Abstract
Introduction:Candida auris is a major threat to public health. Rapid detection is essential for early treatment and transmission control. The use of chromogenic media allows the presumptive identification of this new species. The aim of this study is to describe the morphological characteristics of C. auris colonies on three commercial chromogenic media. Methods: Nineteen C. auris isolates from different countries/clades and 18 isolates of other species were cultivated in CHROMagarTM Candida Plus, HiCromeTM Candida, CHROMagar-Candida, and fluconazole-supplemented (32 mg/L) CHROMagar-Candida media. Results: On CHROMagarTM Candida Plus and HiCromeTM Candida, C. auris isolates from Colombia, Venezuela, India, Korea, and Japan displayed blue-shaded colonies, while isolates from Spain and Germany exhibited light pink shades with a bluish halo. All isolates showed white to pink colonies on CHROMagar-Candida. On CHROMagar Candida supplemented with fluconazole, whilst C. auris, C. glabrata, or C. krusei showed a similar pink color at 48 h incubation, phenotypic differentiation was possible by the rough, paraffin-like texture or the intense purple color acquired by C. krusei and C. glabrata, respectively. Moreover, in this medium, the presence of C. auris in combination with other species of similar color was not limiting for its early identification, due to this medium selecting only strains resistant to this antifungal. Conclusions: The use of chromogenic media such as CHROMagarTM Candida Plus facilitates a presumptive identification of C. auris. However, this identification can be difficult in the presence of mixed cultures. In these cases, the use of CHROMagarTM Candida medium with 32 mg/L fluconazole offers better performance for the identification of C. auris by inhibiting fluconazole-susceptible strains and selecting rare or high fluconazole MIC (>32 mg/L) isolates.
Keywords: Candida auris, chromogenic media, CHROMagar™ Candida Plus, HiCromeTM Candida, fluconazole supplementation, yeast identification
1. Introduction
Since its first description in 2009 [1], Candida auris has been responsible for nosocomial outbreaks in several countries around the world [2]. Distinguishing features of this species include its multi-resistance to antifungal drugs, its potential to cause deep-seated infections with elevated mortality rates [3], its high transmissibility and persistence in the hospital environment [3,4], and the difficulty of identification by conventional biochemical and phenotypic techniques. These characteristics have led the CDC to classify C. auris as a global public health threat [5,6].
Phenotypically, C. auris is difficult to distinguish from other Candida species. On Sabouraud agar, C. auris forms white to cream colonies, and on chromogenic media such as CHROMagar Candida, it forms light pink colonies [7,8].
Available commercial identification systems generally misidentify C. auris as many other species with greater or lesser phylogenetic closeness, such as Candida haemulonii, Rhodotorula glutinis, Saccharomyces cerevisiae, or, less frequently, Candida famata, Candida dobushaemulonii, Candida sake, Candida lusitaniae, Candida albicans, Candida guilliermondii, or Candida parapsilosis [5].
Since the characterization of this yeast as an emerging global threat to public health and the acknowledgment of the difficulties for its identification using conventional biochemical techniques, such as API ID20C (bioMérieux) and AuxaColor™ 2 (BioRad-Laboratories, Marnes-la-Coquette, France), efforts have been driven to develop rapid detection methods with advanced techniques capable of avoiding the limitations of phenotypic tests, such as mass spectrometry (MALDI-TOF MS), or molecular techniques such as internal transcribed spacer sequencing (ITS). Despite their accuracy, these techniques are expensive and require qualified personnel, hampering their use at non-specialized centers or especially resource-limited settings. This fact limits not only the early initiation of appropriate treatment, but also the early detection of cases and contacts [9,10]. Furthermore, all suspected cases of C. auris colonization or infection should be immediately isolated owing to its high transmissibility in the hospital environment and contact cases must be promptly identified and screened for C. auris. In this matter, it is essential to avoid the delay in the microbiological identification of C. auris, limit or control the generation of hospital outbreaks, and diminish patient morbimortality as well as the healthcare-associated burden [11]. The still-present lack of knowledge on easily reproducible, low-cost but sensitive techniques for prompt C. auris identification widens the gap in equity between developing countries [12].
