Abstract
Protein homeostasis (proteostasis) is a dynamic balance of protein synthesis and degradation. Because of the endosymbiotic origin of chloroplasts and the massive transfer of their genetic information to the nucleus of the host cell, many protein complexes in the chloroplasts are constituted from subunits encoded by both genomes. Hence, the proper function of chloroplasts relies on the coordinated expression of chloroplast- and nucleus-encoded genes. The biogenesis and maintenance of chloroplast proteostasis are dependent on synthesis of chloroplast-encoded proteins, import of nucleus-encoded chloroplast proteins from the cytosol, and clearance of damaged or otherwise undesired “old” proteins. This review focuses on the regulation of chloroplast proteostasis, its interaction with proteostasis of the cytosol, and its retrograde control over nuclear gene expression. We also discuss significant issues and perspectives for future studies and potential applications for improving the photosynthetic performance and stress tolerance of crops.
Key words: chloroplast, proteostasis, interaction, retrograde signaling, stress
This review summarizes the current understanding and recent new insights on the biogenesis, maintenance, and remodeling of chloroplast proteostasis, highlighting outstanding questions and offering perspectives in the field.
Introduction
Chloroplasts provide oxygen and food for most life forms through photosynthesis. Moreover, the biosynthesis of many primary and secondary metabolites, such as fatty acids, amino acids, vitamins, pigments, and phytohormone precursors, takes place in chloroplasts (Neuhaus and Emes, 2000; Nelson and Ben-Shem, 2004). The chloroplast originated from a singular endosymbiotic event when a prokaryotic photosynthetic bacterium ancestor was engulfed by a eukaryotic cell approximately 1.5 billion years ago (Archibald, 2009; Keeling, 2013). During coevolution with the host cell, the genome of the endosymbionts (cyanobacteria) has been dramatically reduced through: (1) gene loss, whereby a large number of genes from the endosymbionts have been lost because of their redundancy with nucleus-encoded genes of the host cell; and (2) gene transfer, whereby massive gene transfer events have occurred that relocated essential genes to the nuclear genome for stable inheritance (Timmis et al., 2004; Bock and Timmis, 2008; Bock, 2017). Hence, a protein import machinery needed to be created to reroute the protein products of these genes back to the chloroplasts. Indeed, many protein complexes in chloroplasts are composed of chloroplast- and nucleus-encoded subunits, such as protein complexes from the photosynthetic electron transfer (PET) chain, ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), the chloroplast 70S ribosome, and plastid-encoded RNA polymerase (PEP, which contains many nucleus-encoded cofactors). Thus, the proper function of chloroplasts relies on the harmonious expression of genes from both chloroplast and nuclear genomes.
Coordinated gene expression, referred to as genome-coupled expression, guarantees the successful assembly of these mosaic complexes in correct stoichiometry (Jung and Chory, 2010). The host eukaryotic cells invoke anterograde and retrograde signaling to ensure harmonized gene expression between endosymbiotic organelles and the nucleus. Anterograde signals convey information about cell type and developmental stage to the chloroplasts, synchronizing their function with the whole cell. By contrast, retrograde signals are emitted by chloroplasts, feeding back the status of the chloroplasts to the nucleus and thus reprogramming the expression of nuclear genes for chloroplast function (Chan et al., 2016; Hernández-Verdeja and Strand, 2018; Wu and Bock, 2021).
Protein homeostasis (proteostasis) is the dynamic balance between the generation of correctly folded proteins and the elimination of misfolded/damaged proteins (Webster et al., 2020). Under normal conditions, the proteostasis network rapidly senses and amends disturbances in the proteome to restore basal homeostasis. During stress, cells reprogram the expression of a number of genes, including, e.g., induction of a set of molecular chaperones and repression of general protein translation, to bring proteostasis to a new state that is suitable for battling against challenges (Kim et al., 2013b; Kaushik and Cuervo, 2015). Because of its semiautonomous nature, the regulation of chloroplast proteostasis is more complicated. The biogenesis and maintenance of chloroplast proteostasis involve: (1) synthesis and folding of chloroplast-encoded proteins; (2) import of nucleus-encoded chloroplast proteins (chloroproteins); (3) assembly of protein complexes in their correct stoichiometry from subunits synthesized inside the chloroplasts and imported from the cytosol; and (4) elimination of damaged or at this moment “unwanted” proteins (Sun et al., 2020).
In recent years, chloroplasts have been suggested to be environmental sensors, participating in plant responses to various abiotic and biotic stresses (de Zabala et al., 2015; Kachroo et al., 2021; Schwenkert et al., 2022). An imbalance of chloroplast proteostasis will generate retrograde signals to regulate the expression of development- and stress-related nuclear genes. Further studies on chloroplast proteostasis and its interaction with the proteostasis of other subcellular compartments will provide essential clues toward understanding the role of chloroplasts in stress signaling and plant stress tolerance. This review discusses our current understanding of the biogenesis, maintenance, and remodeling of chloroplast proteostasis. We also highlight recent progress on the coregulation of proteostasis between chloroplasts and the cytosol and on the retrograde control of nuclear gene expression, providing perspectives for future studies toward a complete understanding of these pathways.
Translational control of chloroplast-encoded proteins
The genome of the cyanobacterium ancestor of the chloroplast has been dramatically trimmed by gene loss and gene transfer during evolution. The present-day chloroplast genome typically encodes 100–250 genes in eukaryotic algae and embyrophytes and approximately 130 genes in seed plants, among which fewer than 100 genes are protein-coding genes (Bock, 2007; Bock and Timmis, 2008). According to their functions, chloroplast genes can be sorted into three classes: (1) photosynthetic genes, including subunits of photosystem II (PSII), the cytochrome b6f complex (Cyt b6f), photosystem I (PSI), ATP synthase, NAD(P)H dehydrogenase-like (NDH) complex, and the large subunit of Rubisco (rbcL); (2) genetic (housekeeping) genes, for example, subunits of PEP, ribosomal proteins, rRNA and tRNA genes, and the matK gene, which encodes a putative splicing factor; and (3) other genes with diverse functions, including the accD gene of the fatty acid biosynthesis pathway, clpP of Clp protease, and hypothetical chloroplast open reading frame1 (ycf1) and ycf2, with putative functions in chloroplast protein import. Because of the availability of genetic transformation methods, the green alga Chlamydomonas reinhardtii (Chlamydomonas) and the seed plant Nicotiana tabacum (tobacco) have been developed as model species for investigating the functions of chloroplast genes (Bock, 2015).
Owing to the prokaryotic nature of chloroplasts, many chloroplast genes are organized in clusters with an operon-like structure. However, chloroplast operon-like gene clusters and their regulation are largely distinct from bacterial operons. For example, unlike bacterial operons, which normally encode genes of related function, chloroplast genes in the same operon have more diverse functions (Sugiura, 1995); for example, 50S ribosome protein l20 (rpl20) and 30S ribosome protein s12 (rps12) are in the same operon with clpP, the rpoA gene is located in an operon that contains mainly ribosomal proteins, and so on. The polycistronic transcripts of the chloroplast undergo complicated processing steps (splicing, editing, intercistronic processing) to give rise to mono-, oligo-, and polycistronic RNA species (Barkan, 2011). However, chloroplast transcripts are relatively stable compared with those of bacteria, with half-lives ranging from hours to even days, thus disabling fast transcriptional regulation (Mullet and Klein, 1987; Klaff and Gruissem, 1991). It was already observed in the 1980s that the dark–light transition enhances synthesis of RbcL, an effect that results from translational stimulation without obvious upregulation of rbcL mRNA levels (Berry et al., 1988). Moreover, the stability of chloroplast transcripts seems to be uncorrelated with the coverage of translating ribosomes (Link et al., 2012; Zoschke et al., 2012; Moreno et al., 2017). These studies indicated that, unlike the gene expression of its cyanobacterium ancestor, which is largely regulated at the transcriptional level, chloroplast gene expression is mainly controlled post-transcriptionally, especially at the translational level.
Compared with transcriptional regulation, translational regulation can respond more rapidly to internal or external stimuli. It enables fast modulation of protein abundance, leading to a quick remodeling of the chloroplast proteome, which seems to fit well with the role of chloroplasts as environmental sensors. More importantly, cotranslational translocation was shown to be a common means of protein targeting in chloroplasts, especially for thylakoid membrane-localized proteins (Zoschke and Barkan, 2015; Gawronski et al., 2018). This ensures the production of proteins directly at their functional sites, reduces the risk of misfolding (especially of hydrophobic transmembrane helices) and mistargeting, and saves energy from transport and chaperone binding. Early studies showed that the ribosome-nascent chain complex (RNC, representing translating ribosomes) was associated with the thylakoid membrane (Yamamoto et al., 1981). Chloroplast subfractionation coupled with polysome analysis revealed the cotranslational targeting of the core subunits of PSI (PsaA and PsaB), PSII (D1/PsbA, D2/PsbD, CP43/PsbC, and CP47/PsbB), and Cyt b6f (PetA) to the thylakoid membrane (Friemann and Hachtel, 1988; van Wijk et al., 1996). However, because coupled translation can take place for genes in unprocessed polycistronic chloroplast transcripts, one cotranslationally inserted polypeptide is sufficient to target the RNC to the thylakoid fraction during suborganelle fractionation, which argues for the cotranslational insertion of gene products from a part of polycistronic transcripts (e.g., PsaB and PsbC). This difficulty has been overcome by the successful application of the ribosome profiling approach combined with suborganelle fractionation, which revealed that 19 out of the 37 intrinsic transmembrane domain-containing thylakoid proteins in maize insert into the membrane cotranslationally (Zoschke and Barkan, 2015). Thus, cotranslational insertion of these membrane proteins effectively prevents their aggregation in the stroma. It represents an efficient way to maintain proteostasis at a low cost and is not only employed by chloroplasts but also commonly used for proteins targeted to the endoplasmic reticulum (ER) or mitochondria (Jan et al., 2014; Williams et al., 2014).
Chloroplast protein translation is predominantly controlled at the initiation step. Most identified factors (such as ATP4 [required for normal accumulation of the chloroplast ATP synthase], PENTATRICOPEPTIDE REPEAT10 [PPR10], HIGH CHLOROPHYLL FLUORESCENCE107 [HCF107], HCF173, and HCF244) that promote translation of specific chloroplast genes act at the initiation step (Felder et al., 2001; Schult et al., 2007; Prikryl et al., 2011; Link et al., 2012; Zoschke et al., 2012). Similar to bacteria, the preinitiation complex, which contains the 30S ribosome subunit, three initiation factors (IFs), and the initiator tRNA (charged with N-formylmethionine), scans along the mRNA until the start codon of AUG is located (GUG and UUG can also be used as start codons in chloroplasts), triggering the binding of the 50S subunit to complete the functional 70S ribosome (Kozak, 1999; Yamamoto et al., 2016). It has been shown that the differential expression of IF3 paralogs may participate in the translational control of chloroplast genes (Nesbit et al., 2015). Sequence elements in the translation initiation region of the transcripts are important for the regulation of translation initiation. The Shine–Dalgarno (SD) sequence upstream of the start codon plays a crucial role in translation initiation in Escherichia coli and other bacteria (Shine and Dalgarno, 1974; Osada et al., 1999); it interacts with the anti-SD (aSD) at the 3′ end of 16S rRNA by base pairing and initiates translation (McCarthy and Brimacombe, 1994; Scharff et al., 2011). Approximately two-thirds of chloroplast genes contain an SD sequence in their 5′ untranslated region. A recent study at the translatome level that involved ribosome profiling in tobacco transplastomic lines with disrupted aSD revealed the SD dependence of SD-containing chloroplast genes for translation initiation (Scharff et al., 2017). The translation of chloroplast genes is also regulated at the elongation step as well as the initiation step. For example, the translational elongation of psbC is reduced under heat stress in Chlamydomonas (Trösch et al., 2022). The expression of chloroplast elongation factors is regulated by environmental stimuli, such as light, temperature, and developmental cues (Akkaya and Breitenberger, 1992; Albrecht et al., 2006; Liu et al., 2010b), suggesting their considerable contribution to translational regulation.
Chloroplast translation is regulated by many environmental cues, especially light illumination and temperature shifts. Light-regulated translation actually coordinates chloroplast protein synthesis with photosystem biogenesis. Moreover, chlorophyll biosynthesis is light dependent, as the reduction of protochlorophyllide (Pchlide) to chlorophyllide catalyzed by Pchlide reductase is dependent on light (Gabruk and Mysliwa-Kurdziel, 2015; Vedalankar and Tripathy, 2019). This enables additional coordination of light illumination with the accumulation of chlorophyll-binding apoproteins in PSI and PSII, as folding of these subunits and assembly of the photosynthetic complexes rely on the incorporation of chlorophyll molecules (Nickelsen and Rengstl, 2013; Zabret et al., 2021), which protects the freshly synthesized apoproteins from immediate degradation (Eichacker et al., 1996). The translation of several chloroplast proteins was shown to be regulated by light illumination (Kim and Mullet, 1994; Muhlbauer and Eichacker, 1998). For example, the translation elongation rate of rbcL and psbA is stimulated by dark-to-light transition and is dependent on PET, as inhibition of PET by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) represses the light stimulation of their translation (Berry et al., 1988; Klein et al., 1988; Edhofer et al., 1998). The light perception of plants is independent of photosynthesis by photoreceptors located outside of chloroplasts. Light-stimulated translation was also observed in isolated chloroplasts, providing further evidence that it is dependent on photosynthesis but not photoreceptor-mediated light signaling (Muhlbauer and Eichacker, 1998).
Temperature is another major external factor that acts on translational regulation. The elongation factor EF-Tu is induced by heat stress, conferring heat tolerance to maize (Bhadula et al., 2001). Chloroplast protein synthesis undergoes fast and global reorganization during heat acclimation in the green alga Chlamydomonas and the seed plant tobacco (Trösch et al., 2022). Recently, translational regulation of chloroplasts was shown to play an important role in plant responses to chilling stress (Gao et al., 2022). Mutations in chloroplast translation initiation factor FUG1 (IF2) and ribosomal proteins reduce chilling tolerance in Arabidopsis and tobacco (Rogalski et al., 2008; Marino et al., 2019). Thus, through translational regulation, chloroplasts can rapidly adjust their proteome to achieve a new balance of proteostasis, quickly adapting to environmental and developmental changes.
Import of nucleus-encoded chloroplast proteins
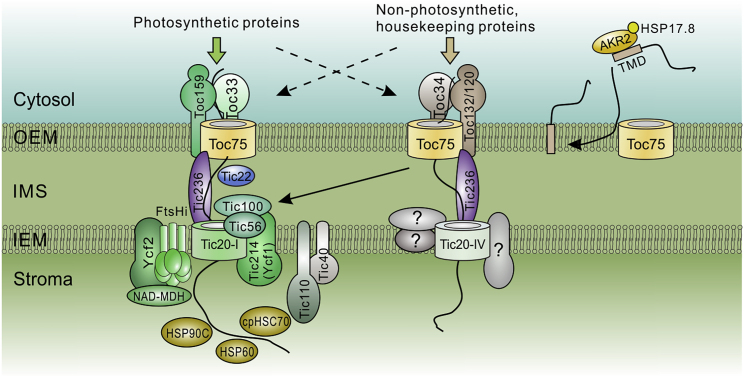
As discussed above, the chloroplast genome typically contains fewer than 100 protein-coding genes. However, the chloroplast proteome has been shown to contain more than 2000 proteins by mass spectrometry analysis (PPDB, http://ppdb.tc.cornell.edu/; van Wijk and Baginsky, 2011). A large number of plastid genes have been transferred to the host nucleus, and their protein products, synthesized in the cytosol, need to be rerouted and imported into the chloroplasts (Jarvis and López-Juez, 2013; Shi and Theg, 2013). Except for insertion of outer envelope membrane (OEM)-localized proteins directly into the OEM mediated by cytosolic sorting factors (AKR2 and HSP17.8; Bae et al., 2008; Lee et al., 2017) and the largely uncharacterized system that mediates chloroplast import of proteins that lack cleavable targeting signals (Nada and Soll, 2004; Miras et al., 2007), the translocons at the outer (TOC) and inner (TIC) envelope membranes of the chloroplasts mediate the import of most chloroproteins (Figure 1; Li and Chiu, 2010; Paila et al., 2015).
Figure 1.
The chloroplast protein import machineries.
