Abstract
Background
Circular RNAs (circRNAs) generated by back‐splicing of precursor mRNAs (pre‐mRNAs) are often aberrantly expressed in cancer cells. Accumulating evidence has revealed that circRNAs play a critical role in the progression of several cancers, including colorectal cancer (CRC). However, the current understandings of the emerging functions of circRNAs in CRC lipid metabolism and the underlying molecular mechanisms are still limited. Here, we aimed to explore the role of circCAPRIN1 in regulating CRC lipid metabolism and tumorigenesis.
Methods
circRNA microarray was performed with three pairs of tumor and non‐tumor tissues from CRC patients. The expression of circRNAs were determined by quantitative PCR (qPCR) and in situ hybridization (ISH). The endogenous levels of circRNAs in CRC cells were manipulated by transfection with lentiviruses overexpressing or silencing circRNAs. The regulatory roles of circRNAs in the occurrence of CRC were investigated both in vitro and in vivo using gene expression array, RNA pull‐down/mass spectrometry, RNA immunoprecipitation assay, luciferase reporter assay, chromatin immunoprecipitation analysis, and fluorescence in situ hybridization (FISH).
Results
Among circRNAs, circCAPRIN1 was most significantly upregulated in CRC tissue specimens. circCAPRIN1 expression was positively correlated with the clinical stage and unfavorable prognosis of CRC patients. Downregulation of circCAPRIN1 suppressed proliferation, migration, and epithelial‐mesenchymal transition of CRC cells, while circCAPRIN1 overexpression had opposite effects. RNA sequencing and gene ontology analysis indicated that circCAPRIN1 upregulated the expressions of genes involved in CRC lipid metabolism. Moreover, circCAPRIN1 promoted lipid synthesis by enhancing Acetyl‐CoA carboxylase 1 (ACC1) expression. Further mechanistic assays demonstrated that circCAPRIN1 directly bound signal transducer and activator of transcription 2 (STAT2) to activate ACC1 transcription, thus regulating lipid metabolism and facilitating CRC tumorigenesis.
Conclusions
These findings revealed the oncogenic role and mechanism of circCAPRIN1 in CRC. circCAPRIN1 interacted with STAT2 to promote CRC tumor progression and lipid synthesis by enhancing the expression of ACC1. circCAPRIN1 may be considered as a novel potential diagnostic and therapeutic target for CRC patients.
Keywords: circRNA, colorectal cancer, circCAPRIN1, lipid metabolism, STAT2, ACC1
List of abbreviations
- CRC
colorectal cancer
- circRNA
circular RNA
- miRNA
microRNA
- ACC
acetyl‐CoA carboxylase
- FASN
fatty acid synthase
- CPT1A
carnitine palmitoyltransferase 1A
- EMT
epithelial‐mesenchymal transition
- TOFA
5‐(Tetradecyloxy)‐2‐furoic acid
- lncRNA
long noncoding RNA
- TG
triglyceride
- STAT
signal transducer and activator of transcription
- JAK
Janus kinase
- PTK
protein tyrosine kinases
- IFN
interferon
- FUSCC
Fudan University Shanghai Cancer Center
- qPCR
quantitative polymerase chain reaction
- TMA
tissue microarrays
- FISH
fluorescence in situ hybridization
- IF
immunofluorescence
- TEM
transmission electron microscopy
- RIP
RNA immunoprecipitation
- ChIP
chromatin immunoprecipitation
- IHC
immunohistochemistry
- HE
hematoxylin‐eosin
- OS
overall survival
- DFS
disease free survival
- CCK‐8
Cell Counting Kit‐8
- GO
Gene Ontology
- FH
fumarate hydratase
- SDH
succinate dehydrogenase
- IDH
isocitrate dehydrogenase
- TGFβ1
transforming growth factor beta 1
- ACSL3
acyl‐CoA synthetase long‐chain 3
- FAO
fatty acid β‐oxidation
- ATP
adenosine triphosphate
- NADPH
nicotinamide adenine dinucleotide phosphate
- ZEB1
Zinc finger E‐box binding homeobox 1
- HFD
high‐fat diet
- CPT2
carnitine palmitoyltransferase 2
- ACBD3
acyl‐coenzyme A binding domain containing 3
- MBL
muscleblind
- FUBP1
far upstream element binding protein 1
- PRDX2
peroxiredoxin 2
- CDK2
cyclin‐dependent kinase 2
1. BACKGROUND
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer‐related mortality among 36 cancer types worldwide [1, 2]. Although oxaliplatin‐based chemotherapy is recommended as a standard treatment for CRC, the development of chemoresistance can cause recurrence and metastasis, leading to poor prognosis of CRC patients, which needs to be addressed urgently. The pathogenesis of CRC includes genetic alterations, epigenetic changes, metabolic disorders, and aberrant signaling pathways [3, 4, 5, 6]. Thus, elucidating the underlying molecular mechanisms and identifying the precise predictive biomarkers for CRC is essential for early diagnosis and optimized treatment of CRC, as well as for monitoring cancer recurrence and metastasis.
Circular RNAs (circRNAs) are non‐coding RNAs that are produced by back‐splicing of pre‐mRNA transcripts to form a covalently closed continuous loop without a 5’cap or a 3’poly‐A tail [7]. Compared to traditional linear RNAs, circRNAs are more stable in expression and resistant to RNase R treatment. The biological functions of circRNAs have been widely reported [8, 9, 10, 11, 12]. Accumulating evidence has indicated that circRNAs participate in tumor proliferation, invasion, migration, apoptosis, and differentiation [13, 14]. Several circRNAs exert their regulatory effects in CRC progression by sponging with specific microRNAs (miRNAs) or binding to specific proteins. For instance, circ3823 promotes CRC growth, metastasis, and angiogenesis through the circ3823/miR‐30c‐5p/ transcription factor 7 axis and may serve as a new diagnostic marker or treatment target for CRC patients [15]. circMEMO1 affects the expression of transcription factor 21 gene by modulating promoter methylation gene and regulates hepatocellular carcinoma progression by acting as a sponge for miR‐106b‐5p [16]. circMAPK1 encodes a tumor suppressor protein MAPK1‐109aa, which can inhibit the malignant biological behavior of gastric cancer [17]. circSPARC regulates the Janus kinase 2/signal transducer and activator of transcription 3 pathway involved in CRC development and may serve as a potential diagnostic and prognostic biomarker and a therapeutic target of CRC [18]. A previous study delineated a global reduction of circRNA abundance in CRC cell lines and a negative correlation between global circRNA abundance and CRC cell proliferation [19]. Emerging evidence has indicated that dysregulation of circRNAs can lead to metabolic disturbance by disrupting glucose and lipid homeostasis [20]. Recently, the role of circRNAs in glycolysis has been widely discussed [21, 22, 23, 24, 25]. However, the effect of circRNAs on lipid metabolism in CRC remains unclear.
Cancer cells have been well recognized to exhibit distinct metabolic patterns [26] and can utilize distinct metabolic pathways to support aberrant cellular replication, distant tumor metastasis, and immune evasion [27, 28]. Notably, in addition to aerobic glycolysis, tumor cells gain energy through lipid metabolism including fatty acid uptake [29], de novo lipid synthesis [26], and fatty acid β‐oxidation [30] to thrive in and challenge the tumor microenvironment. Additionally, hyperproliferative cancer cells can utilize fatty acids for post‐translational modification of oncogenic proteins and synthesizing cellular membrane during replication [31, 32]. Fatty acid biosynthesis from glucose in cancer cells, which is associated with cell growth, epithelial‐mesenchymal transition (EMT), and chemoresistance, is mainly controlled by the rate‐limiting enzymes acetyl‐CoA carboxylase (ACC) and fatty acid synthase (FASN) [33, 34, 35, 36]. 5‐(Tetradecyloxy)‐2‐furoic acid (TOFA), an allosteric inhibitor of ACC, can promote the carboxylation of acetyl‐CoA to malonyl‐CoA, thereby inhibiting fatty acid synthesis, lactate and pyruvate accumulation and CO2 release in isolated rat adipocytes [37]. Cerulenin, a non‐competitive inhibitor of FAS, prevents the condensation of malonyl‐CoA with acetyl‐CoA to elongate fatty acids, inhibits fatty acid synthesis, and induces apoptosis in cancer cells [38, 39]. Targeting the lipid metabolism pathways via inhibition of acetyl‐CoA carboxylase 1 (ACC1) significantly reduces cell proliferation and induces apoptosis in prostate tissue explants [35]. In addition, knockdown of long non‐coding RNA (lncRNA) TSPEAR‐AS2 reduced the triglyceride (TG) content and the expression of FASN and ACC1 and modulated the fatty acid metabolism in CRC cells [40]. Hitherto, the regulatory role of circRNAs in fatty acid biosynthesis and the correlation between circRNAs and ACC in CRC have not yet been elucidated.
The signal transducer and activator of transcription (STAT) family consists of transcription factors (STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6) which regulate the proliferation, apoptosis, angiogenesis and differentiation of cancer cells [41]. STATs are activated by receptors with intrinsic tyrosine kinase activity or by receptor‐associated tyrosine kinases like Janus kinases (JAKs) [42]. The JAK/STAT pathway always constitutes a rapid membrane‐to‐nucleus signaling module and regulates the expression of various critical mediators of cancer and inflammation. Dysregulation of the JAK/STAT pathway is associated with various cancers and autoimmune diseases [43]. STATs could be also activated by constitutively active non‐receptor protein tyrosine kinases (PTKs), including Brk, Bcr‐Abl, and c‐Src [44, 45]. Functionally, STAT3 and STAT5 play a role in cancer occurrence, progression and prognosis [46]. STAT1 is an essential component of interferon (IFN) signaling, mediating cellular functions in response to stimulations with cytokines such as IFNs and interleukin (IL)‐6, growth factors, and hormones [47]. STAT4 specifically mediates IL‐12 signaling, affecting a variety of immune cells [48]. STAT signaling plays an important role in modulating multiple upstream oncogenic pathways, and suppressing this signaling affects cancer cell growth [49, 50, 51]. STAT2 is necessary for the anti‐viral, immunomodulatory, anti‐apoptotic, and anti‐proliferative activities of IFN‐I, and exhibits tumor‐suppressive effects in mouse tumor models and cancer cell lines [52, 53].
