Abstract
Pseudomonas aeruginosa invades various epithelial cell types in vitro and in vivo. The P. aeruginosa genome possesses a gene (flhA) which encodes a protein that is believed to be part of the export apparatus for flagellum assembly and which is homologous to invA of Salmonella spp. Because invA is required for invasion of Salmonella spp., a role for flhA in P. aeruginosa invasion was explored using cultured rabbit corneal epithelial cells. An flhA mutant of P. aeruginosa strain PAO1 was constructed and was shown to be nonmotile. Complementation with flhA in trans restored motility. Corneal cells were infected for 3 h with the wild type (PAO1), the flhA mutant, the flhA mutant complemented with flhA in trans, an flhA mutant containing the plasmid vector control, or an fliC mutant (nonmotile mutant control). Invasion was quantified by amikacin exclusion assays. Both the flhA and the fliC mutants invaded at a lower level than the wild-type strain did, suggesting that both fliC and flhA played roles in invasion. However, loss of motility was not sufficient to explain the reduced invasion by flhA mutants, since centrifugation of bacteria onto cells did not restore invasion to wild-type levels. Unexpectedly, the flhA mutant adhered significantly better to corneal epithelial cells than wild-type bacteria or the fliC mutant did. The percentage of adherent bacteria that invaded was reduced by ∼80% for the flhA mutant and ∼50% for the fliC mutant, showing that only part of the role of flhA in invasion involves fliC. Invasion was restored by complementing the flhA mutant with flhA in trans but not by the plasmid vector control. Intracellular survival assays, in which intracellular bacteria were enumerated after continued incubation in the presence of antibiotics, showed that although flhA and fliC mutants had a reduced capacity for epithelial cell entry, they were not defective in their ability to survive within those cells after entry. These results suggest that the flagellum assembly type III secretion system plays a role in P. aeruginosa invasion of epithelial cells. Since the flhA mutants were not defective in their ability to adhere to corneal epithelial cells, to retain viability at the cell surface, or to survive inside epithelial cells after entry, the role of flhA in invasion of epithelial cells is likely to occur during the process of bacterial internalization.
Pseudomonas aeruginosa is an opportunistic gram-negative pathogen that can cause sight-threatening corneal infection in contact lens wearers and life-threatening infections among neutropenic patients, individuals with severe burns, and those with cystic fibrosis (3, 21). Although P. aeruginosa is traditionally thought to be an extracellular pathogen, evidence is accumulating to implicate corneal epithelial cell invasion in the pathogenesis of corneal disease caused by this bacterium (9, 10). Blocking the cystic fibrosis transmembrane conductance regulator, which has been shown to be a corneal cell factor involved in P. aeruginosa invasion, reduces corneal pathology caused by P. aeruginosa (27). Lipopolysaccharide (LPS) deep rough mutants of P. aeruginosa demonstrate reduced capacity for corneal epithelial cell invasion (26) and are not virulent in vivo (20). Bacterial factors other than LPS are also likely to be involved in epithelial cell entry, since LPS mutants demonstrate low levels of invasion (26) and rpoN mutants of invasive strains, which have normal LPS, are noninvasive (S. M. J. Fleiszig et al., unpublished data). Therefore, an RpoN-regulated factor(s) may also be involved in invasion by this organism.
Type III secretion systems are used by gram-negative bacteria to translocate bacterial proteins directly into host cells during infection. Effector molecules of these systems play diverse roles: some mediate bacterial invasion, while others paralyze or kill host cells. P. aeruginosa is known to possess two type III systems. One system is regulated by the transcriptional activator ExsA (24) and is essential for acute cytotoxic activity and resistance to phagocytosis by eukaryotic cells (4a, 13). The other system belongs to the family of type III secretion systems involved in flagellum biogenesis in many bacteria (16, 17).
In Salmonella spp., InvA is part of the machinery for a type III secretion system that secretes proteins affecting host cell signal transduction leading to bacterial internalization (4), without affecting bacterial adhesion to host cells. InvA is homologous to MxiA of Shigella spp. and LcrD of Yersinia spp. (14), which are also involved in the surface presentation or secretion of invasion factors. A search of the P. aeruginosa genome sequence (www.pseudomonas.com) revealed a gene (flhA) that is homologous to invA of Salmonella spp.