Most microbiology laboratories perform presumptive identification of Candida spp. using chromogenic media [13,14]. Previous reports on CHROMagarTM Candida medium have shown that it is not useful for differentially identifying C. auris and C. parapsilosis colonies [15]. Consequently, other selective media such as CHROMagarTM Candida Plus medium (CHROMagar, Paris, France) and HiCromeTM Candida have been developed to further discriminate C. auris colonies from other species [16,17,18]. Despite recent progress in this area, microbiological data with these modified media for presumptive identification of C. auris are still limited to studies with a relatively low number of species and strains tested, discrepancies between morphotypes, or lack of information on mixed cultures.
Therefore, the aim of this study was to describe the phenotypic characteristic of C. auris colonies on four commercially available chromogenic media, as well as to evaluate their performance for presumptive identification of C. auris compared to other non-C. auris species and mixed cultures, including a modified fluconazole-supplemented CHROMagarTM Candida.
2. Materials and Methods
A total of 35 suspensions of pathogenic yeast species were inoculated in CHROMagarTM Candida Plus (CHROMagar, Paris, France), HiCromeTM Candida (HIMEDIA LBS, Mumbai, India), CHROMagarTM- Candida (Biolife, Milan, Italy), and CHROMagar Candida supplemented with 32 mg/L of fluconazole (CHROMagar Candida plus fluconazole) (MAIM, Barcelona, Spain). Of these, 19 C. auris strains from different countries were selected to represent the four main global clonal lineages. Thirteen of these strains were from the Westerdijk Fungal Biodiversity Institute (CBS-KNAW): CBS-12766, CBS-12882, CBS-12766 (clade I; India); CBS-12372, CBS-12373 (clade II; Korea); CBS-10913, JM-15448 (clade II: Japan); CBS-15603, CBS-15604, CBS-15605, CBS-15606, CBS-15607, CBS-15610 (clade III: Spain); COL-01, COL-02, COL-03 (clade IV: Colombia), 10-05-15-23, 10-05-15-19, 10-05-15-22 (clade IV: Venezuela). Also, 5 reference Candida strains were analyzed (C. albicans ATCC-90028, Candida glabrata ATCC-90030, Candida tropicalis ATCC-200956, C. lusitaniae ATCC-135, and Candida krusei ATCC-6258), as well as one each of Candida utilis, Candida rugosa, Candida orthopsilosis, Candida metapsilosis, Candida boidinii, Candida nivariensis, C. guillermondii, S. cerevisiae, Geotrichum capitatum, Trichosporon asahii, Kodamaea ohmeri, and Rodothorula glutinis from clinical samples. Identification of all strains were confirmed by MALDI-TOF VITEK-MS® (bioMérieux, Marcy-l`Étoile, France) and sequencing of ITS 1-2 [19].
For monocultures, inoculum suspensions were prepared at 0.5 McFarland, adjusted to a concentration of 104 colony-forming units (CFU), and inoculated on three chromogenic media using a calibrated 5 µL pipette. All strains in monoculture were incubated at 37 °C for 48 h. Four mixed cultures containing a C. auris strain with 3 other non-C. auris species were performed to assess their ability to differentiate among Candida species. For all mixed cultures, inoculum suspensions were prepared at 0.5 McFarland and adjusted to 104 CFU. Then, the inoculum was adjusted to 103 CFU, and suspensions were inoculated on four chromogenic media using a calibrated 100 µL pipette. Mixed cultures were incubated at 37 °C and read at 24 h and 48 h by a clinical mycologist. Recording of color, size, and texture of colonies in each medium was performed both visually and using a magnifying lens (Nikon SMZ1500).