In this study, we aimed to explore the clinical relevance of circCAPRIN1 (hsa_circ_0021639) and demonstrated its regulatory role in CRC tumorigenesis and the underlying mechanism, particularly its involvement in fatty acid biosynthesis.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
A total of 91 pairs of human CRC and adjacent non‐tumor tissues were collected from Fudan University Shanghai Cancer Center (FUSCC, Shanghai, China) between September 2020 and April 2021 for quantitative PCR (qPCR) analysis. Tissue microarrays (TMA) were collected for in situ hybridization (ISH) analysis from patients with CRC admitted to FUSCC between January 2007 and November 2009. This TMA was constructed as previously described [54]. All samples were obtained from surgical resections for CRC patients and reviewed by two independent pathologists from FUSCC. The clinicopathological data of TMA are summarized in Table 1. Overall survival (OS) was defined as the duration from surgery date to death due to any cause or the recent follow‐up. Disease‐free survival (DFS) was calculated from the date of surgery to disease relapse or the recent follow‐up.
TABLE 1.
Comparison of clinicopathological features according to circCAPRIN1 expression in 259 colorectal cancer patients
| circCAPRIN1 expression, n (%) | ||||
|---|---|---|---|---|
| Clinicopathological feature | Low expression (n = 106) | High expression (n = 153) | P value | |
| Age (years) | 0.900 | |||
| ≤60 | 65 (61.3) | 95 (62.1) | ||
| >60 | 41 (38.7) | 58 (37.9) | ||
| Sex | 0.746 | |||
| Male | 63 (59.4) | 94 (61.4) | ||
| Female | 43 (40.6) | 59 (38.6) | ||
| Location | 0.276 | |||
| Colon | 53 (50.0) | 66 (43.1) | ||
| Rectum | 53 (50.0) | 87 (56.9) | ||
| Histologic type | 0.179 | |||
| Adenocarcinoma | 103 (97.2) | 143 (93.5) | ||
| Mucinous | 3 (2.8) | 10 (6.5) | ||
| Differentiation | 0.042 | |||
| Poor | 14 (13.2) | 38 (24.8) | ||
| Moderate/ Well | 88 (83.0) | 106 (69.3) | ||
| Unknown | 4 (3.8) | 9 (5.9) | ||
| CEA status | 0.532 | |||
| Normal | 69 (65.1) | 89 (58.2) | ||
| Elevated | 33 (31.1) | 57 (37.3) | ||
| Unknown | 4 (3.8) | 7 (4.6) | ||
| T stage | 0.074 | |||
| T1* | 0 (0) | 0 (0) | ||
| T2 | 21 (19.8) | 20 (13.1) | ||
| T3 | 24 (22.6) | 24 (15.7) | ||
| T4 | 61 (57.5) | 109 (71.2) | ||
| N stage | 0.002 | |||
| N0 | 58 (54.7) | 55 (35.9) | ||
| N1 | 31 (29.2) | 46 (30.1) | ||
| N2 | 17 (16.0) | 52 (34.0) | ||
| M stage | 0.026 | |||
| M0 | 96 (90.6) | 123 (80.4) | ||
| M1 | 10 (9.4) | 30 (19.6) | ||
| Stage | 0.006 | |||
| I | 13 (12.3) | 8 (5.2) | ||
| II | 38 (35.8) | 36 (23.5) | ||
| III | 45 (42.5) | 79 (51.6) | ||
| IV | 10 (9.4) | 30 (19.6) | ||
| Perineural invasion | 0.197 | |||
| Absent | 94 (88.7) | 126 (82.4) | ||
| Present | 12 (11.3) | 26 (17.0) | ||
| Unknown | 0 (0) | 1 (0.6) | ||
| Vascular invasion | 0.059 | |||
| Absent | 80 (75.5) | 94 (61.4) | ||
| Present | 25 (23.6) | 56 (36.6) | ||
| Unknown | 1 (0.9) | 3 (2.0) | ||
T1 patients were excluded from analysis due to limited tumor size.
Abbreviations: CEA, carcinoembryonic antigen.
2.2. Cell lines and cell culture
Human CRC cell lines LoVo, HCT116, SW480, SW620, DLD1, RKO, and HCT8 were purchased from purchased from Cell Bank of Type Culture Collection of the Chinese Academy of Sciences in Shanghai, China and cultured in DMEM medium (Cytiva, Marlborough, USA) supplemented with 10% fetal bovine serum (Gibco, Invitrogen, USA) and 1% penicillin/streptomycin (Gibco, Invitrogen, USA), while the intestinal epithelial cell line NCM460 was purchased from purchased from Cell Bank of Type Culture Collection of the Chinese Academy of Sciences in Shanghai, China and cultured in RPMI 1640 medium (Cytiva, Marlborough, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. All cells were incubated at 37°C with 5% CO2.
siRNAs were transfected into CRC cells using Lipofectamine 3000 reagent (Invitrogen, California, USA) according to the manufacturer's protocol. The siRNA sequences targeting STAT2 were listed in the Supplementary Table S1.
HEK293T cells were co‐transfected with pLKO.1 (lncbio, Shanghai, China) or pLenti ciR6024 (lncbio, Shanghai, China) and the packaging plasmids psPAX2 (lncbio, Shanghai, China) and pMD2G (lncbio, Shanghai, China). The lentiviral particles were collected and filtered via a 0.45‐μm membrane (Merck Millipore, Darmstadt, Germany) after 72 h post‐transfection. The lentiviral particles and polybrene (10 μg/mL; Beyotime Biotechnology, Shanghai, China) were added to CRC cells, and infected cells were selected under puromycin (Beyotime Biotechnology, Shanghai, China) before harvest or subsequent experiments.
2.3. CCK‐8 and colony formation assays
For CCK‐8 assays, 1000 cells/well from different groups were seeded in 96‐well plates. CCK‐8 (Meilunbio, Dalian, China) reagent was added to each well at a fixed time each day for five days, and the cells were incubated at 37°C for 2 h. Then, the absorbance was measured in the single‐wavelength mode (450nm) by microplate reader (Biotek, Synergy H1, Vermont, USA).
For colony formation assays, 500 cells/well were seeded in 6‐well plates for each group. When visible colonies were formed, the cells were washed with phosphate‐buffered saline (PBS) and stained with 0.1% crystal violet for 5 min. Subsequently, the visible colonies were counted.
2.4. Cell migration assays
The wound healing and transwell assays were used to detect the migration ability of CRC cells. In the wound healing assay, RKO and LoVo cells (5 × 105) cells were cultured in six‐well plates and scratched with a sterile pipette tip when each well was filled with cells. After that, the cells were cultured in serum‐free DMEM medium and the wound size was measured at 0 and 24 h. In transwell assay, RKO and LoVo cells (5 × 104) suspending in 200 μL serum‐free DMEM medium were cultured in upper chambers, and 500 μL 10% FBS‐medium was filled in the lower chamber. After 24 h incubation, 0.1% crystal violet were used to stain the cells covered in the bottom chamber. After removing the cells in the upper chamber, the residual cells adhered to the bottom chambers were photographed.
2.5. RNA extraction and real‐time qPCR
Total RNAs were extracted from tissues and cells using TRIzol (Invitrogen, California, USA) and Isopropanol (Aladdin, Shanghai, China), and reverse transcription was performed using PrimeScript RT Master Mix (Takara, Osaka, Japan). Real‐time qPCR was performed using SYBR Premix (EZBioscience, Roseville, USA). The primers are listed in Supplementary Table S1.
2.6. Plasmid construction
HcircCAPRIN1 was PCR‐amplified and inserted into pLenti ciR6024 vector (lncbio, Shanghai, China). shRNA targeting circCAPRIN1 were generated and inserted into pLKO.1 vector (lncbio, Shanghai, China). The primers and shRNA sequences used in this study are summarized in Supplementary Table S1.
2.7. Actinomycin D and RNase R treatment
CRC cell lines were seeded in a 24‐well plate and cultured overnight. Total RNAs were extracted from cells using TRIzol (Invitrogen, California, USA) and Isopropanol (Aladdin, Shanghai, China) after actinomycin D (2 mg/mL; MedChemExpress, Shanghai, China) or DMSO (Solarbio, Beijing, China) treatment for different periods (0, 12, and 24 h). An equivalent of 5 μg RNA was incubated with or without RNase R (20 U; Thermo Fisher Scientific, Waltham, USA) at 37°C for different periods (0, 10, 20, 30, and 40 min). After treatment with actinomycin D or RNase R, the relative expression of circCAPRIN1 or CAPRIN1 mRNA was detected by PCR or qPCR. The sequences of indicated primer used in qPCR are summarized in Supplementary Table S1.
2.8. Oil Red O and Nile Red staining
CRC cells grown on coverslips were fixed with 4% (wt/vol) paraformaldehyde (PFA; Sangon Biotech, Shanghai, China) for 15 min and stained with 3 mg/mL Oil Red O solution (Yuanye, Shanghai, China) for 30 min. After nuclear counterstaining with hematoxylin (Yuanye, Shanghai, China), lipid droplet was visualized under a microscopy (Olympus, DP73, Tokyo, Japan) and evaluated using Image J software (National Institutes of Health, Bethesda, USA).
Similarly, CRC cells grown on coverslips were fixed with 4% (wt/vol) PFA for 15 min and stained with 2 μmol/L Nile Red solution (Solarbio, Beijing, China) for 30 min. Subsequently, the nuclei were stained with 4,6‐diamidino‐2‐phenylindole (DAPI; Thermo Fisher Scientific, Waltham, USA), and immunofluorescence signals were visualized under confocal microscopy (ZEISS, LSM880, Oberkochen, Germany).
