Yuling Li et al. identify selective expression of BCL6 in CCR6+ ILC3 among ILC family. BCL6 expression, promoted by intestinal microbiota, restrains the effector function and plasticity of CCR6+ ILC3.
Abstract
Innate lymphoid cells (ILC) are similar to T helper (Th) cells in expression of cytokines and transcription factors. For example, RORγt is the lineage-specific transcription factor for both ILC3 and Th17 cells. However, the ILC counterpart for BCL6-expressing T follicular helper (Tfh) cells has not been defined. Here, we report that in the ILC compartment, BCL6 is selectively co-expressed with not only CXCR5 but also RORγt and CCR6 in ILC3 from multiple tissues. BCL6-deficient ILC3 produces enhanced levels of IL-17A and IL-22. More importantly, phenotypic and single-cell ATAC-seq analysis show that absence of BCL6 in mature ILC3 increases the numbers of ILC1 and transitional cells co-expressing ILC3 and ILC1 marker genes. A lineage-tracing experiment further reveals BCL6+ ILC3 to ILC1 trans-differentiation under steady state. Finally, microbiota promote BCL6 expression in colonic CCR6+ ILC3 and thus reinforce their stability. Collectively, our data have demonstrated that CCR6+ ILC3 have both Th17 and Tfh programs and that BCL6 expression in these cells functions to maintain their lineage identity.
Introduction
Innate lymphoid cells (ILC), residing in multiple tissues and organs, especially at the mucosal barriers, provide first-line immune responses to alarmins (Spits et al., 2013). Their functions, including activation of immune cell networks, tissue repairs post damage, and regulation of host-microbe interactions, significantly contribute to innate host defense and maintenance of tissue homeostasis (Artis and Spits, 2015). One key feature of ILC is their similarity with CD4+ T helper (Th) subsets in expression of cytokines and transcription factors. ILC develop from common lymphoid progenitor in bone marrow, regulated by of IL-7 and IL-2 (Mebius et al., 2001; Yang et al., 2015), resulting in three main subsets of ILC mirroring Th cells. Group 1 ILC consists of conventional natural killer (NK) cells and the non-cytotoxic ILC1 that are similar to Th1 cells (Spits et al., 2013) in expression of transcription factor T-bet and cytokines IFN-γ and TNF (Fuchs et al., 2013; Klose et al., 2014). Group 2 ILC, namely, ILC2, requires GATA3 and RORα for development and secrete Th2-associated cytokines like IL-4, IL-5, and IL-13 (Halim et al., 2012; Monticelli et al., 2011). Group 3 ILC highly express Th17 signature transcription factor RORγt and secrete type 3 cytokines upon IL-23 stimulation (Cella et al., 2009; Chen et al., 2015).
ILC3 are heterogeneous and generally characterized into subsets based on the expression of CCR6 and NKP46 in mice (Eberl et al., 2004; Klose et al., 2013). CCR6+ ILC3, also called lymphoid tissue inducer (LTi) or NCR− ILC3, developed from GATA3loPLZF− LTi precursors (LTiP) and consisting of both CD4− and CD4+ subgroups (Constantinides et al., 2014; Eberl et al., 2004; Zhong et al., 2020). The fetal CCR6+ ILC3, also called LTi, require CXCR5 in response to CXCL13-mediated recruitment by stromal cells, which further activates adhesion between integrin α4β1 and its ligand VCAM-1 during the initial development of secondary lymphoid organs (Finke et al., 2002; Zhong et al., 2018). CCR6+ ILC3 in adult mice are enriched in gut, providing innate source of IL-17A, IL-17F, IL-22, and GM-CSF as a result of pathogen infection and IL-23 stimulation (Zhong et al., 2018). These cells also express MHC-II for antigen presentation to Th cells (Hepworth et al., 2015). NKP46+ or NCR+ ILC3 are derived from GATA3hiPLZF+ ILC precursor cells (Constantinides et al., 2014; Zhong et al., 2020). Although negative for CCR6, NKP46+ ILC3 are similar to CCR6+ ILC3 in expression of RORγt and IL-22; however, they are also positive for T-bet and IFN-γ, making them a hybrid of both type 1 and type 3 features (Klose et al., 2013; Reynders et al., 2011).
ILC are plastic and mature ILC have been shown to transform into other subsets under certain circumstances (DuPage and Bluestone, 2016; Locksley, 2009). In particular, the mixed-lineage feature of NKP46+ ILC3 suggests it may be an intermediate stage between ILC3 and ILC1. This was confirmed by an RORγt fate-mapping study, which showed ILC3 transition from CCR6+ to NKP46+, and finally to the so-called “ex-ILC3” ILC1, accompanied by gradual loss of RORγt expression with increase in that of T-bet (Klose et al., 2013; Vonarbourg et al., 2010), although it was not clear whether CCR6+ ILC3 becomes ILC1 under normal circumstances. It appears that the conversion between ILC3 and ILC1 is reversible (Bernink et al., 2015), and factors that promote RORγt (i.e., IL-7, IL-23, commensal microbiota) or T-bet (i.e., IL-12, IL-15, IL-18, Notch signaling) may provide extrinsic control of cellular trans-differentiation (Chea et al., 2016; Klose et al., 2013; Vonarbourg et al., 2010). Other factors such as inflammation (Bernink et al., 2015), tumor micro-environment (Goc et al., 2021; Shen et al., 2021), or transcription factors (TFs) like c-Maf (Tizian et al., 2020) and IKZF3/Aiolos (Mazzurana et al., 2019) were also indicated to regulate ILC3 plasticity.
Although ILC1, 2, and 3 share common features with Th1, 2, 17 cells, respectively, thus far in the ILC compartment, there is no equivalent for T follicular helper (Tfh) cells characterized by selective expression of CXCR5 and BCL6 and with critical roles in germinal center reactions (Dong, 2021; Nurieva et al., 2009). Interestingly, LTi/CCR6+ ILC3 express the Tfh signature marker CXCR5 (Marchesi et al., 2009) and play similar roles as Tfh in lymphoid follicle development. However, a recent study predicted high levels of BCL6 expression in ILC1 but not CCR6+ ILC3 based on bulk mRNA and Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-seq) analysis of total intestinal ILCs (Pokrovskii et al., 2019). These investigators showed mildly decreased NK cells in NcrcreBcl6fl/fl mice and claimed that BCL6 functioned to promote ILC1 and NK program but repress that of ILC3.
In the current study, we attempted to better characterize BCL6 expression in ILC. Contradictory to the previous report (Pokrovskii et al., 2019), we discovered instead selective expression of BCL6 in CCR6+ ILC3, which also expressed CXCR5. Loss of BCL6 in ILC3 increased ILC3-to-ILC1 conversion. Thus, we have identified that CCR6+ ILC3 have both Th17 and Tfh programs, and that BCL6 expression in these cell functions to maintain their lineage identity.
Results
BCL6 is selectively expressed by CCR6+ ILC3
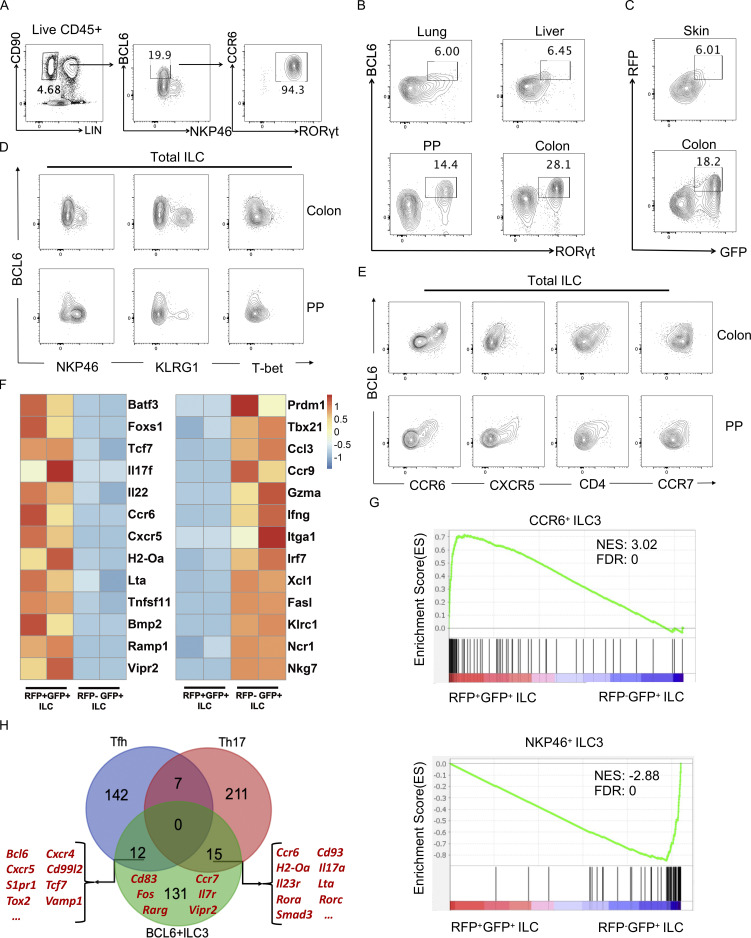
To better characterize the expression of BCL6 in ILC, we used a BCL6 antibody to analyze ILC from multiple organs of C57BL/6 (B6) mice. From intestinal lamina propria lymphocytes (LPL), where a significant portion of ILC reside, a group of ILC were found to express highest levels of BCL6, which also co-expressed RORγt and CCR6, but not NKP46 (Fig. 1 A), suggesting that BCL6 is expressed by a portion of ILC3. BCL6 expression in RORγt+ ILC3 were also observed in other tissues including lung, liver, Peyer’s patches (PP), and colon (Fig. 1 B). Consistently in Bcl6RFP/RFPRorcGFP/+ double reporter mice, RFP+GFP+ ILC3 were found in skin and colon tissues (Fig. 1 C). Moreover, BCL6 was not co-expressed with KLRG1 or NKP46, markers for ILC2 and ILC1, respectively (Fig. 1 D). Thus, BCL6 is selectively expressed in ILC3 at steady state in multiple organs in mice.
Figure 1.
BCL6 is expressed in ILC3 in multiple tissues. (A) Gating strategy for ILC in colon LPLs from B6 mice. Total ILC were gated on Lin−CD90+CD45+ live cells, in which a group of BCL6+NKP46− population was further gated and analyzed for CCR6 and RORγt expression by flow cytometry. (B) BCL6 and RORγt expression in total ILC isolated from B6 mice among indicated tissues at steady state. (C) BCL6 and RORγt expression in total ILC isolated from Bcl6RFP/RFPRorcGFP/+ mice among indicated tissues at steady state. (D) Co-staining of BCL6 versus NKP46, KLRG1, and T-bet in total ILCs isolated from the colon LP and PP of B6 mice. (E) Co-staining of BCL6 versus CCR6, CXCR5, CD4, and CCR7 in total ILCs isolated from the colon LP and PP of B6 mice. (F) Heat map of RNA-seq data for selected key signature genes of CCR6+ ILC3 (Schleussner et al., 2018; Talbot et al., 2020; Zhong et al., 2016; Zhong et al., 2018) and NKP46+ ILC3 (Gury-BenAri et al., 2016) in RFP+GFP+ vs. RFP−GFP+ ILCs obtained from the LPLs of Bcl6RFP/RFPRorcGFP/+ mice. The processed RNA-seq data was listed in Table S1, and raw data are available through GSE220438 on GEO DataSets. (G) GSEA analysis of literature signature genes of CCR6+ ILC3 (top panel) and NKP46+ ILC3 (bottom panel) enriched in RFP+GFP+ (BCL6+RORγt+) ILC versus RFP−GFP+ (BCL6−RORγt+) ILC gene sets. Gene sets were originated from RNA-seq data in F. See also Table S2 for detailed list of literature signature genes. NES, normalized enrichment score; FDR, false discovery rate. (H) Veen graph of top expressed signature genes in BCL6+ ILC3s (RNA-seq data of RFP+GFP+ ILC3 in F) and Tfh and Th17 cells (Liu et al., 2016; Tanaka et al., 2018). The listed signature genes were determined by overlapping top expressed genes from the three indicated gene pools and removing pseudogenes and housekeeping genes. Representative Tfh, Th17, and BCL6+ ILCs genes were highlighted in red (please also see Table S3 for detailed list of genes). Data are a representative of three (A, D, and E) or two (B and C) independent experiments.
ILC3s are heterogeneous and can be further divided into subsets using two surface markers, CCR6 and NKP46, with mutual exclusive expression. Since not all RORγt+ ILC3 expressed BCL6 (Fig. 1 B), we further analyzed the expression of BCL6 with a series of ILC markers in total ILC and found BCL6 expression positively correlated with that of CCR6, CXCR5, and CCR7, with a portion of BCL6+ ILC3 also expressed CD4 (Fig. 1 E). Consistent results were obtained from tissues including PP, colon, and small intestine (SI) LPL of B6 mice and also colon LPL of Bcl6RFP/RFP reporter mice (Fig. 1 E; and Fig. S1, A and B). Therefore, BCL6+ ILC seem to match CCR6+ ILC3.
Figure S1.
