Abstract
Dystrophinopathies are X-linked recessive muscle disorders caused by mutations in the dystrophin (DMD) gene that include deletions, duplications, and point mutations. Correct diagnosis is important for providing adequate patient care and family planning, especially at this time when mutation-specific therapies are available. We report a large single-centre study on the spectrum of DMD gene variants observed in 750 patients analyzed for suspected Duchenne (DMD) or Becker (BMD) muscular dystrophy, over the past 30 years, at the Cardiomyology and Medical Genetics of the University of Campania. We found 534 (71.21%) large deletions, 73 (9.73%) large duplications, and 112 (14.93%) point mutations, of which 44 (5.9%) were small ins/del causing frame-shifts, 57 (7.6%) nonsense mutations, 8 (1.1%) splice site and 3 (0.4%) intronic mutations, and 31 (4.13%) non mutations. Moreover, we report the prevalence of the different types of mutations in patients with DMD and BMD according to their decade of birth, from 1930 to 2020, and correlate the data to the different techniques used over the years. In the most recent decades, we observed an apparent increase in the prevalence of point mutations, probably due to the use of Next-Generation Sequencing (NGS). In conclusion, in southern Italy, deletions are the most frequent variation observed in DMD and BMD patients followed by point mutations and duplications, as elsewhere in the world. NGS was useful to identify point mutations in cases of strong suspicion of DMD/BMD negative on deletions/duplications analyses. In the era of personalized medicine and availability of new causative therapies, a collective effort is necessary to enable DMD and BMD patients to have timely genetic diagnoses and avoid late implementation of standard of care and late initiation of appropriate treatment.
Keywords: DMD gene variants, DMD gene variants in southern Italy, deletions, duplications, point mutations, gene therapy
1. Introduction
Duchenne Muscular Dystrophy (DMD) and Becker Muscular Dystrophy (BMD) are progressive degenerative muscle disorders falling in the group of dystrophinopathies, which also includes X-linked Dilated Cardiomyopathy (XL-DCM) and Carrier Dystrophinopathy (CD) [1,2,3]. Dystrophinopathies are caused by mutations in DMD gene, which is located on the short arm of the X chromosome at Xp21.1 locus and represents one of the largest genes in humans. The gene encodes a protein called dystrophin, which has a molecular weight of 427 kDa and 3685 amino acids [4,5]. These diseases show heterogeneous clinical phenotypes and genetic characteristics [6,7]. Mutations causing disease include deletions, duplications, and point mutations. The phenotype, BMD or DMD, depends on whether these mutations allow the preservation of the DNA reading frame or not. Usually, out-of-frame mutations result in the total absence of the protein and lead to DMD, the most severe form of dystrophinopathy, while in frame mutations that can determine partial, reduced or localized absence of dystrophin lead to more benign forms [8]. Dystrophin acts as a bridge between sarcolemma and actin cytoskeleton. Though representing only 4% of all muscle proteins, it plays a fundamental role in protecting the membrane from the stress induced by muscle contraction, and in regulating the function of dozens of muscle proteins on the sarcolemma [9]. Dystrophinopathies are inherited as X-linked recessive disorders. In about 2/3 of cases, the transmission occurs through mothers who are carriers of the defective gene, while in the remaining 1/3 of cases the disease arises from de novo mutations [10].
1.1. Clinical Pictures of Dystrophinopathies
1.1.1. Rapidly Evolving Dystrophinopathy
Duchenne or rather Conte-Duchenne Muscular Dystrophy, from the name of the Neapolitan clinician Gaetano Conte, who in 1836, 32 years before Duchenne [11] described in two brothers what he called “scrofola muscolorum” (muscle degeneration) [12], is the most frequent and more severe muscular dystrophy in children. Its current incidence is estimated at 1:6000 live birth males. The age of onset is between 3 and 5 years, with delay in the motor skills (difficulty in getting up from the ground with “Gowers maneuver” and climbing stairs, frequent falls, waddling gait) [13]. As the disease progresses, muscles of the upper limbs are also compromised. Since adolescence, the heart is also involved in the dystrophic process with evolution towards severe dilated cardiomyopathy [14,15]. The transition to the wheelchair causes the involvement of the respiratory muscles and the progressive decrease in forced vital capacity (FVC) [16]. In the vast majority of cases, the patient’s mental abilities remain unaffected, though the presence of mental retardation or attentional/autism spectrum disorders has been reported in some studies in up to 30% of patients [17]. Creatine kinase (CK) values can increase up to 100 times the maximum normal value [18].
1.1.2. Benign Evolving Dystrophinopathy
Becker Muscular Dystrophy (BMD), first described by E. Becker in 1955 [19] shows high phenotypic variability with pictures ranging from asymptomatic hyperCKemia, cramps, and myoglobinuria to mild or moderate muscle involvement characterized by progressive symmetrical muscle weakness and atrophy (proximal greater than distal). Calf hypertrophy is often observed though quadriceps femoral weakness may be the only sign [20,21]. The mean age of onset is approximately 12 years (range 1–70 years). Patients with symptoms and/or signs before age five are indistinguishable from those with DMD. About 50% of patients become symptomatic by the age of 10 and most by the age of 20. However, some patients may not experience muscle symptoms until 50–60 years [22,23]. Respiratory involvement is rare and usually seen in the end stages of the disease. Cardiac involvement is common even before the age of 30 and is always present after this age. In a few patients, heart dilation caused by diffuse fibrosis [15,24], may be the first manifestation of heart involvement and underlying myopathy. Cardiomyopathy is characterized by qualitative defects and/or quantitative abnormalities of dystrophin in the myocardium [24]. Its severity increases with age towards stages of dilated cardiomyopathy and congestive heart failure, passing through a stage of transient compensatory myocardial hypertrophy, sometimes associated with arrhythmias [15,24]. The presence of cardiomyopathy, closely related to the type of dystrophin gene mutation [15,25,26,27,28], strongly influences the life expectancy. Increasingly often, these patients benefit from heart transplantation with excellent long-term results [29,30].
1.1.3. X-Linked Dilated Cardiomyopathy
In XL-DCM [31], dystrophin deficiency is limited to the myocardium. The clinical picture in males between the age of 20 and 50 is characterized by early onset congestive heart failure requiring heart transplantation. However, the disease can similarly affect both boys, with rapid and fatal evolution within 2 years of the onset of symptoms, and older patients. Cardiac involvement, in the form of cardiomegaly or ECG abnormalities is sometimes detected in asymptomatic subjects, during a routine medical examination. Patients may present with signs/symptoms of low cardiac output, or report a history of impaired exercise tolerance and exertional dyspnea in the previous months. The absence of muscle weakness is the rule, but can be associated with increased CK values. Mutations responsible for XL-DCM are located in the 5’ end of the gene and in the spectrin domain [32,33]. Mutations in N-terminal actin binding domain (N-ABD) appear to be associated with a more severe phenotype [32] because the lack of M isoform in the heart is not compensated by alternative mechanisms (upregulation of dystrophin B and P isoforms, exon skipping or alternative splicing), which instead are present in the muscle. Cardiomyopathy in female carriers is slowly progressive.
1.1.4. Carrier’s Dystrophinopathy
Females with dystrophinopathy are usually asymptomatic; however, about 8–10% of them may experience muscle symptoms [34,35,36], while a severe myocardial involvement (16–18% in DMD and 7% in DMB) and in particular dilated cardiomyopathy can occur after the age of 40 [37,38,39].
1.1.5. Treatment of Dystrophinopathies
At present, there are no effective treatments for dystrophinopathies. Glucocorticosteroids (GCS) are regarded as the gold standard of care in DMD. Common practice is treatment with GCS between age 3 and 6 years, until the loss of ambulation, with the aim of delaying cardiac and respiratory involvement and preventing the development of scoliosis. Pharmacological treatment for cardiac manifestations includes the standard treatments of dilated cardiomyopathy and arrhythmias such as the use of angiotensin converting enzyme (ACE) inhibitors, β-blockers and diuretics. Patients with BMD and DMD/BMD carriers may benefit from supportive treatment such as pacemaker (PM) or implantable cardioverter defibrillators (ICD) implant. Heart transplantation is also performed, with excellent long-term results [29,30]. Noninvasive ventilatory support associated with cough assisting techniques, has significantly improved the survival in DMD. In the last years, an increasing number of strategies were studied aimed to ameliorate the DMD phenotype by restoring dystrophin production and preserving muscle mass. Therapeutic approaches that aim to restore partially functioning muscle dystrophinin patients with DMD focus on one of these three approaches: 1. Gene delivery using viral vectors [40]; 2. Stop codon read-through [41]; 3. Converting out-of-frame mutations to in-frame mutations (exon skipping, multiple approaches) [42].
The purpose of this large single-centre retrospective study was to report the spectrum of DMD gene variants observed in 750 patients from southern Italy with suspicion of dystrophinopathy, in the last 30 years. As a second step, to report the prevalence of the different types of mutations in patients with DMD and BMD according to their decade of birth, from 1930 to 2020, by correlating the data to the different techniques used over the years.
2. Patients and Methods
2.1. Patients
A systematic full DMD gene mutation analysis was carried out at the Cardiomyology and Medical Genetics of Naples University (now Luigi Vanvitelli Campania University) Hospital in the period 1991–2021 in patients with the suspicion of muscular dystrophy. They prevalently came from southern Italy (Abruzzo, Apulia, Basilicata, Calabria, Campania and Molise) and Sicily. The informed consent for genetic analysis was obtained from patients or parents/guardians if minors, before blood sampling at the time of the first visit, as per established practice in the University Hospital. Only Duchenne and Becker patients still alive at the time in which genetic analysis was available were included in the study.
2.2. Genomic Analysis
Several different techniques were used to perform the genetic analysis, depending on the period in which they were carried out. In the 1990s, the Southern blotting (SB) technique was the gold standard for detecting deletions, replaced by PCR and quantitative PCR [43,44,45] as they became available. Currently, multiplex ligation-dependent probe amplification (MLPA) analysis is the most reliable test to identify deletions or duplications [46,47,48], also due to its cost- and labor-efficiency. Since 2014, Next-Generation Sequencing (NGS) was introduced as the prominent strategy [49,50,51], to provide a reliable genetic diagnosis in patients who were negative to MLPA analysis. Overall, targeted NGS can bolster a more precise understanding of ambiguous mutations [49,50], and identify pathogenic small mutations also in DMD/BMD patients without a large deletion/duplication, especially in non-coding regions [51,52].
