Abstract
Group B streptococci (GBS) contain a family of protective surface proteins characterized by variable numbers of repeating units within the proteins. The prototype alpha C protein of GBS from the type Ia/C strain A909 contains a series of nine identical 246-bp tandem repeat units. We have previously shown that deletions in the tandem repeat region of the alpha C protein affect both the immunogenicity and protective efficacy of the protein in animal models, and these deletions may serve as a virulence mechanism in GBS. The molecular mechanism of tandem repeat deletion is unknown. To determine whether RecA-mediated homologous recombination is involved in this process, we identified, cloned, and sequenced the recA gene homologue from GBS. A strain of GBS with recA deleted, A909ΔrecA, was constructed by insertional inactivation in the recA locus. A909ΔrecA demonstrated significant sensitivity to UV light, and the 50% lethal dose of the mutant strain in a mouse intraperitoneal model of sepsis was 20-fold higher than that of the parent strain. The spontaneous rate of tandem repeat deletion in the alpha C protein in vitro, as well as in our mouse model of immune infection, was studied using A909ΔrecA. We report that tandem repeat deletion in the alpha C protein does occur in the absence of a functional recA gene both in vitro and in vivo, indicating that tandem repeat deletion in GBS occurs by a recA-independent recombinatorial pathway.
Group B Streptococcus (GBS) is a leading cause of meningitis, pneumonia, and sepsis in neonates. There is some evidence that heavy maternal colonization by GBS contributes to premature birth (3, 39). GBS has also been frequently noted to cause invasive infections in women as a complication of childbirth and in other adults who are immunologically impaired by liver disease, diabetes, or malignancy (15, 61). Recent Centers for Disease Control and Prevention surveillance data demonstrate that although the widespread use of intrapartum antibiotic prophylaxis has decreased the incidence of early-onset neonatal GBS disease, this practice has not impacted late-onset neonatal disease or adult disease. Premature infants and adults over age 65 continue to show high case fatality rates from GBS disease (52).
The alpha C protein is a surface protein present in the majority of non-type III GBS. Similar proteins have recently been identified in type III and type V strains (30, 31, 54). Antibodies to these surface proteins have been demonstrated to be protective against infection in animal models (18). The gene for the alpha C protein (bca) from the prototype type Ia/C strain A909 includes a series of nine tandem repeats of 246 bp that are identical at the nucleotide level (40).
The molecular weight of the alpha C protein in clinical isolates has been shown to vary widely, with the size differences due to different numbers of tandem repeat units within the protein (35, 36). Significantly, alpha C protein size has been noted to vary in paired mother/infant isolates (20). Furthermore, we have demonstrated in a mouse model of infection that in the presence of antibody to one size of alpha C protein, mutant GBS strains arise that express smaller-size alpha C proteins on the surface and thus escape from the immune response. The genes for the smaller proteins have precise deletions of repeat elements (36). The change in the number of repeats and in the size of the protein alters the antigenicity of the alpha C protein and the virulence of the strain in the presence of antibody (19). Tandem repeat deletion in the alpha C protein allows antigenic variation and may thus serve as a virulence mechanism in GBS.
The mechanism by which excision of tandem repeat units in the bca gene is accomplished is unknown. Identification of the molecular factors involved in tandem repeat deletion is of broader interest, since tandem repeat sequences of DNA are found in both prokaryotic and eukaryotic genomes. Tandem repeat sequences in eukaryotic genomes are often found in noncoding regions, and variation in these sequences has been associated with a variety of human diseases, such as myotonic dystrophy, Huntington's disease, fragile X syndrome, and colon cancer (58, 59). In prokaryotes, variation in short sequence tandem repeats, such as those present in the promoter region of the Haemophilus influenzae hifA/B fimbriae genes (60), in the 5′ coding region of the H. influenzae lic1 genes (63), which are involved in lipopolysaccharide synthesis, and in the Neisseria gonorrhoeae opa gene, which encodes opacity proteins (55), can regulate gene expression. In addition, an increasing number of bacterial, fungal, and protozoal antigens have been identified that have larger tandem repeat motifs within the coding regions of the associated genes, such as the alpha-like proteins of GBS (30, 62), the Esp protein of Enterococcus faecalis (53), the M proteins of group A Streptococcus (GAS) (22), the 120-kDa major glycoprotein of Blastomyces dermatitidis (25), the 190-kDa surface antigen of Rickettsia rickettsi (4), the ALS proteins of Candida albicans (24), the P270 protein of Trichomonas vaginalis (43), and the shed proteins of Trypanosoma cruzi (11).
Variation in short sequence repeats, such as in the CAAT motif found in the H. influenzae lic gene and in the CTCTT motif found in the Neisseria meningitidis opa gene, is attributed to a RecA-independent strand-slippage mechanism (23, 32, 42). Intermolecular crossover events between identical or near-identical longer stretches of DNA in bacteria, however, are highly dependent on RecA-mediated homologous recombination, and it is possible that tandem repeat deletion within the alpha C protein takes place by a similar RecA-mediated intramolecular crossover mechanism. RecA is a highly conserved ATP-binding protein that mediates pairing of homologous stretches of DNA and catalyzes the formation of heteroduplex structures (48). RecA gene homologues have been identified in more than 60 bacterial species. Although RecA is thought to be essential in all pathways of bacterial homologous recombination, both RecA-dependent and RecA-independent tandem repeat deletion has been observed in Escherichia coli (5, 6, 33, 34).
To begin to understand the mechanism of tandem repeat deletion in GBS, we identified and cloned the recA homologue in GBS. We constructed A909ΔrecA, a strain in which the recA gene is functionally deleted by insertional inactivation. We compared the rate of spontaneous deletion in A909 with that in A909ΔrecA in vitro and compared the strains in our mouse model of GBS infection. We demonstrate that tandem repeat deletion occurs in the absence of intact recA both in vitro and in vivo.
MATERIALS AND METHODS
Amplification of a portion of the GBS recA gene.
To identify the recA gene, degenerate PCR primers designed to anneal to highly conserved regions of the recA gene from other bacterial species (14) were used to amplify a 320-bp fragment from GBS genomic DNA by PCR. The following primers were used: primer A, 5′ CGTAAGCTTYATHGAYGCNGARCAYGC 3′; primer B, 5′ CTCAAGCTTGRTTDATRAADATNGC 3′. The amplified fragment was cloned in vector pCR2 (Invitrogen) and sequenced. The fragment was identified as a portion of the GBS recA gene by sequence homology with the previously identified recA gene in GAS (57).
Creation of GBS cosmid library.
Genomic DNA was prepared from type Ia/C strain A909 as previously described (36). To create the cosmid library, genomic DNA was partially digested with Sau3AI at 37°C for 15 min, followed by heating to inactivate the enzyme. Cosmid vector pHC79 (21) was digested with BamHI and ligated to the partially digested genomic DNA. The ligation mixture was packaged with a commercial lambda packing extract (Gigapack III Gold; Stratagene). The cosmid library titer was 3 × 105/ml.
Isolation of the recA gene from the cosmid library.