2.9. Western blotting
Total proteins were extracted using Radio Immunoprecipitation Assay lysis buffer (Beyotime Biotechnology, Shanghai, China) supplemented with protease inhibitors (Selleck, Shanghai, China) and phosphatase inhibitors (Selleck, Shanghai, China). An aliquot of protein (40 μg) was separated by Sodium dodecyl‐sulfate polyacrylamide gel electrophoresis (SDS‐PAGE; 10%) and transferred to a PVDF membrane (Bio‐Rad Laboratories, Shanghai, China). The membranes were blocked with 5% milk and probed with the primary antibodies at 4°C overnight, followed by incubation with the secondary antibodies (1/5000; #7074 or #7076; Cell Signaling Technology) at room temperature for 1 h. The primary antibodies used in this study were as follows: CPT1A (1/1000; ab220789; Abcam), STAT2 (1/500; sc‐1668; Santa Cruz Biotechnology), Snail (1/1000; #3879; Cell Signaling Technology), N‐Cadherin (1/1000; #14215; Cell Signaling Technology), E‐Cadherin (1/1000; #14472; Cell Signaling Technology), ACC1 (1/1000; 21923‐1‐AP; Proteintech), ACC2 (1/1000; #8578; Cell Signaling Technology), GAPDH (1/1000; #2118; Cell Signaling Technology), p‐ACC (1/1000; #11818; Cell Signaling Technology), Vimentin (1/1000; #5741; Cell Signaling Technology), and FAS (1/1000; #4233; Cell Signaling Technology)., p‐STAT2 (1/1000; #ab191601; Abcam); TOFA (3.3 μg/mL; #S6690; Selleck); Cerulenin (10 μg/mL; #HY‐A0210; MedChemExpress). Then, the membranes were exposed with ECL reagent (Beyotime, Shanghai, China) for chemiluminescent detection.
2.10. Fluorescence in situ hybridization (FISH) and immunofluorescence (IF) staining
FISH was conducted to detect circCAPRIN1 in CRC cells using the FISH Probe Mix (green; GenePharma, Shanghai, China). First, RKO and LoVo cells were immobilized by 4% formaldehyde for 15 min, followed by incubation with FAM‐labeled probe against circCAPRIN1 at 37°C overnight. The sequence of the probe is listed in Supplementary Table S1. Second, the cells were incubated with antibodies specific for ACC1 (1/200; 21923‐1‐AP; Proteintech) and STAT2 (1/50; Santa Cruz Biotechnology, sc‐1668) at 4°C overnight, and then with Alexa Fluor 594 AffiniPure goat anti‐rabbit IgG (1:200; 111‐585‐003; Jackson ImmunoResearch, West Grove, PA, USA) and Alexa Fluor 647 AffiniPure goat anti‐mouse IgG (1:200; 115‐645‐003; Jackson ImmunoResearch, West Grove, PA, USA) at 37°C for 2 h. Subsequently, the slides were washed with PBS. Finally, the nuclei were stained with DAPI (Thermo Fisher Scientific, Waltham, USA), and the IF signal was visualized under a confocal microscopy (ZEISS, LSM880, Oberkochen, Germany).
2.11. In situ hybridization (ISH)
TMA was dewaxed in xylene and rehydrated sequentially with 100%, 95%, 85%, and 75% alcohol. Then, the tissues were hybridized with a specific digoxin‐labeled circCAPRIN1 probe (Servicebio, Wuhan, China). The sequence of the probe is listed in Supplementary Table S1. The expression of circCAPRIN1 was quantified and analyzed. The positive staining intensity (0, negative; 1, weak; 2, moderate; 3, strong) was multiplied by the percentage of positive staining cells (0, < 10%; 1, 10%‐25%; 2, 26%‐50%; 3, 51%‐75%; 4, > 75%) to calculate the score for ISH staining. The ISH score ≤ 8 defined low expression of circCAPRIN1, while > 8 indicated high expression of circCAPRIN1.
2.12. Transmission electron microscopy (TEM)
For TEM, CRC cells were washed with PBS and fixed with 2.5% pentadiol (Servicebio, Wuhan, China) before analysis at 120 kV using TEM (Hitachi HT7700). Briefly, CRC cell lines were fixed with 2.5% pentadiol (Servicebio, Wuhan, China) at room temperature for about 5 min. Then the cells were detached gently with cell scrapers and were centrifuged (no more than 3000 rpm/min) for about 2 min. Finally, CRC cells were resuspended with 2.5% pentadiol and were observed under TEM.
2.13. RNA pull‐down, silver staining, and mass spectrometry analysis
Biotin‐labeled probes were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). The sequence of the probe is listed in Supplementary Table S1. RNA pull‐down assay was performed using the Pierce Magnetic RNA‐Protein Pull‐Down Kit (Thermo Fisher Scientific, USA) according to the manufacturer's protocols. circCAPRIN1‐binding proteins were resolved on SDS‐PAGE (10%) and stained using Fast Silver Stain Kit (P0017S; Beyotime), following the manufacturer's instructions. Subsequently, the proteins were identified by mass spectrometry (Shanghai Applied Protein Technology Co., Ltd) and verified by Western blotting analysis. Liquid chromatography‐mass spectrometry analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific, Waltham, USA) coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 120 min. The mass spectrometer was operated in positive ion and peptide recognition modes. Mass spectrometry data was acquired using a data‐dependent top 10 method dynamically choosing the most abundant precursor ions from the survey scan (300‐1800 m/z) for high‐energy collisional dissociation HCD fragmentation. Automatic gain control (AGC) target was set to 3e6, and maximum inject time to 10 ms. Dynamic exclusion duration was 40.0 s. Survey scans were acquired at a resolution of 70,000 at 200 m/z and resolution for HCD spectra was set to 17,500 at 200 m/z, and isolation width was 2 m/z. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value that to could be reached at the maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
2.14. RNA immunoprecipitation (RIP) assay
STAT2 primary antibody (2 μg; sc‐1668; Santa Cruz Biotechnology) was selected for RIP assay using EZ‐Magna RIP Kit (Merck Millipore, Darmstadt, Germany). In brief, cell extracts were mixed with protein A/G beads conjugated to an antibody against STAT2 or IgG (negative control). Then, the precipitated RNAs were analyzed by qPCR analysis. The sequences of the primers used are summarized in Supplementary Table S1.
2.15. Dual‐luciferase reporter assay
The 2000 bp upstream of the ACC1 gene promoter was amplified using PCR and cloned into the pGL3‐basic vector (Promega, Madison, USA). The sequences of the primers used are summarized in Supplementary Table S1. An equivalent of 200 ng luciferase vector pGL3‐ACC1 or pGL3‐mut‐ACC1, 200 ng STAT2 overexpressing vector or negative control (NC) vector, and 15 ng plasmid expressing a renilla luciferase gene (pRL, Promega, Madison, USA) were transiently co‐transfected into HEK293T cells and the cells were cultured in a 24‐well plate. Then, the cells were lysed to detect the luciferase activity after 48 h post‐transfection using Dual‐Luciferase Reporter Assay System (Promega, Madison, USA) as instructions. The value of luciferase activity was detected by Synergy H1 microplate reader (Biotek, Vermont, USA). The relative firefly luciferase activity was normalized to renilla luciferase activity, and the fold changes in the reporters were calculated.
2.16. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using Pierce Agarose ChIP Kit (ThermoFisher Scientific, USA) to enrich 2000 bp upstream of ACC1 gene promoter region along with specific binding proteins. Primers were designed to amplify every 200 bp sequence (Supplementary Table S1) and detect the exact protein binding sites on or close STAT2 and ACC1 gene promoters. Briefly, RKO cells were fixed with 37% formaldehyde. The chromatin was fragmented by sonication on ice and immunoprecipitation was performed using antibodies against normal immunoglobulin G and STAT2 (sc‐1668; Santa Cruz Biotechnology). PCR products were separated and then visualized on a 1.5% agarose gel and then visualized under an UV light.
2.17. Immunohistochemistry (IHC) and hematoxylin‐eosin (HE) staining
For IHC assay, tissues were fixed with 4% PFA, embedded in paraffin, and cut into 4‐μM‐thick slices. The slices were baked for 24h at 58°C, deparaffinized in xylene, rehydrated in graded ethanol, quenched for endogenous peroxidase activity in 0.3% hydrogen peroxide for 15 minutes at 37°C, and subjected to antigen retrieval. Then, slices sections were blocked with 1% BSA/PBS (Sangon, Shanghai, China). Primary antibodies against ACC1 (1/400; 21923‐1‐AP; Proteintech), CPT1A (1/2000; ab220789; Abcam), and Ki‐67 (1/600, GB121141; Servicebio) were incubated overnight at 4°C, followed by secondary antibody incubation, and incubation with horseradish peroxidase‐labeled streptavidin for 15 min ‐. The immunocomplex was visualized with DAB, and the nucleus was counterstained with hematoxylin. For HE staining, a hematoxylin‐eosin staining kit (Beyotime, Shanghai, China) was employed to directly stain the nucleus and cytoplasm of cells.
2.18. Mice and animal experiments
The protocols of animal experiments involved in this study were reviewed and approved by the Animal Ethics Committee of FUSCC (permit number: 2019FUSCCJS‐092). Nude mice (4‐6 weeks old) weighing 14‐16 g were purchased from Shanghai Jie Si Jie Laboratory Animal Co., Ltd., (Shanghai, China) and bred under specific pathogen‐free conditions. LoVo and RKO cells transfected with the above‐mentioned lentiviruses were subcutaneously injected into 4‐week‐old nude mice to establish a mice xenograft tumor model. Each nude mouse was subcutaneously inoculated with 1 × 106 cells and measured the tumor volume was measured every one week. The mice were euthanized with carbon dioxide (CO2) 4 weeks later, then the subcutaneous tumors were harvested, weighed, and stained.
For in vivo metastasis assay, 1 × 106 HCT116 cells were injected into 5‐week‐old nude mice via tail vein. The bioluminescent signals of liver metastasis were observed using a specific imaging system (Caliper Life Sciences, USA) after 4‐5 weeks of injection. The metastatic lesions were assessed by HE staining.
2.19. circRNA microarray and RNA‐sequencing (RNA‐seq)
circRNA expression profiles of 3 paired CRC and adjacent cancer tissues were compared by using circRNA microarray. Total RNA from each sample was extracted using TRIzol (Invitrogen, CA, USA) following the manufacturer's instructions. The concentration and quality of total RNA were evaluated using NanoDrop ND‐1000 (Thermo Fisher Scientific, Waltham,USA). Total RNA was digested with RNase R (Thermo Fisher Scientific, Waltham, USA) to remove linear RNAs and enrich circRNAs, followed by circRNAs amplification. Arraystar Human circRNA Array (8 × 15K; Arraystar, Rockville, USA) was used to hybridized the circRNAs. The arrays were then scanned by the Agilent Scanner G2505C (Agilent Technologies, California, USA). Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, California, USA) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the R software limma package.
