Abstract
Brucella spp. can establish themselves and cause disease in humans and animals. The mechanisms by which Brucella spp. evade the antibacterial defenses of their host, however, remain largely unknown. We have previously reported that live brucellae failed to induce tumor necrosis factor alpha (TNF-α) production upon human macrophage infection. This inhibition is associated with a nonidentified protein that is released into culture medium. Outer membrane proteins (OMPs) of gram-negative bacteria have been shown to modulate macrophage functions, including cytokine production. Thus, we have analyzed the effects of two major OMPs (Omp25 and Omp31) of Brucella suis 1330 (wild-type [WT] B. suis) on TNF-α production. For this purpose, omp25 and omp31 null mutants of B. suis (Δomp25 B. suis and Δomp31 B. suis, respectively) were constructed and analyzed for the ability to activate human macrophages to secrete TNF-α. We showed that, in contrast to WT B. suis or Δomp31 B. suis, Δomp25 B. suis induced TNF-α production when phagocytosed by human macrophages. The complementation of Δomp25 B. suis with WT omp25 (Δomp25-omp25 B. suis mutant) significantly reversed this effect: Δomp25-omp25 B. suis-infected macrophages secreted significantly less TNF-α than did macrophages infected with the Δomp25 B. suis mutant. Furthermore, pretreatment of WT B. suis with an anti-Omp25 monoclonal antibody directed against an epitope exposed at the surface of the bacteria resulted in substancial TNF-α production during macrophage infection. These observations demonstrated that Omp25 of B. suis is involved in the negative regulation of TNF-α production upon infection of human macrophages.
Members of the genus Brucella are gram-negative, facultatively intracellular bacteria that can induce chronic infections in humans. Following invasion of the reticuloendothelial system, the bacteria develop intracellularly within mononuclear phagocytes. Chronic infection generally results in the fixation of infected macrophages at specific locations within the body (spleen, brain, heart, bones), and the human disease is characterized by undulant fever, endocarditis, arthritis, and osteomyelitis (42). Brucellae are also pathogenic for animals, but the pathophysiology of the human infection differs in many respects from the illness induced in animals. In domestic ruminants, infection results mainly in abortion in females and orchitis in males (15) whereas in mice, infection resembles septicemia and does not become truly chronic (18). These observations therefore suggest a species-specific interaction of Brucella organisms with the immune systems of their different hosts. To survive and multiply within the host, one of the major strategies of pathogens is to affect the expression of cytokines, which is necessary for the normal protective function of the immune response (26).
In previous papers (6, 7) we have reported that brucellae can adopt the following strategy. (i) In human monocytic phagocytes (but not in mouse macrophages), Brucella spp. impair the production of tumor necrosis factor alpha (TNF-α) induced either by their phagocytosis or by exogenously added lipopolysaccharide (LPS). (ii) The defect in TNF-α production results from specific modulation of macrophage stimulation by a protein factor(s) that is produced by the bacteria and is present in the bacterial culture supernatant. Inhibition of TNF-α production may favor the intracellular development of brucellae at different levels, since this proinflammatory cytokine activates the antibacterial activities of macrophages, stimulates antigen-presenting cells, and participates in the initiation of a specific immune response.
This strategy is not particular to brucellae, as other gram-negative bacteria, such as Ehrlichia risticii (35) or Yersinia spp. (2, 30), are also able to inhibit the production of TNF-α which might result from their interaction with macrophages. The molecular mechanism linked to Yersinia inhibition of TNF-α production was recently characterized by our group (29, 30) and involves the injection of a Yersinia-specific protein (Yop) into host cells through a type III transport system (3, 28). In contrast to yersiniae the Brucella entity (or entities) involved in inhibition of TNF-α production by host cells is still unknown. Its identification should constitute an important step toward the understanding of the virulence of these bacteria. Until now, our efforts to identify this molecule by direct fractionation of Brucella supernatants were unsuccessful. Nevertheless, we hypothesized that a protein that can directly interact with the macrophage membrane during the phagocytic process and can be easily released from the bacterial cell would be a good candidate. In addition to LPS and phospholipids, the membrane of gram-negative bacteria contains outer membrane proteins (OMPs), such as the well-characterized protein OmpA, and porins (OmpC and -F) of Enterobacteriaceae. The major Brucella OMPs are identified and classified according to their apparent molecular masses and include the 36- to 38-kDa OMPs (or group 2 porin proteins) and the 31- to 34-kDa and 25- to 27-kDa OMPs, which belong to the group 3 proteins (34). Two genes, named omp25 and omp31, code for the group 3 OMPs. Omp25 is highly conserved in Brucella species, biovars, and strains (9) and exhibits some sequence homology and antigenic relationship with Escherichia coli OmpA (8, 9, 37). In Proteus mirabilis (41) and more recently in Klebsiella pneumoniae (33), OmpA was shown to modulate cytokine production in LPS-activated macrophages.
We thus examined the possibility that in brucellae, Omp25 and/or Omp31 could be involved in the regulation of TNF-α production by infected macrophages. For this purpose, Δomp25 Brucella suis and Δomp31 B. suis mutants were constructed and analyzed for the ability to activate human macrophages to secrete TNF-α. We report here convergent data demonstrating that the expression of Omp25 correlated with the unusual absence of TNF-α release observed in human macrophages infected with Brucella spp. Finally, we show that Brucella Omp25 is involved in the negative regulation of TNF-α production upon infection of human macrophages.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. suis 1330 (ATCC 23444) and derived mutants were all grown in tryptic soy broth at 37°C. Mutant strains containing a kanamycin or chloramphenicol resistance cassette were cultured in the presence of the respective antibiotic at 50 or 25 μg ml−1. Plasmid pAC2507 carried the omp25 gene of B. suis cloned in pCRII (10). For the complementation assay with B. suis, this native omp25 gene was prepared by codigestion with restriction enzymes KpnI and XbaI and recloned into plasmid pBBR1MCS (24), a broad-host-range vector. In the resulting construct, the omp25 gene is under the control of the Plac promoter. E. coli strain DH5α was used as the recipient strain and was routinely grown in Luria-Bertani medium. Recombinant clones were selected on agar supplemented with chloramphenicol in combination with kanamycin at the concentrations indicated above. Plasmid pNV3151 is derived from pBBR1MCS4 (ampicillin resistant) and maintained in E. coli strain JM109. It contained the native omp31 gene of B. melitensis 16M under the control of its own promoter (22).
DNA manipulations and Southern blots.
