Abstract
The kidney and brain expressed protein (KIBRA) plays an important role in synaptic plasticity. Carriers of the T allele of the KIBRA (WWC1) gene rs17070145 C/T polymorphism have been reported to have enhanced spatial ability and to outperform individuals with the CC genotype in working memory tasks. Since ability in chess and science is directly related to spatial ability and working memory, we hypothesized that the KIBRA T allele would be positively associated with chess player status and PhD status in science. We tested this hypothesis in a study involving 2479 individuals (194 chess players, 119 PhD degree holders in STEM fields, and 2166 controls; 1417 males and 1062 females) from three ethnicities (236 Kazakhs, 1583 Russians, 660 Tatars). We found that frequencies of the T allele were significantly higher in Kazakh (66.9 vs. 55.1%; p = 0.024), Russian (44.8 vs. 32.0%; p = 0.0027), and Tatar (51.5 vs. 41.8%; p = 0.035) chess players compared with ethnically matched controls (meta-analysis for CT/TT vs. CC: OR = 2.05, p = 0.0001). In addition, none of the international chess grandmasters (ranked among the 80 best chess players in the world) were carriers of the CC genotype (0 vs. 46.3%; OR = 16.4, p = 0.005). Furthermore, Russian and Tatar PhD holders had a significantly higher frequency of CT/TT genotypes compared with controls (meta-analysis: OR = 1.71, p = 0.009). Overall, this is the first study to provide comprehensive evidence that the rs17070145 C/T polymorphism of the KIBRA gene may be associated with ability in chess and science, with the T allele exerting a beneficial effect.
Keywords: DNA, memory, education, PhD, intelligence, chess, mind sports, analytical ability, scientists, athletes
1. Introduction
The kidney and brain expressed protein (KIBRA), together with its binding partners (dendrin, synaptopodin, dynein-complex and others), plays an important role in synaptic plasticity [1], which enables the brain to store and recall information. KIBRA is considered a postsynaptic scaffold protein connecting cytoskeletal and signaling molecules, with high expression in memory- and spatial learning-related regions of the brain (cortex, hippocampus), as well as in the cerebellum and hypothalamus [2].
KIBRA attracted the attention of behavioral geneticists following the publication of a 2006 genome-wide association study (GWAS) of memory performance and executive function [3]. In that study, the T allele of the KIBRA (also known as WWC1) gene rs17070145 C/T polymorphism was positively associated with episodic memory performance in three independent cohorts [3]. Subsequent studies have examined the rs17070145 single nucleotide polymorphism (SNP) with respect to cognitive performance in different populations [1,4]. Here, KIBRA T allele carriers have been shown to outperform individuals with the CC genotype in episodic and working memory tasks [4,5,6]. Furthermore, the T allele has been linked with increased cognitive performance [7] and better scores in spatial ability [8], spatial learning [9], and navigational success [10].
Spatial ability is a highly heritable (84%) trait [11] and is a predictor of success in science, technology, engineering, and mathematics (i.e., STEM subjects) and other mentally demanding fields [12,13]. Studies suggest that a high level of spatial ability, working-memory capacity, and other cognitive abilities are necessary to reach a high standard of performance in chess [14,15,16]. Working memory is also a heritable (31–72%) trait [17,18] and is positively associated with analytic problem-solving [19] and fluid intelligence [20], suggesting that working memory capacity may be a contributor to achievement in a scientific career.
Because ability in chess and science both appear to be related to spatial ability and working memory, we hypothesized that the KIBRA rs17070145 T allele might be positively associated with chess player status and PhD status in science. Thus, in the present and first study of its type, our aim was to compare allelic frequencies of the KIBRA rs17070145 C/T polymorphism between chess players, PhD holders, and controls. Through this novel focus, and by comprehensively examining these comparisons in three independent cohorts, combined with follow-up meta-analyses of our results, we sought to make a unique contribution to the field.
2. Materials and Methods
2.1. Ethics Statement
The Ethics Committee of the Federal Research and Clinical Center of Physical-Chemical Medicine of the Federal Medical and Biological Agency of Russia (Approval number 2017/04) and the Ethics Committee of the Al-Farabi Kazakh National University (Approval numbers: IRB-A172 and IRB-A267) approved the protocols for the research. The studies were conducted according to the guidelines of the Declaration of Helsinki and Strengthening The Reporting of Genetic Association Studies (STREGA): An extension of the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) statement recommendations [21].
2.2. Participants
The study involved 2479 individuals (194 chess players, 119 PhD degree holders, and 2166 controls; 1417 males and 1062 females) from Kazakh, Russian and Tatar ethnicities. The Russian subjects were all Caucasians of Eastern European descent, while Kazakhs and Tatars were descendants of Turkic origin living in the Republic of Kazakhstan and Republic of Tatarstan (region of the Russian Federation), respectively. Age and sex characteristics of all cohorts are presented in Table 1. Chess players were classified as non-elite (51 Kazakhs, 42 Russians, 42 Tatars), sub-elite (8 Kazakhs, 16 Russians, 26 Tatars) and elite (9 Russian international chess grandmasters ranked among the 80 best chess players in the world). All PhD (Candidates of Sciences, according to the Russian system) holders earned their degrees in the STEM fields, including medicine (n = 10), biology (n = 49), chemistry (n = 39), technology (n = 8), and physics/mathematics (n = 13).