Analysis of amino acid homology and function indicates that P. aeruginosa flhA encodes a component of the flagellum export apparatus. Recent studies with both Yersinia spp. and Salmonella spp. have shown that in addition to exporting flagellar proteins, the flagellum export apparatus can transport other proteins extracellularly to influence bacterium-host interactions (6, 25). Thus, we explored whether FlhA of P. aeruginosa is involved in surface interactions with epithelial cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains, plasmid vectors, and their derivatives are shown in Table 1. The bacteria were propagated in liquid Luria (L) broth or on L agar plates (1.7% agar) with or without antibiotics. The antibiotics (and concentrations) used were as follows: for Escherichia coli, ampicillin (200 μg/ml), gentamicin (10 μg/ml), and chloramphenicol (30 μg/ml); for P. aeruginosa, carbenicillin (300 μg/ml), gentamicin (100 μg/ml), and chloramphenicol (400 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli DH5α | hsdR recA lacZYA φ80 lacZ ΔM15 | GIBCO-BRL |
| P. aeruginosa | ||
| PAO1 | Laboratory strain, motile | M. Vasil |
| PAO-flhA | PAO1 flhA::Cmr, nonmotile | This study |
| PAO-fliC | PAO1 fliC::Gmr, nonmotile | This study |
| Plasmids | ||
| pBluescript KS(+) | E. coli cloning vector, Ampr | Stratagene Inc. |
| pBSflhA | pBluescript containing the 3-kb HindIII/SstI fragment with the complete flhA gene | This study |
| pUC18Cm | Chloramphenicol resistance gene cassette cloned as a Sau3A fragment into the unique BamHI site of pUC18 | S. Lory |
| pSP72 | Cloning vector, Ampr | Promega Inc. |
| pSP72Cm | Chloramphenicol resistance gene cassette cloned as a Sau3A fragment into the unique BamHI site of pSP72 | This study |
| pBS6970 | pBluescript containing the 1.4-kb HindIII/SstI fragment with the complete fliC gene | This study |
| pUC7G | pUC7 plasmid containing a gentamicin resistance gene cassette excisable with restriction enzymes PstI, SalI, BamHI, and EcoRI | S. Lory |
| pBS6970G | pBS6970 with a gentamicin resistance gene cassette inserted in the BglII site of the fliC gene | This study |
| pPZ375 | φriV in pGEM3Z | 23 |
| pPZ375flhA | pPZ375 containing the HindIII/SstI fragment with the complete flhA gene | This study |
For invasion and adherence assays, bacteria were grown overnight at 37°C on L agar plates with antibiotic as required. The following antibiotics (and concentrations) were used: carbenicillin (300 μg/ml), gentamicin (100 μg/ml), and chloramphenicol (150 μg/ml). Bacterial colonies were resuspended in Eagle's minimal essential medium (MEM) containing Hanks balanced salt solution (Sigma Chemical Co., St. Louis, Mo.), 20 mM HEPES buffer (pH 7.4), 3.5% sodium bicarbonate, and 0.6% bovine serum albumin. The appropriate optical density was determined by spectrophotometry (optical density at 650 nm) and confirmed by viable count.
Enzymes and chemicals.
T4 DNA ligase, Taq DNA polymerase, and all restriction enzymes were purchased from GIBCO-BRL Inc., Gaithersburg, Md. The chemicals were purchased either from Sigma Chemical Co. or from Amresco, Inc., Solon, Ohio.
PCR amplification and primers.
PCR amplification was utilized for specific amplification of a 3-kb PCR product from P. aeruginosa PAO1 containing the flhA gene and some flanking sequence and a 1.4-kb PCR product containing the P. aeruginosa PAO1 fliC gene. PCR was performed with a DNA Thermal Cycler 480 (Perkin-Elmer Cetus). The reactions were performed in 100-μl volumes using Taq polymerase. Each reaction mixture contained final concentrations of 50 ng of DNA template, 2.5 Unit of Taq polymerase, 1.5 mM MgCl2, 0.1 mM deoxyaucleoside triphosphate mix, and 0.2 μM primers. For certain amplifications, dimethyl sulfoxide was added to a 2% (vol/vol) final concentration. Thirty-five cycles were run, each consisting of incubations for 1 min at 94°C, 1 min at 55°C, and 5 or 10 min at 72°C. The primers used for PCRs were purchased from GIBCO-BRL Inc. Restriction enzyme recognition sites were added at the ends of primers to facilitate subsequent cloning of the PCR products if desired. Six or more additional nucleotides were added 5′ to the restriction sites to ensure efficient cleavage. The primers that were used to amplify the flhA region of P. aeruginosa PAO1 included RER49 (with an HindIII site) and RER50 (with an SstI site). Primers RER69 (with an HindIII site) and RER70 (with an SstI site) were used for the amplification of the fliC gene from PAO1 DNA. The PCR products were cut with restriction enzymes to verify that the products were correct and then used for the subsequent construction of a mutation in these genes.