3. Results
3.1. Distribution of Dystrophin Gene Pathogenic Variants in the Study Population
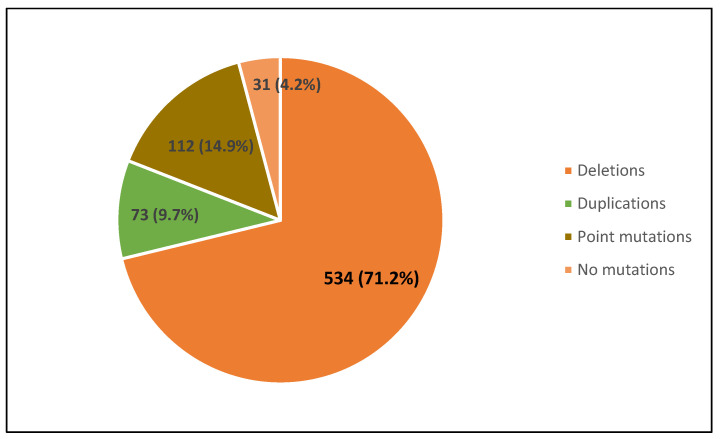
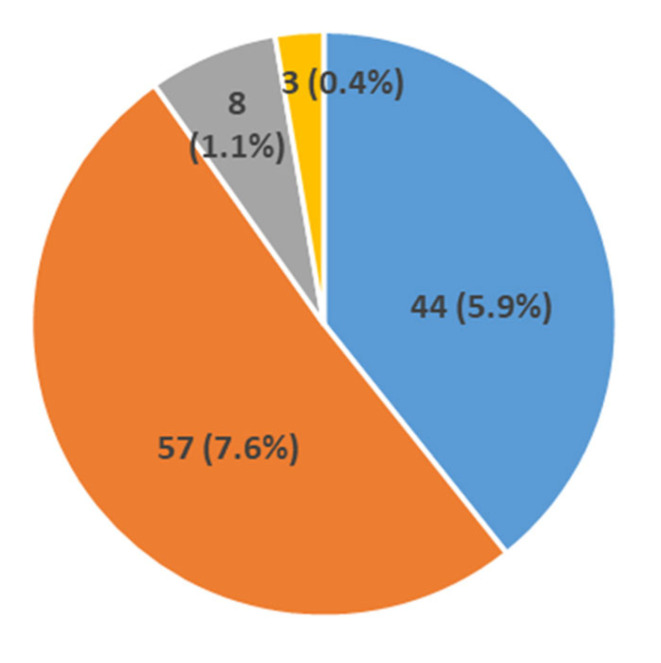
A pathogenic variant in the dystrophin gene was detected in 719 (95.87%) patients, 467 with the Duchenne phenotype and 252 with the Becker phenotype. Taken together, 534 (71.21%) of them had large deletions, 73 (9.73%) large duplications and 112 (14.93%) point mutations. No mutation was found in 31/750 patients (4.13%) (Figure 1).
Figure 1.
Distribution of the pathogenic DMD variants identified in the study population.
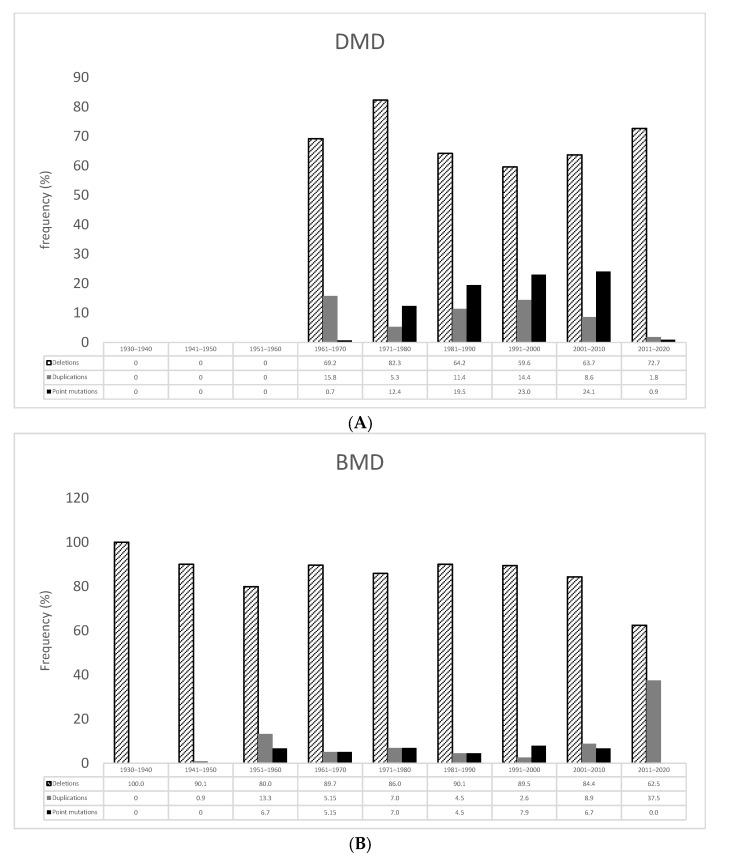
Figure 2 shows the distribution of the type of mutations in Duchenne (Figure 2A) and Becker (Figure 2B) patients according to their decade of birth.
Figure 2.
Frequency of mutations in DMD (A) or BMD (B) analyzed patients according to their decade of birth.
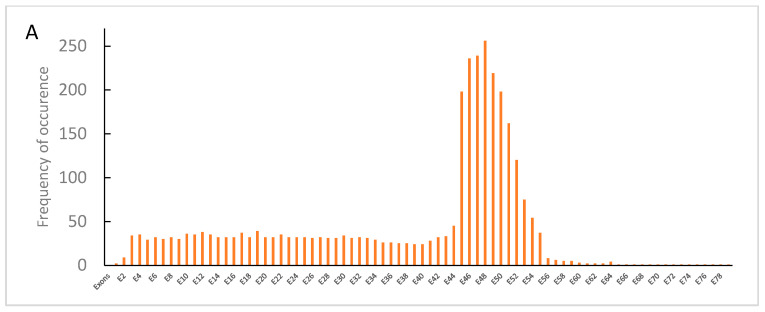
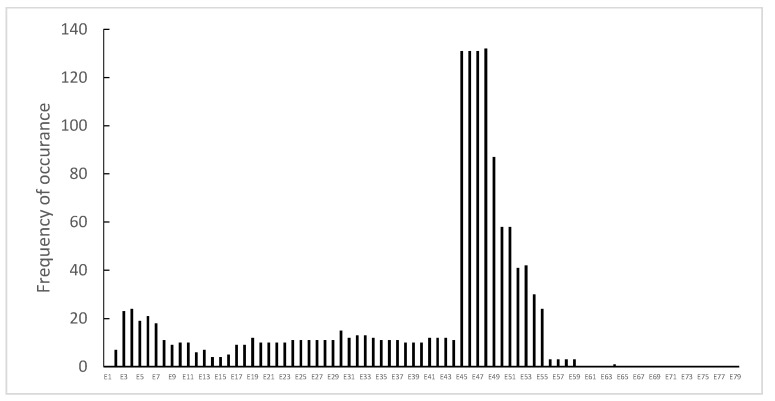
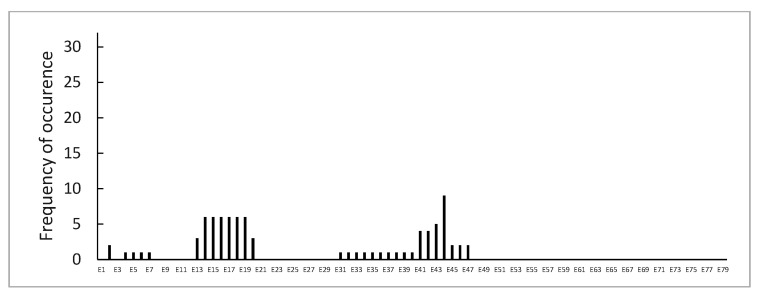
The most frequent hot spot of deletions was between E45–E55 (Figure 3A), while the most frequent hot spots of duplications involved exons E2–E23, and less frequently, exons E41–E60 (Figure 3B).
Figure 3.
Number of times each individual exon of the DMD gene was involved in a deletion (A) or duplications (B) in the 719 patients with DMD or BMD.
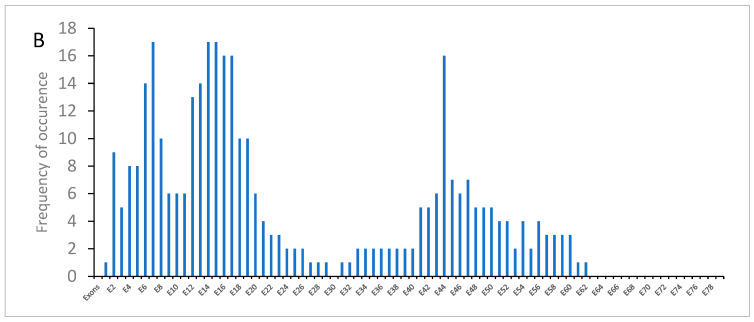
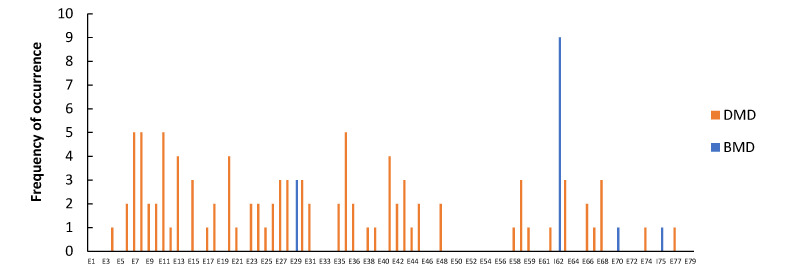
Point mutations were identified in 112 (14.9%) patients, without preferential hotspots (Figure 4). They were distributed as follows: 44 (5.9%) were small ins/del causing a frame-shift, 57 (7.6%) were nonsense mutations, 8 (1.1%) were splice sites, and 3 (0.4%) were intronic mutations (Figure 5).
