and red bars: refractoriness. * p < 0.05, ** p < 0.01, *** p < 0.001. ITD: internal tandem duplication; TKD: tyrosine kinase domain.
3.6. Age–Related Mutational Profile
NGS studies revealed distinct mutational profile in young (<65 years–old) and elderly (≥65 years–old) AML patients. The mean number of gene mutations at diagnosis was higher in older patients than younger (2.9 ± 0.04 vs. 2.5 ± 0.04; p < 0.001). Older patients also had a higher frequency of TET2 (p < 0.001), RUNX1 (p < 0.001), TP53 (p < 0.001), IDH2 (p < 0.01), ASXL1 (p < 0.001), SRSF2 (p < 0.001), U2AF1 (p < 0.01), SF3B1 (p = 0.028), JAK2 (p < 0.001) and EZH2 (p < 0.001). In contrast FLT3 (p < 0.001), NPM1 (p < 0.001), DNMT3A (p = 0.032), NRAS (p < 0.01), PTNP11 (p < 0.001) and WT1 (p < 0.001), were frequently mutated in young AML. Mutational frequencies and p–values are shown in Supplementary Table S6B, Figure 4.
Figure 4.
Age–related mutational profile. Bar chart representing mutational frequencies according to age at diagnosis. Green; <65 years old, red; ≥65 years old. * p < 0.05, ** p < 0.01, *** p < 0.001. ITD: internal tandem duplication; TKD: tyrosine kinase domain.
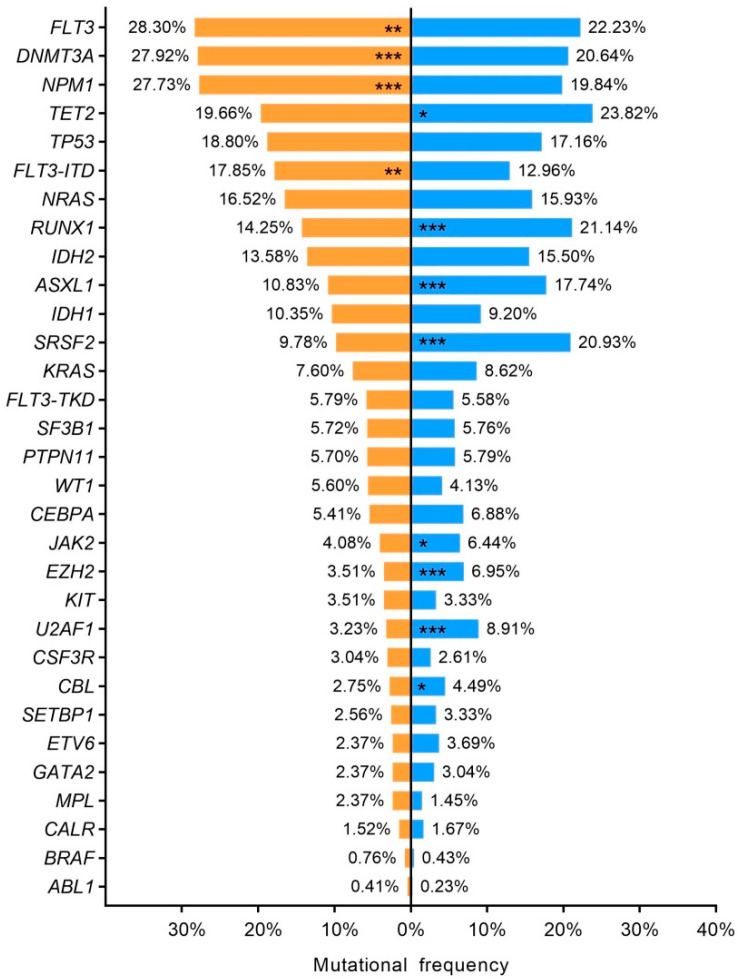
3.7. Sex–Related Mutational Profile
Our cohort showed a well–balanced distribution between male (56.2%) and female (43.8%) patients, similar to that described in previous AML cohorts [1,10]. Sex–specific mutational profiles were observed. Females harbored lower number of mutations than male patients (2.8 ± 0.04 vs. 2.6 ± 0.04; p < 0.01). Mutations in DNMT3A (p < 0.001), FLT3 (p < 0.01) and NPM1 (p < 0.001) were overrepresented in females and TET2 (p = 0.014), RUNX1 (p < 0.001), ASXL1 (p < 0.001), SRSF2 (p < 0.001), EZH2 (p < 0.001), U2AF1 (p < 0.001), JAK2 (p = 0.01) and CBL (p = 0.025) mutations in male patients (Supplementary Table S6C, Figure 5).
Figure 5.
Sex–related mutational profile. Bar chart representing mutational frequencies. Orange; female, blue; male. * p < 0.05, ** p < 0.01, *** p < 0.001. ITD: internal tandem duplication; TKD: tyrosine kinase domain.
3.8. Paired Samples and Mutation Stability
Molecular profiling of paired samples was conducted in 97 patients at diagnosis–relapse (DX–REL) and 59 patients at diagnosis–refractoriness (DX–RES). In DX–REL comparison, loss events were more frequently observed than gain events (43.7% vs. 26.4%) in relapse samples. Interestingly, 17.2% of patients showed simultaneous mutation loss and gain events, while 24.1% maintain the diagnosis’ mutational profile. The most stable mutated genes were TP53, WT1 and NPM1, with stability rates of 81.3%, 80% and 77.8%, respectively. In contrast, signaling activating genes were found to be highly unstable: KIT, FLT3–ITD and FLT3–TKD mutations, NRAS, KRAS and PTPN11 showed stability rates below 50%. Moreover, while mutations in KIT, FLT3–TKD, KRAS and PTPN11 were almost equally lost and acquired, mutations in NRAS and FLT3–ITD were predominantly lost (Supplementary Figure S3).
In refractory AML, 49.2% of patients retained the mutational status found at diagnosis. NPM1 mutations were also the most stable gene at refractory AML (83.3%). Signaling activating genes were highly unstable: NRAS, KRAS, PTPN11 and FLT3–TKD showed stability rates below 45%, being more frequent the loss of these mutations (Supplementary Figure S4).
3.9. New Genomic Classification Applied to PETHEMA–NGS Cohort
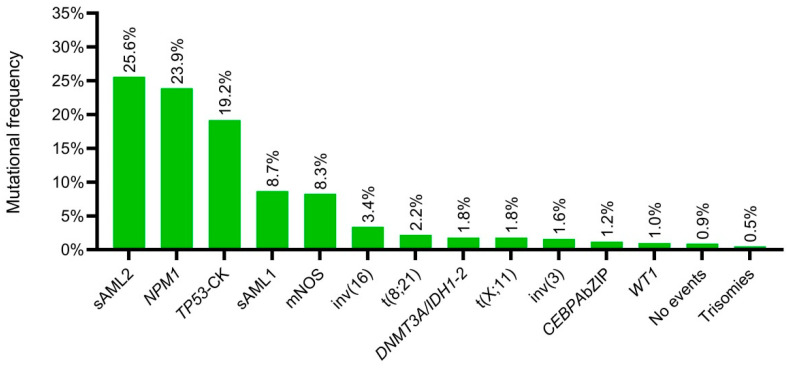
Based on the availability of clinical, cytogenetic and mutational data, 954 patients were eligible to assess the updated genomic classification by Tazi et al. [2]. The most frequently mutated classes were: “sAML2” (25.4%), “NPM1” (23.9%), and “TP53–CK” (19.2%). Other less frequent classes were: “sAML1” (8.7%) and “Not class defining mutations” (8.3%). Molecular classes’ distribution is shown in Figure 6. The “CEBPAbi” category was underrepresented in the PETHEMA cohort (0.8%). Therefore, based on novel diagnosis and prognosis classifications of AML, patients with in–frame mutations in basic leucine zipper domain of CEBPA (CEBPA bZIP) were assessed as a biological AML subgroup (1.2%). In this regard, WHO entities [inv(16), t(8;21), NPM1, CEBPA bZIP, t(11;x), t(6;9) and inv(3)] represented the 34% of PETHEMA–cohort.
