Abstract
Our understanding of diabetes mellitus has benefited from a combination of clinical investigations and work in model organisms and cell lines. Organoid models for a wide range of tissues are emerging as an additional tool enabling the study of diabetes mellitus. The applications for organoid models include studying human pancreatic cell development, pancreatic physiology, the response of target organs to pancreatic hormones and how glucose toxicity can affect tissues such as the blood vessels, retina, kidney and nerves. Organoids can be derived from human tissue cells or pluripotent stem cells and enable the production of human cell assemblies mimicking human organs. Many organ mimics relevant to diabetes mellitus are already available, but only a few relevant studies have been performed. We discuss the models that have been developed for the pancreas, liver, kidney, nerves and vasculature, how they complement other models, and their limitations. In addition, as diabetes mellitus is a multi-organ disease, we highlight how a merger between the organoid and bioengineering fields will provide integrative models.
Subject terms: Diabetes, Biological models
Organoid systems have great potential to improve the study of diseases such as diabetes mellitus. This Review assesses the progress in developing pancreatic organoids and bioengineered systems for modelling diabetes mellitus and its complications.
Key points
Pancreas and islet organoids enable the study of monogenic forms of diabetes mellitus, notably neonatal diabetes mellitus.
For type 1 diabetes mellitus, platforms mixing patient-relevant islets and immune cells are expanding.
Organoid models are emerging that combine cell types derived from multiple organs relevant to diabetes mellitus.
A merger between bioengineering techniques and organoid technology is improving functional and screening assays.
For assays of organ interactions, organ-on-a-chip platforms are moving from cell lines to organoids.
While organoid models for diabetes mellitus complications are being added to the toolbox, the long periods over which complications develop mean that studying these complications in vitro will be challenging.
Introduction
Diabetes mellitus is a disorder in which the body does not produce enough insulin or responds abnormally to this hormone, causing atypically high blood levels of glucose. Diabetes mellitus is traditionally classified into type 1 diabetes mellitus (T1DM), which is autoimmune, and type 2 diabetes mellitus (T2DM), both of which result from a combination of environmental and multigenic factors. However, there are also less common monogenic forms of diabetes mellitus. Although there are excellent drugs available to lower blood levels of glucose, the proportion of patients with well-controlled diabetes mellitus is not increasing, so that a better dissemination of glucose-lowering drugs and the development of new drugs are required for patients whose diabetes mellitus is difficult to control1. Notably, T2DM is increasingly being recognized as a heterogeneous disorder, and so designing drugs targeting different groups of patients will be important, as will designing drugs with fewer and less-severe adverse effects than those currently available. Identifying and testing drugs involving endocrine cell types other than β-cells, including α-cells and δ-cells, and enteroendocrine cells, as well as insulin target organs, are promising avenues for research. With regard to T1DM, an important challenge is to understand the early steps of the disease and the interactions of β-cells with the immune system. For all types of diabetes mellitus, better treatments of the adverse effects of long periods of glucose excursions, notably targeting the macrovasculature and microvasculature, are crucial.
Multiple complementary approaches have led to our current understanding of diabetes mellitus and its treatment, including clinical investigations, work in multiple model organisms and more reductionist cell line models. To address the challenges outlined above, in addition to the existing toolbox, organoid models of a variety of complexities and readiness for the different organs involved in the aetiology of diabetes mellitus and its complications can be used. Organoids are 3D in vitro culture systems that make use of progenitors or stem cells to produce cell assemblies recapitulating aspects of organ structure and function2,3. Notably, organoids enable human cells to be grown in a 3D setting, as tissues do, enabling research and testing of drugs on human cells. Although organoids are not intended to replace animal studies, as it is difficult to recapitulate systemic disorders in vitro, they allow early tests in human cells. Organoids have been developed for the pancreas, a central player in the aetiology of diabetes mellitus, as well as for organs with which the pancreas is metabolically coupled, such as the liver, muscle and adipose tissue. Organoid models also exist for organs involved in diabetes mellitus complications, such as the blood vessels, retina, kidney and nerves4. In addition to these organoids stricto sensu (defined in the glossary (Box 1)), other 3D culture systems not developed from stem cells or progenitors, such as the pseudoislets, are gaining momentum5–7.
In this Review, we discuss the current models available to study the pathogenesis, progression and complications of diabetes mellitus. We first assess how organoid models can be used to study how genetic and environmental factors during pregnancy can predispose the offspring to diabetes mellitus. While these models mimicking pancreas development in humans are already available and their level of functionality can be exploited to study T1DM and T2DM, improvements are ongoing to obtain more mature endocrine cells and improve culture formats. As diabetes mellitus is a multi-organ disease, we next discuss the microfluidic systems enabling exchange between organs and how these systems could help study tissue communication, notably between the pancreas, liver, adipose tissue and brain and immune cells to study T1DM. Finally, we discuss the organoid models, including models of the vasculature, kidney, retina and neurons, that would be amenable to studying diabetes mellitus complications. We do not discuss 3D culture systems for the purpose of producing cells for diabetes mellitus therapy, even though this is a promising therapeutic avenue, and refer the reader to other informative reviews on the topic8,9.
Box 1 Defining terms in organoid research.
Pancreas progenitors 9
Pancreatic cells present in the prenatal developmental period that can give rise to endocrine and ductal cells (bipotent progenitors) or endocrine, ductal and acinar cells (multipotent progenitors).
Organoids 2
3D in vitro culture systems making use of progenitors or stem cells to produce cell assemblies recapitulating aspects of organ structure and function. In this Review, we use organoids as an umbrella term that also encompasses spheres and spheroids, but when no specification of spheres or spheroids are made, ‘organoids’ will usually refer to complex cell assemblies, including branched structures for pancreas organoids. Organoids can form in Matrigel, in suspension culture or on filters at the air–liquid interface.
Spheres or cysts 3
Specific types of organoids with a simple spherical structure made of one cell layer (or less commonly a few cell layers) surrounding a single large lumen. Pancreatospheres are an example of these types of organoid. Spheres are the most common structures found in Matrigel.
Spheroids or pancreatoids 3
Specific types of organoid with a simple spherical structure without a large central lumen. Spheroids commonly form in suspension culture or can be generated by embedding cancer cells or non-epithelial cells in Matrigel.
Dissociated islet cells re-aggregated in defined proportions and sizes. These 3D structures are not commonly referred to as organoids as islet cells are not considered stem cells. However, the same re-aggregation principle can be applied to endocrine cells produced from pluripotent stem cells.
Modelling pancreas development
Historically, many organoid systems were developed using a roadmap recapitulating development to produce the tissues of interest. As a consequence, many organoids still represent a developmental stage rather than a mature organ10. As there is genetic evidence that perturbed development of the pancreas can predispose to diabetes mellitus11 and numerous studies have shown that the environment of the fetus during pregnancy programmes adult metabolic activity12, organoids mimicking pancreas development are useful to study these mechanisms. Here we focus on the pancreas, a central organ in diabetes mellitus, and briefly introduce its development. We then provide an overview of organoids derived from the fetal pancreas or differentiated from pluripotent stem cells (PSCs). We discuss examples illustrating how these organoids have been used to study how genetic mutations and environmental factors predispose to diabetes mellitus.
Pancreas organoids from fetal cells
The pancreas emerges from two buds in the gut tube on embryonic day (E) 9.5 in mice and 30 days after conception in humans. The multipotent pancreas progenitors in these buds will differentiate into either endocrine cells or exocrine cells13,14. By E14.5, progenitors lose the ability to differentiate into acinar cells, and thus become bipotent. These bipotent progenitors continue to form endocrine cells until birth. In terms of morphogenesis, the exocrine part of the pancreas will form a network of tubes ending distally at acini while endocrine cells partially delaminate from the tubes and will assemble into islets of Langerhans9. The currently available organoid systems recapitulate these steps of development, or a subset of them.
Anchored in a long tradition of in vitro culture of tissue explants and primary cells, the development of the 3D matrix Matrigel represented a crucial step in the evolution of organoid systems. Matrigel enables the extended culture of early fetal pancreatic progenitors derived during E10.5–E11.5, which form pancreatospheres (Box 1) after dissociation to single cells15,16. These pancreatospheres are composed of epithelial cells surrounding a central lumen either in co-culture with mesenchymal cells15 or without mesenchyme16. These spheres can be expanded and passaged, although their capacity for differentiation into endocrine cells decreases after three passages15. Purified pancreatic progenitors from later stages of embryonic development (E12–E13 or E14.5) were also later shown to form spheres when embedded in Matrigel17,18. These pancreatic spheres transform into duct-like structures in the presence of epidermal growth factor (EGF) and differentiate towards endocrine cells in its absence. While these systems do not recapitulate the morphogenetic complexity of the pancreas, they are convenient to study endocrine cell formation and partial delamination, which can be done using live imaging18.
More complex organoids were also generated from multipotent progenitors (E10.5–E11.5) in media activating the WNT pathway15,16. These organoids recapitulate the hallmarks of the developing pancreas, including the formation of a ductal network as well as branches with acinar cells at the tips and a few endocrine cells. While these organoids form proportionally fewer endocrine cells compared with spheres16, they are more appropriate to study endocrine cells in the context of acinar cells than are spheres. In these early studies, comparisons of the pancreatic spheres or organoids to the cells of the fetal pancreas in vivo were limited, and the degree of similarity and maturity of the produced cells deserves deeper scrutiny.
In addition to Matrigel-based cultures, suspension cultures of embryonic and fetal pancreatic cells have also been established from E10.5 pancreatic progenitors and have been shown to form pancreatoids comprising all exocrine and endocrine lineages. Notably, 22% of the cells in these cultures differentiated into endocrine-like cells, but only when in the presence of the native mesenchyme, making this system unique for studying the role of the mesenchyme in pancreas development19.
Pancreatic organoids were also derived from human fetuses from 7–11 weeks after conception. Small fragments dissociated from the human pancreas formed hollow spheres that expressed pancreatic progenitor markers17. In the presence of EGF, these organoids could be expanded for 5 months while removing EGF reduced proliferation but promoted differentiation of the organoids towards an endocrine phenotype. Using a different culture medium, a 2021 study derived pancreatic organoids composed of a mixture of hollow spheres and dense spheroids containing ducts with narrow lumens but very few endocrine cells. These organoids could be passaged, indicating the maintenance of pancreas progenitors and single-cell RNA sequencing suggested that a subset of cells acquired a ductal identity in vitro20. Human pancreata from 9–22 weeks after conception were also used to grow organoids harbouring compact but branched structures, as well as cystic areas expressing progenitor and/or ductal markers20. Transcriptomic analyses revealed greater similarity of these organoids to the fetal human pancreas than to adult pancreata or islets.
The fetus-derived organoids described above are progressively becoming a tool to understand the development of the pancreas and its endocrine cells. They notably enable clonal assays to be conducted, which can assess the potency — that is, the ability of the cell to differentiate into other cell types — of individual cells15,21, the morphogenesis of the organ16,18,22 and the function of specific genes in endocrine cell differentiation15,23.
Organoids from pluripotent stem cells
The progressive gain of knowledge of the embryonic developmental cues in model organisms has paved the way to establishing multistep differentiation protocols that efficiently direct mouse PSCs or human PSCs (hPSCs) towards the pancreatic lineage14,24. These protocols, initially making use of 2D cultures, were further developed into 3D culture methods to upscale production and improve the maturity of the β-cells produced, mimicking the in vivo setting where endocrine cells are assembled into islets. The 3D methods used culture at the air–liquid interface after initial production of pancreas progenitors in 2D25, suspension culture from the onset26–31 or culture from later steps32–34, as well as embedding in Matrigel after pancreas progenitor production in 2D20,35 (Fig. 1).