3. Results
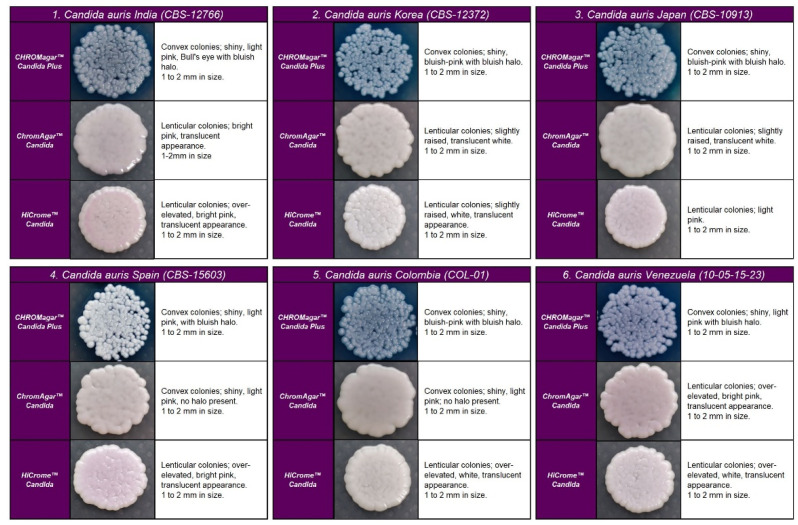
In the four chromogenic media analyzed, the most specific morphology was observed at 48 h in all strains (Figure 1). Recovered colonies were presumptively identified according to the colony color described by the manufacturer.
Figure 1.
Appearance of Candida auris colonies of the four clades described on CHROMagar™ Candida Plus medium, CHROMagar™ Candida medium, and HiCromeTM Candida after 48 h incubation at 35 °C.
C. auris strains developed similar colonies in size, color, and texture in each medium. In CHROMagarTM Candida Plus and HiCromeTM Candida, C. auris strains from Colombia, Venezuela, India, Korea, and Japan exhibited blue-shaded colonies, while strains from Spain exhibited light pink with a bluish halo that diffuses into the surrounding agar. By analysis using HiCromeTM Candida, C. auris showed pale pink shades without a halo (see Figure 1).
On CHROMagarTM-Candida, all C. auris strains showed white and pink colonies at 24 and 48 h, respectively. Colonies of C. auris, C. glabrata, or C. krusei on CHROMagar Candida plus fluconazole showed a similar pink color after 48 h incubation; however, phenotypic differentiation was possible by the rough, paraffin-like texture or the intense purple color acquired by C. krusei and C. glabrata, respectively ( Figure 1 and Figure 2).
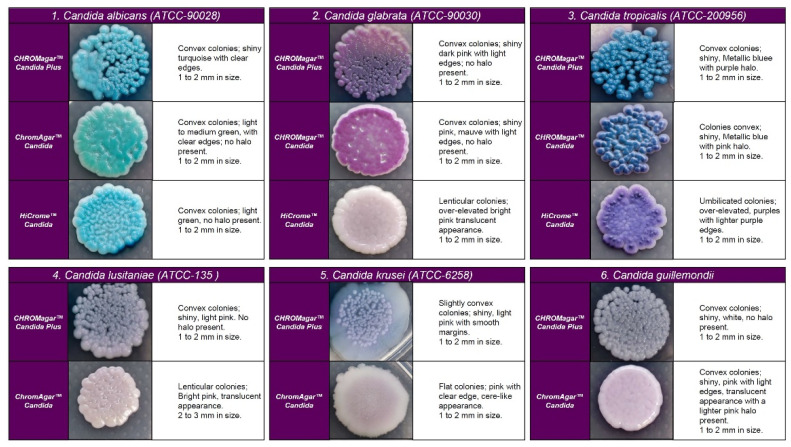
Figure 2.
Appearance of colonies of most pathogenic Candida species on CHROMagar™ Candida Plus medium, CHROMagar™ Candida medium, and HiCromeTM Candida after 48 h incubation at 35 °C.
After 24 h incubation on CHROMagar Candida plus fluconazole, C. auris showed smooth, convex, shiny, and light pink colonies (colony size 0.7–3 mm). After 48 h, they acquired an egg yolk morphology (white periphery and purple center). Although C. auris, C. glabrata, or C. krusei showed a similar pink color at 48 h incubation, phenotypic differentiation was possible due to the same characteristics observed for C. krusei and C. glabrata on CHROMagarTM-Candida. All C. auris strains but COL-01 and CBS-10913 were able to grow (these two isolates were susceptible to fluconazole), while S. cerevisiae, C. albicans, and C. famata showed inhibition of growth in this fluconazole-supplemented medium ( Figure 2 and Figure 3).