Table 1.
Characteristics of studied groups.
| Groups | Males | Females | ||
|---|---|---|---|---|
| n | Age, Years | n | Age, Years | |
| Kazakh chess players | 46 | 17.0 (3.9) | 13 | 16.3 (4.4) |
| Kazakh controls | 111 | 27.5 (4.6) | 66 | 25.0 (3.6) |
| Russian chess players | 44 | 22.2 (3.2) | 23 | 22.8 (3.1) |
| Russian PhD holders | 34 | 27.6 (2.8) | 43 | 30.4 (4.9) |
| Russian controls | 831 | 27.1 (4.3) | 608 | 27.2 (4.4) |
| Tatar chess players | 48 | 21.9 (3.8) | 20 | 21.4 (3.2) |
| Tatar PhD holders | 25 | 30.2 (4.7) | 17 | 32.2 (6.5) |
| Tatar controls | 278 | 25.7 (4.8) | 272 | 25.1 (4.3) |
Data are Mean (SD).
2.3. Genetic Analysis
2.3.1. Russian and Tatar Samples
DNA samples from Russian and Tatar individuals were obtained from leukocytes (venous blood) or epithelial mouth cells. Four ml of venous blood were collected in tubes containing EDTA (Vacuette EDTA tubes, Greiner Bio-One, Kremsmünster, Austria). DNA samples were transported to the laboratory at 4 °C, and DNA was extracted on the same day. DNA extraction and purification from blood samples were performed using commercial kits (Techno-sorb), according to the manufacturer’s instructions (Technoclon, Moscow, Russia), and included chemical lysis, selective DNA binding on silica spin columns, and ethanol washing. DNK-sorb-A kit (Central Research Institute of Epidemiology, Moscow, Russia) was used for the DNA extraction and purification from epithelial mouth samples, according to the manufacturer’s instructions [22].
Genotyping of the KIBRA gene rs17070145 polymorphism was performed using three different methods, including microarray technology [23,24], single-stage PCR followed by hybridization on a hydrogel biochip [25], and a newly designed Restriction Fragment Length Polymorphism method (RFLP). The assay for the microarray analysis required 200 ng of DNA sample as input with a concentration of at least 50 ng/µL. Exact concentrations of DNA in each sample were measured using a Qubit Fluorometer (Invitrogen, Waltham, MA, USA). The genotyping process was performed using HumanOmni1-Quad BeadChips (Illumina, San Diego, CA, USA) to genotype the KIBRA gene polymorphism. All further procedures were performed according to the instructions of the Infinium High-Density Assay.
PCR primers for the RFLP analysis were forward 5′-CTAATGCCAGGACGTCATGGCAG-3′ and reverse 5′-ATCTCTTGACCCAGTATAAAAGGA-3′ (Litech, Moscow, Russia), generating a fragment of 214 bp. PCR was conducted on a multicanal amplificator Tercyk (DNA Technology, Moscow, Russia). PCR products were digested with Tru9 I (SibEnzyme, Novosibirsk, Russia) for 12 h at 65 °C, producing 173- and 41-bp fragments for allele T and a 214-bp fragment for allele C. Fragments were then separated by 8% polyacrylamide gel electrophoresis, stained with ethidium bromide, and visualized in UV light. All genotyping analyses were conducted blind to participant identity.
2.3.2. Kazakh Samples
Genomic DNA was extracted from a buccal swab using a QIAamp DNA Mini kit (Cat No. 51306; Qiagen, Germany) according to the manufacturer’s instructions. Briefly, ari-dried cotton buccal swabs post-collection were cut into a 2 mL tube and resuspended into 400 µL PBS followed by lysis with Proteinase K and AL lysis buffer at 56 °C for 10 min. The digested samples were treated with AL buffer and 100% ethanol followed by washes with wash buffer on the QIAamp mini spin column and eluted with 30 µL nuclease-free water. DNA extracted was quantified using NanodropTM (Thermo Fisher Scientific, Waltham, MA, USA). Approximately, 10 ng of DNA was used for the targeted sequencing using Fluidigm Access Array.