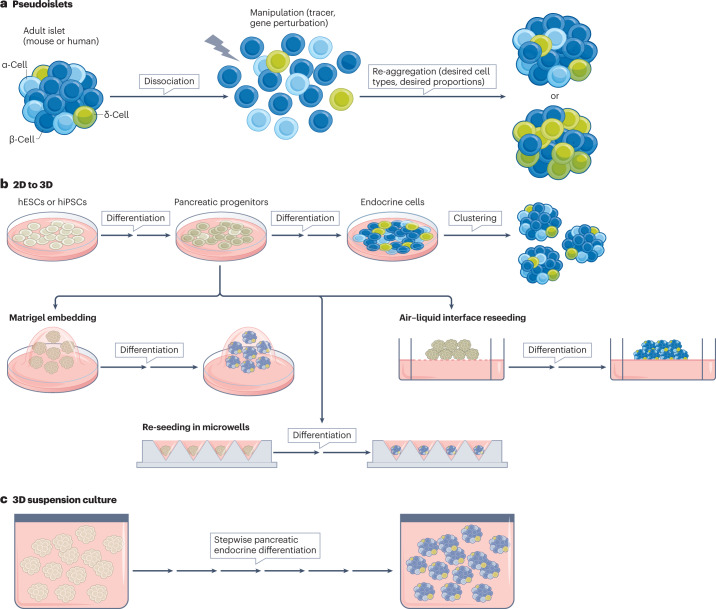
Fig. 1. Types of 3D cultures modelling pancreas and islet development, physiology and disease.
a, Dissociation and re-aggregation of islet cells to form pseudoislets. This process can include a sorting step. The ratio of endocrine cell types and the numbers of cells can be controlled. Genetic manipulations to the cells, such as gene inactivation or gain-of-function, can be applied. b, 3D methods applied to pluripotent stem cell-derived cultures. These methods include clustering of endocrine cells after 2D differentiation, Matrigel culture after pancreas progenitor or endoderm production in 2D20,35, re-seeding in microwells and aggregation after endoderm or pancreas progenitor production32–34 and culture at the air–liquid interface after initial production of pancreas progenitors in 2D25. c, Pancreatic organoids in suspension culture from the onset of differentiation from PSCs26–31. hESCs, human embryonic stem cells; hiPSC, human induced pluripotent stem cells.
Although many of these methods have not been referred to as organoids they are similar to models developed for other organs such as the kidney (air–liquid interface) or brain (steps of suspension culture), which are referred to as organoids (Box 1). Some protocols enabled a step for the expansion of progenitors prior to their differentiation20,33. Variability of efficiency between cell lines and protocols was reported26,36. Although most work focused on hPSCs, mouse PSCs could also be coaxed to form 3D islet-like structures that contained bona fide endocrine cells37,38. Early protocols produced immature cells with some basal insulin secretion in the presence of low levels of glucose and lower levels of insulin secretion in response to glucose than adult β-cells. However, several protocols from the past 5 years have resulted in more mature β-cells that are capable of glucose-stimulated insulin secretion with first-phase and second-phase dynamic insulin secretion (See ‘Improving β-cell maturity in vitro’). While most methods described above result in a mix of pancreas progenitors, pancreatic endocrine cells, enterochromaffin cells and ductal cells, protocols involving cell sorting and re-aggregation form purer islet-like organoids, without progenitors29.
While the relevance of these organoid systems to adult β-cell function needs to be improved (See Improving PSC-derived endocrine cell maturity), the available models are suitable for studying how mutations and environmental factors (Box 2) affect β-cell development, and thereby predispose to diabetes mellitus.
Box 2 Fetal pancreas organoids to study fetal programming of diabetes mellitus.
Organoids can shed light on the molecular effect of maternal health and environment on the developing fetal pancreas. In particular, they can be used to study how early-life exposure to nutrients and environmental toxins can contribute to the vulnerability of the offspring to later-life metabolic disorders, such as diabetes mellitus. Several studies performed in mice and rats have drawn attention to the importance of maternal diet on the fetal pancreas and the health of the progeny. Whether it is malnutrition184, dietary restrictions185, low protein intake186–189 or a high-fat diet190–193, the in utero environment can substantially alter the pancreas development of the offspring. This alteration is associated with lower β-cell production and function, altered islet composition and morphology and glucose intolerance when the offspring reach adulthood, compared with offspring that benefited from an optimal in utero environment. Exposure to endocrine disruptors, such as bisphenol A194–199, air pollution200 and Δ9-THC201, in utero can also predispose to diabetes mellitus.
These findings are also true in humans, where poor maternal nutrition is associated with incident metabolic diseases such as T2DM in the offspring202–204. Exposure to environmental pollutants can induce epigenetic dysregulation and promote the onset of adult metabolic disorders205–207. The mechanisms that might link early life developmental conditions to increased risk of offspring metabolic disease onset are not understood, mainly due to the inability to access the developing organ. While no such study has made use of pancreas organoids yet, a study performed on intestinal organoids is a valuable example addressing the consequences of a high-fat diet on stem cell function208. It showed how a high-fat diet acts via PPARδ to promote the self-renewal of intestinal organoids. Similar strategies can be applied to pancreatic organoids that mimic the fetal developmental stages. These organoids can be used to identify the precise cascade of events that disturb endocrine differentiation and so predispose to glucose intolerance and disease onset in adulthood.
Monogenic diabetes mellitus
There have been tremendous advances in the field of human genetics in the past 20 years that have given insights into the genetic contribution to diabetes mellitus. Monogenic forms of diabetes mellitus are the most straightforward scenario for personalized modelling39. Maturity-onset diabetes of the young (MODY) and neonatal diabetes mellitus (NDM) are the two main forms of monogenic diabetes mellitus40. NDM develops mainly in the first 6 months of life and results from mutations causing abnormal insulin production and secretion, islet cell destruction or abnormal pancreas development11. While some genes are specifically associated with either MODY or NDM, different mutations in the same gene can cause MODY or NDM41. An increasing number of studies have taken advantage of in vitro pancreatic development systems to investigate the impact of these mutations on β-cell formation and function, thereby overcoming challenges such as tissue availability and the limitations of mouse models to recapitulate the human disease phenotype42.
The initial strategy relied on differentiating patient-derived induced PSCs (iPSCs) into β-cells in 2D and comparing them to CRISPR–Cas9 genetically corrected autologous iPSC-derived β-cells41. Focusing here on 3D models, such studies uncovered that mutations in the insulin locus the can cause NDM do so by altering β-cell differentiation43 and impairing β-cell proliferation associated with endoplasmic reticulum (ER) stress44,45. Using a combination of patient mutations and knockout strategy, it was also found that the YIPF5 gene, which is involved in trafficking between the ER and the Golgi, increases β-cell sensitivity to ER stress-induced apoptosis46. Similarly, mutations in the STAT3 locus lead to premature endocrine differentiation during the differentiation protocol through direct induction of NEUROG3 expression47. Other mutations causing functional impairment of the coupling between glucose sensing and insulin secretion via KCNQ1 were identified and studied in a 2021 study (currently available as a preprint)48.
Another approach involves using hPSCs to study the developmental consequences of either naturally occurring mutations found within patients with monogenic diabetes mellitus or loss-of-function mutations generated through gene editing using the non-edited hPSC parental line as a reference. In this approach, the mutation is created in a genetic background that might be different from that found in patients as the starting hPSC lines are generally not characterized for the sensitivity of the background to diabetes mellitus. Studies following this approach have been conducted for PDX1 (refs. 49,50), NEUROG3 (refs. 50,51), RFX6 (ref. 50) and ONECUT1 (ref. 52), which are required at different developmental stages for the production of endocrine cells and the formation of glucose-responsive β-cells. While all studies mentioned above used 3D suspension culture or culture at the air–liquid interface, organoids in extracellular matrix hydrogels such as Matrigel would represent a model of choice to study the diabetes mellitus forms associated with exocrine phenotypes. An example of this application would be studying cystic ducts and their relationship to β-cell defects that can cause diabetes mellitus, as these organoids form ductal-like cells. Moreover, the sorting of endocrine cells and their re-aggregation could be exploited to study isolated islet-like interactions between controlled proportions of endocrine cell types29.
MODY, like NDM, is characterized by abnormally high blood levels of glucose. However, MODY develops later in life than NDM, although usually before 30 years of age. Several studies have generated human 2D in vitro models of monogenic mutations over the years, most of which rely on the generation of iPSC lines from patients and their differentiation into the pancreatic lineage to study genes such as HNF1A, HNF1B and HNF4A53–57. Among those genes, only HNF1A mutations have been studied in 3D48. These studies showed that deleting one or two alleles of HNF1A de-repressed an α-cell gene expression signature, reduced endocrine cell function and changed cellular metabolism54. Studying mutations introduced in human embryonic stem cells (hESCs) and patient-derived iPSCs, similar observations were made for homozygous hypomorphic HNF1A mutations. This study further revealed that heterozygous hypomorphic mutations produce β-cells that are initially normal but develop a reduced insulin secretion phenotype progressively in the 2–8 weeks after their transplantation in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice58. Finally, a study using patient iPSCs and CRISPR–Cas9-engineered hESCs with HNF1αp291fsinsC truncations, the most common mutation associated with MODY3, further showed that this mutation reduces progenitor and β-cell differentiation by inhibiting HNF1B function59. This example shows that comparing different mutations in organoid models enables us to further our understanding of the pathological mechanisms underlying MODY. 3D models recapitulate cell arrangements better than 2D systems; they include interactions between different endocrine cell types, the polarity of β-cells, their maturation and, for some organoids, the association with exocrine cells. It is therefore probable that organoids will be utilized more heavily in the future, and moving from studying the effects of the mutations to devising better treatments would be an essential step forward.
3D culture methods have also been applied to syndromic forms of monogenic diabetes mellitus (Box 3). Beyond the study of genes causing or predisposing to diabetes mellitus, the culture environment of organoids can be easily manipulated, which should, in principle, enable the study of fetal programming of diabetes mellitus by nutrients and environmental toxins. While organoids have not been used for this purpose the background for such studies and possible implementations are discussed in Box 2.
Box 3 3D culture methods applied to syndromic forms of monogenic diabetes mellitus.
There are many forms of monogenic diabetes mellitus that affect multiple organs, leading to syndromic forms of diabetes mellitus. Many of these have been studied in the pancreas or in β-cell models derived from human pluripotent stem cells41. For example, correcting a diabetes mellitus-causing mutation in the WFS1 gene in induced pluripotent stem cells (iPSCs) derived from a patient with Wolfram syndrome improved the differentiation and maturation of the in vitro-produced β-cells209. Another study from 2021 revealed how mutations in the gene MANF (which encodes mesencephalic astrocyte-derived neurotrophic factor) causes a form of diabetes mellitus associated with a neurodevelopmental disorder by promoting endoplasmic reticulum stress in β-cells210. Gene-corrected iPSC-derived β-cells from patients with monogenic diabetes mellitus were also shown to reverse diabetes mellitus in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice following their transplantation211. Moreover, pancreas-on-a-chip models have been used to study cystic fibrosis-related diabetes mellitus by utilizing organoids or spheres that combine pancreatic ducts and endocrine cells35,212.
Improved formats for studying diabetes mellitus
The organoids discussed in the previous section harbour immature, fetal-stage β-cells. These organoids have revealed disease mechanisms for monogenic forms of diabetes mellitus and validated genes found in genome-wide association studies (GWAS) that predispose to diabetes mellitus. However, it would be valuable to develop systems that better mimic postnatal and adult β-cells, in addition to the development of pseudoislet models5–7 (Box 4). Such systems would notably be useful for studying the genes predisposing to diabetes mellitus identified in GWAS60. Moreover, culture methods can be improved for better phenotyping.
Box 4 Islet cells reassembled into pseudoislets.
The importance of 3D structure on endocrine cell function was discovered in the 1980s, owing to the realization that the re-aggregation of rat islet cells regulates insulin release213. Further work focused on developing systems that allowed the spontaneous assembly of pseudoislets with permissive extracellular matrix conditions5,6,72. These early 3D islet systems restored cellular composition, spatial organization and hormone release to levels similar to those of primary islets, establishing them as viable systems for in vitro tests related to diabetes mellitus, drug discovery and pathological investigations. While these pseudoislets are not organoids because they are not derived from progenitor cells or stem cells and do not proliferate, these systems share similarities with organoids owing to their 3D nature, recapitulation of islet functions and ability to be genetically and compositionally manipulated. The pseudoislet system (Fig. 1a) has also been extended to human cells, both human islet-derived endocrine cells7 and human β-cell lines214. These pseudoislets exhibit similar morphological properties to their in vivo counterparts and similar functionality after transplantation into diabetic or non-diabetic immunodeficient mice7,214.
The dissociation and re-aggregation steps required to generate pseudoislets allow these culture systems to study islet cells with or without the effects of neighbouring cells and in desired cellular microenvironments with set cell type ratios215. Moreover, combining genetic manipulation with 3D culture systems provides a robust and adaptable model to study gene function within human islet cells while preserving hormone secretion dynamics216–218. Such a system involving successive steps of dissociation, genetic manipulation and re-aggregation of primary adult human islet cells was used to study the role of SIX2 and SIX3 in diabetes mellitus219. A common regulatory region of these genes harbours variants promoting type 2 diabetes mellitus. Gene inactivation of SIX2 and SIX3 in pseudoislets was shown to regulate β-cell maturity, notably insulin processing and glucose sensing219.