Plasmid constructions.
The 3-kb amplification product containing_the PAO1 flhA gene (www.pseudomonas.com) and some flanking sequence was cloned into the HindIII and SstI sites of pBluescript KS(+) to yield pBSflhA. A chloramphenicol resistance gene cassette was excised from pUC18Cm as a 1-kb Sau3A fragment and was cloned into the unique BamHI site of pSP72 (Promega Inc., Madison, Wis.), giving rise to pSP72Cm. The chloramphenicol resistance gene cassette was then moved from pSP72Cm to pBSflhA as a 1.5-kb AatII/Bgl/II fragment. As a result of this cloning, a 0.2-kb internal AatII/BglII fragment was deleted from the flhA gene. The resulting construct (7.3 kb) was named pBSflhACm and was used for allelic exchange in PAO1. The 1.4-kb amplification product containing the PAO1 fliC gene and some flanking sequence was cloned into the HindIII and SstI sites of pBluescript KS(+) to yield pBS6970. The plasmid pBS6970 was linearized at the unique BglII site in the PAO1 fliC gene. A 1.8-kb gentamicin resistance gene cassette having BamHI ends was excised from pUC7G. The gentamicin resistance gene cassette was inserted at the BglII site in the fliC gene, leading to the construction of pBS6970G. This construct was utilized to generate a chromosomal mutation in the PAO1 fliC gene by marker exchange. The plasmid used for the complementation of the flhA mutation in PAO-flhA (pPZ375flhA) was obtained by cloning the 3-kb DNA fragment carrying the complete PAO1 flhA gene into the shuttle vector pPZ375 (23). This 3-kb fragment was excised from pBSflhA with HindIII and SstI.
Electroporations.
Electroporations were performed using a modification of the protocol of Smith and Iglewski (22). The plasmid DNA used for the electroporations was prepared by the alkaline lysis procedure (2). For gene replacement experiments involving chromosomal recombinations, the plasmid DNA was linearized by a restriction enzyme recognizing a site in the vector and the DNA was gel purified. About 1 μg of linear DNA fragment was electroporated into the electrocompetent P. aeruginosa cells. For complementation experiments, 50 to 100 ng of supercoiled or covalently closed circular plasmid DNA was electroporated into the target strains.
Motility assay.
Motility assays were used to study the motility function of different P. aeruginosa strains. Bacterial strains were grown overnight at 37°C on fresh L agar with or without antibiotics. The cells were then transferred with a sterile toothpick to 0.3% agar plates with or without antibiotics. These plates were incubated at 37°C for 16 h, and motility was assessed qualitatively by examining the circular swarm formed by the growing motile bacterial cells.
Preparation of cell cultures.
Both immortalized rabbit corneal epithelial cells (12) and primary cultures of rabbit corneal epithelial cells (11) were used in these experiments. Epithelial cells were grown in 24-well plates, in 3.5-cm tissue culture dishes (Becton Dickinson Labware, Franklin Lakes, N.J), or on 0.4-μm-pore-size, 12-mm Transwell filters (Corning Costar Corp., Cambridge, Mass.) as previously described (13). Cells were fed with SHEM and were used 3 to 6 days after passaging (11). Results presented here were obtained from cells grown between passages 6 and 11 for immortalized cells.
Invasion of cells grown in plastic tissue culture plates.
Since the fliC mutant was gentamicin resistant, amikacin rather than gentamicin was used to quantify the extent of bacterial invasion of corneal epithelial cells (15). These assays were performed as previously described (12), with minor modifications as described below. An inoculum of 2 × 105 CFU in 200 μl of MEM was used for each well of cells in 24-well tissue culture plates. Following a 3-h incubation at 37°C, bacterial cell growth was enumerated by bacterial cell counts of the supernatant to determine whether mutants were able to survive and grow as efficiently as the wild type, PAO1, in the presence of epithelial cells. Cells were then washed with phosphate-buffered saline prior to incubation of cells with 400 μg of amikacin (Sigma)/ml for 1 h to kill extracellular bacteria. Survivors of amikacin treatment were enumerated by viable bacterial cell counts after the cells were washed to remove the antibiotic, followed by cell lysis with a 15-min treatment with 0.25% Triton X-100 (Sigma). At least six wells were used for each bacterial strain and mutant, and experiments were repeated at least three times. In control experiments in which epithelial cells were omitted, amikacin was shown to be effective at killing extracellular bacteria for all strains tested. In experiments using the wild type, PAO1, amikacin survival assays were shown to yield results for quantification of intracellular bacteria that were similar to those of gentamicin survival assays.