Plasmid DNA was isolated from E. coli according to standard procedures (32). B. suis chromosomal DNA was prepared as previously described (1). DNA treatments with restriction and modification enzymes were performed according to the manufacturer's instructions. Restriction fragments were purified after separating bands on low-melting-point agarose gels (Life Technologies) by the Wizard DNA clean-up system (Promega, Madison, Wis.). DNA labeling was carried out with [α-32P]dCTP (3,000 Ci mmol−1; NEN) and a random priming kit (Appligène). Southern blotting was performed with Biodyne B nylon membranes (Pall, Port Washington, N.Y.). The membranes were washed twice at 68°C for 15 min each time in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.0115 M sodium citrate) with 0.1% sodium dodecyl sulfate (SDS).
Inactivation of B. suis omp25 and omp31 by homologous recombination.
Plasmid pUC19, pAC2553, and pNV3153 constructs were used for homologous recombinations. Plasmid pAC2553 containing the omp25 gene of B. melitensis 16M was interrupted by replacement of the 314-bp StyI fragment with the kanamycin resistance gene from plasmid pUC4K. Plasmid pNV3153 harboring the omp31 gene of B. melitensis 16M was mutated by insertion of the chloramphenicol resistance cassette from plasmid pBlueCM-2 into the StyI restriction site. The omp25-Kan and omp31-Cm inserts were excised as 1.9- and 1.8-kb XbaI-SacI fragments, respectively, and recloned into pCVD442 (11) containing the sacB gene which codes for sucrose sensitivity (19). B. suis was transformed with this suicide vector by electroporation as previously described (23). Mutants of B. suis that integrated the inactivated omp25 or omp31 gene into the chromosome by double-recombination events were selected by their resistance to sucrose and kanamycin or chloramphenicol as reported elsewhere (14).
Analysis of Brucella OMPs by SDS-PAGE and immunoblotting.
Equal volumes of stationary-phase cultures of each Brucella strain (i.e., wild-type [WT] B. suis, the omp25 null mutant [Δomp25 B. suis], the omp31 null mutant [Δomp31 B. suis], and the complemented mutants [Δomp25-omp25 B. suis or Δomp31-omp31 B. suis], respectively) were centrifuged. The bacterial pellets were resuspended in Laemmli sample buffer and the bacterial proteins were separated by SDS–15% polyacrylamide gel electrophoresis (PAGE). Proteins were transferred onto a polyvinylidene difluoride membrane (Millipore, Saint-Quentin, France) by a semidry transfer procedure. Transferred Omp25 and Omp31 proteins were detected by using mouse anti-Omp25 mononoclonal antibody (MAb) A19/12B10/F04 (10) and mouse anti-Omp31 MAb A59/10F09/G10, respectively (38). Bound antibodies were visualized with an anti-mouse horseradish peroxidase-conjugated secondary antibody (Amersham, Les Ulis, France) and revealed by enhanced chemiluminescence assay (Amersham).
Preparation and analysis of Brucella supernatants.
Bacteria from a 10-ml stationary-phase culture of each Brucella strain were pelleted by centrifugation. The bacteria were washed twice in phosphate-buffered saline (PBS), suspended in the same volume (10 ml) of RPMI 1640 medium (Gibco BRL, Les Ulis, France) and incubated in this medium at 37°C for 2.5 h with shaking. Bacteria were then discarded by centrifugation, and the supernatants were filtered through a 0.22-μm-pore-size filter (Steritop; Millipore).
When indicated, the supernatant was prepared from a 400-ml stationary-phase culture of WT B. suis, the supernatant being concentrated 400 times in two steps, (i) reduction to a volume of 20 ml by ultrafiltration with an Amicon Concentration Cell using a membrane with a cutoff of 10 kDa and (ii) reduction to a volume of 1 ml by centrifugation at 2,200 × g and 4°C using 10-kDa Centriprep. Supernatants (generally 75 μl) were analyzed by SDS-PAGE and immunoblotting as reported above. Transferred Omp25 or Omp31 protein or LPS was detected by using anti-Omp25 MAb A19/12B10/F04, anti-Omp31 MAb A59/10F09/G10 (38), or anti-LPS MAb 12G12, respectively (10). The bacterial proteins from concentrated supernatants were visualized by Coomassie brilliant blue staining.
Brucella infection of human THP-1 macrophage-like cells and intracellular survival assay.
Human macrophage-like THP-1 cells (ATCC TIB 202) differentiated for 72 h with 10−7 M 1,25-dihydroxyvitamin D3 (VD3) (6) were infected in 24-well plates (Falcon; Becton Dickinson, Meylan, France) with the different B. suis strains as previously described (6). Briefly, cells (8 × 105 ml−1) cultured for 1 night in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (Gibco BRL) at 37°C and 5% CO2 were washed and incubated in the same medium for 30 min with a bacterial suspension corresponding to a multiplicity of infection (MOI) of 20. After three washes with PBS, the cells were reincubated in RPMI 1640 medium–10% fetal calf serum with gentamicin (30 μg ml−1) to kill any remaining extracellular bacteria. At 1.5, 7, 24, and 48 h postinfection, the cells were washed twice with PBS and lysed in 0.2% Triton X-100. CFU counts were determined by plating serial dilutions on tryptic soy agar. Experiments were performed twice in triplicate.
Detection of TNF-α and IL-8 in supernatants of infected VD3-differentiated THP-1 cells.
Culture supernatants from the infection experiments described above were harvested at different times postinfection, centrifuged, and stored at −20°C for TNF-α measurement. As a control, the ability of the cells to produce TNF-α was measured by stimulation with LPS from E. coli O128:B12 (Sigma) at 100 ng ml−1 for 7 h.
The quantification of TNF-α in supernatants was evaluated by a cytotoxicity assay performed with the TNF-α-sensitive murine fibroblast cell line L929 as previously described (6). This method demonstrated the bioactivity of the TNF-α produced by THP-1 cells. Results were evaluated by comparison with a human recombinant TNF-α standard 87/650 from the National Institute for Biological Standards and Controls (Potter Bar, United Kingdom) and expressed as picograms per milliliter.
Quantification of TNF-α by enzyme-linked immunosorbent assay (ELISA) was also performed as previously described (25), by using the OptEIA set (human TNF-α; Pharmigen, San Diego, Calif.). Interleukin-8 (IL-8) was quantified by ELISA (human IL-8 Endogen; Perbio Science, Bezons, France) by following the instructions of the manufacturer. For every condition tested, the TNF-α (or IL-8) concentration was calculated as the mean ± the standard deviation (SD) of three different determinations.
Binding of anti-Omp25 and anti-Omp31 antibodies to WT B. suis.
