Abstract
The nickel-containing enzyme urease is an essential colonization factor of the gastric pathogen Helicobacter pylori, as it allows the bacterium to survive the acidic conditions in the gastric mucosa. Although urease can represents up to 10% of the total protein content of H. pylori, expression of urease genes is thought to be constitutive. Here it is demonstrated that H. pylori regulates the expression and activity of its urease enzyme as a function of the availability of the cofactor nickel. Supplementation of brucella growth medium with 1 or 100 μM NiCl2 resulted in up to 3.5-fold-increased expression of the urease subunit proteins UreA and UreB and up to 12-fold-increased urease enzyme activity. The induction was specific for nickel, since the addition of cadmium, cobalt, copper, iron, manganese, or zinc did not affect the expression of urease. Both Northern hybridization studies and a transcriptional ureA::lacZ fusion demonstrated that the observed nickel-responsive regulation of urease is mediated at the transcriptional level. Mutation of the HP1027 gene, encoding the ferric uptake regulator (Fur), did not affect the expression of urease in unsupplemented medium but reduced the nickel induction of urease expression to only twofold. This indicates that Fur is involved in the modulation of urease expression in response to nickel. These data demonstrate nickel-responsive regulation of H. pylori urease, a phenomenon likely to be of importance during the colonization and persistence of H. pylori in the gastric mucosa.
Helicobacter pylori is a gram-negative, microaerophilic human pathogen, which colonizes the gastric mucosa. Infection with H. pylori leads to gastritis and is associated with the development of peptic ulcer disease and gastric cancer (16, 33). Since approximately half of the world population is infected with H. pylori (16), it constitutes a major public health problem.
H. pylori expresses large quantities of the enzyme urease (3), an essential colonization factor of H. pylori (17, 18, 56). This enzyme converts urea, which is present in millimolar concentrations in the gastric mucosa, into ammonia and bicarbonate. The ammonia protects H. pylori against the acidic microenvironment (31, 44, 47, 58), causes damage to the gastric epithelium (50), is essential for chemotactic behavior (39), and serves as a nitrogen source (23). The bicarbonate protects H. pylori against the bactericidal activity of peroxynitrite, a nitric oxide metabolite (34).
Urease is a multimeric, nickel-containing enzyme which consists of six UreA and six UreB subunits (15, 25, 28). The UreA and UreB subunits have molecular masses of 27 and 62 kDa, respectively, and are encoded by the ureA and ureB genes which are organized in an operon structure (35). The gene products of a second operon, containing the ureIEFGH genes and located downstream of the ureAB genes, are required for the production of active urease. The UreEFGH accessory proteins probably function in subunit assembly and in the incorporation of nickel in the active sites of urease (8, 11, 38). This second operon also contains the ureI gene, which encodes a putative acid-activated urea transporter (44, 46, 49, 58). Transcription of the H. pylori urease gene cluster occurs from two promoters: one upstream of the ureA gene (PureA) (1, 48) and one in the intergenic region between ureB and ureI (PureI) (1). Transcription from these two promoters, followed by pH-dependent differential mRNA decay, leads to the formation of mRNAs containing ureAB, ureABIE′, ureIE′, and ureF′GH (1).
Urease is expressed by a wide variety of bacteria but not usually at the very high levels found in H. pylori (8, 38). Urease activity in other bacteria is seldom constitutive but is regulated in response to environmental changes, such as changes in pH, urea availability, nitrogen availability, or growth phase (8, 9, 12, 38, 40). H. pylori produces large amounts of urease, and it has been estimated that up to 10% of the total protein content of H. pylori consists of urease (3). The production of such large amounts of urease must constitute a heavy metabolic burden, and therefore it is likely that H. pylori regulates the expression of urease. However, the published H. pylori genome sequences lack homologs of the urease regulators UreR and NtrC (2, 8, 40, 55), and urease expression in H. pylori is not transcriptionally regulated by urea availability or pH (3). The activity of urease is increased at low pH, but expression of the UreA and UreB subunits is unchanged (1, 44, 46, 49, 58). Several loci have been demonstrated to affect either urease expression or activity: the nickel transporter genes nixA and abcCD (4, 26), the heat shock gene hspA (30) and the heat shock regulator gene hspR (51), the heavy metal P-type ATPase gene cadA (27), the RNA helicase gene deaD (7) and helicase genes hp0511 and hp0548 (36), the flagellar biosynthesis gene flbA (36), and the hydrogenase subunit genes hypA and hypB (41). However, these loci have not been demonstrated to directly regulate ureAB transcription.
