Summary
Mesenchymal stem cells (MSCs) are gaining increasing prominence as an effective regenerative cellular therapy. However, ensuring consistent and reliable effects across clinical populations has proved to be challenging. In part, this can be attributed to heterogeneity in the intrinsic molecular and regenerative signature of MSCs, which is dependent on their source of origin. The present work uses integrated omics-based profiling, at different functional levels, to compare the anti-inflammatory, immunomodulatory, and angiogenic properties between MSCs from neonatal (umbilical cord MSC [UC-MSC]) and adult (adipose tissue MSC [AD-MSC], and bone marrow MSC [BM-MSC]) sources. Using multi-parametric analyses, we identified that UC-MSCs promote a more robust host innate immune response; in contrast, adult-MSCs appear to facilitate remodeling of the extracellular matrix (ECM) with stronger activation of angiogenic cascades. These data should help facilitate the standardization of source-specific MSCs, such that their regenerative signatures can be confidently used to target specific disease processes.
Keywords: transcriptomics, proteomics, secretome, integrative, multi-omics, mesenchymal stem cell, immunomodulatory
Graphical abstract
Highlights
-
•
MSCs from different sources exhibit molecular and functional heterogeneity
-
•
Omics-based functional analysis defines source-specific regenerative signature
-
•
UC-MSCs promote robust immediate anti-inflammatory responses
-
•
Adult-MSCs facilitate ECM remodeling and promote angiogenesis
In a recently published study, Thakor et al. used integrative transcriptomic-proteomic and secretome analyses to identify differences in the molecular and functional “signature” between human mesenchymal stem cells (MSCs) derived from different tissue sources. These data will help to facilitate the correct MSC choice for tissue regeneration in different diseases.
Introduction
Regenerative medicine is characterized by the ability to replace or repair injured cells. Mesenchymal stem cells (MSCs) are a promising cellular therapy that can orchestrate tissue regeneration and can be used as an allogenic cellular therapy given their immunoevasive nature due to their low expression of major histocompatibility complex (MHC)-II antigens and other co-stimulatory molecules (Wood and Sakaguchi, 2003). Further, given their wide distribution throughout the human body, MSCs can be isolated from both neonatal (i.e., components of the umbilical cord) and adult (i.e., bone marrow and adipose tissue) tissue.
Recently, MSCs derived from neonatal sources (i.e., the different components of the umbilical cord [UC-MSC]), have gained increasing prominence given these embryonic cell populations have been shown to have enhanced differentiation, migratory, and secretory abilities compared with MSCs derived from adult tissues (Kwon et al., 2016; Mebarki et al., 2021; Nagamura-Inoue and He, 2014; Ullah et al., 2015). In addition, umbilical cord tissue is considered medical waste and retrieved at delivery using a painless, safe, and ethical approach, with cells maintaining their neonatal origin based on human leukocyte antigen (HLA) typing and karyotype analysis (Wu et al., 2018). However, despite its many advantages, the use of UC-MSCs in clinical trials is still limited, especially compared to its two main adult counterparts,MSCs derived from bone marrow (BM-MSCs) and adipose tissue (AD-MSCs), which can be attributed to a lack of understanding of the differences in regenerative potential among the different sources.
In the past, studies that have examined differences between UC-MSCs and BM-MSCs/AD-MSC, have focused on their (1) ability to form colonies, (2) expansion potential, (3) differentiation capacity, and (4) cell surface marker expressions (Fabre et al., 2019; Maleki et al., 2014; Montesinos et al., 2009; Sullivan et al., 2016). However, routine phenotypic characterizations alone cannot be used to identify differences in the functional properties of these cells, which has been reflected in the lack of consistent and reproducible success in many MSC-related clinical trials. Recently, several groups have attempted to compare molecular and functional similarities among multiple sources of MSCs using independent omics-based methods; i.e., transcriptional profiling (Elahi et al., 2016; Merimi et al., 2021; Sargent et al., 2018), proteomics (Billing et al., 2016; Kehl et al., 2019), and secretome analysis (Mizukami et al., 2019; Petrenko et al., 2020; Shin et al., 2021; Wangler et al., 2021). However, while data gleaned from these studies somewhat complement conclusions drawn from phenotypic analysis, one major drawback is that each of these studies focus on a different profiling methodology, which makes it difficult to establish a molecular equivalency among the various datasets. Furthermore, a comprehensive understanding of the regulatory behavior of cells should include an interaction among its various regulomes (i.e., mRNA, protein, and secretory molecules). Hence, an integrated analysis of the transcriptomic, proteomic, and secretory profiles would provide extremely useful insights that may not be deciphered from individual analysis of mRNA expressions or protein levels alone.
In the present study, we have therefore undertaken an integrative -omics comparison between UC-MSCs and the two main adult-MSCs used in preclinical and clinical studies: BM-MSCs and AD-MSCs. These data will help provide greater in-depth insight into the molecular heterogeneity between different sources of MSCs such that standardization can occur to allow the selection of source-specific MSCs that have specific properties that can used to develop targeted cellular therapy for certain diseases.
Results
To eliminate bias, all primary isolated MSCs (i.e., AD-MSCs, BM-MSCs, and UC-MSCs) were obtained from three independent donors (n = 3). For the adult-MSCs (AD-MSC and BM-MSC), the donors were chosen to include both male and female donors from different ethnicities (Table S1). An analysis of the MSC surface expression and differentiation potential did not reveal any differences among the different sources as well as the different donors within each source (Figure S1; Table S2).
Evaluating the transcriptomic heterogeneity between UC-MSCs and adult-MSCs
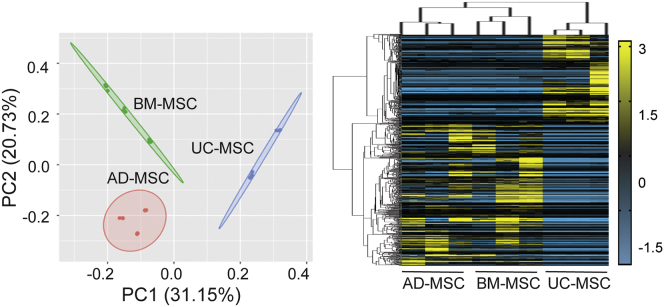
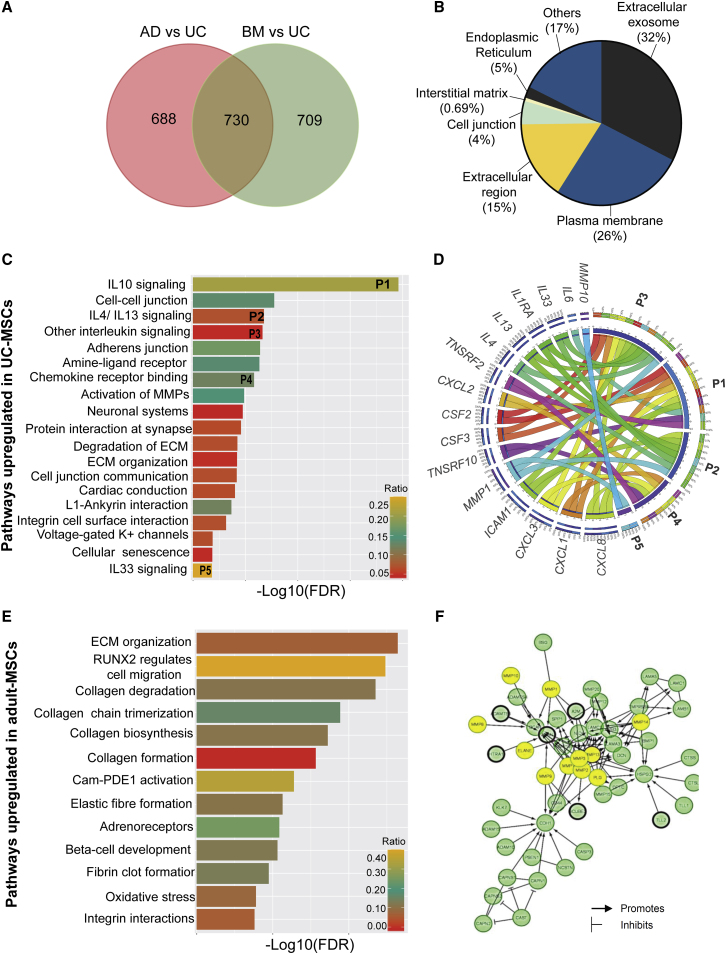
Principal-component analysis (PCA) demonstrated greater than 30% difference between UC-MSC and adult-MSC clusters, while differential expression analysis identified a total of 2,127 significant differentially expressed genes (DEGs) (Figure 1; Table S4) among all three MSC sources (Table S4), with 1,418 DEGs between UC-MSCs and AD-MSCs, and 1,439 DEGs between UC-MSCs and BM-MSCs. The phylogeny of the heatmap also confirmed that adult-MSCs had greater similarity in gene expressions (Figure 1) compared with UC-MSCs. Taken together, the data indicate a consistent difference in the molecular signature between UC-MSCs and adult-MSCs. An overlap of DEGs between UC-MSCs and adult-MSCs identified as many as 730 DEGs (∼51%) (Figure 2A) to be common between adult-MSCs. Gene Ontology (GO) analysis indicated that the common adult-MSC DEGs were predominantly associated with gene clusters localized to extracellular exosomes (32%), followed by plasma membrane (26%), extracellular space (15%), ER (5%), and cell-cell junction (4%) (Figure 2B; Table S4).