Figure 4.
Distribution of point mutations in DMD (red) or BMD (blue) analyzed patients.
Figure 5.
Distribution of the DMD gene sub-type point mutations in the analyzed patients with DMD or BMD.
3.2. Dystrophin Gene Variant Analysis in Duchenne Population
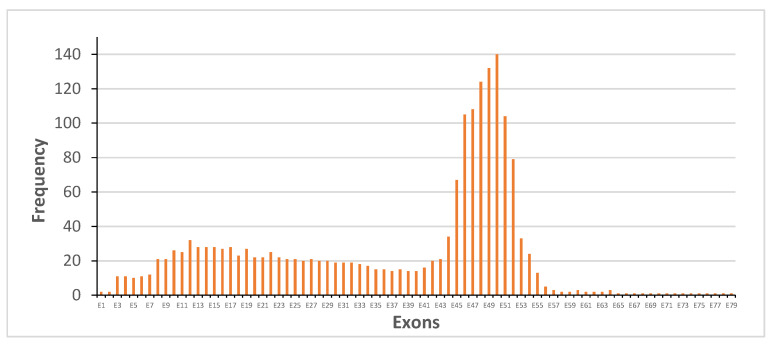
Three-hundred-fourteen DMD patients (68.1%) showed large deletions. The groups of exons most frequently deleted were E48–E50 (6.4%), E45–E50 and E46–E47 (5.1%), followed by E46–E48 (4.1%) and E49–E50 (3.8%). The largest deletions involved the entire dystrophin gene in one patient, 35 (E9–E43; E10–E44), 34 (E10–E43), or 30 (E3–E32) exons, respectively, in another four patients. Deletions of only one exon occurred in 80 patients (25.47%), the most frequent of them involved the exons E51 (63.7%), E44 (17.5%) or E45 (13.75%), followed by exons E52 (10.0%) or E50 (10.9%). Exon deletions prevalently occurred in one hot spot, at the 3′ end (exons E44–E55) of the DMD gene (Figure 6).
Figure 6.
Frequency of single exon deletions in the dystrophin gene, observed in patients with DMD.
Fifty-five DMD patients (11.8%) had large duplications, the largest of them involving 37 exons (E33–E60). Eighteen (32.7%) patients presented a single exon duplication, which involved the exons E2, E12, E44, E50, E51, E52, E54, or E56.
The exons most frequently duplicated were E2 and E44 in 27.8% and 22.2% of cases, respectively. Duplications prevalently occurred at the 5′end in the exons E2–E23 and with a minor frequency at the 3′ end, in exons E44–E60 of the DMD gene (Figure 7).
Figure 7.
Frequency of each single exon duplication in the dystrophin gene observed in patients with DMD.
Ninety-eight DMD patients (20.98%) showed point mutations, which were randomly distributed along the DMD gene, without preferential hotspots.
3.3. Dystrophin Gene Variant Analysis in the Becker Population
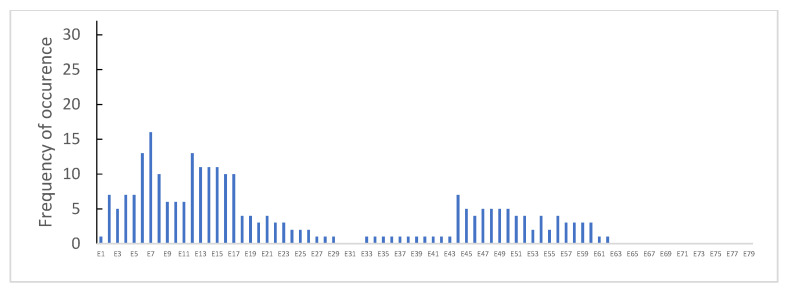
Two-hundred-twenty (87.3%) BMD patients had large deletions. The groups of exons most frequently deleted were E45–E47 (15.9%), E45–E48 (13.2%) or E45–E49 (10.9%). The deletion of these three groups of exons plus the deletion of exon 48 alone (7.3.%) accounts for about half (47.3%) of all deletions in patients with BMD. The largest deletions involved 43 (E17–E59) or 11 exons (E45–E55). Deletions of a single exon were found in 25 patients (11.4%), and beyond exon E48, they involved exons E13 or E30. Deletions occurred in two hot spots, at the 5′ end (E2–E11) and 3′ end (E45–E55) of the DMD gene (Figure 8).
Figure 8.
Frequency of single exon deletions in the dystrophin gene observed in patients with BMD.
Eighteen BMD patients (6.7%) had large duplications, the largest of which involved 15 exons (E31–E44). Four patients (22.2%) had single exon duplications, which involved exons E2 (two patients) or E44 (two patients). The preferential hot spots of duplications were in exons E13–E20 at the 5′end, and in exons E41–E44 at the 3′end of the gene (Figure 9).
Figure 9.
Frequency of single exon duplications in the dystrophin gene observed in BMD patients.
Fourteen patients with BMD (5.2%) had point mutations that were localized in exons E29 (3), E70 (1) or in introns I62 (9) or I75 (1). The pathogenic variant located in intron 62 affected nine members of the same family, in three generations.
4. Discussion
Dystrophinopathies are X-linked progressive muscle disorders caused by mutations in the DMD gene. The mutations causing the diseases include deletions, duplications, and point mutations.
Having a correct diagnosis of DMD/BMD is important for providing proper care to patients and for family planning, but also for assessing whether patients are eligible for mutation-specific treatments [53].
In this retrospective study, we report the genetic profile of 719 DMD/BMD patients from southern Italy (Abruzzo, Apulia, Basilicata, Campania, Calabria, Molise) and Sicily, detected in the last 30 years in the same laboratory with several techniques.
In our laboratory, the genetic testing for mutations in the dystrophin gene began in the second half of 1991, using the SB analysis and the multiplexes PCR of Beggs [43] and Chamberlain [44], which covered at that time about 98% of all deletions. In the following years, the DMD genetic approach in our laboratory evolved according to the progress of techniques, passing from SB and PCR to the MLPA technique [46,47,48] and the more recent Next-Generation Sequencing (NGS) technology [49,50,51,52], obtaining an increase in the detection rate results [54,55,56,57,58,59,60]. This was also possible because the Naples Human Mutation gene Biobank (NHMB)—currently a member of the EuroBioBank (EBB) [61] and the Telethon Network of Genetic Biobanks (TNGB) [62]—were established in our service in 1991, allowing the storage of patients’ blood samples and the re-analysis with NGS of patients’ DNA samples that were negative using previous techniques.
Despite these improvements, a diagnostic delay of up to 4–5 years between the clinical presentation of symptoms and molecular analysis, is still reported in literature. In Italy, the mean age at first medical contact, which raises the suspicion of a DMD, is 31 months, while the mean age at molecular diagnosis is 41 months, 10 months later. In southern Italy, the time for delayed diagnosis is 9 months [63].
We observed over time, in our population, a decrease in the number of patients affected by DMD/BMD, with a parallel reduction in the percentage of point mutations, especially in the last decade. This trend may be the result of genetic counseling and prenatal diagnosis offered to all women at risk of carrying a DMD gene defect, or the fact that patients without DMD gene deletions/duplications arrive later in tertiary centers and labs, and that patients with BMD are usually diagnosed after age 12, regardless of the type of the molecular variation. However, while genetic counseling and prenatal diagnosis of affected males will certainly contribute to a steady reduction in the number of familial cases in the coming years, no strategy will be able to prevent the occurrence of new cases due to “de novo” mutations, which represent about 1/3 of the total.
Deletions represent the highest percentage of mutations in our patients, followed by point mutations, mainly nonsense variants, in patients with DMD and by duplications in patients with BMD. However, when comparing our percentages with those previously reported by Neri et al. in the Italian population [64], we found that point mutations are overestimated in that study (24.6% versus 14.9% in this study). This difference may lie in several factors, such as the origin of the patients (from all over Italy rather than limited to southern Italy), the number of patients analyzed, or the period of analysis. Since deletions (and duplications) are variants more easily identifiable even with less sophisticated techniques, we believe that the sample reported in Neri’s article was particularly enriched by the enrolment (or referral) of patients for whom no deletions/duplications were detected in the peripheral laboratories. An overestimation of point mutations has also been reported by Flanigan et al. [65], who similarly explain the finding of such a large number of point mutations in their study.
Our data are in agreement with previous studies reporting the frequency of mutations in the DMD gene, according to geographical areas in the world (Table 1, and references herein). We considered seven macro-areas—Western and East Europe, Asia, North and South Africa, North and Central-South America—that include 40 Countries.
Table 1.
