Abstract
Yield is a complex parameter of rice due to its polygonal nature, sometimes making it difficult to coat the selection process in the breeding program. In the current study, 34 elite rice genotypes were assessed to evaluate 3 locations for the selection of desirable rice cultivars suitable for multiple environments based on genetic diversity. In variance analysis, all genotypes have revealed significant variations (p ≤ 0.001) for all studied characters, signifying a broader sense of genetic variability for selection purposes. The higher phenotypic coefficient of variation (PCV) and genotypic coefficient of variation (GCV) were found for yield-associated characteristics such as the number of grains panicle−1 (GP), panicles hill−1 (PPH), and tillers hill−1 (TILL). All of the characters had higher heritability (greater than 60%) and higher genetic advance (greater than 20%), which pointed out non-additive gene action and suggested that selection would be effective. The most significant traits causing the genotype variants were identified via principal component analysis. In the findings of the cluster analysis, 34 elite lines were separated into 3 categories of clusters, with cluster II being chosen as the best one. The relationship matrix between each elite cultivar and traits was also determined utilizing a heatmap. Based on multi-trait genotype-ideotype distance index (MGIDI), genotypes Gen2, Gen4, Gen14, Gen22, and Gen30 in Satkhira; Gen2, Gen6, Gen7, Gen15, and Gen30 in Kushtia; and Gen10, Gen12, Gen26, Gen30, and Gen34 in Barishal were found to be the most promising genotypes. Upon validation, these genotypes can be suggested for commercial release or used as potential breeding material in crossing programs for the development of cultivars suitable for multiple environments under the future changing climate.
Keywords: genetic diversity, trait association, principal component analysis, clustered heatmap, MGIDI
1. Introduction
Rice (Oryza sativa L.) is a unique, particularly crucial cereal; over half of the world’s population relies on rice. In Bangladesh, around 158.9 million individuals rely mostly on rice as their main food source [1]. The absolute rice region in Bangladesh is 11.52 million ha and accounts for 75% of the entire crop area and 93% of the entire area cultivated for cereals. The overall rice production in Bangladesh was 36.4 MMT (Million Metric Tons), of which Boro, Aman, and Aus rice occupied 19.5 MMT (53.8%), 14.1 MMT (38.6%), and 2.8 MMT (7.6%) respectively, during 2018–2019 [2]. In contrast with many other rice-producing countries, Bangladesh also has ‘rice security’ as being synonymous with ‘food security’ [3]. The safety of rice production is not only a socio-economic problem but also an essential indicator of the stability of political and cultural harmony [4]. It is estimated that the individuals of Bangladesh will reach 215.4 million by the year 2050, and at the same time, 44.6 MMT of rough rice would be needed to secure the food sufficiency of the rising population [5]. Therefore, the main objectives of plant breeders should be to develop climate-smart varieties as well as yield improvement for the main food crop. Hence, the improvement of high-yielding rice genotypes is expected to be met with agronomic characteristics and the need to cope with the growing demand associated with adaptability in different environments. However, the complex polygenetic trait of the yield, which is influenced by its inherent properties, confuses plant breeders for its direct selection [6,7]. It is crucial to understand how yield relates to its other inherent characteristics [8]. The vast spectrum assortment of agricultural climates, physiological, soil, and hydrological circumstances makes this country a great source of local land racing, and especially, an excessive source of rice with distinguishing phenotypic and genetic characteristics [9]. Discovering this existing diversity in the rice germplasm certainly leads to identifying the fancy genes and boosting the progress in rice breeding [10]. Before selecting the applied breeding method in rice, it is indispensable to know the morpho-genetic dissimilarity in diverse yield-related characters [11,12]. Although there are high environmental effects, assessments based on morphological characteristics are still possible for selection [13,14]. Attributes such as the GCV, PCV, heritability, and genetic advance are convenient biometric aspects for evaluating genetic variability and adaptability [15,16]. By utilizing more genetic varieties and employing efficient selection techniques to boost yield through yield attributes, breeding projects seek to increase rice production. The correlation studies help plant breeders make more accurate selections. Due to the rising importance of crop development, current knowledge of trait connections aids in the proper selection process [17].
Diversity analysis is a tool that helps to divide germplasms into several clusters based on their similar performance in various characteristics [18,19]. Previous literature suggests that there is more genetic dissimilarity in rice for yield traits and its associated characteristics in Bangladesh [20,21]. Nevertheless, even thousands of significant novel genes with economic value remain undiscovered. Therefore, these dissimilarities need to be explored to face current and future challenges in rice breeding. The MGIDI [22] has become one of the most innovative techniques for choosing the best genotypes that might perform much better under various favourable and unfavourable conditions with high yield stability and desired characteristics. Thus, the current investigation was carried out to uncover the genetic divergence and genetic diversity of 34 elite rice genotypes across qualities and agronomic traits for future rice breeding programs for the development of superior and stable cultivars which are suitable for multiple environments under the future changing climate.
2. Materials and Methods
2.1. Experimental Locations and Weather Data
To assess the genotypes, the research was conducted in three representative environments of Bangladesh, namely Barishal (22°53′ N and 90°34′ E), Kushtia (23°55′ N and 89°03′ E), and Sathkira (22°76′ N and 89°11′ E). The monthly mean temperatures, wind speed, and precipitation data during the experimental period from February to May 2017 were presented in Table 1.
Table 1.
Monthly mean agro-climatic data during the field experiment period in Barishal, Kushtia, and Satkhira.
| Locations | Month | Maxi. (°C) | Min. (°C) | Wind (km/h) | Rainfall (mm) | Humidity (%) | Cloud (%) | Pressure (mb) |
|---|---|---|---|---|---|---|---|---|
| Barishal | February | 27.0 | 17.7 | 10.5 | 0 | 43.7 | 0 | 1014.5 |
| March | 31.7 | 21.5 | 9.5 | 0 | 54.0 | 9.7 | 1013.5 | |
| April | 31.5 | 22.7 | 11.7 | 1.0 | 61.0 | 13.7 | 1013.2 | |
| May | 35.2 | 26.7 | 13.7 | 2.4 | 67.7 | 37.5 | 1007.7 | |
| Kushtia | February | 28.0 | 16.5 | 10.0 | 0 | 39.5 | 0 | 1014.7 |
| March | 33.0 | 20.5 | 9.5 | 0 | 38.0 | 3.7 | 1011.7 | |
| April | 35.2 | 21.7 | 9.2 | 1.2 | 42.5 | 5.2 | 1009.7 | |
| May | 38.5 | 26.7 | 12.2 | 7.0 | 55.5 | 33.0 | 1005.5 | |
| Satkhira | February | 27.5 | 17.0 | 12.2 | 0 | 38.5 | 0 | 1015.5 |
| March | 33.7 | 20.7 | 8.5 | 0.0 | 43.7 | 8.7 | 1013.2 | |
| April | 34.5 | 22.5 | 10.2 | 0.3 | 50.2 | 4.7 | 1010.7 | |
| May | 37.2 | 26.2 | 15.2 | 0.1 | 63.5 | 36.2 | 1007.0 |
2.2. Plant Genetic Materials
Thirty-four genotypes containing twenty-six elite breeding lines and eight BRRI-released varieties such as BR16 (released in 1983), BRRI dhan28 (1994), BRRI dhan29 (1994), BRRI dhan50 (2008), BRRI dhan58 (2012), BRRI dhan63 (2014), BRRI dhan74 (2015), and one International Rice Research Institute (IRRI) developed variety; IRBB60 were used. All genetic materials were collected from the Plant Breeding Division, Bangladesh Rice Research Institute (www.brri.gov.bd; accessed on 22 July 2022). The details of those genotypes are illustrated in Table 2.
Table 2.
The designation and parentages of 34 elite rice cultivars utilized in the current investigation.
| Code | Designation | Parentage |
|---|---|---|
| Gen1 | BR7671-37-2-2-37-3-P3 | BRRI dhan29/IR68144 |
| Gen2 | BR8626-19-5-1-2 | BR7166-5B-6/SHEW WAR TUN//BRRI dhan47 |
| Gen3 | BR8626-10-5-1 | BR7166-5B-6/SHEW WAR TUN//BRRI dhan47 |
| Gen4 | BR8109-29-2-2-3 | BM9821/Parija |
| Gen5 | BR(Bio)8333-BC5-1-1 | BRRI dhan29/IRBB60 |
| Gen6 | BR(Bio)8333-BC5-1-20 | BRRI dhan29/IRBB60 |
| Gen7 | BR(Bio)8333-BC5-2-16 | BRRI dhan29/IRBB60 |
| Gen8 | BR(Bio)8333-BC5-2-22 | BRRI dhan29/IRBB60 |
| Gen9 | BR(Bio)8333-BC5-3-10 | BRRI dhan29/IRBB60 |
| Gen10 | BR8631-12-3-5-P2 | BR7166-5B-5/BG305//BRRI dhan29 |
| Gen11 | BR8631-12-3-6-P3 | BR7166-5B-5/BG305//BRRI dhan29 |
| Gen12 | BR7831-59-1-1-4-5 | BR7166-5B-5/BG305//BRRI dhan29 |
| Gen13 | BR8253-9-3-3-1 | BR7305-21-6-1/BRRI dhan29//BR7305-21-6-1 |
| Gen14 | BR8609-2-B-9-1-B5 | BRRIdhan29/BR7166-5B-1 |
| Gen15 | BR7815-18-1-3-2-1 | BRRI dhan36/BR6725-15-1-2 |
| Gen16 | BR7671-37-2-2-3-7-3-P10 | BRRI dhan29/IR68144 |
| Gen17 | BR7671-37-2-2-3-7-3-P11 | BRRI dhan29/IR68144 |
| Gen18 | BR8079-19-1-5-1 | IR71730-51-2/Nayanmoni |
| Gen19 | BR8590-5-2-5-2-2 | BR6902-51-3-2-1/BRRI dhan HR7 |
| Gen20 | BR8590-5-3-3-4-2 | BR6902-51-3-2-1/BRRI dhan HR7 |
| Gen21 | BR8608-39-2-1 | BRRI dhan 29/Shaheen Basmati/BR6902-51-3-2-1 |
| Gen22 | BRC266-5-1-1-1 | BR16/90060-TR1252-8-2-1 |
| Gen23 | BRC266-5-1-2-1 | BR16/90060-TR1252-8-2-1 |
| Gen24 | BR8523-36-2-2-6 | IR 77512-128-2-1-2/BR 6817-25-2-2 |
| Gen25 | BR8938-19-4-3-1-1 | IRBB60/BRRI dhan29 |
| Gen26 | BR8333-15-3-2-2 | BRRI dhan29/IRBB60 |
| Gen27 | IRBB60 | Near isogenic line of IR24 |
| Gen28 | BRRI dhan28 | BR6(IR28)/Purbachi |
| Gen29 | BRRI dhan58 | Somaclonal line of BRRI dhan29 |
| Gen30 | BRRI dhan29 | BG90-2/BR51-46-5 |
| Gen31 | BR16 | IR1416-131-5/IR1364-37-3-1//IR1544A-E666 |
| Gen32 | BRRI dhan50 | BR30/IR67684B |
| Gen33 | BRRI dhan63 | Amol-3/BRRI dhan28 |
| Gen34 | BRRI dhan74 | BRRI dhan29/IR68144 |
2.3. Design of Experiment and Agronomic Practices
The plant material utilized in the current investigation is made up of 34 genotypes of rice accessed from the BRRI (www.brri.gov.bd; accessed on 22 July 2022). All the tested breeding lines were grown in dry seasons in the three experimental fields at BRRI Barishal, BRRI Khustia, and BRRI Satkhira, Bangladesh. The study was evaluated by utilizing the randomized complete block design with three blocks in each experimental site. Thirty-five to forty-day-old seedlings were placed in the experimental field in a 10.8 m2 plot utilizing one seedling at 20 cm × 15 cm spacing. The fertilizers were supplied at 260:100:120:110:10 kg ha−1 of urea, TSP, MP, gypsum, and ZnSO4, respectively. Standard cultural management was practiced and plant protection measures were taken as required according to the suggestion of BRRI [23]. The extra two lines were kept in the plot to lessen the effects of plant growth as well as yield production.
