Abstract
Background
Despite the importance of domesticated animals in the generation and transmission of antibiotic‐resistant Staphylococcus aureus, the role of wild animals, specifically rodents, in the ecology of S. aureus remains unclear. We recovered and genotyped S. aureus isolates from wild Norway rats (Rattus norvegicus) in Boston, Massachusetts to examine genetic relationships between common human and animal S. aureus isolates in a large US metropolitan area.
Methods
We collected and necropsied 63 rats from June 2016 to June 2017. Nasal, foot pad, fur, and fecal swabs were collected. Staphylococcus aureus was isolated using culture‐based methods and polymerase chain reaction confirmation. S. aureus isolates were spa typed, tested for antibiotic susceptibility, and whole genome sequenced. Assembled sequences were uploaded to the Comprehensive Antibiotic Resistance Database to identify antibiotic resistance elements. A phylogenetic tree was constructed using the neighbor‐joining method with the maximum composite likelihood distance in MEGA7.
Results
We recovered 164 Gram‐positive bacterial isolates from Norway rats. Nineteen isolates from eight individual rats were confirmed as S. aureus (prevalence: 12.9% (8/63)). All S. aureus isolates were methicillin‐susceptible S. aureus (MSSA), pvl‐negative, and resistant to penicillin. Two isolates displayed resistance to erythromycin. Four different S. aureus spa types were detected (t933, t10751, t18202, and t189). Thirteen unique antibiotic resistance elements were identified, and all isolates shared genes mepR, mgrA, arlR, and S. aureus norA. Phylogenetic analysis if the 19 S. aureus isolates revealed they were genetically similar to four clades of S. aureus with similar resistance gene profiles isolated from both human‐ and animal‐derived S. aureus, as well as formed a distinct phylogenetic cluster composed only of rat isolates.
Conclusions
Wild rodents may serve as a reservoir or vector of antibiotic resistance genes in the urban environment with relevance for human and animal health.
Keywords: drug resistance, Rattus norvegicus, Staphylococcus aureus, urban health, wild rodent, zoonoses
We identified S. aureus among wild rodents in Boston, MA with antimicrobial resistance genes. Our findings indicate that wild rodents may serve as a under‐studied reservoir of S. aureus and antimicrobial resistance genes with relevance for human exposure and human health.
1. INTRODUCTION
Staphylococcus aureus is a common Gram‐positive bacterium that exists commensally in most mammalian species, residing in the nares, throat, and on the skin of the host (Tong et al., 2015). In humans, S. aureus colonizes approximately 30% of the population (Gorwitz, 2008). S. aureus can also cause a wide range of infections in humans and is the leading cause of bacterial infections globally in both the healthcare and the community setting (Lee et al., 2018). An individual colonized with S. aureu but who does not have an infection, is characterized as an asymptomatic carrier. Carriers are an important reservoir and can be involved in transmission to susceptible individuals through direct personal contact or indirectly through fomites (Senn et al., 2016; Vonberg et al., 2006) or the environment (Katale et al., 2020; Zieliński et al., 2020). S. aureus is also a zoonotic pathogen, and direct contact between humans and animals and animal products can transmit the pathogen to humans and vice versa (Algammal et al., 2020; Pirolo et al., 2019; Kadariya et al., 2014). In the environment, S. aureus has been recovered in animal manure, water, on surfaces and from the air, highlighting the relevance and diversity of the animal reservoir (Smith et al., 2010).
Studies that characterize S. aureus collected from livestock, domestic, and wild animals demonstrate the breadth of animal species that serve as reservoirs of S. aureus with relevance to public health, including over 40 species of wildlife (Heaton et al., 2020; Lozano et al., 2016). Furthermore, S. aureus collected from humans and animals often share virulence factors, antibiotic susceptibility profiles, and spa‐strain types, indicating the bidirectional transmission of bacteria and genes (Huijsdens et al., 2006; van Duijkeren et al., 2016). The Panton Valentine leucocidin (pvl) pro‐toxin subunits lukS and lukF are markers of increased human virulence and pvl is commonly detected in the genomes of human‐associated MRSA strains and has also been detected in animal‐associated strains (Melles et al., 2006). Despite notable research on S. aureus among domesticated animals, particularly those raised for food production, there is less research on S. aureus carriage among wild animals.
Rodents, particularly rats, are well‐adapted to cohabitation with humans in densely populated areas (Santini et al., 2019). Rodent and ectoparasite infestation in urban housing, particularly public housing and homeless shelters, increases risk of pathogen exposure to vulnerable city residents (Byers et al., 2019; Himsworth et al., 2014b; Leibler et al., 2018; McVea et al., 2018). The presence of rodents and rodent exposure among persons living in substandard housing is a well‐established health risk, particularly urban youth with asthma (Perry et al., 2003). Rodents may acquire and transmit these pathogens to/from the environment and humans via their scavenging activities in human‐generated refuse and urban environments (Himsworth et al., 2014). As such, addressing the role of rodent exposures in public health is a key priority in addressing health disparities.
Recent studies of pathogen carriage in wild urban rodents indicate that these animals harbor bacterial pathogens that can cause human foodborne and hospital‐based infections, such as Escherichia coli and methicillin‐resistant Staphylococcus aureus (MRSA) (Jahan et al., 2021; Lee et al., 2019; Rothenburger et al., 2018). Prior studies have recovered human MRSA strains within wild rodents, indicating that these animals may serve as a vector for human staphylococcal disease (Himsworth et al., 2014). Investigators have recovered both the mecA and mecC genes, which code for methicillin resistance, in wild rodents (Mrochen et al., 2018; Silva et al., 2021). Due to the rise in antimicrobial resistance, there is a need to better characterize stains of S. aureus that circulate in wild rodent populations in urban areas and the risks posed by these pathogens to humans.
In this study, we recovered and analyzed S. aureus isolates from wild Norway rats in Boston, MA, a large metropolitan area in the United States, to assess the prevalence of S. aureus carriage among rats, to describe the antibiotic susceptibility profile of recovered isolates, and to determine genetic relatedness between S. aureus circulating in an urban wild rodent reservoir and profiles of strains commonly detected as human pathogens. The goal of this research was to inform a deeper understanding of the potential role of wild rodents in the ecology of S. aureus.