Although urease is essential for gastric colonization (17, 18, 56) and plays a central role in the pathogenesis of H. pylori infection, the expression of urease has so far not been recognized to be regulated by environmental stimuli. In this study, nickel-responsive induction of expression and activity of H. pylori urease is described and is shown to be mediated at the transcriptional level via the ureA promoter.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The H. pylori and Escherichia coli strains and the plasmids used in this study are listed in Table 1. H. pylori was routinely cultured on Dent plates (14), consisting of Columbia agar (Oxoid) supplemented with 7% saponin-lysed defibrinated horse blood, 0.004% triphenyltetrazolium chloride (Sigma), and Dent Selective Supplement (Oxoid), at 37°C under microaerophilic conditions (10% CO2, 5% O2, and 85% N2). H. pylori was grown in broth cultures in brucella broth (Oxoid) supplemented with 3% Newborn Calf Serum (BBN, Gibco). Metal chlorides (ACS quality) were purchased from Sigma, dissolved in distilled water, filter sterilized, and used at the indicated concentrations. The total nickel content of BBN was determined with inductively coupled plasma-mass spectrometry (53) at the Chemische Landesuntersuchungsanstalt, Freiburg, Germany, and was approximately 0.2 μM. E. coli was grown aerobically in Luria-Bertani medium (45) at 37°C. When appropriate, growth media were supplemented with kanamycin or chloramphenicol to final concentrations of 20 or 10 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| H. pylori | ||
| 1061 | Wild-type strain | 7 |
| 26695 | Wild-type strain | 55 |
| NCTC 11638 | Wild-type strain | National Collection of Type Cultures |
| ATCC 43504 | Wild-type strain | American Type Culture Collection |
| N6 | Wild-type strain | 22 |
| J99 | Wild-type strain | 2 |
| AV433 | 1061 ureA::lacZ Kmr | This study |
| 1061fur | 1061 fur::Cmr | J. J. E. Bijlsma et al.a |
| E. coli ER1793 | Host for pBW-derived vectors | 43 |
| Plasmids | ||
| pBW | Insertional vector, Kmr, containing a promoterless lacZ gene in pBCα3 (7) | N. de Vries et al.b |
| pBJD3.3 | pBW containing an H. pylori ureA::lacZ transcriptional fusion | This study |
J. J. E. Bijlsma, S. Bereswill, A. H. M. van Vliet, B. Waidner, C. M. J. E. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters, unpublished data.
N. de Vries, E. J. Kuipers, N. E. Kramer, A. H. M. van Vliet, J. J. E. Bijlsma, M. Kist, S. Bereswill, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters, unpublished data.
Recombinant DNA techniques.
Restriction enzymes and modifying enzymes were purchased from New England Biolabs and were used according to the manufacturer's instructions. Standard protocols were used for manipulation of DNA and the transformation of E. coli (45) and H. pylori (7). Plasmid DNA was prepared using Qiaprep spin columns (Qiagen).
Protein analysis.
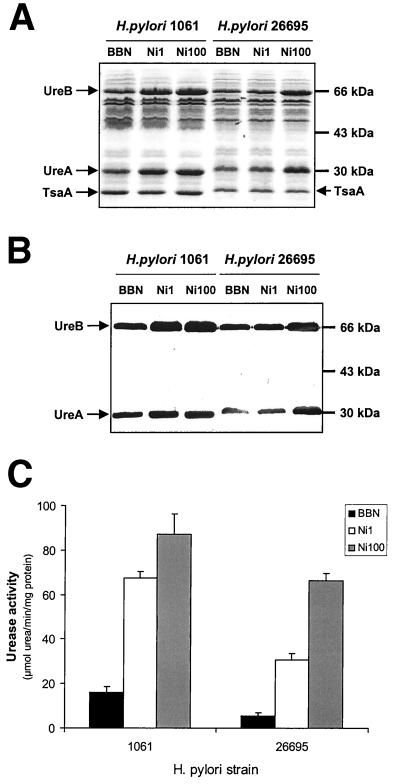
H. pylori cultures were grown in unsupplemented or NiCl2-supplemented BBN for 20 to 24 h with moderate shaking to an optical density at 600 nm (OD600) of 0.4 to 0.8, centrifuged for 10 min at 4,000 × g at 4°C, and resuspended in ice-cold phosphate-buffered saline to a final OD600 of 10. H. pylori cells were lysed by sonication for 15 s on ice with an MSE Soniprep 150 set at amplitude 10. Protein concentrations were determined with the bicinchoninic acid method (Pierce) using bovine serum albumin as the standard. Samples containing 15 μg of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel (45) and stained with Coomassie brilliant blue or subjected to Western immunoblotting with a urease-specific antiserum (7). Protein gels were scanned at 300 dots per inch using a Microtek scanner and analyzed by densitometry using the Kodak 1D Image Analysis Software, version 3.5. Samples were normalized using a total protein content of between 35 and 60 kDa, and expression patterns of the urease subunits were compared to an internal control consisting of the 26-kDa H. pylori TsaA protein (29, 42) (indicated in Fig. 1).
FIG. 1.
Expression and activity of H. pylori urease upon supplementation of growth medium with nickel. H. pylori strains were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1) and 100 μM (Ni100). Changes in urease expression were monitored by SDS-PAGE followed by Coomassie brilliant blue staining (A), immunoblotting with H. pylori urease-specific antiserum (B), or quantitative urease enzyme assay (C). H. pylori strains, UreA and UreB proteins, relevant molecular mass markers, and the 26-kDa TsaA protein used as an internal control for densitometry are indicated.
Urease activity.
