Abstract
The cytochrome b6f (Cyt b6f) complex is a multisubunit protein complex in chloroplast thylakoid membranes required for photosynthetic electron transport. Here we report the isolation and characterization of the new tiny albino 1 (nta1) mutant in Arabidopsis, which has severe defects in Cyt b6f accumulation and chloroplast development. Gene cloning revealed that the nta1 phenotype was caused by disruption of a single nuclear gene, NTA1, which encodes an integral thylakoid membrane protein conserved across green algae and plants. Overexpression of NTA1 completely rescued the nta1 phenotype, and knockout of NTA1 in wild-type plants recapitulated the mutant phenotype. Loss of NTA1 function severely impaired the accumulation of multiprotein complexes related to photosynthesis in thylakoid membranes, particularly the components of Cyt b6f. NTA1 was shown to directly interact with four subunits (Cyt b6/PetB, PetD, PetG, and PetN) of Cyt b6f through the DUF1279 domain and C-terminal sequence to mediate their assembly. Taken together, our results identify NTA1 as a new and key regulator of chloroplast development that plays essential roles in assembly of the Cyt b6f complex by interacting with multiple Cyt b6f subunits.
Key words: albino mutant, chloroplast development, cytochrome b6f complex, assembly factor, Arabidopsis thaliana
The Cyt b6f complex is critical for photosynthetic electron transport, but the mechanisms of its biogenesis and assembly have not been fully revealed. This study reports that the nucleus-encoded thylakoid membrane protein NTA1 mediates Cyt b6f complex assembly by directly interacting with its four subunits. Loss of NTA1 results in albino plants with greatly reduced accumulation of the Cyt b6f complex.
Introduction
Plant photosynthesis takes place in two major phases: light-dependent and light-independent reactions. Light-dependent reactions involve the capture of solar energy by thylakoid membrane–localized pigment–protein complexes and coupled electron transfer reactions, through which the captured light energy is converted to chemical energy in the form of ATP and NADPH (Allen, 2002; Blankenship, 2014). The photosynthetic electron transfer chain is made up of photosystem II (PSII), the cytochrome b6f complex (Cyt b6f), photosystem I (PSI), and the electron carriers plastoquinone and plastocyanin (Joliot and Johnson, 2011). The activity of the Cyt b6f complex is a key determinant of the rate of electron transport (Yamori et al., 2011; Malone et al., 2021).
Cyt b6f is a multisubunit pigment–protein complex that contains one molecule of chlorophyll a and one molecule of β-carotene. Cyt b6f occupies an electrochemically central position in the linear electron transport chain responsible for electron transport and energy transduction between PSII and PSI (Cramer et al., 2006; Joliot and Johnson, 2011; Malone et al., 2021). Cyt b6f is also involved in cyclic electron transport around PSI, in which electrons can be recycled from either reduced ferredoxin or NADPH to plastoquinone and subsequently to Cyt b6f to drive formation of ATP, providing it to the Calvin cycle (Joliot et al., 2006; Dumas et al., 2016; Buchert et al., 2020). Apart from its significant role in electron transport, Cyt b6f also participates in regulating the processes of state transition, redox regulation, and photoprotection (Bellafiore et al., 2005; Dumas et al., 2017; Buchert et al., 2022). In addition, an increase in Cyt b6f activity can improve the efficiency of photosynthesis and stimulate plant growth (Simkin et al., 2017; Ermakova et al., 2019).
The resolved crystal structure of Cyt b6f reveals a highly conserved organization and a dimer structure in the native form, which is the functional form of Cyt b6f and can be converted into a monomer with detergents (Kurisu et al., 2003; Zhang and Cramer, 2004; Baniulis et al., 2008; Malone et al., 2019). The monomeric complex comprises four large subunits (cytochrome f [Cyt f], cytochrome b6 [Cyt b6], the Rieske–FeS protein [PetC], and PetD) and four small subunits (PetG, PetL, PetM, and PetN) and binds several co-factors, the heme c of cytochrome f and the newly discovered heme ci′ covalently attached to Cyt b6 in the quinone binding site Qi (Kurisu et al., 2003; Stroebel et al., 2003). PetC and PetM are encoded by the nuclear genome, and the others are encoded by plastid genes.
Despite the detailed knowledge about the structure and composition of the Cyt b6f complex, much less is known about the mechanisms of its biogenesis and assembly. The biogenesis of Cyt b6f is a sophisticated process regulated at the transcriptional, translational, and post-translational levels and requires coordinated gene expression in the nucleus and chloroplast (Bruce and Malkin, 1991; Saint-Marcoux et al., 2009; Boulouis et al., 2011; Wang and Grimm, 2021). In addition to the eight known subunits, the biogenesis and assembly of the Cyt b6f complex also requires other co-factors and auxiliary proteins. For example, several proteins in the co-factor assembly, complex C, subunit B (PetB) pathway are required for heme binding of Cyt b6 in both Arabidopsis and the green algae Chlamydomonas, and their loss-of-function mutants show severe defects in Cyt b6f accumulation and growth phenotypes (Lezhneva et al., 2008; Saint-Marcoux et al., 2009). HIGH CHLOROPHYLL FLUORESCENCE 164 (HCF164), a thioredoxin-like protein, and HCF222, a DnaJ-like zinc-finger protein, modulate Cyt b6f levels through their disulfide reductase activities (Lennartz et al., 2001; Hartings et al., 2017), and the thylakoid membrane protein DEFECTIVE ACCUMULATION OF Cyt b6f (DAC) is likely involved in the assembly/stabilization of Cyt b6f by interacting with the PetD subunit (Xiao et al., 2012). These nuclear-encoded protein factors therefore have important roles in Cyt b6f biogenesis or accumulation, and some are involved in the assembly of Cyt b6f.
Very recently, another nuclear-encoded protein, DE-ETIOLATION-INDUCED PROTEIN 1 (DEIP1), was reported to play an important role in the assembly of Cyt b6f components. The deip1 mutant in Arabidopsis showed a specific loss of Cyt b6f, and the DEIP1 protein interacts with PetA and PetB, two subunits of Cyt b6f, and mediates the assembly of intermediates in Cyt b6f biogenesis (Sandoval-Ibáñez et al., 2022). In this study, we report the isolation and characterization of the nta1 mutant and the causal protein NTA1. nta1 was identified from a transfer DNA (T-DNA) activation tagging–mediated genetic mutant screen in Arabidopsis and is defective in the same gene locus as the deip1 mutant, AT2G27290. Similar to deip1, nta1 had severe defects in chloroplast development and plant growth, including albinism, seedling lethality, and collapse of thylakoid membranes. Functional characterization indicated that NTA1 localizes in the thylakoid membrane and interacts with four subunits of Cyt b6f (Cyt b6/PetB, PetD, PetG, and PetN) to affect their assembly. Our study provides new molecular, genetic, and biochemical evidence confirming that NTA1/DEIP1 is essential for chloroplast development during the seedling greening process and for accumulation of multiprotein complexes related to photosynthesis in thylakoid membranes, particularly Cyt b6f. In addition, we demonstrate that the C terminus is critical for maintaining the normal function of NTA1.
Results
Isolation and characterization of the nta1 mutant
The nta1 mutant was identified from a T-DNA activation tagging–based genetic mutant screen in Arabidopsis. Activation tagging usually causes gain-of-function mutations (Weigel et al., 2000); however, nta1 turned out to be a recessive mutant. Mutant segregation and genotyping by PCR analysis indicated that the albino nta1 phenotype segregated in a recessive way and was caused by a single nuclear gene (Figure 1A and Supplemental Figure 1A and 1B). When grown on half-strength Murashige and Skoog (½ MS) medium supplemented with 1% sucrose, the mutant terminated its growth quickly after germination and produced only pale-green true leaves (Figure 1B and Supplemental Figure 1C). After transplant to soil, these seedlings could not grow further and eventually died, suggesting that this mutation cannot support photoautotrophic growth and is seedling lethal. Because some reported albino mutants, such as sco (Ruppel and Hangarter, 2007) and spd1 (Ruppel et al., 2011), can produce green true leaves and develop into normal adult plants when supplied with higher concentrations of sucrose (2%–5%) in the growth medium, we tested whether nta1 phenotypes could also be rescued by higher concentrations of sucrose. On 3% and 5% sucrose, the mutant indeed grew better and for a longer time (Supplemental Figure 1D and 1E), but it still did not survive after transfer to soil, indicating that higher sucrose can improve nta1 growth but cannot rescue its lethality. To test whether the albino phenotype of nta1 can be corrected by any plant hormones, we examined its responses to several plant hormones at different concentrations. None of the tested hormones, including auxin, gibberellin, brassinolide (the most active form of brassinosteroid), cytokinin, and 1-aminocyclopropane-1-carboxylic acid (a precursor of ethylene), could rescue the mutant phenotype (Supplemental Figure 1F). All these results suggest that the nta1 mutant is different from any previously known albino mutants; therefore, we named it new tiny albino 1 (nta1).
Figure 1.
nta1 mutant phenotypes and cloning of the NTA1 gene.
(A)nta1 is a recessive albino mutant that segregates in a 1:3 ratio with wild-type-looking plants. The white arrowheads denote the albino plants segregated from a T2 generation. The inset on the upper left shows an enlarged image of the nta1 mutant. Scale bar, 0.1 cm. Plants were grown on half-strength MS medium containing 1% sucrose under a 16-h light (100 μmol photons m−2 s−1)/8-h dark cycle at 22°C.
(B) The nta1 mutant grew very slowly on ½ MS + 1% sucrose medium compared with the wild-type Col-0 plants.
(C) Identification of T-DNA insertion sites in the nta1 genome by genome resequencing using the Illumina HiSeq 2500 platform. The colored bars in the outer circle represent the five chromosomes of Arabidopsis. The dark gray histogram in the inner circle indicates the sequencing depth (0–150×) of the whole-genome resequencing reads in each 10-kb window. Four T-DNA insertion sites (a, b, c, and d) were identified in chromosome 2.
(D) A schematic diagram of the NTA1 gene structure showing the translation initiation codon (ATG), exons (black boxes), introns (black lines between exons), and 5′ and 3′ UTRs (white boxes). One of the T-DNA insertions (d) was inserted in the second intron of the NTA1 (AT2G27290) gene, which caused the nta1 phenotype. NTA1-S1 and NTA1-N1 indicate the two sequences mutated by CRISPR-Cas9, and NTA1-WT shows the wild-type NTA1 sequences selected for CRISPR targeting. The three-letter protospacer adjacent motif is highlighted in blue. P1 and P2 show the primer positions used for NTA1 (AT2G27290) gene expression analysis in NTA1-related mutants (Supplemental Figures 3B and 4B).