BCL6 co-staining data and BCL6 KO-induced cytokine alterations in ILC3 (relative to Fig. 1 and Fig. 2). (A) Co-staining of BCL6 versus CCR6, CD4, CCR7, NKP46, and KLRG1 in total ILC isolated from the colon LPL of Bcl6RFP/RFP reporter mice. (B) Co-staining of BCL6 versus CCR6, CD4, CXCR5, NKP46, and KLRG1 in total ILC isolated from the SI LPL of B6 mice. (C) Expression and statistic data of IL-17A and IL-22 in total ILC3 obtained from the SI LPL of Vav1creBcl6fl/fl and Bcl6fl/fl littermates at steady state. Left: Gating strategy for total ILC3, IL-17A+ ILC3 and IL-22+ ILC3. Right: Statistic data for the percentages of IL-17A+ and IL-22+ ILC3. (D) Staining (left) and statistic data (right) of IFN-γ in total ILC3 (gated on Lin−CD90+CD45+RORγt+ live cells) between Vav1creBcl6fl/fl mice and Bcl6fl/fl littermates from colon LPL at steady state in Fig. 2 A. (E) Staining (left) and statistic data (right) of IFN-γ in total ILC3 between Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates from colon LPL under 2.5% DSS-induced colitis in Fig. 2 C. (F and G) Staining data of total ILC, ILC, IL-17A+, and IL-22+ ILC3s of indicated donor origins (left) in the colon (F) and SI (G) LPL obtained from the lethally irradiated NSG mice reconstituted with Lin− BM precursors from B6 (CD45.1+/+) and Vav1creBcl6fl/fl (CD45.2+/+) mice. Statistic data for the percentages of IL-17A+ and IL-22+ ILC3 of each strain were shown (right). Data are a representative of three (A and B) or two (C–G) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired t test (C–G).
Since BCL6+ ILC3 are abundant in intestine LPL, we focused our research on these cells. To better characterize them, we isolated BCL6+ and BCL6- ILC3 cells from intestinal LPL of Bcl6RFP/RFPRorcGFP/+ mice and performed RNA sequencing (RNA-seq) analysis. Consistent with the flow cytometry data, enhanced expression of CCR6+ ILC3 marker genes (Schleussner et al., 2018; Talbot et al., 2020; Zhong et al., 2016; Zhong et al., 2018) including TFs Batf3, Foxs1, and Tcf7, cytokines/chemokines and receptors Il17f, Il22, Ccr6, Cxcr5, neuropeptide receptors, and growth-related factors Ramp1, Vipr2, Bmp2, and other LTi-specific markers Lta, H2-Oa, and Tnfsf11 were observed specifically in RFP+GFP+ ILC, whereas ILC1-related marker genes (Gury-BenAri et al., 2016) like Tbx21, Ncr1, Ifng, Itga1, Gzma, and Xcl1 were found to be highly expressed in RFP−GFP+ ILC (Fig. 1 F). Gene set enrichment analysis (GSEA) also revealed that the expression profile of RFP+GFP+ ILC was correlative to that of CCR6+ ILC3; in contrast, RFP-GFP+ ILC resembled NKP46+ ILC3 or the classical ILC1 in expression profiles (Fig. 1 G; and Tables S1 and S2).
Thus, BCL6 is selectively expressed by CCR6+ ILC3 in mouse intestine. Considering the similarities between ILC and Th cells, ILC3s were previously considered to resemble Th17 cells. However, since Bcl6 and Cxcr5, the signature genes for Tfh cells, were highly expressed by CCR6+ ILC3, these cells may be analogous to both Th17 and Tfh cells. To assess this idea, we analyzed the expression profiles of BCL6+ ILC3 (Fig. 1 F) with our previously published Tfh and Th17 data (Liu et al., 2016; Tanaka et al., 2018; Fig. 1 H). BCL6+ ILC3 were found to highly express both Tfh signature genes such as Bcl6, Cxcr5, Tcf7, and Tox2, as well as Th17-related genes including Rorc, Il17a, Il23r, Ccr6, and Cd93, in addition to many genes specifically expressed by ILC3 only, i.e., Cd83, Ccr7, and Vipr2 (Fig. 1 H).
Collectively, BCL6 is expressed by CCR6+ ILC3, which exhibit Tfh and Th17 dual expression programs.
Increased cytokine expression by ILC3 in the absence of BCL6
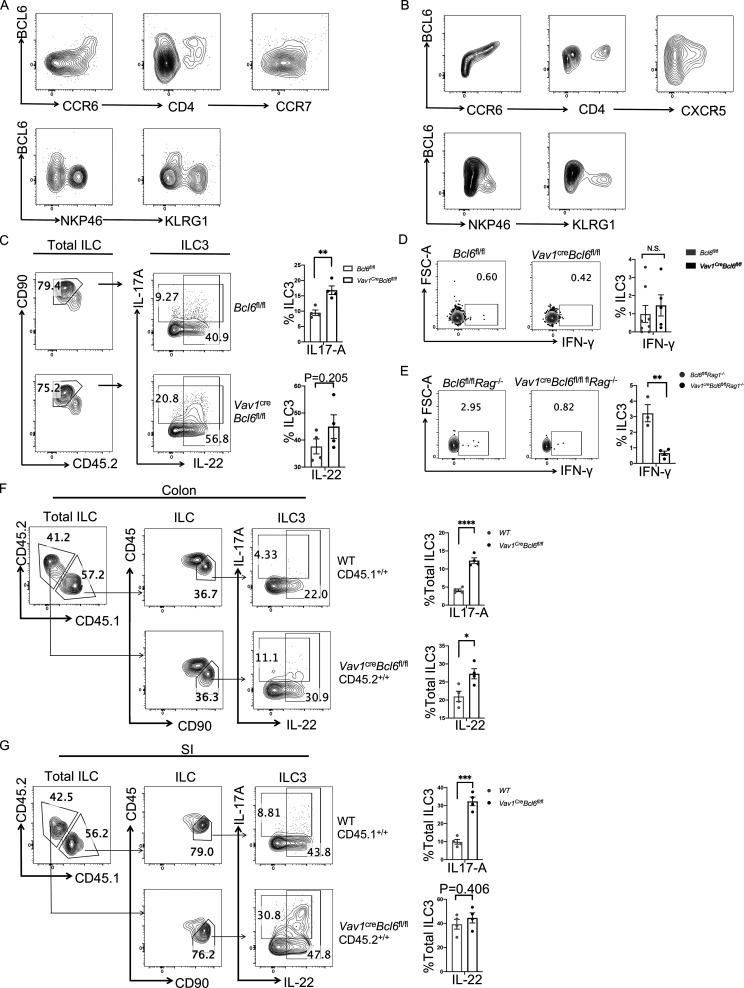
CCR6+ ILC3 produce type 3 cytokines such as IL-17A and IL-22 in response to inflammatory cues. To analyze the function of BCL6 in ILC3, we examined the impact of Bcl6 deficiency by generating Vav1creBcl6fl/fl mice. At steady state, we found that Vav1creBcl6fl/fl mice had similar colonic ILC3 percentages with those of Bcl6fl/fl littermates (Fig. 2 A), indicating that BCL6 is not required for the development of ILC3. However, ILC3 from colon LPL of Vav1creBcl6fl/fl mice secreted significantly higher levels of type 3 cytokines IL-17A and IL-22 than those from control mice, with ILC3 from SI LPL showing similar results (Fig. 2 A and Fig. S1 C). We also constructed bone marrow (BM) chimeric mice by restituting irradiated mice with 1:1 ratio of B6 (CD45.1+/+) versus Vav1creBcl6fl/fl (CD45.2+/+) Lin− BM cells, and noticed that ILC3s of Vav1creBcl6fl/fl strain consistently produced significantly increased levels of IL-17A and IL-22 in colon and SI LPL of the chimeric mice (Fig. S1, F and G). Such results clearly indicate an intrinsic effect of BCL6 in containing the expression of IL-17-A and IL-22 in ILC3, independent of microbiota. On the other hand, production of IFN-γ by BCL6-deficient ILC3 showed no significant difference (Fig. S1 D). Hence, BCL6, selectively expressed in CCR6+ ILC3, may negatively regulate the production of type 3 cytokines such as IL-17A and IL-22 at steady state.
Figure 2.
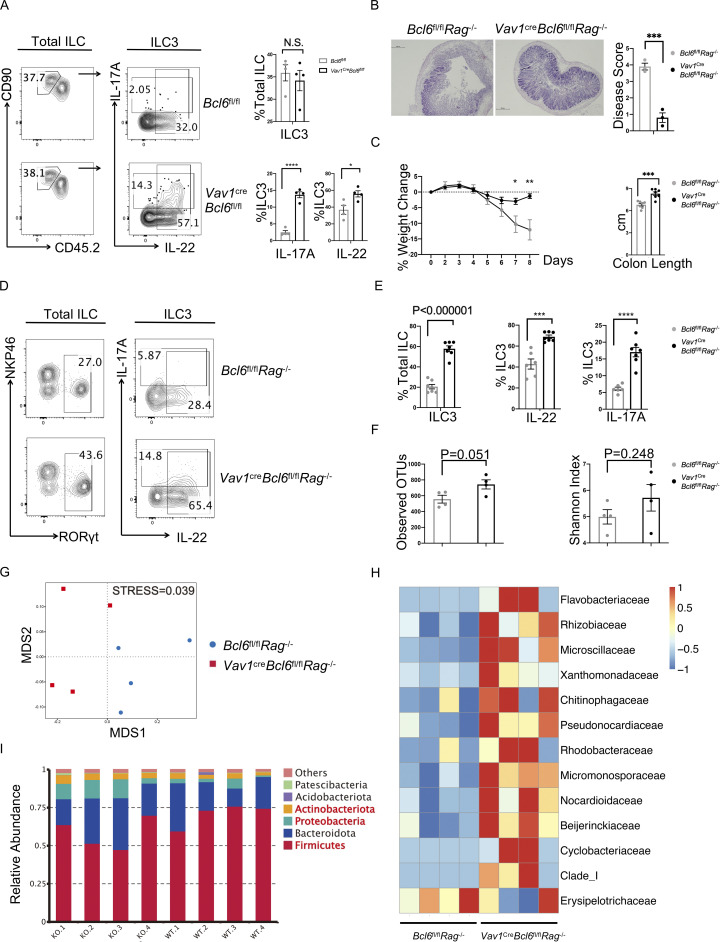
Bcl6 deficiency in ILC3 enhances their cytokine expression and protect to DSS-induced colitis. (A) Expression and statistic data of IL-17A and IL-22 in total ILC3 obtained from the colon LPL of Vav1creBcl6fl/fl and Bcl6fl/fl littermates at steady state. Left: Gating strategy for total ILC3, IL-17A+ ILC3, and IL-22+ ILC3. Right: Statistic data for the percentages of ILC3 and IL-17A+ and IL-22+ ILC3. (B) H&E (left) and estimated disease scores (right) on colon sections from Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates in DSS-induced colitis model. The scale bar indicates length of 200 μm. Also see Table S7 for details on disease score evaluation. (C) Weight changes of Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates in DSS-induced colitis model (left). The statistics of colon length were shown on the right. (D) Expression of IL-17A and IL-22 in total ILC3 obtained from the colon LPL of Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− littermates in the DSS-induced colitis model in C. Left: Gating strategy for total ILC3. Right: IL-17A and IL-22 expression in ILC3. (E) Statistic data for the percentages of ILC3s (left) and IL-17A+ and IL-22 + ILC3 (right) in D. (F) The observed OTU numbers (left) and Shannon index of alpha diversity (right) for the 16 s rDNA-seq data derived from colonic fecal samples from Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates. Raw data of 16s rDNA-seq are available through GSE220438 on GEO DataSets. (G) Non-metric Multi-dimensional Scaling (NMDS) plot for the 16 s rDNA-seq data in F. (H and I) Taxonomic analysis on relative beta diversity abundance for the 16 s rDNA-seq data in F. Comparison of enriched families with >2 significantly changed genera (H) and top 6 enriched phyla (I) between Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates in each sample described in F and G. See also Table S4 for detailed list for differentially enriched phyla and families. Data are representatives of three (A and C–E) or two (B and F–I) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001 by unpaired t test (A, B, C, E, and F).
To further evaluate the role of BCL6+ ILC3 in vivo, we applied dextran sulfate sodium (DSS)–induced colitis model on BCL6-deficient mice. To rule out possible complication by T and B cell alterations in these animals, we generated Vav1creBcl6fl/flRag1−/− mice and found they showed significant resistance to 2.5% DSS treatment, in terms of disease score, weight loss, and colon shortening, compared with Bcl6fl/flRag1−/− littermate controls (Fig. 2, B and C). In the meantime, BCL6-deficient ILC3 were expanded substantially in numbers, and secreted significantly increased amounts of IL-17A and IL-22 at day 8 after DSS treatment (Fig. 2, D and E), which may underscore the protection against DSS-induced colitis (Cao et al., 2019; Ogawa et al., 2004; Zepp et al., 2017; Zhou et al., 2021). Moreover, Vav1creBcl6fl/flRag1−/− mice showed decreased IFN-γ secretion in ILC3, associated with impaired colitis progression (Fig. S1 E). Therefore, Bcl6 deficiency led to increased production of IL-17A and IL-22 in total ILC3.
