Abstract
Background: breast cancer (BC) is the world’s most prevalent cancer in the female population, with 2.3 million new cases diagnosed worldwide in 2020. The great efforts made to set screening campaigns, early detection programs, and increasingly targeted treatments led to significant improvement in patients’ survival. The Full-Field Digital Mammograph (FFDM) is considered the gold standard method for the early diagnosis of BC. From several previous studies, it has emerged that breast density (BD) is a risk factor in the development of BC, affecting the periodicity of screening plans present today at an international level. Objective: in this study, the focus is the development of mammographic image processing techniques that allow the extraction of indicators derived from textural patterns of the mammary parenchyma indicative of BD risk factors. Methods: a total of 168 patients were enrolled in the internal training and test set while a total of 51 patients were enrolled to compose the external validation cohort. Different Machine Learning (ML) techniques have been employed to classify breasts based on the values of the tissue density. Textural features were extracted only from breast parenchyma with which to train classifiers, thanks to the aid of ML algorithms. Results: the accuracy of different tested classifiers varied between 74.15% and 93.55%. The best results were reached by a Support Vector Machine (accuracy of 93.55% and a percentage of true positives and negatives equal to TPP = 94.44% and TNP = 92.31%). The best accuracy was not influenced by the choice of the features selection approach. Considering the external validation cohort, the SVM, as the best classifier with the 7 features selected by a wrapper method, showed an accuracy of 0.95, a sensitivity of 0.96, and a specificity of 0.90. Conclusions: our preliminary results showed that the Radiomics analysis and ML approach allow us to objectively identify BD.
Keywords: breast cancer, mammography, breast density, machine learning, radiomics
1. Introduction
Breast cancer (BC) is the most common cancer among women in the world; about one in eight women will develop breast cancer in their lifetime. Breast cancer survival rates have increased in recent years, and the number of deaths associated with this disease has been steadily declining, largely due to factors such as early detection and personalized treatment approaches. Screening for the early detection of breast cancer is of great interest, as it significantly increases the patient’s chances of survival. It is important to develop techniques to identify key risk factors to benefit from further screening and preventive therapies [1,2,3]. An important role is played by mammography, introduced in the 1960s, which remains the gold standard for breast cancer screening [4]. Over the years, we have witnessed the total replacement of radiographic films with solid-state detectors capable of transforming X-rays into electrical signals to produce the final image directly in a digital format. This technique is called full-field digital mammography (FFDM). Processing and analyzing digital mammograms are significant challenges and can provide quantitative results on breast characteristics. In particular, indices derived from textural patterns of the breast parenchyma can be associated with risk factors. It is established that women with dense breasts have a double risk of developing BC [5,6]. Furthermore, for women with dense breasts, mammography suffers from a loss of sensitivity [7,8]. Breast density (BD) is currently assessed only on a qualitative method by radiologist, therefore based only on a visual assessment, according to the international standard Breast Imaging—Reporting and Data System (BI-RADS) in four classes [9]. However, this qualitative evaluation is subject to intra and inter observer variability. Most of the inter-reader agreement is in the two classifications at the extreme ends of the spectrum (almost entirely fatty and extremely dense), with considerable variation in the agreement between the two middle-density classes.
Therefore, it is desirable to implement a radiomics approach [10], to obtain a quantitative evaluation in order to increase diagnostic accuracy.
The radiomic analysis includes several phases: segmentation of the target region or volume of interest; feature extraction; reduction in the number of characteristics extracted; analysis with the construction of a predictive model and the validation of the results [11,12,13,14,15,16,17,18,19]. The characteristics can be morphological and of a first, second, and higher statistical order.
Such radiomic feature extraction approaches, widely applied in the field of medical imaging [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60], can be combined with the use of regression and classification techniques, with particular regard to machine learning (ML) algorithms, which have been widely used in the medical and biomedical fields for various purposes, ranging from the study of bio-signals and bioimaging [61,62,63,64,65,66,67,68,69] to the analysis of health processes [70,71,72,73].
Today, the main scenario is the oncological environment, since the characteristics of radiomics provide data on the tumor microenvironment that could be related to the risk of developing cancer, histological grade, prognosis, response to treatment and survival in countless tumors [20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60].
Given the importance of breast density as a risk factor for BC and increasing attention to screening strategies based on density and considering the low accuracy that could be correlated with a qualitative assessment, due to the inter- and intra-reader variability in radiologists’ interpretation, the aim of this study was to combine radiomics and ML in order to explore the possibility of correlating statistical and textural characteristics of digital images (such as interactions between pixels or gray levels) with BD [74,75,76,77], to obtain a more objective evaluation. The accuracy of the classifiers on ML algorithms was then evaluated to estimate the breast density class.
2. Methods
2.1. Study Population
This observational retrospective study was approved by the ethics committee of the University “Luigi Vanvitelli”, Naples, Italy, with deliberation n. 469 of 23/07/2019 and, informed consent was waived by the ethics committee. All methods were carried out according to National regulations and guidelines.
We enrolled all consecutive women, who underwent mammography for breast cancer screening programs, at the Breast Unit of the University Hospital “Luigi Vanvitelli”, Naples, from June 2020 to November 2020.
A validation cohort, consisting of a total of 51 patients, obtained from the Breast Team of “Villa Fiorita”, Capua, Italy, was considered in this study and enrolled.
Moreover, we assessed breast structures according to BI-RADS 5th [7].
2.2. Digital Mammograms Acquisition
All the women from the internal cohort underwent two-view bilateral full-field digital mammography according to the standard protocol, using Giotto Class (IMS GIOTTO S.p.A., Sasso Marconi, Bologna, Italy). All the women from the external cohort underwent two-view bilateral full-field digital mammography according to the standard protocol, using GE SENOGRAPHE ESSENTIAL (General Electric Company, Milano, Italy).
For both the internal and external cohort, patient images were acquired both in cranio-caudal (CC) and medio-lateral-oblique (MLO) views, on the right and left sides.
The processing is carried out on the “FOR PRESENTATION” images, i.e., raw images, because being proportional to the attenuation of the X-rays on the breast tissue, they retain the original information on the attenuation of the X-rays and therefore are more suitable for quantitative analyzes.
In total, four images are obtained for each patient. The images have a resolution of 2729 × 3580 (on average for all images) and a laptop with a 2.40 GHz Intel Core i5-9300H CPU, 8 GB of RAM, and the NVIDIA GeForce GTX 1650 graphics card was used for the image processing, training, and machine testing Learning algorithms.
2.3. Image Pre-Processing
Raw images undergo two main stages of preprocessing: image enhancement, to improve the quality and information content of the original data, and breast segmentation, to identify the air-tissue interface and remove the pectoral muscle, mainly in the MLO configuration.
In the first phase, the equalization of the histogram, which consists in applying a logarithmic transformation in order to enhance the pixels of the image and display the different structures of which the breast is composed, and a gray scale normalization with a is used as a range of values between [0,1]. After seeing the image of the breast, three areas can be distinguished: pectoral muscle, which appears whiter than the rest of the breast; mammary tissues, consisting mainly of adipose and fibroglandular tissue; and the region occupied by the air surrounding the breast, also called the bottom.
The purpose of the second step is to isolate only the breast tissue, i.e., the Region of Interest (ROI), eliminating both the pectoral muscle and the background.
Segmentation was achieved using a software tool implemented in MATLAB R2007a (The MathWorks, Inc., Natik, MA, USA), manually drawn by two expert radiologists (22 and 15 years of breast imaging experience, respectively); the segmentation was performed by the two radiologists first separately and then together and in accordance with each other.