A76/02C12/C11 and A59/10F09/G10 are two MAbs of the immunoglobulin G2a (IgG2a) subtype that, respectively, recognize Omp25 and Omp31 on the external surface of intact brucellae. Their characteristics have been published elsewhere (4, 5, 10). As described previously (4, 5), they were used as hybridoma supernatants throughout this study. Twenty-five microliters of a suspension of WT B. suis (optical density, 0.9) was washed, suspended in 500 μl of PBS, and incubated with subagglutinating dilutions (1/100 to 1/5) of anti-Omp25 or anti-Omp31 MAbs or with medium alone. As a control, bacteria were treated with 2 μg of an irrelevant MAb of the IgG2a subtype (anti-CD14 hybridoma RM052; Beckman) under similar conditions. The bacteria were then washed and incubated for a further 30 min with a fluorescein isothiocyanate (FITC)-labeled anti-mouse F(ab)′2 fragment (Beckman). After extensive washings, their fluorescence was analyzed by flow cytometry as previously described (4).
In some experiments, WT B. suis treated for 30 min with different dilutions of anti-Omp25 or anti-Omp31 MAbs or with medium alone, followed by washing, was used to infect differentiated THP-1 cells (MOI, 20). Seven hours later, the amount of TNF-α present in the supernatant was measured as mentioned above.
Statistical analysis.
P values were calculated by using the paired Student t test.
RESULTS
Inactivation of B. suis omp25 and omp31 genes.
To analyze the effect of Omp25 and Omp31 on TNF-α production by infected macrophages, we constructed omp25 and omp31 null mutants of B. suis as an alternative to purifying the OMPs, which is impossible in the absence of detergents. For this reason, the omp25 and omp31 genes of WT B. suis were independently inactivated. Suicide plasmid pCVD442, carrying either the omp25 gene or the omp31 gene interrupted by the kanamycin or chloramphenicol resistance gene, respectively, was transformed into WT B. suis, allowing exchange between chromosomal omp25 or omp31 and the corresponding mutant gene. In both cases, successful allelic exchange was confirmed by Southern blot analysis. HindIII digests of chromosomal DNAs prepared from parent and mutant B. suis strains were hybridized with the omp25 or omp31 probe obtained from plasmid pAC2553 or pNV3153, respectively (see Materials and Methods). A 620-bp fragment that can be deduced from the construction appeared in the omp25 mutant DNA but was absent from the WT DNA; furthermore, instead of the 1.6-kb fragment detected by the omp31 probe in DNA from B. suis (38), two bands of 2,140 and 413 bp were revealed in the omp31 mutant DNA (data not shown).
Western blot analysis using anti-Omp25 or anti-Omp31 MAbs confirmed the absence of Omp25 and Omp31 in the corresponding mutants strains, Δomp25 B. suis and Δomp31 B. suis, respectively (Fig. 1). We were unsuccessful in obtaining a double mutant with both the omp25 and omp31 genes inactivated despite conducting multiple experiments with either the Δomp25 B. suis or the Δomp31 B. suis genetic background.
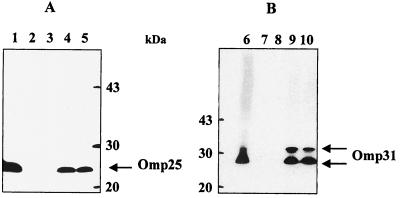
FIG. 1.
Expression of Omp25 and Omp31 by different B. suis mutants. (A) Total proteins of WT B. suis (lane 1) and Δomp25 B. suis transcomplemented (lanes 4 and 5, respectively) or not transcomplemented (lanes 2 and 3) were separated by SDS-PAGE, blotted, and analyzed with an anti-Omp25 antibody. (B) Total proteins of WT B. suis (lane 6) and Δomp31 B. suis transcomplemented (lanes 9 and 10) or not transcomplemented (lanes 7 and 8) were separated by SDS-PAGE, blotted, and analyzed with an anti-Omp31 antibody.
Δomp25 B. suis complemented in trans with the native omp25 gene from B. suis and cloned into pBBR1MCS recovered the expression of Omp25 (Fig. 1) (mutant Δomp25-omp25 B. suis). trans complementation of the omp31 mutant with plasmid pNV3151, containing the intact omp31 gene of B. melitensis led to the production of the Omp31 protein (mutant Δomp31-omp31 B. suis). The profile obtained (Fig. 1) was very similar to the multiple-band pattern observed with the WT B. melitensis strain (38), ranging from 28 to 34 kDa, depending on the sample treatment used before electrophoresis.
TNF-α is produced upon macrophage infection by Δomp25 B. suis, whereas Δomp31 B. suis does not induce any release of this cytokine.
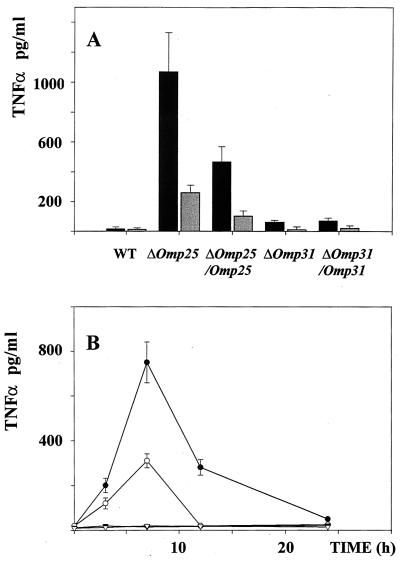
We have previously observed that human macrophagic cells (i.e., human monocytes, VD3- and retinoic acid-differentiated U-937 cells, or VD3-differentiated THP-1 cells) infected with WT B. suis do not produce any TNF-α (6). In a set of similar experiments, we have assayed for the presence of TNF-α in supernatants of THP-1 macrophage-like cells infected with Δomp25 B. suis or Δomp31 B. suis and compared the results to those obtained with WT B. suis. Data from 10 experiments are summarized in Fig. 2A. As expected, there was no TNF-α detectable in supernatants of WT B. suis-infected cells while controls with E. coli LPS showed that the cells were very sensitive to activation (3.0 ± 0.4 ng/ml). In contrast, Δomp25 B. suis-infected cells produced significant concentrations of TNF-α, ranging from 700 to 1,200 pg/ml as measured by ELISA or from 280 to 340 pg/ml as measured with the bioassay. The differences between the values obtained by the two methods were probably due to the different standards used in the assays. Δomp31 B. suis behaved the same way as WT B. suis and did not induce any significant production of the cytokine (<40 pg/ml by ELISA, undetectable by the bioassay). Measurements of the phagocytosis of both mutants were comparable and could therefore not account for the differences in TNF-α production observed (Fig. 3). As in LPS-activated cells, the production of TNF-α induced by the Δomp25 B. suis mutant was transient and optimal 6 to 7 h after infection (Fig. 2B). These results strongly suggested that Omp25 of B. suis could be involved in the previously reported absence of TNF-α production in infected human macrophages. In order to verify if the phenomenon was exclusively due to Omp25, experiments were done with Δomp25-omp25 B. suis. The data presented in Fig. 2 show a significant decrease in TNF-α production when Δomp25-omp25 B. suis was used instead of Δomp25 B. suis. No effect resulted from the complementation of Δomp31 B. suis by the omp31 gene (Δomp31-omp31 B. suis mutant).