The rs17070145 SNP (C/T) was genotyped using Targeted Next Generation Sequencing. First, primers were designed to cover the SNP rs17070145 (chr5: 167845791, GRCh37/hg19) in the KIBRA gene. The primer sequences used for genotyping are F: 5′-TACTCCCAGCACACACCTC-3′ and R: 5′-GTTGGCAGATGGAACCCGT-3′. The primers were evaluated using control DNA samples and the expected size was validated using agarose gel electrophoresis. Then, the primers were tagged with Fluidigm-specific tag sequences CS1: ACACTGACGACATGGTTCTACA for the forward’s primer and CS2: TACGGTAGCAGAGACTTGGTCT for the reverse’s primer. PCR was performed with at least 5 ng of DNA from each sample using Fast Start High Fidelity master mix (Roche, Switzerland). The amplified PCR products were further purified using ExoSAP-IT (Invitrogen, USA) and amplified with the tagged target-specific primers on the 48.48 Access Array integrated Fluidic circuit (IFC) using a Fast start high fidelity master mix (Roche). The prepared amplicons were allied with 48.48 Access Array IFC barcodes, purified with AMPure XP beads (Beckman Coulter, Brea, CA, USA) and quantified using High sensitivity DNA assay kit on Bioanalyzer (Agilent, Santa Clara, CA, USA) following the manufacturer’s instructions. The libraries were further proceeded using emulsion PCR with Ion SphereTM particles with Ion Template OT2 kit in Ion OneTouch™ ES system following the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). All samples were pooled and sequenced using the Ion 520™ Chip on the Ion S5 XL Semiconductor sequencer following the manufacturer’s instructions (Thermo Fischer).
The resulting reads were processed and quality control was assessed using the Ion Torrent Software Suite version 5.12.3. The burrows wheeler aligner (BWA) was used to align the raw sequencing reads to the reference genome GRCh37/hg19. SNP calling and genotyping of the rs17070145 SNP was performed using an in-house bioinformatics pipeline that incorporates SAMtools mpileup commands. Samples showing low coverage depth were excluded from the downstream analysis.
2.4. Statistical Analyses
Statistical analyses were conducted using GraphPad InStat (GraphPad Software, Inc., San Diego, CA, USA) software. Genotype frequencies of cases (chess players and PhD holders) and controls were tested for compatibility with Hardy–Weinberg equilibrium (HWE). Genotype distribution and allelic frequencies between cases and controls were compared using χ2 or Fisher’s exact tests. To perform the meta-analyses the Cochrane Review Manager (RevMan) (London, UK) version 5.3 was used. Random effects models were applied. Odds ratios (OR) with 95% confidence intervals (CI) were estimated using the Mantel-Haenszel method. The heterogeneity degree between the studies was assessed with the I2 statistic. p values < 0.05 were considered statistically significant. Where appropriate, Bonferroni’s correction for multiple testing was performed by dividing the p value (0.05) by the number of tests.
3. Results
3.1. Association of the KIBRA Gene Variant with Chess Player Status
In Kazakh, Russian, and Tatar groups of chess players and controls, the KIBRA gene rs17070145 polymorphism met Hardy-Weinberg expectations (p > 0.05 in all groups tested separately). Frequencies of the rs17070145 T allele were significantly higher in Kazakh (66.9 vs. 55.1%; OR = 1.65, 95% CI 1.07–2.56; p = 0.024, χ2 = 4.64), Russian (44.8 vs. 32.0%; OR = 1.73, 95% CI 1.22–2.45; p = 0.0027, χ2 = 9.00), and Tatar (51.5 vs. 41.8%; OR = 1.48, 95% CI 1.03–2.11; p = 0.035, χ2 = 4.22) chess players compared with ethnically matched controls (Table 2). However, only in Russian chess players did the results remain statistically significant after correction for multiple testing. Across all groups, sub-elite/elite chess players tended to have higher frequencies of the T allele compared with non-elite chess players (Figure 1). In addition, none of the Russian international chess grandmasters (0 vs. 46.3%; OR = 16.4; p = 0.005, χ2 = 6.0) were carriers of the CC genotype (which was the most prevalent genotype in the Russian controls).
Table 2.
KIBRA rs17070145 genotype distribution and T allele frequency in different cohorts.
| Cohort | n | KIBRA Genotypes | T Allele, % | p | ||
|---|---|---|---|---|---|---|
| CC | CT | TT | ||||
| Kazakh chess players | 59 | 4 | 31 | 24 | 66.9 | 0.024 * |
| Kazakh controls | 177 | 34 | 91 | 52 | 55.1 | - |
| Russian chess players | 67 | 20 | 34 | 13 | 44.8 | 0.0027 * |
| Russian PhD holders | 77 | 27 | 37 | 13 | 40.9 | 0.026 * |
| Russian controls | 1439 | 666 | 626 | 147 | 32.0 | - |
| Tatar chess players | 68 | 16 | 34 | 18 | 51.5 | 0.035 * |
| Tatar PhD holders | 42 | 9 | 25 | 8 | 48.8 | 0.252 |
| Tatar controls | 550 | 197 | 246 | 107 | 41.8 | - |
* p < 0.05, statistically significant differences in allelic frequencies between cases (chess players or PhD holders) and controls.
Figure 1.
KIBRA gene rs17070145 T allele frequency in controls and chess players stratified by the level of achievement.
