Summary
In the brain, the complement system plays a crucial role in the immune response and in synaptic elimination during normal development and disease. Here, we sought to identify pathways that modulate the production of complement component 4 (C4), recently associated with an increased risk of schizophrenia. To design a disease-relevant assay, we first developed a rapid and robust 3D protocol capable of producing large numbers of astrocytes from pluripotent cells. Transcriptional profiling of these astrocytes confirmed the homogeneity of this population of dorsal fetal-like astrocytes. Using a novel ELISA-based small-molecule screen, we identified epigenetic regulators, as well as inhibitors of intracellular signaling pathways, able to modulate C4 secretion from astrocytes. We then built a connectivity map to predict and validate additional key regulatory pathways, including one involving c-Jun-kinase. This work provides a foundation for developing therapies for CNS diseases involving the complement cascade.
Keywords: human induced pluripotent stem cells, iPSCs, human embryonic stem cells, ESCs, astrocytes, stem cell-derived astrocytes, disease modeling, complement system, complement component C4, C4, ELISA-based screen, screen, connectivity map, CMap, L1000
Highlights
-
•
New method for 3D differentiation of astrocytes from pluripotent stem cells
-
•
Transcriptional profiling identified these cells as dorsal fetal-like astrocytes
-
•
ELISA-based small-molecule screen identified regulators of C4 secretion
-
•
Connectivity map analysis identified additional pathways that regulate C4
Rapino and colleagues develop a 3D protocol for producing a homogeneous population of astrocytes from pluripotent cells. Combining an ELISA-based screen and a connectivity map assay, they identified compounds and signaling pathways that modulate secretion of complement C4. This work provides a foundation for developing therapies for CNS diseases involving the complement cascade.
Introduction
The complement system is part of the first line of defense against harmful pathogens (Dunkelberger and Song, 2010). While the primary site of complement synthesis is the liver, production of complement also happens in the central nervous system (CNS) (Morgan and Gasque, 1997). In addition to its role in CNS inflammation, the complement system shapes the developing brain by controlling synaptic refinement to ensure proper brain wiring and function (Magdalon et al., 2020). In the mature human brain, aberrant activation of the complement has been observed in the CNS of patients with neurodegeneration, autoimmune diseases, and aging. Excessive complement activation gives rise to early synaptic loss, correlating with cognitive impairment in Alzheimer’s disease (AD) and Tauopathy (Hong et al., 2016; Wu et al., 2019). Blocking the complement system in vivo rescues aberrant synaptic pruning and attenuates neuroinflammation and neurodegeneration in mouse models of AD (Dejanovic et al., 2018; Hong et al., 2016). These examples provide a strong rationale for targeting the complement system as a therapeutic approach to improving brain function.
Genetic variations in complement component 4 (C4) copy number have been linked to the increased risk of schizophrenia. In addition, schizophrenia patients exhibit elevated C4 expression in the cerebral cortex (Rey et al., 2020; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Sekar et al., 2016). Although the exact mechanism underlying schizophrenia is unknown, the neuropathology of patients’ brains is characterized by reduced thickness and synaptic density in the cortex (Glantz and Lewis, 2000; Thompson et al., 2001) consistent with the idea that C4 overexpression leads to enhanced microglia-mediated synaptic engulfment. (Comer et al., 2020; Yilmaz et al., 2020).
Astrocytes play a critical role in synapse formation, function, and elimination (Chaboub and Deneen, 2013). In the last decade, the contribution of astrocytes to neuropsychiatric and neurodegenerative diseases has been increasingly recognized (Druart and Le Magueresse, 2019; Seifert et al., 2006). Astrocytes and microglia are considered the immune cells of the brain due to their ability to secrete chemokines and cytokines, complement proteins, and for their phagocytic function. In particular, astrocytes express and secrete complement components, such as C1r, C1s, C2, C3, and C4, which, therefore, may act cell non-autonomously (Barnum, 1995; Gasque et al., 1995; Gordon et al., 1992; Guttikonda et al., 2021; Lian et al., 2015). Importantly, it has been shown that astrocyte-conditioned media (ACM) can increase neuronal levels of C4 and may participate in complement-mediated synaptic pruning (Sellgren et al., 2017). Despite the importance of mouse experiments, mice and human differ. For example, mice only have one C4 gene, whereas humans have two distinct genes, C4A and C4B (Carroll et al., 1990). Recent work has begun to shed light on the connection between C4AL copy number and microglia engulfment in vitro and in vivo (Sellgren et al., 2019; Yilmaz et al., 2020).
Together, these observations suggest that identifying factors that reduce the secretion of C4 from human astrocytes might be a viable approach to lowering synaptic C4 and preserving synapses in CNS diseases. Restricted accessibility to primary human astrocyte samples limits their use for in vitro studies that require defined genetics and the availability of large numbers of cells, such as small-molecule or genetic screens. Human induced pluripotent stem cells (hiPSCs) offer the unique ability to generate large numbers of patient-specific differentiated cells. However, many standard protocols for differentiating astrocytes from pluripotent stem cells require an extensive culture time (up to 6 months) (Dezonne et al., 2017; Krencik et al., 2011; Palm et al., 2015; Sloan et al., 2017) and/or shorter protocols that rely on the isolation of intermediate cells, such as neural and oligodendrocyte progenitor cells (Barbar et al., 2020; Jiang et al., 2013), or repeated cell passages (Byun et al., 2020; Jovanovic et al., 2021; Leng et al., 2021; Lundin et al., 2018; Peteri et al., 2021; Santos et al., 2017; Soubannier et al., 2020; Tcw et al., 2017). To successfully develop valuable preclinical models for high-throughput screening of astrocytes, it would be preferable to have differentiation protocols that can rapidly and reproducibly generate large numbers of those cells.
To better understand the regulation of C4 in human astrocytes, we developed a new bioreactor-based protocol for the rapid, reproducible, and large-scale production of human astrocytes (hASTROs). Single-cell RNA sequencing (scRNA-seq) confirmed that this protocol produces almost exclusively cells expressing pan-astrocytic markers, albeit exhibiting different levels of maturity. Importantly, a comparison between and within cell lines confirmed the robustness and reproducibility of this differentiation method. Applying a sensitive ELISA-based method, we screened an annotated small-molecule library, identifying several pathways that modulate C4 secretion. By analyzing connectivity map (CMap) datasets, we were also able to discover additional C4 regulators. Combining these two approaches led to the discovery and validation of sets of compounds, such as chromatin remodeling inhibitors (BRD and HDAC inhibitors), NF-κB, JAK, and JNK inhibitors, among others, that reduce C4 secretion from human astrocytes. This type of platform may lead to the discovery of drugs capable of reducing levels of CNS complement potentially diminishing excessive synaptic elimination in neurodegenerative and neuropsychiatric diseases.
