Abstract
Plants have evolved multiple strategies to cope with rapid changes in the environment. During high light (HL) acclimation, the biosynthesis of photoprotective flavonoids, such as anthocyanins, is induced. However, the exact nature of the signal and downstream factors for HL induction of flavonoid biosynthesis (FB) is still under debate. Here, we show that carbon fixation in chloroplasts, subsequent export of photosynthates by triose phosphate/phosphate translocator (TPT), and rapid increase in cellular sugar content permit the transcriptional and metabolic activation of anthocyanin biosynthesis during HL acclimation. In combination with genetic and physiological analysis, targeted and whole-transcriptome gene expression studies suggest that reactive oxygen species and phytohormones play only a minor role in rapid HL induction of the anthocyanin branch of FB. In addition to transcripts of FB, sugar-responsive genes showed delayed repression or induction in tpt-2 during HL treatment, and a significant overlap with transcripts regulated by SNF1-related protein kinase 1 (SnRK1) was observed, including a central transcription factor of FB. Analysis of mutants with increased and repressed SnRK1 activity suggests that sugar-induced inactivation of SnRK1 is required for HL-mediated activation of anthocyanin biosynthesis. Our study emphasizes the central role of chloroplasts as sensors for environmental changes as well as the vital function of sugar signaling in plant acclimation.
Keywords: flavonoid biosynthesis, acclimation, sugar signaling, high light, anthocyanin, SnRK1
In plants, the flavonoid biosynthesis pathway essential for anthocyanin accumulation is transcriptionally activated during high light acclimation. This study reports that triose phosphate export from the chloroplast functions to stimulate the expression of flavonoid biosynthesis genes through increased cellular sugar content in high light. Induction of pathway genes and anthocyanin accumulation require the inactivation of SnRK1 acting downstream of the chloroplast-derived metabolic retrograde signal.
Introduction
Due to their sessile lifestyle, plants evolved mechanisms to adjust cellular processes and metabolism to cope with sudden environmental changes. Among them, light intensity or temperature variations can occur within seconds to minutes and can persist for hours or several days during different seasons of the year. Whereas adaptation imposes genome changes inherited by the offspring and is involved in speciation, acclimation refers to the capacity of plants to adjust cellular processes at all levels as a response to sudden environmental changes (Kleine et al., 2021). Besides rapid adjustments at the metabolic and post-translational levels, tuning gene expression activity is vital for the acclimation response, which depends on substantial transcriptome reprogramming (e.g., Alsharafa et al., 2014; Calixto et al., 2018; Garcia-Molina et al., 2020; Glasser et al., 2014; Huang et al., 2019). During the last decades, it has become apparent that chloroplasts function as sensors of changes in the environment and emitters of signals necessary for establishing a new cellular homeostasis when plants face adverse conditions (Nott et al., 2006; Pogson et al., 2008; Chan et al., 2016; Dietz et al., 2016; Kleine and Leister, 2016; de Souza et al., 2017; Kleine et al., 2021; Schwenkert et al., 2022).
One common trait of plants acclimating to high light (HL) is the accumulation of (colored) flavonoids. The precursors of these secondary metabolites (phenylpropanoids) and flavonoids themselves play a protective role by absorbing excessive amounts of UV light and by providing high antioxidant capacity to the cell in adverse environmental conditions (Gould, 2004; Emiliani et al., 2013; Gould et al., 2018; Agati et al., 2020, 2021). Flavonoids are end products of a combined pathway of cytosolic phenylpropanoid and flavonoid biosynthesis (FB) that produces a great diversity of polyphenolic plant secondary metabolites (Vogt, 2010; Petrussa et al., 2013). Plastid-derived phenylalanine is first converted to p-coumaroyl coenzyme A (CoA) within the phenylpropanoid pathway. Subsequently, chalcone synthase (CHS) catalyzes the initial step of FB, providing chalcones to serve as precursors for various flavonoids, such as (dihydro)flavonols (Figure 1A). Within the anthocyanin biosynthesis branch, dihydroflavonol 4-reductase (DFR) and leucoanthocyanidin dioxygenase (LDOX) produce anthocyanidins, the aglycones of anthocyanins, which are further decorated by glycosyl-, methyl-, and acyltransferases (Appelhagen et al., 2014).
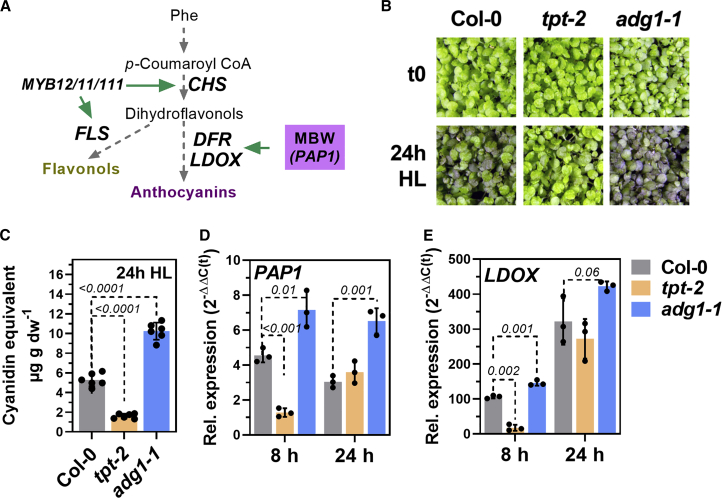
Figure 1.
Flavonoid biosynthesis is differentially regulated in starch-deficient adg1-1 and the triose phosphate/phosphate translocator mutant (tpt-2).
(A) Schematic presentation of the biochemical pathway that leads to the formation of flavonoids. Further information is provided in the main text.
(B) Phenotypes of the wild type (WT; Col-0) and adg1-1 and tpt-2 mutants before (t0) and after exposure to continuous high light (HL) for 24 h.
(C) Anthocyanin accumulation (expressed as cyanidin equivalents) in WT (Col-0), adg1-1, and tpt-2 after exposure to continuous HL for 24 h. Plants were shifted to HL 2 h after the onset of light. Statistical significance of differences between the wild type and mutants was calculated using 1-way ANOVA (Dunnett’s multiple comparisons test) relative to Col-0. Values are means ± standard deviations (n = 6), and the p values are shown.
(D and E) Relative expression of PAP1(D) and LDOX(E) in WT, adg1-1, and tpt-2 after HL treatment. Changes in gene expression were calculated using the 2−ΔΔCt method with SAND as the reference gene and are expressed relative to Col-0 before the HL shift. Statistical significance of differences between Col-0 and mutants was analyzed using Student’s t test. Values are means ± standard deviations (n = 3), and the p values are shown. For all experiments, seedlings from at least 3 different pots shifted at the same time were harvested, and each sample consisted of approximately 10–15 seedlings.
Based on co-expression and function within FB, but not on temporal expression differences during acclimation, pathway genes are grouped into early biosynthetic genes (EBGs), such as CHS and chalcone isomerase. In a side branch flavonol synthase1 (FLS1) catalyses the formation of flavonols which are further glycosylated.Late biosynthetic genes (LBGs), such as DFR and LDOX are essential for the formation of anthocyanidins. These aglycones are decorated by methyl, acyl and glycosyl transferases resulting in the accumulation of anthocyanins (Figure 1). Whereas the expression of EBGs is mostly under the control of the partially redundant transcription factors (TFs) MYB11, MYB12, and MYB111 (Stracke et al., 2007), LBGs are regulated by an MBW complex formed by MYB, basic helix-loop-helix (bHLH), and WD40 components. Together with one WD40 variant (transparent testa glabra 1 [TTG1]), a group of MYB TFs and the bHLH TFs TT8, glabra3 (GL3), and enhancer of GL3 (EGL3) regulate the composition and activity of the MBW complex and the overall activity of the FB pathway (Zhang et al., 2003; Lloyd et al., 2017; LaFountain and Yuan, 2021). MYB components that act as positive regulators of FB are encoded by MYB75 (production of anthocyanin pigment 1 [PAP1]), MYB90 (PAP2), and MYB113 and 114 (Borevitz et al., 2000; Gonzalez et al., 2008). Knockout of PAP1 abolishes the expression of LBGs and anthocyanin accumulation (Teng et al., 2005; Li et al., 2016). LBGs are also repressed by the concurrent interaction between stimulating and repressive MYBs, such as MYB-like 2, with the MBW complex (Dubos et al., 2008; LaFountain and Yuan, 2021).
Enzymes and regulators of FB are targets for post-translational protein modifications, particularly ubiquitination and subsequent proteasomal degradation (Maier et al., 2013; Zhang et al., 2017). In this context, some factors are regulated by light-signaling pathways involving UV (UVR8), blue-light (cryptochromes [CRY]), and red-light (phytochromes (PHY) photoreceptors and the downstream component elongated hypocotyl 5 (HY5) (Kleine et al., 2007; Stracke et al., 2010; Warnasooriya et al., 2011; Heijde et al., 2013; Shin et al., 2013; Maier and Hoecker, 2015; Job et al., 2018; Ponnu et al., 2019; Bursch et al., 2020). Also, various plastid-derived signals or signals that depend on chloroplast function exhibit a function in FB regulation. For example, ascorbate or phytohormones such as jasmonate, gibberellic acid, auxin, abscisic acid, and ethylene are associated with the regulation of FB (Dombrecht et al., 2007; Loreti et al., 2008; Jeong et al., 2010; Qi et al., 2011; Das et al., 2012; Page et al., 2012; Li et al., 2014; Xie et al., 2016; Plumb et al., 2018; Wang et al., 2020; An et al., 2021). Although it remains a subject of controversy, reactive oxygen species accumulation (during HL) positively or negatively affects FB genes and anthocyanin accumulation (Vandenabeele et al., 2004; Vanderauwera et al., 2005; Miller et al., 2007; Viola et al., 2016; Xu et al., 2017). FB genes are also stimulated by exogenous sugar supply (Creasy, 1968; Teng et al., 2005; Solfanelli et al., 2006), and starch-deficient mutants with high sugar content accumulate more anthocyanins than do wild-type (WT) plants (Ragel et al., 2013; Schmitz et al., 2014). Although no change in FB transcript accumulation was observed, knockout of triose phosphate (TP)/phosphate translocator (TPT), essential for the immediate export of photoassimilates (Schneider et al., 2002), resulted in low anthocyanin levels after HL treatment (Schmitz et al., 2014). In addition, blocking photosynthetic electron transfer with 3-(3,4-dichlorophenyl)-1,1-dimethyl urea (DCMU) suppresses FB (Creasy, 1968; Akhtar et al., 2010; Jeong et al., 2010). These findings may directly connect photosynthetic activity and plastid-derived carbohydrates with FB regulation, particularly when light intensity increases. In this context, the analysis of plants with altered activity or content of central components of the sugar-signaling network suggests a function for these factors in FB regulation. For example, perturbation of the target of rapamycin (TOR) complex, which is involved in growth-promoting glucose signaling (Xiong et al., 2013; Chen et al., 2018), results in high anthocyanin contents (Wang et al., 2017; Salem et al., 2018). Also, the function of SNF1-related protein kinase 1 (SnRK1; Baena-Gonzalez et al., 2007), which is activated under low-energy stresses such as sugar starvation or hypoxia, is connected to FB. When cellular sugar levels are high, it is proposed that SnRK1 activity is repressed by regulatory sugar phosphates such as trehalose-6 phosphate (T6P) (Zhang et al., 2009; Nunes et al., 2013; Zhai et al., 2018). Repression of SnRK1 activity by increased T6P level or genetic perturbation activates FB even under normal growth conditions, but reduced anthocyanin contents were observed when T6P phosphatase or the catalytic SnRK1 subunit KIN10 was overexpressed (Schluepmann et al., 2003; Baena-Gonzalez et al., 2007; Zhang et al., 2009; Wingler et al., 2012; Nukarinen et al., 2016; Wang et al., 2021). SnRK1 also functions in the post-translational regulation of phenylalanine ammonia-lyase (PAL), the first enzyme in the cytosolic phenylpropanoid pathway (Wang et al., 2021).
Previous research revealed a complex network for the transcriptional regulation of FB that depended on or interacted with various plastid-derived molecules and known components of other signaling pathways. However, the exact nature of the signal that stimulates FB genes during HL acclimation is still unknown, and molecular mechanisms and components for perception, integration, and translation of the signal to changes in FB gene expression in response to increased light intensity are not well understood.
Results
Perturbation of TP export diminishes HL induction of FB
To analyze the effects of altered photoassimilate partitioning on the regulation of FB, we used Arabidopsis thaliana WT (Col-0), starch-deficient adg1-1 (Supplemental Figure 1A), and the tpt-2 knockout mutant for TPT (Supplemental Figure 1B) in HL shift experiments. For HL treatment, plants were exposed to continuous HL (500 μmol photons m−2 s−1, light-emitting diode [LED]) for up to 24 h without a night phase. WT plants showed a two-fold increase in glucose (GLC), fructose (FRC) and sucrose (SUC) contents after short-term HL treatment, whereas the deficiency of TPT prevented HL-induced accumulation of all three sugars (Supplemental Figure 1C–1E). Blockage of starch biosynthesis in adg1-1 led to marked overaccumulation of sugars 8 h after the HL shift. After 24 h HL, adg1-1 accumulated two-fold more anthocyanins. However, lack of TPT resulted in only 20% of the WT anthocyanin content (Figure 1C). Transcript analysis revealed higher expression of the TF PAP1 (Figure 1D) and its regulated target involved in anthocyanin biosynthesis (LDOX, Figure 1E) in adg1-1 compared with WT after 8 h HL (Figure 1D and 1E). Expression of PAP1 and LDOX was significantly diminished in tpt-2 compared with WT after 8 h, but not 24 h, of HL (Figure 1D and 1E). Except for a stronger absolute increase in PAP1 transcripts, similar results were obtained with plants shifted to HL at the end of the night (Supplemental Figure 1F and 1G). To monitor the maximum induction of FB genes and for better comparison with experiments that required pre-incubation in darkness (see below), plants were shifted to HL at the end of the night in subsequent experiments. In contrast to tpt-2, sugar (phosphate) transporter mutants for glucose 6-phosphate/phosphate translocator 2 (gpt2-1), xylulose 5-phosphate/phosphate translocator (xpt-1 and -2), and plastidic glucose translocator (pglct-2) showed WT-like accumulation of anthocyanins after 24 h HL (Supplemental Figure 1H and 1I). These results indicate specific and dynamic changes in the expression of FB genes and, ultimately, the potential to accumulate anthocyanins during HL treatment, which was significantly reduced in TPT-deficient mutants.