To isolate the recA gene, the library was plated on 20 Luria-Bertani plates containing 100 μg of ampicillin/ml at a density of 200 to 500 colonies per plate, and the colonies were transferred to nylon membranes (Genescreen; Dupont). Colony blotting was carried out with standard methods (49), using a rotisserie oven and a hybridization temperature of 42°C. The 320-bp PCR product was labeled with [32P]dCTP with a random primer kit (Gibco-BRL) and added to the hybridization solution at a final concentration of 106 cpm/ml. The membranes were washed once at low stringency and then twice in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate (SDS) at 42°C for 20 min and exposed to X-ray film at −80°C for 20 h. Thirteen positive colonies were identified, and cosmid DNA was prepared from each with plasmid miniprep columns (Qiagen). The cosmid DNA was digested with HindIII, transferred to a nylon membrane, and probed with the radiolabeled PCR fragment. A 3-kb HindIII fragment from one cosmid showed strong reactivity with the probe, and this fragment was subcloned into pCR2 and named pKPRA3.
Identification of the full-length recA gene.
Comparison of the sequence contained within pKPRA3 with the recA sequence of GAS revealed significant homology but indicated that the cosmid fragment did not contain the full-length recA. PCR primer K1 (5′ CAAATCAAGGGGACTGGTGAGCAT 3′) was designed for the sequence at the 3′ end of the recA cosmid sequence, and primer K2 (5′TTAATCTTCAATTTCAATACCATTATCTAAAT 3′) was designed for the sequence immediately outside the stop codon of the GAS recA sequence (58). An annealing temperature of 45°C was used to amplify an approximately 400-bp fragment from GBS genomic DNA. To confirm the entire recA sequence, a full-length recA PCR product was generated with primer 5′ CGTCGAAAAGCCTTAGATGATGCT 3′ and primer 5′ TAAGTTCCAACCACTGTCAGAACT 3′ with an annealing temperature of 55°C and extension time of 90 s. Pooled PCR products from both reactions were sequenced as described below.
Cloning of an internal fragment of the recA gene into pJRS233.
An internal 603 bp of the recA sequence was amplified from GBS genomic DNA with primers corresponding to the amino acid sequences HALDPAYA (5′ CACGCTCTTGACCCAGCCTATGC 3′) and AGAWYSYN (5′ CATTATATGAGTACCATGCACCTGC 3′). The PCR product was ligated into vector pCR2 by the Topo-cloning method (Invitrogen), following the manufacturer's instructions. The recombinant plasmid was digested with BamHI and XhoI, and the insert was gel purified using a gel extraction column (Qiagen) and ligated into pJRS233, which had been digested with BamHI and XhoI. The recombinant plasmid was named pKP600.
Transformation of pKP600 into GBS.
Competent GBS were prepared as previously described (13). The bacteria were transformed with pKP600 by electroporation (Bio-Rad Gene Pulser; settings, 1.5 kV, 400 Ω, 25 μF), placed on ice for 30 min, incubated at 30°C for 90 min, and finally plated on erythromycin (ERY)-containing sheep's blood agar (ERY/BAP) at 30°C for 48 h. ERY-resistant colonies were streaked on ERY/BAP and grown at 30°C for 48 h. Liquid cultures of ERY-resistant strains were grown in Todd-Hewitt broth (THB) with ERY (THB/ERY) at 30°C overnight and then diluted 1:10 into THB/ERY and grown for 8 to 16 h at 37°C. These cultures were then diluted at 1:100 to 1:10,000, plated on BAP/ERY, and grown overnight at 37°C. ERY-resistant colonies growing at 37°C were passaged a second time in THB/ERY medium at 37°C and again plated at 37°C on ERY/BAP, to decrease any illegitimate persistence of free plasmid.
UV light sensitivity assay.
Single colonies of bacteria were inoculated into THB in the presence or absence of ERY and grown overnight at 37°C. The cultures were diluted 1:100 in fresh THB and grown to an optical density at 650 nm of 0.4. The cultures were diluted 1:10,000, and 0.1 ml was plated on BAP. Plates of wild-type A909 GBS and potential mutants were simultaneously placed on a shortwave UV light box (Fotodyne) for 5, 10, 20, and 30 s. The plates were incubated at 37°C for 16 h, and the CFU were counted at each exposure time and compared with control plates that were not exposed to UV light.
Immune mouse model of infection.
CD-1 outbred mice (adult female, 6 to 8 weeks old; Harlan Laboratory) were given 0.25 ml of rabbit antiserum raised to the nine-repeat alpha C protein, or nonimmune serum as a control, by intraperitoneal injection. Twenty-four hours after injection, the mice were given 106 to 107 CFU of bacteria suspended in 0.5 ml of THB by intraperitoneal injection. Twenty-four hours after bacterial injection, the mice were sacrificed in CO2, and their spleens were immediately removed and homogenized in phosphate-buffered saline, pH 7.0. Mice given nonimmune serum were found dead at 24 h after bacterial injection. The splenic homogenates were directly plated on BAP and grown at 37°C overnight.
Sequencing of bca from splenic isolates.
Sequencing of the repeat region of bca, the gene encoding the alpha C protein, was accomplished by PCR amplification of the complete alpha C protein using primers 5′ GGTGGACAAGAAAAAGTTCTC 3′ and 5′ TGTTCACACCAATAAATGGTGA 3′, with an annealing temperature of 60°C and extension time of 90 s, using DNA isolated from a single colony of each isolate by the protoplast method (12). The pooled product was purified and sequenced as described below. Sequencing primers used were 5′ GTAAAATTGTTGAGGTTAAT 3′ and 5′ GGTAGTTTATTTCCTTTACCG 3′.
Southern blots.
Southern blotting was performed as previously described (36). Probes were prepared using the ECL chemiluminescence direct nucleic acid labeling and detection kit (Amersham) following the manufacturer's instructions.
Western blots.
Bacteria were grown in THB (plus ERY for the A909ΔrecA strain) overnight at 37°C, pelleted, and resuspended in one-third-volume phosphate-buffered saline. Resuspended bacteria were mixed with Laemmli gel sample buffer (plus dithiothreitol), boiled, spun briefly to pellet debris, and immediately run on a 10% Tris-glycine SDS gel. Western blotting was performed as previously described (19) using rabbit antiserum raised to the one-repeat form of the alpha C protein at 1:500 to 1:2,000 dilutions.
Screening for spontaneous tandem repeat deletion in the alpha C protein.
A909 and A909ΔrecA were plated on BAP (with or without ERY) at a density of 200 to 400 colonies/plate. Forty plates were screened for each strain (12,213 colonies for A909; 9,457 colonies for A909ΔrecA.) Colony immunoblotting was performed as previously described (36). Lightly stained colonies were retrieved from the corresponding primary plate and restreaked on BAP with or without ERY. Each potential alpha C protein mutant was examined by Western blotting as described above to determine the size of the alpha C protein. Several A909ΔrecA-derived isolates with alpha C protein deletions were tested for UV sensitivity, and all remained UV sensitive, indicating that the deletions did not arise from reversion to a recA+ phenotype.
PCRs.
