Abstract
Malfunction of the sialic acid transporter caused by various genetic mutations in the SLC17A5 gene encoding Sialin leads to a spectrum of neurodegenerative conditions called free sialic acid storage disorders. Unfortunately, how Sialin transports sialic acid/proton (H+) and how pathogenic mutations impair its function are poorly defined. Here, we present the structure of human Sialin in an inward-facing partially open conformation determined by cryo–electron microscopy, representing the first high-resolution structure of any human SLC17 member. Our analysis reveals two unique features in Sialin: (i) The H+ coupling/sensing requires two highly conserved Glu residues (E171 and E175) instead of one (E175) as implied in previous studies; and (ii) the normal function of Sialin requires the stabilization of a cytosolic helix, which has not been noticed in the literature. By mapping known pathogenic mutations, we provide mechanistic explanations for corresponding functional defects. We propose a structure-based mechanism for sialic acid transport mediated by Sialin.
Sialin structure provides insights into the molecular mechanism of lysosomal sialic acid transport.
INTRODUCTION
Sialic acids are a group of nine-carbon carboxylated monosaccharides synthesized in animals, some bacterial species, and humans, generally found at the terminal end of glycans that are conjugated with proteins and lipids (1, 2). Sialic acids comprise more than 50 natural derivatives, the most widespread form being N-acetylneuraminic acid (Neu5Ac) (fig. S1). The structural diversity and strong negative charge (pKa = ~2) of sialic acids suggest their importance in a wide range of biological processes, such as mediating cell-cell interactions, modulating immune responses, controlling the stability of proteins, and many more (3–5). Thus, the imbalance of sialic acid metabolism is implied in many pathological conditions, including but not limited to cardiovascular diseases (6), cancer (7), immunological disorders (8), and diabetes (9).
Sialic acid metabolism involves a critical recycling process in the endolysosomal system. Various enzymes degrade the glycans to produce free monosaccharides (including sialic acids) that are transported back into the cytosol (10). Thus, genetic mutations of these enzymes and related transporters lead to defective glycan degradation, glycan salvage, and abnormal accumulation of metabolites in lysosomes, resulting in various lysosomal storage disorders (11). For example, multiple mutations in the SLC17A5 gene encoding the lysosomal sialic acid/proton (H+) symporter Sialin could cause free sialic acid storage disorders (12). These disorders are characterized by enlarged cellular lysosomes and elevated levels of free sialic acids in urine (13). They cover a spectrum of pathological forms from a mild, slowly progressive symptom in patients living to adulthood (Salla disease) (14), to an intermediate form resulting in severe developmental delays (15), to a severe condition that is often lethal in early childhood (infantile sialic acid storage disorder) (16). Unfortunately, no approved therapy for these disorders or ongoing clinical trials is listed on clinicaltrials.gov (accessed August 2022). One possible reason for this is that the molecular mechanism of sialic acid/H+ cotransport mediated by Sialin is unclear. Although the transport deficiency of some pathogenic mutants has been characterized in cells and proteoliposomes, how such mutations affect the structure of Sialin remains unknown.
Sialin is a member of the solute carrier 17 (SLC17) family. SLC17 members are responsible for the translocation of various organic anions (phosphate, glutamate, aspartate, sialic acids, and more) driven by an electrochemical gradient with contributions from either electrical potential (Δψ) or H+ gradient (ΔpH) or both (17, 18). Sialin can transport multiple substrates other than sialic acids, depending on where the transporter is located. For example, in rodents, Sialin expressed in synaptic vesicles of hippocampal neurons and pinealocytes is responsible for the vesicular accumulation of aspartate (19). However, this vesicular aspartate accumulation might not be physiologically relevant, as the aspartate released is at a concentration too low to activate postsynaptic N-Methyl-D-aspartic acid (NMDA)–type glutamate receptors (20). In vitro experiments using reconstituted proteoliposomes have confirmed that Sialin could act as an anion transporter for aspartate, glutamate, and neuropeptide NAAG driven by membrane potential (21, 22). In human salivary glands, when localized in the basolateral membrane of the epithelial cells, Sialin is responsible for the active accumulation of nitrate (NO3−), acting like an electrogenic 2NO3−/H+ cotransporter (23). Such function suggests that Sialin plays a critical role in the delicate balance of the nitrate–nitrite–nitric oxide pathway (24). Despite the extensive functional characterization, structural information about Sialin is still missing in the literature, which delays our attempt to reveal the molecular mechanism of this transporter.
Here, using cryo–electron microscopy (cryo-EM) single-particle reconstruction, we determined a 3.4-Å-resolution structure of human Sialin in the apo state. The structure adopts an inward-facing conformation with two domains: N-domain with transmembrane helices (TM) 1 to 6 and C-domain with TM7 to TM12. Using computational docking, we identified the putative substrate-binding pocket in Sialin. Although the pocket is solvent accessible from the cytosolic side, it is not accessible by major substrates such as Neu5Ac because the central pathway is spatially constricted around H183. Thus, the conformation is designated as partially open inward-facing. Nevertheless, the computationally docked Neu5Ac in Sialin reveals a network of critical polar and charged residues in the substrate-binding pocket. In the N-domain of Sialin, there is a small tunnel connecting the lumen with protonatable residues E171 and E175 deeply positioned in the center of the transporter, both of which are critical for H+ coupling and absolutely conserved in the SLC17 family. We mapped known pathogenic mutations onto the Sialin structure and explained how they could cause defective sialic acid transport in patients. In summary, our structural and functional studies reveal the first high-resolution structure of human Sialin and provide essential insights into understanding the role of pathogenic Sialin mutations leading to free sialic acid storage disorders.