Distribution of dystrophin gene pathogenic variants in the different countries of the world, based on their geographical area. The countries were subdivided into seven macro-areas: Western Europe, East Europe, Asia, North Africa, South Africa, North America, and Central-South America.
| Countries | Suspected Dystrophinopathies | Large Deletions | Large Duplications | Point Mutations/Small Rearrangments | Small Ins/Del | Nonsense | Frame-Shift | Splice Site | Missense | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n. | n. | % | n. | % | n. | % | n. | % | n. | % | n. | % | n. | % | n. | % | ||
| EUROPE | ||||||||||||||||||
| Denmark | 182 | 87 | 47.8 | 14 | 7.7 | 4 | 2.2 | 3 | 1.6 | 1 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [48] |
| France | 2411 | 1404 | 58.2 | 362 | 25.6 | 465 | 19.2 | n.r. | n.r. | 187 | 7.6 | 148 | 6.1 | 125 | 8.8 | 7 | 0.4 | [66] |
| France | 2898 | 1901 | 65.5 | 323 | 11.1 | 633 | 21.8 | 201 | 6.9 | 254 | 8.7 | n.r. | n.r. | 158 | 5.4 | 20 | 0.7 | [67] |
| Spain | 284 | 131 | 46.1 | 56 | 19.7 | 97 | 34.1 | 34 | 12.0 | 49 | 17.2 | n.r. | n.r. | 10 | 3.5 | 2 | 0.7 | [68] |
| Spain | 53 | 34 | 64.2 | 4 | 7.5 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [69] |
| Netherlands | 462 | 212 | 45.8 | 42 | 9.0 | n.r. | n.r. | n.r. | n.r. | 49 | 10.6 | n.r. | n.r. | 18 | 3.8 | n.r. | n.r. | [70] |
| Italy | 1902 | 1242 | 65.2 | 190 | 9.9 | 469 | 24.6 | n.r. | n.r. | 200 | 10.5 | 139 | 11.1 | 67 | 5.4 | 38 | 1.9 | [64] |
| South Italy and Sicily | 750 | 534 | 71.2 | 73 | 9.7 | 112 | 14.9 | 44 * | 5.9 | 57 | 7.6 | 44 * | 5.9 | 8 | 1.1 | 0 | 0 | This study |
| EAST EUROPE | ||||||||||||||||||
| Bosnia, Bulgaria, Cyprus, Croatia, Hungary, Lithuania, Poland, Romania, Russia, Serbia, and Ukraine | 260 | 52 | 20 | 17 | 6.5 | n.r. | n.r. | n.r. | n.r. | 49 | 18.8 | 29 | 11.1 | 16 | 9 | 4 | 1.5 | [71] |
| Poland | 180 | 110 | 61.1 | 21 | 4.7 | 2 | 1.1 | n.r. | n.r. | 1 | 0.5 | 1 | 0.5 | n.r. | n.r. | n.r. | n.r. | [72] |
| Turkey | 260 | 128 | 49.2 | 25 | 9.6 | 60 | 23 | n.r. | n.r. | 32 | 12.3 | 17 | 6.5 | 1 | 0.3 | 4 | 1.5 | [73] |
| Czech Republic and Slovakia | 126 | 77 | 61.1 | 12 | 9.5 | 18 | 14.3 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [74] |
| ASIA | ||||||||||||||||||
| Saudi Arabia | 45 | 21 | 46.7 | 8 | 17.8 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [75] |
| Saudi Arabia | 15 | 6 | 40 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [76] |
| Kuwait | 68 | 45 | 66.1 | 3 | 4.4 | 6 | 8.8 | n.r. | n.r. | 4 | 5.8 | n.r. | n.r. | n.r. | n.r. | 2 | 2.9 | [77] |
| China | 1051 | 740 | 70.4 | 87 | 8.2 | 201 | 19.1 | 58 | 5.5 | 102 | 9.7 | n.r. | n.r. | 30 | 2.8 | 15 | 1.4 | [78] |
| China | 67 | 23 | 41.8 | 5 | 7.5 | 23 | 34.3 | 6 | 8.9 | 13 | 19.4 | n.r. | n.r. | 4 | 5.9 | n.r. | n.r. | [79] |
| China | 613 | 360 | 60.2 | 59 | 9.6 | 143 | 23.3 | 52 | 8.4 | 70 | 11.4 | n.r. | n.r. | 21 | 3.4 | n.r. | n.r. | [80] |
| China | 662 | n.r | n.r. | n.r | n.r. | 115 | 18.5 | n.r. | n.r. | 60 | 9 | 20 | 3 | 28 | 4.2 | 6 | 0.9 | [81] |
| China | 1400 | 752 | 53.7 | 92 | 6.5 | 197 | 14 | 45 | 3.2 | 124 | 8.8 | n.r. | n.r. | 22 | 1.5 | 6 | 0.4 | [82] |
| China | 401 | 238 | 59.3 | n.r. | n.r | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [83] |
| East China | 229 | 153 | 66.8 | 22 | 9.6 | 51 | 22.2 | 23 | 10 | 26 | 11.3 | 18 | 7.8 | n.r. | n.r. | 1 | 0.4 | [84] |
| China | 100 | 69 | 69 | 9 | 9 | 22 | 22 | 6 | 6 | 13 | 13 | n.r. | 2 | 2 | 1 | 1 | [85] | |
| Korea | 218 | 147 | 67.4 | 31 | 14.2 | 40 | 18.3 | n.r. | n.r. | 18 | 8.2 | 9 | 4.1 | 11 | 5 | 1 | 0.4 | [86] |
| Korea | 507 | 319 | 65.3 | 65 | 13.3 | 104 | 21.3 | n.r. | n.r. | 60 | 12.3 | 15 | 3.1 | 22 | 4.5 | 6 | 1.2 | [87] |
| Japan | 442 | 270 | 61 | 38 | 9 | n.r. | n.r. | 34 | 7.6 | 69 | 15.6 | n.r. | n.r. | 25 | 5.4 | n.r. | n.r. | [88] |
| Japan | 1497 | 901 | 61 | 188 | 13 | 371 | 24.7 | 116 | 7.7 | 189 | 12.4 | n.r. | n.r. | 61 | 4 | 8 | 0.53 | [89] |
| Iran | 314 | 251 | 79.9 | 17 | 5.4 | 43 | 16.6 | n.r. | n.r. | 24 | 7.6 | 15 | 4.7 | 3 | 0.9 | 1 | 0.3 | [90] |
| Singapore | 145 | 95 | 65.5 | 14 | 9.6 | 31 | 21.3 | n.r. | n.r. | 18 | 12.4 | 8 | 5.5 | 4 | 2.7 | 1 | 0.6 | [91] |
| Malaysia | 35 | 27 | 77.1 | 2 | 5.7 | n.r. | n.r. | 2 | 5.7 | 3 | 8.5 | n.r. | n.r. | n.r. | n.r. | 2 | 5.7 | [92] |
| India | 88 | 65 | 73.8 | n.r | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [93] |
| India | 1660 | 1090 | 65.6 | 98 | 5.9 | 61 | 3.6 | 25 | 1.5 | 34 | 2 | n.r. | n.r. | 5 | 0.3 | 4 | 0.2 | [94] |
| India | 606 | 492 | 81.2 | 33 | 5.4 | 70 | 11.6 | n.r. | n.r. | 40 | 6.6 | 17 | 2.8 | 12 | 1.98 | 1 | 0.16 | [95] |
| India | 83 | 60 | 72.2 | 5 | 6.5 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [96] |
| India | 317 | 285 | 89.9 | 26 | 8.2 | n.r. | n.r. | 317 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [97] |
| India | 961 | 642 | 66.8 | 55 | 5.7 | 101 | 10.5 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [98] |
| Southern India | 510 | 342 | 67.1 | 30 | 5.9 | 10 | 1.9 | 5 | 0.9 | 4 | 0.7 | n.r. | n.r. | n.r. | n.r. | 1 | 0.19 | [99] |
| Sri Lanka | 50 | 40 | 80 | 4 | 8 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [100] |
| North Africa | ||||||||||||||||||
| Algeria | 68 | 36 | 52.9 | 2 | 2.9 | n.r. | n.r. | n.r. | n.r. | 4 | 5.8 | 2 | 2.9 | 2 | 2.9 | 3 | 4.4 | [71] |
| Egypt | 152 | 78 | 51.3 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [101] |
| Egypt | 41 | 22 | 53.6 | 2 | 4.8 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [102] |
| Egypt | 100 | 55 | 55 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [103] |
| Morocco | 72 | 37 | 51.3 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [104] |
| South Africa | ||||||||||||||||||
| South Africa | 128 | 54 | 42.1 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [105] |
| South Africa | 261 | 90 | 34.4 | 34 | 13 | 3 | 1.1 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [106] |
| North America | ||||||||||||||||||
| Canada | 573 | 366 | 63.8 | 64 | 11 | 143 | 25 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [107] |
| USA and Canada | 436 | 256 | 79 | 23 | 7.1 | 45 | 13.9 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [108] |
| USA | 1014 | 460 | 45.3 | 112 | 11 | 442 | 43.5 | 113 | 10.1 | 256 | 25.2 | n.r. | 50 | 4.9 | 15 | 1.4 | [65] | |
| USA | 68 | 45 | 66.1 | 4 | 5.8 | 12 | 17.6 | n.r. | n.r. | 9 | 13.2 | 2 | 2.9 | n.r. | n.r. | 3 | 2.9 | [109] |
| Central-South America | ||||||||||||||||||
| Argentina | 81 * | 40 | 49.3 | 8 | 9.8 | 2 | 2.4 | n.r. | n.r. | 2 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [110] | |
| Colombia | 69 | 40 | 58.8 | 10 | 4.5 | 11 | 15.9 | n.r. | n.r. | 8 | 11.6 | n.r. | n.r. | 3 | 4.3 | n.r. | n.r. | [111] |
| Puerto Rico | 141 | 56 | 66.7 | 2 | 2.4 | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | n.r. | [112] |
* Females at risk were not included.
The differences in the frequency of the different type of mutations observed between our cohort of patients and other parts of the world, but also among the seven considered macro-areas may depend on several factors, such as:
-
-
selection bias, as some studies analyzed subjects from different institutions [79];
-
-
inclusion in the study of a high number of patients still undiagnosed and/or negative when using standard techniques [52,68,71,79];
-
-
failure to sequence all patients negative for deletions or duplications, due to the cost problem [93];
- -
- -
- -
-
-
higher rate of inbreeding in some countries, such as the Middle East and Turkey, leading to a higher incidence of autosomal recessive diseases;
-
-
limitations of the techniques used, which could explain the low frequency of point mutations in some studies [48,75].
The average results from the seven macro-areas are summarized in Table 2. Overall, a total of 25,705 individuals with the suspicion of dystrophinopathies were analyzed.
Table 2.