Invasion by bacteria after centrifugation onto cells.
In other experiments, bacteria were centrifuged onto the cells prior to the 3-h incubation to complement the effects of motility defects on invasion. For this purpose, an inoculum of 106 CFU of each bacterial strain in 1 ml of MEM was added to 3.5-cm tissue culture dishes containing confluent corneal epithelial cells. One group of dishes for each strain was centrifuged at 3,000 rpm for 5 min with a clinical centrifuge (IEC, Needham, Mass.). An identical group of samples was allowed to remain static under the same conditions. At least four samples were used for each group. Invasion assays were then performed as described above for cells grown in 24-well plates. Viable counts of the culture supernatants were performed after centrifugation and again after the 3-h invasion assay, prior to addition of amikacin, to monitor the effects of centrifugation on bacterial viability and growth.
Quantification of invasion efficiency.
In other experiments, the efficiency at which cells internalized adherent bacteria was determined. For this purpose, simultaneous invasion and adherence assays were performed and the percentage of adherent bacteria that had invaded cells was calculated. Since P. aeruginosa is able to adhere in large numbers to the sides of plastic tissue culture wells, these assays were performed using cells grown on semipermeable filters, which could then be removed from their plastic inserts prior to enumeration of adherent bacteria by viable counts. Using this method, only those bacteria that were actually attached to corneal cells were counted. Cells were grown on Transwell filters in 12-well plates and inoculated with 5 × 105 CFU of bacteria in 500 μl of MEM. After 3 h of incubation with bacteria, half of the samples were treated with amikacin to quantify invasion. The other half were used to quantify adherence, so amikacin was omitted. Instead, these samples were washed three times with MEM to remove nonadherent bacteria. The continued confluence of the epithelial cell layer during exposure to bacteria was confirmed by measuring transepithelial resistance across the monolayer using an EVOM meter (World Precision Instruments, Sarasota, Fla.). Each filter was then cut away from the respective plastic holder prior to lysis of cells on the filter with Triton X-100 and enumeration of bacteria in the homogenate by viable counts.
Intracellular survival assays.
An inoculum of 2 × 106 CFU in 200 μl of MEM was added to each well of cells grown in 24-well tissue culture plates. After a 1-h infection at 37°C, the cell samples were divided into two groups. One group was treated with amikacin for 1 h, while the other group was allowed to incubate with amikacin for a total of 4 h. Trypan blue staining and observation of epithelial cell morphology showed that the longer amikacin treatment did not kill or injure cells. The number of bacteria surviving within cells was enumerated by viable counts after cell lysis. The number of bacteria found within cells after 1 h of incubation with amikacin was compared to the number of intracellular bacteria recovered after 4 h of incubation with amikacin to determine the ability of bacteria to survive and grow within epithelial cells after cell entry.
Statistics.
For analysis of the significance of differences in invasion or adherence levels, the Student t test was used for comparison of two groups of data; otherwise, analysis of variance was used. All experiments were repeated at least three times.
RESULTS
Characterization of P. aeruginosa flhA.
The flhA gene of P. aeruginosa was localized in the flagellar regulon in the P. aeruginosa PAO1 genome sequence (www.pseudomonas.com). The flhA gene is flanked by the flhF gene at the 5′ end and the flhB gene at the 3′ end. The deduced amino acid sequence of this gene showed significant similarity and identity to FlhA of other organisms, InvA of Salmonella enterica serovar Typhimurium, LcrD of Yersinia enterocolitica, and HrpI of Pseudomonas syringiae (Table 2), which are all involved in type III secretion. Since many of the P. aeruginosa flagellar genes have been shown to be regulated by RpoN (5) and an RpoN consensus sequence was located in the intergenic region between flhB and flhA, we examined the promoter region of the flhA gene for RpoN dependency. A 600-bp DNA fragment containing the flhA promoter region was obtained by PCR amplification and was cloned into a promoter probe vector, pDN191acΩ. The promoter activity of the flhA promoter fused with the promoterless lacZ gene was estimated by measurement of β-galactosidase activity of this fusion. The promoter activity of the flhA promoter was significantly reduced in the RpoN mutant (data not shown), indicating that the flhA gene expression was directly or indirectly RpoN dependent.
TABLE 2.
Homology of P. aeruginosa FlhA with proteins in other organisms
Construction and characterization of flhA and fliC mutants of PAO1.