RESULTS
Structure determination of Sialin
Previous studies have shown that expression of Sialin could be redirected to the plasma membrane by mutating the N-terminal double Leu (L22 and L23) to either Ala or Gly in multiple human cell lines, such as human embryonic kidney (HEK) 293 (25), HeLa (26), and submandibular gland cells (23). Here, we kept the double Leu unchanged, cloned the full-length human Sialin with an N-terminal His-tag, and mutated three Asn residues (N71A, N77A, and N95A) to abolish potential glycosylation. We expressed the construct in High Five insect cells using a Bac-to-Bac baculovirus expression system (27, 28). The result shows that most of the expressed Sialin is located in the plasma membrane when immunostained with an anti-Sialin monoclonal antibody 8B1 (Fig. 1A). In addition, Sialin is only detectable when the cell is permeated by Triton X-100 and followed by immunostaining with an anti–His-tag antibody, suggesting that the N-terminal His-tag of Sialin is located on the cytosolic side. Thus, Sialin is correctly oriented (topologically equivalent as in lysosomes) (fig. S2). Next, we asked whether the expressed Sialin in High Five cells is functional or not by a cellular transport assay (Fig. 1B). We incubated High Five cells (with or without Sialin expression) with radioactively labeled sialic acid [3H]Neu5Ac in the buffer of pH 5.6 and then quantified the accumulated amount of [3H]Neu5Ac in the cells. The result shows that Sialin can actively uptake Neu5Ac in High Five cells at a level comparable to previous reports using HEK293 and HeLa cells (25, 26).
Fig. 1. Overall architecture of functional Sialin.
(A) Confocal images of High Five cells. Unpermeabilized cells with or without Sialin expression were immunostained using anti-Sialin monoclonal antibody 8B1 (red) and DAPI (blue). (B) Cellular transport of sialic acids using High Five cells with and without Sialin expression. (C) Cryo-EM reconstruction of Sialin-nanodisc-Fab at 3.4-Å resolution colored by local resolution estimation. (D) Cartoon representation of the Sialin model. All helices (12 TMs and a cytosolic helix) are rainbow-colored.
To carry out cryo-EM analysis, we first incorporated the detergent-purified Sialin into nanodiscs made of soybean extract lipids and membrane scaffold protein MSP1D1 (Sialin-nanodisc) (29). The sample was imaged on a 200-keV Talos Arctica microscope. Data analysis showed two-dimensional (2D) averages of an excellent top/bottom view (perpendicular to the membrane bilayer) with apparent densities accounting for the TMs in Sialin but poor side views (parallel to the membrane bilayer) without any sharp features (fig. S3, A and B). Unfortunately, we could not generate a meaningful high-resolution structure using this sample. Thus, we developed a mouse monoclonal antibody 8B1 against purified Sialin. The final sample is the complex of Sialin-nanodisc and Fab fragment from 8B1, which has a size of ~160 kDa, well above the current size limit for cryo-EM studies (fig. S3, A and C). With the Sialin-nanodisc-Fab complex, we obtained a final reconstruction at 3.4-Å resolution and carried out the de novo model building (Fig. 1, C and D, and figs. S4 and S5). There are three patches of residues unresolved in Sialin: N-terminal residues 1 to 31, long luminal loop (L2) between TM1 and TM2 containing residues 69 to 101, and C-terminal residues 489 to 496, because of their poor experimental densities. The overall architecture of Sialin resembles a canonical fold for the major facilitator superfamily with 12 TMs. These TMs are grouped into two distinct domains: N-domain with TM1 to 6 and C-domain with TM7 to 12. The N- and C-domains are connected by a cytosolic loop (L7) between TM6 and TM7. Notably, in this loop, a well-defined small cytosolic helix is sitting right beneath the N-domain with delicate interactions that will be discussed later.
Putative substrate-binding pocket
We inspected the central translocation pathway between the N- and C-domains using the software MoleOnline (30). The path is open to the cytosolic side and closed at the center around residues Y301, N302, F305, Y306, Y54, and R57 (Fig. 2, A and B). Upward from here to the luminal side (or extracellular side), the pathway is entirely sealed by L309 as the pore radius drops to 0.1 Å. Even considering the side chain flexibility, the free radius at this position is narrower than 1.2 Å. Thus, the determined structure adopts an inward-facing conformation. To understand the substrate binding mechanism, we tried to obtain the complex of Neu5Ac and Sialin but failed. Using microscale thermophoresis, we measured the binding between Neu5Ac and purified Sialin in both detergent and nanodisc made of soybean polar extract lipids. The result shows no detectable binding under any pH conditions. We tested the transporter function in proteoliposomes to rule out the possibility of somehow damaging the purified Sialin. Specifically, we reconstituted the purified Sialin into liposomes at pH 7.4. Then, we added [3H]Neu5Ac and dropped the pH outside the proteoliposomes to 5.6. We detected strong substrate transport, confirming that the purified Sialin is functional (fig. S6). This result is consistent with previous studies by other groups (19, 21, 22, 31). Then, why does Sialin not bind Neu5Ac in vitro? We carefully examined the central channel and found a constriction site on the cytosolic side surrounded by residues H183, R195, L415, and A422 (Fig. 2, A and C). At this position, the diameter of the central pathway is constricted to ~3.5 or ~5.5 Å, when considering side-chain flexibility, while the size of Neu5Ac is roughly 6 Å by 12 Å by 12 Å, which is too big to go through the constriction site to reach the substrate-binding pocket. This fact has two implications: (i) After each transport, the substrate will not be able to reenter the substrate-binding pocket from the cytosol to trigger a reversed translocation event. (ii) Since Sialin in proteoliposome can transport Neu5Ac, it is reasonable to suggest that the pH gradient, H+ cotransport, or both are critical to promoting subtle conformational changes at the constriction site to allow Neu5Ac to go through and be released to the cytosol.
Fig. 2. Putative substrate-binding pocket in the partially open inward-facing Sialin.
(A) Sialin (brown) is in the inward-facing conformation with the central pathway shown (blue tube). The pathway’s diameter (blue) and free diameter (orange, treating side chains as flexible) are plotted side-by-side. (B) The narrowest position around L309 in the path. (C) The cytosolic constriction site around H183. (D) The top-scored pose of docked Neu5Ac (ball and stick) in the putative substrate-binding pocket interacts with surrounding residues. The dashed lines are pseudo bonds with a length of 2.5 to 3.3 Å.