2.4. Data Collection of Studied Agronomic Traits
A standard evaluation system (SES, 2013) developed from IRRI was followed to collect data on agronomic characters [24]. Data were gathered on 50% flowering (X50% F, days), plant height (PH, cm), tillers per plant (TILL, no.), number of panicles per hill (PPH), panicle length (PL, cm), grains per panicle (GP, no.), fertility (Ferti, %), grain yield per plant (YPP), 1000-grain weight (TGW, gm), and yield ton per hectare (YTH, t/ha). Days to flowering have been collected as early as when 50% of the panicles appear. The five plants were haphazardly counted, eliminating border plants and plants adjacent to the missing hill, for plant height in terms of centimetre units. The measurement of a plant’s height was taken from the base to the top of the panicle. The number of tillers was collected when the grain had been set and the total number of fertile panicles emerged from each plant. The panicles were harvested after maturity and placed in an envelope separately. The harvested panicles were dried on a threshing floor utilizing sunlight. After that, the number of filled and unfilled grains was counted, and thousand grain-weight and grain yield per plant were measured from harvested panicles. Each unit plot was harvested and then dried on the floor by sunlight. Finally, the plot grain yield was weighed and calculated as a ton per hectare, where moisture was adjusted at 14%.
2.5. Statistical Analysis
2.5.1. Estimations of Variance Components
The variation that exists among the germplasm was measured by measuring mean, phenotypic, and genotypic variance. The mean values of each genotype were utilized for combined analysis of variance following STAR software (Version 2.0.1; http://bbi.irri.org/products; accessed on 20 July 2022).
To evaluate the phenotypic and genotypic variance, GCV and PCV were assessed and marked to utilize the formula suggested by Johnson et al. [25]. Broad sense heritability (h2bs) of all desired characters was determined using the equation as depicted by Allard [26]. Heritability was categorized with a value of 0–10% as low, 10–20% as moderate, and 20% to above as high, which was reported by Burton and DeVane [27]. Genetic Advance (GA) was calculated as interpreted in the equation by Singh and Chaundry [28]:
2.5.2. Correlation Coefficients Analysis
The correlation matrix was created from chart.Correlation()with the “PerformanceAnalytics” package in R.
2.5.3. Multivariate Analysis
PCA was executed to calculate eigenvalues, association between cultivars and/or traits, and contributions with quality of traits and genotypes towards principal components of the total variation. The multivariate analysis was performed utilizing R (v4.1.1) and R Studio software (v1.4.1717). The PCA–biplot was assembled using “ggplot2”, “Factoextra”, and “FactomineR” packages of R. A package, namely NbClust, which was utilized to measure the optimal cluster number in a dataset. NbClust contains 30 different valid indices, namely “KI”, “CH”, “Hartigan”, “CCC”, “Scott”, “Marriot”, “TrCovW”, “TraceW”, “Friedman”, “Rubin”, Cindex”, “DB”, “Silhouette”, “Duda”, Pseudot2”, “Beale”, “Ratkowsky”, “Ball”, “Ptibiserial”, “Gap”, “Frey”, “McClain”, “γ”, “Gplus”, “Tau”, “Dunn”, “Hubert”, “SDindex”, “Dindex”, and “SDbw”. The cluster plot with heatmap was visualized with the “heatmaply” package of R.
2.5.4. Analysis of MGIDI Selection Index
The statistical analysis for the multi-trait genotype–ideotype distance index (MGIDI) was performed utilizing R Package ‘metan’ version 1.16.0 (https://github.com/TiagoOlivoto/metan; accessed on 20 July 2022) in R version 4.0.2 (http://www.r-project.org/; accessed on 20 July 2022) [29].
3. Results
3.1. Genetic Variability Analysis
The variability analysis for different locations, as well as pooled data, exhibited significant (p ≤ 0.01) variation along the rice genotypes for all the traits evaluated (Table 3). The interaction variance between genotypes × locations was found to be significant for the traits, illustrating the consistent performance of the genotypes across the year with a significant effect of environmental factors.
Table 3.
Combined analysis of variance, mean sum of square (MSS) and F-value of the ten traits, and their interaction in elite rice breeding lines in studied locations.
| Traits a | Barishal | Kushtia | Satkhira | Pooled b | Interaction c | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MSS | F-Value | MSS | F-Value | MSS | F-Value | MSS | F-Value | MSS | F-Value | |
| X50F | 196.2 | 160.3 ** | 66.9 | 62.7 ** | 51.7 | 49.2 ** | 224.2 | 203.9 ** | 44.7 | 40.7 ** |
| PH | 391.3 | 23.1 ** | 225.8 | 21.2 ** | 258.2 | 26.8 ** | 783.4 | 63.1 ** | 45.9 | 3.7 ** |
| TILL | 18.7 | 5.9 ** | 31.4 | 4.3 ** | 10.2 | 11.2 ** | 36.3 | 9.5 ** | 12 | 3.2 ** |
| PPH | 21.8 | 7.3 ** | 8.4 | 2.6 ** | 12.3 | 8.4 ** | 16.3 | 6.4 ** | 13 | 5.2 ** |
| PL | 4.8 | 5.7 ** | 8.8 | 3.7 ** | 5.4 | 6.1 ** | 11.3 | 8.2 ** | 3.8 | 2.8 ** |
| GP | 27,549.9 | 7.5 ** | 9620.6 | 11.6 ** | 7675.9 | 7.9 ** | 31,784.3 | 17.4 ** | 6531 | 3.6 ** |
| Ferti | 258.1 | 14.1 ** | 161.6 | 6.4 ** | 124.7 | 5.4 ** | 171.3 | 7.7 ** | 186.8 | 8.4 ** |
| YPP | 27.2 | 12.2 ** | 43.7 | 7.9 ** | 16.2 | 10.4 ** | 48.9 | 15.8 ** | 19 | 6.2 ** |
| TGW | 37.2 | 19.2 ** | 27.3 | 36.8 ** | 20.7 | 6.3 ** | 72.7 | 36.5 ** | 6.2 | 3.2 ** |
| YTH | 1.9 | 10.5 ** | 2.9 | 9.3 ** | 1.8 | 16.8 ** | 4.8 | 22.6 ** | 1 | 5.2 ** |
** Significant at a probability level of 1%, a abbreviation of traits is as per given under material and method, b data of three locations pooled and analysed; c genotype × Location interaction, MSS = Mean sum of square. Genotypes df is 33 and rep df is 2. Notes: X50F = days to 50% flowering, PH = plant height (cm), TILL = number of tillers per hill, PPH = number of panicles per hill, PL = panicle length (cm), GP = grains per panicle, Ferti = fertility (%), YPP = grain yield per plant (gm), TGW = thousand grain weight (gm), and YTH = grain yield (t/ha).
The estimations of range, mean value, variance components, heritability, and genetic advance for various traits with other statistical indicators are shown in Table 4. The days of 50% flowering varied from 103 to 135 with an average of 118.2 ± 0.9 in Barishal, 110 to 129 with an average of 121 ± 0.8 in Kushtia, while in Satkhira, it varied from 106 to 124 days with an average of 114.7 ± 0.8. The range of plant height varied from 80 to 148 cm with an average of 105.7 ± 3.4 in Barishal, 77 to 131 cm with an average of 99 ± 2.6 in Kushtia, while it ranged from 81 to 124 cm with an average of 101.2 ± 2.5 in Satkhira (Table 4).
Table 4.
Estimation of variance components, broad sense heritability, and genetic advance in elite rice genotypes.