Reactions were carried out with PCR Supermix (Gibco-BRL) with a 0.4 μM primer concentration in a thermocycler (Perkin-Elmer) using a cycle program as follows: initial denaturation at 94°C for 5 min, 35 cycles with denaturation at 94°C for 30 s, annealing temperature as indicated for 30 s and extension at 72°C for 30 to 90 s as indicated, and a final extension step at 72°C for 7 min, followed by cooling to 4°C. Template DNA was either purified genomic DNA or DNA prepared from single bacterial colonies by the protoplast method reaction (12).
Reverse transcriptase PCR (RT-PCR).
Bacterial cultures were grown to an approximate optical density at 650 nm of 0.400 in THB, pelleted, and resuspended in 25% glucose–Tris-EDTA (pH 8.0) containing 2.5 mg of lysozyme/ml and 167 μg of mutanolysin/ml, and incubated at room temperature for 10 min. The bacteria were then pelleted, and total RNA isolation was carried out using the RNeasy kit (Qiagen). After treatment with DNase I (Gibco-BRL), the integrity of the RNA was assessed by denaturing agarose gel to ensure that no significant degradation of the RNA had occurred. RT-PCRs were carried out using the One-Step RT-PCR kit (Qiagen), using 0.8 to 1.0 μg of total RNA per reaction. All reactions were carried out with a 0.6 μM primer concentration in a thermocycler (Perkin-Elmer) using a cycle program as follows: reverse transcription at 50°C for 30 min and denaturation at 95°C for 15 min, followed by 35 cycles as described above for PCRs.
Sequencing reactions and facilities.
Direct sequencing from pooled PCR product or sequencing from cloned DNA was performed with an automated fluorescent dideoxy sequencing system (Applied Biosystems) through the Brigham and Women's Hospital Automatic Sequencing and Genotyping Facility and the Beth Israel-Deaconess Medical Center Molecular Medicine Sequencing Facility. Direct PCR product sequencing was carried out from product pooled from 5 to 8 separate PCRs; the pooled product was purified over a PCR purification column (Qiagen) to remove unincorporated primer and deoxynucleoside triphosphates.
Statistical analyses.
Alpha C protein Western blot phenotype data were obtained on 24 mice, with 1 to 124 isolates obtained per mouse. The isolate data were classified into nine-repeat, deletion, and null phenotypes. The primary analysis concerned the association between the A909ΔrecA genotype and the frequency of each isolate phenotype. Special statistical analysis was required to account for mouse-to-mouse variation in the number of isolates obtained. Overdispersed binomial logistic regression (2) was performed to test the null hypothesis that the A909ΔrecA genotype and the nonnull, deletion phenotype were not associated. Colony blot data on alpha C protein repeat number were obtained from 12,213 colonies of A909 and from 9,457 colonies of A909ΔrecA and were analyzed using a two-tailed Fisher's exact test.
Nucleotide sequence accession number.
The complete recA sequence has been deposited in the GenBank database under accession number AF307982.
RESULTS
Identification and cloning of the recA gene of GBS.
Using PCR primers designed to anneal to highly conserved regions of recA from other bacterial species (14), a 320-bp fragment was amplified from GBS genomic DNA. Using the PCR product as probe, a single band was seen on a Southern blot of GBS DNA digested with several restriction enzymes, indicating the presence of a single recA gene in GBS. To isolate the entire GBS recA gene, the PCR product was used to screen a cosmid library of GBS genomic DNA. A cosmid containing a partial recA sequence was identified; PCR methods were used to obtain the full-length sequence. An alignment of the deduced RecA amino acid sequences from GBS, GAS, Streptococcus pneumoniae, and E. coli reveals that the GBS sequence is 88% identical to that of GAS, 79% identical to that of the pneumococcus, and 55% identical to that of E. coli (44, 50, 57).
To assess the functional similarity of the GBS RecA protein to the E. coli RecA protein, we compared the amino acid sequences at the putative active sites of the proteins. E. coli RecA has been crystallized, and the functional residues of the protein have been characterized by site-directed mutagenesis (50). Residues with putative roles in ATP binding, in the ATP-induced conformational switch, in monomer-monomer interactions, and in DNA binding are listed in Table 1. Each of these residues is well conserved in the GBS RecA deduced amino acid sequence; 57 (83%) of the 69 amino acids are identical, and 64 (93%) are identical or conservatively substituted, as shown in Table 1.
TABLE 1.
Comparison of E. coli recA active sites with the GBS recA sequencea
| Function | Sequence (residues) | No. of identical residues (conserved residues)/total |
|---|---|---|
| ATP binding P-loop | GPESSGKT (66–73) | 8/8 |
| GPESSGKT (40–47) | ||
| ATP binding Walker B box | VIVVD (140–144) | 3/5 |
| LVVVD (114–118) | (5/5) | |
| ATP binding | E (96) | 1/1 |
| E (70) | ||
| DNA binding | GEIGDSHM (157–164) | 6/8 |
| GDIGDSHV (131–138) | (8/8) | |
| DNA binding | IRMKIGVMFGNPETT (195–209) | 12/15 |
| LREKVGVMFGNPETT (169–183) | (14/15) | |
| Double-stranded DNA binding | K, W, K, G, K (286, 290, 297, 301, 302) | 4/5 |
| K, W, K, G, S (264, 268, 275, 279, 280) | ||
| Monomer-monomer interactions | L, I, A (114, 128, 148) | 3/3 |
| L, I, A (102, 148, 122) | ||
| MAW motifb | TGSLSLDIALGAGGLPMGRIVEIY (42–65) | 20/24 |
| SGSLAIDIALGAGGYPKGRIVEIY (16–39) | (21/24) |
Creation of a strain of GBS with recA deleted.
pJRS233 is an E. coli/streptococcal shuttle vector that contains a gram-negative replicon, a temperature-sensitive gram-positive replicon, and an ERY resistance gene (45). Persistence of an ERY resistance phenotype at 37°C in a gram-positive strain can be accomplished by integration of this plasmid into the genomic DNA of the organism. An internal fragment of GBS recA was cloned into pJRS233, creating vector pKP600. Integration of pKP600 at the recA locus can occur by homologous recombination, interrupting the recA gene, as shown schematically in Fig. 1. Competent GBS were transformed with the recombinant plasmid, passaged at 30°C, and then shifted to growth at 37°C. After serial passaging at 37°C, five isolates were chosen for further study.
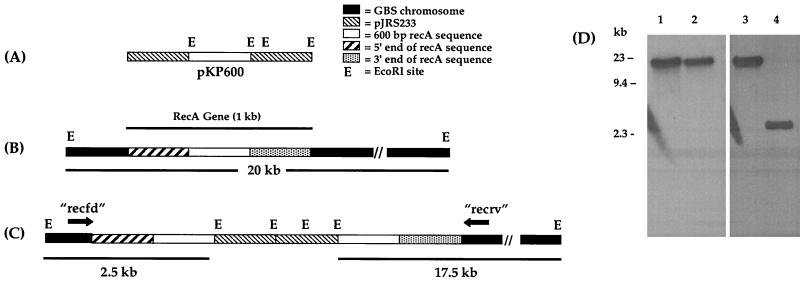
FIG. 1.