(E) Phenotypes of two representative nta1 lines complemented by overexpression of AT2G27290 (Com1 and Com2).
(F) Phenotypes of NTA1-S1 and NTA1-N1 mutants generated by CRISPR-Cas9. The targeting sequences in the NTA1-S1 and NTA1-N1 plants are shown in (D). The deleted nucleotide is shown as a solid dash mark, and the inserted nucleotide is displayed with a red capital letter.
Consistent with their albino phenotype, the mutant plants had significantly lower chlorophyll and carotenoid contents compared with the wild-type (WT) plants, and the degree of decrease was correlated with the developmental stage (Supplemental Figure 2A–2D). The mutants were also demonstrated to be photosynthetically defective (Supplemental Table 1 and Supplemental Figure 2E). The ratio of variable to maximum fluorescence, Fv/Fm, an indicator of the maximum quantum yield of PSII, was reduced to 0.404 in nta1 from the WT value of 0.775. The effective quantum yield of PSII (ΦPSII) was reduced to 0.0363 in nta1 from 0.467 in WT. In addition, the photochemical quenching (qP) dropped to nearly zero in nta1, indicating that the electrons from PSII accumulated in the plastoquinone pool and could not be immediately transported via the electron transport chain. A clear difference was also observed in the quantum yield of non-regulated non-photochemical energy loss in PSII (Y(NO)). The Y(NO) value in WT was 0.306, whereas it was more than doubled in nta1 (0.809). Thus, nta1 is severely disturbed in photosynthetic activity, and most of the energy absorbed is lost in an uncontrolled manner in nta1.
NTA1 gene cloning, nta1 phenotype complementation, and recapitulation
To identify the gene that caused the nta1 phenotype, both whole-genome resequencing and adapter ligation-mediated PCR cloning (O’Malley et al., 2007) were performed in nta1. A total of four T-DNA insertions in the nta1 genome were identified and were all located on chromosome 2, with two in the intergenic region, one in the second exon of the AT2G23980 gene, and one in the last intron of the AT2G27290 gene (Figure 1C and 1D and Supplemental Figure 3). Because a previous study reported that mutants of AT2G23980 have no growth phenotypes (Gao et al., 2012), we focused our attention on AT2G27290. Expression of AT2G27290 was barely detectable at both the RNA and protein levels in the nta1 mutant (Supplemental Figure 4B and 4D). In GenBank, AT2G27290 was annotated as encoding a chloroplast-localized uncharacterized protein. All these data and information suggest that AT2G27290 is likely to be the gene that causes the nta1 phenotype.
To confirm that AT2G27290 is the causal gene of the nta1 mutant phenotype, we first performed nta1 phenotype complementation. The expression of AT2G27290 in nta1 completely rescued its albino phenotype and other associated abnormalities, including dwarfism and lethality (Figure 1E and Supplemental Figure 4A). Consistent with the restored phenotype, the chlorophyll and carotenoid contents of the complementation lines were also recovered to WT levels (Supplemental Figure 2A–2D). All these results suggest that AT2G27290 is the gene that causes the nta1 phenotype.
To further confirm that AT2G27290 is the NTA1 gene, we performed phenotype recapitulation of the nta1 mutant by CRISPR-Cas9 gene editing. Two 19-bp sequences were chosen as the target sites, one in exon 1 (S site) and the other in exon 2 (N site) of AT2G27290 (Figure 1D). Two homozygous lines, NTA1-S1 and NTA1-N1, corresponding to the two target sites, showed the same albino phenotype as the original nta1 mutant, with tiny and albino rosettes, suggesting that the nta1 phenotype was successfully copied by disrupting the AT2G27290 gene (Figure 1D and 1F and Supplemental Figure 4E). The nta1 mutant complementation and recapitulation experiments thus confirmed that AT2G27290 is the NTA1 gene whose mutation causes the nta1 mutant phenotype.
Characterization of the NTA1 protein
The NTA1 protein is made of 201 amino acids with a chloroplast transit peptide (cTP) at the N terminus (amino acids 1–39) and an uncharacterized domain, DUF1279 (amino acids 104–184) (Figure 3A). Amino acid sequence analysis revealed that NTA1 orthologs are widespread in green algae and lower and higher plant species (Supplemental Figure 5A). Domain analysis revealed that all putative NTA1 orthologs contain a highly conserved DUF1279 domain (Supplemental Figure 5B). Thus, the NTA1-homologous gene family may have been conserved during evolution from ancestral green algae to higher plants.
Figure 3.
Subcellular localization and topology of the NTA1 protein.
(A) Domain organization of NTA1 protein in Arabidopsis. The chloroplast transit peptide (cTP) and DUF1279 domain are shown in light-green and orange boxes, respectively. aa, amino acid.
(B) Subcellular localization of YFP-tagged full-length NTA1 (NTA1:YFP), NTA1 lacking the transit peptide (NTA1ΔcTP:YFP), transit peptide (cTP:YFP), and YFP alone (YFP) in protoplasts made from the corresponding transgenic plants. YFP fluorescence (YFP), chlorophyll autofluorescence (chlorophyll), bright-field, and merged images are shown. Scale bars, 20 μm.
(C) NTA1 localizes to the thylakoid fraction. Immunoblots of total protein (total) and protein fractions isolated from chloroplasts, stroma, and thylakoids of wild-type (Col-0) plants were analyzed with antibodies against NTA1, OEC33 (thylakoid marker, luminal protein of PSII), Lhcb1 (thylakoid marker, integral membrane protein), RbcL (stroma marker), and FNR (thylakoid and stroma markers).
(D) Thermolysin treatment of thylakoid membranes. Thylakoids of Col-0 plants were treated with thermolysin for 15, 30, and 45 min on ice and then separated by SDS–PAGE and immunoblotted with anti-OEC33, anti-PsaD, and anti-NTA1 antibodies.
(E) The association of NTA1 with the thylakoid membrane. Wild-type thylakoid membranes were incubated with 1 M NaCl, 1 M CaCl2, and 0.1 M Na2CO3 (pH 11.5) for 30 min on ice. Then the membrane pellet and supernatant (Sup.) were separated by SDS–PAGE and immunoblotted with antibodies against NTA1, OEC33, and CP47 (a PSII core protein). Untreated thylakoid membranes were used as a control.
(F and G) Two possible topologies of NTA1. Predicted localization of the N terminus in the stroma (F) or lumen (G). The short purple lines next to the red line indicate the protein region for NTA1 antibody production.
NTA1 protein accumulates in all tested tissues, with the highest levels in leaves (Supplemental Figure 5C). The NTA1 level decreased markedly when the seedlings entered the senescence stage (10 weeks of age) (Supplemental Figure 5D). In addition, the level of NTA1 increased during the process of light-induced seedling greening, consistent with changes in the levels of the reference proteins Cyt b6f subunit Cyt f, PSII protein OEC33, and light-harvesting chlorophyll a/b binding protein complex (LHC) subunit Lhcb1 involved in photosynthesis (Supplemental Figure 5E). In conclusion, NTA1 accumulates in all photosynthetic tissues and is induced by light during seedling greening.
NTA1 is crucial for chloroplast development during the seedling greening process
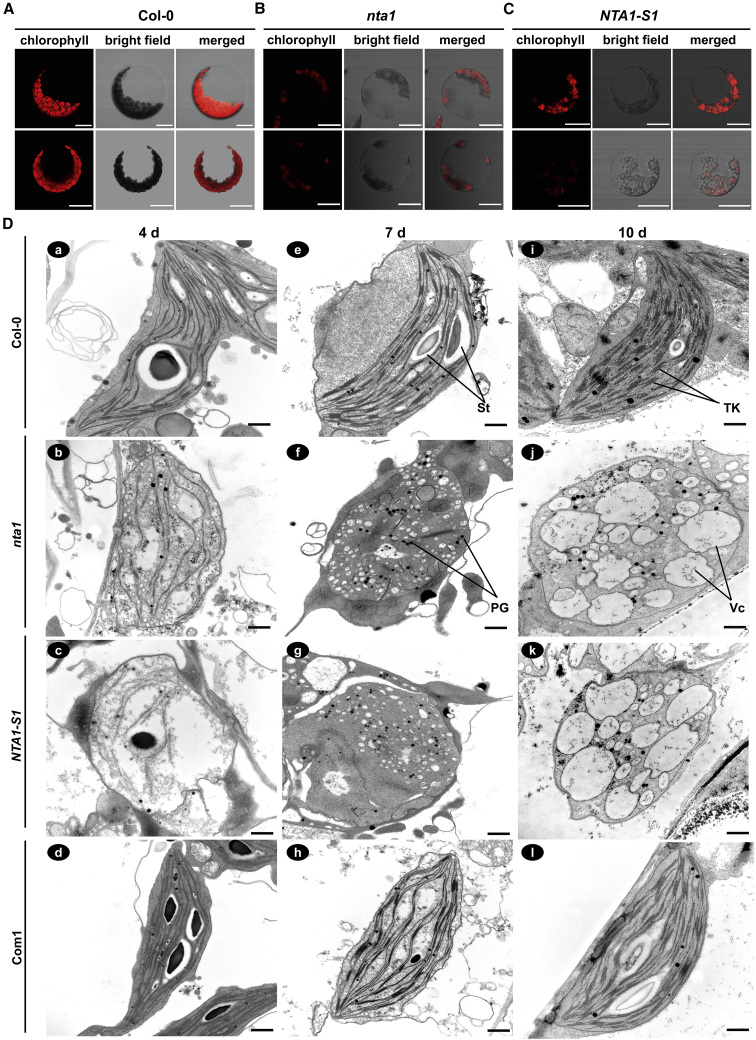
We next investigated whether the chloroplasts were developmentally malformed in NTA1-related mutants. Detailed microscopy observations showed that the cotyledon cells of 10-day-old nta1 and NTA1-S1 mutants exhibited chloroplast abnormalities, including reduced chloroplast number, abnormal chloroplast structure, and lower intensity of chlorophyll autofluorescence, compared with the WT (Figure 2A–2C). To further clarify whether NTA1-related mutants had defects in chloroplast ultrastructure, chloroplasts from cotyledons of 4-, 7-, and 10-day-old mutants were observed under a transmission electron microscope (TEM). In 4-day-old nta1 and NTA1-S1 cotyledons, the chloroplasts had few and abnormal internal membranes, and the thylakoids were considerably less pigmented; some chloroplasts were broken, and some lacked stacked grana compared with those of the WT and the complemented Com1 line (Figure 2D, b and c vs. a and d; Supplemental Figure 6A, d vs. c). No starch granules were observed in the chloroplasts of the mutant plants (Figure 2D, b and c vs. a and d; Supplemental Figure 6A, d vs. c). Similarly, chloroplasts in 7-day-old mutant plants contained very few thylakoid membrane systems but were filled with large vesicles and many densely stained globule structures, similar to plastoglobuli (Figure 2D, f and g vs. e and h). Strikingly, at the 10-day-old seedling stage, chloroplasts of nta1 and NTA1-S1 mutants were completely devoid of thylakoids and more vacuolated (Figure 2D, j and k vs. i and l). Together, these results demonstrate that NTA1 is essential for the maintenance of normal chloroplast structure during seedling development.