| Traits | Location | X50F | PH | TILL | PPH | PL | GP | Ferti | YPP | TGW | YTH |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Barishal | 103–135 | 80–148 | 8–23 | 8–23 | 20–27.9 | 151–607 | 51–87.3 | 16.9–32.5 | 15.4–33 | 4.5–8.6 |
| Kushtia | 110–129 | 77–131 | 8–29 | 5–18 | 19–30 | 160–484 | 57.4–94.9 | 18–37.5 | 16.9–31.2 | 4.4–9.3 | |
| Satkhira | 106–124 | 81–124 | 7–18 | 6.3–16.5 | 18.5–29.8 | 79–375 | 54.3–94.5 | 17.6–28.9 | 13.8–28.9 | 4.6–7.9 | |
| Mean ± SE | Barishal | 118.2 ± 0.9 | 105.7 ± 3.4 | 13.8 ± 1.5 | 12.5 ± 1.4 | 23.9 ± 0.7 | 307.3 ± 49.6 | 66.1 ± 3.4 | 24.2 ± 1.2 | 21.8 ± 1.2 | 6.41 ± 0.4 |
| Kushtia | 121.0 ± 0.8 | 99.0 ± 2.67 | 15.7 ± 2.2 | 10.4 ± 1.5 | 24.4 ± 1.3 | 267.5 ± 23.5 | 79.7 ± 4.2 | 28.6 ± 1.9 | 22.1 ± 0.7 | 7.04 ± 0.5 | |
| Satkhira | 114.7 ± 0.8 | 101.2 ± 2.5 | 11.2 ± 0.7 | 10.7 ± 0.9 | 23.8 ± 0.7 | 210 ± 25.5 | 80.2 ± 3.9 | 22.7 ± 1.1 | 20.7 ± 1.5 | 6.24 ± 0.2 | |
| ẟ2g | Barishal | 64.9 | 124.7 | 5.2 | 6.3 | 1.3 | 7951.9 | 88.9 | 8.30 | 11.7 | 0.6 |
| Kushtia | 21.9 | 71.7 | 8.1 | 1.7 | 2.2 | 2929.7 | 45.4 | 12.7 | 8.8 | 0.8 | |
| Satkhira | 16.8 | 82.8 | 3.1 | 3.6 | 1.5 | 2234.5 | 33.9 | 4.8 | 5.8 | 0.5 | |
| ẟ2p | Barishal | 65.4 | 130.4 | 6.2 | 7.3 | 1.6 | 9183.3 | 95.0 | 9.1 | 12.4 | 0.6 |
| Kushtia | 22.3 | 75.3 | 10.5 | 2.8 | 2.9 | 3206.8 | 53.8 | 14.6 | 9.1 | 0.9 | |
| Satkhira | 17.3 | 89.0 | 3.4 | 4.1 | 1.8 | 2558.6 | 41.5 | 5.4 | 6.9 | 0.6 | |
| ẟ2e | Barishal | 1.3 | 16.9 | 3.2 | 3.0 | 0.8 | 3694.2 | 18.3 | 2.3 | 1.9 | 0.2 |
| Kushtia | 1.1 | 10.7 | 7.4 | 3.2 | 2.4 | 831.5 | 25.5 | 5.5 | 0.7 | 0.3 | |
| Satkhira | 1.1 | 9.6 | 0.9 | 1.5 | 0.8 | 972.4 | 22.9 | 1.5 | 3.3 | 0.1 | |
| GCV (%) | Barishal | 54.9 | 117.9 | 37.3 | 50.3 | 5.6 | 2588.0 | 134.5 | 34.4 | 53.6 | 9.3 |
| Kushtia | 3.8 | 8.5 | 17.9 | 12.7 | 5.9 | 20.3 | 8.5 | 12.5 | 13.4 | 13.2 | |
| Satkhira | 3.5 | 8.9 | 15.7 | 17.6 | 5.2 | 22.5 | 7.3 | 9.7 | 11.6 | 11.9 | |
| PCV (%) | Barishal | 6.8 | 10.5 | 16.5 | 20.0 | 4.8 | 29.0 | 13.5 | 11.9 | 15.6 | 12.0 |
| Kushtia | 3.96 | 9.2 | 24.8 | 8.7 | 22.9 | 10.5 | 14.9 | 13.9 | 15.4 | 8.7 | |
| Satkhira | 3.6 | 9.5 | 17.8 | 6.4 | 26.9 | 9.4 | 11.1 | 14.5 | 13.0 | 6.4 | |
| ECV (%) | Barishal | 6.8 | 11.3 | 20.9 | 6.1 | 35.1 | 14.9 | 13.4 | 16.9 | 13.7 | 6.1 |
| Kushtia | 0.8 | 3.3 | 17.2 | 17.1 | 6.3 | 10.7 | 6.3 | 8.1 | 3.8 | 7.9 | |
| Satkhira | 0.8 | 3.0 | 8.5 | 11.2 | 3.9 | 14.8 | 5.9 | 5.4 | 8.7 | 5.2 | |
| RD (%) | Barishal | 0.9 | 3.9 | 13.0 | 13.8 | 3.8 | 19.7 | 6.4 | 6.1 | 6.3 | 6.7 |
| Kushtia | 1.6 | 4.7 | 23.3 | 37.5 | 27.2 | 8.6 | 15.7 | 12.5 | 2.7 | 10.7 | |
| Satkhira | 2.2 | 6.9 | 8.8 | 11.9 | 16.4 | 12.7 | 18.4 | 9.5 | 16.7 | 5.9 | |
| h2b (%) | Barishal | 99.4 | 95.6 | 83.2 | 86.2 | 82.6 | 86.6 | 93.5 | 91.7 | 94.8 | 90.5 |
| Kushtia | 98.4 | 95.3 | 76.7 | 62.5 | 72.8 | 91.4 | 84.2 | 87.4 | 97.3 | 89.2 | |
| Satkhira | 97.9 | 93.0 | 91.2 | 88.0 | 83.6 | 87.3 | 81.6 | 90.5 | 84.1 | 94.0 | |
| GA (%) | Barishal | 14.0 | 21.3 | 30.7 | 38.4 | 9.1 | 55.6 | 28.4 | 23.5 | 31.4 | 23.5 |
| Kushtia | 7.9 | 17.2 | 32.5 | 20.7 | 10.5 | 39.8 | 15.9 | 24.1 | 27.3 | 25.7 | |
| Satkhira | 7.3 | 17.8 | 30.9 | 34.0 | 9.7 | 43.3 | 13.5 | 19.0 | 21.9 | 23.9 |
X50F = days to 50% flowering, PH = plant height (cm), TILL = number of tillers per hill, PPH = number of panicles per hill, PL = panicle length (cm), GP = grains per panicle, Ferti = fertility (%), YPP = grain yield per plant (gm), TGW = thousand grain weight (gm), and YTH = grain yield (t/ha).
The range of TILL varied from 8 to 23 with a mean of 13.8 ± 1.5 in Barishal; in Kushtia, it varied from 5 to 18 with an average of 15.7 ± 2.2; and in Satkhira, it ranged from 7 to 18 with a mean 11.2 ± 0.7. The PP varied from 8 to 23 (average 12.5 ± 1.4), 5 to 18 (average 10.4 ± 1.5), and from 6.3 to 16.5 (average 10.7 ± 0.9) in Barishal, Kushtia, and Satkhira, respectively. The panicle length varied from 20 to 27.9 cm with a mean value of 23.9 ± 0.7 in Barishal, 19 to 30 cm with an average value of 24.4 ± 1.3 in Kushtia, and 18.5 to 29.8 cm with a mean value of 23.8 ± 0.7 at the Satkhira location. The grains per panicle were found to be variable between 151 and 607 (average 307.3 ± 49.6) in Barishal, between 160 and 484 (average 267.5 ± 23.5) in Kushtia, and between 79 and 375 (average 210 ± 25.5) in Satkhira. The yield per plant was found to range from 16.9 to 32.5 gm with an average of 24.2 ± 1.2 in Barishal, and a range from 18 to 37.5 gm with an average of 28.6 ± 1.9 in Kushtia, while the range was from 17.6 to 28.9 gm with an average of 22.7 ± 1.1 in Satkhira. The range of 1000-grain weight varied from 15.7 to 33 gm with an average of 21.8 ± 1.2 in Barishal, 16.9 to 31.2 gm with an average of 22.1 ± 0.7 in Kushtia, while it ranged from 13.8 to 28.9 gm with an average of 20.7 ± 1.5 in Satkhira. The yield tons per hectare varied from 4.5 to 8.6 (average 6.41 ± 0.4), 4.4 to 9.3 (average 7.04 ± 0.5), and from 4.6 to 7.9 (average 6.24 ± 0.5) in Barishal, Kushtia, and Satkhira, respectively. It was noticed that the average value of yield (t/ha) for Kushtia was higher than in the other two locations.
Various genetic factors were evaluated to determine the genetic variability for specific features that already existed among the genotypes (Table 4). The phenotypic variance was divided into genotypic and error variance to evaluate the heritable component of overall variability. These results clearly showed that variability in the genotypes was primarily because of the genotypic variance, as the error variance units were low in three locations. The PCV values for all the characters were found to be higher than the corresponding GCV. The GP, TILL, and PPH recorded high values of GCV and PCV, while days to X50F, PL, and PH showed the lowest values in three locations (Table 4). Broad sense heritability was high for the days to X50F (99.4%), followed by PH (95.6%), TGW (94.8%), and fertility percentage (93.5%) in Barishal. At the Kushtia and Satkhira locations, these traits also showed higher heritability. The traits PP (62.5%), PL (72.8%), and TILL (76.7%) showed the lowest heritability in Kushtia. In Barishal and Satkhira, the genetic advance was found to be highest for GP at (55.6%) and (43.3%), followed by PP at (58.4%) and (34%), respectively. The highest number of GP (39.8%) was followed by the TILL (32.5%) in Kushtia. The days to 50F and PL had the lowest GA in the three locations.
3.2. Correlation Matrix among the Traits over Locations
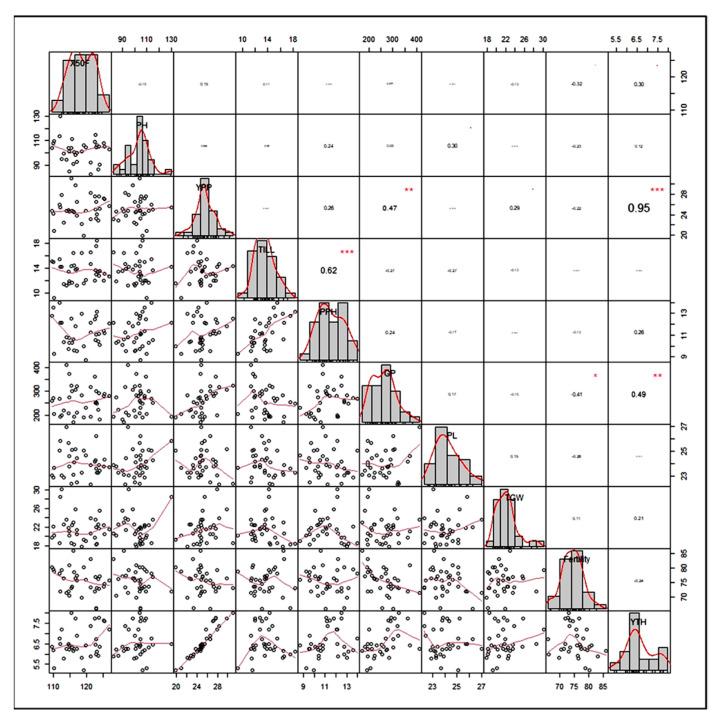
To comprehend the degree of relationship among the investigated variables and to highlight their significance in rice breeding programs, Pearson’s correlation coefficient matrix was displayed in Figure 1.
Figure 1.
Correlation coefficient matrix, scatter plot, and phenotypic frequency distribution among grain yield and yield-related traits over the locations. Each variable’s distribution is displayed diagonally. The bivariate scatter plots with a trend line are shown at the bottom of the diagonal. The correlation coefficient and the level of significance are displayed as stars at the top of the diagonal. * p ≤ 0.05, ** p ≤ 0.01, and *** p > 0.001 show significance level; Note: X50F = days to 50% flowering, PH = plant height (cm), TILL = number of tillers per hill, PPH = number of panicles per hill, PL = panicle length (cm), GP = grains per panicle, Fertility = fertility (%), YPP = grain yield per plant (gm), TGW = thousand grain weight (gm), and YTH = grain yield (t/ha).
The findings exhibited that YTH showed a positive and highly significant correlation along with YPP (0.95 ***) and GP (0.49 **). Fertility (%) was positively associated with TILL (0.02) and TGW (0.11), but there was a negative and significant association with GP (−0.41 *). The TGW had a positive association with YPP (0.29) and PL (0.15). The PL was positively correlated with PH (0.30) and GP (0.17). The GP showed a highly significant positive association with YPP (0.47 **), but a negative correlation with the TILL (−0.27). The PPH revealed a highly significant positive correlation with the TILL (0.62 ***), but only a positive correlation with YPP (0.26) and PH (0.24).