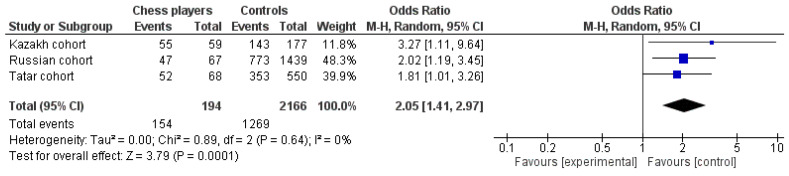
Given that allelic frequencies between three populations were significantly different (p < 0.0001), we performed a meta-analysis. The pooled OR favoring chess players compared with controls was 2.05 (95% CI 1.41–2.97, p = 0.0001 for the random effect model of meta-analysis) for the carriage of the T allele (i.e., rs17070145 CT/TT genotypes). No heterogeneity between studies (I2 = 0%; p = 0.64) was observed (Figure 2). These results indicate that carriage of the KIBRA CT/TT genotypes is strongly associated with chess player status.
Figure 2.
Meta-analysis for association studies for the KIBRA gene and chess player status. The prevalence of the rs17070145 CT/TT genotypes has been shown in chess players. The purple squares represent the ratio of CT/TT genotypes in chess players for each individual study. The black diamond represents the pooled ratio of CT/TT genotypes in all chess players and its 95% CI.
3.2. Association of the KIBRA Gene Variant with PhD Status
The KIBRA gene rs17070145 polymorphism met Hardy-Weinberg expectations in Russian and Tatar PhD holders (p > 0.05). The frequency of the rs17070145 T allele was significantly higher in Russian PhD holders (40.9 vs. 32.0%; OR = 1.47, 95% CI 1.06–2.05; p = 0.026, χ2 = 4.94) compared with Russian controls, but not in Tatar PhD holders (48.8 vs. 41.8%; OR = 1.33, 95% CI 0.85–2.07; p = 0.252, χ2 = 1.29) (Table 2). The pooled OR favoring PhD holders compared with controls was 1.71 (95% CI 1.14–2.57, p = 0.009 for the random effect model of meta-analysis) for carriage of the T allele. No heterogeneity between studies (I2 = 0%; p = 0.59) was observed (Figure 3).
Figure 3.
Meta-analysis for association studies for the KIBRA gene and PhD status. The prevalence of the rs17070145 CT/TT genotypes has been shown in PhD holders. The purple squares represent the ratio of CT/TT genotypes in PhD holders for each individual study. The black diamond represents the pooled ratio of CT/TT genotypes in all PhD holders and its 95% CI.
4. Discussion
The aim of the present study was to compare allelic frequencies of the KIBRA rs17070145 C/T polymorphism between chess players, PhD holders, and controls. As hypothesized, the favorable T allele was associated with chess player status and PhD status in science, and this finding was replicated across three cohorts. The latter replications are essential to confirm associations in genetic studies, when there are a limited number of individuals, such as in the case of elite athletes [26,27,28]. To our knowledge, then, this is the first study demonstrating that ability in chess and science may depend on a specific gene variant—the KIBRA rs17070145 SNP, previously associated with working memory, spatial ability, and other cognitive traits.
Although the rs17070145 C/T polymorphism is intronic, it may be functional. Indeed, according to the GTEx portal [29], this SNP is a splicing quantitative trait locus (sQTL), with the T allele associated with significantly (p = 1.3 × 10−10) altered changes in the splicing ratios of transcripts in brain tissue. Furthermore, Piras et al. [30] reported that the TT genotype was associated with overexpression of genes involved in the MAPK signaling pathway (MAPK8IP1, GADD45B, DUSP2, DUSP5, PRKCG, NR4A1, MAPK11, HSPA1A, HSPA1B and HSPB1) in the human hippocampus of 22 neuropathologically normal individuals. Interestingly, the MAPK signaling pathway has been previously linked with memory and learning processes [31,32].
Studies indicate that the KIBRA rs17070145 SNP is associated with brain metabolism and structure. Accordingly, Palombo et al. [33] demonstrated that T allele carriers had a larger hippocampal volume (cornu ammonis and dentate gyrus subregions) relative to noncarriers (i.e., CC genotype) among young people. This observation was confirmed in another study of older adults [34]. In addition, the KIBRA T allele has been linked with greater gray matter volume in the prefrontal cortex and parahippocampal cortex [35]. Furthermore, T allele carriers have been shown to exhibit higher glucose metabolism than carriers of the CC genotype in posterior cingulate and precuneus brain regions [36]. Lower cerebral glucose metabolism is considered one of pre-symptomatic endophenotypes associated with dementia [36,37]. Indeed, the T allele has been reported to protect against cognitive decline with age [38,39] and Alzheimer’s disease [36,40].
Chess is a board game that falls under the family of mind sports and requires a high level of spatial visual processing, attention, and working memory [41]. The heritability of skill in chess has been estimated at 48% [42]. Consequently, our findings that the KIBRA T allele and CT/TT genotypes are over-represented in three independent cohorts of chess players (and notably in all 9 international chess grandmasters) are supported by the fact that the T allele is associated with better attention/concentration [43], working memory [4] and spatial-related traits [8,9,10]. It should be noted that psychogenetic research in sport until now has been limited to studies involving power, endurance, combat, and game sports athletes only, with a focus on stress resilience (COMT rs4680), reaction time (APOE ε2/ε3/ε4, KIF27 rs10125715, APC rs518013, TMEM229A rs7783359, LRRN3 rs80054135), memory (APOE ε3/ε4, BDNF rs6265), sensation seeking (DRD3 rs167771, DRD4 rs1800955), anxiety (ACE I/D, COMT rs4680, MAOA VNTR polymorphism), aggressiveness (MAOA VNTR polymorphism), and motivation (MAOA VNTR polymorphism) [44,45,46]. Therefore, the present study, the first genetic research into mind sports athletes, represents an important step forward in the field.