Figure 1.
Transcriptomic analysis between UC-MSCs and adult-MSCs
Principal-component analysis (PCA) of transcriptomic datasets for each MSC source (n = 3 donors per source) and corresponding heatmap of normalized fragments per kilobase of exon per million mapped fragments (FPKM) values from 2,127 statistically significant DEGs (Log |FC| > 2; FDR <0.05) between all sources of MSCs.
Figure 2.
Transcriptomic enrichment analysis
(A) Venn diagram showing the distribution of 2,127 DEGs in adult-MSCs compared with UC-MSCs.
(B) GO enrichment analysis indicates the cellular localization of the 730 common adult-MSC DEGs.
(C) All significant (FDR < 0.05) pathways upregulated in UC-MSCs compared with adult-MSCs.
(D) Circos plot depicts the relationship between enriched immune-regulatory pathways and their corresponding genes. Clockwise from top: pathways and genes are ordered by their number of interactions. Ribbon size encodes cell value associated with row/column segment pair. Column segment value and ribbon color are decided by the number of interactions.
(E) All significant (FDR < 0.05) pathways upregulated in adult-MSCs compared with UC-MSCs.
(F) Protein network of genes involved in extracellular matrix (ECM) organization pathway identified in adult-MSCs. Yellow circles indicate ECM proteolysis enzymes (MMPs, ELANE, and PLG) identified in our genomic screen. All other significant genes identified in our screen are highlighted with a black border. No fill circles indicate genes that are not validated experimentally.
UC-MSCs and adult-MSCs differ in their mode of cell-cell signaling
While the gene set enrichment analysis provided a weighted analysis of all genes that were differentially expressed between UC-MSCs and adult-MSCs, it did not provide any data related to the biological pathways that these DEGs affect and ultimately their functional outcome. A pathway analysis was performed separately with the up- and downregulated DEGs (http://www.reactome.org) (Gillespie et al., 2021) (Table S4). Pathways upregulated in UC-MSCs were predominantly related to paracrine signaling via interleukins and chemotactic cytokines (C-X-C-L) (Figure 2C). A Circos plot was used to probe the relationship between enriched chemokine pathways and the genes involved in these pathways (Figure 2D), which identified the activation of anti-inflammatory molecules (i.e., interleukin [IL]4, IL13, IL10, and IL6) that suppress the responses of myeloid cells and lymphocytes via tumor necrosis factor (TNF) receptors (TNFRSF10, TNFFRSF2) and STAT6 signaling (Figure 2D). The same analysis also indicated that, apart from inducing the expression of canonical anti-inflammatory genes, UC-MSCs also induced higher expression of several immune mediators (i.e., CXCL1, CXCL8, CSF2I, and CSF3; Figure 2D) that are canonically associated with activating cells of the innate immune system via their cognate CXCR2 receptors. Interestingly, UC-MSCs were also shown to promote cell migration and cell junction-mediated communications that facilitate ion transport (Figure 2C), all of which have been implicated in neuromuscular and cognitive functions (Könnecke and Bechmann, 2013).
In contrast, adult-MSCs were found to prefer signaling exclusively via extracellular matrix (ECM) remodeling, organization, and degradation (Figure 2E). A large number of ECM proteolytic enzymes, including matrix metalloproteinases (MMPs), elastogens (ELANE), and plasminogens (PLG), which facilitate cell-cell communications and angiogenesis, were found to be enriched in our screen. In particular MMP1, MMP3, MMP8, MMP10, and MMP13 identified in our screen (Figure 2F) are known to facilitate various growth factor signaling. Furthermore, collagen molecules (COL15A1, COL16A1, COL18A1) that constitute the structural framework of the basement membrane were also identified in our screen (Figure S3A), providing evidence toward upregulation of the angiogenic cascade in adult-MSCs. On the other hand, although ECM organization was identified as one of the significant pathways in UC-MSC transcriptomes, a closer look at the genes involved indicated a role for inter-cellular adhesion molecules ICAM1 and ICAM5 (Figure S3B), with strong protective roles in neurodegeneration (Birkner et al., 2019), as opposed to canonical angiogenesis related functions.
Evaluating the proteomic heterogeneity between UC-MSCs and adult-MSCs
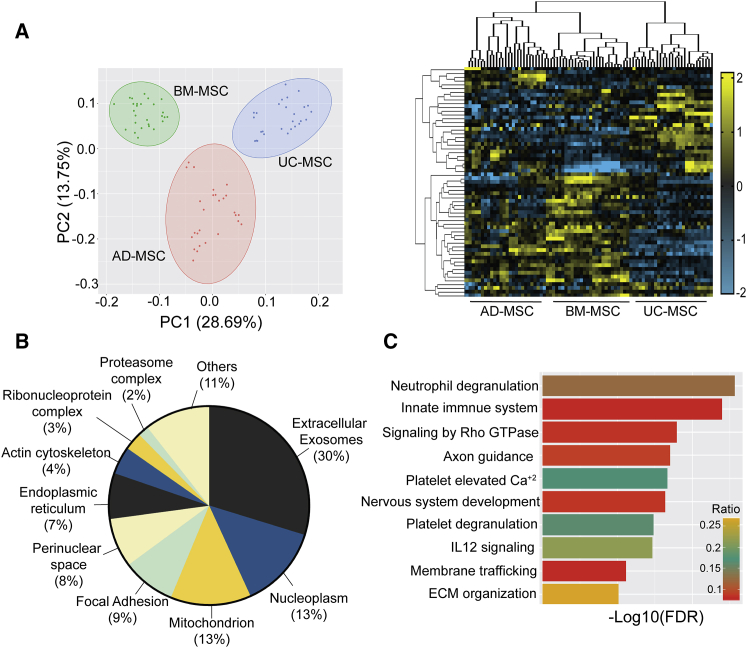
To complement the gene expression comparisons between adult-MSCs and UC-MSCs, we also compared protein levels among all three MSCs. After quality validation, 3,137 proteins were quantified and a PCA multivariate analysis indicated ∼30% difference between the proteome of BM-MSCs and UC-MSCs (Figure 3A), which is similar to that observed for the mRNA transcriptomes (Figure 1). Due to the high heterogeneity observed among the AD-MSC donors, it was difficult to compute a statistically relevant difference for AD-MSCs and other MSCs sources. A Z score-based statistical cutoff (Table S5) identified a total of 393 proteins in AD-MSCs, 610 proteins in BM-MSCs, and 705 proteins in UC-MSCs (Figure S4A). Hierarchical clustering of the 1,341 statistically significant proteins further validated the results obtained from our PCA analysis. Gene set enrichment analysis indicated that major differences among the proteome were clustered in the extracellular fraction of the proteome (30%) (Figure 3B; Table S5), similar to the subcellular localization identified in our transcriptomes (Figure 2B). Hence, in order to differentiate the functionally relevant proteome from that involved in intracellular crosstalk, we performed a pathway analysis exclusively with the extracellular fraction. Significant pathways important in all three sources of MSC signaling include innate immune system (i.e., neutrophil degranulation), anti-inflammatory pathways (i.e., IL12 signaling, VEGFA signaling), nervous system development (i.e., axon guidance, ECM organization), and anti-senescence pathways (i.e., packaging of telomere ends, meiotic recombination) (Figure 3C). Interestingly, other significant differences in proteome distribution were related to the mitochondrion (13%) and nucleoplasm (13%) (Figure 3B and Table S5).