3.3. Analysis of Principal Components of the Studied Traits
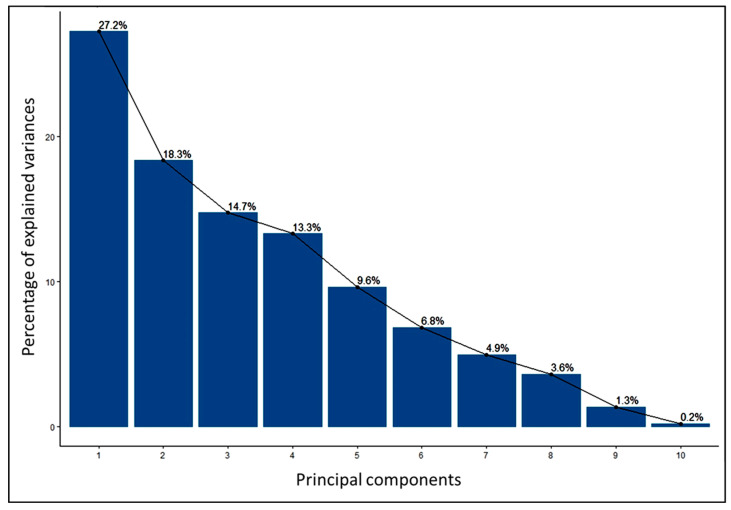
3.3.1. Graphical Presentation of the Scree Plot
The Scree plot described the percentage of variation for the respective principal components (Figure 2). PC1 to PC10 explained 100% of the variance. PC1 showed the maximum amount (27.2%) of the total variance, whereas PC2 depicted 18.3%. The other PC3 to PC10 exhibited 14.7, 13.3, 9.6, 6.8, 3.6, 1.3, and 0.2% of the total variance, respectively. The first five PCs contributed above 80% of the total variability. As a result, five PCs can be used to demonstrate the contribution and quality of top-performer genotypes and variables to total variability.
Figure 2.
Scree plot on variability explained by each component of 34 elite rice genotypes.
3.3.2. PCA-Biplot Analysis
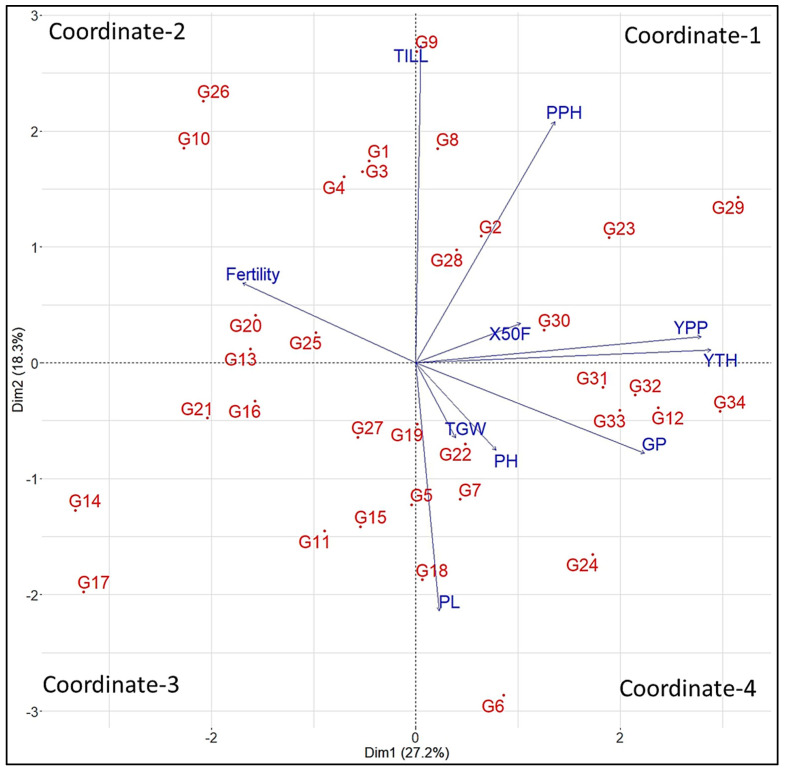
The biplot analysis shows the relationship among the various traits and genotypes concerning the first two principal components, which exhibited 45.5% of divergence in the dataset (Figure 3). In the biplot, the two first principal components were visualized into four coordinates along the x- and y-axis. Coordinate-1 is comprised of seven genotypes, namely, Gen2, Gen8, Gen9, Gen23, Gen28, Gen29, and G30, with the positive value of the first and second PCs. This coordinate’s genotypes were associated with important variables such as the TILL, PPH, X50F, YPP, and YTH. Gen29 is located far from the origin and has the greatest diversity, followed by Gen23, whereas other genotypes are located near the centre. The grain yield (t/ha) exhibited the longest vector, followed by grain yield per plant, which reflected the greater variance.
Figure 3.
Biplot for the 34 rice genotypes and 10 agronomic traits along the first 2 principal components. Note: X50F = days to 50% flowering, PH = plant height (cm), TILL = number of tillers per hill, PPH = number of panicles per hill, PL = panicle length (cm), GP = grains per panicle, Fertility = fertility (%), YPP = grain yield per plant (gm), TGW = thousand grain weight (gm), and YTH = grain yield (t/ha).
The genotypes such as Gen1, Gen3, Gen4, Gen10, Gen13, Gen20, Gen25, and Gen26 are plotted in the direction of negative values of PCA1 and positive values of PCA2 in coordinate-2, where the percent of the fertility trait is strongly associated with those genotypes. The first two principal components were negative in the eight genotypes placed at coordinate 3. The maximum number of genotypes (11) is located in coordinate-4, which had positive and negative values of the first and second PCs, respectively. The characteristics such as PL, GP, TGW, and PH are largely contributed to by the 11 genotypes. Among the four variables, GP showed the longer line that reflected the greater variance. The distribution of genotypes in the biplot represented that there was some genetic variability among the different rice breeding lines.
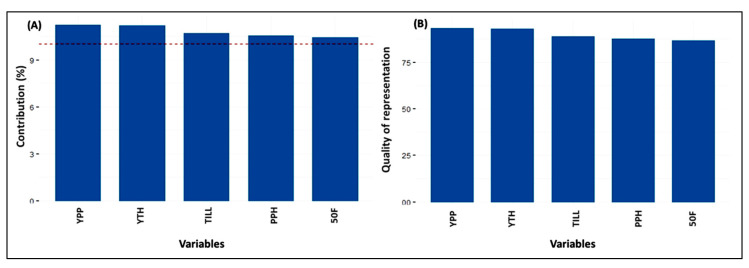
3.3.3. Contribution along with the Quality of Variables and Genotypes
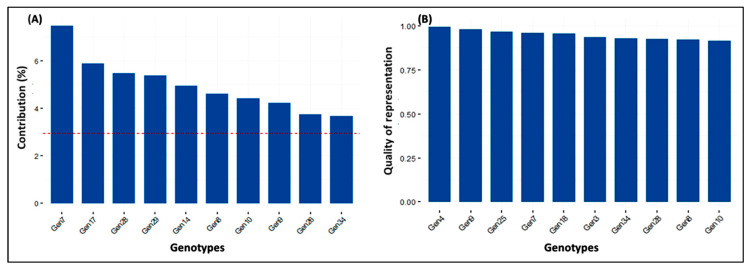
Among the ten variables, the maximum % of contribution and quality of representation towards the total variability was exhibited in the YPP, succeeded by YTH, TILL, PPH, and X50F to principal components 1-2-3-4-5 (Figure 4). In the 34 genotypes, the highest percentage of contribution towards the entire diversity was revealed in the genotype Gen7, followed by genotypes Gen17, Gen28, Gen29, Gen14, Gen8, Gen10, Gen9, Gen26, and Gen34, respectively, to principal components 1-2-3-4-5. However, the genotypes of Gen4 showed the maximum quality divergence, followed by Gen9, Gen25, Gen7, Gen18, Gen3, Gen34, Gen28, Gen8, and Gen10, respectively (Figure 5).
Figure 4.
Graphical representation on (A) percent of contribution and (B) quality of representation of top 5 variables to principal components 1-2-3-4-5.
Figure 5.
Graphical representation of (A) percent of contribution and (B) quality of representation of top 10 genotypes to principal components 1-2-3-4-5.
3.4. Analysis of Clusters among Traits
3.4.1. Choice of the Best Cluster Number
A total of 30 indices were utilized to identify the best cluster number during NbClust analysis. A total of 28 indices represented the number of clusters based on index value (Table 5).
Table 5.
List of the best number of clusters and their index value of 30 indices implemented in the NbClust package in R.
| KL | CH | Hartigan | CCC | Scott | Marriot | |
| No. of clusters | 8 | 3 | 3 | 10 | 3 | 3 |
| Index value | 2.1197 | 7.8206 | 3.051 | −0.7325 | 87.9885 | 2.49 × 1012 |
| TrCovW | TraceW | Friedman | Rubin | Cindex | DB | |
| No. of clusters | 3 | 3 | 8 | 8 | 8 | 10 |
| Index value | 310.342 | 23.6767 | 61.487 | −0.1678 | 0.4 | 1.0447 |
| Silhouette | Duda | PseudoT2 | Beale | Ratkowsky | Ball | |
| No. of clusters | 10 | 2 | 2 | 2 | 4 | 3 |
| Index value | 0.2796 | 1.0678 | −1.6513 | −0.4058 | 0.3152 | 61.7704 |
| PtBiserial | Gap | McClain | γ | Gplus | Tau | |
| No. of clusters | 5 | 2 | 2 | 10 | 10 | 5 |
| Index value | 0.5448 | −0.7636 | 0.8023 | 0.8943 | 2.8324 | 79.1515 |
| Frey | Hubert | Dindex | Dunn | SDindex | SDbw | |
| No. of clusters | 6 | 9 | 3 | 10 | 7 | 10 |
| Index value | 0.6385 | 0.0111 | 2.4227 | 0.5084 | 0.8697 | 0.3125 |
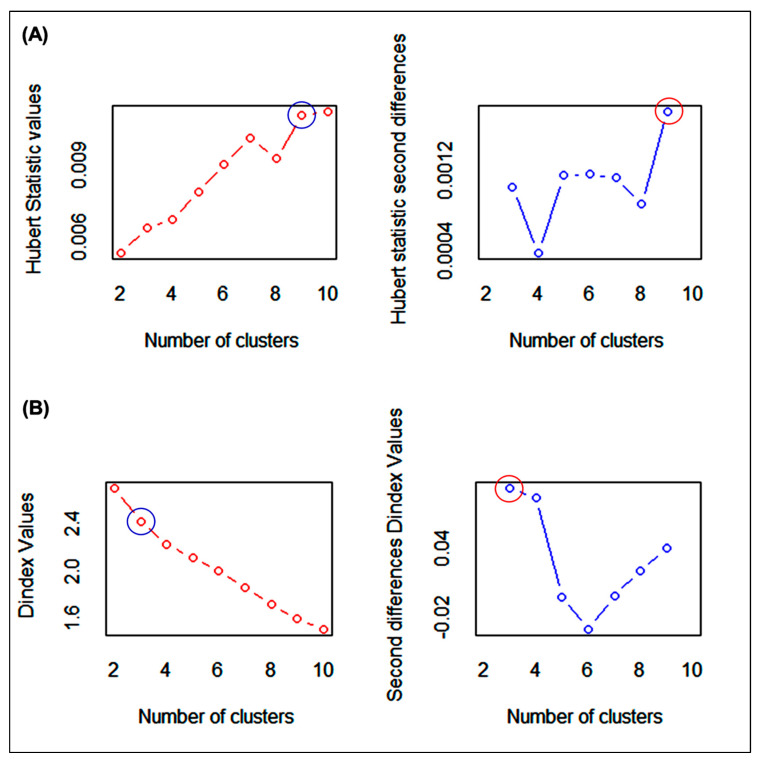
The other two methods, Huber and Dindex, are graphical methods that consider a significant knee for index value and a significant peak for second differences plotted. According to significant knee and peak value, nine is the optimal number of clusters in the Hubert index and three is the best number of clusters in the Dindex method (Figure 6). The results showed that five indices suggested two as the best number of clusters, eight indices recommended three as the best number of clusters, one index recommended four as the best number of clusters, two indices suggested five as the best number of clusters, one index recommended six as the best number of clusters, one index recommended seven as the best number of clusters, four indices suggested eight as the best number of clusters, one index recommended nine as the best number of clusters, and seven indices recommended ten as the best number of clusters. The best number of clusters, as determined by the majority rule, is three (Figure 6). Therefore, the initial value of k = 3 would be utilized to execute the K-means cluster analysis.