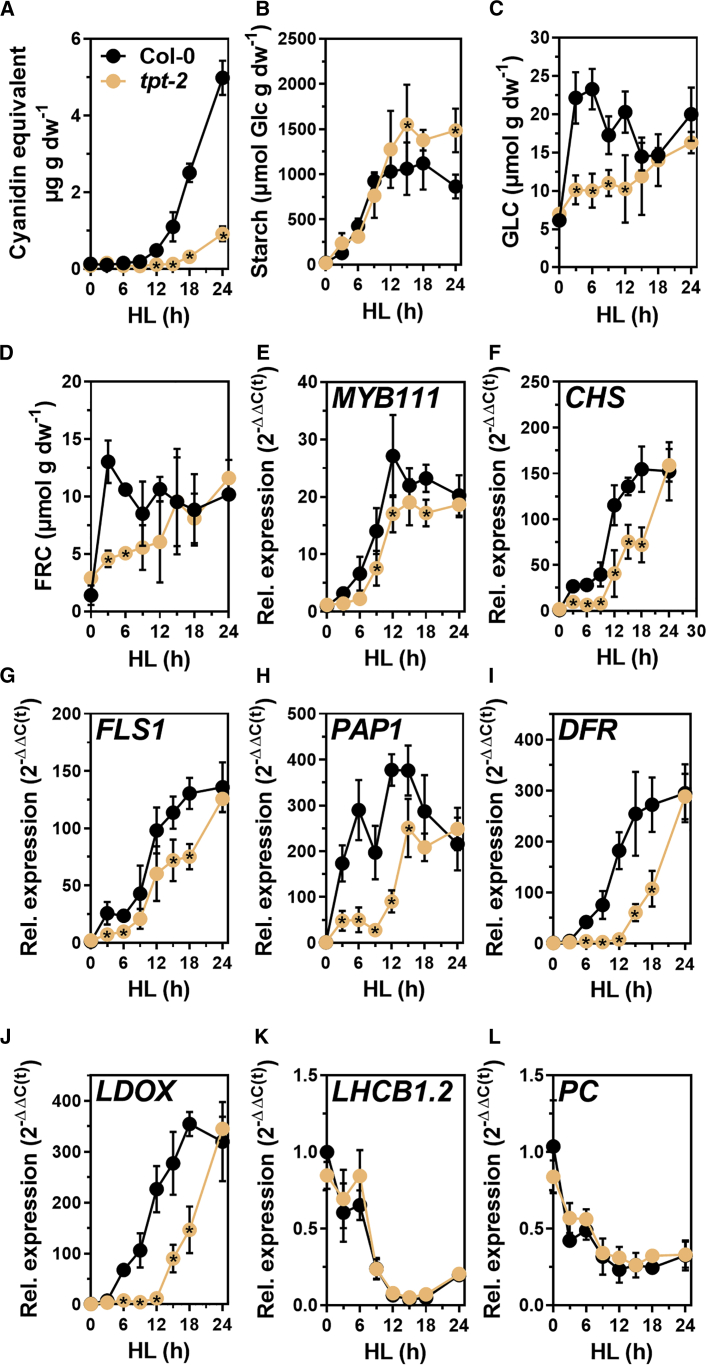
To analyze the induction of FB during HL in more detail, a time-resolved analysis of anthocyanin, starch, sugar, and transcript abundances in HL-shifted Col-0 and tpt-2 was performed (Figure 2). Anthocyanin contents increased after 6–9 h HL in WT, but they began to increase only after 15–18 h HL in tpt-2 (Figure 2A). Starch accumulation was similar between tpt-2 and WT plants until 9–12 h. The tpt-2 mutant continued to synthesize starch, resulting in significantly higher levels at the end of the HL experiment (Figure 2B). HL exposure led to the accumulation of GLC and FRC within 3 h in WT leaves (Figure 2C and 2D), but sugar content increased only gradually in tpt-2 (Figure 2C and 2D). The TF MYB111 and its targets, CHS and FLS1, were strongly induced in WT from the beginning of the HL shift. Although induced with a similar trend, EBGs expression was diminished in tpt-2 compared with WT (Figure 2E–2G). Intriguingly, HL induction of LBGs involved in anthocyanin biosynthesis was significantly delayed in tpt-2 by more than 8 h. Whereas PAP1, DFR, and LDOX markedly increased within 6 h of HL treatment in the WT, they remained low in tpt-2 within the first hours of HL and only began to increase after 9 h of HL (Figure 2H–2J). In tpt-2, LBG expression reached WT levels only after 24 h of HL (Figure 2I and 2J). Notably, the kinetics of LBG induction in tpt-2 after 12 h of HL treatment resembled the induction in WT between 3 and 15 h of HL, and delayed stimulation of LBGs agreed with the late and diminished anthocyanin accumulation (Figures 1C–2A). WT-like repression of LHCB1.2 and PC (Figure 2K and 2L) excluded a general perturbation of the HL response in tpt-2. Also, no differences in reactive oxygen species (ROS) level between tpt-2 and WT were observed using specific stains (Supplemental Figure 2C–2F). In addition, we also found WT-like expression of stress- or ROS-responsive genes such as glutathione peroxidase 7 (GPX7) or ascorbate peroxidase 1 (APX1) in tpt-2 during HL (Supplemental Figure 2A and 2B).
Figure 2.
The tpt-2 mutant showed a delayed HL-response.Fig. 6: Overexpression of KIN10 diminished HL-induction of FB.Fig. 7: Inactivation of SnRK1 induced FB in HL
Kinetic analysis of (A) anthocyanin (represented as cyanidin equivalents), (B) starch (represented as GLC equivalents), (C) GLC, (D) FRC content and gene expression (E–L) in Col-0 (black) and tpt-2 (orange) throughout a 24-h HL shift experiment. Plants were shifted at the end of the night (t0) to the HL condition, and samples were collected at the indicated time points. Gene expression was calculated using the 2−ΔΔCt method relative to Col-0 (t0) with SAND as the reference gene. Values are means ± standard deviations (n = 4). Statistical significance of differences between WT and tpt-2 at each time point was analyzed using Student’s t test (p < 0.05). Significantly different values are indicated with asterisks inside the tpt-2 symbols. (E–G) Expression levels of representative early biosynthetic genes (EBGs) and (H–J) late biosynthetic genes (LBGs) involved in FB and (K and L) light harvesting chlorophyll A/B binding protein 1.2 (LHCB1.2) and plastocyanin (PC) were analyzed.
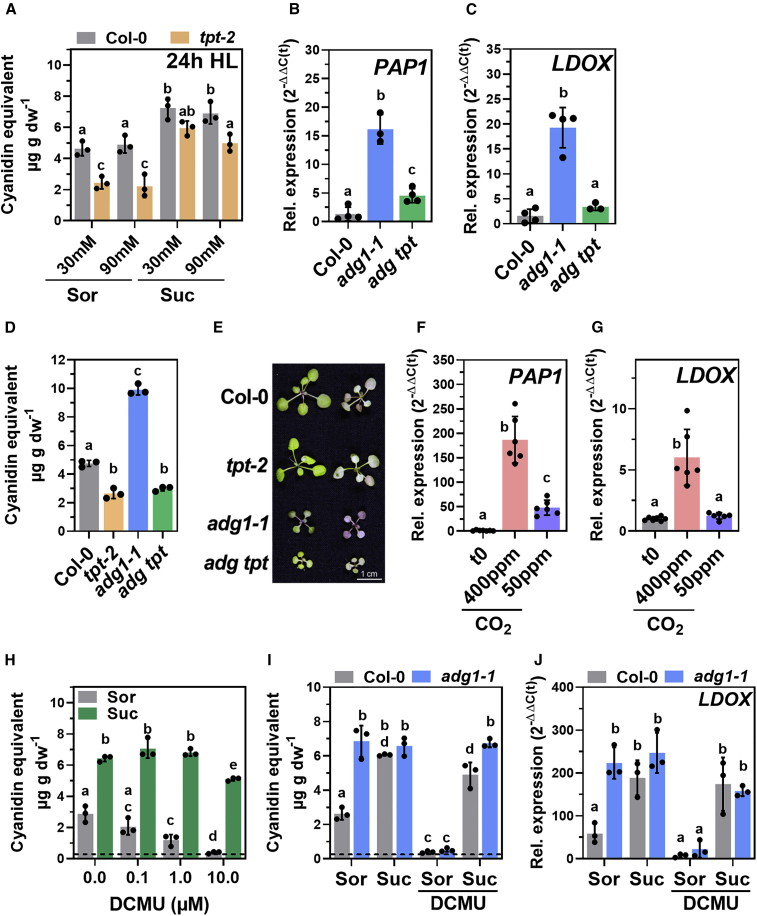
Because TPT deficiency caused a substantial delay in the accumulation of sugars in HL, we hypothesized that the lack of a sugar signal originating from chloroplasts was causative for the low anthocyanin accumulation in tpt-2. To test this, Col-0 and tpt-2 were incubated in a buffer supplemented with SUC or sorbitol (SOR) as an osmotic control and shifted to HL (Figure 3). In the control buffer, tpt-2 showed the same low anthocyanin content compared with WT as was observed for soil-grown plants. However, sucrose feeding significantly increased anthocyanin content in tpt-2 to a WT-like level (Figure 3A). By contrast, the introduction of the tpt-2 allele into the adg1-1 mutant suppressed the more pronounced activation of FB in the adg1-1 single mutant (Figure 3B–3E). To analyze the impact of TP biosynthesis in chloroplasts on FB regulation, carbon fixation within the Calvin cycle was restricted by adjusting the CO2 concentration using a LI-COR device. PAP1 and LDOX were significantly induced after HL treatment at 400 ppm CO2 (Figure 3F and 3G), and the expression changes were comparable to those observed in young WT plants treated with bright LED light in a growth cabinet (Figure 2H). By contrast, a limited CO2 supply of 50 ppm suppressed the HL induction of PAP1 and abolished LDOX induction (Figure 3F and 3G). Likewise, the suppression of photosynthetic activity by DCMU abolished anthocyanin accumulation in Col-0 and adg1-1 in HL (Figure 3H–3J), and this effect could be fully rescued at the metabolic and transcriptional levels by feeding of exogenous SUC to both genotypes (Figure 3H–3J). We also found that plants shifted to 24 h of HL at different time points during short-day (SD) or long-day (LD) growth conditions accumulated more anthocyanins the later they were shifted during the day. These experiments revealed a positive correlation of FB activation with starch content before the HL shift (Supplemental Figure 3A and 3B). Moreover, PAP1 and LDOX expression after 8 h of HL and anthocyanin contents after 24 h of HL were also positively correlated with sugar content before the HL shift (t0) (Supplemental Figure 3C–3K).
Figure 3.
Induction of FB in HL depends on carbon fixation, TP export, and carbohydrates.
(A) Anthocyanin accumulation in Col-0 (gray) and tpt-2 (yellow) in buffer containing different concentrations of SOR or SUC. Seedlings were transferred to the buffer at the end of the night and exposed to HL for 24 h.
(B and C) Relative expression of PAP1(B) and (C)LDOX in Col-0 (gray), adg1-1 (blue), and adg1-1 tpt-2 (green) after 8 h HL treatment. Gene expression was calculated using the 2−ΔΔCt method relative to Col-0 after 8 h HL with SAND as the reference gene.
(D and E) Anthocyanin content (expressed as cyanidin equivalents) (D) and (E) phenotype of WT and mutant plants exposed to 24 h HL. Left column shows the adaxial view and right column the abaxial view of 2 individual plants.
(F and G) Relative expression of PAP1(F) and (G)LDOX in Col-0 before incubation (EoN, t0, gray) in 400 ppm CO2 (light pink) or 50 ppm CO2 (purple). Leaves were mounted in a LI-COR device and incubated for 4 h at 500 μmol photons m−2 s−1. Gene expression was calculated using the 2−ΔΔCt method relative to expression at t0 with SAND as the reference gene. Five-week-old rosette plants were used, and each data point represents 1 leaf sample from independent plants.
(H) Anthocyanin content (expressed as cyanidin equivalents) after 24 h HL in Col-0 incubated in buffer with 90 mM SOR or SUC in the absence or presence of increasing amounts of DCMU (0–10 μM).
(I) Anthocyanin content (expressed as cyanidin equivalents) after 24 h HL in Col-0 and adg1-1 incubated in buffer with 90 mM SOR or SUC in the absence or presence of 10 μM DCMU.
(J) Same conditions as in (I), but samples were harvested after 8 h HL exposure. Gene expression was calculated using the 2−ΔΔCt method relative to Col-0 before the transfer to incubation buffer, and SAND was used as the reference gene. The dashed lines in (J) and (K) show the mean anthocyanin content at t(0) in Col-0. All values are means ± standard deviations.
Statistical significance of differences between WT/control and mutants/treatment was calculated using 1-way ANOVA relative to Col-0 after 8 h HL (in B and C, Dunnett’s multiple comparisons test, n ≥ 3) or 24 h HL (in D, Tukey’s multiple comparisons test, n = 3) or to expression values before incubation at different CO2 concentrations (t0 in F and G, Dunnett’s multiple comparisons test, n = 6) or 2-way ANOVA (Tukey’s multiple comparisons test in A and H–J, n = 3). Letters indicate significant differences at p ≤ 0.01. Values are means ± standard deviations.