For RNA‐seq, strand‐specific RNA‐seq libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Beverly, MA) according to the manufacturer's instructions. The raw sequencing reads were filtered by FastQC, and compared using the spliced read aligner TopHat2. RNA‐sequencing data were analyzed by spliced read aligner HISAT2 (Dusseldorf, Germany), and gene ontology (GO) analysis was carried out for gene functional annotation. Genes with a fold change of > 2 and a P value of < 0.05 was considered to be significantly differentially expressed between human colorectal cancer tissues and adjacent tumor tissues, and between RKO control and RKO shcircCAPRIN1‐2/RKO shcircCAPRIN1‐3 colorectal cancer cell lines.
2.20. Statistical analysis
Statistical data were analyzed using SPSS 24.0 software (IBM, Armonk, NY, USA). Categorical variables were analyzed using chi‐squared test. Experimental data were analyzed using GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA, USA) and presented as mean ± standard deviation (SD). Student's t test was used to compare the statistical difference between two groups. Kaplan‐Meier curves were plotted using R (version 3.6.3), and the log‐rank test was employed to compare the survival differences. P < 0.05 indicated statistical significance.
3. RESULTS
3.1. circCAPRIN1 was associated with CRC
To identify the circRNAs involved in the tumorigenesis of CRC, circRNA microarray was performed using three pairs of tumor and non‐tumor tissues from CRC patients. Differentially expressed circRNAs were identified, with a fold‐change ≤ 0.5 and ≥ 2, and a P value < 0.05 (Figure 1A). Among these, circCAPRIN1 (hsa_circ_0021639) was most significantly upregulated in CRC tissues. CircCAPRIN1 derived from exons 8‐12 of the CAPRIN1 gene on human chromosome (chr) 11 was a sense‐overlapping circular transcript with 467 bp (Figure 1B). The head‐to‐tail splicing of the amplified circCAPRIN1 was verified by Sanger sequencing (Figure 1C). To substantiate that the loop structure of circCAPRIN1 was generated from back‐splicing of pre‐mRNA, we designed divergent and convergent primers for circCAPRIN1 amplification using complementary DNA (cDNA), and genomic DNA (gDNA) extracted from RKO cells as templates. As expected, circCAPRIN1 could be amplified only by using divergent primers and cDNA rather than gDNA as template (Figure 1D). FISH assay revealed that circCAPRIN1 was predominantly located in the nucleus but not cytoplasm in RKO and LoVo cells and upregulated in CRC tissues compared to normal tissues (Figure 1E). Next, the stability of circCAPRIN1 was evaluated. After treating with a transcription inhibitor actinomycin D, the half‐life of circCAPRIN1 was significantly prolonged compared with linear CAPRIN1, as shown by PCR and qPCR results (Figure 1F‐G). Similarly, circCAPRIN1 was more stable than linear CAPRIN1 following RNase R treatment (Figure 1H‐I).
FIGURE 1.
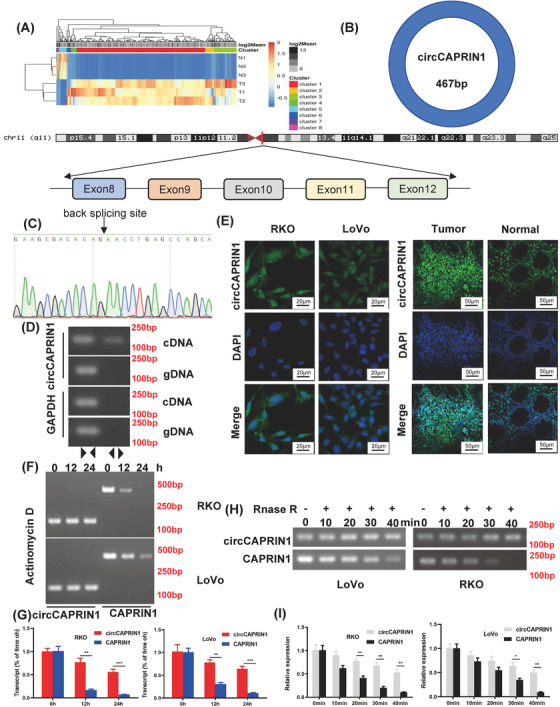
CircCAPRIN1 was identified and verified in CRC cells.
(A) Heatmap showed differentially dysregulated circRNAs in CRC tissues compared to adjacent normal tissues. Red and green represent upregulated and downregulated circRNAs, respectively. (B) Schematic of circCAPRIN1 arose from exon 8‐12 of the CAPRIN1 gene.
(C) Sanger sequencing was conducted to confirm head‐to‐tail splicing.
(D) The existence of circCAPRIN1 was detected with divergent and convergent primers from cDNA and gDNA, GAPDH was used as a negative control.
(E) The localization of circCAPRIN1 was explored in CRC cells and tissues with FISH.
(F‐G) Expression of circCAPRIN1 and CAPRIN1 was measured by RT‐PCR (F) and qPCR (G) at different time points in CRC cells treated with Actinomycin D.
(H‐I) Expression of circCAPRIN1 and CAPRIN1 was measured by RT‐PCR (H) and qPCR (I) at different time points in CRC cells treated with RNase R.
Abbreviations: CRC, colorectal cancer; FISH, fluorescence in situ hybridization.
3.2. Upregulated circCAPRIN1 in CRC predicted poor prognosis
To further investigate the correlation between circCAPRIN1 expression and CRC, we first tested circCAPRIN1 expression in CRC tissues and cell lines using qPCR. The results showed that circCAPRIN1 expression was upregulated in CRC tissues compared to the paired adjacent normal tissues, which also verified our previous microarray findings (Figure 2A). Similarly, circCAPRIN1 expression in cultured CRC cell lines (LoVo, HCT116, SW480, SW620, DLD1, and RKO) was significantly higher than that in intestinal epithelial cell line (NCM460) (Figure 2B). Subsequently, the expression of circCAPRIN1 in CRC tissues was determined by ISH using human TMA (Figure 2C). The circCAPRIN1 ISH score in CRC tissues was significantly higher than that in normal tissues (Figure 2D), and was positively correlated with tumor‐node‐metastasis (TNM) stage and tumor grade (Figure 2E and Table 1). Patients were divided into the high‐ and low‐expression groups according to the circCAPRIN1 ISH score (low expression: ISH score ≤ 8; high expression: ISH score > 8) (Figure 2F). Patients with high circCAPRIN1 expression had unfavorable OS (P = 0.006) and DFS (P = 0.012) (Figure 2G). These results indicated an oncogenic role of circCAPRIN1 in promoting CRC progression.
FIGURE 2.
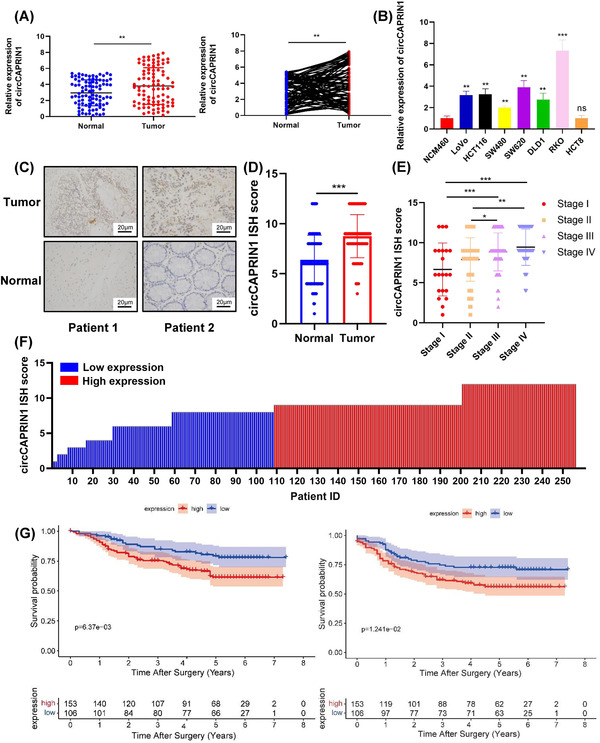
CircCAPRIN1 is upregulated in CRC, and the expression is correlated with tumor progression and patient prognosis.
(A) Relative expression of circCAPRIN1 was detected in 91 paired CRC and adjacent tissues by qPCR.
(B) Relative expression of circCAPRIN1 was determined in the intestinal epithelial cell line (NCM460) and CRC cell lines.
(C) Representative images revealed circCAPRIN1 expression in CRC and adjacent tissues detected by ISH.
(D) The ISH staining scores were compared between CRC and adjacent tissues.
(E) The ISH staining scores were compared among patients at different stages.
(F) The ISH staining score was presented individually. ISH staining score ≤ 8 was regarded as low expression, while a score ≥ 9 was regarded as high expression.
(G) Kaplan–Meier survival curve was plotted to compare over the OS (left panel, P = 0.006) and DFS (right panel, P = 0.012) between patients with low and high expression.
Abbreviations: CRC, colorectal cancer; ISH, In situ hybridization; OS, overall survival; DFS, disease‐free survival.
3.3. circCAPRIN1 promoted the proliferation, migration, and EMT of CRC cells in vitro
In order to investigate the biological function of circCAPRIN1 in CRC development, we used shRNAs to knock down circCAPRIN1 expression in RKO and LoVo cells. The results of qPCR analysis showed that sh‐circCAPRIN1‐2 (sh‐circ‐2) and sh‐circCAPRIN1‐3 (sh‐circ‐3) significantly downregulated the expression of circCAPRIN1 instead of CAPRIN1 (Figure 3A). FISH assay confirmed the reduction in circCAPRIN1 amount in CRC cells (Supplementary Figure S1). Subsequent Cell Counting Kit‐8 (CCK‐8) and colony formation assays demonstrated that decreased circCAPRIN1 expression could lead to significant inhibition of RKO and LoVo cell proliferation (Figure 3B‐C). Furthermore, wound healing and transwell assays showed that the deletion of circCAPRIN1 impaired CRC cell migration (Figure 3D‐E). Additionally, Western blotting analysis confirmed that downregulation of circCAPRIN1 was accompanied with increased expression of E‐cadherin protein but decreased expression of N‐cadherin, Vimentin, and Snail proteins (Figure 3F), indicating that circCAPRIN1 could promote EMT of CRC cells. As expected, the overexpression of circCAPRIN1 did not affect CAPRIN1 expression (Supplementary Figure S2A‐B) but facilitated the proliferation ability and EMT of CRC cells (Supplementary Figure S2C‐D).
FIGURE 3.