Figure 6.
Graphical method (A) Hubert index and (B) Dindex for detecting the optimum number of clusters.
3.4.2. Grouping of Genotypes by Heatmap Oriented Clustering Pattern
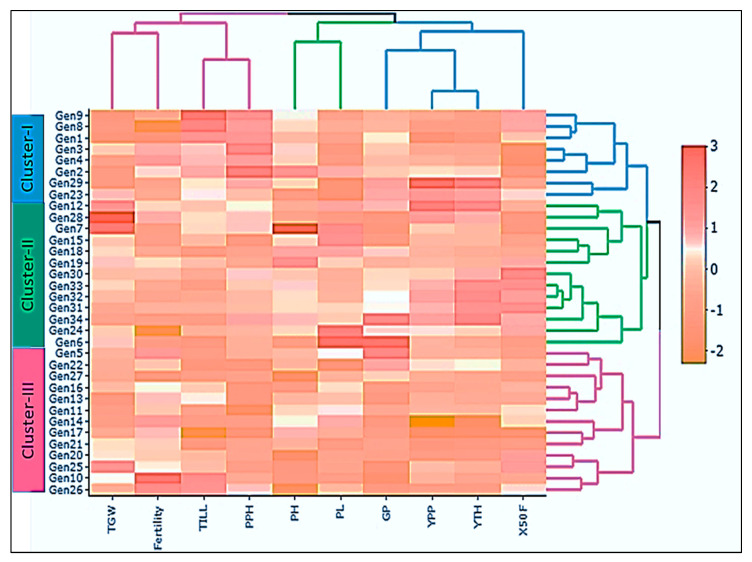
A heatmap is a two-dimensional data visualization approach that employs the use of colour to depict a phenomenon’s extent. The reader can see how the phenomenon is grouped or changes over space by looking at colour variation by intensity. It shows the relative distribution of highly expressed characteristics over a background of predominantly small features (Figure 7). As a result, the heatmap analysis generated two dendrograms: one in the horizontal direction reflecting the features that triggered the diffusion and one in the vertical direction representing the germplasm accessions.
Figure 7.
The heatmap displays the clustering pattern of 34 rice genotypes with 10 quantitative characters. Each column indicates a character, whereas each row represents a genotype. The various colours and intensities (−2 to 3) were adjusted based on the genotype–characters relationship. The orange colour represents a lower value, the white colour for mid value, and the dark red indicates a higher value.
In the present study, heatmap-oriented cluster analysis using average values of all the studied traits of 34 rice genotypes was conducted, and genotypes were classified into 3 clusters. The frequency pattern of clusters exhibited that a large number of genotypes (13) was found in cluster II and cluster III, whereas the lowest number of genotypes (8) was situated in cluster I (Table 6 and Figure 7).
Table 6.
Clustering of 34 elite rice breeding lines.
| Cluster | Genotypes Number | Name of Genotypes Code |
|---|---|---|
| I | 8 | Gen1, Gen2, Gen3, Gen4, Gen8, Gen9, Gen23, Gen29 |
| II | 13 | Gen6, Gen7, Gen12, Gen15, Gen18, Gen19, Gen24, Gen28, Gen30, Gen31, Gen32, Gen33 Gen34 |
| III | 13 | Gen5, Gen10, Gen11, Gen13, Gen14, Gen16, Gen17, Gen20, Gen21, Gen22, Gen25, Gen26, Gen27 |
Cluster I genotypes are BR7671-37-2-2-37-3-P3 (Gen1), BR8626-19-5-1-2 (Gen2), BR8626-10-5-1(Gen3), BR8109-29-2-2-3 (Gen4), BR(Bio)8333-BC5-2-22 (Gen8), BR(Bio)8333-BC5-3-10 (Gen9), BRC266-5-1-2-1 (Gen23), and BRRI dhan58 (Gen29). Cluster II contains 13 genotypes: BR(Bio)8333-BC5-1-20 (Gen6), BR(Bio)8333-BC5-2-16 (Gen7), BR7831-59-1-1-4-5 (Gen12), BR7815-18-1-3-2-1 (Gen15), BR8079-19-1-5-1 (Gen18), BR8590-5-2-5-2-2 (Gen19), BR8523-36-2-2-6 (Gen24), BRRI dhan28 (Gen28), BRRI dhan29 (Gen30), BR16 (Gen31), BRRI dhan50 (Gen32), BRRI dhan63 (Gen33), and BRRI dhan74 (Gen34). Cluster III contains 13 genotypes, including BR (Bio) 8333-BC5-1-1 (Gen5), BR8631-12-3-5-P2 (Gen10), BR8631-12-3-6-P3 (Gen11), BR8253-9-3-3-1 (Gen13), BR8609-2-B-9-1-B5 (Gen14), BR7671-37-2-2-3-7-3-P10 (Gen16), BR7671-37-2-2-3-7-3-P11 (Gen17), BR8590-5-3-3-4-2 (Gen20), BR8608-39-2-1 (Gen21), BRC266-5-1-1-1 (Gen22), BR8938-19-4-3-1-1 (Gen25), BR8333-15-3-2-2 (Gen26), and IRBB60 (Gen27). Other dendrograms revealed three distinct groups. Group-1 is linked with four characters (TGW, Fertility, TILL, and PPH). Group-2 is connected with two characters (PH and PL). Group-3 is allied with four characters (GP, YPP, YTH, and X50F) (Figure 7).
3.5. Identification of the Best Performing Genotypes through Multi-Trait Genotype–Ideotype Distance Index (MGIDI)
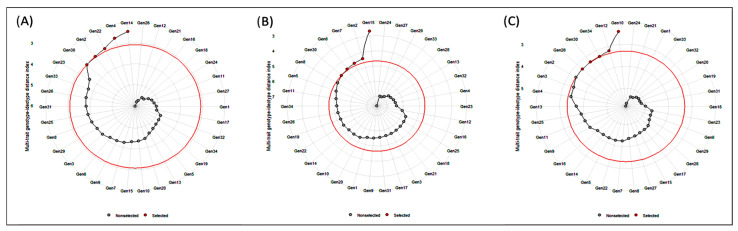
The multi-trait genotype–ideotype distance index (MGIDI) was used to select the best-performing genotypes utilizing the MGIDI selection index. The selection outputs of different locations are presented accordingly. A selection difference in percent (DS) of 16.3% for GP is highlighted followed by 12.4% for YPP in Satkhira (Table 7), 11.9% for YTH is compared to 11.4% for YPP in Kushtia (Table 8), and 12.6% for PPH is highlighted followed 8.31% for TILL in Barishal in the selection gain analysis results (Table 9) from MGIDI analysis, which revealed gains in the desired sense in 34 genotypes of the 10 characters. A total of 34 genotypes, 5 genotypes in each location, e.g., genotypes Gen2, Gen4, Gen14, Gen22, and Gen30 in Satkhira; Gen2, Gen6, Gen7, Gen15, and Gen30 in Kushtia; and Gen10, Gen12, Gen26, Gen30, and Gen34 in Barishal were selected. These genotype performances went well, which is equivalent to the ideotype utilized in MGIDI. However, because of their superior performance, genotypes Gen28 and Gen12 in Satkhira, Gen24 and G27 in Kushtia, and Gen24 and Gen21 in Barishal displayed greater sensitivity (Figure 8).
Table 7.
Original value (Xo), selected value (Xs), selection differential in percentage (SDperc), heritability (h2), and selection gain in percent (SGperc) for the MGIDI in 34 rice genotypes in Satkhira.
| Variable | Factor | Xo | Xs | SDperc | h2 | SGperc | Sense | Goal |
|---|---|---|---|---|---|---|---|---|
| PPH | FA1 | 10.7 | 11.8 | 9.88 | 0.88 | 8.7 | increase | 100 |
| GP | FA1 | 210 | 244 | 16.3 | 0.873 | 14.3 | increase | 100 |
| TGW | FA1 | 20.7 | 20.4 | −1.88 | 0.841 | −1.58 | increase | 0 |
| X50F | FA2 | 115 | 115 | −0.117 | 0.98 | −0.115 | increase | 0 |
| PH | FA2 | 101 | 101 | −0.276 | 0.963 | −0.266 | increase | 0 |
| TILL | FA2 | 11.2 | 11.5 | 2.82 | 0.91 | 2.57 | increase | 100 |
| YPP | FA3 | 22.8 | 25.6 | 12.4 | 0.904 | 11.2 | increase | 100 |
| YTH | FA3 | 6.24 | 6.67 | 6.84 | 0.941 | 6.43 | increase | 100 |
| PL | FA4 | 23.8 | 25.1 | 5.29 | 0.836 | 4.42 | increase | 100 |
| Fertility | FA4 | 80.2 | 76.9 | −4.12 | 0.816 | −3.36 | increase | 0 |
Table 8.