2.4. Features Extraction and Features Selection
A total of 229 features were extracted: 1–112 features of Haralick; 113–157 features of Law; 158–185 features of Run Length; 186–215 features of Wavelet; 216–227 features of the histogram; 228 Fractal dimensions; 229 Local binary patterns. The textural features were calculated according to the Image Biomarker Standardization Initiative [78].
To eliminate redundant features, both a selection of filtering features based on correlation and a selection of wrapper features were used in this study. Filter feature selection is a method of applying statistical measures by assigning a certain score to each feature. The characteristics are ranked by score and selected or removed from the dataset according to the classification. Therefore, a filter characteristics selection method that searches for correlation between characteristics using functions cor and find Correlation of the caret package [79] with a cutoff set to 0.9 [80] was a performed.
The wrapper’s feature selection method is to set up the problem of selecting a suitable set of features as a search problem between sets of different combinations of features. To select the best set, you can use a predictive model in which you assign a score to each combination. The research process can be carried out following different approaches, some of these are for example methodical algorithms, such as “best-first” or stochastic methods such as “random-hill climbing” or heuristic methods. An example of a wrapper method is Recursive Feature Elimination (RFE).
The implementation of the wrapper features selection was achieved using the rfe and rfeControl functions of the caret package [79] by choosing an algorithm as a model to evaluate the accuracy of a Random Forest (RF) and choosing equal to 100 the number of features to be selected among the starting point. This value was chosen empirically, evaluating that the maximum accuracy in relation to the number of features chosen is already reached before the selected value, making it unnecessary to select multiple features.
2.5. Statistical and Machine Learning Analysis
A nonparametric Mann Whitney test was performed to identify statistically significant differences in radiomic metrics among groups for age and BMI.
A p-value < 0.05 was considered significant.
The data obtained through the extraction of features and the application of a feature selection algorithm are classified through different ML algorithms (support vector machine (SVM), linear discrimination analysis (LDA), artificial neural network (ANN), decision tree (DT), Random Forest Tree) [81].
To solve the problem of the unbalanced dataset, we transformed it into a binary classification problem by associating patients with classes A and B and patients with classes C and D [77]. The division into training and testing on the dataset was made by partitioning the data and leaving 80% for training and 20% for model testing.
The performance of a classification can be summarized using the Confusion Matrix, while for binary classification, it is also possible to add the ROC curves (Receiver operation features).
Sensitivity, specificity, and accuracy were calculated. Sensitivity is the portion of correctly identified true positives. Specificity is the portion of true negatives correctly identified. Accuracy is the parameter that summarizes the effectiveness of the classification by evaluating the proportion of correct predictions (both positive and negative) on the total number of cases examined.
The classification analysis was cross-validated using the 10-fold cross-validation approach, and median values of AUC, accuracy, sensitivity, and specificity were obtained.
The best model was chosen considering the highest area under the ROC curve and the highest accuracy. The McNemar test was used to assess the statistical significance of dichotomous tables. A p-value < 0.05 was considered significant.
All analyses were performed with the R-Studio Version 1.3.959 (https://www.rstudio.com/ Accessed date 14 March 2022) [82].
3. Results
A total of 168 patients from the internal cohort aged 38 to 84 were enrolled over a 5-month period from June 2020 to November 2020.
A total of 51 patients from the validation cohort aged 37 to 80 were enrolled over a 4-month period from January 2021 to April 2021. Patients’ characteristics are reported in Table 1 and Table 2.
Table 1.
Number of patients across breast density and age group.
| Age Group (yrs) | Breast Density | |||
|---|---|---|---|---|
| A | B | C | D | |
| Internal Cohort | ||||
| 15–44 | 0 | 6 | 2 | 7 |
| 45–54 | 4 | 18 | 25 | 9 |
| 55–64 | 8 | 41 | 5 | 15 |
| 65–78 | 5 | 14 | 0 | 5 |
| >78 | 0 | 3 | 1 | 0 |
| Sum | 17 | 82 | 33 | 36 |
| Validation Cohort | ||||
| 15–44 | 0 | 1 | 1 | 2 |
| 45–54 | 1 | 2 | 2 | 1 |
| 55–64 | 1 | 4 | 3 | 3 |
| 65–78 | 2 | 8 | 5 | 2 |
| >78 | 2 | 6 | 3 | 2 |
| Sum | 6 | 21 | 14 | 10 |
Table 2.
Number of patients across breast density and BMI.
| Age Group (yrs) | Breast Density | |||
|---|---|---|---|---|
| A | B | C | D | |
| Internal Cohort | ||||
| Normal (<25 kg/m2) | 12 | 15 | 9 | 8 |
| Overweight (25–29 kg/m2) | 9 | 17 | 16 | 10 |
| Obese (≥30 kg/m2) | 11 | 35 | 13 | 13 |
| Sum | 32 | 67 | 38 | 31 |
| Validation Cohort | ||||
| Normal (<25 kg/m2) | 4 | 2 | 3 | 4 |
| Overweight (25–29 kg/m2) | 6 | 3 | 4 | 5 |
| Obese I (≥30 kg/m2) | 6 | 6 | 3 | 5 |
| Sum | 16 | 11 | 10 | 14 |
To reduce the computational problem in the classification analysis considering the small size for the four classes, we transformed the dataset into a binary dataset.
According to the binary classification, the internal dataset is transformed into 99 patients with densities A and B: fat breast; and 69 patients with densities C and D: dense breast.
While the external validation cohort included 27 fat breast and 24 dense breast.
The results related to the features selection correlation method was a subclass of 35 textural characteristics, as shown in Table 3. No statistically significant difference (p-value > 0.05 at Kruskal Wallis test) was found in the median values of these 35 selected textural features for age subgroups (15–44; 45–54; 55–64; 65–78; ≥78 years). Moreover, no statistically significant difference (p-value > 0.05 at Kruskal Wallis test) was found in median values of these 35 selected textural features for BMI subgroups (Normal (<25 kg/m2); Overweight (25–29 kg/m2); Obese (≥30 kg/m2)).
Table 3.
35 features selected with the filter features selection method.
| Variable Number | Variable Name |
|---|---|
| V687 | Fractal dimensions |
| V685 | 95th percentile Hist feat |
| V684 | mean 5th percentile Hist feat |
| V682 | Entropy Hist feat |
| V679 | Std Hist feat |
| V678 | Min histogram value |
| V677 | Max histogram value |
| V674 | Kurtosis Diagonal comp. Wavelet 5° iteration |
| V672 | Kurtosis Vertical comp. Wavelet 5° iteration |
| V671 | Variance Vertical comp. Wavelet 5° iteration |
| V670 | Kurtosis Horizontal comp. Wavelet 5° iteration |
| V669 | Variance Horizontal comp. Wavelet 5° iteration |
| V669 | Kurtosis Diagonal comp. Wavelet 4° iteration |
| V668 | Kurtosis Vertical comp. Wavelet 4° iteration |
| V666 | Variance Vertical comp. Wavelet 4° iteration |
| V665 | Kurtosis Horizontal comp. Wavelet 4° iteration |
| V664 | Kurtosis Diagonal comp. Wavelet 3° iteration |
| V662 | Kurtosis Vertical comp. Wavelet 3° iteration |
| V660 | Kurtosis Horizontal comp. Wavelet 3° iteration |
| V656 | Kurtosis Diagonal comp. Wavelet 2° iteration |
| V654 | Kurtosis Vertical comp. Wavelet 2° iteration |
| V652 | Kurtosis Horizontal comp. Wavelet 2° iteration |
| V646 | Kurtosis Horizontal comp. Wavelet 1° iteration |
| V641 | Run Percentage RL 180 degrees |
| V636 | Low Gray Level Run Emphasys RL 90 degrees |
| V633 | Run Percentage RL 90 degrees |
| V631 | Short Run Emphasys RL 90 degrees |
| V618 | Short Run Emphasys RL 0 degrees |
| V607 | Mean Map8 Law |
| V598 | Standard Deviation Map6 Law |
| V569 | Haralick D120 Measure of correlation I |
| V530 | Haralick D30 Energy |
| V518 | Haralick D15 Correlation |
| V473 | Maximal Correl. Coeff. (Mean 0°, 45°, 90°, 135°) |
| V469 | Haralick Entropy (Mean 0°, 45°, 90°, 135°) |
Following the application of the Wrapper feature selection, a subclass of 7 features was obtained and reported in Table 4. No statistically significant difference (p value > 0.05 at Kruskal Wallis test) was found in the median values of these 7 selected textural features for age subgroups (15–44; 45–54; 55–64; 65–78; ≥78 years). Moreover, no statistically significant difference (p value > 0.05 at Kruskal Wallis test) was found in the median values of these 7 selected textural features for BMI subgroups (Normal (<25 kg/m2); Overweight (25–29 kg/m2); Obese I (≥30 kg/m2)).