Figure 6.
Molecular classes’ distribution according to the Tazi et al., 2022 genomic classification. CK: Complex karytotype. mNOS: Not class defining mutations.
3.9.1. Prognosis Value of Molecular Classes
Molecular classes have also been associated with different prognostic values. The previously established WHO categories, “inv(16)” (Median OS not reached at 42 months), “CEBPA bZIP” (Median OS not reached at 32 months) and “NPM1” [29.0 months (95%CI 19.9–38.0)] had the best outcomes while inv(3) had the worst prognostic value [4.9 months (95%CI 0.8–9.1)]. Among new molecular classes, those patients without driver mutations “No events” (Median OS not reached at 33 months) or “Not class defining mutations” [23.3 months (95%CI 11.0–35.6)] showed the best prognostic value while “TP53/CK” [5.3 months (95%CI 2.9–7.6)], “sAML2” [12.1 months (95%CI 9.9–14.2)] and WT1 [4.0 months (95%CI 0.0–18.4)] classes had the worst OS (p < 0.001) (Table 2; Supplementary Figure S5). When we evaluated the prognostic value according to the mono–allelic (N = 47; 32.9%) or multi–hit (N = 96; 67.1%) status of TP53 mutations, we did not find a different outcome between both groups [mono–allelic: 5.4 months (95%CI 0.007–10.7); multi–hit: 4.1 months (95%CI 2.9–5.3) (p = 0.088)] (Supplementary Figure S6).
Table 2.
Median overall survival and 95% CI for molecular AML classes.
| Molecular Classes | OS | (95% CI) | p | |
|---|---|---|---|---|
| Lower IC | Upper IC | |||
| inv(16) | NR | <0.001 | ||
| CEBPA bZIP | NR | |||
| No events | NR | |||
| NPM1 | 29.0 | 19.9 | 38.0 | |
| Not class defining mutations | 23.3 | 11.0 | 35.6 | |
| DNMT3A/IDH1–2 | 18.5 | 1.7 | 35.3 | |
| sAML1 | 18.1 | 12.5 | 23.7 | |
| t(8;21) | 17.5 | 3.7 | 31.3 | |
| Trisomies | 14.4 | |||
| t(X;11) | 13.2 | 0.0 | 31.3 | |
| sAML2 | 12.1 | 9.9 | 14.2 | |
| TP53–CK | 5.3 | 2.9 | 7.6 | |
| inv(3) | 4.9 | 0.8 | 9.1 | |
| WT1 | 4.0 | 0.0 | 18.4 | |
NR: Mean overall survival not reached at: “inv(16)”: 42 months; “No events”: 33 months and “CEBPA bZIP”: 31 months.
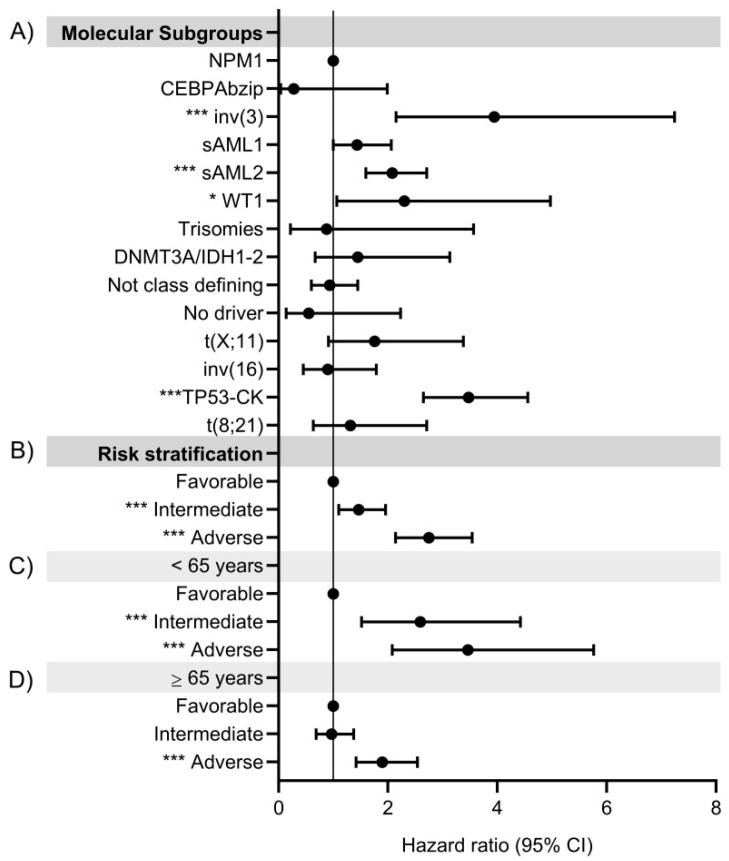
In terms of risk of death, the molecular classes “inv(3)” [3.9 (95%CI 2.1–7.2) (p < 0.001)] and “TP53/CK” [3.5 (95%CI 2.6–4.6) (p < 0.001)] showed the highest risk of death compared to “NPM1” class. Remarkably, the new established “sAML2” [2.1 (95%CI 1.6–2.7) (p < 0.001)] and “WT1” [2.3 (95%CI 1.1–5) (p < 0.05)] categories were the next with higher risk of death (Figure 7A, Supplementary Table S7A).
Figure 7.
Hazard ratio for death according to (A) New molecular subgroups in the global cohort; and new genomic risk score for: (B) Global cohort, (C) Patients < 65 years and (D) Patients ≥65 years. * p < 0.05, *** p < 0.001.
3.9.2. Integrated Risk Score
We also assessed the integrated risk score based on cytogenetic and gene mutations proposed in the new genomic classification [2]. In the evaluable cohort (N = 954), 23.8% of patients were classified to a favorable risk group, 27.1% were included in the intermediate risk group and 49.1% in the adverse risk group. We also evaluated the impact of age at diagnosis in the new genomic classification. Risk stratification distribution was significantly different between age groups: <65 years (intensive = 416; non–intensive = 10) vs. ≥65 years (intensive = 150; non–intensive = 378). Young patients showed homogeneous distribution of the different risk groups (Favorable: 31.2%, Intermediate: 32.4% and adverse: 36.4%), while older patients were predominantly included in the adverse risk group (Favorable: 17.8%, Intermediate: 22.9% and adverse: 59.3%; p < 0.001).
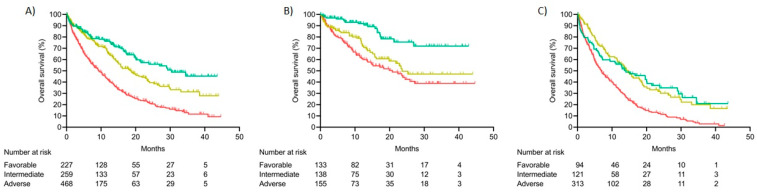
In terms of outcomes, median OS was 30.8 months (95%CI NR) in the favorable risk group, 18.5 months (95%CI 14.3–22.7) in the intermediate group and 9.4 months (95%CI 7.8–11.00) in the adverse risk group (p < 0.001) (Figure 8A). Intermediate and adverse risk patients had 1.5 (95%CI 1.1–2.0; p < 0.01) and 2.7 (95%CI 2.1–3.5; p < 0.001) increased risk of death relative to favorable risk group (Figure 7B, Supplementary Table S7B).