Results
Rapid and efficient 3D differentiation of a pure population of astrocytes from pluripotent cells
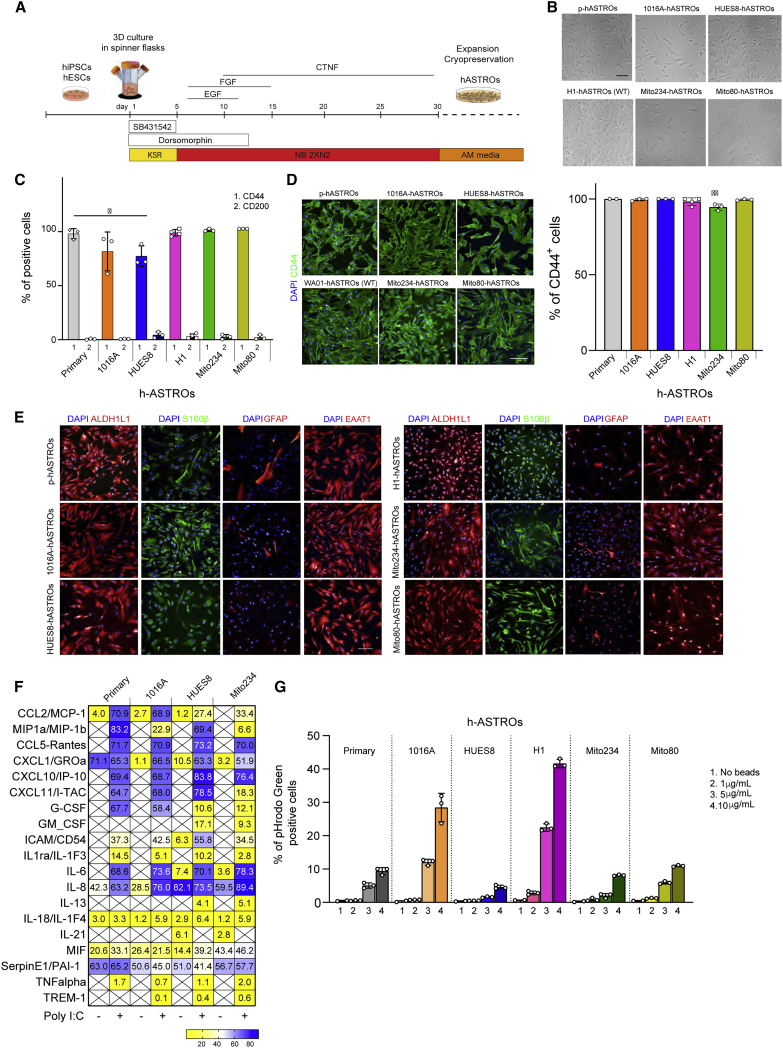
To generate large numbers of human astrocytes in a short period of time, we optimized a previously published protocol (Emdad et al., 2012). In brief, the embryonic stem cell (ESC) lines HUES8 and H1 and induced pluripotent stem cell (iPSC) lines 1016A, Mito234, and Mito80, were grown as spheres in 3D spinner flasks (Rigamonti et al., 2016). Pluripotent spheres were patterned toward a neuroectodermal fate using dual SMAD inhibition and further differentiated into astrocytes utilizing a combination of neurobasal medium and cytokines. After 30 days, spheres could be dissociated and cryopreserved or expanded in astrocyte medium (Figure 1A). Bright-field images of the differentiated astrocytes (hASTROs) show that their morphology closely resembled that of primary human astrocytes (p-hASTROs) (Figures 1B and S1A). To evaluate the purity of these cultures, dissociated cells (P0) were grown until 80% confluent and assessed by flow cytometry for the astrocyte- or neuron-specific cell surface markers (CD44 and CD200) (Lundin et al., 2018; Turac et al., 2013). All cell lines tested expressed high levels of CD44 (70%–100%), with only a negligible percentage (<1%) of the cells expressing CD200 (Figures 1C and S1B). Moreover, after the first passage (P1), nearly all the cells expressed CD44 (Figures 1D and S1C). For the different lines tested, 100% of the cells expressed CD44, except for Mito234 hASTROs in which 95% of the cells expressed CD44 after the first passage (Figure 1D). Antibody labeling demonstrated that these cells expressed other canonical astrocyte markers, such as ALDH1L1, S100β, CX43, and the glutamate transporter EAAT1, at levels comparable with those of p-hASTROs. However, only a small percentage of the astrocytes differentiated using this protocol expressed the intermediate filament protein GFAP (Figures 1E, S1D, and S1E).
Figure 1.
Differentiation and characterization of hASTROs
(A) Schematic of hASTRO differentiation method.
(B) Representative bright-field images of primary human astrocytes (p-hASTROs) and iPSC- and ESC-hASTROs, showing astrocyte-like morphology. Scale bar, 100 μm.
(C) Flow cytometry analysis of CD44 and CD200 expression in hASTROs compared with p-hASTROs. Data are means ± SD of biological differentiations. Each symbol represents an independent differentiation (n = 3). In the H1 line, the circles represent C4 WT hASTROs (n = 2), the squares represent C4 KO hASTROs (n = 2). For primary astrocytes each symbol represents a different passage of the same batch of primary human astrocytes (n = 3). One-way ANOVA, Dunnett’s multiple comparison test mean of each column versus primary human astrocytes. ∗Adjusted p = 0.0356.
(D) Representative immunofluorescence and quantification of CD44 expression in different cell lines. Scale bar, 100 μm. Data are means ± SD of biological differentiations (n = 3 per cell line). For H1, squares and circles represent C4 WT and C4 KO hASTROs, respectively. One-way ANOVA, Dunnett’s multiple comparison test mean of each column versus primary human astrocytes. ∗∗Adjusted p = 0.0097.
(E) Representative immunocytochemistry images of astrocyte markers ALDH1L1, S100β, GFAP, and EAAT1. Blue, DAPI staining. Scale bar, 100 μm.
(F) Human cytokine array quantification of supernatants from p-hASTROs, 1016A, HUES8, and Mito234-hASTROs in basal conditions or after stimulation with poly(I:C). Heatmaps represent averages of technical duplicates.
(G) Percentage of phagocytic astrocytes (GFP-positive cells) after incubation with different concentrations of pHrodo Green Zymosan A Bioparticle conjugate.
To evaluate the immune capability of hASTROs, supernatants were analyzed for the expression of 36 cytokines and chemokines in basal conditions or after stimulation with a TLR3 ligand (polyinosinic-polycytidylic acid [poly(I:C)]). We found that all astrocyte lines tested secreted a variety of factors, including CXCL1, MIF, and Serpin (Figures 1F and S1F). Moreover, stimulation with poly(I:C), induced secretion of pro-inflammatory cytokines and chemokines, such as IL-6, IL-8, MIP1a, and IL1RA, as described previously (Barbar et al., 2020; Choi et al., 2014; Tcw et al., 2017). Finally, we tested the functional phagocytic potential of hASTROs by measuring the engulfment of pHrodo green Zymosan particles (Figures 1G and S1G). We found that cell lines that were at slightly higher density when the assay was performed displayed higher percentages of GFP-positive cells (1016A and H1), suggesting that the phagocytic capacity of astrocytes may be influenced by cell density (Figure S1H).
Transcriptional profiling of stem cell-derived astrocytes
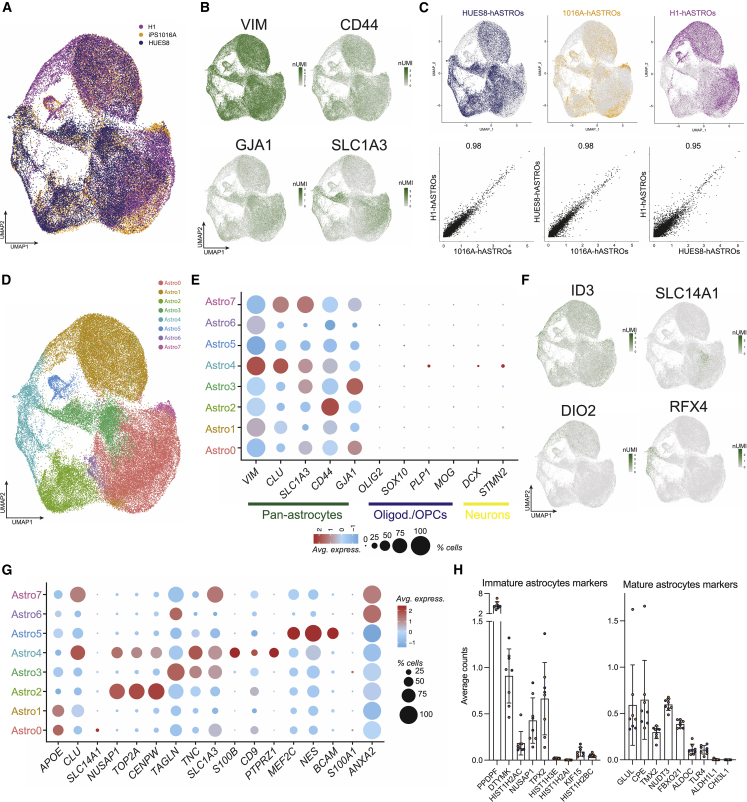
To further characterize the astrocytes to assess the reproducibility of our method, we performed scRNA-seq of three hASTRO lines (1016A, HUES8, and H1). UMAP analysis revealed equal distribution of the three lines in the populations of cells (Figure 2A). These cells expressed canonical markers of astrocytes, such as vimentin (VIM), CD44, gap junction protein alpha 1 (GJA1), and the SLC1A3 gene coding for excitatory amino acid transporter 1 (EATT1) (Figure 2B). Correlation of gene expression between the different lines confirmed that there was very low inter- and intra-line variability (Figures 2C and S2A). We next evaluated the heterogeneity of these astrocytes and the potential presence of subpopulations. We were able to distinguish eight different subgroups of cells (Figure 2D). All clusters expressed pan-astrocytic markers, but none expressed oligodendrocyte or oligodendrocyte precursor (OPC) (OLIG2, SOX10, PLP1, and MOG) or neuronal markers (DCX and STMN2). We identified a group of cells (Astro2) that expressed markers of immature astrocytes or cycling cells (such as NUSAP1, TOP2A, and CENPW), a group of cells (Astro5) containing more progenitor-like cells (expressing MEF2C, NES, and BCAM), and a group (Astro4) characterized by the expression of more mature markers (S100B, CD9, and PTPRZ1) (Figures 2G and S2B). To determine the maturation stage of the cells, we performed hierarchical clustering of our data and compared them with available datasets of mature (adult astrocytes) and fetal primary samples (fetal cortex and fetal pre-frontal cortex [PFC GW25], as well as astrocytes and excitatory neurons [iPSC-ExN]) produced from iPSCs (Figure S2C). As expected, hASTROs clustered farther from primary adult human astrocytes, total fetal cortex, and excitatory neurons. Interestingly, iPSC-derived astrocytes clustered together and closer to PFC GW25. A further analysis of recently identified markers for mature and immature astrocytes revealed that the three cell lines analyzed expressed markers of both immature (PPDPF, DTYMK, NUSAP1, TPX2, and others) and mature astrocytes (GLUL, CPE, NUDT3, and others) (Figure 2H).
