Abstract
The metabolic interdependence, interactions, and coordination of functions between chloroplasts and mitochondria are established and intensively studied. However, less is known about the regulatory components that control these interactions and their responses to external stimuli. Here, we outline how chloroplastic and mitochondrial activities are coordinated via common components involved in signal transduction pathways, gene regulatory events, and post-transcriptional processes. The endoplasmic reticulum emerges as a point of convergence for both transcriptional and post-transcriptional pathways that coordinate chloroplast and mitochondrial functions. Although the identification of molecular components and mechanisms of chloroplast and mitochondrial signaling increasingly suggests common players, this raises the question of how these allow for distinct organelle-specific downstream pathways. Outstanding questions with respect to the regulation of post-transcriptional pathways and the cell and/or tissue specificity of organelle signaling are crucial for understanding how these pathways are integrated at a whole-plant level to optimize plant growth and its response to changing environmental conditions.
Key words: mitochondria, chloroplast, co-regulation
Although the interdependence between mitochondria and chloroplasts has been studied extensively at the metabolic level, less is known about the components and signaling pathways that coordinate their functions. This review summarizes emerging and potential routes and molecular components that affect or regulate the functions of the two organelles.
Introduction
Mitochondria are defining structures in eukaryotic cells. Mitochondrial endosymbiosis has been proposed as the triggering event for the establishment of eukaryotic cells (Martin and Müller, 1998; Gray, 2017). Except in some specialized lineages of life, largely parasitic in nature, mitochondria are essential for viability, and even in those specialized lineages, vestigial mitochondria are present in the form of hydrogenosomes or mitosomes (Makiuchi and Nozaki, 2014; Lewis et al., 2020). The mitochondrial endosymbiosis took place over 1 billion years ago and was followed by a second endosymbiosis that led to the formation of plastids (chloroplasts) and the plant lineage. Thus, chloroplasts evolved in a cellular environment where mitochondria were already established. It is not surprising that mitochondria and chloroplasts have become interdependent in their >500 million years of co-evolution. It is well established from over 50 years of research that essential processes such as photorespiration are performed by interconnected metabolic pathways within mitochondria and chloroplasts (and peroxisomes) (Eisenhut et al., 2019). However, there are a variety of other interdependences between chloroplast and mitochondria. For example, both organelles are central players in carbon and nitrogen metabolism (Smith et al., 2019; Medeiros et al., 2021), synthesis of FeS co-factors (Balk and Pilon, 2011), and fatty acid synthesis (Guan et al., 2020); alterations in galactolipid synthesis in chloroplasts are rescued by mutations in a protein located in the mitochondrial contact site and cristae organizing system (MICOS) complex (Li et al., 2019); mitochondrial alternative electron transport is induced in extended darkness (Pedrotti et al., 2018); and protease systems/proteostasis are coordinated in some manner between both organelles (Kmiec et al., 2018).
In contrast to the well-studied exchange between mitochondria and chloroplasts at the metabolic level, much less is known about the molecular components that co-regulate mitochondrial and chloroplast function. Both mitochondria and chloroplasts signal their functional state to the nucleus to modify gene expression and optimize function in a process called retrograde signaling (Figure 1). Thus, their functional state has a sensory function (Kmiecik et al., 2016; Crawford et al., 2018; Mielecki et al., 2020; Mackenzie and Mullineaux, 2022). Perturbations of mitochondrial or chloroplast signaling often result in greater sensitivity to environmental factors, e.g., high light or drought, demonstrating that organelle retrograde signaling is required for optimal growth (Dahal et al., 2017; Pornsiriwong et al., 2017; Dahal and Vanlerberghe, 2018; Duan et al., 2020; Barreto et al., 2022; Zeng et al., 2022). Given the coordination between these organelles at a metabolic level, it is presumed that their signaling pathways are also integrated by shared regulatory components.
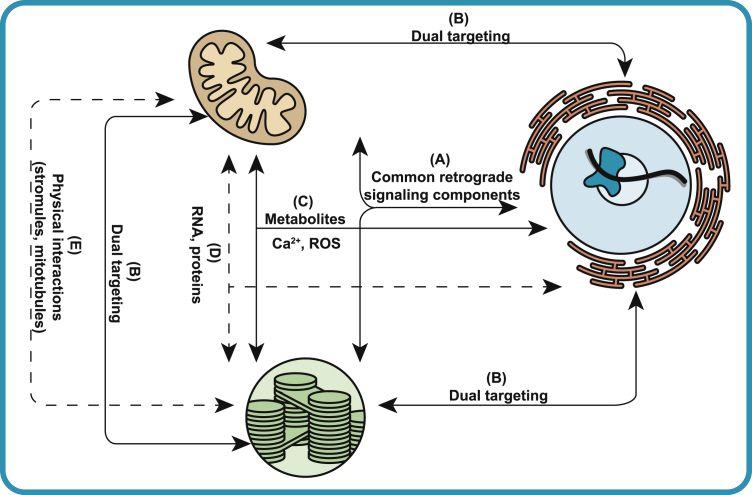
Figure 1.