Figure 8.
Overall survival according to new genomic risk score: (A) Global cohort, (B) Patients < 65 years, (C) Patients ≥ 65 years.
We also found differences in terms of OS in both age groups. Median OS in favorable–risk group was not reached at 42.8 months in patients < 65 years, while intermediate and adverse risk groups reached a median OS of 23.6 (95%CI NR) months and 20.8 (95%CI 13.2–28.3), respectively (p < 0.001) (Figure 8B). In contrast, median OS of older patients was significantly decreased for all risk groups: Favorable (14.0; 95%CI 6.3–21.8), intermediate (14.6; 95%CI 11.9–17.4) and adverse (6.9; 95%CI 5.6–8.3) (p < 0.001) (Figure 8C).
Regarding risk score, young patients classified in the intermediate and adverse risk group showed 2.6 (95%CI 1.5–4.4; p < 0.001) and 3.5 (95%CI 2.1–5.8; p < 0.001) higher risk of death relative to those patients classified in the favorable risk group (Figure 7C, Supplementary Table S7C). On the other hand, patients >65 years classified in the adverse risk group showed an increased risk of death of 1.9 (95%CI 1.4–2.5; p < 0.001) compared to those classified as favorable risk. No statistically significant results were found in the risk of death of intermediate risk patients: 1.0 (95%CI 0.7–1.4; p = 0.864) (Figure 7D, Supplementary Table S7D). In general, patients >65 years had a dismal prognosis and higher risk of death compared to younger ones for all risk groups: Favorable: 4.7 (95%CI 2.8–8.0; p < 0.001), intermediate: 1.8 (95%CI 1.2–2.6; p < 0.01) and adverse: 2.6 (95%CI 2.0–3.5; p < 0.001).
Comparison between the Integrated Risk Score and 2022 ELN Risk Classification
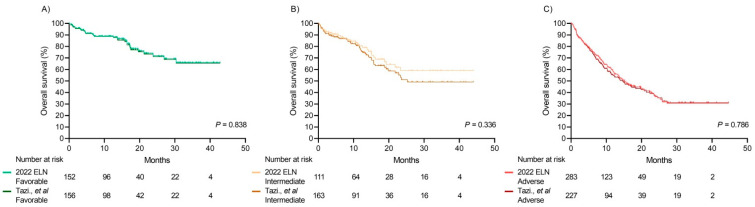
According to current 2022 ELN risk stratification, we selected 546 fit patients for a tentative comparison with the AML genomic classification risk score. We did not find a distinct OS (2022 ELN vs. Tazi et al.) for favorable (Median OS not reached in both groups; p = 0.839), intermediate (Median OS not reached vs. 25.3 months; p = 0.336) or adverse–risk patients (Median OS 15.2 months vs. 14.7 months; p = 0.786) according to both classifications (Figure 9).
Figure 9.
Overall survival curves according to 2022 ELN risk classification and Tazi et al. [2] genomic AML classification: (A) Favorable, (B) Intermediate and (C) Adverse risk groups.
4. Discussion
NGS has become the preferred technology to capture the heterogeneous molecular landscape of AML [2,3,4,5]. These approaches have been rapidly adopted as a potential routine tool for molecular diagnosis in AML patients [11]. However, its translation into clinical practice is hampered by specific requirements, such as the necessity of highly skilled laboratories, the increased cost compared to single–gene assays and the expected high turnaround time [7,12]. In this scenario, our results show that an NGS diagnostic platform, established by the PETHEMA cooperative group, was able to provide NGS reliable results with all relevant molecular data currently required for diagnosis and prognosis stratification and therapy choice. In addition, this kind of cooperative study allows for the assessment of the genetic heterogeneity of AML in large cohorts of patients and provides an extensive quality molecular data to evaluate current genomic knowledge in “real–life” AML cohorts.
Due to the rapid NGS implementation into routine molecular diagnosis of AML, NGS workflows and quality specifications are heavily reliant on laboratory specific procedures [13,14]. Therefore, current NGS analyses are characterized by the lack of standardized procedures, the diversity of quality metrics criteria, and the high variability of the assessed genes and variant reporting criteria [15]. To address the need for harmonization procedures, the PETHEMA cooperative group implemented the first Spanish nationwide NGS testing strategy. Regular rounds of cross–validation were planned in order to identify weaknesses and to establish consensus quality metrics criteria for variant reporting among seven central laboratories in AML molecular diagnosis.
Networking for NGS studies allowed us to identify challenges in its clinical implementation [9]. The first CV round revealed the absence of AML key genes in some NGS approaches, and consequently a consensus set of relevant genes for the clinical management was defined. Next steps of the diagnostic platform enabled us to unify variant reporting according to the role of genes in diagnostic classification, prognostic utility and targeted therapy. Cooperative studies have also allowed us to address technical challenges. Comparison of results between centers facilitates discrimination of polymorphic variants and sequencing artifacts from real AML–related variants [7].
Our results demonstrated that NGS standardization in the context of a cooperative group is possible with a concordance of 96.6% in variant detection (VAF > 5%). Noteworthy, the detection of low VAF (<5%) variants (concordance 41.2%) was also consistent with previous studies, which report that accurate detection of low VAF variants by NGS could be compromised. CV rounds results reflected the improvement of the diagnostic platform performance as the error rate decreased in variants VAF > 5% as a result of the experience gained in NGS studies. Although the second CV round included 5 variants with a mean VAF of 3.3%, in the third CV round we aim to explore the performance of NGS studies in very low VAF variants. For this purpose, we included 11 variants with a mean VAF of 1.2%. Data analysis revealed an increase of the ER between both CV rounds due to the very low VAF variants of the third CV round. Consequently, we established VAF 5% cut–off for variant reporting, although according to other specific studies, variants in hotspot regions were reported up to 1% VAF even at borderline technical quality [16,17,18].
Our results reflect that a comprehensive NGS approach is suitable for defining the molecular profile of AML as over 96% of patients of the “real–life” PETHEMA cohort harbored at least one mutated gene. Our study also reflects the genomic heterogeneity that encompasses AML (13,14) as several gene–to–gene interactions but also co–occurrence and mutual exclusivity patterns across functional categories were described. Recently, published studies have highlighted the impact of co–mutations in modulating treatment response [19,20].
Similarly, distinct mutational profiles were detected among disease stages, reflecting the clonal evolution of AML [1,21]. In fact, when compared to the diagnosis’ molecular profile we detected fewer mutational changes in resistance to induction therapy (51%) than relapse AML patients (87%) which may suggest different mechanisms underlying both moments [22,23,24,25]. Clonal evolution of the disease could be especially relevant for treatment response [26,27], and in relapsed AML patients, our results supported that molecular testing should be conducted again in order to identify targetable abnormalities such as FLT3 mutations, which showed a stability rate of 43.3% in our cohort [28].
In addition to mutational changes according to disease stages, specific mutational profiles have been associated to patient’s clinical characteristics, such as sex and age, with significant impact on prognosis, therapeutic allocation and disease monitoring [29]. In fact, we report that young patients show a mutational profile very similar to female AML patients, which allows them to benefit more from targeted therapies due to higher frequency of FLT3 mutations [30,31,32,33]. In contrast, male and elderly patients show a molecular profile with a higher frequency of adverse risk genetic abnormalities and limited targeted therapy opportunities [34,35,36]. Indeed, some studies suggest that these features should be considered as an essential variable in clinical trials to deepen understanding of the disease and to identify new treatment opportunities [37,38].