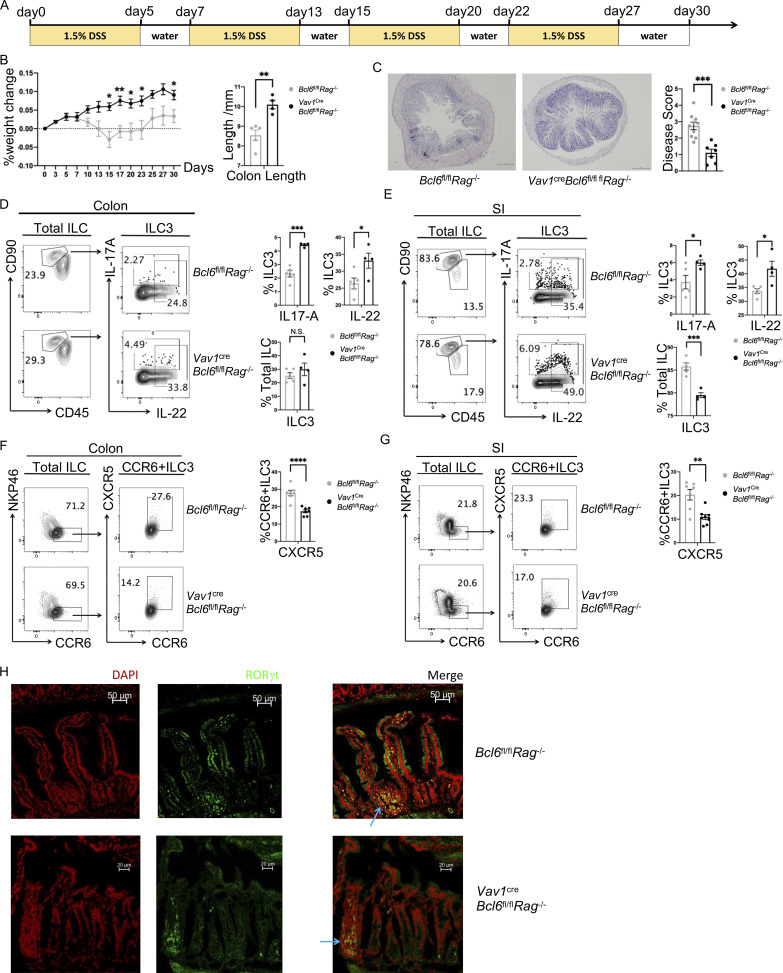
We also established a 30-d chronic DSS-induced colitis model using Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates. Similar with what was observed under 2.5% DSS-induced acute colitis (Fig. 2, C and E), secretion of IL-17A and IL-22 were still increased in BCL6-deficient ILC3 from both colon and SI (Fig. S2, D and E), associated with elevated resistance against long-term 1.5% DSS challenge. Consistently, Bcl6-deficient mice showed improved weight loss and colon shortening with less severe disease scores (Fig. S2, B and C). Of note, ILC3 in colon or especially in SI were not increased in Vav1creBcl6fl/flRag1−/− mice (Fig. S2, D and E), different from the acute model where the percentages of ILC3s were expanded almost three times (from around 20 to 60% of total ILC; Fig. 2, D and E). This is possible due to a lower DSS concentration that activated ILC3 to a lesser extent in the chronic model.
Figure S2.
Chronic DSS-induced colitis and CXCR5-related phenotypes in BCL6-deficient mice (relative to Fig. 2). (A) 30-d 1.5% DSS treatment protocol of the chronic DSS-induced colitis model. (B) Weight changes (left) of Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates in 30-d chronic DSS-induced colitis model. The statistics of colon length were shown on the right. (C) H&E (left) and accordingly estimated disease score (right) of colon sections from Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates in the chronic DSS-induced colitis model in B. Scale bar indicates length of 200 μm. Also see Table S7 for details on disease score evaluation. (D and E) Expression and statistic data of IL-17A and IL-22 in total ILC3 obtained from the colon (D) and SI (E) LPL of Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− littermates in the chronic DSS-induced colitis model in B. Left: Gating strategy for total ILC3, IL-17A+ ILC3 and IL-22+ ILC3. Right: Statistic data for the percentages of total ILC3, IL-17A+ and IL-22+ ILC3. (F and G) Expression and statistic data of CXCR5 in CCR6+ ILC3 from the colon (F) and SI (G) LPL of Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− littermates at steady state. Left: Gating strategy for CCR6+ ILC3 and CXCR5+CCR6+ ILC3. Right: Statistic data for the percentages of CXCR5+CCR6+ ILC3. (H) Immunofluorescence staining of proximal SI sections of Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− littermates at steady state. DAPI staining shown in color red and RORγt staining shown in color green; blue arrows pointing at structures of RORγt+ aggregates that represent atypical SILTs in merged photos; scale bars represented length with indicated value. Data are representative (B–E and H) or combined (F and G) of two independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired t test (B–G).
Since intestinal microbiota and ILC are known to cross-regulate each other (Goc et al., 2021; Gury-BenAri et al., 2016), we conducted bacterial 16S rDNA sequencing (rDNA-seq) analysis on fecal matters from Vav1creBcl6fl/flRag1−/− and control Bcl6fl/flRag1−/− mice at steady state. Our data suggested increased abundance of the microbiota in Vav1creBcl6fl/flRag1−/− samples: increased operational taxonomic unit (OTU) numbers with rather separated Non-metric Multi-dimensional Scaling locations from the Bcl6fl/flRag1−/− group, despite comparable alpha diversity (Shannon index; Fig. 2, F and G). Deficiency in cytokines such as IL-22 usually leads to dysbiosis of microbiota with reduced quantity and diversity (Zenewicz et al., 2013). So, in Vav1creBcl6fl/flRag1−/− mice, with elevated secretion of IL-22 and IL-17A by BCL6-deficient ILC3s (Fig. 2 A), likely resulted in the moderately expanded OTUs (Fig. 2, F and G). Taxon-based analysis provided more evidences. Amplicon Sequence Variants in samples from Vav1creBcl6fl/flRag1−/− mice were significantly increased in Proteobacteria and Actinobacteriota while significantly decreased in Firmicutes among the top six abundant phyla (Fig. 2 I). Specifically, in the Vav1creBcl6fl/flRag1−/− samples, there existed increased colonization of families including Rhizobiaceae, Rhodobacteraceae, Beijerinckiaceae, and Clade_I (Fig. 2 H), all members of class Alphaproteobacteria, which were reported to have positive correlations with expression levels of IL-22 in gut (Gaudino et al., 2021). These four subtypes of Alphaproteobacteria belong to phylum Proteobacteria and may add to increase of this phylum. Conversely, the colitis-promoting family Erysipelotrichaceae under phylum Firmicutes (Chen et al., 2017) was the rare one found with decreased Amplicon Sequence Variants in Vav1creBcl6fl/flRag1−/− samples (Fig. 2 H). Such decrease in the colitogenic bacteria may contribute to enhanced protection in the BCL6-deficient strain following DSS treatment. Collectively, the compositional changes of microbiota in Vav1creBcl6fl/flRag1−/− mice were consistent with increased cytokine production by ILC3.
Interestingly, a previous study showed that CXCR5-deficient CCR6+ ILC3 also exhibited enhanced IL-17A and IL-22 secretion (Sécca et al., 2021). We, therefore, analyzed CXCR5 expression and found that under steady state, there was a significant reduction (nearly 50%) of CXCR5 expression in CCR6+ ILC3 from both colon and SI LPL of Vav1creBcl6fl/flRag1−/− mice (Fig. S2, F and G). It was shown that unlike the Rag1−/− mice where CCR6+ ILC3 included solitary intestinal lymphoid tissues (SILTs) restrictedly developed within the crypt base of the proximal SI, Cxcr5−/−Rag1−/− mice developed atypical RORγt+ aggregates that extended up from crypt to villus (Sécca et al., 2021). Consistently, we observed that BCL6-deficient strain, similar as Cxcr5−/−Rag1−/− mice, also exhibited lost restriction of SILT development in the proximal SI (Fig. S2 H). Therefore, our findings thus suggest a strong possibility that CXCR5 is a functional downstream factor regulated by BCL6 in controlling positioning as well as cytokine secretion of ILC3.
Loss of BCL6 reduced CCR6+ but increased NKP46+ ILC3
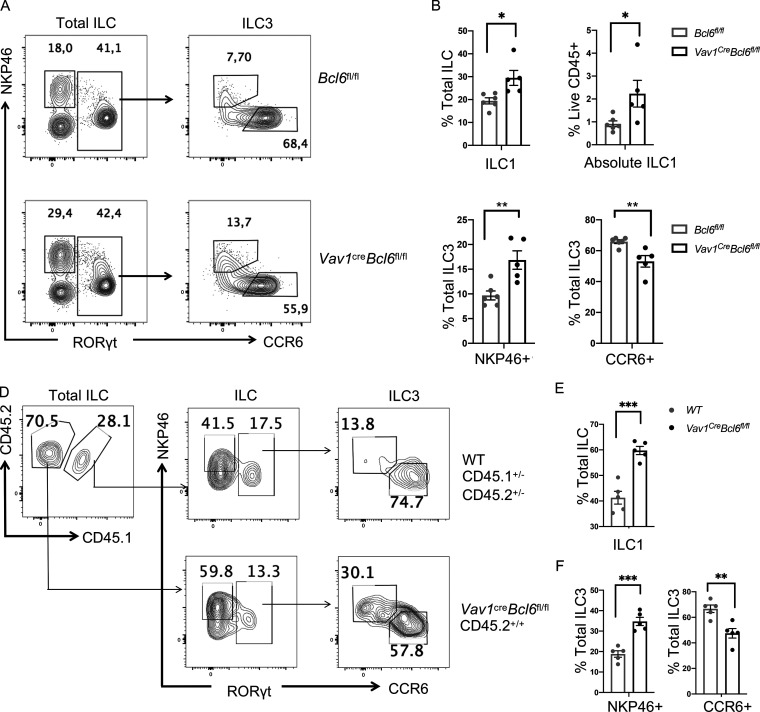
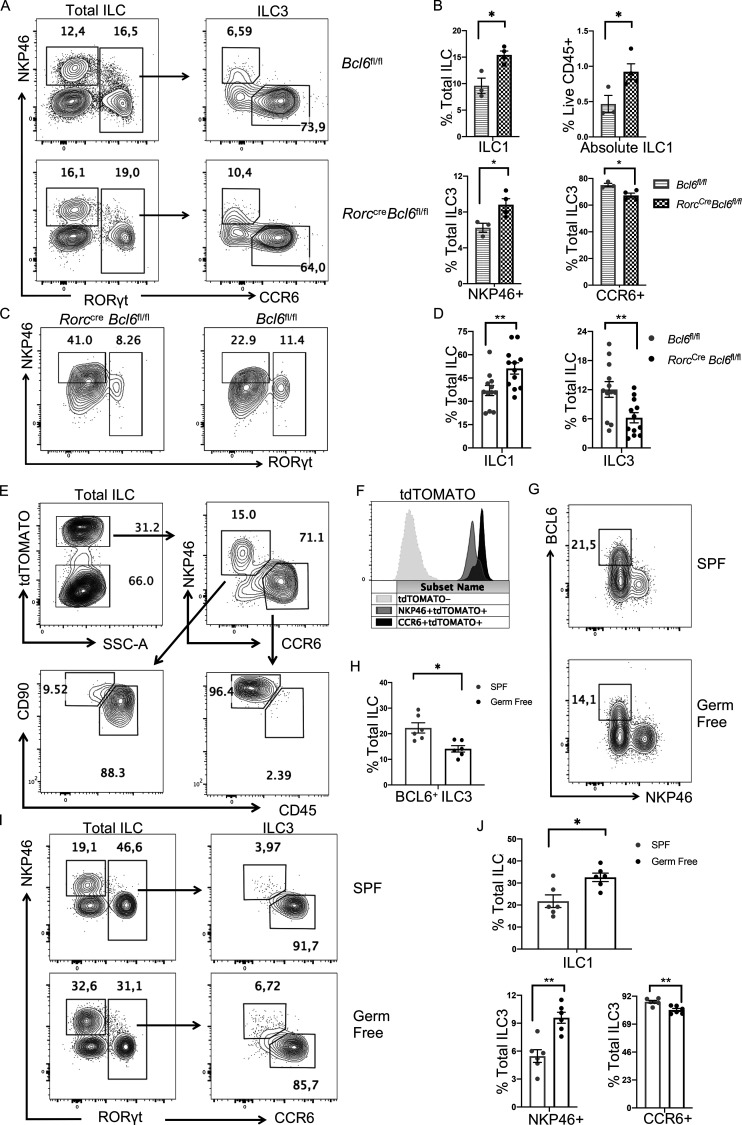
To further understand the function of BCL6 in ILC3, we analyzed their major subsets, CCR6+ and NKP46+. Surprisingly, although total ILC3 were unchanged in percentages at steady state (Fig. 2 A), we observed that in both colon and SI LPL of Vav1creBcl6fl/fl mice, CCR6+ ILC3 were significantly reduced, whereas NKP46+ ILC3 significantly increased, compared with Bcl6fl/fl mice (Fig. 3, A and C; and Fig. S3, A and B). Following this trend, NKP46+ ILC1 were also found to expand in both relative percentages and absolute numbers (Fig. 3, A and B; and Fig. S3 B). Within T-bet+ ILC1, we further examined Eomes expression and found the T-bet+Eomes− cNK subset was not affected by Bcl6 deficiency (data not shown).
Figure 3.
Bcl6 deficiency alters ILC3 vs. ILC1 ratio. (A) Staining data of total ILCs (left) and ILC3s (right) in the colon LP of Vav1creBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. (B and C) Statistic data for the percentages of ILC1 (B), NKP46+ ILC3, and CCR6+ ILC3 (C) in A. Gating on RORγt- NKP46+ ILC as ILC1, RORγt+NKP46+ ILC as NKP46+ ILC3, and RORγt+CCR6+ ILC as CCR6+ ILC3. (D) Staining data of ILC and ILC3 of indicated donor origins in the colon LPL obtained from the lethally irradiated B6 (CD45.1+/+) mice reconstituted with Lin− BM precursors from B6 (CD45.1+/−CD45.2+/−) and Vav1creBcl6fl/fl (CD45.2+/+) mice. (E and F) Statistic data for the percentages of ILC1 (E), NKP46+ ILC3, and CCR6+ ILC3 (F) from the chimera mice in D. Data are representatives of three (A, B, and C) and two (D, E, and F) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data with significance are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired t test (B, C, E, and F).
Figure S3.