Schematic diagram of insertional inactivation at the recA locus. (A) pKP600; internal 603 bp of the GBS recA nucleotide sequence cloned into the pJRS233 shuttle vector at the BamHI and XhoI sites. The white area represents the 603-bp cloned fragment. EcoRI sites on the vector are noted with “E.” (B) recA chromosomal locus located on the approximately 20-kb EcoRI fragment. Striped shading represents the 5′ end of recA exclusive of the cloned fragment. Grey shading represents the 3′ end of recA exclusive of the cloned fragment. Black shading represents chromosome. (C) recA chromosomal locus after insertion of pKP600. Digestion with EcoRI should generate approximately 2.5-kb and 17.5-kb fragments; use of the 5′-specific probe should identify the smaller fragment, and use of the 3′-specific probe should identify the larger fragment. (D) Southern blot of A909 (lanes 1 and 3) and A909ΔrecA (lanes 2 and 4) chromosomal DNA, digested with EcoRI and probed with 3′-specific recA probe (lanes 1 and 2) or 5′-specific recA probe (lanes 3 and 4).
Bacterial strains with recA deleted are markedly more sensitive to the killing effects of UV light due to the reduced efficiency of DNA repair that results from deletion of recA. (28). The potential recA mutants were tested for growth sensitivity to UV light. One of the five isolates, which was markedly more sensitive to exposure to UV light than wild-type A909, was designated A909ΔrecA. This isolate demonstrated essentially complete killing after only 5 s of exposure to UV light (data not shown).
Molecular analysis of A909ΔrecA.
To confirm disruption of recA in A909ΔrecA, EcoRI restriction digests of genomic DNA from wild-type and mutant bacteria were analyzed by a Southern blot with fragments of the recA sequence. The Southern blot shown in Fig. 1D reveals the generation of a new fragment of the predicted size in the mutant strain when it was probed with sequence from the 5′ portion of recA and a slight shift downward of the large fragment when probed with sequence at the 3′ end of the recA gene, consistent with integration of the plasmid at the recA locus. The remaining four erythromycin-resistant isolates were also analyzed by a Southern blot (data not shown). One of these (which also showed enhanced susceptibility to UV killing) showed a pattern identical to that of A909ΔrecA. The others were not UV sensitive and showed patterns incompatible with interruption of the recA gene.
The insertion of pKP600 into recA should prevent the transcription of full-length recA mRNA and thus prevent synthesis of the RecA protein. To confirm that intact recA mRNA is absent in A909ΔrecA, total RNA from strains A909 and A909ΔrecA was analyzed by RT-PCR. Primer 15 and primer 16 were used as a positive control, since this primer pair should amplify a 603-bp recA fragment from both A909 and A909ΔrecA. The primer pair recfd and recrv correspond to sequences immediately outside recA (Fig. 1) and amplify an 1,155-bp fragment that contains the entire recA coding region. RT-PCR was carried out as described in Materials and Methods; control reaction mixtures were held on ice during the reverse transcription step to ensure that products were not the result of contaminating genomic DNA. The 603-bp product was amplified as predicted from both the wild-type strain and the recA mutant, but the 1,155-bp product was amplified only from the wild-type strain, indicating that full-length recA mRNA is not made in the mutant strain. There was no product amplified from any of the control reactions (data not shown).
Attenuation of A909ΔrecA in a mouse model of sepsis.
The 50% lethal dose (LD50) of the mutant strain was compared with that of the wild-type strain A909 in our mouse model of intraperitoneal sepsis. The LD50 of A909ΔrecA was 9.4 × 104, and that of wild-type A909 was 3.5 × 103, as calculated by the method of Reed and Muench (47) (data not shown), demonstrating that A909ΔrecA was attenuated in this model. To determine if the difference in virulence of A909ΔrecA was due to a growth defect in the mutant strain, we compared the in vitro growth rates of the wild-type and mutant strains. The slope of the growth curves during log-phase growth was essentially identical between the strains, indicating that there was not a significant growth defect in the mutant strain. The spleen of one of the mice used in the LD50 study was removed, homogenized, and plated on blood agar plates. Single bacterial colonies were isolated and were observed to retain both the ERY resistance marker and the UV sensitivity phenotype.
Comparison of alpha C protein tandem repeat deletion in the wild-type and recA mutant strains in a mouse model of infection.
In our mouse model of GBS infection, when mice are infected with wild-type GBS after receiving a protective antibody to the alpha C protein, bacterial colonies with truncated forms of the alpha C protein are frequently isolated. The genes encoding the truncated proteins contain precise deletions of tandem repeat elements (36). To determine whether the process of tandem repeat deletion in the alpha C protein of GBS involves recA-mediated homologous recombination, we compared the occurrence and frequency of isolating mutants with tandem repeat deletions in mice challenged with A909ΔrecA and those challenged with wild-type A909.
Rabbit antiserum raised to the nine-repeat alpha C protein was given to mice by intraperitoneal injection. Nonimmune serum was used as a control. Twenty-four hours after antiserum was given, the mice were challenged by intraperitoneal injection with doses of either wild-type A909 or A909ΔrecA GBS, at or above the LD50 for each strain. Mice given nonimmune serum were found dead at 24 h, but mice given immune serum were generally alive and appeared well after 24 h. At 24 h, the surviving mice were sacrificed and the spleens from all mice were immediately removed. Individual bacterial colonies were isolated from the spleens, and the alpha C protein from each isolate was assessed by a Western blot. Rabbit antiserum to the one-repeat alpha C protein was used because it recognizes all forms of the alpha C protein on a Western blot.
Western blot analysis of the alpha C protein from splenic isolates.
The Western blot shown in Fig. 2A shows representative bacterial splenic isolates obtained from a mouse given nine-repeat immune serum and challenged with the wild-type strain A909.
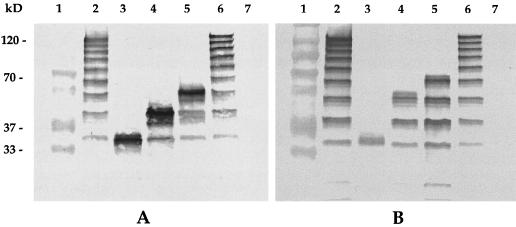
FIG. 2.
Western blots of splenic isolates from mice challenged with A909 (A) or A909ΔrecA (B). Bacterial samples were separated by SDS-PAGE, transferred to nitrocellulose, and probed with rabbit antiserum to one-repeat alpha C protein. Blot A: lane 1, SDS-PAGE prestained molecular mass markers; lane 2, A909 control; lanes 3 to 6, individual splenic isolates with alpha C proteins containing one repeat (lane 3), two repeats (lane 4), three repeats (lane 5), or nine repeats (lane 6). Lane 7, isolate not expressing alpha C protein (null mutant). Blot B: lane 1, SDS-PAGE prestained molecular mass markers; lane 2, A909ΔrecA control; lanes 3 to 6, individual A909ΔrecA splenic isolates with alpha C proteins containing one repeat (lane 3), three repeats (lane 4), four repeats (lane 5), or nine repeats (lane 6). Lane 7, A909ΔrecA-derived isolate not expressing alpha C protein (null mutant).