In a study of the personality traits of 2015 scientists and 78,753 nonscientists, it was demonstrated that scientists scored higher than nonscientists on tough mindedness (a disposition to be analytical—drawing conclusions, and making decisions, based on logic, facts, and data) and openness (to innovation, novel experiences, and new learning) [47]. Scientists also need to be creative and have spatial ability [12,48]. Analytical ability and openness to experience positively correlate with intelligence and creativity [49,50]. There is also a positive relationship between working memory and other cognitive traits, such as creativity, analytic problem-solving, fluid intelligence, and spatial ability [19,20,51,52]. Given the above, it is, therefore, unsurprising that the PhD holders in our study had a greater frequency of the favorable KIBRA T allele and CT/TT genotypes compared with the general population. Indeed, in line with our findings that possessing the CT/TT genotype was associated with a greater disposition to earn a PhD in Russian and Tatar participants, bioinformatic analysis revealed that the KIBRA T allele was positively associated (p = 0.00089) with educational attainment in the UK Biobank cohort (n = 240,547) [53]. Importantly, there are at least 3952 known genome-wide significant SNPs associated with educational attainment [54], which may be considered in future research as potential candidate genetic markers related to achievement in science.
Against the backdrop of this study’s novel findings, there are some limitations that should be acknowledged. First, the size of the sample of PhD holders and chess players was moderate. Thus, although our results were further supported through meta-analyses, extension to and replication within groups of differing geographic ancestry is needed to translate these findings more broadly. Second, although our hypothesis—that the T allele of the KIBRA rs17070145 SNP would be associated with chess player status and PhD status in science—was based on evidence for the importance of spatial ability and working memory, we did not actually assess the latter traits in this study. Given the preceding lines, a third limitation is then that our comparison was between chess players and PhD holders with presumably high levels of key cognitive traits and controls, whose cognitive traits were essentially unknown. A fruitful (and potentially more powerful) future design would thus be to compare allelic frequencies of the KIBRA gene between individuals with known (measured) high and low cognitive abilities.
5. Conclusions
In conclusion, our results suggest that the rs17070145 C/T polymorphism of the KIBRA gene is associated with ability in chess and science, with the T allele exerting a beneficial effect. Nevertheless, given the substantial heritable component of cognitive and personality traits, and that each DNA locus may explain only a very small proportion of phenotypic variance (e.g., ~0.01% to ~1%), there are clearly more—and probably many more—genetic variants associated with ability in chess and science that remain to be identified. We thus caution that more evidence is needed before application of findings such as ours (e.g., via genetic testing) in talent identification in chess and science.
Acknowledgments
The authors would like to thank Ilya Alexeev and Anna Danilushkina for their help with data collection and genotyping.
Author Contributions
Conceptualization, I.I.A.; formal analysis, I.I.A., E.V.V., M.B.Y., G.K.D. and E.A.S.; investigation, I.I.A., E.V.V., M.B.Y., G.K.D., L.M.S., E.S.E., E.A.S., A.B., P.M.B., L.J.G., R.A.Y., A.K.L., N.A.K., E.V.G., R.H., A.M.K. and T.R.; writing—original draft preparation, I.I.A.; writing—review and editing, T.R.; project administration, I.I.A., E.V.G. and A.M.K.; funding acquisition, A.M.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the Al-Farabi Kazakh National University (Approval numbers: IRB-A172 and IRB-A267) and the Ethics Committee of Federal Research and Clinical Center of Physical-chemical Medicine of the Federal Medical and Biological Agency of Russia (Approval number 2017/04).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The Kazakhstan part of the study was funded by the Ministry of Education and Science of the Republic of Kazakhstan (grant no. AP08856595: “EEG/MRI studies of brain development, emotional-cognitive functions and their genetic markers in different age groups”). The UAE part of the study (targeted sequencing) was funded by University of Sharjah (grant no. AI-foL-2201) and ASPIRE AD PM ARI Project VRI-20-10 grant.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Schneider A., Huentelman M.J., Kremerskothen J., Duning K., Spoelgen R., Nikolich K. KIBRA: A new gateway to learning and memory? Front. Aging Neurosci. 