Urease activity of fresh lysates was determined by measuring ammonia production from urea hydrolysis with the Berthelot reaction (11). Briefly, lysates of freshly sonicated cells (0.6 to 1.0 μg of protein) were incubated for 10 min at room temperature in 1 ml of PEB buffer (100 mM sodium phosphate, 10 mM EDTA; pH 7.5) containing 50 mM urea. Subsequently, 90-μl aliquots were mixed with 150 μl of phenol-nitroprusside (Sigma Diagnostics 640-1), 150 μl of alkaline hypochlorite (Sigma Diagnostics 640-3), and 750 μl of distilled water and then incubated for 10 min at 37°C. The ODs of the samples were determined at 570 nm. The amount of ammonia present in the 90-μl samples was then inferred from a standard NH4Cl concentration curve. Urease activity was expressed as micromoles of urea hydrolyzed per minute per milligram of protein.
RNA analysis.
Total RNA was isolated from 4 × 109 freshly grown bacteria with RNeasy spin columns (Qiagen), according to the manufacturer's instructions. RNA was separated on 2% formaldehyde–1.5% agarose gels in 20 mM sodium phosphate buffer (pH 7), transferred to positively charged nylon membranes (Roche Diagnostics), and covalently bound to the membrane by cross-linking with 0.120 J of UV light of 254-nm wavelength per cm2. RNA was visualized by methylene blue staining (1), and RNA samples were normalized based on 16S and 23S rRNA band intensities. The sizes of the hybridizing RNA species were calculated from comparison with a digoxigenin-labeled RNA marker (RNA Marker I; Roche Diagnostics). Internal fragments of the ureA, ureI, and ureG genes were PCR amplified with the primers listed in Table 2. The resulting PCR fragments contained a T7 promoter sequence and were used for the production of antisense RNA probes labeled with digoxigenin by in vitro transcription using T7 RNA polymerase (Roche Diagnostics). Northern hybridization and stringency washes were done at 68°C, and bound probe was visualized with the DIG-Detection Kit (Roche Diagnostics) and the chemiluminescent substrate CPD-Star (Amersham Pharmacia).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| UreA-F1 | ATGAAACTCACCCCAAAAGA |
| UreA-R1-T7a | ctaatacgactcactatagggagaGGAAGTGTGAGCCGATTTGA |
| UreI-F2 | AAGCACTGCGGTGATGAACT |
| UreI-R2-T7a | ctaatacgactcactatagggagaACCAATCGCCTTCAGTGATG |
| UreG-F1 | TGACATGGCGGTCATCACTA |
| UreG-R-T7a | ctaatacgactcactatatagggagaCAAGTCTGAGCGCGTGATTC |
| BJD3.3-F1b | cgcggatccTTTTTGAAGGGCATTTGTGC |
| BJD3.3-R1b | cgcggatccAACTTGTCTAACTCTTTTGG |
Primers contained a 5′ extension with T7 promoter sequence (in lowercase letters) for the creation of an antisense RNA probe.
Primers contained a 5′ extension for cloning purposes, indicated in lowercase letters. A BamHI restriction site is underlined.
Construction and characterization of a H. pylori ureA::lacZ transcriptional fusion.
Plasmid pBJD3.3 (Table 1) was constructed by PCR amplification of the H. pylori 1061 ureA promoter with primers BJD3.3-F1 and BJD3.3-R1 (Table 2), subsequent digestion with BamHI, and cloning into the unique BglII site upstream of the promoterless lacZ gene of vector pBW (Table 1). The ureA PCR fragment was sequenced to verify the fidelity of the amplification. pBJD3.3 was transformed to H. pylori 1061 as described previously (7), resulting in the kanamycin-resistant H. pylori strain AV433 (Table 1). The β-galactosidase activity in H. pylori AV433 grown in either unsupplemented or in nickel-supplemented BBN medium was determined in lysates from freshly sonicated cells. A portion (0.1 ml) of lysate (corresponding to approximately 109 cells) was mixed with 0.9 ml of Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, 0.002% SDS) and warmed to 37°C. Reactions were started by the addition of 0.2 ml of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml), and terminated by the addition of 0.5 ml of Na2CO3 (1 M). The OD420, the OD550, and the time required for the solution to become yellow were recorded. The β-galactosidase activity was expressed in Miller units (45).
RESULTS
Nickel supplementation of growth medium increases urease expression and activity.
We tested the effect of medium supplementation with the urease cofactor nickel on protein expression patterns in one clinical isolate and one reference strain of H. pylori (1061 [7] and 26695 [55], respectively). Strains were grown either in unsupplemented BBN or in BBN supplemented with NiCl2 to final concentrations of 1 and 100 μM. These NiCl2 concentrations represent a significant increase in the total nickel content of BBN, which is approximately 0.2 μM, but are well below the nickel toxicity levels for H. pylori, which are between 1 and 6 mM (27, 37, 57). Supplementation of BBN with 1 or 100 μM NiCl2 did not have a significant effect on the growth of H. pylori (not shown) but did result in the increased expression of two proteins with molecular masses of approximately 30 and 67 kDa, respectively (Fig. 1A). The molecular mass, amount, and recognition of these two proteins by antibodies to H. pylori urease (Fig. 1B) verified that these proteins were the urease subunit proteins UreA and UreB. BBN supplementation with 500 μM NiCl2 did not give rise to higher levels of urease expression, and BBN supplementation with 1 and 5 mM NiCl2 started to adversely affect growth of H. pylori 1061 and 26695 (not shown).