When the alpha C protein is visualized on SDS-polyacrylamide gel electrophoresis (PAGE), a laddering pattern is seen that corresponds to the number of repeat elements in the protein. The pattern is thought to reflect posttranslational proteolysis of the protein at the repeat site—so that, for example, nine bands are seen with nine-repeat alpha C protein, whereas two bands are seen with a two-repeat form of the alpha C protein (41, 54). Wild-type A909 is shown in Fig. 2A, lane 2. Lanes 3 to 6 show alpha C protein from individual splenic isolates, consistent with the one-repeat (lane 3), two-repeat (lane 4), three-repeat (lane 5) and nine-repeat (lane 6) forms of the alpha C protein. Lane 7 exhibits no alpha C protein. Many isolates of this type were obtained that apparently produce no alpha C protein; they were termed null mutants. Control experiments with representative null mutants demonstrate that these mutants produce overall adequate amounts of protein, and specifically, these mutants do produce another antigenic surface protein, the beta C protein (data not shown). Some isolates appeared to produce intact nine-repeat alpha C protein at markedly lower levels than the wild type. For the purposes of statistical analysis, these were also considered to be null mutants since they did not appear to have undergone tandem repeat deletion (see PCR amplification below). In control mice that received nonimmune serum, 56 isolates were tested and all had intact nine-repeat alpha C protein as determined by a Western blot, whether the challenge strain was A909 or A909ΔrecA.
Similar results were obtained when mice received nine-repeat immune serum and were challenged with the mutant strain A909ΔrecA. A Western blot of representative bacterial splenic isolates is shown in Fig. 2B, with A909ΔrecA in lane 2. Lanes 3 to 6 show alpha C protein from individual splenic isolates from a mouse challenged with A909ΔrecA, consistent with one-repeat (lane 3), three-repeat (lane 4), four-repeat (lane 5) and nine-repeat (lane 6) protein. In addition, a number of null mutants were also observed, as shown in lane 7. Each of the splenic isolates obtained from mice challenged with A909ΔrecA retained their resistance to erythromycin; representative null and deletion isolates were further tested for sensitivity to the killing effects of UV light and were observed to retain the UV sensitivity phenotype of A909ΔrecA.
PCR amplification and sequencing of the alpha C protein gene in splenic isolates.
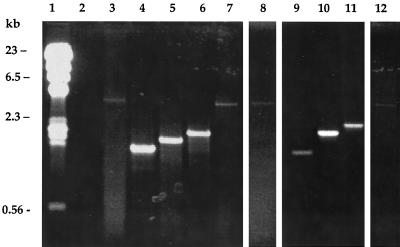
To ensure that the phenotypes seen on Western blots in these experiments corresponded to gene deletions, the alpha C protein genes from the wild-type strain, the A909ΔrecA mutant, and several splenic isolates were amplified by PCR. The products obtained from the wild-type isolates are shown in Fig. 3. In lane 3, a product of the predicted size, approximately 3.5 kb, is observed with amplification from the wild-type A909 strain. In lanes 4, 5, and 6, the band sizes correspond to products from isolates with one, two, and three repeats on a Western blot. The null mutants have not been previously genetically characterized; amplification of DNA from a null mutant, shown in lane 7, yielded a band close to that of the wild type. Similar results were observed with truncated and null mutants isolated from spleens of mice challenged with A909ΔrecA. Products consistent with a nine-repeat alpha C protein are observed in lane 8 (A909ΔrecA parent strain) and lane 12, a null mutant by a Western blot. In lanes 9, 10, and 11, the band sizes correspond to products from isolates with one, three, and four repeats on a Western blot.
FIG. 3.
PCR amplification of the alpha C protein gene (bca) from bacterial splenic isolates obtained from mice challenged with A909 (lanes 4 to 7) or A909ΔrecA (lanes 9 to 12). DNA was prepared from each isolate by the protoplast method as outlined in Materials and Methods. PCR products were separated on 1% agarose gels. Lane 1, λHindIII markers; lane 2, blank; lane 3, wild-type A909; lanes 4 to 6, individual splenic isolates with alpha C proteins containing one repeat (lane 4), two repeats (lane 5), or three repeats (lane 6); lane 7, isolate not expressing the alpha C protein (null mutant); lane 8, A909ΔrecA; lanes 9 to 11, individual A909ΔrecA splenic isolates with alpha C proteins containing one repeat (lane 9), three repeats (lane 10), or four repeats (lane 11); lane 12, A909ΔrecA-derived isolate not expressing the alpha C protein (null mutant).
The repeat region of the alpha C protein from wild-type isolates with two and three repeats (shown in Fig. 3, lanes 5 and 6) and recA deletion isolates with 3 and four repeats (shown in Fig. 3, lanes 10 and 11) was sequenced from pooled PCR product, as described in Materials and Methods. The number of repeat sequences found in all four isolates corresponded exactly to that predicted by Western blotting and PCR. The sequencing revealed precise deletions of repeat sequences; there was no evidence of illegitimate or partial deletions in the isolates with recA deleted.
Statistical analysis of the splenic isolate data.
A summary of the Western blot analyses of the splenic isolates is shown in Table 2. Non-nine-repeat mutants were obtained from 7 of 10 mice challenged with the wild-type A909 strain, and 293 of these isolates were analyzed by Western blots. Non-nine-repeat mutants were obtained from 7 of 13 mice challenged with the recA deletion strain A909ΔrecA, and 341 isolates were analyzed by Western blots. If fewer than 125 isolates were recovered from an individual spleen, 50 to 100% of the isolates were analyzed. If more than 250 isolates were recovered from an individual spleen, a representative proportion of the isolates was randomly chosen for analysis. The number of isolates obtained from a spleen ranged from 1 to more than 1,000 and did not depend on the challenge strain (P = 0.64 by the Wilcoxon test).
TABLE 2.
Summary of Western blot analysis of splenic isolatesa
| Strain | No. of mice | Total no. of isolates | No. of isolates with alpha C protein phenotype (%)
|
||
|---|---|---|---|---|---|
| 9-repeat | Deletion | Null | |||
| A909 | 10 | 293 | 86 (29) | 73 (25) | 134 (46) |
| ΔrecA | 13 | 341 | 184 (54) | 108 (32) | 49 (14) |
The probability of obtaining the non-9-repeat alpha C protein phenotype is 0.71 for A909 isolates and 0.46 for A909 ΔrecA (P = 0.12). Eliminating the null mutants, the probability of obtaining a deletion mutant is 0.46 for A909 and 0.37 for A909ΔrecA (P = 0.57).
We sought to determine whether there was a difference in the rate at which bacterial isolates with mutant forms of the alpha C protein were obtained from the wild-type and recA mutant challenged mice. The probability of obtaining any non-nine-repeat alpha C protein phenotype is lower in the recA mutant isolates: 0.46 versus 0.71 for A909 isolates. However, statistical analysis of the data using overdispersed binomial logistic regression (2) to account for both the number of mice and the number of isolates from each mouse revealed that this difference is not significant (P = 0.12). To determine whether the rate of deletion mutants alone differed between the two strains, the data were analyzed with elimination of the null mutants from the data set. The probability of obtaining a deletion mutant is 0.46 for A909 and 0.37 for A909ΔrecA, with a nonsignificant P value of 0.57.