Table 4.
7 features selected with the wrapper features selection method.
| Variable Number | Variable Name |
|---|---|
| V558 | Energy Haralick D120 |
| V552 | Entropy Haralick D60 |
| V544 | Energy Haralick D60 |
| V566 | Sum Entropy Haralick D120 |
| V530 | Energy Haralick D30 |
| V614 | Skewness Map9 Law |
| V516 | Energy Haralick D15 |
Only the Energy Haralick D30 structural feature was identified as a significant predictor by the two different feature selection methods.
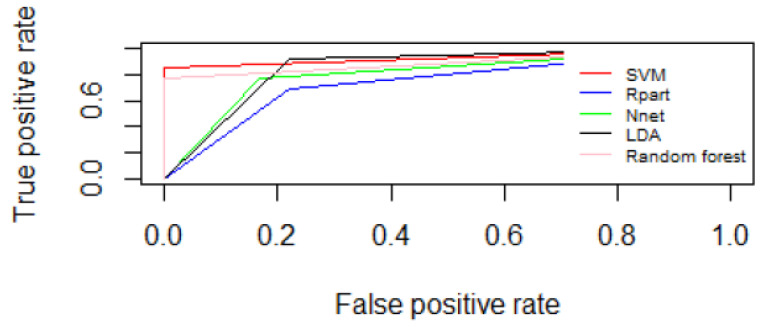
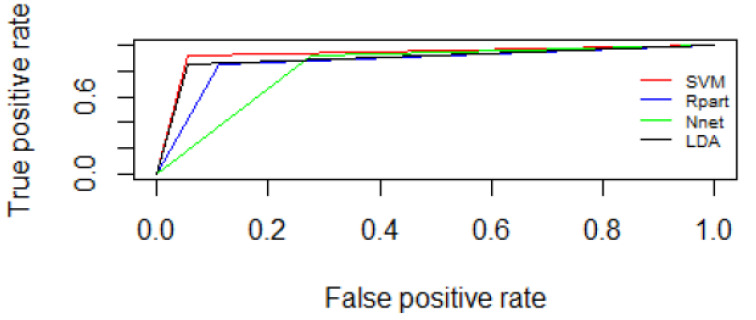
Table 5 and Table 6 reported the preliminary results of the ML algorithms tested using the two feature selection approaches: the filter feature selection correlation method and the feature selection wrapper method. Figure 1 and Figure 2 report ROC curves between fat and dense breasts considering the 35 selected textural features by means of filter feature selection correlation method and ROC curves between fat and dense breasts considering the 7 features selected with the wrapper features selection method.
Table 5.
Classification evaluation metrics between fat and dense breasts with elimination of correlate features (filter feature selection correlation method).
| Method | Accuracy | 95% CI | Kappa | Sensitivity | Specificity | p Value |
|---|---|---|---|---|---|---|
| SVM | 0.93 | 0.79–0.99 | 0.86 | 1.00 | 0.85 | <0.001 |
| Random Forest Tree | 0.91 | 0.79–0.99 | 0.86 | 1.00 | 0.85 | <0.001 |
| LDA | 0.84 | 0.66–0.94 | 0.68 | 0.778 | 0.92 | <0.01 |
| ANN using the nnet package | 0.74 | 0.55–0.88 | 0.47 | 0.778 | 0.69 | <0.01 |
| Decision Tree using the Rpart package | 0.74 | 0.55–0.88 | 0.47 | 0.778 | 0.69 | <0.01 |
Note. SVM: support vector machine, LDA: linear discrimination analysis, ANN: artificial neural network.
Table 6.
Classification evaluation metrics between fat and dense breasts with RFE.
| Method | Accuracy | 95% CI | Kappa | Sensitivity | Specificity | p Value |
|---|---|---|---|---|---|---|
| SVM | 0.93 | 0.79–0.99 | 0.87 | 0.94 | 0.92 | <0.001 |
| LDA | 0.90 | 0.74–0.98 | 0.80 | 0.94 | 0.85 | <0.01 |
| ANN using the nnet package | 0.68 | 0.49–0.83 | 0.34 | 0.83 | 0.46 | <0.01 |
| Decision Tree using the Rpart package | 0.87 | 0.70–0.96 | 0.73 | 0.89 | 0.85 | <0.01 |
Note. SVM: support vector machine, LDA: linear discrimination analysis, ANN: artificial neural network.
Figure 1.
ROC curves between fat and dense breasts considering the 35 selected textural features by means of filter feature selection correlation method.
Figure 2.
ROC curves between fat and dense breasts considering the 7 features selected with the wrapper features selection method.
The accuracy varied between 74.15% and 93.55% using the filter feature selection correlation method; while using a feature selection wrapper method, the range of accuracy was 87.10% and 93.55%. In both cases, the SVM algorithm, with k = 0.8675, reached the best results (accuracy of 93.55% and a percentage of true positives and negatives equal to TPP = 94.44% and TNP = 92.31%). The F1 score for the SVM algorithm was equal to 0.86. The training and prediction time was a few seconds.
Considering the external validation cohort, the SVM, including the 35 predictors obtained from the feature selection correlation method, showed an accuracy of 0.94, a sensitivity of 0.97, and a specificity of 0.88.
Considering the external validation cohort, the SVM, as the best classifier with the 7 features selected by the wrapper method, showed an accuracy of 0.95, a sensitivity of 0.96, and a specificity of 0.90. The best SVM classifier had the following configuration setting: Linear SVM; kernel function: linear; kernel scale: automatic; box constraint level: 1; multiclass method: one-vs-one; standardize data: true; optimizer options; and hyperparameter options disabled.
4. Discussion
Breast density is reflected as an independent increased risk factor for breast cancer, and denser breasts can mask the presence of cancer, especially in the early stages, when it is critically important to make the diagnosis to ensure a positive prognosis for the patient. The purpose of the study is to define a quantitative analysis of mammographic images to support the qualitative assessment of breast density by radiologists. Although evaluation of BD has shown several values for estimating BC risk, a qualitative or BI-RADS approach is highly subjective to inter- and intra-observer variability [83,84,85]. In contrast, radiomics evaluation of mammographic data could allow a more complex characterization of the intricacy and morphologic organization of the parenchymal patterns.
Radiomics analysis is based on the conversion of medical images to higher dimensional data by using computer-based algorithms. Several authors showed that Radiomics data, when correlated with clinical-pathological outcomes, could allow a noninvasive and cost-effective approach to favor precision medicine [86,87,88,89,90,91,92,93,94,95,96]. Preliminary studies showed that radiomics features were correlated with BC independent of BD and, therefore, had the potential to augment BD in assessing a woman’s risk of developing cancer [97,98,99].
