Abstract
Maintaining genomic stability is crucial for embryonic stem cells (ESCs). ESCs with unrepaired DNA damage are eliminated through differentiation and apoptosis. To date, only tumor suppressor p53 is known to be implicated in this quality control process. Here, we identified a p53-independent quality control factor lncRNA NONMMUT028956 (Lnc956 for short) in mouse ESCs. Lnc956 is prevalently expressed in ESCs and regulates the differentiation of ESCs after DNA damage. Mechanistically, Ataxia telangiectasia mutated (ATM) activation drives m6A methylation of Lnc956, which promotes its interaction with Krüppel-like factor 4 (KLF4). Lnc956-KLF4 association sequestrates the KLF4 protein and prevents KLF4’s transcriptional regulation on pluripotency. This posttranslational mechanism favors the rapid shutdown of the regulatory circuitry of pluripotency. Thus, ATM signaling in ESCs can activate two pathways mediated by p53 and Lnc956, respectively, which act together to ensure robust differentiation and apoptosis in response to unrepaired DNA damage.
DNA damage induces ESC differentiation via Lnc956.
INTRODUCTION
Pluripotent stem cells (PSCs) including embryonic stem cells (ESCs) or induced PSCs (iPSCs) are capable of unlimited proliferation and differentiation into all cell types in the body. PSCs have wide applications in biomedical researches and in cell-based regenerative medicine. However, genomic instability and tumorigenicity hamper their full application. Studying the underlying mechanisms is critical to address this issue (1–4). Because of the fundamental roles in organism development, PSCs have super competence to maintain stable genome. For instance, mouse ESCs display 100-fold lower mutation rates than their isogenic embryonic fibroblasts (5). Although the underpinning mechanisms remain largely unknown, previous studies including ours suggested that PSCs use unique strategies to secure high level of genomic integrity. Compared to differentiated somatic cells, PSCs are more efficient to resolve DNA replication stress and prevent the replication-associated DNA damage (6, 7). The DNA damage response and repair processes are also robust and accurate in PSCs (8–10). In addition, telomere lengthening and protection strategies are distinct between PSCs and somatic cells (11–13). In particular, PSCs can undergo rapid apoptosis or differentiation to eliminate cells with unrepaired DNA damage and prevent DNA damage from spreading onto progenies. This genome quality control is regulated by tumor suppressor protein TRP53 (also known as p53). However, the mechanism by which p53 safeguards genomic stability in PSCs is distinct to that in somatic cells. In response to DNA damage in PSCs, the primary function of p53 is to induce stem cell differentiation (14–16). A global examination of p53 targets in mouse ESCs revealed that p53, which acts as a transcription factor, directly activated and suppressed more than 3600 genes in response to DNA damage. Among the p53-activated genes, many are associated with stem cell differentiation. Meanwhile, most of the PSC core transcription factors are suppressed by p53 (e.g., Pou5f1, Nanog, Sox2, Zic3, Jmjd1c, Esrrb, Tcfcp2l1, Utf1, and n-Myc) (14). Thus, through its dual functions to activate differentiation genes and suppress PSC core regulatory factors at transcriptional level, p53 switches off pluripotency on differentiation to control PSC quality and safety.
In addition to p53, whether there is p53-independent genome guardian in PSCs remains unexplored. Pluripotent state is regulated not only at transcriptional level but also at posttranscriptional level of the core pluripotency factors. For instance, the core pluripotency transcription factor Krüppel-like factor 4 (KLF4) shows very high protein stability. Complete depletion of Klf4 transcripts only induces subtle decrease in protein level (17). Notably, protein and transcript levels usually become decoupled at the early phase of dynamic cell fate transition (18), suggesting a vital role of the posttranscriptional regulation in initiating cell fate transition. On the basis of the above knowledge, we speculated that there might be p53-independent posttranscriptional mechanism to facilitate the rapid pluripotency switching off in response to DNA damage. In this study, we report a posttranslational mechanism which acts downstream of Ataxia telangiectasia mutated (ATM) kinase but is parallel to the p53-mediated transcriptional pathway. This posttranslational regulation is mediated by an ESC-specific lncRNA NONMMUT028956 (Lnc956 for short). Upon DNA damage, Lnc956 undergoes m6A methylation that drives Lnc956 to interact with KLF4. Lnc956-KLF4 association sequestrates the KLF4 protein and disrupts its transcriptional regulations on pluripotency, thereby favoring the rapid shutdown of the regulatory circuitry of pluripotency before the protein levels of the core pluripotent transcription factors go decline.
RESULTS
Lnc956 loss compromises ESC differentiation and apoptosis after DNA damage
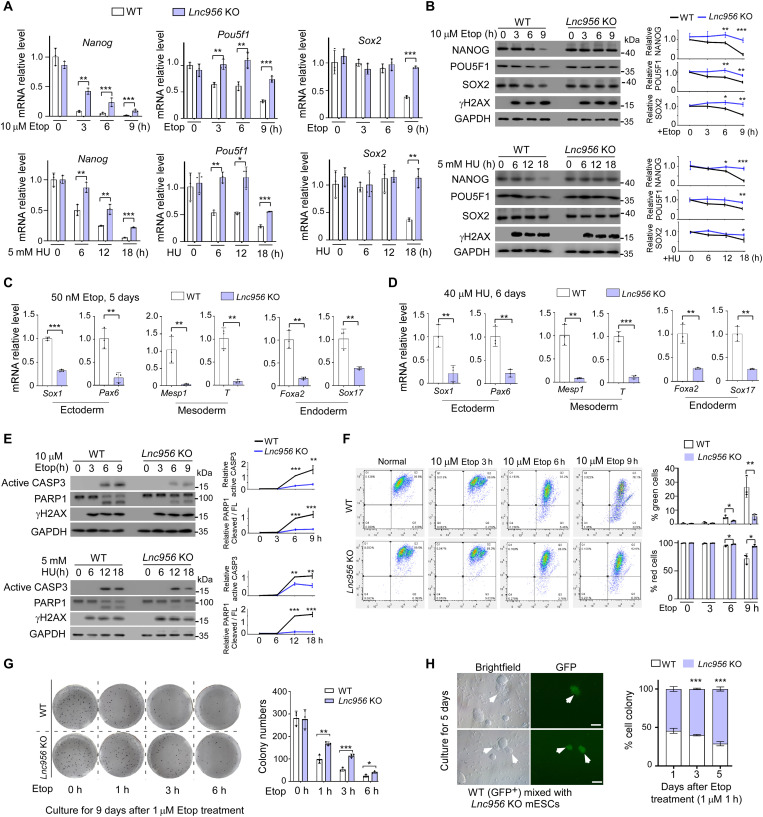
We identified an ESC-specific lncRNA Lnc956, which promoted the stalled replication fork restart and ensured stem cell genomic stability (bioRxiv doi: https://doi.org/10.1101/2022.03.13.484185). Because of the DNA replication deficit, Lnc956 knockout (KO) mouse ESCs, derived from Lnc956 KO mouse blastocysts (fig. S1A), contained more endogenous DNA double-strand breaks (DSBs) (fig. S1B) and proliferated slightly slower than wild-type (WT) counterparts (fig. S1C). However, they showed normal clonal morphology (fig. S1A) and pluripotency marker expressions under unperturbed culture condition (fig. S1D). Considering that DNA damage can induce ESC differentiation, we speculated that Lnc956 KO might render ESCs tolerance to DNA damage. To test this hypothesis, we treated the WT and Lnc956 KO ESCs with high concentration of etoposide (19) and hydroxyurea (HU) to induce severe DNA DSBs and DNA replication stress, respectively (fig. S2, A and B). Under DNA damage condition, ESCs exit pluripotency before initiating differentiation. We first examined the pluripotency markers expression after DNA damage treatment. The mRNA expressions of pluripotency markers Nanog, Pou5f1, and Sox2 decreased in slower kinetics in KO ESCs than in WT ESCs under all treatment conditions (Fig. 1A). We reduced the concentrations of DNA damaging agents and observed the consistent cellular response kinetics (fig. S3A). Similarly, the protein levels of Nanog homeobox (NANOG), Pou domain class 5, transcription factor 1 (POU5F1), and SRY (sex determining region Y)-box 2 (SOX2) decreased slower in Lnc956 KO ESCs under severe (Fig. 1B) or mild treatment conditions (fig. S3B). Treatment with other DNA damaging agents zeocin (20) and melphalan (21) obtained similar results (fig. S3, C and D). We then examined the ESC differentiation after DNA damage by assessing the expression of three germ layer markers (Sox1 and Pax6 for ectoderm, Mesp1 and T for mesoderm, and Foxa2 and Sox17 for endoderm) (22). Because high concentration of DNA damaging agents caused acute cell death, which prevented the investigation on ESC differentiation, we used mild DNA damage conditions by treating ESCs with 50 nM etoposide for 5 days (Fig. 1C) or 40 μM HU for 6 days (Fig. 1D). Under both conditions, WT ESCs expressed significantly higher level of three germ layer marker genes than Lnc956 KO ESCs. Together, these observations suggested that Lnc956 KO ESCs were resistant to differentiation after genotoxic insults.
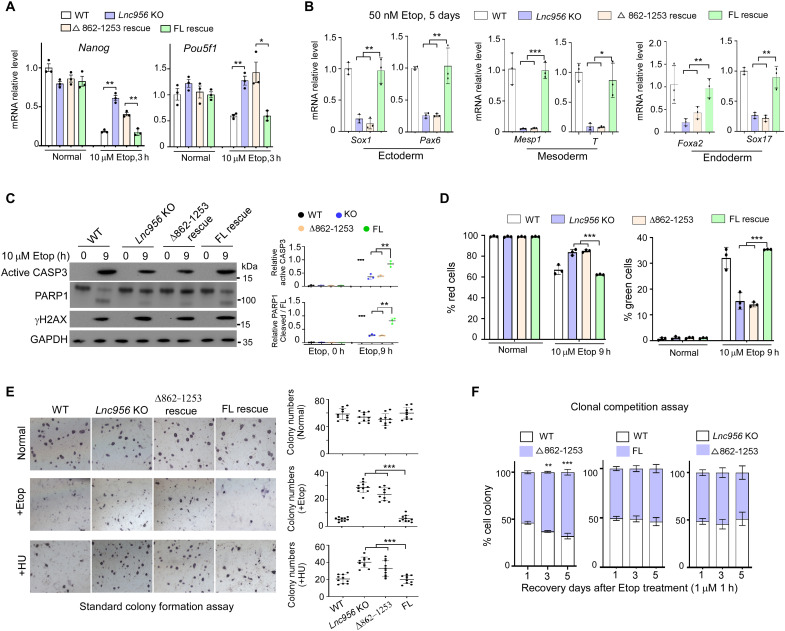
Fig. 1. Lnc956 depletion compromises ESC differentiation and apoptosis after DNA damage.