Figure 2.
Transcriptomic analysis of hASTROs
(A) UMAP projection of three hASTRO lines.
(B) UMAP projections representing pan-astrocytic marker expression.
(C) UMAP projections of hASTROs from three lines with corresponding gene expression correlations.
(D) UMAP projections of identified subclusters.
(E) Dot plot for markers of different cell types in the brain.
(F) UMAP projections for expression of markers associated with cortical astrocytes.
(G) Dot plots representing expression of markers characteristic of each subgroup.
(H) Expression of markers associated with astrocyte maturation. Each dot represents a biological replicate; different colors represent the different cell lines (blue HUES8, orange 1016A and purple H1).
We assessed the regionality of hASTROs by plotting the expression of canonical dorsal and ventral markers confirming cortical identity (RFX4) and identifying both protoplasmic (ID3 and SLC14A1) and fibrous astrocytes (DI O 2) (Hodge et al., 2019; Schirmer et al., 2019) (Figures 2F and S2D). Gene expression correlated with differentiation patterning, suggesting that hASTROs have a rostral, rather than caudal, anatomical localization (Figure S2D). In summary, using this 3D differentiation protocol, we can generate a homogeneous population of human astrocytes with high reproducibility between and within cell lines.
Human astrocyte secretion complement C4 is regulatable
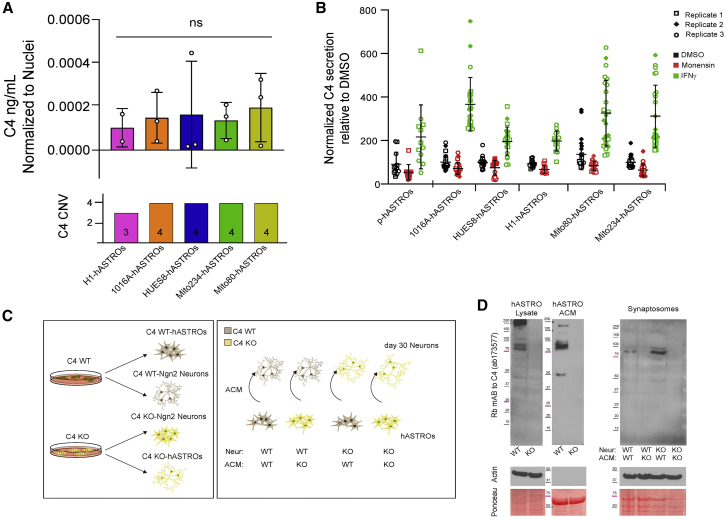
To measure C4 secretion from hASTROs and establish a reliable screening platform, we developed an ELISA-based assay. To ensure the specificity of the antibody used, we confirmed the absence of C4 secretion from a knockout (KO) ESC line (C4 KO-hASTROs) (Figure S3A). Using this assay, we found that iPSC and ESC hASTROs secreted detectable levels of C4 in basal conditions, as did pASTROs (Figures 3A and 3B). The amount of C4 secretion did not correlate with C4 copy number, since there was no significant difference observed between the lines tested (Figure 3A). C4 secretion was reduced by monensin (an ionophore that causes protein accumulation in the Golgi) and increased by treatment with interferon-γ (IFN-γ) (Walker et al., 1998) (Figure 3B).
Figure 3.
Human astrocytes secrete C4
(A) C4 secretion normalized to number of nuclei. Each circle represents the average of eight wells per biological replicate per line; n = 3 independent differentiations for 1016A, Hues8, Mito234, and Mito80, and n = 2 for H1. The lower panel represents the total copy number variation of C4 per line. Ordinary one-way ANOVA, Tukey’s multiple comparisons test. n.s., non-significant.
(B) Normalized C4 secretion measured by ELISA comparing the effects of monensin and IFN-γ in hASTROs. Data represent individual wells (n = 8) of three biological triplicates (squares, circles, and rhombi) per line. Statistics available in Table S1.
(C) Schematic of experimental design to assess C4 detection at synaptosomes using C4 KO and C4 WT-hASTROs and Ngn2 differentiated neurons.
(D) Western blots of C4 expression in C4WT and KO-hASTROs, total lysates, and concentrated astrocyte-conditioned media (ACM) (left panel); C4 expression on synaptosomes purified from C4-WT and KO-Ngn2 neurons incubated with C4 WT or C4 KO ACM in different combinations (right panel). Ponceau and actin are used as loading controls.
To verify that the ELISA was only detecting human C4, we assayed the supernatant of astrocytes cultured with two different media with or without FBS. We found no differences in the amounts of C4 secreted in basal conditions or with treatment. We did observe a stronger response to IFN-γ when the cells were cultured in NB media with or without FBS (Figure S3B). To elucidate the contribution of astrocyte-derived C4 to that present at synapses, we differentiated hASTROs and Ngn2 neurons from C4-WT and KO ESCs (Figure 3C). Western blots of total lysates and concentrated ACM from hASTROs showed specific recognition of C4 bands in the WT C4-hASTRO and total absence of signal in the C4 KO-ASTROs lanes (Figures 3D and S3C). ACM collected from C4-WT or KO hASTROs was incubated with WT or KO Ngn2 neurons in different combinations, as illustrated in Figure 3C. After 24 h, synaptosomes were prepared from each of the conditions and analyzed by western blotting. As expected, no C4 bands were detected when KO ACM was incubated with KO neurons. Furthermore, no C4 was detected when WT neurons were treated with KO ACM. However, when KO neurons were incubated with WT ACM, synaptosome-bound C4 was readily detected. This indicates that, at least under these experimental conditions, C4 released from astrocytes can bind to synaptic regions and that astrocytes are the primary source of synaptic C4 protein (Figures 3D and S3C).
Small-molecule screening identifies diverse pathways able to decrease C4 secretion
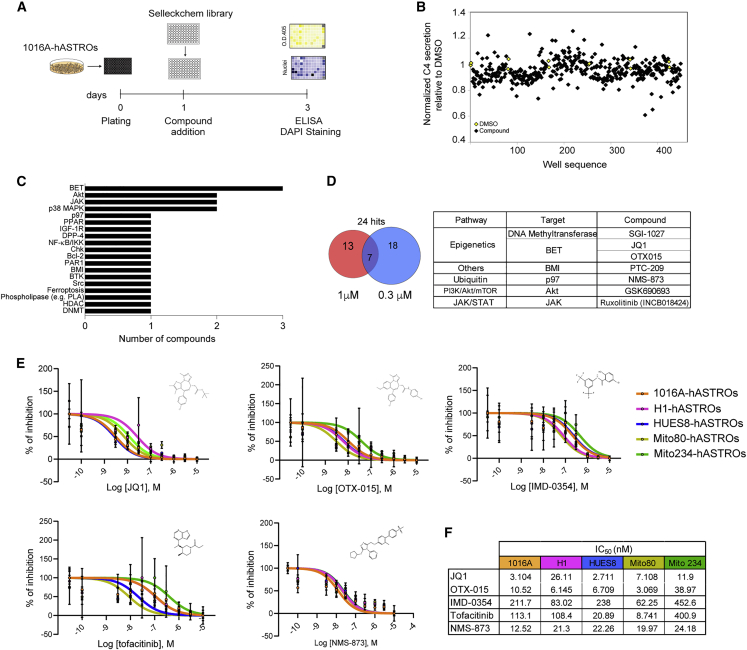
To better understand the molecular mechanisms that regulate astrocyte C4 release, we used the ELISA-based assay just described, with monensin and IFN-γ as controls, to perform a small-molecule screen of 464 unique inhibitors, covering a wide variety of signaling pathways, each tested at two different concentrations. ELISAs were performed on supernatants from each well, and plates were stained with DAPI to quantify the number of nuclei per well (Figure 4A). Absorbance values were normalized for cell number; triplicates were averaged and normalized relative to DMSO controls (Figure 4B). Hits were ranked based on the percentage decrease of C4 release compared with DMSO. The top 24 compounds that were active at either one or both concentrations were selected for further analysis. An examination of the set of hit compounds confirmed that there was no bias due to the extent of representation of the compounds in the library (Figure S4A). The highest represented hit target was the bromodomain extra terminal (BET) subfamily of chromatin readers (3 compounds), followed by AKT, JAK, and p38 MAPK inhibitors (Figure 4C). Of the 24 hits, 7 compounds scored at both concentrations tested. Of those, 4 regulate gene expression and the others are inhibitors of ubiquitination, PI3K/AKT/mTOR, or JAK/STAT signaling (Figure 4D).