2. MATERIALS AND METHODS
2.1. Sample collection and isolation
During the period of July 2016 and June 2017, we collected wild rodents in Boston in collaboration with the City of Boston Inspectional Services Department (ISD). A convenience sampling design was used. Rodents were trapped using Professional Expanded Trigger snap traps with approximately 2 cm pieces of beef jerky for bait at locations with histories of rodent infestations (Back Bay, Chinatown, Long Wharf, Dorchester, Boston Common, North End, Roxbury) in the city of Boston per the existing schedule of ISD. Traps were set and animals collected within 6 h. Specimens were transported under refrigerated conditions for aseptic necropsy and sample collection according to prior protocol (Cummings et al., 2019).
We collected data on rat weight and sex. We acquired nasal, foot pad, fur, and descending colon fecal swabs using Henry Schein 6 dacron‐tipped sterile applicators and swabs were immediately streaked for isolation on Remel Columbia Naladixic Acid with 5% sheep's blood (CNA) agar plates (37°C for 24 h) for the isolation of Gram‐positive organisms on receipt at the laboratory. One of each distinguishable colony type grown on the CNA agar plates was individually Gram‐stained and observed under a light microscope. Colonies of Gram‐positive cocci were subsequently plated on Remel™ Blood Agar (37°C for 24 h) and then subcultured onto Mannitol Salt Agar (MSA) and Vogel‐Johnson Agar (VJA) (37°C for 24 h) for selective isolation of salt tolerant, coagulase‐positive, mannitol fermenting staphylococci (Gómez et al., 2014; Kato et al., 1995). Colonies that were phenotypically consistent with S. aureus on MSA and VJA agars were cryopreserved for future analysis. A single colony of each isolated sample was inoculated into 10 ml of Brain Heart Infusion broth and placed into a benchtop incubating shaker for 18 h at 37°C. These colonies were subsequently resuspended in 50% glycerol in a 50 ml conical tube and vortexed with three 2 s bursts at 2000 rpm. One ml aliquots were stored at −80°C in 2 ml cryogenic storage vials for subsequent characterization.
2.2. Characterization of isolates
S. aureus isolates were subcultured from glycerol stock on Columbia colistin and nalidixic acid (CNA) plates (Hardy Diagnostics). Isolates grown on CNA plates were confirmed by a series of biochemical tests including the catalase test, coagulase test, and the S. aureus latex agglutination assay (Pastorex Staph‐plus, Bio‐Rad, Hercules, CA). Isolates positive for the catalase test, coagulase test, and S. aureus latex agglutination assay were classified as S. aureus.
Genomic DNA from S. aureus isolates was extracted using the Wizard Genomic DNA preparation kit (Promega, Madison, WI). Polymerase chain reaction (PCR) was carried out on all S. aureus isolates to detect the presence of the methicillin resistance gene mecA, which is commonly found on methicillin‐resistant S. aureus (MRSA) strains that infect humans (Wielders et al., 2002), and the Panton Valentine leukocidin (pvl) genes (lukS, lukF). PCR was used to amplify spa genes using Spa2F (5′‐GAACAA‐CGTAACGGCTTCATCC‐3′) and 1514R (5′‐CAGCAGTAGTGCCGTTTGCCT‐3′) according to previously published methods (Koreen et al., 2004). PCR was also used to identify seven housekeeping genes (arcC, aroE, glp, gmk, pta, tpi, yqiL) associated with multilocus sequence typing (MLST) (Enright et al., 2000). MLST remains incomplete due to time restraints and quality of reads. Isolates with complete reads were assigned sequence types (ST) using the specific MLST database for S. aureus (http://saureus.mlst.net).
We used Ridom StaphType software to assign spa types (version2.2.1; Ridom GmbH, Würzburg, Germany). The Based Upon Repeat Pattern (BURP) algorithm was applied to group spa types based on their genetic proximity (Mellmann et al., 2007).
2.3. Antibiotic susceptibility testing
S. aureus isolates were tested for antibiotic susceptibility with the VITEK 2 System (bioMérieux, Durham, NC) using AST‐GP76 cards according to manufacturer's instructions, in accordance with the Clinical Laboratory Standards Institute standards (CLSI, 2012). Isolates were tested for susceptibility to penicillin, oxacillin, tetracycline, erythromycin, ciprofloxacin, moxifloxacin, minocycline, clindamycin, trimethoprim‐sulfamethoxazole, quinupristin/dalfopristin, gentamicin, levofloxacin, linezolid, daptomycin, vancomycin, rifampin, minocycline, tigecycline, and nitrofurantoin. Any isolates resistant to three or more classes of antibiotics were considered multi‐drug resistant (Magiorakos et al., 2012).
Assembled genomic sequences were uploaded to the Comprehensive Antibiotic Resistance Database (CARD) and run against the Resistance Gene Identifier (RGI) to identify antibiotic resistance elements, including genes and nucleotide polymorphisms. The following parameters were selected: DNA sequence, perfect and strict hits only, exclude nudge, and high quality/coverage.
2.4. Whole‐genome sequencing and assembly
Whole‐genome sequencing (WGS) was performed on 20 S. aureus isolates. Isolate genomic DNA was prepared using Nextera DNA Flex library prep and WGS was performed using paired‐end (2×250 bp) sequencing on a MiSeq instrument (Illumina) to an average sequencing depth of at least 50. No custom primers were used. Staphylococcus aureus subsp. aureus NCTC 8325 (RefSeq NC_007795.1) was used as the reference genome for mapping trimmed reads (https://www.ncbi.nlm.nih.gov/genome/?term=NC_007795) using CLC Genomics (ver. 12). Once mapped, consensus genomic sequences were inferred using the cut‐off thresholds of at least five reads per site. We further focused our analyses on protein‐coding regions, using blastn to capture the respective sequences (Camacho et al., 2009). Because there were regions of missing coverage in each isolate, to maximize the number of shared sites used in the phylogenetic analysis, we only included genes that were present across all isolates and reference genomes. This resulted in a subset of 1730 protein‐coding genes (out of a total of 2767 genes in NC_007795.1) included in the concatenated multiple sequence alignment. The list of genes and their respective names, and the list of genomic reference strains are available in Table 2, respectively. Isolates with the same R number (i.e., Rat R51HC2‐3‐1 and Rat R51HC2‐2) were recovered from the same rat.
TABLE 2.