The possibility of objectively identifying risk factors, such as BD, allows for better risk stratification of the patient and better management. We assessed 168 patients, 99 patients with fat breast and 69 patients with dense breast. A total of 229 features were extracted, and after a selection correlation method, we obtained a subclass of 35 textural characteristics. Thanks to the extraction of the texture characteristics from the image dataset, it was possible to evaluate a binary classification using ML algorithms, which led to results that show a good agreement with the radiologist’s report; a maximum accuracy of 93.55% was obtained in the internal cohort with an SVM method and percentage of true positives and negatives equal to TPP = 94.44% and TNP = 92.31%. Considering the external validation cohort, the SVM, as the best classifier with the 7 features selected by the wrapper method, showed an accuracy of 0.95, a sensitivity of 0.96, and a specificity of 0.90.
In line with our results, also Kantos et al. [99] assessed phenotypes of mammographic parenchymal complexity by using radiomic features and evaluated their associations with breast density and other breast cancer risk factors. They showed that an unsupervised clustering identified four phenotypes with increasing parenchymal complexity that were reproducible between training and test sets. Breast density was not strongly correlated with the phenotype category. The low- to intermediate-complexity phenotype had the lowest proportion of dense breasts, whereas similar proportions were observed across other phenotypes. In the independent case-control sample, phenotypes showed a significant association with BC, resulting in a higher discriminatory capacity when added to a model with BD and body mass index.
Li et al. [100] combined the characteristics of normal parenchyma from the contralateral breast with radiomic features of breast tumors to improve the accuracy in the diagnosis of BC. They showed that the performance of the combined lesion and parenchyma classifier was better than that of the lesion features alone. Overall, six radiomic features, such as spiculation, margin sharpness, size, circularity from the tumor feature set, and skewness and power law beta from the parenchymal feature set, were selected [100].
Being able to objectively assess the risk of BC would be an important step to reducing the effects of this disease, along with its mortality rate. Yala et al. [101] tried to develop a model that used mammograms as risk factors (BD and patient age) to predict the risk of BC. They used 71,689 images for training (2732 positive vs. 68,957 negative), 8554 for validation (316 positive vs. 8238 negative), and 8869 for testing (269 positive vs. 8282). The authors proposed 3 models. The first is based on a logistic regression, containing only the risk factors; the second is based only on images, using a ResNet18 architecture. The last, a hybrid DL model that combines data from the previous two models. The hybrid model achieved an AUC of 0.70 [101].
Our results may be considered preliminary, on a still limited dataset, but they highlight the possibility of obtaining promising results with a larger dataset.
A future endpoint could be to correlate imaging metrics with clinical features, such as the age of a patient that could influence breast tissue and the possibility to obtain a hybrid model to predict the risk of BC.
The present study has several limitations. It was performed in a retrospective manner at a single academic institution, and the results may not generalize to other practice settings. We did not assess the interreader agreement for the BD assessment. In addition, we did not evaluate the BC rate considering BD categories. Future studies should be aimed at evaluating better BC detection in the context of dense breasts.
5. Conclusions
Our results showed that the Radiomics analysis and ML approach can objectively identify BD. Thanks to the extraction of the texture characteristics from the image dataset and a binary classification using ML algorithms we obtained an accuracy of 93.55% with an SVM method and a percentage of true positives and negatives equal to TPP = 94.44% and TNP = 92.31%.
Author Contributions
Methodology: M.S., F.G., G.G., R.G. (Roberta Galdiero), R.G. (Roberta Grassi), V.G. and R.G. (Roberto Grassi); Formal analysis: M.S. and R.F.; Investigation: M.S., R.F., F.G., G.G., M.P.B., F.A. (Francesca Angelone), C.R., A.M.P., F.A. (Francesco Amato), R.G. (Roberta Galdiero), V.G. and R.G. (Roberto Grassi); Writing—original draft: M.S., V.G. and R.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study has been conducted in the framework of a wider study (“Senologia per te”) promoted by SIRM (Italian Society of Medical Radiology) approved by the Ethical Committee of University “Luigi Vanvitelli” (prot. N 469 of 23/07/2019).
Informed Consent Statement
Informed consent was waived by the ethics committee.
Data Availability Statement
The data presented in this study is available in this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study has been partially funded by IMS GIOTTO S.p.A. Sasso Marconi (BO), Italy. Special thanks to Sara Vecchio at IMS for useful discussions.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hall P., Easton D. Breast cancer screening: Time to target women at risk. Br. J. Cancer. 2013;108:2202–2204. doi: 10.1038/bjc.2013.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howell A., Astley S., Warwick J., Stavrinos P., Sahin S., Ingham S., McBurney H., Eckersley B., Harvie M., Wilson M., et al. Prevention of breast cancer in the context of a national breast screening programme. J. Intern. Med. 2012;271:321–330. doi: 10.1111/j.1365-2796.2012.02525.x. [DOI] [PubMed] [Google Scholar]
- 3.Li H., Giger M.L., Olopade O.I., Margolis A., Lan L., Chinander M.R. Computerized texture analysis of mammographic parenchymal patterns of digitized mammograms. Acad. Radiol. 2005;12:863–873. doi: 10.1016/j.acra.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 4.Heywang-Köbrunner S., Viehweg P., Heinig A., Küchler C. Contrast-enhanced MRI of the breast: Accuracy, value, controversies, solutions. Eur. J. Radiol. 1997;24:94–108. doi: 10.1016/S0720-048X(96)01142-4. [DOI] [PubMed] [Google Scholar]
- 5.McCormack V.A., dos Santos Silva I. Breast Density and Parenchymal Patterns as Markers of Breast Cancer Risk: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006;15:1159–1169. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 6.Boyd N.F., Martin L.J., Yaffe M.J., Minkin S. Mammographic density and breast cancer risk: Current understanding and future prospects. Breast Cancer Res. 2011;13:223. doi: 10.1186/bcr2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spak D.A., Plaxco J.S., Santiago L., Dryden M.J., Dogan B. BI-RADS ® fifth edition: A summary of changes. Diagn. Interv. Imaging. 2017;98:179–190. doi: 10.1016/j.diii.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Carney P.A., Miglioretti D.L., Yankaskas B.C., Kerlikowske K., Rosenberg R., Rutter C.M., Geller B.M., Abraham L.A., Taplin S.H., Dignan M., et al. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann. Intern. Med. 2003;138:168–175. doi: 10.7326/0003-4819-138-3-200302040-00008. [DOI] [PubMed] [Google Scholar]