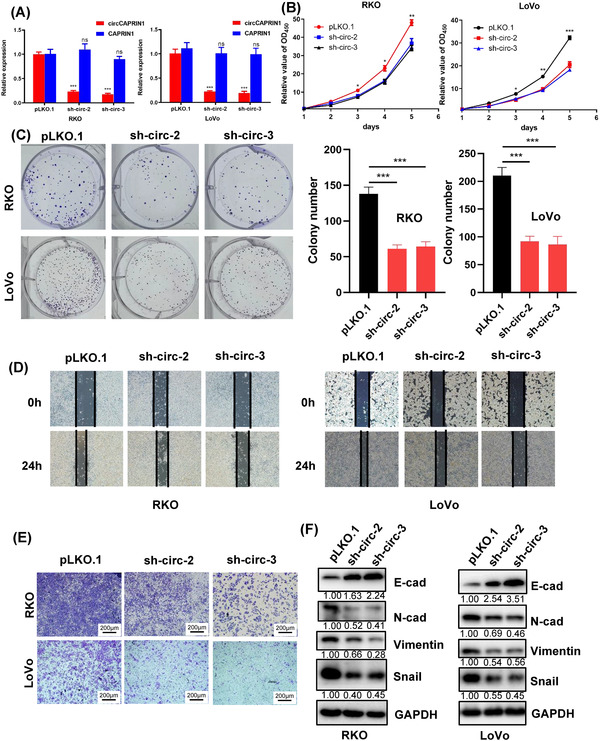
CircCAPRIN1 knockdown in CRC cells impairs the ability of cell proliferation and migration.
(A) Relative expressions of circCAPRIN1 and CAPRIN1 were determined by qPCR in CRC cells transfected with shcircCAPRIN1 or control.
(B‐C) CCK‐8 assay (B) and colony formation assay (C) (left panel) were performed to evaluate the proliferation ability of CRC cells after circCAPRIN1 knockdown, and the number of colonies was counted (right panel).
(D‐E) Wound healing assay (D) and transwell assay (E) were performed to evaluate the migration ability of CRC cells after circCAPRIN1 knockdown.
(F) Expression of EMT‐related proteins was detected by Western blotting in CRC cells transfected with shcircCAPRIN1 or control.
Abbreviations: qPCR, quantitative PCR; CRC, colorectal cancer; CCK‐8, Cell Counting Kit‐8; EMT, epithelial‐mesenchymal transition.
3.4. circCAPRIN1 knockdown inhibited the expression of genes involved in adipogenesis
To gain an in‐depth insight into the molecular function of circCAPRIN1 in CRC progression, RNA sequencing was performed to depict the gene expression profile of RKO cells following circCAPRIN1 knockdown. We then conducted GO analysis for mRNA‐encoding genes to identify the potential functions of circRNAs in the occurrence and development of CRC. The enriched GO terms including biological process, cellular component, and molecular function are shown in Figure 4A. Intriguingly, adipogenesis was regulated by circCAPRIN1. According to the results of TEM and Oil Red O and Nile Red staining, downregulation of circCAPRIN1 was accompanied with decreased lipid droplets (Figure 4B‐D). Overexpression of circCAPRIN1 resulted in enhanced lipid droplets, as shown by Nile Red staining (Supplementary Figure S2E). To demonstrate the relationship between circCAPRIN1‐dependent lipogenesis and EMT in CRC, we treated circCAPRIN1‐overexpressed RKO and LoVo cells with TOFA (3.3 μg/mL) or Cerulenin (10 μg/mL), respectively. Colony formation assay and transwell assays indicated that overexpression of circCAPRIN1 could promote proliferation and migration ability of CRC cells, and inhibition of ACC or FASN reversed this promoting effect (Supplementary Figure S3A‐B). Similarly, Western blotting conformed that inhibition of ACC or FASN increased the expression of E‐cadherin protein but decreased the expression of N‐cadherin, Vimentin, and Snail proteins in circCAPRIN1‐overexpressed cells (Supplementary Figure S3C).
FIGURE 4.

CircCAPRIN1 knockdown inhibits adipogenesis by downregulating ACC1 expression.
(A) GO analysis showed that adipogenesis was regulated by CircCAPRIN1.
(B‐D) The lipid droplets in RKO and LoVo cells was detected by electron microscopy (B), Oil Red O (C), and Nile Red (D) (left panel), and the lipid droplet areas of different groups were counted (right panel).
(E) Expression of ACC1, ACC2, p‐ACC, and FAS was detected by Western blotting in CRC cells transfected with shcircCAPRIN1 or control.
Abbreviations: GO, gene ontology.
Next, we searched for the lipid metabolism enzymes that mediated the role of circCAPRIN1 in adipogenesis. The data of RNA sequencing revealed that ACC1 expression was positively correlated with circCAPRIN1 level (Supplementary Figure S4). Western blotting showed that ACC1 was significantly downregulated following circCAPRIN1 depletion, while the expressions of ACC2, p‐ACC, and FAS were not affected (Figure 4E), indicating that ACC1 might be a key target gene of circCAPRIN1. Moreover, the expression of CPT1A, a downstream protein of ACC1 signaling, was decreased as a result of circCAPRIN1 knockdown (Supplementary Figure S5A). To further confirm whether circCAPRIN1 promotes fatty acid synthesis via upregulating ACC1 expression, RKO and LoVo cells overexpressing ACC1 and circCAPRIN1 knockdown were established (Supplementary Figure S5B). Functional assays showed that ACC1 overexpression reversed the suppressive effects of circCAPRIN1 knockdown on CRC cell proliferation and fatty acid synthesis in these tumor cells (Supplementary Figure S6).
3.5. circCAPRIN1 interacted with STAT2 to transcriptionally activate ACC1
Since circCAPRIN1 is mainly localized in the nucleus, it might regulate transcription via interaction with specific transcription factors, thereby affecting ACC1 expression and tumor progression. Next, we conducted RNA pull‐down assay, silver staining, and mass spectrometry analysis to identify circCAPRIN1‐binding proteins (Figure 5A). Consistent with our hypothesis, STAT2, an interferon‐regulated transcription factor, was significantly enriched in the circCAPRIN1 group instead of the lgG group (Figure 5B). FISH assay showed that circCAPRIN1 and STAT2 were colocalized in the nucleus (Figure 5C). Furthermore, RIP assay substantiated that STAT2 specifically bound to circCAPRIN1 in CRC cells, as indicated by both PCR and qPCR analyses (Figure 5D). Notably, circCAPRIN1 knockdown did not affect STAT2 expression (Figure 5E). Additionally, siRNA‐mediated STAT2 knockdown resulted in decreased ACC1 level in CRC cells, suggesting that ACC1 expression was regulated by STAT2 (Figure 5F‐G). Moreover, according to the GTEx database, a positive correlation between ACC1 and STAT2 levels was established in colon tissues (Supplementary Figure S7A). As expected, ACC1 expression was upregulated in colon cancer tissues compared with normal tissues according to the results from the TCGA and GTEx databases (Supplementary Figure S7B).
FIGURE 5.
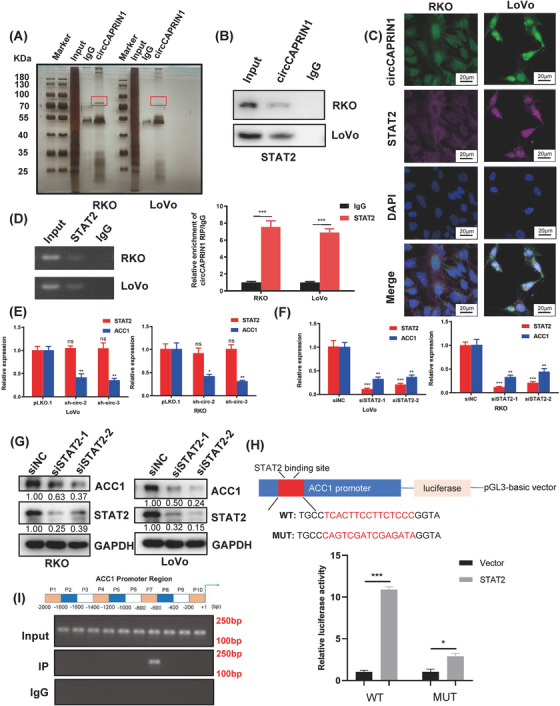
CircCAPRIN1 recruits STAT2 to activate ACC1 transcriptionally.
(A) RNA pull‐down assay was conducted using the specific biotin‐labeled circCAPRIN1 probe in RKO and LoVo cells, followed by silver staining and mass spectrometry.
(B) Western blotting proved that circCAPRIN1 interacts with STAT2.
(C) FISH assay of CRC cells demonstrated that circCAPRIN1 and STAT2 are colocalized in the nucleus.
(D) RIP assay was performed in RKO and LoVo cells using anti‐STAT2 or anti‐IgG, and the enrichment of RNA was detected by PCR (left panel) and qPCR (right panel).
(E) Relative expression of STAT2 and ACC1 was detected by qPCR in CRC cells transfected with shcircCAPRIN1 or control.
(F) Relative expressions of STAT2 and ACC1 were detected by qPCR in CRC cells transfected with siSTAT2 or control.
(G) Expression of ACC1 and STAT2 was detected by Western blotting in CRC cells transfected with siSTAT2 or control.
(H) The binding site of the ACC1 promoter was predicted (upper panel), and a luciferase reporter assay was performed to confirm the correlation between STAT2 and ACC1 (lower panel).
(I) PCR results of ChIP analysis showed that STAT2 binds to the specific region of the ACC1 promoter region.
Abbreviations: qPCR, quantitative PCR; CRC, colorectal cancer; FISH, fluorescence in situ hybridization; RIP, RNA immunoprecipitation; ChIP, chromatin immunoprecipitation.
Next, the binding site of ACC1 was predicted, and a luciferase reporter assay was performed to confirm the correlation between STAT2 and ACC1 (Figure 5H). Compared with the control group, STAT2 overexpression significantly increased the luciferase activity of wild‐type ACC1 and had a minimal effect on mutant ACC1, wherein STAT2 binding site was mutated. To identify the precise binding site of STAT2 on ACC1 gene promoter, we performed a ChIP assay in RKO cells. One STAT2 binding site was located at approximately 600‐800 bp upstream of the open reading frame of ACC1 (Figure 5I). Moreover, the overexpression of STAT2 partially reversed the downregulation of ACC1 gene expression upon circCAPRIN1 knockdown and counteracted the inhibitory effect of circCAPRIN1 knockdown on the proliferation, migration, and EMT of CRC cells (Supplementary Figure S7C‐E). Collectively, these data suggested that circCAPRIN1 recruits STAT2 to transcriptionally activate ACC1 gene expression in CRC cells.