Most nucleus-encoded chloroplast proteins (chloroproteins) are imported into chloroplasts through the OEM-localized TOC and IEM-localized TIC complexes. The core TOC complex comprises Toc159 and Toc34 transit peptide receptors and the channel-forming Toc75. The receptors are GTPases, providing energy at the early stage of import by hydrolyzing GTP. In Arabidopsis (and other plants), the receptors have multiple isoforms. The Toc159 family member Toc159 and the Toc34 family member Toc33-containing TOC complex is mainly in charge of importing highly abundant photosynthetic proteins. Meanwhile, Toc132/Toc120 together with Toc34 form an alternative TOC complex that is responsible for the import of non-photosynthetic/housekeeping proteins. However, the clients of Toc159/Toc33 and Toc132/120/Toc34 receptors may largely overlap and/or complement one another when the activity of one receptor is reduced. The nature of the TIC complex and its associated molecular motors remains controversial. In the model plant Arabidopsis, the newly identified 1-MD TIC complex contains the channel-forming Tic20 (Tic20-I), the chloroplast-encoded Tic214/Ycf1, Tic100, and Tic56. The molecular motor that hydrolyzes ATP to provide energy for the passage of preproteins through the TIC complex has been identified. It includes another chloroplast-encoded large open reading frame, Ycf2, the IEM-localized FtsHi, and NAD-malate dehydrogenase (NAD-MDH). The previously identified chaperones (cpHSC70, HSP90C, and HSP60) that associate with the Tic110 scaffold, may act downstream at the late stage of the cotranslocational folding process. An alternative TIC complex may exist, which comprises the Tic20 isoform, Tic20-IV, and other unidentified components without Tic214/Ycf1, as the plastids still contain non-photosynthetic chloroproteins when plastid translation is fully blocked, which abolishes chloroplast-encoded Tic214/Ycf1 and Ycf2. It should be noticed that Tic214/Ycf1 is absent in some monocots such as Poaceae. The cytosolic factor AKR2 and the small HSP, HSP17.8, facilitate the targeting of OEM-localized proteins. Toc75 may participate in this process.
Proteins imported by the TOC–TIC machinery contain an N-terminal targeting sequence, termed the chloroplast transit peptide (cTP), which is generally necessary and sufficient for chloroplast targeting (Lee et al., 2019). However, unlike signal peptides for other subcellular compartments such as the nucleus, ER, or peroxisome, which comprise a relatively conserved sequence motif, the length of the cTP can vary from 13 to more than 100 amino acids. Some amino acids are overrepresented in the cTP but do not seem to be conserved among species (Lee et al., 2006, 2018; Zybailov et al., 2008; Holbrook et al., 2016). In Arabidopsis, the sequence motif in the cTP has been shown to contain the information necessary for determining the tissue-specific and developmental-stage-specific import of different chloroproteins (Teng et al., 2012; Chu et al., 2020), suggesting that the cTP could possess detailed plastid-type-specific targeting information. The sequence motif of the cTP has also been shown to direct protein targeting to different types of chloroplasts in the same cell in Bienertia sinuspersici, a plant species that performs C4 photosynthesis within individual chlorenchyma cells (Wimmer et al., 2017).
Recognition of the cTP from chloroprotein precursors is mediated by two families of OEM-integrated preprotein receptors, Toc159 and Toc34 (Wallas et al., 2003; Smith et al., 2004; Andres et al., 2010). Precursors can bind directly to the Toc159 and Toc34 receptors, and this is the major pathway of chloroprotein targeting to the TOC complex (Paila et al., 2015). Cytosolic chaperones have been shown to participate in sorting of chloroproteins to the TOC complex. HSP90, HSP90/HSP70 organizing protein (HOP), and FKBP73 interact with the cTP of some chloroproteins and stimulate their import in vitro (Fellerer et al., 2011). The HSP90–preprotein complex may dock at the OEM surface through Toc64, which was hypothesized to transfer the precursors to TOC receptors (Qbadou et al., 2007). Besides the HSP90–Toc64 pathway, HSP70 together with 14-3-3 protein, designated as the cytosolic guidance complex, represents another cytosolic sorting route of the chaperone-assisted pathway for precursor targeting (May and Soll, 2000; Lee et al., 2013). However, knockout of Toc64 in Arabidopsis and the moss Physcomitrium patens, or mutations to eliminate the 14-3-3 phosphorylation site in cTP, did not result in visible import defects (Nakrieko et al., 2004; Aronsson et al., 2007). The Arabidopsis toc64 mutant displays only a very mild import phenotype under specific conditions, such as osmotic or cold stress (Sommer et al., 2013). Thus, the function of these cytosolic chaperones in precursor targeting requires further clarification. They may act as complementary routes that rescue escaped preproteins to prevent their mistargeting, especially under some stress conditions that reduce the accuracy of precursor targeting.
In Arabidopsis, the Toc159 and Toc34 receptor families both contain multiple members. The Toc159 family contains four homologs (Toc159, Toc132, Toc120, and Toc90), and the Toc34 family contains two (Toc33 and Toc34). The null mutation of Toc159 in the ppi2 mutant results in an albino phenotype with a dramatic reduction in photosynthetic proteins. However, the ppi2 mutant is viable on sucrose-containing medium, indicating that Toc159 is preferentially in charge of importing photosynthetic proteins (Bauer et al., 2000; Smith et al., 2004; Agne et al., 2009). Although single mutation of Toc132 or Toc120 does not produce visible phenotypes compared with the wild type, the double mutant is paler or even lethal. Importantly, the non-green plastids of roots are specifically affected in the toc132 toc120 double mutant but not in the toc159 mutant (Kubis et al., 2004), suggesting that Toc132 and Toc120 are major receptors for non-photosynthetic and housekeeping proteins. Toc159 was shown to be in the same complex with Toc33, excluding the Toc132/120 receptors, which coexist with Toc34 (Ivanova et al., 2004). The Toc33 mutant ppi1 displays a chlorotic phenotype with reduced accumulation of photosynthetic proteins (Jarvis et al., 1998; Kubis et al., 2003), providing further evidence for its role in the import of photosynthetic proteins (Figure 1).
The different substrate preferences of the TOC receptors play a critical role in the biogenesis and regulation of chloroplast proteostasis. Toc159 is highly expressed in green tissues, where the bulk of photosynthetic chloroprotein import occurs. By contrast, Toc132/120 begin to accumulate very early during germination, and their expression appears to be comparable in green or etiolated aerial tissues and in roots, consistent with their function in the import of non-photosynthetic/housekeeping chloroproteins (Ivanova et al., 2004; Inoue et al., 2010). The separation of less abundant housekeeping proteins from highly abundant photosynthetic proteins by distinct import machinery can prevent the out-competition of import machinery by photosynthetic proteins. In fact, Toc159 and Toc132/120 do represent distinct import pathways, as the overexpression of Toc132 cannot rescue the albino phenotype of the ppi2 mutant, and vice versa (Ivanova et al., 2004; Kubis et al., 2004). It is worth noting that large-scale proteomic analysis with Toc159-deficient lines and the null allele ppi2 demonstrated that, in addition to photosynthetic proteins, chloroproteins from various metabolic pathways are also affected by Toc159 depletion (Bischof et al., 2011), indicating a broad substrate import activity of Toc159 (Figure 1). However, the less accumulated chloroproteins revealed by the proteomic approach in these Toc159-deficient lines should be interpreted with caution as substrates of Toc159, as they could result from the strong secondary effects of the albino mutant phenotype. Disruption of Toc90 does not result in visible phenotypes in Arabidopsis (Kubis et al., 2004); however, its overexpression can partially complement the albino phenotype of ppi2, indicating that Toc90 may play a minor role in protein import and is redundant in Toc159 (Infanger et al., 2011).
In addition to the most abundant photosynthetic plastids (chloroplasts), the plastid family of organelles also includes the leucoplast, etioplast, chromoplast, amyloplast, elaioplast, and gerontoplast, among others. All of these different plastid types have a unique proteome, and thus a new proteostasis needs to be established when plastids change form. During chloroplast biogenesis (from proplastids or etioplasts to chloroplasts upon illumination), large amounts of photosynthetic chloroproteins must be imported into the plastids. Proteostasis needs to be remodeled when plastid form changes, for example, from chloroplast to chromoplast during fruit ripening or to gerontoplast during senescence. Meanwhile, the plastids need to quickly adjust their proteome in response to environmental stimuli. How plastids regulate the proteins that need to be imported to reach a new state of proteostasis during these processes remains unclear. However, the regulation of TOC complex abundance, especially the abundance and/or type of import receptors, seems to play a crucial role (Nakai, 2015; Richardson and Schnell, 2020). The phytohormone gibberellic acid (GA) controls the abundance of Toc159 during germination, leading to accumulation of photosynthetic chloroproteins (Shanmugabalaji et al., 2018). Under low GA, the DELLA protein RGA-LIKE2 (RGL2), a negative regulator of GA signaling, targets Toc159 (possibly prior to the OEM integration of Toc159) for degradation via the ubiquitin proteasome system (UPS). The increasing GA concentration during germination leads to degradation of RGL2 and consequently to accumulation of Toc159, thus promoting chloroplast biogenesis (Figure 2A). Interestingly, the control of Toc159 abundance by GA and DELLA proteins is independent of the identified chloroplast-associated protein degradation (CHLORAD) pathway, as mutation of the E3 ligase gene SP1 (SUPPRESSOR OF ppi1 LOCUS1) in the CHLORAD pathway has no effect on the reduction of Toc159 when GA synthesis is inhibited (Ling et al., 2012, 2019; Shanmugabalaji et al., 2018). In addition, the stability of TOC components has also been shown to be regulated by the small ubiquitin-like modifier (SUMO) system, thus participating in the establishment of plastid proteostasis during chloroplast biogenesis (Accossato et al., 2020; Watson et al., 2021).
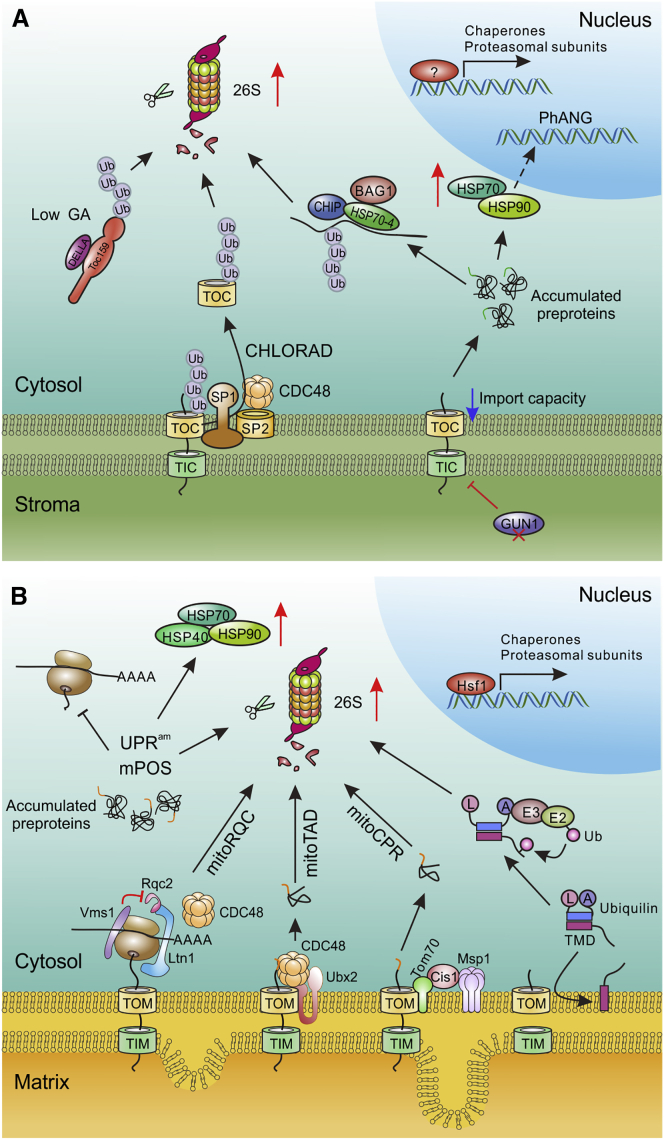
Figure 2.
Precursor accumulation response of chloroplasts and mitochondria.
(A) Chloroplast precursor accumulation response (PARcp). Impaired protein import into chloroplasts causes overaccumulation of chloroprotein precursors in the cytosol. In the Arabidopsis Toc159 mutant ppi2, the cytosolic HSP70 chaperone cyHSP70-4 (HSP70-4) and the E3 ubiquitin ligase CHIP were shown to participate in the degradation of overaccumulated chloroprotein precursors. The nucleotide exchange factor of HSP70, BAG1, recruits the proteasome to the substrates for degradation. Overaccumulation of chloroprotein precursors upregulates the cytosolic chaperones cyHSP70 and cyHSP90 and components of the 26S proteasome in gun1 mutants (which reduces plastid protein import capacity when chloroplast biogenesis is blocked, e.g., upon lincomycin or norflurazon treatment) to maintain cytosolic proteostasis. The chloroplast-associated protein degradation (CHLORAD) pathway is involved in the UPS-dependent turnover of components from the TOC complex during development and in response to stress. In the CHLORAD pathway, TOC subunits are ubiquitinated by the E3 ligase SP1 and extracted from the OEM by AAA+ ATPase CDC48 and Omp85 protein SP2 for proteasomal degradation. The DELLA protein RGL2 also participates in UPS-dependent TOC159 degradation at the early stage of germination when the GA concentration is low. However, whether the CHLORAD and DELLA pathways are involved in PARcp for degradation of clogged precursors in the import channel and/or overaccumulated precursors in the cytosol remains to be determined.
(B) Mitochondrial precursor accumulation response (PARmt). Multiple pathways have been identified in yeast, which surveils mitochondrial protein import and eliminates overaccumulated precursors in the cytosol. mitoRQC (ribosome quality control pathway for mitochondrial polypeptides): Listerin1 (Ltn1) gains access to precursors that stall in the ribosome, followed by ubiquitination, CDC48-mediated extraction, and proteasomal degradation. Vms1 (a 60S ribosome-binding protein) recognizes those stalled precursors that fail to be ubiquitinated by Ltn1 in close proximity to the translocase of outer membrane (TOM) complex at the OEM and prevents them from being CAT-tailed (C-terminal alanyl/threonyl sequences that mediate the aggregation of stalled polypeptides after import into mitochondria) by Rqc2. These precursors are imported into the mitochondrial matrix and either folded or degraded by the mitochondrial quality control system. mitoTAD (mitochondrial protein translocation-associated degradation): the mitoTAD pathway continuously monitors import under non-stress conditions to prevent clogging of the TOM channel by precursors. UBX domain protein Ubx2 (a component of ERAD) binds to the TOM complex to recruit CDC48 to extract the arrested precursors for proteasomal degradation. mitoCPR (mitochondrial compromised protein import response): inhibition of mitochondrial protein import induces the expression of Cis1, which associates with the TOM complex by interacting with Tom70. Cis1 recruits the AAA+ ATPase Msp1, which mediates the extraction and proteasomal degradation of arrested precursor proteins. Overaccumulation of mitochondrial precursor proteins (mPOS) in the cytosol causes an unfolded protein response activated by mistargeting of proteins (UPRam). mPOS and UPRam upregulate the expression of chaperones and proteasomal components through the action of heat-shock factor 1 (Hsf1) and inhibit translation to rebalance proteostasis. In mammals, ubiquilins bind to the transmembrane domain (TMD) of OEM-localized proteins in the cytosol to prevent their aggregation. The C-terminal ubiquitin-associating (UBA) domain (A) interacts with its N-terminal ubiquitin-like (UBL) domain (L) to stabilize the client–ubiquilin complex. Over time, the UBA domain recruits an unknown E3 ligase to ubiquitinate the substrates, while the free UBL domain directs the proteasomal degradation of the client proteins.
Although the regulation of plastid proteostasis at the import level during plastid form transition has been less investigated, the CHLORAD pathway-regulated turnover of TOC components (such as Toc159, Toc75, and Toc33) seems to participate in the transition of chloroplasts to chromoplasts during tomato fruit ripening (Thomson et al., 2020; Ling et al., 2021). However, the role of different TOC complexes in the proteome shift during the transition from chloroplasts to chromoplasts in flowers and fruits and, to an extent, from chloroplasts or proplastids to other plastid forms such as amyloplasts in cereals and elaioplasts in oilseeds, remains to be explored (Sadali et al., 2019).
The CHLORAD pathway also plays a pivotal role in the biogenesis and remodeling of chloroplast proteostasis under abiotic stress conditions (Ling and Jarvis, 2015, 2016). Moreover, a recent study indicated that the SUMO modification of various chloroproteins is essential for their precise chloroplast localization under heat stress. At least the Toc33-containing TOC complex is involved in the import of SUMOylated preproteins (Zheng et al., 2022), further indicating that import regulation makes a large contribution to the shifting and rebalancing of chloroplast proteostasis in response to environmental stimuli.
Preproteins are translocated across the OEM through the TOC channel formed by Toc75, a β-barrel channel-forming protein, and reach the intermembrane space (IMS). The polypeptide transport-associated (POTRA) domain of Toc75, the IMS-localized small TIC protein Tic22, and the DUF490 domain of Tic236 may form a chaperoning conduit that facilitates passage of the preproteins through the IMS (by preventing their misfolding) and their arrival at the TIC complex (Figure 1; Kasmati et al., 2013; Paila et al., 2016; O'Neil et al., 2017; Chen et al., 2018b). However, the nature of the TIC complex and its associated molecular motors, which provide energy for preprotein translocation through ATP hydrolysis, remains controversial (Kikuchi et al., 2009, 2013; de Vries et al., 2015; Nakai, 2015, 2020; Bolter and Soll, 2017; Shingo et al., 2018; Li et al., 2020). The early-identified typical TIC components Tic110 and Tic40 (Chou et al., 2003; Inaba et al., 2005; Kovacheva et al., 2005) were excluded from the new 1-MD (megadalton) TIC complex, which is composed of the channel-forming subunit Tic20 and the newly identified Tic100, Tic56, and chloroplast-encoded subunit Tic214/Ycf1 (Kikuchi et al., 2013). The new import motor of the Ycf2–FtsHi complex, other than ClpC1, cpHSC70, and HSP90C, was later suggested by the same laboratory (Figure 1; Shingo et al., 2018). However, it should be noticed that some components of the 1-MD complex are not ubiquitous in higher plants; for example, Tic214/Ycf1 is absent in Poaceae (de Vries et al., 2015), giving rise to questions about the extent to which this new complex is conserved (de Vries et al., 2015). The connection of Tic110/Tic40 with the 1-MD TIC complex and the function of the Ycf2–FtsHi motor require further clarification in future studies.