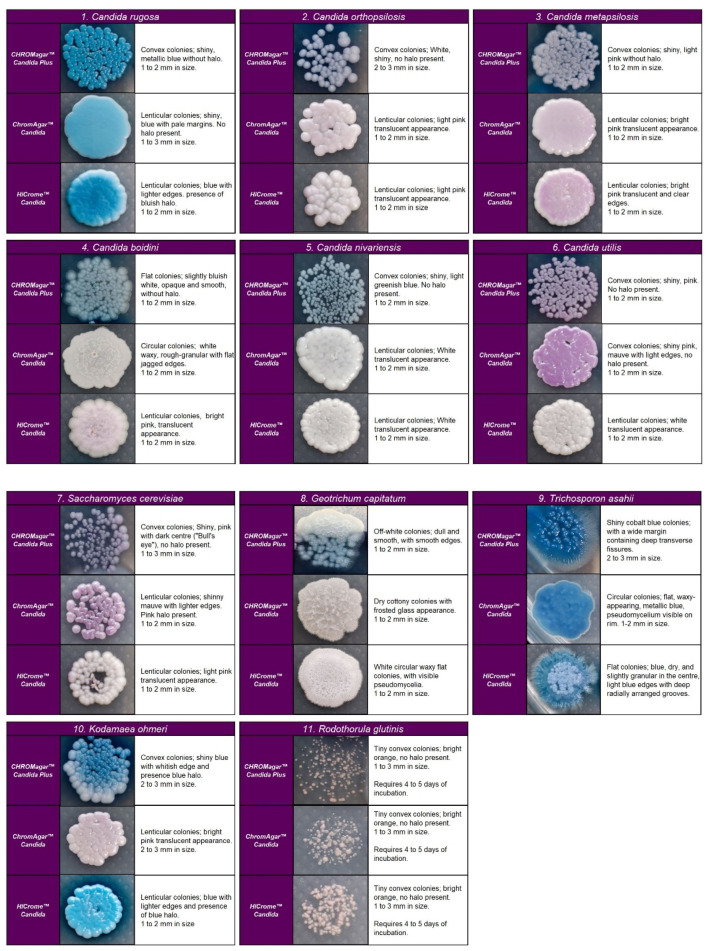
Figure 3.
Colony appearance of five non-Candida species on CHROMagar™ Candida Plus, CHROMagar™ Candida, and HiCromeTM Candida after 48 h incubation at 35 °C.
Regarding the colony morphology, the strains grown on CHROMagarTM Candida Plus developed individualized colonies, in contrast to the other three media where the growth is agglomerated at 48 h ( Figure 2 and Figure 3). On CHROMagarTM Candida and HiCromeTM Candida, after this incubation time, differentiation between the mauve/light pink species (C. auris, C. ortopsilosis, C. metapsilosis, C. nivariensis, C. guillermondii, and C. lusitaniae) was difficult. In CHROMagarTM Candida Plus, some species (C. albicans, C. tropicalis, C. rugose, and K. ohmeri) developed a similar metallic blue color, the distinction of which was difficult without significant changes with incubations over 48 h (Figure 2). Similar results were reported for species such as C. auris, C. albicans, C. parapsilosis/orthopsilosis, or C. tropicalis that presented a bluish color. However, differentiation was possible through the variety of shades or halos with different colors [17].