- 9.Timmers J.M., van Doorne-Nagtegaal H.J., Zonderland H.M., van Tinteren H., Visser O., Verbeek A.L., den Heeten G.J., Broeders M.J. The Breast Imaging Reporting and Data System (BI-RADS) in the Dutch breast cancer screening programme: Its role as an assessment and stratification tool. Eur Radiol. 2012;22:1717–1723. doi: 10.1007/s00330-012-2409-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Timmeren J.E., Cester D., Tanadini-Lang S., Alkadhi H., Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santone A., Brunese M.C., Donnarumma F., Guerriero P., Mercaldo F., Reginelli A., Miele V., Giovagnoni A., Brunese L. Radiomic features for prostate cancer grade detection through formal verification. Radiol. Med. 2021;126:688–697. doi: 10.1007/s11547-020-01314-8. [DOI] [PubMed] [Google Scholar]
- 12.Granata V., Fusco R., Costa M., Picone C., Cozzi D., Moroni C., La Casella G., Montanino A., Monti R., Mazzoni F., et al. Preliminary Report on Computed Tomography Radiomics Features as Biomarkers to Immunotherapy Selection in Lung Adenocarcinoma Patients. Cancers. 2021;13:3992. doi: 10.3390/cancers13163992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granata V., Fusco R., Barretta M.L., Picone C., Avallone A., Belli A., Patrone R., Ferrante M., Cozzi D., Grassi R., et al. Radiomics in hepatic metastasis by colorectal cancer. Infect. Agent Cancer. 2021;16:39. doi: 10.1186/s13027-021-00379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fusco R., Piccirillo A., Sansone M., Granata V., Rubulotta M.R., Petrosino T., Barretta M., Vallone P., Di Giacomo R., Esposito E., et al. Radiomics and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography in the Breast Lesions Classification. Diagnostics. 2021;11:815. doi: 10.3390/diagnostics11050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusco R., Granata V., Mazzei M.A., Di Meglio N., Del Roscio D., Moroni C., Monti R., Cappabianca C., Picone C., Neri E., et al. Quantitative imaging decision support (QIDSTM) tool consistency evaluation and radiomic analysis by means of 594 metrics in lung carcinoma on chest CT scan. Cancer Control. 2021;28:1073274820985786. doi: 10.1177/1073274820985786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granata V., Fusco R., Avallone A., De Stefano A., Ottaiano A., Sbordone C., Brunese L., Izzo F., Petrillo A. Radiomics-Derived Data by Contrast Enhanced Magnetic Resonance in RAS Mutations Detection in Colorectal Liver Metastases. Cancers. 2021;13:453. doi: 10.3390/cancers13030453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granata V., Fusco R., Risi C., Ottaiano A., Avallone A., De Stefano A., Grimm R., Grassi R., Brunese L., Izzo F., et al. Diffusion-Weighted MRI and Diffusion Kurtosis Imaging to Detect RAS Mutation in Colorectal Liver Metastasis. Cancers. 2020;12:2420. doi: 10.3390/cancers12092420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurgitano M., Angileri S.A., Rodà G.M., Liguori A., Pandolfi M., Ierardi A.M., Wood B.J., Carrafiello G. Interventional Radiology ex-machina: Impact of Artificial Intelligence on practice. Radiol. Med. 2021;126:998–1006. doi: 10.1007/s11547-021-01351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scapicchio C., Gabelloni M., Barucci A., Cioni D., Saba L., Neri E. A deep look into radiomics. Radiol. Med. 2021;126:1296–1311. doi: 10.1007/s11547-021-01389-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlo R., Renato C., Giuseppe C., Lorenzo U., Giovanni I., Domenico S., Valeria R., Elia G., Maria C.L., Mario C. Mediterranean Conference on Medical and Biological Engineering and Computing. Springer; Berlin/Heidelberg, Germany: 2019. Distinguishing functional from non-functional pituitary macroadenomas with a machine learning analysis; pp. 1822–1829. [Google Scholar]
- 21.Donisi L., Cesarelli G., Castaldo A., De Lucia D.R., Nessuno F., Spadarella G., Ricciardi C. A Combined Radiomics and Machine Learning Approach to Distinguish Clinically Significant Prostate Lesions on a Publicly Available MRI Dataset. J. Imaging. 2021;7:215. doi: 10.3390/jimaging7100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benedetti G., Mori M., Panzeri M.M., Barbera M., Palumbo D., Sini C., Muffatti F., Andreasi V., Steidler S., Doglioni C., et al. CT-derived radiomic features to discriminate histologic characteristics of pancreatic neuroendocrine tumors. Radiol. Med. 2021;126:745–760. doi: 10.1007/s11547-021-01333-z. [DOI] [PubMed] [Google Scholar]
- 23.Nardone V., Reginelli A., Grassi R., Boldrini L., Vacca G., D’Ippolito E., Annunziata S., Farchione A., Belfiore M.P., Desideri I., et al. Delta radiomics: A systematic review. Radiol. Med. 2021;126:1571–1583. doi: 10.1007/s11547-021-01436-7. [DOI] [PubMed] [Google Scholar]
- 24.Granata V., Fusco R., Avallone A., Cassata A., Palaia R., Delrio P., Grassi R., Tatangelo F., Grazzini G., Izzo F., et al. Abbreviated MRI protocol for colorectal liver metastases: How the radiologist could work in pre surgical setting. PLoS ONE. 2020;15:e0241431. doi: 10.1371/journal.pone.0241431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granata V., Fusco R., Setola S.V., Raso M.M., Avallone A., De Stefano A., Nasti G., Palaia R., Delrio P., Petrillo A., et al. Liver radiologic findings of chemotherapy-induced toxicity in liver colorectal metastases patients. Eur. Rev. Med Pharmacol. Sci. 2019;23:9697–9706. doi: 10.26355/eurrev_201911_19531. [DOI] [PubMed] [Google Scholar]
- 26.Granata V., Fusco R., Maio F., Avallone A., Nasti G., Palaia R., Albino V., Grassi R., Izzo F., Petrillo A. Qualitative assessment of EOB-GD-DTPA and Gd-BT-DO3A MR contrast studies in HCC patients and colorectal liver metastases. Infect. Agents Cancer. 2019;14:1–9. doi: 10.1186/s13027-019-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fusco R., Granata V., Petrillo A. Introduction to Special Issue of Radiology and Imaging of Cancer. Cancers. 2020;12:2665. doi: 10.3390/cancers12092665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caruso D., Pucciarelli F., Zerunian M., Ganeshan B., De Santis D., Polici M., Rucci C., Polidori T., Guido G., Bracci B., et al. Chest CT texture-based radiomics analysis in differentiating COVID-19 from other interstitial pneumonia. Radiol. Med. 2021;126:1415–1424. doi: 10.1007/s11547-021-01402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satake H., Ishigaki S., Ito R., Naganawa S. Radiomics in breast MRI: Current progress toward clinical application in the era of artificial intelligence. Radiol. Med. 2021;127:39–56. doi: 10.1007/s11547-021-01423-y. [DOI] [PubMed] [Google Scholar]
- 30.Fusco R., Granata V., Sansone M., Rega D., Delrio P., Tatangelo F., Romano C., Avallone A., Pupo D., Giordano M., et al. Validation of the standardized index of shape tool to analyze DCE-MRI data in the assessment of neo-adjuvant therapy in locally advanced rectal cancer. Radiol. Med. 2021;126:1044–1054. doi: 10.1007/s11547-021-01369-1. [DOI] [PubMed] [Google Scholar]
- 31.Qin H., Que Q., Lin P., Li X., Wang X.-R., He Y., Chen J.-Q., Yang H. Magnetic resonance imaging (MRI) radiomics of papillary thyroid cancer (PTC): A comparison of predictive performance of multiple classifiers modeling to identify cervical lymph node metastases before surgery. Radiol. Med. 2021;126:1312–1327. doi: 10.1007/s11547-021-01393-1. [DOI] [PubMed] [Google Scholar]