To elucidate residues critical for the substrate interactions, we computationally docked a Neu5Ac molecule into the inward-facing partially open Sialin structure using Autodock Vina (32, 33). The program’s top 5 scored poses were found at the narrowest position in the central cavity as previously described (Figs. 2, A and B), suggesting a putative substrate-binding pocket in Sialin (Fig. 2D and fig. S7). The pocket is surrounded by a network of polar and charged residues, including Y54, R57, Y119, H298, Y301, N302, F305, Y306, S411, and N430, that can form hydrogen bonds and salt bridges with Neu5Ac. Specifically, using relaxed constraints of 0.4-Å distance and 20° angle, the top-scored pose forms multiple hydrogen bonds with surrounding polar residues (Y54, N302, Y306, Y119, and N430) as well as salt bridges with R57 (Fig. 2D). Most of these residues are highly conserved in Sialin homologs (fig. S8). For example, in two known structures of Sialin homologs, d-galactonate/H+ symporter from Escherichia coli (EcDgoT) (34) and vesicular glutamate transporter 2 from Rattus norvegicus (rVGLUT2) (35), R47 in EcDgoT and R88 in rVGLUT2 are equivalent to R57 in Sialin (figs. S8 and S9). These arginines are known to interact with a carboxyl group in respective substrates electrostatically. As expected, for Sialin, the transport efficiency of the R57A mutant is only ~10% compared to the wild type, confirming its substrate-binding function (Fig. 3B and fig. S10).
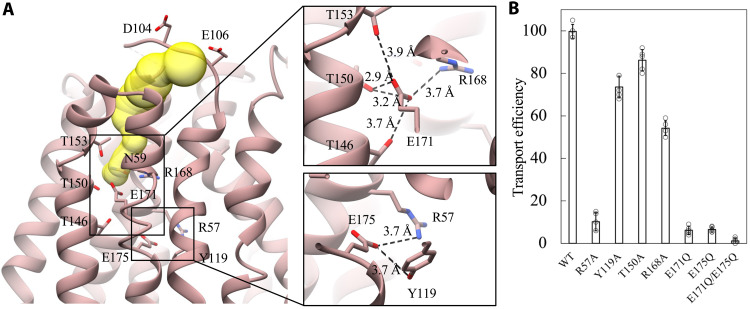
Fig. 3. Critical H+ coupling residues E171 and E175.
(A) The lumen-accessible tunnel in N-domain (yellow, same view as in Fig. 2A). The hydrogen bonds and charged interactions around E171 and E175 are zoomed in. (B) The transport efficiency of mutants measured in High Five cells using [3H]Neu5Ac is compared with that of wild-type Sialin.
Proton coupling
Secondary transporters move substrates across the cellular membrane using energy stored in the electrochemical gradient, such as the H+ gradient (36). To sense or cotransport H+, acidic residues (Asp and Glu) are often required and delicately positioned in membrane transporters. In EcDgoT, both D46 and E133 (equivalent to L56 and E175 in Sialin, respectively) are exposed to the luminal solution and capable of reversible protonation to carry out the symport of H+/D-galactonate (figs. S8 and S11A) (34), while in rVGLUT2, H128 and E191 (equivalent to L112 and E175 in Sialin) are proposed H+ binding sites to activate substrate transport without H+ efflux allosterically (figs. S8 and S11B) (35). These residues are all located in a lumen-accessible tunnel in the N-domains of EcDgoT and rVGLUT2. As expected, in Sialin, there is a similar tunnel that starts from residues on L2 (D104 and E106), goes through multiple polar residues (T153, N59, and R168) on TMs, and ends around T150 and E171 (Fig. 3A). Thus, E175 is completely buried and not solvent accessible to the tunnel, which is locally different from that of homologs, because, in N-domains of EcDgoT and rVGLUT2, the residues equivalent to Sialin’s E175 (E133 and E191, respectively) are readily solvent accessible to the tunnel. In Sialin, the diameter of the lumen-accessible tunnel is 3 to 4.8 Å, allowing full access to water molecules. There is a network of interactions built around E171 and E175. Specifically, E171 seems to be stabilized by interacting with T146, T150, T153, and R168, while E175 is stabilized by interacting with Y119 and R57. Both R168 and R57 are along the central substrate transport pathway. We hypothesize that the protonation/deprotonation status of both E171 and E175 is essential for sensing and cotransporting H+ during sialic acid translocation. To test that, we carried out the cellular transport assay (as in Fig. 1B) with various Sialin mutants. The transport efficiency of each mutant was normalized by its expression level in the High Five cells. The result (Fig. 3B and fig. S10) shows that Y119A, T150A, and R168A still retain most of their substrate transport function, suggesting the moderate importance of these residues. However, R57A is seriously dysfunctional, which is consistent with the fact that R57C is a pathogenic mutation found in patients with infantile sialic acid storage disorder (37). There are two reasons why R57 is functionally essential: (i) It is responsible for substrate recognition, and (ii) its interaction with E175 is critical for H+ coupling (in Discussion). In addition, we found that both E171Q and E175Q lose ~95% of their transport function, while the E171Q/E175Q double mutant has no detectable substrate transport. Thus, we concluded that E171 and E175 are equally critical for H+ coupling and therefore for the substrate transport mediated by Sialin.