Summary of average frequencies of DMD gene variants identified in the seven considered macro-areas in the world.
| World Macro-Areas | Suspected Dystrophinopathies | DMD/BMD Patients | Detection Rate | Large Deletions | Large Duplications | Point Mutations/Small Rearrangements | |||
|---|---|---|---|---|---|---|---|---|---|
| N | N | N | % | N | % | N | % | ||
| Western Europe | 8922 | 7670 | 86.0 | 5011 | 65.3 | 991 | 12.9 | 1668 | 21.7 |
| East Europe | 826 | 620 | 75.1 | 367 | 59.2 | 75 | 12.1 | 178 | 28.7 |
| Asia | 12084 | 9913 | 82.0 | 7433 | 74.9 | 891 | 9.0 | 1589 | 16.0 |
| North Africa | 433 | 236 | 54.5 | 228 | 96.6 | 4 | 0.02 | 4 | 0.02 |
| South Africa | 389 | 178 | 45.8 | 144 | 80.9 | 34 | 19.1 | n.a. | n.a. |
| North America | 2091 | 1972 | 94.3 | 1127 | 57.2 | 203 | 10.3 | 642 | 32.5 |
| Central-South America | 210 | 169 | 80.4 | 136 | 80.5 | 20 | 11.8 | 13 | 7.7 |
| Total or percentages | 25.7 | 20.7 | 80.8 | 14.4 | 69.6 | 2218 | 10.7 | 4094 | 19.7 |
| This report | 750 | 719 | 95.8 | 534 | 71.2 | 73 | 10.1 | 112 | 15.6 |
The highest number of DMD/BMD patients were investigated in Asia and Europe, while only 1/4 of the patients analyzed in Europe were analyzed in North America, despite a population 25% higher than Europe. This finding is most probably due to a complex mix of different factors, such as:
-
-
the geographic size of North America, which limits the access to diagnostic procedures;
-
-
the large difference in urban and rural medical care;
-
-
the lack of knowledge in primary care providers about DMD, resulting in less CK and transaminase testing;
-
-
insurance coverage issues, especially for genetic testing procedures.
In this study—the largest, by number of patients, performed to our knowledge in a single-center and over such a long period—the detection rate of DMD gene variations is 95.8%, the highest among the seven macro-areas. This result was possible because, in our lab, we usually re-analyze all patients with suspected dystrophinopathy, who were negative to previous techniques, using new generation techniques.
It is intriguing to note that dystrophinopathies, with their higher frequency of deletions, differ from other genetic diseases, in which point mutations are the most frequent pathogenic variants (http://www.hgmd.org (accessed on 6 November 2022)), and among them missense variants usually prevail [113]. A possible explanation for the high percentage of deletions could rely on the giant size of the DMD gene and/or on the presence of numerous repeat sequences, that can favor deleterious crossover events during meiosis. The low percentage of point mutations, on the other hand, may be related to the difficulty of their detection using standard techniques, or to the fact that they may cause non-severe (and therefore undiagnosed) phenotypes, leading, consequently, to their underestimation.
5. Conclusions
The life expectancy of patients with DMD has doubled, and more and more patients reach and exceed the fourth decade of life [114] thanks to a multifaceted approach that makes use of pharmacological treatment, cardiac and respiratory support, and rehabilitation.
Therapeutic approaches that aim to restore partially functional muscle dystrophin in patients with DMD are now available for patients with deletions of one or more exons [41], or with stop-codon (nonsense) point mutations [42]. However, the possibility to have access to genetic testing for DMD is not homogeneous in the different countries of the world, causing not only a delay in diagnosis and timely treatment, but also a delay in genetic counseling, implementation of standard care, and participation in clinical trials.
In this study, we found that the major hot spot for deletions is in exons E45–E55, which are currently the most suitable for gene therapy via exon skipping [40,41].
The data reported here also show that, in industrialized countries, the evolution of molecular strategies was able to improve the detection rate of DMD gene variants and the achievement of an early and precise genetic diagnosis in patients with BMD/DMD.
In the era of personalized medicine and the recent availability of causative therapies, a collective effort is necessary to enable patients with DMD or BMD to have timely genetic diagnoses, even in economically disadvantaged countries, to avoid delayed genetic counseling and problems in family planning, late implementation of the standard of care, and late initiation of treatment.
Acknowledgments
We thanks Stefania Aurino, Francesca D’Amico, Maria Grazia Esposito, Arcomaria Garofalo, Teresa Giugliano, Amelia Trimarco, Vega Maria Ventriglia, Roberta Zeuli and the technical staff of the Medical Genetics Laboratory who, over the years, have alternated in the molecular analysis of the DMD gene. We also thank Rosaria Di Martino of the Luigi Vanvitelli University Library who supported us in providing most of the papers we consulted.
Author Contributions
Conceptualization, L.P. (Luisa Politano); Data curation, G.P., A.T. and V.N.; Formal analysis, E.P., M.E.O. and A.T.; Investigation, L.P. (Luigia Passamano) and M.S.; Methodology, E.V. and L.P. (Luisa Politano); Supervision, L.P. (Luisa Politano); Validation, G.P. and V.N.; Writing—original draft, E.V. and L.P. (Luisa Politano); Writing—review and editing, E.V. and L.P. (Luisa Politano). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. As a current practice in our hospital, EC approval is not required when data are reported in an aggregate manner.
Informed Consent Statement
Patients and guardians in the case of minors give their informed consent for DNA analysis and study research at the time of the blood sampling.
Data Availability Statement
Data are available on request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Emery A.E.H. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 2.Mercuri E., Bönnemann C.G., Muntoni F. Muscular dystrophies. Lancet. 2019;394:2025–2038. doi: 10.1016/S0140-6736(19)32910-1. [DOI] [PubMed] [Google Scholar]
- 3.Brandsema J.F., Darras B.T. Dystrophinopathies. Semin. Neurol. 2015;35:369–384. doi: 10.1055/s-0035-1558982. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman E.P., Kunkel L.M. Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron. 1989;2:1019–1029. doi: 10.1016/0896-6273(89)90226-2. [DOI] [PubMed] [Google Scholar]
- 6.Koeks Z., Bladen C.L., Salgado D., van Zwet E., Pogoryelova O., McMacken G., Monges S., Foncuberta M.E., Kekou K., Kosma K., et al. Clinical Outcomes in Duchenne Muscular Dystrophy: A Study of 5345 Patients from the TREAT-NMD DMD Global Database. J. Neuromuscul. Dis. 2017;4:293–306. doi: 10.3233/JND-170280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman E.P. Causes of clinical variability in Duchenne and Becker muscular dystrophies and implications for exon skipping therapies. Acta Myol. 2020;39:179–186. doi: 10.36185/2532-1900-020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leturcq F., Kaplan J.-C. Bases moléculaires des dystrophinopathies [Molecular bases of dystrophinopathies] J. Soc. Biol. 2005;199:5–11. doi: 10.1051/jbio:2005001. (In French) [DOI] [PubMed] [Google Scholar]
- 9.Blake D.J., Weir A., Newey S.E., Davies K.E., Spaulding H.R., Quindry T., Hammer K., Quindry J.C., Selsby J.T., Amirouche A., et al. Function and Genetics of Dystrophin and Dystrophin-Related Proteins in Muscle. Physiol. Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 10.Haldane J.B. The rate of spontaneous mutation of a human gene. J. Genet. 2004;83:235–244. doi: 10.1007/BF02717892. [DOI] [PubMed] [Google Scholar]
- 11.Duchenne G.B.A. Recherches sur la paralysie musculaire pseudohypertrophique ou paralysie myo-sclerosique. Arch. Gen. Med. 1868;11:5–25. [Google Scholar]
- 12.Conte G., Gioia L. Scrofola del sistema muscolare. Ann. Clin. Osp. Incurabili Napoli. 1836;2:66–79. [Google Scholar]
- 13.Thangarajh M. The Dystrophinopathies. Continuum. 2019;25:1619–1639. doi: 10.1212/CON.0000000000000791. [DOI] [PubMed] [Google Scholar]
- 14.Nigro G., Comi L.I., Politano L., Bain R.J. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int. J. Cardiol. 1990;26:271–277. doi: 10.1016/0167-5273(90)90082-G. [DOI] [PubMed] [Google Scholar]
- 15.Nigro G., Comi L.I., Politano L., Nigro G. Engel & Franzini-Armstrong: Myology. McGraw-Hill; New York, NY, USA: 2004. Cardiomyopathies associated with Muscular Dystrophies; pp. 1239–1256. [Google Scholar]
- 16.LoMauro A., Romei M., Gandossini S., Pascuzzo R., Vantini S., D’Angelo M.G., Aliverti A. Evolution of respiratory function in Duchenne muscular dystrophy from childhood to adulthood. Eur. Respir. J. 2018;51:1701418. doi: 10.1183/13993003.01418-2017. [DOI] [PubMed] [Google Scholar]
- 17.Banihani R., Smile S., Yoon G., Dupuis A., Mosleh M., Snider A., McAdam L. Cognitive and Neurobehavioral Profile in Boys With Duchenne Muscular Dystrophy. J. Child Neurol. 2015;30:1472–1482. doi: 10.1177/0883073815570154. [DOI] [PubMed] [Google Scholar]
- 18.Kiessling W.R., Beckmann R. Serum levels of myoglobin and creatine kinase in duchenne muscular dystrophy. Klin. Wochenschr. 1981;59:347–348. doi: 10.1007/BF01525003. [DOI] [PubMed] [Google Scholar]
- 19.Becker P.E., Kiener F. A new X-chromosomal muscular dystrophy. Arch. Psychiatr. Nervenkr Z Gesamte Neurol. Psychiatry. 1955;193:427–448. doi: 10.1007/BF00343141. [DOI] [PubMed] [Google Scholar]