(A) The mRNA expressions of Nanog, Pou5f1, and Sox2 decreased in slower kinetics in Lnc956 KO ESCs compared to WT ESCs under etoposide (Etop) or HU treatment condition. (B) The protein expressions of NANOG, POU5F1, and SOX2 decreased faster in WT than in Lnc956 KO ESCs after Etop or HU treatment. (C and D) The mRNA expressions of three germ layer markers were statistically higher in WT ESCs than in Lnc956 KO ESCs after Etop (C) or HU (D) treatment. (E) The CASP3 activation and PARP1 cleavage were more robust in WT ESCs than in Lnc956 KO cells treated with Etop or HU. (F) JC-1 staining revealed that more WT ESCs showed green fluorescence indicative of apoptosis when compared to Lnc956 KO ESCs after Etop treatment. Reversely, less WT ESCs showed red fluorescence. (G) In standard colony formation assay, WT and KO ESCs formed similar numbers of alkaline phosphatase–positive (AP+) colonies under normal condition. After Etop treatment, more AP+ colonies were formed in KO ESCs than in WT counterparts. (H) In clonal competition assay, WT ESCs formed fewer colonies than KO ESCs. Moreover, WT GFP+ colonies lost dome-like morphology indicative of ESC differentiation (white arrows). All experiments were repeated three times with consistent results. In (B) and (E), the relative protein levels were normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data were shown as mean ± SEM from three independent experiments. Two-tailed Student’s t test. Scale bars, 100 μm. *P < 0.05, **P < 0.01, and ***P < 0.001.
DNA damage induces ESC differentiation, and the differentiated cells consequently undergo cell death. We then treated the ESCs with high concentration of DNA damaging agents and examined the apoptotic response of ESCs in a time course manner. The apoptosis, monitored by caspase 3 (CASP3) activation and poly (ADP-ribose) polymerase family, member 1 (PARP1) cleavage, occurred with slower kinetics in KO ESCs than in WT ESCs (Fig. 1E and fig. S3E), indicating that Lnc956 KO rendered ESCs resistant to DNA damage–induced apoptosis. To verify this observation, we further examined the mitochondrial membrane permeabilization, which acts upstream to induce apoptosis. The mitochondrial membrane potential was measured by sensitive JC-1 fluorescent probe assay, in which the green fluorescence indicates the low membrane potential, whereas the red fluorescence reflects high membrane potential. Consistently, beginning at the 6 hours of etoposide treatment, the ESCs with green fluorescence (green cells) were detected in both groups, and the percentages were statistically higher in WT than in KO group (Fig. 1F). Meanwhile, the ESCs with red fluorescence (red cells) showed reverse pattern (Fig. 1F). Intriguingly, neither WT nor Lnc956 KO ESCs underwent cell senescence (fig. S3F), suggesting that senescence may not be the major cell fate in response to DNA damages in ESCs.
To evaluate the long-term effect of Lnc956 KO on cellular resistance to DNA damage, we performed standard colony formation assay in which ESCs were treated without or with 1 μM etoposide for different times and cultured for up to 9 days to allow colony formation from single cell. Under normal culture condition, the numbers of colony positive for alkaline phosphatase–positive (AP+) staining did not differ between WT and Lnc956 KO ESCs. However, more AP+ colonies were formed in Lnc956 KO ESCs than in WT ESCs under etoposide treatment conditions (Fig. 1G). This result suggested that Lnc956 depletion rendered ESCs resistant to DNA damage. We also carried out the clonal competition assay in which green fluorescent protein (GFP)–labeled WT ESCs were mixed with Lnc956 KO cells in the same numbers, treated with 1 μM etoposide for 1 hour, and cocultured for 5 days as described previously (8). Consistently, Lnc956 KO ESCs displayed much higher survival rate than WT ESCs (Fig. 1H). Moreover, Lnc956 KO cell colonies showed dome-shaped undifferentiated status, whereas WT cell colonies exhibited flattened morphology indicative of differentiation (Fig. 1H). To further validate the function of Lnc956, we efficiently knocked down (KD) Lnc956 via two independent doxycycline (Dox)–inducible short-hairpin RNAs (fig. S4A). Lnc956 KD reproducibly delayed ESC differentiation (fig. S4, B to D) and apoptosis (fig. S4E) and enhanced colony formation (fig. S4F) after DNA damage treatment. These results together support that loss of Lnc956 compromises the ESC differentiation and apoptosis in response to DNA damage.
We also investigated whether Lnc956 depletion hampered the physiological differentiation of ESCs. During the spontaneous differentiation via embryoid body formation, the expression levels of three germ layer markers were significantly higher in WT ESCs than in KO ESCs (fig. S4G). Similar results were obtained when the ESC differentiation was induced by retinoid acid (fig. S4H). Thus, Lnc956 depletion also compromised the physiological differentiation of ESCs.
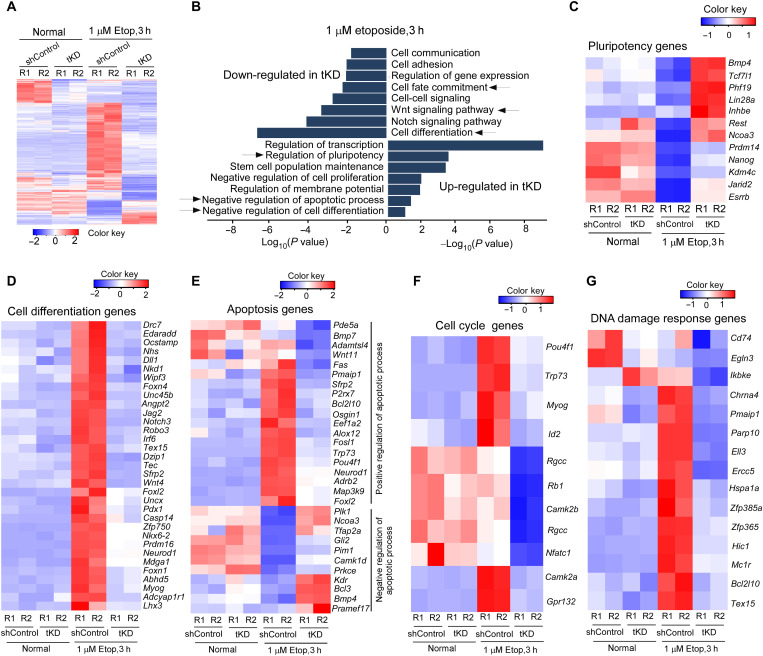
Last, we performed RNA sequencing (RNA-seq) analysis to view the global molecular changes associated with Lnc956 depletion. To avoid the long-term effect of Lnc956 KO on gene expression, we used Dox-inducible KD ESCs to transiently knock down (tKD) Lnc956 for 2 days. Under normal culture condition, 228 genes were down-regulated and 32 genes were up-regulated in Lnc956 tKD ESCs when compared to WT ESCs (Fig. 2A, fig. S5A, and dataset S1). Gene ontology (GO) enrichment analysis revealed that the down-regulated genes were enriched in the processes including cell differentiation and positive regulation of apoptosis (fig. S5, B to D). Under 3-hour etoposide treatment condition, we identified 812 down-regulated genes and 195 up-regulated genes, respectively, in Lnc956 tKD ESC compared to WT counterparts (Fig. 2A, fig. S5A, and dataset S1). The processes of “regulation of pluripotency,” “stem cell population maintenance,” “negative regulation of apoptosis,” and “negative regulation of cell differentiation” were enriched in the up-regulated genes in tKD ESCs after etoposide treatment. Meanwhile, the processes including “cell differentiation” and “cell fate commitment” were enriched in down-regulated genes in tKD ESCs (Fig. 2B). We further looked closely at the differentially expressed genes (DEGs) involved in pluripotency maintenance (Fig. 2C), cell differentiation (Fig. 2D), apoptosis (Fig. 2E), cell cycle (Fig. 2F), and DNA damage responses (Fig. 2G). Specifically, DEGs relevant to pluripotency maintenance included Esrrb, Nanog, and Prdm14 (Fig. 2C). DEGs related to cell differentiation included Neurod1, Nkx6-2, and Tex15 (Fig. 2D). DEGs relevant to apoptosis regulation included Fas, Bcl2l10, and Trp73 (Fig. 2E). It should be mentioned that in our experimental setting, ESCs were subject to 3-hour etoposide treatment. Under this circumstance, ESCs were at early stage of differentiation, and the down-regulation of some key pluripotency genes was not initiated due to the distinct expression kinetics during ESC differentiation (23, 24). Together, these transcriptional changes were in accordance with the above phenotypic observations.
Fig. 2. DEGs in Lnc956 KD ESCs.
(A) Heatmap of DEGs between Lnc956 transient knockdown (tKD) ESCs and shControl counterparts under normal culture and Etop treatment conditions in two replicates (R). (B) GO analysis revealed the biological processes enriched in down-regulated and up-regulated genes, respectively, in tKD ESCs under Etop treatment condition. (C to G) Heatmaps of DEGs relevant to pluripotency maintenance (C), cell differentiation (D), apoptosis (E), cell cycle (F), and DNA damage responses (G).
p53 does not mediate the regulations of Lnc956 on ESC differentiation and apoptosis
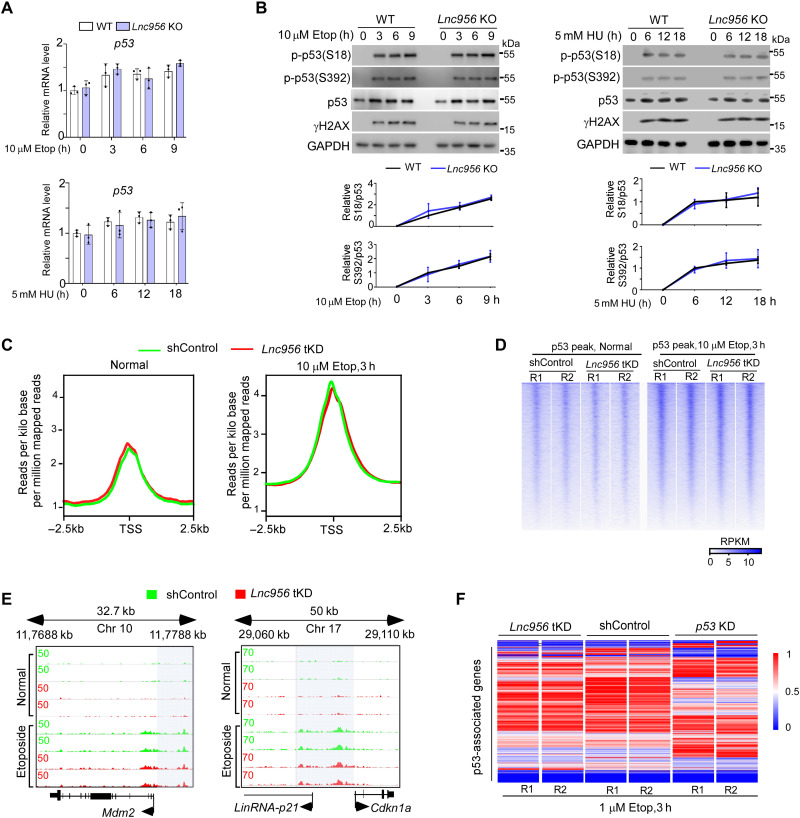
Upon DNA damage, cells initiate DNA damage response to arrest cell cycle, to repair DNA damages, and to determine the cell fate. We wondered whether the DNA damage response signaling was affected in Lnc956 KO ESCs, which underlined the compromised cell fate determination. Like WT ESCs, Lnc956 KO ESCs were able to normally activate ATM-CHK2 (fig. S6A) and ATR-CHK1 signals (fig. S6B). Similarly, the cell cycle distribution and G2/M progression were not altered by Lnc956 KO (fig. S6, C and D). p53, a downstream target of ATM-CHK2 and ATR-CHK1 signaling, plays crucial role in regulating cell fate determination after DNA damage in ESCs (25, 26). In response to genotoxic stress, p53 is activated and acts as a transcription factor to directly suppress Nanog expression and induce ESC differentiation (16). p53 mRNA (Fig. 3A) and protein expressions, as well as its phosphorylation at Ser18 (Ser15 in human) and Ser392, which monitors the p53 activation after DNA damage (14), did not differ between WT and Lnc956 KO ESCs after different damage treatments (Fig. 3B). Similar results were obtained between WT and Lnc956 KD ESCs (fig. S6E). Thus, the overall DNA damage response signaling was normally activated in Lnc956-depleted ESCs after DNA damage.