Interestingly, the TIC complex also seems to vary among different tissues and at different developmental stages, thus participating in regulation of the substrates to be imported. In Arabidopsis, Tic20-I is highly expressed in green tissues and is thought to be responsible for importing photosynthetic and non-photosynthetic chloroproteins in these tissues, whereas Tic20-IV is the major form in non-green tissues such as roots (Hirabayashi et al., 2011; Nakai, 2015). In addition, plastids can still import chloroproteins in seedlings established in the presence of spectinomycin (Bolter and Soll, 2017), a specific inhibitor of chloroplast protein translation that blocks the synthesis of plastid-encoded Tic214/Ycf1. This result indicates that an alternative TIC complex without Tic214 may exist, which is particularly necessary for the import of chloroproteins under specific conditions (Figure 1; Nakai, 2015).
The turnover of TIC components has been shown to be regulated under heat-stress conditions by IEM-localized FtsH11 protease, which plays a putative role in the regulation of chloroplast proteostasis in response to heat stress (Adam et al., 2019). GUN1, an integrator of multiple retrograde signaling pathways, was shown to interact with cpHSC70-1 to participate in the regulation of protein import and the maintenance of chloroplast proteostasis under lincomycin or norflurazon (an inhibitor of carotenoid synthesis, which thus blocks chloroplast biogenesis) treatment conditions (Wu et al., 2019a, 2019b). Recently, the giant TIC component, Tic236, together with the OEM-localized CRL protein, was shown to participate in the regulation of chloroplast division by facilitating the import of plastid division machinery components (FTSZ1, FTSZ2; Chen et al., 2018b; Fang et al., 2022). These studies indicate that specific TIC components may have specialized roles in the maintenance of plastid proteostasis and may function at given developmental stages or under specific environmental conditions.
Chloroplast protein import stress and its interaction with the cytosolic protein quality control system
Various stress conditions, especially light and temperature stresses that inhibit chloroplast function, will reduce the chloroplast protein import capacity and result in an aberrant accumulation of chloroprotein precursors in the cytosol (Ling and Jarvis, 2015; Eisa et al., 2019). The unfolded precursors are highly proteotoxic to the cell because their exposed hydrophobic regions will disrupt cytosolic proteostasis, resulting in arrested cell growth and plant development. The cytosolic HSP70 chaperone (cyHSP70-4/HSC70-4), Bcl-associated athanogene 1 (BAG1), and the E3 ubiquitin ligase CHIP have been shown to participate in the degradation of chloroprotein precursors in the ppi2 mutant (Lee et al., 2009, 2016), as well as remodeling the stoichiometry of the Clp complex in ClpP4 knockdown and overexpression lines by CHIP-dependent ubiquitination of ClpP3 and ClpP5 (Wei et al., 2015). The transcript level of GLK1 (a key regulator of photosynthetic gene expression and chloroplast biogenesis; Waters et al., 2009) remains high in the gun1 mutant compared with the wild type under lincomycin or norflurazon treatment (because of the GUN phenotype). However, the GLK1 protein does not accumulate in either the wild type or the gun1 mutant under these conditions. The uncoupling of transcript level from protein accumulation of GLK1 is also involved in UPS-dependent GLK1 protein degradation (Tokumaru et al., 2017). These observations indicate the crucial role of cytosolic UPS in the chloroplast related protein degradation and precursor accumulation response (PARcp; Hristou et al., 2020; Thomson et al., 2020). A recent study on GUN1 indicated that cyHSP90.1, cyHSP70-2/cyHSP70-4, and the UPS are involved in PARcp induced by the reduced import capacity of the gun1 mutant (Wu et al., 2019b). However, how the clogged precursors in the TOC machinery are extracted from the import channel, the subcellular sites in the cell that deal with the aberrantly accumulated precursors, the response of nuclear gene expression to the PARcp, and whether there are other components of the cytosolic protein quality control (PQC) system that are involved in this process, are poorly understood (Figure 2A and Table 1).
Table 1.
Identified pathways of PARcp and PARmt.
| Pathway | Key factors | Brief description | References |
| PARcp | |||
| cyHSP70-CHIP | cyHSP70-4, CHIP, BAG1 | cyHSP70-4 interacts with chloroprotein precursors, E3 ubiquitin ligase CHIP ubiquitinates the substrates, BAG1 recruits the 26S proteasome to the substrates | Lee et al., 2009, 2016 |
| GUN1-mediated retrograde signaling | GUN1, cyHSP90, cyHSP70 | Depletion of GUN1 reduces the import capacity of plastids under stress conditions and causes overaccumulation of chloroprotein precursors, which upregulates cyHSP90, cyHSP70, and 26S proteasomal subunits to rebalance cytosolic proteostasis | Wu et al., 2019b |
| CHLORAD | SP1, SP2, CDC48 | Turnover of TOC components by UPS. E3 ubiquitin ligase SP1 ubiquitinates the substrates, AAA+ ATPase CDC48 together with the Omp85 protein SP2 extracts substrates from the OEM for degradation by UPS. Unclear whether the CHLORAD pathway is involved in precursor degradation | Ling et al., 2012, 2019 |
| GA-DELLA | RGL2 | RGL2 interacts with Toc159 and promotes its degradation through the UPS, thus repressing chloroplast biogenesis under low GA. Unclear whether this pathway is involved in precursor degradation | Shanmugabalaji et al., 2018 |
| PARmt | |||
| mitoRQC | Rqc2, Ltn1, Vms1, CDC48 | A ribosome quality control pathway to prevent the import of ribosome-stalled mitochondrial polypeptides. Rqc2 adds CAT-tails to the C terminus of ribosome-stalled polypeptides, which are ubiquitinated by E3 ligase Ltn1 and degraded by the UPS. Vms1 antagonizes the Rqc2-dependent CAT-tailing and allows these proteins to be imported into mitochondria for folding or degradation | Izawa et al., 2017 |
| mitoTAD | Ubx2, CDC48 | A quality control pathway under non-stress conditions that continuously monitors mitochondrial import. Ubx2 binds to the TOM complex and recruits CDC48, which extracts precursors that are arrested in the TOM channel for UPS-dependent degradation | Martensson et al., 2019 |
| mitoCPR | Pdr3, Cis1, Msp1, Tom70 | Under import-stress conditions, transcription factor Pdr3 induces the upregulation of Cis1, which binds to the TOM complex via Tom70 and recruits Msp1 for extraction of clogged precursors from the import channel. The extracted precursors are degraded by the UPS | Weidberg and Amon, 2018 |
| UPRam | – | Overaccumulation of mitochondrial protein precursors inhibits protein synthesis and activates proteasome activity | Wrobel et al., 2015 |
| mPOS | Gis2, Nog2 | Overaccumulation of mitochondrial protein precursors modulates ribosomal biogenesis, messenger RNA decapping, transcript-specific translation, protein chaperoning, and turnover. Gis2 stimulates Cap-independent translation. Nog2 inhibits nuclear export of the 60S ribosomal subunit | Wang and Chen, 2015 |
| Ubiquilin pathway | Ubiquilins (UBQLN1, 2, and 4) | Ubiquilins bind to the transmembrane domain of OEM-localized proteins for their membrane targeting. Over time, the C-terminal UBA domain of the ubiquilin disassociates with its N-terminal UBL domain. The UBA domain recruits an unknown E3 ligase for the ubiquitination of the substrate. The freed UBL domain engages the proteasome for substrate degradation | Itakura et al., 2016 |
However, cellular responses to mitochondrial precursor accumulation (PARmt) have been intensively studied in yeast (Boos et al., 2020) and may provide informative clues for future studies of the PARcp in plants. These global adaptive responses include: (1) upregulation of the cytosolic PQC system (chaperones and UPS) to clear proteotoxic precursors (Wrobel et al., 2015; Boos et al., 2019); (2) inhibition of cellular translation to reduce import loads of the mitochondria (Wang and Chen, 2015; Boos et al., 2019); and (3) extraction of the clogged precursors from the import channel to release import capacity, thus enabling mitochondrial function (Izawa et al., 2017; Weidberg and Amon, 2018; Martensson et al., 2019). Interestingly, in addition to the cytosolic clearance pathway, mitochondrial precursors were also shown to be relocated to the nucleus and ER for degradation (Laborenz et al., 2021; Shakya et al., 2021). These studies demonstrate that the host cell makes use of a sophisticated mechanism to prevent the accumulation of mitochondrial precursors and safeguard cytosolic proteostasis (Figure 2B and Table 1). Whether plant cells have a conserved network for response to aberrantly accumulated precursors of evolutionarily related chloroplasts will need to be resolved in future studies.
The function of the CHLORAD pathway in the turnover of TOC components (such as Toc159, Toc75, and Toc33) is well documented. The OEM-localized SP2 and the cytosolic AAA+ (ATPase associated with diverse cellular activities) chaperone CDC48 of the CHLORAD pathway cooperate to retrotranslocate TOC components from the OEM, thus enabling their ubiquitin-dependent proteasomal degradation (Ling et al., 2019). However, whether the CHLORAD pathway also participates in the extraction of clogged precursors from the TOC channel is still unknown. The CDC48-mediated retrotranslocation of proteins from the ER and their subsequent UPS-dependent degradation have also been identified and extensively studied in the ER-associated degradation (ERAD) pathway (Meusser et al., 2005; Liu and Howell, 2016), suggesting a conserved mechanism of protein retrotranslocation from the chloroplasts, mitochondria (where the extraction of clogged preproteins is also dependent on CDC48; Figure 2B), and ER. Surprisingly, the intra-chloroplast proteins RbcL and AtpB have recently been demonstrated to be retrotranslocated from the chloroplast by CDC48, then ubiquitinated and degraded through the cytosolic UPS (Li et al., 2022a). However, much less is known about this mechanism; in particular, how intra-chloroplast proteins pass the IEM to reach the retrotranslocation channel on the OEM is not clear.
It was observed in yeast that mitochondrial protein import stress triggers mitophagy to eliminate mitochondria that undergo dysfunctional protein import (Killackey et al., 2022). The U-box 4 (PUB4) E3 ubiquitin ligase was demonstrated to mediate the clearance of 1O2-damaged chloroplasts by chlorophagy (Woodson et al., 2015). However, whether chloroplast protein import stress results in chlorophagy-mediated chloroplast degradation is still unknown. Interestingly, it has recently been observed that mild elevation of chloroprotein precursors in the cytosol caused by attenuation of cytosolic UPS increases the accumulation of functional photosynthetic complexes, resulting in enhanced photosynthetic performance (Grimmer et al., 2020). This finding indicates that precursors may have signaling functions: cells may monitor the precursor level and use it as a signal for proper coordination between endosymbionts and host cells.
Elimination of chloroplast proteins
Protein abundance is determined by the rates of protein synthesis and degradation. The maintenance of functional organelles requires the effective sweeping of damaged or otherwise “unwanted” proteins during developmental or environmental transitions. The proteolytic machineries play vital roles in plastid proteome biogenesis, maintenance, and remodeling. More than 20 proteases of prokaryotic origin have been identified in chloroplasts through various bioinformatic, genetic, biochemical, and proteomic approaches (Kato and Sakamoto, 2010; van Wijk, 2015). These proteolytic machineries include the ATP-dependent Clp, FtsH, and Lon proteases, ATP-independent Deg endopeptidase, and peptidases that function in transit peptide cleavage and further degradation of peptide fragments, including cleaved transit peptides and degradation products from other proteases (Figure 3 and Table 2; Nishimura et al., 2016, 2017).
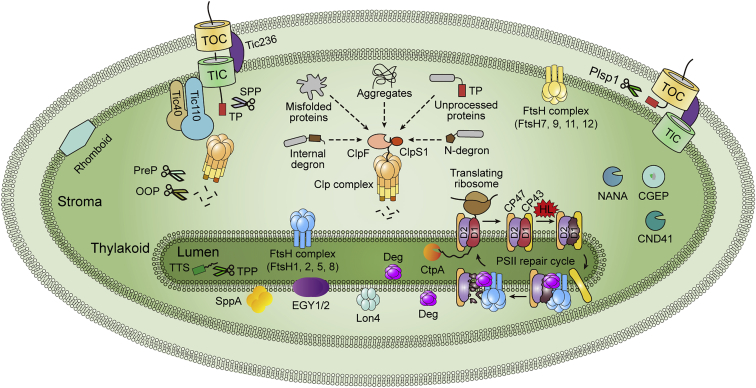
Figure 3.
Proteolytic machineries in chloroplasts of land plants.
The Clp protease complex is the major protease in the stroma. The adaptor subunit ClpS1 interacts with chloroplast-specific subunit ClpF, participating in substrate recognition. The substrates of Clp protease include: (1) proteins with internal or N-degrons; (2) misfolded and/or aggregated proteins; and (3) unprocessed proteins. Moreover, a portion of Clp proteases closely associate with the TIC complex through the interaction of ClpC1 with Tic110, where they function as a checkpoint for newly imported proteins. Thylakoid membrane-localized FtsH metalloprotease (FtsH1, 2, 5, 8) plays a central role in D1 turnover during the PSII repair cycle and is crucial for thylakoid biogenesis. IEM-anchored FtsH (FtsH7, 9, 11, 12) may participate in the turnover of IEM proteins and protein import. The Deg endopeptidases localize either at the stromal side (Deg2, 7) or the lumenal side (Deg1, 5, 8) of thylakoids and participate in D1 degradation by cleaving the inter-loops that connect the five transmembrane helices of D1. The C terminus of newly synthesized D1 proteins must be processed by the C-terminal processing enzyme (CtpA) in the lumen. Other chloroplast proteases include the thylakoid-localized Lon4, EGY1/2 (ethylene-dependent gravitropism-deficient and yellow-green 1/2), SppA, the stroma-localized NANA, CGEP (chloroplast glutamyl peptidase), CND41 (41-kDa chloroplast nucleoid DNA-binding protein), and the IEM-localized Rhomboid (Table 2). The short peptide fragments from protease degradation products and cleaved TPs are further processed by peptidases and recycled. These peptidases include TPP (Plsp1) in the thylakoid lumen, SPP, PreP, and OPP in the stroma, and Plsp1 in the IMS. TP, transit peptide; TTS, thylakoid targeting sequence; TPP, thylakoidal processing peptidase (Plsp1); SPP, stromal processing peptidase; PreP, presequence peptidase; OOP, organellar oligopeptidase; HL, high light.
Table 2.