Figure 3.
Proteomic analysis for AD-MSCs, BM-MSCs, and UC-MSCs
(A) PCA of all identified proteins for each MSC source (n = 3 donors per source) and the corresponding heatmap of normalized Z scores from 1,341 statistically significant proteins (Z > 2; FDR < 0.05) enriched among all three MSC sources.
(B) GO enrichment analysis for 1,341 significant proteins demonstrating their cellular localization.
(C) Top 10 pathways identified from gene enrichment analysis of proteins identified in the extracellular fraction.
Integrated analysis of the functional overlap between transcriptome and proteome
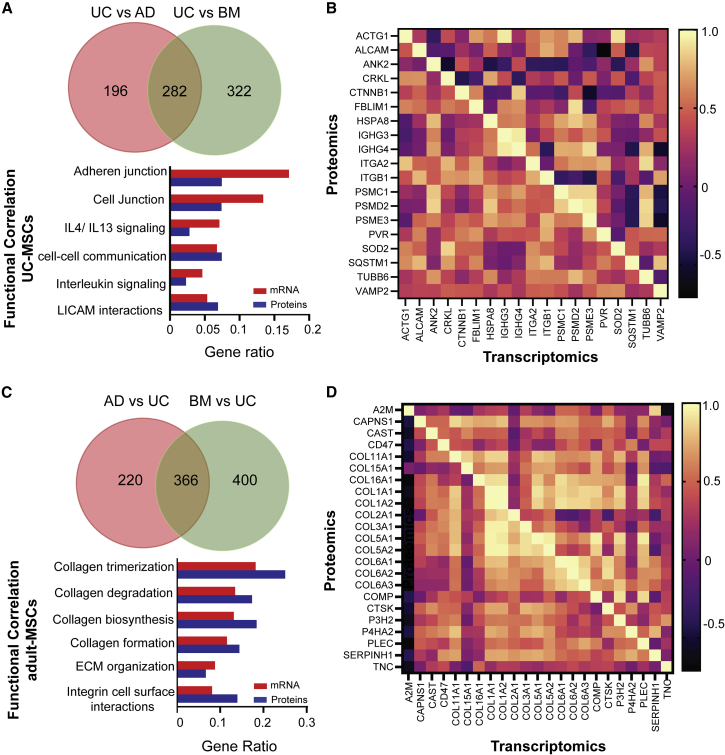
One of the objectives of this study was to correlate the regenerative signature among different sources of MSCs at different components of the regulome. In order to compare the expression level between these two datasets, a Z score for the mRNA transcripts was calculated in the same way as Z score for protein abundance quantification for each MSC source (Table S6), which indicated a high correlation between expression values of transcripts and proteins for each of the three MSC sources (Figure S4B). However, the observed read expression counts in the genomic screen were 50 times higher than that observed by mass spectrometry (Figure S5A), while the dynamic variability of deep sequencing had a 20-fold lower distribution than those obtained from proteomics (Figure S5B). Thus, considering these inherent statistical biases for higher abundance genes and to prevent loss of information arising from post-transcriptional regulatory events, we adopted an integrative approach wherein we extracted and compared the common functional processes enriched in both the transcriptomic and proteomic datasets, using significant differentially abundant proteins (DAPs) between UC-MSCs and adult-MSCs (Table S6).
Using this approach, we identified 478 proteins that were upregulated in UC-MSCs compared with AD-MSCs and 604 proteins were upregulated in UC-MSC compared with BM-MSCs. Among them, as many as 282 genes were found to be common among adult-MSCs (Figure 4A). Similarly, 586 proteins were downregulated in UC-MSCs compared with AD-MSCs, while 766 proteins were downregulated in UC-MSCs compared with BM-MSCs, with as many as 366 genes common among the adult-MSCs (Figure 4C). A functional overlap between the enriched pathways obtained from gene set enrichment analysis of the DEGs and DAPs (Figure S6; Table S6), identified as many as 6 significant pathways related to immune cell regulation that were upregulated in UC-MSCs (Figure 4A) among the Top10 category. However, the Pearson correlation coefficients for genes involved in the functionally correlated pathways were higher for the translated proteins as opposed to their mRNA counterpart (Figure 4B). Similarly, significant pathways that were consistently downregulated in UC-MSCs compared with adult-MSCs were related to the ability of MSCs to ensure cell-cell communication via ECM remodeling (Figure 4C). For adult-MSCs, all functionally relevant pathways were among the Top10 enriched pathways in both transcriptomic and proteomic over-enrichment studies, which was also reflected in the Pearson correlation coefficients (Figure 4D). This underscores the inherent heterogeneity that underlies mRNA transcript abundance and protein level variability, especially in cytokines as opposed to structural proteins.
Figure 4.
Functional correlation analysis between transcriptomics and proteomics
(A) Functional correlation analysis for UC-MSCs. Venn diagram shows the distribution of differentially abundant proteins (DAPs) upregulated in UC-MSCs compared with adult-MSCs. Bar plot shows the overlap of pathways between transcriptomics and proteomics datasets upregulated in UC-MSCs.
(B) Correlation heatmap between transcriptomics and proteomics of genes for functionally overlapped pathways for UC-MSCs.
(C) Functional correlation analysis for adult-MSCs. Venn diagram shows the distribution of DAPs upregulated in adult-MSCs compared with UC-MSCs. Bar plot shows the overlap of pathways between transcriptomics and proteomics datasets upregulated in adult-MSCs.
(D) Correlation heatmap between transcriptomics and proteomics of genes for functionally overlapped pathways for adult-MSCs.
Analysis of secretome for UC-MSCs and adult-MSCs
One of the paradigms in MSC-therapies is that their regenerative effect is mainly exerted through their ability to secrete various soluble factors (Joseph et al., 2020; Sagaradze et al., 2019). This is supported by the evidence that the actual engraftment and differentiation of MSCs at the damage site is very low and not persistent (von Bahr et al., 2012). Indeed, our integrated transcriptomic-proteomic analysis also identified significant functional differences among the cargo that constitutes the extracellular fraction of MSCs (Figures 2B and 3B). In order to correlate the secretory proteome with the biological pathways identified through our integrated transcriptomic-proteomic study, we also performed a targeted secretome analysis on the MSC-derived culture media (CM), and individual expression pattern of analytes was visualized using unsupervised hierarchical clustering (Table S7).