Fig. 3. Lnc956 depletion does not affect the expression and transcriptional activity of p53.
(A) The mRNA expression of p53 was not affected by Lnc956 KO. (B) p53 protein expression and phosphorylation activation were normal in Lnc956 KO ESCs under normal and DNA damage conditions. (C) Average p53 binding peak intensities were comparable in shControl and Lnc956 transient KD (tKD) ESCs under normal and Etop treatment conditions. (D) Heatmap of p53 binding around transcription start sites (TSS) ±2.5 kb in ESCs in two replications (R), RPKM, Reads Per Kilobase per Million mapped reads. (E) ChIP-seq genome views of p53 binding peaks at Mdm2 and p21 loci. (F) Lnc956 tKD did not affect the overall mRNA expressions of p53-associated genes. As a control, p53 KD drastically changed their expressions. All experiments in (A) and (B) were repeated three times with similar results. In (B), the relative protein levels were normalized by GAPDH. Data were shown as mean ± SEM. Two-tailed Student’s t test.
Although p53 protein expression and activation were not affected in Lnc956 deficient ESCs, we wondered whether its transcriptional activity was dysregulated. To comprehensively evaluate the transcriptional regulation of p53 on its target genes, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) to analyze the genome-wide chromatin binding of p53 in WT and Lnc956 tKD (KD for 2 days) ESCs under normal and etoposide treatment conditions. Globally, Lnc956 transient loss had no substantial influence on p53 (Ser18P) chromatin binding under either normal or treatment condition (Fig. 3, C and D, and dataset S2). Specifically, we looked closely at the promoter binding of its well-known target genes Mdm2 and p21. Binding of p53 to their promoters did not differ between WT and Lnc956 tKD ESCs under normal or etoposide treatment condition (Fig. 3E). Therefore, the binding of p53 to its genome-wide target sites was not affected by Lnc956 depletion. Concordantly, the majority of p53-associated genes (dataset S3) located nearby the p53-binding peaks displayed comparable expression levels in Lnc956 tKD and shControl ESCs after etoposide treatment, whereas KD of p53 in ESCs (fig. S6, F and G) affected their expressions (Fig. 3F). Together, these data suggested that the transcriptional activity of p53 was not affected by Lnc956 depletion, and p53 did not lay downstream of Lnc956 to mediate Lnc956’s function.
Interaction with KLF4 mediates the regulation of Lnc956 on ESC differentiation after DNA damage
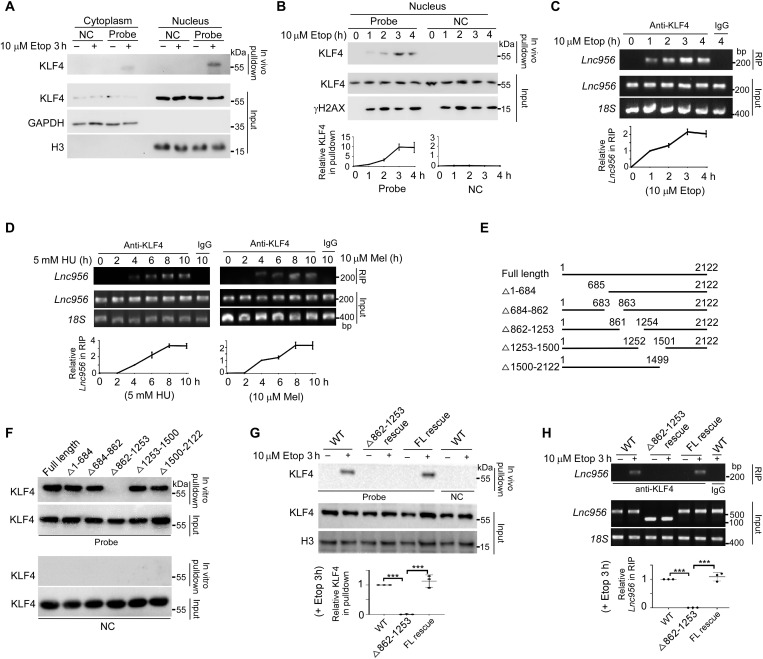
To investigate the molecular mechanism underlying the regulation of Lnc956 on cell fate determination of ESCs after DNA damage, we performed Lnc956 in vitro pulldown assay combined with mass spectrometry analysis. Intriguingly, the pluripotency transcription factor KLF4 was in the list with relatively high count score (fig. S7A). Binding of Lnc956 to KLF4 was validated by the immunoblotting analysis of in vitro RNA pulldown sample (fig. S7B). Considering that in vitro pulldown assay may yield false-positive interaction, we performed in vivo cross-linking followed by nucleus-cytoplasm fractionation and RNA pulldown to verify their spatial interaction. Immunoblotting of pulldown samples revealed that Lnc956 and KLF4 barely interacted under normal culture condition in nucleus or cytoplasm (Fig. 4A). Notably, obvious association was detected in the nucleus after etoposide treatment (Fig. 4A). We then performed time course analysis of Lnc956-KLF4 association in nucleus. The interaction was weak at the early stage of etoposide treatment but was gradually enhanced at the later stage (Fig. 4B). Consistent result was obtained by RNA immunoprecipitation (RIP) analysis using KLF4 antibody (Fig. 4C). Further, we treated the ESCs with other types of DNA damaging agents and examined Lnc956-KLF4 association by RIP. All treatments evoked the Lnc956-KLF4 interaction (Fig. 4D), supporting that Lnc956-KLF4 association is a general response to DNA damages. Thus, we conclude that Lnc956 interacts with KLF4 in the nucleus and Lnc956-KLF4 association is strengthened by DNA damage accumulation.
Fig. 4. Lnc956 interacts with KLF4.
(A) In vivo RNA pulldown analysis detected strong Lnc956-KLF4 association in nucleus after Etop treatment. NC, negative control. (B) Time course in vivo RNA pulldown analysis revealed that Lnc956-KLF4 interaction increased at later stage of Etop treatment. (C) RIP confirmed that KLF4 pulled down more Lnc956 at later stage of Etop treatment. IgG, immunoglobulin. (D) KLF4 pulled down more Lnc956 at later stage of HU or melphalan (Mel) treatment. (E) Schematic representation of FL and truncated Lnc956 mutants. (F) Δ862-1253 Lnc956 did not interact with KLF4 in vitro. (G) In vivo RNA pulldown assay did not detect the KLF4-Lnc956 association in Δ862-1253 Lnc956–rescued ESCs. However, KLF4-Lnc956 interaction was fully restored in FL Lnc956–rescued ESCs. (H) RIP confirmed the KLF4-Lnc956 association in WT and FL Lnc956–rescued ESCs but not in Δ862-1253 Lnc956–rescued ESCs. All experiments were repeated three times with similar results. The relative protein levels in (B) and (G) were normalized by input KLF4, and the relative RNA levels in (C), (D), and (H) were normalized by input Lnc956. Data in (B), (C), (D), (G), and (H) were shown as mean ± SEM. Two-tailed Student’s t test. ***P < 0.001.
Next, we moved on to map the KLF4-binding sites on Lnc956. On the basis of the reported KLF4 nucleotide–binding motif, we predicted seven putative binding sites and generated a series of truncated Lnc956 mutants. These mutants included Δ1-684 [deletion of 1 to 684 base pairs (bp) containing one putative binding site], Δ684-862 (deletion of 684 to 862 bp), Δ862-1253 (deletion of 862 to 1253 bp containing two putative binding sites), Δ1253-1500 (deletion of 1253 to 1500 bp containing two putative binding sites), and Δ1500-2122 (deletion of 1500 to 2122 bp containing two putative sites) (Fig. 4E). In vitro RNA pulldown assay revealed that Δ862-1253 truncate Lnc956 failed to pull down KLF4, suggesting that 862-1253 fragment was essential to interact with KLF4 (Fig. 4F). We further introduced full-length (FL) Lnc956 and Δ862-1253 truncate Lnc956 into Lnc956 KO ESCs and established stable cell lines (fig. S7C). Concordantly, in vivo RNA pulldown assay on nuclear fraction validated the inability of Δ862-1253 truncate to interact with KLF4. As a control, re-expression of FL Lnc956 restored the Lnc956-KLF4 association after etoposide treatment (Fig. 4G). Consistent result was obtained by RIP using KLF4 antibody (Fig. 4H).
KLF4 is one of the core pluripotency maintenance transcription factors and functions in a highly integrated way with other transcription factors to maintain pluripotency (27). It also regulates the pluripotency-associated three-dimensional enhancer networks (28). We speculated that binding to KLF4 might mediate the regulation of Lnc956 on ESC differentiation and apoptosis in response to DNA damage. To test this hypothesis, we performed rescue experiments. Re-expression of FL Lnc956 in KO ESCs successfully restored the cellular sensitivity to exit pluripotency (Fig. 5A), initiate differentiation (Fig. 5B), and to undergo apoptosis (Fig. 5, C and D) in response to DNA damage. The recovery of cellular survival in DNA damages was also validated by standard colony formation assay (Fig. 5E) and clonal competition assay (Fig. 5F). However, Δ862-1253 truncate Lnc956, which failed to interact with KLF4, had no rescue effect on ESC pluripotency exit (Fig. 5A), differentiation (Fig. 5B), apoptosis (Fig. 5, C and D), or colony formation (Fig. 5, E and F). These results suggested that Lnc956-KLF4 association was essential to initiate ESC differentiation and apoptosis after DNA damage.
Fig. 5. Lnc956-KLF4 interaction is required for ESC differentiation after DNA damage.