2010;2:4. doi: 10.3389/neuro.24.004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johannsen S., Duning K., Pavenstädt H., Kremerskothen J., Boeckers T. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155:1165–1173. doi: 10.1016/j.neuroscience.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Papassotiropoulos A., Stephan D.A., Huentelman M.J., Hoerndli F.J., Craig D.W., Pearson J.V., Huynh K.-D., Brunner F., Corneveaux J., Osborne D., et al. Common Kibra Alleles Are Associated with Human Memory Performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 4.Milnik A., Heck A., Vogler C., Heinze H.-J., de Quervain D.J.-F., Papassotiropoulos A. Association of KIBRA with episodic and working memory: A meta-analysis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2012;159B:958–969. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 5.Muse J., Emery M., Sambataro F., Lemaitre H., Tan H.-Y., Chen Q., Kolachana B.S., Das S., Callicott J., Weinberger D.R., et al. WWC1 Genotype Modulates Age-Related Decline in Episodic Memory Function Across the Adult Life Span. Biol. Psychiatry. 2013;75:693–700. doi: 10.1016/j.biopsych.2013.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlomuzica A., Preusser F., Roberts S., Woud M.L., Lester K.J., Dere E., Eley T.C., Margraf J. The role of KIBRA in reconstructive episodic memory. Mol. Med. 2018;24:7. doi: 10.1186/s10020-018-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T.A., Bowers P., Sidorenko J., Karlsson Linnér R., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazantseva A.V., Enikeeva R.F., Davydova Y.D., Mustafin R.N., Takhirova Z.R., Malykh S.B., Lobaskova M.M., Tikhomirova T.N., Khusnutdinova E.K. The role of the KIBRA and APOE genes in developing spatial abilities in humans. Vavilov J. Genet. Breed. 2021;25:839–846. doi: 10.18699/VJ21.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuck N.W., Doeller C.F., Schjeide B.M., Schröder J., Frensch P.A., Bertram L., Li S.C. Aging and KIBRA/WWC1 genotype affect spatial memory processes in a virtual navigation task. Hippocampus. 2013;23:919–930. doi: 10.1002/hipo.22148. [DOI] [PubMed] [Google Scholar]
- 10.Rovira E., Mackie R.S., Clark N., Squire P.N., Hendricks M.D., Pulido A.M., Greenwood P.M. A role for attention during wilderness navigation: Comparing effects of BDNF, KIBRA, and CHRNA4. Neuropsychology. 2016;30:709–719. doi: 10.1037/neu0000277. [DOI] [PubMed] [Google Scholar]
- 11.Malanchini M., Rimfeld K., Shakeshaft N.G., McMillan A., Schofield K.L., Rodic M., Rossi V., Kovas Y., Dale P.S., Tucker-Drob E.M., et al. Evidence for a unitary structure of spatial cognition beyond general intelligence. NPJ Sci. Learn. 2020;5:9. doi: 10.1038/s41539-020-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kell H.J., Lubinski D., Benbow C.P., Steiger J.H. Creativity and technical innovation: Spatial ability’s unique role. Psychol. Sci. 2013;24:1831–1836. doi: 10.1177/0956797613478615. [DOI] [PubMed] [Google Scholar]
- 13.Rodic M., Tikhomirova T., Kolienko T., Malykh S., Bogdanova O., Zueva D.Y., Gynku E.I., Wan S., Zhou X., Kovas Y. Spatial complexity of character-based writing systems and arithmetic in primary school: A longitudinal study. Front. Psychol. 2015;6:333. doi: 10.3389/fpsyg.2015.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frydman M., Lynn R. The general intelligence and spatial abilities of gifted young Belgian chess players. Br. J. Psychol. 1992;83:233–235. doi: 10.1111/j.2044-8295.1992.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 15.Burgoyne A.P., Sala G., Gobet F., Macnamara B.N., Campitelli G., Hambrick D.Z. The relationship between cognitive ability and chess skill: A comprehensive meta-analysis. Intelligence. 2016;59:72–83. doi: 10.1016/j.intell.2016.08.002. [DOI] [Google Scholar]
- 16.Smith E.T., Bartlett J.C., Krawczyk D.C., Basak C. Are the advantages of chess expertise on visuo-spatial working-memory capacity domain specific or domain general? Mem. Cognit. 2021;49:1600–1616. doi: 10.3758/s13421-021-01184-z. [DOI] [PubMed] [Google Scholar]
- 17.Wilson R.S., Barral S., Lee J.H., Leurgans S.E., Foroud T.M., Sweet R.A., Graff-Radford N., Bird T.D., Mayeux R., Bennett D.A. Heritability of different forms of memory in the Late Onset Alzheimer’s Disease Family Study. J. Alzheimers Dis. 2011;23:249–255. doi: 10.3233/JAD-2010-101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogler C., Gschwind L., Coynel D., Freytag V., Milnik A., Egli T., Heck A., de Quervain D.J., Papassotiropoulos A. Substantial SNP-based heritability estimates for working memory performance. Transl. Psychiatry. 2014;4:e438. doi: 10.1038/tp.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis D.M., Ball B.H., Kimpton N., Brewer G.A. The role of working memory capacity in analytic and multiply-constrained problem-solving in demanding situations. Q. J. Exp. Psychol. 2020;73:920–928. doi: 10.1177/1747021820909703. [DOI] [PubMed] [Google Scholar]