Figure 2.
Effects of NTA1 mutations on chloroplast development.
(A–C) Protoplasts isolated from the cotyledons of 10-day-old wild-type (A), nta1(B), and NTA1-S1(C) plants observed under a confocal microscope. Chloroplasts are shown by the chlorophyll autofluorescence. Two representative types of chloroplasts with different distributions are shown (top and bottom images). Scale bars, 20 μm.
(D) Ultrastructure of chloroplasts in cotyledons of 4-day-old (a–d), 7-day-old (e–h), and 10-day-old (i–l) wild type (Col-0), nta1, NTA1-S1, and complementation line (Com1). St, starch granule; TK, thylakoid; Vc, vesicle; PG, plastoglobule. Scale bars, 500 nm.
Because the mutant shows albinism in cotyledons, which are embryonic tissues, we hypothesized that NTA1 mutations might affect plastid development or differentiation during certain stages of embryogenesis. To test this hypothesis, we first examined developing ovules in immature siliques of the self-fertilizing heterozygous nta1 and NTA1-S1 mutants. No difference in the appearance of ovules was observed between the WT and the mutants (Supplemental Figure 6B). Embryo development was further examined by manually dissecting embryos from these plants at different developmental stages to visually track embryo development under a stereomicroscope. From 5 days post-anthesis to maturity at 17 days post-anthesis, embryos of WT, nta1, and NTA1-S1 were identical in appearance with respect to developmental rate, morphology, and color (Supplemental Figure 6C). Therefore, the chloroplast defects most likely manifested after embryogenesis.
Next, we examined the chloroplast morphology at different time points during the 24 h of seedling greening under light (100 μmol photons m−2 s−1). First, in the dark, the etioplasts of homozygous mutants showed no differences from the WT and Com1 chloroplasts in either the prolamellar bodies or the plastid structures (Supplemental Figure 7A–7D). In WT and Com1, the first sign of chloroplast development was detected after 4 h of illumination, when the prolamellar bodies start to disappear and the first batch of thylakoid membranes are formed (Supplemental Figure 7E and 7H). A similar pattern of chloroplast development was observed in 4-h-illuminated mutants (Supplemental Figure 7F and 7G). After 9 and 13 h of illumination, the chloroplasts of all plants were already fully developed, and the basic internal thylakoid membranes and grana stacks were formed (Supplemental Figure 7I–7P). However, after 24 h of illumination, chloroplasts in the mutants displayed less evolved thylakoid membranes that lacked grana structures and starch granules compared with those of WT and complemented Com1 plants (Supplemental Figure 7R and 7S vs. 7Q and 7T). Collectively, these studies confirm that NTA1 mutations affect chloroplast development during the seedling greening process.
NTA1 is a chloroplast thylakoid transmembrane protein with its C terminus facing the stroma
To understand the function of NTA1 in chloroplast development, we first determined the subcellular localization of this protein. Analysis with the web-based program CHLOROP1.1 suggested that the NTA1 protein localizes in the chloroplast (Supplemental Figure 8A). By expressing yellow fluorescent protein (YFP)–tagged NTA1 (NTA1-YFP) in Arabidopsis protoplasts and tobacco leaves, we found that the NTA1-YFP fluorescence overlapped completely with the autofluorescence signal of chlorophylls (Figure 3B and Supplemental Figure 8D), indicating that NTA1 is localized in chloroplasts. Furthermore, the chloroplast localization of NTA1 was dependent on its N-terminal sequence (1–39 aa), predicted to be the cTP. Full-length NTA1 and the cTP alone can directly target to chloroplasts, whereas NTA1 without the cTP (ΔcTP) aggregated in the cytoplasm (Figure 3B).
To further determine the sublocalization of NTA1 within the chloroplast, chloroplasts were fractionated into soluble (stromal) and membrane (thylakoid) fractions. Figure 3C shows that NTA1 co-localized with the thylakoids, consistent with a previous study of the thylakoid proteome in which peptides corresponding to NTA1 were detected only in the thylakoids of chloroplasts (Peltier et al., 2004). As a comparison, the Rubisco large subunit RBCL was exclusively located in the stromal fractions, whereas the PSII subunit OEC33 and the light-harvesting chlorophyll A/B binding protein Lhcb1 were detected only in the thylakoid fractions. However, ferredoxin NADP+ oxidoreductase (FNR) was detected in both the stromal fractions and the thylakoid membranes.
To verify whether NTA1 is a thylakoid transmembrane protein, purified thylakoids from WT plants were treated with NaCl, CaCl2, or Na2CO3. As shown in Figure 3E, NTA1 could not be extracted from thylakoid membranes incubated with any of these salts, similar to the thylakoid integral protein CP47. However, the peripherally associated luminal protein OEC33 could be extracted from the membrane. These results clearly indicate that NTA1 is an integral thylakoid membrane protein. Analysis with Membranome software (Lomize et al., 2017) also suggests that NTA1 is a transmembrane bitopic protein (Supplemental Figure 8B).
To determine the topology of NTA1 in thylakoid membranes, isolated thylakoid membranes from WT and nta1-complementation plants were treated with the protease thermolysin, followed by immunoblotting with an NTA1-specific antibody raised against the C-terminal portion of NTA1 (14 amino acids; Supplemental Figure 8C). It was anticipated that if the NTA1 protein could be detected in the thylakoid after thermolysin treatment, the C-terminal region of NTA1 must be located on the lumenal side of the membrane as shown in Figure 3F; otherwise, the topology would be opposite, as indicated in Figure 3G. The results showed that NTA1 was cleaved and undetectable in thylakoid membranes in the presence of thermolysin (Figure 3D and Supplemental Figure 8E). In comparison, the stromal-exposed thylakoid protein PsaD was clearly cleaved, whereas the luminal protein OEC33 was unaffected. Together, these results strongly suggest that the C terminus of NTA1 faces the chloroplast stromal side, as illustrated in Figure 3G.
NTA1 is required for accumulation of the Cyt b6f complex
To further understand the function of NTA1, we examined the effect of NTA1 deficiency on transcription of genes encoding key chloroplast proteins. The expression of plastidic genes is dependent on the activity of two RNA polymerases, plastid-encoded RNA polymerase and nucleus-encoded RNA polymerase (Hedtke et al., 1997; Sakamoto et al., 2008). Transcript levels of representative plastid-encoded RNA polymerase and nucleus-encoded RNA polymerase genes showed no noticeable differences between the WT and the mutant, except for RNA polymerase B subunit (RPOB), whose expression was slightly higher in 4-day-old mutants (Supplemental Figures 9A and 9B). Moreover, there were no differences in the accumulation of chloroplast and cytosolic rRNAs (Supplemental Figure 9C). Hence, NTA1 mutation appears to have no effect on the transcription of genes encoding the major chloroplast proteins.
We next investigated the influence of NTA1 on the accumulation of chloroplast proteins. To obtain a global and quantitative view of proteins that were altered in NTA1-related mutants, a comparative proteomic analysis was performed. Four hundred forty-eight differentially expressed proteins were detected in nta1 and 543 in NTA1-S1, with a fold change of at least 1.5. Among the differentially expressed proteins (P < 0.05), 88 were upregulated and 118 were downregulated in both mutants (Supplemental Figure 10A and Supplemental Data 1). Most of the upregulated proteins are localized to the membrane or extracellular region and are stress-related or transport proteins (Supplemental Figure 10B and 10C and Supplemental Data 1). By contrast, 89 of 118 downregulated proteins were localized to chloroplasts, and the majority (68 of 89) were thylakoid membrane proteins involved in photosynthesis (Supplemental Figure 10D–10L and Supplemental Data 1). These results indicate that loss of NTA1 had a strong effect on the accumulation of photosynthetic thylakoid membrane proteins.
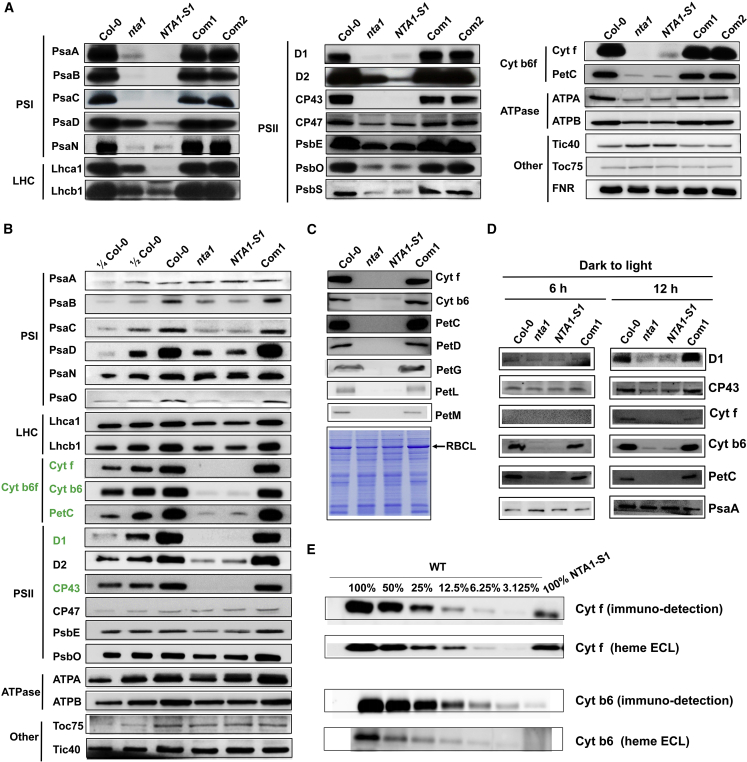
The protein profiles of nta1 and NTA1-S1 showed a significant reduction in the large subunit of Rubisco based on Coomassie brilliant blue staining (Supplemental Figure 9D–9E). Immunoblot analyses further confirmed that most of the tested thylakoid membrane proteins involved in photosynthesis (e.g., PSI, PSII, and Cyt b6f subunits) were nearly undetectable in 10-day-old nta1 and NTA1-S1 mutants (Figure 4A), in which the chloroplasts were completely devoid of thylakoids (Figure 2D). By contrast, proteins located in other regions of the chloroplasts were present at comparable levels in WT and mutant plants (Figure 4A). Because the TEM results showed that chloroplasts of 4-day-old mutant plants contained some thylakoids (Figure 2D), we next examined which photosynthetic complex was most affected by the NTA1 mutations in 4-day-old nta1 and NTA1-S1 mutants. The results indicated that the subunits of Cyt b6f and the PSII core subunits D1 and CP43 were barely detectable in the mutants (Figure 4B and 4C), whereas the abundance of other thylakoid membrane proteins was less affected (Figure 4B vs. 4A). When dark-grown plants were transferred to the light, levels of Cyt b6f subunits, including the nuclear-encoded PetC, were almost undetectable at 6 h of illumination in the mutants, whereas levels of D1 and CP43 of PSII remained comparable to those of the WT and complemented Com1 plants (Figure 4D). A reduction in D1 and CP43 became evident after 12 h of illumination. Therefore, the reduction in D1 and CP43 was most likely a secondary effect of NTA1 mutation. We therefore conclude that NTA1 particularly perturbed the accumulation of Cyt b6f.