Mapping the pathogenic mutations
With the high-resolution structure of Sialin, we start to understand why pathogenic mutations result in dysfunctional proteins. According to the ClinVar database, more than 380 mutations in the SLC17A5 gene are documented in patients with free sialic acid storage disorders (accessed August 2022). In this study, we do not discuss genetic mutations that either result in premature termination of protein translation or do not affect protein translation at all. Instead, we will focus on mutations of specific amino acids that still produce Sialin, particularly the 10 mutations that are clinically important (designated as “pathogenic”) (table S2). These pathogenic mutations (Fig. 4A) could be roughly grouped into four classes. The first class contains G328E, P334R, and G409E (fig. S12A). G328 on TM8 is positioned directly against G218 on TM5 and surrounded by hydrophobic side chains of V62, I219, L214, and P215. P334 on TM8 fits tightly in the small cavity among L396, T397, and T400. G409 is the kink on TM10 surrounded by F367, T368 from TM9, F474 from TM12, and A349 from TM8. Structurally, it is understandable that replacing any G328, P334, and G409 with large, charged residues (Glu and Arg) is energetically unfavorable for the tight helical packing in Sialin. Thus, as demonstrated in previous functional studies, G328E, P334R, and G409E are all loss-of-function pathogenic mutants (table S2). K136E represents the second class of pathogenic mutations that interferes with transporter-lipid interactions (fig. S12B). In our structure, the side chain of K136 is very well defined by the experimental density and does not form any hydrogen bond or salt bridge with surrounding residues in Sialin. However, K136 seems to connect with a lipid-like density and most likely interacts with the negatively charged phosphate in phospholipids. K136E reverses the charge of the side chain from positive to negative, resulting in destabilized transporter-lipid interactions that likely impair the transporter function. Previous studies have shown that K136E has only 10 to 55% of the wild-type activity (table S2). It is worth noting that different cells and lipid systems were used in these studies. We speculate that subtle differences in the lipid composition may be why the measured activity of K136E covers a relatively large range. The third class includes R57C and H183R, which completely abolishes sialic acid transport (table S2). We hypothesize that these mutations directly impair the substrate-transporter interactions (Figs. 2 and 4). For R57, it is likely the most critical residue for substrate recognition. Thus, in R57C, the positively charged side chain is lost, resulting in lower substrate binding efficiency. Meanwhile, R57C loses the ability to interact with E175, resulting in less efficient H+ coupling. For H183, it forms the cytosolic constriction site together with R195, L415, and A422. H183 is most likely neutral at physiological pH as the side chain pKa is predicted to be ~3.6 (38). Thus, in H183R, the constriction site would be more positively charged and even narrower as Arg is slightly larger than His. The two effects could cooperatively trap negatively charged substrate Neu5Ac around the constriction site and effectively block the substrate release. The fourth class of mutations includes R39H, R39C, E262D, and ΔSSLRN (268 to 272) (Fig. 4B). It seems to be the most interesting class, as these mutants affect the relative structural stability between the cytosolic helix and N-domain. In our structure, the cytosolic helix (residues 264 to 272) is stably localized below the N-domain by strong interactions between three charged residues: R39, E262, and E264. In addition, P191 and P192 may also contribute to the stable localization with CH/π interaction with Y265 in the helix (39). However, in the cryo-EM map, the side chain of Y265 is not visible due to the quality of experimental density. The fixed localization of the cytosolic helix is essential, as it ensures that the following loop (between the cytosolic helix and TM7) is physically away from the center of the translocation pathway to maintain the solvent accessibility (Fig. 4C). In the ΔSSLRN mutant, the cytosolic helix is shortened but may still be correctly anchored under the N-domain, because the essential interactions remain. However, the distance between TM7 (and C-domain) and the N-domain needs to be closer to accommodate the shortened amino acid sequence, resulting in a narrower central translocation pathway that completely abolishes substrate transport (table S2). Here, we performed a cellular transport assay with R39H and E262D mutants. The result shows that R39H retains ~28 ± 3%, and E262D has only ~19 ± 2% of the transport activity compared to wild-type Sialin (table S2). The side chains in R39H, R39C, and E262D mutants are likely too small to form correct hydrogen bonds to stabilize the cytosolic helix. Thus, the whole L7 loop becomes flexible and could potentially cover the central pathway for substrate translocation, which explains the mild loss of transport function.
Fig. 4. Pathogenic mutations mapped onto Sialin.
(A) Nine residues (purple) corresponding to 10 pathogenic mutations are marked in the cartoon structure of Sialin. (B) The cytosolic helix is stably localized by interactions among corresponding residues. (C) L7 between TM6 and TM7 is physically away from the central pathway (marked by Δ).
DISCUSSION
As a secondary active transporter, Sialin functions through an “alternating access mechanism” in which the substrate-binding pocket is solvent accessible from one side of the membrane or the other when the transporter conformation alters between inward-facing and outward-facing (40, 41). The cryo-EM structure of Sialin presented in this study shows an inward-facing conformation with a cytosolic gate around H183 partially open. The gate does not allow substrate Neu5Ac to go through, which is advantageous to prevent Neu5Ac in the cytosol from being transported back into lysosomes. To understand the conformational change, we compared our Sialin structure with the AlphaFold predicted version (AF-Q9NRA2-F1) (42). As expected, the N-domain and C-domain of the predicted structure could be superimposed with their counterparts in our structure well with an RMSD of 0.737 and 0.698 Å, respectively (fig. S13A). However, as a whole, the two structures do not overlap with each other, indicating that AF-Q9NRA2-F1 may adopt a different conformation. We analyzed the central pore in AF-Q9NRA2-F1 by MoleOnline and attempted computational docking with Autodock Vina. The result shows that (i) the docked Neu5Ac does not sit in the putative substrate-binding pocket; instead, it remains close to the luminal opening; (ii) the substrate-binding pocket around R57 in the middle of the transporter is solvent accessible to the luminal side; and (iii) the translocation pathway on the cytosolic side is entirely closed by H183, R195, L415, and L199 (fig. S13B). Thus, we concluded that the AlphaFold predicted Sialin structure AF-Q9NRA2-F1 likely in the outward-facing partially open conformation. It remains to be seen whether this conformation is physiologically relevant or not. However, it confirms the existence of the cytosolic gate around H183 and, thus, the pathological effect of the H183R mutant.