Proteases and peptidases in chloroplasts.
| Name | AGI | Features | Localization | Mutant phenotype | References |
|---|---|---|---|---|---|
| ATP-dependent proteases | |||||
| Clp | |||||
| ClpP1 | ATCG00670 | Clp complex core (R ring), chloroplast-encoded | Stroma | Essential, homoplasmic knockout not available | Shikanai et al., 2001; Kuroda and Maliga, 2003 |
| ClpP3 | AT1G66670 | Clp complex core (P ring), essential for early development | Stroma | Arrest at the cotyledon stage in autotrophic conditions | Kim et al., 2013a |
| ClpP4 | AT5G45390 | Clp complex core (P ring), essential for embryogenesis | Stroma | Embryo lethal | Zheng et al., 2006; Kim et al., 2013a |
| ClpP5 | AT1G02560 | Clp complex core (P ring), essential for embryogenesis | Stroma | Embryo lethal | Kim et al., 2009 |
| ClpP6 | AT1G11750 | Clp complex core (P ring), chloroplast biogenesis | Stroma | Knockdown lines show pale-green leaves at early developmental stages | Sjögren et al., 2006 |
| ClpR1 | AT1G49970 | Clp complex core (R ring), essential for early development | Stroma | Knockdown and point mutation lines show pale-green and retarded growth phenotype | Koussevitzky et al., 2007a; Kim et al., 2009 |
| ClpR2 | AT1G12410 | Clp complex core (R ring), essential for early development | Stroma | Arrest at the cotyledon stage in autotrophic conditions and death after several weeks | Kim et al., 2009 |
| ClpR3 | AT1G09130 | Clp complex core (R ring) | Stroma | Substitution of ClpR1 | Kim et al., 2009 |
| ClpR4 | AT4G17040 | Clp complex core (R ring), essential for early development | Stroma | Arrest at the cotyledon stage in autotrophic conditions and death after several weeks | Kim et al., 2009 |
| ClpT1 | AT4G25370 | Assembly and stabilization of Clp complex | – | No visible phenotypes (single mutant), delayed development and virescent leaves (double mutant) | Sjögren and Clarke, 2011; Kim et al., 2015 |
| ClpT2 | AT4G12060 | ||||
| ClpC1 | AT5G50920 | Molecular chaperone, leaf development | Stroma | Virescent leaves and retarded growth | Sjögren et al., 2004 |
| ClpC2 | AT3G48870 | Molecular chaperone | Stroma | Indistinguishable from the wild type | Park and Rodermel, 2004; Nishimura et al., 2013 |
| ClpD | AT5G51070 | Molecular chaperone | Stroma | Indistinguishable from the wild type, induced by abiotic stresses | Nakashima et al., 1997; Zheng et al., 2002; Nishimura et al., 2013 |
| ClpS | AT1G68660 | Adaptor subunit, substrate recognition and delivery | Stroma | Indistinguishable from the wild type; reduced accumulation of chlorophyll a and b under short-day conditions | Nishimura et al., 2013 |
| ClpF | AT2G03390 | Binding to ClpS, substrate recognition and delivery | Stroma | Indistinguishable from the wild type | Nishimura et al., 2015 |
| FtsH | |||||
| FtsH1 | AT1G50250 | Forming the thylakoid FtsH hexamer (type A), functioning in PSII repair | Thylakoid membrane | Indistinguishable from the wild type | Sakamoto et al., 2003 |
| FtsH2 | AT2G30950 | VAR2, forming the thylakoid FtsH hexamer (type B), functioning in PSII repair and PSI assembly | Thylakoid membrane | Variegated leaves and sensitivity to photoinhibition | Sakamoto et al., 2003; Kato et al., 2012; Järvi et al., 2016 |
| FtsH5 | AT5G42270 | VAR1, forming the thylakoid FtsH hexamer (type A), functioning in PSII repair and PSI assembly | Thylakoid membrane | Variegated leaves and sensitivity to photoinhibition | Sakamoto et al., 2002, 2003; Järvi et al., 2016 |
| FtsH8 | AT1G06430 | Forming the thylakoid FtsH hexamer (type B), functioning in PSII repair | Thylakoid membrane | Indistinguishable from the wild type | Sakamoto et al., 2003 |
| FtsH6 | AT5G15250 | Degradation of LHCB1and LHCB3 in darkness or high light treatment, thermomemory | Thylakoid membrane | Indistinguishable from the wild type, sensitive to heat stress | Sakamoto et al., 2003; Zelisko et al., 2005; Sedaghatmehr et al., 2016 |
| FtsH7 | AT3G47060 | – | IEM | Indistinguishable from the wild type | Wagner et al., 2011 |
| FtsH9 | AT5G58870 | – | IEM | Indistinguishable from the wild type | Wagner et al., 2011 |
| FtsH11 | AT5G53170 | TIC protein turnover, thermotolerance | IEM | Virescent leaves and retarded growth, sensitive to heat stress | Chen et al., 2006, 2018a; Adam et al., 2019 |
| FtsH12 | AT1G79560 | Protein import, early chloroplast development | IEM | Embryo lethal | Wagner et al., 2011; Schreier et al., 2018; Shingo et al., 2018; Mielke et al., 2021 |
| Lon | |||||
| Lon1 | AT5G26860 | Removal of damaged proteins, organelle biogenesis | Chloroplast and mitochondria | Shorter root and stem, smaller leaves, and retarded growth | Rigas et al., 2009; Daras et al., 2014 |
| Lon4 | AT3G05790 | Removal of damaged proteins, drought tolerance | Thylakoid, dual-localized to mitochondria | Sensitivity to drought stress | Ostersetzer et al., 2007; Li et al., 2010 |
| ATP-independent proteases | |||||
| Deg | |||||
| Deg1 | AT3G27925 | Cleaving the lumen transmembrane loop of D1, PSII repair | Lumenal side of thylakoid membrane | Early flowering; fewer, thinner, and pale-green leaves; sensitivity to photoinhibition | Kapri-Pardes et al., 2007; Kley et al., 2011 |
| Deg2 | AT2G47940 | Degradation of LHCB6, chaperone-like activity, possibly cleaving the stroma transmembrane loop of D1 | Stromal side of thylakoid membrane | Indistinguishable from the wild type | Huesgen et al., 2006; Lucinski et al., 2011a; Sun et al., 2012 |
| Deg5 | AT4G18370 | Degradation of PsbF and synergistic function in the degradation of D1 (cleaving the lumenal transmembrane loop of D1) with Deg8 | Lumenal side of thylakoid membrane | Slightly reduced growth rate | Sun et al., 2007; Lucinski et al., 2011b |
| Deg7 | AT3G03380 | PSII (D1, D2, CP43, and CP47) repair, possibly cleaving the stromal transmembrane loop of D1 | Stromal side of thylakoid membrane | Indistinguishable from the wild type | Sun et al., 2010 |
| Deg8 | AT5G39830 | Synergistic function in the degradation of D1 (cleaving the lumenal transmembrane loop of D1) with Deg5 | Lumenal side of thylakoid membrane | Slightly reduced growth rate | Sun et al., 2007 |
| Peptidases | |||||
| SPP | AT5G42390 | TP cleavage of newly imported proteins | Stroma | Embryo lethal | Trösch and Jarvis, 2011 |
| PreP1 | AT3G19170 | Degradation of peptides with length of 10–65 amino acids | Stroma | Very weak chlorotic phenotype for prep1 mutant, no visible phenotypes for prep2 mutant, virescent leaves with reduced thylakoid grana stacks for prep1 prep2 double mutant | Nilsson Cederholm et al., 2009 |
| PreP2 | AT1G49630 | ||||
| OOP | AT5G65620 | Degradation of peptides with length of 8–23 amino acids | Stroma | Indistinguishable from the wild type, may be involved in salicylic acid signaling | Kmiec et al., 2013; Moreau et al., 2013 |
| Plsp1/TPP | AT3G24590 | Degradation of TT in the lumen and maturation of TOC75 | Thylakoid membrane and envelope | Albino and death before development of complete true leaves | Inoue et al., 2005; Shipman and Inoue, 2009; Shipman-Roston et al., 2010 |
| Others | |||||
| CtpA | AT4G17740 | C-terminal processing of D1 | Lumen | Essential for autotrophic growth and chloroplast development | Che et al., 2013 |
| EGY1 | AT5G35220 | Metalloprotease activity, chloroplast development | Thylakoid membrane | Pigmentation deficiency | Chen et al., 2005 |
| EGY2 | AT5G05740 | Metalloprotease activity, hypocotyl elongation, regulation of chloroplast gene expression | Thylakoid membrane | Shorter hypocotyls and larger rosette leaves at flowering time | Chen et al., 2012; Adamiec et al., 2018 |
| SppA | AT1G73990 | High light acclimation | Thylakoid membrane | Indistinguishable from the wild type, sensitive to high light stress | Wetzel et al., 2009 |
| NANA | AT3G12700 | Aspartyl protease, regulates carbohydrate metabolism, starch accumulation | Chloroplast | Shorter hypocotyl and roots at seedling stages, dwarf phenotypes in soil | Paparelli et al., 2012 |
| CND41 | AT5G10770 | Aspartyl protease, DNA binding, degradation of Rubisco holoproteins during leaf senescence | Stroma | Tobacco antisense lines show delayed-senescence phenotype in nitrogen-depleted conditions | Nakano et al., 1997; Kato et al., 2004 |
| CGEP | AT2G47390 | Glutamyl peptidase, may be involved in starch metabolism | Stroma | Indistinguishable from the wild type, genetically interacts with Clp mutants | Bhuiyan et al., 2020 |
Clp is an ATP-dependent Ser-type protease complex that functions mainly in the turnover of stromal proteins (Olinares et al., 2011; Nishimura and van Wijk, 2015). It is composed of a chaperone complex and a proteolytic core complex. The chaperone complex includes ClpC1 (also known as HSP93V), ClpC2 (HSP93III), and ClpD. They presumably assemble into homohexamers and are responsible for substrate docking, unfolding, and translocation to the core complex (Montandon et al., 2019; Rei Liao et al., 2022). The protein abundance of ClpC1 is much higher than that of ClpC2 and ClpD (Zybailov et al., 2008; Sjögren et al., 2014), and knockout of ClpC1 results in a pale-green phenotype. By contrast, clpc2 and clpd mutants are indistinguishable from the wild type, indicating that ClpC1 is the major isomer of the Clp chaperone (Park and Rodermel, 2004; Sjögren et al., 2004; Nishimura et al., 2013). However, the expression of ClpD (ERD1) has been shown to be induced by various abiotic stress conditions and senescence, suggesting more specific roles for ClpD in response to environmental stimuli or during specific developmental stages (Nakashima et al., 1997; Zheng et al., 2002). The small Clp adaptor protein ClpS1, together with ClpF, has been demonstrated to participate in the recognition of a set of substrate proteins (Nishimura et al., 2013, 2015).
The proteolytic core of Clp comprises two asymmetric rings. The R ring is composed of catalytically inactive ClpR1, R2, R3, R4, and the plastid-encoded proteolytically active ClpP1, whereas the P ring includes the catalytically active subunits ClpP3, P4, P5, and P6 (Kim et al., 2009). The small subunits ClpT1 and T2 participate in the assembly and stabilization of the core complex (Sjögren and Clarke, 2011; Kim et al., 2015). Extensive studies have been carried out to identify the Clp substrates. These experimentally verified substrates include, for example, key enzymes of the tetrapyrrole (glutamyl-tRNA reductase) and carotenoid (phytoene synthase) biosynthesis pathways (Apitz et al., 2016; Welsch et al., 2018), the rate-limiting enzyme of the methylerythritol 4-phosphate (MEP) pathway (deoxyxylulose 5-phosphate synthase [DXS]; Pulido et al., 2016), and the central integrator of the plastid retrograde signaling pathway, GUN1 (Wu et al., 2018). To survey the Clp substrates at the proteome-wide level, comparative proteomic studies of various Clp subunit mutants (clpr2, clpr4, clpp3, clps1, clpf, clpc1, and the clpc1 clps1 double mutant; Kim et al., 2009; Zybailov et al., 2009; Kim et al., 2013a; Nishimura et al., 2013; Nishimura et al., 2015) and in vivo substrate trapping of ClpC1 (Montandon et al., 2019; Rei Liao et al., 2022) were performed, revealing a broad range of putative Clp substrates and a vital role for Clp protease in the regulation of stromal proteostasis (Nishimura and van Wijk, 2015). However, the overaccumulated proteins (putative substrates) observed in clp mutants could also be secondary effects, consequences of the strong chlorotic or even albino phenotype of the mutants, and they must thus be further verified by other approaches (Figure 3 and Table 2).
Interestingly, ClpC has been shown to dually localize in the stroma and associated to IEM (∼30% of total ClpC associated to IEM; Sjögren et al., 2014). The IEM-associated ClpC, through interaction with Tic110 and presumably Tic40, tethers the Clp core complex in close proximity to the import channel, which was believed to function as a checkpoint for newly imported proteins (Flores-Pérez et al., 2016). This quality control mechanism ensures fast inspection and elimination of damaged, misfolded, or currently unneeded proteins immediately upon their release from the TIC complex. Consistent with this hypothesis, mutation of the Clp core subunit also results in reduced import capacity (Sjögren et al., 2014).
In the thylakoid membrane, FtsH and Deg are the major proteases that play critical roles in the biogenesis of thylakoid membranes and the PQC of thylakoid membrane proteins. FtsH is an ATP-dependent zinc metalloprotease complex. There are 12 FtsH-encoding genes in the model plant Arabidopsis, nine of which are localized in chloroplasts. FtsH1, 2, 5, 6, and 8 are anchored into the thylakoid membrane by their N-terminal single transmembrane domains. The C-terminal region that protrudes into the stroma contains the ATPase and proteolytic domains (Kato and Sakamoto, 2018). The thylakoid-localized FtsH forms a heterohexameric complex composed of type A (FtsH1 and FtsH5) and type B (FtsH2 and FtsH8) subunits with a mole ratio of 2:4 in Arabidopsis and 3:3 in cyanobacteria (Boehm et al., 2012; Moldavski et al., 2012). This hexametric FtsH complex plays a central role in the degradation of unassembled photosynthetic proteins and the so-called PSII repair cycle to renew the photodamaged PSII reaction center protein D1, a process that has been extensively studied in the last decades (Lindahl et al., 2000; Bailey et al., 2002; Sakamoto et al., 2003; Zaltsman et al., 2005a, 2005b; reviewed in Järvi et al., 2015; Theis and Schroda, 2016). In this process, the photodamaged PSII migrates to the thylakoid grana margins, partially disassembled (release of CP43 and oxygen evolving complex). The damaged D1 protein is then pulled out from the membrane and degraded by FtsH. The Deg endopeptidases facilitate D1 degradation by cleaving the inter-loops connecting the five transmembrane helices of the D1 protein, either at the lumenal side (Deg1, 5, 8) or the stromal side (Deg2, 7) of the thylakoid membrane (Kapri-Pardes et al., 2007; Kato et al., 2012; Butenko et al., 2018). In addition to D1 repair, thylakoid FtsH also participates in the turnover of various photosynthetic proteins (Ostersetzer and Adam, 1997; Malnoe et al., 2014; Bujaldon et al., 2017) and is involved in the biogenesis of PSI (Figure 3 and Table 2; Järvi et al., 2016).
The loss of function of FtsH2 (VAR2), the most abundant FtsH isomer, and FtsH5 (VAR1), the second most abundant FtsH isomer, results in a strong and weak leaf-variegation phenotype, respectively, even under normal light conditions, indicating their vital role in thylakoid biogenesis (Takechi et al., 2000; Sakamoto et al., 2002). Interestingly, an intensive suppressor screening of var1/var2 mutants identified many components of the chloroplast translation machineries (Miura et al., 2007; Yu et al., 2008; Liu et al., 2010a, 2010b). Although the precise mechanism of the suppression remains elusive, a “threshold” model has been suggested in which these mutations rebalance chloroplast proteostasis in the var mutants by reducing the loads of the proteome. The other thylakoid-localized isomer, FtsH6, is not involved in the PSII repair cycle. It was shown to participate in thermomemory in Arabidopsis by negatively regulating the abundance of HSP21 (Sedaghatmehr et al., 2016; Chen et al., 2017), indicating the diverse roles of FtsH in plants during environmental challenges.
Another set of FtsH proteases is localized in the IEM, including FtsH7, 9, 11, 12, and five proteolytically inactivated FstHs (FtsHi1–5). FtsH11 may participate in the regulation of TIC protein turnover and plays a role in plant thermotolerance (Chen et al., 2006; Adam et al., 2019). Interestingly, as discussed above, FtsH12 and four FtsHi proteins (FtsHi1, 2, 4, 5), together with NAD-malate dehydrogenase and the chloroplast-encoded Ycf2, form a 2-MD complex that serves as an import motor involved in the ATP-dependent translocation of preproteins through the TIC channel (Schreier et al., 2018; Shingo et al., 2018). FtsHi1, also known as ARC1 (ACCUMULATION AND REPLICATION OF CHLOROPLAST1), was identified early through its role in chloroplast division (Kadirjan-Kalbach et al., 2012; Mishra and Funk, 2021). The connection of its functions in protein import and chloroplast division is still not clear. Interestingly, a recent study showed that Tic236, a large TIC component constantly linking the TIC and TOC complex, also plays a role in chloroplast division (Chen et al., 2018b; Fang et al., 2022), providing a hint for the connection of these two processes. The depletion of FtsHi5 results in hypersensitivity to decreased ambient temperature (Li et al., 2021), which has also been observed in many mutants with disrupted chloroplast proteostasis (fug1, prps17, prpl11, and var2; Marino et al., 2019), indicating a close link between chloroplast proteostasis and temperature stress.
The aberrant accumulation of peptides generated by precursor processing and protein degradation can affect chloroplast function, and such peptides must be further degraded and recycled. Transit peptides are cleaved by stromal processing peptidase (SPP) and by thylakoidal processing peptidase (TPP) for cleavage of the second peptide that directs the targeting of thylakoid lumen proteins (Chaal et al., 1998; Richter and Lamppa, 1998). The resulting transit peptide fragments and protein degradation products (short peptides) from Clp, FtsH, and other proteases are further degraded by SPP, presequence peptidase (PreP, degrading peptides with a length of 10–65 amino acids), and/or organellar oligopeptidase (OOP, degrading peptides with a length of 8–23 amino acids) to avoid their proteotoxic effects on chloroplast proteostasis (Figure 3 and Table 2; Richter and Lamppa, 1999; Stahl et al., 2005; Kmiec et al., 2013; Kmiec et al., 2014).
The lifetimes of different proteins can vary from hours to days or even months. Radiolabeling was widely used in early studies to estimate protein turnover rate by following the decreased kinetics of isotope pulse-labeled proteins in the chasing phase. These studies provided insights mainly into the protein lifetimes of enzymes involved in primary metabolism, with a strong focus on photosynthetic machineries (Kemp and Sutton, 1971; Trewavas, 1972). Development of approaches that combined isotope labeling with mass spectrometry analysis enabled a comprehensive view of protein turnover at the proteome level (Nelson et al., 2014a, 2014b; Ishihara et al., 2015; Li et al., 2017). Contrary to conventional knowledge, highly abundant proteins, e.g., subunits of Rubisco, have been shown to turn over very slowly. This could be explained by the fact that protein synthesis and degradation are highly energy-consuming processes. It has been estimated that the synthesis of new proteins can account for 38% of total respiratory ATP, half of these ATP are used for the synthesis of Rubisco (Li et al., 2017). It would be energy-saving to maintain these highly abundant proteins with long lifetimes.