Transcriptome analysis revealed dynamic regulation of the HL acclimation response in tpt-2
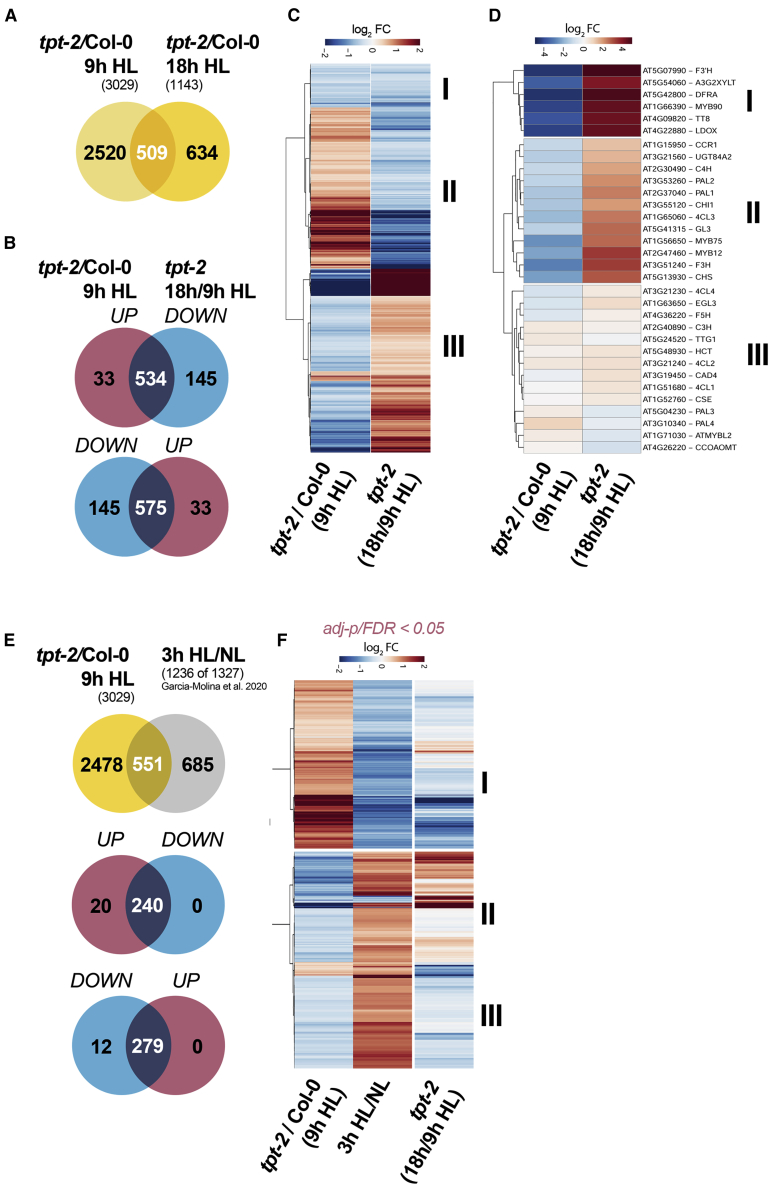
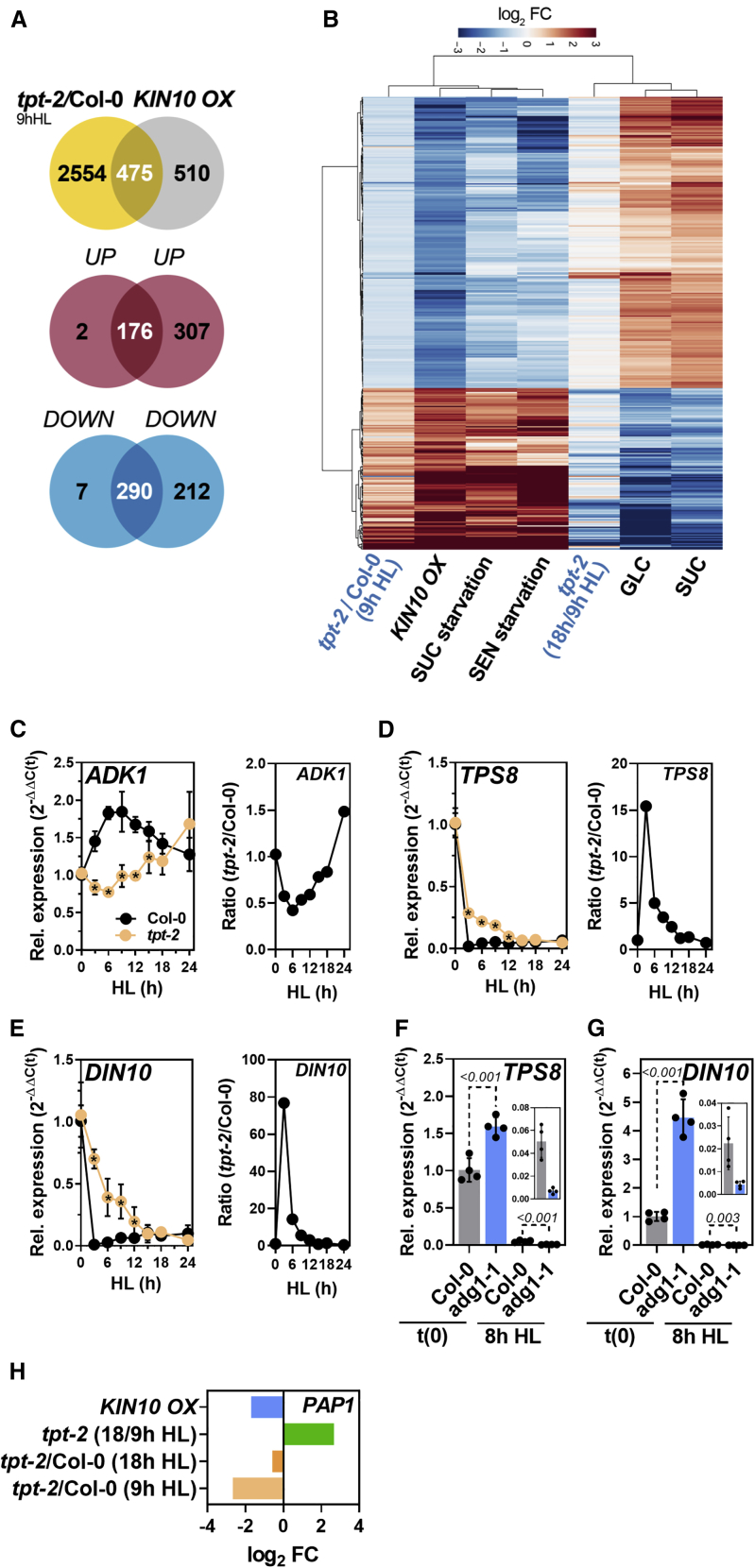
To gain further insights into the HL acclimation response and its dependence on TP export, we performed a whole-transcriptome sequencing analysis (mRNA sequencing [mRNA-seq]) of WT and tpt-2 after 9 and 18 h of HL. We selected these time points because the anthocyanin branch of FB was not induced in tpt-2 compared with WT at 9 h of HL treatment. By contrast, after 18 h of HL, LBG expression reached a maximum in WT but was induced in tpt-2 relative to 9 h HL (Figure 2). Hence, ratiometric analysis of gene expression changes between these time points would also disclose other dynamic changes in the tpt-2 transcriptome. From the initial comparison of WT and tpt-2, we found only a minor overlap of commonly differentially expressed genes (DEGs, adjusted p < 0.05) between 9 and 18 h of HL (Figure 4A and Supplemental Figure 4A; Supplemental Data 2). Although this already indicated a dynamic transcript expression in tpt-2, relative changes between the mutant and WT were also influenced by changes in the WT between 9 and 18 h of HL treatment (Supplemental Figure 4A). Gene Ontology term (GO term) enrichment analysis revealed phenylpropanoid/flavonoid/anthocyanin biosynthesis and regulation, starch metabolic process, and cellular carbohydrate metabolic process as the most strongly downregulated gene groups in tpt-2 versus WT at 9 h of HL (Supplemental Data 2, log2 FC [fold change] <−1). Among the downregulated genes in tpt-2 at 9 h of HL, we found GPT2, whose expression was previously reported to depend on TP export from chloroplasts during HL (Weise et al., 2019). Furthermore, the large subunits of ADP-glucose pyrophosphorylase 3 (APL3) and granule-bound starch synthase 1 (GBSS1), both involved in starch biosynthesis (regulation), were repressed in tpt-2. It is worth noting that components of light-signaling pathways, such as CRYs, PHYs, or components of the E3 ubiquitin ligase COP1/SPA, and the downstream factors HY5 and its interactor BBX20-22, were not repressed in tpt-2 relative to WT throughout the HL kinetics (Supplemental Data 1). Transcripts related to trehalose metabolism (trehalose-6-phosphate synthase [TPS8,9,10,11]) were upregulated in tpt-2 at 9 h of HL (log2 FC > 1). Upregulated and downregulated transcripts in tpt-2/WT at 9 h of HL were also associated with the terms cellular response to oxygen levels/hypoxia (Supplemental Data 2).
Figure 4.
mRNA-seq analysis of HL-treated plants revealed a delayed HL response in tpt-2.
(A) Venn diagram depicting the overlap of significant DEGs (adjusted p < 0.05) between Col-0 and tpt-2 incubated for 9 h (red) and 18 h (blue) in HL.
(B and C) Venn diagram (B) and (C) heatmap showing transcripts differentially expressed in tpt-2/Col-0 at 9 h HL and their change in expression after 18 h relative to 9 h HL in tpt-2 (tpt-2 18 h/9 h HL) (adjusted p < 0.05). A significant number of up- or downregulated DEGs in tpt-2 relative to Col-0 after 9 h HL were induced or repressed in tpt-2 after 18 h of HL treatment (in total, 1109). DEGs in cluster I were downregulated in tpt-2/Col-0 after 9 h HL and were further repressed in tpt-2 18 h/9 h HL. Cluster II encompasses DEGs upregulated in tpt-2/Col-0 after 9 h HL but significantly downregulated in tpt-2 18 h/9 h HL. Cluster III contains DEGs downregulated at 9 h HL in tpt-2 relative to Col-0 but induced at 18 h HL treatment in tpt-2 relative to 9 h HL.
(D) Expression of genes encoding enzymes and transcriptional regulators of phenylpropanoid and flavonoid biosynthesis in tpt-2/Col-0 at 9 h HL and relative changes in tpt-2 at 18 h compared with 9 h HL. Clusters I and II contain transcripts that were strongly suppressed in tpt-2/Col-0 at 9 h HL but were induced after 18 h HL in tpt-2 relative to 9 h HL. Cluster III contains transcripts that showed only minor (relative) changes between 9 and 18 h HL treatment in tpt-2.
(E and F) Comparison of DEGs in tpt-2/Col-0 (9 h HL) with DEGs after a short-term (3 h) HL shift of Arabidopsis WT plants. DEGs after 3 h HL relative to NL conditions were extracted from Garcia-Molina et al. (2020) (FDR < 0.05). Of the 1327 DEGs after 3 h of HL treatment, transcripts for 1236 genes were found in the tpt-2 dataset and used for comparison (E).
(F) Heatmap comparing relative expression changes of transcripts in Col-0 treated with 3 h HL (Garcia-Molina et al., 2020), tpt-2/Col-0 (9 h HL), and tpt-2 (18 h HL/9 h HL). Changes in gene expression are given as log2 FC relative to the control, and hierarchical row clustering (Euclidean distance) using the ward.D method was used for all heatmaps.
We next focused on the dynamic changes in the tpt-2 transcriptome by analyzing the DEGs at 9 h HL for their expression changes at 18 h HL. Among the deregulated transcripts at 9 h HL, approximately 35% (1109 transcripts) showed opposite expression at 18 h HL in tpt-2 (i.e., they were induced or repressed; adjusted p < 0.05, Figure 4B and 4C, clusters II and III). Transcripts that we classified as late-induced in tpt-2 were assigned to GO terms related to flavonoid biosynthesis and carbohydrate metabolic process (Figure 4D and Supplemental Data 3, filtered for −1 > log2 FC > 1). In addition to APL3, GBSS1, and GPT2, transcripts encoding FB enzymes and regulators (e.g, LBGs, MYB75 [PAP1], MYB90 [PAP2], MYB113, MYB114, TT8) were markedly induced after 18 h of HL treatment in tpt-2 (Figure 4D, clusters I and II; Supplemental Data 5). By contrast, dark inducible 1 (DIN1/SEN1/STR15), DIN10, and TPS8 and 11, regulated by SnRK1 signaling, were induced relative to WT at 9 h of HL but showed late repression in tpt-2 after 18 h of HL.
In addition, we compared the tpt-2 dataset (9 h HL) with the result of a previous transcriptome analysis of WT plants shifted to 3 h HL under similar conditions (Supplemental Data 6; Garcia-Molina et al., 2020), and we found a significant overlap of DEGs in both sets (Figure 4E and 4F). Of the common DEGs, 50% were repressed in WT plants at 3 h HL but almost exclusively upregulated in tpt-2/WT after 9 h HL (Figure 4E). Nearly all of the transcripts in this cluster showed late repression in tpt-2 at 18 h HL (cluster I, Figure 4F) and belonged to, for example, carbohydrate metabolism (including trehalose metabolism) and response to hypoxia (Supplemental Data 6). By contrast, 279 transcripts induced in WT plants after 3 h of HL were downregulated at 9 h of HL in tpt-2, and some of them showed late induction in tpt-2 after 18 h of HL (Figure 4F, cluster II). These transcripts belonged to carbohydrate metabolic process, protein folding, and ribosome biogenesis and function (Supplemental Data 6). In summary, plants that lack TPT cannot induce transcriptome adjustments during the early phases of HL acclimation as observed for WT plants.