- 20.Shelton J.T., Elliott E.M., Matthews R.A., Hill B.D., Gouvier W.D. The relationships of working memory, secondary memory, and general fluid intelligence: Working memory is special. J. Exp. Psychol. Learn. Mem. Cogn. 2010;36:813–820. doi: 10.1037/a0019046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little J., Higgins J.P.T., Ioannidis J.P.A., Moher D., Gagnon F., von Elm E., Khoury M.J., Cohen B., Davey-Smith G., Grimshaw J., et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE Statement. Hum. Genet. 2009;125:131–151. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- 22.Akhmetov I.I., Astranenkova I.V., Rogozkin V.A. Association of PPARD gene polymorphism with human physical performance. Mol. Biol. 2007;41:852–857. [PubMed] [Google Scholar]
- 23.Al-Khelaifi F., Yousri N.A., Diboun I., Semenova E.A., Kostryukova E.S., Kulemin N.A., Borisov O.V., Andryushchenko L.B., Larin A.K., Generozov E.V., et al. Genome-Wide Association Study Reveals a Novel Association Between MYBPC3 Gene Polymorphism, Endurance Athlete Status, Aerobic Capacity and Steroid Metabolism. Front. Genet. 2020;11:595. doi: 10.3389/fgene.2020.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikuchi N., Moreland E., Homma H., Semenova E.A., Saito M., Larin A.K., Kobatake N., Yusupov R.A., Okamoto T., Nakazato K., et al. Genes and Weightlifting Performance. Genes. 2021;13:25. doi: 10.3390/genes13010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fesenko D.O., Ivanovsky I.D., Ivanov P.L., Zemskova E.Y., Agapitova A.S., Polyakov S.A., Fesenko O.E., Filippova M.A., Zasedatelev A.S. A Biochip for Genotyping Polymorphisms Associated with Eye, Hair, Skin Color, AB0 Blood Group, Sex, Y Chromosome Core Haplogroup, and Its Application to Study the Slavic Population. Mol. Biol. 2022;56:860–880. doi: 10.1134/S0026893322050053. [DOI] [PubMed] [Google Scholar]
- 26.Eynon N., Nasibulina E.S., Banting L.K., Cieszczyk P., Maciejewska-Karlowska A., Sawczuk M., Bondareva E.A., Shagimardanova R.R., Raz M., Sharon Y., et al. The FTO A/T polymorphism and elite athletic performance: A study involving three groups of European athletes. PLoS One. 2013;8:e60570. doi: 10.1371/journal.pone.0060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmetov I.I., Hakimullina A.M., Popov D.V., Lyubaeva E.V., Missina S.S., Vinogradova O.L., Williams A.G., Rogozkin V.A. Association of the VEGFR2 gene His472Gln polymorphism with endurance-related phenotypes. Eur. J. Appl. Physiol. 2009;107:95–103. doi: 10.1007/s00421-009-1105-7. [DOI] [PubMed] [Google Scholar]
- 28.Semenova E.A., Zempo H., Miyamoto-Mikami E., Kumagai H., Larin A.K., Sultanov R.I., Babalyan K.A., Zhelankin A.V., Tobina T., Shiose K., et al. Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition. Cells. 2022;11:3910. doi: 10.3390/cells11233910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Portal. [(accessed on 25 November 2022)]. Available online: https://gtexportal.org.
- 30.Piras I.S., Krate J., Schrauwen I., Corneveaux J.J., Serrano G.E., Sue L., Beach T.G., Huentelman M.J. Whole transcriptome profiling of the human hippocampus suggests an involvement of the KIBRA rs17070145 polymorphism in differential activation of the MAPK signaling pathway. Hippocampus. 2017;27:784–793. doi: 10.1002/hipo.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng S., Zhang Y., Zhang J., Wang H., Ren B. ERK in learning and memory: A review of recent research. Int. J. Mol. Sci. 2010;11:222–232. doi: 10.3390/ijms11010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciccarelli A., Giustetto M. Role of ERK signaling in activity-dependent modifications of histone proteins. Neuropharmacology. 2014;80:34–44. doi: 10.1016/j.neuropharm.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 33.Palombo D.J., Amaral R.S., Olsen R.K., Müller D.J., Todd R.M., Anderson A.K., Levine B. KIBRA polymorphism is associated with individual differences in hippocampal subregions: Evidence from anatomical segmentation using high-resolution MRI. J. Neurosci. 2013;33:13088–13093. doi: 10.1523/JNEUROSCI.1406-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witte A.V., Köbe T., Kerti L., Rujescu D., Flöel A. Impact of KIBRA Polymorphism on Memory Function and the Hippocampus in Older Adults. Neuropsychopharmacology. 2016;41:781–790. doi: 10.1038/npp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R., Wan W., Li J. KIBRA polymorphism modulates gray matter volume to influence cognitive ability in the elderly. Brain Imaging Behav. 2020;14:1388–1394. doi: 10.1007/s11682-019-00047-1. [DOI] [PubMed] [Google Scholar]