Secretome of adult-MSCs support angiogenesis while UC-MSCs mediate stronger immune responses
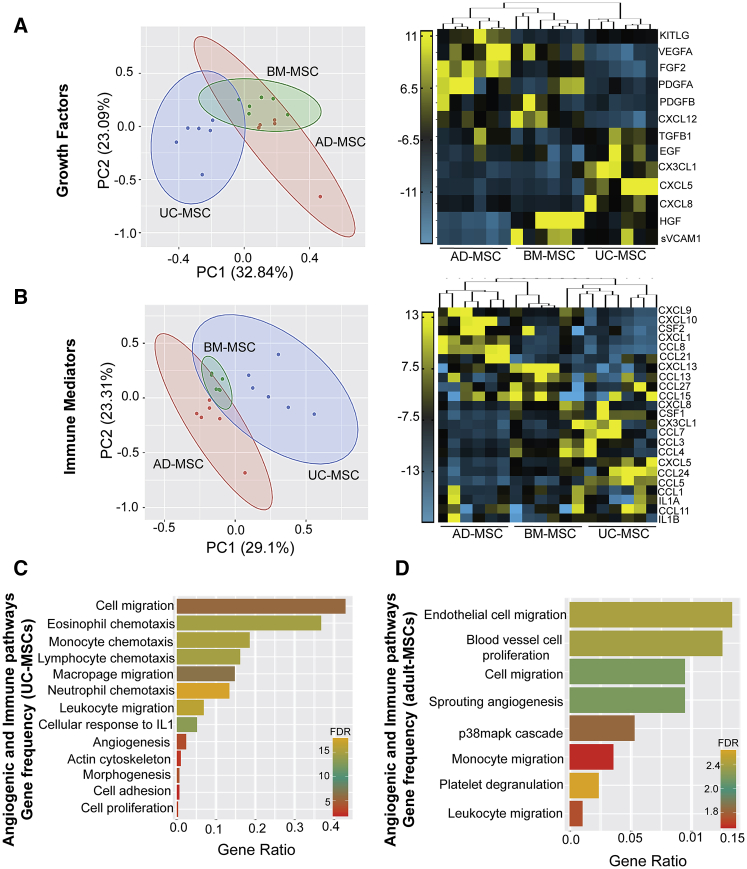
Consistent with our integrated transcriptomic-proteomic analysis, UC-MSCs were found to diverge significantly from adult-MSCs in the secretion of canonical growth factors (∼32%) (Figure 5A) and immune mediators (∼30%) (Figure 5B). Compared with UC-MSCs, adult-MSCs were found to secrete higher amounts of growth factors (i.e., FGF, PDGF, VEGF, and CXCL12) (Figure 5A), associated with homing, vascular permeability, and stability following tissue injury (Shibuya, 2011). On the other hand, several factors involved in the crosstalk between immune cells and angiogenesis were found to be highly secreted by UC-MSCs (Figure 5B). Prominent among them are CXCL3, CXCL5, and CXCL8, which act on the CXCR2 receptor found on neutrophils and endothelial and epithelial cells. Myeloid cell growth factor CSF1, which helps in differentiation and proliferation of neutrophil and monocytes but also potentiates differentiation to anti-inflammatory M2 macrophage phenotype (Figure 5B), were also found to be highly expressed by UC-MSCs. Surprisingly among all the MSCs, only AD-MSCs were found to secrete high levels of CXCL1, CXCL9, and CXCL10 ligands, which promote angiostasis (Figure 5B) via its CXCR3 receptor (Cui et al., 2010; Fulkerson and Rothenberg, 2006). Further, the colony-stimulating growth factor CSF2, which promotes pro-inflammatory M1 macrophage response, was also highly expressed in AD-MSCs. Among the adult-MSCs, BM-MSCs had the lowest secretory profile for both angiostatic factors and immune mediators, while showing higher expression for growth factors.
Figure 5.
Comparison of growth factors and immune mediators in the secretome of UC-MSCs and adult-MSCs
(A) PCA and heatmap of mean fluorescent intensity (MFI) values of (A) growth factors and (B) immune mediators secreted in CM from all three MSC sources (n = 3 donors per source). Comparison of gene ratio frequency (number of genes in our screen versus all background genes associated with that pathway) between significant (FDR < 0.05) angiogenic pathways (i.e., canonical and immune modulatory) enriched in (C) UC-MSCs and (D) adult-MSCs.
Interestingly, pathway enrichment analysis for UC-MSCs found the highest gene ratio values (0.4–0.1) with pathways related to positive regulation of microglial cell proliferation followed by activation of the innate immune system, including neutrophil chemotaxis, eosinophil chemotaxis, and macrophage migration (Figure 5C). Pathways related to angiogenesis, including positive regulation of angiogenesis, positive regulation of cytoskeleton organization, and anatomical structure morphogenesis, had the lowest gene ratio values (0.0002–0.01). Similarly, adult-MSC enriched pathways included endothelial cell chemotaxis, endothelial cell proliferation, and sprouting angiogenesis with the highest gene ratio values, while leukocyte chemotaxis-related terms had the lowest (Figure 5D).
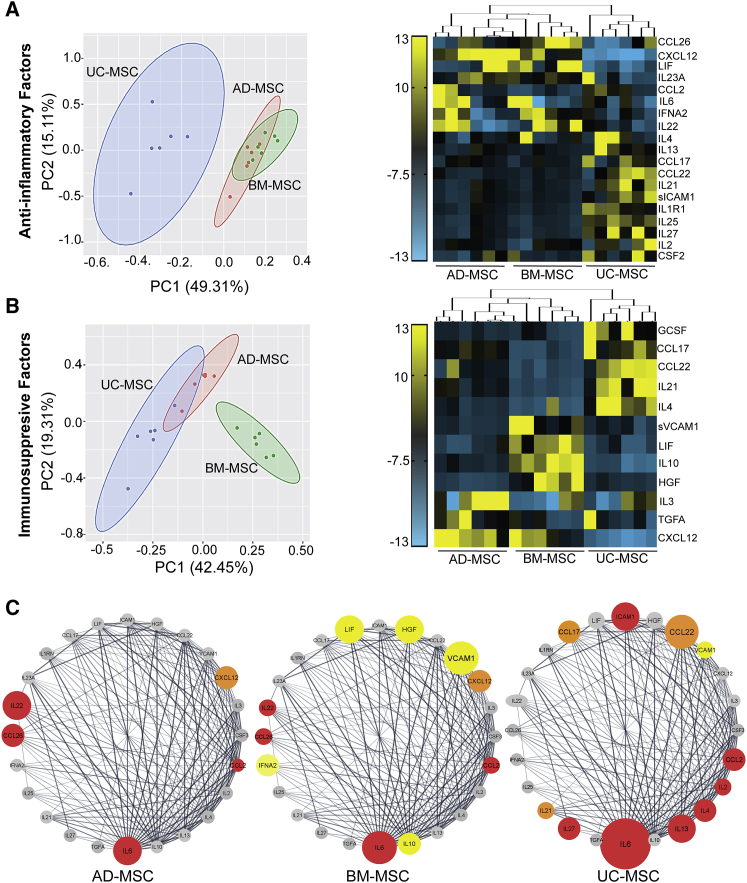
Immunomodulatory potential of UC-MSCs is lower than BM-MSCs but higher than AD-MSCs
Although activation of immune cells and angiogenesis cascade is essential to prevent further tissue damage and allow tissue renewal to begin, there are immunomodulatory checkpoints that regulate the continuous and premature activation of these signaling pathways in order to prevent long-term chronic inflammatory responses. Analytes in the CM of different sources of MSCs were categorized based on their immunomodulatory functions: (1) anti-inflammatory, inhibiting proliferation and differentiation of immune cells, and (2) immunosuppressive, including regulatory T cells for maintaining immune tolerance and limiting chronic inflammatory signals (Table S3). Interestingly, two-dimensional PCA indicated as much as 49% difference in the anti-inflammatory secretome between UC-MSCs and adult-MSCs (Figure 6A). The UC-MSCs had a predominantly anti-inflammatory secretome that was found to be enriched in T cell inhibitory interleukins (IL4, IL13, IL6, IL35, IL2, IL22, IL1R1, and IL25) as well as the colony-stimulating growth factor CSF3, which promote anti-inflammatory M2 macrophage polarization (Figure 6A).
Figure 6.
Comparison of immunomodulatory factors in the secretome of UC-MSCs and adult-MSCs
(A and B) PCA and heatmap of MFI values of (A) anti-inflammatory factors; and (B) immunosuppressive factors, secreted in culture media (CM) from all three MSC sources (n = 3 donors per source).
(C) Protein-protein interaction maps (PPI score >1.0 × 10−12) of 24 immunomodulatory factors were identified using the String database and ordered using the number of edges associated with each gene (degree sorted) in Cytoscape for AD-MSCs, BM-MSCs, and UC-MSCs. Genes with significant MFI values are color coded based on their anti-inflammatory (red), immunosuppressive (yellow), and anti-inflammatory/immunosuppressive dual roles (orange). Gray represents no change in MFI.
In contrast, PCA analysis for the immunosuppressive profile of MSCs indicated that BM-MSCs cluster away from both UC-MSCs and AD-MSCs with as much as 42% difference (Figure 6B). Indeed, BM-MSCs were found to be enriched specifically with immunomodulatory factors, including CCL2, CCL22, LIF, IL10, IL4, CD274, VCAM1, and HGF, associated with maintaining the Th1/Th2 balance. A protein-protein interaction (PPI) network analysis among all the identified immunomodulatory factors further validated the exclusive ability of BM-MSCs to secrete high amounts of immunosuppressive modulators (Figure 6C), although the mean expression level for immunomodulatory factors was found to be higher in UC-MSCs (Figure 6C). This was further confirmed using a gene enrichment analysis, which highlighted that immune-regulatory pathways upregulated in UC-MSCs were related to activation of the host innate immune response (Figure S6).