- 20.Bushby K.M., Gardner-Medwin D. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. I. Natural history. J. Neurol. 1993;240:98–104. doi: 10.1007/BF00858725. [DOI] [PubMed] [Google Scholar]
- 21.Bushby K.M., Gardner-Medwin D., Nicholson L.V., Johnson M.A., Haggerty I.D., Cleghorn N.J., Harris J.B., Bhattacharyal S.S. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. II. Correlation of phenotype with genetic and protein abnormalities. J. Neurol. 1993;240:105–112. doi: 10.1007/BF00858726. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki D., Yoshida K., Fukushima K., Nakamura A., Suzuki K., Sato T., Takeda S., Ikeda S.-I. Characterization of deletion breakpoints in patients with dystrophinopathy carrying a deletion of exons 45–55 of the Duchenne muscular dystrophy (DMD) gene. J. Hum. Genet. 2009;54:127–130. doi: 10.1038/jhg.2008.8. [DOI] [PubMed] [Google Scholar]
- 23.Taglia A., Petillo R., D’Ambrosio P., Picillo E., Torella A., Orsini C., Ergoli M., Scutifero M., Passamano L., Palladino A., et al. Clinical features of patients with dystrophinopathy sharing the 45–55 exon deletion of DMD gene. Acta Myol. 2015;34:9–13. [PMC free article] [PubMed] [Google Scholar]
- 24.Nigro G., Comi L.I., Politano L., Limongelli F.M., Nigro V., De Rimini M.L., Giugliano M.A., Petretta V.R., Passamano L., Restucci B., et al. Evaluation of the cardiomyopathy in becker muscular dystrophy. Muscle Nerve. 1995;18:283–291. doi: 10.1002/mus.880180304. [DOI] [PubMed] [Google Scholar]
- 25.Arbustini E., Diegoli M., Morbini P., Bello B.D., Banchieri N., Pilotto A., Magani F., Grasso M., Narula J., Gavazzi A., et al. Prevalence and characteristics of the dystrophin defects in adult male patients with dilated cardiomyopathy. J. Am. Coll. Cardiol. 2000;35:1760–1768. doi: 10.1016/S0735-1097(00)00650-1. [DOI] [PubMed] [Google Scholar]
- 26.Nicolas A., Raguénès-Nicol C., Ben Yaou R., Ameziane-Le Hir S., Chéron A., Vié V., Claustres M., Leturcq F., Delalande O., Hubert J.F., et al. French network of clinical reference centres for neuromuscular diseases (CORNEMUS). Becker muscular dystrophy severity is linked to the structure of dystrophin. Hum. Mol. Genet. 2015;24:1267–1279. doi: 10.1093/hmg/ddu537. [DOI] [PubMed] [Google Scholar]
- 27.Nigro G., Politano L., Nigro V., Petretta V.R., Comi L.I. Mutation of dystrophin gene and cardiomyopathy. Neuromuscul. Disord. 1994;4:371–379. doi: 10.1016/0960-8966(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 28.Kaspar R.W., Allen H.D., Ray W.C., Alvarez C.E., Kissel J.T., Pestronk A., Weiss R.B., Flanigan K.M., Mendell J.R., Montanaro F. Analysis of Dystrophin Deletion Mutations Predicts Age of Cardiomyopathy Onset in Becker Muscular Dystrophy. Circ. Cardiovasc. Genet. 2009;2:544–551. doi: 10.1161/CIRCGENETICS.109.867242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papa A.A., D’Ambrosio P., Petillo R., Palladino A., Politano L. Heart transplantation in patients with dystrophinopathic cardiomyopathy: Review of the literature and personal series. Intractable Rare Dis. Res. 2017;6:95–101. doi: 10.5582/irdr.2017.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascencio-Lemus M.G., Barge-Caballero E., Paniagua-Martín M.J., Barge-Caballero G., Couto-Mallón D., Crespo-Leiro M.G. Is Becker Dystrophinopathy a Contraindication to Heart Transplant? Experience in a Single Institution. Rev. Esp. Cardiol. 2019;72:584–585. doi: 10.1016/j.recesp.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 31.Towbin J.A., Hejtmancik J.F., Brink P., Gelb B., Zhu X.M., Chamberlain J.S., McCabe E.R., Swift M. X-linked dilated cardiomyopathy. Molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87:1854–1865. doi: 10.1161/01.CIR.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 32.Muntoni F., Cau M., Ganau A., Congiu R., Arvedi G., Mateddu A., Marrosu M.G., Cianchetti C., Realdi G., Cao A., et al. Brief report: Deletion of the Dystrophin Muscle-Promoter Region Associated with X-Linked Dilated Cardiomyopathy. N. Engl. J. Med. 1993;329:921–925. doi: 10.1056/NEJM199309233291304. [DOI] [PubMed] [Google Scholar]
- 33.Ferlini A., Sewry C., Melis M.A., Mateddu A., Muntoni F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul. Disord. 1999;9:339–346. doi: 10.1016/S0960-8966(99)00015-2. [DOI] [PubMed] [Google Scholar]
- 34.Zhong J., Xie Y., Bhandari V., Chen G., Dang Y., Liao H., Zhang J., Lan D. Clinical and genetic characteristics of female dystrophinopathy carriers. Mol. Med. Rep. 2019;19:3035–3044. doi: 10.3892/mmr.2019.9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norman A., Harper P. A survey of manifesting carriers of Duchenne and Becker muscular dystrophy in Wales. Clin. Genet. 1989;36:31–37. doi: 10.1111/j.1399-0004.1989.tb03363.x. [DOI] [PubMed] [Google Scholar]
- 36.Seemann N., Selby K., McAdam L., Biggar D., Kolski H., Goobie S., Yoon G., Campbell C., Canadian Pediatric Neuro-muscular Group Symptomatic dystrophinopathies in female children. Neuromuscul. Disord. 2011;21:172–177. doi: 10.1016/j.nmd.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Politano L., Nigro V., Nigro G., Petretta V.R., Passamano L., Papparella S., Di Somma S., I Comi L. Development of cardiomyopathy in female carriers of Duchenne and Becker muscular dystrophies. JAMA. 1996;275:1335–1338. doi: 10.1001/jama.1996.03530410049032. [DOI] [PubMed] [Google Scholar]
- 38.Hoogerwaard E.M., Bakker E., Ippel P.F., Oosterwijk J.C., Majoor-Krakauer D.F., Leschot N.J., Van Essen A.J., Brunner H.G., van der Wouw P.A., Wilde A.A., et al. Signs and symptoms of Duchenne muscular dystrophy and Becker muscular dystrophy among carriers in the Netherlands: A cohort study. Lancet. 1999;353:2116–2119. doi: 10.1016/S0140-6736(98)10028-4. [DOI] [PubMed] [Google Scholar]
- 39.Grain L., Cortina-Borja M., Forfar C., Hilton-Jones D., Hopkin J., Burch M. Cardiac abnormalities and skeletal muscle weakness in carriers of Duchenne and Becker muscular dystrophies and controls. Neuromuscul. Disord. 2001;11:186–191. doi: 10.1016/S0960-8966(00)00185-1. [DOI] [PubMed] [Google Scholar]
- 40.Elangkovan N., Dickson G. Gene Therapy for Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021;8((Suppl. S2)):S303–S316. doi: 10.3233/JND-210678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda S., Clemens P.R., Hoffman E.P. Exon-Skipping in Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021;8((Suppl. S2)):S343–S358. doi: 10.3233/JND-210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landfeldt E., Sejersen T., Tulinius M. A mini-review and implementation model for using ataluren to treat nonsense muta-tion Duchenne muscular dystrophy. Acta Paediatr. 2019;108:224–230. doi: 10.1111/apa.14568. [DOI] [PubMed] [Google Scholar]
- 43.Beggs A.H., Koenig M., Boyce F.M., Kunkel L.M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum. Genet. 1990;86:45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- 44.Chamberlain J.S., Farwell N.J., Chamberlain J.R., Cox G.A., Caskey C.T. PCR analysis of dystrophin gene mutation and expression. J. Cell. Biochem. 1991;46:255–259. doi: 10.1002/jcb.240460309. [DOI] [PubMed] [Google Scholar]
- 45.Dunnen J.T.D., Beggs A.H. Multiplex PCR for Identifying DMD Gene Deletions. Curr. Protoc. Hum. Genet. 2006;49:9.3.1–9.3.22. doi: 10.1002/0471142905.hg0903s49. [DOI] [PubMed] [Google Scholar]
- 46.Lalic T., Vossen R.H., Coffa J., Schouten J.P., Guc-Scekic M., Radivojevic D., Djurisic M., Breuning M.H., White S.J., den Dunnen J.T. Deletion and duplication screening in the DMD gene using MLPA. Eur. J. Hum. Genet. 2005;13:1231–1234. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 47.Okizuka Y., Takeshima Y., Awano H., Zhang Z., Yagi M., Matsuo M. Small Mutations Detected by Multiplex Ligation–Dependent Probe Amplification of the Dystrophin Gene. Genet. Test. Mol. Biomark. 2009;13:427–431. doi: 10.1089/gtmb.2009.0002. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz M., Dunø M. Improved Molecular Diagnosis of Dystrophin Gene Mutations Using the Multiplex Ligation-Dependent Probe Amplification Method. Genet. Test. 2004;8:361–367. doi: 10.1089/gte.2004.8.361. [DOI] [PubMed] [Google Scholar]
- 49.Slatko B.E., Gardner A.F., Ausubel F.M. Overview of Next Generation Sequencing technologies (and bioinformatics) in cancer. Mol. Biol. 2018;122:e59. doi: 10.1002/cpmb.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volk A.E., Kubisch C. The rapid evolution of molecular genetic diagnostics in neuromuscular diseases. Curr. Opin. Neurol. 2017;30:523–528. doi: 10.1097/WCO.0000000000000478. [DOI] [PubMed] [Google Scholar]
- 51.Zhang K., Yang X., Lin G., Han Y., Li J. Molecular genetic testing and diagnosis strategies for dystrophinopathies in the era of next generation sequencing. Clin. Chim. Acta. 2019;491:66–73. doi: 10.1016/j.cca.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Lim B.C., Lee S., Shin J.-Y., Kim J.-I., Hwang H., Kim K.J., Hwang Y.S., Seo J.-S., Chae J.H. Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: Comprehensive mutational search in a single platform. J. Med. Genet. 2011;48:731–736. doi: 10.1136/jmedgenet-2011-100133. [DOI] [PubMed] [Google Scholar]