Antibiotic susceptibility and associated genetic profile of recovered S. aureus isolates (n = 19) from wild Norway rats in Boston, MA a
| Rodent ID and location | Isolate ID | Sex, weight (g) | Source | spa | MLST | Antibiotic resistance identified |
|---|---|---|---|---|---|---|
| R7, DT | R7F1 | M, 150 | Fecal | t10751 | nd | PEN |
| R28, BB | R28N4 | F, 225 | Nasal | t933 | 1094 | PEN |
| R44, BB | R44NC4 | F, 200 | Nasal | t933 | 1094 | PEN |
| R44F6‐C1 | Nasal | t933 | 1094 | PEN | ||
| R44F6‐C2 | Nasal | t933 | nd | PEN | ||
| R44P12 | Foot pad | t933 | 1094 | PEN | ||
| R44P4‐C1 | Foot pad | t933 | 1094 | PEN | ||
| R44P4‐C2 | Foot pad | t933 | 1094 | PEN | ||
| R46, BB | R46FC1‐1 | M, 100 | Hair | t933 | nd | PEN |
| R267C1‐2 | Hair | t933 | nd | PEN | ||
| R47, BB | R47NC1‐1 | M, 300 | Nasal | t933 | 1094 | PEN |
| R47NC1‐2 | Nasal | t933 | nd | PEN | ||
| R47NC3‐1 | Nasal | t933 | nd | PEN | ||
| R47NC3‐2 | Nasal | t933 | nd | PEN | ||
| R47P12 | Foot pad | t933 | nd | PEN | ||
| R50, BB | R50HC3‐1 | F, 100 | Hair | t933 | nd | PEN |
| R51, BB | R51HC2‐2 | F, 400 | Hair | t18202 (new) | nd | PEN, ERY |
| R51HC2‐3‐1 | Hair | t18202 (new) | nd | PEN, ERY | ||
| R52, BB | R52NC3 | M, 325 | Nasal | t189 | nd | PEN |
Abbreviations: PEN: penicillin; ERY: erythromycin; BB: Back Bay, DT: Downtown (BB and DT are sampling locations in Boston, MA). “nd” indicates missing data. spa type t18202 first identified in this study. Positive control used was mecA and pvl positive, spa type t9683, and resistant to PEN, OXA, ERY, CIP and LVX.
All recovered isolates were mecA and pvl negative.
2.5. Phylogenetic analysis
To determine whether S. aureus rat isolates are all closely related to each other or to isolates from other species, we reconstructed a phylogenetic tree using 29 commonly used reference strains (Supporting Information 1). Using the alignment of 1507253 nucleotide positions shared among 49 sequences, a phylogenetic tree was reconstructed using the neighbor‐joining method with the maximum composite likelihood distance as implemented in MEGAX (Kumar et al., 2018). Five hundred bootstrap replicas were used to evaluate the reliability of internal branches . We also reconstructed a maximum likelihood (ML) tree using PhyML 3.0 (Guindon et al., 2010) with the substitution model (GTR) selected using the SMS method with the AIC (Lefort et al., 2017), with branch support evaluated using aLRT (Anisimova & Gascuel, 2006). To reduce computational burden, the ML tree was based on a subset of variable sites only (a total of 46930 positions). Both methods yielded essentially the same topologies; thus, we will only show the NJ tree here.
3. RESULTS
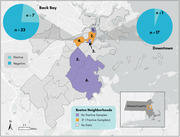
We collected 63 individual Norway rats in this study (n = 63). Average rat weight was 188.5 g, and slightly more than half were female (36/63; Table 2). A total of 164 Gram‐positive isolates were obtained (Table 1). Nineteen isolates from eight individual rats were identified as S. aureus, yielding a prevalence among Norway rats of 12.9% (8/63), based on positive results to the catalase test, coagulase test, and latex agglutination assay (Figure 2). Half of the rats with positive isolates were male and half were female. Rats with a positive isolate were heavier than rats without a positive isolate (225 g vs. 183 g), although this difference was not statistically significant. Four rats had a single positive isolate recovered while four rats had more than one positive isolate recovered (Table 2). The majority of rats (seven of eight) with a positive isolate were recovered in the Back Bay (BB) neighborhood of Boston, while one positive rat was recovered from Downtown (DT) (Figure 1). VITEK 2 identification revealed 22 non‐S. aureus Gram‐positive species, 13 of which were coagulase‐negative staphylococci (CoNS) (Table 1). All S. aureus isolates were resistant to penicillin, and two displayed resistance to erythromycin (spa type t18202). All isolates were pvl‐negative and mecA‐negative.
TABLE 1.
Characterization of isolates recovered from wild Norway rats in Boston, MA (n = 164 isolates recovered from 63 rodents)
| Gram‐positive isolates | N (%) |
|---|---|
| Staphylococcus aureus | 19 (11.6%) |
| Coagulase negative staphylococci (CoNS) | |
| Staphylococcus arlettae | 1 (1.6) |
| Staphylococcus auricularis | 1 (1.6) |
| Staphylococcus chromogenes | 1 (1.6) |
| Staphylococcus cohnii ssp urealyticus | 34 (20.7) |
| Staphylococcus equorum | 9 (5.5) |
| Staphylococcus gallinarum | 9 (5.5) |
| Staphylococcus hominis ssp hominis | 2 (1.2) |
| Staphylococcus lentus | 1 (1.6) |
| Staphylococcus sciuri | 9 (5.5) |
| Staphylococcus simulans | 1 (1.6) |
| Staphylococcus warneri | 5 (3.0) |
| Staphylococcus vitulinus | 2 (1.2) |
| Staphylococcus xylosus | 10 (16.4) |
| Other species | |
| Gardnerella vaginalis | 4 (2.4) |
| Aerococcus viridians | 8 (4.9) |
| Kocuria varians | 1 (1.6) |
| Kocuria rosea | 1 (1.6) |
| Leuconostoc mesenteroides ssp cremoris | 1 (1.6) |
| Enterococcus avium | 2 (1.2) |
| Enterococcus faecalis | 18 (11.0) |
| Enterococcus faecium | 1 (1.6) |
| Enterococcus hirae | 2 (1.2) |
| Unidentified (no growth) | 22 (13.4) |
FIGURE 2.