Supplementation of BBN with 1 and 100 μM NiCl2 gave a significant induction of up to 3.5-fold of UreA and UreB expression in H. pylori 1061 (Table 3, Fig. 1A). H. pylori 26695 did not significantly induce UreA and UreB expression in response to supplementation with 1 μM NiCl2 (Table 3, Fig. 1A) but did give a significant 3.3-fold induction at 100 μM NiCl2 (Table 3, Fig. 1A). Expression of the 26-kDa TsaA protein (29, 42), which was used as an internal control, was not affected by nickel supplementation (Table 3, Fig. 1A). Four other reference strains (NCTC11638, ATCC43504, J99, and N6) also showed two- to threefold-induced expression of UreA and UreB upon medium supplementation with 100 μM NiCl2. Nickel supplementation of BBN also led to increased urease activity in strains 26695 and 1061, albeit to different levels (Fig. 1C). Urease activity increased approximately 5-fold in H. pylori 1061 and almost 12-fold in H. pylori 26695 (Fig. 1C). The induction of urease expression was specific for nickel, since medium supplementation with 25 μM CoCl2 or with 100 μM CdCl2, CuCl2, FeCl3, MnCl2, or ZnCl2 did not affect urease expression in H. pylori 26695 (data not shown).
TABLE 3.
Quantitative determination of nickel-responsive induction of H. pylori urease expression
| Strain and protein | Change in relative level of protein expression ± SDa after supplementation with:
|
|
|---|---|---|
| 1 μM NiCl2 | 100 μM NiCl2 | |
| H. pylori 26695 | ||
| UreA | 1.3 ± 0.4 | 3.3 ± 0.9* |
| UreB | 1.4 ± 0.3 | 3.4 ± 0.2* |
| TsaA | 1.0 ± 0.1 | 1.0 ± 0.1 |
| H. pylori 1061 | ||
| UreA | 2.8 ± 0.6* | 3.5 ± 0.6* |
| UreB | 2.8 ± 0.6* | 3.4 ± 0.4* |
| TsaA | 1.1 ± 0.2 | 1.1 ± 0.2 |
| H. pylori 1061fur | ||
| UreA | 1.9 ± 0.1* | 2.0 ± 0.2* |
| UreB | 1.7 ± 0.2* | 1.9 ± 0.5* |
| TsaA | 1.1 ± 0.1 | 1.0 ± 0.1 |
Relative levels of protein were determined by densitometry as described in Materials and Methods and were compared to the expression in unsupplemented BBN medium. The expression in unsupplemented BBN was set to 1. Samples were derived from three or four independent growth experiments. Asterisks indicate a significant change in expression compared to the level in unsupplemented medium (P < 0.01; Student's t test). TsaA served as an internal control, as indicated in Fig. 1A and Fig. 4.
Nickel induction of urease expression is mediated at the transcriptional level.
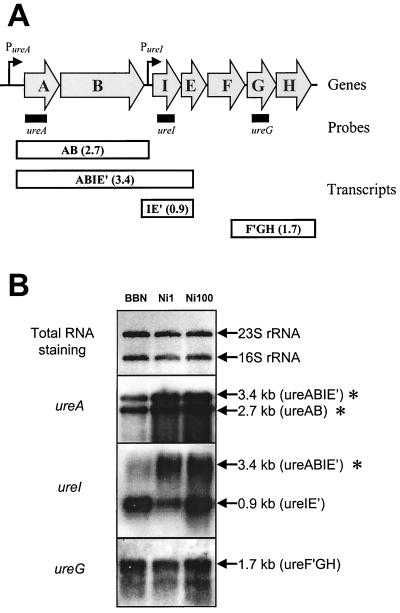
The levels of the mRNA species coding for urease subunit or accessory genes in H. pylori strain 1061 were analyzed with probes specific for the ureA, ureI, and ureG genes (Fig. 2A). The ureA-specific probe hybridized with a ureAB-containing mRNA of approximately 2.7 kb (1) and a ureABIE′-containing mRNA of 3.4 kb (1). The amounts of both mRNAs were clearly increased upon nickel supplementation of the medium (Fig. 2B). The ureI-specific probe hybridized with mRNAs of approximately 3.4 kb (ureABIE′) and 0.9 kb (ureIE′) (1). The amount of the 3.4-kb mRNA was clearly induced upon nickel supplementation, while the amount of the 0.9-kb mRNA did not change significantly upon nickel supplementation (Fig. 2B). Finally, the ureG-specific probe hybridized with a single mRNA, with a length of approximately 1.7 kb (ureF′GH) (1), whose amount did not change significantly upon nickel supplementation (Fig. 2B). In summary, only transcripts originating from the promoter upstream of the ureA gene were induced upon nickel supplementation.
FIG. 2.