Figure 4.
Small-molecule screen to identify C4 modulators
(A) Schematic of the screen.
(B) Scatterplot of C4 secretion (black squares represent the average of triplicates) compared with DMSO (yellow squares are averages of triplicates) at 1 μM.
(C) Bar graph showing the targets of the identified compounds.
(D) Venn diagram showing the number of hits selected from 1 or 0.3 μM concentration and their overlap. Table shows overlapping compound targets and pathways.
(E) Normalized dose-response curves showing the percent inhibition of selected compounds tested on hASTROs. Individual points represent mean ± SD. n = 3 replicate wells for each dose; different colors represent the different cell lines tested.
(F) Table with calculated IC50 for JQ1, OTX-015, IMD-0354, tofacitinib, and NMS-873 in the five cell lines tested.
Selected compounds were validated using 4 different concentrations (1, 0.3, 0.1, and 0.03 μM). Of the 24 compounds tested, 20 showed a decrease of C4 in a dose-dependent manner (Figure S4B; Table S2). We then tested these compounds on hASTROs produced from an iPSC line obtained from a patient diagnosed with schizophrenia (Mito234-hASTROs). This line contains the same C4 copy number (CNV = 4) as the 1016A-hASTRO line; however, it has a different ratio of C4AL to C4BL (Figure S4C). A few compounds were somewhat more toxic to Mito234-hASTROs, as measured by a larger decrease in nuclei number after treatment (Figure S4D; Table S3). We do not yet know if this was related to the donor’s disease state or to line-to-line variability. Hence, we decided to focus on compounds, including kinase inhibitors (JAK, p38 MAPK, Akt, and Src), a nuclear receptor inhibitor (PPAR antagonist), an ATPase (VCP/p97) inhibitor, and epigenetic regulators/modifiers (BET inhibitors and a BMI inhibitor), that had consistent effects on both lines.
To assess whether the difference observed in the potency of the compounds was biologically relevant, dose-response assays were conducted to identify effective concentrations for the hits in hASTROs differentiated from ESCs (HUES8 and H1) or iPSCs from healthy controls (1016A) and disease patients (Mito80 and Mito234) (Figure 4E). Log half-maximal inhibitory concentration (IC50) calculation revealed that all compounds tested were effective in the nanomolar range, with BRD inhibitors (JQ1 and OTX-015) and NMS-873 on average showing higher potencies (Figure 4F). A closer look at the IC50 results did not reveal any correlation between the potency of the compound, the C4 copy number, and the origin of the cells (iPSC versus ESCs or heathy versus disease).
Three different pathways decrease hASTRO C4 secretion and block the response to pro-inflammatory stimuli
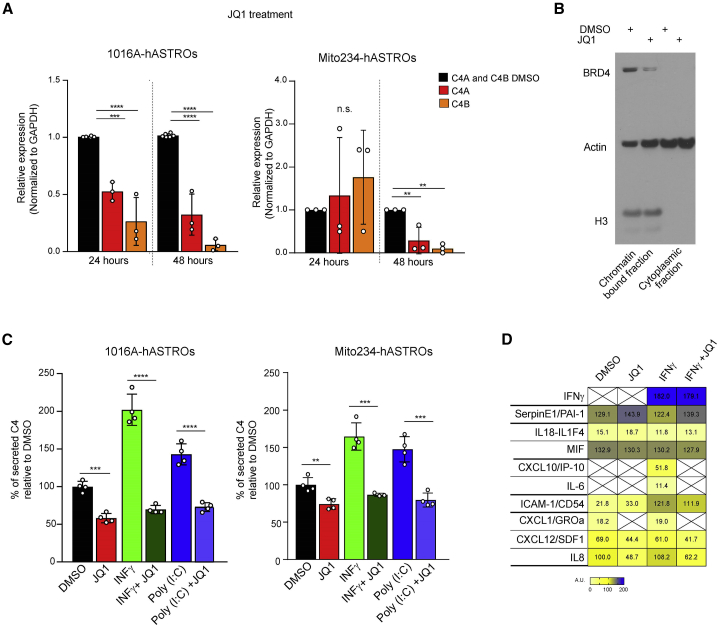
To explore the mechanism of C4 regulation in hASTROs, we studied the effects of compounds belonging to three different pathways: JQ1, a BET inhibitor; IMD-0354, an NF-κB inhibitor; and tofacitinib (Xeljanz), a clinically approved JAK inhibitor. These compounds are known to have anti-inflammatory properties, but their specific roles in modulating C4 secretion in human astrocytes had not been explored. We first treated 1016A and Mito234-hASTROs with JQ1 for 24 and 48 h and found a reduction in C4A and C4B mRNA at 48 h in both lines, consistent with the known effects of JQ1 on gene transcription (Figure 5A). TaqMan probes used to detect C4A and C4B were validated using the C4-KO hASTRO line (Figure S5A). To assess whether JQ1 was working as a BET inhibitor, we quantified the amount of BRD4 in the chromatin fraction of cells treated for 24 h with JQ1. We confirmed that BRD4 was displaced from chromatin, leading to a decrease in mRNA transcription (Figures 5B, S5B, and S5C). JQ1 also downregulated the mRNA levels of other key complement components, such as C1s, C2, C3, and C5 (Figure S5D).
Figure 5.
JQ1 represses C4 transcription and interferes with pro-inflammatory signaling
(A) qPCR of C4A and C4B in hASTROs treated with DMSO or JQ1 in 1016A and Mito234-hASTROs. Data are presented as biological triplicates. One-way ANOVA, ∗∗∗p < 0.0001, ∗∗∗∗p ≤ 0.0001, ∗∗p = 0.0026; n.s., non-significant.
(B) BRD4 displacement from chromatin in astrocytes treated with JQ1 at 1 μM for 24 h compared with DMSO control. The nuclear marker histone H3 was used as a control for the nucleus/cytoplasm fractionation.
(C) C4 ELISA of 1016A-hASTROs and Mito234-hASTROs treated with IFN-γ or poly(I:C) with or without JQ1. Data are presented as means of technical replicates ± SD relative to DMSO control (100% secretion) n = 4 wells per condition. Unpaired t test, ∗∗p = 0.005, ∗∗∗p = 0.0001, ∗∗∗∗p < 0.0001.
(D) Heatmap showing cytokine measurement in supernatants of 1016A-hASTROs treated with DMSO, IFN-γ, JQ1, or IFN-γ and JQ1.
We then asked if JQ1 could reduce the effects of two different inflammatory stimuli, IFN-γ and poly(I:C), both of which increased the secretion of C4. C4 secretion was blocked and returned below baseline when 1016A- and Mito234-hASTROs were co-treated with pro-inflammatory stimuli and JQ1 (Figure 5C). To expand our investigation of the effects of JQ1 on cytokine secretion, we used the cytokine array already described and quantified levels of cytokines secreted from hASTROs with or without additional IFN-γ treatment. Nuclear counts were used to exclude the contribution of cell number changes (Figure S5F). Under normal culture conditions, JQ1 decreased the expression of CXCL10 and IL-8 and completely abolished the secretion of the chemokine CXCL1. Stimulation with IFN-γ promoted the secretion of CXCL10, IL-6, and ICAM-1. JQ1 treatment entirely blocked the secretion of CXCL10 and IL-6 but had a much smaller effect on ICAM-1. No effect of JQ1 on Serpin E1, IL18, MIF, or IFN-γ secretion was observed (Figures 5D and S5E).
Next, we carried out additional studies of IMD-0354. To validate the mechanism of action (MoA) of this NF-κB inhibitor, we treated cells with poly(I:C) and measured p65 nuclear translocation (Figure S5G). As expected, quantification of the percentage of p65-positive nuclei confirmed that IMD-0345 strongly inhibits this process (Figure S5H). We next investigated the effect of IMD-0354 on C4 transcription, finding that the C4 decrease at 48 h could be mainly attributed to reducing C4B mRNA, whereas C4A was transiently upregulated after 24 h of treatment, subsequently returning to baseline levels (Figure S5I). Although similar trends were observed with Mito234-hASTROs, no statistically significant decrease of either C4A or C4B was found at 48 h. When we challenged the astrocytes with inflammatory stimuli and co-treated with IMD-0354, we observed a significant decrease in C4 secretion under all conditions tested (Figure S5J). We then looked at the effect of the JAK-STAT inhibitor tofacitinib on C4 modulation. hASTROs treated with tofacitinib showed a robust decrease in both C4A and C4B mRNA levels in 1016A- and Mito234-hASTROs (Figure S5K). Moreover, tofacitinib treatment decreased C4 secretion both under basal conditions and upon treatment with pro-inflammatory stimuli (Figure S5L).