It has been shown that HL acclimation also involves and depends on the transcriptional adjustment of phytohormone biosynthesis (Huang et al., 2019), and phytohormones are involved in the transcriptional and post-translational regulation of FB. Hence, we analyzed the RNA-seq dataset for the expression of gene products involved in phytohormone biosynthesis and signaling (Supplemental Data 4, −1 > log2 FC > 1, adjusted p < 0.05). From the group of auxin-responsive genes analyzed, only small auxin-up RNA (SAUR) 21, 20, 22, and 64 were moderately upregulated in tpt-2 throughout the HL treatment. Among the ABA-related transcripts tested, PYL5 and PYL7 were moderately induced at 9 h HL in tpt-2/WT (two-fold), and PYL7 was repressed after 18 h HL in tpt-2 (Supplemental Data 4). Two transcripts related to jasmonate (JA) biosynthesis were deregulated in tpt-2/WT at 9 h HL. Acyl-CoA oxidase 4 (ACX4) was significantly upregulated, jasmonic acid carboxyl methyltransferase (JMT) was downregulated, and both transcripts showed opposite expression in tpt-2/WT relative to values published for HL acclimation kinetics of WT plants (Supplemental Figure 4B). In addition, we found that JA biosynthesis genes, particularly gene products for precursor biosynthesis inside the chloroplast (lipoxygenase, allene oxide synthase, and allene oxide cyclase), were strongly induced in tpt-2 at 18 h HL relative to 9 h HL (Supplemental Figure 4C; Supplemental Data 4). Although JA promotes anthocyanin biosynthesis and FB was induced in tpt-2 after long-term HL, the JA-deficient aos1-1 and jassy mutants (Guan et al., 2019) showed WT-like anthocyanin accumulation during HL treatment (Supplemental Figures 4D–4F).
The impact of SnRK1 on HL induction of FB
The gene expression analyses identified a large number of DEGs strongly upregulated (9 h HL) but later repressed (18 h HL) in tpt-2 that were also reported to be regulated by SnRK1 (Figure 5; Supplemental Data 7; Baena-Gonzalez et al., 2007). These transcripts were almost exclusively deregulated in the same direction by the overexpression of KIN10 in protoplasts (KIN10ox) and tpt-2 after 9 h HL, but they were overall repressed or induced, respectively, after 18 h HL in tpt-2 (Figure 5B). Furthermore, the tpt-2 transcriptome at 9 h HL showed a positive correlation with changes in gene expression induced by KIN10ox and starvation conditions. After prolonged HL treatment, expression changes in tpt-2 were positively correlated with those observed after SUC and GLC feeding (Figure 5B and Supplemental Figure 5A). The late-repressed DEGs in tpt-2 were related to GO terms for carbohydrate metabolism, catabolic process, starvation, and absence of light (Supplemental Data 7), and late-induced transcripts were mainly associated with ribosome assembly and function and protein translation (Supplemental Data 7). We also performed a direct comparison of transcripts deregulated in KIN10ox (Baena-Gonzalez et al., 2007) and WT plants exposed to 0.5 and 3 h HL (Garcia-Molina et al., 2020) and found that approximately 50% of the DEGs in KIN10ox showed expression changes in HL-treated WT (Supplemental Figure 5B). Overall, these DEGs showed an opposite trend in expression (i.e., they were induced in HL-treated WT but repressed in KIN10ox and vice versa; Pearson correlation coefficient r = −0.71, p < 0.05). In summary, this analysis revealed dynamic changes in the expression of transcripts responsive to carbohydrate availability and pointed to SnRK1 as a potential downstream component of sugar signaling during HL treatment.
Figure 5.
Comparative transcriptome analysis indicated deregulation of SnRK1 signaling in tpt-2 during HL acclimation.
(A) Venn diagram showing the overlap between DEGs in tpt-2/Col-0 after 9 h HL and overexpression of KIN10 in protoplasts (KIN10 OX) extracted from Baena-Gonzalez et al. (2007) (Supplemental Data 7). Of the 985 reported transcripts for KIN10 OX, 475 were deregulated in the same direction in tpt-2 after 9 h HL (compare Venn diagrams for UP/UP and DOWN/DOWN).
(B) Heatmap comparing the relative expression of DEGs in tpt-2/Col-0 9 h HL (adjusted p < 0.05) and in tpt-2 (18 h/9 h HL) with the expression changes induced by KIN10 overexpression in protoplasts, SUC and senescence (SEN) starvation, GLC, and SUC feeding (Supplemental Data 7). Changes in gene expression are given as log2 FC relative to the control in each dataset. Hierarchical column and row clustering (Euclidean distance) using the ward.D method were applied.
(C–E) Relative expression of SnRK1-repressed (C)ADK1, and SnRK1-induced (D)TPS8 and (E)DIN10 during the HL shift kinetics of Col-0 and tpt-2.
(F and G) Expression of (F)TPS8 and (G)DIN10 in Col-0 and adg1-1. Gene expression was calculated using the 2−ΔΔCt method relative to Col-0 (t0, end of night) with SAND as the reference gene. Insets show the expression of TPS8 and DIN10 after 8 h HL. Statistical significance of differences between WT and tpt-2 at each time point was analyzed using Student’s t test (p < 0.05, asterisk inside the tpt-2 symbols) or 1-way ANOVA (Dunnett’s multiple comparisons test); adjusted p values are shown. Values are means ± standard deviations (n = 4).
(H) Expression of PAP1 in the indicated datasets/comparisons. Expression values for PAP1 in protoplasts overexpressing KIN10 were extracted from previously published data (Baena-Gonzalez et al., 2007). Changes in gene expression are given as log2 FC relative to the control in each dataset. PAP1 was significantly deregulated in KIN10 OX relative to control protoplasts, tpt-2/WT (9 h HL), and tpt-2 18 h/9 h HL.
To test whether SnRK1-dependent signaling was compromised in tpt-2 during HL, a targeted analysis of transcripts regulated by KIN10ox was performed using the HL kinetics sample set. Adenosine kinase 1 (ADK1), repressed by KIN10ox, was significantly induced in WT plants within the first hours but was not induced in tpt-2 until 12 h of HL treatment (Figure 5C). Two SnRK1-induced transcripts, TPS8 and DIN10, were repressed within 3 h of HL treatment in WT plants, but this repression was markedly delayed in tpt-2 (Figure 5D and 5E). By contrast, SnRK1 marker genes were upregulated in adg1-1 at the end of the night, but repression of TPS8 and DIN10 was more pronounced compared with WT after 8 h of HL (Figure 5F and 5G), reflecting low and high sugar content in darkness and HL, respectively, in the starch-deficient mutant adg1-1. The catalytic SnRK1 subunit KIN10 and other subunits in tpt-2 were not differentially expressed between WT and tpt-2 (Supplemental Figure 5C and Supplemental Data 1). Because the overexpression of KIN10 represses PAP1 in protoplast assays (Baena-Gonzalez et al., 2007) and PAP1 demonstrated dynamic changes in expression in tpt-2 throughout the HL treatment (Figures 2H and 5H), we hypothesized that SnRK1 was also involved in the regulation of FB during HL treatment.
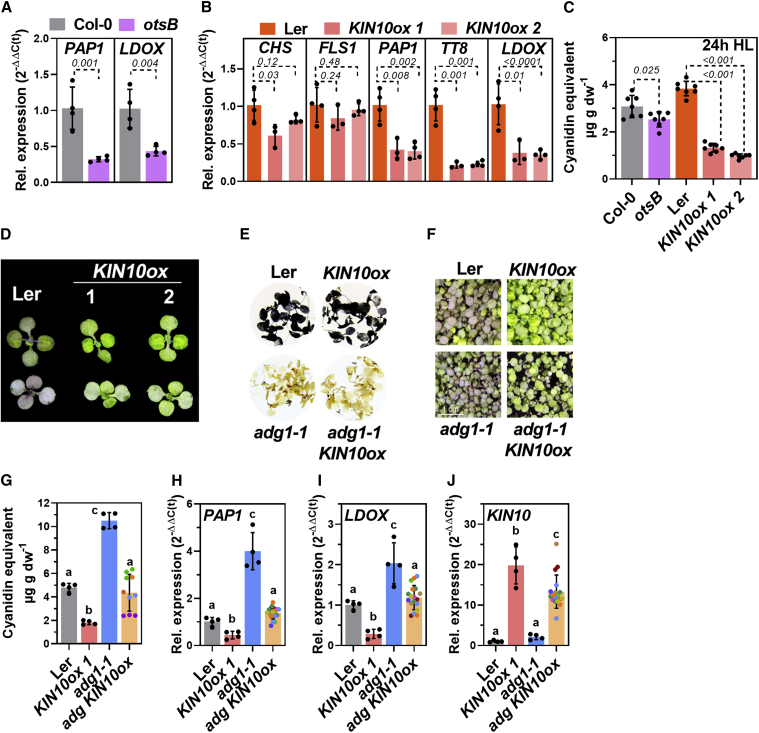
Previous studies have suggested the repression of SnRK1 activity by regulatory sugar phosphates such as T6P. Overexpression of a T6P phosphatase (encoded by otsB, Supplemental Figure 6A) led to diminished expression of PAP1 and LDOX during early time points of HL treatment (Figure 6A) and significantly fewer anthocyanins compared with WT seedlings after 24 h of HL (Figure 6C). HL induction of LBGs (PAP1, TT8, and LDOX) but not of EBGs (CHS and FLS1) was perturbed in two KIN10ox lines compared with WT plants (Figure 6B and Supplemental Figure 6B). This result indicated that an excess of KIN10 particularly repressed the anthocyanin branch of FB during short-term HL exposure. Consequently, the anthocyanin content was reduced by approximately 70% in the KIN10ox lines compared with the WT after 24 h of HL exposure (Figure 6C and 6D). The overexpression of KIN10 in the adg1-1 mutant background (partially) suppressed the more robust activation of anthocyanin biosynthesis observed for the adg1-1 single mutant (Figure 6E–6J), indicating that SnRK1 acts downstream of the chloroplast-derived sugar signal.
Figure 6.
Overexpression of KIN10 diminished HL-induction of FB
(A) Expression of PAP1 and LDOX in Col-0 and Col-0 expressing otsB (light purple) after 8 h HL.
(B) Expression of CHS, FLS1, PAP1, TT8, and LDOX in Landsburg erecta (Ler) (orange) and 2 independent overexpression lines for KIN10 in the Ler background (ox1 and ox2, pink and light pink) after 8 h HL. Plants were shifted at the end of the night. Gene expression was calculated relative to the corresponding WT background after 8 h HL using the 2−ΔΔCt method with SAND as the reference gene. Values are means ± standard deviations (n ≥ 3), and p values for Student’s t test between mutant and WT background are shown.
(C) Anthocyanin accumulation (expressed as cyanidin equivalents) in the lines shown in (A) and (B) after 24 h HL determined from 2 independent HL shift experiments. Values are means ± standard deviations (n = 7 from 2 independent shifts), and p values for Student’s t test between the mutant and the corresponding WT background are shown.
(D) Phenotypes of Ler, KIN10ox #1, and #2 after 24 h HL. Top: adaxial; bottom: abaxial.
(E and F) Staining of starch in seedlings exposed to 24 h HL (E) and (F) phenotypes of Ler, KIN10ox 1, adg1-1, and the adg1-1 KIN10ox double mutant after 24 h HL.
(G–J) Anthocyanin content after 24 h HL (G) and (H)PAP1, (I)LDOX, and (J)KIN10 expression after 8 h HL in the different genotypes. Note that for adg1-1 KIN10ox, seeds from different plants obtained after crossing and selection were used (same color of circle, see section “methods”). Each data point (circle) represents the result from 10–15 seedlings per “line” analyzed as 1 sample. Gene expression was calculated relative to Ler using the 2−ΔΔCt method with SAND as the reference gene. Values are means ± standard deviations. In (G)–(J), for Col-0, KIN10ox 1, and adg1-1, n = 4. For adg1-1 KIN10ox in (G), n = 10, and in (H)–(J), n = 18. Statistical significance of differences was analyzed using 1-way ANOVA (Tukey’s multiple comparisons test), and letters indicate significant differences at p < 0.05.
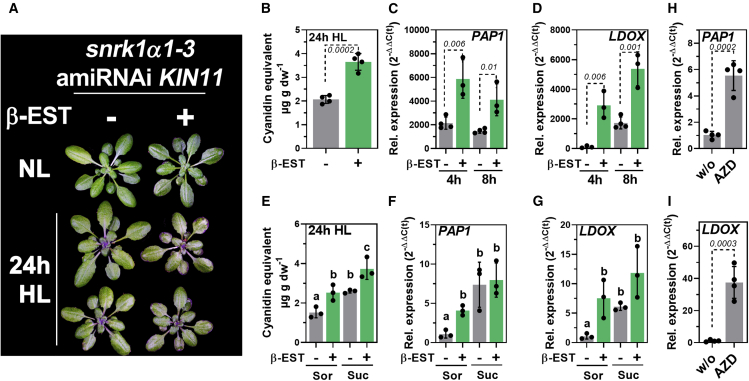
To study the effect of SnRK1 suppression, an inducible RNAi line for KIN11 (SnRK1α2), the second catalytic subunit of SnRK1, in the background of a KIN10 (SnRK1α1) knockout mutant was analyzed (snrk1α1-3 amiRNAi KIN11; Pedrotti et al., 2018). This line had to be used because constitutive snrk1α1/α2 mutants are not viable, and single knockout results in no obvious phenotype (Baena-Gonzalez et al., 2007; Nukarinen et al., 2016; Belda-Palazon et al., 2020). After spraying β-estradiol (β-Est), KIN11 was efficiently suppressed but still detectable (Supplemental Figure 6C), and SnRK1-induced DIN10 was downregulated compared with control plants at the end of the night (Supplemental Figure 6D). After being subjected to HL, +β-Est plants had a visibly darker appearance (Figure 7A), which was attributed to higher anthocyanin contents (Figure 7B) and more pronounced induction of PAP1 and LDOX compared with control plants (Figure 7C and 7D). However, the suppression of SnRK1 had no impact on the HL induction of EBGs (Supplemental Figure 6E and 6F). In the presence and absence of SUC, β-Est-induced snrk1α1-3 amiRNAi KIN11 accumulated more anthocyanins in HL compared with the non-induced control (Figure 7E), and the knockdown of SnRK1 resulted in the same anthocyanin level as in the non-induced plants fed with SUC. Compared with the non-induced control, expression of PAP1 and LDOX was strongly induced in β-Est-induced snrk1α1-3 amiRNAi KIN11 after 8 h of HL in SOR-containing buffer (Figure 7F and 7G). Whereas SUC stimulated the expression of both transcripts by a factor of 7–8 when KIN11 was still present, the suppression of SnRK1 resulted in a less pronounced SUC induction of PAP1 and LDOX (approximately 0.5- to 0.75-fold) (Figure 7F and 7G).