Molecular diagnosis of AML has shifted towards a comprehensive mutational study driven by NGS, yielding large amounts of data. Updated diagnostic (WHO and ICC) and prognostic (ELN) classifications [3,4,5] include an increasing number of genetic abnormalities, which may challenge its applicability in many countries and institutions who cannot afford molecular data integration. However, comprehensive molecular analyses, as the recent revision of the genomic classification of AML are needed to understand the clinical relevance of molecular biomarkers to define novel clusters with prognostic value. In our comparison between genomic AML classification and 2022 ELN risk stratification we did not find differences in OS for favorable, intermediate and adverse risk groups. However, for a comprehensive comparison it would be necessary to evaluate the prognosis impact of individual genetic abnormalities in larger cohorts. Mutations in sAML genes are considered in both risk classification proposals with different prognostic impact [2,5]. Although ELN guidelines associate mutations in “myelodisplasia–related genes” as adverse risk regardless of the number of mutations, our results found significant differences in the OS between sAML1 and sAML2 patients, as described by Tazi et al., (p = 0.018). In this sense, further studies are needed in order to certainly clarify the impact of sAML mutations on prognosis.
Regarding TP53 mutations, in the PETHEMA cohort, 47 patients were included in the mono–allelic group (32.9%), while 96 were included in the multi–hit group (67.1%). Similar results were described in the Tazi et al. cohort: mono–allelic (31.1%) vs. multi–hit: (68.9%). We did not find a distinct outcome between these groups, although the sample size was limited to draw solid conclusions. Our results are concordant with Tazi et al., who concluded that the allelic state of TP53 (mono allelic or multi–hit) provide no further prognostic information in AML.
On the other hand, recent studies reported that instead of biallelic mutations, only in–frame mutations in the bZIP domain of CEBPA should be considered as a favorable prognosis marker [39]. In our cohort, the percentage of patients with CEBPA bZIP mutations is similar to the CEBPAbi subgroup reported by Tazi et al., (1.2% vs. 1.8%) and in both cases, OS analysis includes them within a favorable risk subgroup. However, CEBPAbi and CEBPA bZIP may be overlapping categories in some patients as several studies suggest that CEBPA double mutated frequently includes mutations in the C–terminal region which allocated bZIP domain [40]. Moreover, we believe that the recent recognition of in–frame mutations in the bZIP domain of CEBPA as a prognostic biomarker may allow for more homogeneous analysis by reducing the variability in the interpretation of the biallelic character of these mutations [41].
5. Conclusions
This report reflects the efforts of the PETHEMA cooperative scientific group to adopt a nationwide strategy network of reliable and consistent NGS analyses. The establishment of consensus subset genes and the periodic CV rounds have strengthened the diagnostic network by unifying analysis criteria and decreasing reporting variability. Molecular analyses through NGS are routinely performed for AML patients and a comprehensive molecular profile of the disease is offered to clinicians in order to individualize the therapeutic strategy. Moreover, NGS results have provided a large amount of molecular data that has revealed the molecular complexity of the disease. In this cohort, mutual exclusion and mutational co–occurrences among genes and functional categories have been deciphered and a distinct molecular profile between age groups at diagnosis and sex has been detected. Moreover, clinical validation of the current genomic classification in the “real–life” PETHEMA cohort has demonstrated the correlation of the molecular subgroups with clinical prognosis, reflecting the utility of the cooperative NGS studies in routine molecular diagnostics.
Acknowledgments
The authors would like to thank María D. García, Carlos Pastorini, Rafael Vianney, Yolanda Mendizabal and Alvaro Fernández for data collection and management.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15020438/s1, Table S1: Description of the sequencing technology, NGS panel and NGS platform used by each reference laboratory; Table S2: Detailed gene panel composition; Table S3: Genomic groups and integrated risk score based on 16 molecular classes defined by Tazi et al., 2022 in the updated genomic classification of AML; Table S4: Third Cross–validation round results; Table S5: Molecular alterations pertaining to described functional categories. Table S6: Mutational frequency distribution according to moment disease, age and sex; Table S7: Hazard ratio for (A) molecular classes, (B) Risk score in global cohort, (C) Risk score in <65 years–old, (D) Risk score in ≥65 years–old; Figure S1: Third Cross–validation round results; Figure S2: Mutational frequency of PETHEMA consensus genes in the global cohort; Figure S3: Stability rates in diagnosis–relapse samples. Figure S4: Stability rates in diagnosis–refractoriness samples; Figure S5: Overall survival curves according to AML molecular classes; Figure S6: Overall survival curves of TP53 mutated patients according to mono–allelic vs. multi–hit configurations.
Appendix A
Institutions and clinicians participating in the PETHEMA epidemiologic registry of acute myeloid leukemia and acute promyelocytic leukemia.