Comparison of the spontaneous rate of alpha C protein tandem repeat deletion in the wild-type and recA mutant strains.
The mouse model of immune infection provides a powerful selection for mutations in the alpha C protein and has the advantage of illustrating the biological relevance of mutations in this bacterial surface protein. We thought it possible that the strength of this in vivo selection could obscure some effect of the recA mutation on the spontaneous rate of tandem repeat deletion in the alpha C protein. To address this, we compared the rate of spontaneous deletion in the alpha C proteins in the wild-type and mutant strains by colony immunoblots using a monoclonal antibody, 4G8, which specifically recognizes the repeat region of the protein (26). This monoclonal antibody is highly reactive with intact nine-repeat alpha C protein but only weakly reactive with lower-repeat-number forms of the protein (37). Based on our previous work that indicated the rate of spontaneous deletion was 6 × 10−4 (36), we prospectively determined that screening of approximately 10,000 colonies of each strain would allow us to detect a 10-fold decrease in the rate of mutation with a P value of <0.05. As described in Materials and Methods, each potential mutant identified by colony immunoblotting was isolated and analyzed by a Western blot to confirm the deletion phenotype. We screened 12,213 wild-type colonies and obtained 22 mutants for a rate of deletion of 1.8 × 10−3 (1 mutant per 555 colonies); we screened 9,457 mutant colonies and obtained 16 mutants for a rate of deletion of 1.7 × 10−3 (1 mutant per 591 colonies). The difference is not significant (P = 0.872), indicating that the recA mutation does not affect the rate of spontaneous tandem repeat deletion of the alpha C protein.
DISCUSSION
Single nucleotide changes that result in differences in enzyme activity or protein expression can confer a survival advantage to bacteria that are subject to adverse environmental conditions. Tandem repeat deletion within the alpha C protein gene confers a survival advantage to GBS bearing this surface protein by resulting in antigenic change that allows escape from humoral immunity, thereby serving as a pathogenicity trait in GBS. Similar to the deletions found in the M proteins of GAS (22), deletions in the alpha C protein occur by homologous recombination between intragenic repeats, in that the deletions are precise and occur between identical sequences, but the molecular mechanisms involved in the process are unknown.
Tandem repeat deletion has been most extensively studied with E. coli with both plasmid-based and chromosomal reporter constructs, and the dependence on RecA in these systems is variable, ranging from no RecA dependence for deletion of small (<100-bp) repeats to modest reductions (2- to 40-fold) in deletion rates for larger repeats (5, 6, 33, 34). Nontandem repeat deletion appears to be more dependent on RecA, with increasing distance between repeats (5, 34). We sought to determine whether RecA-mediated homologous recombination was involved in deletion of the 246-bp tandem repeats found in the alpha C protein during passage in an immune host.
Nothing is known about homologous recombination or recombinatorial mechanisms in general in GBS, and we report here the identification of a recA homologue in this species. The sequence of this gene is highly related to those of other reported streptococcal species and to the well-studied recA gene of E. coli. As evidenced by the high degree of conservation with putative active sites in the E. coli RecA protein (Table 1) and by the phenotype of sensitivity to UV light in the recA deletion mutant strain, it is expected that the GBS RecA protein plays a role in homologous recombination similar to that of RecA in E. coli. Deletion of recA in other bacterial species has a variable effect on the virulence of the organism (10, 16, 17, 46, 56); A909ΔrecA is significantly attenuated in our mouse model of sepsis, with an LD50 that is approximately 20-fold higher than that with wild-type A909. Although it has been shown that recA mutant E. coli bacteria grow poorly (48), our in vitro growth studies of A909ΔrecA demonstrate that during log phase, the mutant strain grows similarly to A909, suggesting that the attenuation of the strain is not due simply to a growth rate defect in the mutant. In addition to its role in homologous recombination, E. coli RecA is involved in LexA repressor cleavage and the induction of the SOS response, in chromosome partitioning, and in induced stable DNA replication (28). In S. pneumoniae, coordinate expression of recA and cinA is induced during competence (38, 44). GBS RecA may also be involved in bacterial processes aside from its role in homologous recombination, and the decreased virulence observed in our recA mutant strain could be due to the loss of another RecA function.
We found that tandem repeat deletion in the alpha C protein in vitro and in our mouse model of immune infection occurs in the absence of RecA. Deletion mutants were found with the wild-type GBS strain and with the strain of GBS with recA deleted, indicating that intramolecular recombination between tandem repeats in GBS does not require functional RecA. This observation is of particular importance, since we have been able to study this process on the chromosome with a protective surface antigen in a pathogenic species, rather than with an experimental plasmid construct. The mutant construct A909ΔrecA was created by insertional inactivation at the recA locus; thus, a partial recA sequence does remain on the chromosome. This partial sequence lacks the C-terminal 68 amino acid residues; in E. coli, the C-terminal region of RecA functions in double-stranded DNA binding (1, 29, 65). Although we cannot completely exclude the possibility that a truncated and partially functional form of the protein is present, the extreme sensitivity of this strain to the killing effects of UV light argue that the strain functionally has a deletion of RecA with respect to DNA repair, which requires the same processes—binding of single-stranded DNA, pairing of homologous sequences, and promotion of DNA strand exchange—as does RecA-mediated homologous recombination.
RecA-independent tandem repeat deletion in GBS may be explained by a number of alternate recombination mechanisms. It is possible that forms of RecA-like homologous recombination are involved in tandem repeat deletion in GBS; such a RecA-like function has been attributed to the RecE and RecT proteins of E. coli (28). Alternatively, other recombination mechanisms may be involved in tandem repeat deletion in GBS, including site-specific recombination, which involves highly specific sequences and unique proteins, and illegitimate recombination, which does not require DNA homology and is relatively inefficient, occurring at low frequency. RecA-independent tandem repeat deletion in E. coli, however, does not seem to be supported by any of these mechanisms, in that the process is not sequence specific, does involve homologous stretches of DNA, and occurs with some efficiency between repeats (6). It is also apparently influenced by replication factors such as DNA polymerases and helicases. Deletions of several genes in E. coli have been shown to result in increased levels of RecA-independent tandem repeat deletion, including deletion of dnaE (9) and dnaQ (51), which encode subunits of DNA polymerase III, and rep, which encodes a replicative helicase (9). Deletion of dnaB, which encodes a replication fork helicase, and deletion of uvrD, which encodes helicase II, appear to increase RecA-dependent tandem repeat deletion (8, 64).
In light of these observations, several models of RecA-independent tandem repeat deletion have been suggested in E. coli, including sister-strand exchange, replication slippage, rolling-circle replication, and misalignment exchange (6, 7, 33). The different models proposed for RecA-independent tandem repeat deletion are essentially all replication-driven models that invoke the formation of recombinogenic intermediates to result in intramolecular recombination. Each of these models would predict involvement of some elements of the RecA pathways distal to RecA, as well as predicting a prominent role for replication enzymes.