- 36.Corneveaux J.J., Liang W.S., Reiman E.M., Webster J.A., Myers A.J., Zismann V.L., Joshipura K.D., Pearson J.V., Hu-Lince D., Craig D.W., et al. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol. Aging. 2010;31:901–909. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiman E.M., Chen K., Alexander G.E., Caselli R.J., Bandy D., Osborne D., Saunders A.M., Hardy J. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc. Natl. Acad. Sci. USA. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter T., Burnham S.C., Doré V., Savage G., Bourgeat P., Begemann K., Milicic L., Ames D., Bush A.I., Maruff P., et al. KIBRA is associated with accelerated cognitive decline and hippocampal atrophy in APOE ε4-positive cognitively normal adults with high Aβ-amyloid burden. Sci. Rep. 2018;8:2034. doi: 10.1038/s41598-018-20513-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stickel A., Kawa K., Walther K., Glisky E., Richholt R., Huentelman M., Ryan L. Age-Modulated Associations between KIBRA, Brain Volume, and Verbal Memory among Healthy Older Adults. Front. Aging Neurosci. 2018;9:431. doi: 10.3389/fnagi.2017.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling J., Huang Y., Zhang L., Wei D., Cheng W. Association of KIBRA polymorphism with risk of Alzheimer’s disease: Evidence based on 20 case-control studies. Neurosci. Lett. 2018;662:77–83. doi: 10.1016/j.neulet.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Zuo C., Wang D., Tao S., Hao L. Reduced Thalamus Volume and Enhanced Thalamus and Fronto-Parietal Network Integration in the Chess Experts. Cereb. Cortex. 2020;30:5560–5569. doi: 10.1093/cercor/bhaa140. [DOI] [PubMed] [Google Scholar]
- 42.Vinkhuyzen A.A., van der Sluis S., Posthuma D., Boomsma D.I. The heritability of aptitude and exceptional talent across different domains in adolescents and young adults. Behav. Genet. 2009;39:380–392. doi: 10.1007/s10519-009-9260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda Y., Hashimoto R., Ohi K., Fukumoto M., Takamura H., Iike N., Yoshida T., Hayashi N., Takahashi H., Yamamori H., et al. Association study of KIBRA gene with memory performance in a Japanese population. World J. Biol. Psychiatry. 2010;11:852–857. doi: 10.3109/15622971003797258. [DOI] [PubMed] [Google Scholar]
- 44.Valeeva E.V., Ahmetov I.I., Rees T. Sports, Exercise, and Nutritional Genomics. Academic Press; Cambridge, MA, USA: 2019. Psychogenetics and sport; pp. 147–165. [DOI] [Google Scholar]
- 45.Boulygina E.A., Borisov O.V., Valeeva E.V., Semenova E.A., Kostryukova E.S., Kulemin N.A., Larin A.K., Nabiullina R.M., Mavliev F.A., Akhatov A.M., et al. Whole genome sequencing of elite athletes. Biol. Sport. 2020;37:295–304. doi: 10.5114/biolsport.2020.96272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmetov I.I., Hall E.C.R., Semenova E.A., Pranckevičienė E., Ginevičienė V. Advances in sports genomics. Adv. Clin. Chem. 2022;107:215–263. doi: 10.1016/bs.acc.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Lounsbury J.W., Foster N., Patel H., Carmody P., Gibson L.W., Stairs D.R. An investigation of the personality traits of scientists versus nonscientists and their relationship with career satisfaction. R&D Manag. 2012;42:47–59. doi: 10.1111/j.1467-9310.2011.00665.x. [DOI] [Google Scholar]
- 48.Webb R.M., Lubinski D., Benbow C.P. Spatial ability: A neglected dimension in talent searches for intellectually precocious youth. J. Educ. Psychol. 2007;99:397–420. doi: 10.1037/0022-0663.99.2.397. [DOI] [Google Scholar]
- 49.Harris J.A. Measured intelligence, achievement, openness to experience, and creativity. Personal. Individ. Differ. 2004;36:913–929. doi: 10.1016/S0191-8869(03)00161-2. [DOI] [Google Scholar]
- 50.Anglim J., Dunlop P.D., Wee S., Horwood S., Wood J.K., Marty A. Personality and intelligence: A meta-analysis. Psychol. Bull. 2022;148:301–336. doi: 10.1037/bul0000373. [DOI] [Google Scholar]
- 51.Remoli T.C., Santos F.H. Interactions between working memory and creativity: A systematic review. Psicol. Estud. 2017;22:53–65. doi: 10.4025/psicolestud.v22i1.32518. [DOI] [Google Scholar]
- 52.Mitolo M., Gardini S., Caffarra P., Ronconi L., Venneri A., Pazzaglia F. Relationship between spatial ability, visuospatial working memory and self-assessed spatial orientation ability: A study in older adults. Cogn. Process. 2015;16:165–176. doi: 10.1007/s10339-015-0647-3. [DOI] [PubMed] [Google Scholar]
- 53.UK Biobank GWAS round 2 results [released 1st August 2018] [(accessed on 25 November 2022)]. Available online: http://www.nealelab.is/uk-biobank/
- 54.Okbay A., Wu Y., Wang N., Jayashankar H., Bennett M., Nehzati S.M., Sidorenko J., Kweon H., Goldman G., Gjorgjieva T., et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat. Genet. 2022;54:437–449. doi: 10.1038/s41588-022-01016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.