On the basis of the functional and structural analysis, here, we propose a sialic acid/H+ cotransport mechanism (Fig. 5). Let us start the transport cycle with Sialin in the outward-facing conformation (state 1). This state is likely similar to the predicted AlphaFold structure. However, since the AlphaFold structure is only partially open, it has to fully open the luminal side to allow the substrate to enter the central pathway. So far, we do not know if such an opening is regulated by a luminal gate or a more substantial structural change within the transporter. Because the N-domain most likely functions as a rigid body, E171 shall interact with R168, and E175 shall interact with R57, as found in our inward-open conformation. Here, to probe the protonation/deprotonation status of E171 and E175, we used multiple pKa prediction servers (38, 43, 44). The pKa of E171 is predicted to be between 5.6 and 8.2, while the pKa of E175 is ~3.0. Considering the physiological environment (pH ~5.6 in the lysosomal lumen and ~7.4 in the cytosol), it is most likely that E171 balances between the protonated and deprotonated states. At the same time, E175 strongly favors the deprotonated form. Sialin changes from state 1 to state 2 when sialic acid is recognized and moves into the central pathway. Because sialic acid has a strong negative charge (pKa ~2.6), it will likely interact with the R168 side chain and thus break the E171-R168 interaction. Then, the free E171 side chain can be easily protonated. As the sialic acid moves down and reaches the substrate-binding pocket, Sialin changes from state 2 to state 3. Sialic acid interacts with R57 and breaks the E175-R57 interaction to free E175. Here, H+ on the E171 side chain can be freely transferred onto E175 because of the proximity of the two residues (one helical turn away). The deprotonated E171 restores its interaction with R168, destabilizing the transient R168–sialic acid interaction and promoting the R57–sialic acid interaction in the pocket. Next, Sialin will change from state 3 to state 4 to release both H+ and sialic acid into the cytosol. For the transferred H+, the release process shall be easy because E175’s low pKa favors a deprotonated state under cytosolic pH. For sialic acid, the release process may be complicated and involve many residues around the binding pocket. For example, polar residues, such as N430, N302, Y54, and Y119 (Fig. 2D), may provide electrostatic force to move the sialic acid away from R57. The sugar ring in sialic acid could interact with nearby hydrophobic residues. In addition, deprotonated E175 will likely rebind R57. All these possible interactions probably act coordinately to remove the sialic acid from the substrate-binding pocket. Last, Sialin could reset itself to state 1, the outward-facing conformation. The structure determined here represents Sialin at state 3 but without sialic acid.
Fig. 5. Proposed model of sialic acid/H+ cotransport by Sialin.
In the outward-facing conformation (state 1), R168 interacts with E171, and R57 interacts with E175. When the sialic acid is recognized (state 2), it likely interacts with R168 and frees E171 to be pronated. As the substrate moves down the translocation pathway (state 3), it interacts with R57 and frees E175. E171 then transfers the H+ to E175. As E171 rebonds with R168, the substrate moves further down. In the inward-facing conformation (state 4), since E175 prefers a deprotonation state (pKa ~3.0), the H+ is quickly released to the cytosol. E175 rebonds with R57, which promotes the substrate release from the binding pocket. Sialin is then reset to the outward-facing conformation for another cycle.
In summary, our structure provides an excellent start to understanding the sialic acid/H+ cotransport mediated by Sialin. However, there are still many open questions regarding the molecular mechanism. For example, how does Sialin open the H183 constriction site to release the substrate? How does the transporter switch from inward-facing to outward-facing? Future studies in biophysics, structural biology, and molecular dynamics simulations will be necessary to answer these questions.
MATERIALS AND METHODS
Protein expression and purification
The full-length human Sialin (UniProt: Q9NRA2) was cloned into a pFastBac vector (Thermo Fisher Scientific) with an N-terminal His-tag and a thrombin digestion site. Three predicted N-linked glycosylation sites were abolished by site-directed mutagenesis (N71A, N77A, and N95A). Sialin was overexpressed in High Five cells using the Bac-to-Bac Baculovirus Expression System (Thermo Fisher Scientific). Cells were harvested by centrifugation 72 hours after virus infection and lysed by passing through a microfluidizer M110P (Microfluidics Corporation). The membrane fraction was collected by centrifugation at 150,000g for 1 hour and resuspended in buffer A [20 mM tris (pH 7.4) and 150 mM NaCl]. Lauryl maltose neopentyl glycol (LMNG; 1%) (Anatrace) was used to solubilize the membrane at 4°C for 2 hours. The supernatant was isolated by centrifugation at 150,000g for 1 hour and incubated with TALON IMAC resin (Clontech) in buffer A with 5 mM imidazole. The resin was washed with buffer B [20 mM tris (pH 7.4), 150 mM NaCl, and 0.003% LMNG] with 10 mM imidazole, and then the protein was eluted in buffer B with 200 mM imidazole. The eluted protein was digested with thrombin (Enzyme Research Laboratories) at a molar ratio of 1:50 overnight at 4°C. Sialin was further purified by gel filtration chromatography with a Superdex 200 column (Sigma-Aldrich) in buffer B and concentrated to ~5 mg/ml for storage.
Fab generation
Monoclonal antibodies against Sialin were generated in mice using purified protein in detergent as the antigen in the Monoclonal Antibody Core Laboratory at the Oregon Health & Science University (45). Antibody 8B1 was selected as it had the strongest binding affinity to Sialin when tested by Western blot and ELISA. The Fab was produced by papain digestion and purified by protein A affinity chromatography (Thermo Fisher Scientific).
Cryo-EM sample preparation
To overcome the challenge presented by the small size of Sialin (~55 kDa), we first reconstituted the purified protein into lipid nanodiscs. We then formed a larger complex with Fab generated from the 8B1 antibody. Briefly, membrane scaffold protein MSP1D1 (Addgene) was purified as described previously (46). Purified Sialin in detergent was mixed with MSP1D1 and soybean polar extract lipids (Anatrace) at a molar ratio of 1:8:400 on ice for 30 min. Prewetted Bio-Beads (0.2 mg; Bio-Rad Laboratories) was added for overnight incubation at 4°C. The mixture was purified by gel filtration with a Superose 6 column in buffer A, incubated with Fab at a molar ratio of 1:2, and then purified by gel filtration. The final complex of Sialin-nanodisc-Fab was concentrated to ~2.5 mg/ml.