Nickel induction of urease expression is mediated at the transcriptional level. (A) Transcriptional organization of the H. pylori urease operon, as modified from Akada et al. (1). The position of the probes used in this study are indicated, together with the different mRNA transcripts of the urease operon. The size (in kilobases) and the genes present on the respective mRNA species are indicated. An apostrophe indicates a truncated transcript; genes and the direction of transcription are indicated by arrows. PureA, ureA promoter; PureI, ureI promoter. (B) Analysis of urease transcription using Northern hybridization. H. pylori strains were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1) and 100 μM (Ni100). Staining of transferred RNA by methylene blue is included for comparison of the RNA amounts. The probes used are indicated on the left, whereas the rRNA and the sizes and content of the hybridizing mRNA species are indicated on the right. Nickel-induced mRNA species are marked with an asterisk.
Nickel induction of urease expression is mediated via the ureA promoter.
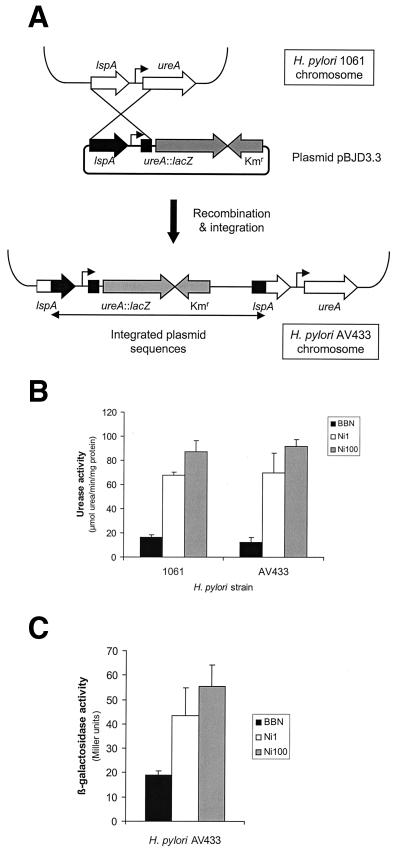
A chromosomal ureA::lacZ fusion was used to measure the effect of nickel supplementation on ureAB transcription. This transcriptional fusion would also allow us to determine whether the increase in ureAB mRNA resulted from true transcriptional induction (increased de novo mRNA synthesis) or increased mRNA stability. To construct the chromosomal ureA::lacZ fusion, plasmid pBJD3.3, which contains the ureA promoter in front of a promoterless lacZ gene (Table 1, Fig. 3A), was transformed to H. pylori 1061. The integration of the pBJD3.3 vector into the H. pylori 1061 chromosome by single homologous recombination leads to kanamycin resistance (7). The resulting transformant, named AV433, contains a duplicated ureA promoter (Fig. 3A). One ureA promoter is fused to the promoterless lacZ gene, whereas the other copy is still preceding the intact urease operon (Fig. 3A). Correct chromosomal integration of pBJD3.3 and the presence of the ureA::lacZ and wild-type urease operons in H. pylori AV433 was confirmed by PCR, sequencing, and Southern hybridization. The insertion of the pBW vector did not have a major effect on the expression, activity, and nickel induction of urease, since there were no significant differences in urease activity between strain AV433 and its parent strain 1061 (Fig. 3B). The β-galactosidase activity in H. pylori AV433 increased approximately threefold when the growth medium was supplemented with NiCl2 compared to unsupplemented growth medium (Fig. 3C).
FIG. 3.
Nickel induction of urease transcription is mediated via the ureA promoter, as determined by using a transcriptional ureA::lacZ fusion. (A) Schematic representation of H. pylori AV433, containing a single genomic copy of both the ureA::lacZ transcriptional fusion and the wild-type ureA promoter and downstream urease operon. lspA is the gene upstream of ureA, and Kmr indicates the kanamycin resistance determinant. (B) Activity and nickel-responsive induction of urease in H. pylori AV433 is not altered by duplication of the ureA promoter, compared to the parent strain 1061. (C) Induction of the ureA::lacZ transcriptional fusion in H. pylori AV433, as determined by β-galactosidase activity. BBN, unsupplemented medium; Ni1, medium supplemented with NiCl2 to a final concentration of 1 μM; Ni100, medium supplemented with NiCl2 to a final concentration of 100 μM. β-Galactosidase and urease activities were determined from three to eight independent cultures.
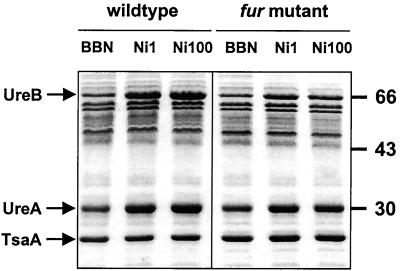
The H. pylori Fur homolog modulates nickel-responsive induction of urease.
The role of the H. pylori metal-responsive regulator Fur (HP1027) (6) in the nickel-responsive induction of urease was established by insertional mutagenesis of the fur gene in H. pylori 1061. Mutation of fur did not affect basal levels of urease expression (Fig. 4), nor did it affect transcription of the different mRNA species of the urease operon (not shown). However, nickel-responsive induction of urease was clearly diminished in the H. pylori fur mutant. Where wild-type H. pylori 1061 showed up to 3.5-fold induction of UreA and UreB expression (Table 3), in the fur mutant this was only to a maximum of 2-fold induction at 100 μM NiCl2 (Table 3). This difference was significant (P < 0.05) at both 1 and 100 μM NiCl2. Expression of the 26-kDa TsaA protein was again independent from the NiCl2 supplementation or the fur mutation (Fig. 4, Table 3). Increasing the NiCl2 concentration to 500 μM did not result in an additional induction of urease expression in the fur mutant, indicating that maximum levels of expression of UreA and UreB are lower in the H. pylori 1061 fur mutant than in wild-type H. pylori 1061.