Figure 4.
Analysis of major subunits of multiprotein complexes related to photosynthesis.
(A and B) Immunoblot analyses of chloroplast proteins on the basis of equal total proteins (20 μg) from the cotyledons of 10-day-old (A) or 4-day-old (B) wild-type, nta1, NTA1-S1, and complementation lines (Com1 and Com2). Proteins were separated by SDS–PAGE and analyzed by immunoblotting using antibodies specific for the indicated subunits.
(C) Immunoblot analysis of Cyt b6f subunits of 4-day-old Col-0, nta1, NTA1-S1, and Com1. Total proteins (20 μg per lane) were separated by SDS–PAGE or SDS–urea–PAGE and analyzed by immunoblotting using antibodies specific for the indicated subunits. The top shows the immunoblot, and the bottom shows the gel image stained with Coomassie brilliant blue.
(D) Light-induced accumulation of major components of PSI, PSII, and Cyt b6f in the Col-0, nta1, NTA1-S1, and Com1 plants. Seeds were germinated and grown in the dark for 4 days and then transferred to light (100 μmol photons m−2 s−1) and sampled after 6, 9, and 12 h. Total proteins were extracted and separated by SDS–PAGE, followed by immunoblotting with specific antisera against Cyt f, Cyt b6, PetC, D1, and CP43. Twenty micrograms of protein was loaded in each lane.
(E) Detection of heme peroxidase activity of Cyt f and Cyt b6 by chemiluminescence (heme ECL) and immunodetection.
To determine whether the significant reduction in abundance of Cyt b6f subunits observed in NTA1-related mutants was due to reduced gene transcription, transcript levels of Cyt b6f subunits were measured in 4-day-old and 10-day-old mutant seedlings. As shown in Supplemental Figure 9F and 9G, transcripts of the eight Cyt b6f subunits were comparable among the mutants, WT, and Com1 plants. Hence, reduced accumulation of Cyt b6f components in NTA1-related mutants is not the consequence of a transcriptional defect. Defects in heme binding have been reported to result in low accumulation of Cyt b6f (Lennartz et al., 2001; Lezhneva et al., 2008). We therefore compared the heme-binding ability of Cyt f and Cyt b6 in the WT and the NTA1-S1 mutant by assaying their heme peroxidase activity. The peroxidase activity of the mutant associated with Cyt f was about 25%–30% that of the WT, proportional to its relative protein level in the loaded leaf total proteins, whereas that of Cyt b6 could not be detected in the mutant owing to its extremely low level (∼1% of the WT) (Figure 4E). These results suggest that the heme-binding activity of the remaining Cyt f and Cyt b6 holoproteins was not affected in the mutant.
NTA1 directly interacts with four subunits of Cyt b6f through its DUF1279 domain and C-terminal region
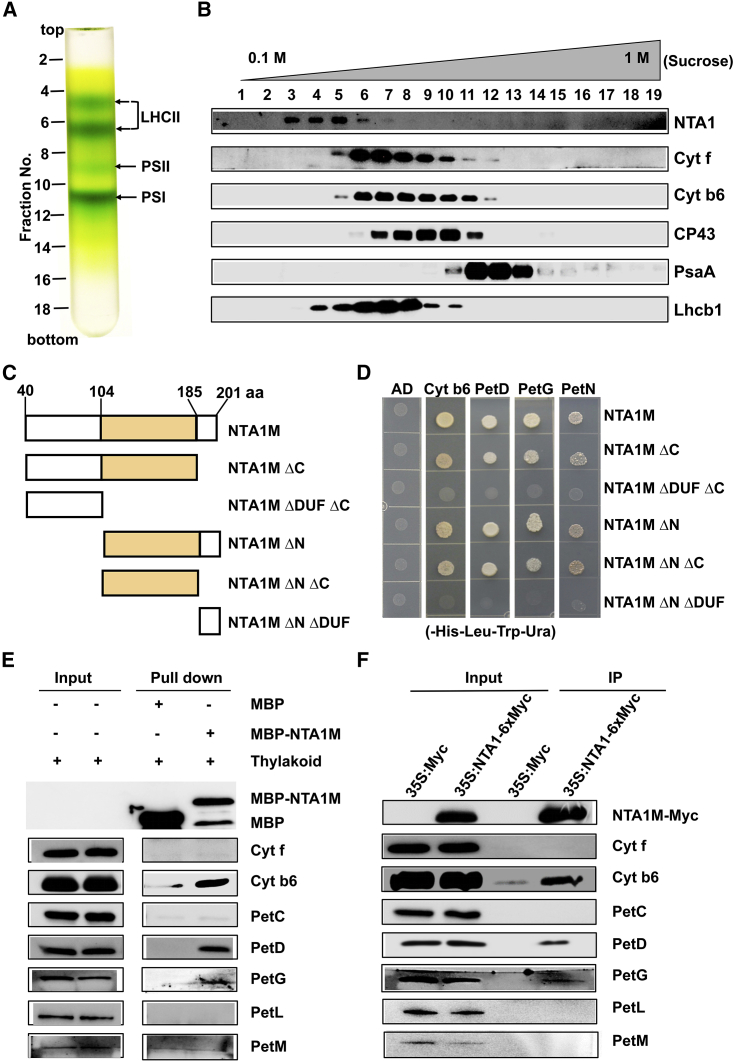
Next, we assayed whether NTA1 is a component of the Cyt b6f or functions as an assembly factor to modulate the assembly of Cyt b6f components. To determine whether NTA1 is part of the Cyt b6f complex, thylakoid membranes were isolated from the WT chloroplasts, solubilized with n-dodecyl-β-D-maltoside (DM), and then subjected to separation by a continuous sucrose gradient. After centrifugation, 19 fractions were collected from the gradients and subjected to immunoblot analysis (Figure 5A). Based on the migration patterns of protein fractions, the NTA1 protein did not show co-migration with Cyt b6f or other protein complexes (Figure 5B). Instead, it was present in a less dense fraction that contained free LHCs (released from PSI-LHCI or PSII-LHCII supercomplexes as a consequence of solubilization). These results suggest that NTA1 is not part of the fully assembled Cyt b6f complex. However, we cannot rule out the possibility that NTA1 interacts with certain subunits of Cyt b6f to mediate their assembly. As shown by the sucrose gradient fractionation experiment, the NTA1 protein fraction overlapped with the intermediates of Cyt b6f (Figure 5B). Therefore, we next tested its interaction with the eight subunits of Cyt b6f. In yeast two-hybrid assays, clear interactions were detected between the mature NTA1 protein (NTA1M) and the two large subunits (apocytochrome b6/PetB and PetD) and two small subunits (PetG and PetN) of Cyt b6f (Figure 5D and Supplemental Figure 11A and 11B). Because NTA1-related mutants also had reduced accumulation of other photosynthetic proteins in thylakoid membranes (Figure 4 and Supplemental Figure 10F–10L), we next examined the possible association of NTA1 with selected key subunits of PSI, PSII, and LHC. The results showed that NTA1 had no interactions with any of the selected subunits (Supplemental Figure 11G–11I). Therefore, we conclude that NTA1 specifically interacts with Cyt b6f subunits before they are fully assembled into the complex. To confirm the interactions of NTA1 with the four subunits of Cyt b6f, a semi-in vivo pull-down experiment was performed. Solubilized thylakoid membranes were incubated with recombinant maltose-binding protein (MBP)-tagged NTA1M that was immobilized on the amylose resin, and the precipitated proteins were then eluted and analyzed by immunoblotting. Consistent with the results of the yeast two-hybrid assays, Cyt b6, PetD, and PetG were detected in precipitates of MBP-NTA1M but not in solubilized thylakoid membranes incubated with MBP (Figure 5E) (here, PetN was not detected because its antibody was not available). To further investigate these interactions in a plant system, a co-immunoprecipitation experiment was performed using 35S:NTA1-Myc transgenic plants. The results showed that NTA1 indeed interacted with Cyt b6, PetD, and PetG in vivo (Figure 5F). Taken together, these results suggest that NTA1 has an essential function in the assembly of Cyt b6f by directly interacting with its four subunits.
Figure 5.
Protein–protein interaction studies of NTA1 with Cyt b6f subunits.
(A) Separation of major photosynthetic protein complexes on a 0.1–1 M sucrose density gradient. Thylakoid membranes from wild-type Col-0 chloroplasts were solubilized by 1% DM and then loaded onto the gradient. The relative positions of the fractions and LHCII, PSI, and PSII are indicated.
(B) Nineteen fractions were collected from the gradients, and proteins from each fraction were separated by SDS–PAGE. After electrophoresis, the separated proteins were immunodetected with the NTA1-specific antibody. To identify the positions of major photosynthetic complexes, blots were also probed with antibodies against Cyt f, Cyt b6, CP43, PsaA, and Lhcb1.
(C) Schematic diagram of truncated NTA1 mature protein (NTA1M) structures. aa, amino acid.
(D) Yeast two-hybrid assays of the interactions between four subunits of the Cyt b6f complex and NTA1M truncated versions. No interactions were detected between NTA1M ΔDUF ΔC or NTA1M ΔN ΔDUF and the indicated preys on the selection medium (−His−Leu−Trp−Ura). All experiments were repeated at least three times with similar results.
(E)In vitro pull-down assay of the interactions between NTA1 and Cyt b6f subunits. Recombinant MBP-NTA1M or MBP control bound to the amylose resin was incubated with solubilized thylakoid membranes, and the bound proteins were eluted and analyzed by immunoblotting with specific antibodies shown on the right.