(A) After Etop treatment, the mRNA expressions of Nanog and Pou5f1 were down-regulated in WT and FL Lnc956–rescued ESCs. However, the down-regulation was compromised in Lnc956 KO and Δ862-1253 Lnc956–rescued ESCs, in which Lnc956-KLF4 interaction was lost. (B) After Etop treatment, the expression levels of three germ layer markers were significantly higher in WT and FL Lnc956–rescued ESCs than in Lnc956 KO or Δ862-1253 Lnc956–rescued ESCs. (C and D) Immunoblotting (C) and JC-1 staining (D) showed that Etop-induced apoptosis was compromised in Lnc956 KO and Δ862-1253 Lnc956–rescued ESCs. (E and F) Standard colony formation (E) and clonal competition assays (F) revealed that Lnc956 KO and Δ862-1253 Lnc956–rescued ESCs had higher potential to form colonies after Etop treatment. All experiments were repeated three times with similar results. The relative protein levels in (C) were normalized by GAPDH. Data were shown as mean ± SEM. Two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Lnc956-KLF4 interaction sequestrates KLF4 and suppresses KLF4’s transcriptional activity
LncRNA can serve as a “bait” to sequester proteins from binding target genomic loci (29). We speculated that Lnc956-KLF4 interaction might sequestrate KLF4 and prevent it from binding to the promoters of its target genes, thereby promoting ESC differentiation under DNA damage condition. The protein levels of KLF4 in nucleus did not differ between WT and Lnc956 KO ESCs under normal or 3-hour etoposide treatment condition (fig. S8A). We isolated insoluble chromatin and measured the amounts of chromatin-bound KLF4 before and after DNA damage. Intriguingly, DNA damage evoked a decrease in chromatin-bound KLF4 level in WT ESCs. However, the amount of chromatin-bound KLF4 remained comparable before and after etoposide treatment in Lnc956 KO ESCs (Fig. 6A). This observation implicated that Lnc956-KLF4 association sequestrated KLF4 and prevented it from binding to chromatin.
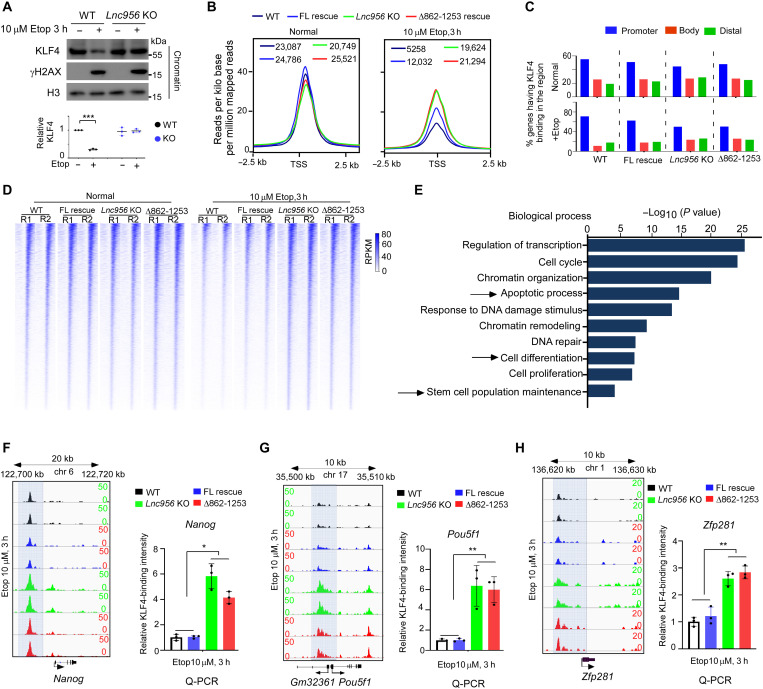
Fig. 6. Lnc956-KLF4 interaction sequestrates KLF4 and suppresses KLF4’s transcriptional regulation on pluripotencyABCDEF H.
(A) After Etop treatment, the level of chromatin-bound KLF4 decreased in WT ESCs but remained constant in Lnc956 KO ESCs. The relative protein levels were normalized by histone H3. (B) Average KLF4 binding peak intensities were comparable among four ESC types under normal condition. After Etop treatment, the peak intensities in ESCs with Lnc956-KLF4 interaction (WT and FL Lnc956 rescued) were lower than in ESCs without Lnc956-KLF4 interaction (Lnc956 KO and Δ862-1253 Lnc956 rescued). (C) The distribution of KLF4 binding regions in the four ESC types. Genes that contain at least one KLF4 peak in each region were calculated. (D) Heatmap of KLF4 binding around TSS ± 2.5 kb in four types of ESCs under normal and Etop treatment conditions. Data were from two independent replications (R). (E) GO enrichment of differential KLF4-targeting genes among the four types of ESCs under Etop treatment condition. (F to H) ChIP-seq genome views of KLF4 binding peaks at loci of Nanog (F), Pou5f1 (G), and Zfp281 (H) in four types of ESCs. Right panel showed the qPCR validation of ChIP samples. Data were shown as mean ± SEM from three independent replications. Two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Further, we performed ChIP-seq analysis in different ESCs (WT, Lnc956 KO, Δ862-1253 Lnc956 rescued, and FL Lnc956 rescued) to examine in detail the influence of Lnc956-KLF4 association on promoter binding of KLF4. Using a peak-finding algorithm (30), we reproducibly identified 23,087, 20,749, 24,786, and 25,521 KLF4 binding sites, respectively, in WT, Lnc956 KO, FL Lnc956–rescued, and Δ862-1253 Lnc956–rescued ESCs under normal culture condition in two replicates. Following etoposide treatment for 3 hours, the number of KLF4 consensus binding sites common in two replicates dropped substantially in WT (from 23,087 to 5258, ~77.2% reduction) and FL Lnc956–rescued ESCs (from 24,786 to 12032, ~51.5% reduction). However, only slight reduction was observed in Lnc956 KO (from 20,749 to 19,624, ~5.4% reduction) and Δ862-1253 Lnc956–rescued ESCs (from 25,521 to 21,294, ~16.6% reduction) (Fig. 6B). KLF4 predominantly bound at the promoter-proximal regions. The other binding sites included gene body regions or distal regions. This binding pattern was not substantially altered by loss of Lnc956-KLF4 association in Lnc956 KO or Δ862-1253 Lnc956–rescued ESCs (Fig. 6C). We also performed heatmap analysis of KLF4 binding around the transcription start site (TSS). Under etoposide treatment condition, ESCs with proficient Lnc956-KLF4 association (WT and FL Lnc956–rescued cells) had weaker KLF4 binding peaks than ESCs without Lnc956-KLF4 association (Lnc956 KO and Δ862-1253 Lnc956–rescued cells) (Fig. 6D). GO analysis on these differentially bound promoters between ESCs with and without Lnc956-KLF4 association identified the enrichment of biology processes, including apoptosis, stem cell differentiation, and maintenance (Fig. 6E). Looking closely, we validated the binding of KLF4 to the promoters of several well-known target genes. KLF4 binds to the promoters of core pluripotency maintenance genes including Nanog, Pou5f1, and Zfp281 (31). ChIP-quantitative polymerase chain reaction (PCR) showed that binding of KLF4 to these gene promoters did not differ between ESCs with and without Lnc956-KLF4 association under normal culture condition (fig. S8, B to D). After etoposide treatment, KLF4 binding intensity was significantly lower in ESCs with Lnc956-KLF4 association (WT and FL Lnc956–rescued cells) than in ESCs without Lnc956-KLF4 association (Lnc956 KO and Δ862-1253 Lnc956–rescued cells) (Fig. 6, F to H). Together, these results demonstrate that Lnc956-KLF4 interaction sequestrates KLF4 and suppresses its transcriptional activity.
Lnc956-KLF4 axis and p53 function independently to regulate cell fate determination of ESCs in response to DNA damage
We have shown that Lnc956-KLF4 axis regulated cell fate determination of ESCs after DNA damage. Depletion of Lnc956 had no influence on the overall p53 expression and its transcriptional activity, suggesting that the function of Lnc956-KLF4 axis did not rely on p53 (Fig. 3). To further verify this conclusion, we examined whether KLF4 could regulate p53. We did not find the unambiguous binding of KLF4 to p53 promoter in mouse ESCs (fig. S8E). Concordantly, KD of Klf4 by small interfering RNAs (siRNA) (fig. S8F) had no influence on the transcription of p53 under normal or DNA damage condition (Fig. 7A). This was in line with the observation that Lnc956 KO did not affect the p53 expression (Fig. 3, A and B). RNA-seq analysis of siControl and siKlf4 ESCs also showed that Klf4 KD had marginal disturbance on the global expression of p53-associated genes located nearby its binding peaks (Fig. 7B). Thus, these data collectively support that Lnc956-KLF4 axis does not regulate p53 function.
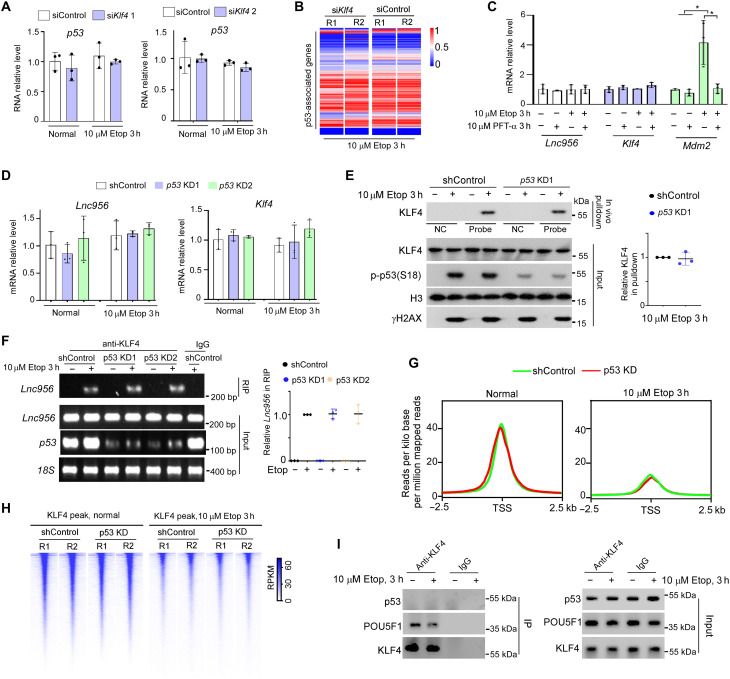
Fig. 7. Lnc956-KLF4 axis and p53 function independently.
(A) Klf4 KD by siRNA did not influence the mRNA expression of p53. (B) Klf4 KD had no impact on the overall expression of p53-associated genes under DNA damage condition. (C) Modulation of p53 activity by etoposide treatment or its inhibitor PFT-α affected Mdm2 expression but had no influence on the expression of Lnc956 or Klf4. (D) Similarly, p53 KD did not affect the expression of Lnc956 or Klf4. (E and F) p53 KD had no influence on the Lnc956-KLF4 association as indicated by in vivo RNA pulldown (E) and RIP analysis (F). (G and H) The KLF4 binding to chromatin was not affected by p53 KD under normal or Etop treatment condition. In (G), the average KLF4 binding peak intensities were comparable between shControl and p53 KD ESCs. The heatmap of KLF4 binding around TSS ± 2.5 kb was shown in (H). (I) Co-immunoprecipitation (IP) did not detect the association of KLF4 with p53. As a control, POU5F1 was co-immunoprecipitated with KLF4. Except in (B), (G), and (H), all experiments were repeated three times with similar results. The relative protein levels in (E) were normalized by input KLF4. The relative RNA levels in (F) were normalized by input Lnc956. Data were shown as mean ± SEM. Two-tailed Student’s t test. *P < 0.05.