Improving β-cell maturity in vitro
Improving PSC-derived endocrine cell maturity
Ensuring that β-cells produced from in vitro pancreatic differentiation of PSCs display similar functionality to adult β-cells has become an important focus in the past 5 years. Progress was made in 2019 by isolating and re-aggregating in vitro-derived immature β-like cells29. These clusters displayed dynamic insulin secretion and responded to multiple secretagogues through calcium signalling. Clustering the immature β-like cells also promoted their mitochondrial metabolic maturation. Fine-tuning the medium composition and controlling the cellular cluster size through spontaneous re-aggregation without endocrine enrichment also improved insulin secretion in response to glucose30. Improvements in culture media and extension of culture time also resulted in high-level functionality, similar to that of human islets. This functionality was assessed by dynamic insulin secretion assays, respirometry measurements, electrophysiological assays, assessment of Ca2+ levels, and cAMP and insulin granule exocytosis. Targeted metabolite-tracing studies of glucose flux into the major pathways coupling metabolism to insulin secretion, together with single-cell transcriptomic profiling, showed that a remarkably high level of β-cell functionality was achieved in vitro, although some metabolic and transcriptomic differences persisted between stem cell-derived islet β-cells and primary β-cells34. In the past 10 years, transcriptomic20,61,62, metabolic29,34,63 and functional30,34,64 investigations of β-cells and their progenitors have been an important focus in the field. Together, these studies have provided a more holistic view of the similarities between stem cell-derived β-cells and endogenous β-cells and of the limitations of in vitro pancreatic models. Such studies are invaluable for addressing the relevance of the models, especially when providing comparisons with high-quality endogenous cells. Further improvements are ongoing to recapitulate the different stages of postnatal maturation, including the changes in transcription factors and silencing of ‘disallowed genes’ (a set of genes that prevent adult β-cell function) during development. Additional improvements include recapitulation in the medium of the major changes in metabolites in the environment of β-cells during postnatal maturation, which are linked to changes in food sources and therefore affect nutrients such as lipids and amino acids65,66.
Organoids from potential islet stem cells
In addition to hPSC-derived islets, a promising report from 2020 suggested that adult stem cells expressing the marker Procr could be isolated from adult mouse islets and could be used to seed the formation of islet organoids composed of multiple endocrine cell types67. While the existence of stem cells in the adult pancreas, whether in ducts or in islets, is highly debated68, further follow-up studies will be worthwhile, particularly with human islets. Several populations of the adult pancreas69, notably ALDHhi cells, which are probably exocrine cells, and CD51+ (human) or CD24+ (mouse) ductal cells have been suggested to contain progenitors. These populations indeed appear to re-express progenitor markers while expanding in culture and can form some endocrine cells when triggered by differentiation cocktails21,70. The efficiency and nature of these cells deserves more thorough investigation.
Improved formats for phenotyping
Developing gel scaffolds that improve organoid culture
While suspension cultures of hESC-derived or iPSC-derived β-cells were initially designed to be high-throughput for transplantation, the batch format of these culture systems is not ideal for any large-scale standardized screening. Suspension cultures in microwells (Fig. 1) enabling the assembly of a single large islet in each well offer both reproducibility and consistency in size as well as a convenient format to screen multiple conditions34. Culture at the air–liquid interface is also not high-throughput25. While pancreatic spheroid cultures in Matrigel have been used in screening20, Matrigel is produced from the secretion of mouse sarcoma cells, making it an animal-based, undefined product with observed batch-to-batch variation71. While more defined matrices based on collagen were used as early as the 1980s72, they have remained less effective at promoting endocrine differentiation and insulin secretion compared with Matrigel73. Although mixing collagen with Matrigel enabled the production of islet organoids from hESCs in 3D74, a more systematic comparison of efficiency between collagen-based matrices with Matrigel and Matrigel alone is required.
Hydrogel-based scaffolds are being tested as a more defined environment than Matrigel that is amenable to systematic variation of physical and chemical properties75. Work conducted in 2013 in mice explored soft hydrogels and revealed that pancreas organoids could grow in polyethylene-glycol-based gels, provided that these gels were covalently functionalized with laminin 1 (ref. 16). In 2018, a hydrogel-based platform called Amikagel was used to promote the aggregation of hESC-derived pancreatic progenitors into spheroids, with controlled size and cellular heterogeneity. They were then further differentiated into cells capable of insulin secretion, although in vitro culture beyond 7 days duration was not possible in this extracellular matrix76. Further work in the field towards more defined culture scaffolds appears necessary to enable stabilization of pancreas organoids for high-throughput sampling towards disease modelling and therapeutic applications focused on T2DM75,77.
Pancreas-on-a-chip models and high-throughput analyses
Despite being a relatively new field78, pancreas-on-a-chip systems are advancing swiftly, enabling the culture of 3D clusters of islet cells in microwells where perfusion and collection of samples can be coupled via microfluidic chips79 (Fig. 2). These systems can allow multiparametric assessments, for example, continuous measurements of oxygen consumption rate, insulin secretion and calcium imaging, as well as precise spatiotemporal stimulation with glucose and potentially other molecules79,80. Pancreas-on-a-chip systems also enable the cell clusters to be lysed inside the device to analyse gene expression, DNA methylation and micro-RNA secretion81. To date, these systems have been used to model oxidative stress-induced diabetes mellitus with human β-cell line spheroids81, study intra-islet communication based on rodent islets or pseudoislets82,83, improve functionality and glucose sensitivity of iPSC-derived human islets80,84,85, study insulin kinetics in single human pseudoislets86, and maintain islet physiology in vitro87. In a 2020 study, a microperfusion system in which genetically modified and re-aggregated native human islet cells were cultured was developed to allow the synchronous acquisition of several functional parameters88. Monitoring calcium signalling and hormone secretory profiles demonstrated differences in GPCR signalling pathways between human β-cells and α-cells88. Such studies give new insights into human islet biology and allow novel therapeutic approaches in diabetes mellitus research. A few challenges remaining for the widespread use of pancreas-on-a-chip systems including their complicated handling procedure and the suboptimal biophysical properties of the materials used in their fabrication (as, for example, the propensity of polydimethylsiloxane to absorb small molecules)79.
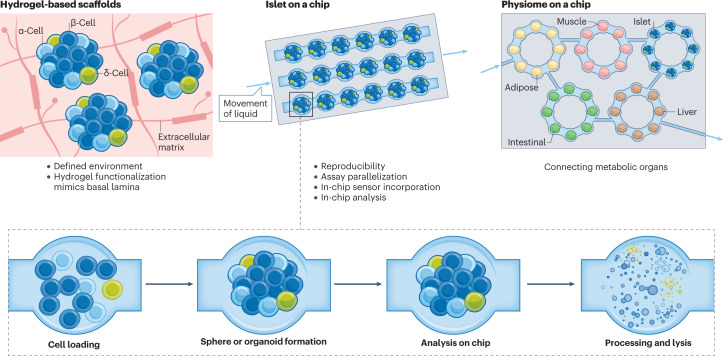
Fig. 2. Novel and emerging organoid culture systems using microfluidics.
Synergies between the bioengineering and organoid fields are enabling the development of standardization and miniaturization of assays, as well as the ability to collect biological information in real time and at the end point of culture. Bioengineered hydrogels have been synthesized and tested for their ability to sustain organoid 3D cultures, although they are often not as effective at promoting growth as extracellular matrix-derived gels. Hydrogels are water-insoluble, 3D networks of polymer chains capable of holding large amounts of water. They include natural polymers, such as alginate, chitosan, fibrin, heparin and hyaluronic acid fibrin, and synthetic polymers, such as acrylate polymers, polyethylene glycol, polyvinyl alcohol, sodium polyacrylate and their copolymers. Microwells of a defined size enable more reproducibility of organoid or pseudoislet size. Islets-on-a-chip assemble islets or pseudoislets, ideally of homogeneous size and composition in each single well, which can be used as an individual test tube. In the same spirit other organoids can be arrayed on a chip. Microfluidic systems can be designed to control the mixing and ratios of different cell types. Many different samples can be imaged at the same time, and functional imaging assays can be conducted. The system can also be designed to collect cells or their contents at the experimental end point. In the physiome-on-a-chip format, organoids of different types can be seeded in different wells and a microfluidic system enables the connection of different types of organoids to observe their crosstalk, attempting to reproduce physiological organ interactions.
As pancreas-on-a-chip systems have the potential for scalable manufacturing80, they would be an excellent tool for diabetic drug testing, especially if combined with the high number of β-cells that can be produced from hPSCs. Bioprinting is another promising technology that is currently being developed; it allows the positioning of living cells in a controllable pattern using biomaterials. There are several technical challenges associated with this technology, including limitations in resolution and cellular distribution, as well as the lack of vascularization and innervation for use in complex organs such as the endocrine pancreas89; however, such technologies could also provide new avenues for higher-scale drug and medical testing. Indeed, in principle they enable cells to be associated in defined numbers and patterns, allowing, for example, better islet mimicry and reproducibility. A few reports of high-throughput screening have indicated that islet spheroids can be predictive of the in vivo response to diabetes mellitus drugs90,91. Moving forward, challenges in modelling T2DM and developing new treatments will be the long time periods over which the disease develops as it will be difficult and costly to maintain organoids in culture for months or years.
Connecting organs and cell types
Because diabetes mellitus is a disease that perturbs the crosstalk of multiple tissues, models incorporating multiple pancreatic cell types and organs are needed to recapitulate certain aspects of the disease (Figs. 2 and 3).
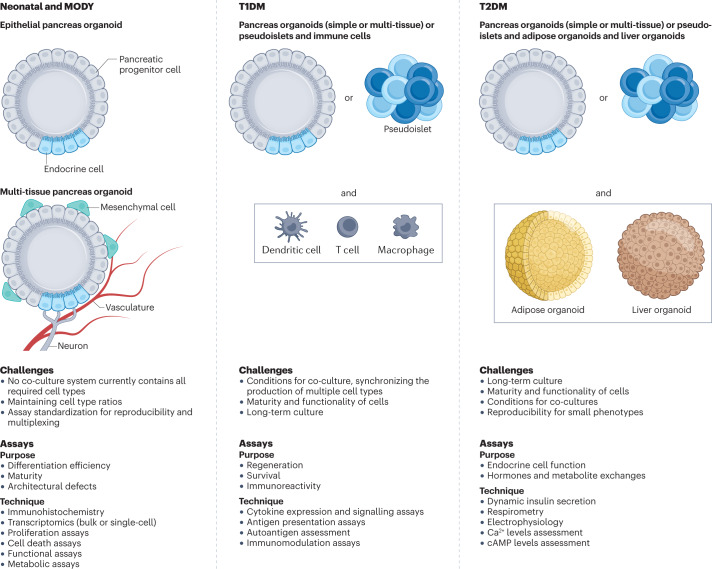
Fig. 3. Organoid systems of different complexity, enabling interactions between cells and organs in the course of diabetes mellitus to be studied.
Different 3D culture models exist for monogenic diabetes mellitus, type 1 diabetes mellitus (T1DM) and type 2 diabetes mellitus (T2DM). The figure outlines the challenges associated with modelling these diseases and the assays that are available for each system. Simple epithelial pancreas organoid models for neonatal diabetes mellitus and maturity-onset diabetes of the young (MODY) are available. These models can be composed of either progenitors and endocrine cells or aggregated pluripotent stem cell-derived endocrine cells alone. Co-culture of epithelial cells with mesenchyme and/or endothelial cells to form blood vessels is possible but difficult to stabilize and has not yet been achieved with pancreatic mesenchyme and endothelial cells. The maturity of the cells corresponds to embryonic cells and so can be problematic for modelling monogenic diabetes mellitus. The first models for the study of T1DM and T2DM are emerging. These models combine pancreatic organoids with immune cells or organoids from other metabolic organs for the study of T1DM and T2DM, respectively.
Pancreas organoids beyond the epithelium
Integrating non-endocrine cell types that are part of the native islets of Langerhans into organoid cultures can create more complex and mature systems, better recapitulating the in vivo conditions. In this regard, there are numerous ongoing efforts to produce endothelial, mesenchymal, neuronal and exocrine co-culture systems with pancreatic organoids.
The vasculature is an indispensable part of mature pancreatic islets in vivo and supports islet architecture, survival and secretion. To date, several studies have been conducted with co-cultures of endothelial and pancreatic cells in organoid systems, encompassing several goals (Fig. 3). The aim of some 3D co-cultures is to improve the differentiation of hESC-derived β-cells76 and the maturity of these cells92–94. The aim of others is to study the islet microenvironment more faithfully in hESC-derived or mouse cell line-based systems95,96, notably the impact of the spatial distribution of different cell types on islet or pseudoislet functionality94. Finally, the aim of some 3D co-cultures is to improve vascularization in both mouse organoids and isolated human islets prior to transplantation97,98. Adding bone marrow-derived vasculogenic endothelial progenitors92, islet-derived endothelial cells93 or human umbilical vein endothelial cells (HUVECs)94 to mouse or human β-cell pseudoislets was shown to improve glucose-stimulated insulin secretion and β-cell function. The degree of connectivity of blood vessels and the extent to which they penetrate 3D islet cultures is currently being investigated and improved, as the fenestration of the islet-associated microvasculature is of major importance for endocrine function97,99. Furthermore, there are efforts in the endothelial field to produce vasculogenic human endothelial cells that can directly interact with cells of the 3D co-cultured organoids, a promising and exciting line of research that could provide a perfusable and physiological platform for vascularized organoids, including the pancreas100.