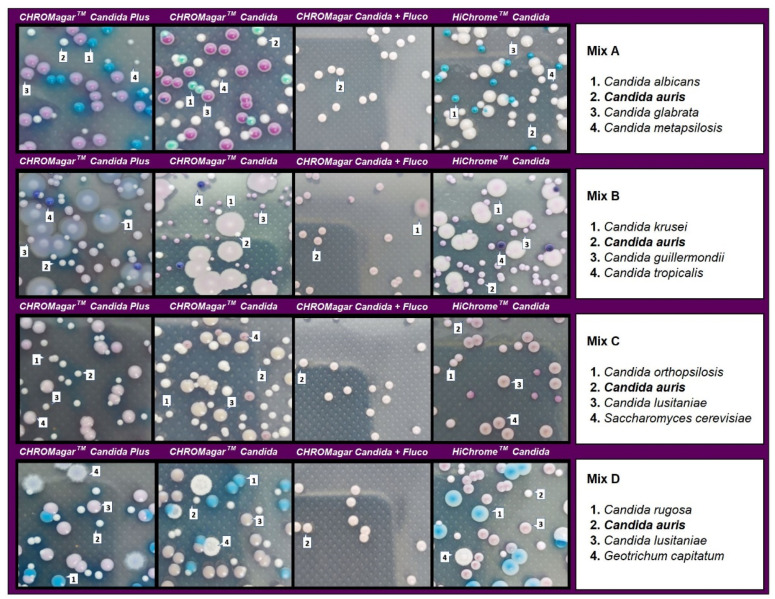
In the evaluation of mixed cultures, on CHROMagar Candida plus fluconazole, C. auris showed smooth, convex, shiny, light purple colonies with an egg yolk morphology (white periphery and purple center). After 48 h of incubation, in cultures including C. auris in combination with C. albicans, C. metapsilosis, and C. glabrata, identification was difficult in the three tested media due to the whitish to pinkish coloration of the colonies. However, after 48 h incubation, C. auris colonies acquired a turquoise blue halo in CHROMagarTM Candida Plus that encouraged differentiation. All C. albicans, C. metapsilosis, and C. glabrata isolates were completely inhibited on this medium (Figure 4).
Figure 4.
Appearance of Candida species and other yeast species colonies from mixed cultures on CHROMagar™ Candida Plus, CHROMagar™ Candida, CHROMagar™ Candida plus fluconazole, and HiCromeTM Candida after 48 h incubation at 35 °C.
In mixed cultures, the number of colonies was similar for CHROMagarTM Candida Plus and CHROMagarTM Candida with some differences depending on the species assayed. CHROMagar Candida plus fluconazole was able to recover 97–100% of C. auris colonies before 48 h of incubation. Differentiation was not possible with HiCromeTM Candida in mixed cultures containing C. auris and C. guillermondii (Table 1).
Table 1.
Comparison of CHROMagarTM Candida Plus, HiCromeTM Candida, CHROMagarTM Candida, and CHROMagar Candida supplemented with 32 mg/L of fluconazole for the recovery (CFU) of mixed cultures after 48 h of incubation at 35 °C.
| Mixed Culture/Organism | Recovered Colonies (n) | |||
|---|---|---|---|---|
| CHROMagarTM Candida Plus | HiCromeTM Candida | CHROMagarTM Candida | CHROMagar Candida Plus Fluconazole | |
| Mix A | ||||
| C. auris | 142 | 74 | 144 | 112 |
| C. glabrata | 137 | 134 | 137 | NG |
| C. metapsilosis | 32 | 85 | 30 | NG |
| C. albicans | 65 | 65 | 74 | NG |
| Total | 376 | 358 | 385 | 112 |
| C. auris recovered vs. total (%) | 37.77 | 20.67 | 37.40 | 100 |
| Mix B | ||||
| C. auris | 100 | * 325 | 81 | 124 |
| C. guillermondii | 213 | 166 | NG | |
| C. tropicalis | 21 | 37 | 13 | NG |
| C. krusei | 89 | 77 | 67 | 3 |
| Total | 423 | 114 | 327 | 127 |
| C. auris recovered vs. total (%) | 23.64 | ND | 24.77 | 97.64 |
| Mix C | ||||
| C. auris | 105 | 122 | 107 | 103 |
| C. lusitaniae | 108 | 107 | 108 | NG |
| S. cerevisiae | 39 | 35 | 28 | NG |
| C. orthopsilosis | 85 | 20 | 70 | NG |
| Total | 337 | 284 | 313 | 103 |
| C. auris recovered vs. total (%) | 31.16 | 42.96 | 34.19 | 100 |
| Mix D | ||||
| C. auris | 105 | 85 | 72 | 74 |
| C. rugosa | 57 | 57 | 35 | NG |
| C. lusitaniae | 96 | 49 | 130 | NG |
| G. capitatum | 37 | 24 | 19 | NG |
| Total | 295 | 215 | 256 | 74 |
| C. auris recovered vs. total (%) | 35.59 | 39.53 | 28.13 | 100 |
NG = Non-growth; ND = not determined; * uncountable.