Which, if any, of the proposed mechanisms of RecA-independent tandem repeat deletion in gram-negative organisms might be functional in GBS is not known. Neither the baseline rate of spontaneous tandem repeat deletion in the alpha C protein nor the relatively high rate at which we recover deletion mutants in our immune mouse model of infection was significantly affected by the recA mutation. The spontaneous rate of deletion we report here is somewhat higher than previously reported (36); we attribute this difference to the fact that in the previous work only a relatively small number of colonies was screened. The rate of approximately 1.8 × 10−3 is much higher than the single base DNA replication error rate (estimated in prokaryotes to be 10−8 to 10−12) but is within the range of tandem repeat deletion rates reported for E. coli with plasmid-based reporter systems (ranging from 3 × 10−3 to 6 × 10−5) (27, 33, 34). It is likely that these deletions occur in vivo at an ongoing baseline rate and are biologically amplified in our immune mouse model by the survival advantage of the shift in antigenicity conferred by the deletions. Whether this process is mediated by alternate forms of homologous recombination, by site-specific recombination, by illegitimate recombination, or by replicational error may be best addressed by the identification of mutations in the recA-deleted background which significantly decrease the rate of tandem repeat deletion. The high degree of sequence conservation at the junctional regions between repeats suggests that these specific sequences may be important in facilitating recombination (30). The specificity and frequency of these deletions argue against a role for illegitimate recombination and support a role for site-directed recombination or replicational error. Replicational error, which under some circumstances might be disadvantageous to the species, may in the immune environment confer a survival advantage by resulting in antigenic variation.
It is also possible, but far less likely, that the binding of effective antibody to the alpha C protein transmits a signal that results in a targeted deletion process. This model would invoke a more site-specific process, which is attractive given the quite precise nature of these deletions. Yet it does not explain the rate of spontaneous tandem repeat deletion in the absence of immune pressure, unless it is assumed that the presence of effective antibody results in upregulation of a baseline rate of deletion. Further study of the recombination and replication processes of GBS, and in particular the identification of gene mutations that result in both increased and decreased rates of tandem repeat deletion, may lead to a better understanding of this process and other bacterial strategies for antigenic variation and immune evasion.
ACKNOWLEDGMENTS
This work was supported by National Research Service Award grant HD070466, National Institutes of Health grants AI38424 and AI33963, and National Institutes of Health contract N01-AI-75326.
We thank Beth Lurvey and Anne Behrens for technical assistance, Dennis Kasper for many helpful discussions, Eric Eichenwald for critical review of the manuscript, and Merton Bernfield, Steven Ringer, and Gary Silverman for their ongoing support of our work.
REFERENCES
- 1.Aihara H, Ito Y, Kurumizaka H, Terada T, Yokoyama S, Shibata T. An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J Mol Biol. 1997;274:213–221. doi: 10.1006/jmbi.1997.1403. [DOI] [PubMed] [Google Scholar]
- 2.Aitkin M, Anderson D, Francis B, Hinde J, editors. Statistical modelling in GLIM. Oxford, United Kingdom: Oxford University Press; 1989. [Google Scholar]
- 3.Alger L S, Lovchik J C, Hebel J R, Blackmon L R, Crenshaw M C. The association of Chlamydia trachomatis, Neisseria gonorrhoeae, and group B streptococci with preterm rupture of the membranes and pregnancy outcome. Am J Obstet Gynecol. 1988;159:397–404. doi: 10.1016/s0002-9378(88)80093-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson B E, McDonald G A, Jones D C, Regnery R L. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun. 1990;58:2760–2769. doi: 10.1128/iai.58.9.2760-2769.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi X, Liu L F. recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J Mol Biol. 1994;235:414–423. doi: 10.1006/jmbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 6.Bi X, Liu L F. recA-independent DNA recombination between repetitive sequences: mechanisms and implications. Prog Nucleic Acid Res Mol Biol. 1996;54:253–292. doi: 10.1016/s0079-6603(08)60365-7. [DOI] [PubMed] [Google Scholar]
- 7.Bi X, Liu L F. A replicational model for DNA recombination between direct repeats. J Mol Biol. 1996;256:849–858. doi: 10.1006/jmbi.1996.0131. [DOI] [PubMed] [Google Scholar]
- 8.Bierne H, Seigneur M, Ehrlich S D, Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol. 1997;26:557–567. doi: 10.1046/j.1365-2958.1997.6011973.x. [DOI] [PubMed] [Google Scholar]
- 9.Bierne H, Vilette D, Ehrlich S D, Michel B. Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol Microbiol. 1997;24:1225–1234. doi: 10.1046/j.1365-2958.1997.4381795.x. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier N A, Lipps C J, So M Y, Heffron F. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 11.Buscaglia C A, Alfonso J, Campetella O, Frasch A C. Tandem amino acid repeats from Trypanosoma cruzi shed antigens increase the half-life of proteins in blood. Blood. 1999;93:2025–2032. [PubMed] [Google Scholar]
- 12.Chaffin D O, Rubens C E. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- 13.Dunny G M, Lee L, LeBlanc D J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dybvig K, Hollingshead S K, Heath D G, Clewell D B, Sun F, Woodard A. Degenerate oligonucleotide primers for enzymatic amplification of recA sequences from gram-positive bacteria and mycoplasmas. J Bacteriol. 1992;174:2729–2732. doi: 10.1128/jb.174.8.2729-2732.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farley M M, Harvey R C, Stull T, Smith J D, Schuchat A, Wenger J D, Stephens D S. A population-based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N Engl J Med. 1993;328:1807–1811. doi: 10.1056/NEJM199306243282503. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher J J, Gordon R C. Perinatal transmission of bacterial sexually transmitted diseases. Part II: group B streptococcus and Chlamydia trachomatis. J Fam Pract. 1990;30:689–696. [PubMed] [Google Scholar]
- 17.Fuchs S, Muhldorfer I, Donohue-Rolfe A, Kerenyi M, Emody L, Alexiev R, Nenkov P, Hacker J. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb Pathog. 1999;27:13–23. doi: 10.1006/mpat.1999.0279. [DOI] [PubMed] [Google Scholar]
- 18.Gravekamp C, Kasper D L, Michel J L, Kling D E, Carey V, Madoff L C. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infect Immun. 1997;65:5216–5221. doi: 10.1128/iai.65.12.5216-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravekamp C, Rosner B, Madoff L C. Deletion of repeats in the alpha C protein enhances the pathogenicity of group B streptococci in immune mice. Infect Immun. 1998;66:4347–4354. doi: 10.1128/iai.66.9.4347-4354.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hervas J A, Gonzalez L, Gil J, Paoletti L C, Madoff L C, Benedi V J. Neonatal group B streptococcal infection in Mallorca, Spain. Clin Infect Dis. 1993;16:714–718. doi: 10.1093/clind/16.5.714. [DOI] [PubMed] [Google Scholar]