(F) Co-immunoprecipitation assay of the interactions between NTA1 and Cyt b6f subunits. Solubilized thylakoid membranes from transgenic plants overexpressing NTA1-Myc or the Myc epitope alone were incubated with protein A/G-coupled anti-Myc antiserum. The immunoprecipitants were probed with the antibodies as in (E).
To determine which part of NTA1 is responsible for its interaction with the four Cyt b6f subunits, deletion derivatives of NTA1M were generated and tested for interactions with the potential partners (Figure 5C). As shown in Figure 5D and Supplemental Figure 11C–11F, the N or C terminus of NTA1M alone failed to interact with the four subunits of Cyt b6f. However, the N-terminal or C-terminal region combined with the DUF1279 domain could interact with them, indicating that the DUF1279 domain is essential for the interaction. Consistent with this result, the DUF1279 domain alone was able to interact with all four interaction partners. In addition, the interactions became weaker without the C terminus. Therefore, both the DUF1279 domain and the C-terminal region are required for the interaction of NTA1 with the four subunits of Cyt b6f.
The C terminus of NTA1 is indispensable for its physiological function in plant development
To further understand the importance of the DUF1279 domain and C-terminal region to NTA1 function, truncated versions of NTA1, including deletion of the C-terminal region (NTA1 ΔC) or deletion of both the DUF1279 domain and the C-terminal region (NTA1 ΔDUF ΔC), were introduced into the NTA1-S1 mutant to determine whether they could rescue the mutant phenotype (Figure 6A). The results indicated that expression of these truncated NTA1 genes in NTA1-S1 could not rescue the mutant phenotype, whereas the full-length NTA1 could completely rescue the phenotype (Figure 6B). This result suggests that the C-terminal part of NTA1 is essential for maintaining its normal functions in chloroplast development.
Figure 6.
Transgenic plants expressing truncated versions of NTA1 show abnormal growth phenotypes.
(A) Schematic of NTA1 gene truncations.
(B) Morphology of the NTA1-S1 mutants transformed with the 35S:NTA1ΔDUFΔC, 35S:NTA1ΔC, or 35S:NTA1 construct. Overexpression of these truncated gene fragments could not rescue the NTA1-S1 mutant phenotype. Scale bars, 0.2 cm.
(C) Western blot analysis of endogenous NTA1 level in NTA1ΔC-overexpressing plants. Amido black–stained blot image is shown for the loading control.
(D) Morphology of wild-type (Col-0) plants transformed with the 35S:NTA1ΔC or 35S:NTA1ΔDUFΔC plasmid. An abnormal pale-green phenotype was observed in the heterozygous 35S:NTA1ΔC/Col+/− plants, whereas the homozygous plants (35S:NTA1ΔC/Col+/+) showed an albino phenotype. Scale bars, 0.2 cm.
To test whether the DUF1279 domain and C terminus of NTA1 are essential for the interactions between NTA1 and Cyt b6f subunits in vivo and thus affect Cyt b6f formation and chloroplast development, we overexpressed the truncated NTA1s in the Col-0 WT background and examined the phenotypes of 35S:NTA1 ΔC and 35S:NTA1 ΔDUF ΔC transgenic plants. Overexpression of NTA1 ΔC resulted in a pale-green phenotype in heterozygous plants and an albino phenotype in homozygous plants, whereas we did not observe any abnormal phenotypes from 35S:NTA1 ΔDUF ΔC (Figure 6C and 6D). These results suggest that the DUF1279 domain is essential for the interaction of NTA1 with Cyt b6f subunits, whereas the C-terminal portion is crucial for NTA1 function.
Discussion
The biogenesis and assembly of Cyt b6f is a complicated process that must be highly regulated and coordinated with additional nucleus-encoded factors (Lennartz et al., 2001, 2006; Xiao et al., 2012; Sandoval-Ibáñez et al., 2022). However, factors and mechanisms involved in the assembly of this complex have not yet been fully unraveled. In this study, we demonstrated the involvement of NTA1, a thylakoid membrane–localized protein also named DEIP1 in a recent study (Sandoval-Ibáñez et al., 2022), in the biogenesis and assembly of Cyt b6f. Arabidopsis plants deficient in NTA1 are albino and seedling lethal. Moreover, NTA1 plays a critical role in maintaining photosynthetic activity and accumulation of major thylakoid membrane proteins, particularly Cyt b6f. The nta1 mutations not only caused significant reduction in Cyt b6f accumulation but also reduced the abundance of PSI and PSII complexes. However, during the first 6 h of illumination, unlike the Cyt b6f subunits, the PSI and PSII core subunits showed comparable abundance in NTA1-related mutants and WT plants (Figure 4D). Therefore, Cyt b6f is the primary complex affected in the NTA1-related mutants. Because NTA1 does not co-migrate with PSI and PSII and has no interaction with their core subunits (Figure 5B and Supplemental Figure 11G and 11H), we reason that the reduction in PSI and PSII complexes is likely to be a secondary effect of NTA1 mutation caused by the altered photosynthetic redox control associated with modified retrograde signaling. The importance of the redox states of photosynthetic electron transport components (e.g., plastoquinone, the Cyt b6f complex, and reactive oxygen species) as a source of chloroplast signals to the nuclear compartment has been corroborated and stressed by earlier studies (Fey et al., 2005; Nott et al., 2006). Therefore, nta1 can be classified as a mutant deficient in Cyt b6f accumulation, and the changes in stacked thylakoid membranes in chloroplasts of NTA1-related mutants can be attributed to the loss of Cyt b6f and the accompanying physiological effects.
The assembly of Cyt b6f subunits has been explored in previous studies. For example, Wollman (1998) revealed that Cyt b6 and PetD can form a relatively protease-resistant subcomplex that serves as a template for assembly of Cyt f and PetG (Wollman, 1998). Other subunits such as PetC and PetL then join to form the functional dimer (Schwenkert et al., 2007). It has been demonstrated that PetD will become unstable if Cyt b6 is absent, and the synthesis of Cyt f is also greatly reduced when Cyt b6 or PetD is inactivated (Kuras and Wollman, 1994). All these studies indicate a prerequisite role of Cyt b6 and PetD in assembly of the Cyt b6f complex. Our protein–protein interaction experiments revealed that NTA1 interacted not only with Cyt b6 and PetD but also with the two small subunits PetG and PetN of Cyt b6f (Figure 5D–5F and Supplemental Figure 11A–11F), and Sandoval-Ibáñez et al. (2022) recently reported that DEIP1/NTA1 could also interact with PetA (Cyt f) and PetB (Cyt b6) subunits. All these results indicate the important role of NTA1 in assembly of the Cyt b6f complex. The importance of the physical interactions of assembly factors with specific subunits has been demonstrated in the assembly of other photosynthetic complexes (Peng et al., 2006; Lu et al., 2011; Xiao et al., 2012; Bhuiyan et al., 2015; Wittkopp et al., 2018; Zhang et al., 2018). Our sucrose gradient experiment showed that NTA1 does not co-migrate with the Cyt b6f complex but overlaps with its assembly intermediates (Figure 5B); thus, it ought to function before the full assembly of Cyt b6f subunits. Based on protein–protein interaction and sucrose gradient results, we propose that NTA1 functions as an assembly factor that mediates the assembly of Cyt b6f subunits by directly interacting with multiple subunits. When NTA1 is not available, the subunits Cyt b6, PetD, PetG, and PetN become unstable, and the stability of other subunits will also be affected. Proteolytic degradation has been shown to modulate the abundance of Cyt b6f subunits via degradation of those that remain unassembled. For instance, an early study on a mutant of Lemna perpusilla observed that when the Cyt b6f complex is not fully assembled, its subunits are degraded by proteases (Bruce and Malkin, 1991). Through later in vitro studies, the major thylakoid membrane protease FtsH was shown to participate in degradation of unassembled PetC (Ostersetzer and Adam, 1997). Similarly, a recent study in Chlamydomonas reinhardtii demonstrated that FtsH-dependent degradation is responsible for loss of the Cyt b6f complex under dark and sulfur-starvation conditions (Malnoë et al., 2014; Wei et al., 2014). These studies indicate that when Cyt b6f is not fully assembled, its subunits will be degraded by proteases. Whether loss of NTA1 in NTA1-related mutants will lead to activation of certain proteases such as FtsH is unknown; thus, the instability and degradation of Cyt b6f subunits are worthy of further study.
Very recently, a role for DEIP1/NTA1 in assembly of the Cyt b6f complex was also reported by Sandoval-Ibáñez et al. (2022). They showed that DEIP1/NTA1 mediates the assembly of Cyt b6f subunits by physically interacting with the PetA/Cyt f and PetB/Cyt b6 subunits. The reason for the different interaction results in our study and that of Sandoval-Ibáñez et al. is unclear but may be due to the different methods used for detection of protein–protein interactions. We used three different approaches, and no interaction was observed for NTA1 and Cyt f/PetA. In addition to DEIP1/NTA1, several other nuclear-encoded proteins in Arabidopsis, maize, and a green alga (C. reinhardtii) have been claimed to play a role in the assembly of Cyt b6f subunits, including DAC, HCF153, and CPLD49 (Voelker and Barkan, 1995; Lennartz et al., 2006; Xiao et al., 2012; Wittkopp et al., 2018). Whether these factors work together to coordinate the assembly of the Cyt b6f complex awaits clarification by future studies.
Our study revealed that the DUF1279 domain and the C-terminal part of NTA1 mediate the interaction of NTA1 with Cyt b6f subunits (Figure 5D and Supplemental Figure 11C–11F). The importance of these interacting domains was further demonstrated by transgenic studies. Transgenic plants that expressed a truncated NTA1 lacking the DUF1279 and C-terminal domains (NTA1 ΔDUF ΔC) or the C terminus alone (NTA1 ΔC) could not rescue the NTA1-S1 mutant phenotype (Figure 6B). More interestingly, overexpression of the NTA1 ΔC fragment alone in the Col-0 WT background could induce a pale green (heterozygous) or albino (homozygous) phenotype (Figure 6D), indicating the essential role of the C-terminal domain in NTA1 function.