FIG. 2.
The Δomp25 B. suis mutant induces TNF-α production in human macrophages. (A) THP-1 cells were infected with WT or mutant B. suis and cultured for 7 h in complete culture medium supplemented with gentamicin at 30 μg/ml. The cell supernatants were then harvested, and their TNF-α contents were determined by ELISA (black bar) or a bioassay (grey bar). Each experiment included infection with WT B. suis and the following mutants: Δomp25 B. suis, the complemented Δomp25 strain of B. suis (Δomp25-omp25 B. suis), Δomp31 B. suis, and the complemented Δomp31 strain of B. suis (Δomp31-omp31 B. suis). Each experiment also included a control cell activation with E. coli LPS at 100 ng/ml. In these experiments, LPS-stimulated cells produced 3,000 ± 400 pg of TNF-α per ml. Values represent the mean ± SD of 10 different experiments. (B) THP-1 cells were infected with WT B. suis (▿), Δomp25 B. suis (●), Δomp25-omp25 B. suis (○), or Δomp31 B. suis (▾). At different times postinfection, supernatants were harvested and TNF-α production was measured by ELISA. Each value represents the mean ± SD of four similar samples.
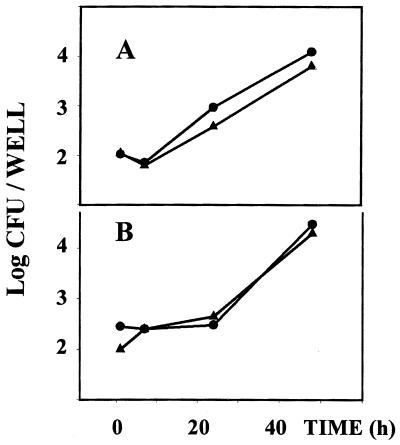
FIG. 3.
Intracellular behavior of Δomp25 B. suis and Δomp31 B. suis within THP-1 cells compared to that of WT B. suis. VD3–THP-1 cells (7 × 105 per well) were infected with Δomp25 B. suis (A, ▴) or with Δomp31 B. suis (B, ▴) (MOI, 20) in 24-well plates. For each mutant, a control infection was performed with WT B. suis (●). At different time periods, cells were lysed and the number of viable bacteria was then determined. Panels A and B report the results of one typical experiment representative of three.
Upon infection with WT B. suis, human macrophages which did not produce TNFα secreted other inflammatory cytokines like IL-1, IL-6 (6), or IL-8 (unpublished results). We thus compared the production of IL-8 in VD3-differentiated THP-1 cells infected with WT and Δomp25 B. suis and found no significant difference. In three different experiments, the IL-8 concentrations measured in supernatants of cells infected for 24 h with WT, Δomp25, and Δomp25-omp25 B. suis were 280 ± 35, 320 ± 40, and 290 ± 25 pg/ml, respectively.
Our previous data have shown that activation of macrophage-like U-937 cells by TNF-α results in reduced multiplication of WT B. suis inside the cells (7). We therefore measured the multiplication of Δomp25 and Δomp31 B. suis in VD3-differentiated THP-1 cells. No significant difference was noted in the development of WT, Δomp25, or Δomp31 B. suis within VD3-differentiated THP-1 cells (Fig. 3).
Omp25 is released into the supernatants of WT B. suis cultures.
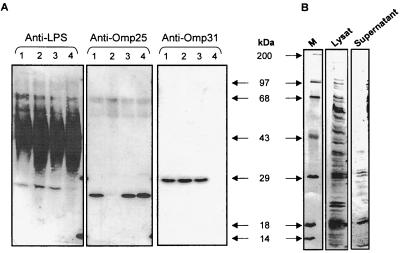
In addition to the absence of TNF-α production in WT B. suis-infected macrophages under conditions in which other gram-negative bacteria are active (7), it was also observed that Brucella culture supernatants contain a protein factor(s) that is able to inhibit the secretion of TNF-α in LPS-activated macrophages (6). We therefore analyzed the expression of Omp25 in the supernatants of the different Brucella strains studied as described above. Western blot analysis with an anti-Omp25 MAb revealed that Omp25 was present in the supernatants of WT B. suis and Δomp31 B. suis (Fig. 4A). On the contrary, Omp25 was not observed in the supernatants of Δomp25 B. suis and as expected, the protein reappeared in the supernatant of the complemented Δomp25-omp25 B. suis strain. Experiments performed in parallel with an anti-Omp31 MAb demonstrated the presence of the protein in WT B. suis, Δomp25 B. suis, or Δomp25-omp25 B. suis supernatant but not in Δomp31 B. suis supernatant (Fig. 4A). Figure 4 also shows the presence of Brucella LPS in all of the bacterial supernatants studied. Analyses of concentrated supernatants and bacterial lysats by SDS-PAGE revealed that beside Omp25, several proteins were present in bacteria supernatants; this is shown for WT B. suis in Fig. 4B. The release of outer membrane vesicles (blebs) by exponentially growing WT B. suis could explain the presence of Omp25 in bacterial supernatants (17). Indeed, high-speed centrifugation revealed the presence of blebs in all of the Brucella supernatants analyzed (R.-A. Boigegrain et al., unpublished results).
FIG. 4.
The inhibitory capacity of the bacterial supernatants correlates with the presence of Omp25. (A) WT B. suis or B. suis mutants (2.5 × 1010/ml) were cultured for 150 min in RPMI 1640 medium, the bacteria were centrifuged, and 75 μl of each supernatant was separated by SDS-PAGE and analyzed by Western blotting with an anti-Omp25, an anti-Omp31, or an anti-LPS antibody. Lanes: 1, WT B. suis; 2, Δomp25 B. suis; 3, Δomp25-omp25 B. suis; 4, Δomp31 B. suis). (B) Supernatants of WT B. suis cultured as reported above were concentrated 400-fold and analyzed by Coomassie blue staining after electrophoresis and blotting. Lanes: M, molecular size markers; Lysat, bacterial lysate; Supernatant, proteins in supernatant of B. suis cultured in RPMI 1640 medium (see Materials and Methods).