4. Discussion
Rapid and reliable diagnosis of C. auris infection or colonization is essential for prompt and accurate antifungal therapy as well as for transmission control. Classical yeast identification techniques still available such as API-20C, AuxaColorTM 2, VITEK-2 YST, BD-PhoenixTM, and MicroScan misidentify C. auris [20,21]. To avoid the identification problems of available commercial techniques, the use of commercial media such as CHROMagar Candida® supplemented with Pal agar (sunflower seed extract) has been suggested as a simple and low-cost method to distinguish between C. auris isolates and the C. haemulonii complex [22].
New media such as HiCromeTM Candida and CHROMagarTM Candida Plus have been previously studied with identification rates between 0% and 33%, respectively [21]. Our results using CHROMagarTM Candida Plus agree with those described by other authors who reported good results in the detection of C. auris in both artificial cultures and surveillance samples after 36 h of incubation, respectively [17,21]. Identification of C. auris was difficult in the presence of white to mauve growing species at 24–36 h of incubation [17]. In our study, after 24 h of incubation, inhibition of susceptible species in CHROMagarTM Candida supplemented with 32 mg/L fluconazole allows a rapid recovery of C. auris colonies, although those were small and not colored enough.
In studies carried out with CHROMagarTM Candida Plus, it has been described that C. auris, C. albicans, C. parapsilosis/orthopsilosis, or C. tropicalis species can have a similar bluish color before 36 h [17,21,23]; in our study, we found similar results for C. auris, C. orthopsilosis, C. metapsilosis, C. nivariensis, and C. glabrata in this medium.
According to our results, in culture assays performed on CHROMagar Candida with 32 mg/L of fluconazole, the presence of C. auris in combination with other species of similar color was not limiting for its early identification, due to this medium selecting only fluconazole-resistant strains. Although C. auris isolation in this medium requires further confirmation, it allows for a prompt response that enables rapid isolation of patients and thus improved control of its transmission.
Currently, molecular methods or mass spectrophotometry (MALDI-TOF MS) are recommended for the definitive identification of C. auris. Among the molecular methods, sequencing of genetic loci (D1/D2, RPB1 and RPB2), rRNA internal transcribed spacers (ITS), and commercialized molecular technologies are the most commonly used [24,25]. These techniques are reliable but their availability outside reference laboratories is limited, especially in developing countries [26,27]. In these settings, the use of CHROMagarTM Candida agar with 32 mg/L of fluconazole as the primary culture medium in patients with suspected C. auris colonization allows fast presumptive identification of C. auris (24–48 h). This achievement is similar to that observed by other authors who used CHROMagarTM Candida Plus for the isolation and identification of C. auris from clinical samples in monoculture [17,21]. Although a presumptive identification can be achieved in 36 h, in all the media studied, a minimum incubation of 48 h is necessary to facilitate the differentiation of the species, and definitive identification must be done by spectrophotometric techniques such as MALDI-TOF or by molecular methodology sequencing the ITS regions [28,29,30].
In conclusion, the use of low-cost and accessible chromogenic media could allow the presumptive identification of C. auris infection or colonization in areas with a high prevalence of this species. This advantage is more evident with the use of CHROMagarTM Candida agar with 32 mg/L of fluconazole: this medium offers better performance for the identification of C. auris by inhibiting other species susceptible to this antifungal and selecting rare strains or those with high fluconazole MIC (>32 mg/L). Nevertheless, in all cases, definitive confirmation is required.