3.6. circCAPRIN1 promoted CRC cell growth and metastasis in vivo
To determine the role of circCAPRIN1 in CRC in vivo, RKO and LoVo cells with stable knockdown of circCAPRIN1 were inoculated subcutaneously into nude mice. Tumor volume was measured every week. circCAPRIN1 silencing dramatically inhibited tumor growth (Figure 6A‐C). IHC staining for xenograft tumors displayed weaker Ki67 staining of the tumor tissues derived from stable knockdown of circCAPRIN1 compared to the controls. Moreover, we determined the effects of circCAPRIN1 knockdown on ACC1 signaling and its downstream molecular CPT1A by IHC staining. As expected, ACC1 and CPT1A labeling was weaker after knocking circCAPRIN1 (Figure 6D).
FIGURE 6.

Oncogenic role of circCAPRIN1 in vivo.
(A) The indicated tumor cells were injected subcutaneously into nude mice, and xenograft tumors were collected after 4 weeks.
(B) Tumor weight was measured in different groups.
(C) Tumor volume was measured every week in different groups.
(D) The tumor tissues were stained with anti‐Ki67, ACC1, and CPT1A antibodies.
(E) Bioluminescence imaging of metastases after injecting tumor cells into the tail vein of nude mice.
(F) Representative macroscopic (upper panel) and H&E staining (lower panel) images of the liver derived from different groups.
Abbreviations: H&E, hematoxylin‐eosin.
Next, we explored the metastasis‐promoting potential of circCAPRIN1 in vivo. HCT116 cell lines stably transfected with a luciferase‐encoding plasmid were and injected into nude mice via tail vein. Metastatic tumors of mice were assessed using the IVIS Spectrum Imaging System. The results demonstrated that circCAPRIN1 knockdown impaired the metastatic ability of CRC cells (Figure 6E‐F). Taken together, it could be deduced that circCAPRIN1 inhibited CRC cell growth and metastasis in vivo. Schematic of biogenesis, function, and regulatory mechanism of circCAPRIN1 in CRC cells is shown in Figure 7.
FIGURE 7.

Schematic of biogenesis, function, and regulatory mechanism of circCAPRIN1 in CRC cells.
Abbreviations: CRC, colorectal cancer.
4. DISCUSSION
Recent advances in high‐throughput sequencing and integrated bioinformatics analysis have enabled the dissection of a large amount of circRNAs in mammalian cells. Emerging evidence has suggested that abnormal expression of circRNAs plays a key role in tumorigenesis. Herein, we identified hsa_circ_0021639, also termed circCAPRIN1, as a CRC‐related circRNA through circular RNA microarray analysis. The expression of circCAPRIN1 was abnormally upregulated in CRC and positively correlated with the pathological stage of CRC. High expression of circCAPRIN1 was associated with poor prognosis, and upregulated circCAPRIN1 promoted the proliferation and migration of CRC cells both in vitro and in vivo. Mechanistically, circCAPRIN1 might interact with STAT2 to promote the expression of ACC1, thus enhancing lipid synthesis in CRC cells and promoting tumorigenesis.
Based on RNA‐Seq and GO analysis, we found that circCAPRIN1 could target ACC1 gene to regulate adipogenesis. We also observed that ACC1 overexpression could partially reverse the suppressive effect of silencing circCAPRIN1 on fatty acid synthesis in CRC cells. Metabolic alterations in cancer cells are well‐documented and regarded as the hallmark of carcinogenesis. EMT is considered as an important process during organ development and embryogenesis and is causally linked to cancer invasion and metastasis [55, 56, 57]. Cancer cells always rewire their metabolic pathways to satisfy their requirement for (adenosine triphosphate) ATP production. Many studies have indicated that metabolism and EMT are intertwined [58, 59]. While metabolic alterations possibly induce EMT, EMT may also lead to metabolic changes [60]. Except for glycolysis, TCA cycle, one of the main metabolic pathways within mitochondria, plays a key role in tumorigenesis, providing important intermediates for biosynthesis of lipids, nucleic acid and amino acids [61]. Several enzymes involved in the TCA cycle including fumarate hydratase (FH), succinate dehydrogenase (SDH), and isocitrate dehydrogenase (IDH), have been shown to be mutated or functionally impaired in cancer patients [62, 63, 64, 65]. Breast cancer cell lines with low SDH expression and hepatocellular carcinoma cell lines with reduced SDH level are characterized by increased expression of EMT markers, indicating a linkage between EMT and SDH [66, 67]. In colorectal cancer cell lines, 2‐hydroxyglutarate induced (Zinc finger E‐box binding homeobox 1) ZEB1 expression by modifying trimethylation of lysine 4 on histone H3 (H3K4me3) in the promoter region of ZEB1 gene [68].
Little is known about the regulatory role of lipogenesis genes in EMT. Transforming growth factor beta 1 (TGFβ1) has been reported to induce the up‐regulation of acyl‐CoA synthetase long‐chain 3 (ACSL3) expression promoting metabolic reprogramming in colorectal carcinoma (CRC) cells, ACSL3 also mediates EMT and metastasis of CRC cells by activating the fatty acid β‐oxidation (FAO) pathway, which stimulates ATP production and inhibits the level of nicotinamide adenine dinucleotide phosphate (NADPH) [69]. Previous study has shown that TGFβ and leptin can inhibit the activity of ACC1 through AMPK‐catalyzed phosphorylation of ACC1 at Ser79, thereby promoting EMT, ACC1 inhibition combined with concomitantly increased acetylation of SMAD2, which is triggered by accumulated acetyl‐CoA, mediates TGFβ‐induced EMT [70]. Furthermore, enhanced FASN expression in cisplatin resistant non‐small cell lung cancer cells promoted EMT through TGFβ signaling [71].
Enhanced fatty acid synthesis and lipid metabolism support the growth and division of cancer cells, interfering with diverse cellular processes. In our study, we examined the effects of TOFA and Cerulenin treatment on CRC cells. Inhibited lipogenesis has been reported to cause the suppressing of proliferation and migration in CRC cells. For instance, high‐fat diet (HFD) drove colorectal tumorigenesis through inducing gut microbial dysbiosis, metabolomic dysregulation with elevated lysophosphatidic acid, and gut barrier dysfunction in mice [72]. FASN is a key enzyme responsible for de novo fatty acid synthesis and is often highly expressed in cancer cells. FASN regulates colorectal cancer invasion and metastasis by influencing lipid droplet accumulation via Wnt signaling pathway [73]. Consistently, inhibition of FASN suppresses the malignant phenotype of colorectal cancer cells by down‐regulating lipid droplet accumulation and inhibiting mTOR signaling pathway [74]. ACC1 is a key lipogenic enzyme linked to EMT and invasion‐promoting pathways in cancer cells [70]. Moreover, ACC1 together with FASN control cell growth and apoptosis in HCT‐116 cells, suggesting that the pharmacologic inhibitors of either enzyme maybe useful in eradicating cancer cells [75]. Recently, the crosstalk between circRNAs and lipid metabolism has attracted increasing attention. For instance, circRNA_0046367 sponges miR‐34a to protect PPARα, degrading lipids via upregulating expressions of CPT2 and ACBD3 [76]. circNF1‐419,which regulates glycerophospholipid metabolic pathway and retrogrades endocannabinoid signaling and promotes cell apoptosis, is a potential therapeutic target for treating astroglioma [77]. The current findings indicated that the circCAPRIN1 regulated to CRC cell proliferation and migration via ACC1‐dependent metabolic reprogramming.
Strikingly, a large number of circRNAs are located in the cell cytoplasm, deeming as miRNA sponges [78]. CiRS‐7 is a well‐characterized circRNA with more than 70 conserved binding sites with miR‐7, rendering it the potential capability to regulate the target genes of miR‐7 [79]. Additionally, a small subset of endogenous circRNAs, primarily located in the cytoplasm, are translated into proteins. For instance, circ‐ZNF609 is translated in a splicing‐dependent and cap‐independent manner, based on its association with heavy polysomes [80]. A proportion of circRNAs are expressed in the nucleus and act as protein sponges to interact with RNA‐binding proteins, serving as decoys or scaffolds to mediate protein complex formation and regulate protein function. Accumulating evidence has demonstrated that circRNAs regulate gene expression. circMbl, a circRNA derived from muscleblind (MBL) gene that and harbors a binding site for MBL, regulates gene expression by competing with linear splicing [81]. Similarly, Wang et al. [82] reported that circACTN4 activated MYC transcription by recruiting FUBP1 to disrupt the binding between FUBP1 and FIR. Besides, circRNAs also serve as protein scaffolds to facilitate or block protein complex functions. circ‐Amotl1 physically recruits both PDK1 and AKT1, facilitating PDK1‐dependent phosphorylation of AKT1 [83]. circDIDO1 specifically binds to PRDX2 and promotes RBX1‐mediated ubiquitination and degradation of PRDX2, thereby inactivating PRDX2‐dependent signaling pathways [84]. circ‐Foxo3 is a protein scaffold that binds to CDK2 and p21 to form a circ‐Foxo3‐p21‐CDK2 ternary complex, which arrests the function of CDK2 and blocks the cell cycle progression [85]. Furthermore, circRNAs may exert biological functions by recruiting specific proteins to certain cellular locations. For instance, circRNAFECR1 promotes demethylation of CpG sites of FLI1 gene and to activate its transcription by recruiting TET1 to its promoter region [86]. The present study revealed that circCAPRIN1 was primarily located in the nucleus, although less expression was observed in cell the cytoplasm. The mechanisms of the action of circRNA in CRC were investigated by determining their subcellular locations. Herein, we speculated that circCAPRIN1 upregulated ACC1 expression at the transcriptional level. Using RNA pull‐down assay and mass spectrometry analysis, we demonstrated that STAT2 as a circCAPRIN1‐binding protein was recruited to the promoter region of ACC1 to regulate its expression.