NTA1 is encoded by the same gene (AT2G27290) as the recently reported DEIP1 protein (Sandoval-Ibáñez et al., 2022). The nta1 and deip1 mutants show similar dwarf and albino phenotypes, and both studies implicated NTA1/DEIP1 in the assembly of the Cyt b6f complex, which further suggests the importance of this protein. Although both our study and that of Sandoval-Ibáñez et al. address the same protein and have reached similar conclusions, there are several major differences between our study and their work. First, our nta1 mutant was identified by a different approach (T-DNA activation-tagging mutant screen), and we have provided additional details about the mutant phenotype, including the defects in chloroplast development and the physiological responses to sucrose and phytohormones. Second, CRISPR-Cas9 genome-editing technology was used to recapitulate the nta1 mutant phenotype, and we raised an NTA1-specific antibody to study its function. Third, to determine the steps of chloroplast biogenesis in which nta1 mutant defects occur, we performed time-course experiments on the chloroplast ultrastructure by TEM and demonstrated that chloroplasts of NTA1-related mutants begin to display defects during the seedling greening process. Fourth, we performed comparative proteomic analyses among nta1, NTA1-S1, and WT plants to identify global changes in protein expression caused by the NTA1 mutation and found that loss of NTA1 primarily affects the abundance of photosynthesis-related proteins in thylakoid membranes. Fifth, we performed a thorough detection of NTA1 interactions with all eight subunits of Cyt b6f and revealed by both in vitro and in vivo studies (Figure 5D–5F) that NTA1 interacts with the four subunits Cyt b6/PetB, PetD, PetG, and PetN. Sandoval-Ibáñez et al. (2022) tested the interaction of DEIP1/NTA1 with only the four large subunits and detected interactions with the PetA and PetB subunits and a weak interaction with PetD. Finally, we demonstrated that the DUF1279 and C-terminal domains are required for the interaction of NTA1 with the four Cyt b6f components, and the significance of this interaction was confirmed by transgenic studies using truncated versions of NTA1.
In summary, our results provide more and solid evidence that NTA1/DEIP1 is a crucial regulator of chloroplast development and that it functions as an assembly factor for the Cyt b6f complex in the thylakoid membrane by directly interacting with the four Cyt b6f subunits (Cyt b6, PetD, PetG, and PetN) through its DUF1279 domain and C terminus. Once assembly is complete, NTA1 leaves the Cyt b6f complex by a yet unknown mechanism. Based on our results, we propose a model for NTA1 function in Cyt b6f assembly (Figure 7). Future studies should be performed to elucidate the detailed biochemical mechanisms by which NTA1/DEIP1 mediates assembly of the Cyt b6f complex.
Figure 7.
A hypothetical model of NTA1 function in the assembly of Cyt b6f subunits.
Here we provide a highly simplified outline of the proposed Cyt b6f assembly process, emphasizing the role of NTA1 in mediating the assembly of Cyt b6f by interacting with its four subunits Cyt b6, PetD, PetG, and PetN through the DUF1279 domain and C terminus (with the C terminus facing the stroma). NTA1 is believed to interact with these four subunits before they are assembled into a complex; once assembly is complete, NTA1 leaves the b6f complex by an unknown mechanism. Without NTA1, these subunits become unstable and will be degraded quickly, resulting in greatly reduced accumulation of Cyt b6f components, as seen in the NTA1-related mutants.
Methods
Plant growth and plasmid construction
Arabidopsis thaliana plants were grown in soil or on ½ Murashige and Skoog (MS) medium plates (pH 5.6–5.8) containing 1%, 3%, or 5% sucrose at 22°C under a 16-h light (100 μmol photons m−2 s−1)/8-h dark cycle. For hormone treatment, seeds were germinated on solid ½ MS medium with the indicated concentrations of hormones for 6 days. The ½ MS medium supplemented with corresponding solvents was used as a control. The construct used for complementation experiments was created by cloning the PCR-amplified cDNA of NTA1 into the pCanG vector backbone. To generate the constructs for NTA1 knockout by CRISPR-Cas9, two 19-bp sequences from the first exon (nucleotide positions 182–201) and the second exon (nucleotide positions 326–345) of NTA1 were inserted into the pBGK01 vector with an AtU6 promoter (Ma et al., 2015; Wang et al., 2015). To analyze the subcellular localization of the NTA1 protein, full-length and truncated coding sequences of NTA1 were amplified and cloned into the Gateway binary vector pEG101, generating the NTA1-YFP, ΔcTP-YFP, and cTP-YFP fusions. For the construction of 35S:NTA1 ΔC and 35S:NTA1 ΔDUF ΔC, each truncated NTA1 cDNA was amplified using the full-length NTA1 cDNA as a template. To generate the transgenic plants for these different NTA1 sequences, all the resulting constructs were transformed into Arabidopsis Col-0 by the Agrobacterium-mediated floral dipping method (Clough and Bent, 1998). The primers used for plasmid construction and plant genotyping are listed in Supplemental Data 2.
NTA1 gene cloning by adapter ligation–mediated PCR
An adapter ligation–mediated PCR was used for NTA1 gene cloning following the procedure of O’Malley et al. (2007). The CTAB method (Voelker and Barkan, 1995) was used to isolate genomic DNA from the homozygous mutant nta1. Genomic DNA (100 ng) was digested with HindIII, EcoRI, or BamHI and then ligated with adapters to create adapter-flanked templates. The target T-DNA/gDNA junction was exclusively amplified using T-DNA primers (DSKLB1, DSKLB2, and DSKLB3) and adapters (AP2 and AP4). After amplification, the moiety of the T-DNA/gDNA junction was sequenced with T-DNA sequencing primers. The nucleotide sequences of adapters and specific T-DNA primers are available in Supplemental Data 2.
Measurement of chlorophylls, carotenoids, and chlorophyll fluorescence
Chlorophylls and carotenoids were measured as described previously (Lichtenthaler and Wellburn, 1983), with small modifications. Fresh leaves obtained from 4-, 7-, and 10-day-old Arabidopsis seedlings grown on ½ MS medium with 1% sucrose were first homogenized in 80% (v/v) acetone. Absorbance (A) of the final solution was measured at 663, 647, and 470 nm, and concentrations of total chlorophylls, chlorophyll a, chlorophyll b, and carotenoids were calculated. Chlorophyll fluorescence parameters were measured with the MAXI version of the Imaging-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments) as described previously (Lu, 2011; Jin et al., 2014). Data were analyzed with Imaging PAM software. Fv/Fm, the ratio of variable to maximum fluorescence for the maximum quantum yield of PSII, was measured under dark-adapted conditions and calculated using the equation Fv/Fm = (Fm − F0)/Fm, in which F0 is the minimum fluorescence and Fm is the maximum fluorescence in the dark-adapted state. The effective quantum yield of PSII was measured at 100 μmol photons m−2 s−1 light intensity and was calculated as ΦPSII = (Fm′ − F′)/Fm′, where Fm′ is the maximum fluorescence at a given actinic light intensity and F′ is the fluorescence at a given actinic light intensity. The non-photochemical quenching parameter NPQ was calculated using the following equation: NPQ = (Fm − Fm′)/Fm′. qP was calculated from the equation qP = (Fm′ − Fs)/(Fm′ − F0′), where Fs is the steady-state fluorescence. Y(NO) was calculated from the equation Y(NO) = F′/Fm.
Transmission electron microscopy
TEM experiments were performed according to Ruppel et al. (2011) with small modifications. Leaves of seedlings in different stages were cut into small pieces and placed into a fixation solution containing 2% glutaraldehyde solution and 0.1 ml picric acid in 25 mM Sodium cacodylate buffer (pH 7.2) and fixed overnight at 4°C. The fixed samples were washed and post-fixed in 2% OsO4 at room temperature for 2 h. The samples were then washed, dehydrated, and embedded in soft Spurr’s resin (Electron Microscopy Sciences). Cotyledon pieces were sectioned using an automated ultramicrotome with a diamond knife. The sections were stained with uranyl acetate solution and lead citrate. Stained sections were observed and imaged under a Hitachi H7650 TEM (Hitachi High-Technologies) with a MacroFire monochrome CCD camera (Optronics) operating at 80 kV.
RNA isolation and quantitative RT-PCR assays
Total RNA was isolated from plant tissues using the Total RNA Kit Ⅱ (Omega Bio-Tek), followed by treatment with RNase-free DNase I (Thermo Scientific) at room temperature for 15 min to remove genomic DNA. One microgram of treated RNA was used as a template for first-strand cDNA synthesis using a RevertAid Strand cDNA Synthesis Kit (Thermo Scientific) with oligo(dT) or random hexamer primers. Quantitative RT-PCR was performed using SYBR-Green Supermix (Bio-Rad) and the CFX96 Touch real-time PCR detection system (Bio-Rad). Primer sequences of genes tested in quantitative RT-PCR and RT-PCR are listed in Supplemental Data 2. For RNA gel electrophoresis, 8 μg of total RNA was denatured by heating at 70°C for 10 min in 47.5% formamide, 0.25 mM EDTA, and 0.0125% SDS and fractionated on a 1.2% formaldehyde gel.
Protein extraction, immunoblot analysis, and heme staining
Total proteins were isolated from Arabidopsis tissues by pulverizing the tissues in liquid nitrogen. SDS sample buffer (2×) was added in the ratio of 2:1 (2 μl of buffer/1 mg of tissue powder) to extract the proteins. The extracted proteins were then heated at 70°C for 10 min, followed by centrifugation at 12 000 g for 10 min. The resulting supernatants were transferred to a new microfuge tube. Protein concentrations were determined by the Bradford method using bovine serum albumin (BSA) as the standard (Bradford, 1976). Extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using a minigel system (Bio-Rad). After electrophoresis, the separated proteins were visualized by Coomassie Brilliant Blue G-250 or transferred electrophoretically onto a polyvinylidene fluoride membrane (Bio-Rad). After 2 h of blocking with 1% BSA solution, the membranes were incubated with a primary antibody for 3 h and a secondary antibody (horseradish peroxidase IgG [H + L]) for 1 h at room temperature. Immunodetection was carried out using the Western Blotting Luminol Reagent (Thermo Scientific). Heme peroxidase activity of Cyt f and Cyt b6 on blots was detected using chemiluminescence as previously described (Feissner et al., 2003; Kuras et al., 2007). Total proteins from shoots of 22-day-old plants (on 1% sucrose plates) were loaded, and immunolabeled bands were visualized by chemiluminescence and autoradiography on X-ray films.
Proteomics analysis
Total proteins were extracted from 14-day-old WT, nta1, and NTA1-S1 seedlings using the phenol extraction method (Yang et al., 2007). Isobaric tags for relative and absolute quantification (iTRAQ)-based proteomic analysis, including protein purification, digestion, iTRAQ labeling, protein identification, and quantification, was performed as described previously (Yang et al., 2011). Functional classification of identified proteins was performed on the basis of the bin codes from MapMan. In silico predictions of protein subcellular localizations were carried out using TAIR10 software (https://www.arabidopsis.org/).