The regulation of protein lifetime (stability) in chloroplasts is not yet fully understood. However, the first N-terminal amino acid (after excision of the starting Met for chloroplast-encoded proteins or cleavage of the TP for nucleus-encoded proteins) is an important determinant of protein stability, referred to as N-end rules, in both prokaryotes and eukaryotes (Mogk et al., 2007; Varshavsky, 2011). For tobacco plastid proteins, it has been shown that N-terminal Glu, Met, and Val residues result in high accumulation of plastid genome-expressed GFP protein, whereas N-terminal His and Cys-GFP are very unstable (Apel et al., 2010). The adaptor subunit of Clp protease, ClpS, appears to participate in substrate recognition by binding directly to the N-terminal destabilizing residues (N-degron) for substrate delivery in E. coli (Roman-Hernandez et al., 2009; Schmidt et al., 2009). A homolog of ClpS (ClpS1) has been identified in Arabidopsis (Nishimura et al., 2013). ClpS1 interacts with the chloroplast-specific subunit ClpF, forming a binary adaptor complex that directly binds to Clp substrates for their degradation (Figure 3; Nishimura et al., 2015). However, analysis of the degradation rate of Arabidopsis proteins by isotope labeling and mass spectrometry shows that the degradation rate seems to be uncorrelated with the rules of N-terminal stable/unstable amino acids; it is instead determined by their intrinsic properties (Li et al., 2017), adding more complexities to our understanding of the regulation of chloroplast proteostasis. Moreover, protein degradation rate is strongly regulated by environmental stimuli, enabling fast remodeling of the proteome for new growth conditions (Li et al., 2022b).
Chloroplast chaperones and the unfolded protein response
Newly synthesized or imported proteins must be efficiently folded for their functionality, at the same time preventing the proteotoxicity of unfolded proteins to the cell. Chaperones play a central role in protein folding/refolding processes and the delivery of proteins that cannot be correctly folded to proteases for degradation (Kim et al., 2013b; Balchin et al., 2016). Several chloroplast chaperones have been demonstrated to play roles in protein import in association with Tic110, including HSP93/ClpC, HSP90C, cpHSC70, and the chaperonin CPN60 (Flores-Perez and Jarvis, 2013). The Tic110–Tic40 complex has been excluded from the 1-MD TIC complex; however, Tic110 with its associated chaperones may act at the later stage of the import process that is closely linked to post-import folding. HSP93/ClpC, as discussed above, has dual roles in Clp-mediated protein degradation and the PQC of protein import (Flores-Pérez et al., 2016). Early studies identified cpHSC70 and HSP90C as import motors. Although the new Ycf2–FtsHi motor has been suggested, a role for cpHSC70 and HSP90C in protein import cannot be excluded, as loss of function of cpHSC70-1, the major form of cpHSC70, and pharmacological inhibition of HSP90C activity by radicicol produce strong import defects (Shi and Theg, 2010; Su and Li, 2010; Inoue et al., 2013). Their role in protein import may be as post-import chaperones that bind to preproteins to prevent their misfolding/aggregation and participate in the follow-up folding process. In fact, other than a TIC-associated chaperone, cpHSC70 plays a critical role in the maintenance of stromal proteostasis by participating in the folding/refolding process of stromal proteins. In Arabidopsis, cpHSC70 has been shown to work together with HSP100 chaperone ClpB3 and ClpC1 to determine the refolding/reactivation (through ClpB3) or degradation (through Clp protease) of DXS, the rate-limiting enzyme of the MEP pathway (Pulido et al., 2016).
In addition to the HSP70 and HSP100 families, group I chaperonin CPN60s and their co-chaperonin CPN10/20 form another major machinery that assists in chloroplast protein folding (Vitlin Gruber et al., 2013; Zhao and Liu, 2017). A well-studied protein substrate of CPN60 chaperonins is RbcL. Indeed, chloroplast chaperonin was initially identified as a Rubisco-binding protein (Hemmingsen and Ellis, 1986; Hemmingsen et al., 1988). Recently, CPN60 has been shown to participate in the thylakoid membrane integration of Plsp1 (thylakoid-processing peptidase) by capturing Plsp1 and releasing it to the SEC translocon on the thylakoid membrane (Klasek et al., 2020), suggesting that CPN60s also function in intra-chloroplast protein targeting. Moreover, CPN60s also participate in chloroplast protein import and chloroplast division (Kessler and Blobel, 1996; Suzuki et al., 2009), indicating the multiple functions of these chaperonins.
When misfolded proteins accumulate and exceed the buffering capacity of PQC, an unfolded protein response (UPR) will be induced to upregulate genes encoding PQC components and rebalance proteostasis. The UPRs in the ER (UPRer) and mitochondria (UPRmt) have been well studied in mammals and plants (Howell, 2013; Anderson and Haynes, 2020). Inhibition of chloroplast translation by lincomycin (which induces a rapid accumulation of aggregated proteins in chloroplasts), conditional depletion of Clp protease, and mutation of FtsH2 has been shown to trigger a UPR-like response in chloroplasts (UPRcp; Ramundo and Rochaix, 2014; Ramundo et al., 2014; Llamas et al., 2017; Dogra et al., 2019a). As in the UPRer and UPRmt, a group of nucleus-encoded proteins involved in chloroplast PQC are induced in response to UPRcp, including ClpB3, cpHSC70, HSP21, and reactive oxygen species (ROS) detoxifiers (in the var2 mutant), and this appears to be mediated by the heat-shock factor HSFA2 (Llamas et al., 2017; Dogra et al., 2019a). A similar UPRcp was observed in Chlamydomonas. Interestingly, a cytosolic protein kinase named Mars1 (for mutant affected in chloroplast-to-nucleus retrograde signaling) has been identified and shown to participate in the retrograde signaling pathway of the UPRcp to mediate HSP induction (Kessler and Longoni, 2019; Perlaza et al., 2019). However, the cytosolic components that relay UPRcp signaling in higher plants remain elusive.
Retrograde signaling related to chloroplast proteostasis
Chloroplasts have evolved critical roles in sensing physiological and environmental cues to tune nuclear gene expression by retrograde signaling (Chan et al., 2016; Schwenkert et al., 2022). In the 1970s, it had already been noticed that cytosolic synthesis of chloroproteins was inhibited by defective chloroplast gene expression in the “albostrains” barley mutant (Bradbeer et al., 1979). Since then, multiple retrograde signaling pathways have been identified that relay the status of the chloroplast to the nucleus to control the expression of nuclear genes and, hence, the sets of chloroproteins to be imported into chloroplasts. These retrograde signals include tetrapyrrole intermediates and signals from plastid gene expression (known as biogenetic control), ROS and redox signals from PET, and other chloroplast metabolites (termed operational control; de Souza et al., 2017; Hernández-Verdeja and Strand, 2018; Li and Kim, 2022).
The imbalance of chloroplast proteostasis was shown to trigger a nuclear response to upregulate components of chloroplast PQC, indicating the existence of proteostasis-related chloroplast retrograde signaling. Mutation of FtsH2 (var2 mutant) disturbs thylakoid proteostasis and induces a UPR-like response that upregulates the expression of chloroplast chaperones, proteases, and ROS detoxifiers (Dogra et al., 2019a). FtsH2 was shown to be involved in the turnover of EXECUTOR1 (EX1) and EX2 proteins (putative singlet oxygen [1O2] sensors) at the granal margins, thus participating in the 1O2 retrograde signaling pathway that is functionally linked to PSII repair (Wang et al., 2016; Dogra et al., 2019b, 2022). The stress hormone salicylic acid (SA) seems to take part in FtsH2-related retrograde signaling, as SA-responsive genes were largely upregulated in the var2 mutant. Meanwhile, the inactivation of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1, a vital immune component that positively regulates SA synthesis and signaling; Cui et al., 2017) in the flu mutant (conditional fluorescent [flu] mutant that generates 1O2 in chloroplasts upon a dark-to-light shift; Meskauskiene et al., 2001) compromises 1O2-triggered stress responses (Ochsenbein et al., 2006; Duan et al., 2019). However, a direct link between SA and plastid ROS retrograde signaling needs to be established in future studies.
GUN1 was initially identified as a central integrator of multiple retrograde signaling pathways, as its mutation uncoupled nuclear gene expression under conditions that altered different retrograde signaling pathways (Koussevitzky et al., 2007b). Recent studies suggested that GUN1 plays a critical role in the maintenance of chloroplast proteostasis. Inactivation of GUN1 only weakly delays seedling greening at early germination stages, with no obvious phenotypes in mature plants in which chloroplast biogenesis is completed. However, introduction of the gun1 allele into single mutants with reduced chloroplast proteostasis capacity, including mutants of plastid translation initiation factor (fug1; Marino et al., 2019), a ribosomal protein (prpl11; Tadini et al., 2016), proteases (var1 and var2; Tadini et al., 2020), and subunits of the protein import apparatus (clpc1 and cphsc70-1; Wu et al., 2019b), severely aggravates their mutant phenotype. Hence, GUN1 emerges as a putative harmonizer of chloroplast proteostasis (Wu and Bock, 2021). GUN1 seems to regulate chloroplast proteostasis by controlling the import of housekeeping proteins under conditions that alter retrograde signaling (Wu et al., 2019a, 2019b). Depletion of GUN1 results in aberrant accumulation of chloroprotein precursors in the cytosol, induction of cyHSP90 and cyHSP70 chaperones, and, hence, expression of the GUN phenotype, as the expression of photosynthesis-associated nuclear genes (PhANGs) is positively correlated with HSP90 activity (Wu et al., 2019b). It is worth mentioning that although PhANG transcripts are accumulated in the gun1 mutant, their protein products are not. This post-transcriptional regulation involves both translational repression and UPS-dependent degradation of these proteins (Tokumaru et al., 2017; Wu et al., 2019a).
However, it should be noticed that the GUN phenotype of gun1 is based on a comparison of PhANG expression between gun1 and the wild type under the same conditions that block chloroplast biogenesis (treated with lincomycin or norflurazon), but not between gun1 and the untreated wild type in which normal chloroplast biogenesis proceeds. Indeed, PhANG expression in gun1 is only higher than that of the lincomycin/norflurazon-treated wild type; it is still much lower than that of the untreated wild type or gun1 mutants. Thus, this model cannot be used to explain the reduced expression of PhANGs in ppi2, tic100cue8, or any other virescent mutants (with import defects) in comparison with the healthy wild type (Kakizaki et al., 2009; Loudya et al., 2022). The reduced protein import in the ppi2 or tic100cue8 mutant represses the biogenesis of chloroplasts, thus triggering downregulation of PhANG expression, further indicating that chloroplasts with disturbed proteostasis emit a retrograde signal to tune nuclear gene expression for rebalancing proteostasis. However, the nature of this retrograde signal and the mechanism by which it regulates nuclear gene expression remain elusive.
Concluding remarks
The biogenesis, maintenance, and remodeling of chloroplast proteostasis require orchestration of the synthesis, import, assembly, and turnover of proteins expressed from both chloroplast and nuclear genomes. Significant progress has been made in the last decades in understanding the regulation of chloroplast proteostasis. However, our knowledge of these regulatory processes, their underlying mechanisms, and the factors involved is still limited. For example, the influence of different environmental factors on chloroplast translation and the trans-factors involved in these regulations need to be identified. Chloroplast translation seems to strongly interact with protein synthesis in the cytosol. Likewise, import defects will result in aberrant accumulation of chloroprotein precursors in the cytosol, which strongly affects cytosolic proteostasis and triggers PARcp. However, the cellular network of PARcp remains largely elusive. Future studies will gradually reveal how plant cells coordinate protein synthesis with the maintenance of proteostasis between the chloroplast and cytosol, and also with proteostasis in other subcellular compartments such as the mitochondria and ER.
Thousands of chloroproteins need to be imported into plastids during their biogenesis and/or transition into different types of plastids. TOC receptors must decode the information embedded in the cTP, as the cTP seems to contain detailed developmental-stage-specific and tissue-specific targeting signals. Different TOC complexes have been shown to prefer the import of different sets of proteins, i.e., photosynthetic proteins for Toc159/Toc33 and housekeeping proteins for Toc132/Toc120/Toc34. Identification of new factors involved in the specificity of substrate selection will provide new insights into the spatiotemporal regulation of chloroplast protein import. In addition, most of our knowledge of protein import has been obtained from model species such as Arabidopsis and pea. Future studies may focus more effort on crop plants such as the important monocot crops rice, wheat, and maize and on regulation during plastid type transitions, as in tomato fruits.
Chloroplasts with imbalanced proteostasis will emit signals to modulate nuclear gene expression, especially by upregulating the genes that encode chloroplast PQC components. However, this retrograde signaling pathway is still poorly understood. Moreover, the chloroplast signals, such as 1O2 and methylerythritol cyclodiphosphate (MEcPP), seem to have a close connection with other organelles, for example, modulating the UPRer (Walley et al., 2015). Understanding the cooperation mechanism between chloroplast proteostasis and the proteostasis of other subcellular compartments will be essential for future improvement of stress tolerance in crops.
Funding
Research on chloroplast biology in the authors’ laboratory has been funded by the National Natural Science Foundation of China (NSFC; 32070299, 32270285), the Shanghai Pujiang Program (20PJ1405600), the Shanghai Collaborative Innovation Center of Agri-Seeds (ZXWH2150201/014), and the Partner Group program of the Max Planck Society to G.-Z.W.
Author contributions
G.-Z.W. conceived the review. L.-L.G., Z.-H.H., Y.W., and G.-Z.W. wrote the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgments
We apologize to those authors whose research could not be cited owing to space limitations. No conflict of interest is declared.