In summary, we confirmed that three compounds targeting different signaling pathways and cellular mechanisms reduce C4 secretion from human astrocytes under baseline conditions and block the increase in secretion that was produced by treatment with two inflammatory stimuli. Interestingly, JQ1, as might have been expected, reduced the secretion of additional complement components and cytokines.
Building a CMap to predict novel C4 modulatory pathways
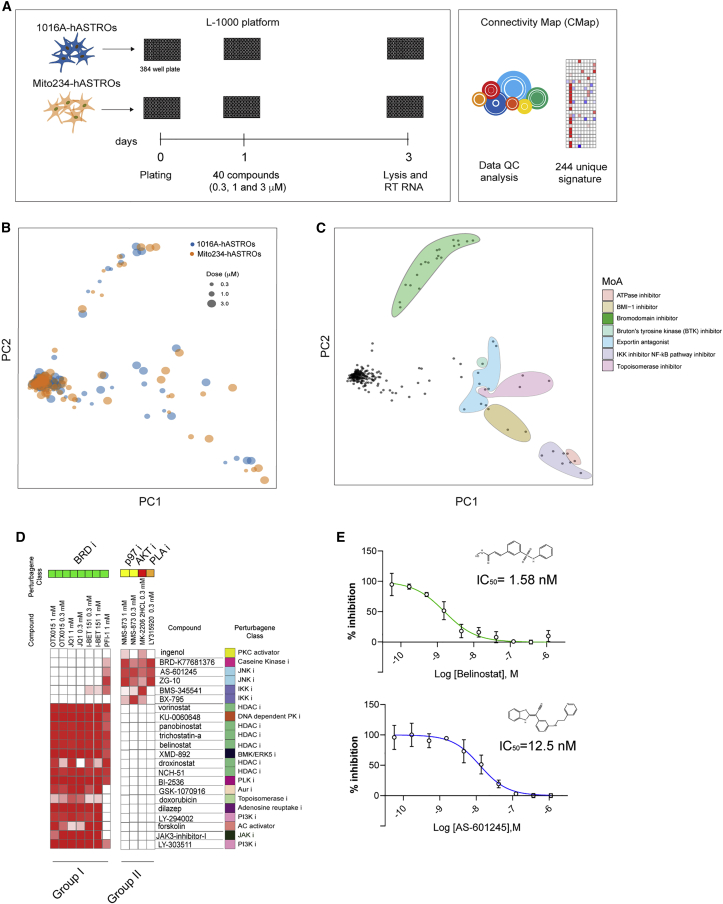
To broaden our ability to discover new modulatory pathways for astrocyte C4, we built a C4 CMap (C4-CMap). Drug-induced gene expression signature was generated using the high-throughput and cost-effective L1000 Luminex bead-based assay platform (Subramanian et al., 2017). In brief, L1000 measures the transcript abundance of 978 landmark genes and computational analysis can infer the expression of non-measured genes. Gene-wide robust Z scores are then computed across all samples in the same plate to calculate differential gene expression signatures for each sample. These signatures are used to explore relationships—similarities or dissimilarities—among diseases, drugs, genes, and pathways, by comparing their associated differential expression signatures. To generate C4-CMap data, we selected a list of perturbagens belonging to diverse pathways, such as inflammation, neuroactive compounds, chromatin remodeling, and others, based on our primary screening results. When possible, we included multiple compounds against the same target (Figure S6A). In total, we compared hASTROs and Mito234-hASTROs using 40 compounds at 3 different concentrations, generating 244 gene expression signatures (including untreated and DMSO-treated cells) (Figure 6A). Principal-component analysis of the C4-CMap signature showed no clustering bias resulting from cell origin or compound dosage (Figures 6B, S6B, and S6C). To assess what might be driving the observed separation among signatures, we used the Broad Institute’s Drug Repurposing Hub (Corsello et al., 2017) to label signatures based on known MoA and targets. Closely clustered compounds are highlighted in colored groups based on the compounds’ on-target mechanisms (Figure 6C). We identified seven clusters with different mechanisms of action, the largest being the bromodomain inhibitor group containing JQ1, OTX015, I-BET151, and PFI-1. Among the other identified groups, each contained one compound identified from the primary screen: IMD-0354, an IKK inhibitor; NMS-873, an ATPase inhibitor; PTC-209, a BMI inhibitor; AVL-292, a BTK inhibitor; KPT330, an Exportin-1 inhibitor; and the Topoisomerase inhibitor, mitoxantrone (used as a calibrator control for its strong and distinguishable transcriptional signature) (Figure 6C).
Figure 6.
Building a C4-CMap
(A) Schematic of experimental and analytic pipeline.
(B) Principal-component analysis (PCA) representing healthy control and patient cell lines treated with selected compounds. Size of circles indicates the dose of treatment.
(C) PCA projection of the two lines treated with 40 compounds at 3 doses. Clusters are colored according to the compounds’ mechanisms of action (MoA).
(D) Heatmap showing the connectivity of two groups of compounds clustered by the similarity of their signatures.
(E) Dose-response curve of HDAC inhibitor (belinostat) and JNK inhibitor (AS-601245) at 72 h after treatment. Individual points represent the mean ± SD of triplicate wells.
We then sought to leverage the CMap resource to predict other compound classes capable of affecting C4 secretion. To do this, we computed connectivity scores between the 244 C4-CMap compound signatures and the >2,400 compounds in the CMap Touchstone reference dataset. We then identified Touchstone compounds connecting preferentially to C4-CMap compounds that had previously been observed to reduce C4 secretion without decreasing cell number. This analysis revealed two groups of compounds: group I contained all the BRD inhibitors (OTX-015, JQ1, I BET, and PFI 1), and group II consisted of a p97 APTase inhibitor (NMS-873), an AKT inhibitor (MK-2206), and a PLA phospholipase inhibitor (LY315920) (Figure 6D). Connectivity results suggested that BRD inhibitors generated signatures similar to those of PLK inhibitors, PI3K inhibitors, and HDAC inhibitors. Indeed, HDAC inhibitors, such as Belinostat, were a validated hit class in our screen. The effects of Belinostat are shown in Figure 6E. The second group of compounds identified by connectivity contained multiple JNK inhibitors. Since there were no JNK inhibitors present in our original screening library, we tested an inhibitor of JNKs 1,2,3 (AS-601245) and found that it reduces C4 in a dose-dependent manner with an IC50 of 12.5 nM (Figure 6E).
Discussion
Despite the growing interest in the role that astrocytes play in human disease, there is still a great deal to be discovered about human glial biology. In this study, we developed a relatively rapid protocol for differentiating human pluripotent cells into astrocytes in large numbers. Compared with most existing protocols for astrocyte differentiation, our 3D method provides a straightforward and reproducible system to generate a large number of cells that can be either passaged or cryopreserved. Importantly, the cells produced by this protocol contain nearly 100% astrocytes, based on expression of canonical-astrocyte markers. Of note, this protocol is robust and reproducible when applied across pluripotent iPSC and ESC lines. Only a small percentage of differentiated cells expressed glial fibrillary acidic protein (GFAP) and primarily appear to be “resting astrocytes.” As reported previously, GFAP is heterogeneously expressed in the human brain and is primarily associated with reactive states in disease (DeSilva et al., 2012; Liddelow et al., 2017; Lundin et al., 2018; Macikova et al., 2009; Tcw et al., 2017).
Crucially, astrocytes derived following this protocol secrete C4. To understand the role of C4 secretion from astrocytes and its connection to synaptic pruning, we used C4-KO lines and showed that C4 secreted from astrocytes can be a source of C4 located at synapses. This is consistent with previous data and with our scRNA-seq data on C4 expression in the mouse brain (Ximerakis et al., 2019). Although we did not detect C4 in neurons differentiated using Ngn2 overexpression, this does not exclude the possibility that other types of neurons can express and release C4. Nonetheless, these experiments suggest that decreasing secreted C4 from astrocytes can influence synaptic C4 and modulate the synaptic over-pruning observed in neuropsychiatric and neurodegenerative diseases.