FIG. 4.
Effect of a fur mutation in H. pylori 1061 on nickel-responsive induction of urease expression. H. pylori 1061 (wild type) and its isogenic fur mutant (fur mutant) were grown in unsupplemented medium (BBN) or in BBN supplemented with NiCl2 to final concentrations of 1 μM (Ni1) and 100 μM (Ni100). Changes in UreA and UreB expression were monitored by SDS-PAGE, followed by Coomassie brilliant blue staining. H. pylori strains, UreA and UreB proteins, relevant molecular mass markers, and the 26-kDa TsaA protein used as internal control for densitometry are indicated.
DISCUSSION
Urease activity is an essential factor in the colonization of the gastric mucosa by H. pylori (17, 18, 56). It is therefore not surprising that H. pylori expresses high levels of urease, up to 10% of its total protein content (3). This must constitute a heavy metabolic burden for this fastidious pathogen, but since none of the regulatory mechanisms described for other bacterial ureases apply to H. pylori urease, the expression of H. pylori urease was thought to be constitutive (3, 8, 38). As demonstrated here, H. pylori regulates urease transcription via the availability of the urease cofactor nickel, representing a novel type of transcriptional regulation for bacterial ureases.
Supplementation of growth media with nickel resulted in increased expression of the urease subunit proteins UreA and UreB and increased urease activity (Fig. 1). There were differences in the induction levels quantified in the two strains tested, since H. pylori 26695 expressed lower amounts of urease subunits and had a lower urease activity than strain 1061 but demonstrated much higher levels of induction in response to nickel supplementation of the growth medium (Fig. 1). The increase in urease expression and activity was accompanied by increased amounts of mRNA species containing the ureA and ureB genes (Fig. 2). The increase in ureA- and ureB-containing mRNAs is likely to be mediated exclusively through increased transcription from the ureA promoter (Fig. 3), since the observed threefold increase in β-galactosidase activity in H. pylori AV433 (ureA::lacZ) upon nickel supplementation seems to match the increase in the amounts of urease proteins and transcripts. In contrast to our findings, Olson et al. (41) did not find induction of urease activity when H. pylori strain ATCC43504 was grown with medium supplementation of 1 or 5 μM NiCl2. This is likely to be due to differences in medium composition and growth conditions. Olson et al. (41) used brucella agar plates supplemented with 10% blood, and agar and blood components are likely to chelate nickel. Also, plate-grown cultures usually contain a mix of cells in different growth phases, and this might also influence urease activity and nickel availability. We have tested strain ATCC 43504 in BBN medium supplemented with 100 μM NiCl2 and, under these conditions, it demonstrated a clear nickel-responsive induction of urease expression, to similar levels as H. pylori 1061.
The increase in urease activity resulting from nickel supplementation is higher than the observed increase in UreA and UreB subunit proteins (Fig. 1A and C, Table 3). This suggests that not only the amount of urease protein but also the amount of the cofactor nickel is a limiting factor for urease activity of H. pylori. Apparently, when grown in unsupplemented growth medium, H. pylori contains inactive urease resulting from the limited availability of nickel. Assuming that in the natural niche of H. pylori (i) the bioavailability of nickel is limited and (ii) the availability of nickel is positively affected by a decrease in pH (32), as is, for example, the case for iron, then the increased availability of nickel at the low pH would provide a physiological mechanism for a rapid increase of urease activity. This would result in an instantaneous increase in acid resistance resulting from activation of inactive urease apoenzyme due to increased nickel availability, without the need for de novo synthesis of the urease apoenzyme. While the concentration of nickel in the gastric lumen or gastric mucosa is unknown, the daily nickel intake in industrialized countries averages 150 μg/day but can vary significantly since some food sources, such as coffee, tea, nuts, and chocolate, are rich in nickel (52).
The nickel-responsive induction of urease expression observed in this study seems to be mediated through more than one regulatory system. The iron-regulatory protein Fur is involved in the nickel-responsive regulation of urease expression, since urease expression is only induced to a maximum of twofold in H. pylori 1061fur, whereas in wild-type H. pylori 1061 it is induced more than threefold. Fur is not involved in the basal levels of urease expression, however, since urease expression in wild-type and fur mutant strains is very similar (Fig. 4, Table 3). Whether Fur exerts its regulatory function directly at the urease promoter or through other pathways is unknown. Although the ureA promoter region contains a stem-loop structure (48) that somewhat resembles the structure used by Fur (20), it does not contain significant sequence homology to the Fur binding sequence (20) and was not identified in screening of H. pylori DNA in the original and H. pylori-optimized Fur titration assays (21). This indicates that the Fur protein plays an important role in metal-responsive gene regulation in H. pylori, since it is also involved in metal-responsive repression of ferritin synthesis in H. pylori (5).