Original value (Xo), selected value (Xs), selection differential in percentage (SDperc), heritability (h2), and selection gain in percent (SGperc) for the MGIDI in 34 rice genotypes in Kushtia.
| Variable | Factor | Xo | Xs | SDperc | h2 | SGperc | Sense | Goal |
|---|---|---|---|---|---|---|---|---|
| YPP | FA1 | 28.6 | 31.9 | 11.4 | 0.874 | 9.96 | increase | 100 |
| YTH | FA1 | 7.04 | 7.88 | 11.9 | 0.892 | 10.6 | increase | 100 |
| TILL | FA2 | 15.7 | 16 | 1.89 | 0.767 | 1.45 | increase | 100 |
| PPH | FA2 | 10.4 | 10.4 | 0.0471 | 0.624 | 0.0294 | increase | 100 |
| GP | FA2 | 267 | 267 | −0.108 | 0.914 | −0.0989 | increase | 0 |
| X50F | FA3 | 121 | 124 | 2.31 | 0.984 | 2.28 | increase | 100 |
| PH | FA3 | 99.1 | 105 | 6.01 | 0.952 | 5.72 | increase | 100 |
| TGW | FA4 | 22.2 | 24.1 | 8.6 | 0.973 | 8.36 | increase | 100 |
| PL | FA5 | 24.4 | 25.2 | 3.48 | 0.728 | 2.53 | increase | 100 |
| Fertility | FA5 | 79.8 | 78.1 | −2.03 | 0.842 | −1.71 | increase | 0 |
Table 9.
Original value (Xo), selected value (Xs), selection differential in percentage (SDperc), heritability (h2), and selection gain in percent (SGperc) for the MGIDI in 34 rice genotypes in Barishal.
| Variable | Factor | Xo | Xs | SDperc | h2 | SGperc | Sense | Goal |
|---|---|---|---|---|---|---|---|---|
| YPP | FA1 | 24.2 | 26 | 7.75 | 0.918 | 7.11 | increase | 100 |
| YTH | FA1 | 6.41 | 6.93 | 8.20 | 0.905 | 7.42 | increase | 100 |
| TILL | FA2 | 13.9 | 15 | 8.31 | 0.827 | 6.87 | increase | 100 |
| PPH | FA2 | 12.5 | 14.1 | 12.6 | 0.863 | 10.8 | increase | 100 |
| PH | FA3 | 106 | 112 | 5.92 | 0.956 | 5.66 | increase | 100 |
| GP | FA3 | 307 | 323 | 5.07 | 0.866 | 4.39 | increase | 100 |
| PL | FA3 | 23.9 | 24.9 | 4.15 | 0.826 | 3.43 | increase | 100 |
| Fertility | FA3 | 66.1 | 60.1 | −9.06 | 0.929 | −8.41 | increase | 0 |
| X50F | FA4 | 118 | 114 | −3.79 | 0.994 | −3.77 | increase | 0 |
| TGW | FA4 | 21.9 | 23.4 | 6.96 | 0.948 | 6.6 | increase | 100 |
Figure 8.
The circular preview indicates the ranking of genotypes based on the MGIDI selection index as well as the best rice genotypes with the associated locations (A) Satkhira, (B) Kushtia, and (C) Barishal. The selected ones are marked in red colour.
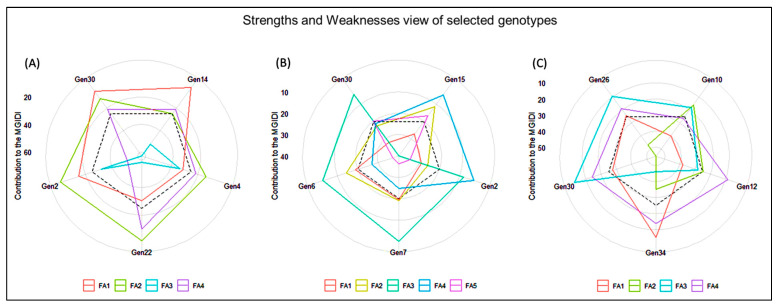
In the Satkhira region, regarding the strengths and weaknesses of the five selected genotypes (Figure 9A and Table 7), we found the Gen4 lineage with a strong contribution to factor 1 (FA1: PPH, GP and TGW) and a weak contribution of Gen14. For factor 2 (FA2: X50F, PH, and TILL), Gen14 lineage observed strong and weak contributions of Gen2. In factor 3 (FA3: YPP, and YTH), the Gen30 lineages were observed as strong and Gen2 presented a weak contribution, and in factor 4 (FA4: PL and Fertility), the Gen2 lineage revealed a strong contribution in this factor.
Figure 9.
The strengths and weaknesses view of the selected genotypes represent the proportion of each factor on the computed MGIDI index. (A) Satkhira, (B) Kushtia, and (C) Barishal.
In Kushtia, for factor 1 (FA1: YPP and YTH), Gen30 were strong and Gen6 were a weak contributor (Figure 9B and Table 8). To factor 2 (FA2: TILL, PPH, and GP), the Gen30 lineage presented strong and weak participation of Gen15. For factor 3 (FA3: X50F and PH), the Gen15 lineage noticed strong and weak points of Gen7. For factor 4 (FA4: TGW), the genotypes Gen6 and Gen15 have strong and weak points, respectively. The Gen6 genotypes were strong and the Gen15 genotypes were weak contributions in factor 5 (FA5: PL and Fertility).
In Barishal, for factor 1 (FA1: YPP and YTH), Gen10 were strong and Gen34 was a weak contributor (Figure 9C and Table 9). For factor 2 (FA2: TILL and PPH), the Gen30 lineage presented strong and weak participation of Gen10. For factor 3 (FA3: PH, PL, Fertility and GP), the Gen12 lineage revealed strong and weak points of Gen30. In factor 4 (FA4: X50F and TGW), the genotypes Gen10 and Gen12 have strong and weak points.
4. Discussion
4.1. Variability Analysis among the Studied Traits
Genetic variations in crop gene-pool resources can be displayed using morphological markers easily and straightforwardly [30]. The assessment of genetic variability and genetic relationships among the major morphological and yield-associated traits paves the first step for any breeding program [31]. The agronomic features and yield attributes are primarily quantitative characters influenced by both genetic and environmental factors. The interactions between genotype and environment have been found in several crops [32,33,34]. Therefore, we have evaluated 34 potential genotypes for 3 locations for variability, principal component, and cluster studies. In general, the phenotypic and genotypic variances were often comparable for all the characters in three locations, illustrating that all the attributes were under governance by genotypic variance. The trait had higher PCV values than corresponding GCV values with very small differences (RD), which implies that genetic factors primarily control these traits rather than environmental factors [35,36]. The results showed that the PCV value was greater than the GCV value for all the studied traits, implying that all the traits were influenced by the environment to some extent among rice genotypes. GCV and PCV values were high for GP, TILL, and PPH compared to the other variables. The closer PCV and GCV values were found in X50F, PL, and PH in three locations. The result reflected that those traits were slightly influenced by environmental factors, indicating that the selection would be rewarding for future breeding programs. This type of result was recorded in rice by other researchers [37,38,39].
The higher value of ECV for the traits TILL, PPH, GP, YPP, and YTH showed that the environment had more impact on the expression of these traits. However, X50F, PH, and Fertility percentage were found to have the very lowest ECV values, indicating lower environmental effects. A similar statement was reported in the findings of [40]. The estimation of heritability with genetic advances would be an effective approach in a selective breeding program [41]. The presence of a high genetic advance value and a high heritability indicates the trait is governed by additive gene action and would be particularly effective in terms of selection and accuracy. The GP, PPH, and TILL have high heritability and GA, suggesting that these variables are controlled by additive genes with little environmental influence. On the other hand, the smaller heritability and GA indicated that traits were influenced by non-additive genes (with more contribution from the environment). For future breeding programs, traits exhibiting high GCV, heritability, and GA should be prioritized [42].
4.2. Trait Association
Significant selection promoting genotypes only marked on yield may not be beneficial due to having some environmental impact on polygenic characteristics. As a result, the selection must be made through interrelated traits to improve yield or plant morphology. Thus, correlation analysis was performed among the yield, with the yield contributing ten characters. Yield is assumed to play an essential role in crop production, and several studies have been initiated to examine the association between yield and different yield-related attributes in various crops. In this study, YTH had a positive and significant association (p ≤ 0.01 and p ≤ 0.001) with GP and YPP, but no significant association with TGW. The results were consistent with the report of [6,43] in rice. There was no correlation between 100-seed weight and yield in Adzuki beans [44], which was in accordance with our results. Debsharma et al. [6] examined 16 germplasms of elite rice and found that PL and TILL are negatively associated with yield, which agreed with these findings. There was a positive correlation between X50F and YTH, indicating that the longer-duration varieties generate higher yield. It has been noted that a positive relationship is observed between PL and PH [45]. The X50F was found to be negatively associated with PH, which corroborated the report of Prasad et al. [46]. It has been reported that YPP exhibited a significantly positive association (p ≤ 0.01) with GP, which was also noticed by Debsharma et al. [6]. As a result of their significant impact on yield, GP and YTH should receive priority attention in any rice improvement efforts.
4.3. Principal Component Analysis among Traits
Principal Component Analysis (PCA) is a widely used technique to explore the maximum variability through dimensionality reduction from a large number of components [47]. The PCA1, PCA2, PCA3, and PCA4 had eigen values of 27.2%, 18.3%, 14.7%, and 13.3% respectively. Together, they accounted for 73.5% variability of the genotypes used for the diversity analysis. By analysing 31 rice germplasms, Pokhrel et al. [48] found that the first 4 components were responsible for 73.8% of the total variation, which is quite comparable with our findings. PCA-biplots integrate characteristics and objects in two dimensions and reduce conflicting variations, facilitating the identification of the primary characters in datasets [49]. The YTH, YPP, TILL, PPH, and X50F were the top contributors, and the quality of representations elaborates on the total variations because of the highest positive values in the first five PCAs, which is the line in the studies of [50,51]. Those characters might be given keen emphasis when choosing parents.
4.4. Heatmap-Oriented Cluster Analysis among Genotypes and Traits
In general, an analysis of clustering is performed to choose the best parents for exploiting high heterotic gain. Based on agro-morphological characteristics, cluster analysis has been previously reported in numerous studies for rice [52,53,54]. In the present study, heatmap-oriented cluster analysis resulted in 3 clusters among 34 elite rice cultivars. The heatmap displays the maximum and minimum values for each genotype for all comparable features in a variety of colours ranging from light hues to deeper intensities. A heatmap scattering analysis showed that cluster II and cluster III had the most genotypes (13), whereas cluster I had the fewest cultivars (8) (Table 6). The rice breeding lines were classified into four clusters, which are depicted in earlier reports by [11,50]. The mean assessment of various clusters for the traits manifested in cluster II genotypes with the PH and PL were accumulated in Group 2, whereas genotypes with TGW, Fertility, TILL, and PPH were more divergent in Group 1. On the other hand, again, cluster II genotypes having GP, YPP, YTH, and X50F were aggregated into Group-3 with high red colour compactness (Figure 7). The cluster-oriented mean values of the mentioned characters were used to estimate the supremacy of the cluster, and therefore, could be assumed in the enhancement of different traits [11,55]. To maximize heterosis and produce a wide variety of segregating populations, Nisar et al. [56] and Ashok et al. [57] advised that genotypes that are used for hybridization be selected from the farthest clusters with good mean accomplishment for studied traits. From cluster analysis, it was recommended that, regarding yield effectiveness and other yield-associated characteristics, elite rice genotypes grouped in clusters II should have more obvious potential for further rice breeding programs.