Published: August 12, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Accossato S., Kessler F., Shanmugabalaji V. SUMOylation contributes to proteostasis of the chloroplast protein import receptor TOC159 during early development. Elife. 2020;9:e60968. doi: 10.7554/eLife.60968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z., Aviv-Sharon E., Keren-Paz A., Naveh L., Rozenberg M., Savidor A., Chen J. The chloroplast envelope protease FTSH11 - interaction with CPN60 and identification of potential substrates. Front. Plant Sci. 2019;10:428. doi: 10.3389/fpls.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamiec M., Misztal L., Kosicka E., Paluch-Lubawa E., Luciński R. Arabidopsis thaliana egy2 mutants display altered expression level of genes encoding crucial photosystem II proteins. J. Plant Physiol. 2018;231:155–167. doi: 10.1016/j.jplph.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Agne B., Infanger S., Wang F., Hofstetter V., Rahim G., Martin M., Lee D.W., Hwang I., Schnell D., Kessler F. A Toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J. Biol. Chem. 2009;284:8670–8679. doi: 10.1074/jbc.M804235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya M.S., Breitenberger C.A. Light regulation of protein synthesis factor EF-G in pea chloroplasts. Plant Mol. Biol. 1992;20:791–800. doi: 10.1007/BF00027150. [DOI] [PubMed] [Google Scholar]
- Albrecht V., Ingenfeld A., Apel K. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 2006;60:507–518. doi: 10.1007/s11103-005-4921-0. [DOI] [PubMed] [Google Scholar]
- Anderson N.S., Haynes C.M. Folding the mitochondrial UPR into the integrated stress response. Trends Cell Biol. 2020;30:428–439. doi: 10.1016/j.tcb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrès C., Agne B., Kessler F. The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta. 2010;1803:715–723. doi: 10.1016/j.bbamcr.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Apel W., Schulze W.X., Bock R. Identification of protein stability determinants in chloroplasts. Plant J. 2010;63:636–650. doi: 10.1111/j.1365-313X.2010.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz J., Nishimura K., Schmied J., Wolf A., Hedtke B., van Wijk K.J., Grimm B. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 2016;170:2040–2051. doi: 10.1104/pp.15.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J.M. The puzzle of plastid evolution. Curr. Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Aronsson H., Boij P., Patel R., Wardle A., Töpel M., Jarvis P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007;52:53–68. doi: 10.1111/j.1365-313X.2007.03207.x. [DOI] [PubMed] [Google Scholar]
- Bae W., Lee Y.J., Kim D.H., Lee J., Kim S., Sohn E.J., Hwang I. AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 2008;10:220–227. doi: 10.1038/ncb1683. [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Berry J.O., Carr J.P., Klessig D.F. mRNAs encoding ribulose-1, 5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc. Natl. Acad. Sci. USA. 1988;85:4190–4194. doi: 10.1073/pnas.85.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadula S.K., Elthon T.E., Habben J.E., Helentjaris T.G., Jiao S., Ristic Z. Heat-stress induced synthesis of chloroplast protein synthesis elongation factor (EF-Tu) in a heat-tolerant maize line. Planta. 2001;212:359–366. doi: 10.1007/s004250000416. [DOI] [PubMed] [Google Scholar]
- Bhuiyan N.H., Rowland E., Friso G., Ponnala L., Michel E.J.S., van Wijk K.J. Autocatalytic processing and substrate specificity of Arabidopsis chloroplast glutamyl peptidase. Plant Physiol. 2020;184:110–129. doi: 10.1104/pp.20.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P.A., Roschitzki B., Hirsch-Hoffmann M., Hennig L., Kessler F., Gruissem W., et al. Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell. 2011;23:3911–3928. doi: 10.1105/tpc.111.092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Structure, function, and inheritance of plastid genomes. Top. Curr. Genet. 2007;19:29–63. [Google Scholar]
- Bock R. Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015;66:211–241. doi: 10.1146/annurev-arplant-050213-040212. [DOI] [PubMed] [Google Scholar]
- Bock R. Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu. Rev. Genet. 2017;51:1–22. doi: 10.1146/annurev-genet-120215-035329. [DOI] [PubMed] [Google Scholar]
- Bock R., Timmis J.N. Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- Boehm M., Yu J., Krynicka V., Barker M., Tichy M., Komenda J., Nixon P.J., Nield J. Subunit organization of a synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell. 2012;24:3669–3683. doi: 10.1105/tpc.112.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B., Soll J. Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol. Plant. 2017;10:219–221. doi: 10.1016/j.molp.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Boos F., Labbadia J., Herrmann J.M. How the mitoprotein-induced stress response safeguards the cytosol: a unified view. Trends Cell Biol. 2020;30:241–254. doi: 10.1016/j.tcb.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Boos F., Krämer L., Groh C., Jung F., Haberkant P., Stein F., Wollweber F., Gackstatter A., Zöller E., van der Laan M., et al. Mitochondrial protein-induced stress triggers a global adaptive transcriptional programme. Nat. Cell Biol. 2019;21:442–451. doi: 10.1038/s41556-019-0294-5. [DOI] [PubMed] [Google Scholar]
- Bradbeer J.W., Atkinson Y.E., Börner T., Hagemann R. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature. 1979;279:816–817. [Google Scholar]
- Bujaldon S., Kodama N., Rappaport F., Subramanyam R., de Vitry C., Takahashi Y., Wollman F.A. Functional accumulation of antenna proteins in chlorophyll b-less mutants of Chlamydomonas reinhardtii. Mol. Plant. 2017;10:115–130. doi: 10.1016/j.molp.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Butenko Y., Lin A., Naveh L., Kupervaser M., Levin Y., Reich Z., Adam Z. Differential roles of the thylakoid lumenal Deg protease homologs in chloroplast proteostasis. Plant Physiol. 2018;178:1065–1080. doi: 10.1104/pp.18.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaal B.K., Mould R.M., Barbrook A.C., Gray J.C., Howe C.J. Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana. Implications for the origin and catalytic mechanism of the enzyme. J. Biol. Chem. 1998;273:689–692. doi: 10.1074/jbc.273.2.689. [DOI] [PubMed] [Google Scholar]
- Chan K.X., Phua S.Y., Crisp P., McQuinn R., Pogson B.J. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- Che Y., Fu A., Hou X., McDonald K., Buchanan B.B., Huang W., Luan S. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:16247–16252. doi: 10.1073/pnas.1313894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Bi Y.R., Li N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 2005;41:364–375. doi: 10.1111/j.1365-313X.2004.02308.x. [DOI] [PubMed] [Google Scholar]
- Chen G., Law K., Ho P., Zhang X., Li N. EGY2, a chloroplast membrane metalloprotease, plays a role in hypocotyl elongation in Arabidopsis. Mol. Biol. Rep. 2012;39:2147–2155. doi: 10.1007/s11033-011-0962-4. [DOI] [PubMed] [Google Scholar]
- Chen J., Burke J.J., Xin Z. Chlorophyll fluorescence analysis revealed essential roles of FtsH11 protease in regulation of the adaptive responses of photosynthetic systems to high temperature. BMC Plant Biol. 2018;18:11. doi: 10.1186/s12870-018-1228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Burke J.J., Velten J., Xin Z. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J. 2006;48:73–84. doi: 10.1111/j.1365-313X.2006.02855.x. [DOI] [PubMed] [Google Scholar]
- Chen S.T., He N.Y., Chen J.H., Guo F.Q. Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5-mediated retrograde pathway in Arabidopsis. Plant J. 2017;89:1106–1118. doi: 10.1111/tpj.13447. [DOI] [PubMed] [Google Scholar]
- Chen Y.L., Chen L.J., Chu C.C., Huang P.K., Wen J.R., Li H.M. TIC236 links the outer and inner membrane translocons of the chloroplast. Nature. 2018;564:125–129. doi: 10.1038/s41586-018-0713-y. [DOI] [PubMed] [Google Scholar]
- Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M., Levin J.Z., Keegstra K., Li H.M. Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 2003;22:2970–2980. doi: 10.1093/emboj/cdg281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.-C., Swamy K., Li H.-M. Tissue-specific regulation of plastid protein import via transit-peptide motifs. Plant Cell. 2020;32:1204–1217. doi: 10.1105/tpc.19.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Gobbato E., Kracher B., Qiu J., Bautor J., Parker J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2017;213:1802–1817. doi: 10.1111/nph.14302. [DOI] [PubMed] [Google Scholar]
- Daras G., Rigas S., Tsitsekian D., Zur H., Tuller T., Hatzopoulos P. Alternative transcription initiation and the AUG context configuration control dual-organellar targeting and functional competence of Arabidopsis Lon1 protease. Mol. Plant. 2014;7:989–1005. doi: 10.1093/mp/ssu030. [DOI] [PubMed] [Google Scholar]
- de Souza A., Wang J.-Z., Dehesh K. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 2017;68:85–108. doi: 10.1146/annurev-arplant-042916-041007. [DOI] [PubMed] [Google Scholar]
- de Vries J., Sousa F.L., Bölter B., Soll J., Gould S.B. YCF1: a green TIC? Plant Cell. 2015;27:1827–1833. doi: 10.1105/tpc.114.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M., Littlejohn G., Jayaraman S., Studholme D., Bailey T., Lawson T., Tillich M., Licht D., Bölter B., Delfino L., et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants. 2015;1:15074. doi: 10.1038/nplants.2015.74. [DOI] [PubMed] [Google Scholar]
- Dogra V., Duan J., Lee K.P., Kim C. Impaired PSII proteostasis triggers a UPR-like response in the var2 mutant of Arabidopsis. J. Exp. Bot. 2019;70:3075–3088. doi: 10.1093/jxb/erz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V., Li M., Singh S., Li M., Kim C. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun. 2019;10:2834. doi: 10.1038/s41467-019-10760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V., Singh R.M., Li M., Li M., Singh S., Kim C. EXECUTER2 modulates the EXECUTER1 signalosome through its singlet oxygen-dependent oxidation. Mol. Plant. 2022;15:438–453. doi: 10.1016/j.molp.2021.12.016. [DOI] [PubMed] [Google Scholar]
- Duan J., Lee K.P., Dogra V., Zhang S., Liu K., Caceres-Moreno C., Lv S., Xing W., Kato Y., Sakamoto W., et al. Impaired PSII proteostasis promotes retrograde signaling via salicylic acid. Plant Physiol. 2019;180:2182–2197. doi: 10.1104/pp.19.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edhofer I., Mühlbauer S.K., Eichacker L.A. Light regulates the rate of translation elongation of chloroplast reaction center protein D1. Eur. J. Biochem. 1998;257:78–84. doi: 10.1046/j.1432-1327.1998.2570078.x. [DOI] [PubMed] [Google Scholar]
- Eichacker L.A., Müller B., Helfrich M. Stabilization of the chlorophyll binding apoproteins, P700, CP47, CP43, D2, and D1, by synthesis of Zn-pheophytin a in intact etioplasts from barley. FEBS Lett. 1996;395:251–256. doi: 10.1016/0014-5793(96)01026-5. [DOI] [PubMed] [Google Scholar]
- Eisa A., Malenica K., Schwenkert S., Bölter B. High light acclimation induces chloroplast precursor phosphorylation and reduces import efficiency. Plants. 2019;9:24. doi: 10.3390/plants9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J., Li B., Chen L.J., Dogra V., Luo S., Wu W., Wang P., Hwang I., Li H.M., Kim C. TIC236 gain-of-function mutations unveil the link between plastid division and plastid protein import. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2123353119. e2123353119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Meierhoff K., Sane A.P., Meurer J., Driemel C., Plücken H., Klaff P., Stein B., Bechtold N., Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellerer C., Schweiger R., Schöngruber K., Soll J., Schwenkert S. Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant. 2011;4:1133–1145. doi: 10.1093/mp/ssr037. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Jarvis P. Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta. 2013;1833:332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Bédard J., Tanabe N., Lymperopoulos P., Clarke A.K., Jarvis P. Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol. 2016;170:147–162. doi: 10.1104/pp.15.01538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friemann A., Hachtel W. Chloroplast messenger RNAs of free and thylakoid-bound polysomes from Vicia faba L. Planta. 1988;175:50–59. doi: 10.1007/BF00402881. [DOI] [PubMed] [Google Scholar]
- Gabruk M., Mysliwa-Kurdziel B. Light-dependent protochlorophyllide oxidoreductase: phylogeny, regulation, and catalytic properties. Biochemistry. 2015;54:5255–5262. doi: 10.1021/acs.biochem.5b00704. [DOI] [PubMed] [Google Scholar]
- Gao Y., Thiele W., Saleh O., Scossa F., Arabi F., Zhang H., Sampathkumar A., Kühn K., Fernie A., Bock R., et al. Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell. 2022;34:2056–2079. doi: 10.1093/plcell/koac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawroński P., Jensen P.E., Karpiński S., Leister D., Scharff L.B. Pausing of chloroplast ribosomes is induced by multiple features and is linked to the assembly of photosynthetic complexes. Plant Physiol. 2018;176:2557–2569. doi: 10.1104/pp.17.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmer J., Helm S., Dobritzsch D., Hause G., Shema G., Zahedi R.P., Baginsky S. Mild proteasomal stress improves photosynthetic performance in Arabidopsis chloroplasts. Nat. Commun. 2020;11:1662. doi: 10.1038/s41467-020-15539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen S.M., Ellis R.J. Purification and properties of ribulosebisphosphate carboxylase large subunit binding protein. Plant Physiol. 1986;80:269–276. doi: 10.1104/pp.80.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen S.M., Woolford C., van der Vies S.M., Tilly K., Dennis D.T., Georgopoulos C.P., Hendrix R.W., Ellis R.J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hernández-Verdeja T., Strand Å. Retrograde signals navigate the path to chloroplast development. Plant Physiol. 2018;176:967–976. doi: 10.1104/pp.17.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Kikuchi S., Oishi M., Nakai M. In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant Cell Physiol. 2011;52:469–478. doi: 10.1093/pcp/pcr010. [DOI] [PubMed] [Google Scholar]
- Holbrook K., Subramanian C., Chotewutmontri P., Reddick L.E., Wright S., Zhang H., Moncrief L., Bruce B.D. Functional analysis of semi-conserved transit peptide motifs and mechanistic implications in precursor targeting and recognition. Mol. Plant. 2016;9:1286–1301. doi: 10.1016/j.molp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Howell S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Hristou A., Grimmer J., Baginsky S. The secret life of chloroplast precursor proteins in the cytosol. Mol. Plant. 2020;13:1111–1113. doi: 10.1016/j.molp.2020.07.004. [DOI] [PubMed] [Google Scholar]
- Huesgen P.F., Schuhmann H., Adamska I. Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett. 2006;580:6929–6932. doi: 10.1016/j.febslet.2006.11.058. [DOI] [PubMed] [Google Scholar]
- Inaba T., Alvarez-Huerta M., Li M., Bauer J., Ewers C., Kessler F., Schnell D.J. Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infanger S., Bischof S., Hiltbrunner A., Agne B., Baginsky S., Kessler F. The chloroplast import receptor Toc90 partially restores the accumulation of Toc159 client proteins in the Arabidopsis thaliana ppi2 mutant. Mol. Plant. 2011;4:252–263. doi: 10.1093/mp/ssq071. [DOI] [PubMed] [Google Scholar]
- Inoue H., Rounds C., Schnell D.J. The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell. 2010;22:1947–1960. doi: 10.1105/tpc.110.074328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Li M., Schnell D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA. 2013;110:3173–3178. doi: 10.1073/pnas.1219229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Baldwin A.J., Shipman R.L., Matsui K., Theg S.M., Ohme-Takagi M. Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 2005;171:425–430. doi: 10.1083/jcb.200506171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Obata T., Sulpice R., Fernie A.R., Stitt M. Quantifying protein synthesis and degradation in Arabidopsis by dynamic 13CO2 labeling and analysis of enrichment in individual amino acids in their free pools and in protein. Plant Physiol. 2015;168:74–93. doi: 10.1104/pp.15.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Zavodszky E., Shao S., Wohlever M.L., Keenan R.J., Hegde R.S. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y., Smith M.D., Chen K., Schnell D.J. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell. 2004;15:3379–3392. doi: 10.1091/mbc.E03-12-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Park S.H., Zhao L., Hartl F.U., Neupert W. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell. 2017;171:890–903.e18. doi: 10.1016/j.cell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., Weissman J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Aro E.M. Photosystem II repair in plant chloroplasts-regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta. 2015;1847:900–909. doi: 10.1016/j.bbabio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Tadini L., Ivanauskaite A., Rantala S., Allahverdiyeva Y., Leister D., Aro E.M. Thylakoid-bound FtsH proteins facilitate proper biosynthesis of photosystem I. Plant Physiol. 2016;171:1333–1343. doi: 10.1104/pp.16.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Jung H.S., Chory J. Signaling between chloroplasts and the nucleus: can a systems biology approach bring clarity to a complex and highly regulated pathway? Plant Physiol. 2010;152:453–459. doi: 10.1104/pp.109.149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P., Burch-Smith T.M., Grant M. An emerging role for chloroplasts in disease and defense. Annu. Rev. Phytopathol. 2021;59:423–445. doi: 10.1146/annurev-phyto-020620-115813. [DOI] [PubMed] [Google Scholar]
- Kadirjan-Kalbach D.K., Yoder D.W., Ruckle M.E., Larkin R.M., Osteryoung K.W. FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J. 2012;72:856–867. doi: 10.1111/tpj.12001. [DOI] [PubMed] [Google Scholar]
- Kakizaki T., Matsumura H., Nakayama K., Che F.S., Terauchi R., Inaba T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009;151:1339–1353. doi: 10.1104/pp.109.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapri-Pardes E., Naveh L., Adam Z. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell. 2007;19:1039–1047. doi: 10.1105/tpc.106.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmati A.R., Töpel M., Khan N.Z., Patel R., Ling Q., Karim S., Aronsson H., Jarvis P. Evolutionary, molecular and genetic analyses of Tic22 homologues in Arabidopsis thaliana chloroplasts. PLoS One. 2013;8:e63863. doi: 10.1371/journal.pone.0063863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. New insights into the types and function of proteases in plastids. Int. Rev. Cell Mol. Biol. 2010;280:185–218. doi: 10.1016/S1937-6448(10)80004-8. [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. FtsH protease in the thylakoid membrane: physiological functions and the regulation of protease activity. Front. Plant Sci. 2018;9:855. doi: 10.3389/fpls.2018.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sun X., Zhang L., Sakamoto W. Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 2012;159:1428–1439. doi: 10.1104/pp.112.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Murakami S., Yamamoto Y., Chatani H., Kondo Y., Nakano T., Yokota A., Sato F. The DNA-binding protease, CND41, and the degradation of ribulose-1, 5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta. 2004;220:97–104. doi: 10.1007/s00425-004-1328-0. [DOI] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. Proteostasis and aging. Nat. Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- Keeling P.J. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- Kemp J.D., Sutton D.W. Protein metabolism in cultured plant tissues. Calculation of an absolute rate of protein synthesis, accumulation, and degradation in tobacco callus in vivo. Biochemistry. 1971;10:81–88. doi: 10.1021/bi00777a013. [DOI] [PubMed] [Google Scholar]