Inhibition of TNF-α production by the supernatant of B. suis cultures.
To confirm the regulation of TNF-α production by Omp25, the effect of the culture supernatants from B. suis strains on TNF-α production was assessed in LPS-induced macrophages. It was observed that the LPS-induced secretion of TNF-α was inhibited by the supernatants of the WT B. suis strain but not by those of Δomp25 B. suis (Table 1), the LPS-induced production of TNF-α in the presence of both supernatants being significantly different (P < 0.005). Furthermore, the complementation in trans of the Δomp25 mutant by omp25 significantly restored the ability of the bacterial culture supernatant to impair TNF-α production (P < 0.01).
TABLE 1.
TNF-α production by THP-1 cells activated by LPS in the presence of supernatants of B. suis mutantsa
| Expt no. | TNF-α concn (pg/ml)
|
|||
|---|---|---|---|---|
| Medium | WT supernatant | Δomp25 B. suis supernatant | Δomp25-omp25 B. suis supernatant | |
| 1 | 2,580 ± 230 | 1,320 ± 90* | 1,940 ± 212† | 1,120 ± 110‡ |
| 2 | 658 ± 120 | 90 ± 12* | 600 ± 100† | 300 ± 30‡ |
| 3 | 790 ± 95 | 142 ± 21* | 700 ± 50† | 342 ± 20‡ |
VD3-differentiated THP-1 cells were preincubated for 30 min in complete medium supplemented with a 1/10 dilution of supernatant from WT B. suis, Δomp25 B. suis, or Δomp25-omp25 B. suis or in medium alone. They were then stimulated by addition of E. coli LPS at 100 ng/ml. Seven hours later, the TNF-α concentration in each supernatant was measured by ELISA. Three experiments were performed, and each value shown represents the mean ± SD of four different determinations in one experiment. Paired Student t tests showed that the differences between values bearing the symbols * and † (P = 0.0031) and between those bearing the symbols † and ‡ (P = 0.01) are significant, while those between values bearing the symbols ∗ and ‡ (P = 0.65) are not significant.
TNF-α production by macrophages infected with anti-Omp25-treated WT B. suis.
In smooth bacteria, LPS affects the accessibility of outer membrane protein antigens to antibodies. Nevertheless, two MAbs secreted by the hybridomas A76/02C12/C11 and A59/10F09/G10, respectively, recognize Omp25 and Omp31 exposed on the intact Brucella surface (4, 10). Figure 5A confirmed these data and showed that WT B. suis bound the anti-Omp25 and anti-Omp31 antibodies. For both antibodies, optimal binding was observed for dilutions of hybridoma supernatants ranging from 1/10 to 1/5, with the anti-Omp25 MAb being less effective than the anti-Omp31 MAb. The specificity of the antibodies has been previously established (4, 5); it was confirmed by using omp25 and omp31 null mutants of B. suis. Δomp25 B. suis bound the anti-Omp31 MAb but not the anti-Omp25 MAb (Fig. 5B), while Δomp31 B. suis, which did not interact with the anti-Omp31 MAb, bound the anti-Omp25 MAb (data not shown).
FIG. 5.
Anti-Omp25 antibodies specifically reverse the inhibition of TNF-α in human macrophage infection. (A and B) WT B. suis (A) and Δomp25 B. suis (B) were incubated for 30 min with anti-Omp25 or anti-Omp31 (dilution, 1/5) or with an irrelevant IgG2a. The bacteria were then extensively washed and incubated for 30 min with an anti-mouse FITC-F(ab′)2 fragment. Their fluorescence was then measured by flow cytometry. 1, WT B. suis plus anti-CD14 (irrelevant IgG2a) plus anti-mouse FITC-F(ab′)2; 2, WT B. suis plus anti-Omp25 plus anti-mouse FITC-F(ab′)2; 3, WT B. suis plus anti-Omp31 plus anti-mouse FITC-F(ab′)2. FL1-H, fluorescence arbitrary units. (C) WT B. suis was incubated for 30 min with different dilutions of anti-Omp25 (●) or anti-Omp31 (○) antibodies or with medium alone. (x axis, percentages of hybridoma supernatants [HS] during incubation.) The bacteria were then washed and used to infect THP-1 cells as described in Materials and Methods (MOI, 20). Seven hours later, macrophage culture supernatants were harvested and TNF-α concentrations in cell supernatants were measured (y axis). The values shown are the means ± SD of four different experiments. In this set of experiment, noninfected cells released 73 ± 10 pg of TNF-α per ml. Differences in TNF-α production between cells infected with WT B. suis, anti-Omp25-treated WT B. suis, and anti Omp31-treated B. suis were analyzed by paired t tests.
The hypothesis that a blockade of Omp25 affects TNF-α production was tested. In parallel experiments, WT B. suis was preincubated with different dilutions of anti-Omp25 or anti-Omp31 antibodies. Both antibodies were of the IgG2a isotype. Macrophages were then infected with anti-Omp-treated bacteria or with WT B. suis (Fig. 5C), and 7 h later, TNF-α concentrations were measured in cell supernatants. As expected, no TNF-α production was induced by WT B. suis infection and the slight levels of TNF-α measured in the supernatants of controls (noninfected cells) and WT B. suis-infected cells were similar. On the contrary: a relatively large amount of this cytokine was found in the supernatants of cells infected with anti-Omp25-treated B. suis and TNF-α production increased with antibody binding to the bacteria. The optimal effect was observed for bacteria preincubated with a 1/10 dilution of anti-Omp25 hybridoma supernatant, and TNF-α production was 15-fold higher than that which occurred in a noninfected cell culture (P < 0.0005). The bacteria treated with the anti-Omp31 MAb induced only weak production of TNF-α, two- to threefold higher than that of the noninfected cells (P < 0.025). The TNF-α production promoted by anti-Omp25-treated bacteria was thus much higher than that promoted by anti-Omp31-treated bacteria. (P < 0.0024). Since both the anti-Omp25 and anti-Omp31 MAbs are of the IgG2a subtype, the participation of the Fc portion of the antibodies could not explain the differences in TNF-α production observed in this experiment. Moreover, anti-Omp25-treated and anti-Omp31-treated B. suis bacteria were phagocytosed at very similar levels (data not shown).