STAT2 is a member of the STAT family that plays a critical role in immune responses to extracellular and intracellular stimuli, including cancer initiation, inflammatory reactions, and tumor cell invasion [87]. IFN‐I‐activated transcriptional factors, including STAT1 and STAT2, drive increased PRL‐3‐induced glycolysis in multiple myeloma [88]. LncRNA DLX6‐AS1 regulates cell proliferation, cell cycle, and glycolysis in vitro and tumor growth in vivo. It promotes neuroblastoma development by upregulating STAT2 expression via targeting miR‐506‐3p [89]. STAT2 is also identified as a novel contributor to colorectal and skin carcinogenesis, and upregulates the expression and secretion of proinflammatory mediators that activate the oncogenic STAT3 signaling pathway [90]. In the present study, we found that circCAPRIN1 did not affect STAT2 expression, while STAT2 directly regulated ACC1 level. Moreover, overexpression of STAT2 partially reversed the inhibitory effect of knocking down circCAPRIN1 on the proliferation and migration of CRC cells in an ACC1‐dependent manner. Taken together, our results demonstrated that circCAPRIN1 interacted with STAT2 to activate ACC1 gene transcription, thereby enhancing lipid metabolism and promoting CRC progression.
circRNAs play a vital role in CRC tumorigenesis and progression, and are involved in CRC cell proliferation, metastasis, immune escape and drug resistance [91, 92, 93, 94, 95]. Recent studies have indicated that circRNAs can not only be detected in tumor tissues but also in exosomes, blood, saliva and urine [96, 97, 98]. Differential expressions of circRNAs in various pathological states indicated that circRNAs could be considered as promising diagnostic and therapeutic biomarker for diverse cancer types [99, 100, 101]. Like other circRNAs, circCAPRIN1 holds great potential as a novel attractive diagnostic biomarker for CRC due to its resistance to RNase R digestion and their stability during circulation. Recently, circRNA‐based therapy has attracted increasing attention. Compared with conventional drugs, such as monoclonal antibodies or small molecule inhibitors, circRNA‐based therapy has several advantages, including a long half‐life of circRNAs, less drug resistance, and more precise delivery to specific lesions [102]. In the present study, the regulatory function of circCAPRIN1 in CRC was dissected. Inhibiting lipid synthesis via targeting circCAPRIN1‐regulated pathways will be expected to improve the poor prognosis of CRC patients with high circCAPRIN1 expression.
Studies have suggested that the generation of circRNAs from precursor‐mRNAs requires spliceosomal machinery and is dependent on cis‐regulatory elements and trans‐acting factors [7]. However, little is known about the regulation of back‐splicing circularization to form circCAPRIN1. Thus, further studies are needed to clarify the exact molecular mechanism underlying circCAPRIN1 biogenesis.
5. CONCLUSIONS
We identified a novel circRNA, termed circCAPRIN1, that was upregulated in human CRC tissues and CRC cell lines, and a high level of circCAPRIN1 was associated with poor prognosis of CRC patients. Herein, we illustrated that the circCAPRIN1/STAT2 complex promoted CRC progression by facilitating lipid synthesis in an ACC1‐dependent manner. These findings implicated circCAPRIN1 to be a novel prognostic biomarker and a promising therapeutic target for CRC.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Xinxiang Li, Ziliang Wang and Qingguo Li designed research concept. Yufei Yang conducted experiments and drafted the manuscript. Dakui Luo and Yang Shao analyzed data. Zezhi Shan, Qi Liu, Junyong Weng, Weijing He and Ruoxin Zhang offered technical support. Xinxiang Li, Ziliang Wang and Qingguo Li supervised the study and edited the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
FUNDING
This work was supported by the National Natural Science Foundation of China (81972260 and 82103259), Shanghai Municipal Natural Science Foundation (21ZR1414400), and Shanghai Medical Innovation Research Project (22Y11907600).
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study and the plasmids used in this study are available from the corresponding author upon reasonable request.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Ethical Committee and Institutional Review Board of Fudan University Shanghai Cancer Center (permit number: 050432‐4‐2108). All subjects in our study have written informed consent. The animal study was carried out in compliance with the guidance suggestion of Animal Care Committee of Fudan University Shanghai Cancer Center (permit number: 2019FUSCCJS‐092).
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
Not applicable.
Yang Y, Luo D, Shao Y, Shan Z, Liu Q, Weng J, et al. circCAPRIN1 interacts with STAT2 to promote tumor progression and lipid synthesis via upregulating ACC1 expression in colorectal cancer. Cancer Commun. 2023;43:100–122. 10.1002/cac2.12380
Yufei Yang, Dakui Luo and Yang Shao contributed equally to this work.
Contributor Information
Qingguo Li, Email: qingguoli@fudan.edu.cn.
Ziliang Wang, Email: huf_zlwang@126.com.
Xinxiang Li, Email: xinxiangli@fudan.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209‐49. [DOI] [PubMed] [Google Scholar]
- 2. Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time‐trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond). 2021;41(10):1037‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li C, Sun YD, Yu GY, Cui JR, Lou Z, Zhang H, et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell. 2020;38(5):734‐47 e9. [DOI] [PubMed] [Google Scholar]
- 4. Vuletic A, Mirjacic Martinovic K, Tisma Miletic N, Zoidakis J, Castellvi‐Bel S, Cavic M. Cross‐Talk Between Tumor Cells Undergoing Epithelial to Mesenchymal Transition and Natural Killer Cells in Tumor Microenvironment in Colorectal Cancer. Front Cell Dev Biol. 2021;9:750022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53‐dependent manner. Mol Cancer. 2021;20(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silva VR, Santos LS, Dias RB, Quadros CA, Bezerra DP. Emerging agents that target signaling pathways to eradicate colorectal cancer stem cells. Cancer Commun (Lond). 2021;41(12):1275‐313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428‐42. [DOI] [PubMed] [Google Scholar]
- 8. Zhang S, Sun J, Gu M, Wang G, Wang X. Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Med. 2021;10(24):8725‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue C, Li G, Lu J, Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct Target Ther. 2021;6(1):400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y, Mao Q, Xia Q, Cheng J, Huang Z, Li Y, et al. Noncoding RNAs link metabolic reprogramming to immune microenvironment in cancers. J Hematol Oncol. 2021;14(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He L, Man C, Xiang S, Yao L, Wang X, Fan Y. Circular RNAs' cap‐independent translation protein and its roles in carcinomas. Mol Cancer. 2021;20(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD‐1/PD‐L1 pathway in cancer immunotherapy. Mol Cancer. 2021;20(1):116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205‐11. [DOI] [PubMed] [Google Scholar]
- 14. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675‐91. [DOI] [PubMed] [Google Scholar]
- 15. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR‐30c‐5p/TCF7 axis. Mol Cancer. 2021;20(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong ZR, Ke AW, Li T, Cai JB, Yang YF, Zhou W, et al. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer. 2021;20(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S, et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer. 2021;20(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachmayr‐Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner‐Hofmann T, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu YL, Li HF, Chen HH, Lin H. Emergent Roles of Circular RNAs in Metabolism and Metabolic Disorders. Int J Mol Sci. 2022;23(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J, Zhang X, Cao J, Xu P, Chen Z, Wang S, et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3‐mediated autophagy and glycolysis. Cell Death Dis. 2021;12(10):910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin C, Lu R, Yuan M, Zhao R, Zhou H, Fan X, et al. Circular RNA 0006349 Augments Glycolysis and Malignance of Non‐small Cell Lung Cancer Cells Through the microRNA‐98/MKP1 Axis. Front Cell Dev Biol. 2021;9:690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zeng Z, Zhao Y, Chen Q, Zhu S, Niu Y, Ye Z, et al. Hypoxic exosomal HIF‐1alpha‐stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40(36):5505‐17. [DOI] [PubMed] [Google Scholar]
- 25. Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids. 2021;23:355‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐74. [DOI] [PubMed] [Google Scholar]
- 27. Wu Q, Yu X, Li J, Sun S, Tu Y. Metabolic regulation in the immune response to cancer. Cancer Commun (Lond). 2021;41(8):661‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang Z, Xu Z, Zhu X, Zhang J. New insights into molecules and pathways of cancer metabolism and therapeutic implications. Cancer Commun (Lond). 2021;41(1):16‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang PB, Hou PP, Liu FY, Hong WB, Chen HZ, Sun XY, et al. Blocking PPARgamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression. Proc Natl Acad Sci U S A. 2020;117(44):27412‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garland PB, Shepherd D, Nicholls DG, Ontko J. Energy‐dependent control of the tricarboxylic acid cycle by fatty acid oxidation in rat liver mitochondria. Adv Enzyme Regul. 1968;6:3‐30. [DOI] [PubMed] [Google Scholar]
- 31. Suburu J, Chen YQ. Lipids and prostate cancer. Prostaglandins Other Lipid Mediat. 2012;98(1‐2):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Broadfield LA, Pane AA, Talebi A, Swinnen JV, Fendt SM. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev Cell. 2021;56(10):1363‐93. [DOI] [PubMed] [Google Scholar]
- 33. Schlaepfer IR, Rider L, Rodrigues LU, Gijon MA, Pac CT, Romero L, et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol Cancer Ther. 2014;13(10):2361‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanagasabai T, Li G, Shen TH, Gladoun N, Castillo‐Martin M, Celada SI, et al. MicroRNA‐21 deficiency suppresses prostate cancer progression through downregulation of the IRS1‐SREBP‐1 signaling pathway. Cancer Lett. 2022;525:46‐54. [DOI] [PubMed] [Google Scholar]
- 35. Butler LM, Mah CY, Machiels J, Vincent AD, Irani S, Mutuku SM, et al. Lipidomic Profiling of Clinical Prostate Cancer Reveals Targetable Alterations in Membrane Lipid Composition. Cancer Res. 2021;81(19):4981‐93. [DOI] [PubMed] [Google Scholar]
- 36. Soukupova J, Malfettone A, Bertran E, Hernandez‐Alvarez MI, Penuelas‐Haro I, Dituri F, et al. Epithelial‐Mesenchymal Transition (EMT) Induced by TGF‐beta in Hepatocellular Carcinoma Cells Reprograms Lipid Metabolism. Int J Mol Sci. 2021;22(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Halvorson DL, McCune SA. Inhibition of fatty acid synthesis in isolated adipocytes by 5‐(tetradecyloxy)‐2‐furoic acid. Lipids. 1984;19(11):851‐6. [DOI] [PubMed] [Google Scholar]