Intrinsity-related analysis of ILC1/ILC3 ratio alterations in Vav1creBcl6fl/fl mice (relative to Fig. 3). (A) Staining data of total ILCs (left) and ILC3s (right) in the SI LPL of Vav1creBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. (B) Statistic data for the percentages of ILC1, NKP46+ ILC3, and CCR6+ ILC3 in A. (C) Staining data of ILC and ILC3 of indicated donor origins in the SI LPL obtained from the lethally irradiated B6 (CD45.1+/+) mice mentioned in Fig. 3, D−F. (D) Statistic data for the percentages of ILC1, NKP46+ ILC3, and CCR6+ ILC3 from the chimeric mice in C. (E) Staining pattern of total ILC and ILC3 populations obtained from colon LPL of Cd2CreBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. (F) Statistic data for percentages of indicated ILC subsets in A. Data are a representative of three (A and B) and two (C–F) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01 by unpaired t test (B, D, and F).
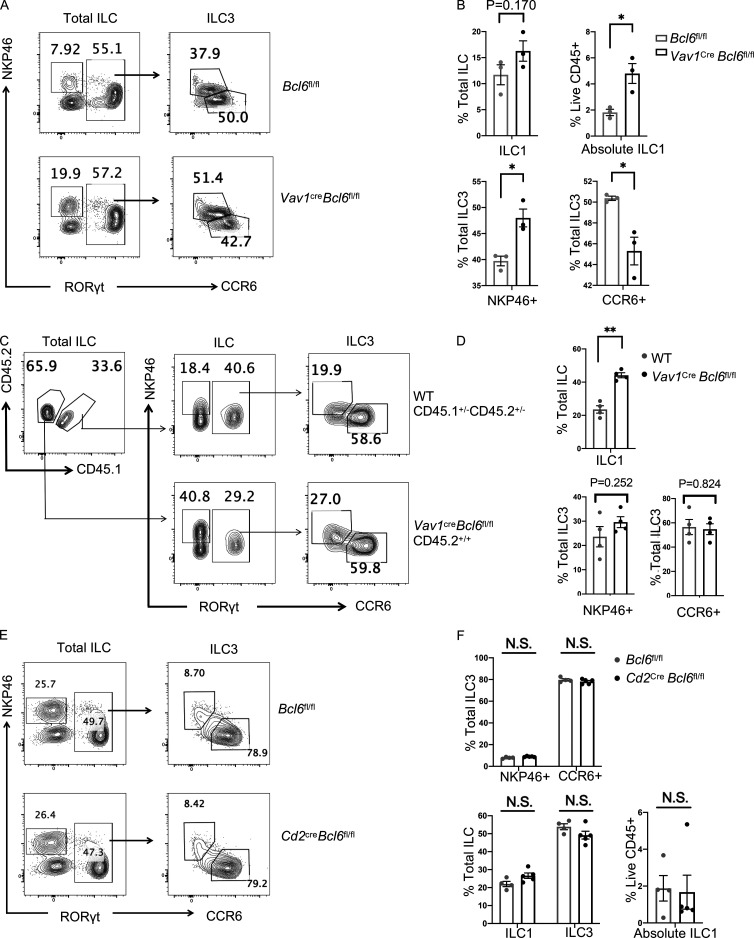
Thus, Bcl6 deficiency in Vav1creBcl6fl/fl mice decreases CCR6+ ILC3 but increases NKP46+ ILC including NKP46+ ILC3 and ILC1. To determine whether this change was cell intrinsic, we constructed mixed BM chimeras in which 1:1 mix of ILC progenitors-containing Lin− BM cells from Vav1creBcl6fl/fl mice (CD45.2+/+ background) and CD45.1+/−CD45.2+/− B6 background mice were co-transferred into lethally irradiated CD45.1+/+ recipient mice (Fig. 3, D−F). Similar as above, in both colon and SI LPL of the chimeric mice, we observed decreased CCR6+ ILC3, whereas increased NKP46+ ILC3 and ILC1 in BCL6-deficient CD45.2+/+ cells, as compared with CD45.1+/−CD45.2+/− donor-derived cells (Fig. 3, D−F; and Fig. S3, C and D). Next, to rule out the complication by other cell types, particularly T cells, we analyzed Cd2creBcl6fl/fl mice, where BCL6 was deficient in T cells but not ILC, and found no alteration in ILC subsets compared with their control littermates (Fig. S3, E and F).
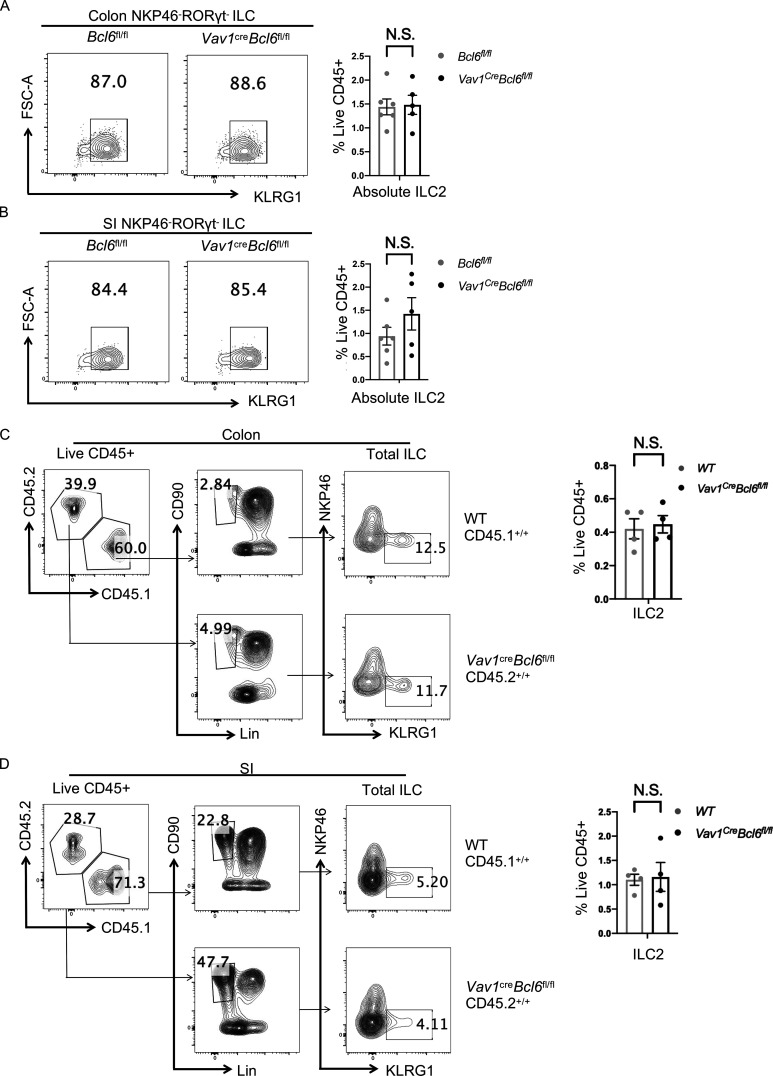
On the other hand, although BCL6 is not expressed in ILC2 at steady state (Fig. 1, A−D), the absolute numbers of ILC2s in both colon and SI LPL in Vav1creBcl6fl/fl mice under steady status were examined, and we found them not affected by Bcl6 deletion (Fig. S4, A and B). To further validate these results, we also examined ILC2 cells in the BM chimeric mice restituted with 1:1 ratio of B6 (CD45.1+/+) versus Vav1creBcl6fl/fl (CD45.2+/+) Lin− BM cells. After re-constitution, the absolute numbers of ILC2 remained similar between the two strains of BM cells (Fig. S4, C and D), suggesting that BCL6 does not regulate ILC3 plasticity toward ILC2.
Figure S4.
ILC2-related alteration in Vav1creBcl6fl/fl mice (relative to Fig. 3). (A and B) Expression and statistic data of ILC2 from the colon (A) and SI (B) LPL of Vav1creBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. Left: Gating strategy for ILC2 (NKP46−RORγt− KLRG1+ ILC). Right: Statistic data for the percentages of ILC2. (C and D) Staining data of ILC and ILC2 of indicated donor origins (left) in the colon (C) and SI (D) LPL obtained from the lethally irradiated NSG mice indicated in (Fig. S1, F and G). Statistic data for the percentages of ILC2s of each strain were shown on the right. Data are a representative of two (A–D) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM by unpaired t test (A–D).
Thus, Bcl6 deficiency in ILC results in decreased CCR6+ ILC3 and increased NKP46+ ILC3 and ILC1 in a cell-intrinsic manner, without affecting ILC2.
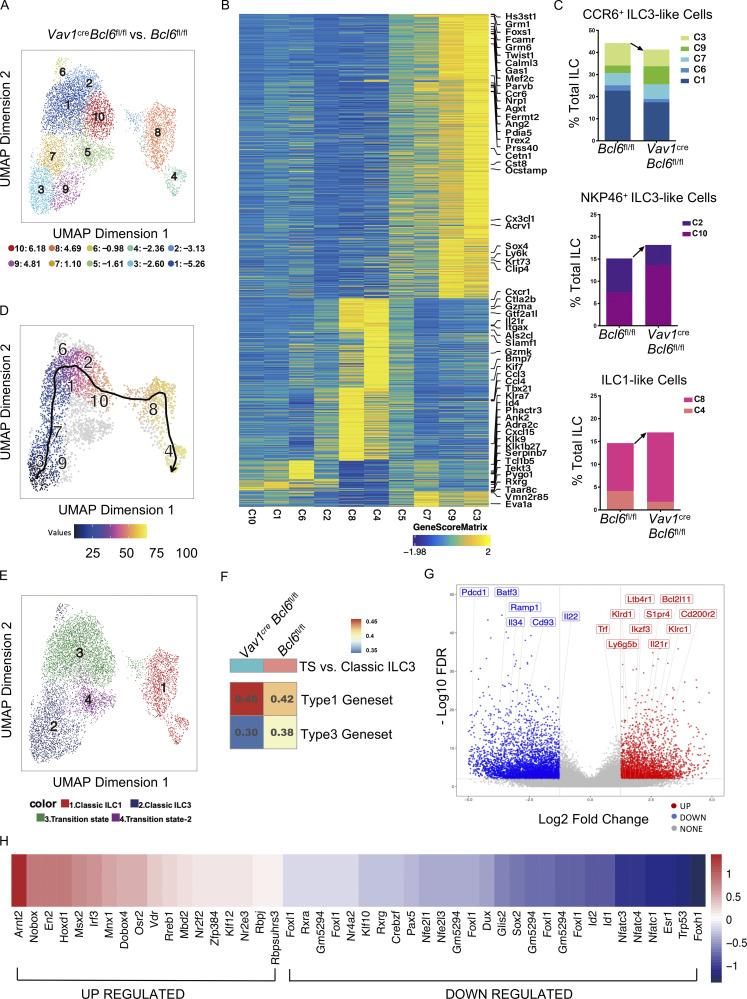
Single-cell analysis reveals increased ILC3 plasticity in the absence of BCL6
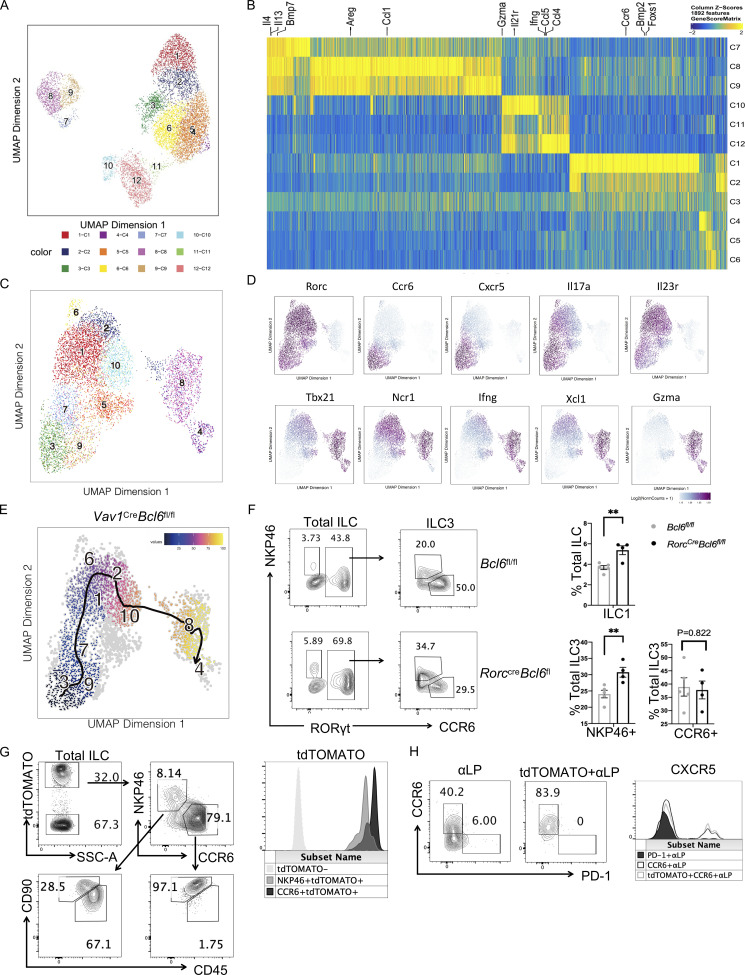
Our data indicate that Bcl6 deficiency decreased CCR6+ but increased NKP46+ ILC (both ILC3 and ILC1), suggestive of enhanced ILC3 plasticity toward ILC1. To test this hypothesis, we applied single-cell ATAC-seq (scATAC-seq) analysis on total ILC from colon and SI LPL of Vav1creBcl6fl/fl and their control littermate mice. Since ILC3s and ILC1s were of our interests, we eliminated cells with ILC2 type in our analysis based on preliminary classification (Fig. S5, A and B), and obtained 10 clusters from the remaining cells with selective highly accessible lineage-related marker genes (Fig. S5 C and Fig. 4 B). Cluster 5 did not possess gene score patterns specific to any ILC lineages and thus was excluded from further analysis. Post evaluation of the marker gene-based lineage-specific trend shown by genescore heatmap (Fig. 4 B) and “geographic” distributions (Fig. S5 D), as well as the proportional changes of Vav1creBcl6fl/fl vs. Bcl6fl/fl source ILCs (Fig. 4 A), we predicted expression profiles of the nine clusters and characterized these ILC3 or ILC1 feature cells into three subgroups: the decreased “CCR6+ ILC3-like cells” including cluster 3, 9, 7, 6, and 1, the increased “NKP46+ ILC3-like cells” containing cluster 2 and 10, and the also increased “ILC1-like cells” consisting of clusters 8 and 4 (Fig. 4 C). Proportional alteration on the three subgroups defined by their chromatin accessibility patterns (Fig. 4 C) was consistent to the ILC3 and ILC1 ratio changes in BCL6-deficient ILCs found by flow cytometry (Fig. 3, A−C).