- 32.Karmazanovsky G., Gruzdev I., Tikhonova V., Kondratyev E., Revishvili A. Computed tomography-based radiomics approach in pancreatic tumors characterization. Radiol. Med. 2021;126:1388–1395. doi: 10.1007/s11547-021-01405-0. [DOI] [PubMed] [Google Scholar]
- 33.Gregucci F., Fiorentino A., Mazzola R., Ricchetti F., Bonaparte I., Surgo A., Figlia V., Carbonara R., Caliandro M., Ciliberti M.P., et al. Radiomic analysis to predict local response in locally advanced pancreatic cancer treated with stereotactic body radiation therapy. Radiol. Med. 2021;127:100–107. doi: 10.1007/s11547-021-01422-z. [DOI] [PubMed] [Google Scholar]
- 34.Fusco R., Granata V., Maio F., Sansone M., Petrillo A. Textural radiomic features and time-intensity curve data analysis by dynamic contrast-enhanced MRI for early prediction of breast cancer therapy response: Preliminary data. Eur. Radiol. Exp. 2020;4:1–7. doi: 10.1186/s41747-019-0141-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrillo A., Fusco R., Granata V., Filice S., Sansone M., Rega D., Delrio P., Bianco F., Romano G.M., Tatangelo F., et al. Assessing response to neo-adjuvant therapy in locally advanced rectal cancer using Intra-voxel Incoherent Motion modelling by DWI data and Standardized Index of Shape from DCE-MRI. Ther. Adv. Med Oncol. 2018;10:1758835918809875. doi: 10.1177/1758835918809875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fusco R., Sansone M., Granata V., Grimm R., Pace U., Delrio P., Tatangelo F., Botti G., Avallone A., Pecori B., et al. Diffusion and perfusion MR parameters to assess preoperative short-course radiotherapy response in locally advanced rectal cancer: A comparative explorative study among Standardized Index of Shape by DCE-MRI, intravoxel incoherent motion- and diffusion kurtosis imaging-derived parameters. Abdom. Radiol. 2018;44:3683–3700. doi: 10.1007/s00261-018-1801-z. [DOI] [PubMed] [Google Scholar]
- 37.Petrillo A., Fusco R., Granata V., Setola S.V., Sansone M., Rega D., Delrio P., Bianco F., Romano G.M., Tatangelo F., et al. MR imaging perfusion and diffusion analysis to assess preoperative Short Course Radiotherapy response in locally advanced rectal cancer: Standardized Index of Shape by DCE-MRI and intravoxel incoherent motion-derived parameters by DW-MRI. Med. Oncol. 2017;34:198. doi: 10.1007/s12032-017-1059-2. [DOI] [PubMed] [Google Scholar]
- 38.Petrillo A., Fusco R., Petrillo M., Granata V., Delrio P., Bianco F., Pecori B., Botti G., Tatangelo F., Caracò C., et al. Standardized Index of Shape (DCE-MRI) and Standardized Uptake Value (PET/CT): Two quantitative approaches to discriminate chemo-radiotherapy locally advanced rectal cancer responders under a functional profile. Oncotarget. 2016;8:8143–8153. doi: 10.18632/oncotarget.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrillo A., Fusco R., Petrillo M., Granata V., Sansone M., Avallone A., Delrio P., Pecori B., Tatangelo F., Ciliberto G. Standardized Index of Shape (SIS): A quantitative DCE-MRI parameter to discriminate responders by non-responders after neoadjuvant therapy in LARC. Eur. Radiol. 2015;25:1935–1945. doi: 10.1007/s00330-014-3581-3. [DOI] [PubMed] [Google Scholar]
- 40.Van der Lubbe M.F.J.A., Vaidyanathan A., de Wit M., Burg E.L.V.D., Postma A.A., Bruintjes T.D., Bilderbeek-Beckers M.A.L., Dammeijer P.F.M., Bossche S.V., Van Rompaey V., et al. A non-invasive, automated diagnosis of Menière’s disease using radiomics and machine learning on conventional magnetic resonance imaging: A multicentric, case-controlled feasibility study. Radiol. Med. 2021;127:72–82. doi: 10.1007/s11547-021-01425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunese L., Brunese M.C., Carbone M., Ciccone V., Mercaldo F., Santone A. Automatic PI-RADS assignment by means of formal methods. Radiol. Med. 2021;127:83–89. doi: 10.1007/s11547-021-01431-y. [DOI] [PubMed] [Google Scholar]
- 42.Vicini S., Bortolotto C., Rengo M., Ballerini D., Bellini D., Carbone I., Preda L., Laghi A., Coppola F., Faggioni L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: Focus on the three most common cancers. Radiol. Med. 2022;127:819–836. doi: 10.1007/s11547-022-01512-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang F.-H., Zheng H.-L., Li J.-T., Li P., Zheng C.-H., Chen Q.-Y., Huang C.-M., Xie J.-W. Prediction of recurrence-free survival and adjuvant therapy benefit in patients with gastrointestinal stromal tumors based on radiomics features. Radiol. Med. 2022;127:1085–1097. doi: 10.1007/s11547-022-01549-7. [DOI] [PubMed] [Google Scholar]
- 44.De Robertis R., Geraci L., Tomaiuolo L., Bortoli L., Beleù A., Malleo G., D’Onofrio M. Liver metastases in pancreatic ductal adenocarcinoma: A predictive model based on CT texture analysis. Radiol. Med. 2022;127:1079–1084. doi: 10.1007/s11547-022-01548-8. [DOI] [PubMed] [Google Scholar]
- 45.Chiti G., Grazzini G., Flammia F., Matteuzzi B., Tortoli P., Bettarini S., Pasqualini E., Granata V., Busoni S., Messserini L., et al. Gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs): A radiomic model to predict tumor grade. Radiol. Med. 2022;127:928–938. doi: 10.1007/s11547-022-01529-x. [DOI] [PubMed] [Google Scholar]
- 46.Yao F., Bian S., Zhu D., Yuan Y., Pan K., Pan Z., Feng X., Tang K., Yang Y. Machine learning-based radiomics for multiple primary prostate cancer biological characteristics prediction with 18F-PSMA-1007 PET: Comparison among different volume segmentation thresholds. Radiol. Med. 2022;127:1170–1178. doi: 10.1007/s11547-022-01541-1. [DOI] [PubMed] [Google Scholar]
- 47.Fiaschetti V., Ubaldi N., De Fazio S., Ricci A., Maspes F., Cossu E. Digital tomosynthesis spot view in architectural distortions: Outcomes in management and radiation dose. Radiol. Med. 2022 doi: 10.1007/s11547-022-01570-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue K., Liu L., Liu Y., Guo Y., Zhu Y., Zhang M. Radiomics model based on multi-sequence MR images for predicting preoperative immunoscore in rectal cancer. Radiol. Med. 2022;127:702–713. doi: 10.1007/s11547-022-01507-3. [DOI] [PubMed] [Google Scholar]
- 49.Granata V., Fusco R., De Muzio F., Cutolo C., Setola S.V., Dell’Aversana F., Grassi F., Belli A., Silvestro L., Ottaiano A., et al. Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. Radiol. Med. 2022;127:763–772. doi: 10.1007/s11547-022-01501-9. [DOI] [PubMed] [Google Scholar]
- 50.Cozzi D., Bicci E., Cavigli E., Danti G., Bettarini S., Tortoli P., Mazzoni L.N., Busoni S., Pradella S., Miele V. Radiomics in pulmonary neuroendocrine tumours (NETs) Radiol. Med. 2022;127:609–615. doi: 10.1007/s11547-022-01494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Granata V., Fusco R., De Muzio F., Cutolo C., Setola S.V., Grassi R., Grassi F., Ottaiano A., Nasti G., Tatangelo F., et al. Radiomics textural features by MR imaging to assess clinical outcomes following liver resection in colorectal liver metastases. Radiol. Med. 2022;127:461–470. doi: 10.1007/s11547-022-01477-6. [DOI] [PubMed] [Google Scholar]