Argentina (Grupo Argentino para el Tratamiento de la Leucemia Aguda—GATLA)—Hospital de Clínicas, Buenos Aires: F. Rojas; H. Longoni; Fundaleu, Buenos Aires: G. Milone, I. Fernández, Clínica Conciencia, Neuquén: R. Ramirez; Hospital Rossi, La Plata: C. Canepa, S. Saba, G. Balladares, Hospital General San Martin, Parana: G. Milone, C. Ventiurini, R. Mariano, P. Negri; Hospital Italiano de La Plata, La Plata: M. V. Prates, J. Milone; Hospital General San Martín, La Plata:P. Fazio, M. Gelemur; Hospital Clemente Alvarez, Rosario: G. Milone, S. Ciarlo, F. Bezares; Hospital de Córdoba, Córdoba: L. López, Hospital Privado de Córdoba, Córdoba: J. J.García; Instituto Privado Hematología, Paraná: P. Negri, M. Giunta, G. Milone; Hospital Teodoro Alvarez, Buenos Aires: M. Kruss; Hospital Tornú, Buenos Aires: D. Lafalse, G. Milone; Hospital Gobernador Centeno, La Pampa: E. Marquesoni, M. F. Casale; Hospital Italiano de Buenos Aires, Buenos Aires: A. Gimenez, E. B. Brulc, M. A. Perusini; Complejo Médico Policía Federal, La Plata: G. Milone, L. Palmer; Colombia (Asociación Colombian de Hematología y Oncología—ACHO)—Clínica La Estancia, Popayán: M. E. Correa; Fundación Valle del Lili, Cauca: F.J. Jaramillo, J. Rosales; Instituto FOSCAL, Bucaramanga: C. Sossa, J. C. Herrera; Hospital Pablo Tobón Uribe, Antioquia: M. Arango; Poland (Polish Adult Leukemia Group—PALG)—City Hospital Legnica, Baja Silesia: J. Holojda; IHIT Hematology and transfusiology institute, Warszawa: A. Golos, A. Ejduk; Wojewódzki Szpital Specjalistyczny w Olsztynie, Olsztyn: B. Ochrem; WIM (Military Institute of Medicine in Warsaw), Warszawa: G. Małgorzata; Poland Medical University of Warsaw Banacha, Warszawa:A. Waszczuk-Gajda, J. Drozd-Sokolowska, M. Czemerska, M. Paluszewska; Medical University School Gdansk, Gdansk: E. Zarzycka; Wojewódzki Szpital Specjalistyczny im. Św.. Jadwigi Śląskiej, Opole: A. Masternak; Hospital Brzozow, Brzozow: Dr. Hawrylecka; Medical University Lublin, Lublin: M. Podhoreka, K. Giannopoulos, T. Gromek; Medical University Bialystok, Bialystok: J. Oleksiuk; Silesian Medical University Katowice, Katowice:bA. Armatys, G. Helbig; Universitary Hospital Wroclaw, Wroclaw: M. Sobas; Poznan University of Medical Sciences, Pozna: A. Szczepaniak; Rydigier City Hospital Krakow, Krakow: E. Rzenno, M. Rodzaj; Collegium Medicum Jagiellonian University Krakow, Krakow: B. Piatkowska-Jakubas; City Hospital Rzeszów, Rzeszów: A. Skret; Medical University Lodz, Lodz: A. Pluta, M. Czemerska; Center of Oncology Kielce, Kielce: E. Barańska; Medical University of Warsaw, Warsaw: M. Paluszewska; Portugal—Hospital de Santa Maria-Lisboa, Lisboa:G. Vasconcelos, J. Brioso; IPOFG Lisboa, Lisboa: A. Nunes, I. Bogalho; Centro Hospitalar e Universitário de Coimbra, Coimbra: A. Espadana, M. Coucelo, S. Marini, J. Azevedo, A. I. Crisostomo, L. Ribeiro, V. Pereira; Centro Hospitalar de Lisboa Central E. P. E, Lisboa: A. Botelho; Instituto Português Oncologia do Porto Francisco Gentil, Porto: J. M. Mariz; Centro Hospitalar São João, Porto: J. E. Guimaraes, E. Aguiar; Centro Hospitalar do Porto E.P.E. Porto: J. Coutinho; Spain (Programa Español de Tratamiento de las Hemopatías Malignas, PETHEMA)—Complejo Hospitalario Universitario A Coruña, A Coruña: V. Noriega, L. García, C. Varela, G. Debén, M. R. González; Hospital Clínico Universitario de Santiago, A Coruña: M. Encinas, A. Bendaña, S. González, J.L. Bello, M. Albors; Hospital General de Albacete, Albacete: L. Algarra, J.R.Romero, J.S.Bermon, M.J. Varo; Hospital Vinalopó, Alicante: V. López, E. López; Hospital Virgen de los Lirios, Alcoy: C. Mora, C. Amorós; Hospital General Elche, Alicante: E. López, A. Romero; Hospital Torrevieja Salud, Alicante: A. Jaramillo, N. Valdez, I. Molina, A. Fernández, B. Sánchez; Hospital de la Marina Baixa Villajoyosa, Alicante: A. García; Hospital General de Elda, Alicante: V. Castaño, T. López, J. Bernabeu; Hospital de Denia-Marina Salud, Alicante: M.J. Sánchez; Hospital de la Vega Baja de Orihuela, Alicante: C. Fernández; Hospital General de Alicante, Alicante: C. Gil, C. Botella, P. Fernández, M. Pacheco, F. Tarín; J.J. Verdú; Complejo Hospitalario Torrecardenas, Almeria: M.J. García, A. Mellado, M.C. García, J. González; Hospital Central de Asturias, Asturias: T. Castillo, E. Colado, S. Alonso; Complejo Asistencial Ávila, Ávila: I. Recio, M. Cabezudo, J. Davila, M. J. Rodríguez, A. Barez, B. Díaz; Hospital Don Benito-Villanueva, Badajoz: J. Prieto; Institut Catala d’Oncologia L’Hospitalet, Barcelona: M. Arnan, C. Marín, M. Mansilla; Hospital de Cruces, Bizkaia: A. Balaberdi, M. E. Amutio, R. A. del Orbe, I. Ancin, J. C. Ruíz; Hospital Galdakao-Usansolo, Bizkaia: M. Olivalres, C. Gómez, I. gonzález, M. Celis, K. Atutxa, T. Carrascosa, T. Artola, M. Lizuain; Basurtuko Ospitalea, Bizkaia: J.I. Rodriguez, O. Arce, J. A. Márquez, J. Atuch, F. Marco de Lucas, Z. Díez, B. Dávila; Hospital Santos Reyes, Burgos: R. Cantalejo, M. Díaz; Hospital Universitario de Burgos, Burgos: J. Labrador, F. Serra, G. Hermida, F. J. Díaz, P. de Vicente, R. Álvarez: Hospital Santiago Apóstol, Burgos: C. Alonso, Hospital San Pedro de Alcántara, Cáceres: J. M. Bergua; Hospital Campo Arañuelo, Cáceres: N. Ugalde; Hospital Virgen del Puerto, Cáceres: E. Pardal; Hospital General Jerez de la Frontera, Cádiz: R. Saldaña, F. Rodríguez, E. Martín, L. Hermosín; Hospital Universitario Puerta del Mar, Cádiz: M. P. Garrastazul, I. Marchante, J. A. Raposo, F. J. Capote; Hospital U. Marqués de Valdecilla, Cantabria: M. Colorado, A. Batlle, L. Yañez, S. García, P. González, E. M. Ocio, M. Briz, A. Bermúdez, S. García; Consorcio Hospitalario Provincial de Castellón, Castellón: C. Jiménez, S. Beltrán; Hospital de Vinaroz: M. Montagud; Hospital Universitario de La Plana, Castellón: I. Castillo; Hospital General de Castellón, Castellon: R. García, A. Gascón, J. Clavel, A. Lancharro, L. Lnares; Hospital Santa Bárbara, Ciudad Real: M. M. Herráez, A. Milena; Hospital Virgen de Altagracia, Ciudad Real: M. J. Romero, Hospital General de Ciudad Real, Ciudad Real: B. Hernández, C. Calle, R. Benegas; Hospital Gutierrez Ortega de Valdepeñas, Ciudad Real: Dr. Bolívar; Hospital General La Mancha Centro, Ciudad Real: M. A. Pozas; Hospital Reina Sofia, Córdoba: J. Serrano, F. J. Dorado, J. Sánchez, M. C. Martínez; Hospital Virgen de la Luz, Cuenca: C. J. Cerveró, M. J. Busto; Hospitales HUVN-HC San Cecilio de Ganada, Granada: M. Bernal, E. López, L. Moratalla, Z. Mesa, M. Jurado, A. Romero, P. González; Complejo Hospitalario Universitario Granada, Granada: L. Moratalla, A. Romero, L. López; Hospital Universitario de Guadalajara, Guadalajara: M. Díaz, D. De Miguel, A. B. Santos, J. Arbeteta; Hospital Donostia, Donosti: E. Pérez, N. Caminos, N. Uresandi, N. Argoitiaituart, T. Artola, J. Swen, A. Uranga, I. Olazaba, M. Lizuain, E. Gainza, P. Romero; Hospital Juan Ramón Jimenez Huelva, Huelva: E. Gil, A. J. Palma, K. G. Gómez, M. Solé, J. N. Rodríguez; Hospital San Jorge, Huesca: I. M. Murillo, J. Marco, J. Serena, V. Marco; Hospital de Barbastro, Huesca: M. Perella, L. Costilla; Hospital General Ciudad de Jaen, Jaén: J. A. López, A. Baena, P. Almagro; Hospital San Pedro de Logroño, La Rioja: M. Hermosilla, A. Esteban, B. A. Campeny, M. J. Nájera, P. Herrra; Hospital Insular de Las Palmas, Las Palmas: R. Fernández, J. D. González, L. Torres; Hospital Dr. Negrín, Las Palmas: S. Jiménez; M. T. Gómez, C. Bilbao, C. Rodríguez; Hospital Doctor