Although the inhibition of complement is recognized as a potentially valuable therapeutic approach, developing useful clinical CNS complement-inhibiting drugs has been challenging, due to the limited permeability of the blood-brain barrier (BBB) (Ricklin and Lambris, 2016), although antibodies that block C1q can lessen microglia-mediated synapse loss in animal models of AD (Dejanovic et al., 2018; Hong et al., 2016). Using an ELISA-based screen we aimed to identify compounds and pathways that reduce secreted C4 from cultured human astrocytes. We focused our attention on BRD, NF-kB, and JAK inhibitors for their strong effects, and known involvement in the inflammatory response. Previous work is consistent with our observations. For example, two studies highlighted the ability of JQ1 to cross the BBB, reduce inflammation, and improve brain plasticity in both WT and mouse models of AD (Benito et al., 2017; Magistri et al., 2016). The transcription factor NF-kB is a well-known master regulator of the inflammatory response and has been implicated in neurodegenerative disease (Flood et al., 2011). Moreover, NF-κB has been shown to be activated in a mouse model of AD and to lead to increased release of C3 and subsequent effects on dendritic morphology and synaptic function (Lian et al., 2015). Finally, the JAK/STAT pathway is activated by IFN-γ, a well-known inflammatory mediator that modulates C3 and C4 levels in different cell types, including astrocytes (Kitamura et al., 1999; Mitchell et al., 1996). Our data provide evidence that JAK inhibition can prevent C4 secretion from astrocytes both under basal conditions and in response to pro-inflammatory stimuli.
To identify additional pathways that can regulate C4 in human astrocytes, we built an in silico CMap. As our data clearly show, CMap is a valuable and cost-effective tool to predict potentially therapeutic drug candidates in neuronal cell types, even though this dataset was generated using primarily cancer cell lines. Among the targets predicted by this analysis, HDAC inhibitors were excellent positive controls. One of the HDAC inhibitors was present in our initial hit collection and was confirmed in a secondary assay. While it might seem counterintuitive that BRD inhibitors generate a signature similar to HDAC inhibitors, it has already been shown in other cellular systems that HDAC and BRD modulate similar genes through global acetylation rearrangements, pause of RNA elongation, and selective depletion of BRD-containing proteins (Bhadury et al., 2014; Mackmull et al., 2015). Most importantly, the CMap analysis allowed us to identify and validate new targets, such as JNK, that were not covered by compounds in our screening library. Prior to this study, very little was known about the regulation of C4 in human astrocytes. Our approaches help identify multiple pathways that regulate C4 secretion and that can be targeted in different ways. One of the advantages of our study may be the possibility of repurposing drugs for serious CNS indications.
Using the CMap analysis, we also explored the transcriptional differences between hASTROs differentiated from healthy donors or patients affected by schizophrenia. We did not detect statistically meaningful differences between the lines in response to the selected perturbations. More in-depth analysis (such as RNA-seq) comparing a larger number of hASTROs lines treated with more perturbagens, may help to reveal interesting differences between healthy and disease lines.
The recent association of C4A with elevated risk of schizophrenia and its role in controlling synapse numbers motivated us to investigate the modulation of astrocyte-secreted C4 using small molecules. Understanding the mechanisms of C4 regulation and discovering C4-modulating compounds in human astrocytes is a first step toward the development of CNS penetrant complement-modulating drugs with a broad application in neuroinflammatory, neurodegenerative, and neuropsychiatric diseases. Our data provide an increased understanding of pathways that regulate C4 and that may themselves be targeted for therapeutic application. In addition, our system permits the identification of compounds that might differentially affect C4A or C4B in human cells. The ability to distinguish between the two C4 genes might help guide the choice of the optimal C4-acting compounds.
It will come as no surprise that our hit compounds, in addition to regulating C4, modulate multiple cytokines and other complement components. The general reduction in inflammation may, therefore, synergize with the effects of decreased complement, allowing single compounds to have synergistic effects in treating serious neurodegenerative and neuropsychiatric diseases.
In conclusion, we developed a robust protocol for the differentiation of astrocytes from stem cells that can be used to interrogate human glial biology in vitro. We explored the biology of the regulation of C4 in human astrocytes, identifying multiple pathways that modulate its secretion. Furthermore, we built a C4-CMap that allowed us to explore and confirm new pathways able to regulate C4. The tools and knowledge that we accumulated will stimulate further research exploring C4 as a potential target for modulating synapse number and function.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors Francesca Rapino (francesca_rapino@harvard.edu or francescarapino82@gmail.com) and Lee L. Rubin (lee_rubin@harvard.edu).
Materials availability
The H1 C4 KO line generated in this study will be made available upon request, following appropriate institutional guidelines for cell line use and distribution. The 1016A and the HUES8 lines are available for request through Divvly from the HSCI iPS Core Facility. Additional cell-line-specific restrictions apply for the sharing of Mito80 and Mito234.
Data and code availability
The raw scRNA-seq data are available in NCBI’s Gene Expression Omnibus (GEO) under the accession number SE213352. The processed datasets can be downloaded at https://singlecell.broadinstitute.org/single_cell/study/SCP1960/hpsc-derived-astrocytes-from-rapino-et-al-2022#study-summary. Codes used for the analysis of the RNA-seq are described in detail in supplemental experimental procedures.
hiPSC culture and astrocyte differentiation
Stem cells were cultured on Matrigel-coated plates in StemFlex medium. When PSCs reached confluency, they were dissociated to single cells using Accutase. Single suspensions of stem cells were seeded into 125 mL spinner flasks in 100 mL of mTeSR medium at a concentration of 1 × 106 cells/mL in 10 μM ROCK inhibitor Y-27632. After 48 h, the medium was changed to KSR medium with SB431542 and dorsomorphin (10 and 1 μM, respectively). The medium was changed every day for the first 5 days. From days 6 to 12, the medium was changed every 2 days with complete neurobasal supplemented with dorsomorphin, and different cytokines as specified. From day 16 onward, the medium was changed every 2 days (NB 2× N2 CTNF 20 ng/mL). At day 30, spheres were dissociated and either cryopreserved or expanded on PLL-coated plates. For details about stem cell culture, 3D astrocyte differentiation, sphere dissociation, astrocytes culture, and cryopreservation see supplemental experimental procedures.
scRNA-seq
For scRNA-seq experiments, cells were harvested and run through the 10X Chromium Single Cell 3′ Reagents V3 system according to the vendor’s instructions (10X Genomics, San Francisco, CA). Samples were then sequenced on a NovaSeq 6000 system (Illumina) using an S2 flow cell at 2× 100 bp. Details about data normalization and analysis can be found in the supplemental experimental procedures.
ELISA
Ninety-six-well plates were coated with goat anti-human C4 antibody in PBS. The next day, the plates were washed and incubated with a blocking solution (1% BSA in PBS) for 1 h. After eliminating the blocking solution, astrocyte supernatant was added to each well and incubated for 1 h and 30 min. Following washes, the samples were incubated with a capture antibody to detect C4. Plates were washed three times with PBS and incubated for 30 min with goat-anti-rabbit alkaline phosphatase. In the last step, after additional washes, the plates were developed using a diethanolamine buffer. The reaction was stopped and read at 405 nm using a Molecular Devices SpectraMax M5 Reader. For antibody dilution and details on ELISA see supplemental experimental procedures and Table S4.
Screen, hit selection, secondary screen, and dose response
1016A-derived astrocytes were plated on poly-L-lysine-coated 96-well plates. The next day, the medium was replaced with fresh medium, and compounds were added at two different concentrations (1 and 0.3 μM). The screen was performed in triplicate plates using the highly selective inhibitor Library (464 compounds) from Sellckchem. Two days after compound addition, the supernatant was used to perform ELISA and plates were stained for nuclei count. Absorbance (OD405) was divided by nuclei number and normalized on DMSO control (100%). Compounds were ranked by the percentage decrease of C4. The top 24 compounds were selected for a secondary validation screen. The secondary screen was performed as described previously. Cherry-picked compounds from the stock library or freshly purchased compounds were tested at four concentrations (3, 1, 0.3, and 0.1 μM) in triplicate in 1016A-hASTRO 10%. The same criteria were applied when testing compounds on Mito234-hASTROs. Twelve-point dose responses (starting at 10 μM with one to three dilutions) were performed with selected compounds in stem cell-derived astrocytes. For details about primary screen, secondary validation, and dose-response check supplemental experimental procedures.
L1000 data generation, processing, and analysis
1016A-hASTROs and Mito234-hASTROs were plated in AM complete medium in poly-L-lysine-coated 384-well plates. The nest day, cells were treated with 40 compounds at three different concentrations in triplicate with each cell line on a separate 384-well plate. Cells were lysed after 48 h and were then subject to L1000 profiling as described in Subramanian et al. (2017). The C4-CMap signatures were converted into gene sets and queried in the CMap Touchstone (TS). C4-CMap signatures were identified as C4-reducing according to the corresponding compound’s effect on C4 and lack of effect on cell fitness, using data from the C4 ELISA experiment. These signatures were then manually grouped according to the compounds’ canonical MoA, resulting in two groups. We calculated the frequency with which each TS signature connected to the C4-CMap signatures in groups I and II as well as to the un-interesting C4-CMap signatures, using a threshold of tausummary ≥ 90. C4 preferential connectors were those that frequently connected to the C4-reducing signatures and infrequently to the un-interesting signatures.