Mesenchymal cells have also been included together with endothelial cells in pancreatic 3D cultures, from hESC-derived pancreatic cells or isolated rat islets99,101 (Fig. 3). These experiments showed that human mesenchymal cells drive the condensation of endothelial, mesenchymal and specific parenchymal cell types to form vascularized organ buds, including a vascularized pancreatic islet-like tissue with a mouse β-cell line102. Later studies in 2018 expanded on this idea using intact mouse and human islets103,104.
In a 2018 report, simulated cholinergic re-innervation of mouse β-cells through 2D and 3D co-culture with cholinergic neurons revealed that islet function is increased by re-innervation in a 3D pseudoislet environment105. Additional models to study the innervation of pancreatic organoids are underway, with improved protocols to generate innervated mouse pancreas organoids that also allow the role of neurogenesis in pancreas development and disease to be assessed106 (Fig. 3).
Finally, there are emerging studies highlighting the interaction of pancreatic exocrine tissue with the endocrine cells, and showing the effect of the exocrine tissue on diabetes mellitus prevalence107–109. The organoid system could also provide a valuable platform to study the communication and crosstalk between the exocrine and endocrine compartments of the pancreas in healthy and diseased conditions. Generation and long-term maintenance of human acinar organoids from stem cells has been achieved in the past 10 years, although these acinar cells had immature gene expression patterns and zymogen granules35,110,111. Optimization of culture conditions could enable further maturation of human acinar cells for co-culture studies with β-cells.
Co-culture systems for studying T1DM
T1DM occurs by autoimmune-mediated destruction of pancreatic β-cells. Although systemic readouts such as autoantibodies or circulating immune cell types can be monitored in patients or their siblings who are susceptible to developing T1DM, the early stages of the disease go undetected. Mouse models such as non-obese diabetic (NOD) mice have provided important information on T1DM mechanisms but fail to recapitulate certain aspects of the human disease, notably by being more biased towards females and showing more insulitis than human diabetes mellitus112. Moreover, several T1DM treatments that were promising in NOD mice have failed to translate to humans113. While recapitulating a systemic disease is difficult in an in vitro setting, organoid models could provide a means to recreate in vitro interactions between immune cells and β-cells (Fig. 3).
While models combining β-cells and immune cells from healthy individuals can be informative, patient cells can be used to test how dysfunction in either β-cells or immune cells can lead to T1DM. GWAS have revealed that most genetic loci associated with T1DM occur in genes expressed in cells of the immune system, while only a few affect genes expressed in β-cells114. However, the function of these genes needs to be tested. As T1DM is multigenic, iPSCs derived from patients could enable the testing of genetic or epigenetic predispositions towards developing the disease115. In this spirit, iPSCs were derived from skin fibroblasts from patients with T1DM, from which β-cells were differentiated in vitro and assembled in 3D116–118. A model of exposure to inflammatory cytokines was developed in which iPSC-derived β-cells were exposed to inflammatory cytokines. In this model, β-cells from individuals without T1DM and β-cells derived from patients with T1DM were shown to be similarly susceptible to the treatment by cytokines such as IL-1β, TNF and IFNγ, which reduced the proportion of β-cells as compared with the proportion in untreated controls111. In another study, β-cells derived from patients with fulminant T1DM were shown to be a little more sensitive to treatment with a cocktail of cytokines than β-cells derived from healthy individuals. These β-cells exhibited higher levels of cleaved caspase 3 than cells from healthy individuals, although apoptosis was not increased in the patient-derived cells119. While the scope of these models is still limited, so far they have not provided strong evidence that β-cells derived from patients with T1DM or a genetic susceptibility to T1DM are more sensitive to destruction by the immune system than β-cells from healthy individuals.
Following this work, additional human iPSC-derived β-cells were generated from patients with T1DM to test defects in immune cell function or the interactions between patient immune cells and β-cells120. The human iPSC-derived β-cells were exposed to peripheral blood mononuclear cells (PBMCs) isolated from autologous donors. The human iPSC-derived islets were observed to upregulate HLA class I (HLA-ABC) molecules as well as the immunomodulatory proteins PDL1 and FasR upon addition of IFNγ to the culture medium. HLA-ABC induction occurred to a similar extent in hPSC-derived β-cells from individuals without T1DM, patients with T1DM and healthy primary human islets, suggesting that patient β-cells are not intrinsically more sensitive to cytokines than β-cells from healthy individuals. The PBMCs from patients with T1DM expressed similar levels of mRNA for six autoantigens compared to the levels expressed by PBMCs from healthy donors without T1DM. When exposed to hPSC-derived β-cells pretreated with molecules to induce ER stress, autologous T cells were activated regardless of whether the hPSC-derived β-cells originated from a patient with T1DM or a healthy individual120. This finding is in line with reports that the numbers of islet-reactive T cells are similar between individuals with T1DM and individuals without T1DM121. The use of Transwell cultures in this study could be adapted to future studies and contribute to showing the need for engagement of HLA–T cell receptor interactions.
On a practical level, these co-culture experiments remain challenging due to the challenges in obtaining large numbers of PBMCs and the need to synchronize the production of hPSC-derived β-cells in lengthy protocols to the collection of patient blood samples. This synchronization could be facilitated by methods to maintain both populations in culture or from cryopreservation of hPSC-derived β-cells. Exposure to specific immune cell types would also be needed to gain depth in the immunological responses. In this spirit, differentiation of isogenic dendritic cells, macrophages, endothelial cells and β-cells was achieved in 2021 (ref. 122), and antigen presentation from iPSC-derived macrophages from patients with T1DM to T cell receptors was tested123. Immune responses in hPSC lines harbouring mutations identified in GWAS have not been studied yet. As for β-cell defects, inactivation of GLIS3, a gene that predisposes to T1DM and T2DM, was shown to promote β-cell death124. Similar experiments with patient-relevant variants rather than full inactivation would be valuable in the future.
Modelling other organs contributing to diabetes mellitus
Liver organoids
The liver is an important organ in the development of T2DM, and human liver organoids composed of hepatocytes are potentially valuable. Indeed, liver organoids recapitulate fundamental aspects of the liver such as the architecture, certain functions and genetic signature125, attesting to their relevance as models for T2DM pathogenesis. Liver organoids could be used to study glucose metabolism in the liver, how insulin resistance develops over time as well as non-alcoholic fatty liver disease, a disease that increases the risk of T2DM, and its progression to steatosis. Liver organoids are produced either from PSCs or from adult or embryonic liver tissue. The first experiments were conducted using PSCs to produce hepatocytes, which improved in functionality over time126. These cells exhibit features of fetal hepatocytes127 and protocols are being further developed to improve the maturity of these cells128,129. The PSC-derived hepatic organoids are generally not composed purely of epithelial cells but tend to include mesodermal derivatives such as stellate cells and immune cells, which can be exploited to model inflammation and fibrosis130. Assays enabling the screening of drugs or toxic compounds have been developed, making use of these systems130. Methods to add the vasculature to these liver organoid systems have been developed, although perfusion of the vascular compartment has not yet been achieved130,131. Starting from embryonic tissue has shown that, while some hepatoblasts commit early to the cholangiocyte lineage, bipotent cells that can give rise to both hepatocytes and cholangiocytes remain throughout life132. In adults, such progenitors can be expanded and subsequently differentiated into hepatocytes133. The ability to isolate and maintain hepatocytes in vitro was achieved only recently, in 2018 (refs. 134,135). These epithelial organoids have already been used to model steatosis, accompanied by fibrosis and inflammation in vitro136,137.
Intestinal organoids
Intestinal organoids are one of the earliest and most extensively studied organoid models138. These self-organizing 3D structures with regenerative ability simulate the phenotypic structure, cellular composition and partial function of the small intestine in vitro. They can be generated either from adult intestinal stem cells139 or hPSCs140. In the context of diabetes mellitus research, small intestinal organoids harbouring enteroendocrine cells are useful for investigating nutrient and drug transport, incretin hormone sensing and secretion, and intracellular signalling processes, as demonstrated in mice141. Intestinal enteroendocrine cells have also been used in 3D mouse co-culture systems with pancreatic endocrine cells, such as β-cells, to measure the dynamic effect of enteroendocrine hormones on insulin production in a controlled environment142. In humans, intestinal organoids have been shown to be composed of around 1% enteroendocrine cells140. Despite making up a small percentage of cells in intestinal organoids, these cells could be exploited with the help of reporter lines or genetic manipulations. For example, enteroendocrine cells in organoid culture could be used to further study the mechanism of endogenous GLP1 secretion to improve incretin-based diabetes mellitus therapy, such as for T2DM. Studies based on intestinal organoids and co-culture systems can be used for transcriptomic, peptidomic and functional analyses that can lead to the development of new anti-diabetic drugs targeting the hormone-secreting intestinal enteroendocrine system, thus aiding research into metabolism and diabetes mellitus143.
Muscle organoids
Primary cell-derived 3D skeletal muscle cultures have been in use in the field for more than 25 years to model function and disease, notably in the form of a human bio-artificial muscle model144. In the past 5 years, several groups have developed hPSC-derived 3D human skeletal muscle cultures, some of which have already been employed for various disease modelling approaches145,146. Insulin resistance in skeletal muscle is an important contributor to T2DM development and is considered to be predictive of the disease147. For this reason, investigating the mechanisms underlying skeletal muscle insulin resistance and its prevention and reversion is highly desirable for diabetes mellitus research, particularly for T2DM. There have been a few studies modelling insulin resistance in skeletal muscle differentiated from human iPSCs, from either individuals with mutations in the insulin receptor or patients with T2DM148,149. However, a thorough analysis in a 3D culture system is currently lacking. It would be highly informative to use cells derived from patients with diabetes mellitus to develop similar organoid models. This is feasible using 3D culture of human iPSCs derived from patients with T2DM146, which recapitulate muscle tissue functional units and niches.
Adipose organoids
In T2DM, adipocyte dysfunction can cause insulin resistance. Furthermore, the accumulation of lipid intermediates in other organs such as the liver and the skeletal muscle can lead to insulin resistance150. While adipose tissue cultures have been achieved in 2D, recapitulating 3D architectures could enhance adipose tissue models, as seen in 3D models of other organs. Adipose organoids could be useful in drug discovery studies to elucidate the progression of T2DM, as well as the complications related to the disease. A few research groups have been able to culture adipose spheroids in suspension using the adipose stromal vascular fraction derived from both mice151,152 and humans151,153. Supplementing the adipose organoids with endogenous endothelial cells153 or co-culture with immune cells152,154 broadens the complexity of the analyses that can be performed with these systems, such as the chronic inflammatory phenotype seen in relation to T2DM. Adapting the adipose spheroids to high-throughput formats such as multiwell plates would allow large-scale drug and toxin screening complemented by ongoing adipose-on-a-chip approaches154,155. These systems also have the potential to model healthy adipose tissue in comparison to diabetic adipose tissue, to better predict responses in a clinical setting.
Building multi-organ systems
Organ-on-a-chip approaches have great potential as they allow multiparametric assessments and scalable manufacturing, and hence permit high-throughput culture and drug testing. Especially for complex metabolic diseases that affect multiple tissues, such as diabetes mellitus, being able to mimic the interaction of different organs in an in vitro context is invaluable (Figs. 2 and 3).
In 2017, an early approach to modelling human pancreatic islets and liver together made use of re-aggregated human pancreatic pseudoislets and liver spheroids in a multi-organ-on-a-chip platform156 (Fig. 2). In this study, the perfusable setup enabled the researchers to investigate islet responses to glucose and insulin secretion as well as glucose uptake by the liver spheroids. Such platforms are immensely valuable for drug testing approaches; for example, the effect of diabetic drugs on glucose regulation and pancreas–liver crosstalk, as well as the effect of specific pharmaceuticals on inter-organ communication, could readily be studied. Further studies incorporating more organs to create a ‘physiome-on-a-chip’ platform relevant to T2DM are in the pipeline157–160 (Fig. 2).