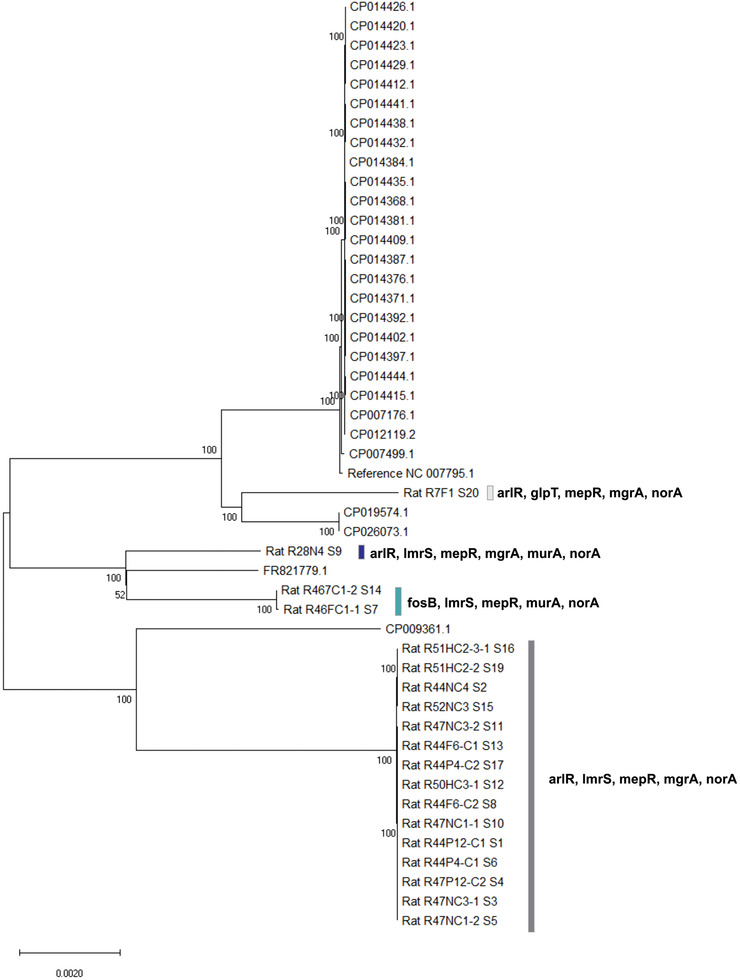
Neighbor‐joining tree based on the maximum composite likelihood distance that shows clustering of isolates from rats and other reference isolates. Antimicrobial genes recovered from isolates are noted. Isolates from rats are designated with “rat” and a sample number, while other isolates are denoted with the respective GenBank accession numbers, with the exception of reference genome NC_007795.1, used in mapping and assembling. Bootstrap values above 50% are shown as numbers at respective nodes
FIGURE 1.
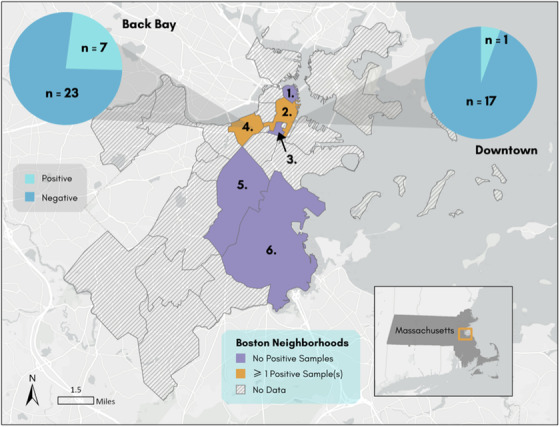
Map of neighborhoods in Boston, MA
We detected five different spa types (t18202, t18292, t933, t10751, and t189). Of the 19 isolates, 79% (15/19) were spa type t933, and MLST indicated sequence type (ST) 1094. Other spa types detected were t10751 and t189. The BURP algorithm identified no linkages between spa types, classifying all strains as singletons.
The CARD database and RGI application identified 13 unique antibiotic resistance elements in the genomes of the 19 S. aureus isolates; all isolates shared genes mepR, mgrA, arlR, and S. aureus norA (Figure 2). Of these antibiotic resistance genes identified, some genes and their functions were correlated with phenotypic results.
3.1. Phylogenetic analysis
As depicted in the midpoint rooted tre, the isolates from rats split into three distinct clades, with the bootstrap support of 100% each. The majority of isolates from rats were closely related to each other and appeared to be closest to CP009361 isolate (cc30). This clade was composed of 15 isolates recovered from five individual rats. The second clade consisted of three rat isolates, two of which were highly proximal and were recovered from the same rat (Figure 2), and clustered with the LGA251 strain (FR821779). The third clade included a single isolate from a rat that clustered with reference isolates USA400 and with ST1. The observed clustering pattern roughly corresponded with the distribution of antibiotic resistance genes (Figure 2, Supporting Information 2) across isolates, where isolates that shared the same resistance genes were clustered together.
4. DISCUSSION
The molecular and bioinformatics analyses performed in this study indicate that Boston city wild rodents carry S. aureus. These isolates predominately reflected distinct clusters of S. aureus within the rodent reservoir and lack the human virulence gene pvl, suggesting animal adaptation. However, some recovered rat isolates were similar to strains of potential concern for human health (USA400/ST1) (de Matos et al., 2016) as well as S. aureus strains identified in domesticated animal species (Brody et al., 2008; Ko et al., 2019). Additionally, wild rodents also carried non‐S. aureus, Gram‐positive bacterial species capable of causing human disease, including CoNS and enterococci, further highlighting public health risk.
The most frequently observed spa type among the S. aureus isolates was t933/ST1094, accounting for 79% (15/19) of the isolates. While t933/ST1094 has not been commonly identified in human or animal studies in the past, nor reported in communities or hospitals, it was previously documented in ewes in Tunisia (Said et al., 2017). In addition, this strain was reported in China, Germany, and the Netherlands, according to the Staph Ridom database (http://spa.ridom.de), although not previously in the United States. Although these strains may be independent, it is possible that these isolates were carried and seeded in a number of geographical regions by human, animal, or other transport. Our findings here indicate that the majority of S. aureus isolates recovered from our wild rat population are genetically distinct from strains that typically infect humans.
Among the other spa types identified, only t189 is reported in publicly available data. Spa type t189 was identified in Ohio and Iowa, recovered in environmental samples and carried by daycare employees, respectively (Thapaliya et al., 2017; Moritz et al., 2015). These observations suggest that while there may be limited similarity to some, more isolated spa types of S. aureus circulating in humans, the rodent S. aureus is relatively distinct, with clear genotypic and phenotypic differences from S. aureus circulating among humans.