DISCUSSION
We have previously reported that in human macrophage infection, Brucella impairs TNF-α production and that this inhibitory effect results from the action of a protein factor(s) of the bacteria (6). In this report, we present data demonstrating that one major OMP of Brucella spp., Omp25, is involved in the inhibition of TNF-α production that normally occurs when gram-negative bacteria are phagocytosed by human macrophages. Different sets of experiments based on the effects of OMP null mutants of B. suis led to this conclusion.
(i) When they were infected with Δomp25 B. suis, macrophages secreted active TNF-α. Furthermore, the inhibition of TNF-α production was partially recovered when Δomp25 B. suis was complemented in trans with the intact omp25 gene. The differences observed in the levels of TNF-α secretion were possibly due to the quantitative differences in Omp25 expression between WT B. suis and Δomp25-omp25 B. suis (Fig. 1). It should be noted that the complementation was performed with the B. suis omp25 gene under the control of the E. coli Plac promoter, which is probably less active than the genuine promoter of WT B. suis (see Materials and Methods).
(ii) No effect was linked to omp31 deletion. This result was, in fact, foreseeable, as this molecule is absent from B. abortus (39) and no TNF-α is detected upon phagocytosis of this bacterium by human macrophages (7).
(iii) The absence of Omp25, which promoted the secretion of TNF-α in Brucella-infected macrophages, did not modify the production of other inflammatory cytokines, such as IL-8.
(iv) In contrast to the WT B. suis supernatants, those derived from Δomp25 B. suis cultures did not inhibit the secretion of TNF-α by LPS-activated macrophages. The data obtained with the different bacteria showed that, in fact, the inhibitory property of the supernatants paralleled the presence of Omp25 in the medium.
(v) Macrophages infected with anti-Omp25-treated WT B. suis produced TNF-α, an observation which is in line with a blockade of Omp25, since the anti-Omp25 antibody bound to an epitope of the protein which, in spite of LPS, was directly accessible on the bacteria and the comparison of the binding of anti-Omp25 and anti-Omp31 (two IgG2a MAbs) to Brucella excluded the participation of the anti-Omp25 Fc group in TNF-α induction, as the levels of bound anti-Omp31 were significantly higher, yet the binding of anti-Omp31 exerted only a slight effect on the production of the cytokine during infection.
Together, these data demonstrate that Omp 25 is specifically involved in the inhibition of TNF-α production in Brucella-infected macrophages and that an Omp25-induced effect accounts for our previous observations on cytokine release during macrophage infection by brucellae.
OMPs of Brucella spp. have been identified several years ago (12, 13); however, interest in these proteins has focused on their potential as protein antigens, and to date, no specific function has been attributed to them. The involvement of bacterial OMPs in the modulation of the interaction of the protein with the host is not an uncommon phenomenon. Indeed, for other gram-negative bacteria, reports have been published claiming that, in addition to the maintenance of membrane structural integrity, OMPs affect macrophage functions by directly interacting with the host cell membrane. For instance, OmpC from Salmonella typhimurium mediates adherence to macrophages (27), OmpF from E. coli enhances macrophage cytotoxicity (40), and purified K. pneumoniae OmpA was recently shown to induce cytokine production in macrophages (33). Thus, it seems possible that Brucella Omp25 interacts with a macrophage receptor(s) and induces negative signals that specifically modify the pathway leading to TNF-α secretion, while the nature of the receptor(s) and the mechanisms remain unidentified. Omp25 could also modulate the release of bacterial proteins antagonistic to macrophage activation. To our knowledge, there is no evidence that Omp25, which is not a porin, is involved in a protein secretion pathway. In fact, outer membrane fragments (blebs) produced by exponentially growing brucellae explain the presence of Omp25, Omp31, and other proteins in bacterial supernatants (17). It could be that the expression of Omp25 regulates bleb formation and, in this way, protein release and TNF-α production. However, we did not observe any significant difference in Omp31 levels in supernatants of WT B. suis, Δomp25 B. suis, and Δomp25-omp25 B. suis. (Fig. 4A). This observation argued against the control of bleb release by Omp25, even if it is awaiting confirmation by analysis of proteins present in the blebs produced by the different mutants.
Alternatively, Omp25 could act through a modification of the interaction between macrophages and bacterial LPS. Experiments using complexes of LPS and OmpA have indeed shown that P. mirabilis OmpA evokes enhancement of LPS-induced transcription of the TNF-α-encoding gene but inhibition of the transcription of the gene for IL-1β (41). This effect is due to the modulation of LPS responses following its strong interaction with OmpA. Omp25 from Brucella spp. is tightly associated with LPS, and so it is possible that such an interaction specifically impairs the Brucella LPS signaling leading to TNF-α production while not affecting the messages linked to IL-1β, IL-6, and/or IL-8 production (6). In this case, a competition between the Brucella and E. coli LPSs would explain the results presented in Table 1. However, different observations argue against this possibility. Brucella LPS is a poor inducer of macrophage activation (100- to 1,000-fold less potent than E. coli LPS [20]), and there is no direct evidence that macrophagic stimulation is due to bacterial LPS in a Brucella infection (7). Moreover, Brucella supernatants do not inhibit TNF-α production in LPS-induced murine macrophages (6) and in human cells, Brucella supernatants impair the production of TNF-α triggered by opsonized zymosan (6). Finally, Omp31, the other major OMP found in the culture supernatant, which shares 34% identity with Omp25 and is also tightly bound to LPS (9, 10), is not involved in the inhibition of TNF-α production.
We have previously shown that pretreatment of U-937 cells with TNF-α results in an activation that partially inhibits the intracellular multiplication of brucellae (7). In the present study, it was observed that the development of Δomp25 B. suis was not significantly affected compared to WT B. suis development. It is possible that in THP-1 cells, the kinetics and amount of TNF-α released during the period of infection is not consistent with efficient microbicidal activation of the host cells.
Nevertheless, it is clear that Brucella Omp25 is involved in the production of a key factor of the host immune response. The levels of TNF-α produced with Δomp25 B. suis were of the same order of magnitude as those produced by E. coli LPS and appeared to be biologically significant, since E. coli LPS at 100 ng/ml induced the same production of TNF-α as macrophage infection by nonvirulent E. coli (MOI, 20) (J. Dornand et al., unpublished results). Thus, deletion of the omp25 gene might affect Brucella virulence in a more appropriate model.