In order to test the function of flhA in P. aeruginosa, a chromosomal mutant of PAO1 flhA was constructed by gene replacement. The P. aeruginosa PAO1 flhA gene located on a PCR-generated 3.0-kb HindIII/SstI fragment was inactivated by inserting a chloramphenicol resistance gene cassette as an AatII/BglII fragment. The insertionally inactivated flhA gene on a nonreplicating, multicopy plasmid (pBSflhACm) was introduced into PAO1 by electroporation, where it replaced the corresponding chromosomal copy of the flhA gene by double reciprocal recombination, giving rise to an flhA mutant strain, PAO-flhA. The replacement of the wild-type flhA gene in PAO-flhA was confirmed by PCR (data not shown). This strain was nonmotile on 0.3% L agar plates and did not make a flagellum, as observed under the electron microscope (data not shown). Pili appeared normal in number and length. PAO-C (fliC mutant strain) was constructed using the same strategy and was used as a nonmotile control in the invasion assays.
Complementation of motility.
Plasmid pPZ375flhA, carrying the PAO1 flhA gene, and its vector control without the flhA insert were electroporated into the flhA mutant strain PAO-flhA. The resulting strains were tested for motility on 0.3% agar plates. As expected, pPZ375flhA fully complemented the motility defect of PAO-flhA, while the vector control remained nonmotile (data not shown). The complemented mutant was shown to have flagella by electron microscopy (data not shown).
Effect of fliC and flhA mutations on epithelial cell invasion.
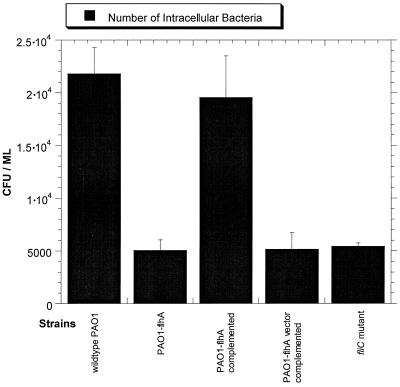
The invasion level of the nonmotile flhA mutant was compared to those of the wild-type and control strains. Since this was a study of invasion rather than motility, an fliC mutant was included as a control. flhA and fliC mutants are nonmotile for similar reasons; i.e., neither mutant assembles flagella. In the case of the fliC mutant, the mutation is in the gene encoding flagellin, the flagellar protein. The flhA mutant was found to be less invasive than the wild-type strain, PAO1 (Fig. 1). Complementation of flhA in trans restored invasion. The fliC mutant was also found to be less invasive than the wild type. There was no significant difference between the fliC and the flhA mutants, suggesting that loss of motility may be responsible for reduced invasion levels by the flhA mutant.
FIG. 1.
Effects of flhA and fliC mutations on invasion of corneal epithelial cells.
Since the loss of motility of the flhA mutant might be expected to affect the ability of bacteria to gain access to the cell surface, experiments were performed in which bacteria were centrifuged onto cells at the beginning of the invasion assay. Centrifugation did not restore flhA mutant invasion to wild-type levels (Table 3). Although flhA mutant invasion was enhanced by centrifugation (approximately twofold), a similar increase was found to occur with the wild-type, PAO1. Thus, the flhA mutant remained markedly less invasive than the wild-type strain (P = 0.0017). Bacterial viable counts of the media were performed before and after centrifugation and also after the 3-h incubation of cells with bacteria. Those data showed that reduced levels of invasion by the flhA mutant were not explained by any loss in bacterial cell viability as a result of centrifugation, nor were they due to a reduced ability of mutants to grow or survive in the presence of corneal epithelial cells (data not shown).
TABLE 3.
Invasion assays comparing the wild type, PAO1, to the flhA mutant in static assays and after bacteria have been centrifuged onto cells
| Strain | Mean no. of surviving CFU ± SD in:
|
||
|---|---|---|---|
| Inoculum | Invasion assay
|
||
| Before centrifugation | After centrifugation | ||
| PAO1 | (2.8 ± 0.3) × 106 | (3.0 ± 0.7) × 104 | (5.4 ± 1.7) × 104 |
| flhA mutant | (3.4 ± 0.1) × 106 | (0.8 ± 0.3) × 104 | (1.6 ± 0.4) × 104 |
Effect of the flhA mutation on bacterial association with cells.