- 52.Autorino R., Gui B., Panza G., Boldrini L., Cusumano D., Russo L., Nardangeli A., Persiani S., Campitelli M., Ferrandina G., et al. Radiomics-based prediction of two-year clinical outcome in locally advanced cervical cancer patients undergoing neoadjuvant chemoradiotherapy. Radiol. Med. 2022;127:498–506. doi: 10.1007/s11547-022-01482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gitto S., Bologna M., Corino V.D.A., Emili I., Albano D., Messina C., Armiraglio E., Parafioriti A., Luzzati A., Mainardi L., et al. Diffusion-weighted MRI radiomics of spine bone tumors: Feature stability and machine learning-based classification performance. Radiol. Med. 2022;127:518–525. doi: 10.1007/s11547-022-01468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao W., Wang W., Song D., Yang C., Zhu K., Zeng M., Rao S.-X., Wang M. A predictive model integrating deep and radiomics features based on gadobenate dimeglumine-enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. Radiol. Med. 2022;127:259–271. doi: 10.1007/s11547-021-01445-6. [DOI] [PubMed] [Google Scholar]
- 55.Nicosia L., Bozzini A.C., Palma S., Montesano M., Signorelli G., Pesapane F., Latronico A., Bagnardi V., Frassoni S., Sangalli C., et al. Contrast-Enhanced Spectral Mammography and tumor size assessment: A valuable tool for appropriate surgical management of breast lesions. Radiol. Med. 2022;127:1228–1234. doi: 10.1007/s11547-022-01561-x. [DOI] [PubMed] [Google Scholar]
- 56.Tsuchiya M., Masui T., Terauchi K., Yamada T., Katyayama M., Ichikawa S., Noda Y., Goshima S. MRI-based radiomics analysis for differentiating phyllodes tumors of the breast from fibroadenomas. Eur. Radiol. 2022;32:4090–4100. doi: 10.1007/s00330-021-08510-8. [DOI] [PubMed] [Google Scholar]
- 57.Palatresi D., Fedeli F., Danti G., Pasqualini E., Castiglione F., Messerini L., Massi D., Bettarini S., Tortoli P., Busoni S., et al. Correlation of CT radiomic features for GISTs with pathological classification and molecular subtypes: Preliminary and monocentric experience. Radiol. Med. 2022;127:117–128. doi: 10.1007/s11547-021-01446-5. [DOI] [PubMed] [Google Scholar]
- 58.Chiloiro G., Cusumano D., de Franco P., Lenkowicz J., Boldrini L., Carano D., Barbaro B., Corvari B., Dinapoli N., Giraffa M., et al. Does restaging MRI radiomics analysis improve pathological complete response prediction in rectal cancer patients? A prognostic model development. Radiol. Med. 2021;127:11–20. doi: 10.1007/s11547-021-01421-0. [DOI] [PubMed] [Google Scholar]
- 59.Bracci S., Dolciami M., Trobiani C., Izzo A., Pernazza A., D’Amati G., Manganaro L., Ricci P. Quantitative CT texture analysis in predicting PD-L1 expression in locally advanced or metastatic NSCLC patients. Radiol. Med. 2021;126:1425–1433. doi: 10.1007/s11547-021-01399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ricciardi C., Cuocolo R., Verde F., Improta G., Stanzione A., Romeo V., Maurea S., D’Armiento M., Sarno L., Guida M., et al. European Medical and Biological Engineering Conference. Springer; Berlin/Heidelberg, Germany: 2020. Resolution resampling of ultrasound images in placenta previa patients: Influence on radiomics data relia- bility and usefulness for machine learning; pp. 1011–1018. [Google Scholar]
- 61.Ponsiglione A.M., Ricciardi C., Scala A., Fiorillo A., Sorrentino A., Triassi M., Orabona G.D., Improta G. Application of DMAIC Cycle and Modeling as Tools for Health Technology Assessment in a University Hospital. J. Health Eng. 2021;2021:8826048. doi: 10.1155/2021/8826048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponsiglione A.M., Cesarelli G., Amato F., Romano M. Optimization of an artificial neural network to study accelerations of foetal heart rhythm; Proceedings of the 2021 IEEE 6th International Forum on Research and Technology for Society and Industry (RTSI); Naples, Italy. 6–9 September 2021; pp. 159–164. [Google Scholar]
- 63.Donisi L., Cesarelli G., Balbi P., Provitera V., Lanzillo B., Coccia A., D’Addio G. Positive impact of short-term gait rehabilitation in Parkinson patients: A combined approach based on statistics and machine learning. Math. Biosci. Eng. 2021;18:6995–7009. doi: 10.3934/mbe.2021348. [DOI] [PubMed] [Google Scholar]
- 64.Ponsiglione A.M., Amato F., Romano M. Multiparametric Investigation of Dynamics in Fetal Heart Rate Signals. Bioengineering. 2021;9:8. doi: 10.3390/bioengineering9010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponsiglione A.M., Cosentino C., Cesarelli G., Amato F., Romano M. A Comprehensive Review of Techniques for Processing and Analyzing Fetal Heart Rate Signals. Sensors. 2021;21:6136. doi: 10.3390/s21186136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jang S.-K., Chang J.Y., Lee J.S., Lee E.-J., Kim Y.-H., Han J.H., Chang D.-I., Cho H.J., Cha J.-K., Yu K.H., et al. Reliability and Clinical Utility of Machine Learning to Predict Stroke Prognosis: Comparison with Logistic Regression. J. Stroke. 2020;22:403–406. doi: 10.5853/jos.2020.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leite A.F., de Faria Vasconcelos K., Willems H., Jacobs R. Radiomics and machine learning in oral healthcare. Proteom. Clin. Appl. 2020;14:1900040. doi: 10.1002/prca.201900040. [DOI] [PubMed] [Google Scholar]
- 68.Recenti M., Ricciardi C., Edmunds K., Gislason M.K., Gargiulo P. Machine learning predictive system based upon radiodensitometric distributions from mid-thigh CT images. Eur. J. Transl. Myol. 2020;30:121–124. doi: 10.4081/ejtm.2019.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scrutinio D., Ricciardi C., Donisi L., Losavio E., Battista P., Guida P., Cesarelli M., Pagano G., D’Addio G. Machine learning to predict mortality after rehabilitation among patients with severe stroke. Sci. Rep. 2020;10:20127. doi: 10.1038/s41598-020-77243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trunfio T.A., Ponsiglione A.M., Ferrara A., Borrelli A., Gargiulo P. A comparison of different regression and classification methods for predicting the length of hospital stay after cesarean sections; Proceedings of the 2021 5th International Conference on Medical and Health Informatics; Kyoto Japan. 14–16 May 2021; pp. 63–67. [Google Scholar]
- 71.Ricciardi C., Valente A.S., Edmund K., Cantoni V., Green R., Fiorillo A., Picone I., Santini S., Cesarelli M. Linear discriminant analysis and principal com- ponent analysis to predict coronary artery disease. Health Inform. J. 2020;26:2181–2192. doi: 10.1177/1460458219899210. [DOI] [PubMed] [Google Scholar]
- 72.Profeta M., Ponsiglione A.M., Ponsiglione C., Ferrucci G., Giglio C., Borrelli A. Comparison of machine learning algorithms to predict length of hospital stay in patients undergoing heart bypass surgery; Proceedings of the 2021 International Symposium on Biomedical Engineering and Computational Biology; Nanchang, China. 13–15 August 2021; pp. 1–5. [Google Scholar]
- 73.Ge L., Chen Y., Yan C., Zhao P., Zhang P., Runa A., Liu J. Study Progress of Radiomics with Machine Learning for Precision Medicine in Bladder Cancer Management. Front. Oncol. 2019;9:1296. doi: 10.3389/fonc.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gastounioti A., Oustimov A., Keller B.M., Pantalone L., Hsieh M.-K., Conant E.F., Kontos D. Breast parenchymal patterns in processed versus raw digital mammograms: A large population study toward assessing differences in quantitative measures across image representations. Med. Phys. 2016;43:5862–5877. doi: 10.1118/1.4963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Y., Keller B.M., Ray S., Wang Y., Conant E.F., Gee J.C., Kontos D. Parenchymal texture analysis in digital mammography: A fully automated pipeline for breast cancer risk assessment. Med. Phys. 2015;42:4149–4160. doi: 10.1118/1.4921996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 77.Sacchetto D., Morra L., Agliozzo S., Bernardi D., Bjorklund T., Brancato B., Bravetti P., Carbonaro L.A., Correale L., Fanto C., et al. Mammographic density: Comparison of visual assessment with fully automatic calculation on a multivendor dataset. Eur. Radiol. 2016;26:175–183. doi: 10.1007/s00330-015-3784-2. [DOI] [PubMed] [Google Scholar]