Antimicrobial susceptibility testing revealed that all isolates were resistant to penicillin. Spa type t18202 was additionally resistant to erythromycin (Supporting Information 1). RGI analysis confirmed that all isolates shared mepR, mgrA, arlR, and norA. MepR, part of the gene cluster, mepRAB, encodes mepR, a substrate‐binding mepA regulatory protein. MepA is a MATE family multidrug efflux pump, and its presence in S. aureus has been shown to decrease tigecycline susceptibility (McAleese et al., 2005). All the S. aureus isolates in this study had mepR, but none presented the mepA gene, which is commonly expressed by MRSA, perhaps reflecting greater adaptation to antimicrobial susceptibility among MRSA compared to MSSA. MgrA is a global regulator, controlling many genes, and also constitutes a regulatory pathways with ArlRS; together these proteins regulate the expression of large surface proteins, like Ebh, a protein that interferes with fibrinogen and prevents clotting and in turn, clumping of bacteria to host tissue. ArlS, a protein histidine kinase, phosphorylates ArlR, and influences norA expression. Together, arlRS activates expression of mgrA, which represses ebh (Crosby et al., 2016).
Resistance among rodent‐recovered S. aureus isolates to antibiotics used in hospital settings, such as benzylpenicillin and erythromycin, may suggest that these animals may play a role in transmission or perpetuation of antibiotic resistance genes in the urban environment with relevance to human health. Phylogenic analyses highlighted a small number of recovered isolates clustered alongside reference isolates for USA400/ST1, reflecting potential overlap between rodent and clones associated with epidemic MRSA. The rodent‐recovered isolates clustered near USA400/ST1 recovered in this study were methicillin sensitive and pvl‐negative, indicating a clonal departure from this prior epidemic MRSA strain. While our sample size was too small to draw comprehensive conclusions, the phylogenic similarity may suggest a role for wild rodents in the transmission and/or preservation of pathogenic isolates within the urban environment.
Norway rats in our study carried CoNS, a pathogen of increasing relevance to global health as a driver of hospital‐acquired infections (Becker et al., 2014). Further genetic analysis of CoNS identified here is needed to examine shared genes to the 19 S. aureus isolates. MRSA and CoNS surveillance among wild animal populations in urban areas is necessary to better understand transmission and risk to susceptible humans and animals (Abdel‐Moein & Zaher, 2020; Algammal et al., 2020).
Generalizability from our study is hampered due to small sample size, given the pilot nature of our research. A larger sample size would provide greater diversity of strain types and may also recover more characteristics of community and hospital‐associated S. aureus in the wild rodent reservoir. Due to limitations of short‐read sequencing, some genomic regions of our isolate draft assemblies—such as those representing repeated sequences (Ben Khedher et al., 2022)—were left unresolved. Our use of latex agglutination assays in support of S. aureus identification may be limited by high level of false positives in specimens recovered from some animal species. Our study was likewise limited by the convenience sampling strategy, which relied on active trapping efforts by the City of Boston to acquire rodents. The number of complete whole genomes for S. aureus available for analyses provides an additional limitation. On one hand, the number of completed genomes is now quite large and is growing, leading to computational challenges of reconstructing large‐scale phylogenetic histories (Aanensen et al., 2016). On the other hand, many published sequences were derived from clinical settings rather than animal surveillance efforts and/or lack comprehensive metadata that describe the origin and source of the sample and what is known about its resistance genes. Such details and background information is essential to provide a comprehensive comparative analysis of the S. aureus strains that have been described. Additionally, further research is needed to investigate mechanisms through which rodents may transmit S. aureus to humans, or vice versa, in the context of shared environments. Close attention to rodent behavior may elucidate mechanisms of bacterial transmission. How often rodents surface, where and when they search for food, and how they defend themselves from animal/human interaction are questions that may provide clarification towards bacterial dissemination in urban environments.
5. CONCLUSIONS
Our findings suggest that wild rodents in the City of Boston carry S. aureus of relevance to human health as well as distinct strains not commonly reported in the United States that may be specific to the rodent population. Our study contributes to the growing literature on the urban ecology of S. aureus, suggesting that wild urban mammals play a role alongside humans and domesticated animals in the evolution and ecology of this pathogen within shared urban environments. These pathogens may pose risks for vulnerable populations in urban areas and our study warrants continued study of the animal:human interface in urban centers.
AUTHOR CONTRIBUTIONS
Formal analysis (equal), visualization (supporting), writing–original draft preparation (lead): Gracen R. Gerbig. Data curation (lead), formal analysis (equal), visualization (lead), writing–review and editing (equal): Helen Piontkivska. Resources (supporting), supervision (supporting), writing–review and editing (supporting): Tara C. Smith. Conceptualization (supporting), formal analysis (equal), funding acquisition (supporting), investigation (equal): Ruairi White. Formal analysis (supporting): Jean Mukherjee; Data curation (supporting), project administration (supporting), writing–review and editing (supporting): Hayley Benson. Conceptualization (equal), formal analysis (lead), investigation (lead), project administration (equal), supervision (equal), writing – review and editing (equal): Marieke Rosenbaum. Conceptualization (equal), funding acquisition (lead), supervision (equal), investigation (supporting), project administration (equal), supervision (equal), writing – review and editing (lead): Jessica H. Leibler.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
ETHICS APPROVAL STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The US National Research Council's guidelines for the Care and Use of Laboratory Animals were followed.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.1020
Supporting information
Supporting Information 1
Supporting Information 2
ACKNOWLEDGEMENTS
The authors extend their gratitude to the City of Boston's Inspectional Services Department for their participation in this study. In particular, we would like to thank John Ulrich, Leo Boucher, Christopher McNally, Brian Oliveri, Brendan Sheehan, Eric McGevna, and Pedro Torres for their tireless efforts and good spirits. This research was funded by an Early Career Catalyst Award from the Boston University School of Public Health to JHL. Marieke Rosenbaum and Hayley Benson's contributions were supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), award number KL2TR002545. Ruairi White's contributions were supported by NIH Short Term Training, grant number T35OD010963. Gracen R. Gerbig received funding from the American Society for Microbiology Undergraduate Research Fellowship and the SURE program at Kent State University.