- 21.Hohn B, Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980;11:291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- 22.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 23.Hood D W, Deadman M E, Jennings M P, Bisercic M, Fleischmann R D, Venter J C, Moxon E R. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyer L L, Payne T L, Hecht J E. Identification of Candida albicans ALS2 and ALS4 and localization of als proteins to the fungal cell surface. J Bacteriol. 1998;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein B S, Hogan L H, Jones J M. Immunologic recognition of a 25-amino acid repeat arrayed in tandem on a major antigen of Blastomyces dermatitidis. J Clin Investig. 1993;92:330–337. doi: 10.1172/JCI116571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kling D E, Gravekamp C, Madoff L C, Michel J L. Characterization of two distinct opsonic and protective epitopes within the alpha C protein of the group B Streptococcus. Infect Immun. 1997;65:1462–1467. doi: 10.1128/iai.65.4.1462-1467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong D, Masker W. Deletion between direct repeats in T7 DNA stimulated by double-strand breaks. J Bacteriol. 1994;176:5904–5911. doi: 10.1128/jb.176.19.5904-5911.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurumizaka H, Aihara H, Ikawa S, Kashima T, Bazemore L R, Kawasaki K, Sarai A, Radding C M, Shibata T. A possible role of the C-terminal domain of the RecA protein. A gateway model for double-stranded DNA binding. J Biol Chem. 1996;271:33515–33524. doi: 10.1074/jbc.271.52.33515. [DOI] [PubMed] [Google Scholar]
- 30.Lachenauer C, Creti R, Michel J, Madoff L. Mosaicism in the alpha-like protein genes of group B streptococci. Proc Natl Acad Sci USA. 2000;97:9630–9635. doi: 10.1073/pnas.97.17.9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachenauer C S, Madoff L C. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect Immun. 1996;64:4255–4260. doi: 10.1128/iai.64.10.4255-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 33.Lovett S T, Drapkin P T, Sutera V A, Jr, Gluckman-Peskind T J. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993;135:631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovett S T, Gluckman T J, Simon P J, Sutera V A, Jr, Drapkin P T. Recombination between repeats in Escherichia coli by a recA-independent, proximity-sensitive mechanism. Mol Gen Genet. 1994;245:294–300. doi: 10.1007/BF00290109. [DOI] [PubMed] [Google Scholar]
- 35.Madoff L C, Hori S, Michel J L, Baker C J, Kasper D L. Phenotypic diversity in the alpha C protein of group B streptococcus. Infect Immun. 1991;59:2638–2644. doi: 10.1128/iai.59.8.2638-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madoff L C, Michel J L, Kasper D L. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect Immun. 1991;59:204–210. doi: 10.1128/iai.59.1.204-210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin B, Garcia P, Castanie M P, Glise B, Claverys J P. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls an SOS regulon. Dev Biol Stand. 1995;85:293–300. [PubMed] [Google Scholar]
- 39.McDonald H, Vigneswaran R, O'Loughlin J A. Group B streptococcal colonization and preterm labour. Aust N Z J Obstet Gynaecol. 1989;29:291–293. doi: 10.1111/j.1479-828x.1989.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 40.Michel J L, Madoff L C, Kling D E, Kasper D L, Ausubel F M. Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infect Immun. 1991;59:2023–2028. doi: 10.1128/iai.59.6.2023-2028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michel J L, Madoff L C, Olson K, Kling D E, Kasper D L, Ausubel F M. Large identical tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci USA. 1992;89:10060–10065. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy G L, Connell T D, Barritt D S, Koomey M, Cannon J G. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- 43.Musatovova O, Alderete J F. The Trichomonas vaginalis phenotypically varying P270 immunogen is highly conserved except for numbers of repeated elements. Microb Pathog. 1999;27:93–104. doi: 10.1006/mpat.1999.0281. . (Erratum, 28:191, 2000.) [DOI] [PubMed] [Google Scholar]
- 44.Pearce B J, Naughton A M, Campbell E A, Masure H R. The rec locus, a competence-induced operon in Streptococcus pneumoniae. J Bacteriol. 1995;177:86–93. doi: 10.1128/jb.177.1.86-93.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Casal J, Price J, Maguin E, Scott J. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 46.Pogson C A, Simmons C P, Strugnell R A, Hodgson A L. Cloning and manipulation of the Corynebacterium pseudotuberculosis recA gene for live vaccine vector development. FEMS Microbiol Lett. 1996;142:139–145. doi: 10.1111/j.1574-6968.1996.tb08421.x. [DOI] [PubMed] [Google Scholar]
- 47.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 48.Roca A I, Cox M M. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sancar A, Stachelek C, Konigsberg W, Rupp W D. Sequences of the recA gene and protein. Proc Natl Acad Sci USA. 1980;77:2611–2615. doi: 10.1073/pnas.77.5.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saveson C J, Lovett S T. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics. 1997;146:457–470. doi: 10.1093/genetics/146.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrag S J, Zywicki S, Farley M M, Reingold A L, Harrison L H, Lefkowitz L B, Hadler J L, Danila R, Cieslak P R, Schuchat A. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N Engl J Med. 2000;342:15–20. doi: 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 53.Shankar V, Baghdayan A S, Huycke M M, Lindahl G, Gilmore M S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67:193–200. doi: 10.1128/iai.67.1.193-200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stalhammar-Carlemalm M, Stenberg L, Lindahl G. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J Exp Med. 1993;177:1593–1603. doi: 10.1084/jem.177.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 56.Stroeher U H, Lech A J, Manning P A. Gene sequence of recA+ and construction of recA mutants of Vibrio cholerae. Mol Gen Genet. 1994;244:295–302. doi: 10.1007/BF00285457. [DOI] [PubMed] [Google Scholar]
- 57.Tao L, Hollingshead S K, Suvorov A N, Ferretti J J, McShan W M. Construction of a Streptococcus pyogenes recA mutant via insertional inactivation, and cloning and sequencing of the complete recA gene. Gene. 1995;162:59–62. doi: 10.1016/0378-1119(95)00273-9. [DOI] [PubMed] [Google Scholar]
- 58.Tapscott S J, Klesert T R, Widrow R J, Stoger R, Laird C D. Fragile-X syndrome and myotonic dystrophy: parallels and paradoxes. Curr Opin Genet Dev. 1998;8:245–253. doi: 10.1016/s0959-437x(98)80148-2. [DOI] [PubMed] [Google Scholar]
- 59.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Ham S M, van Alphen L, Mooi F R, van Putten J P. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73:1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- 61.Verghese A, Mireault K, Arbeit R D. Group B streptococcal bacteremia in men. Rev Infect Dis. 1986;8:912–917. doi: 10.1093/clinids/8.6.912. [DOI] [PubMed] [Google Scholar]
- 62.Wastfelt M, Stalhammar-Carlemalm M, Delisse A, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem. 1996;271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- 63.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 64.Yamashita T, Hanada K, Iwasaki M, Yamaguchi H, Ikeda H. Illegitimate recombination induced by overproduction of DnaB helicase in Escherichia coli. J Bacteriol. 1999;181:4549–4553. doi: 10.1128/jb.181.15.4549-4553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu C, Ferretti J. Molecular characterization of new group A streptococcal bacteriophages containing the gene for streptococcal erythrogenic toxin A (speA) Mol Gen Genet. 1991;231:161–168. doi: 10.1007/BF00293833. [DOI] [PubMed] [Google Scholar]