Chloroplast isolation and stroma and thylakoid fractionation
Isolation and subfractionation of chloroplasts into stroma and thylakoids were carried out according to Bals and Schünemann (2011) with minor modifications. Chloroplasts were isolated from leaves of 4-week-old WT Arabidopsis (Col-0) grown in soil with a 12-h light (100 μmol photons m−2 s−1) (22°C)/12-h dark (22°C) cycle. Leaves were first homogenized in homogenization buffer (450 mM sorbitol, 20 mM Tricine–KOH [pH 8.4], 10 mM EDTA, 10 mM Na2CO3, and 0.1% [w/v] BSA) and filtered through two layers of Miracloth. Crude chloroplasts were then collected by a 5-min spin at 1500 g and 4°C and further purified on a Percoll gradient by mixing 15 ml of cold 100% Percoll and 15 ml of cold 2× RB buffer (300 mM sorbitol, 20 mM Tricine–KOH [pH 7.6], 5 mM MgCl2, and 2.5 mM EDTA) by ultracentrifuging for 20 min at 53 000 g and 4°C. Intact chloroplasts were subsequently incubated in TE buffer (10 mM Tris–HCl [pH 8.0] and 1 mM EDTA) on ice for 30 min. For further fractionation of the lysed chloroplasts, sucrose gradients were generated in ultracentrifugation tubes in the following order: 2.5 ml of 1.2 M sucrose, 3.5 ml of 1 M sucrose, and 3.5 ml of 0.46 M sucrose. Then, 1 ml of the lysed chloroplasts was carefully loaded onto the sucrose gradient and ultracentrifuged for 2 h at 41 000 g and 4°C. Stromal proteins (upper 0.8 ml) from the top of the gradient were collected. Thylakoid pellets in the bottom were resuspended in cold HM buffer (10 mM HEPES–KOH [pH 8.0] and 5 mM MgCl2) and then stored on ice for direct use or at −80°C for later use.
Thermolysin treatment and salt washing of thylakoids
The topology of the NTA1 protein was tested by a protease protection assay (Bhuiyan et al., 2015). Isolated thylakoids were resuspended in HM buffer, adjusted to a concentration of 0.3 mg chlorophyll/ml, and treated with thermolysin (0.1 mg/ml final concentration) for 15, 30, or 45 min on ice. Proteolysis was stopped by adding EGTA (10 mM final concentration), and then protection of NTA1 against degradation by thermolysin was tested by immunoblotting using anti-NTA1, anti-OEC33, and anti-PsaD antibodies.
For high-salt washes, the purified thylakoid fraction (30 μg of protein) was incubated for 30 min on ice in a salt solution consisting of 1 M NaCl, 1 M CaCl2, and 0.1 M Na2CO3 (pH 11.5). The mixtures were centrifuged, and the pellet (integral membrane proteins) and the resulting supernatant (peripheral membrane proteins) were resuspended in 2× SDS sample buffer, analyzed by SDS–PAGE, and immunoblotted using anti-NTA1, anti-OEC33, and anti-CP47 antibodies.
Sucrose gradient fractionation of thylakoid membranes
Purified thylakoids were resuspended in 5 mM MgCl2, 10 mM NaCl, and 25 mM MES–NaOH (pH 6.0) and adjusted to a concentration of 0.5 mg/ml chlorophyll. Then the thylakoids were solubilized in a final concentration of 1% (w/v) DM on ice for 5 min. After 10-min centrifugation at 14 000 g and 4°C, the supernatant (500 μl) was loaded onto a linear 0.1–1 M sucrose gradient in 5 mM MgCl2, 10 mM NaCl, 0.06% DM, and 25 mM MES–NaOH (pH 5.7) (Xiao et al., 2012). The gradient was centrifuged (SW41-Ti rotor; Beckman) at 180 000 g for 22 h at 4°C. Then, 19 fractions were collected and subjected to immunoblot analyses as described previously (Peng et al., 2006).
Yeast two-hybrid assays
Yeast two-hybrid assays were carried out using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). Full-length and truncated versions of NTA1 and coding sequences of Cyt b6f subunits and representative subunits of PSI, PSII, and LHC complexes were cloned into both the bait vector pGBKT7 and the prey vector pGADT7 (see Supplemental Data 2 for primer information). Bait and prey vector pairs were co-transformed into the AH109 yeast strain. Co-transformants were selected on synthetic dropout medium lacking leucine and tryptophan (−L−T). To detect protein–protein interactions, yeast with each pair of transformants were grown on synthetic dropout medium lacking leucine, tryptophan, histidine, and uracil (−L−T−H−U).
Semi-in vivo pull-down and co-immunoprecipitation assays of protein–protein interactions
MBP and MBP-tagged full-length NTA1 (MBP-NTA1M) were expressed in Escherichia coli strain BL21(DE3) by induction with 1 or 0.4 mM isopropyl β-D-1-thiogalactopyranoside. The MBP and MBP-NTA1M proteins were purified using amylose resin (BioLabs) according to a reported protocol (Riggs, 1994). A semi-in vivo pull-down assay was performed as described by Sun et al. (2007). MBP or MBP-NTA1M (0.1 mg) was incubated with 200 μl of a 50% suspension (v/v) of amylose resin in column buffer (20 mM Tris–HCl [pH 7.4], 0.2 M NaCl, and 1 mM EDTA) for 1 h at 4°C in Pierce spin columns. After the thylakoid membranes (100 μg chlorophyll) were solubilized in 1% (w/v) DM in 20% glycerol (v/v) and 25 mM Bis–Tris–HCl [pH 7.0] for 20 min on ice, they were centrifuged at 12 000 g for 10 min at 4°C. The supernatant was then incubated with MBP or NTA1M-MBP for 4–6 h with constant rotation at 4°C. After being washed five times with ice-cold 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, and 1 mM EDTA buffer, resin-bound proteins were eluted with 2× SDS sample buffer, resolved by SDS–PAGE, and subjected to immunoblot analyses.
Immunoprecipitation of Cyt b6f subunits was performed as described previously (Jin et al., 2014) with minor modifications. Thylakoid membrane proteins (200 μl, 0.5 mg chlorophyll/ml) were solubilized with 2% (w/v) DM in 20% (v/v) glycerol and 25 mM Bis–Tris–HCl (pH 7.0) for 10 min on ice. After centrifugation, the supernatant was diluted in an equal volume of the same buffer without DM and incubated with anti-Myc antibody at 4°C for 3–4 h in Pierce spin columns. The agarose was then washed five times with ice-cold PBS buffer (pH 7.8), and bound proteins were eluted with 2× SDS sample buffer, resolved by SDS–PAGE, and subjected to immunoblot analysis.
Bioinformatic analysis
Protein sequences predicted from the Arabidopsis NTA1 gene and their orthologs were aligned using the ClustalX program (Larkin et al., 2007). A phylogenetic tree was constructed using MrBayes 3.2 (Ronquist and Huelsenbeck, 2003) and drawn with FigTree 1.4.0. Protein localization and transit peptides were predicted using ChloroP1.1 (Emanuelsson et al., 1999). Differentially expressed proteins in the comparative proteomic analysis were categorized into functional groups using MapMan (Thimm et al., 2004).
Antiserum production
The NTA1 antibody was raised against synthetic peptides corresponding to the C terminus of NTA1 (CANWIGKKVDKEKDD) in rabbit by Agrisera (Vännäs, Sweden). Other antibodies were purchased from Agrisera and PhytoAB (San Jose, CA, USA).
Data availability
The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038714. Other datasets used and/or analyzed in this study are available from the corresponding author upon reasonable request.
Funding
This work is supported by the General Research Fund (CUHK codes 14121915, 14148916, and 14104521) and the Area of Excellence Scheme (AoE/M-403/16 and AoE/M-05/12) of the Research Grants Council (RGC) of Hong Kong, a grant from the National Natural Science Foundation of China (NSFC)-RGC Joint Scheme (N_CUHK452/17), and direct grants from the Chinese University of Hong Kong (CUHK).
Author contributions
Conceptualization, J.-X.H.; methodology, N.L., W.W., L.J., and J.-X.H.; investigation, N.L., W.W., L.F., C.W., K.W., N.Z., and W.Y.; writing – original draft, N.L. and J.-X.H.; writing – review & editing, N.L., Y.J., L.J., and J.-X.H.; funding acquisition, J.-X.H.; resources & supervision, Y.J., L.J., and J.-X.H.
Acknowledgments
We thank Professor Hong-Bin Wang (Sun Yat-Sen University) for assisting in measurement of chlorophyll fluorescence. No conflict of interest is declared.
Published: December 22, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
Sequence data for Arabidopsis genes in this article can be found at The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: NTA1 (AT2G27290), PetA (ATCG00540), PetB (ATCG00720), PetC (AT4G03280), PetD (ATCG00730), PetG (ATCG00600), PetL (ATCG00590), PetM (AT2G26500), and PetN (ATCG00210).