Acknowledgments
We thank Jacques Meis (Centre of Expertise in Mycology Radboudumc/Canisius-Wilhelmina Hospital, Nijmegen, The Netherlands), and Indira Berrio (Medical and Experimental Mycology Group, Biological Research Corporation, Medellín, Colombia) for the supply of clinical isolates recovered from Indian, Korean, Japanese, Venezuelan and Colombian isolates.
Author Contributions
Conceptualization, A.R.-G., I.S.-G. and J.P.; Literature search, V.G.-B., M.G.-H. and E.V.-G.; Writing—original draft preparation, A.R.-G., J.P. and V.G.-B.; editing, S.G.A. and P.W.J.d.G.; Elaboration of Tables and Figures, A.R.-G., I.S.-G. and J.M.P.-R.; Supervision, A.R.-G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. Candida aurissp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Jeffery-Smith A., Taori S.K., Schelenz S., Jeffery K., Johnson E.M., Borman A., Manuel R., Brown C.S. Candida auris: A review of the literature. Clin. Microbiol. Rev. 2017;31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz-Gaitán A., Moret A.M., Tasias-Pitarch M., Aleixandre-López A.I., Martínez-Morel H., Calabuig E., Salavert-Lletí M., Ramírez P., López-Hontangas J.L., Hagen F., et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61:498–505. doi: 10.1111/myc.12781. [DOI] [PubMed] [Google Scholar]
- 4.Welsh R.M., Bentz M.L., Shams A., Houston H., Lyons A., Rose L.J., Litvintseva A.P. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 2017;55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Bustos V., Cabanero-Navalon M.D., Ruiz-Saurí A., Ruiz-Gaitán A.C., Salavert M., Tormo M.Á., Pemán J. What Do We Know about Candida auris? State of the Art, Knowledge Gaps, and Future Directions. Microorganisms. 2021;9:2177. doi: 10.3390/microorganisms9102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control . Antibiotic Resistance Threats in the United States, 2019. CDC; Atlanta, Georgia: 2019. [Google Scholar]
- 7.Wang X., Bing J., Zheng Q., Zhang F., Liu J., Yue H., Tao L., Du H., Wang Y., Wang H., et al. The first isolate of Candida auris in China: Clinical and biological aspects. Emerg. Microbes Infect. 2018;7:1–9. doi: 10.1038/s41426-018-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidaud A., Chowdhary A., Dannaoui E. Candida auris: An emerging drug resistant yeast—A mini-review. J. Med. Mycol. 2018;28:568–573. doi: 10.1016/j.mycmed.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Prakash A., Sharma C., Singh A., Singh P.K., Kumar A., Hagen F., Govender N., Colombo A., Meis J., Chowdhary A. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin. Microbiol. Infect. 2016;22:277.e1–277.e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Perry J.D. A Decade of Development of Chromogenic Culture Media for Clinical Microbiology in an Era of Molecular Diagnostics. Clin. Microbiol. Rev. Am. Soc. Microbiol. 2017;30:449–479. doi: 10.1128/CMR.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fasciana T., Cortegiani A., Ippolito M., Giarratano A., Di Quattro O., Lipari D., Graceffa D., Giammanco A. Candida auris: An Overview of How to Screen, Detect, Test and Control This Emerging Pathogen. Antibiotics. 2020;9:778. doi: 10.3390/antibiotics9110778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S., Singh S., Tawde Y., Chakrabarti A., Rudramurthy S.M., Kaur H., Shankarnarayan S.A., Ghosh A. A Selective Medium for Isolation and Detection of Candida auris, an Emerging Pathogen. J. Clin. Microbiol. 2021;59:e00326-20. doi: 10.1128/JCM.00326-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orenga S., James A.L., Manafi M., Perry J.D., Pincus D.H. Enzymatic substrates in microbiology. J. Microbiol. Methods. 2009;79:139–155. doi: 10.1016/j.mimet.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 14.De Angelis G., Menchinelli G., Torelli R., De Carolis E., Posteraro P., Sanguinetti M., Posteraro B. Different detection capabilities by mycological media for Candida isolates from mono- or dual-species cultures. PLoS ONE. 2020;15:e0226467. doi: 10.1371/journal.pone.0226467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura T., Alshahni M.M., Makimura K. Evaluation of CHROMagarTM Candida Plus chromogenic agar for the presumptive identification of Candida auris. Microbiol. Immunol. 2022;66:292–298. doi: 10.1111/1348-0421.12973. [DOI] [PubMed] [Google Scholar]