Assessing the immunogenic and angiogenic response of UC-MSCs and adult-MSCs in vitro
In order to assess the differential immunogenic and angiogenic responses exerted by different sources of MSCs and the variability associated with the donors within each source, we established in vitro models for activation of immune and angiogenic cascade.
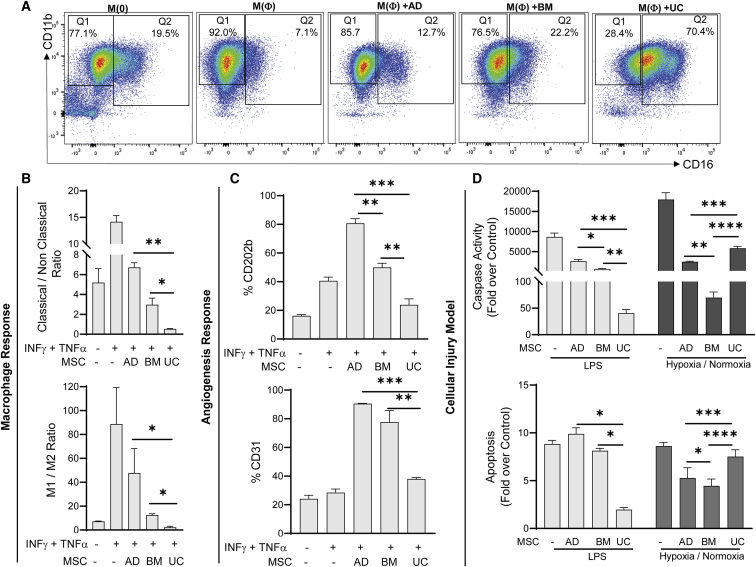
UC-MSCs induce non-classical monocyte and anti-inflammatory M2 macrophage differentiation
The ability of MSCs to induce different subpopulations of monocytes associated with different immune functions was assessed using macrophage/monocyte cells (RAW 264.7), where the cells were activated with inflammatory stimuli and co-cultured with different sources of MSCs. The expression of surface markers associated with classical (CD11b+CD16−) and non-classical (CD11b+CD16+) subsets were measured (Figure 7A). While adult-MSCs favored the migratory classical monocyte, UC-MSCs were found to promote a population shift toward the non-migratory and non-classical subset, which is associated with clearance of apoptotic cells as well as anti-viral and anti-bacterial immune surveillance roles in the vasculature (Figure 7B). In addition, we estimated the ability of different MSCs to induce polarization of differentiated macrophages into pro-inflammatory (M1) or anti-inflammatory (M2) phenotypes. Our data show that UC-MSCs were also able to induce a stronger polarization to the M2 phenotype (Figure 7B), which is in concordance with the higher expression of immune mediators observed in the UC-MSC secretome (Figure 5B). Furthermore, the ability of each source of MSCs to elicit a specific immune response was found to be consistent across all donors (Figure S7B).
Figure 7.
Differences in immune and angiogenic response between UC-MSCs and adult-MSCs
(A) Representative flow cytometry figures showing classical (CD11b+CD16−) and non-classical (CD11b+CD16+) monocytes populations in RAW 264.7 macrophages under resting (M(0)) and activated (M(Φ)) states (INFγ (150 ng/mL) + TNFα (50 ng/mL)) and following treatment with different MSC sources.
(B) Changes in population of monocytes (CD11b+CD16+/−) (top) and Macrophages (M1, CD86+; M2, CD206+) (bottom) following activation with INFγ (150 ng/mL) + TNFα (50 ng/mL) and treatment with different MSC sources.
(C) Changes in expression of CD202b (top) and CD31(PECAM1) (bottom) in MS-1 endothelial cells following activation with INFγ (150 ng/mL) + TNFα (50 ng/mL) and treatment with different MSC sources.
(D) Changes in Caspase-3/7 activity (top) and apoptosis (bottom) for HK-2 cellular injury models using LPS or hypoxia (ischemia)/normoxia (reperfusion). For all experiments, three different donors (n = 3) for each MSC source were used as replicates.
Data represents mean ± SEM. Statistical analysis was computed using one-way anova or Fisher's t-test; p∗ < 0.05, p∗∗ < 0.01 and p∗∗∗ < 0.001.”
Adult-MSCs stimulate angiogenesis signaling pathways
To assess the angiogenic potential of the three different MSC sources, pancreatic endothelial (MS-1) cells were exposed to inflammatory stimuli, and the ability of adult-MSCs to stimulate stronger angiogenic signaling cascades was assessed by computing the surface marker expression of adhesion molecules Angiopoietin-1 receptor (CD202b) and platelet endothelial cell adhesion molecule (PECAM1/CD31), both of which are known to play a critical role in vessel maturation and endothelial cell migration (Brindle et al., 2006; Cao et al., 2002). Furthermore, the variability in expression among the donors within each source was found to be negligible (Figure 7C).
In vitro models confirm differences in the regenerative signature between UC-MSCs and adult-MSCs
Although we validated the individual differences in the immunogenic and angiogenic potential between UC-MSCs and adult-MSCs, these effects may not reflect the systemic regenerative effect exerted by MSCs at sites of injury. As a proof of concept, we also developed two different cellular injury models by exposing proximal renal tubular epithelial cells (HK-2) to (1) Lipopolysaccharide (LPS), used to represent sepsis induced injury, and (2) hypoxia followed by normoxia, used to represent ischemia/reperfusion injury. Injured HK-2 cells were co-cultured with three different MSC sources and assayed for caspase activity as well as release of cytoplasmic nucleosomes as a surrogate for cell death (Figure 7D). For LPS-mediated injury, we expect an increased damage to cellular components and release of inflammatory cytokines via Toll-like receptor (TLR) signaling. Here, UC-MSCs were able to significantly reduce caspase activity and cell apoptosis compared with adult-MSCs. Similarly, following ischemia/reperfusion injury, which is characterized by direct mitochondrial damage and enhanced mitochondrial Reactive Oxygen Species (mitoROS, associated with inhibition of angiogenesis, adult-MSCs were found to reduce apoptosis more significantly than UC-MSCs. However, when comparing adult-MSCs, BM-MSCs were found to have a better therapeutic signature in attenuating cell death compared with AD-MSCs in both our model systems. In part, this may be attributed to the lower expression of angiostatic factors and a higher expression of immunosuppressive modulators in BM-MSCs compared with AD-MSCs (Figure 6C).
Discussion
This study demonstrated that the origin from which MSCs are derived plays a significant role in influencing their regenerative potential. In particular, we explored source-specific differences among MSCs in response to the different phases of tissue regeneration, including (1) suppression of the acute inflammatory cascade, and (2) initiation of cellular proliferation with concordant angiogenesis. Our integrative-omics study explored these differences at the genomic and proteomics levels with further validation using targeted secretome analysis and proof-of-concept in vitro models.
In our studies, UC-MSCs were found to have low expression of canonical growth factors, but were enriched in expression of immune mediators. Our proof-of-concept in vitro model studies showed that UC-MSCs can cause a shift in the population of classical monocytes toward their non-classical counterpart. Non-classical monocytes are crucial for activating the innate host defense against invading pathogens, including HIV (Saito et al., 2017) and parasites such as Leishmania (Oghumu et al., 2010), and in clearance of apoptotic endothelial cells from the vasculature (Narasimhan et al., 2019). Ironically, this immune surveillance function is also associated with secretion of pro-inflammatory cytokines such as TNFα and IL1β in response to pathogen detection (Kapellos et al., 2019). Further, UC-MSCs were also found to secrete high amounts of CC-chemokine ligands that promote differentiation of a subcategory of regulatory T cells (Tregs) via alternate M2 macrophage polarization (Ruytinx et al., 2018). Overall levels of anti-inflammatory molecules (i.e., IL4, IL13, IL12, IL35) were also found to be higher in UC-MSCs, which acts in different ways to prevent T cell proliferation (Garlanda and Mantovani, 2013). Taken together, activation of these pathways would be crucial in treating any acute inflammatory responses at sites of tissue injury or infection. Further evidence to support this finding comes from a large prospective, double-blind, multi-center, randomized phase 2a trial examining the effect of MSCs in ARDS (NCT02097641), in which BM-MSCs were shown to have no effect on patient mortality (Laffey and Matthay, 2017), whereas, in a recent double-blind, single-center, randomized, phase 1/2a trial, UC-MSCs were shown to improve the outcome in patients with ARDS from COVID-19 (Sharma and Zhao, 2021). This unique biological characteristic of UC-MSCs can possibly be attributed to the critical role that the umbilical cord plays in the maintenance of a healthy pregnancy. Recent longitudinal studies with blood samples of pregnant women have shown that, although induction of pregnancy induces an immunosuppressive state in the maternal placental system, the umbilical cord has an enhanced activation of neutrophils, natural killer (NK) cells, and mast cells to prevent infection to the fetus (Cornish et al., 2020).