Possible communication pathways between chloroplasts and mitochondria optimize chloroplast/photosynthetic function.
A number of pathways, both transcriptional and post-transcriptional/translational, coordinate mitochondrial and chloroplast function.
(A) Both chloroplasts and mitochondria signal to the nucleus to alter nuclear gene expression by sharing components in these pathways to coordinate functions.
(B) Dual targeting (including proteins dually targeted to mitochondria and chloroplasts and retro-translocation of proteins from one organelle to another or to the nucleus) enables coordination of organelle function, but how or whether it is regulated remains unknown.
(C) Metabolites, ions, and ROS are all known to be produced by, exchanged between, or capable of affecting chloroplasts and mitochondria. Clearly, they are involved in coordinating function, but an understanding of how this is mediated still requires identification of molecular components.
(D) Although there are no documented cases of protein or RNA transport from mitochondria to chloroplasts, or vice versa, this represents a powerful hypothetical mechanism for coordination of organelle function.
(E) Organelle extensions (stromules, mitotubules) have been documented, but any role in exchange between different organelle species has yet to be demonstrated. Paths illustrated with solid lines represent cases for which there is experimental evidence of communication, and dotted lines indicate hypothetical, as yet undemonstrated communication routes.
This review will focus on the direct co-regulation of mitochondrial and chloroplast function by transcription factors (transcriptional regulation) and post-transcriptional mechanisms that affect protein components or abundance in both organelles (Figure 1A). The potential role for dual-targeted proteins will be reviewed, as this provides a direct mechanism for co-regulation (Figure 1B). Possible and emerging routes of regulation such as direct exchange of RNA or protein molecules between mitochondria and chloroplasts and physical interactions between organelles will be discussed (Figure 1E). We will not focus on the function of metabolite signaling molecules such as reactive oxygen species (ROS), oxidation-reduction (redox) equivalents, or Ca2+ unless their direct target(s) have been identified or they have been shown to co-regulate chloroplast and mitochondrial functions. The involvement and importance of such metabolite signaling molecules in organelle signaling has been extensively documented (Bittner et al., 2022; Castro et al., 2021; Foyer and Hanke, 2022; Leister, 2019; Li and Kim, 2022; Noctor and Foyer, 2016; Pollastri et al., 2021). Overexpression or knockout of a variety of genes encoding mitochondrial or chloroplast proteins often results in changes in the other organelle. Although such experiments are valuable in elucidating links, they often identify secondary responses rather than the initial regulatory event(s). An example of the difficulty in identification of regulatory components and mechanisms is seen with mitochondrial succinate dehydrogenase. An RNA-interference approach that reduced succinate dehydrogenase activity by 30% resulted in plants with higher CO2 assimilation rates and enhanced growth (Fuentes et al., 2011). Furthermore, two reports strongly suggest a role for succinate dehydrogenase in salicylic acid (SA)-mediated signaling via ROS (Belt et al., 2017; Gleason et al., 2011), but the direct link between the two has remained elusive. The large number of such reports, and their limitations in generating a mechanistic understanding, is beyond the scope of this review.