Reversely, we investigated whether p53 could regulate the Lnc956-KLF4 axis. To this end, we first examined whether p53 regulated the transcription of Lnc956 and Klf4. Manipulating p53 activity by DNA damage treatment or p53 inhibitor pifithrin-α (PFT-α) (32) had no influence on RNA expression of Lnc956 or Klf4. As a control, manipulation of p53 activity drastically affected the expression of its target gene Mdm2 (Fig. 7C). KD of p53 obtained similar results (Fig. 7D). In addition, KLF4 protein expression and Lnc956-KLF4 association was not altered by p53 KD, as indicated by in vivo RNA pulldown (Fig. 7E) and RIP analyses (Fig. 7F). Further, we examined whether p53 KD affected the KLF4’s transcriptional activity by ChIP-seq analysis of shControl and p53 KD ESCs under normal and etoposide treatment conditions. Notably, the KLF4 binding intensity was comparable between shControl and p53 KD ESCs under either normal or etoposide treatment condition (Fig. 7, G and H, and dataset S4). Together, these data demonstrated that p53 did not regulate the Lnc956-KLF4 axis.
Previous study on other cellular context reported that KLF4-p53 formed protein complex to regulate gene transcription (33). We examined whether KLF4 associated with p53 in mouse ESCs. Coimmunoprecipitation did not detect the KLF4-p53 interaction. As a control, KLF4 successfully pulled down its known interaction protein POU5F1 (Fig. 7I).
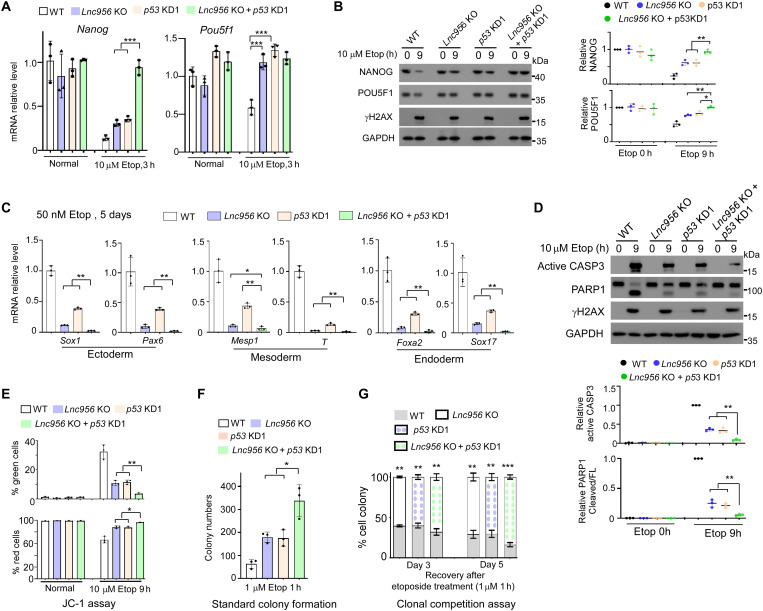
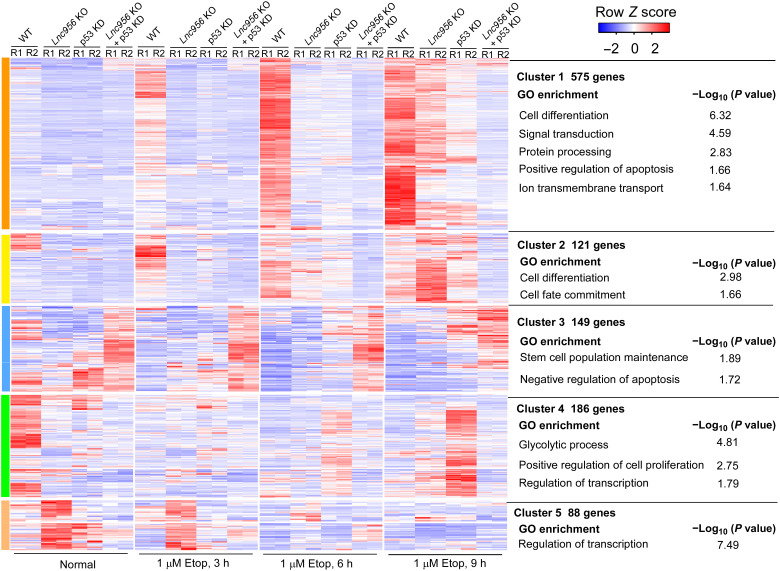
The above lines of evidence suggested that Lnc956-KLF4 axis and p53 function independently. To further verify the conclusion, we knocked down p53 in Lnc956 KO ESCs (fig. S9A). Simultaneous depletion of p53 and Lnc956 displayed additive effects on the pluripotency genes expression (Fig. 8, A and B, and fig. S9, B and C), the three germ layer markers induction (Fig. 8C and fig. S9D), the apoptosis (Fig. 8, D and E, and fig. S9E), and the colony formation rate in response to DNA damage (Fig. 8, F and G, and fig. S9F). We also performed RNA-seq to analyze the global gene expressions in WT, Lnc956 KO, p53 KD, and Lnc956 KO + p53 KD ESCs cultured normally or treated with etoposide for different time. The majority of DEGs (75%, 845 of 1119) showed additive change pattern in Lnc956 KO + p53 KD ESCs when compared to Lnc956 KO or p53 KD ESCs. The DEGs in clusters 1 and 2 were up-regulated by etoposide treatment in WT ESCs. However, Lnc956 KO or p53 KD suppressed the up-regulation, simultaneous depletion of p53, and Lnc956 displayed additive influence. These genes were enriched in the processes of cell differentiation and positive regulation of apoptosis (Fig. 9). In contrast, DEGs in cluster 3 showed reverse change pattern and were enriched in the processes of stem cell maintenance and negative regulation of apoptosis (Fig. 9). Thus, these data together support that Lnc956 and p53 function independently to regulate cell fate determination of ESCs in response to DNA damage.
Fig. 8. Simultaneous depletion of p53 and Lnc956 displays additive effects on cell differentiation and apoptosis.
(A) Lnc956 or p53 depletion compromised the down-regulation of Nanog mRNA expression after Etop treatment. Simultaneous depletion displayed additive effect. (B) Lnc956 or p53 depletion compromised the down-regulation of NANOG and POU5F1 protein expressions after Etop treatment. Simultaneous depletion displayed additive effect. (C) Lnc956 or p53 depletion suppressed the three germ layer markers expression during Etop-induced differentiation. Simultaneous depletion had additive effect. (D and E) Lnc956 or p53 depletion compromised the Etop-induced apoptosis measured by immunoblotting (D) and JC-1 probe assay (E). Simultaneous depletion had additive effect. (F and G) Standard colony formation (F) and clonal competition (G) assays showed that Lnc956 or p53 depletion enhanced the colony formation of ESCs after Etop treatment. Simultaneous depletion had additive effect. All experiments were repeated three times with similar results. In (B) and (D), the relative protein levels were normalized by GAPDH. Data were shown as mean ± SEM. Two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Fig. 9. Simultaneous depletion of p53 and Lnc956 has additive effect on gene expression changes.
RNA-seq analysis of the gene expressions in WT, Lnc956 KO, p53 KD, and Lnc956 KO + p53 KD ESCs showed that the majority of DEGs (75%) showed additive change pattern in Lnc956 KO + p53 KD ESCs when compared to Lnc956 KO or p53 KD ESCs after DNA damage treatment.
ATM-driven m6A modification of Lnc956 regulates Lnc956-KLF4 association
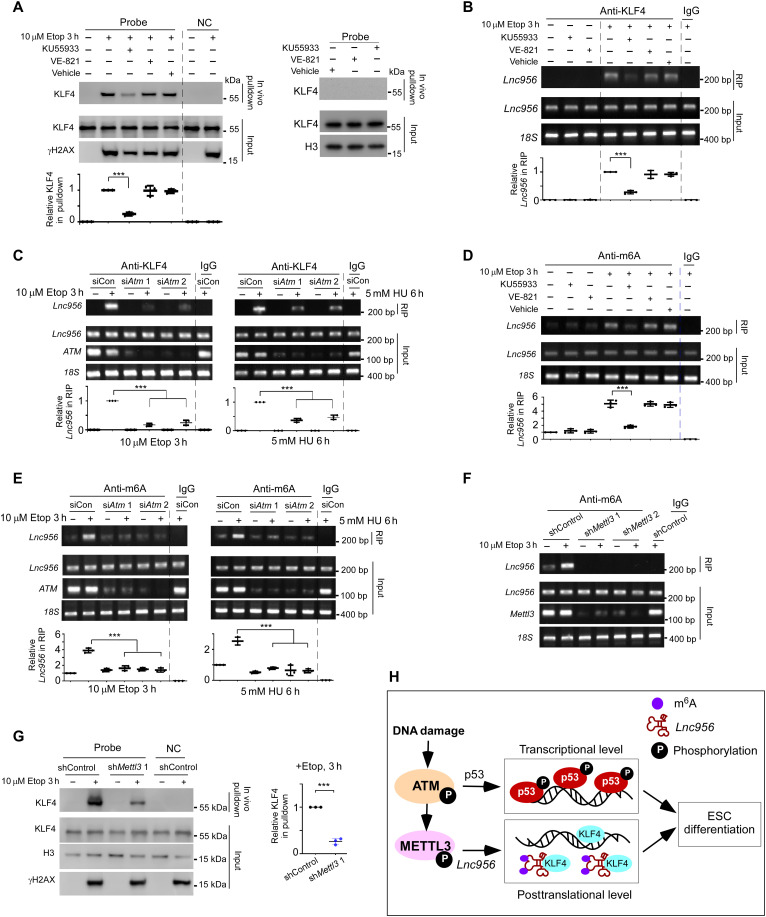
Considering that Lnc956-KLF4 association was promoted under DNA damage condition, we wondered whether DNA damage signaling regulated the interaction. ATM and ATR kinases are central coordinators of DNA damage response and trigger a cascade of numerous downstream effects (34). To examine whether ATM or ATR signaling regulated the Lnc956-KLF4 association, we inhibited ATM activity with a specific inhibitor KU55933 (35) and ATR activity with VE-821 (36), respectively (fig. S10A). Intriguingly, in vivo RNA pulldown showed that blockage of ATM activation compromised Lnc956-KLF4 association under etoposide treatment condition, whereas inhibiting ATR activity had no effect (Fig. 10A). Consistent result was obtained by RIP with KLF4 antibody (Fig. 10B). To further verify the regulatory role of ATM on Lnc956-KLF4 association, we efficiently knocked down ATM in ESCs by two independent siRNAs (fig. S10B) and examined its influence on Lnc956-KLF4 interaction under distinct genotoxic treatments. RIP analysis confirmed that ATM KD compromised the Lnc956-KLF4 association under different damaging conditions (Fig. 10C). Thus, ATM, but not ATR kinase, regulated the Lnc956-KLF4 association after genotoxic insult.
Fig. 10. ATM-driven m6A modification of Lnc956 is required for interaction with KLF4.