- 38. Funabashi H, Kawaguchi A, Tomoda H, Omura S, Okuda S, Iwasaki S. Binding site of cerulenin in fatty acid synthetase. J Biochem. 1989;105(5):751‐5. [DOI] [PubMed] [Google Scholar]
- 39. Jeong NY, Lee JS, Yoo KS, Oh S, Choe E, Lee HJ, et al. Fatty acid synthase inhibitor cerulenin inhibits topoisomerase I catalytic activity and augments SN‐38‐induced apoptosis. Apoptosis. 2013;18(2):226‐37. [DOI] [PubMed] [Google Scholar]
- 40. Peng Y, Xu C, Wen J, Zhang Y, Wang M, Liu X, et al. Fatty Acid Metabolism‐Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Patients With Colorectal Cancer. Front Oncol. 2021;11:704038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol. 2020;60:41‐56. [DOI] [PubMed] [Google Scholar]
- 42. Xin P, Xu X, Deng C, Liu S, Wang Y, Zhou X, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol. 2020;80:106210. [DOI] [PubMed] [Google Scholar]
- 43. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther. 2021;6(1):402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945‐54. [PubMed] [Google Scholar]
- 45. Weaver AM, Silva CM. Signal transducer and activator of transcription 5b: a new target of breast tumor kinase/protein tyrosine kinase 6. Breast Cancer Res. 2007;9(6):R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Catlett‐Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10(1):105‐15. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24(130):19‐29. [PubMed] [Google Scholar]
- 48. Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunol Res. 2005;31(3):231‐42. [DOI] [PubMed] [Google Scholar]
- 49. Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97‐105. [DOI] [PubMed] [Google Scholar]
- 50. Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 2011;1815(1):104‐14. [DOI] [PubMed] [Google Scholar]
- 51. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789‐99. [DOI] [PubMed] [Google Scholar]
- 52. Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13(6):795‐804. [DOI] [PubMed] [Google Scholar]
- 53. Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, et al. A cytomegaloviral protein reveals a dual role for STAT2 in IFN‐{gamma} signaling and antiviral responses. J Exp Med. 2005;201(10):1543‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sauter G, Simon R, Hillan K. Tissue microarrays in drug discovery. Nat Rev Drug Discov. 2003;2(12):962‐72. [DOI] [PubMed] [Google Scholar]
- 55. Kriz W, Kaissling B, Le Hir M. Epithelial‐mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin Invest. 2011;121(2):468‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thiery JP. Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442‐54. [DOI] [PubMed] [Google Scholar]
- 57. Yang J, Weinberg RA. Epithelial‐mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818‐29. [DOI] [PubMed] [Google Scholar]
- 58. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kang X, Li C. A Dimension Reduction Approach for Energy Landscape: Identifying Intermediate States in Metabolism‐EMT Network. Adv Sci (Weinh). 2021;8(10):2003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hua W, Ten Dijke P, Kostidis S, Giera M, Hornsveld M. TGFbeta‐induced metabolic reprogramming during epithelial‐to‐mesenchymal transition in cancer. Cell Mol Life Sci. 2020;77(11):2103‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sciacovelli M, Frezza C. Metabolic reprogramming and epithelial‐to‐mesenchymal transition in cancer. FEBS J. 2017;284(19):3132‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Aspuria PP, Lunt SY, Varemo L, Vergnes L, Gozo M, Beach JA, et al. Succinate dehydrogenase inhibition leads to epithelial‐mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab. 2014;2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grassian AR, Lin F, Barrett R, Liu Y, Jiang W, Korpal M, et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)‐200‐dependent epithelial‐mesenchymal transition (EMT). J Biol Chem. 2012;287(50):42180‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Castro‐Vega LJ, Buffet A, De Cubas AA, Cascon A, Menara M, Khalifa E, et al. Germline mutations in FH confer predisposition to malignant pheochromocytomas and paragangliomas. Hum Mol Genet. 2014;23(9):2440‐6. [DOI] [PubMed] [Google Scholar]
- 65. Renkema GH, Wortmann SB, Smeets RJ, Venselaar H, Antoine M, Visser G, et al. SDHA mutations causing a multisystem mitochondrial disease: novel mutations and genetic overlap with hereditary tumors. Eur J Hum Genet. 2015;23(2):202‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li J, Liang N, Long X, Zhao J, Yang J, Du X, et al. SDHC‐related deficiency of SDH complex activity promotes growth and metastasis of hepatocellular carcinoma via ROS/NFkappaB signaling. Cancer Lett. 2019;461:44‐55. [DOI] [PubMed] [Google Scholar]
- 67. Rosland GV, Dyrstad SE, Tusubira D, Helwa R, Tan TZ, Lotsberg ML, et al. Epithelial to mesenchymal transition (EMT) is associated with attenuation of succinate dehydrogenase (SDH) in breast cancer through reduced expression of SDHC. Cancer Metab. 2019;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Colvin H, Nishida N, Konno M, Haraguchi N, Takahashi H, Nishimura J, et al. Oncometabolite D‐2‐Hydroxyglurate Directly Induces Epithelial‐Mesenchymal Transition and is Associated with Distant Metastasis in Colorectal Cancer. Sci Rep. 2016;6:36289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Quan J, Cheng C, Tan Y, Jiang N, Liao C, Liao W, et al. Acyl‐CoA synthetase long‐chain 3‐mediated fatty acid oxidation is required for TGFbeta1‐induced epithelial‐mesenchymal transition and metastasis of colorectal carcinoma. Int J Biol Sci. 2022;18(6):2484‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rios Garcia M, Steinbauer B, Srivastava K, Singhal M, Mattijssen F, Maida A, et al. Acetyl‐CoA Carboxylase 1‐Dependent Protein Acetylation Controls Breast Cancer Metastasis and Recurrence. Cell Metab. 2017;26(6):842‐55 e5. [DOI] [PubMed] [Google Scholar]
- 71. Visca P, Sebastiani V, Botti C, Diodoro MG, Lasagni RP, Romagnoli F, et al. Fatty acid synthase (FAS) is a marker of increased risk of recurrence in lung carcinoma. Anticancer Res. 2004;24(6):4169‐73. [PubMed] [Google Scholar]
- 72. Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin Y, et al. High‐Fat Diet Promotes Colorectal Tumorigenesis Through Modulating Gut Microbiota and Metabolites. Gastroenterology. 2022;162(1):135‐49 e2. [DOI] [PubMed] [Google Scholar]
- 73. Wang H, Xi Q, Wu G. Fatty acid synthase regulates invasion and metastasis of colorectal cancer via Wnt signaling pathway. Cancer Med. 2016;5(7):1599‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chang L, Wu P, Senthilkumar R, Tian X, Liu H, Shen X, et al. Loss of fatty acid synthase suppresses the malignant phenotype of colorectal cancer cells by down‐regulating energy metabolism and mTOR signaling pathway. J Cancer Res Clin Oncol. 2016;142(1):59‐72. [DOI] [PubMed] [Google Scholar]
- 75. Zhan Y, Ginanni N, Tota MR, Wu M, Bays NW, Richon VM, et al. Control of cell growth and survival by enzymes of the fatty acid synthesis pathway in HCT‐116 colon cancer cells. Clin Cancer Res. 2008;14(18):5735‐42. [DOI] [PubMed] [Google Scholar]
- 76. Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 Prevents Hepatoxicity of Lipid Peroxidation: An Inhibitory Role against Hepatic Steatosis. Oxid Med Cell Longev. 2017;2017:3960197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li R, Tang X, Xu C, Guo Y, Qi L, Li S, et al. Circular RNA NF1‐419 Inhibits Proliferation and Induces Apoptosis by Regulating Lipid Metabolism in Astroglioma Cells. Recent Pat Anticancer Drug Discov. 2022;17(2):162‐77. [DOI] [PubMed] [Google Scholar]
- 78. Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67‐79. [DOI] [PubMed] [Google Scholar]
- 79. Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q, et al. ciRS‐7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma. Mol Cancer. 2021;20(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ‐ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22‐37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell. 2014;56(1):55‐66. [DOI] [PubMed] [Google Scholar]
- 82. Wang X, Xing L, Yang R, Chen H, Wang M, Jiang R, et al. The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto‐oncogene MYC. Mol Cancer. 2021;20(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7(16):3842‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1‐529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lee CJ, An HJ, Cho ES, Kang HC, Lee JY, Lee HS, et al. Stat2 stability regulation: an intersection between immunity and carcinogenesis. Exp Mol Med. 2020;52(9):1526‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vandsemb EN, Rye MB, Steiro IJ, Elsaadi S, Ro TB, Slordahl TS, et al. PRL‐3 induces a positive signaling circuit between glycolysis and activation of STAT1/2. FEBS J. 2021;288(23):6700‐15. [DOI] [PubMed] [Google Scholar]
- 89. Hu Y, Sun H, Hu J, Zhang X. LncRNA DLX6‐AS1 Promotes the Progression of Neuroblastoma by Activating STAT2 via Targeting miR‐506‐3p. Cancer Manag Res. 2020;12:7451‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gamero AM, Young MR, Mentor‐Marcel R, Bobe G, Scarzello AJ, Wise J, et al. STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev Res (Phila). 2010;3(4):495‐504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR‐193a‐5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39(1):283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF‐H1/RhoA axis. Theranostics. 2020;10(18):8211‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wu M, Kong C, Cai M, Huang W, Chen Y, Wang B, et al. Hsa_circRNA_002144 promotes growth and metastasis of colorectal cancer through regulating miR‐615‐5p/LARP1/mTOR pathway. Carcinogenesis. 2021;42(4):601‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xu YJ, Zhao JM, Gao C, Ni XF, Wang W, Hu WW, et al. Hsa_circ_0136666 activates Treg‐mediated immune escape of colorectal cancer via miR‐497/PD‐L1 pathway. Cell Signal. 2021;86:110095. [DOI] [PubMed] [Google Scholar]
- 96. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. The landscape of microRNA, Piwi‐interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kolling M, Haddad G, Wegmann U, Kistler A, Bosakova A, Seeger H, et al. Circular RNAs in Urine of Kidney Transplant Patients with Acute T Cell‐Mediated Allograft Rejection. Clin Chem. 2019;65(10):1287‐94. [DOI] [PubMed] [Google Scholar]
- 98. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1‐7. [DOI] [PubMed] [Google Scholar]
- 101. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365(2):141‐8. [DOI] [PubMed] [Google Scholar]
- 102. Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi L, et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study and the plasmids used in this study are available from the corresponding author upon reasonable request.