Bacterial adherence assays were performed to determine whether the reduced invasion by the flhA mutant was due to a reduced ability to adhere to cells. Simultaneous invasion and adherence assays using cells grown on filters were used to calculate the percentage of adherent bacteria that invaded, i.e., to determine the efficacy of internalization (Table 4). Similar to the invasion of cells grown in plastic tissue culture wells, invasion of these filter-grown cells was reduced by both fliC and flhA mutations. Surprisingly, the flhA mutant was better at adhering to cells than the wild type was (P = 0.007). This enhanced adherence capacity was not noted with the fliC mutant (P = 0.6). Complementation of flhA mutants with flhA in trans reduced adherence to levels similar to those noted with wild-type bacteria or with the fliC mutant (P = 0.3). Although the absolute levels of invasion by flhA and fliC mutants were similar, the percentage of adherent bacteria that invaded (efficacy of internalization) was more reduced with the flhA mutation (81% reduction) than with the fliC mutation (55% reduction). This finding suggested that fliC was involved in internalization but that mutation of flhA played an additional role and that this was associated with differences in binding to epithelial cells.
TABLE 4.
P. aeruginosa adherence to and invasion of corneal epithelial cells grown on filters
| Strain | Mean no. of
surviving CFU ± SD during:
|
% Invasion of adherent bacteria | |
|---|---|---|---|
| Adherence | Invasion | ||
| PAO1 | (7.4 ± 1.1) × 105 | (6.1 ± 1.2) × 104 | 8.2 |
| flhA mutant | (18.6 ± 4.4) × 105 | (2.5 ± 0.8) × 104 | 1.3 |
| flhA mutant, complemented | (7.0 ± 1.2) × 105 | (4.2 ± 0.1) × 104 | 5.9 |
| flhA mutant (vector) | (15.9 ± 2.5) × 105 | (3.1 ± 0.6) × 104 | 1.9 |
| fliC mutant | (7.4 ± 1.9) × 105 | (2.5 ± 0.3) × 104 | 3.4 |
Bacteria did not reduce transepithelial resistance in any of these experiments, eliminating the possibility that enumerated bacteria in adherence assays included a population that had bound directly to exposed filter surfaces.
Effect of the flhA mutation on intracellular survival.
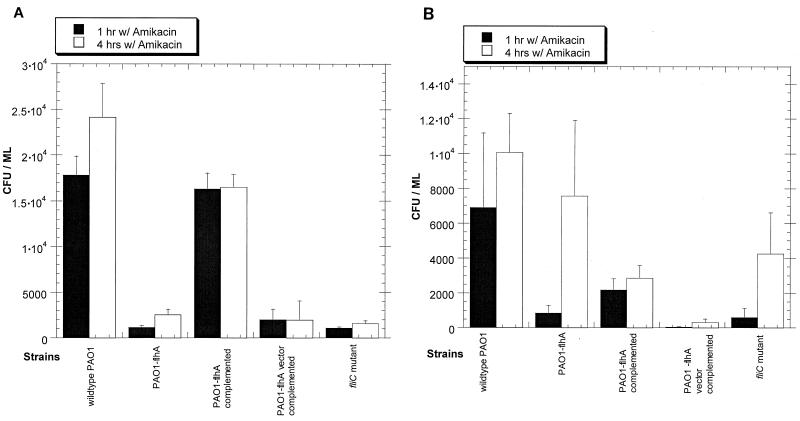
The flhA mutants had a reduced capacity for epithelial cell entry, but they were not entirely noninvasive. Intracellular survival assays showed that although flhA was involved in epithelial cell entry, it was not required for survival or growth within cells after entry (Fig. 2). A comparison of the number of intracellular bacteria recovered from cells after 1 and 4 h of incubation with antibiotic revealed that both the fliC and the flhA mutants could survive inside epithelial cells after entry as effectively as the wild type did (P > 0.05). Interestingly, experiments with primary cultures consistently showed a tendency for these mutants to survive and grow even more efficiently inside cells than the wild type did (Fig. 2B). These experiments also demonstrated that the reduced ability of flagellar mutants to enter cells could be detected as early as 1 h after incubation with bacteria (Fig. 2).
FIG. 2.
Intracellular survival assays show that flhA and fliC mutants are not defective in their ability to survive within corneal epithelial cells. (A) Immortalized rabbit corneal epithelial cells, (B) primary cultures of rabbit corneal epithelial cells. To monitor intracellular survival, samples incubated with amikacin for 4 h were compared to samples incubated with amikacin for 1 h.