(A) Inhibition of ATM activity with KU55933 compromised Etop-induced Lnc956-KLF4 association detected by in vivo RNA pulldown. Inhibition of ATR activity with VE-821 had no influence (left). No Lnc956-KLF4 association was detected under normal culture condition (right). (B) RIP confirmed that inhibition of ATM but not ATR activity attenuated Etop-induced Lnc956-KLF4 interaction. (C) ATM KD by siRNA compromised Lnc956-KLF4 interaction induced by Etop or HU treatment. (D) m6A modification of Lnc956 was enhanced by Etop treatment. Inhibition of ATM but not ATR activity suppressed the damage-induced m6A modification of Lnc956. (E) ATM KD by siRNAs suppressed the m6A modification of Lnc956 induced by Etop or HU treatment. (F) Mettl3 KD abolished m6A modification of Lnc956 under normal and Etop treatment conditions. (G) Mettl3 KD compromised Etop-induced Lnc956-KLF4 association. (H) Working model for quality control of ESCs. Downstream of ATM activation, p53 pathway, and Lnc956-KLF4 axis work independently to ensure robust elimination of ESC population with unrepaired DNA damages. All experiments were repeated three times with similar results. The relative protein levels in (A) and (G) were normalized by input KLF4. The relative RNA levels in (B) to (E) were normalized by input Lnc956. Data were shown as mean ± SEM. Two-tailed Student’s t test. ***P < 0.001.
We next asked how ATM regulated the Lnc956-KLF4 association. Previous study reported that ATM could phosphorylate and activate methyltransferase like 3 (METTL3), which methylates the adenosine at the N6 position (m6A) of DNA damage–associated RNAs (37). We thus wondered whether ATM stimulated m6A modification of Lnc956, which, in turn, modulated the Lnc956-KLF4 interaction. To this end, we first examined the potential m6A modification of Lnc956 and its regulation by ATM. Computational prediction of m6A modification by m6A site predictor sequence-based RNA adenosine methylation site predictor (38) suggested that Lnc956 contained nine potential m6A methylation sites (fig. S10C). RIP through m6A antibody showed that Lnc956 could be immunoprecipitated in ESCs under normal culture condition, and the pulldown efficiency was robustly enhanced by etoposide treatment (Fig. 10D). The result implicated that Lnc956 was subject to m6A methylation, and DNA damages potentiated this modification. In line with this result, previous study using m6A immunoprecipitation coupled with high-throughput sequencing in mESCs also detected Lnc956 (39). Of note, blockage of ATM, but not ATR kinase activity, drastically attenuated the m6A immunoprecipitation with Lnc956 under etoposide treatment (Fig. 10D), suggesting that ATM kinase signaling stimulated the m6A modification of Lnc956. The regulation of ATM on m6A modification of Lnc956 in response to DNA damages was further confirmed in ATM KD ESCs treated with different DNA damaging agents (Fig. 10E). Intriguingly, Lnc956 also underwent m6A modification during physiological ESC differentiation (fig. S10D), although the ATM or ATR activation was not detected (fig. S10E). This observation suggested that Lnc956 was modified through different mechanisms in different contexts.
Next, we examined whether m6A methylation of Lnc956 regulated its association with KLF4. We knocked down METTL3, the core RNA methyltransferase responsible for methyl group transfer during m6A modification, in ESCs and examined its effects. Efficient knockdown of Mettl3 in ESCs (fig. S10F) robustly decreased the m6A modification of Lnc956 (Fig. 10F). As a result, Lnc956-KLF4 association was significantly reduced (Fig. 10G and fig. S10G). Thus, m6A methylation of Lnc956 induced Lnc956-KLF4 interaction. The m6A nuclear reader YTH domain containing 1 (YTHDC1) was reported to play a role in DNA damage response (37). We wondered whether YTHDC1 participated in the regulation of Lnc956-KLF4 interaction. To this end, we knocked down YTHDC1 via two independent siRNAs (fig. S10H) and examined its effect. YTHDC1 KD affected neither the Lnc956-KLF4 interaction (fig. S10I) nor the DNA damage-induced pluripotency exit (fig. S10J). Thus, unlike the involvement of METTL3-m6A-YTHDC1 axis in DNA DSB repair (37), YTHDC1 did not mediate the regulations of m6A modification on Lnc956-KLF4 association and cell fate determination after DNA damage in ESCs. In summary, we proposed a working model in which ATM-driven m6A modification of Lnc956 promotes Lnc956-KLF4 interaction, which rapidly sequestrates KLF4 and attenuates its transcriptional activity. This pathway functions independent of p53 pathway to control ESC’s quality and safety. By integrating regulations at the posttranslational level (Lnc956 pathway) and the transcriptional level (p53 pathway), ESCs can robustly eliminate populations with unrepaired DNA damages (Fig. 10H).
DISCUSSION
Efficient elimination of cell populations with unrepaired DNA damage is crucial to maintain genomic stability of stem cell pool. p53 plays key roles in this process by inducing stem cell differentiation and apoptosis. Except p53, whether there exist other independent regulators remains unknown. In this study, we reported a stem cell–specific player Lnc956, which functions independent of p53 to regulate the cell fate determination of ESCs after DNA damage. Lnc956 functions through binding and sequestrating the core pluripotency maintenance factor KLF4, thereby attenuating KLF4’s transcriptional activity to induce ESC differentiation. Several lines of evidence supported that Lnc956-KLF4 axis and p53 were independent of regulating cell fate determination. First, depletion of Lnc956 or KLF4 did not affect the expression of p53 nor did it influence the p53’s transcriptional activity as revealed by ChIP-seq and RNA-seq analyses. Second, p53 KD or activity inhibition had no influence on the expression of Lnc956 or KLF4. Moreover, p53 depletion did not impair the Lnc956-KLF4 association and the chromatin binding profile of KLF4. Third, p53 and KLF4 did not physically interact, indicating that they do not work as a whole. Fourth, simultaneous depletion of p53 and Lnc956 exhibited additive effects on ESC differentiation, apoptosis, and cell survival in DNA damage treatment. The gene expression changes also showed additive effect. Thus, Lnc956-KLF4 axis added a regulatory layer. This posttranslational regulation has an advantage over p53-mediated transcriptional regulation in evoking fast cell fate transition. Thus, unlike in somatic cells, ATM signaling in ESCs can activate two independent pathways, which work together to ensure robust differentiation and apoptosis to eliminate damaged cells and maintain genome stability of the cell population.
Our present data suggested that Lnc956-KLF4 association was regulated by m6A modification of Lnc956, which was controlled by ATM signaling. On the one hand, RIP using m6A antibody could pull down Lnc956. On the other hand, blockage of m6A modification by knocking down the catalytic enzyme METTL3 significantly suppressed the m6A modification of Lnc956 and the Lnc956-KLF4 interaction. However, it was unclear which sites in Lnc956 underwent m6A methylation and how the m6A modification facilitated the interaction of Lnc956 with KLF4. Although our result showed that the nuclear m6A reader YTHDC1 was not involved in the regulation process, it remained unclear whether other readers are involved or no reader is required. In addition, the results and conclusion were obtained by using murine ESCs in this study. Whether the human ortholog of Lnc956 has conserved role and the similar mechanism operates in genome quality surveillance of human ESCs need to be confirmed in the future study.
MATERIALS AND METHODS
Derivation and culture of mouse ESCs
Mouse ESCs were derived as previously reported (40). Briefly, embryonic day 3.5 blastocysts were recovered from Lnc956+/− females mated with Lnc956−/− male mice. After removal of zona pellucida with acid tyrode solution (100 ml; Sigma-Aldrich, T1788), blastocysts were plated on mitomycin C–treated mouse embryonic fibroblasts (MEFs) and cultured for 5 days. The outgrowths were then picked with a mouth pipette and transferred into 50-μl droplet of 0.05% TrypLE solution (Gibco, 12604021) for 3 min. Dissociated cells were pipetted into 48-well culture plates with a MEF feeder for further culture. After routine clonal expansion, genotype was confirmed by PCR with specific primers (table S1).
Mouse ESCs were maintained in Dulbecco’s modified Eagle's medium (DMEM)/F12 supplemented with 20% KO serum replacement (Thermo Fisher Scientific, A3181502), recombinant mouse leukemia inhibitory factor (104 U/ml; Millipore, ESG1107), 2 mM l-glutamine (Sigma-Aldrich, G8540-25g), penicillin (100 U/ml)/streptomycin (100 μg/ml) (Gibco, 15140-122), 100 μM β-mercaptoethanol (Sigma-Aldrich, M7522), and 1× MEM nonessential amino acids solution (Gibco, 11140-035).
Neutral comet assay
The neutral comet assay was carried out as previously described (6). Briefly, ESCs were resuspended with ice-cold phosphate-buffered saline [PBS; (pH 7.4) Ca2+ and Mg2+ free] at a concentration of 2 × 105 cells/ml, and then an aliquot of 30 μl suspension was mixed with 70 μl of 0.8% low-melting agarose kept at 37°C. The mixture was immediately and evenly spread onto the comet slides. Then, the slides were placed at 4°C for 10 min and transferred into lysis buffer [2.5 M NaCl, 100 mM Na2EDTA, 10 mM tris, 1% N-lauroylsarcosine, and 1% Triton X-100 (pH 8.3)] at room temperature for 1 hour away from light. The slides were washed twice and placed in cold electrophoresis buffer [300 mM sodium acetate and 100 mM tris (pH 8.3)]. Electrophoresis was performed at 1 V/cm, 300 mA for 25 min at 4°C away from light. After electrophoresis, The slides were washed in deionized water twice and immersed in ice-cold 100% ethanol at room temperature for 5 min. After air dry, DNA was stained with DAPI (4′,6-diamidino-2-phenylindole; 10 ng/ml; Thermo Fisher Scientific) and immediately analyzed with a fluorescence microscope. Two hundred cells per slides were captured for analysis, and each experiment was independently repeated three times. CASP comet assay software (Andor Technology) was used to analyze comet.
Western blotting
Cells were lysed with radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitor cocktail (Roche, 4693132001) and phosphatase inhibitors (NaF and Na3VO4). Protein samples were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto the polyvinylidene difluoride membrane. After blocking with 5% bovine serum albumin, the primary antibodies were added and incubated at 4°C overnight. Then, the membrane was washed with PBS and incubated with secondary antibodies at room temperature for 1 hour. Images were captured by the ProteinSimple FluorChem system. The antibody information was listed in table S2.
Quantitative RT-PCR
Total RNA was isolated using TRIzol (Invitrogen) based on the protocols of manufacture. The concentration of RNA was measured by NanoDrop 2000 (Thermo Fisher Scientific). One microgram of total RNA per sample was transcribed into complementary DNA (cDNA) using the PrimeScript RT Reagent Kit (TaKaRa, catalog no. RR037A). Quantitative PCR was carried out with a SYBR Premix Ex Taq kit (DRR041S, TaKaRa) on the LightCycler 480 II (Roche). Gapdh was used to normalize the gene expression. The method of 2–ΔΔCt was used for data analysis. All primers for reverse transcription PCR (RT-PCR) were listed in table S1.
Mitochondrial membrane potential measurement with JC-1 fluorescent probe
Mitochondrial membrane potential assay was carried out on the basis of manufacturer’s instructions (Beyotime,C2003S). Briefly, cell samples with or without DNA damage treatment were harvested and washed twice with ice-cold PBS, followed by incubation with JC-1 probe for 20 min at 37°C. After incubation, cells were washed and resuspended with ice-cold PBS and sorted using a fluorescence-activated cell sorting LSRFortessa flow cytometer (Becton Dickinson). Data were analyzed with FlowJo software (version 7.6).