- 78.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H.J.W.L., Andrearczyk V., Apte A., Ashrafinia S., Bakas S., Beukinga R.J., Boellaard R., et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Im-age-based Phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuhn M., Wing J., Weston S., Williams A. The caret package. Gene Expr. 2007 [Google Scholar]
- 80.Pietropaolo A., Geraghty R.M., Veeratterapillay R., Rogers A., Kallidonis P., Villa L., Boeri L., Montanari E., Atis G., Emiliani E., et al. A machine learning predictive model for post-ureteroscopy urosepsis needing intensive care unit admission: A case–control yau endourology study from nine euro- pean centres. J. Clin. Med. 2021;10:3888. doi: 10.3390/jcm10173888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fusco R., Sansone M., Filice S., Carone G., Amato D.M., Sansone C., Petrillo A. Pattern Recognition Approaches for Breast Cancer DCE-MRI Classification: A Systematic Review. J. Med. Biol. Eng. 2016;36:449–459. doi: 10.1007/s40846-016-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Copyright 2000–2020, R-Tools Technology Inc. [(accessed on 15 May 2020)]. Available online: https://www.r-tt.com/
- 83.Pinker K. Beyond Breast Density: Radiomic Phenotypes Enhance Assessment of Breast Cancer Risk. Radiology. 2019;290:50–51. doi: 10.1148/radiol.2018182296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arslan A., Aktas E., Sengul B., Tekin B. Dosimetric evaluation of left ventricle and left anterior descending artery in left breast radiotherapy. Radiol. Med. 2020;126:14–21. doi: 10.1007/s11547-020-01201-2. [DOI] [PubMed] [Google Scholar]
- 85.Tagliafico A.S., Campi C., Bianca B., Bortolotto C., Buccicardi D., Francesca C., Prost R., Rengo M., Faggioni L. Blockchain in radiology research and clinical practice: Current trends and future directions. Radiol. Med. 2022;127:391–397. doi: 10.1007/s11547-022-01460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qin H., Wu Y., Lin P., Gao R., Li X., Wang X., Chen G., He Y., Yang H. Ultrasound Image–Based Radiomics: An Innovative Method to Identify Primary Tumorous Sources of Liver Metastases. J. Ultrasound Med. 2020;40:1229–1244. doi: 10.1002/jum.15506. [DOI] [PubMed] [Google Scholar]
- 87.Li N., Wakim J., Koethe Y., Huber T., Schenning R., Gade T.P., Hunt S.J., Park B.J. Multicenter assessment of augmented reality registration methods for image-guided interventions. Radiol. Med. 2022;127:857–865. doi: 10.1007/s11547-022-01515-3. [DOI] [PubMed] [Google Scholar]
- 88.Wei S., Han Y., Zeng H., Ye S., Cheng J., Chai F., Wei J., Zhang J., Hong N., Bao Y., et al. Radiomics diagnosed histopathological growth pattern in prediction of response and 1-year progression free survival for colorectal liver metastases patients treated with bevacizumab containing chemotherapy. Eur. J. Radiol. 2021;142:109863. doi: 10.1016/j.ejrad.2021.109863. [DOI] [PubMed] [Google Scholar]
- 89.Spinelli M.S., Balbaa M.F., Gallazzi M.B., Eid M.E.-E., Kotb H.T., El Shafei M., Ierardi A.M., Daolio P.A., Barile A., Carrafiello G. Role of percutaneous CT–guided radiofrequency ablation in treatment of intra-articular, in close contact with cartilage and extra-articular osteoid osteomas: Comparative analysis and new classification system. Radiol. Med. 2022;127:1142–1150. doi: 10.1007/s11547-022-01542-0. [DOI] [PubMed] [Google Scholar]
- 90.Giannini V., Rosati S., DeFeudis A., Balestra G., Vassallo L., Cappello G., Mazzetti S., De Mattia C., Rizzetto F., Torresin A., et al. Radiomics predicts response of individual HER2-amplified colorectal cancer liver metastases in patients treated with HER2-targeted therapy. Int. J. Cancer. 2020;147:3215–3223. doi: 10.1002/ijc.33271. [DOI] [PubMed] [Google Scholar]
- 91.Caruso D., Polici M., Rinzivillo M., Zerunian M., Nacci I., Marasco M., Magi L., Tarallo M., Gargiulo S., Iannicelli E., et al. CT-based radiomics for prediction of therapeutic response to Everolimus in metastatic neuroendocrine tumors. Radiol. Med. 2022;127:691–701. doi: 10.1007/s11547-022-01506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han D., Yu N., Yu Y., He T., Duan X. Performance of CT radiomics in predicting the overall survival of patients with stage III clear cell renal carcinoma after radical nephrectomy. Radiol. Med. 2022;127:837–847. doi: 10.1007/s11547-022-01526-0. [DOI] [PubMed] [Google Scholar]
- 93.Donati O.F., Hany T.F., Reiner C.S., von Schulthess G.K., Marincek B., Seifert B., Weishaupt D. Value of Retrospective Fusion of PET and MR Images in Detection of Hepatic Metastases: Comparison with 18F-FDG PET/CT and Gd-EOB-DTPA–Enhanced MRI. J. Nucl. Med. 2010;51:692–699. doi: 10.2967/jnumed.109.068510. [DOI] [PubMed] [Google Scholar]
- 94.Masci G.M., Ciccarelli F., Mattei F.I., Grasso D., Accarpio F., Catalano C., Laghi A., Sammartino P., Iafrate F. Role of CT texture analysis for predicting peritoneal metastases in patients with gastric cancer. Radiol. Med. 2022;127:251–258. doi: 10.1007/s11547-021-01443-8. [DOI] [PubMed] [Google Scholar]
- 95.Viganò L., Arachchige V.S.J., Fiz F. Is precision medicine for colorectal liver metastases still a utopia? New perspectives by modern biomarkers, radiomics, and artificial intelligence. World J. Gastroenterol. 2022;28:608–623. doi: 10.3748/wjg.v28.i6.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sansone M., Marrone S., Di Salvio G., Belfiore M.P., Gatta G., Fusco R., Vanore L., Zuiani C., Grassi F., Vietri M.T., et al. Comparison between two packages for pectoral muscle removal on mammographic images. Radiol. Med. 2022;127:848–856. doi: 10.1007/s11547-022-01521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gastounioti A., Conant E.F., Kontos D. Beyond breast density: Are view on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Res. 2016;18:91. doi: 10.1186/s13058-016-0755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pinker-Domenig K., Chin J., Melsaether A.N., Morris E.A., Moy L. Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology. 2018;287:732–747. doi: 10.1148/radiol.2018172171. [DOI] [PubMed] [Google Scholar]
- 99.Kontos D., Winham S.J., Oustimov A., Pantalone L., Hsieh M.-K., Gastounioti A., Whaley D.H., Hruska C.B., Kerlikowske K., Brandt K., et al. Radiomic Phenotypes of Mammographic Parenchymal Complexity: Toward Augmenting Breast Density in Breast Cancer Risk Assessment. Radiology. 2019;290:41–49. doi: 10.1148/radiol.2018180179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H., Mendel K.R., Lan L., Sheth D., Giger M.L. Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma. Radiology. 2019;291:15–20. doi: 10.1148/radiol.2019181113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yala A., Lehman C., Schuster T., Portnoi T., Barzilay R. A Deep Learning Mammography-based Model for Improved Breast Cancer Risk Prediction. Radiology. 2019;292:60–66. doi: 10.1148/radiol.2019182716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study is available in this article.