4.5. Multi-Trait Selection Index Based on Factor and MGIDI Index Analysis
Plant breeders typically evaluate a variety of attributes throughout the selection process. A plant ideotype that symbolizes the selection of high-performing plants is kept in mind by plant breeders. By using a stepwise trial-and-error approach, an ideotype gives breeders an ultimate aim for selection, ultimately improving plant performance. The recently introduced MGIDI (multi-trait genotype–ideotypes distance index) is a new approach for selecting genotypes based on multiple trait information [22]. It was simple to identify the genotype’s strengths and weaknesses based on the multiple-trait approach using the MGIDI’s view (Figure 6). The chosen genotypes (Gen2, Gen4, Gen14, Gen22, and Gen30 in Satkhira; Gen2, Gen6, Gen7, Gen15, and Gen30 in Kushtia; and Gen10, Gen12, Gen26, Gen30, and Gen34 in Barishal) had great productivity potential in terms of various yield-contributing traits in addition to grain yield. According to Gabriel et al. [58] and Olivoto and Nardino [22], who evaluated thirteen strawberry genotypes, the Albion cultivar had the highest productivity. In contrast to the selected genotypes of Gen28 and Gen12 in Satkhira, Gen24 and G27 in Kushtia, and Gen24 and Gen21 in Barishal were quite near the cut point (Figure 8), suggesting that this genotype may have advantageous traits. Therefore, when evaluating genotypes, the investigator should pay special attention to those that are quite close to the cut point [22].
5. Conclusions
A total of thirty-four elite rice breeding lines were assessed based on ten agronomic and yield-related traits in three locations, revealing that sufficient genetic variation occurred in those studied traits. The majority of the traits under investigation showed significant genetic variability. The GP, TILL, and PPH of all locations exhibited high values of PCV and GCV coupled with GA and heritability. The correlation studies exposed a highly significant positive interrelation between YPP and GP and might be effectively applied as a selection parameter for rice breeding development. The heatmap-oriented cluster analysis exposed a group of 34 genotypes into 3 clusters, displaying significant genetic variability among them. The genotypes accumulated in cluster II performed well in terms of agro-morphological traits. Thus, genotypes Gen2, Gen4, Gen14, Gen22, and Gen30 in Satkhira; Gen2, Gen6, Gen7, Gen15, and Gen30 in Kushtia; and Gen10, Gen12, Gen26, Gen30, and Gen34 in Barishal might be assumed as superior parents based on the MGIDI selection index. Additionally, a heatmap was used to estimate the association matrix between each elite cultivar and its attributes. With the help of the diversity analysis, it will be possible to select the finest recombinants for various traits and to modify these traits in subsequent segregates, which will provide valuable information for further trait-oriented breeding programs or use these selected germplasms as potential breeding material in crossing programs for the development of cultivars suitable for multiple environments under the future changing climate.
Acknowledgments
The authors acknowledge the Bangladesh Rice Research Institute, Regional stations Satkhira, Barishal, and Kushtia for providing facilities to accomplish the research work. The authors extend their gratitude to the Taif University Researchers Supporting Project number (TURSP-2020/39), Taif University, Taif, Saudi Arabia and the ‘Slovak University of Agriculture’, Nitra, Tr. A. Hlinku 2, 949 01 Nitra, the Slovak Republic for partially supporting the study.
Author Contributions
Conceptualization, S.K.D.; design of the experiment, S.M, S.K.D. and M.H.A.; methodology, S.M., S.K.D. and M.H.A.; data analysis, S.K.D., M.A.S., A.G. and A.H.; data curation, S.K.D. and M.A.S., writing—original draft preparation, S.K.D., M.A.S., and P.R.R.; critical review and editing, S.K.D., M.A.S., A.G., M.B. and A.H.; funding, M.B. and A.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The research work was technically and financially supported by Bangladesh Rice Research Institute (BRRI), Gazipur 1701, Bangladesh. This research was also partially funded by the ‘Slovak University of Agriculture’, Nitra, Tr. A. Hlinku 2, 949 01 Nitra, Slovak Republic under the projects APVV-20-0071 and partially supported by Taif University Researchers Supporting Project number (TURSP-2020/39), Taif University, Taif, Saudi Arabia.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.BBS . Bangladesh Bureau of Statistics. Statistics and Informatics Division (SID), Ministry of Planning, Government of the People’s Republic of Bangladesh; Dhaka, Bangladesh: 2017. Yearbook of Agricultural Statistics; p. 42. [Google Scholar]
- 2.BBS . Bangladesh Bureau of Statistics. Statistics and Informatics Division (SID), Ministry of Planning, Government of the People’s Republic of Bangladesh; Dhaka, Bangladesh: 2020. Yearbook of Agricultural Statistics; p. 39. [Google Scholar]
- 3.Brolley M. Rice Today. International Rice Research Institute (IRRI); Metro Manila, Philippines: 2015. Rice security is food security for much of the world; pp. 30–32. [Google Scholar]
- 4.Nath N.C. Food security in Bangladesh: Status, challenges and strategic policy options; Proceedings of the 19th Biennial Conference of the Bangladesh Economic Association (BAE); Dhaka, Bangladesh. 8–10 January 2015. [Google Scholar]
- 5.Kabir M.S., Salam M.U., Chowdhury A., Rahman N.M.F., Iftekharuddaula K.M., Rahman M.S., Rashid M.H., Dipti S.S., Islam A., Latif M.A., et al. Rice vision for Bangladesh: 2050 and beyond. Bangladesh Rice J. 2015;19:1–18. doi: 10.3329/brj.v19i2.28160. [DOI] [Google Scholar]
- 6.Debsharma S.K., Disha R.F., Ahmed M.M.E., Khatun M., Ibrahim M., Aditya T.L. Assessment of Genetic Variability and Correlation of Yield Components of Elite Rice Genotypes (Oryza sativa L.) Bangladesh Rice J. 2020;24:21–29. doi: 10.3329/brj.v24i1.53237. [DOI] [Google Scholar]
- 7.Debsharma S.K., Roy P.R., Begum R.A., Iftekharuddaula K.M., Roy K.K., Hossain M.Z. Distinctness, Uniformity and Stability (DUS) characterization for BRRI developed rice varieties of Bangladesh. Int. J. Sustain. Crop Prod. 2020;15:13–19. [Google Scholar]
- 8.Akinwale M.G., Gregorio G., Nwilene F., Akinyele B.O., Ogunbayo S.A., Odiyi A.C. Heritability and correlation coefficient analysis for yield and its components in rice (Oryza sativa L.) Afr. J. Plant Sci. 2011;5:207–212. doi: 10.5897/AJPS.9000137. [DOI] [Google Scholar]
- 9.Sweeney M.T., Thomson M.J., Cho Y.G., Park Y.J., Williamson S.H., Bustamante C.D., McCouch S.R. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson M.J., Septiningsih E.M., Suwardjo F., Santoso T.J., Silitonga T.S., McCouch S.R. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2007;114:559–568. doi: 10.1007/s00122-006-0457-1. [DOI] [PubMed] [Google Scholar]
- 11.Hannan A., Rana M.R.I., Hoque M.N., Sagor G.H.M. Genetic variability, character association and divergence analysis for agro-morphological traits of local rice (Oryza sativa L.) germplasm in Bangladesh. J. Bangladesh Agric. Univ. 2020;18:289–299. doi: 10.5455/JBAU.84906. [DOI] [Google Scholar]
- 12.Debsharma S.K., Roy P.R., Begum R.A., Iftekharuddaula K.M. Elucidation of Genotype × Environment interaction for identification of stable genotypes to grain yield of rice (Oryza sativa L.) varieties in Bangladesh rainfed condition. Bangladesh Rice J. 2020;24:59–71. doi: 10.3329/brj.v24i1.53240. [DOI] [Google Scholar]
- 13.Megloire N. Ph.D. Dissertation. University of Free State; Bloemfontein, South Africa: 2005. The Genetic, Morphological and Physiological Evaluation of African Cowpea Genotypes. [Google Scholar]
- 14.Quddus M.R., Rahman M.A., Jahan N., Debsharma S.K., Disha R.F., Hasan M.M., Aditya T.L., Iftekharuddaula K.M., Collard B.C.Y. Estimating pedigree-based breeding values and stability parameters of elite rice breeding lines for yield under salt stress during the Boro season in Bangladesh. Plant Breed. Biotechnol. 2019;7:257–271. doi: 10.9787/PBB.2019.7.3.257. [DOI] [Google Scholar]
- 15.Akter F., Islam M.Z., Akter A., Debsharma S.k., Shama A., Khatun M. Genetic diversity of bacterial blight resistant rice (Oryza sativa L.) genotypes from INGER. Bangladesh Rice J. 2019;23:59–64. doi: 10.3329/brj.v23i2.48248. [DOI] [Google Scholar]
- 16.Aditya J.P., Bhartiya P., Bhartiya A. Genetic variability, heritability and character association for yield and component characters in soybean (Glycine. max L. Merrill) J. Cent. Eur. Agric. 2011;12:27–34. doi: 10.5513/JCEA01/12.1.877. [DOI] [Google Scholar]
- 17.Gonçalves D.D.L., Barelli M.A.A., Oliveira T.C.D., Santos P.R.J.D., Silva C.R.D., Poletine J.P., Neves L.G. Genetic correlation and path analysis of common bean collected from Caceres Mato Grosso state, Brazil. Ciênc. Rural. 2017;47:1–7. doi: 10.1590/0103-8478cr20160815. [DOI] [Google Scholar]
- 18.Vural H., Karasu A. Variability studies in cowpea (Vigna unguiculata L. Walp.) varieties grown in Isparta, Turkey. Revista Científica UDO Agrícola. 2007;1:29–34. [Google Scholar]
- 19.Debsharma S.K., Nath U.K., Azad A.K., Alam M.J., Das P.B. Selection of groundnut lines for genetic diversity and high yield performance. Bangladesh J. Seed Sci. Tech. 2011;15:173–180. [Google Scholar]