Figure S5.
Supplemental data of scATAC-seq as well as phenotypes in RorccreBcl6fl/fl mice and Bcl6ERT2creRosatdtomato mice (relative to Fig. 4 and Fig. 5). (A) UMAP visualization of the scATAC-seq (Fig. 4) clustering pattern of the total intestinal ILC from colon and SI LPL of Vav1creBcl6fl/fl mice or Bcl6fl/fl littermates. (B) Heatmap of marker genes (determined by gene score matrix) of each cluster (vertically labeled) of the total ILC in A. Selected ILC lineage specific genes (Gury-BenAri et al., 2016) with high gene scores in the 12 clusters were horizontally labeled on top (see also Table S5). (C) UMAP visualization of clustering pattern of the ILC post deletion of ILC2 form cells from original scATAC-seq data pool in A. (D) UMAP distribution of relative chromatin accessibility of indicated genes for cells in C, represented by color with scale bar (Log2 = 1.15–1.30). (E) Trajectory analysis of Vav1creBcl6fl/fl source clusters shown in C. (F) Staining data of total ILC and ILC3 (left) in the SI LPL of RorccreBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. Statistic data of indicated ILC populations were shown (right). (G) Staining pattern (left) of CCR6+ and NKP46+ ILCs in tdTOMATO fated ILCs obtained from SI LPL of Bcl6ERT2creRosatdtomato fate mapping mice mentioned in Fig. 5, E and F. Expression of tdTOMATO for indicated ILC subsets were shown (right). (H) Staining pattern (left) of CCR6+ and PD-1+ αLP (Lin− IL-7R+α4β7+) in total or tdTOMATO fated BMs obtained from Tamoxifen treated Bcl6ERT2creRosatdtomato fate-mapping mice. Expression of CXCR5 for indicated cell types were shown (right). Data are a representative of three (F–G) or two (H) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data are shown as mean ± SEM. **P < 0.01 by unpaired t test (F).
Figure 4.
scATAC-seq data reveals increased ILC3 plasticity in BCL6-deficient mice. The total intestinal ILC were obtained from the SI and colon LPL of Vav1creBcl6fl/fl or Bcl6fl/fl littermates (combination of three mice for each strain) by FACS sorting (gated on Lin−CD90+CD45+ live cells), and used for scATAC-seq analysis. UMAP visualizations for both Vav1creBcl6fl/fl and Bcl6fl/fl samples are visually identical, so all UMAP figures shown are representative data of one sample unless specifically indicated. Raw data of scATAC-seq are available through GSE221341 on GEO DataSets. (A) UMAP visualization of clustering pattern of the ILCs post deletion of ILC2 form cells from original scATAC-seq data pool (Fig. S5 A). Proportional changes of each cluster of Vav1creBcl6fl/fl versus Bcl6fl/fl littermate samples were labeled by blue-red colors with scales shown below. (B) Heatmap of marker genes (determined by gene score matrix) of each cluster (vertically labeled) of the total ILC2-depleted ILC in A. Selected ILC lineage specific genes (Gury-BenAri et al., 2016) with high gene scores in the 10 clusters were horizontally labeled on top (see also Table S5). (C) Frequencies of three subgroups from the total ILC2-depleted ILCs in A between Bcl6fl/fl and Vav1creBcl6fl/fl samples. Each subgroup is consisted of specified clusters (labeled by different colors) indicated by chromatin level characterization from B and Fig. S5 D. Arrow indicated direction of cell number changes. (D) Trajectory analysis for Bcl6fl/fl source clusters from A. See also Fig. S5 E for trajectory analysis result for Vav1creBcl6fl/fl sample. (E) Representative UMAP visualization of broad rename of cells from A. (F) GSVA of differential genesets generated by comparing chromatin accessibility of genes from TS vs. those from classic ILC3 for Vav1creBcl6fl/fl and Bcl6fl/fl samples, respectively. The overlapped genes between the two genesets were excluded from analysis (see also Table S5). (G) Volcano plot for Vav1creBcl6fl/fl source TS vs. classic ILC3 differential geneset generated in F. Selective regulated peaks (labeled by nearest gene) were Vav1creBcl6fl/fl-specific representatives matching literature ILC1 marker genes (right red, up-regulated) or CCR6+ ILC3 marker genes (left blue, down-regulated). See also Table S2 for literature marker genes and Table S6 for detailed information of selected peaks and nearby genes. (H) Transcription factor motif enrichment scores of Vav1creBcl6fl/fl TS vs. those of Bcl6fl/fl TS. The motifs shown here are selected by enrichment score of mlog10Padj >0.1. Value of score is labeled by blue-red color scale.
Trajectory analysis at single-cell level is a powerful tool for lineage tracing. So in our case, the whole process of transformation from CCR6+ ILC3 to NKP46+ ILC3 and finally to ILC1 could be captured at the chromatin accessibility level using this method. Indeed, we found an identical order following the direction of ILC3 to ILC1 transdifferentiation from clusters 3, 9, 7, 6, 1, 2, 10, and 8 to cluster 4 for both Vav1creBcl6fl/fl and Bcl6fl/fl source ILCs (Fig. 4 D and Fig. S5 E), which was consistent with the predicted expression-driven characterization (Fig. 4 C).
To better analyze the ILC3 to ILC1 plasticity, we re-grouped the nine clusters based on lineage steadiness: cells in clusters 6, 1, 2, and 10 were selected as “transition state” (TS) exhibiting high scores of both ILC3 and ILC1 marker genes, while single lineage-trending cells including clusters 3, 9, and 7 were termed as “classic ILC3” and clusters 4 and 8 as “classic ILC1” (Fig. 4, B and E; and Fig. S5 D).
Since BCL6 is selectively expressed in CCR6+ ILC3, its loss should directly impact the initial step of ILC3 to ILC1 transdifferentiation: from classic ILC3 to TS. We generated lists of genes with top modulated chromatin accessibility in TS relative to classic ILC3 for both Vav1creBcl6fl/fl and Bcl6fl/fl mice samples, separately (Table S2), and deleted overlapped genes between the two genesets to focus on differential modulations. Gene set variation analysis (GSVA) of these gene lists revealed modulations with enhanced trend toward type 1 features while impaired type 3 signatures during transformation of especially Vav1creBcl6fl/fl sample (Fig. 4 F). More precisely, Bcl6 deficiency resulted in specifically upregulated chromatin accessibility of type 1 marker genes such as Ikzf3, Klrc1, Il21r, and Trf, and downregulation of type 3 marker genes including Batf3, Ramp1, Cd93, and Il22 through classic ILC3–to-TS transition (Fig. 4 G). Interestingly, Ikzf3 (encodes Aiolos) was also previously recognized to favor ILC3-to-ILC1 transformation in human (Cella et al., 2019).
Given the dynamic analysis, TS from Vav1creBcl6fl/fl mice might locate at a stage of transition that is closer to ILC1. We then examined motif enrichment in TS and found that TS of Vav1creBcl6fl/fl sample displayed upregulation of type 1 driving motifs including the Rorc-inhibiting Arnt2 (Dong et al., 2014), the type 1–promoting ones Irf3 and Klf12 (James et al., 2018; Lam et al., 2019), and the plasticity promoting Rbpj (Cella et al., 2019), whereas reduced enrichment of type 3 signatures inducing motifs containing the Rorc-promoting Nfatc1, Nfatc3 (Yahia-Cherbal et al., 2019), the Il17a-related Esr1 (Fuseini et al., 2019), and ILC development regulating factors such as Id2, Sox2, and Rxrg (Cherrier et al., 2012; Ishizuka et al., 2016) were observed (Fig. 4 H). Hence, the epigenetic regulation at TS in Vav1creBcl6fl/fl mice favored an overall pro-Tbx21 while anti-Rorc direction.
Taken together, at the chromatin level, Bcl6 deficiency led to alteration at the initial step of ILC3 to ILC1 transition with enhanced plasticity in CCR6+ ILC3.
Microbiota promotes BCL6 expression to restrict ILC3 plasticity
As scATAC-seq analysis suggested, BCL6 might function to suppress ILC3-to-ILC1 transformation. To confirm this and to locate the development step at which BCL6 functions in the ILC3 lineage, we generated RorccreBcl6fl/fl mice, in which Bcl6 is specifically deleted in ILC3 (Constantinides et al., 2014; Walker et al., 2019). Similar to Vav1creBcl6fl/fl mice (Fig. 3, A−C), decreased CCR6+ and expanded NKP46+ ILC3 and ILC1 were observed in both colon and SI LPL of RorccreBcl6fl/fl mice (Fig. 5, A and B; and Fig. S5 F), supporting that Bcl6 deficiency in ILC3 promotes CCR6+-to-NKP46+ transition.
Figure 5.
Microbiota restrain ILC3 to ILC1 plasticity through enhancing BCL6 expression. (A) Staining data of total ILC (left) and ILC3 (right) in the colon LPL of RorccreBcl6fl/fl mice and Bcl6fl/fl littermates at steady state. (B) Statistic data of indicated ILC populations in A. (C) Staining data of total ILCs showing ILC1 and ILC3 populations for donor CCR6+ ILC3s obtained from RorccreBcl6fl/fl and Bcl6fl/fl littermates 25 d following transfer into M-NSG recipient mice. (D) Statistic data of indicated ILC populations in C. (E) Staining pattern of CCR6+ and NKP46+ ILCs in tdTOMATO fated ILCs obtained from colon LPL of Tamoxifen treated Bcl6ERT2creRosatdtomato fate mapping mice. (F) Representative expression of tdTOMATO for indicated ILC subsets in E. (G) BCL6 staining data in total ILC obtained from colon LPL from SPF and germ-free mice. (H) Statistic data for the percentages of BCL6+ ILC3 within total ILCs from SPF and germ-free mice in G. (I) Staining pattern of total ILC and ILC3 in SPF and germ-free mice in G. (J) Statistic data for the percentages of ILC1, NKP46+ ILC3, and CCR6+ ILC3 in SPF and germ-free mice in G. Data are representative of three (A, B, E, and F), or combined from three (C and D) or two (G, H, I, and J) independent experiments. Each dot in the bar charts represents a sample generated from an individual mouse. Data with significance are shown as mean ± SEM. *P < 0.05, **P < 0.01 by unpaired t test (B, D, H, and J).
Since not all RORγt+ ILC3 are CCR6+, we transferred CCR6+ ILC3s from intestinal LPL of either RorccreBcl6fl/fl mice or Bcl6fl/fl mice to the ILC-null M-NSG (NOD-PrkdcscidIl2rgem1/Smoc) recipient mice. Previous work (Chea et al., 2016) showed that the rate for CCR6+ ILC3 to lose their RORγt and upregulate NKP46 and T-bet expression post transfer represents the degree of ILC3 plasticity. At 25 d after transfer, we noticed that CCR6+ ILC3 from RorccreBcl6fl/fl mice exhibited significantly increased differentiation into ILC1, compared with those from Bcl6fl/fl mice (Fig. 5, C and D). Hence, BCL6 expression in ILC3 restrains their plasticity and trans-differentiation into ILC1.
To understand whether BCL6 is involved in the spontaneous transition of ILC3 toward ILC1 in vivo, we generated a Bcl6ERT2creRosatdtomato fate-mapping mouse and analyzed their ILC from both colon and SI LPL after 1 mo injection of Tamoxifen. Using a ILC lineage-specific gating strategy using surface markers CD45 and CD90 (Guo et al., 2015), we found that in tdTOMATO+ ILC, which had at least historically expressed BCL6, there existed both CCR6+ ILC3, as well as NKP46+ ILC, consisting of both NKP46 + ILC3 (tdTOMATO+NKP46+CD45medCD90hi) and ILC1 (tdTOMATO+NKP46+CD45hiCD90med; Fig. 5 E and Fig. S5 G). tdTOMATO+CCR6+ cells, with dominant percentages, expressed higher levels of tdTOMATO, whereas tdTOMATO+NKP46+ cells, with reduced percentages, had lower tdTOMATO expression (Fig. 5, E and F; and Fig. S5 G). Since NKP46+ ILC do not express BCL6 (Fig. 1, A−D), existence of tdTOMATO+NKP46+ ILC in Bcl6ERT2creRosatdtomato mice indicates conversion of BCL6-expressing CCR6+ into NKP46+ ILC physiologically.
We also examined the expression of BCL6 in BM progenitors of ILC using Bcl6ERT2creRosatdtomato fate-mapping mice. Results showed tdTOMATO+ αLP mostly in LTiP-containing CCR6+ progenitors, but not in common ILC progenitor–containing PD-1+ progenitors (Yu et al., 2016; Fig. S5 H). Since CCR6+ ILC3s developed from LTiP while other ILC subsets from common ILC progenitor, our result indicates selective expression of BCL6 in CCR6+ ILC3 lineage before its maturation. Therefore, except for its function in CCR6+ ILC3 maintenance at mature stage (Fig. 5, A−D), BCL6 might also regulate ILC3, but not other ILC types, at early developmental stage.