Gerbig, G. R. , Piontkivska, H. , Smith, T. C. , White, R. , Mukherjee, J. , Benson, H. , Rosenbaum, M. , & Leibler, J. H. (2023). Genetic characterization of Staphylococcus aureus isolated from Norway rats in Boston, Massachusetts. Veterinary Medicine and Science, 9, 272–281. 10.1002/vms3.1020
Marieke Rosenbaum and Jessica Leibler should be considered joint senior authors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abdel‐Moein, K. A. , & Zaher, H. M. (2020). The nasal carriage of coagulase‐negative staphylococci among animals and its public health implication. Vector‐Borne and Zoonotic Diseases, 20(12), 897–902. 10.1089/vbz.2020.2656 [DOI] [PubMed] [Google Scholar]
- Anisimova, M. , & Gascuel, O. (2006). Approximate likelihood‐ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology, 55(4), 539–552. [DOI] [PubMed] [Google Scholar]
- Algammal, A. M. , Hetta, H. F. , Elkelish, A. , Alkhalifah, D. H. H. , Hozzein, W. N. , Batiha, G. E.‐S. , El Nahhas, N. , & Mabrok, M. A. (2020). Methicillin‐resistant Staphylococcus aureus (MRSA): One health perspective approach to the bacterium epidemiology, virulence factors, antibiotic‐resistance, and zoonotic impact. Infection and Drug Resistance, 13, 3255–3265. 10.2147/IDR.S272733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aanensen, D. M. , Feil, E. J. , Holden, M. T. , Dordel, J. , Yeats, C. A. , Fedosejev, A. , Goater, R. , Castillo‐Ramírez, S. , Corander, J. , Colijn, C. , & Chlebowicz, M. A. (2016). Whole‐genome sequencing for routine pathogen surveillance in public health: A population snapshot of invasive Staphylococcus aureus in Europe. MBio, 7(3), e00444–16. https://journals.asm.org/doi/full/10.1128/mBio.00444‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, K. , Heilmann, C. , & Peters, G. (2014). Coagulase‐negative staphylococci. Clinical Microbiology Reviews, 27(4), 870–926. 10.1128/CMR.00109-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khedher, M. , Ghedira, K. , Rolain, J. M. , Ruimy, R. , & Croce, O. (2022). Application and challenge of 3rd generation sequencing for clinical bacterial studies. International Journal of Molecular Sciences, 23(3), 1395. https://www.mdpi.com/1422‐0067/23/3/1395/htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Said, M. , Abbassi, M. S. , Gómez, P. , Ruiz‐Ripa, L. , Sghaier, S. , El Fekih, O. , Hassen, A. , & Torres, C. (2017). Genetic characterization of Staphylococcus aureus isolated from nasal samples of healthy ewes in Tunisia. High prevalence of CC130 and CC522 lineages. Comparative Immunology, Microbiology and Infectious Diseases, 51, 37–40. 10.1016/j.cimid.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Brody, T. , Yavatkar, A. S. , Lin, Y. , Ross, J. , Kuzin, A. , Kundu, M. , Fann, Y. , & Odenwald, W. F. (2008). Horizontal gene transfers link a human MRSA pathogen to contagious bovine mastitis bacteria. PLoS One, 3(8), e3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, K. A. , Cox, S. M. , Lam, R. , & Himsworth, C. G. (2019). “They're always there”: Resident experiences of living with rats in a disadvantaged urban neighbourhood. BMC Public Health, 19(1), 853. 10.1186/s12889-019-7202-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , & Madden, T. L. (2009). BLAST+: architecture and applications. BMC Bioinformatics, 10(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2012). Performance standards for antimicrobial susceptibility testing (22nd ed.). Clinical and Laboratory Standards Institute. [Google Scholar]
- Crosby, H. A. , Schlievert, P. M. , Merriman, J. A. , King, J. M. , Salgado‐Pabon, W. , & Horswill, A. R. (2016). The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathogens, 12(5), e1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, C. O. , Hill, N. J. , Puryear, W. B. , Rogers, B. , Mukherjee, J. , Leibler, J. H. , Rosenbaum, M. H. , & Runstadler, J. A. (2019). Evidence of influenza A in wild Norway rats (Rattus norvegicus) in Boston, Massachusetts. Frontiers in Ecology and Evolution, 7. 10.3389/fevo.2019.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Matos, P. D. M. , de Oliveira, T. L. R. , Cavalcante, F. S. , Ferreira, D. C. , Iorio, N. L. P. , Pereira, E. M. , Chamon, R. C. , & Dos Santos, K. R. N. (2016). Molecular markers of antimicrobial resistance in methicillin‐resistant Staphylococcus aureus SCC mec IV presenting different genetic backgrounds. Microbial Drug Resistance, 22(8), 700–706. [DOI] [PubMed] [Google Scholar]
- Enright, M. C. , Day, N. P. , Davies, C. E. , Peacock, S. J. , & Spratt, B. G. (2000). Multilocus sequence typing for characterization of methicillin‐resistant and methicillin‐susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology, 38(3), 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, P. , González‐Barrio, D. , Benito, D. , García, J. T. , Viñuela, J. , Zarazaga, M. , Ruiz‐Fons, F. , & Torres, C. (2014). Detection of methicillin‐resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. The Journal of Antimicrobial Chemotherapy, 69(8), 2061–2064. 10.1093/jac/dku100 [DOI] [PubMed] [Google Scholar]
- Gorwitz, R. J. , Kruszon‐Moran, D. , McAllister, S. K. , McQuillan, G. , McDougal, L. K. , Fosheim, G. E. , Jensen, B. J. , Killgore, G. , Tenover, F. C. , & Kuehnert, M. J. (2008). Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. The Journal of Infectious Diseases, 197(9), 1226–1234. 10.1086/533494 [DOI] [PubMed] [Google Scholar]
- Guindon, S. , Dufayard, J. F. , Lefort, V. , Anisimova, M. , Hordijk, W. , & Gascuel, O. (2010). New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3), 307–321. [DOI] [PubMed] [Google Scholar]