Diabetes mellitus complications
The increasing incidence of diabetes mellitus worldwide is accompanied by coincident increases in diabetic complications, including retinopathy, neuropathy and nephropathy (Fig. 4). The complications arise from the systemic nature of diabetes mellitus and are therefore not easy to study in vitro. Moreover, these complications develop over years. Organoids present an opportunity to study the cell types affected the first and the most, as well as the effect of glucose and other metabolites in the context of these diabetic complications. Organoid-based studies would possibly complement mouse models to inspect human-specific aspects of these complications.
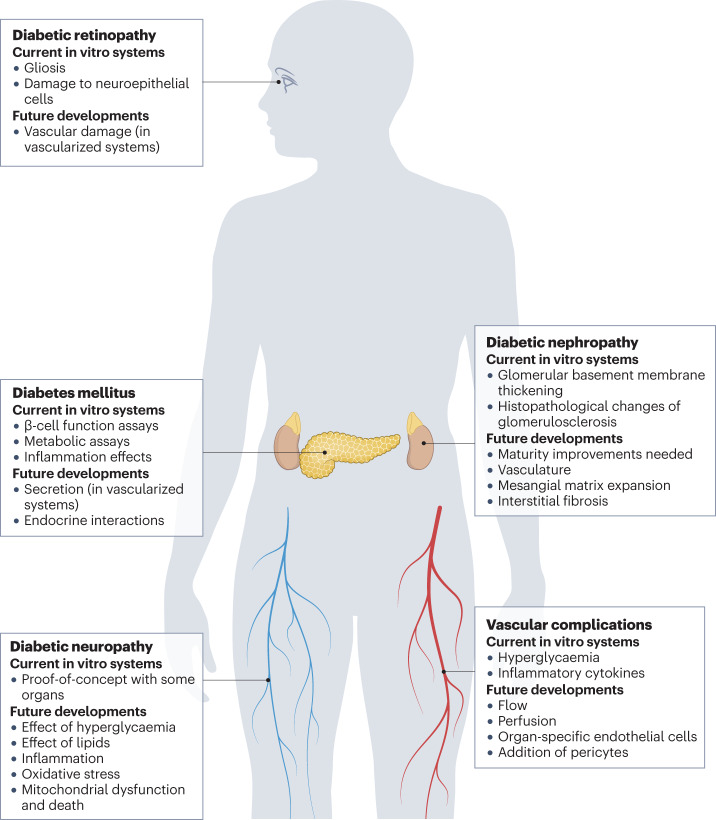
Fig. 4. Diabetes mellitus complication problems that might be solved with organoids.
Examples of complications that can be studied with organoids. Organoid systems for different organs are at different stages of development, the pancreas, kidney and retina being the more advanced and organoids recapitulating nerves the least advanced. The disease-relevant assays that are already available (current in vitro systems) are summarized and interesting assays that might become testable (future developments) pending improvements of models are proposed.
Vascular complications
In the past 10 years, progress has been made in the production of 3D vascular networks differentiated from hPSCs, moving away from similar systems traditionally using HUVECs161. Organoids composed of endothelial cells and pericytes can be generated and exposed to hyperglycaemic conditions or inflammatory cytokines in vitro. These conditions were shown to induce a thickening in the vascular basement membrane, similar to the effect observed in patients with diabetes mellitus161 (Fig. 4). DLL4 and NOTCH3 were further identified as drivers of diabetic vasculopathy using this system. With the realization that the vasculature is different molecularly and morphologically in different organs, protocols are emerging to mimic the vasculature within specific organs162. Moreover, while the systems above are self-organized, bioprinting can be used to create designer blood vessels by subtractive (removing material) or additive (adding layer-by-layer) methods163,164. Bioprinting can also be designed to impose a flow that can be used to present growth factors or therapeutic drugs on the apical side of endothelial cells and to promote their maturation, which is promoted by shear stress sensing. While these systems have mostly been tested with vascular networks derived from HUVECs, similar methods can, in principle, be applied to endothelial cells for specific tissues, including those derived from hPSCs. Such platforms are amenable to the screening of drugs to decrease vascular complications, in the spirit of what was done in 2D for diabetic cardiomyopathy165.
Nephropathies
Diabetic nephropathy affects about one-third of patients with T1DM and patients with T2DM, and there is a strong familial contribution to this risk166. Diabetic nephropathy has been studied in mouse models, which recapitulate the early stages of the disease well, including mild albuminuria, mesangial matrix expansion and glomerular hypertrophy166. However, mouse models fall short in recapitulating the late progression of diabetic nephropathy such as overt albuminuria, histopathological changes of glomerulosclerosis, glomerular basement membrane thickening, interstitial fibrosis and progressive renal insufficiency166.
Kidney organoids can be derived from large aggregates of hPSCs but diabetic nephropathy has not yet been studied using kidney organoids. However, a 2022 study made use of organoids to reconstitute the diabetic milieu in vitro by altering the concentration of glucose in the medium from 5 mM to 25 mM in 24-h oscillatory cycles. This model showed that these oscillating concentrations of glucose induce metabolic programming in the kidney that increases SARS-CoV-2 infection susceptibility as compared with a culture with a constant glucose concentration of 5 mM, mimicking healthy individuals167. Kidney organoids can be derived from patient cells to recapitulate the genetic susceptibility to diabetes mellitus, or from genetically modified hPSCs that can be used to test the function of specific genes168. Most kidney cell types can currently be cultured in organoid format, including podocytes, several types of tubules and interstitial stromal populations, including mesangial cells (Fig. 4). A limitation is that the current kidney organoids represent a fetal rather than a mature organ168. For this reason, kidney organoids have so far been exploited to study diseases with fetal onsets, such as cystic diseases. In these studies, sensitized conditions were used to reveal disease phenotypes in cells harbouring disease-causing mutations as compared with cells without the mutation or variant of interest168. In the same spirit, mouse models of diabetic nephropathy have been improved by sensitive genetic backgrounds, such as endothelial nitric oxide synthase deficiency, and this approach might be adaptable to a human organoid setting. The current kidney organoid models have been shown to be amenable to screening for molecules that improve or worsen phenotypes of different diseases, although assays relevant to diabetic nephropathy remain to be established. These avascular organoids would be sufficient to test the effects of glucose as well as other metabolites and proteins such as insulin on kidney cells, particularly on podocytes, while many other diabetic nephropathy symptoms would require vascularized organoids.
An important early component of diabetic nephropathy is endothelial damage. Therefore, models including endothelial cells and blood-like perfusion would be very useful, particularly to study podocyte foot effacement and glomerular basement membrane thickening. The glomeruli produced in vitro in organoids can be vascularized upon in vivo transplantation or chicken chorioallantoic membrane engraftment169–171. While a few endothelial cells were co-developed in the early kidney organoids produced around 2015 (ref. 172), these cells were not able to establish the filtration barrier, even after endothelial cell production was boosted168,173. However, culturing kidney organoids under flow on microfluidic chips resulted in the expansion of perfusable vascular networks, the formation of podocyte feet and a primitive glomerular basement membrane, which will provide useful early readouts for diabetic nephropathy174. However, the absence of glomerular filtration will be a limitation to studying diabetic nephropathy. Previous experience with modelling other kidney diseases also suggests that proteinuria will not be an easy phenotype to model. Another limitation in the current organoids is the absence of a fully interconnected branched network of glomerular filtration units, although this limitation might be less important for diabetic nephropathy modelling168.
In vivo, endothelial damage is also strongly involved in promoting the expression of inflammatory molecules such as cytokines that affect podocytes and increase immune cell infiltration in diabetic kidneys. Immune cells could be another component worth integrating into future models.
The limited lifespan of kidney organoids will be an important limitation to overcome for diabetic nephropathy studies as these organoids undergo fibrosis and expansion of off‐target populations after 30 days in culture168. This background fibrosis would also make it more difficult to detect the late-stage fibrosis found in diabetic nephropathy. Hypertension and glomerular filtration rate are two important features of late-stage diabetic nephropathy that would probably be difficult to recapitulate in organoids as it would require vascularized filtration units reconstituting the architecture of glomeruli and a perfusion system where pressure can be precisely controlled over weeks. More bioengineered approaches, including bioprinting, have been designed to model glucose reabsorption in patients with diabetes mellitus175.
Neuropathies
Diabetic neuropathies affect multiple nerves, both sensory and autonomous. The most prevalently affected neurons are the longest, innervating the feet and, secondarily, the hands176. Organoids are emerging to serve as models of peripheral sensory neurons177 (Fig. 4) and their connection to peripheral organs such as the skin178. However, given the long periods during which complications of diabetes mellitus develop and the contribution of genetic, metabolic, neurovascular and autoimmune factors in their aetiology176, neuropathies will be very challenging to mimic in vitro.
Retinopathies
Diabetic retinopathies affect 80% of patients with diabetes mellitus for more than 20 years179. The first steps of diabetic retinopathy are traditionally thought to target the vasculature, although there is also evidence that multiple neuronal cell types of the retina are affected early and that gliosis can also be a precocious event179. Ideal models would thus recapitulate the retina and the perfused vasculature. Mimicking the specificities of the eye vasculature would enable addressing why these blood vessels seem particularly affected in diabetes mellitus. Retinal organoids mimicking the neural retina were initially generated from mouse embryonic stem cells180 and subsequently from hESCs181, and protocols have since evolved to generate more functional and diverse neuronal cell types and more faithful cell organization182. In 2019, a microfluidic system combining the retinal epithelium and retinal pigmented epithelium was developed that could be used to easily apply media to simulate the diabetic environment or to test drugs that could protect neuronal or glial cells from the effects of diabetic retinophathy183 (Fig. 4). However, systems combining retinal vasculature and the retina are not yet available.
Conclusions
So far, organoid models have mostly been applied to monogenic forms of diabetes mellitus and, rarely, to T1DM or T2DM. However, the field is moving forward, adapting assays developed for cell lines or in vivo models to organoids. Several organoid models are available for the pancreas, as are related systems using 3D aggregates of endocrine cells, such as pseudoislets. The most advanced organoid systems for other organs are the kidney and the liver. All systems derived from hPSCs easily provide us with millions of cells to work with and enable genetic manipulations to compare patient cells to cells from people without diabetes mellitus. However, progress towards a more mature adult structure and function would be needed to best model the adult stages of diabetes mellitus. Even for organoids derived from adult cells, the retention of function in vitro requires careful benchmarking. This limitation has been a long-standing problem and although 3D culture has enabled in vitro systems to retain more similarity to their in vivo counterparts, culture media are simpler than in vivo physiological fluids.
Technological evolutions are also expected to provide growth devices enabling more flexibility, multiwell formats amenable to screening and microfluidic systems. Microfluidic systems would enable retrieving multiple physiological parameters in parallel and connecting organoid models of different human organs. These systems could enable the recapitulation of inter-organ communications, although challenges remain in finding stable conditions to maintain the functionality of all organoid systems, especially for long periods of time. Bioengineering and bioprinting are also developing, providing devices that improve organ architecture. Given the links between architecture, differentiation and function, hybrid culture methods providing guiding geometrical constraints while enabling self-organization properties of organoids are emerging. Although the field raises a lot of hope, we have to be careful, as even though organoids enable disease mechanisms and potential drugs to be tested directly in human cells, their relevance and whether they do better than animal models remain open questions. Diabetes mellitus research has so far benefited from complementary approaches, and organoids are a new type of weapon in the fight against the disease.
Author contributions
A.G.-B. and B.S.B.-T. researched data for the article, contributed substantially to discussion of content, wrote the article and reviewed/edited the article before submission. S.Y. researched data for the article, contributed substantially to discussion of content and wrote the article.