Cryo-EM data collection
Three microliters of the freshly purified complex was applied to a plasma-cleaned C-flat holy carbon grid (1.2/1.3, 400 mesh, Electron Microscopy Sciences) and prepared using a Vitrobot Mark IV (Thermo Fisher Scientific) with the environmental chamber set at 100% humidity and 4°C. The grid was blotted for ~3 s and then flash-frozen in liquid ethane. The data were collected on a Titan Krios (Thermo Fisher Scientific) operated at 300 keV and equipped with a K3 direct detector (Gatan). A total of 10,235 movies were recorded with a calibrated pixel size of 0.826 Å, a defocus range of −1 to −2.5 μm, and 50 frames with a total dose of ~60 electrons/Å2.
Image processing, model building, and refinement
The data were processed with cryoSPARC (47). Patch motion correction was used to correct the beam-induced movement, and patch contrast transfer function was used to estimate contrast transfer function parameters for each movie. A total of ~3 million particles were automatically picked using “Blob Picker” and extracted with a box size of 320 × 320 pixels. After two rounds of reference-free 2D classification, ab initio reconstruction, and heterogeneous refinement, ~800,000 particles were selected for further processing. 3D classification without alignment was performed with the following parameters: five classes, 6-Å target resolution, and principal component analysis initiation mode. The final reconstruction was obtained with a particular class of 394,078 particles. After homogenesis refinement and nonuniform refinement, the map reached a resolution of ~3.7 Å. A soft mask around Sialin and half of the Fab was generated for a final round of local refinement to produce the 3.4-Å-resolution map. The model was built in Coot (48). The AlphaFold-predicted model of Sialin was used as a guide together with several secondary structure prediction programs: Jpred (49) and PSSpred (50). The final model contains residues 32 to 68 and 102 to 488, missing flexible N-termini, C-termini, and the luminal loop between TM2 and TM3. The model was refined in PHENIX (51). The quality of the model was assessed by MolProbity (52). Statistical details can be found in table S1. All superpositions of structures were calculated in the program UCSF Chimera with the alignment algorithm of Needleman-Wunsch and BLOSUM-62 matrix (53).
Cellular transport assay
Sialic acid uptake was measured in High-Five cells expressing wild-type and various mutations of Sialin. Specifically, 200 μl of High-Five cells (0.4 × 106 count/ml) was seeded in 24-well plates in Grace’s Insect Medium supplemented with fetal bovine serum and infected with the corresponding baculoviruses for 24 hours. Cells were washed twice with buffer C [20 mM MES (pH 5.6), 5 mM glucose, 150 mM NaCl, and 1 mM MgSO4] and then incubated with [3H]Neu5Ac (0.05 μCi, assumed to be 10 nM in 250 μl of buffer C) for 15 min at room temperature. After washing twice with ice-cold buffer at pH 7.4, the radioactivity in the cells was counted by liquid scintillation counting using a PerkinElmer Tri-Carb 2910 TR machine. Each transport was measured five times independently. The expression level of wild-type and mutant Sialin in High-Five cells was determined by Western blot analysis, which was used to normalize the substrate transport measurements. The transported amount of [3H]Neu5Ac was plotted in Fig. 1B, and the transport efficiency of mutants was calculated against that of wild type in percentage in Fig. 3B.
Proteoliposome transport assay
The experiment followed a previously published protocol (54). Briefly, 10 mg of Soybean polar extract lipids (Avanti Polar Lipids) were dissolved in chloroform, dried by nitrogen gas, and immediately resuspended in Buffer A to a final concentration of 10 mg/ml. The large unilamellar liposome vesicles were made by extruding the suspension through a 400-nm polycarbonate membrane filter using a mini extruder (Avanti Polar Lipids). To reconstitute proteoliposomes, the liposomes were destabilized by 0.015% Triton X-100 and then incubated with purified Sialin at a ratio of 150:1 (wt/wt) for 20 min at room temperature. Bio-Beads SM-2 (150 mg; Bio-Rad Laboratories) were added to the mixture to absorb all detergents overnight at 4°C. The reconstituted proteoliposomes were harvested by ultracentrifugation at 180,000g for 20 min and then resuspended in buffer D [20 mM MES (pH 5.6) and 150 mM NaCl]. Proteoliposome solution (100 μl) for each transport was incubated with 10 nM [3H]Neu5Ac at room temperature. At different time points, the proteoliposomes were filtered and washed with ice-cold Buffer A. The filter membrane was scintillation-counted. Each transport was repeated five times. The transported amount was calculated with the assumption that the orientation of reconstituted Sialin was 50:50.
Immunofluorescence staining and microscopy imaging
High Five cells with or without His-tagged Sialin expression were fixed in 2% paraformaldehyde for 30 min at room temperature. After washing three times with PBS, cells were permeabilized with 0.2% Triton X-100 for 30 min. Unpermeabilized or permeabilized cells were blocked in PBS containing 10% horse serum for 30 min at room temperature. Cells were then incubated with primary antibodies against 6×His (1:500; UBPBio) or anti-Sialin monoclonal antibody 8B1 (1:2000) in 5% horse serum in PBS for 1 hour at room temperature. Cells were washed three times in PBS before costained with secondary antibody (1:800; Molecular Probes Alexa Fluor 555) and nuclei dye Hoechst (1:5000; Molecular Probes) or 4′,6-diamidino-2-phenylindole (Sigma-Aldrich; 1 μg/ml) for 1 hour at room temperature. Stained cells were washed three times in PBS, followed by microscopy imaging with an Olympus FV1000 FCS/RICS confocal microscope or a Keyence BZ-X710 All-in-One Fluorescence Microscope.
Acknowledgments
We thank the staff in the Pacific Northwest Center, especially T. Humphreys, for the cryo-EM data collection. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. We also thank J. Kieft for the critical reading and suggestions.
Funding: This work was supported by NIH (R01 GM126626, R21 AG064572, R01HL133230, and R01HL159086).