Standard colony formation assay
Cells were plated at a density of 350 cells per 3.8-cm2 petri dish (12-well plate, Corning 3336). One day after inoculation, cells were treated with dimethyl sulfoxide (DMSO) or 1 μM etoposide for 1, 3, and 6 hours, respectively, and washed three times with prewarming PBS. Cells were then cultured for 9 days followed by fixation and staining for AP (Beyotime, C3206). The numbers of AP+ colonies in each well were counted. Triplicate wells were calculated for each condition.
Clonal competition assay
Clonal competition assay was performed as previously described (8). Briefly, the same numbers of two types of ESCs, of which one was labeled with GFP or red fluorescent protein, were mixed and plated on the dish. After attachment, the mixed ESCs were treated with 1 μM etoposide for 1 hour and cocultured till analysis. The clonal growth and state were monitored daily and counted at each time point for statistics.
Cell senescence assay
Cell senescence was evaluated using a senescence β-galactosidase staining kit (Beyotime, C0602). Briefly, cells were plated at a density of 104 cells per 9.5-cm2 petri dish (six-well plate, Corning 3335). One day after inoculation, cells were treated with DMSO or 50 nM etoposide for 2 days. Following treatment, cells were cultured for 2 days and then were subject to staining by following the manufacturer’s instructions. Triplicate wells were performed for each condition.
In vitro RNA pulldown and mass spectrometry analysis
Biotin-labeled RNA was in vitro transcribed with the Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific, 20163) according to the manufacturer’s instructions. RNA secondary structure was recovered by heating to 95°C for 3 min in RNA structure buffer [0.1 M KCl, 10 mM tris (pH 7.0), and 10 mM MgCl2], followed by incubating samples on ice for 5 min and returning to room temperature. Folded RNAs were subject to in vitro RNA pulldown using a Pierce Magnetic RNA-protein pull-down kit (Thermo Fisher Scientific, 20164) according to the manufacturer’s instructions. The pulldown proteins were denatured and separated on SDS-PAGE for mass spectrometry analysis as previously described (8).
In vivo RNA pulldown assay
In vivo RNA pulldown assay was performed as previously described (41). Briefly, biotin-labeled DNA probes for target LncRNA were synthesized by RiboBio Co. Ltd. Cells (2 × 108) with or without DNA damage treatment were washed twice with ice-cold PBS and cross-linked at the energy of 400 mJ using 265-nm ultraviolet light. Then, cross-linked samples were lysed with CSKT solution [10 mM Pipes, 0.5% Triton X-100 (pH 6.8), 3 mM MgCl2, 100 mM NaCl, and 0.3 M sucrose] containing proteinase inhibitors (Roche) and SUPERaseIn (Thermol Fisher Scientific, AM2694) at 4°C for 10 min. After centrifuge at 1200g for 10 min at 4°C, the pellet was suspended with 3 ml of deoxyribonuclease I (DNase I) buffer [50 mM tris (pH 7.5), 0.1% sodium lauroyl sarcosine, SUPERaseIn, 0.5% NP-40, 1 × protease inhibitors, 10 mM vanadyl ribonucleoside complex, and 600 U of riboxynuclease (RNase)–free DNase I] and incubated at 37°C with rotation. The supernatant was precleared by adding 50 μl of M-280 streptavidin Dynabeads (Thermo Fisher Scientific, 00781251) for 30 min at room temperature with rotation. After discarding preclear beads, 100 pmol of DNA probes and 150 μl of beads were mixed and added into the supernatant and incubated for 20 min at room temperature with rotation. Then, the samples with beads were heated to 65°C for 15 min and cooled down slowly to 37°C for 1 hour. Beads were then washed three times with wash buffer I [50 mM tris (pH 7.5), 0.3 M LiCl, 1% SDS, 0.5% NP-40, and 1 mM dithiothreitol (DTT)] containing protease inhibitors. Beads were incubated with 300 μl of DNase buffer containing 20 U of DNase at 37°C for 10 min. Beads were washed twice with wash buffer I containing protease inhibitors at 37°C and once with wash buffer 2 (1% SDS, 1 mM DTT, 5 mM EDTA, and 150 mM NaCl) containing protease inhibitors. Proteins were eluted with elution buffer [10 mM tris (pH 7.5) and 1 mM EDTA] at 70°C for 15 min and boiled in protein loading buffer at 100°C for 15 min.
RNA immunoprecipitation
RIP was performed as previously described (42, 43). Briefly, 1 × 107 cells were lysed in 1 ml of polysome lysis buffer [100 mM KCl, 5 mM MgCl2, 10 mM Hepes (pH 7.0), 0.5% NP-40, 1 mM DTT, and 400 μM Ribonucleoside vanadyl complex (VRC)] supplemented with RNase inhibitors and protease inhibitors. After incubation on ice for 5 min, 200 μl of preclear protein A/G beads was added into lysis and incubated at 4°C for 30 min. Then, 2.5 to 5 μg of antibody was added into lysis beads complex and incubated at 4°C overnight. The antibody-coated beads were then washed with 1 ml of ice-cold NT2 buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, and 0.05% NP-40] containing RNase inhibitors and protease inhibitors for five times at 4°C. After washing, the beads were resuspended with 850 μl of ice-cold NT2 buffer supplemented with 1 μl of DTT (1 M), 35 μl of EDTA (0.5 M, pH 8.0), RNase inhibitors, and protease inhibitors and incubated at 4°C for 12 hours. The beads were then washed four to five times (5 min for each time) with ice-cold NT2 buffer. After washing, the beads were resuspended with ice-cold NT2 buffer containing proteinase K and 1% SDS and incubated at 55°C for 30 min. The RNAs were then extracted using TRIzol (Invitrogen) and dissolved in 10 to 20 μl of diethyl pyrocarbonate–treated water.
ChIP-seq and data analysis
ChIP was performed as previously described (44). Briefly, 5 × 106 cells were cross-linked with 1.5% formaldehyde for 10 min at room temperature. Cross-linking was then quenched by adding 0.125 M glycine for 5 min. After washing twice with ice-cold PBS, cells were lysed with 1 ml of cytoplasm lysis buffer [5 mM Pipes (pH 8.0), 85 mM KCl, and 0.5% NP-40 with protease inhibitors] and incubated on ice for 5 min. After centrifuge at 5000g for 5 min at 4°C, the pellet was resuspended with 0.5 ml of nuclear lysis buffer [50 mM tris-HCl (pH 8.1), 10 mM EDTA, and 1% SDS with protease inhibitors]. Chromatin was subject to sonication to a size of 0.2 to 0.5 kb using Bioruptor 200. The supernatant was diluted with nine volumes of dilution buffer [0.01% SDS, 1.1% Trition X-100, 2 mM EDTA, 50 mM tris-HCl (pH 8.1), and 150 mM NaCl] containing protease inhibitors. Chromatin samples were incubated with specific antibodies at 4°C overnight. Then, the chromatin-protein supernatant and protein A/G beads were coincubated for 12 hours at 4°C. The beads were then washed with low salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.1), and 150 mM NaCl], high-salt buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.1), and 500 mM NaCl] and LiCl salt buffer [0.25 M LiCl, 1% IGEPAL-CA630 (NP-40), 1% deoxycholic acid (sodium salt), 1 mM EDTA, and 10 mM tris (pH 8.1)], respectively. After wash twice with Tris-EDTA (TE) buffer, cross-link was reversed in TE buffer at 65°C for 12 hours. The samples were then digested by proteinase K and RNase A. DNA was purified with phenol:chloroform:isoamyl alcohol. The precipitated DNA samples were either performed for DNA deep sequencing or analyzed by RT-PCR.
For ChIP-seq data analysis, clean reads were mapped to the mouse genome (mm10) using Bowtie 2 (version 2.1.0). PCR duplicates were removed using SAMtools (version 0.1.19). The binding sites of KLF4 and p53 were called using the MACS2 (version 2.2.7.1) as described (30). Differential binding analyses of ChIP-seq peak data were performed using DiffBind package (version 3.2.6) (Stark R, Brown GD. DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. http://bioconductor.org/packages/release/bioc/html/DiffBind.html). Enriched peaks were annotated using ChIPseeker package (version 1.20.0) (45). The genome-wide average of peak intensity for each sample was calculated using deepTools computeMatrix (version 3.5.0) and illustrated using deepTools plotHeatmap (version 3.5.0) (46). The ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) database (accession number: GSE188431).
Nucleus and cytoplasm fragmentation
Nucleus and cytoplasm fragmentation was performed following the standard protocol (47). Briefly, 2 × 107 cells were washed with PBS. Cells were collected by centrifugation at 1000 rpm for 10 min and resuspended with cytoplasm extract buffer [10 mM Hepes, 50 mM KCl, 1 mM EDTA, 0.1% (v/v) NP-40, and 1 mM DTT supplemented with Roche complete protease inhibitor (Roche Applied System)]. Cells were incubated on ice for 5 min and centrifuged at 3000 rpm for 5 min at 4°C. Supernatants were harvested for cytoplasm fraction. The pellet was washed three times with cytoplasm extract buffer without NP-40. Then, the pellet was resuspended with nucleus extract buffer [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, and 0.3% NP-40 supplemented with Roche complete protease inhibitor (Roche Applied System)] and incubated on ice for 10 min. Centrifuge for 5 min at 12,000 rpm was done, and supernatants were harvested for nucleus fraction.
Lentivirus package and cell transfection
Human embryonic kidney 293T cells were grown in 15-cm2 culture dishes and prepared for transfection at the time of 80% confluency. Twelve micrograms of expression vector (pLKO.1 or pTRIPZ), 6 μg of pSPAX, and 6 μg of pMD 2.0 plasmid were cotransfected with opti-MEM containing polyethyleneimine (Sigma-Aldrich). At 18 hours after transfection, opti-MEM was replaced with DMEM containing 5% fetal bovine serum. The viral supernatant was harvested at 48 and 72 hours after transfection. For Lnc956, p53, and Mettl3 knockdown, ESCs were transducted with virus and polybrene (10 μg/ml).
RNA-seq and data analyses
Total RNA was isolated from cultured cells by TRIzol (Tiangen, DP424). The cDNA libraries were constructed with the TruSeq RNA Sample Preparation Kit (Illumina) and sequenced on the HiSeq X Ten platform. After sequencing, the clean reads were mapped to mouse reference genome (mm10) using TopHat2 software (version 2.0.8). The values of gene expression and DEGs were calculated by Cufflinks (version 2.1.1) and Cuffdiff (Version 2.1.1) software, respectively. GO enrichment was carried out using DAVID. The heatmaps were constructed by the “gplots” R packages with default parameter. The RNA-seq data have been deposited in the GEO database (accession number: GSE188431).
Statistical analyses
All statistical analyses were two-tailed test and carried out using Prism (GraphPad). P < 0.05 was used to set a cutoff of statistical analysis. An unpaired Student’s t test was applied for two-group comparisons when variance between them was equal (determined by the F test). For the normally distributed data with unequal variance, unpaired t test with Welch’s correction was used. All the data were presented as means ± SEM.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (31930027), National Key Research & Developmental Program of China (2021YFA1102002), and Yunnan Fundamental Research Projects (2021000051 and 202201AS070041).