Author contributions
F.R. and L.L.R. designed the project, the experiments, and wrote the manuscript. E.N. and J.N. differentiated stem cells into ASTROs, helped with the small-molecule screen, and performed follow-up experiments. A.O’N. helped perform the primary screen and U.K.B. differentiated KO-hASTROs. T.N. performed all analyses on CMap data, developed the algorithm for the analysis of C4 connectivity data, and helped write the manuscript. F.L. and M.T. performed the 10X Genomic analysis. W.C. perform the phagocytosis assay. J.B. and E.O’C. helped with cell culture and performed the FACS and the immunofluorescences, respectively. D.H., G.A., and L.B. generated the C4 KO lines. H.d.R. helped develop the ELISA protocol. J.M.T., S.D.S., R.H.P., B.S., A.S., S.A.M., and R.N. commented on the experiments and the manuscript.
Acknowledgments
We thank M. Ximerakis, N. Rodriguez-Muela, J. LaLonde, F.D. Price, L. Davidow, K. Pfaff, G. Rizki, D.J. Selkoe, M. Johnson, A. Carey, J. Presumey, and Soyon Hong for helpful advice in different aspects of our work and/or for reviewing the manuscript. We also thank the staff members of the Connectivity Map (CMAP) at the Broad Institute for their technical advice and assistance. This work was supported by the Stanley Center for Psychiatric Research, the Broad Institute of Harvard and MIT, and National Institute of Mental Health grant U01MH115727 (to S.A.M. and L.L.R.), R01-MH120227 (to R.H.P., S.D.S., and J.M.T.) and P50 MH112491 (to S.A.M. and B.S.).
Conflict of interests
B.S. serves on the scientific advisory board of Annexon LLC and is a minor shareholder of Annexon LLC; however, this is unrelated to the submitted work. L.L.R. is a founder of Elevian, Rejuveron, and Vesalius Therapeutics, a member of their scientific advisory boards, and a private equity shareholder. All are interested in formulating approaches intended to treat diseases of the nervous system and other tissues. He is also on the advisory board of Alkahest, a Grifols company, focused on the plasma proteome. None of these companies provided any financial support for the work in this paper. The current work is unrelated to work carried out at any of these companies. All other authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.11.018.
Contributor Information
Francesca Rapino, Email: francesca_rapino@harvard.edu.
Lee L. Rubin, Email: lee_rubin@harvard.edu.
Supplemental information
References
- Barbar L., Jain T., Zimmer M., Kruglikov I., Sadick J.S., Wang M., Kalpana K., Rose I.V.L., Burstein S.R., Rusielewicz T., et al. CD49f is a novel marker of functional and reactive human iPSC-derived astrocytes. Neuron. 2020;107:436–453.e412. doi: 10.1016/j.neuron.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnum S.R. Complement biosynthesis in the central nervous system. Crit. Rev. Oral Biol. Med. 1995;6:132–146. doi: 10.1177/10454411950060020301. [DOI] [PubMed] [Google Scholar]
- Benito E., Ramachandran B., Schroeder H., Schmidt G., Urbanke H., Burkhardt S., Capece V., Dean C., Fischer A. The BET/BRD inhibitor JQ1 improves brain plasticity in WT and APP mice. Transl. Psychiatry. 2017;7:e1239. doi: 10.1038/tp.2017.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadury J., Nilsson L.M., Muralidharan S.V., Green L.C., Li Z., Gesner E.M., Hansen H.C., Keller U.B., McLure K.G., Nilsson J.A. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc. Natl. Acad. Sci. USA. 2014;111:E2721–E2730. doi: 10.1073/pnas.1406722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun J.S., Lee C.O., Oh M., Cha D., Kim W.K., Oh K.J., Bae K.H., Lee S.C., Han B.S. Rapid differentiation of astrocytes from human embryonic stem cells. Neurosci. Lett. 2020;716:134681. doi: 10.1016/j.neulet.2019.134681. [DOI] [PubMed] [Google Scholar]
- Carroll M.C., Fathallah D.M., Bergamaschini L., Alicot E.M., Isenman D.E. Substitution of a single amino acid (aspartic acid for histidine) converts the functional activity of human complement C4B to C4A. Proc. Natl. Acad. Sci. USA. 1990;87:6868–6872. doi: 10.1073/pnas.87.17.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub L.S., Deneen B. Astrocyte form and function in the developing central nervous system. Semin. Pediatr. Neurol. 2013;20:230–235. doi: 10.1016/j.spen.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.S., Lee H.J., Lim I., Satoh J.i., Kim S.U. Human astrocytes: secretome profiles of cytokines and chemokines. PLoS One. 2014;9:e92325. doi: 10.1371/journal.pone.0092325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer A.L., Jinadasa T., Sriram B., Phadke R.A., Kretsge L.N., Nguyen T.P.H., Antognetti G., Gilbert J.P., Lee J., Newmark E.R., et al. Increased expression of schizophrenia-associated gene C4 leads to hypoconnectivity of prefrontal cortex and reduced social interaction. PLoS Biol. 2020;18:e3000604. doi: 10.1371/journal.pbio.3000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M., et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejanovic B., Huntley M.A., De Mazière A., Meilandt W.J., Wu T., Srinivasan K., Jiang Z., Gandham V., Friedman B.A., Ngu H., et al. Changes in the synaptic proteome in Tauopathy and rescue of Tau-induced synapse loss by C1q antibodies. Neuron. 2018;100:1322–1336.e7. doi: 10.1016/j.neuron.2018.10.014. [DOI] [PubMed] [Google Scholar]
- DeSilva T.M., Borenstein N.S., Volpe J.J., Kinney H.C., Rosenberg P.A. Expression of EAAT2 in neurons and protoplasmic astrocytes during human cortical development. J. Comp. Neurol. 2012;520:3912–3932. doi: 10.1002/cne.23130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezonne R.S., Sartore R.C., Nascimento J.M., Saia-Cereda V.M., Romão L.F., Alves-Leon S.V., de Souza J.M., Martins-de-Souza D., Rehen S.K., Gomes F.C.A. Derivation of functional human astrocytes from cerebral organoids. Sci. Rep. 2017;7:45091. doi: 10.1038/srep45091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart M., Le Magueresse C. Emerging roles of complement in psychiatric disorders. Front. Psychiatry. 2019;10:573. doi: 10.3389/fpsyt.2019.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkelberger J.R., Song W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- Emdad L., D'Souza S.L., Kothari H.P., Qadeer Z.A., Germano I.M. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 2012;21:404–410. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- Flood P.M., Qian L., Peterson L.J., Zhang F., Shi J.S., Gao H.M., Hong J.S. Transcriptional factor NF-kappaB as a target for therapy in Parkinson's disease. Parkinsons Dis. 2011;2011:216298. doi: 10.4061/2011/216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasque P., Fontaine M., Morgan B.P. Complement expression in human brain. Biosynthesis of terminal pathway components and regulators in human glial cells and cell lines. J. Immunol. 1995;154:4726–4733. [PubMed] [Google Scholar]
- Glantz L.A., Lewis D.A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Gordon D.L., Avery V.M., Adrian D.L., Sadlon T.A. Detection of complement protein mRNA in human astrocytes by the polymerase chain reaction. J. Neurosci. Methods. 1992;45:191–197. doi: 10.1016/0165-0270(92)90076-p. [DOI] [PubMed] [Google Scholar]
- Guttikonda S.R., Sikkema L., Tchieu J., Saurat N., Walsh R.M., Harschnitz O., Ciceri G., Sneeboer M., Mazutis L., Setty M., et al. Fully defined human pluripotent stem cell-derived microglia and tri-culture system model C3 production in Alzheimer's disease. Nat. Neurosci. 2021;24:343–354. doi: 10.1038/s41593-020-00796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Chen C., Wang R., Chechneva O.V., Chung S.H., Rao M.S., Pleasure D.E., Liu Y., Zhang Q., Deng W. hESC-derived Olig2+ progenitors generate a subtype of astroglia with protective effects against ischaemic brain injury. Nat. Commun. 2013;4:2196. doi: 10.1038/ncomms3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic V.M., Malley C., Tristan C.A., Ryu S., Chu P.-H., Barnaeva E., Ormanoglu P., Mercado J.C., Michael S., Ward M.E., et al. Directed differentiation of human pluripotent stem cells into radial glia and astrocytes bypasses neurogenesis. bioRxiv. 2021 doi: 10.1101/2021.08.23.457423. Preprint at. [DOI] [Google Scholar]