- Kessler F., Blobel G. Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Longoni P. How chloroplasts protect themselves from unfolded proteins. Elife. 2019;8:e51430. doi: 10.7554/eLife.51430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Oishi M., Hirabayashi Y., Lee D.W., Hwang I., Nakai M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell. 2009;21:1781–1797. doi: 10.1105/tpc.108.063552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339:571–574. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]
- Killackey S.A., Bi Y., Soares F., Hammi I., Winsor N.J., Abdul-Sater A.A., Philpott D.J., Arnoult D., Girardin S.E. Mitochondrial protein import stress regulates the LC3 lipidation step of mitophagy through NLRX1 and RRBP1. Mol. Cell. 2022;82:2815–2831.e5. doi: 10.1016/j.molcel.2022.06.004. [DOI] [PubMed] [Google Scholar]
- Kim J., Mullet J.E. Ribosome-binding sites on chloroplast rbcL and psbA mRNAs and light-induced initiation of D1 translation. Plant Mol. Biol. 1994;25:437–448. doi: 10.1007/BF00043872. [DOI] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D.B., van Wijk K.J. Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell. 2009;21:1669–1692. doi: 10.1105/tpc.108.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Olinares P.D., Oh S.H., Ghisaura S., Poliakov A., Ponnala L., van Wijk K.J. Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol. 2013;162:157–179. doi: 10.1104/pp.113.215699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kimber M.S., Nishimura K., Friso G., Schultz L., Ponnala L., van Wijk K.J. Structures, functions, and interactions of ClpT1 and ClpT2 in the Clp protease system of Arabidopsis chloroplasts. Plant Cell. 2015;27:1477–1496. doi: 10.1105/tpc.15.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Klaff P., Gruissem W. Changes in chloroplast mRNA stability during leaf development. Plant Cell. 1991;3:517–529. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasek L., Inoue K., Theg S.M. Chloroplast chaperonin-mediated targeting of a thylakoid membrane protein. Plant Cell. 2020;32:3884–3901. doi: 10.1105/tpc.20.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.R., Mason H.S., Mullet J.E. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kley J., Schmidt B., Boyanov B., Stolt-Bergner P.C., Kirk R., Ehrmann M., Knopf R.R., Naveh L., Adam Z., Clausen T. Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure. Nat. Struct. Mol. Biol. 2011;18:728–731. doi: 10.1038/nsmb.2055. [DOI] [PubMed] [Google Scholar]
- Kmiec B., Teixeira P.F., Glaser E. Shredding the signal: targeting peptide degradation in mitochondria and chloroplasts. Trends Plant Sci. 2014;19:771–778. doi: 10.1016/j.tplants.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Kmiec B., Teixeira P.F., Berntsson R.P.A., Murcha M.W., Branca R.M.M., Radomiljac J.D., Regberg J., Svensson L.M., Bakali A., Langel U., et al. Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc. Natl. Acad. Sci. USA. 2013;110:E3761–E3769. doi: 10.1073/pnas.1307637110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjögren L.L.E., Zhao Y., Clarke A.K., Chory J. An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 2007;63:85–96. doi: 10.1007/s11103-006-9074-2. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Kovacheva S., Bédard J., Patel R., Dudley P., Twell D., Ríos G., Koncz C., Jarvis P. In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 2005;41:412–428. doi: 10.1111/j.1365-313X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell. 2003;15:1859–1871. doi: 10.1105/tpc.012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Ríos G., Koncz C., et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Maliga P. The plastid clpP1 protease gene is essential for plant development. Nature. 2003;425:86–89. doi: 10.1038/nature01909. [DOI] [PubMed] [Google Scholar]
- Laborenz J., Bykov Y.S., Knöringer K., Räschle M., Filker S., Prescianotto-Baschong C., Spang A., Tatsuta T., Langer T., Storchová Z., et al. The ER protein Ema19 facilitates the degradation of nonimported mitochondrial precursor proteins. Mol. Biol. Cell. 2021;32:664–674. doi: 10.1091/mbc.E20-11-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Jung C., Hwang I. Cytosolic events involved in chloroplast protein targeting. Biochim. Biophys. Acta. 2013;1833:245–252. doi: 10.1016/j.bbamcr.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Lee J., Hwang I. Sorting of nuclear-encoded chloroplast membrane proteins. Curr. Opin. Plant Biol. 2017;40:1–7. doi: 10.1016/j.pbi.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Yoo Y.J., Razzak M.A., Hwang I. Prolines in transit peptides are crucial for efficient preprotein translocation into chloroplasts. Plant Physiol. 2018;176:663–677. doi: 10.1104/pp.17.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Kim S.J., Oh Y.J., Choi B., Lee J., Hwang I. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol. Plant. 2016;9:1428–1431. doi: 10.1016/j.molp.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 2006;140:466–483. doi: 10.1104/pp.105.074575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee J., Woo S., Razzak M.A., Vitale A., Hwang I. Molecular mechanism of the specificity of protein import into chloroplasts and mitochondria in plant cells. Mol. Plant. 2019;12:951–966. doi: 10.1016/j.molp.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee D.W., Lee Y., Mayer U., Stierhof Y.-D., Lee S., Jürgens G., Hwang I. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 2009;21:3984–4001. doi: 10.1105/tpc.109.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Chiu C.C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Li H.M., Schnell D., Theg S.M. Protein import motors in chloroplasts: on the role of chaperones. Plant Cell. 2020;32:536–542. doi: 10.1105/tpc.19.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yuan J., Li Y., Sun H., Ma T., Huai J., Yang W., Zhang W., Lin R. The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022;39:110664. doi: 10.1016/j.celrep.2022.110664. [DOI] [PubMed] [Google Scholar]
- Li J.Y., Sun J.L., Tian Y.Y., Liu J.X. The FtsH-inactive protein FtsHi5 is required for chloroplast development and protein accumulation in chloroplasts at low ambient temperature in Arabidopsis. Front. Plant Sci. 2021;12:830390. doi: 10.3389/fpls.2021.830390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Nelson C.J., Trösch J., Castleden I., Huang S., Millar A.H. Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell. 2017;29:207–228. doi: 10.1105/tpc.16.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Duncan O., Ganguly D.R., Lee C.P., Crisp P.A., Wijerathna-Yapa A., Salih K., Trösch J., Pogson B.J., Millar A.H. Enzymes degraded under high light maintain proteostasis by transcriptional regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2121362119. e2121362119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Kim C. Chloroplast ROS and stress signaling. Plant Commun. 2022;3:100264. doi: 10.1016/j.xplc.2021.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mu Y., Sun X., Zhang L. Increased sensitivity to drought stress in atlon4 Arabidopsis mutant. Chin. Sci. Bull. 2010;55:3668–3672. [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A.B., Adam Z., Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q., Jarvis P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 2015;25:2527–2534. doi: 10.1016/j.cub.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q., Jarvis P. Plant signaling: ubiquitin pulls the trigger on chloroplast degradation. Curr. Biol. 2016;26:R38–R40. doi: 10.1016/j.cub.2015.11.022. [DOI] [PubMed] [Google Scholar]
- Ling Q., Huang W., Baldwin A., Jarvis P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science. 2012;338:655–659. doi: 10.1126/science.1225053. [DOI] [PubMed] [Google Scholar]
- Ling Q., Broad W., Trösch R., Töpel M., Demiral Sert T., Lymperopoulos P., Baldwin A., Jarvis R.P. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science. 2019;363:eaav4467. doi: 10.1126/science.aav4467. [DOI] [PubMed] [Google Scholar]
- Ling Q., Sadali N.M., Soufi Z., Zhou Y., Huang B., Zeng Y., Rodriguez-Concepcion M., Jarvis R.P. The chloroplast-associated protein degradation pathway controls chromoplast development and fruit ripening in tomato. Nat. Plants. 2021;7:655–666. doi: 10.1038/s41477-021-00916-y. [DOI] [PubMed] [Google Scholar]
- Link S., Engelmann K., Meierhoff K., Westhoff P. The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol. 2012;160:2202–2218. doi: 10.1104/pp.112.205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016;211:418–428. doi: 10.1111/nph.13915. [DOI] [PubMed] [Google Scholar]
- Liu X., Yu F., Rodermel S. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 2010;154:1588–1601. doi: 10.1104/pp.110.164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Rodermel S.R., Yu F. A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 2010;10:287. doi: 10.1186/1471-2229-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas E., Pulido P., Rodriguez-Concepcion M. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet. 2017;13:e1007022. doi: 10.1371/journal.pgen.1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudya N., Bédard J., Ali S.M., Devlin P.F., Jarvis R.P., López-Juez E. Mutations in the chloroplast inner envelope protein TIC100 impair and repair chloroplast protein import and impact retrograde signalling. Plant Cell. 2022;34:3028–3046. doi: 10.1093/plcell/koac153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciński R., Misztal L., Samardakiewicz S., Jackowski G. The thylakoid protease Deg2 is involved in stress-related degradation of the photosystem II light-harvesting protein Lhcb6 in Arabidopsis thaliana. New Phytol. 2011;192:74–86. doi: 10.1111/j.1469-8137.2011.03782.x. [DOI] [PubMed] [Google Scholar]
- Luciński R., Misztal L., Samardakiewicz S., Jackowski G. Involvement of Deg5 protease in wounding-related disposal of PsbF apoprotein. Plant Physiol. Biochem. 2011;49:311–320. doi: 10.1016/j.plaphy.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Malnoë A., Wang F., Girard-Bascou J., Wollman F.A., de Vitry C. Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell. 2014;26:373–390. doi: 10.1105/tpc.113.120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Naranjo B., Wang J., Penzler J.F., Kleine T., Leister D. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 2019;99:521–535. doi: 10.1111/tpj.14342. [DOI] [PubMed] [Google Scholar]
- Mårtensson C.U., Priesnitz C., Song J., Ellenrieder L., Doan K.N., Boos F., Floerchinger A., Zufall N., Oeljeklaus S., Warscheid B., et al. Mitochondrial protein translocation-associated degradation. Nature. 2019;569:679–683. doi: 10.1038/s41586-019-1227-y. [DOI] [PubMed] [Google Scholar]
- May T., Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.E., Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K., den Camp R., Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Mielke K., Wagner R., Mishra L.S., Demir F., Perrar A., Huesgen P.F., Funk C. Abundance of metalloprotease FtsH12 modulates chloroplast development in Arabidopsis thaliana. J. Exp. Bot. 2021;72:3455–3473. doi: 10.1093/jxb/eraa550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras S., Salvi D., Piette L., Seigneurin-Berny D., Grunwald D., Reinbothe C., Joyard J., Reinbothe S., Rolland N. Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J. Biol. Chem. 2007;282:29482–29492. doi: 10.1074/jbc.M611112200. [DOI] [PubMed] [Google Scholar]
- Mishra L.S., Funk C. The FtsHi enzymes of Arabidopsis thaliana: pseudo-proteases with an important function. Int. J. Mol. Sci. 2021;22:5917. doi: 10.3390/ijms22115917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E., Kato Y., Matsushima R., Albrecht V., Laalami S., Sakamoto W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell. 2007;19:1313–1328. doi: 10.1105/tpc.106.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Schmidt R., Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Moldavski O., Levin-Kravets O., Ziv T., Adam Z., Prag G. The hetero-hexameric nature of a chloroplast AAA+ FtsH protease contributes to its thermodynamic stability. PLoS One. 2012;7:e36008. doi: 10.1371/journal.pone.0036008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montandon C., Friso G., Liao J.Y.R., Choi J., van Wijk K.J. In vivo trapping of proteins interacting with the chloroplast CLPC1 chaperone: potential substrates and adaptors. J. Proteome Res. 2019;18:2585–2600. doi: 10.1021/acs.jproteome.9b00112. [DOI] [PubMed] [Google Scholar]
- Moreau M., Westlake T., Zampogna G., Popescu G., Tian M., Noutsos C., Popescu S. The Arabidopsis oligopeptidases TOP1 and TOP2 are salicylic acid targets that modulate SA-mediated signaling and the immune response. Plant J. 2013;76:603–614. doi: 10.1111/tpj.12320. [DOI] [PubMed] [Google Scholar]
- Moreno J.C., Tiller N., Diez M., Karcher D., Tillich M., Schöttler M.A., Bock R. Generation and characterization of a collection of knock-down lines for the chloroplast Clp protease complex in tobacco. J. Exp. Bot. 2017;68:2199–2218. doi: 10.1093/jxb/erx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer S.K., Eichacker L.A. Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J. Biol. Chem. 1998;273:20935–20940. doi: 10.1074/jbc.273.33.20935. [DOI] [PubMed] [Google Scholar]
- Mullet J.E., Klein R.R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada A., Soll J. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci. 2004;117:3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- Nakai M. YCF1: a green TIC: response to the de Vries et al. commentary. Plant Cell. 2015;27:1834–1838. doi: 10.1105/tpc.15.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. REPLY: the revised model for chloroplast protein import. Plant Cell. 2020;32:543–546. doi: 10.1105/tpc.19.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Murakami S., Shoji T., Yoshida S., Yamada Y., Sato F. A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell. 1997;9:1673–1682. doi: 10.1105/tpc.9.9.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 1997;12:851–861. doi: 10.1046/j.1365-313x.1997.12040851.x. [DOI] [PubMed] [Google Scholar]
- Nakrieko K.A., Mould R.M., Smith A.G. Fidelity of targeting to chloroplasts is not affected by removal of the phosphorylation site from the transit peptide. Eur. J. Biochem. 2004;271:509–516. doi: 10.1046/j.1432-1033.2003.03950.x. [DOI] [PubMed] [Google Scholar]
- Nelson C.J., Li L., Millar A.H. Quantitative analysis of protein turnover in plants. Proteomics. 2014;14:579–592. doi: 10.1002/pmic.201300240. [DOI] [PubMed] [Google Scholar]
- Nelson C.J., Alexova R., Jacoby R.P., Millar A.H. Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiol. 2014;166:91–108. doi: 10.1104/pp.114.243014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Ben-Shem A. The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 2004;5:971–982. doi: 10.1038/nrm1525. [DOI] [PubMed] [Google Scholar]
- Nesbit A.D., Whippo C., Hangarter R.P., Kehoe D.M. Translation initiation factor 3 families: what are their roles in regulating cyanobacterial and chloroplast gene expression? Photosynth. Res. 2015;126:147–159. doi: 10.1007/s11120-015-0074-4. [DOI] [PubMed] [Google Scholar]
- Neuhaus H.E., Emes M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B. Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 2013;64:609–635. doi: 10.1146/annurev-arplant-050312-120124. [DOI] [PubMed] [Google Scholar]
- Nilsson Cederholm S., Bäckman H.G., Pesaresi P., Leister D., Glaser E. Deletion of an organellar peptidasome PreP affects early development in Arabidopsis thaliana. Plant Mol. Biol. 2009;71:497–508. doi: 10.1007/s11103-009-9534-6. [DOI] [PubMed] [Google Scholar]
- Nishimura K., van Wijk K.J. Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta. 2015;1847:915–930. doi: 10.1016/j.bbabio.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kato Y., Sakamoto W. Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol. 2016;171:2280–2293. doi: 10.1104/pp.16.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Kato Y., Sakamoto W. Essentials of proteolytic machineries in chloroplasts. Mol. Plant. 2017;10:4–19. doi: 10.1016/j.molp.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Apitz J., Friso G., Kim J., Ponnala L., Grimm B., van Wijk K.J. Discovery of a unique Clp component, ClpF, in chloroplasts: a proposed binary ClpF-ClpS1 adaptor complex functions in substrate recognition and delivery. Plant Cell. 2015;27:2677–2691. doi: 10.1105/tpc.15.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Asakura Y., Friso G., Kim J., Oh S.H., Rutschow H., Ponnala L., van Wijk K.J. ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell. 2013;25:2276–2301. doi: 10.1105/tpc.113.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil P.K., Richardson L.G.L., Paila Y.D., Piszczek G., Chakravarthy S., Noinaj N., Schnell D. The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proc. Natl. Acad. Sci. USA. 2017;114:E4868–E4876. doi: 10.1073/pnas.1621179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein C., Przybyla D., Danon A., Landgraf F., Göbel C., Imboden A., Feussner I., Apel K. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. Plant J. 2006;47:445–456. doi: 10.1111/j.1365-313X.2006.02793.x. [DOI] [PubMed] [Google Scholar]