Peer review
Peer review information
Nature Reviews Endocrinology thanks Mostafa Bakhti, Françoise Carlotti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nauck MA, Wefers J, Meier JJ. Treatment of type 2 diabetes: challenges, hopes, and anticipated successes. Lancet Diabetes Endocrinol. 2021;9:525–544. doi: 10.1016/S2213-8587(21)00113-3. [DOI] [PubMed] [Google Scholar]
- 2.Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The hope and the hype of organoid research. Development. 2017;144:938–941. doi: 10.1242/dev.150201. [DOI] [PubMed] [Google Scholar]
- 3.Marsee A, et al. Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell. 2021;28:816–832. doi: 10.1016/j.stem.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Tsakmaki A, Fonseca Pedro P, Bewick GA. Diabetes through a 3D lens: organoid models. Diabetologia. 2020;63:1093–1102. doi: 10.1007/s00125-020-05126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopcroft DW, Mason DR, Scott RS. Insulin secretion from perifused rat pancreatic pseudoislets. Vitr. Cell. Dev. Biol. 1985;21:421–427. doi: 10.1007/BF02620828. [DOI] [PubMed] [Google Scholar]
- 6.Halban PA, Powers SL, George KL, Bonner-Weir S. Spontaneous reassociation of dispersed adult rat pancreatic islet cells into aggregates with three-dimensional architecture typical of native islets. Diabetes. 1987;36:783–790. doi: 10.2337/diab.36.7.783. [DOI] [PubMed] [Google Scholar]
- 7.Hilderink J, et al. Controlled aggregation of primary human pancreatic islet cells leads to glucose-responsive pseudoislets comparable to native islets. J. Cell. Mol. Med. 2015;19:1836–1846. doi: 10.1111/jcmm.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velazco-Cruz L, Goedegebuure MM, Millman JR. Advances toward engineering functionally mature human pluripotent stem cell-derived β cells. Front. Bioeng. Biotechnol. 2020;8:786. doi: 10.3389/fbioe.2020.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusko TM, Russ HA, Stabler CL. Strategies for durable β cell replacement in type 1 diabetes. Science. 2021;373:516–522. doi: 10.1126/science.abh1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frum T, Spence JR. hPSC-derived organoids: models of human development and disease. J. Mol. Med. 2021;99:463–473. doi: 10.1007/s00109-020-01969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrand J, et al. Neonatal diabetes mellitus. Front. Pediatr. 2020;8:602. doi: 10.3389/fped.2020.540718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hara SE, Gembus KM, Nicholas LM. Understanding the long-lasting effects of fetal nutrient restriction versus exposure to an obesogenic diet on islet-cell mass and function. Metabolites. 2021;11:514. doi: 10.3390/metabo11080514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen HL, Grapin-Botton A. The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 2017;66:51–68. doi: 10.1016/j.semcdb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Petersen MBK, Gonçalves CAC, Kim YH, Grapin-Botton A. Recapitulating and deciphering human pancreas development from human pluripotent stem cells in a dish. Curr. Top. Dev. Biol. 2018;129:143–190. doi: 10.1016/bs.ctdb.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Sugiyama T, et al. Reconstituting pancreas development from purified progenitor cells reveals genes essential for islet differentiation. Proc. Natl Acad. Sci. USA. 2013;110:12691–12696. doi: 10.1073/pnas.1304507110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greggio C, et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonfanti P, et al. Ex vivo expansion and differentiation of human and mouse fetal pancreatic progenitors are modulated by epidermal growth factor. Stem Cell Dev. 2015;24:1766–1778. doi: 10.1089/scd.2014.0550. [DOI] [PubMed] [Google Scholar]
- 18.Bakhti M, et al. Establishment of a high-resolution 3D modeling system for studying pancreatic epithelial cell biology in vitro. Mol. Metab. 2019;30:16–29. doi: 10.1016/j.molmet.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scavuzzo MA, Yang D, Borowiak M. Organotypic pancreatoids with native mesenchyme develop insulin producing endocrine cells. Sci. Rep. 2017;7:10810. doi: 10.1038/s41598-017-11169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonçalves CA, et al. A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion. Nat. Commun. 2021;12:3144. doi: 10.1038/s41467-021-23295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loomans CJM, et al. Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Rep. 2018;10:712–724. doi: 10.1016/j.stemcr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahl-Jensen SB, et al. Deconstructing the principles of ductal network formation in the pancreas. PLoS Biol. 2018;16:e2002842. doi: 10.1371/journal.pbio.2002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scavuzzo MA, Teaw J, Yang D, Borowiak M. Generation of scaffold-free, three-dimensional insulin expressing pancreatoids from mouse pancreatic progenitors in vitro. J. Vis. Exp. 2018;2018:e57599. doi: 10.3791/57599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair GG, Tzanakakis ES, Hebrok M. Emerging routes to the generation of functional β-cells for diabetes mellitus cell therapy. Nat. Rev. Endocrinol. 2020;16:506–518. doi: 10.1038/s41574-020-0375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rezania A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 26.Nostro MC, et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagliuca FW, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russ HA, et al. Controlled induction of human pancreatic progenitors produces functional beta‐like cells in vitro. EMBO J. 2015;34:1759–1772. doi: 10.15252/embj.201591058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair GG, et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat. Cell Biol. 2019;21:263–274. doi: 10.1038/s41556-018-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velazco-Cruz L, et al. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Rep. 2019;12:351–365. doi: 10.1016/j.stemcr.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyoda T, et al. Cell aggregation optimizes the differentiation of human ESCs and iPSCs into pancreatic bud-like progenitor cells. Stem Cell Res. 2015;14:185–197. doi: 10.1016/j.scr.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J, et al. Generation of insulin‐producing islet‐like clusters from human embryonic. Stem Cell. Stem Cells. 2007;25:1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- 33.Zhu S, et al. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 2016;7:10080. doi: 10.1038/ncomms10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balboa D, et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 2022;40:1042–1055. doi: 10.1038/s41587-022-01219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hohwieler M, et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66:473–486. doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wesolowska-Andersen A, et al. Analysis of differentiation protocols defines a common pancreatic progenitor molecular signature and guides refinement of endocrine differentiation. Stem Cell Rep. 2020;14:138–153. doi: 10.1016/j.stemcr.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Ye K. Three-dimensional differentiation of embryonic stem cells into islet-like insulin-producing clusters. Tissue Eng. Part A. 2009;15:1941–1952. doi: 10.1089/ten.tea.2008.0181. [DOI] [PubMed] [Google Scholar]
- 38.Saito H, Takeuchi M, Chida K, Miyajima A. Generation of glucose-responsive functional islets with a three-dimensional structure from mouse fetal pancreatic cells and iPS cells in vitro. PLoS ONE. 2011;6:e28209. doi: 10.1371/journal.pone.0028209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braverman-Gross C, Benvenisty N. Modeling maturity onset diabetes of the young in pluripotent stem cells: challenges and achievements. Front. Endocrinol. 2021;12:20. doi: 10.3389/fendo.2021.622940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riddle MC, et al. Monogenic diabetes: from genetic insights to population-based precision in care. Reflections from a diabetes care editors’ expert forum. Diabetes Care. 2020;43:3117–3128. doi: 10.2337/dci20-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balboa D, Iworima DG, Kieffer TJ. Human pluripotent stem cells to model islet defects in diabetes. Front. Endocrinol. 2021;12:149. doi: 10.3389/fendo.2021.642152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelalim EM. Modeling different types of diabetes using human pluripotent stem cells. Cell. Mol. Life Sci. 2021;78:2459–2483. doi: 10.1007/s00018-020-03710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma S, et al. β Cell replacement after gene editing of a neonatal diabetes-causing mutation at the insulin locus. Stem Cell Rep. 2018;11:1407–1415. doi: 10.1016/j.stemcr.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balboa D, et al. Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. Elife. 2018;7:e38519. doi: 10.7554/eLife.38519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panova AV, et al. Aberrant splicing of INS impairs beta-cell differentiation and proliferation by ER stress in the isogenic iPSC model of neonatal diabetes. Int. J. Mol. Sci. 2022;23:8824. doi: 10.3390/ijms23158824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Franco E, et al. YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J. Clin. Invest. 2020;130:6338–6353. doi: 10.1172/JCI141455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saarimäki-Vire J, et al. An activating STAT3 mutation causes neonatal diabetes through premature induction of pancreatic differentiation. Cell Rep. 2017;19:281–294. doi: 10.1016/j.celrep.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Z, et al. A missense KCNQ1 mutation impairs insulin secretion in neonatal diabetes. bioRxiv. 2021 doi: 10.1101/2021.08.24.457485v1. [DOI] [Google Scholar]
- 49.Wang X, et al. Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Mol. Metab. 2019;24:80–97. doi: 10.1016/j.molmet.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Z, et al. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell. 2016;18:755–768. doi: 10.1016/j.stem.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, et al. A comprehensive structure-function study of neurogenin3 disease-causing alleles during human pancreas and intestinal organoid development. Dev. Cell. 2019;50:367–380.e7. doi: 10.1016/j.devcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philippi A, et al. Mutations and variants of ONECUT1 in diabetes. Nat. Med. 2021;27:1928–1940. doi: 10.1038/s41591-021-01502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teo AKK, et al. Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Rep. 2016;6:357–367. doi: 10.1016/j.stemcr.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vethe H, et al. Probing the missing mature β-cell proteomic landscape in differentiating patient iPSC-derived cells. Sci. Rep. 2017;7:4780. doi: 10.1038/s41598-017-04979-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braverman-Gross C, et al. Derivation and molecular characterization of pancreatic differentiated MODY1-iPSCs. Stem Cell Res. 2018;31:16–26. doi: 10.1016/j.scr.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 56.Yabe SG, et al. Expression of mutant mRNA and protein in pancreatic cells derived from MODY3- iPS cells. PLoS ONE. 2019;14:e0217110. doi: 10.1371/journal.pone.0217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cardenas-Diaz FL, et al. Modeling monogenic diabetes using human ESCs reveals developmental and metabolic deficiencies caused by mutations in HNF1A. Cell Stem Cell. 2019;25:273–289.e5. doi: 10.1016/j.stem.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González BJ, et al. Reduced calcium levels and accumulation of abnormal insulin granules in stem cell models of HNF1A deficiency. Commun. Biol. 2022;5:779. doi: 10.1038/s42003-022-03696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cujba A-M, et al. An HNF1α truncation associated with maturity-onset diabetes of the young impairs pancreatic progenitor differentiation by antagonizing HNF1β function. Cell Rep. 2022;38:110425. doi: 10.1016/j.celrep.2022.110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeForest N, Majithia AR. Genetics of type 2 diabetes: implications from large-scale studies. Curr. Diab. Rep. 2022;22:227–235. doi: 10.1007/s11892-022-01462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veres A, et al. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Augsornworawat P, Maxwell KG, Velazco-Cruz L, Millman JR. Single-cell transcriptome profiling reveals β cell maturation in stem cell-derived islets after transplantation. Cell Rep. 2020;32:108067. doi: 10.1016/j.celrep.2020.108067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis JC, et al. Glucose response by stem cell-derived β cells in vitro is inhibited by a bottleneck in glycolysis. Cell Rep. 2020;31:107623. doi: 10.1016/j.celrep.2020.107623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basford CL, et al. The functional and molecular characterisation of human embryonic stem cell-derived insulin-positive cells compared with adult pancreatic beta cells. Diabetologia. 2012;55:358–371. doi: 10.1007/s00125-011-2335-x. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez-Dominguez JR, Melton DA. Cell maturation: hallmarks, triggers, and manipulation. Cell. 2022;185:235–249. doi: 10.1016/j.cell.2021.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helman A, et al. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metab. 2020;31:1004–1016.e5. doi: 10.1016/j.cmet.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang D, et al. Long-term expansion of pancreatic islet organoids from resident Procr+ progenitors. Cell. 2020;180:1198–1211.e19. doi: 10.1016/j.cell.2020.02.048. [DOI] [PubMed] [Google Scholar]
- 68.Domínguez-Bendala J, Qadir MMF, Pastori RL. Pancreatic progenitors: there and back again. Trends Endocrinol. Metab. 2019;30:4–11. doi: 10.1016/j.tem.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rezanejad H, et al. Heterogeneity of SOX9 and HNF1β in pancreatic ducts is dynamic. Stem Cell Rep. 2018;10:725–738. doi: 10.1016/j.stemcr.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beşikcioğlu, H. E. et al. Organoids: questions and answers. STAR Protocolshttps://star-protocols.cell.com/categories/organoids/qa# (2022).