Interestingly, adult-MSCs were found to be exclusively enriched in factors related to ECM remodeling, which provides the spatial localization for the dissemination of all inter-cellular signaling (Caley et al., 2015). This signaling is crucial in treating diseases associated with diabetic wound healing, re-vascularization following islet transplant, or ischemia-related tissue injury. In fact, based on a recent review by Wang et al. in 2021, despite the higher proliferation and differentiation ability of neonatal sources of MSCs, adult-MSCs still remain the preferred choice for the treatment of acute burn wound related injuries. Further, our transcriptomic pathway analysis also identified a critical role of adult-MSCs in β cell differentiation, which is transcriptionally linked (Bensellam et al., 2018) to islet re-vascularization. Similarly, most studies related to the treatment of ischemia-related diseases, including acute kidney injury (AKI) and myocardial infraction, have observed favorable results with the use of BM-MSCs as opposed to other sources (Pires et al., 2016), similar to our cellular model of ischemia/reperfusion.
In contrast to the significant difference observed between different sources of MSCs, intra-donor heterogeneity within the sources, for both transcriptomic and proteomic data were not found to be significant. Furthermore, the donor-associated variability was higher for the transcriptome, as opposed to the proteome. Previous studies of cellular economics have shown that genes with higher protein abundance not only express high mRNA but also stimulate higher expression of regulatory factors (Vogel et al., 2010), which can account for this observation. The highest donor variability was observed among AD-MSCs. In part, this can be attributed to the distinct metabolic profile that characterizes AD-MSCs based on their diverse anatomical distributions (Morais et al., 2021). However, the variability among donors did not affect the functional biological pathways, through which MSCs exert their regenerative effect, as validated by our in vitro cellular injury models.
Although multiple methods can be employed for developing a joint model to compare the different omics platforms, the ultimate objective of any study will dictate the manner of the integrative platform chosen (Haider and Pal, 2013). Despite the high correlation in expression values between transcriptome and proteome levels, mRNA abundance as a proxy to protein measurement often results in discrepancy due to factors impacting translational efficiency, protein degradation and clearance (Eldad and Arava, 2008; Hargrove and Schmidt, 1989; Shine and Dalgarno, 1974). This would affect the degree of overlap between the gene sets identified. Thus, a direct integration of the transcriptomic and proteomic datasets would potentially limit the number of identified protein-coding genes in the master list, leading to loss of genes related to signaling and metabolic pathways, whose expressions are tightly regulated via transcriptional feedback loops (Du et al., 2019). Additionally, signaling molecules are also characterized by low half-lives. Since the main goal of the present work is to compare the functional attributes between the different sources of MSCs and their associated regenerative signatures, we decided to extract and compare the common functional processes between the two omics platforms (Perco et al., 2010). One drawback of this method is that it does not allow for creating a dynamic dependency model between transcripts and proteins (Piruzian et al., 2010). However, this is only relevant for the identification of upstream transcriptional regulators as a surrogate for downstream protein functions, and was beyond the scope of this study. We also acknowledge our in vitro secretome study may not fully represent the differences between secreted proteins from different sources of MSCs in stimulated or adverse conditions, relative to the basal conditions used in the present study. Thus, a complete investigation of MSCs and MSC-derived extracellular vesicles and their effects in vivo is still necessary to validate our hypothesis-driven targeted treatment model.
Overall, our study integrates various omics platforms at multiple functional levels in order to decipher the molecular mechanisms associated with differences in regenerative signature observed among the different sources. Phenotypic characterizations cannot be solely used to standardize MSC therapies with the present data providing proof-of-concept validation for the need to use molecular signatures to fully understand and characterize MSC therapies for particular clinical indications. This understanding will hopefully help researchers and clinicians select source-specific MSCs targeted for specific disease indication. Further, it also opens up the possibility of using source-specific MSCs in combination, or even sequentially, depending on the time point when they are administered during the disease process.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources should be directed to and will be fulfilled by the corresponding author, Dr. Avnesh S Thakor (asthakor@stanford.edu).
Materials availability
This study did not generate any new cell lines. All human mesenchymal stem cell donors used were obtained from StemBioSys, USA.
Cell culture
All MSCs used in this study were obtained from StemBioSys, USA (https://www.stembiosys.com/), at passage (P) 1 and characterized by the minimal criteria defined by the International Society for Cell & Gene Therapy (ISCT) (Dominici et al., 2006). MSCs were expanded in vitro using the same culture conditions (supplemental experimental procedures) to ensure standardization.
Transcriptomics analysis and gene enrichment pathway visualization
Following sequencing, quality assessment, read filtering and genome mapping (see supplemental experimental procedures), DESeq2 (v3.14) (Love et al., 2014) was used to perform the differential gene expression analysis. DEGs were determined using a significance threshold of adjusted p <0.05 and log-fold change ≥2. Gene enrichment analyses of DEG (Gene Ontology Consortium, 2021; Ashburner et al., 2000) were performed using the Bioconductor packages (Gentleman et al., 2004) GO.db (v3.14 https://doi.org/10.18129/B9.bioc.GO.db). Functional pathway analysis was also performed using Reactome.db (https://doi.org/10.18129/B9.bioc.reactome.db) (Gillespie et al., 2021). Hierarchical clustering was performed using Cluster 3.0 using correlation uncentered data and single linkage clustering. The relationship between the biological pathways and corresponding genes was visualized using Circos (Krzywinski et al., 2009). Protein-network analysis was performed using the String database (Szklarczyk et al., 2015) and visualized using Cytoscape (v 3.9.0) (Shannon et al., 2003).
Peptide identification, quantification, and statistical analysis
The raw data files were searched using Byonic 2.11.0 software (Protein Metrics) against the Swiss-Prot human proteome database (2020, 20,385 entries) and peptide identification was filtered with false discovery rate (FDR) <0.01 and those identified in two or more spectra. All quantitative information was expressed in terms of Z scores at protein level (Navarro et al., 2014). Only proteins having a p value less than 5% and fold change higher than two were considered for further analysis. For identifying DAPs, we first computed a differential using the formula log(p value) × [mean(X)-mean(Y)]. Only proteins having a p value less than 5% and differential levels higher than 2 were considered for further analysis.
Luminex cytokine assays
Concentrations of cytokines in cell supernatants were measured using a custom Human Cytokine 80-plex Assay HCP2MAG-62K-PX23 (EMD, Millipore). Samples were prepared following the manufacturer’s protocol and normalized using both internal bead-based standards as well as by using only media as a negative control. Raw mean fluorescence intensity values (MFI) obtained were normalized across samples using MFI per 1 × 106 cells.
Immune and angiogenesis assays
Mouse macrophage/monocyte cell line RAW 264.7 and mouse pancreatic endothelial cell line MS-1 were used for the immunogenic and angiogenic assays respectively (see supplemental experimental procedures). RAW 264.7 cells and MS-1 cell lines were treated with inflammatory cocktail [INFγ (150 ng/mL) + TNFα (50 ng/mL)] in serum-free media for 24 h. After 24h, cells were co-incubated with different MSCs sources (n = 3 donors) in a contact-independent manner using a 0.6 μM Transwell in a ratio of 1:10 (MSC:cell) for an additional 24 h. Cells were then detached, washed with PBS, and stained with fluorochrome-labeled monoclonal for flow cytometry. Data were analyzed with FlowJo v10.2 software (TreeStar, United States).