Nickel-responsive regulation has only been characterized at the molecular level in a few bacterial species (19). Nickel induces the cnr cobalt and nickel resistance determinant of Ralstonia sp. via the CnrY, CnrX, and CnrH proteins (24, 54), whereas it represses the E. coli nickel acquisition system nik via the NikR protein (10, 13). H. pylori does not contain clear homologs of the CnrY, CnrX, or CnrH proteins, but it does contain a NikR homolog, designated HP1338 (55). We are currently assessing the role of the HP1338 protein in metal-responsive regulation of H. pylori.
In conclusion, H. pylori regulates the transcription and production of its virulence factor urease in response to nickel, and the H. pylori Fur homolog is involved in this regulation. Since urease expression is essential for colonization by H. pylori, it might be of interest to investigate the effect of a low-nickel diet on the colonization ability of H. pylori in animal models. Nickel-responsive induction of urease levels may also play a role in other urease-producing bacteria. If so, this may allow the development of new or improved strategies to prevent or control infection with urease-positive bacteria.
ACKNOWLEDGMENTS
We thank Jetta Bijlsma and Ben Appelmelk for helpful discussions.
This study was supported by grants from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO 901-14-206) to A.H.M.V.V., the Universitair Stimulerings Fonds (USF) of the Vrije Universiteit to E.J.K., the Deutsche Forschungsgemeinschaft (Ki201/9-1) to M.K., and the Biotechnology and Biological Sciences Research Council (BBSRC) and National Institute for Biological Standards and Control (NIBSC) to B.J.D.
REFERENCES
- 1.Akada J K, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol Microbiol. 2000;36:1071–1084. doi: 10.1046/j.1365-2958.2000.01918.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Bauerfeind P, Garner R, Dunn B E, Mobley H L. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauerfeind P, Garner R M, Mobley L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bereswill S, Greiner S, van Vliet A H M, Waidner B, Fassbinder F, Schiltz E, Kusters J G, Kist M. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J Bacteriol. 2000;182:5948–5953. doi: 10.1128/jb.182.21.5948-5953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bereswill S, Lichte F, Vey T, Fassbinder F, Kist M. Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol Lett. 1998;159:193–200. doi: 10.1111/j.1574-6968.1998.tb12860.x. [DOI] [PubMed] [Google Scholar]
- 7.Bijlsma J J E, Vandenbroucke-Grauls C M J E, Phadnis S H, Kusters J G. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect Immun. 1999;67:2433–2440. doi: 10.1128/iai.67.5.2433-2440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burne R A, Chen Y M. Bacterial ureases in infectious diseases. Microbes Infect. 2000;2:533–542. doi: 10.1016/s1286-4579(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y Y, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chivers P T, Sauer R T. Regulation of high-affinity nickel uptake in bacteria: Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem. 2000;275:19735–19741. doi: 10.1074/jbc.M002232200. [DOI] [PubMed] [Google Scholar]
- 11.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Koning-Ward T F, Robins-Browne R M. A novel mechanism of urease regulation in Yersinia enterocolitica. FEMS Microbiol Lett. 1997;147:221–226. doi: 10.1111/j.1574-6968.1997.tb10245.x. [DOI] [PubMed] [Google Scholar]
- 13.De Pina K, Desjardin V, Mandrand-Berthelot M A, Giordano G, Wu L F. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dent J C, McNulty C A. Evaluation of a new selective medium for Campylobacter pylori. Eur J Clin Microbiol Infect Dis. 1988;7:555–558. doi: 10.1007/BF01962615. [DOI] [PubMed] [Google Scholar]
- 15.Dunn B E, Campbell G P, Perez-Perez G I, Blaser M J. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. 1990;265:9464–9469. [PubMed] [Google Scholar]
- 16.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eitinger T, Mandrand-Berthelot M A. Nickel transport systems in microorganisms. Arch Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- 20.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron-box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fassbinder F, van Vliet A H M, Gimmel V, Kusters J G, Kist M, Bereswill S. Identification of iron-regulated Genes of Helicobacter pylori by a modified Fur titration assay (FURTA-Hp) FEMS Microbiol Lett. 2000;184:225–229. doi: 10.1111/j.1574-6968.2000.tb09018.x. [DOI] [PubMed] [Google Scholar]
- 22.Ferrero R L, Cussac V, Coureoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner R M, Fulkerson J, Jr, Mobley H L. Helicobacter pylori glutamine synthetase lacks features associated with transcriptional and posttranslational regulation. Infect Immun. 1998;66:1839–1847. doi: 10.1128/iai.66.5.1839-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grass G, Grosse C, Nies D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia sp. strain CH34. J Bacteriol. 2000;182:1390–1398. doi: 10.1128/jb.182.5.1390-1398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawtin P R, Delves H T, Newell D G. The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol Lett. 1991;61:51–54. doi: 10.1016/0378-1097(91)90012-y. [DOI] [PubMed] [Google Scholar]