Author contributions: H.M. and Y.N. performed most of the experiments. L.W. analyzed the RNA-seq and ChIP-seq data. W.Z. prepared the in vitro RNA pulldown samples for mass spectrometry analysis. H.M., Y.N., and L.W. prepared the figures. H.M. and P.Z. wrote the manuscript. P.Z. supervised the study.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq and ChIP-seq data are available through NCBI GEO datasets (access number GSE188431). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 and S2
Dataset S1 to S5
Other Supplementary Material for this : manuscript includes the following:
Datasets S1 to S5
REFERENCES AND NOTES
- 1.I. Vitale, G. Manic, R. De Maria, G. Kroemer, L. Galluzzi, DNA damage in stem cells. Mol. Cell 66, 306–319 (2017). [DOI] [PubMed] [Google Scholar]
- 2.S. P. Wyles, E. B. Brandt, T. J. Nelson, Stem cells: The pursuit of genomic stability. Int. J. Mol. Sci. 15, 20948–20967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U. Weissbein, N. Benvenisty, U. Ben-David, Quality control: Genome maintenance in pluripotent stem cells. J. Cell Biol. 204, 153–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. Tapia, H. R. Scholer, Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell 19, 298–309 (2016). [DOI] [PubMed] [Google Scholar]
- 5.E. D. Tichy, P. J. Stambrook, DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 314, 1929–1936 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B. Zhao, W. Zhang, Y. Cun, J. Li, Y. Liu, J. Gao, H. Zhu, H. Zhou, R. Zhang, P. Zheng, Mouse embryonic stem cells have increased capacity for replication fork restart driven by the specific Filia-Floped protein complex. Cell Res. 28, 69–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.X. Q. Ge, J. Han, E. C. Cheng, S. Yamaguchi, N. Shima, J. L. Thomas, H. F. Lin, Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Reports 5, 185–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. Zhao, W. D. Zhang, Y. L. Duan, Y. Q. Lu, Y. X. Cun, C. H. Li, K. Guo, W. H. Nie, L. Li, R. Zhang, P. Zheng, Filia is an ESC-specific regulator of DNA damage response and safeguards genomic stability. Cell Stem Cell 16, 684–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Xiong, D. Todorova, N. Y. Su, J. Kim, P. J. Lee, Z. Shen, S. P. Briggs, Y. Xu, Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J. Cell Biol. 208, 513–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. D. Tichy, R. Pillai, L. Deng, L. Liang, J. Tischfield, S. J. Schwemberger, G. F. Babcock, P. J. Stambrook, Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 19, 1699–1711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.P. Ruis, D. Van Ly, V. Borel, G. R. Kafer, A. McCarthy, S. Howell, R. Blassberg, A. P. Snijders, J. Briscoe, K. K. Niakan, P. Marzec, A. J. Cesare, S. J. Boulton, TRF2-independent chromosome end protection during pluripotency. Nature 589, 103–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Markiewicz-Potoczny, A. Lobanova, A. M. Loeb, O. Kirak, T. Olbrich, S. Ruiz, E. Lazzerini Denchi, TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 589, 110–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. Zalzman, G. Falco, L. V. Sharova, A. Nishiyama, M. Thomas, S. L. Lee, C. A. Stagg, H. G. Hoang, H. T. Yang, F. E. Indig, R. P. Wersto, M. S. Ko, Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. Li, Y. He, W. Dubois, X. Wu, J. Shi, J. Huang, Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell 46, 30–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.X. Fu, S. Wu, B. Li, Y. Xu, J. Liu, Functions of p53 in pluripotent stem cells. Protein Cell 11, 71–78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. Lin, C. Chao, S. Saito, S. J. Mazur, M. E. Murphy, E. Appella, Y. Xu, p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7, 165–171 (2005). [DOI] [PubMed] [Google Scholar]
- 17.N. K. Dhaliwal, L. E. Abatti, J. A. Mitchell, KLF4 protein stability regulated by interaction with pluripotency transcription factors overrides transcriptional control. Genes Dev. 33, 1069–1082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. R. Kristensen, J. Gsponer, L. J. Foster, Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 9, 689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Z. Nagy, E. Soutoglou, DNA repair: Easy to visualize, difficult to elucidate. Trends Cell Biol. 19, 617–629 (2009). [DOI] [PubMed] [Google Scholar]
- 20.S. G. Chankova, E. Dimova, M. Dimitrova, P. E. Bryant, Induction of DNA double-strand breaks by zeocin in Chlamydomonas reinhardtii and the role of increased DNA double-strand breaks rejoining in the formation of an adaptive response. Radiat. Environ. Biophys. 46, 409–416 (2007). [DOI] [PubMed] [Google Scholar]
- 21.M. van Kan, K. E. Burns, P. Browett, N. A. Helsby, A higher throughput assay for quantification of melphalan-induced DNA damage in peripheral blood mononuclear cells. Sci. Rep. 9, 18912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. C. Poh, J. Chen, Y. Hong, H. Yi, S. Zhang, J. Chen, D. C. Wu, L. Wang, Q. Jia, R. Singh, W. Yao, Y. Tan, A. Tajik, T. S. Tanaka, N. Wang, Generation of organized germ layers from a single mouse embryonic stem cell. Nat. Commun. 5, 4000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.I. Chambers, J. Silva, D. Colby, J. Nichols, B. Nijmeijer, M. Robertson, J. Vrana, K. Jones, L. Grotewold, A. Smith, Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 (2007). [DOI] [PubMed] [Google Scholar]
- 24.A. Filipczyk, C. Marr, S. Hastreiter, J. Feigelman, M. Schwarzfischer, P. S. Hoppe, D. Loeffler, K. D. Kokkaliaris, M. Endele, B. Schauberger, O. Hilsenbeck, S. Skylaki, J. Hasenauer, K. Anastassiadis, F. J. Theis, T. Schroeder, Network plasticity of pluripotency transcription factors in embryonic stem cells. Nat. Cell Biol. 17, 1235–1246 (2015). [DOI] [PubMed] [Google Scholar]
- 25.A. Sigal, V. Rotter, Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer Res. 60, 6788–6793 (2000). [PubMed] [Google Scholar]
- 26.D. P. Lane, Cancer. p53, guardian of the genome. Nature 358, 15–16 (1992). [DOI] [PubMed] [Google Scholar]
- 27.X. Chen, H. Xu, P. Yuan, F. Fang, M. Huss, V. B. Vega, E. Wong, Y. L. Orlov, W. Zhang, J. Jiang, Y. H. Loh, H. C. Yeo, Z. X. Yeo, V. Narang, K. R. Govindarajan, B. Leong, A. Shahab, Y. Ruan, G. Bourque, W. K. Sung, N. D. Clarke, C. L. Wei, H. H. Ng, Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 28.D. C. Di Giammartino, A. Kloetgen, A. Polyzos, Y. Liu, D. Kim, D. Murphy, A. Abuhashem, P. Cavaliere, B. Aronson, V. Shah, N. Dephoure, M. Stadtfeld, A. Tsirigos, E. Apostolou, KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat. Cell Biol. 21, 1179–1190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Han, W. Li, C. H. Lin, J. Yang, C. Shang, S. T. Nurnberg, K. K. Jin, W. H. Xu, C. Y. Lin, C. J. Lin, Y. Q. Xiong, H. C. Chien, B. Zhou, E. Ashley, D. Bernstein, P. S. Chen, H. S. V. Chen, T. Quertermous, C. P. Chang, A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Kim, J. L. Chu, X. H. Shen, J. L. Wang, S. H. Orkin, An extended transcriptional network for pluripotency of embryonic stem cells. Cell 133, 1049–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.P. G. Komarov, E. A. Komarova, R. V. Kondratov, K. Christov-Tselkov, J. S. Coon, M. V. Chernov, A. V. Gudkov, A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737 (1999). [DOI] [PubMed] [Google Scholar]
- 33.G. Xi, X. Shen, C. Wai, M. F. White, D. R. Clemmons, Hyperglycemia induces vascular smooth muscle cell dedifferentiation by suppressing insulin receptor substrate-1-mediated p53/KLF4 complex stabilization. J. Biol. Chem. 294, 2407–2421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Marechal, L. Zou, DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.I. Hickson, Y. Zhao, C. J. Richardson, S. J. Green, N. M. Martin, A. I. Orr, P. M. Reaper, S. P. Jackson, N. J. Curtin, G. C. Smith, Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64, 9152–9159 (2004). [DOI] [PubMed] [Google Scholar]
- 36.P. M. Reaper, M. R. Griffiths, J. M. Long, J. D. Charrier, S. Maccormick, P. A. Charlton, J. M. Golec, J. R. Pollard, Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 7, 428–430 (2011). [DOI] [PubMed] [Google Scholar]
- 37.C. Zhang, L. Chen, D. Peng, A. Jiang, Y. He, Y. Zeng, C. Xie, H. Zhou, X. Luo, H. Liu, L. Chen, J. Ren, W. Wang, Y. Zhao, METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 79, 425–442.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Y. Zhou, P. Zeng, Y. H. Li, Z. Zhang, Q. Cui, SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 44, e91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Y. Wang, Y. Li, J. I. Toth, M. D. Petroski, Z. Zhang, J. C. Zhao, N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Czechanski, C. Byers, I. Greenstein, N. Schrode, L. R. Donahue, A. K. Hadjantonakis, L. G. Reinholdt, Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 9, 559–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.H. P. Chu, C. Cifuentes-Rojas, B. Kesner, E. Aeby, H. G. Lee, C. Wei, H. J. Oh, M. Boukhali, W. Haas, J. T. Lee, TERRA RNA antagonizes ATRX and protects telomeres. Cell 170, 86–101.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Bierhoff, Analysis of lncRNA-protein interactions by RNA-protein pull-down assays and RNA immunoprecipitation (RIP). Methods Mol. Biol. 1686, 241–250 (2018). [DOI] [PubMed] [Google Scholar]
- 43.R. Jain, T. Devine, A. D. George, S. V. Chittur, T. E. Baroni, L. O. Penalva, S. A. Tenenbaum, RIP-Chip analysis: RNA-binding protein immunoprecipitation-microarray (Chip) profiling. Methods Mol. Biol. 703, 247–263 (2011). [DOI] [PubMed] [Google Scholar]
- 44.J. Huang, R. Sengupta, A. B. Espejo, M. G. Lee, J. A. Dorsey, M. Richter, S. Opravil, R. Shiekhattar, M. T. Bedford, T. Jenuwein, S. L. Berger, p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007). [DOI] [PubMed] [Google Scholar]
- 45.G. Yu, L. G. Wang, Q. Y. He, ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 46.F. Ramirez, D. P. Ryan, B. Gruning, V. Bhardwaj, F. Kilpert, A. S. Richter, S. Heyne, F. Dundar, T. Manke, deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. F. Pendle, P. J. Shaw, Isolation of nuclei and nucleoli. Methods Mol. Biol. 1511, 31–44 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 and S2
Dataset S1 to S5
Datasets S1 to S5