Since microbiota have been found to shape the compositions of ILC (Gury-BenAri et al., 2016), we analyzed BCL6 expression in germ-free mice. BCL6+ ILC were significantly reduced in colonic LPL of these mice, compared with specific pathogen–free (SPF) animals (Fig. 5, G and H). Thus, BCL6 expression in ILC is independent, while under modulation by microbiota. Consistently, in germ-free mice, there existed reduced percentages of CCR6+ and expanded portions of NKP46+ ILC3 and ILC1, compared with SPF animals (Fig. 5, I and J), suggesting increased ILC3 to ILC1 plasticity.
Collectively, our data indicate that microbiota limit ILC3-ILC1 trans-differentiation, at least in part through regulation of BCL6 expression.
Discussion
ILC, similar to Th cells in their cytokine production, provides acute host protection in infection and tissue damage. In the current study, we started by analyzing ILC expression of BCL6, the master transcription factor in Tfh cells. We found that CCR6+ ILC3 have both Tfh and Th17 programs featured by co-expression of BCL6 and RORγt. More importantly, we showed that BCL6 functions to maintain ILC3 identity and repress their conversion toward ILC1.
Our findings on BCL6 expression and function in ILC differ sharply from a previous report, where BCL6 was reported to be expressed at highest level in ILC1 post-paralleled RNA-seq and ATAC-seq analysis on NK and major ILC subsets (Pokrovskii et al., 2019). However, this transcriptional and epigenetic analysis lacked validation by authentic BCL6 protein data among ILCs. Here, we thoroughly analyzed BCL6 expression pattern in total ILC both at protein level through FACS staining and at transcription level using Bcl6RFP/RFPRorcGFP/+ reporter mice, and we found that statistically all (94.3%) BCL6+ ILC expressed CCR6 and RORγt but not the ILC1 marker NKP46. Moreover, the sorted BCL6+ RORγt+ ILC possess an expression profile highly overlapping with that of CCR6+ ILC3 relative to NKP46+ ILC3 or ILC1 as indicated by GSEA. Our results were in line with established studies showing BCL6-inducing factor STAT3 is essential for ILC3 development (Guo et al., 2014), and that BCL6 inhibitor STAT5 is enriched in ILC1 (Choi et al., 2013; Villarino et al., 2017). In addition, BCL6 expression patterns defined by us are consistent with several previous studies, in which the Bcl6 mRNA was higher in CCR6+ ILC3/LTi than type 1 NKP46+ ILC3 or NK cells (Tizian et al., 2020; GSE184841). According to human research, existence of BCL6+ ILC was found in human tonsil with an expression profile overlapped mainly with that of CCR6+ ILC3, featured by high levels of CXCR5, CCR6, and CCR7 expressions (O’Connor et al., 2021). But these human “ILCFR” do not express other lineage markers including RORγt, likely due to variations between human and mice.
To prove the function of BCL6 as NK or ILC1-promoting factor in vivo, the previous report (Pokrovskii et al., 2019) generated NcrcreBcl6fl/fl mice and found ILC1 in these mice were decreased from 4.28 to 1.93%. Such in vivo phenotype alone was not sufficient in making a conclusion on BCL6 function. Whether the BCL6-dependent “ILC1-promoting function” was cell-intrinsic, or the mechanisms underlying BCL6 function were not addressed in this report. Since we found BCL6 selective expression only in CCR6+ ILC3, we first generated Vav1creBcl6fl/fl mice and detected decreased CCR6+ ILC3 from relatively 70 to 50%, but increased ILC1 from relatively 20 to 30%. Analysis of mixed BM chimeric mice, RorccreBcl6fl/fl mice, BCL6 lineage tracing model as well as transfer experiments all provide supportive evidences that deficiency of Bcl6 in CCR6+ ILC3 greatly elevates ILC3-to-ILC1 plasticity and thus causes the aforementioned phenotype. We further applied scATAC-seq to WT or BCL6-deficient ILC and found that BCL6 helped maintain Rorc-promoting genes (Batf3, Cd93, Il22, Nfatc1, and Esr1) but repressed Tbx21-promoting genes (Ikzf3, Klrc1, Il21r, Klf12, and Rbpj) to restrict ILC3-to-ILC1 transition. In addition, functions of BCL6 in CD4+ T cells are very similar with our findings in CCR6+ ILC3. In Tfh cells, BCL6 blocks T-bet expression through inhibitory binding of Tbx21, Ifng, and Il12rb1 (Liu et al., 2016) and represses Tfh cells plasticity toward Th1 cells (Alterauge et al., 2020). In Th17 cells, BCL6 restricts Rorc function by reducing the expression of Il17a and Il17f cytokines (Hatzi et al., 2015; Nurieva et al., 2009), without affecting Rorc expression (Nurieva et al., 2009). Consistent with its function in Th17 cells, we found that BCL6 in CCR6+ ILC3 restrains secretion of IL-17A and IL-22 while retaining expression of RORγt. Interestingly, Vav1creBcl6fl/flRag1−/− mice and Cxcr5−/−Rag1−/− mice showed similar phenotypes in terms of atypical SILT development, and enhanced type 3 cytokine secretion in CCR6+ ILC3, indicating a strong possibility that CXCR5 is a functional downstream factor for BCL6 in regulating ILC3. Although there exists evidence supporting this hypothesis, for instance, BCL6-deficient Tfh cells also exhibited decreased CXCR5 expression (Nurieva et al., 2009), the exact mechanism underlying BCL6 regulation of CXCR5 expression in ILC3 requires further investigation. In short, we concluded that BCL6 is a canonical marker, an effector function repressor as well as a lineage maintaining contributor in CCR6+ ILC3, likely through CXCR5.
BCL6 regulation of CCR6+ ILC3 might start before their maturation, since selective expression of BCL6 in CCR6+ ILC3 lineage was observed early during its developmental stage (LTiP). In this study, evidences regarding the altered ILC3/ILC1 ratios and the transfer experiments using the RorccreBcl6fl/fl strain have sufficiently proved the maintenance of CCR6+ ILC3 lineage identity by BCL6. Nevertheless, whether BCL6 could affect the differentiation of CCR6+ ILC3 precursors and how require further research in the future.
Regulation of ILC plasticity is associated with pathology of inflammation. For example, the rates of ILC3-to-ILC1 transformation were found to increase in Crohn’s disease patients, reflecting physiological necessity for elevated immune responses to pathogens (Bernink et al., 2015). It is also witnessed in the same study that there exists ILC1-to-ILC3 transition, which was promoted by IL-23 and IL-1β in vitro. The reconstitution of steady state ILC3-ILC1 balance likely occurs spontaneously for tissue repair post inflammation, but lack in vivo verification and mechanistic analysis. Given the BCL6-promoting cytokine IL-6 (Choi et al., 2013) was reported to augment the ILC3-driving IL-23/IL-1α secretion in intestinal ILCs (Powell et al., 2015), it is possible that the ILC1-to-ILC3 transition could be modulated through targeting BCL6-involved pathways.
ILC plasticity is also environmentally sensitive and can be affected by microbiota (Goc et al., 2021; Gury-BenAri et al., 2016). We found that microbiota regulate BCL6 expression in ILC. Although the bacteria species that modulate BCL6 expression ILC3 remain elusive, there exist candidate bacteria such as Bacteroidales (Kuhn et al., 2018) and Lactobacillus rhamnosus GR-1 (Yeganegi et al., 2010) likely elevate BCL6 expression in gut with positive correlation with BCL6-promoting factor IL-6 (Wan et al., 2021) and JAK/STAT signaling (Liu et al., 2016). Moreover, since Vav1creBcl6fl/flRag1−/− mice possessed a shift in colonic microbiome reflecting high levels of IL-22, BCL6 may in-turn regulate the composition of microbiome through affecting CCR6+ ILC3. A recent study showed that increased ILC3 plasticity changed CD4+ T cell proportion through MHC-II to induce cancer-promoting microbiota dysbiosis (Goc et al., 2021). Such work demonstrated connections among microbiota, adaptive immunity, and ILC homeostasis, while our finding on the microbiota-BCL6 crosstalk showed cytokine-based regulation directly between ILC3 plasticity and host microbiome.
For decades, CCR6+ ILC3/LTi, as part of group 3 ILC, have been only considered to be the functional innate couterpart of Th17 cells due to high expression of canonical Th17-signature genes, such as Rorc, Ccr6, Il17a, and Il22. However, CCR6+ ILC3/LTi also share similarity with Tfh cells in expression of the signature gene Cxcr5. CCR6+ ILC3/LTi require CXCR5 for recruitment of immune cells during secondary lymphoid organ genesis and positional restriction of SILT development at mature stage (Finke et al., 2002; Sécca et al., 2021; Zhong et al., 2018), and Tfh cells also rely on CXCR5–CXCL13 axis for contacting with B cells in germinal center formation (Dong, 2021). Interestingly, our study characterizes BCL6, the master transcription factor of Tfh cells, as a canonical marker for CCR6+ ILC3/LTi cells which maintain their lineage identity. In addition to Bcl6 and Cxcr5, CCR6+ ILC3/LTi also express several key Tfh marker genes including Tox2, Tcf7, S1pr1, and Cxcr4, which thus also defines CCR6+ ILC3/LTi cells as a “Tfh-like” population, a phenomenon not yet reported before. Currently, it is unclear why CCR6+ ILC3/LTi cells express dual signature genes of both Th17 and Tfh cells, but it is possibly caused by their developmental requirement on the IL-6/STAT3 axis (Choi et al., 2013). Considering the current findings of ILCs functionally mimicking each of their corresponding T helper cell counerparts, our studies indicate that CCR6+ ILC3/LTi cells may also play a role in regulating germinal reactions or humoral response in response to complex gut associated microenvironments, an exciting field that remains to be explored.
In summary, we found that as a signature transcription factor of CCR6+ ILC3, BCL6 is required for their lineage maintenance and restriction of effector function at mature stage. CCR6+ ILC3 are both Tfh cells and Th17 cells equivalents in ILC. Together with future research on BCL6 targets and regulation of BCL6/RORγt co-expression in ILC, our findings add to ILC and CD4+ T cell biology, and provide new insights into the regulation of intestinal homeostasis.
Materials and methods
Lead contact and materials availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chen Dong (cdonglab@hotmail.com).
Experimental model and subject details
Mice
B6 (C57BL/6), Rag1−/− and CD45.1, Rosatdtomato,Vav1cre, Cd2cre, Rorccre and RorcGFP/+ mice were all obtained from The Jackson Lab. M-NSG (NOD-PrkdcscidIl2rgem1/Smoc) mice were purchased from Shanghai Model Organisms Center, Inc. Bcl6ERT2cre mice were generated by Biocytogen, Co., Ltd. Briefly, P2A-iCreERT2 was inserted before the stop codon of Bcl6 gene to generate Bcl6ERT2cre mice. Bcl6RFP/RFP, Bcl6fl/fl mice were generated by our lab and described in previous studies (Liu et al., 2016). B6 mice were crossed with CD45.1 mice to generate CD45.2+CD45.1+ congenic mice. Bcl6RFP/RFP mice were crossed with RorcGFP/+ mice to generate double report mice. Vav1cre, Cd2cre, and Rorccre mice were crossed with Bcl6fl/fl mice to generate various conditional knockout mice. All these mice were bred and maintained in the SPF animal facility at Tsinghua University. All animal experiments were performed according to the governmental and institutional guidelines for animal welfare and approved by the Institutional Animal Care and Use Committee, Tsinghua University.
Method details
LPL isolation and flow cytometry
The intestine tissues from indicated mice were longitudinally opened, cut into small pieces, and then incubated in pre-digestion media (RPMI 1640 media containing 20 mM Hepes, 5 mM EDTA, 1% penicillin and streptomycin, 1 mM dithiothreitol) for 30 min at 37°C. After washing, the pieces were incubated in digestion buffer (RPMI 1640 supplemented with 1% penicillin and streptomycin, 20 mM Hepes, 0.5 mg/ml collagenase D, 1 mg/ml Dispase, 0.5 mg/ml DNase) for 30 min at 37°C. After filtering, LPLs were finally isolated by Percoll density-gradient centrifugation. The purified LPLs were stimulated with 5 ng/ml IL-23 (Fig. 2, D and E) or 2 ng/ml IL-1β plus 2 ng/ml IL-23 for IL-22 and IL-17A measurement, and 0.06 ng/ml IL-12 plus 0.05 ng/ml IL-15 for IFN-γ measurement, in the presence of Golgi-Plug (BD) for 4 h prior to staining for flow cytometry analysis by LSR Fortessa (BD). Cell surface staining was performed by incubating cells with antibodies for 30 min at 4°C after blocking with Fc antibody. RORγt, T-bet, IL-22, IFN-γ, IL-17A staining was carried out using intracellular transcription factor kit (eBioscience) or cytokine staining kit (BD Biosciences).
RNA-seq
The BCL6+RORγt+ ILC gated as CD45+Lin−CD90.2+RFP+GFP+ live cells were sorted from intestine LPLs of indicated mice, with BCL6-RORγt+ ILC (CD45+Lin−CD90.2+RFP-GFP+) as control. Total RNAs were isolated with RNeasy Mini Kit (QIAGEN) and the RNA-seq library was built by BGI Genomics. Sequence reads were obtained by BGISEQ-500, and the clean reads were mapped to mouse genome. Gene expression was indicated by Reads Per Kilobases per Million reads, and the differentially expressed genes were carried out by DESeq2 using a cut-off thresh hold of log2 Fold Change >1 and adjusted P value <0.05.
DSS-induced colitis
Age-matched male mice were given drinking water containing 2.5% DSS for 5 d, and then followed by regular water for 3 d. The DSS water was replaced daily. Body weight was monitored throughout the experiments.