DISCUSSION
The results of this study showed that both flhA and fliC contribute to P. aeruginosa invasion, as determined by antibiotic survival assays. The reduced capacity of flagellar mutants to invade cells was not explained by any loss of ability to survive or grow in the assay systems in the presence of the corneal epithelial cells. Centrifugation of bacteria onto cells at the beginning of invasion assays did not complement the invasion defect of flagellar mutants. Adherence assay results confirmed that these mutants, although nonmotile, were not defective in their ability to gain access to the cell surface. Even without centrifugation, the flhA mutant could adhere to epithelial cells more efficiently than could the wild type and the fliC mutant adhered equally as well as the wild-type strain. These results showed that lack of motility, and hence access to the cell surface, was not directly responsible for the reduced invasive capacity of flhA mutants.
It is difficult at this time to conceive of a model encompassing roles for both FliC and FlhA during such an interaction with the cell. Since there is no reduction of contact with the cells in either mutant, motility due to the flagellar filament made up principally of FliC is not important. However, there were differences noted between the flhA and fliC mutants in their invasion efficiency, suggesting that factors other than fliC are also involved. Based on the Escherichia coli-Sralmonella Typhimurium paradigm of flagellum assembly (17) an fliC mutant should retain the flagellar cap and hook but not an intact flagellum. We have noted that the cap and hook are made in an fliC mutant of P. aeruginosa (unpublished data). On the other hand, the flhA mutant, if defective in transport, would be unable to make these structures, raising the possibility that some intermediate structure of the flagellum between the flagellar cap and the cell wall may be the effector molecule for invasion or is used to transport it. Alternatively, flhA-dependent invasion factors might be unrelated to flagellar assembly. For example in Yersinia spp., YlpA, a virulence factor that is involved in bacterium host cell interactions, has been found to be secreted via the flagellar type III secretion apparatus (25). Yersinia flhA mutants do not secrete YlpA. In Salmonella spp., regulators of flagellar assembly also regulate invasion proteins secreted via a separate type III system (SPI-1) (6). By homology to Yersinia spp. and Salmonella spp., it is likely that P. aeruginosa FlhA controls or is used to secrete an invasion protein(s) rather than functioning directly as a ligand for invasion. These data therefore suggest that FlhA may play a dual role as a secretion apparatus for an invasin and for the distal flagellar structures much the same way that FlhB, FlhA, and FlhE in Y. enterocolitica play a dual role of secreting flagellar structures and phospholipase (25).
There are at least five major stages through which bacteria must progress in order to be counted by antibiotic survival assays that are used for quantifying bacterial invasion. Bacteria must be able to (i) gain access to the cell surface; (ii) survive at the cell surface despite the presence of innate antimicrobial epithelial defenses, such as defensins (18); (iii) adhere to the cell membrane; (iv) be internalized; and (v) remain viable or replicate inside the cell for the remainder of the assay. Bacterial mutations that affect any one of these five stages would be expected to reduce bacterial invasion of epithelial cells as quantified by antibiotic survival assays. The flhA mutants adhered more efficiently to cells than did the wild type, PAO1, and although reduced in their capacity for cell entry, they were at least as capable as the wild type of surviving within cells after entry. Thus, the role of flagellar proteins in the invasion process apparently occurs after adherence to the cell and during internalization by the host cell. This is different from the role of LPS in invasion, which is involved in survival after epithelial cell entry (7). The effects of FliC and FlhA on internalization occur relatively quickly, since there was significant loss of invasion ability even with a short (1-h) invasion assay. This is also different from the effect of LPS mutation, which does not affect capacity to invade in short assays (7).
The markedly increased cellular adhesion of the flhA mutant suggests that there is an inverse relationship between expression of a corneal cell adhesin(s) and expression of the flagellar type III secretion system, which will require further exploration. A mutation in flhA may result in up regulation of a cellular nonpilus adhesin, since the pilus number appeared normal. It has been noted that a mutation of one flagellar regulator resulted in up regulation of many different flagellar gene operons (5). Alternatively, the increased adherence levels noted with flhA mutants might be related to evasion of target epithelial cell defenses. Innate epithelial cell defense systems, such as those that lead to defensin and mucin production (18), may recognize and respond to proteins that are secreted by the flagellar assembly apparatus (19). In this case, mutants that lack these proteins might not trigger these defense mechanisms.
In summary, the data presented herein support the growing body of evidence suggesting that the P. aeruginosa flagellum plays various roles in virulence, some of which are unrelated to motility. These functions include modulation of adhesion to mucins (1), mediation of inflammatory responses (8), and a contribution to host cell invasion. Continued studies of the structure, function, and regulation of proteins related to this system in P. aeruginosa might lead to new strategies for preventing or treating infections caused by this opportunistic pathogen.