- 20.Akter N., Khalequzzaman M., Islam M.Z., Mamun M.A.A., Chowdhury M.A.Z. Genetic variability and character association of quantitative traits in Jhum rice genotypes. SAARC J. Agric. 2018;16:193–203. doi: 10.3329/sja.v16i1.37434. [DOI] [Google Scholar]
- 21.Islam M.Z., Akter N., Chakrabarty T., Bhuiya A., Siddique M.A., Khalequzzaman M. Agro-morphological characterization and genetic diversity of similar named aromatic rice (Oryza sativa L.) landraces of Bangladesh. Bangladesh Rice J. 2018;22:45–56. doi: 10.3329/brj.v22i1.41836. [DOI] [Google Scholar]
- 22.Olivoto T., Nardino M. MGIDI: Toward an effective multivariate selection in biological experiments. Bioinformatics. 2021;37:1383–1389. doi: 10.1093/bioinformatics/btaa981. [DOI] [PubMed] [Google Scholar]
- 23.BRRI . Modern Rice Cultivation. 22nd special ed. BRRI; Dhaka, Bangladesh: 2019. p. 96. [Google Scholar]
- 24.Yoshida S., Forno D.A., Cock J.H., Gomez K.A. Laboratory Manual for Physiological Studies of Rice. IRRI83; Los Baños, Philippines: 1976. [Google Scholar]
- 25.Johnson H.W., Robinson H., Comstock R. Estimates of Genetic and Environmental Variability in Soybeans. Agronomy. 1955;47:314–318. doi: 10.2134/agronj1955.00021962004700070009x. [DOI] [Google Scholar]
- 26.Allard R.W. Principles of Plant Breeding. 2nd ed. John Wiley and Sons; New York, NY, USA: 1999. p. 254. [Google Scholar]
- 27.Burton G.W., DeVane E.H. Estimating heritability in tall fescue (Festuca arundinacea) from replicated clonal material. Agron. J. 1953;45:478–481. doi: 10.2134/agronj1953.00021962004500100005x. [DOI] [Google Scholar]
- 28.Singh R.K., Chaundry B.D. Biometrical Methods in Quantitative Genetic Analysis. 2nd ed. Kalayani Publishers; New Delhi, India: Ludhiana, India: 1985. [Google Scholar]
- 29.Olivoto T., Lúcio A.D.C. Metan: An R package for multi environment trial analysis. Methods Ecol. Evol. 2020;11:783–789. doi: 10.1111/2041-210X.13384. [DOI] [Google Scholar]
- 30.Bibi S., Shah A.H., Khan I.A., Jan S.A., Zubair M., Ali N., Khan U. Assessment of genetic diversity in hexaploid wheat of northern areas of Pakistan (Gilgit Baltistan) using morphological marker. Int. J. Agric. Biol. 2019;22:283–289. [Google Scholar]
- 31.Wang Q.J., Jiang Y., Liao Z.Q., Xie W.B., Zhang X.M., Lan H., Hu E.L., Xu J., Feng X.J., Wu F.K., et al. Evaluation of the contribution of teosinte to the improvement of agronomic, grain quality and yield traits in maize (Zea mays) Plant Breed. 2020;139:589–599. doi: 10.1111/pbr.12796. [DOI] [Google Scholar]
- 32.Kovacevic V., Kadar I., Andric L., Zdunic Z., Iljkic D., Varga I., Jovic J. Environmental and genetic effects on cadmium accumulation capacity and yield of maize. Czech. J. Genet. Plant. 2019;55:70–75. doi: 10.17221/5/2018-CJGPB. [DOI] [Google Scholar]
- 33.Mwiinga B., Sibiya J., Kondwakwenda A., Musvosvi C., Chigeza G. Genotype x environment interaction analysis of soybean (Glycine max L. Merrill) grain yield across production environments in Southern Africa. Field Crop Res. 2020;256:107922. doi: 10.1016/j.fcr.2020.107922. [DOI] [Google Scholar]
- 34.Debsharma S.K., Rahman M.A., Quddus M.R., Khatun H., Disha R.F., Roy P.R., Ahmed S., El-Sharnouby M., Iftekharuddaula K.M., Aloufi S., et al. SNP based trait characterization detects genetically important and stable multiple stress tolerance rice genotypes in salt-stress environments. Plants. 2022;11:1150. doi: 10.3390/plants11091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vardhan K.M.V., Rao S.S. Genetic variability for seed yield and its components in linseed (Linum usitatissimum L.) Int. J. Appl. Biol. Pharm. Technol. 2012;3:200–202. [Google Scholar]
- 36.Adjah K.L., Abe A., Adetimirin V.O., Asante M.D. Genetic variability, heritability and correlations for milling and grain appearance qualities in some accessions of rice (Oryza sativa L.) Physiol. Mol. Biol. Plants. 2020;26:1309–1317. doi: 10.1007/s12298-020-00826-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manikya C.M., Reddy T.D. Studies on genetic divergence in medium duration elite rice genotypes (Orzya sativa L.) J. Res. ANGRAU. 2011;39:122. [Google Scholar]
- 38.Rahman M.M., Syed M.A., Akte A., Alam M.M., Ahsan M.M. Genetic variability, correlation and path coefficient analysis of morphological traits in transplanted aman rice (Oryza sativa L.) Am.-Eurasian J. Agric. Environ. Sci. 2014;14:387–391. [Google Scholar]
- 39.Hasan-Ud-Daula M., Sarker U.K. Variability, heritability, character association, and path coefficient analysis in advanced breeding lines of rice (Oryza sativa L.) Genetika. 2020;52:711–726. doi: 10.2298/GENSR2002711H. [DOI] [Google Scholar]
- 40.Mukul M. Elucidation of genotypic variability, character association, and genetic diversity for stem anatomy of twelve Tossa Jute (Corchorus olitorius L.) genotypes. BioMed Res. Int. 2020:9424725. doi: 10.1155/2020/9424725. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Sarker U., Islam M.T., Rabbani M.G., Oba S. Genetic variation and interrelationship among antioxidant, quality and agronomic traits in green amaranth. Turk. J. Agric. For. 2016;40:526–535. doi: 10.3906/tar-1405-83. [DOI] [Google Scholar]
- 42.Kuswantoro H. The role of heritability and genetic variability in estimated selection response of soybean lines on Tidal Swamp land. Pertanika J. Trop. Agric. 2017;40:319–328. [Google Scholar]
- 43.Golam F., Yin Y.H., Masitah A., Afnierna N., Majid N.a., Khalid N., Osman M. Analysis of aroma and yield components of aromatic rice in Malaysian tropical environment. Aust. J. Crop Sci. 2011;5:1318–1325. [Google Scholar]
- 44.Hu L., Luo G., Zhu X., Wang S., Wang L., Cheng X., Chen H. Genetic Diversity and Environmental Influence on Yield and Yield-Related Traits of Adzuki Bean (Vigna angularis L.) Plants. 2022;11:1132. doi: 10.3390/plants11091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rangare N.R., Krupakar A., Ravichandra K., Shukla A.K., Mishra A.K. Estimation of characters association and direct and indirect effects of yield contributing traits on grain yield in exotic and Indian rice (Oryza sativa L.) germplasm. Int. J. Agric. Sci. 2012;2:54–61. [Google Scholar]
- 46.Prasad B., Patwari A.K., Biswas P.S. Genetic variability and selection criteria in fine grain rice (Oryza sativa L.) Pak. J. Bio. Sci. 2001;4:1188–1190. [Google Scholar]
- 47.Morrison D.E. Multivariate Statistical Methods. 2nd ed. McGraw Hill Kogakusta Ltd.; New York, NY, USA: 1978. [Google Scholar]
- 48.Pokhrel A., Dhakal A., Sharma S., Poudel A. Evaluation of physicochemical and cooking characteristics of rice (Oryza sativa L.) landraces of lamjung and Tanahun Districts, Nepal. Int. J. Food Sci. 2020:1589150. doi: 10.1155/2020/1589150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kose A., Oguz O., Ozlem B., Ferda K. Application of multivariate statistical analysis for breeding strategies of spring safflower (Carthamus tinctorius L.) Turkish J. F. Crop. 2018;23:12–19. doi: 10.17557/tjfc.413818. [DOI] [Google Scholar]
- 50.Ahmad F., Hanafi M.M., Hakim M.A., Rafii M.Y., Arolu I.W., Abdullah S.N.A., Barendse W. Genetic divergence and heritability of 42 coloured upland rice genotypes (Oryza sativa L.) as revealed by microsatellites marker and agro-morphological traits. PLoS ONE. 2015;10:e0138246. doi: 10.1371/journal.pone.0138246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saha S.R., Ferdausi A., Hassan L., Haque M.A., Begum S.N., Yasmin F., Akram W. Rice landraces from haor areas of Bangladesh possess greater genetic diversity as revealed by morpho-molecular approaches along with grain quality traits. Cogent Food Agric. 2022;8:2075130. doi: 10.1080/23311932.2022.2075130. [DOI] [Google Scholar]
- 52.Li X., Yan W., Agrama H., Hu B., Jia L., Jia M., Jackson A., Moldenhauer K., McClung A., Wu D. Genotypic and phenotypic characterization on genetic differentiation and diversity in the USDA rice mini-core collection. Genetica. 2010;138:1221–1230. doi: 10.1007/s10709-010-9521-5. [DOI] [PubMed] [Google Scholar]
- 53.Mathure S., Shaikh A., Renuka N., Wakte K., Jawali N., Thengane R., Nadaf A. Characterization of aromatic rice (Oryza sativa L.) germplasm and correlation between their agronomic and quality traits. Euphytica. 2011;179:237–246. doi: 10.1007/s10681-010-0294-9. [DOI] [Google Scholar]
- 54.Nascimento W.F., Silva E.F., Veasey E.A. Agro-morphological characterization of upland rice accessions. Sci. Agric. 2011;68:652–660. doi: 10.1590/S0103-90162011000600008. [DOI] [Google Scholar]
- 55.Ahmed A., Shaon S.G., Islam M.S., Saha P.S., Islam M.M. Genetic divergence analysis in HRDC rice (Oryza sativa L.) hybrids in Bangladesh. Bangladesh J. Plant Breed. Genet. 2014;27:25–32. doi: 10.3329/bjpbg.v27i2.27842. [DOI] [Google Scholar]
- 56.Nisar M., Kumar A., Pandey V.R., Singh P.K., Verma O.P. Studies on genetic divergence analysis in rice (Oryza sativa L.) under sodic soil. Int. J. Curr. Microb. Appl. Sci. 2017;6:3351–3358. doi: 10.20546/ijcmas.2017.612.390. [DOI] [Google Scholar]
- 57.Ashok S., Jyothula D.P.B., Ratnababu D. Genetic divergence studies for yield, yield components and grain quality parameters in rice (Oryza sativa L.) Electron. J. Plant Breed. 2017;8:1240–1246. doi: 10.5958/0975-928X.2017.00178.8. [DOI] [Google Scholar]
- 58.Gabriel A., De Resende J.T., Zeist A.R., Resende L.V., Resende N., Zeist R.A. Phenotypic stability of strawberry cultivars based on physicochemical traits of fruits. Hortic. Bras. 2019;37:75–81. doi: 10.1590/s0102-053620190112. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.