Author contributions: W.H., C.C., K.S., and H.Z. designed the experiments, collected and analyzed the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The cryo-EM map of Sialin was deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/emdb/) under the accession code EMD-27755. Coordinates of the atomic model were deposited in the Protein Data Bank (https://www.rcsb.org/) under the accession code 8DWI. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.D. Adams, M. Wasserstein, in GeneReviews((R)), M. P. Adam, H. H. Ardinger, R. A. Pagson, S. E. Wallace, L. J. H. Bean, K. W. Gripp, G. M. Mirzaa, A. Amemiya, Eds. (University of Washington, Seattle, 1993). [PubMed]
- 2.T. Angata, A. Varki, Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary perspective. Chem. Rev. 102, 439–470 (2002). [DOI] [PubMed] [Google Scholar]
- 3.C. Traving, R. Schauer, Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 54, 1330–1349 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. Schauer, J. P. Kamerling, Exploration of the sialic acid world. Adv. Carbohydr. Chem. Biochem. 75, 1–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Ghosh, in Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease, S. Ghosh, Ed. (Academic Press, 2020), pp. 1–61. [Google Scholar]
- 6.O. Basaran, A. Dei Giudici, M. Federici, F. Versaci, Sialic acid: An important contributor to cardiovascular risk. Minerva Cardiol. Angiol. 69, 477–479 (2021). [DOI] [PubMed] [Google Scholar]
- 7.C. Bull, M. A. Stoel, M. H. den Brok, G. J. Adema, Sialic acids sweeten a tumor’s life. Cancer Res. 74, 3199–3204 (2014). [DOI] [PubMed] [Google Scholar]
- 8.M. S. Macauley, P. R. Crocker, J. C. Paulson, Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Mazzanti, R. A. Rabini, E. Salvolini, M. Tesei, D. Martarelli, B. Venerando, G. Curatola, Sialic acid, diabetes, and aging: A study on the erythrocyte membrane. Metabolism 46, 59–61 (1997). [DOI] [PubMed] [Google Scholar]
- 10.H. H. Freeze, G. W. Hart, R. L. Schnaar, in Essentials of Glycobiology, A. Varki, R. D. Cummings, P. Stanley, M. Aebi, A. G. Darvill, Taroh Kinoshita, N. H. Packer, J. H. Prestegard, P. H. Seeberger, Eds. (Cold Spring Harbor Laboratory Press, ed. 3, 2015), pp. 51–63. [PubMed] [Google Scholar]
- 11.C. Reily, T. J. Stewart, M. B. Renfrow, J. Novak, Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.F. W. Verheijen, E. Verbeek, N. Aula, C. E. M. T. Beerens, A. C. Havelaar, M. Joosse, L. Peltonen, P. Aula, H. Galjaard, P. J. van der Spek, G. M. S. Mancini, A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat. Genet. 23, 462–465 (1999). [DOI] [PubMed] [Google Scholar]
- 13.M. Huizing, M. E. Hackbarth, D. R. Adams, M. Wasserstein, M. C. Patterson, S. U. Walkley, W. A. Gahl, D. R. Adams, K. Dobrenis, J. Foglio, W. A. Gahl, B. Gasnier, M. Hackbarth, M. Huizing, M. Lek, M. C. V. Malicdan, L. E. Paavola, M. C. Patterson, R. Reimer, S. U. Walkley, M. Wasserstein, R. Y. Wang, R. Zoncu, Free sialic acid storage disorder: Progress and promise. Neurosci. Lett. 755, 135896 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.P. Aula, K. Raivio, S. Autio, C. E. Thoden, J. Rapola, S. L. Koskela, I. Yamashina, Four patients with a new lysosomal storage disorder (Salla Disease). Monogr. Hum. Genet. 10, 16–22 (1978). [DOI] [PubMed] [Google Scholar]
- 15.R. Kleta, R. P. Morse, E. Orvisky, D. Krasnewich, J. Alroy, A. A. Ucci, I. Bernardini, D. A. Wenger, W. A. Gahl, Clinical, biochemical, and molecular diagnosis of a free sialic acid storage disease patient of moderate severity. Mol. Genet. Metab. 82, 137–143 (2004). [DOI] [PubMed] [Google Scholar]
- 16.E. Lemyre, P. Russo, S. B. Melançon, R. Gagné, M. Potier, M. Lambert, Clinical spectrum of infantile free sialic acid storage disease. Am. J. Med. Genet. 82, 385–391 (1999). [PubMed] [Google Scholar]
- 17.R. J. Reimer, SLC17: A functionally diverse family of organic anion transporters. Mol. Aspects Med. 34, 350–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. P. Alexander, E. Kelly, A. Mathie, J. A. Peters, E. L. Veale, J. F. Armstrong, E. Faccenda, S. D. Harding, A. J. Pawson, C. Southan, J. A. Davies, L. Amarosi, C. M. H. Anderson, P. M. Beart, S. Broer, P. A. Dawson, B. Hagenbuch, J. R. Hammond, K.-I. Inui, Y. Kanai, S. Kemp, G. Stewart, D. T. Thwaites, T. Verri, The concise guide to pharmacology 2021/22: Transporters. Br. J. Pharmacol. 178, S412–S513 (2021). [DOI] [PubMed] [Google Scholar]
- 19.T. Miyaji, N. Echigo, M. Hiasa, S. Senoh, H. Omote, Y. Moriyama, Identification of a vesicular aspartate transporter. Proc. Natl. Acad. Sci. U.S.A. 105, 11720–11724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. E. Herring, K. Silm, R. H. Edwards, R. A. Nicoll, Is aspartate an excitatory neurotransmitter? J. Neurosci. 35, 10168–10171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.T. Miyaji, H. Omote, Y. Moriyama, Functional characterization of vesicular excitatory amino acid transport by human sialin. J. Neurochem. 119, 1–5 (2011). [DOI] [PubMed] [Google Scholar]
- 22.J. Lodder-Gadaczek, V. Gieselmann, M. Eckhardt, Vesicular uptake of N-acetylaspartylglutamate is catalysed by sialin (SLC17A5). Biochem. J. 454, 31–38 (2013). [DOI] [PubMed] [Google Scholar]
- 23.L. Qin, X. Liu, Q. Sun, Z. Fan, D. Xia, G. Ding, H. L. Ong, D. Adams, W. A. Gahl, C. Zheng, S. Qi, L. Jin, C. Zhang, L. Gu, J. He, D. Deng, I. S. Ambudkar, S. Wang, Sialin (SLC17A5) functions as a nitrate transporter in the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 13434–13439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. O. Lundberg, E. Weitzberg, M. T. Gladwin, The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167 (2008). [DOI] [PubMed] [Google Scholar]