ACKNOWLEDGMENTS
This work was supported by NIH grants EY11221 (S.M.J.F.); and AI145014 and HL33622 (R.R.), a UC Berkeley Faculty Research Grant (S.M.J.F.), and a Hellman Family Award (S.M.J.F.).
REFERENCES
- 1.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosaflagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 4.Callazo C M, Galan J E. The invasion-associated type III system of Salmonella typhimuriumdirects the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 4a.Cowell B A, Chen D Y, Frank D W, Vallis A J, Fleiszig S M J. ExoT of cytotoxic Pseudomonas aeruginosaprevents uptake by corneal epithelial cells. Infect Immun. 2000;68:403–406. doi: 10.1128/iai.68.1.403-406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dasgupta N, Arora S K, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichelberg K, Galán J E. The flagellar sigma factor FliA (ς28) regulates the expression of Salmonellagenes associated with the centisome 63 type III secretion system. Infect Immun. 2000;68:2735–2743. doi: 10.1128/iai.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans D J, Tuo T C, Van R, Fleiszig S M J. The role of lipopolysaccharide in Pseudomonas aeruginosacorneal epithelial cell invasion includes intracellular survival. Investig Ophthalmol Vis Sci. 2000;41:S153. [Google Scholar]
- 8.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosapulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleiszig S M J, Zaidi T S, Fletcher E L, Preston M J, Pier G B. Pseudomonas aeruginosainvades corneal epithelial cells during experimental infection. Infect Immun. 1994;62:3485–3493. doi: 10.1128/iai.62.8.3485-3493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleiszig S M J, Zaidi T S, Pier G B. Pseudomonas aeruginosainvasion of and multiplication within corneal epithelial cells in vitro. Infect Immun. 1995;63:4072–4077. doi: 10.1128/iai.63.10.4072-4077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleiszig S M J, Zaidi T S, Preston M J, Grout M, Evans D J, Pier G B. Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:2288–2294. doi: 10.1128/iai.64.6.2288-2294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Yen T S B, Frank D W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleiszig S M J, Evans D J, Do N, Shin S, Vallas V, Mostov K E. Epithelial cell polarity affects susceptibility to Pseudomonas aeruginosainvasion and cytotoxicity. Infect Immun. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginocchio C C, Galán J E. Functional conservation among members of the Salmonella typhimuriumInvA family of proteins. Infect Immun. 1995;63:729–732. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser A R, Fleiszig S, Kang P J, Mostov K, Engel J N. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosaby epithelial cells. Infect Immun. 1998;66:1413–1420. doi: 10.1128/iai.66.4.1413-1420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNamara N A, Van R, Tuchin O S, Fleiszig S M J. Ocular surface epithelia express mRNA for human beta defensin-2. Exp Eye Res. 1999;69:483–490. doi: 10.1006/exer.1999.0722. [DOI] [PubMed] [Google Scholar]
- 19.McNamara N A, Khong A, Caterina M, Prince A, Julius D, Basbaum C. Putative identification of the purinergic receptor P2Y5 as a co-receptor for bacterial flagellin: role in bacterial induced gene expression. Mol Biol Cell. 1999;10:118a. [Google Scholar]
- 20.Preston M J, Fleiszig S M J, Zaidi T S, Goldberg J B, Shortridge V D, Vasil M L, Pier G B. Rapid and sensitive method for evaluating Pseudomonas aeruginosavirulence factors during corneal infections in mice. Infect Immun. 1995;63:3497–3501. doi: 10.1128/iai.63.9.3497-3501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C.: ASM Press; 1994. pp. 260–268. [Google Scholar]
- 22.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosaby electroporation. Nucleic Acids Res. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Temple L, Sage A, Christie G E, Phibbs P V., Jr Two genes for carbohydrate catabolism are divergently transcribed from a region of DNA containing the hexC locus in Pseudomonas aeruginosaPAO1. J Bacteriol. 1994;176:4700–4709. doi: 10.1128/jb.176.15.4700-4709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yahr T L, Mende-Mueller L M, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosaexoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagella export apparatus functions as a protein secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidi T S, Fleiszig S M J, Preston M J, Goldberg J B, Pier G B. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Investig Ophthalmol Vis Sci. 1996;37:976–986. [PubMed] [Google Scholar]
- 27.Zaidi T S, Lyczak J, Preston M, Pier G B. Cystic fibrosis transmembrane conductance regulator-mediated corneal epithelial cell ingestion of Pseudomonas aeruginosais a key component in the pathogenesis of experimental murine keratitis. Infect Immun. 1999;67:1481–1492. doi: 10.1128/iai.67.3.1481-1492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]