José Molina Orosa, Las Palmas: A. Hong, Y. Ramos de Laón, V. Afonso; Hospital Universitario de León, León: F. Ramos, M. Fuertes; Hospital Comarcal del Bierzo, León: E. de Cabo, C. Aguilera, M. Megido; Hospital Universitari Arnau de Vilanova de Lleida, Leida: T. García; Hospital Lucus Augusti, Lugo: E. Lavilla, M. Varela, S. Ferrero, M. J. Sánchez, L. López, J. Arias, L. Vizcaya; Hospital Infanta Sofía, Madrid: A. Roldán, A. Vilches, M. J. Penalva, J. Vázquez; Hospital Central de la Defensa Gómez Ulla, Madrid: M. T. Calderón, A. Matilla, C. Serí, M. J. Otero, N. García, E. Sandoval; Hospital de Fuenlabrada, Madrid: C. Franco, R. Flores, P. Bravo, A. López; Hospital Fundación Jiménez Díaz, Madrid: J. L. López, C. Blas, A. Díez, J. M. Alonso, C. Soto, A. Arenas; Hospital U. Príncipe de Asturias, Madrid: J. García, Y. Martín, P. S. Villafuerte, E. Magro; Hospital Puerta de Hierro, Madrid: G. Bautista; A. De Laiglesia; Hospital Gregorio Marañón, Madrid: G. Rodríguez, L. Solán, M. Chicano, P. Balsalobre, S. Monsalvo, P. Font, D. Carbonell, C. Martínez; Hospital U. La Paz, Madrid: K. Humala, A. E. Kerguelen, D. Hernández, M. Gasior, P. Gómez, I. Sánchez; Hospital Madrid Norte Sanchinarro, Madrid: S. Redondo, L. Llorente, M. Bengochea, J. Pérez; Hospital Sanitas Torrejón, Madrid: A. Sebrango, M. santero, A. Morales; Hospital La Princesa, Madrid: A. Figuera, P. Villafuerte, A. Alegre, E. Fernández; Hospital Ruber Internacional, Madrid: A. Alonso; Hospital 12 de Octubre, Madrid: M. P. Martínez, J. Martínez, M. T. Cedena, L. Moreno; MD Anderson Cancer Center, Madrid: A. De la Fuente; Hospital Sanitas La Zarzuela, Madrid: D. García; Hospital Universitario Quiron, Madrid: C. Chamorro, V. Pradillo, E. Martí, J. M. Sánchez, I. Delgado, A. Alonso; Hospital Rey Juan Carlos, Madrid: B. Rosado, A. Velasco, C. Miranda, G. Salvatierra, J. M. Alonso,, J. L. López; Hospital Infanta Leonor, Madrid: M. Foncillas, J. A. Hernández; Hospital Universitario de Getafe, Madrid: C. Escolano, L. García, I. Delgado; Hospital Clínico San Carlos, Madrid: C. Benabente, R. Martínez, M. Polo, E. Anguita; Hospital Universitario Severo Ochoa, Madrid: R. Riaza, G. Amores, M. J. Requena; Hospital Universitario Fundación Alcorcón, Madrid: F. Javier, L. Villaloón; Hospital Universitario Moncloa, Madrid: C. Aláez, V. Pradillo, S. Nistal, B. Navas; Hospital Universitario de Móstoles, Madrid: J. Sánchez, M. A. Andreu; Hospital Ramon y Cajal, Madrid: P. Herrera, J. López; Hospital U. Virgen de la Victoria, Málaga: M. García, M. J. Moreno, A. Fernández, M. P. Queipo; Hospital Quirónsalud Málaga, Málaga: A. Hernández; Hospital Regional de Málaga, Málaga: M. Barrios, A. Heiniger, A. Jiménez, A. Contento, F. López, M. Alcalá; Hospital Vithas Xanit Internacional, Málaga: S. Lorente, M. González, E. M. Morales, J. Gutierrez; Hospital Virgen del Castillo, Murcia: M. J. Serna, V. Beltrán; Hospital Santa Lucía de Cartagena, Murcia: M. Romera, M. Berenguer, A. Martínez, A. Tejedor; Hospital Morales Meseguer, Murcia: M. L. Amigo, F. Ortuño, L. García, A. Jerez, O. López; Hospital U. Virgen de la Arrixaca, Murcia: J. M. Moraleda, P. Rosique, J. Gómez, M. C. Garay; Hospital Los Arcos Mar Menor, Murcia: P. Cerezuela, C. Martínez, A. B. MArtínez, A. González; Hospital STª Mª del Rosell, Murcia: J. Ibáñez; Clínica San Miguel, Navarra: M. J. Alfaro; Complejo Hospitalario de Navarra, Navarra: M. Mateos, M. A. Goñi, M. A. Araiz, A. Gorosquieta, M. Zudaire, M. Viguria, A. Zabala, M. Alvarellos, I. Quispe, M. P. Sánchez, G. Hurtado, M. Pérez, Y. Burguete, N. Areizaga, T. Galicia; Clínica Universitaria de Navarra, Navarra: J. Rifón, A. Alfonso, F. Prósper, M. Marcos, L. E. Tamariz, V. Riego. A. Manubens, M. J. Larrayoz, M. J. Calasanz, A. Mañú, B. Paiva, I. Vázquez, L. Burgos; Complejo Hospitalario de Ourense (CHOU), Ourense: M. Pereiro, M. Rodríguez, M. C. Pastoriza, J. A. Mendez, J. L. Sastre, M. Iglesias, C. Ulibarrena, F. Campoy; Hospital Valdeorras, Ourense: D. Jaimes; Hospital Rio Carrión, Palencia: J. M. Alonso, B. Albarrán, J. Solano, A. Silvestre; Complexo Hospitalario Universitario de Vigo, Vigo: C. Albo, S. Suarez, C. Loureiro, I. Figueroa, M. Rodríguez, M. A. Fernández, A. Martínez, C. Poderós, J. Vazquez, L. Iglesias, A. Nieto, T. Torrado, A. M. Martínez; Hospital Provincial de Pontevedra, Pontevedra: M.L. Amador, P. Oubiña, E. Feijó, A. Dios, I. Loyola, R. Roreno; Hospital POVISA, Pontevedra: A. Simiele, L. Álvarez, V. Turcu; Hospital U. Salamanca, Salamanca: B. Vidriales, M. González, R. García, A. Avendaño, C. Chillón, E. Pérez, V. González; Hospital General La Palma, Santa Cruz de Tenerife: J. V. Govantes, S. Rubio, M. Tapia; Hospital General de Segovia, Segovia: C. Olivier, J. A. Queizán; Hospital U. Virgen Macarena, Sevilla: O. Pérez, J. A. Vera, C. Muñoz, A. rodriguez, N. González; Hospital U. Virgen del Rocio, Sevilla: J. A. Pérez, E. Soria, I.Espigado, J. Falantes, I. Montero, P. García, E. Rodríguez, E. Carrillo, T. Caballero, C. García; Hospital Virgen de Valme, Sevilla: C. Couto, I. Simón, M. Gómez; Hospital Virgen del Mirón de Soria, Soria: C. Aguilar; Hospital Universitario Canarias, Tenerife: B. J. González, S. Lakhwani, A. Bienert, B. González; Hospital Universitario Nuestra Señora de Candelaria, Tenrife: A. Cabello, A. Y. Oliva, H. González; Hospital Obispo Polanco, Teruel: N. González, Hospital de Alcañiz, Teruel: L. Sancho, M. Paricio, L. Perdiguer; Hospital General Nuestra Señora del Prado, Toledo: F. Solano, A. Lerma, M. D. Martínez; Hospital Virgen de la Salud de Toledo, Toledo: M. I. Gómez, A. Yeguas; Hospital U. La Fe, Valencia: P. Montesinos, E. Barragán, C. Sargas, R. Amigo, D. Martinez, B. Boluda, R. Rodríguez, E. Acuña, I. Cano; Hospital de Requena, Valencia: A. Escrivá, M. Pedreño; Hospital de Lluis Alcanyis de Xativa, Valencia: R. Renart; IVO (Instituto Valenciano de Oncología), Valencia: A. Navalón; Hospital de Sagunto, Valencia: I. Castillo, M. Orts; Hospital Dr. Peset, Valencia: M. J. Sayas, M. J. Fernández, M. L. Juan, E. Gómez, M. Gimeno, E. Donato, M. Cejalvo, J. Marco; Hospital Clínico Universitario, Valencia: M. Tormo, M. Calabuig, B. Navarro, I. Martin, E. Villamont, A. Miralles; Hospital de La Ribera, Valencia: R. Lluch; Hospital Casa de la Salud, Valencia: J. García; Hospital de Gandía, Valencia: M. Moragues, M. A. Ruiz; Hospital Arnau de Vilanova, Valencia: A. López, C. Benet, M. Valero; Hospital General de Valencia, Valencia: M. Linares, R. Collado, M. Orero, P. Ibañez, M. J. Lis, P. L. Pérez, M. Roig, M. López, A. V. Mena; Hospital