- Heaton, C. J. , Gerbig, G. R. , Sensius, L. D. , Patel, V. , & Smith, T. C. (2020). Staphylococcus aureus epidemiology in wildlife: A systematic review. Antibiotics, 9(2), 89. 10.3390/antibiotics9020089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himsworth, C. G. , Miller, R. R. , Montoya, V. , Hoang, L. , Romney, M. G. , Al‐Rawahi, G. N. , Kerr, T. , Jardine, C. M. , Patrick, D. M. , Tang, P. , & Weese, J. S. (2014). Carriage of methicillin‐resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLOS ONE, 9(2), Article e87983. 10.1371/journal.pone.0087983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himsworth, C. G. , Parsons, K. L. , Feng, A. Y. T. , Kerr, T. , Jardine, C. M. , & Patrick, D. M. (2014b). A mixed methods approach to exploring the relationship between Norway rat (Rattus norvegicus) abundance and features of the urban environment in an inner‐city neighborhood of Vancouver, Canada. PLOS ONE, 9(5), e97776. 10.1371/journal.pone.0097776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijsdens, X. W. , van Dijke, B. J. , Spalburg, E. , van Santen‐Verheuvel, M. G. , Heck, M. E. , Pluister, G. N. , Voss, A. , Wannet, W. J. , & de Neeling, A. J. (2006). Community‐acquired MRSA and pig‐farming. Annals of Clinical Microbiology and Antimicrobials, 5(1), 26. 10.1186/1476-0711-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan, N. A. , Lindsey, L. L. , & Larsen, P. A. (2021). The role of peridomestic rodents as reservoirs for zoonotic foodborne pathogens. Vector‐Borne and Zoonotic Diseases, 21(3), 133–148. 10.1089/vbz.2020.2640 [DOI] [PubMed] [Google Scholar]
- Kadariya, J. , Smith, T. C. , & Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food‐borne disease: An ongoing challenge in public health. BioMed Research International, 2014, e827965. 10.1155/2014/827965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katale, B. Z. , Misinzo, G. , Mshana, S. E. , Chiyangi, H. , Campino, S. , Clark, T. G. , Good, L. , Rweyemamu, M. M. , & Matee, M. I. (2020). Genetic diversity and risk factors for the transmission of antimicrobial resistance across human, animals and environmental compartments in East Africa: A review. Antimicrobial Resistance & Infection Control, 9(1), 127. 10.1186/s13756-020-00786-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y. , Matsunaga, S. , Misuna, Y. , Ushioda, H. , Yamamoto, T. , & Kaneuchi, C. (1995). Isolation and characterization of Staphylococcus aureus in rats trapped at restaurants in buildings in downtown Tokyo. The Journal of Veterinary Medical Science, 57(3), 499–502. 10.1292/jvms.57.499 [DOI] [PubMed] [Google Scholar]
- Ko, D. S. , Kim, D. , Kim, E. K. , Kim, J. H. , & Kwon, H. J. (2019). Evolution of a major bovine mastitic genotype (rpoB sequence type 10‐2) of Staphylococcus aureus in cows. Journal of Microbiology, 57(7), 587–596. [DOI] [PubMed] [Google Scholar]
- Koreen, L. , Ramaswamy, S. V. , Graviss, E. A. , Naidich, S. , Musser, J. M. , & Kreiswirth, B. N. (2004). Spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro‐ and macrovariation. Journal of Clinical Microbiology, 42(2), 792–799. 10.1128/JCM.42.2.792-799.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A. S. , de Lencastre, H. , Garau, J. , Kluytmans, J. , Malhotra‐Kumar, S. , Peschel, A. , & Harbarth, S. (2018). Methicillin‐resistant Staphylococcus aureus . Nature Reviews. Disease Primers, 4, 18033. 10.1038/nrdp.2018.33 [DOI] [PubMed] [Google Scholar]
- Lee, M. J. , Byers, K. A. , Donovan, C. M. , Zabek, E. , Stephen, C. , Patrick, D. M. , & Himsworth, C. G. (2019). Methicillin‐resistant Staphylococcus aureus in urban Norway rat (Rattus norvegicus) populations: Epidemiology and the impacts of kill‐trapping. Zoonoses and Public Health, 66(3), 343–348. 10.1111/zph.12546 [DOI] [PubMed] [Google Scholar]
- Leibler, J. H. , Robb, K. , Joh, E. , Gaeta, J. M. , & Rosenbaum, M. (2018). Self‐reported animal and ectoparasite exposure among urban homeless people. Journal of Health Care for the Poor and Underserved, 29(2), 664–675. 10.1353/hpu.2018.0050 [DOI] [PubMed] [Google Scholar]
- Lefort, V. , Longueville, J. E. , & Gascuel, O. (2017). SMS: smart model selection in PhyML. Molecular Biology and Evolution, 34(9), 2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano, C. , Gharsa, H. , Ben Slama, K. , Zarazaga, M. , & Torres, C. (2016). Staphylococcus aureus in animals and food: Methicillin resistance, prevalence and population structure. A review in the African Continent. Microorganisms, 4(1), 12. 10.3390/microorganisms4010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos, A.‐P. , Srinivasan, A. , Carey, R. B. , Carmeli, Y. , Falagas, M. E. , Giske, C. G. , Harbarth, S. , Hindler, J. F. , Kahlmeter, G. , Olsson‐Liljequist, B. , Paterson, D. L. , Rice, L. B. , Stelling, J. , Struelens, M. J. , Vatopoulos, A. , Weber, J. T. , & Monnet, D. L. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- McAleese, F. , Petersen, P. , Ruzin, A. , Dunman, P. M. , Murphy, E. , Projan, S. J. , & Bradford, P. A. (2005). A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory‐derived Staphylococcus aureus mutants to tigecycline. Antimicrobial Agents and Chemotherapy, 49(5), 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVea, D. A. , Himsworth, C. G. , Patrick, D. M. , Lindsay, L. R. , Kosoy, M. , & Kerr, T. (2018). Exposure to rats and rat‐associated Leptospira and Bartonella species among people who use drugs in an impoverished, inner‐city neighborhood of Vancouver, Canada. Vector‐Borne and Zoonotic Diseases, 18(2), 82–88. 10.1089/vbz.2017.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melles, D. C. , van Leeuwen, W. B. , Boelens, H. A. M. , Peeters, J. K. , Verbrugh, H. A. , & van Belkum, A. (2006). Panton‐valentine leukocidin genes in Staphylococcus aureus . Emerging Infectious Diseases, 12(7), 1174–1175. 10.3201/eid1207.050865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann, A. , Weniger, T. , Berssenbrügge, C. , Rothgänger, J. , Sammeth, M. , Stoye, J. , & Harmsen, D. (2007). Based upon repeat pattern (BURP): An algorithm to characterize the long‐term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiology, 7(1), 98. 10.1186/1471-2180-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, E. D. , Hanson, B. M. , Kates, A. E. , & Smith, T. C. (2015). Molecular characteristics of Staphylococcus aureus isolated from employees, children, and environmental surfaces in Iowa child daycare facilities. American Journal of Infection Control, 43(5), 482–488. 10.1016/j.ajic.2015.01.022 [DOI] [PubMed] [Google Scholar]