In vitro cellular injury model
For a sepsis injury model, HK-2 cells were incubated with LPS at a concentration of 0.1 μg/mL (Sigma-Aldrich) in serum-free medium for 24 h. After 24 h, the medium was changed to normal medium and HK-2 cells were incubated with MSCs (n = 3 donor) in a contact-independent manner using 0.6 μM Transwell in a ratio of 1:10 for an additional 24 h. For the ischemia/reperfusion-induced model, HK-2 cells were subjected to hypoxia for 48 h, following which they were changed to normoxia conditions with fresh normal medium and incubated with MSC (n = 3 donor) in a contact-independent manner in a ratio of 1:10 for an additional 24 h. Cells were then detached, washed with PBS, and assayed for Caspase-3/7 activity (Promega, United States) and Apoptosis (Roche Diagnostic, Switzerland).
Author contributions
Conceptualization, A.G., G.S., and A.S.T.; methodology, A.G., G.S., FM., S.R., and A.B; investigation, A.G. and F.M; writing, A.G.; supervision, A.S.T. and S.P; editing: R.Y., R.P., and S.C.
Acknowledgments
This work was supported by NIH grant DK129598 and DK119293 and the Kidney for Dane Community, the Akiko Yamzaki and Jerry Yang Faculty Scholar Fund in Pediatric Translational Medicine from the Stanford Maternal and Child Health Research Institute. The authors thank Novogene (China) for mRNA NGS, Ken Lau at for the Canary Center at Stanford proteomics sample preparation, and Stanford Immuno-Phenotyping Core for secretome data acquisition.
Conflict of interests
The authors declare no competing interests.
Published: December 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.11.006.
Supplemental information
Data and code availability
The authors declare that all data supporting the findings of this study are available within the article and its supplementary material files. The accession number for the transcriptomics data reported in this paper is “GEO: GSE199826” and the accession number for the proteomics data is “PRIDE: PDXD032994”.
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensellam M., Jonas J.-C., Laybutt D.R. Mechanisms of β-cell dedifferentiation in diabetes: recent findings and future research directions. J. Endocrinol. 2018;236:R109–R143. doi: 10.1530/JOE-17-0516. [DOI] [PubMed] [Google Scholar]
- Billing A.M., Hamidane H.B., Dib S.S., Cotton R.J., Bhagwat A.M., Kumar P., Hayat S., Yousri N.A., Goswami N., Suhre K.J.S.r. Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Sci. Rep. 2016;6:21507. doi: 10.1038/srep21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkner K., Loos J., Gollan R., Steffen F., Wasser B., Ruck T., Meuth S.G., Zipp F., Bittner S. Neuronal ICAM-5 plays a neuroprotective role in progressive neurodegeneration. Front. Neurol. 2019;10:205. doi: 10.3389/fneur.2019.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle N.P.J., Saharinen P., Alitalo K. Signaling and functions of angiopoietin-1 in vascular protection. Circ. Res. 2006;98:1014–1023. doi: 10.1161/01.RES.0000218275.54089.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caley M.P., Martins V.L.C., O'Toole E.A. Metalloproteinases and wound healing. Adv. Wound Care. 2015;4:225–234. doi: 10.1089/wound.2014.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., O'Brien C.D., Zhou Z., Sanders S.M., Greenbaum J.N., Makrigiannakis A., DeLisser H.M. Involvement of human PECAM-1 in angiogenesis and in vitro endothelial cell migration. Am. J. Physiol. Cell Physiol. 2002;282:C1181–C1190. doi: 10.1152/ajpcell.00524.2001. [DOI] [PubMed] [Google Scholar]
- Cornish E.F., Filipovic I., Åsenius F., Williams D.J., McDonnell T. Innate immune responses to acute viral infection during pregnancy. Front. Immunol. 2020;11:572567. doi: 10.3389/fimmu.2020.572567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui A., Anhenn O., Theegarten D., Ohshimo S., Bonella F., Sixt S.U., Peters J., Sarria R., Guzman J., Costabel U. Angiogenic and angiostatic chemokines in idiopathic pulmonary fibrosis and granulomatous lung disease. Respiration. 2010;80:372–378. doi: 10.1159/000245332. [DOI] [PubMed] [Google Scholar]
- Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Du Y., Clair G.C., Al Alam D., Danopoulos S., Schnell D., Kitzmiller J.A., Misra R.S., Bhattacharya S., Warburton D., Mariani T.J., et al. Integration of transcriptomic and proteomic data identifies biological functions in cell populations from human infant lung. Am. J. Physiol. Lung Cell Mol. Physiol. 2019;317 doi: 10.1152/ajplung.00475.2018. L347–l360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi K.C., Klein G., Avci-Adali M., Sievert K.D., MacNeil S., Aicher W.K. Human mesenchymal stromal cells from different sources diverge in their expression of cell surface proteins and display distinct differentiation patterns. Stem Cells Int. 2016;2016:5646384. doi: 10.1155/2016/5646384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldad N., Arava Y. A ribosomal density-mapping procedure to explore ribosome positions along translating mRNAs. Methods Mol. Biol. 2008;419:231–242. doi: 10.1007/978-1-59745-033-1_16. [DOI] [PubMed] [Google Scholar]
- Fabre H., Ducret M., Degoul O., Rodriguez J., Perrier-Groult E., Aubert-Foucher E., Pasdeloup M., Auxenfans C., McGuckin C., Forraz N., et al. Characterization of different sources of human MSCs expanded in serum-free conditions with quantification of chondrogenic induction in 3D. Stem Cells Int. 2019;2019:2186728. doi: 10.1155/2019/2186728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson P.C., Rothenberg M.E. In: Encyclopedia of Respiratory Medicine. Laurent G.J., Shapiro S.D., editors. Academic Press; Oxford: 2006. Chemokines, CXC | CXCL9 (MIG) pp. 398–402. [Google Scholar]
- Garlanda C., Mantovani A. Ligands and receptors of the interleukin-1 family in immunity and disease. Front. Immunol. 2013;4:396. doi: 10.3389/fimmu.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa1113. D325–d334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S., Pal R. Integrated analysis of transcriptomic and proteomic data. Curr. Genomics. 2013;14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove J.L., Schmidt F.H. The role of mRNA and protein stability in gene expression. FASEB. J. 1989;3:2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- Joseph A., Baiju I., Bhat I.A., Pandey S., Bharti M., Verma M., Pratap Singh A., Ansari M.M., Chandra V., Saikumar G., et al. Mesenchymal stem cell-conditioned media: a novel alternative of stem cell therapy for quality wound healing. J. Cell. Physiol. 2020;235:5555–5569. doi: 10.1002/jcp.29486. [DOI] [PubMed] [Google Scholar]
- Kapellos T.S., Bonaguro L., Gemünd I., Reusch N., Saglam A., Hinkley E.R., Schultze J.L. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front. Immunol. 2019;10:2035. doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl D., Generali M., Mallone A., Heller M., Uldry A.-C., Cheng P., Gantenbein B., Hoerstrup S.P., Weber B. Proteomic analysis of human mesenchymal stromal cell secretomes: a systematic comparison of the angiogenic potential. NPJ Regen. Med. 2019;4:8. doi: 10.1038/s41536-019-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könnecke H., Bechmann I. The role of microglia and matrix metalloproteinases involvement in neuroinflammation and gliomas. Clin. Dev. Immunol. 2013;2013:914104. doi: 10.1155/2013/914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M.I., Schein J.E., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon A., Kim Y., Kim M., Kim J., Choi H., Jekarl D.W., Lee S., Kim J.M., Shin J.-C., Park I.Y. Tissue-specific differentiation potency of mesenchymal stromal cells from perinatal tissues. Sci. Rep. 2016;6:23544. doi: 10.1038/srep23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffey J.G., Matthay M.A. Fifty years of research in ARDS. Cell-Based therapy for acute respiratory distress syndrome. Biology and potential therapeutic value. Am. J. Respir. Crit. Care Med. 2017;196:266–273. doi: 10.1164/rccm.201701-0107CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki M., Ghanbarvand F., Reza Behvarz M., Ejtemaei M., Ghadirkhomi E. Comparison of mesenchymal stem cell markers in multiple human adult stem cells. Int. J. Stem Cells. 