Chronic DSS-induced colitis
Age-matched male mice were given drinking water containing 1.5% DSS during day 0–5, 7–13, 15–20, and 22–27, and replaced by regular water during day 5–7, 13–15, 20–22, and 27–30. Body weights were monitored by every 2–3 d throughout the experiments.
Bacterial 16s rDNA-seq
Fresh mouse fecal samples were collected from stools of indicated mice. Isolation of sample microbial DNA by cetyltriethylammnonium bromide/sodium dodecylsulfate method (Xia et al., 2019). The 16s rDNA library was constructed using NEBNext UltraTM IIDNA Library Prep Kit (Cat No. E7645), and sequenced based on NovaSeq PE250 and analyzed using Consult QII-ME2_EN method by Novogene.
Generation of BM chimera
BM cells from donor mice were collected and flushed with 1× PBS. Lineage depletion of the isolated BM cells was achieved by incubation with Biotin anti-mouse lineage panel (Biolegend) for 20 min, washed and incubated with streptavidin beads (Miltenyi) for 30 min, and then negatively isolated by MACS columns (Miltenyi) following the manufacturer’s instructions. Cell suspension containing ∼2 × 106 Lin− donor BM cells at a 1:1 ratio (WT: Vav1creBcl6fl/fl) were resuspended in 300 ml of 1xPBS, and then intravenously injected into lethally irradiated recipient mice. These mice were sacrificed and analyzed 8 wk after reconstitution.
Histology and immunofluorescence staining
Intestine tissues of indicated mice were dissected and immediately fixed in 10% formalin. H&E staining was applied to paraffin-embedded colon sections. Disease scores of colon tissue were assessed with published scoring system based on indexes evaluating wall thickening, mononuclear cell infiltration, transmural leucocyte infiltration, and goblet cell loss (Wirtz et al., 2007). Immunofluorescence staining was applied to frozen sections of proximal SI, during which SI sections were fixed by iced acetone and then by 0.1% Triton X-100 for around 10 min, after blocked by 5% rat serum for 30 min, and then stained with RORγt antibody (1:50) for 1 h before finally mounted with DAPI. Each step requires three times PBST washing in between, and samples were examined with Zeiss LSM780.
scATAC-seq
Intestinal total ILCs (CD45+Lin−CD90.2+) were isolated from WT Bcl6fl/fl mice and Vav1creBcl6fl/fl mice (two mice combined per group) by FACS sorting. Cells were tested for live conditions and then washed and lysed for 3 min on ice according to the low input protocol recommendations by 10× Genomics (CG000169-Rev C). For single-cell library preparation, the 10× Genomics Chromium Controller and the 10× Genomics Chromium Single-Cell ATAC Library & Gel Bead Kit (1000111) were used according to the manufacturer’s instructions (CG000168-Rev B). Libraries were sequenced on an Illumina NovaSeq PE50 (Noveogene) to a depth of around 260 million reads per sample (∼5,000 cells per sample) with paired-end reads according to the recommendations by 10× Genomics.
Processing of scATAC-seq
Single-cell accessibility counts matrices were generated by aligning reads to mouse ATAC reference (cellranger-atac reference v1.2.0, mm10) using cellranger-atac count. Then, strict quality control of scATAC-seq data was conducted by ArchR (v1.0.1, https://www.archrproject.com/index.html) with default parameters. Cells as outliers in quality control matrix (unique nuclear fragments <1,000 or transcription start site enrichment score <4 or non-ILC cells) were removed from further analysis. The predicted doublets were deleted by setting filterRatio to 1. After preprocessing and filtering, we obtained 4713 ILCs from WT mouse and 4,323 ILCs from KO mouse, and these two samples were aggregated into an ArchRProject. The aggregated ArchRProject achieved dimensionality reduction through iterative Latent Semantic Indexing. Normalization using term frequency-inverse document frequency followed by singular value decomposition was performed twice. Then, each single-cell data were clustered using Seurat’s FindClusters function (resolution was set to 0.8) and were visualized by Uniform Manifold Approximation and Projection (UMAP).
Gene-activity matrix, motif deviation enrichment, and trajectory analysis
A gene-activity score matrix was calculated using addGeneScoreMatrix function, and the detailed algorithm was described in the full manual of ArchR (https://www.archrproject.com/bookdown/index.html). Identification of marker features in each cluster was based on gene score matrix. The peak set was generated using MACS2. The differential peak between TS and ILC3 in Vav1creBcl6fl/fl mice was calculated by getMarkerFeatures function using “wilcoxon” test method, then visualized by ggplot2 R package. Motif deviations enrichment was conducted by ArchR’s wrapper of ChromVAR and visualized by pheatmap R package. GSVA was conducted by GSVA R package. Trajectory of the two samples was calculated using supervised algorithm in ArchR, respectively, then visualized by overlaying the pseudo-time values on corresponding UMAP embedding.
Transfer experiment of CCR6+ ILC3
LPLs were isolated from Bcl6fl/fl and RorccreBcl6fl/fl mice (pool of 6–8 mice) and Lin+ cells depleted according to above-mentioned procedures. In total, ∼3.5 × 105 CCR6+ ILC3 cells (gated on CD45medLin−CD90.2+ CCR6+) were obtained by FACS sorting per group, and intravenously injected to M-NSG mice (∼50,000 CCR6+ ILC3 cells in 300 ml of 1× PBS per recipient mice). The LPLs were isolated from the recipient mice and analyzed 25 d after transfer.
BCL6 fate-mapping
The Bcl6ERT2cre were crossed with Rosatdtomato mice to create fate-mapping reporter mice. To track Bcl6 expression, 200 ml of Tamoxifen (300 nM, dissolved in corn oil) was i.p. injected into Bcl6ERT2creRosatdtomato mice (∼5–6 wk old) every 5 d, lasting for 1 mo, and the mice were then sacrificed for LPL isolation and analysis.
Quantification and statistical analysis
Unless specifically indicated, otherwise, comparison between two different groups was done with unpaired two-tailed Student’s t tests. All P values below 0.05 were considered significant. Statistical analysis was performed with Graphpad Prism 8.
Online supplementary material
Figs. S1, S2, S3, S4, and S5 include supplementary details of BCL6 co-staining data and BCL6 KO-induced cytokine alterations in ILC3 (Fig. S1), chronic DSS-induced colitis and CXCR5-related phenotypes in Vav1creBcl6fl/flRag1−/− mice and Bcl6fl/flRag1−/− littermates (Fig. S2), intrinsity-related analysis of ILC1/ILC3 ratio alterations in Vav1creBcl6fl/fl mice (Fig. S3), ILC2-related alteration in Vav1creBcl6fl/fl mice (Fig. S4), and additional data relative to scATAC-seq as well as phenotypes in RorccreBcl6fl/fl mice and BCL6 fate mapping mice (Fig. S5). Table S1 lists processed RNA-seq data of BCL6+RORγt+ (RFP+GFP+) ILC and BCL6−RORγt+ (RFP−GFP+) ILC. Table S2 lists literature marker genes of CCR6+ ILC3 and NKP46+ ILC. Table S3 lists full gene lists of the Veen graph relative to Fig. 1 H. Table S4 lists detailed beta diversity abundance of differential phyla and families between 16 s rDNA-seq data from Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− mice. Table S5 lists marker gene lists of each clusters of the scATAC-seq data. Table S6 lists differential genesets of TS vs. classic ILC3 from Vav1creBcl6fl/ and Bcl6fl/fl mice scATAC-seq sample in Fig. 4, F and G, respectively, with overlapped genes excluded. Table S7 lists systematic disease score evaluation of histology data relative to Fig. 2 B and Fig. S2 C.
Supplementary Material
shows processed RNA-seq data of BCL6+RORγt+ (RFP+GFP+) ILC and BCL6-RORγt+ (RFP-GFP+) ILC.
shows literature marker genes of CCR6+ ILC3, NKP46+ ILC.
shows full gene lists of the Veen graph relative to Fig. 1 H.
shows detailed beta diversity abundance of differential phyla and families between 16 s rDNA-seq data from Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− mice. Relative to Fig. 2, H and I.
shows marker gene lists of each clusters of the scATAC-seq data in Fig. 4 B.
shows differential genesets of TS vs. classic ILC3 from Vav1creBcl6fl/ and Bcl6fl/fl mice scATAC-seq sample in Fig. 4, F and G, respectively. Overlapped genes excluded.
shows systematic disease score evaluation of histology data relative to Fig. 2 B and Fig. S2 C.
Acknowledgments
We thank the core facility of Institute for Immunology, Tsinghua University for FACS, Dr. Xiaohuan, Guo (Tsinghua University, Beijing) and his lab members for sharing experience on ILC-related experimental skills, and all the Dong lab members for their help.
The work was supported in part by National Natural Science Foundation of China (31821003 and 31991170 to C. Dong, 32070889 to X. Wang), the National Key Research and Development Program of China (2020YFA0707803 to X. Wang), Innovative Research Team of High-level Local Universities in Shanghai (to C. Dong), and Shanghai Science and Technology Commission (21JC1404200 to C. Dong). C. Dong is a FViL Investigator.
Author contributions: C. Dong conceived and supervised the project. Y. Li and C. Dong designed the experiments and analyzed the data. J. Ge analyzed the scATAC-seq data. X. Zhao analyzed the RNA-seq data. M. Gou helped crossed Bcl6ERT2creRosatdtomato mice. Y. Li performed the experiments. M. Xu, J. Huang, B. Xie, and X. Bai provided advice on experimental design. Q. Sun, L. Sun, and S. Tan helped with the experiments. Y. Li, X. Wang, and C. Dong prepared the manuscript.
Data availability
RNA-seq data (relative to Fig. 1) and 16 s rDNA-seq data (relative to Fig. 2) that support the findings of this study have been deposited in GEO DataSets with the accession number GSE220438. scATAC-seq data (relative to Fig. 4) are available through GSE221341. All other relevant data are available from the corresponding author.