- Mrochen, D. M. , Schulz, D. , Fischer, S. , Jeske, K. , El Gohary, H. , Reil, D. , Imholt, C. , Trübe, P. , Suchomel, J. , Tricaud, E. , Jacob, J. , Heroldová, M. , Bröker, B. M. , Strommenger, B. , Walther, B. , Ulrich, R. G. , & Holtfreter, S. (2018). Wild rodents and shrews are natural hosts of Staphylococcus aureus . International Journal of Medical Microbiology, 308(6), 590–597. 10.1016/j.ijmm.2017.09.014 [DOI] [PubMed] [Google Scholar]
- Perry, T. , Matsui, E. , Merriman, B. , Duong, T. , & Eggleston, P. (2003). The prevalence of rat allergen in inner‐city homes and its relationship to sensitization and asthma morbidity. Journal of Allergy and Clinical Immunology, 112(2), 346–352. 10.1067/mai.2003.1640 [DOI] [PubMed] [Google Scholar]
- Pirolo, M. , Visaggio, D. , Gioffrè, A. , Artuso, I. , Gherardi, M. , Pavia, G. , Samele, P. , Ciambrone, L. , Di Natale, R. , Spatari, G. , Casalinuovo, F. , & Visca, P. (2019). Unidirectional animal‐to‐human transmission of methicillin‐resistant Staphylococcus aureus ST398 in pig farming; evidence from a surveillance study in southern Italy. Antimicrobial Resistance & Infection Control, 8(1), 187. 10.1186/s13756-019-0650-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburger, J. L. , Rousseau, J. D. , Weese, J. S. , & Jardine, C. M. (2018). Livestock‐associated methicillin‐resistant Staphylococcus aureus and Clostridium difficile in wild Norway rats (Rattus norvegicus) from Ontario swine farms. Canadian Journal of Veterinary Research = Revue Canadienne De Recherche Veterinaire, 82(1), 66–69. [PMC free article] [PubMed] [Google Scholar]
- Santini, L. , González‐Suárez, M. , Russo, D. , Gonzalez‐Voyer, A. , von Hardenberg, A. , & Ancillotto, L. (2019). One strategy does not fit all: Determinants of urban adaptation in mammals. Ecology Letters, 22(2), 365–376. 10.1111/ele.13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn, L. , Clerc, O. , Zanetti, G. , Basset, P. , Prod'hom, G. , Gordon, N. C. , Sheppard, A. E. , Crook, D. W. , James, R. , Thorpe, H. A. , Feil, E. J. , & Blanc, D. S. (2016). The stealthy superbug: The role of asymptomatic enteric carriage in maintaining a long‐term hospital outbreak of ST228 methicillin‐resistant Staphylococcus aureus . MBio, 7(1), e02039‐15. 10.1128/mBio.02039-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, V. , Gabriel, S. I. , Borrego, S. B. , Tejedor‐ Junco, M. T. , Manageiro, V. , Ferreira, E. , Reis, L. , Caniça, M. , Capelo, J. L. , Igrejas, G. , & Poeta, P. (2021). Antimicrobial resistance and genetic lineages of Staphylococcus aureus from wild rodents: first report of mecC‐positive methicillin‐resistant S. aureus (MRSA) in Portugal. Animals, 11, 1537. 10.3390/ani11061537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T. C. , Moritz, E. D. , Leedom Larson, K. R. , & Ferguson, D. D. (2010). The environment as a factor in methicillin‐resistant Staphylococcus aureus transmission. Reviews on Environmental Health, 25(2), 121–134. 10.1515/REVEH.2010.25.2.121 [DOI] [PubMed] [Google Scholar]
- Thapaliya, D. , Hellwig, E. J. , Kadariya, J. , Grenier, D. , Jefferson, A. J. , Dalman, M. , Kennedy, K. , DiPerna, M. , Orihill, A. , Taha, M. , & Smith, T. C. (2017). Prevalence and characterization of Staphylococcus aureus and methicillin‐resistant Staphylococcus aureus on public recreational beaches in northeast Ohio. GeoHealth, 1(10), 320–332. 10.1002/2017GH000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S. Y. , Davis, J. S. , Eichenberger, E. , Holland, T. L. , & Fowler, V. G., Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinical Microbiology Reviews, 28(3), 603–661. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijkeren, E. , Hengeveld, P. , Zomer, T. P. , Landman, F. , Bosch, T. , Haenen, A. , & van de Giessen, A. (2016). Transmission of MRSA between humans and animals on duck and turkey farms. Journal of Antimicrobial Chemotherapy, 71(1), 58–62. 10.1093/jac/dkv313 [DOI] [PubMed] [Google Scholar]
- Vonberg, R. ‐ P. , Stamm‐Balderjahn, S. , Hansen, S. , Zuschneid, I. , Rüden, H. , Behnke, M. , & Gastmeier, P. (2006). How often do asymptomatic healthcare workers cause methicillin‐resistant Staphylococcus aureus outbreaks? A systematic evaluation. Infection Control & Hospital Epidemiology, 27(10), 1123–1127. 10.1086/507922 [DOI] [PubMed] [Google Scholar]
- Wielders, C. L. C. , Fluit, A. C. , Brisse, S. , Verhoef, J. , & Schmitz, F. J. (2002). MecA gene is widely disseminated in Staphylococcus aureus population. Journal of Clinical Microbiology, 40(11), 3970–3975. 10.1128/JCM.40.11.3970-3975.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, D. M. , Harris, H. W. , Charlebois, E. D. , Chambers, H. , Campbell, A. , Perdreau‐Remington, F. , Lee, C. , Mankani, M. , Mackersie, R. , & Schecter, W. P. (2004). An epidemic of methicillin‐resistant Staphylococcus aureus soft tissue infections among medically underserved patients. Archives of Surgery (Chicago, Ill.: 1960), 139(9), 947–951. 10.1001/archsurg.139.9.947 [DOI] [PubMed] [Google Scholar]
- Zieliński, W. , Korzeniewska, E. , Harnisz, M. , Hubeny, J. , Buta, M. , & Rolbiecki, D. (2020). The prevalence of drug‐resistant and virulent Staphylococcus spp. In a municipal wastewater treatment plant and their spread in the environment. Environment International, 143, 105914. 10.1016/j.envint.2020.105914 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1
Supporting Information 2
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