Figure 7.
Inactivation of SnRK1 induced FB in HL
(A–D) Phenotype (A) and (B) anthocyanin content (expressed as cyanidin equivalents) after 24 h HL and (C)PAP1 and (D)LDOX expression in snrk1α1-3 amiRNAi KIN11 induced (+) or not (–) induced with β-Est (see methods) after 4 and 8 h HL, respectively. Gene expression was calculated relative to β-Est at EoN using the 2−ΔΔCt method with SAND as the reference gene. Statistical significance of differences was analyzed using Student’s t test (n ≥ 3) in (B)–(D), and p values are shown.
(E–G) Anthocyanin content after 24 h HL (E) and PAP1(F) and LDOX(G) expression in leaves incubated with 30 mM SOR or 30 mM SUC. For statistical analysis, 2-way ANOVA (Dunnett’s multiple comparisons test, n = 3) was used, and letters indicate significant differences at p < 0.05.
(H and I) Relative expression of PAP1(H) and (I)LDOX in Col-0 after 6 h of HL treatment in the absence (w/o) or presence of 1 μM AZD8055 (AZD). Seedlings were grown on MS plates for 10 d in SD conditions. Gene expression was calculated relative to the control (w/o) using the 2−ΔΔCt method with SAND as the reference gene. Statistical significance of differences between control and AZD treatment was analyzed using Student’s t test. Values are means ± standard deviations (n = 4), and the p values are shown. For all experiments, plants were shifted to HL at the end of the night.
The HXK1 mutant gin2-1, defective in GLC-mediated regulation of gene expression (Moore et al., 2003), accumulated WT-like amounts of anthocyanins under HL (Supplemental Figure 6G). Inactivation of SnRK1 during HL shifts or increased sugar content may also activate TOR. To test the involvement of TOR, Col-0 was grown in the absence or presence of the TOR inhibitor AZD8055 and shifted to HL. The inhibition of TOR further increased the expression of PAP1 and LDOX in HL-treated Col-0 (Figure 7H and 7I). MAP kinase (MPK) 3 and 6 phosphorylation depends on SnRK1 (Cho et al., 2016), and MPK3, -4, -6, and -11 have been shown to phosphorylate PAP1 (Li et al., 2016; Kreynes et al., 2020). Furthermore, MPK6 functions in the immediate regulation of gene expression downstream of TP export upon HL shifts (Vogel et al., 2014). However, two allelic mpk6 mutants were not affected in anthocyanin accumulation during a 24-h HL shift (Supplemental Figure 6H).
Discussion
Carbon fixation, TP export, and increase in cellular sugar level are essential for HL induction of FB
The time-resolved targeted and global gene expression analysis revealed dynamic regulation of the HL-induced transcriptome response in tpt-2. Based on the results presented here, we propose that TPT deficiency did not render the plants entirely insensitive to HL, but instead, dynamic changes in the signal connected to TP export from chloroplasts caused the delayed stimulation of FB genes in tpt-2. Several lines of evidence support a role for TP export from chloroplasts as a metabolic retrograde signal and for cellular sugar content in the induction of FB pathway genes and accumulation of anthocyanins during HL. First, in WT seedlings, the stimulation of FB genes and anthocyanin accumulation during HL treatment was positively correlated with cellular sugar content before the HL shift. Second, limitation of carbon fixation significantly attenuated the activation of FB during short-term HL exposure (Figure 3). In agreement with this result, mutants impaired in photosynthesis owing to perturbed chlorophyll accumulation showed lower amounts of anthocyanin after HL exposure compared with the WT (Richter et al., 2020). Also, perturbed FB gene expression and anthocyanin accumulation resulting from diminished photosynthetic activity caused by DCMU application were rescued by SUC feeding (Figure 3), corroborating studies with strawberry leaf discs (Creasy, 1968). Third, genetic perturbation of starch biosynthesis resulted in pronounced accumulation of soluble sugars (Supplemental Figure 1C–1E), stimulation of FB pathway gene expression, and anthocyanin production in HL-treated adg1-1 (Figures 1 and 3; Blasing et al., 2005; Ragel et al., 2013; Schmitz et al., 2014). Stronger activation of FB in adg1-1 was suppressed in the adg1-1 tpt-2 double mutant and by DCMU but rescued through exogenous SUC application (Figure 3). Lastly, diminished anthocyanin accumulation in tpt-2 was fully rescued by SUC feeding in HL, providing strong evidence for the lack of a sugar signal in tpt-2. WT-like starch accumulation (Figure 2), ROS response (Supplemental Figure 2), and regulation of photosynthetic genes (Figures 2H, 2L, and 7I) in tpt-2 indicated that photosynthesis, carbon fixation, or plastid-to-nucleus communication were overall not perturbed in the mutant. Therefore, a specific function for TP export from chloroplasts and subsequent conversion in cytosolic sugar biosynthesis pathways to regulate FB genes (and others) during HL is strongly supported. This conclusion agrees with the PAP1-dependent positive impact of sugars on the expression of FB genes even in standard growth conditions (Teng et al., 2005; Solfanelli et al., 2006). Furthermore, transcripts induced under energy-limiting conditions were enriched among the DEGs during early time points of the HL shift, and sugar-responsive transcripts showed a delayed response in tpt-2 (Figure 5 and Supplemental Figure 5). For example, GPT2 was markedly repressed at 9 h of HL relative to WT but strongly upregulated after 18 h of HL in tpt-2. GPT2 is induced in GLC-fed potato leaves (Quick et al., 1995) or by high sugar content in the starch biosynthesis mutants adg1 and pgm (Kunz et al., 2010; Weise et al., 2019) and upon HL treatment of Arabidopsis WT plants (Huang et al., 2019). Hence, the late induction of FB genes is most likely explained by a delayed increase in sugar content in tpt-2 (Figure 2), thereby supporting the suggested model of sugar-dependent regulation of FB during HL.
Given the lack of a primary route for TP export, it remains unclear which alternative pathway/transporter enables cytosolic sugar pools to increase in HL-treated tpt-2. WT-like anthocyanin accumulation in gpt2, pglct, and xpt (Supplemental Figure 1F) suggests that these routes for carbohydrate exchange are not essential for HL induction of FB in the presence of a functional TPT. Nevertheless, we cannot exclude the possibility that one of these transporters partially compensated for the lack of TPT in tpt-2. For example, XPT has also been reported to transport TP (Eicks et al., 2002) and could partially complement the lack of TPT in HL as it does under normal growth conditions in TPT-deficient mutants (Hilgers et al., 2018). Alternatively, starch degradation already occurring in TPT-deficient plants during the day (Hausler et al., 1998; Schneider et al., 2002; Walters et al., 2004), subsequent export of maltose through maltose exporter 1 (MEX1; Niittyla et al., 2004), and conversion in the cytosol could also contribute to the increasing sugar content of tpt-2 exposed to long-term HL. Indeed, a function of MEX1 in cold-induced anthocyanin accumulation has been reported (Purdy et al., 2013), and maltose feeding can stimulate FB (Teng et al., 2005). Future analyses of double mutants deficient in TPT and other plastid-localized sugar transporters could reveal which alternative export pathways are responsible for the late increase in sugar content and sugar-dependent regulation of FB under long-term HL.
Previously, ROS were shown to negatively or positively affect the expression of FB genes through, for example, redox-dependent regulation of transcriptional FB repressors such as TCP14 or -15 (Viola et al., 2016). However, time-resolved analysis of the WT and the tpt-2 mutant enabled separation of the (early) ROS response from a metabolic sugar signal regulating FB during HL acclimation. More precisely, the expression of GPX7 was rapidly stimulated upon HL exposure (3 h HL), then declined and strongly increased again after 9 h HL in WT and tpt-2 (Supplemental Figure 2). Whereas FB gene expression paralleled that of ROS marker genes during early time points in WT, tpt-2 showed a WT-like ROS response, but, at the same time, failed to stimulate crucial positive regulators (PAP1 and TT8) of LBGs (LDOX and DFR). The substantial increase in GPX7 (and others, Supplemental Figure 2) after prolonged HL paralleled the expression of FB genes in tpt-2 but not in WT, in which the expression of FB pathway genes reached a maximum or was even repressed in the case of PAP1. Unchanged expression of ascorbate biosynthesis genes (e.g., VTC2, Supplemental Data 1) and WT-like total ascorbate level in the tpt-2 mutant (Schmitz et al., 2014) may exclude diminished ascorbate level as causative for the deregulation of FB in tpt-2 (Page et al., 2012; Plumb et al., 2018). We do not rule out a function of ROS in the regulation of EBGs (e.g., CHS, FLS1) or LBGs under long-term HL (e.g., several days) or under other and/or more severe abiotic stress conditions, but we concluded that ROS are most likely not involved in the stimulation of, in particular, LBGs at early time points of HL treatment. In addition to ROS, phytohormones such as JA (Shan et al., 2009; Qi et al., 2011) or ethylene (Jeong et al., 2010) also affect FB. Ethylene biosynthesis or ethylene-responsive genes and other phytohormone biosynthesis pathways were not enriched among the DEGs, and only JA biosynthesis genes showed late induction in HL-treated tpt-2 (Supplemental Figure 4B and 4C; Supplemental Data 4). Although some FB genes were responsive to exogenous JA feeding (Dombrecht et al., 2007), JA contents did not change when WT plants were subjected to 7 h of HL (Balfagon et al., 2019), and anthocyanin accumulation was WT-like in JA-deficient aos1-1 and jassy mutants (Supplemental Figure 4). These findings suggest that JA availability is most likely not a determining factor for anthocyanin accumulation after short-term HL treatment, but a potential role for JA in the late induction of FB genes in tpt-2 or WT cannot be ruled out under different conditions.
At this point, we want to highlight that diminished anthocyanin accumulation in tpt-2 during HL exposure has been reported previously by Schmitz et al. (2014). However, using microarray-based transcriptomics, no changes in the expression of FB pathway genes were observed. Although we cannot entirely exclude the possibility that the different experimental conditions (shift to 300 μmol m−2 s−1 and another light source) had an impact on the different outcomes of the experiments, the time point of harvest may explain most of the differences: Schmitz et al. harvested plant material after 4 h and 48 h of the HL shift. However, our time-resolved analysis revealed only minor differences in the expression of FB pathway genes between WT and tpt-2 after 4 h HL and WT-like expression of PAP1, DFR, and LDOX after prolonged HL exposure (i.e., 24 h HL). Hence, owing to the selection of time points, the previous study could not detect the dynamic changes and altered transcriptional response of FB during HL acclimation in tpt-2.
SnRK1 downregulates HL induction of anthocyanin biosynthesis
Transcriptome changes throughout the HL exposure revealed late induction and repression of sugar-responsive genes in tpt-2, and we propose that increasing cellular sugar content serves to induce FB at the transcriptional level upon HL exposure. Changes in sugar content are sensed and translated into adjustments of gene expression by, for instance, HXK1, TOR, and SnRK1 and downstream components (Baena-Gonzalez and Lunn, 2020; Li et al., 2021). Our analysis showed that HXK1-dependent GLC signaling is not essential for FB activation when plants are exposed to HL (Supplemental Figure 6E). Instead, knockdown of SnRK1 activity resulted in faster activation of FB and almost complete insensitivity to SUC. By contrast, overexpression of KIN10 diminished FB induction in HL and suppressed the more robust activation of FB in the adg1-1 background (Figures 6 and 7). Therefore, the results presented here support a model in which SnRK1 functions downstream of the chloroplasts, and inhibition of SnRK1 is a prerequisite for HL-induced anthocyanin biosynthesis. This conclusion is supported by the targeted and global gene expression analysis, which showed altered SnRK1-dependent signaling in tpt-2 over the course of the HL experiment (Figures 4 and 5) and differential expression of SnRK1-regulated transcripts in HL-treated WT (Supplemental Figure 5B). Based on the expression profile of well-established marker genes for SnRK1-dependent signaling, we concluded that the delayed increase in sugar content (Figure 2) led to delayed inactivation of SnRK1, thereby permitting the activation of FB only after prolonged HL treatment in tpt-2. When SUC feeding was applied (Figure 3), anthocyanin accumulation was restored to the WT level in tpt-2. In this regard, more pronounced repression of SnRK1 marker genes (Figure 5) and activation of FB in adg1-1 in HL (Figures 1 and 3) are probably explained by high sugar (Supplemental Figure 1) and T6P contents in starch-deficient mutants (Carillo et al., 2013), resulting in more substantial suppression of SnRK1 activity compared with WT plants. High sugar content most likely also partially suppresses SnRK1 activity even when KIN10 is overexpressed, thereby leading to stronger activation of FB in adg1-1 KIN10ox compared with the KIN10ox line (Figure 6). The newly established connection between FB activation in HL and SnRK1 activity receives strong support from previous studies: PAP1 was repressed by SnRK1 in protoplast assays, and excess of KIN10 diminished sucrose-induced activation of FB in planta (Baena-Gonzalez et al., 2007; Ramon et al., 2019). By contrast, FB was activated and pathway genes were partially de-repressed in partial loss-of-function snrk1α1−/−snrk1α2+/− and snrk1α1-3 amiRNAi KIN11 mutants, even under normal growth conditions (Nukarinen et al., 2016; Peixoto et al., 2021; Wang et al., 2021). Overexpression of T6P phosphatase supposedly activated SnRK1 but repressed FB in stably transformed lines (otsB, Zhang et al., 2009). On the contrary increased T6P level in T6P synthase (otsA) overexpressor plants induced FB (Schluepmann et al., 2003; Zhang et al., 2009; Wingler et al., 2012). In addition to the transcriptional regulation of FB pathway genes, the inactivation of SnRK1 leads to indirect post-translational stabilization of PAL, which catalyzes the initial step of phenylpropanoid biosynthesis upstream of FB (Wang et al., 2021). Based on the new results, we propose that anthocyanin accumulation during HL acclimation is the result of both transcriptional and post-translational activation of FB through SnRK1 inhibition.