Supplemental information
References
- Allen J.F. Photosynthesis of ATP-electrons, proton pumps, rotors, and poise. Cell. 2002;110:273–276. doi: 10.1016/s0092-8674(02)00870-x. [DOI] [PubMed] [Google Scholar]
- Bals T., Schünemann D. Chloroplast Research in Arabidopsis. Springer; 2011. Isolation of Arabidopsis thylakoid membranes and their use for in vitro protein insertion or transport assays; pp. 321–338. [DOI] [PubMed] [Google Scholar]
- Baniulis D., Yamashita E., Zhang H., Hasan S.S., Cramer W.A. Structure-function of the cytochrome b6f complex. Photochem. Photobiol. 2008;84:1349–1358. doi: 10.1111/j.1751-1097.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Bhuiyan N.H., Friso G., Poliakov A., Ponnala L., van Wijk K.J. MET1 is a thylakoid-associated TPR protein involved in photosystem II supercomplex formation and repair in Arabidopsis. Plant Cell. 2015;27:262–285. doi: 10.1105/tpc.114.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship R.E. John Wiley & Sons; 2014. Molecular Mechanisms of Photosynthesis. [Google Scholar]
- Boulouis A., Raynaud C., Bujaldon S., Aznar A., Wollman F.A., Choquet Y. The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell. 2011;23:333–349. doi: 10.1105/tpc.110.078170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruce B.D., Malkin R. Biosynthesis of the chloroplast cytochrome b6f complex: studies in a photosynthetic mutant of Lemna. Plant Cell. 1991;3:203–212. doi: 10.1105/tpc.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert F., Mosebach L., Gäbelein P., Hippler M. PGR5 is required for efficient Q cycle in the cytochrome b6f complex during cyclic electron flow. Biochem. J. 2020;477:1631–1650. doi: 10.1042/BCJ20190914. [DOI] [PubMed] [Google Scholar]
- Buchert F., Scholz M., Hippler M. Electron transfer via cytochrome b6f complex displays sensitivity to antimycin A upon STT7 kinase activation. Biochem. J. 2022;479:111–127. doi: 10.1042/BCJ20210802. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cramer W.A., Zhang H., Yan J., Kurisu G., Smith J.L. Transmembrane traffic in the cytochrome b6f complex. Annu. Rev. Biochem. 2006;75:769–790. doi: 10.1146/annurev.biochem.75.103004.142756. [DOI] [PubMed] [Google Scholar]
- Dumas L., Chazaux M., Peltier G., Johnson X., Alric J. Cytochrome b6f function and localization, phosphorylation state of thylakoid membrane proteins and consequences on cyclic electron flow. Photosynth. Res. 2016;129:307–320. doi: 10.1007/s11120-016-0298-y. [DOI] [PubMed] [Google Scholar]
- Dumas L., Zito F., Blangy S., Auroy P., Johnson X., Peltier G., Alric J. A stromal region of cytochrome b6f subunit IV is involved in the activation of the Stt7 kinase in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 2017;114:12063–12068. doi: 10.1073/pnas.1713343114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova M., Lopez-Calcagno P.E., Raines C.A., Furbank R.T., von Caemmerer S. Overexpression of the Rieske FeS protein of the Cytochrome b6f complex increases C4 photosynthesis. bioRxiv. 2019:574897. doi: 10.1038/s42003-019-0561-9. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feissner R., Xiang Y., Kranz R.G. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal. Biochem. 2003;315:90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- Fey V., Wagner R., Braütigam K., Wirtz M., Hell R., Dietzmann A., Leister D., Oelmüller R., Pfannschmidt T. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J. Biol. Chem. 2005;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- Gao F., Han X., Wu J., Zheng S., Shang Z., Sun D., Zhou R., Li B. A heat-activated calcium-permeable channel-Arabidopsis cyclic nucleotide-gated ion channel 6-is involved in heat shock responses. Plant J. 2012;70:1056–1069. doi: 10.1111/j.1365-313X.2012.04969.x. [DOI] [PubMed] [Google Scholar]
- Hartings S., Paradies S., Karnuth B., Eisfeld S., Mehsing J., Wolff C., Levey T., Westhoff P., Meierhoff K. The DnaJ-like zinc-finger protein HCF222 is required for thylakoid membrane biogenesis in plants. Plant Physiol. 2017;174:1807–1824. doi: 10.1104/pp.17.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B., Börner T., Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–811. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- Jin H., Liu B., Luo L., Feng D., Wang P., Liu J., Da Q., He Y., Qi K., Wang J., et al. HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell. 2014;26:1213–1229. doi: 10.1105/tpc.113.122424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Johnson G.N. Regulation of cyclic and linear electron flow in higher plants. Proc. Natl. Acad. Sci. USA. 2011;108:13317–13322. doi: 10.1073/pnas.1110189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Joliot A., Johnson G. In: Photosystem I: The Light-Driven Plastocyanin: Ferredoxin Oxidoreductase. Golbeck J.H., editor. Springer; Berlin: 2006. Cyclic electron transfer around Photosystem I. [Google Scholar]
- Kuras R., Wollman F.A. The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Saint-Marcoux D., Wollman F.A., de Vitry C. A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:9906–9910. doi: 10.1073/pnas.0702340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G., Zhang H., Smith J.L., Cramer W.A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lennartz K., Bossmann S., Westhoff P., Bechtold N., Meierhoff K. HCF153, a novel nuclear-encoded factor necessary during a post-translational step in biogenesis of the cytochrome b6f complex. Plant J. 2006;45:101–112. doi: 10.1111/j.1365-313X.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- Lennartz K., Plücken H., Seidler A., Westhoff P., Bechtold N., Meierhoff K. HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell. 2001;13:2539–2551. doi: 10.1105/tpc.010245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Kuras R., Ephritikhine G., de Vitry C. A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in arabidopsis chloroplasts. J. Biol. Chem. 2008;283:24608–24616. doi: 10.1074/jbc.M803869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler h.k., Wellburn A.R. Portland Press Limited; 1983. Determinations of Total Carotenoids and Chlorophylls a and B of Leaf Extracts in Different Solvents. [Google Scholar]
- Lomize A.L., Lomize M.A., Krolicki S.R., Pogozheva I.D. Membranome: a database for proteome-wide analysis of single-pass membrane proteins. Nucleic Acids Res. 2017;45:D250–D255. doi: 10.1093/nar/gkw712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. The occurrence of a thylakoid-localized small zinc finger protein in land plants. Plant Signal. Behav. 2011;6:1881–1885. doi: 10.4161/psb.6.12.18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hall D.A., Last R.L. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell. 2011;111:085456. doi: 10.1105/tpc.111.085456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Malnoë A., Wang F., Girard-Bascou J., Wollman F.A., de Vitry C. Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell. 2014;26:373–390. doi: 10.1105/tpc.113.120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone L.A., Proctor M.S., Hitchcock A., Hunter C.N., Johnson M.P. Cytochrome b(6)f-Orchestrator of photosynthetic electron transfer. Biochim. Biophys. Acta. Bioenerg. 2021;1862:148380. doi: 10.1016/j.bbabio.2021.148380. [DOI] [PubMed] [Google Scholar]
- Malone L.A., Qian P., Mayneord G.E., Hitchcock A., Farmer D.A., Thompson R.F., Swainsbury D.J.K., Ranson N.A., Hunter C.N., Johnson M.P. Cryo-EM structure of the spinach cytochrome b(6 )f complex at 3.6 Å resolution. Nature. 2019;575:535–539. doi: 10.1038/s41586-019-1746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- O'Malley R.C., Alonso J.M., Kim C.J., Leisse T.J., Ecker J.R. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2007;2:2910–2917. doi: 10.1038/nprot.2007.425. [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg A.J., Sun Q., van Wijk K.J. New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 2004;279:49367–49383. doi: 10.1074/jbc.M406763200. [DOI] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell. 2006;18:955–969. doi: 10.1105/tpc.105.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs P. Expression and purification of maltose-binding protein fusions. Curr. Protoc. Mol. Biol. 1994;28 doi: 10.1002/0471142727.mb1606s28. 16.16.11-16.16.14. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruppel N.J., Hangarter R.P. Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 2007;7:37. doi: 10.1186/1471-2229-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel N.J., Logsdon C.A., Whippo C.W., Inoue K., Hangarter R.P. A mutation in Arabidopsis SEEDLING PLASTID DEVELOPMENT1 affects plastid differentiation in embryo-derived tissues during seedling growth. Plant Physiol. 2011;155:342–353. doi: 10.1104/pp.110.161414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Marcoux D., Wollman F.A., de Vitry C. Biogenesis of cytochrome b6 in photosynthetic membranes. J. Cell Biol. 2009;185:1195–1207. doi: 10.1083/jcb.200812025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Miyagishima S., Jarvis P. Vol. 6. The Arabidopsis book/American Society of Plant Biologists; 2008. p. e0110. (Chloroplast Biogenesis: Control of Plastid Development, Protein Import, Division and Inheritance). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval-Ibáñez O., Rolo D., Ghandour R., Hertle A.P., Armarego-Marriott T., Sampathkumar A., Zoschke R., Bock R. De-etiolation-induced protein 1 (DEIP1) mediates assembly of the cytochrome b(6)f complex in Arabidopsis. Nat. Commun. 2022;13:4045. doi: 10.1038/s41467-022-31758-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Legen J., Takami T., Shikanai T., Herrmann R.G., Meurer J. Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol. 2007;144:1924–1935. doi: 10.1104/pp.107.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A.J., McAusland L., Lawson T., Raines C.A. Overexpression of the RieskeFeS protein increases electron transport rates and biomass yield. Plant Physiol. 2017;175:134–145. doi: 10.1104/pp.17.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. An atypical haem in the cytochrome b 6 f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- Sun X., Peng L., Guo J., et al. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell. 2007;19:1347–1361. doi: 10.1105/tpc.106.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Voelker R., Barkan A. Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b 6 f complex in maize. Mol. Gen. Genet. 1995;249:507–514. doi: 10.1007/BF00290576. [DOI] [PubMed] [Google Scholar]
- Wang P., Grimm B. Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. 2021;26:484–495. doi: 10.1016/j.tplants.2020.12.005. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Derrien B., Gautier A., Houille-Vernes L., Boulouis A., Saint-Marcoux D., Malnoë A., Rappaport F., de Vitry C., Vallon O., et al. Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell. 2014;26:353–372. doi: 10.1105/tpc.113.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Ahn J.H., Blázquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Ferrándiz C., Kardailsky I., Malancharuvil E.J., Neff M.M., et al. Activation tagging in arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp T.M., Saroussi S., Yang W., Johnson X., Kim R.G., Heinnickel M.L., Russell J.J., Phuthong W., Dent R.M., Broeckling C.D., et al. GreenCut protein CPLD 49 of Chlamydomonas reinhardtii associates with thylakoid membranes and is required for cytochrome b6f complex accumulation. Plant J. 2018;94:1023–1037. doi: 10.1111/tpj.13915. [DOI] [PubMed] [Google Scholar]
- Wollman F.A. The structure, function and biogenesis of cytochrome b6f complexes. Adv Photosynth Res. 1998;7:459–476. [Google Scholar]
- Xiao J., Li J., Ouyang M., Yun T., He B., Ji D., Ma J., Chi W., Lu C., Zhang L. DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis thaliana. Plant Physiol. 2012;112:204891. doi: 10.1104/pp.112.204891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W., Takahashi S., Makino A., Price G.D., Badger M.R., von Caemmerer S. The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 2011;155:956–962. doi: 10.1104/pp.110.168435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Qiang X., Owsiany K., Zhang S., Thannhauser T.W., Li L. Evaluation of different multidimensional LC-MS/MS pipelines for isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis of potato tubers in response to cold storage. J. Proteome Res. 2011;10:4647–4660. doi: 10.1021/pr200455s. [DOI] [PubMed] [Google Scholar]
- Yang Y., Thannhauser T.W., Li L., Zhang S. Development of an integrated approach for evaluation of 2-D gel image analysis: impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis. 2007;28:2080–2094. doi: 10.1002/elps.200600524. [DOI] [PubMed] [Google Scholar]
- Zhang H., Cramer W.A. Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol. Biol. 2004;274:67–78. doi: 10.1385/1-59259-799-8:067. [DOI] [PubMed] [Google Scholar]
- Zhang L., Pu H., Duan Z., Li Y., Liu B., Zhang Q., Li W., Rochaix J.D., Liu L., Peng L. Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell. 2018;30:1770–1788. doi: 10.1105/tpc.18.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038714. Other datasets used and/or analyzed in this study are available from the corresponding author upon reasonable request.