- Kitamura K., Andoh A., Inoue T., Amakata Y., Hodohara K., Fujiyama Y., Bamba T. Sodium butyrate blocks interferon-gamma (IFN-gamma)-induced biosynthesis of MHC class III gene products (complement C4 and factor B) in human fetal intestinal epithelial cells. Clin. Exp. Immunol. 1999;118:16–22. doi: 10.1046/j.1365-2249.1999.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R., Weick J.P., Liu Y., Zhang Z.J., Zhang S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng K., Rose I.V., Kim H., Xia W., Romero-Fernandez W., Rooney B., Koontz M., Li E., Ao Y., Wang S., et al. CRISPRi screens in human astrocytes elucidate regulators of distinct inflammatory reactive states. bioRxiv. 2021 doi: 10.1101/2021.08.23.457400. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Yang L., Cole A., Sun L., Chiang A.C.A., Fowler S.W., Shim D.J., Rodriguez-Rivera J., Taglialatela G., Jankowsky J.L., et al. NFkappaB-activated astroglial release of complement C3 compromises neuronal morphology and function associated with Alzheimer's disease. Neuron. 2015;85:101–115. doi: 10.1016/j.neuron.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Delsing L., Clausen M., Ricchiuto P., Sanchez J., Sabirsh A., Ding M., Synnergren J., Zetterberg H., Brolén G., et al. Human iPS-derived astroglia from a stable neural precursor state show improved functionality compared with conventional astrocytic models. Stem Cell Rep. 2018;10:1030–1045. doi: 10.1016/j.stemcr.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macikova I., Perzelova A., Mraz P., Bizik I., Steno J. GFAP-positive astrocytes are rare or absent in primary adult human brain tissue cultures. Biologia. 2009;64:833–839. doi: 10.2478/s11756-009-0136-1. [DOI] [Google Scholar]
- Mackmull M.T., Iskar M., Parca L., Singer S., Bork P., Ori A., Beck M. Histone deacetylase inhibitors (HDACi) cause the selective depletion of bromodomain containing proteins (BCPs) Mol. Cell. Proteomics. 2015;14:1350–1360. doi: 10.1074/mcp.M114.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalon J., Mansur F., Teles E Silva A.L., de Goes V.A., Reiner O., Sertié A.L. Complement system in brain architecture and neurodevelopmental disorders. Front. Neurosci. 2020;14:23. doi: 10.3389/fnins.2020.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistri M., Velmeshev D., Makhmutova M., Patel P., Sartor G.C., Volmar C.H., Wahlestedt C., Faghihi M.A. The BET-bromodomain inhibitor JQ1 reduces inflammation and Tau phosphorylation at Ser396 in the brain of the 3xTg model of Alzheimer's disease. Curr. Alzheimer Res. 2016;13:985–995. doi: 10.2174/1567205013666160427101832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T.J., Naughton M., Norsworthy P., Davies K.A., Walport M.J., Morley B.J. IFN-gamma up-regulates expression of the complement components C3 and C4 by stabilization of mRNA. J. Immunol. 1996;156:4429–4434. [PubMed] [Google Scholar]
- Morgan B.P., Gasque P. Extrahepatic complement biosynthesis: where, when and why? Clin. Exp. Immunol. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm T., Bolognin S., Meiser J., Nickels S., Träger C., Meilenbrock R.L., Brockhaus J., Schreitmüller M., Missler M., Schwamborn J.C. Rapid and robust generation of long-term self-renewing human neural stem cells with the ability to generate mature astroglia. Sci. Rep. 2015;5:16321. doi: 10.1038/srep16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteri U.K., Pitkonen J., Utami K.H., Paavola J., Roybon L., Pouladi M.A., Castrén M.L. Generation of the human pluripotent stem-cell-derived astrocyte model with forebrain identity. Brain Sci. 2021;11:209. doi: 10.3390/brainsci11020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey R., Suaud-Chagny M.F., Bohec A.L., Dorey J.M., d'Amato T., Tamouza R., Leboyer M. Overexpression of complement component C4 in the dorsolateral prefrontal cortex, parietal cortex, superior temporal gyrus and associative striatum of patients with schizophrenia. Brain Behav. Immun. 2020;90:216–225. doi: 10.1016/j.bbi.2020.08.019. [DOI] [PubMed] [Google Scholar]
- Ricklin D., Lambris J.D. New milestones ahead in complement-targeted therapy. Semin. Immunol. 2016;28:208–222. doi: 10.1016/j.smim.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti A., Repetti G.G., Sun C., Price F.D., Reny D.C., Rapino F., Weisinger K., Benkler C., Peterson Q.P., Davidow L.S., et al. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem Cell Rep. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R., Vadodaria K.C., Jaeger B.N., Mei A., Lefcochilos-Fogelquist S., Mendes A.P.D., Erikson G., Shokhirev M., Randolph-Moore L., Fredlender C., et al. Differentiation of inflammation-responsive astrocytes from glial progenitors generated from human induced pluripotent stem cells. Stem Cell Rep. 2017;8:1757–1769. doi: 10.1016/j.stemcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer L., Velmeshev D., Holmqvist S., Kaufmann M., Werneburg S., Jung D., Vistnes S., Stockley J.H., Young A., Steindel M., et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G., Schilling K., Steinhäuser C. Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat. Rev. Neurosci. 2006;7:194–206. doi: 10.1038/nrn1870. [DOI] [PubMed] [Google Scholar]
- Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren C.M., Sheridan S.D., Gracias J., Xuan D., Fu T., Perlis R.H. Patient-specific models of microglia-mediated engulfment of synapses and neural progenitors. Mol. Psychiatry. 2017;22:170–177. doi: 10.1038/mp.2016.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren C.M., Gracias J., Watmuff B., Biag J.D., Thanos J.M., Whittredge P.B., Fu T., Worringer K., Brown H.E., Wang J., et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat. Neurosci. 2019;22:374–385. doi: 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Paşca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V., Maussion G., Chaineau M., Sigutova V., Rouleau G., Durcan T.M., Stifani S. Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci. Lett. 2020;731:135028. doi: 10.1016/j.neulet.2020.135028. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K., et al. A next generation connectivity map: L1000 platform and the first 1, 000, 000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J., Wang M., Pimenova A.A., Bowles K.R., Hartley B.J., Lacin E., Machlovi S.I., Abdelaal R., Karch C.M., Phatnani H., et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 2017;9:600–614. doi: 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M., Vidal C., Giedd J.N., Gochman P., Blumenthal J., Nicolson R., Toga A.W., Rapoport J.L. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. Proc. Natl. Acad. Sci. USA. 2001;98:11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turaç G., Hindley C.J., Thomas R., Davis J.A., Deleidi M., Gasser T., Karaöz E., Pruszak J. Combined flow cytometric analysis of surface and intracellular antigens reveals surface molecule markers of human neuropoiesis. PLoS One. 2013;8:e68519. doi: 10.1371/journal.pone.0068519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.G., Kim S.U., McGeer P.L. Expression of complement C4 and C9 genes by human astrocytes. Brain Res. 1998;809:31–38. doi: 10.1016/s0006-8993(98)00811-7. [DOI] [PubMed] [Google Scholar]
- Wu T., Dejanovic B., Gandham V.D., Gogineni A., Edmonds R., Schauer S., Srinivasan K., Huntley M.A., Wang Y., Wang T.M., et al. Complement C3 is activated in human AD brain and is required for neurodegeneration in mouse models of amyloidosis and Tauopathy. Cell Rep. 2019;28:2111–2123.e6. doi: 10.1016/j.celrep.2019.07.060. [DOI] [PubMed] [Google Scholar]
- Ximerakis M., Lipnick S.L., Innes B.T., Simmons S.K., Adiconis X., Dionne D., Mayweather B.A., Nguyen L., Niziolek Z., Ozek C., et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019;22:1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- Yilmaz M., Yalcin E., Presumey J., Aw E., Ma M., Whelan C.W., Stevens B., McCarroll S.A., Carroll M.C. Overexpression of schizophrenia susceptibility factor human complement C4A promotes excessive synaptic loss and behavioral changes in mice. Nat. Neurosci. 2020;24:214–224. doi: 10.1038/s41593-020-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw scRNA-seq data are available in NCBI’s Gene Expression Omnibus (GEO) under the accession number SE213352. The processed datasets can be downloaded at https://singlecell.broadinstitute.org/single_cell/study/SCP1960/hpsc-derived-astrocytes-from-rapino-et-al-2022#study-summary. Codes used for the analysis of the RNA-seq are described in detail in supplemental experimental procedures.