TOR activity is stimulated by GLC (Xiong et al., 2013; Li and Sheen, 2016) and repressed by SnRK1-dependent phosphorylation of the RAPTOR component (Nukarinen et al., 2016). Therefore, high sugar content and inactivation of SnRK1 during HL could lead to TOR activation, which may induce FB. However, diminished TOR activity in raptor1B or inducible tor knockdown mutants results in increased flavonoid contents (Caldana et al., 2013; Wang et al., 2017; Salem et al., 2018). Inhibition of TOR by AZD even further stimulated rather than repressed FB pathway genes in HL (Figure 6), suggesting that active TOR is not essential for HL stimulation of FB but may have a repressive function in FB. However, perturbed TOR function leads to starch and sugar accumulation (Caldana et al., 2013; Salem et al., 2018; Zhang et al., 2018). Because of the strong correlation between cellular sugar and T6P content (Carillo et al., 2013; Yadav et al., 2014; Peixoto et al., 2021), FB could be (further) stimulated when TOR activity is suppressed (in HL) because high sugar levels may result in more substantial inactivation of SnRK1 (as in the sugar-overaccumulating starch mutants). The involvement of TOR and its interconnection with SnRK1 in the regulation of FB during HL acclimation should be analyzed in the future using (double) mutants with altered activities of both factors.
Potential downstream components of the sugar signal and the connection with light signaling
MPK6 was shown to be activated by a TP-dependent signal, thereby permitting the adjustment of nuclear gene expression within 10 min of HL exposure (Vogel et al., 2014). Although it is still a matter of debate, phosphorylation of PAP1 by MPK3, -4, -6, or -11 may adjust the stability or activity of the TF and eventually the accumulation of anthocyanins (Li et al., 2016; Kreynes et al., 2020, 2021; Yang et al., 2021). Interestingly, phosphorylation of MPK3 and -6 depends on SnRK1 in response to the submergence of leaves (Cho et al., 2016), which would provide a connection between TP export, SnRK1, MPK activity, and the regulation of FB. However, mpk6 mutants were indistinguishable from WT plants in terms of anthocyanin accumulation (Supplemental Figure 6F; Li et al., 2016), indicating that at least MPK6 is dispensable for HL-induced anthocyanin accumulation.
Components of light-signaling pathways also regulate FB. Hence, the question arises of how SnRK1 may interact with those pathways to regulate FB during HL. CRY- and PHY-dependent inactivation of the constitutive photomorphogenic1/suppressor of phyA-105 (COP1/SPA)-containing E3 ubiquitin ligase complex in the light permits stabilization of HY5 and its interacting B-box-containing proteins (BBX20, BBX21, and BBX22, Bursch et al., 2020) but also PAP1 and PAP2 (Maier et al., 2013; Ponnu et al., 2019), and direct binding of these TFs to their target genes promotes FB (Shin et al., 2013; Bursch et al., 2020). Consequently, cry1, hy5, and bbx202122 mutants showed low expression of FB regulators and pathway genes and diminished anthocyanin contents when exposed to monochromatic light but also HL (Kleine et al., 2007; Stracke et al., 2010; Ponnu et al., 2019; Bursch et al., 2020), thereby resembling mutants with active SnRK1 in terms of FB regulation. Because physical interaction of SnRK1 with components for ubiquitin-mediated protein turnover has been reported (E3 ligases and 26S proteasome; Farras et al., 2001; Nietzsche et al., 2014), it is tempting to speculate that SnRK1 (in)directly represses the function of positive regulators of FB (e.g., HY5, BBX, PAP1, TT8). A potential function of SnRK1 as a repressor of highly energy-demanding FB is supported by, for example, SnRK1-mediated destabilization of WRINKLED1, a TF that regulates fatty acid biosynthesis (Zhai et al., 2018). Future experiments will reveal how and at which point light- and sugar-dependent signaling pathways converge at the molecular level to permit the activation of FB.
Taken together and in conjunction with previous studies revealing a crucial function for TP export in the regulation of nuclear gene expression under (short-term) HL (Vogel et al., 2014; Weise et al., 2019), our study emphasizes the critical role of chloroplasts as sensors for changes in the environment and as emitters of metabolic signals for the adjustment of nuclear gene expression and pathways relevant for acclimation responses, such as FB. Although a shift in light intensity leads to the partial photoinhibition of photosynthetic complexes and, thus, would result in lower photosynthetic electron transfer rates and carbon fixation, sophisticated protection (e.g., non-photochemical quenching, flavonoid accumulation) and repair mechanisms ensure high photosynthetic activity and CO2 fixation rates in HL. Even beyond the light saturation point at approximately 400–500 μmol photons m−2 s−1 in Arabidopsis (Schumann et al., 2017), photosynthesis is still active, producing sufficient amounts of photosynthates for growth and reproduction. Therefore, a (transient) rise in cellular sugar content could serve as a metabolic signal informing the cell about changing light intensities (Schmitz et al., 2014 and results presented above). The accumulation of anthocyanins is a widespread response of plants exposed to adverse environmental conditions. It is of great interest to test whether FB is also regulated by a sugar signal when plants are exposed to other abiotic stress conditions (e.g., low temperatures). In this context, comparative gene expression analysis (Figures 4 and 5) also revealed that other cellular processes and pathways, such as the biogenesis of ribosomes, which function as hubs for plant acclimation (Garcia-Molina et al., 2020), were affected when TP export was perturbed. Hence, the importance of TP as a metabolic retrograde signal (Pfannschmidt, 2010) and the connected cytosolic sugar biosynthesis and signaling pathways for HL acclimation should be analyzed in more detail in the future.
Methods
Genotypes and growth conditions
If not otherwise stated, A. thaliana WT and mutant plants were grown on soil in SD (10 h light) at 100 μmol photons m−2 s−1 at 22°C. Genotypes used in this study are listed in Supplemental Data 8. Homozygous mpk6-2 and mpk6-3 were obtained from K.-J. Dietz (University Bielefeld, Germany); gpt2-1, adg1-1 tpt2-1, xpt-1, xpt-2, and gin2-1 from R. Häusler (University of Cologne, Germany); and jassy from S. Schwenkert (Ludwig-Maximilians-Universität Munich, Germany). Mutant lines overexpressing T6P from Escherichia coli (otsB) were provided by A. Wiese-Klinkenberg (Forschungszentrum Jülich, Germany) and snrk1α1-3 amiRNAi KIN11 by W. Dröge-Laser (University of Würzburg, Germany). The aos1-1 mutant (SALK_017756) was obtained from I. Feußner (University of Göttingen, Germany). The tDNA insertion mutants pglct-2 and tpt-2 and the ethyl methanesulfonate mutant adg1-1 were obtained from the Nottingham Arabidopsis Stock Centre (NASC). Gene knockout was confirmed by qPCR analysis or lack of starch in adg1-1. To induce the knockdown of KIN11, snrk1α1-3 amiRNAi KIN11 lines were grown for 3 weeks in SD and were sprayed with or without 20 μM β-estradiol in water supplemented with 0.05% Tween 20 for another 7 days.
The adg1-1 KIN10 overexpression lines were obtained by crossing KIN10ox 1 with adg1-1. In the T2 generation, phosphinothricin-resistant (selection for 35S:KIN10-HA) and starch-deficient (for adg1-1 mutation) plants were selected. Overexpression of KIN10-HA was confirmed by means of western blotting and hemagglutinin (HA)-specific antibody, and the presence of the point mutation in ADG1 was confirmed by Sanger sequencing. In the T3, seeds of plants homozygous for adg1-1 were sown on soil and sprayed with phosphinothricin. Plants were sprayed 3 times within 7 days starting 7 days after germination and were kept for another 5 days without any treatment before the HL shift was performed. Several adg1-1 KIN10ox lines were obtained and analyzed.
Treatment of plants
Standard HL shift experiments were carried out with 18- to 21-d-old plants exposed to 500 μmol photons m−2 s−1 for the indicated time periods under a constant temperature (22°C) in a Conviron Adaptis A/GEN1000 growth chamber equipped with a white LED light source. Plants were either shifted at the end of the night or 2 h after the onset of light. For young plants, samples contained leaves of approximately 10 seedlings pooled from at least 3 different pots. Experiments were repeated at least one time. For HL shift experiments with snrk1α1-3 amiRNAi KIN11, rosette leaves of 4-week-old plants were harvested for each sample, and each sample contained leaves from two individual plants.
For HL shift experiments in normal (400 ppm) or low (50 ppm) CO2 conditions (21% oxygen), rosette leaves were mounted on a LI-COR LI-6400XT device (LI-COR, USA) and incubated at 500 μmol photons m−2 s−1 (10% blue and 90% red LED light) for 4 h. The flow rate was set to 300 μmol s−1, the block temperature to 22°C, and the relative humidity to 60%. After 4 h, one leaf disc (2 cm2) was punched out from the treated area of the leaf and directly frozen in liquid nitrogen. Six leaves from individual rosette plants were analyzed on consecutive days.
Sugar (SUC) feeding and DCMU application were performed by incubating plants in 20 mM Tris buffer (pH 7.4). SUC was applied at 30 (∼1%; g/v) or 90 mM (∼3%; g/v). As an osmotic control, 30 or 90 mM SOR was used. DCMU stock was prepared in DMSO, and control buffers contained the same amount of DMSO. Plants were transferred to the buffer at the end of the night and pre-incubated for 30 min in darkness before the HL shift.
For AZD treatment, seeds were surface sterilized by incubation in 70% ethanol (plus 0.05% Triton X-100) for 10 min. Subsequently, seeds were washed with 70% ethanol, followed by a washing step in 100% ethanol. After decanting the ethanol, seeds were dried and plated on 0.5× Murashige and Skoog (MS) medium (4.4 g/l (w/v) MS including vitamins, 0.5 g/l (w/v) MES, 0.8% agar, pH 5.7). MS plates were supplemented with 1 μM AZD8055 (MedChemExpress [USA], HY-10422). After stratification at 4°C for 2 d, plants were grown in SD conditions for 10 d before the HL shift.
Detection of ROS
Superoxide radical accumulation was analyzed using nitro blue tetrazolium chloride (NBT, Sigma-Aldrich [USA], 93862). Leaves of half-rosettes from approximately 4-week-old plants were incubated in NBT staining solution (25 mM HEPES/KOH, 1 mg/ml NBT, pH 7.5). After 15–30 min vacuum infiltration, leaves were further incubated for 2 h at room temperature (RT) in the dark. Hydrogen peroxide content was analyzed using 3,3′-diaminobenzidine (DAB, Merck-Millipore [USA], D8001). The DAB staining solution (20 mM Tris/acetate, 1 mg/ml DAB [Sigma-Aldrich], pH 5) was prepared 1 h before use. After vacuum infiltration, the leaves were kept in darkness for 24 h at RT. Chlorophyll was destained with 80% (v/v) ethanol for 20 min at 80°C in a water bath. Hydrogen peroxide contents were also analyzed using 2′,7′-dichlorofluorescein diacetate (H2DCF-DA, Sigma-Aldrich, 35845). To this end, leaves were incubated in 20 mM Tris/HCl (pH 7.4) with or without 10 μM H2DCF-DA. After 15–30 min vacuum infiltration at RT, leaves were left in the solution for another 0.5–2 h at RT in the dark. For the analysis, leaf discs were cut, and the fluorescence signal was detected using a Zeiss (Germany) LSM 800 confocal laser-scanning microscope (63× magnification) at an excitation wavelength of 488 nm. The DCF signal was detected with an emission wavelength of 500–575 nm. Chlorophyll fluorescence emission was recorded between 650 and 680 nm. The settings for analysis were the same for all analyzed samples. The mean fluorescence signals for DCF were quantified using ImageJ (https://imagej.nih.gov/ij/).
Quantification of starch and starch staining
Starch was extracted from dried or frozen leaf material using 80% (v/v) ethanol and incubation at 80°C for 30 min. After centrifugation (10 min, 13 000 rpm, RT), the soluble carbohydrate-containing supernatants were transferred to new tubes and prepared for sugar analysis (see below). The starch-containing pellet was resuspended in 750 μl of 0.5 M NaOH and incubated for 30 min at 95°C before adding 750 μl of 1 M CH3COOH. Starch was digested by mixing 100 μl of the starch suspension with 100 μl of amyloglucosidase solution (1 mg/ml in 200 mM CH3COOH and 100 mM NaOH) for 2 h shaking at 1050 rpm at 55°C. Then, 100 μl of a 1:5, 1:10, or 1:20 dilution of the starch digestion diluted in H2Odd was mixed with 200 μl of glucose oxidase (GlcOx) reagent (1 mg glucose oxidase, 1.5 mg horseradish peroxidase [HRP], 5 mg dianisidine/HCl per 50 ml 0.5 M Tris/HCl [pH 7.0], 40% [v/v] glycerol). For the glucose standard curve, 0–1 mM glucose was mixed with 200 μl of the GlcOx. The standards and samples were incubated at 30°C for 30 min before the reaction was stopped by the addition of 400 μl of 5 M HCl. After a short centrifugation (30 s, RT), the absorbance of the samples at 540 nm was determined using a 96-well plate reader (SpectraMax M2 microplate reader [Molecular Devices, USA]). The amount of starch was determined as glucose equivalents, and the glucose content was calculated using the standard curve.
For the staining of starch in intact seedlings, chlorophyll was destained with 80% (v/v) ethanol for 20 min at 80°C. Subsequently, seedlings were incubated for 1 h with Lugol’s iodine solution. After destaining with water for 2 h at RT, starch accumulation was documented.
Quantification of soluble sugars
Concentrations of the free sugars SUC, GLU, and FRC were quantified photometrically as previously described (Atanasov et al., 2020). Supernatants from the first step of the starch extraction (see above) were collected and dried in a speed vacuum concentrator. Sugars were extracted from dried pellets in H2Odd by constantly shaking at 500 rpm and 22°C. For sucrose quantification, sample extracts were incubated for 10 min with 30% KOH at 95°C, followed by incubation for 30 min at 40°C with an anthrone reagent (0.14% w/v anthrone in 14.6 M H2SO4). Together with a standard calibration curve, sample absorbance was determined at 620 nm. Glucose concentrations were determined from extracts in a coupled hexokinase/glucose-6-phosphate dehydrogenase assay, resulting in NADPH + H+ formation, detectable at 340 nm. Following glucose quantification, phosphoglucose isomerase was added to the mixture to determine fructose concentrations. The absolute amount was calculated using the standard calibration curves.