- 72.Montesano R, Mouron P, Amherdt M, Orci L. Collagen matrix promotes reorganization of pancreatic endocrine cell monolayers into islet-like organoids. J. Cell Biol. 1983;97:935–939. doi: 10.1083/jcb.97.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberg-Welsh C. Long-term culture in matrigel enhances the insulin secretion of fetal porcine islet-like cell clusters in vitro. Pancreas. 2001;22:157–163. doi: 10.1097/00006676-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 74.Wang W, Jin S, Ye K. Development of islet organoids from H9 human embryonic stem cells in biomimetic 3D scaffolds. Stem Cell Dev. 2017;26:394–404. doi: 10.1089/scd.2016.0115. [DOI] [PubMed] [Google Scholar]
- 75.Kozlowski MT, Crook CJ, Ku HT. Towards organoid culture without Matrigel. Commun. Biol. 2021;4:1387. doi: 10.1038/s42003-021-02910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Candiello J, et al. 3D heterogeneous islet organoid generation from human embryonic stem cells using a novel engineered hydrogel platform. Biomaterials. 2018;177:27–39. doi: 10.1016/j.biomaterials.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 77.Jiang K, et al. 3-D physiomimetic extracellular matrix hydrogels provide a supportive microenvironment for rodent and human islet culture. Biomaterials. 2019;198:37–48. doi: 10.1016/j.biomaterials.2018.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohammed JS, Wang Y, Harvat TA, Oberholzer J, Eddington DT. Microfluidic device for multimodal characterization of pancreatic islets. Lab Chip. 2009;9:97–106. doi: 10.1039/B809590F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abadpour S, et al. Pancreas-on-a-chip technology for transplantation applications. Curr. Diab. Rep. 2020;20:72. doi: 10.1007/s11892-020-01357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glieberman AL, et al. Synchronized stimulation and continuous insulin sensing in a microfluidic human islet on a chip designed for scalable manufacturing. Lab Chip. 2019;19:2993–3010. doi: 10.1039/C9LC00253G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee SH, et al. Microphysiological analysis platform of pancreatic islet β-cell spheroids. Adv. Healthc. Mater. 2018;7:1701111. doi: 10.1002/adhm.201701111. [DOI] [PubMed] [Google Scholar]
- 82.Jun Y, et al. In vivo-mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci. Adv. 2019;5:eaax4520. doi: 10.1126/sciadv.aax4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Essaouiba A, et al. Microwell-based pancreas-on-chip model enhances genes expression and functionality of rat islets of Langerhans. Mol. Cell. Endocrinol. 2020;514:110892. doi: 10.1016/j.mce.2020.110892. [DOI] [PubMed] [Google Scholar]
- 84.Tao T, et al. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip. 2019;19:948–958. doi: 10.1039/C8LC01298A. [DOI] [PubMed] [Google Scholar]
- 85.Liu H, et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 2020;7:1903739. doi: 10.1002/advs.201903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zbinden A, et al. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 2020;85–86:205–220. doi: 10.1016/j.matbio.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 87.Patel SN, et al. Organoid microphysiological system preserves pancreatic islet function within 3D matrix. Sci. Adv. 2021;7:eaba5515. doi: 10.1126/sciadv.aba5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walker JT, et al. Integrated human pseudoislet system and microfluidic platform demonstrate differences in GPCR signaling in islet cells. JCI Insight. 2020;5:e137017. doi: 10.1172/jci.insight.137017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar SA, Delgado M, Mendez VE, Joddar B. Applications of stem cells and bioprinting for potential treatment of diabetes. World J. Stem Cell. 2019;11:13–32. doi: 10.4252/wjsc.v11.i1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amin J, et al. A simple, reliable method for high-throughput screening for diabetes drugs using 3D β-cell spheroids. J. Pharmacol. Toxicol. Methods. 2016;82:83–89. doi: 10.1016/j.vascn.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 91.Yang K, et al. A 3D culture platform enables development of zinc-binding prodrugs for targeted proliferation of β cells. Sci. Adv. 2020;6:3207–3225. doi: 10.1126/sciadv.abc3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Penko D, et al. Incorporation of endothelial progenitor cells into mosaic pseudoislets. Islets. 2011;3:73–79. doi: 10.4161/isl.3.3.15392. [DOI] [PubMed] [Google Scholar]
- 93.Spelios MG, Afinowicz LA, Tipon RC, Akirav EM. Human EndoC-βH1 β-cells form pseudoislets with improved glucose sensitivity and enhanced GLP-1 signaling in the presence of islet-derived endothelial cells. Am. J. Physiol. Metab. 2018;314:E512–E521. doi: 10.1152/ajpendo.00272.2017. [DOI] [PubMed] [Google Scholar]
- 94.Urbanczyk M, Zbinden A, Layland SL, Duffy G, Schenke-Layland K. Controlled heterotypic pseudo-islet assembly of human β-cells and human umbilical vein endothelial cells using magnetic levitation. Tissue Eng. Part. A. 2020;26:387–399. doi: 10.1089/ten.tea.2019.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Augsornworawat P, Velazco-Cruz L, Song J, Millman JR. A hydrogel platform for in vitro three dimensional assembly of human stem cell-derived islet cells and endothelial cells. Acta Biomater. 2019;97:272–280. doi: 10.1016/j.actbio.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seo H, Son J, Park J-K. Controlled 3D co-culture of beta cells and endothelial cells in a micropatterned collagen sheet for reproducible construction of an improved pancreatic pseudo-tissue. Apl. Bioeng. 2020;4:046103. doi: 10.1063/5.0023873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nalbach L, et al. Improvement of islet transplantation by the fusion of islet cells with functional blood vessels. EMBO Mol. Med. 2021;13:e12616. doi: 10.15252/emmm.202012616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aghazadeh Y, et al. Microvessels support engraftment and functionality of human islets and hESC-derived pancreatic progenitors in diabetes models. Cell Stem Cell. 2021;28:1936–1949.e8. doi: 10.1016/j.stem.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 99.Rambøl MH, Han E, Niklason LE. Microvessel network formation and interactions with pancreatic islets in three-dimensional chip cultures. Tissue Eng. Part. A. 2020;26:556–568. doi: 10.1089/ten.tea.2019.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palikuqi B, et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature. 2020;585:426–432. doi: 10.1038/s41586-020-2712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soltanian A, et al. Generation of functional human pancreatic organoids by transplants of embryonic stem cell derivatives in a 3D‐printed tissue trapper. J. Cell. Physiol. 2019;234:9564–9576. doi: 10.1002/jcp.27644. [DOI] [PubMed] [Google Scholar]
- 102.Takebe T, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 103.Takahashi Y, Sekine K, Kin T, Takebe T, Taniguchi H. Self-condensation culture enables vascularization of tissue fragments for efficient therapeutic transplantation. Cell Rep. 2018;23:1620–1629. doi: 10.1016/j.celrep.2018.03.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takahashi Y, Takebe T, Taniguchi H. Methods for generating vascularized islet‐like organoids via self‐condensation. Curr. Protoc. Stem Cell Biol. 2018;45:e49. doi: 10.1002/cpsc.49. [DOI] [PubMed] [Google Scholar]
- 105.Jiao A, et al. Simulated cholinergic reinnervation of β (INS-1) cells: antidiabetic utility of heterotypic pseudoislets containing β cell and cholinergic cell. Int. J. Endocrinol. 2018 doi: 10.1155/2018/1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Besikcioglu HE, et al. Innervated mouse pancreas organoids as an ex vivo model to study pancreatic neuropathy in pancreatic cancer. STAR Protoc. 2021;2:100935. doi: 10.1016/j.xpro.2021.100935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chiou J, et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature. 2021;594:398–402. doi: 10.1038/s41586-021-03552-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kahraman S, et al. Abnormal exocrine–endocrine cell cross-talk promotes β-cell dysfunction and loss in MODY8. Nat. Metab. 2022;4:76–89. doi: 10.1038/s42255-021-00516-2. [DOI] [PubMed] [Google Scholar]
- 109.Overton DL, Mastracci TL. Exocrine-endocrine crosstalk: the influence of pancreatic cellular communications on organ growth, function and disease. Front. Endocrinol. 2022;13:1010. doi: 10.3389/fendo.2022.904004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang L, et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell. 2021;28:1090–1104.e6. doi: 10.1016/j.stem.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khosravi-Maharlooei M, et al. Modeling human T1D-associated autoimmune processes. Mol. Metab. 2021;56:101417. doi: 10.1016/j.molmet.2021.101417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skyler JS. Hope vs hype: where are we in type 1 diabetes? Diabetologia. 2018;61:509–516. doi: 10.1007/s00125-017-4530-x. [DOI] [PubMed] [Google Scholar]
- 114.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N. Engl. J. Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 115.Joshi K, et al. Modeling type 1 diabetes using pluripotent stem cell technology. Front. Endocrinol. 2021;12:260. doi: 10.3389/fendo.2021.635662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maehr R, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl Acad. Sci. USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thatava T, et al. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol. Ther. 2013;21:228–239. doi: 10.1038/mt.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Millman JR, et al. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 2016;7:11463. doi: 10.1038/ncomms11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hosokawa Y, et al. Insulin-producing cells derived from ‘induced pluripotent stem cells’ of patients with fulminant type 1 diabetes: vulnerability to cytokine insults and increased expression of apoptosis-related genes. J. Diabetes Investig. 2018;9:481–493. doi: 10.1111/jdi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leite NC, et al. Modeling type 1 diabetes in vitro using human pluripotent stem cells. Cell Rep. 2020;32:107894. doi: 10.1016/j.celrep.2020.107894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Culina S, et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci. Immunol. 2018;3:4013. doi: 10.1126/sciimmunol.aao4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Armitage LH, et al. Use of induced pluripotent stem cells to build isogenic systems and investigate type 1 diabetes. Front. Endocrinol. 2021;12:1376. doi: 10.3389/fendo.2021.737276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Joshi K, et al. Induced pluripotent stem cell macrophages present antigen to proinsulin-specific T cell receptors from donor-matched islet-infiltrating T cells in type 1 diabetes. Diabetologia. 2019;62:2245–2251. doi: 10.1007/s00125-019-04988-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Amin S, et al. Discovery of a drug candidate for GLIS3-associated diabetes. Nat. Commun. 2018;9:2681. doi: 10.1038/s41467-018-04918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Prior N, Inacio P, Huch M. Liver organoids: from basic research to therapeutic applications. Gut. 2019;68:2228–2237. doi: 10.1136/gutjnl-2019-319256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thompson WL, Takebe T. Human liver model systems in a dish. Dev. Growth Differ. 2021;63:47–58. doi: 10.1111/dgd.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Camp JG, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 128.Ardalani H, et al. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes. Acta Biomater. 2019;95:371–381. doi: 10.1016/j.actbio.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 129.Zabulica M, et al. Guide to the assessment of mature liver gene expression in stem cell-derived hepatocytes. Stem Cell Dev. 2019;28:907–919. doi: 10.1089/scd.2019.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thompson WL, Takebe T. Generation of multi-cellular human liver organoids from pluripotent stem cells. in. Methods Cell Biol. 2020;159:47–68. doi: 10.1016/bs.mcb.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 132.Prior N, et al. Lgr5+ stem/progenitor cells reside at the apex of a heterogeneous embryonic hepatoblast pool. Development. 2019;146:dev174557. doi: 10.1242/dev.174557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peng WC, et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell. 2018;175:1607–1619.e15. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu H, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606.e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 136.Kruitwagen HS, et al. Long-term adult feline liver organoid cultures for disease modeling of hepatic steatosis. Stem Cell Rep. 2017;8:822–830. doi: 10.1016/j.stemcr.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ouchi R, et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384.e6. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fair KL, Colquhoun J, Hannan NRF. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. B Biol. Sci. 2018;373:20170217. doi: 10.1098/rstb.2017.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 140.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zietek T, Rath E, Haller D, Daniel H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci. Rep. 2015;5:16831. doi: 10.1038/srep16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nguyen DTT, van Noort D, Jeong I-K, Park S. Endocrine system on chip for a diabetes treatment model. Biofabrication. 2017;9:015021. doi: 10.1088/1758-5090/aa5cc9. [DOI] [PubMed] [Google Scholar]
- 143.Goldspink DA, et al. Labeling and characterization of human GLP-1-secreting L-cells in primary ileal organoid culture. Cell Rep. 2020;31:107833. doi: 10.1016/j.celrep.2020.107833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vandenburgh H, et al. Tissue-engineered skeletal muscle organoids for reversible gene therapy. Hum. Gene Ther. 1996;7:2195–2200. doi: 10.1089/hum.1996.7.17-2195. [DOI] [PubMed] [Google Scholar]