- 16.Borman A.M., Fraser M., Johnson E.M. CHROMagarTM Candida Plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med. Mycol. 2020;11:1–6. doi: 10.1093/mmy/myaa049. [DOI] [PubMed] [Google Scholar]
- 17.Mulet Bayona J.V., Salvador García C., Tormo Palop N., Gimeno Cardona C. Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagarTM Candida for the detection of Candida auris in surveillance samples. Diagn. Microbiol. Infect. Dis. 2020;98:115168. doi: 10.1016/j.diagmicrobio.2020.115168. [DOI] [PubMed] [Google Scholar]
- 18.Bayona J.V.M., García C.S., Palop N.T., Martín A.V., Padrón C.G., Rodríguez J.C., Pemán J., Cardona C.G. Novel Chromogenic Medium CHROMagarTM Candida Plus for Detection of Candida auris and Other Candida Species from Surveillance and Environmental Samples: A Multicenter Study. J. Fungi. 2022;8:281. doi: 10.3390/jof8030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellemain E., Carlsen T., Brochmann C., Coissac E., Taberlet P., Kauserud H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhary A., Sharma C., Meis J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong A.W., Dieleman C., Carbia M., Tap R.M., Hagen F. Performance of Two Novel Chromogenic Media for the Identification of Multidrug-Resistant Candida auris Compared with other Commercially Available Formulations. J. Clin. Microbiol. 2021;59:e03220-20. doi: 10.1128/JCM.03220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A., Sachu A., Mohan K., Vinod V., Dinesh K., Karim S. Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal’s medium. Rev. Iberoam Micol. 2017;34:109–111. doi: 10.1016/j.riam.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Sasoni N., Maidana M., Latorre-Rapela M.G., Morales-Lopez S., Berrio I., Gamarra S., Garcia-Effron G. Candida auris and some Candida parapsilosis strains exhibit similar characteristics on CHROMagarTMCandida Plus. Med. Mycol. 2022;60:myac062. doi: 10.1093/mmy/myac062. [DOI] [PubMed] [Google Scholar]
- 24.Sharma C., Kumar N., Meis J.F., Pandey R., Chowdhary A. Draft Genome Sequence of a Fluconazole-Resistant Candida auris Strain from a Candidemia Patient in India. Genome Announc. 2015;3:e00722-15. doi: 10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhary A., Sharma C., Duggal S., Agarwal K., Prakash A., Singh P.K., Jain S., Kathuria S., Randhawa H.S., Hagen F., et al. New Clonal Strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013;19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkow E.L., Lockhart S.R. Fluconazole resistance in Candida species: A current perspective. Infect. Drug Resist. 2017;10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Colombo A.L., Calvo B., Cuomo C.A., Desjardins C.A., et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pino-Calm B., de Artola D.G.M., Gil-Campesino H., Alcoba-Flórez J. Restriction fragment length polymorphism de las regiones ITS1-ITS2 como método para identificar Candida auris. Rev. Iberoam Micol. 2018;35:167–168. doi: 10.1016/j.riam.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Bao J.R., Master R.N., Azad K.N., Schwab D.A., Clark R.B., Jones R.S., Moore E.C., Shier K.L. Rapid, Accurate Identification of Candida auris by Using a Novel Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) Database (Library) J. Clin. Microbiol. 2018;56:e01700-17. doi: 10.1128/JCM.01700-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmoudi S., Afshari S.A.K., Gharehbolagh S.A., Mirhendi H., Makimura K. Methods for identification of Candida auris, the yeast of global public health concern: A review. J. Med. Mycol. 2019;29:174–179. doi: 10.1016/j.mycmed.2019.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.