- 25.P. Morin, C. Sagne, B. Gasnier, Functional characterization of wild-type and mutant human sialin. EMBO J. 23, 4560–4570 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. C. Wreden, M. Wlizla, R. J. Reimer, Varied mechanisms underlie the free sialic acid storage disorders. J. Biol. Chem. 280, 1408–1416 (2005). [DOI] [PubMed] [Google Scholar]
- 27.T. R. Davis, K. M. Trotter, R. R. Granados, H. A. Wood, Baculovirus expression of alkaline phosphatase as a reporter gene for evaluation of production, glycosylation and secretion. Biotechnology (N. Y) 10, 1148–1150 (1992). [DOI] [PubMed] [Google Scholar]
- 28.T. J. Wickham, T. Davis, R. R. Granados, M. L. Shuler, H. A. Wood, Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus expression system. Biotechnol. Prog. 8, 391–396 (1992). [DOI] [PubMed] [Google Scholar]
- 29.T. K. Ritchie, Y. V. Grinkova, T. H. Bayburt, I. G. Denisov, J. K. Zolnerciks, W. M. Atkins, S. G. Sligar, Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. Pravda, D. Sehnal, D. Toušek, V. Navrátilová, V. Bazgier, K. Berka, R. Svobodová Vařeková, J. Koča, M. Otyepka, MOLEonline: A web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.L. Dubois, N. Pietrancosta, A. Cabaye, I. Fanget, C. Debacker, P. A. Gilormini, P. M. Dansette, J. Dairou, C. Biot, R. Froissart, A. Goupil-Lamy, H. O. Bertrand, F. C. Acher, I. McCort-Tranchepain, B. Gasnier, C. Anne, Amino acids bearing aromatic or heteroaromatic substituents as a new class of ligands for the lysosomal sialic acid transporter sialin. J. Med. Chem. 63, 8231–8249 (2020). [DOI] [PubMed] [Google Scholar]
- 32.O. Trott, A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. Eberhardt, D. Santos-Martins, A. F. Tillack, S. Forli, AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. B. Leano, S. Batarni, J. Eriksen, N. Juge, J. E. Pak, T. Kimura-Someya, Y. Robles-Colmenares, Y. Moriyama, R. M. Stroud, R. H. Edwards, Structures suggest a mechanism for energy coupling by a family of organic anion transporters. PLOS Biol. 17, e3000260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.F. Li, J. Eriksen, J. Finer-Moore, R. Chang, P. Nguyen, A. Bowen, A. Myasnikov, Z. Yu, D. Bulkley, Y. Cheng, R. H. Edwards, R. M. Stroud, Ion transport and regulation in a synaptic vesicle glutamate transporter. Science 368, 893–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O. Boudker, G. Verdon, Structural perspectives on secondary active transporters. Trends Pharmacol. Sci. 31, 418–426 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Ruivo, A. Sharifi, S. Boubekeur, P. Morin, C. Anne, C. Debacker, J. C. Graziano, C. Sagné, B. Gasnier, Molecular pathogenesis of sialic acid storage diseases: Insight gained from four missense mutations and a putative polymorphism of human sialin. Biol. Cell 100, 551–559 (2008). [DOI] [PubMed] [Google Scholar]
- 38.R. Anandakrishnan, B. Aguilar, A. V. Onufriev, H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. J. Zondlo, Aromatic-proline interactions: Electronically tunable CH/π interactions. Acc. Chem. Res. 46, 1039–1049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.P. Mitchell, A general theory of membrane transport from studies of bacteria. Nature 180, 134–136 (1957). [DOI] [PubMed] [Google Scholar]
- 41.O. Jardetzky, Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966). [DOI] [PubMed] [Google Scholar]
- 42.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. H. Olsson, C. R. Sondergaard, M. Rostkowski, J. H. Jensen, PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 44.S. Pahari, L. Sun, S. Basu, E. Alexov, DelPhiPKa: Including salt in the calculations and enabling polar residues to titrate. Proteins 86, 1277–1283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. Zheng, G. Wisedchaisri, T. Gonen, Crystal structure of a nitrate/nitrite exchanger. Nature 497, 647–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Z. Wang, W. Hu, H. Zheng, Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter. Sci. Adv. 6, eaay7997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 48.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. Drozdetskiy, C. Cole, J. Procter, G. J. Barton, JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. Yan, D. Xu, J. Yang, S. Walker, Y. Zhang, A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci. Rep. 3, 2619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkóczi, V. B. Chen, T. I. Croll, B. Hintze, L. W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell, T. C. Terwilliger, A. G. Urzhumtsev, L. L. Videau, C. J. Williams, P. D. Adams, Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.V. B. Chen, W. B. Arendall III, J. J. Headd, D. A. Keedy, R. M. Immormino, G. J. Kapral, L. W. Murray, J. S. Richardson, D. C. Richardson, MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 54.E. R. Geertsma, N. A. Nik Mahmood, G. K. Schuurman-Wolters, B. Poolman, Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008). [DOI] [PubMed] [Google Scholar]
- 55.N. J. Myall, C. C. Wreden, M. Wlizla, R. J. Reimer, G328E and G409E sialin missense mutations similarly impair transport activity, but differentially affect trafficking. Mol. Genet. Metab. 92, 371–374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 and S2
References