- 26.Hendricks J K, Mobley H L. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrmann L, Schwan D, Garner R, Mobley H L, Haas R, Schafer K P, Melchers K. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol Microbiol. 1999;33:524–536. doi: 10.1046/j.1365-2958.1999.01496.x. [DOI] [PubMed] [Google Scholar]
- 28.Hu L T, Mobley H L. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungblut P R, Bumann D, Haas G, Zimny-Arndt U, Holland P, Lamer S, Siejak F, Aebischer A, Meyer T F. Comparative proteome analysis of Helicobacter pylori. Mol Microbiol. 2000;36:710–725. doi: 10.1046/j.1365-2958.2000.01896.x. [DOI] [PubMed] [Google Scholar]
- 30.Kansau I, Guillain F, Thiberge J M, Labigne A. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA) Mol Microbiol. 1996;22:1013–1023. doi: 10.1046/j.1365-2958.1996.01536.x. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamurthy P, Parlow M, Zitzer J B, Vakil N B, Mobley H L, Levy M, Phadnis S H, Dunn B E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect Immun. 1998;66:5060–5066. doi: 10.1128/iai.66.11.5060-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnaswamy R, Wilson D B. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl Environ Microbiol. 2000;66:5383–5386. doi: 10.1128/aem.66.12.5383-5386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuipers E J. Review article: relationship between Helicobacter pylori, atrophic gastritis and gastric cancer. Aliment Pharmacol Ther. 1998;12:25–36. doi: 10.1111/j.1365-2036.1998.00009.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuwahara H, Miyamoto Y, Akaike T, Kubota T, Sawa T, Okamoto S, Madea H. Helicobacter pylori urease suppresses bactericidal activity of peroxynitrite via carbon dioxide production. Infect Immun. 2000;68:4378–4383. doi: 10.1128/iai.68.8.4378-4383.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGee D J, May C A, Garner R M, Himpsl J M, Mobley H L. Isolation of Helicobacter pylori genes that modulate urease activity. J Bacteriol. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobley H L, Garner R M, Chippendale G R, Gilbert J V, Kane A V, Plaut A G. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter. 1999;4:162–169. doi: 10.1046/j.1523-5378.1999.99286.x. [DOI] [PubMed] [Google Scholar]
- 38.Mobley H L, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun. 1998;66:4832–4837. doi: 10.1128/iai.66.10.4832-4837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholson E B, Concaugh E A, Foxall P A, Island M D, Mobley H L. Proteus mirabilis urease: transcriptional regulation by UreR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson J W, Mehta N S, Maier R J. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole P W, Logan S M, Kostrzynska M, Wadstrom T, Trust T J. Isolation and biochemical and molecular analyses of a species-specific protein antigen from the gastric pathogen Helicobacter pylori. J Bacteriol. 1991;173:505–513. doi: 10.1128/jb.173.2.505-513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phadnis S H, Westblom T U, Normark S. Molecular cloning of Helicobacter pylori DNA: important differences between mcrBC deletion host strains. Mol Microbiol. 1993;10:1151. doi: 10.1111/j.1365-2958.1993.tb00984.x. [DOI] [PubMed] [Google Scholar]
- 44.Rektorschek M, Buhmann A, Weeks D, Schwan D, Bensch K W, Eskandari S, Scott D R, Sachs G, Melchers K. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol Microbiol. 2000;36:141–152. doi: 10.1046/j.1365-2958.2000.01835.x. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Scott D R, Marcus E A, Weeks D L, Lee A, Melchers K, Sachs G. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect Immun. 2000;68:470–477. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott D R, Weeks D, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 48.Shirai M, Fujinaga R, Akada J K, Nakazawa T. Activation of Helicobacter pylori ureA promoter by a hybrid Escherichia coli-H. pylori rpoD gene in E. coli. Gene. 1999;239:351–359. doi: 10.1016/s0378-1119(99)00389-3. [DOI] [PubMed] [Google Scholar]
- 49.Skouloubris S, Thiberge J M, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smoot D T, Mobley H L, Chippendale G R, Lewison J F, Resau J H. Helicobacter pylori urease activity is toxic to human gastric epithelial cells. Infect Immun. 1990;58:1992–1994. doi: 10.1128/iai.58.6.1992-1994.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spohn G, Scarlato V. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol Microbiol. 1999;34:663–674. doi: 10.1046/j.1365-2958.1999.01625.x. [DOI] [PubMed] [Google Scholar]
- 52.Sunderman F W. Biological monitoring of nickel in humans. Scand J Work Environ Health. 1993;19:34–38. [PubMed] [Google Scholar]
- 53.Sutton K L, Caruso J A. Liquid chromatography-inductively coupled plasma mass spectrometry. J Chromatogr A. 1999;856:243–258. doi: 10.1016/s0021-9673(99)00580-4. [DOI] [PubMed] [Google Scholar]
- 54.Tibazarwa C, Wuertz S, Mergeay M, Wyns L, van Der Lelie D. Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J Bacteriol. 2000;182:1399–1409. doi: 10.1128/jb.182.5.1399-1409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 56.Tsuda M, Karita M, Morshed M G, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Velayudhan J, Hughes N J, McColm A A, Bagshaw J, Clayton C L, Andrews S C, Kelly D J. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol Microbiol. 2000;37:274–286. doi: 10.1046/j.1365-2958.2000.01987.x. [DOI] [PubMed] [Google Scholar]
- 58.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]