References
- Alterauge, D., Bagnoli J.W., Dahlström F., Bradford B.M., Mabbott N.A., Buch T., Enard W., and Baumjohann D.. 2020. Continued Bcl6 expression prevents the transdifferentiation of established Tfh cells into Th1 cells during acute viral infection. Cell Rep. 33:108232. 10.1016/j.celrep.2020.108232 [DOI] [PubMed] [Google Scholar]
- Artis, D., and Spits H.. 2015. The biology of innate lymphoid cells. Nature. 517:293–301. 10.1038/nature14189 [DOI] [PubMed] [Google Scholar]
- Bernink, J.H., Krabbendam L., Germar K., de Jong E., Gronke K., Kofoed-Nielsen M., Munneke J.M., Hazenberg M.D., Villaudy J., Buskens C.J., et al. 2015. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. 43:146–160. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Cao, Q., Gao X., Lin Y., Yue C., Wang Y., Quan F., Zhang Z., Liu X., Lu Y., Zhan Y., et al. 2019. Thymopentin ameliorates dextran sulfate sodium-induced colitis by triggering the production of IL-22 in both innate and adaptive lymphocytes. Theranostics. 9:7490–7505. 10.7150/thno.35015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K., Doherty J.M., Mills J.C., and Colonna M.. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725. 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M., Gamini R., Sécca C., Collins P.L., Zhao S., Peng V., Robinette M.L., Schettini J., Zaitsev K., Gordon W., et al. 2019. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20:980–991. 10.1038/s41590-019-0425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea, S., Perchet T., Petit M., Verrier T., Guy-Grand D., Banchi E.G., Vosshenrich C.A., Di Santo J.P., Cumano A., and Golub R.. 2016. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci. Signal. 9:ra45. 10.1126/scisignal.aaf2223 [DOI] [PubMed] [Google Scholar]
- Chen, L., He Z., Slinger E., Bongers G., Lapenda T.L.S., Pacer M.E., Jiao J., Beltrao M.F., Soto A.J., Harpaz N., et al. 2015. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol. 8:390–402. 10.1038/mi.2014.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Wilson J.E., Koenigsknecht M.J., Chou W.-C., Montgomery S.A., Truax A.D., Brickey W.J., Packey C.D., Maharshak N., Matsushima G.K., et al. 2017. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 18:541–551. 10.1038/ni.3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier, M., Sawa S., and Eberl G.. 2012. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209:729–740. 10.1084/jem.20111594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.S., Eto D., Yang J.A., Lao C., and Crotty S.. 2013. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 190:3049–3053. 10.4049/jimmunol.120303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C. 2021. Cytokine regulation and function in T cells. Annu. Rev. Immunol. 39:51–76. 10.1146/annurev-immunol-061020-053702 [DOI] [PubMed] [Google Scholar]
- Dong, P., Zhang S., Cai M., Kang N., Hu Y., Cui L., Zhang J., and He W.. 2014. Global characterization of differential gene expression profiles in mouse Vγ1+ and Vγ4+ γδ T cells. PLoS One. 9:e112964. 10.1371/journal.pone.0112964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage, M., and Bluestone J.A.. 2016. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat. Rev. Immunol. 16:149–163. 10.1038/nri.2015.18 [DOI] [PubMed] [Google Scholar]
- Eberl, G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Finke, D., Acha-Orbea H., Mattis A., Lipp M., and Kraehenbuhl J.. 2002. CD4+CD3- cells induce Peyer’s patch development: Role of α4β1 integrin activation by CXCR5. Immunity. 17:363–373. 10.1016/S1074-7613(02)00395-3 [DOI] [PubMed] [Google Scholar]
- Fuchs, A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., and Colonna M.. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 38:769–781. 10.1016/j.immuni.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuseini, H., Cephus J.-Y., Wu P., Davis J.B., Contreras D.C., Gandhi V.D., Rathmell J.C., and Newcomb D.C.. 2019. ERα signaling increased IL-17A production in Th17 cells by upregulating IL-23R expression, mitochondrial respiration, and proliferation. Front. Immunol. 10:2740. 10.3389/fimmu.2019.02740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudino, S.J., Beaupre M., Lin X., Joshi P., Rathi S., McLaughlin P.A., Kempen C., Mehta N., Eskiocak O., Yueh B., et al. 2021. IL-22 receptor signaling in Paneth cells is critical for their maturation, microbiota colonization, Th17-related immune responses, and anti-Salmonella immunity. Mucosal Immunol. 14:389–401. 10.1038/s41385-020-00348-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc, J., Lv M., Bessman N.J., Flamar A.L., Sahota S., Suzuki H., Teng F., Putzel G.G., Eberl G., Withers D.R., et al. 2021. Dysregulation of ILC3s unleashes progression and immunotherapy resistance in colon cancer. Cell. 184:5015–5030.e16. 10.1016/j.cell.2021.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Liang Y., Zhang Y., Lasorella A., Kee B.L., and Fu Y.X.. 2015. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. 42:731–743. 10.1016/j.immuni.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Qiu J., Tu T., Yang X., Deng L., Anders R.A., Zhou L., and Fu Y.X.. 2014. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Infect. Immun. 40:25–39. 10.1016/j.immuni.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri, M., Thaiss C.A., Serafini N., Winter D.R., Giladi A., Lara-Astiaso D., Levy M., Salame T.M., Weiner A., David E., et al. 2016. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 166:1231–1246.e13. 10.1016/j.cell.2016.07 [DOI] [PubMed] [Google Scholar]
- Halim, T.Y., Krauss R.H., Sun A.C., and Takei F.. 2012. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 36:451–463. 10.1016/j.immuni.2011.12.02 [DOI] [PubMed] [Google Scholar]
- Hatzi, K., Nance J.P., Kroenke M.A., Bothwell M., Haddad E.K., Melnick A., and Crotty S.. 2015. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212:539–553. 10.1084/jem.20141380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, M.R., Fung T.C., Masur S.H., Kelsen J.R., McConnell F.M., Dubrot J., Withers D.R., Hugues S., Farrar M.A., Reith W., et al. 2015. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4⁺ T cells. Science. 348:1031–1035. 10.1126/science.aaa4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka, I.E., Chea S., Gudjonson H., Constantinides M.G., Dinner A.R., Bendelac A., and Golub R.. 2016. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17:269–276. 10.1038/ni.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, K.R., Soon M.S.F., Sebina I., Fernandez-Ruiz D., Davey G., Liligeto U.N., Nair A.S., Fogg L.G., Edwards C.L., Best S.E., et al. 2018. IFN regulatory factor 3 balances Th1 and T follicular helper immunity during nonlethal blood-stage infection. J. Immunol. 200:1443–1456. 10.4049/jimmunol.1700782 [DOI] [PubMed] [Google Scholar]
- Klose, C.S., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d’Hargues Y., Göppert N., Croxford A.L., Waisman A., Tanriver Y., and Diefenbach A.. 2013. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature. 494:261–265. 10.1038/nature11813 [DOI] [PubMed] [Google Scholar]
- Klose, C.S.N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356. 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Kuhn, K.A., Schulz H.M., Regner E.H., Severs E.L., Hendrickson J.D., Mehta G., Whitney A.K., Ir D., Ohri N., Robertson C.E., et al. 2018. Bacteroidales recruit IL-6-producing intraepithelial lymphocytes in the colon to promote barrier integrity. Mucosal Immunol. 11:357–368. 10.1038/mi.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, V.C., Folkersen L., Aguilar O.A., and Lanier L.L.. 2019. KLF12 regulates mouse NK cell proliferation. J. Immunol. 203:981–989. 10.4049/jimmunol.1900396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Lu H., Chen T., Nallaparaju K.C., Yan X., Tanaka S., Ichiyama K., Zhang X., Zhang L., Wen X., et al. 2016. Genome-wide analysis identifies bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 14:1735–1747. 10.1016/j.celrep.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley, R.M. 2009. Nine lives: plasticity among T helper cell subsets. J. Exp. Med. 206:1643–1646. 10.1084/jem.20091442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi, F., Martin A.P., Thirunarayanan N., Devany E., Mayer L., Grisotto M.G., Furtado G.C., and Lira S.A.. 2009. CXCL13 expression in the gut promotes accumulation of IL-22-producing lymphoid tissue-inducer cells, and formation of isolated lymphoid follicles. Mucosal Immunol. 2:486–494. 10.1038/mi.2009.113 [DOI] [PubMed] [Google Scholar]
- Mazzurana, L., Forkel M., Rao A., Van Acker A., Kokkinou E., Ichiya T., Almer S., Höög C., Friberg D., and Mjösberg J.. 2019. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur. J. Immunol. 49:1344–1355. 10.1002/eji.201848075 [DOI] [PubMed] [Google Scholar]
- Mebius, R.E., Miyamoto T., Christensen J., Domen J., Cupedo T., Weissman I.L., and Akashi K.. 2001. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3- cells, as well as macrophages. J. Immunol. 166:6593–6601. 10.4049/jimmunol.166.11.6593 [DOI] [PubMed] [Google Scholar]
- Monticelli, L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1031/ni.213110.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor, M.H., Muir R., Chakhtoura M., Fang M., Moysi E., Moir S., Carey A.J., Terk A., Nichols C.N., Metcalf T., et al. 2021. A follicular regulatory Innate lymphoid cell population impairs interactions between germinal center Tfh and B cells. Commun. Biol. 4:563. 10.1038/s42003-021-02079-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, A., Andoh A., Araki Y., Bamba T., and Fujiyama Y.. 2004. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin. Immunol. 110:55–62. 10.1016/j.clim.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Pokrovskii, M., Hall J.A., Ochayon D.E., Yi R., Chaimowitz N.S., Seelamneni H., Carriero N., Watters A., Waggoner S.N., Littman D.R., et al. 2019. Characterization of transcriptional regulatory networks that promote and restrict identities and functions of intestinal innate lymphoid cells. Immunity. 51:185–197.e6. 10.1016/j.immuni.2019.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, N., Lo J.W., Biancheri P., Vossenkämper A., Pantazi E., Walker A.W., Stolarczyk E., Ammoscato F., Goldberg R., Scott P., et al. 2015. Interleukin 6 increases production of cytokines by colonic innate lymphoid cells in mice and patients with chronic intestinal inflammation. Gastroenterology. 149:456–467.e15. 10.1053/j.gastro.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders, A., Yessaad N., Vu Manh T.P., Dalod M., Fenis A., Aubry C., Nikitas G., Escalière B., Renauld J.C., Dussurget O., et al. 2011. Identity, regulation and in vivo function of gut NKp46+RORγt+ and NKp46+RORγt- lymphoid cells. EMBO J. 30:2934–2947. 10.1038/emboj.2011.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleussner, N., Merkel O., Costanza M., Liang H.-C., Hummel F., Romagnani C., Durek P., Anagnostopoulos I., Hummel M., Jöhrens K., et al. 2018. The AP-1-BATF and -BATF3 module is essential for growth, survival and TH17/ILC3 skewing of anaplastic large cell lymphoma. Leukemia. 32:1994–2007. 10.1038/s41375-018-0045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sécca, C., Bando J.K., Fachi J.L., Gilfillan S., Peng V., Di Luccia B., Cella M., McDonald K.G., Newberry R.D., and Colonna M.. 2021. Spatial distribution of LTi-like cells in intestinal mucosa regulates type 3 innate immunity. Proc. Natl. Acad. Sci. USA. 118:e2101668118. 10.1073/pnas.2101668118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L., Ye Y., Sun H., and Su B.. 2021. ILC3 plasticity in microbiome-mediated tumor progression and immunotherapy. Cancer Cell. 39:1308–1310. 10.1016/j.ccell.2021.08.002 [DOI] [PubMed] [Google Scholar]
- Spits, H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N., Mebius R.E., et al. 2013. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149. 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- Talbot, J., Hahn P., Kroehling L., Nguyen H., Li D., and Littman D.R.. 2020. Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature. 579:575–580. 10.1038/s41586-020-2039-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Martinez G.J., Yan X., Long W., Ichiyama K., Chi X., Kim B.S., Reynolds J.M., Chung Y., Tanaka S., et al. 2018. Regulation of pathogenic T helper 17 cell differentiation by steroid receptor coactivator-3. Cell Rep. 23:2318–2329. 10.1016/j.celrep.2018.04.088 [DOI] [PubMed] [Google Scholar]
- Tizian, C., Lahmann A., Hölsken O., Cosovanu C., Kofoed-Branzk M., Heinrich F., Mashreghi M.F., Kruglov A., Diefenbach A., and Neumann C.. 2020. c-Maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. eLife. 9:e52549. 10.7554/eLife.52549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino, A.V., Sciumè G., Davis F.P., Iwata S., Zitti B., Robinson G.W., Hennighausen L., Kanno Y., and O’Shea J.J.. 2017. Subset- and tissue-defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J. Exp. Med. 214:2999–3014. 10.1084/jem.20150907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg, C., Mortha A., Bui V.L., Hernandez P.P., Kiss E.A., Hoyler T., Flach M., Bengsch B., Thimme R., Hölscher C., et al. 2010. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity. 33:736–751. 10.1016/j.immuni.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.A., Clark P.A., Crisp A., Barlow J.L., Szeto A., Ferreira A.C.F., Rana B.M.J., Jolin H.E., Rodriguez-Rodriguez N., Sivasubramaniam M., et al. 2019. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity. 51:104–118.e7. 10.1016/j.immuni.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, S., Ni L., Zhao X., Liu X., Xu W., Jin W., Wang X., and Dong C.. 2021. Costimulation molecules differentially regulate the ERK-Zfp831 axis to shape T follicular helper cell differentiation. Immunity. 54:2740–2755.e6. 10.1016/j.immuni.2021.09.018 [DOI] [PubMed] [Google Scholar]
- Wirtz, S., Neufert C., Weigmann B., and Neurath M.F.. 2007. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2:541–546. 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]
- Xia, Y., Chen F., Du Y., Liu C., Bu G., Xin Y., and Liu B.. 2019. A modified SDS-based DNA extraction method from raw soybean. Biosci. Rep. 39. 10.1042/BSR20182271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahia-Cherbal, H., Rybczynska M., Lovecchio D., Stephen T., Lescale C., Placek K., Larghero J., Rogge L., and Bianchi E.. 2019. NFAT primes the human RORC locus for RORγt expression in CD4+ T cells. Nat. Commun. 10:4698. 10.1038/s41467-019-12680-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Li F., Harly C., Xing S., Ye L., Xia X., Wang H., Wang X., Yu S., Zhou X., et al. 2015. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16:1044–1050. 10.1038/ni.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeganegi, M., Leung C.G., Martins A., Kim S.O., Reid G., Challis J.R., and Bocking A.D.. 2010. Lactobacillus rhamnosus GR-1-induced IL-10 production in human placental trophoblast cells involves activation of JAK/STAT and MAPK pathways. Reprod. Sci. 17:1043–1051. 10.1177/1933719110377237 [DOI] [PubMed] [Google Scholar]
- Yu, Y., Tsang J.C., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L., et al. 2016. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature. 539:102–106. 10.1038/nature20105 [DOI] [PubMed] [Google Scholar]
- Zenewicz, L.A., Yin X., Wang G., Elinav E., Hao L., Zhao L., and Flavell R.A.. 2013. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190:5306–5312. 10.4049/jimmunol.1300016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J.A., Zhao J., Liu C., Bulek K., Wu L., Chen X., Hao Y., Wang Z., Wang X., Ouyang W., et al. 2017. IL-17A-Induced PLET1 expression contributes to tissue repair and colon tumorigenesis. J. Immunol. 199:3849–3857. 10.4049/jimmunol.1601540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C., Cui K., Wilhelm C., Hu G., Mao K., Belkaid Y., Zhao K., and Zhu J.. 2016. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17:169–178. 10.1038/ni.3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C., Zheng M., Cui K., Martins A.J., Hu G., Li D., Tessarollo L., Kozlov S., Keller J.R., Tsang J.S., et al. 2020. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity. 52:83–95.e4. 10.1016/j.immuni.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C., Zheng M., and Zhu J.. 2018. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev. 42:5–12. 10.1016/j.cytogfr.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Wu D., Jawale C., Li Y., Biswas P.S., McGeachy M.J., and Gaffen S.L.. 2021. Divergent functions of IL-17-family cytokines in DSS colitis: Insights from a naturally-occurring human mutation in IL-17F. Cytokine. 148:155715. 10.1016/j.cyto.2021.155715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows processed RNA-seq data of BCL6+RORγt+ (RFP+GFP+) ILC and BCL6-RORγt+ (RFP-GFP+) ILC.
shows literature marker genes of CCR6+ ILC3, NKP46+ ILC.
shows full gene lists of the Veen graph relative to Fig. 1 H.
shows detailed beta diversity abundance of differential phyla and families between 16 s rDNA-seq data from Vav1creBcl6fl/flRag1−/− and Bcl6fl/flRag1−/− mice. Relative to Fig. 2, H and I.
shows marker gene lists of each clusters of the scATAC-seq data in Fig. 4 B.
shows differential genesets of TS vs. classic ILC3 from Vav1creBcl6fl/ and Bcl6fl/fl mice scATAC-seq sample in Fig. 4, F and G, respectively. Overlapped genes excluded.
shows systematic disease score evaluation of histology data relative to Fig. 2 B and Fig. S2 C.
Data Availability Statement
RNA-seq data (relative to Fig. 1) and 16 s rDNA-seq data (relative to Fig. 2) that support the findings of this study have been deposited in GEO DataSets with the accession number GSE220438. scATAC-seq data (relative to Fig. 4) are available through GSE221341. All other relevant data are available from the corresponding author.