Anthocyanin extraction and quantification
Anthocyanins were extracted from ground and frozen leaf material using 1 ml of anthocyanin extraction buffer (18% 1-propanol, 1% HCl in water). Homogenates were incubated for 2 h at RT in darkness. After centrifugation for 10 min at RT (13 000 rpm), supernatants were transferred to cuvettes, and the absorption at 547, 650, and 720 nm was recorded. The absorption of anthocyanins was calculated using the following formula: (A537−A720) − 0.25×(A650−A720). Absorption values were transformed into cyanidin equivalents using cyanidin chloride as a standard (Sigma-Aldrich, 79457) and normalized to either grams fresh weight (fw) or dry weight (dw).
Protein extraction and western blot analysis
Total leaf proteins were extracted from ground and frozen leaf material using protein extraction buffer (56 mM Na2CO3, 56 mM dithiothreitol [DTT], 2% [w/v] SDS, 12% [w/v] SUC, and 2 mM EDTA). After the powder was resuspended, samples were incubated at 90°C for 10 min, and protein extracts were obtained by centrifugation (10 min, 13 000 rpm, RT). Protein extracts were separated on 12% polyacrylamide-SDS gels and blotted onto a nitrocellulose membrane. After blocking for 1 h in 4% milk solution in TBS-T (50 mM Tris/HCl, 150 mM NaCl, pH 7.5; 0.1% [v/v] Tween 20), membranes were incubated overnight at 4°C with the primary antibody in 1% milk solution in TBS (phospho-AMPK, 1:2000, Cell Signaling Technology [USA], #2535). The next day, membranes were washed and incubated with the secondary antibody for 2 h at RT (goat anti-rabbit immunoglobulin G [IgG] coupled with HRP, 1:10 000, 1% milk solution, TBS). Signals were detected using Clarity Western ECL substrate (Bio-Rad, Germany) and an ECL Chemostar CCD camera (Intas, Germany).
RNA extraction and qPCR analysis
Whole-leaf RNA was extracted using a previously published protocol (Oñate-Sánchez and Vicente-Carbajosa, 2008). In brief, frozen and ground leaf material was resuspended in 300 μl cell lysis buffer (2% [w/v] SDS, 68 mM sodium citrate, 132 mM citric acid, 1 mM EDTA). After the addition of 100 μl DNA/protein precipitation solution (4 M NaCl, 16 mM sodium citrate, 32 mM citric acid), samples were vortexed and incubated on ice for 10 min. Subsequently, samples were centrifuged at 13 000 rpm (4°C) for 10 min, and 300 μl of the supernatant was used to precipitate the RNA with 300 μl isopropanol. After precipitation (5 min, 13 000 rpm, RT), pellets were washed with 800 μl 75% ethanol, centrifuged again, and dried. The RNA was resuspended in 20–25 μl of H2Odd (RNAse free) and stored at −80°C until further use.
For qPCR analysis, 1–2 μg of DNase I (Thermo Fisher [USA])-treated RNA was transcribed into cDNA using RevertAid reverse transcriptase (Thermo Fisher) according to the manufacturer’s protocol. qPCR analysis was carried out in a CFX96-C1000 96-well plate thermocycler (Bio-Rad) with ChamQ Universal SYBR qPCR Master Mix (Absource Diagnostics, Germany) in 6-μl reactions containing 1 μl of diluted (1:5) cDNA. Relative gene expression was calculated using the 2−ΔΔCt method with SAND (AT2G28390) as the reference gene. The primers used for qPCR analysis are listed in Supplemental Data 9.
For RNA-seq, RNA was treated with DNase I and purified using a phenol/chloroform/isoamyl alcohol precipitation protocol. After washing with 75% ethanol (v/v), RNA was dried and resuspended in RNase-free water.
RNA-seq and data analysis
RNA-seq was performed by Novogene (UK/China). Three biological replicates for Col-0 and tpt-2 after 9 and 18 h of HL treatment were analyzed. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first-strand cDNA was synthesized using random hexamer primers, followed by second-strand cDNA synthesis. The libraries were tested with a Qubit fluorometer and real-time PCR for quantification and an Agilent Bioanalyzer for detection of size distribution. Quantified libraries were sequenced on the Illumina HiSeq 4000 platform, according to effective library concentration and data amount (4 GB raw reads). Raw data in fastq format were processed using in-house perl scripts to obtain clean data by removing adapter-contaminated, poly-N, and low-quality reads. At the same time, Q20, Q30, and GC content of the clean data were calculated. Paired-end clean reads were aligned to the TAIR10 reference genome and gene model annotation files using HISAT2 version 2.0.5, and featureCounts version 1.5.0-p3 was used to count the number of reads that mapped to each gene. Fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) was calculated for each gene based on its length and mapped read counts. Differential gene expression analysis (three biological replicates per condition) was performed using the DESeq2 R package (1.20.0). The resulting p values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Post-sequencing analysis of data was performed using the Galaxy webserver (Galaxy Europe, https://usegalaxy.eu/), the Venny tool for preparation of Venn diagrams (https://bioinfogp.cnb.csic.es/tools/venny/), and R studio (heatmap package: ‘pheatmap’ and correlation analysis: ‘corrplot’). GO term analysis was performed using GOTermFinder with settings for A. thaliana (https://go.princeton.edu/cgi-bin/GOTermFinder). The transcriptome data were compared with the expression data published in Huang et al. (2019), Baena-Gonzalez et al. (2007), and Garcia-Molina et al. (2020).
Funding
Work in the lab of A.S.R. is supported by a grant from the German Research Foundation (DFG) to A.S.R. (TRR175, project C06). Work in the lab of T.N. was supported by the DFG (TRR175, D03).
Author contributions
M.-E.Z., A.S.R., A.K., K.J., T.R., G.E.A., C.K., and T.N. performed the experiments and analyzed the data. A.S.R. designed, conceived, and supervised the study and wrote the article, with the support of all authors. A.S.R. and T.N. acquired funding.
Acknowledgments
We acknowledge the generous gifts of seeds from the following colleagues: K.-J. Dietz (mpk6), W. Dröge-Laser (snrk1α1-3 amiRNAi KIN11), R. Häusler (gpt2-1, adg tpt, xpt, gin2-1), A. Wiese-Klinkenberg (otsB), S. Schwenkert (jassy), and I. Feussner (aos1-1). We also thank R. Häusler for discussion on the project. No conflict of interest is declared.
Published: August 11, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
GO term analysis was conducted using https://go.princeton.edu/cgi-bin/GOTermFinder without plastome-encoded genes. Only DEGs with log2 FC < 1 and with an adjusted p < 0.05 were considered for GO term analysis. WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value.
GO term analysis was conducted using https://go.princeton.edu/cgi-bin/GOTermFinder. DEGs with log2 fold change <–1 for tpt/WT 9 h HL and log2 FC > 1 for tpt-2 (18 h/9 h HL) and with an adjusted p < 0.05 were used for GO term analysis. WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value.
WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value. Adapted from Huang et al. (2019).
WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value
Data availability
Transcriptome data were deposited at GEO (https://www.ncbi.nlm.nih.gov/geo/info/update.html) under the record GSE196053.
References
- Agati G., Guidi L., Landi M., Tattini M. Anthocyanins in photoprotection: knowing the actors in play to solve this complex ecophysiological issue. New Phytol. 2021;232:2228–2235. doi: 10.1111/nph.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agati G., Brunetti C., Fini A., Gori A., Guidi L., Landi M., Sebastiani F., Tattini M. Are Flavonoids Effective Antioxidants in Plants? Twenty Years of Our Investigation. Antioxidants. 2020;9:1098. doi: 10.3390/antiox9111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar T.A., Lees H.A., Lampi M.A., Enstone D., Brain R.A., Greenberg B.M. Photosynthetic redox imbalance influences flavonoid biosynthesis in Lemna gibba. Plant Cell Environ. 2010;33:1205–1219. doi: 10.1111/j.1365-3040.2010.02140.x. [DOI] [PubMed] [Google Scholar]
- Alsharafa K., Vogel M.O., Oelze M.L., Moore M., Stingl N., König K., Friedman H., Mueller M.J., Dietz K.J. Kinetics of retrograde signalling initiation in the high light response of Arabidopsis thaliana. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130424. doi: 10.1098/rstb.2013.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J.P., Zhang X.W., Liu Y.J., Wang X.F., You C.X., Hao Y.J. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021;72:1460–1472. doi: 10.1093/jxb/eraa525. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Thiedig K., Nordholt N., Schmidt N., Huep G., Sagasser M., Weisshaar B. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta. 2014;240:955–970. doi: 10.1007/s00425-014-2088-0. [DOI] [PubMed] [Google Scholar]
- Atanasov V., Fürtauer L., Nägele T. Indications for a central role of hexokinase activity in natural variation of heat acclimation in Arabidopsis thaliana. Plants. 2020;9:819. doi: 10.3390/plants9070819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Lunn J.E. SnRK1 and trehalose 6-phosphate - two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 2020;55:52–59. doi: 10.1016/j.pbi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Balfagon D., Sengupta S., Gomez-Cadenas A., Fritschi F.B., Azad R.K., Mittler R., Zandalinas S.I. Jasmonic acid is required for plant acclimation to a combination of high light and heat stress. Plant Physiol. 2019;181:1668–1682. doi: 10.1104/pp.19.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B., Adamo M., Valerio C., Ferreira L.J., Confraria A., Reis-Barata D., Rodrigues A., Meyer C., Rodriguez P.L., Baena-González E. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants. 2020;6:1345–1353. doi: 10.1038/s41477-020-00778-w. [DOI] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch K., Toledo-Ortiz G., Pireyre M., Lohr M., Braatz C., Johansson H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants. 2020;6:921–928. doi: 10.1038/s41477-020-0725-0. [DOI] [PubMed] [Google Scholar]
- Caldana C., Li Y., Leisse A., Zhang Y., Bartholomaeus L., Fernie A.R., Willmitzer L., Giavalisco P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73:897–909. doi: 10.1111/tpj.12080. [DOI] [PubMed] [Google Scholar]
- Calixto C.P.G., Guo W., James A.B., Tzioutziou N.A., Entizne J.C., Panter P.E., Knight H., Nimmo H.G., Zhang R., Brown J.W.S. Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant Cell. 2018;30:1424–1444. doi: 10.1105/tpc.18.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo P., Feil R., Gibon Y., Satoh-Nagasawa N., Jackson D., Bläsing O.E., Stitt M., Lunn J.E. A fluorometric assay for trehalose in the picomole range. Plant Methods. 2013;9:21. doi: 10.1186/1746-4811-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J. Learning the Languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- Chen G.H., Liu M.J., Xiong Y., Sheen J., Wu S.H. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA. 2018;115:12823–12828. doi: 10.1073/pnas.1809526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.Y., Wen T.N., Wang Y.T., Shih M.C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016;67:2745–2760. doi: 10.1093/jxb/erw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy L.L. The significance of carbohydrate metabolism in flavonoid synthesis in strawberry leaf disks. Phytochemistry. 1968;7:1743–1749. [Google Scholar]
- Das P.K., Shin D.H., Choi S.B., Yoo S.D., Choi G., Park Y.I. Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells. 2012;34:93–101. doi: 10.1007/s10059-012-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A., Wang J.Z., Dehesh K. Retrograde signals: Integrators of Interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 2017;68:85–108. doi: 10.1146/annurev-arplant-042916-041007. [DOI] [PubMed] [Google Scholar]
- Dietz K.J., Turkan I., Krieger-Liszkay A. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 2016;171:1541–1550. doi: 10.1104/pp.16.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.M., Alboresi A., Weisshaar B., Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Eicks M., Maurino V., Knappe S., Flügge U.I., Fischer K. The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 2002;128:512–522. doi: 10.1104/pp.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani J., Grotewold E., Falcone Ferreyra M.L., Casati P. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol. Plant. 2013;6:1376–1379. doi: 10.1093/mp/sst021. [DOI] [PubMed] [Google Scholar]
- Farrás R., Ferrando A., Jásik J., Kleinow T., Okrész L., Tiburcio A., Salchert K., del Pozo C., Schell J., Koncz C. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001;20:2742–2756. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Molina A., Kleine T., Schneider K., Mühlhaus T., Lehmann M., Leister D. Translational components contribute to acclimation responses to high light, heat, and cold in arabidopsis. iScience. 2020;23:101331. doi: 10.1016/j.isci.2020.101331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläßer C., Haberer G., Finkemeier I., Pfannschmidt T., Kleine T., Leister D., Dietz K.J., Häusler R.E., Grimm B., Mayer K.F.X. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant. 2014;7:1167–1190. doi: 10.1093/mp/ssu042. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Gould K.S. Nature's Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004;2004:314–320. doi: 10.1155/S1110724304406147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K.S., Jay-Allemand C., Logan B.A., Baissac Y., Bidel L.P. When are foliar anthocyanins useful to plants? Re-evaluation of the photoprotection hypothesis using Arabidopsis thaliana mutants that differ in anthocyanin accumulation. Environ. Exp. Bot. 2018;154:11–22. doi: 10.1016/j.envexpbot.2018.02.006. [DOI] [Google Scholar]
- Guan L., Denkert N., Eisa A., Lehmann M., Sjuts I., Weiberg A., Soll J., Meinecke M., Schwenkert S. JASSY, a chloroplast outer membrane protein required for jasmonate biosynthesis. Proc. Natl. Acad. Sci. USA. 2019;116:10568–10575. doi: 10.1073/pnas.1900482116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler R.E., Schlieben N.H., Schulz B., Flügge U.I. Compensation of decreased triose phosphate/phosphate translocator activity by accelerated starch turnover and glucose transport in transgenic tobacco. Planta. 1998;204:366–376. doi: 10.1007/s004250050268. [DOI] [PubMed] [Google Scholar]