- Olinares P.D.B., Kim J., van Wijk K.J. The Clp protease system: a central component of the chloroplast protease network. Biochim. Biophys. Acta. 2011;1807:999–1011. doi: 10.1016/j.bbabio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Osada Y., Saito R., Tomita M. Analysis of base-pairing potentials between 16S rRNA and 5' UTR for translation initiation in various prokaryotes. Bioinformatics. 1999;15:578–581. doi: 10.1093/bioinformatics/15.7.578. [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O., Kato Y., Adam Z., Sakamoto W. Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol. 2007;48:881–885. doi: 10.1093/pcp/pcm052. [DOI] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G.L., Schnell D.J. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 2015;427:1038–1060. doi: 10.1016/j.jmb.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G., Inoue H., Parks E.S., McMahon J., Inoue K., Schnell D.J. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. Elife. 2016;5:e12631. doi: 10.7554/eLife.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparelli E., Gonzali S., Parlanti S., Novi G., Giorgi F.M., Licausi F., Kosmacz M., Feil R., Lunn J.E., Brust H., et al. Misexpression of a chloroplast aspartyl protease leads to severe growth defects and alters carbohydrate metabolism in Arabidopsis. Plant Physiol. 2012;160:1237–1250. doi: 10.1104/pp.112.204016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Rodermel S.R. Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:12765–12770. doi: 10.1073/pnas.0402764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlaza K., Toutkoushian H., Boone M., Lam M., Iwai M., Jonikas M.C., Walter P., Ramundo S. The Mars1 kinase confers photoprotection through signaling in the chloroplast unfolded protein response. Elife. 2019;8:e49577. doi: 10.7554/eLife.49577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P., Llamas E., Llorente B., Ventura S., Wright L.P., Rodríguez-Concepción M. Specific Hsp100 chaperones determine the fate of the first enzyme of the plastidial isoprenoid pathway for either refolding or degradation by the stromal Clp protease in Arabidopsis. PLoS Genet. 2016;12:e1005824. doi: 10.1371/journal.pgen.1005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S., Becker T., Bionda T., Reger K., Ruprecht M., Soll J., Schleiff E. Toc64-a preprotein-receptor at the outer membrane with bipartide function. J. Mol. Biol. 2007;367:1330–1346. doi: 10.1016/j.jmb.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Ramundo S., Rochaix J.D. Chloroplast unfolded protein response, a new plastid stress signaling pathway? Plant Signal. Behav. 2014;9:e972874. doi: 10.4161/15592316.2014.972874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S., Casero D., Mühlhaus T., Hemme D., Sommer F., Crèvecoeur M., Rahire M., Schroda M., Rusch J., Goodenough U., et al. Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell. 2014;26:2201–2222. doi: 10.1105/tpc.114.124842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rei Liao J.Y., Friso G., Forsythe E.S., Michel E.J.S., Williams A.M., Boguraev S.S., Ponnala L., Sloan D.B., van Wijk K.J. Proteomics, phylogenetics, and coexpression analyses indicate novel interactions in the plastid CLP chaperone-protease system. J. Biol. Chem. 2022;298:101609. doi: 10.1016/j.jbc.2022.101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G.L., Schnell D.J. Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids. J. Exp. Bot. 2020;71:1226–1238. doi: 10.1093/jxb/erz517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA. 1998;95:7463–7468. doi: 10.1073/pnas.95.13.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J. Cell Biol. 1999;147:33–44. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S., Daras G., Laxa M., Marathias N., Fasseas C., Sweetlove L.J., Hatzopoulos P. Role of Lon1 protease in post-germinative growth and maintenance of mitochondrial function in Arabidopsis thaliana. New Phytol. 2009;181:588–600. doi: 10.1111/j.1469-8137.2008.02701.x. [DOI] [PubMed] [Google Scholar]
- Rogalski M., Schöttler M.A., Thiele W., Schulze W.X., Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–2237. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Hernández G., Grant R.A., Sauer R.T., Baker T.A. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc. Natl. Acad. Sci. USA. 2009;106:8888–8893. doi: 10.1073/pnas.0903614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadali N.M., Sowden R.G., Ling Q., Jarvis R.P. Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep. 2019;38:803–818. doi: 10.1007/s00299-019-02420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Tamura T., Hanba-Tomita Y., Murata M., Sodmergen The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Gene Cell. 2002;7:769–780. doi: 10.1046/j.1365-2443.2002.00558.x. [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Childs L., Walther D., Bock R. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 2011;7:e1002155. doi: 10.1371/journal.pgen.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff L.B., Ehrnthaler M., Janowski M., Childs L.H., Hasse C., Gremmels J., Ruf S., Zoschke R., Bock R. Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in Tobacco. Plant Cell. 2017;29:3085–3101. doi: 10.1105/tpc.17.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Zahn R., Bukau B., Mogk A. ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol. Microbiol. 2009;72:506–517. doi: 10.1111/j.1365-2958.2009.06666.x. [DOI] [PubMed] [Google Scholar]
- Schreier T.B., Cléry A., Schläfli M., Galbier F., Stadler M., Demarsy E., Albertini D., Maier B.A., Kessler F., Hörtensteiner S., et al. Plastidial NAD-dependent malate dehydrogenase: a moonlighting protein involved in early Chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell. 2018;30:1745–1769. doi: 10.1105/tpc.18.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K., Meierhoff K., Paradies S., Töller T., Wolff P., Westhoff P. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell. 2007;19:1329–1346. doi: 10.1105/tpc.106.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Fernie A.R., Geigenberger P., Leister D., Möhlmann T., Naranjo B., Neuhaus H.E. Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. 2022;27:577–587. doi: 10.1016/j.tplants.2021.12.004. [DOI] [PubMed] [Google Scholar]
- Sedaghatmehr M., Mueller-Roeber B., Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat. Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya V.P., Barbeau W.A., Xiao T., Knutson C.S., Schuler M.H., Hughes A.L. A nuclear-based quality control pathway for non-imported mitochondrial proteins. Elife. 2021;10:e61230. doi: 10.7554/eLife.61230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugabalaji V., Chahtane H., Accossato S., Rahire M., Gouzerh G., Lopez-Molina L., Kessler F. Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 2018;28:2616–2623.e5. doi: 10.1016/j.cub.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell. 2010;22:205–220. doi: 10.1105/tpc.109.071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta. 2013;1833:314–331. doi: 10.1016/j.bbamcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 2001;42:264–273. doi: 10.1093/pcp/pce031. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Asakura Y., Imai M., Nakahira Y., Kotani Y., Hashiguchi Y., Nakai Y., Takafuji K., Bédard J., Hirabayashi-Ishioka Y., et al. A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell. 2018;30:2677–2703. doi: 10.1105/tpc.18.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman-Roston R.L., Ruppel N.J., Damoc C., Phinney B.S., Inoue K. The significance of protein maturation by plastidic type I signal peptidase 1 for thylakoid development in Arabidopsis chloroplasts. Plant Physiol. 2010;152:1297–1308. doi: 10.1104/pp.109.151977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman R.L., Inoue K. Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett. 2009;583:938–942. doi: 10.1016/j.febslet.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Sjögren L.L.E., Clarke A.K. Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell. 2011;23:322–332. doi: 10.1105/tpc.110.082321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L.E., MacDonald T.M., Sutinen S., Clarke A.K. Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 2004;136:4114–4126. doi: 10.1104/pp.104.053835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L.E., Stanne T.M., Zheng B., Sutinen S., Clarke A.K. Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell. 2006;18:2635–2649. doi: 10.1105/tpc.106.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L.E., Tanabe N., Lymperopoulos P., Khan N.Z., Rodermel S.R., Aronsson H., Clarke A.K. Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 2014;289:11318–11330. doi: 10.1074/jbc.M113.534552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.D., Rounds C.M., Wang F., Chen K., Afitlhile M., Schnell D.J. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 2004;165:323–334. doi: 10.1083/jcb.200311074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M., Rudolf M., Tillmann B., Tripp J., Sommer M.S., Schleiff E. Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ. 2013;36:970–983. doi: 10.1111/pce.12030. [DOI] [PubMed] [Google Scholar]
- Ståhl A., Nilsson S., Lundberg P., Bhushan S., Biverståhl H., Moberg P., Morisset M., Vener A., Mäler L., Langel U., et al. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J. Mol. Biol. 2005;349:847–860. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Su P.-H., Li H.-M. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell. 2010;22:1516–1531. doi: 10.1105/tpc.109.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Essays Biochem. 1995;30:49–57. [PubMed] [Google Scholar]
- Sun J.L., Li J.Y., Wang M.J., Song Z.T., Liu J.X. Protein quality control in plant organelles: current progress and future perspectives. Mol. Plant. 2021;14:95–114. doi: 10.1016/j.molp.2020.10.011. [DOI] [PubMed] [Google Scholar]
- Sun R., Fan H., Gao F., Lin Y., Zhang L., Gong W., Liu L. Crystal structure of Arabidopsis Deg2 protein reveals an internal PDZ ligand locking the hexameric resting state. J. Biol. Chem. 2012;287:37564–37569. doi: 10.1074/jbc.M112.394585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., Chi W., Ma J., Lu C., Zhang L. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell. 2007;19:1347–1361. doi: 10.1105/tpc.106.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 2010;152:1263–1273. doi: 10.1104/pp.109.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Nakanishi H., Bower J., Yoder D.W., Osteryoung K.W., Miyagishima S.Y. Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol. 2009;9:38. doi: 10.1186/1471-2229-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadini L., Peracchio C., Trotta A., Colombo M., Mancini I., Jeran N., Costa A., Faoro F., Marsoni M., Vannini C., et al. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 2020;101:1198–1220. doi: 10.1111/tpj.14585. [DOI] [PubMed] [Google Scholar]
- Tadini L., Pesaresi P., Kleine T., Rossi F., Guljamow A., Sommer F., Mühlhaus T., Schroda M., Masiero S., Pribil M., et al. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 2016;170:1817–1830. doi: 10.1104/pp.15.02033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi K., Sodmergen, Murata M., Motoyoshi F., Sakamoto W. The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 2000;41:1334–1346. doi: 10.1093/pcp/pcd067. [DOI] [PubMed] [Google Scholar]
- Teng Y.S., Chan P.T., Li H.M. Differential age-dependent import regulation by signal peptides. PLoS Biol. 2012;10:e1001416. doi: 10.1371/journal.pbio.1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J., Schroda M. Revisiting the photosystem II repair cycle. Plant Signal. Behav. 2016;11:e1218587. doi: 10.1080/15592324.2016.1218587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S.M., Pulido P., Jarvis R.P. Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 2020;48:71–82. doi: 10.1042/BST20190274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Tokumaru M., Adachi F., Toda M., Ito-Inaba Y., Yazu F., Hirosawa Y., Sakakibara Y., Suiko M., Kakizaki T., Inaba T. Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol. 2017;173:524–535. doi: 10.1104/pp.16.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A. Control of the protein turnover rates in Lemna minor. Plant Physiol. 1972;49:47–51. doi: 10.1104/pp.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Jarvis P. The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS One. 2011;6:e23039. doi: 10.1371/journal.pone.0023039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Ries F., Westrich L.D., Gao Y., Herkt C., Hoppstädter J., Heck-Roth J., Mustas M., Scheuring D., Choquet Y., et al. Fast and global reorganization of the chloroplast protein biogenesis network during heat acclimation. Plant Cell. 2022;34:1075–1099. doi: 10.1093/plcell/koab317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 2015;66:75–111. doi: 10.1146/annurev-arplant-043014-115547. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Baginsky S. Plastid proteomics in higher plants: current state and future goals. Plant Physiol. 2011;155:1578–1588. doi: 10.1104/pp.111.172932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J., Andersson B., Aro E.M. Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 1996;271:9627–9636. doi: 10.1074/jbc.271.16.9627. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedalankar P., Tripathy B.C. Evolution of light-independent protochlorophyllide oxidoreductase. Protoplasma. 2019;256:293–312. doi: 10.1007/s00709-018-1317-y. [DOI] [PubMed] [Google Scholar]
- Vitlin Gruber A., Nisemblat S., Azem A., Weiss C. The complexity of chloroplast chaperonins. Trends Plant Sci. 2013;18:688–694. doi: 10.1016/j.tplants.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Wagner R., Aigner H., Pružinská A., Jänkänpää H.J., Jansson S., Funk C. Fitness analyses of Arabidopsis thaliana mutants depleted of FtsH metalloproteases and characterization of three FtsH6 deletion mutants exposed to high light stress, senescence and chilling. New Phytol. 2011;191:449–458. doi: 10.1111/j.1469-8137.2011.03684.x. [DOI] [PubMed] [Google Scholar]
- Wallas T.R., Smith M.D., Sanchez-Nieto S., Schnell D.J. The roles of Toc34 and Toc75 in targeting the Toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 2003;278:44289–44297. doi: 10.1074/jbc.M307873200. [DOI] [PubMed] [Google Scholar]
- Walley J., Xiao Y., Wang J.Z., Baidoo E.E., Keasling J.D., Shen Z., Briggs S.P., Dehesh K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2015;112:6212–6217. doi: 10.1073/pnas.1504828112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim C., Xu X., Piskurewicz U., Dogra V., Singh S., Mahler H., Apel K. Singlet oxygen- and EXECUTER1-mediated signaling is initiated in grana margins and depends on the protease FtsH2. Proc. Natl. Acad. Sci. USA. 2016;113:E3792–E3800. doi: 10.1073/pnas.1603562113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S.J., Li N., Ye Y., Wu F., Ling Q., Jarvis R.P. Crosstalk between the chloroplast protein import and SUMO systems revealed through genetic and molecular investigation in Arabidopsis. Elife. 2021;10:e60960. doi: 10.7554/eLife.60960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B.M., Gildea H.K., Dillin A. Protein homeostasis from the outside in. Nat. Cell Biol. 2020;22:911–912. doi: 10.1038/s41556-020-0534-8. [DOI] [PubMed] [Google Scholar]
- Wei J., Qiu X., Chen L., Hu W., Hu R., Chen J., Sun L., Li L., Zhang H., Lv Z., et al. The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis. J. Exp. Bot. 2015;66:5809–5820. doi: 10.1093/jxb/erv286. [DOI] [PubMed] [Google Scholar]
- Weidberg H., Amon A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science. 2018;360:eaan4146. doi: 10.1126/science.aan4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Zhou X., Yuan H., Álvarez D., Sun T., Schlossarek D., Yang Y., Shen G., Zhang H., Rodriguez-Concepcion M., et al. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol. Plant. 2018;11:149–162. doi: 10.1016/j.molp.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Wetzel C.M., Harmacek L.D., Yuan L.H., Wopereis J.L.M., Chubb R., Turini P. Loss of chloroplast protease SPPA function alters high light acclimation processes in Arabidopsis thaliana L. (Heynh.) J. Exp. Bot. 2009;60:1715–1727. doi: 10.1093/jxb/erp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer D., Bohnhorst P., Shekhar V., Hwang I., Offermann S. Transit peptide elements mediate selective protein targeting to two different types of chloroplasts in the single-cell C4 species Bienertia sinuspersici. Sci. Rep. 2017;7:41187. doi: 10.1038/srep41187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Joens M.S., Sinson A.B., Gilkerson J., Salomé P.A., Weigel D., Fitzpatrick J.A., Chory J. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science. 2015;350:450–454. doi: 10.1126/science.aac7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L., Topf U., Bragoszewski P., Wiese S., Sztolsztener M.E., Oeljeklaus S., Varabyova A., Lirski M., Chroscicki P., Mroczek S., et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- Wu G.Z., Bock R. GUN control in retrograde signaling: how GENOMES UNCOUPLED proteins adjust nuclear gene expression to plastid biogenesis. Plant Cell. 2021;33:457–474. doi: 10.1093/plcell/koaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.Z., Meyer E.H., Wu S., Bock R. Extensive post-transcriptional regulation of nuclear gene expression by plastid retrograde signals. Plant Physiol. 2019;180:2034–2048. doi: 10.1104/pp.19.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.Z., Chalvin C., Hoelscher M., Meyer E.H., Wu X.N., Bock R. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 2018;176:2472–2495. doi: 10.1104/pp.18.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.Z., Meyer E.H., Richter A.S., Schuster M., Ling Q., Schöttler M.A., Walther D., Zoschke R., Grimm B., Jarvis R.P., et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants. 2019;5:525–538. doi: 10.1038/s41477-019-0415-y. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Wittek D., Gupta R., Qin B., Ueda T., Krause R., Yamamoto K., Albrecht R., Pech M., Nierhaus K.H. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc. Natl. Acad. Sci. USA. 2016;113:E1180–E1189. doi: 10.1073/pnas.1524554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Burke J., Autz G., Jagendorf A.T. Bound ribosomes of pea chloroplast thylakoid membranes: location and release in vitro by high salt, puromycin, and RNase. Plant Physiol. 1981;67:940–949. doi: 10.1104/pp.67.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Liu X., Alsheikh M., Park S., Rodermel S. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell. 2008;20:1786–1804. doi: 10.1105/tpc.107.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabret J., Bohn S., Schuller S.K., Arnolds O., Möller M., Meier-Credo J., Liauw P., Chan A., Tajkhorshid E., Langer J.D., et al. Structural insights into photosystem II assembly. Nat. Plants. 2021;7:524–538. doi: 10.1038/s41477-021-00895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Ori N., Adam Z. Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell. 2005;17:2782–2790. doi: 10.1105/tpc.105.035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Feder A., Adam Z. Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts-implications for thylakoid formation and photosystem II maintenance. Plant J. 2005;42:609–617. doi: 10.1111/j.1365-313X.2005.02401.x. [DOI] [PubMed] [Google Scholar]
- Zelisko A., García-Lorenzo M., Jackowski G., Jansson S., Funk C. AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA. 2005;102:13699–13704. doi: 10.1073/pnas.0503472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Liu C. Chloroplast chaperonin: an intricate protein folding machine for photosynthesis. Front. Mol. Biosci. 2017;4:98. doi: 10.3389/fmolb.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A.K. Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol. Plant. 2002;114:92–101. doi: 10.1034/j.1399-3054.2002.1140113.x. [DOI] [PubMed] [Google Scholar]
- Zheng B., MacDonald T.M., Sutinen S., Hurry V., Clarke A.K. A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta. 2006;224:1103–1115. doi: 10.1007/s00425-006-0292-2. [DOI] [PubMed] [Google Scholar]
- Zheng X.T., Wang C., Lin W., Lin C., Han D., Xie Q., Lai J., Yang C. Importation of chloroplast proteins under heat stress is facilitated by their SUMO conjugations. New Phytol. 2022;235:173–187. doi: 10.1111/nph.18121. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Barkan A. Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. USA. 2015;112:E1678–E1687. doi: 10.1073/pnas.1424655112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Kroeger T., Belcher S., Schöttler M.A., Barkan A., Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012;72:547–558. doi: 10.1111/j.1365-313X.2012.05081.x. [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Friso G., Kim J., Rudella A., Rodríguez V.R., Asakura Y., Sun Q., van Wijk K.J. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics. 2009;8:1789–1810. doi: 10.1074/mcp.M900104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]