- 53.Aartsma-Rus A., Ginjaar I.B., Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J. Med. Genet. 2016;53:145–151. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nigro V., Politano L., Nigro G., Romano S.C., Molinari A.M., Puca G.A. Detection of a nonsense mutation in the dystrophin gene by multiple SSCP. Hum. Mol. Genet. 1992;1:517–520. doi: 10.1093/hmg/1.7.517. [DOI] [PubMed] [Google Scholar]
- 55.Nigro V., Nigro G., Esposito M.G., Comi L.I., Molinari A.M., Puca G.A., Politano L. Novel small mutations along the DMD/BMD gene associated with different phenotypes. Hum. Mol. Genet. 1994;3:1907–1908. doi: 10.1093/hmg/3.10.1907. [DOI] [PubMed] [Google Scholar]
- 56.Trimarco A., Torella A., Piluso G., Maria Ventriglia V., Politano L., Nigro V. Log-PCR: A New Tool for Immediate and Cost-Effective Diagnosis of up to 85% of Dystrophin Gene Mutations. Clin. Chem. 2008;54:973–981. doi: 10.1373/clinchem.2007.097881. [DOI] [PubMed] [Google Scholar]
- 57.Torella A., Trimarco A., Blanco F.D.V., Cuomo A., Aurino S., Piluso G., Minetti C., Politano L., Nigro V. One Hundred Twenty-One Dystrophin Point Mutations Detected from Stored DNA Samples by Combinatorial Denaturing High-Performance Liquid Chromatography. J. Mol. Diagn. 2010;12:65–73. doi: 10.2353/jmoldx.2010.090074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torella A., Zanobio M., Zeuli R., Del Vecchio Blanco F., Savarese M., Giugliano T., Garofalo A., Piluso G., Politano L., Nigro V. The position of nonsense mutations can predict the phenotype severity: A survey on the DMD gene. PLoS ONE. 2020;15:e0237803. doi: 10.1371/journal.pone.0237803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savarese M., Qureshi T., Torella A., Laine P., Giugliano T., Jonson P.H., Johari M., Paulin L., Piluso G., Auvinen P., et al. Identification and Characterization of Splicing Defects by Single-Molecule Real-Time Sequencing Technology (PacBio) J. Neuromuscul. Dis. 2020;7:477–481. doi: 10.3233/JND-200523. [DOI] [PubMed] [Google Scholar]
- 60.Onore M.E., Torella A., Musacchia F., D’Ambrosio P., Zanobio M., Blanco F.D.V., Piluso G., Nigro V. Linked-Read Whole Genome Sequencing Solves a Double DMD Gene Rearrangement. Genes. 2021;12:133. doi: 10.3390/genes12020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mora M., Angelini C., Bignami F., Bodin A.-M., Crimi M., Di Donato J.H., Felice A., Jaeger C., Karcagi V., LeCam Y., et al. The EuroBioBank Network: 10 years of hands-on experience of collaborative, transnational biobanking for rare diseases. Eur. J. Hum. Genet. 2014;23:1116–1123. doi: 10.1038/ejhg.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Filocamo M., Baldo C., Goldwurm S., Renieri A., Angelini C., Moggio M., Mora M., Merla G., Politano L., Garavaglia B., et al. Telethon Network of Genetic Biobanks: A key service for diagnosis and research on rare diseases. Orphanet J. Rare Dis. 2013;8:129. doi: 10.1186/1750-1172-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Amico A., Catteruccia M., Baranello G., Politano L., Govoni A., Previtali S.C., Pane M., D’Angelo M.G., Bruno C., Messina S., et al. Diagnosis of Duchenne Muscular Dystrophy in Italy in the last decade: Critical issues and areas for improvements. Neuromuscul. Disord. 2017;27:447–451. doi: 10.1016/j.nmd.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 64.Neri M., Rossi R., Trabanelli C., Mauro A., Selvatici R., Falzarano M.S., Spedicato N., Margutti A., Rimessi P., Fortunato F., et al. The Genetic Landscape of Dystrophin Mutations in Italy: A Nationwide Study. Front. Genet. 2020;11:131. doi: 10.3389/fgene.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flanigan K.M., Dunn D.M., von Niederhausern A., Soltanzadeh P., Gappmaier E., Howard M.T., Sampson J.B., Mendell J.R., Wall C., King W.M., et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: Application of modern diagnostic techniques to a large cohort. Hum. Mutat. 2009;30:1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tuffery-Giraud S., Béroud C., Leturcq F., Ben Yaou R., Hamroun D., Michel-Calemard L., Moizard M.-P., Bernard R., Cossée M., Boisseau P., et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: A model of nationwide knowledgebase. Hum. Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 67.The DMD Mutations Database UMD-DMD France. [(accessed on 4 October 2021)]. Available online: http://www.umd.be/DMD/
- 68.Vieitez I., Gallano P., Gonzalez-Quereda L., Borrego S., Marcos I., Millán J.M., Jairo T., Prior C., Molano J.M., Trujillo-Tiebas M., et al. Mutational spectrum of Duchenne muscular dystrophy in Spain: Study of 284 cases. Neurología. 2017;32:377–385. doi: 10.1016/j.nrl.2015.12.009. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 69.Garcia S., de Haro T., Zafra-Ceres M., Poyatos A., Gomez-Capilla J.A., Gomez-Llorente C. Identification of de novoMutations of Duchénnè/Becker Muscular Dystrophies in Southern Spain. Int. J. Med. Sci. 2014;11:988–993. doi: 10.7150/ijms.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van den Bergen J.C., Ginjaar H.B., van Essen A.J., Pangalila R., de Groot I.J., Wijkstra P.J., Zijnen M.P., Cobben N.A., Kampelmacher M.J., Wokke B.H., et al. Forty-Five Years of Duchenne Muscular Dystrophy in The Netherlands. J. Neuromuscul. Dis. 2014;1:99–109. doi: 10.3233/JND-140005. [DOI] [PubMed] [Google Scholar]
- 71.Selvatici R., Rossi R., Fortunato F., Trabanelli C., Sifi Y., Margutti A., Neri M., Gualandi F., Szabò L., Fekete B., et al. Ethnicity-related DMD Genotype Landscapes in European and Non-European Countries. Neurol. Genet. 2020;7:e536. doi: 10.1212/NXG.0000000000000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimowski J.G., Purzycka J., Pawelec M., Ozdarska K., Zaremba J. Small mutations in Duchenne/Becker muscular dystrophy in 164 unrelated Polish patients. J. Appl. Genet. 2021;62:289–295. doi: 10.1007/s13353-020-00605-0. [DOI] [PubMed] [Google Scholar]
- 73.Toksoy G., Durmus H., Aghayev A., Bagirova G., Rustemoglu B.S., Basaran S., Avci S., Karaman B., Parman Y., Altunoglu U., et al. Mutation spectrum of 260 dystrophinopathy patients from Turkey and important highlights for genetic counseling. Neuromuscul. Disord. 2019;29:601–613. doi: 10.1016/j.nmd.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 74.Brabec P., Vondráček P., Klimeš D., Baumeister S., Lochmüller H., Pavlík T., Gregor J. Characterization of the DMD/BMD patient population in Czech Republic and Slovakia using an innovative registry approach. Neuromuscul. Disord. 2009;19:250–254. doi: 10.1016/j.nmd.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Elhawary N.A., Jiffri E.H., Jambi S., Mufti A.H., Dannoun A., Kordi H., Khogeer A., Jiffri O.H., Elhawary A.N., Tayeb M.T. Molecular characterization of exonic rearrangements and frame shifts in the dystrophin gene in Duchenne muscular dystrophy patients in a Saudi community. Hum. Genom. 2018;12:18. doi: 10.1186/s40246-018-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tayeb M.T. Deletion mutations in Duchenne muscular dystrophy (DMD) in Western Saudi children. Saudi J. Biol. Sci. 2010;17:237–240. doi: 10.1016/j.sjbs.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mohammed F., Elshafey A., Al-Balool H., Alaboud H., Al Ben Ali M., Baqer A., Bastaki L. Mutation spectrum analysis of Duchenne/Becker muscular dystrophy in 68 families in Kuwait: The era of personalized medicine. PLoS ONE. 2018;13:e0197205. doi: 10.1371/journal.pone.0197205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kong X., Zhong X., Liu L., Cui S., Yang Y., Kong L. Genetic analysis of 1051 Chinese families with Duchenne/Becker Muscular Dystrophy. BMC Med. Genet. 2019;20:139. doi: 10.1186/s12881-019-0873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lo I.F., Lai K.K., Tong T.M., Lam S.T. A different spectrum of DMD gene mutations in local Chinese patients with Duchenne/Becker muscular dystrophy. Chin. Med. J. 2006;119:1079–1087. [PubMed] [Google Scholar]
- 80.Guo R., Zhu G., Zhu H., Ma R., Peng Y., Liang D., Wu L. DMD mutation spectrum analysis in 613 Chinese patients with dystrophinopathy. J. Hum. Genet. 2015;60:435–442. doi: 10.1038/jhg.2015.43. [DOI] [PubMed] [Google Scholar]
- 81.Wang L., Xu M., Li H., He R., Lin J., Zhang C., Zhu Y. Genotypes and Phenotypes of DMD Small Mutations in Chinese Patients with Dystrophinopathies. Front. Genet. 2019;10:114. doi: 10.3389/fgene.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma P., Zhang S., Zhang H., Fang S., Dong Y., Zhang Y., Hao W., Wu S., Zhao Y. Comprehensive genetic characteristics of dystrophinopathies in China. Orphanet J. Rare Dis. 2018;13:109. doi: 10.1186/s13023-018-0853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y., Liu Z., OuYang S., Zhu Y., Wang L., Wu J. Distribution of dystrophin gene deletions in a Chinese population. J. Int. Med. Res. 2016;44:99–108. doi: 10.1177/0300060515613223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X., Zhao L., Zhou S., Hu C., Shi Y., Shi W., Li H., Liu F., Wu B., Wang Y. A comprehensive database of Duchenne and Becker muscular dystrophy patients (0–18 years old) in East China. Orphanet J. Rare Dis. 2015;10:5. doi: 10.1186/s13023-014-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y.-M., Yan K., Liu B., Chen M., Wang L.-Y., Huang Y.-Z., Qian Y.-Q., Sun Y.-X., Li H.-G., Dong M.-Y. Comprehensive genetic diagnosis of patients with Duchenne/Becker muscular dystrophy (DMD/BMD) and pathogenicity analysis of splice site variants in the DMD gene. J. Zhejiang Univ. B. 2019;20:753–765. doi: 10.1631/jzus.B1800541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yun U., Lee S.-A., Choi W.A., Kang S.-W., Seo G.H., Lee J.H., Park G., Lee S., Choi Y.-C., Park H.J. Clinical and genetic spectra in patients with dystrophinopathy in Korea: A single-center study. PLoS ONE. 2021;16:e0255011. doi: 10.1371/journal.pone.0255011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho A., Seong M.-W., Lim B.C., Lee H.J., Byeon J.H., Kim S.S., Kim S.Y., Choi S.A., Wong A.-L., Lee J., et al. Consecutive analysis of mutation spectrum in the dystrophin gene of 507 Korean boys with Duchenne/Becker muscular dystrophy in a single center. Muscle Nerve. 2017;55:727–734. doi: 10.1002/mus.25396. [DOI] [PubMed] [Google Scholar]