Upon infection with Listeria monocytogenes or B. abortus, it was reported that mice lacking receptors for TNF-α are severely deficient in IL-12 production and that the earliest infection is exacerbated. These observations show that TNF-α controls early IL-12 production, suggesting a key role for TNF-α in induction of acquired cellular immunity (44). In fact, mice deficient in the TNF-α response finally control a Brucella infection because they are able to produce gamma interferon by a TNF-α-independent mechanism, since the requirement for TNF-α in the induction of acquired cellular immunity is not absolute in the model (43, 44). Nevertheless, it must be kept in mind that these findings arise from mice which were not naturally infected and display a Brucella immunity different from that of humans. For instance, the functions of NK cells, which are inhibited in Brucella-infected patients (31), are not involved in mouse infection (16) and nitric oxide synthase, which has a key role in the elimination of the bacteria in mice (43, 44), is not induced in Brucella-infected human macrophages (21). In humans, it remains possible that the Omp25-induced effect on TNF-α production affects the host defense at different levels, (i) by inhibiting innate immunity and (ii) by impairing the production of IL-12 and the development of a Th1 response, thus changing the immune response to the Th2 type (one of the major features of human chronic focused brucellosis associated with a high titer of antibodies and poor delayed-type hypersensitivity [36]). Moreover, significant production of anti-Omp25 antibodies might block the negative effect of the protein and thus participate in protective immunity against Brucella spp. Such an effect might be more important in rough than in smooth Brucella strains, since Omp25 is more readily accessible to antibodies in rough bacteria because of the steric hindrance by S-LPS (9).
If recognition of the surface of human macrophages by Omp25 is an initiating event in the intracellular development of brucellae, in spite of the lack of effect of Brucella supernatant on TNF-α production in murine macrophages (6), it seems unlikely that the recognition results from a specific evolution. Humans do not transmit brucellosis; they are always contaminated by animals. This means that the property of Omp25 to affect TNF-α production could also be true for the infected primary host and that deletion of the omp25 gene might affect Brucella virulence in this host (swine in the case of B. suis). This proposal must be analyzed to support the general applicability of the proposed mechanism.
In conclusion, the data presented here show that the expression of Omp25 at the surface of Brucella spp. controls TNF-α production in human macrophage infection. This finding, which explains our previous observations (6, 7), is of importance for the analysis of the virulence of Brucella spp. and for the construction of attenuated bacteria that are able to induce a network of interacting cytokines which can result in a protective Th1 response against the intracellular pathogen. Work is now in progress to examine if this effect is due to direct recognition of the OMP by a specific receptor(s) of the macrophage membrane and to determine the molecular pathways linked to this recognition.
ACKNOWLEDGMENTS
V. Jubier-Maurin and R.-A. Boigegrain contributed equally to this work.
We thank S. Ouahrani-Bettache for skillful help with fluorescence-activated cell sorter analysis. We are grateful to J. Oliaro for critical reading of the manuscript.
This work was supported by EC (contract QLK2-1999-00014).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Beuscher H U, Rödel F, Forsberg A, Röllinghoff M. Bacterial evasion of host immune defense: Yersinia enterocolitica encodes a suppressor for tumor necrosis factor alpha expression. Infect Immun. 1995;63:1270–1277. doi: 10.1128/iai.63.4.1270-1277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland A, Cornelis G. Rôle of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect Immun. 1998;66:1878–1884. doi: 10.1128/iai.66.5.1878-1884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden R A, Cloeckaert A, Zygmunt M S, Bernard S, Dubray G. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect Immun. 1995;63:3945–3952. doi: 10.1128/iai.63.10.3945-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowden R A, Cloeckaert A, Zygmunt M, Dubray G. Evaluation of immunogenicity and protective activity in BALB/c mice of the 25-kDa major outer-membrane protein of Brucella melitensis (Omp25) expressed in E. coli. J Med Microbiol. 1998;47:39–48. doi: 10.1099/00222615-47-1-39. [DOI] [PubMed] [Google Scholar]
- 6.Caron E, Gross A, Liautard J P, Dornand J. Brucella spp. release a specific, protease-sensitive, inhibitor of TNF-α expression, active on human but not on murine macrophage-like cells. J Immunol. 1996;156:2885–2893. [PubMed] [Google Scholar]
- 7.Caron E, Peyrard T, Köhler S, Cabane S, Liautard J-P, Dornand J. Live Brucella spp. fail to induce tumor necrosis factor alpha excretion in U937-derived phagocytes. Infect Immun. 1994;62:5267–5274. doi: 10.1128/iai.62.12.5267-5274.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert A, Verger J M, Grayon M, Crepinet O. Restriction site polymorphism of the genes encoding the major 25kDa and 36kDa outer-membrane proteins of Brucella. Microbiology. 1995;141:2111–2121. doi: 10.1099/13500872-141-9-2111. [DOI] [PubMed] [Google Scholar]
- 9.Cloeckaert A, Verger J M, Grayon M, Vizcaino N. Molecular and immunological characterization of the major outer membrane proteins of Brucella. FEMS Microbiol Lett. 1996;145:1–8. doi: 10.1111/j.1574-6968.1996.tb08547.x. [DOI] [PubMed] [Google Scholar]
- 10.Cloeckaert A, Verger J M, Grayon M, Zygmunt M S, Grépinet O. Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect Immun. 1996;64:2047–2055. doi: 10.1128/iai.64.6.2047-2055.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubray G. Protectives antigens in brucellosis. Ann Inst Pasteur/Microbiol (Paris) 1987;138:84–87. doi: 10.1016/0769-2609(87)90080-9. [DOI] [PubMed] [Google Scholar]
- 13.Dubray G, Charriaut C. Evidence of three major polypeptide species and two major polysaccharide species in the Brucella outer membrane. Ann Rech Vet. 1983;14:311–318. [PubMed] [Google Scholar]
- 14.Ekaza E, Guilloteau L, Teyssier J, Liautard J P, Köhler S. Functional analysis of the ClpATPase ClpA of Brucella suis, and persistence of the knockout mutant in BALB/c mice. Microbiology. 2000;146:1605–1616. doi: 10.1099/00221287-146-7-1605. [DOI] [PubMed] [Google Scholar]
- 15.Enright F M. The pathogenesis and pathobiology of Brucella infection in domestic animals. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 301–320. [Google Scholar]