Manises, Valencia: I. Picón, V. Cánovas, A. Palacios, E. Martí; Hospital Clínico de Valladolid, Valladolid: R. Cuello, J. Borrego, M. burgois; Hospital Rio Hortega, Valladolid: A. Cantalapiedra, O. Norberto, E. Angomas, B. Cidoncha; Hospital Universitario Araba, Victoria: L. Cuevas, D. Robles, A. Mendiazabal, I. Oiartzabal, J. M. Guinea de Castro; Hospital Virgen de la Concha, Zamora: C. Montes, M. Pérez, L. García; Hospital Royo Villanova, Zaragoza: V. Carrasco, A. Pérez, L. López, J. J. Moneva; Hospital Clínico U. Lozano Blesa, Zaragoza: M. Olave, E. Bonafonte, L. Mayor, G. Azaceta, L. Palomera; Hospital Ernest Lluch Martin, Zaragoza: M. Malo, M. J. Escobar; Hospital Quiron Salud Zaragoza, Zaragoza: J. M. Grasa; Hospital Miguel Servet, Zaragoza: B. De Rueda, A. Aulés, C. Salvador, V. Ansó, A. Iborra, P. Delagado, A. Rubio; Uruguay—Hospital de Clínicas, Montevideo: M. Stevenazzi, I. Alpire, V. Irigoin, L. Díaz, C. Guillermo, R. Guadagna, S. Grille, C. Oliver, M. Boada, V. Vales; Hospital Maciel, Montevideo: A. I. Prado; COMERO (Cooperativa Medica de Rocha), Rocha: A. P. De los Santos.
Author Contributions
Conceptualization, C.S., E.B. and P.M.; methodology: C.S., R.A., M.J.L., M.C.C., E.C.-C. and C.B.-S.; formal analysis, C.S. and E.B.; data curation, C.S., E.B., D.M.-C., B.B. and P.M.; investigation, C.S., R.A., M.J.L., M.C.C., E.C.-C., C.B.-S., E.P.d.l.T., D.M.-C., R.R.-V., B.B., C.G., T.B., J.M.B., L.A., M.T., P.M.-S., E.S., J.S., J.M.A.-D., R.G.-B., M.L.A., P.H.-P., M.J.S., E.L.-R., J.M.-L., M.J.C., R.G.-S., J.A.P.-S., M.T.G.-C., J.S.-G., E.B. and P.M.; writing—original draft preparation, C.S.; writing—review and editing, E.B. and P.M. visualization, C.S.; supervision, E.B. and P.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Instituto de Investigación Sanitaria La Fe (2017/00304/PI; 13/09/2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was partially funded by Bristol-Myers Squibb/Celgene: PETHEMA-NGS-LMA; Ministry of Economy and Competitiveness|Instituto de Salud Carlos III, Spain, Spain: PI18/01340, PI19/00730, PI19/01518, FI19/00059.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tazi Y., Arango-Ossa J.E., Zhou Y., Bernard E., Thomas I., Gilkes A., Freeman S., Pradat Y., Johnson S.J., Hills R., et al. Unified Classification and Risk-Stratification in Acute Myeloid Leukemia. medRxiv. 2022:2022.03.09.22271087. doi: 10.1038/s41467-022-32103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khoury J.D., Solary E., Abla O., Akkari Y., Alaggio R., Apperley J.F., Bejar R., Berti E., Busque L., Chan J.K.C., et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber D.A., Orazi A., Hasserjian R.P., Borowitz M.J., Calvo K.R., Kvasnicka H.M., Wang S.A., Bagg A., Barbui T., Branford S., et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemia: Integrating Morphological, Clinical, and Genomic Data. Blood. 2022;140:1200–1228. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Döhner H., Wei A.H., Appelbaum F.R., Craddock C., DiNardo C.D., Dombret H., Ebert B.L., Fenaux P., Godley L.A., Hasserjian R.P., et al. Diagnosis and Management of AML in Adults: 2022 ELN Recommendations from an International Expert Panel. Blood. 2022;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PubMed] [Google Scholar]
- 6.Levine R.L., Valk P.J.M. Next-Generation Sequencing in the Diagnosis and Minimal Residual Disease Assessment of Acute Myeloid Leukemia. Haematologica. 2019;104:868. doi: 10.3324/haematol.2018.205955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacher U., Shumilov E., Flach J., Porret N., Joncourt R., Wiedemann G., Fiedler M., Novak U., Amstutz U., Pabst T. Challenges in the Introduction of Next-Generation Sequencing (NGS) for Diagnostics of Myeloid Malignancies into Clinical Routine Use. Blood Cancer J. 2018;8:113. doi: 10.1038/s41408-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haferlach T. Advancing Leukemia Diagnostics: Role of Next Generation Sequencing (NGS) in Acute Myeloid Leukemia. Hematol. Rep. 2020;12:8957. doi: 10.4081/hr.2020.8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sargas C., Ayala R., Chillón M.C., Larráyoz M.J., Carrillo-Cruz E., Bilbao C., Yébenes-Ramírez M., Llop M., Rapado I., García-Sanz R., et al. Networking for Advanced Molecular Diagnosis in Acute Myeloid Leukemia Patients Is Possible: The PETHEMA NGS-AML Project. Haematologica. 2021;106:3079–3089. doi: 10.3324/haematol.2020.263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson G., Hoadley K., Triche T.J., Laird P.W., Baty J.D., et al. Genomic and Epigenomic Landscapes of Adult de Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leisch M., Jansko B., Zaborsky N., Greil R., Pleyer L. Next Generation Sequencing in AML-On the Way to Becoming a New Standard for Treatment Initiation and/or Modulation? Cancers. 2019;11:252. doi: 10.3390/cancers11020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luthra R., Patel K.P., Routbort M.J., Broaddus R.R., Yau J., Simien C., Chen W., Hatfield D.Z., Medeiros L.J., Singh R.R. A Targeted High-Throughput Next-Generation Sequencing Panel for Clinical Screening of Mutations, Gene Amplifications, and Fusions in Solid Tumors. J. Mol. Diagn. 2017;19:255–264. doi: 10.1016/j.jmoldx.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Petrackova A., Vasinek M., Sedlarikova L., Dyskova T., Schneiderova P., Novosad T., Papajik T., Kriegova E. Standardization of Sequencing Coverage Depth in NGS: Recommendation for Detection of Clonal and Subclonal Mutations in Cancer Diagnostics. Front. Oncol. 2019;9:851. doi: 10.3389/fonc.2019.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endrullat C., Glökler J., Franke P., Frohme M. Standardization and Quality Management in Next-Generation Sequencing. Appl. Transl. Genom. 2016;10:2–9. doi: 10.1016/j.atg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandzic T., Ladenvall C., Engvall M., Mattsson M., Hermanson M., Cavelier L., Ljungström V., Baliakas P. Five Percent Variant Allele Frequency Is a Reliable Reporting Threshold for TP53 Variants Detected by Next Generation Sequencing in Chronic Lymphocytic Leukemia in the Clinical Setting. Hemasphere. 2022;6:e761. doi: 10.1097/HS9.0000000000000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jennings L.J., Arcila M.E., Corless C., Kamel-Reid S., Lubin I.M., Pfeifer J., Temple-Smolkin R.L., Voelkerding K.V., Nikiforova M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillermin Y., Lopez J., Chabane K., Hayette S., Bardel C., Salles G., Sujobert P., Huet S. What Does This Mutation Mean? The Tools and Pitfalls of Variant Interpretation in Lymphoid Malignancies. Int. J. Mol. Sci. 2018;19:1251. doi: 10.3390/ijms19041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strom S.P. Current Practices and Guidelines for Clinical Next-Generation Sequencing Oncology Testing. Cancer Biol. Med. 2016;13:3–11. doi: 10.20892/j.issn.2095-3941.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin G., Ilya S., Kim T.-K., Tercan B., Martins T.J., Dai J., Chien S., Carson A., Patay B., Estey E.H., et al. Co-Occurring Mutation Clusters Predict Drug Sensitivity in Acute Myeloid Leukemia. Blood. 2020;136:12–13. doi: 10.1182/blood-2020-142727. [DOI] [Google Scholar]