The malate valve mediates the chloroplast-to-mitochondrion communication pathway
A good example of metabolites directly coordinating activities between mitochondria and chloroplasts and for which the mechanism has been elucidated is the function of the MOSAIC DEATH 1 protein (Zhao et al., 2018). It is a plastid-located enoyl-ACP reductase that is involved in de novo fatty acid synthesis. The mutant shows induced programmed cell death caused by increased mitochondrial ROS production (Wu et al., 2015), and a suppressor screen identified components involved in the malate shuttle (Zhao et al., 2018). A defect in MOSAIC DEATH 1 leads to an increase in chloroplast NADH levels, resulting in conversion of oxaloacetate to malate in the chloroplast stroma and its export to the cytosol via the dicarboxylate carrier. The malate is subsequently imported into mitochondria and oxidized back to oxaloacetate in the tricarboxylic acid cycle with the excessive oxidation of NAD(H) by the mitochondrial electron transport chain, resulting in ROS production that triggers programmed cell death. In addition to mitochondrial mutations in complex I, this study identified key enzymes and carrier proteins in the malate valve that act as suppressors, e.g., the plastid-localized, NAD-dependent malate dehydrogenase, the chloroplast dicarboxylate carrier, and mitochondrial malate dehydrogenase. The external addition of malate also induces ROS and cell death in mammalian cell lines, indicating that this may be a well-conserved communication pathway in eukaryotic cells (Zhao et al., 2018). The localization of the malate transporter in mitochondria (Lee et al., 2021), the difference in metabolism between day and night, and the operation of the tricarboxylic acid cycle and the extent of photorespiration (Lim et al., 2020) together suggest that exported chloroplast redox equivalents may be oxidized directly or indirectly by mitochondria (Zhao et al., 2020). Furthermore, the involvement of the citrate valve indicates that additional metabolites are likely to be involved in such signaling pathways (Igamberdiev, 2020; Lee et al., 2021). Notably, the mitochondrial malate transporter is induced transcriptionally as part of the mitochondrial stress response (Van Aken et al., 2009); thus, transcriptional and post-transcriptional pathways are linked and coordinated (Van Aken et al., 2009) (see below).
Transcription factors co-regulating chloroplast and mitochondrial function
Endoplasmic reticulum (ER)-bound NAC (no apical meristem, transcription activation factors 1/2, and cup-shaped cotyledon 2) transcription factors in Arabidopsis have emerged as key regulators of mitochondrial and chloroplast signaling (Ng et al., 2013a; De Clercq et al., 2013; Shapiguzov et al., 2019). Arabidopsis NAC transcription factor 017 (ANAC017) was identified in a forward genetic screen for regulators of Arabidopsis thaliana Alternative Oxidase 1a (AOX1a) (Ng et al., 2013a), the latter being the pre-eminent marker for mitochondrial signaling in a variety of plants. At the same time, reverse genetic approaches identified ANAC013 as a regulator of the mitochondrial dysfunction response (MDR) (De Clercq et al., 2013), a set of genes that respond when mitochondrial function is perturbed by several different approaches. Both these ER-bound transcription factors are released from the ER upon perturbation of mitochondrial function. The exact mechanism has not been defined, but evidence suggests that proteolytic activation by a rhomboid protease plays a role in releasing transcription factors from the ER (Ng et al., 2013a; De Clercq et al., 2013). ANAC017 is not transcriptionally responsive to stress application (Meng et al., 2019) and appears to be constitutively expressed (see below). It is a direct positive regulator of the expression of ANAC013, which is transcriptionally responsive to stress, and both ANAC017 and ANAC013 are required for mitochondrial signaling. The connection to chloroplast function comes from the interaction of RADICAL CELL DEATH 1 (RCD1) with both ANAC013 and ANAC017 (Shapiguzov et al., 2019). The rcd1 mutant shows greatly altered chloroplast redox responses and is resistant to oxidative stress, but it has a highly retarded growth phenotype. ANAC017 and ANAC013 are required to activate the mitochondrial stress response, which is essential for oxidation of excess chloroplast reducing equivalents (Chadee et al., 2021). ANAC017 directly binds to the promoter of RCD1, positively regulating its expression (He et al., 2022) (Figure 2). RCD1 directly binds ANAC017 and ANAC013 and inhibits their activity in the nucleus (Shapiguzov et al., 2019). Thus, ANAC017 acts as a positive regulator of a negative regulatory loop to suppress ANAC activity, which acts as a convergence point for chloroplast and mitochondrial ROS signaling pathways.
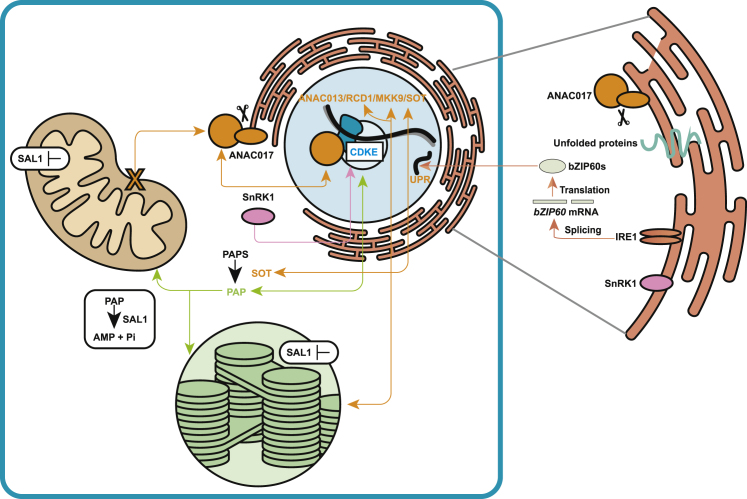
Figure 2.