- 16.Fernandes D M, Benson R, Baldwin C L. Lack of a role for natural killer cells in early control of Brucella abortus 2308 infections in mice. Infect Immun. 1995;63:4029–4033. doi: 10.1128/iai.63.10.4029-4033.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamazo C, Moriyon I. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect Immun. 1987;55:609–615. doi: 10.1128/iai.55.3.609-615.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Carrillo C. Laboratory animal models for brucellosis studies. In: Nielsen K, Duncan J R, editors. Animal brucellosis. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 423–442. [Google Scholar]
- 19.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein J, Hoffman T, Frasch C, Lizzio E F, Beining P R, Hochstein D, Lee Y L, Angus R D, Golding B. Lipopolysaccharide (LPS) from Brucella abortus is less toxic than that from Escherichia coli, suggesting the possible use of B. abortus or LPS from B. abortus as a carrier in vaccines. Infect Immun. 1992;60:1385–1389. doi: 10.1128/iai.60.4.1385-1389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross A, Spiesser S, Terraza A, Rouot B, Caron E, Dornand J. Expression and bactericidal activity of nitric oxide synthase in Brucella suis infected murine macrophages. Infect Immun. 1998;66:1309–1316. doi: 10.1128/iai.66.4.1309-1316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilloteau L A, Loriucau K, Vizcaino N, Jacques I, Dubray G. Immunogenicity of recombinant Escherichia coli expressing the omp31 gene of Brucella melitensis in BALB/c mice. Vaccine. 1999;17:353–361. doi: 10.1016/s0264-410x(98)00205-9. [DOI] [PubMed] [Google Scholar]
- 23.Köhler S, Teyssier J, Cloeckaert A, Rouot B, Liautard J P. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol Microbiol. 1996;20:701–712. doi: 10.1111/j.1365-2958.1996.tb02510.x. [DOI] [PubMed] [Google Scholar]
- 24.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBRIMCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 25.Lafont V, Liautard J, Gross A, Liautard J, Favero J. Tumor necrosis factor-α production is differently regulated in γδ and αβ human T lymphocytes. J Biol Chem. 2000;275:19282–19287. doi: 10.1074/jbc.M910487199. [DOI] [PubMed] [Google Scholar]
- 26.Marrack P, Kappler J. Subversion of the immune system by pathogens. Cell. 1994;76:323–325. doi: 10.1016/0092-8674(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 27.Negm R S, Pistole T G. The porin OmpC of Salmonella typhimurium mediates adherence to macrophages. Can J Microbiol. 1999;45:658–664. [PubMed] [Google Scholar]
- 28.Palmer L E, Hobbie S, Galan J E, Bliska J B. YopJ of Yersinia pseudotuberculosis is required for the inhibition of TNF-α production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 29.Ruckdeschel K, Harb S, Roggenkamp A, Hornef M, Zumbihl R, Köhler S, Heesemann J, Rouot B. Yersinia enterocolitica impairs activation of transcription factor NF-κB: involvement in the induction of programmed cell death and in the supression of the macrophage TNF-α production. J Exp Med. 1998;187:1069–1079. doi: 10.1084/jem.187.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruckdeschel K, Machold J, Roggenkamp A, Schubert S, Zumbhil R, Liautard J P, Heesemann J, Rouot B. Yersinia enterocolitica promotes deactivation of macrophage mitogen-activated protein kinases, extracellular signal-regulated kinase-1/2, p38, and c-Jun NH2-terminal kinase. J Biol Chem. 1997;272:15920–15927. doi: 10.1074/jbc.272.25.15920. [DOI] [PubMed] [Google Scholar]
- 31.Salmeron, I., Rodriguez-Zapata, Salmeron O., Manzano L., Vaquer S., and Alvarez-Mon, M. 1992. Impaired activity of natural killer cells in patients with acute brucellosis. Clin. Infect. Dis. 15:764–770. [DOI] [PubMed]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Soulas C, Baussant T, Aubry J P, Delmeste Y, Barillat N, Caron G, Renno T, Bonnefoy J Y, Jeannin P. Outer membrane protein A (OmpA) binds to and activates human macrophages. J Immunol. 2000;165:2335–2340. doi: 10.4049/jimmunol.165.5.2335. [DOI] [PubMed] [Google Scholar]
- 34.Sowa B A. Membrane proteins of Brucella spp. In: Adams L G, editor. Advances in brucellosis research. College Station: Texas A&M University; 1993. pp. 89–105. [Google Scholar]
- 35.Van Heeckeren A, Rikihisa Y, Park J, Fertel R. Tumor necrosis factor alpha, interleukin-1 alpha, interleukin-6, and prostaglandin E2 production in murine peritoneal macrophages infected with Ehrlichia risticii. Infect Immun. 1993;61:4333–4337. doi: 10.1128/iai.61.10.4333-4337.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vendrell J P, Conge A M, Secondy M, Lombroso S, Huguet M F, Bertand A, Janbon F, Serre A. In vitro antibody secretion by peripheral blood mononuclear cells as an expression of the immune response to Brucella spp. in humans. J Clin Microbiol. 1992;30:2200–2203. doi: 10.1128/jcm.30.8.2200-2203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verstreate D R, Creasy M T, Caveney N T, Baldwin C L, Blab M W, Winter A J. Outer membrane proteins of Brucella abortus: isolation and characterization. Infect Immun. 1982;35:979–989. doi: 10.1128/iai.35.3.979-989.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vizcaíno N, Cloeckaert A, Zygmunt M S, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis opm31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64:3744–3751. doi: 10.1128/iai.64.9.3744-3751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaíno N, Cloeckaert A, Zygmunt M S, Fernández-Lago L. Molecular characterization of Brucella species large DNA fragment deleted in Brucella abortus strains: evidence for a locus involved in the synthesis of a polysaccharide. Infect Immun. 1999;67:2700–2712. doi: 10.1128/iai.67.6.2700-2712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vordermeier H M, Hoffman P, Gombert F O, Jung G, Bessler W G. Synthetic peptide segments from Escherichia coli porin OmpF constitute leukocyte activators. Infect Immun. 1990;58:2719–2725. doi: 10.1128/iai.58.8.2719-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber G, Link F, Ferber E, Munder P G, Zeitter D, Barlett R R, Nixdorff K. Differential modulation of the effects of lipopolysaccharide on macrophages by a major outer membrane protein of Proteus mirabilis. J Immunol. 1993;151:415–424. [PubMed] [Google Scholar]
- 42.Young E J. Human brucellosis. Rev Infect Dis. 1983;5:821–842. doi: 10.1093/clinids/5.5.821. [DOI] [PubMed] [Google Scholar]
- 43.Zhan Y, Liu Z, Cheers C. Tumor necrosis factor alpha and interleukin-12 contribute to resistance to the intracellular bacterium Brucella abortus by different mechanisms. Infect Immun. 1996;64:2782–2786. doi: 10.1128/iai.64.7.2782-2786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan Y, Cheers C. Control of IL-12 and IFN-gamma production in response to live or dead bacteria by TNF and other factors. J Immunol. 1998;161:1447–1453. [PubMed] [Google Scholar]