- 145.Jalal S, Dastidar S, Tedesco FS. Advanced models of human skeletal muscle differentiation, development and disease: three-dimensional cultures, organoids and beyond. Curr. Opin. Cell Biol. 2021;73:92–104. doi: 10.1016/j.ceb.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maffioletti SM, et al. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 2018;23:899–908. doi: 10.1016/j.celrep.2018.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rothman DL, et al. Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc. Natl Acad. Sci. USA. 1995;92:983–987. doi: 10.1073/pnas.92.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Iovino S, Burkart AM, Warren L, Patti ME, Kahn CR. Myotubes derived from human-induced pluripotent stem cells mirror in vivo insulin resistance. Proc. Natl Acad. Sci. USA. 2016;113:1889–1894. doi: 10.1073/pnas.1525665113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Batista TM, et al. A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab. 2020;32:844–859.e5. doi: 10.1016/j.cmet.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019;20:242–258. doi: 10.1038/s41580-018-0093-z. [DOI] [PubMed] [Google Scholar]
- 151.Klingelhutz AJ, et al. Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci. Rep. 2018;8:523. doi: 10.1038/s41598-017-19024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Taylor J, et al. Generation of immune cell containing adipose organoids for in vitro analysis of immune metabolism. Sci. Rep. 2020;10:21104. doi: 10.1038/s41598-020-78015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Muller S, et al. Human adipose stromal-vascular fraction self-organizes to form vascularized adipose tissue in 3D cultures. Sci. Rep. 2019;9:7250. doi: 10.1038/s41598-019-43624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu Y, et al. Adipose-on-a-chip: a dynamic microphysiological in vitro model of the human adipose for immune-metabolic analysis in type II diabetes. Lab Chip. 2019;19:241–253. doi: 10.1039/C8LC00481A. [DOI] [PubMed] [Google Scholar]
- 155.McCarthy M, et al. Fat-on-a-chip models for research and discovery in obesity and its metabolic comorbidities. Tissue Eng. Part. B Rev. 2020;26:586–595. doi: 10.1089/ten.teb.2019.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bauer S, et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 2017;7:14620. doi: 10.1038/s41598-017-14815-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 2016;113:2213–2227. doi: 10.1002/bit.25989. [DOI] [PubMed] [Google Scholar]
- 158.Edington CD, et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 2018;8:4530. doi: 10.1038/s41598-018-22749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rogal J, Zbinden A, Schenke-Layland K, Loskill P. Stem-cell based organ-on-a-chip models for diabetes research. Adv. Drug Deliv. Rev. 2019;140:101–128. doi: 10.1016/j.addr.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 160.Tao T, et al. Microengineered multi‐organoid system from hiPSCs to recapitulate human liver‐islet axis in normal and type 2 diabetes. Adv. Sci. 2022;9:2103495. doi: 10.1002/advs.202103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wimmer RA, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nguyen J, Lin Y-Y, Gerecht S. The next generation of endothelial differentiation: tissue-specific ECs. Cell Stem Cell. 2021;28:1188–1204. doi: 10.1016/j.stem.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 163.Gao G, et al. Construction of a novel in vitro atherosclerotic model from geometry‐tunable artery equivalents engineered via in‐bath coaxial cell printing. Adv. Funct. Mater. 2021;31:2008878. doi: 10.1002/adfm.202008878. [DOI] [Google Scholar]
- 164.Zhang S, Wan Z, Kamm RD. Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip. 2021;21:473–488. doi: 10.1039/D0LC01186J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Drawnel FM, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014;9:810–820. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 166.Azushima K, Gurley SB, Coffman TM. Modelling diabetic nephropathy in mice. Nat. Rev. Nephrol. 2018;14:48–56. doi: 10.1038/nrneph.2017.142. [DOI] [PubMed] [Google Scholar]
- 167.Garreta E, et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab. 2022;34:857–873.e9. doi: 10.1016/j.cmet.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nishinakamura R. Human kidney organoids: progress and remaining challenges. Nat. Rev. Nephrol. 2019;15:613–624. doi: 10.1038/s41581-019-0176-x. [DOI] [PubMed] [Google Scholar]
- 169.Buzhor E, et al. Kidney spheroids recapitulate tubular organoids leading to enhanced tubulogenic potency of human kidney-derived cells. Tissue Eng. Part A. 2011;17:2305–2319. doi: 10.1089/ten.tea.2010.0595. [DOI] [PubMed] [Google Scholar]
- 170.Garreta E, et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 2019;18:397–405. doi: 10.1038/s41563-019-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kaisto S, et al. Optimization of renal organoid and organotypic culture for vascularization, extended development, and improved microscopy imaging. J. Vis. Exp. 2020;2020:e60995. doi: 10.3791/60995. [DOI] [PubMed] [Google Scholar]
- 172.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 173.Ryan AR, et al. Vascular deficiencies in renal organoids and ex vivo kidney organogenesis. Dev. Biol. 2021;477:98–116. doi: 10.1016/j.ydbio.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Homan KA, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin NYC, et al. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl Acad. Sci. USA. 2019;116:5399–5404. doi: 10.1073/pnas.1815208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sloan G, Selvarajah D, Tesfaye S. Pathogenesis, diagnosis and clinical management of diabetic sensorimotor peripheral neuropathy. Nat. Rev. Endocrinol. 2021;17:400–420. doi: 10.1038/s41574-021-00496-z. [DOI] [PubMed] [Google Scholar]
- 177.Pereira JD, et al. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat. Commun. 2021;12:4744. doi: 10.1038/s41467-021-24776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee J, et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Antonetti DA, Silva PS, Stitt AW. Current understanding of the molecular and cellular pathology of diabetic retinopathy. Nat. Rev. Endocrinol. 2021;17:195–206. doi: 10.1038/s41574-020-00451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 181.Zhong X, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.O’Hara-Wright M, Gonzalez-Cordero A. Retinal organoids: a window into human retinal development. Development. 2020;147:dev189746. doi: 10.1242/dev.189746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Achberger K, et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife. 2019;8:e46188. doi: 10.7554/eLife.46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Blondeau B, Avril I, Duchene B, Bréant B. Endocrine pancreas development is altered in foetuses from rats previously showing intra-uterine growth retardation in response to malnutrition. Diabetologia. 2002;45:394–401. doi: 10.1007/s00125-001-0767-4. [DOI] [PubMed] [Google Scholar]
- 185.Reusens B, Theys N, Dumortier O, Goosse K, Remacle C. Maternal malnutrition programs the endocrine pancreas in progeny. Am. J. Clin. Nutr. 2011;94:1824S–1829S. doi: 10.3945/ajcn.110.000729. [DOI] [PubMed] [Google Scholar]
- 186.Chamson-Reig A, Thyssen SM, Arany E, Hill DJ. Altered pancreatic morphology in the offspring of pregnant rats given reduced dietary protein is time and gender specific. J. Endocrinol. 2006;191:83–92. doi: 10.1677/joe.1.06754. [DOI] [PubMed] [Google Scholar]
- 187.Calzada L, et al. Maternal protein restriction during gestation impairs female offspring pancreas development in the rat. Nutr. Res. 2016;36:855–862. doi: 10.1016/j.nutres.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 188.King R, et al. Offspring of mice exposed to a low-protein diet in utero demonstrate changes in mTOR signaling in pancreatic islets of Langerhans, associated with altered glucagon and insulin expression and a lower β-cell mass. Nutrients. 2019;11:605. doi: 10.3390/nu11030605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alejandro EU, et al. Maternal low-protein diet on the last week of pregnancy contributes to insulin resistance and β-cell dysfunction in the mouse offspring. Am. J. Physiol. Integr. Comp. Physiol. 2020;319:R485–R496. doi: 10.1152/ajpregu.00284.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Han J, Xu J, Epstein PN, Liu YQ. Long-term effect of maternal obesity on pancreatic beta cells of offspring: reduced beta cell adaptation to high glucose and high-fat diet challenges in adult female mouse offspring. Diabetologia. 2005;48:1810–1818. doi: 10.1007/s00125-005-1854-8. [DOI] [PubMed] [Google Scholar]
- 191.Bringhenti I, et al. Maternal obesity during the preconception and early life periods alters pancreatic development in early and adult life in male mouse offspring. PLoS ONE. 2013;8:e55711. doi: 10.1371/journal.pone.0055711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ohta T, et al. Maternal high-fat diet promotes onset of diabetes in rat offspring. Anim. Sci. J. 2017;88:149–155. doi: 10.1111/asj.12606. [DOI] [PubMed] [Google Scholar]
- 193.Nicholas LM, et al. Exposure to maternal obesity programs sex differences in pancreatic islets of the offspring in mice. Diabetologia. 2020;63:324–337. doi: 10.1007/s00125-019-05037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bodin J, et al. Transmaternal bisphenol A exposure accelerates diabetes type 1 development in NOD mice. Toxicol. Sci. 2014;137:311–323. doi: 10.1093/toxsci/kft242. [DOI] [PubMed] [Google Scholar]
- 195.Alonso-Magdalena P, García-Arévalo M, Quesada I, Nadal Á. Bisphenol-A treatment during pregnancy in mice: a new window of susceptibility for the development of diabetes in mothers later in life. Endocrinology. 2015;156:1659–1670. doi: 10.1210/en.2014-1952. [DOI] [PubMed] [Google Scholar]
- 196.Whitehead R, Guan H, Arany E, Cernea M, Yang K. Prenatal exposure to bisphenol A alters mouse fetal pancreatic morphology and islet composition. Horm. Mol. Biol. Clin. Investig. 2016;25:171–179. doi: 10.1515/hmbci-2015-0052. [DOI] [PubMed] [Google Scholar]
- 197.García-Arévalo M, et al. Maternal exposure to bisphenol-A during pregnancy increases pancreatic β-cell growth during early life in male mice offspring. Endocrinology. 2016;157:4158–4171. doi: 10.1210/en.2016-1390. [DOI] [PubMed] [Google Scholar]
- 198.La Merrill MA, et al. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 2020;16:45–57. doi: 10.1038/s41574-019-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nadal A, et al. Bisphenol-A alters pancreatic B-cell proliferation and mass in an estrogen receptor beta-dependent manner [abstract] J. Endocr. Soc. 2020;4(Suppl. 1):SAT-715. doi: 10.1210/jendso/bvaa046.791. [DOI] [Google Scholar]
- 200.Malmqvist E, et al. Maternal exposure to air pollution and type 1 diabetes – accounting for genetic factors. Environ. Res. 2015;140:268–274. doi: 10.1016/j.envres.2015.03.024. [DOI] [PubMed] [Google Scholar]
- 201.Gillies R, et al. Maternal exposure to Δ9-tetrahydrocannabinol impairs female offspring glucose homeostasis and endocrine pancreatic development in the rat. Reprod. Toxicol. 2020;94:84–91. doi: 10.1016/j.reprotox.2020.04.070. [DOI] [PubMed] [Google Scholar]
- 202.Moullé VS, Parnet P. Effects of nutrient intake during pregnancy and lactation on the endocrine pancreas of the offspring. Nutrients. 2019;11:2708. doi: 10.3390/nu11112708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lahti-Pulkkinen M, et al. Consequences of being overweight or obese during pregnancy on diabetes in the offspring: a record linkage study in Aberdeen, Scotland. Diabetologia. 2019;62:1412–1419. doi: 10.1007/s00125-019-4891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Perng W, Oken E, Dabelea D. Developmental overnutrition and obesity and type 2 diabetes in offspring. Diabetologia. 2019;62:1779–1788. doi: 10.1007/s00125-019-4914-1. [DOI] [PubMed] [Google Scholar]
- 205.Sandovici I, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc. Natl Acad. Sci. USA. 2011;108:5449–5454. doi: 10.1073/pnas.1019007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vaiserman A, Lushchak O. Prenatal malnutrition-induced epigenetic dysregulation as a risk factor for type 2 diabetes. Int. J. Genomics. 2019;2019:3821409. doi: 10.1155/2019/3821409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kadayifci FZ, et al. Early-life programming of type 2 diabetes mellitus: understanding the association between epigenetics/genetics and environmental factors. Curr. Genomics. 2019;20:453–463. doi: 10.2174/1389202920666191009110724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Beyaz S, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Shang L, et al. β-Cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes. 2014;63:923–933. doi: 10.2337/db13-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Montaser H, et al. Loss of MANF causes childhood-onset syndromic diabetes due to increased endoplasmic reticulum stress. Diabetes. 2021;70:1006–1018. doi: 10.2337/db20-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Maxwell KG, et al. Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 2020;12:eaax9106. doi: 10.1126/scitranslmed.aax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shik Mun K, et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 2019;10:3124. doi: 10.1038/s41467-019-11178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Halban PA, et al. The possible importance of contact between pancreatic islet cells for the control of insulin release. Endocrinology. 1982;111:86–94. doi: 10.1210/endo-111-1-86. [DOI] [PubMed] [Google Scholar]
- 214.Tsonkova VG, et al. The EndoC-βH1 cell line is a valid model of human beta cells and applicable for screenings to identify novel drug target candidates. Mol. Metab. 2018;8:144–157. doi: 10.1016/j.molmet.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Reissaus CA, Piston DW. Reestablishment of glucose inhibition of glucagon secretion in small pseudoislets. Diabetes. 2017;66:960–969. doi: 10.2337/db16-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Broutier L, et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016;11:1724–1743. doi: 10.1038/nprot.2016.097. [DOI] [PubMed] [Google Scholar]
- 217.Harata M, et al. Delivery of shRNA via lentivirus in human pseudoislets provides a model to test dynamic regulation of insulin secretion and gene function in human islets. Physiol. Rep. 2018;6:e13907. doi: 10.14814/phy2.13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu S, et al. Lentiviral mediated gene silencing in human pseudoislet prepared in low attachment plates. J. Vis. Exp. 2019;2019:139–148. doi: 10.3791/59578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bevacqua RJ, et al. SIX2 and SIX3 coordinately regulate functional maturity and fate of human pancreatic β cells. Genes Dev. 2021;35:234–249. doi: 10.1101/gad.342378.120. [DOI] [PMC free article] [PubMed] [Google Scholar]