- 20.Lachowiez C.A., Reville P.K., Kantarjian H., Jabbour E., Borthakur G., Daver N., Issa G., Furudate K., Tanaka T., Pierce S., et al. Contemporary Outcomes in IDH-Mutated Acute Myeloid Leukemia: The Impact of Co-Occurring NPM1 Mutations and Venetoclax-Based Treatment. Am. J. Hematol. 2022;97:1443–1452. doi: 10.1002/ajh.26694. [DOI] [PubMed] [Google Scholar]
- 21.Bullinger L., Döhner K., Dohner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 22.Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., McLellan M.D., et al. Clonal Evolution in Relapsed Acute Myeloid Leukaemia Revealed by Whole-Genome Sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu J., Peng D., Liu L. Drug Resistance Mechanisms of Acute Myeloid Leukemia Stem Cells. Front. Oncol. 2022;12:896426. doi: 10.3389/fonc.2022.896426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Gils N., Denkers F., Smit L. Escape From Treatment; the Different Faces of Leukemic Stem Cells and Therapy Resistance in Acute Myeloid Leukemia. Front. Oncol. 2021;11:659253. doi: 10.3389/fonc.2021.659253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Gu Y., Chen B. Mechanisms of Drug Resistance in Acute Myeloid Leukemia. Onco. Targets Ther. 2019;12:1937–1945. doi: 10.2147/OTT.S191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benard B.A., Leak L.B., Azizi A., Thomas D., Gentles A.J., Majeti R. Clonal Architecture Predicts Clinical Outcomes and Drug Sensitivity in Acute Myeloid Leukemia. Nat. Commun. 2021;12:7244. doi: 10.1038/s41467-021-27472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thol F., Heuser M. Treatment for Relapsed/Refractory Acute Myeloid Leukemia. Hemasphere. 2021;5:E572. doi: 10.1097/HS9.0000000000000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daver N., Schlenk R.F., Russell N.H., Levis M.J. Targeting FLT3 Mutations in AML: Review of Current Knowledge and Evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarlock K., Zhong S., He Y., Ries R., Severson E., Bailey M., Morley S., Balasubramanian S., Erlich R., Lipson D., et al. Distinct Age-Associated Molecular Profiles in Acute Myeloid Leukemia Defined by Comprehensive Clinical Genomic Profiling. Oncotarget. 2018;9:26417–26430. doi: 10.18632/oncotarget.25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimwade D., Ivey A., Huntly B.J.P. Molecular Landscape of Acute Myeloid Leukemia in Younger Adults and Its Clinical Relevance. Blood. 2016;127:29. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.di Nardo C.D., Cortes J.E. Mutations in AML: Prognostic and Therapeutic Implications. Hematol. Am. Soc. Hematol. Educ. Program. 2016;2016:348. doi: 10.1182/asheducation-2016.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J., Jiang P.Y.Z., Sun H., Zhang X., Jiang Z., Li Y., Song Y. Advances in Targeted Therapy for Acute Myeloid Leukemia. Biomark. Res. 2020;8:17. doi: 10.1186/s40364-020-00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ranieri R., Pianigiani G., Sciabolacci S., Perriello V.M., Marra A., Cardinali V., Pierangeli S., Milano F., Gionfriddo I., Brunetti L., et al. Current Status and Future Perspectives in Targeted Therapy of NPM1-Mutated AML. Leukemia. 2022;36:2351–2367. doi: 10.1038/s41375-022-01666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayala R., Rapado I., Onecha E., Martínez-Cuadrón D., Carreño-Tarragona G., Bergua J.M., Vives S., Algarra J.L., Tormo M., Martinez P., et al. The Mutational Landscape of Acute Myeloid Leukaemia Predicts Responses and Outcomes in Elderly Patients from the PETHEMA-FLUGAZA Phase 3 Clinical Trial. Cancers. 2021;13:2458. doi: 10.3390/cancers13102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prassek V.V., Rothenberg-Thurley M., Sauerland M.C., Herold T., Janke H., Ksienzyk B., Konstandin N.P., Goerlich D., Krug U., Faldum A., et al. Genetics of Acute Myeloid Leukemia in the Elderly: Mutation Spectrum and Clinical Impact in Intensively Treated Patients Aged 75 Years or Older. Haematologica. 2018;103:1853. doi: 10.3324/haematol.2018.191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni J., Hong J., Long Z., Li Q., Xia R., Zeng Q. Mutation Profile and Prognostic Relevance in Elderly Patients with de Novo Acute Myeloid Leukemia Treated with Decitabine-Based Chemotherapy. Int. J. Lab. Hematol. 2020;42:849–857. doi: 10.1111/ijlh.13299. [DOI] [PubMed] [Google Scholar]
- 37.Hellesøy M., Engen C., Grob T., Löwenberg B., Valk P.J.M., Gjertsen B.T. Sex Disparity in Acute Myeloid Leukaemia with FLT3 Internal Tandem Duplication Mutations: Implications for Prognosis. Mol. Oncol. 2021;15:2285–2299. doi: 10.1002/1878-0261.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De-Morgan A., Meggendorfer M., Haferlach C., Shlush L. Male Predominance in AML Is Associated with Specific Preleukemic Mutations. Leukemia. 2021;35:867–870. doi: 10.1038/s41375-020-0935-5. [DOI] [PubMed] [Google Scholar]
- 39.Bullinger L. CEBPA Mutations in AML: Site Matters. Blood. 2022;139:6–7. doi: 10.1182/blood.2021013557. [DOI] [PubMed] [Google Scholar]
- 40.Wakita S., Sakaguchi M., Oh I., Kako S., Toya T., Najima Y., Doki N., Kanda J., Kuroda J., Mori S., et al. Prognostic Impact of CEBPA BZIP Domain Mutation in Acute Myeloid Leukemia. Blood Adv. 2022;6:238–247. doi: 10.1182/bloodadvances.2021004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mannelli F., Ponziani V., Bencini S., Bonetti M.I., Benelli M., Cutini I., Gianfaldoni G., Scappini B., Pancani F., Piccini M., et al. CEBPA–Double-Mutated Acute Myeloid Leukemia Displays a Unique Phenotypic Profile: A Reliable Screening Method and Insight into Biological Features. Haematologica. 2017;102:529–540. doi: 10.3324/haematol.2016.151910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.