- 88.Takeshima Y., Yagi M., Okizuka Y., Awano H., Zhang Z., Yamauchi Y., Nishio H., Matsuo M. Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J. Hum. Genet. 2010;55:379–388. doi: 10.1038/jhg.2010.49. [DOI] [PubMed] [Google Scholar]
- 89.Okubo M., Goto K., Komaki H., Nakamura H., Mori-Yoshimura M., Hayashi Y.K., Mitsuhashi S., Noguchi S., Kimura E., Nishino I. Comprehensive analysis for genetic diagnosis of Dystrophinopathies in Japan. Orphanet J. Rare Dis. 2017;12:149. doi: 10.1186/s13023-017-0703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zamani G., Hosseinpour S., Ashrafi M.R., Mohammadi M., Badv R.S., Tavasoli A.R., Akbari M.G., Bereshneh A.H., Malamiri R.A., Heidari M. Characteristics of disease progression and genetic correlation in ambulatory Iranian boys with Duchenne muscular dystrophy. BMC Neurol. 2022;22:162. doi: 10.1186/s12883-022-02687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tomar S., Moorthy V., Sethi R., Chai J., Low P.S., Hong S.T.K., Lai P.S. Mutational spectrum of dystrophinopathies in Singapore: Insights for genetic diagnosis and precision therapy. Am. J. Med. Genet. Part C Semin. Med. Genet. 2019;181:230–244. doi: 10.1002/ajmg.c.31704. [DOI] [PubMed] [Google Scholar]
- 92.Rani A.Q., Sasongko T.H., Sulong S., Bunyan D., Salmi A.R., Zilfalil B.A., Matsuo M., Zabidi-Hussin Z.A. Mutation Spectrum ofDystrophinGene in Malaysian Patients with Duchenne/Becker Muscular Dystrophy. J. Neurogenet. 2013;27:11–15. doi: 10.3109/01677063.2012.762580. [DOI] [PubMed] [Google Scholar]
- 93.Rao M.V., Sindhav G.M., Mehta J.J. Duchenne/Becker muscular dystrophy: A report on clinical, biochemical, and genetic study in Gujarat population, India. Ann. Indian Acad. Neurol. 2014;17:303–307. doi: 10.4103/0972-2327.138508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kohli S., Saxena R., Thomas E., Singh K., BijarniaMahay S., Puri R.D., Verma I.C. Mutation Spectrum of Dystrophinopathies in India: Implications for Therapy. Indian J. Pediatr. 2020;87:495–504. doi: 10.1007/s12098-020-03286-z. [DOI] [PubMed] [Google Scholar]
- 95.Polavarapu K., Preethish-Kumar V., Sekar D., Vengalil S., Nashi S., Mahajan N.P., Thomas P.T., Sadasivan A., Warrier M., Gupta A., et al. Mutation pattern in 606 Duchenne muscular dystrophy children with a comparison between familial and non-familial forms: A study in an Indian large single-center cohort. J. Neurol. 2019;266:2177–2185. doi: 10.1007/s00415-019-09380-3. [DOI] [PubMed] [Google Scholar]
- 96.Manjunath M., Kiran P., Preethish-Kumar V., Nalini A., Singh R.J., Gayathri N. A comparative study of mPCR, MLPA, and muscle biopsy results in a cohort of children with Duchenne muscular dystrophy: A first study. Neurol. India. 2015;63:58–62. doi: 10.4103/0028-3886.152635. [DOI] [PubMed] [Google Scholar]
- 97.Vengalil S., Preethish-Kumar V., Polavarapu K., Mahadevappa M., Sekar D., Purushottam M., Thomas P.T., Nashi S., Nalini A. Duchenne Muscular Dystrophy and Becker Muscular Dystrophy Confirmed by Multiplex Ligation-Dependent Probe Amplification: Genotype-Phenotype Correlation in a Large Cohort. J. Clin. Neurol. 2017;13:91–97. doi: 10.3988/jcn.2017.13.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumar S.H., Athimoolam K., Suraj M., Das ChristuDas M.S., Muralidharan A., Jeyam D., Ashokan J., Karthikeyan P., Krishna R., Khanna-Gupta A., et al. Comprehensive genetic analysis of 961 unrelated Duchenne Muscular Dystrophy patients: Focus on diagnosis, prevention and therapeutic possibilities. PLoS ONE. 2020;15:e0232654. doi: 10.1371/journal.pone.0232654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tallapaka K., Ranganath P., Ramachandran A., Uppin M.S., Perala S., Aggarwal S., Lakshmi D., Meena A.K., Dalal A.B. Molecular and Histopathological Characterization of Patients Presenting with the Duchenne Muscular Dystrophy Phenotype in a Tertiary Care Center in Southern India. Indian Pediatr. 2019;56:556–559. doi: 10.1007/s13312-019-1553-z. [DOI] [PubMed] [Google Scholar]
- 100.Thakur N., Abeysekera G., Wanigasinghe J., Dissanayake V.H.W. The spectrum of deletions and duplications in the dystrophin (DMD) gene in a cohort of patients with Duchenne muscular dystrophy in Sri Lanka. Neurol. India. 2019;67:714–715. doi: 10.4103/0028-3886.263235. [DOI] [PubMed] [Google Scholar]
- 101.Elhawary N.A., Shawky R.M., Hashem N. Frameshift deletion mechanisms in Egyptian Duchenne and Becker muscular dystrophy families. Mol. Cells. 2004;18:141–149. Erratum in Mol. Cells 2005, 19, 155; Nasser A, Elhawary [corrected to Elhawary, Nasser A]. [PubMed] [Google Scholar]
- 102.El Sherif R.M., Fahmy N.A., Nonaka I., A Etribi M. Patterns of dystrophin gene deletion in Egyptian Duchenne/Becker muscular dystrophy patients. Acta Myol. 2007;26:145–150. doi: 10.1016/j.nmd.2006.05.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Effat L.K., El-Harouni A.A., Amr K., El-Minisi T.I., Meguid N., El Awady M. Screening of Dystrophin Gene Deletions in Egyptian Patients with DMD/BMD Muscular Dystrophies. Dis. Markers. 2000;16:125–129. doi: 10.1155/2000/437372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sbiti A., El Kerch F., Sefiani A. Analysis of Dystrophin Gene Deletions by Multiplex PCR in Moroccan Patients. J. Biomed. Biotechnol. 2002;2:158–160. doi: 10.1155/S1110724302205069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ballo R., Viljoen D., Beighton P. Duchenne and Becker muscular dystrophy prevalence in South Africa and molecular findings in 128 persons affected. S.Afr. Med. J. 1994;84Pt 1:494–497. [PubMed] [Google Scholar]
- 106.Kerr R., Robinson C., Essop F.B., Krause A. Genetic testing for Duchenne/Becker muscular dystrophy in Johannesburg, South Africa. S.Afr. Med. J. 2013;103((Suppl. S1)):999. doi: 10.7196/SAMJ.7274. [DOI] [PubMed] [Google Scholar]
- 107.Mah J.K., Korngut L., Dykeman J., Day L., Pringsheim T., Jette N. A systematic review and meta-analysis on the epidemiology of Duchenne and Becker muscular dystrophy. Neuromuscul. Disord. 2014;24:482–491. doi: 10.1016/j.nmd.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 108.Gambetta K.E., McCulloch M.A., Lal A.K., Knecht K., Butts R.J., Villa C.R., Johnson J.N., Conway J., Bock M.J., Schumacher K.R., et al. Diversity of Dystrophin Gene Mutations and Disease Progression in a Contemporary Cohort of Duchenne Muscular Dystrophy. Pediatr. Cardiol. 2022;43:855–867. doi: 10.1007/s00246-021-02797-6. [DOI] [PubMed] [Google Scholar]
- 109.Dent K.M., Dunn D.M., von Niederhausern A.C., Aoyagi A.T., Kerr L., Bromberg M.B., Hart K.J., Tuohy T., White S., den Dunnen J.T., et al. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am. J. Med. Genet. Part A. 2005;134:295–298. doi: 10.1002/ajmg.a.30617. [DOI] [PubMed] [Google Scholar]
- 110.Luce L.N., Dalamon V., Ferrer M., Parma D., Szijan I., Giliberto F. MLPA analysis of an Argentine cohort of patients with dystrophinopathy: Association of intron breakpoints hot spots with STR abundance in DMD gene. J. Neurol. Sci. 2016;365:22–30. doi: 10.1016/j.jns.2016.03.047. [DOI] [PubMed] [Google Scholar]
- 111.Triana-Fonseca P., Parada-Márquez J.F., Silva-Aldana C.T., Zambrano-Arenas D., Arias-Gomez L.L., Morales-Fonseca N., Medina-Méndez E., Restrepo C.M., Silgado-Guzmán D.F., Fonseca-Mendoza D.J. Genetic Profile of the Dystrophin Gene Reveals New Mutations in Colombian Patients Affected with Muscular Dystrophinopathy. Appl. Clin. Genet. 2021;14:399–408. doi: 10.2147/TACG.S317721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ramos E., Conde J.G., Berrios R.A., Pardo S., Gómez O., Rodríguez M.F.M. Prevalence and Genetic Profile of Duchene and Becker Muscular Dystrophy in Puerto Rico. J. Neuromuscul. Dis. 2016;3:261–266. doi: 10.3233/JND-160147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stenson P.D., Mort M., Ball E.V., Chapman M., Evans K., Azevedo L., Hayden M., Heywood S., Millar D.S., Phillips A.D., et al. The Human Gene Mutation Database (HGMD®): Optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020;139:1197–1207. doi: 10.1007/s00439-020-02199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Passamano L., Taglia A., Palladino A., Viggiano E., D’Ambrosio P., Scutifero M., Cecio M.R., Torre V., De Luca F., Picillo E., et al. Improvement of survival in Duchenne Muscular Dystrophy: Retrospective analysis of 835 patients. Acta Myol. 2012;31:121–125. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on request.