- Heijde M., Binkert M., Yin R., Ares-Orpel F., Rizzini L., Van De Slijke E., Persiau G., Nolf J., Gevaert K., De Jaeger G., et al. Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:20326–20331. doi: 10.1073/pnas.1314336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers E.J.A., Schöttler M.A., Mettler-Altmann T., Krueger S., Dörmann P., Eicks M., Flügge U.I., Häusler R.E. The combined loss of triose phosphate and xylulose 5-phosphate/phosphate translocators leads to severe growth retardation and impaired photosynthesis in Arabidopsis thaliana tpt/xpt double mutants. Front. Plant Sci. 2018;9:1331. doi: 10.3389/fpls.2018.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhao X., Chory J. The arabidopsis transcriptome responds specifically and dynamically to high light stress. Cell Rep. 2019;29:4186–4199.e3. doi: 10.1016/j.celrep.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S.W., Das P.K., Jeoung S.C., Song J.Y., Lee H.K., Kim Y.K., Kim W.J., Park Y.I., Yoo S.D., Choi S.B., et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 2010;154:1514–1531. doi: 10.1104/pp.110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job N., Yadukrishnan P., Bursch K., Datta S., Johansson H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018;176:2963–2976. doi: 10.1104/pp.17.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Leister D. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta. 2016;1857:1313–1325. doi: 10.1016/j.bbabio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 2007;144:1391–1406. doi: 10.1104/pp.107.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Nägele T., Neuhaus H.E., Schmitz-Linneweber C., Fernie A.R., Geigenberger P., Grimm B., Kaufmann K., Klipp E., Meurer J., et al. Acclimation in plants - the green hub consortium. Plant J. 2021;106:23–40. doi: 10.1111/tpj.15144. [DOI] [PubMed] [Google Scholar]
- Kreynes A.E., Yong Z., Ellis B.E. Developmental phenotypes of Arabidopsis plants expressing phosphovariants of AtMYB75. Plant Signal. Behav. 2021;16:1836454. doi: 10.1080/15592324.2020.1836454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreynes A.E., Yong Z., Liu X.M., Wong D.C.J., Castellarin S.D., Ellis B.E. Biological impacts of phosphomimic AtMYB75. Planta. 2020;251:60. doi: 10.1007/s00425-020-03350-0. [DOI] [PubMed] [Google Scholar]
- Kunz H.H., Häusler R.E., Fettke J., Herbst K., Niewiadomski P., Gierth M., Bell K., Steup M., Flügge U.I., Schneider A. The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol. 2010;12(Suppl 1):115–128. doi: 10.1111/j.1438-8677.2010.00349.x. [DOI] [PubMed] [Google Scholar]
- LaFountain A.M., Yuan Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021;231:933–949. doi: 10.1111/nph.17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Sheen J. Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 2016;33:116–125. doi: 10.1016/j.pbi.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Liu K.H., Sheen J. Dynamic nutrient signaling networks in plants. Annu. Rev. Cell Dev. Biol. 2021;37:341–367. doi: 10.1146/annurev-cellbio-010521-015047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang W., Gao J., Yin K., Wang R., Wang C., Petersen M., Mundy J., Qiu J.L. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in arabidopsis. Plant Cell. 2016;28:2866–2883. doi: 10.1105/tpc.16.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Van den Ende W., Rolland F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol. Plant. 2014;7:570–572. doi: 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]
- Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., Gonzalez A. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and Co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58:1431–1441. doi: 10.1093/pcp/pcx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Povero G., Novi G., Solfanelli C., Alpi A., Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- Maier A., Hoecker U. COP1/SPA ubiquitin ligase complexes repress anthocyanin accumulation under low light and high light conditions. Plant Signal. Behav. 2015;10:e970440. doi: 10.4161/15592316.2014.970440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., Rubio V., Uhrig J.F., Hülskamp M., Hoecker U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013;74:638–651. doi: 10.1111/tpj.12153. [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Rizhsky L., Hegie A., Koussevitzky S., Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Nietzsche M., Schießl I., Börnke F. The complex becomes more complex: protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Front. Plant Sci. 2014;5:54. doi: 10.3389/fpls.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Messerli G., Trevisan M., Chen J., Smith A.M., Zeeman S.C. A previously unknown maltose transporter essential for starch degradation in leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Nukarinen E., Nägele T., Pedrotti L., Wurzinger B., Mair A., Landgraf R., Börnke F., Hanson J., Teige M., Baena-Gonzalez E., et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016;6:31697. doi: 10.1038/srep31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C., Primavesi L.F., Patel M.K., Martinez-Barajas E., Powers S.J., Sagar R., Fevereiro P.S., Davis B.G., Paul M.J. Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol. Biochem. 2013;63:89–98. doi: 10.1016/j.plaphy.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M., Sultana N., Paszkiewicz K., Florance H., Smirnoff N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2012;35:388–404. doi: 10.1111/j.1365-3040.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- Pedrotti L., Weiste C., Nägele T., Wolf E., Lorenzin F., Dietrich K., Mair A., Weckwerth W., Teige M., Baena-González E., et al. Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell. 2018;30:495–509. doi: 10.1105/tpc.17.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto B., Moraes T.A., Mengin V., Margalha L., Vicente R., Feil R., Höhne M., Sousa A.G.G., Lilue J., Stitt M., et al. Impact of the SnRK1 protein kinase on sucrose homeostasis and the transcriptome during the diel cycle. Plant Physiol. 2021;187:1357–1373. doi: 10.1093/plphys/kiab350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrussa E., Braidot E., Zancani M., Peresson C., Bertolini A., Patui S., Vianello A. Plant flavonoids--biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013;14:14950–14973. doi: 10.3390/ijms140714950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. Plastidial retrograde signalling--a true "plastid factor" or just metabolite signatures? Trends Plant Sci. 2010;15:427–435. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Plumb W., Townsend A.J., Rasool B., Alomrani S., Razak N., Karpinska B., Ruban A.V., Foyer C.H. Ascorbate-mediated regulation of growth, photoprotection, and photoinhibition in Arabidopsis thaliana. J. Exp. Bot. 2018;69:2823–2835. doi: 10.1093/jxb/ery170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Woo N.S., Förster B., Small I.D. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Ponnu J., Riedel T., Penner E., Schrader A., Hoecker U. Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc. Natl. Acad. Sci. USA. 2019;116:27133–27141. doi: 10.1073/pnas.1909181116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy S.J., Bussell J.D., Nunn C.P., Smith S.M. Leaves of the Arabidopsis maltose exporter1 mutant exhibit a metabolic profile with features of cold acclimation in the warm. PLoS One. 2013;8:e79412. doi: 10.1371/journal.pone.0079412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick W.P., Scheibe R., Neuhaus H.E. Induction of hexose-phosphate translocator activity in spinach chloroplasts. Plant Physiol. 1995;109:113–121. doi: 10.1104/pp.109.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragel P., Streb S., Feil R., Sahrawy M., Annunziata M.G., Lunn J.E., Zeeman S., Mérida Á. Loss of starch granule initiation has a deleterious effect on the growth of arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 2013;163:75–85. doi: 10.1104/pp.113.223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M., Dang T.V.T., Broeckx T., Hulsmans S., Crepin N., Sheen J., Rolland F. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell. 2019;31:1614–1632. doi: 10.1105/tpc.18.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A.S., Tohge T., Fernie A.R., Grimm B. The genomes uncoupled-dependent signalling pathway coordinates plastid biogenesis with the synthesis of anthocyanins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190403. doi: 10.1098/rstb.2019.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M.A., Li Y., Bajdzienko K., Fisahn J., Watanabe M., Hoefgen R., Schöttler M.A., Giavalisco P. RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol. 2018;177:565–593. doi: 10.1104/pp.17.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2003;100:6849–6854. doi: 10.1073/pnas.1132018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Heinrichs L., Scossa F., Fernie A.R., Oelze M.L., Dietz K.J., Rothbart M., Grimm B., Flügge U.I., Häusler R.E. The essential role of sugar metabolism in the acclimation response of Arabidopsis thaliana to high light intensities. J. Exp. Bot. 2014;65:1619–1636. doi: 10.1093/jxb/eru027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A., Häusler R.E., Kolukisaoglu U., Kunze R., van der Graaff E., Schwacke R., Catoni E., Desimone M., Flügge U.I. An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J. 2002;32:685–699. doi: 10.1046/j.1365-313x.2002.01460.x. [DOI] [PubMed] [Google Scholar]
- Schumann T., Paul S., Melzer M., Dörmann P., Jahns P. Plant growth under natural light conditions provides highly flexible short-term acclimation properties toward high light stress. Front. Plant Sci. 2017;8:681. doi: 10.3389/fpls.2017.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Fernie A.R., Geigenberger P., Leister D., Möhlmann T., Naranjo B., Neuhaus H.E. Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. 2022;27:577–587. doi: 10.1016/j.tplants.2021.12.004. [DOI] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009;60:3849–3860. doi: 10.1093/jxb/erp223. [DOI] [PubMed] [Google Scholar]
- Shin D.H., Choi M., Kim K., Bang G., Cho M., Choi S.B., Choi G., Park Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 2004;39:45–58. doi: 10.1111/j.1365-313X.2004.02105.x. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inzé D., Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola I.L., Camoirano A., Gonzalez D.H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in arabidopsis. Plant Physiol. 2016;170:74–85. doi: 10.1104/pp.15.01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M.O., Moore M., König K., Pecher P., Alsharafa K., Lee J., Dietz K.J. Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell. 2014;26:1151–1165. doi: 10.1105/tpc.113.121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Walters R.G., Ibrahim D.G., Horton P., Kruger N.J. A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiol. 2004;135:891–906. doi: 10.1104/pp.104.040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhao X., Zhao Y., Shanklin J., Zhao Q., Liu C.J. Arabidopsis SnRK1 negatively regulates phenylpropanoid metabolism via Kelch domain-containing F-box proteins. New Phytol. 2021;229:3345–3359. doi: 10.1111/nph.17121. [DOI] [PubMed] [Google Scholar]
- Wang C.K., Han P.L., Zhao Y.W., Ji X.L., Yu J.Q., You C.X., Hu D.G., Hao Y.J. Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Biophys. Res. Commun. 2020;533:717–722. doi: 10.1016/j.bbrc.2020.09.065. [DOI] [PubMed] [Google Scholar]
- Wang L., Li H., Zhao C., Li S., Kong L., Wu W., Kong W., Liu Y., Wei Y., Zhu J.K., et al. The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ. 2017;40:56–68. doi: 10.1111/pce.12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnasooriya S.N., Porter K.J., Montgomery B.L. Tissue- and isoform-specific phytochrome regulation of light-dependent anthocyanin accumulation in Arabidopsis thaliana. Plant Signal. Behav. 2011;6:624–631. doi: 10.4161/psb.6.5.15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise S.E., Liu T., Childs K.L., Preiser A.L., Katulski H.M., Perrin-Porzondek C., Sharkey T.D. Transcriptional regulation of the glucose-6-phosphate/phosphate translocator 2 is related to carbon exchange across the chloroplast envelope. Front. Plant Sci. 2019;10:827. doi: 10.3389/fpls.2019.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A., Delatte T.L., O'Hara L.E., Primavesi L.F., Jhurreea D., Paul M.J., Schluepmann H. Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 2012;158:1241–1251. doi: 10.1104/pp.111.191908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Tan H., Ma Z., Huang J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant. 2016;9:711–721. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Xiong Y., McCormack M., Li L., Hall Q., Xiang C., Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Mahmood K., Rothstein S.J. ROS induces anthocyanin production via late biosynthetic genes and anthocyanin deficiency confers the hypersensitivity to ROS-generating stresses in arabidopsis. Plant Cell Physiol. 2017;58:1364–1377. doi: 10.1093/pcp/pcx073. [DOI] [PubMed] [Google Scholar]
- Yadav U.P., Ivakov A., Feil R., Duan G.Y., Walther D., Giavalisco P., Piques M., Carillo P., Hubberten H.M., Stitt M., et al. The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 2014;65:1051–1068. doi: 10.1093/jxb/ert457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Ma H., Li Y., Zhang Y., Zhang J., Wu T., Song T., Yao Y., Tian J. Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J. 2021;106:1728–1745. doi: 10.1111/tpj.15267. [DOI] [PubMed] [Google Scholar]
- Zhai Z., Keereetaweep J., Liu H., Feil R., Lunn J.E., Shanklin J. Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell. 2018;30:2616–2627. doi: 10.1105/tpc.18.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhang X., Abrahan C., Colquhoun T.A., Liu C.J. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in arabidopsis. Plant Cell. 2017;29:1157–1174. doi: 10.1105/tpc.16.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L.F., Jhurreea D., Andralojc P.J., Mitchell R.A.C., Powers S.J., Schluepmann H., Delatte T., Wingler A., Paul M.J. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009;149:1860–1871. doi: 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Y., McFarlane H.E., Obata T., Richter A.S., Lohse M., Grimm B., Persson S., Fernie A.R., Giavalisco P. Inhibition of TOR represses nutrient consumption, which Improves greening after extended periods of etiolation. Plant Physiol. 2018;178:101–117. doi: 10.1104/pp.18.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO term analysis was conducted using https://go.princeton.edu/cgi-bin/GOTermFinder without plastome-encoded genes. Only DEGs with log2 FC < 1 and with an adjusted p < 0.05 were considered for GO term analysis. WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value.
GO term analysis was conducted using https://go.princeton.edu/cgi-bin/GOTermFinder. DEGs with log2 fold change <–1 for tpt/WT 9 h HL and log2 FC > 1 for tpt-2 (18 h/9 h HL) and with an adjusted p < 0.05 were used for GO term analysis. WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value.
WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value. Adapted from Huang et al. (2019).
WT, Col-0; tpt, tpt-2 mutant; 9, 9 h HL; 18, 18 h HL; padjust, adjusted p value
Data Availability Statement
Transcriptome data were deposited at GEO (https://www.ncbi.nlm.nih.gov/geo/info/update.html) under the record GSE196053.