2014;7:118–126. doi: 10.15283/ijsc.2014.7.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebarki M., Abadie C., Larghero J., Cras A. Human umbilical cord-derived mesenchymal stem/stromal cells: a promising candidate for the development of advanced therapy medicinal products. Stem Cell Res. Ther. 2021;12:152. doi: 10.1186/s13287-021-02222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merimi M., Buyl K., Daassi D., Rodrigues R.M., Melki R., Lewalle P., Vanhaecke T., Fahmi H., Rogiers V., Lagneaux L., et al. Transcriptional profile of cytokines, regulatory mediators and TLR in mesenchymal stromal cells after inflammatory signaling and cell-passaging. Int. J. Mol. Sci. 2021;22:7309. doi: 10.3390/ijms22147309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami A., Thomé C.H., Ferreira G.A., Lanfredi G.P., Covas D.T., Pitteri S.J., Swiech K., Faça V.M. Proteomic identification and time-course monitoring of secreted proteins during expansion of human mesenchymal stem/stromal in stirred-tank bioreactor. Front. Bioeng. Biotechnol. 2019;7:154. doi: 10.3389/fbioe.2019.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos J.J., Flores-Figueroa E., Castillo-Medina S., Flores-Guzmán P., Hernández-Estévez E., Fajardo-Orduña G., Orozco S., Mayani H. Human mesenchymal stromal cells from adult and neonatal sources: comparative analysis of their morphology, immunophenotype, differentiation patterns and neural protein expression. Cytotherapy. 2009;11:163–176. doi: 10.1080/14653240802582075. [DOI] [PubMed] [Google Scholar]
- Morais T., Seabra A.L., Patrício B.G., Guimarães M., Nora M., Oliveira P.F., Alves M.G., Monteiro M.P. Visceral adipose tissue displays unique metabolomic fingerprints in obesity, pre-diabetes and type 2 diabetes. Int. J. Mol. Sci. 2021;22:5695. doi: 10.3390/ijms22115695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamura-Inoue T., He H. Umbilical cord-derived mesenchymal stem cells: their advantages and potential clinical utility. World J. Stem Cells. 2014;6:195–202. doi: 10.4252/wjsc.v6.i2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan P.B., Marcovecchio P., Hamers A.A.J., Hedrick C.C. Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. [DOI] [PubMed] [Google Scholar]
- Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., Núñez E., Martínez-Acedo P., Pérez-Hernández D., Jorge I., Mesa R., Calvo E., Carrascal M., et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014;13:1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- Oghumu S., Lezama-Dávila C.M., Isaac-Márquez A.P., Satoskar A.R. Role of chemokines in regulation of immunity against leishmaniasis. Exp. Parasitol. 2010;126:389–396. doi: 10.1016/j.exppara.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perco P., Mühlberger I., Mayer G., Oberbauer R., Lukas A., Mayer B. Linking transcriptomic and proteomic data on the level of protein interaction networks. Electrophoresis. 2010;31:1780–1789. doi: 10.1002/elps.200900775. [DOI] [PubMed] [Google Scholar]
- Petrenko Y., Vackova I., Kekulova K., Chudickova M., Koci Z., Turnovcova K., Kupcova Skalnikova H., Vodicka P., Kubinova S. A comparative analysis of multipotent mesenchymal stromal cells derived from different sources, with a focus on neuroregenerative potential. Sci. Rep. 2020;10:4290. doi: 10.1038/s41598-020-61167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires A.O., Mendes-Pinheiro B., Teixeira F.G., Anjo S.I., Ribeiro-Samy S., Gomes E.D., Serra S.C., Silva N.A., Manadas B., Sousa N., et al. Unveiling the differences of secretome of human bone marrow mesenchymal stem cells, adipose tissue-derived stem cells, and human umbilical cord perivascular cells: a proteomic analysis. Stem Cells Dev. 2016;25:1073–1083. doi: 10.1089/scd.2016.0048. [DOI] [PubMed] [Google Scholar]
- Piruzian E., Bruskin S., Ishkin A., Abdeev R., Moshkovskii S., Melnik S., Nikolsky Y., Nikolskaya T. Integrated network analysis of transcriptomic and proteomic data in psoriasis. BMC Syst. Biol. 2010;4:41. doi: 10.1186/1752-0509-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruytinx P., Proost P., Van Damme J., Struyf S. Chemokine-induced macrophage polarization in inflammatory conditions. Front. Immunol. 2018;9:1930. doi: 10.3389/fimmu.2018.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaradze G., Grigorieva O., Nimiritsky P., Basalova N., Kalinina N., Akopyan Z., Efimenko A. Conditioned medium from human mesenchymal stromal cells: towards the clinical translation. Int. J. Mol. Sci. 2019;20:E1656. doi: 10.3390/ijms20071656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Sejima H., Naito T., Ushirogawa H., Matsuzaki T., Matsuura E., Tanaka Y., Nakamura T., Takashima H. The CC chemokine ligand (CCL) 1, upregulated by the viral transactivator Tax, can be downregulated by minocycline: possible implications for long-term treatment of HTLV-1-associated myelopathy/tropical spastic paraparesis. Virol. J. 2017;14:234. doi: 10.1186/s12985-017-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent A., Shano G., Karl M., Garrison E., Miller C., Miller R.H. Transcriptional profiling of mesenchymal stem cells identifies distinct neuroimmune pathways altered by CNS disease. Int. J. Stem Cells. 2018;11:48–60. doi: 10.15283/ijsc17062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Zhao F. Updates on clinical trials evaluating the regenerative potential of allogenic mesenchymal stem cells in COVID-19. NPJ Regen. Med. 2021;6:37. doi: 10.1038/s41536-021-00147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: a crucial target for anti- and pro-angiogenic therapies. Genes Cancer. 2011;2:1097–1105. doi: 10.1177/1947601911423031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Lee J., Kwon Y., Park K.-S., Jeong J.-H., Choi S.-J., Bang S.I., Chang J.W., Lee C. Comparative proteomic analysis of the mesenchymal stem cells secretome from adipose, Bone Marrow, Placenta and Wharton’s Jelly. Int. J. Mol. Sci. 2021;22:845. doi: 10.3390/ijms22020845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.O., Gordon-Evans W.J., Fredericks L.P., Kiefer K., Conzemius M.G., Griffon D.J. Comparison of mesenchymal stem cell surface markers from bone marrow aspirates and adipose stromal vascular fraction sites. Front. Vet. Sci. 2015;2:82. doi: 10.3389/fvets.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Subbarao R.B., Rho G.J. Human mesenchymal stem cells - current trends and future prospective. Biosci. Rep. 2015;35:e00191. doi: 10.1042/BSR20150025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Abreu R.d.S., Ko D., Le S.Y., Shapiro B.A., Burns S.C., Sandhu D., Boutz D.R., Marcotte E.M., Penalva L.O. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bahr L., Batsis I., Moll G., Hägg M., Szakos A., Sundberg B., Uzunel M., Ringden O., Le Blanc K. Analysis of tissues following mesenchymal stromal cell therapy in humans indicates limited long-term engraftment and no ectopic tissue formation. Stem Cell. 2012;30:1575–1578. doi: 10.1002/stem.1118. [DOI] [PubMed] [Google Scholar]
- Wangler S., Kamali A., Wapp C., Wuertz-Kozak K., Häckel S., Fortes C., Benneker L.M., Haglund L., Richards R.G., Alini M., et al. Uncovering the secretome of mesenchymal stromal cells exposed to healthy, traumatic, and degenerative intervertebral discs: a proteomic analysis. Stem Cell Res. Ther. 2021;12:11. doi: 10.1186/s13287-020-02062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.J., Sakaguchi S. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 2003;3:199–210. doi: 10.1038/nri1027. [DOI] [PubMed] [Google Scholar]
- Wu M., Zhang R., Zou Q., Chen Y., Zhou M., Li X., Ran R., Chen Q. Comparison of the biological characteristics of mesenchymal stem cells derived from the human placenta and umbilical cord. Sci. Rep. 2018;8:5014. doi: 10.1038/s41598-018-23396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary material files. The accession number for the transcriptomics data reported in this paper is “GEO: GSE199826” and the accession number for the proteomics data is “PRIDE: PDXD032994”.