Interactions between mitochondrial and chloroplast retrograde signaling pathways.
Release of ANAC017 from the ER is initiated by perturbation of mitochondrial function; after translocation to the nucleus, ANAC017 activates the expression of target genes. ANAC017 binds directly to the promoters of RCD1, MKK9, and SOT, which have been shown to participate directly in chloroplast retrograde signaling pathways (see text for details). SAL1-PAP signaling has been extensively characterized as a chloroplast retrograde pathway, but emerging experimental evidence from complementation of a sal1 mutant with a mitochondria-only targeted protein shows interaction with mitochondrial signaling. CDKE has been shown to regulate AOX1a and a variety of other genes at a transcriptional, and for AOX1a post-transcriptional, level. It interacts with SnRK1, which displays a dynamic subcellular localization, with cytosolic, ER, and nuclear localizations reported. ANAC017 is necessary for the ER UPR response and ER-located RNA splicing enzyme inositol-requiring enzyme 1 that is involved in the alternative splicing of bZIP60.
ER, endoplasmic reticulum; UPR, unfolded protein response; ANAC017, Arabidopsis NAC transcription factor 017; ANAC013, Arabidopsis NAC transcription factor 013; RCD1, radical cell death 1; MKK9, mitogen-activated protein kinase kinase 9; SOT, sulphotransferase; CDKE, cyclin-dependent kinase E1; SnRK1, SNF1-related kinase 1; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; PAP, 3′-phosphoadenosine 5′-phosphate; AMP, adenosine monophosphate; IRE1, inositol requiring 1; bZIP60, basic region/leucine zipper motif 60.
There are several other direct molecular links between the ANAC pathway and chloroplast function. Chromatin immunoprecipitation sequencing analysis of the direct targets of ANAC017 revealed that it binds to the promoter of MITOGEN-ACTIVATED PROTEIN KINASE KINASE 9, which activates the MITOGEN-ACTIVATED PROTEIN KINASE (MPK) 3/MPK6 pathway and ethylene signaling (He et al., 2022) (Figure 2). This acts as a convergence point with the ability of ethylene to repress photomorphogenesis (Gommers et al., 2021). The PAP/SAL1 pathway, which controls the levels of the chloroplast signaling molecule 3′-phosphoadenosine 5′-phosphate (PAP), is also linked to ethylene (Balparda et al., 2020), as are the fast high light retrograde signaling (Vogel et al., 2014) and methylerythritol cyclodiphosphate (MEcPP) chloroplast retrograde signaling pathways (Zeng et al., 2022) (Figure 2). ANAC017 also directly targets BASIC REGION/LEUCINE ZIPPER MOTIF (bZIP) 60, bZIP53, ANAC013, ANAC044, ANAC081, and WRKY DNA-BINDING PROTEIN 25, which are all regulators of the unfolded protein response associated with the ER (Gladman et al., 2016) (Figure 2). Cytosolic protein folding stress has been linked to the chloroplast GENOMES UNCOUPLED 1 (GUN1) signaling retrograde pathway (Wu et al., 2019). Thus, it is possible there is a direct link between ANAC017 and GUN1 signaling converging in the ER stress hub. It has also been reported that ANAC017 is necessary for the ER unfolded protein response induced by the addition of dithiothreitol (DTT) (Fuchs et al., 2022). ANAC017 signaling can be triggered by DTT stress, then induce the expression of AOX1a to alleviate the reductive stress caused by DTT (Fuchs et al., 2022). The impaired alternative splicing of bZIP60 in a mutant of the INOSITOL REQUIRING 1 protein, with both proteins localized to the ER membrane (Nagashima et al., 2011), leads to a greater induction of AOX1a (Ng et al., 2013a), showing an integration of the unfolded protein response and the mitochondrial stress signaling pathway at the ER. The chloroplast signaling metabolite MEcPP also induces bZIP60, providing another convergence point for mitochondrial and chloroplast retrograde signaling in the ER (Walley et al., 2015). It has also been reported that mitochondrial retrograde signaling (MRS) is regulated diurnally, with increased binding of ANAC017 to promoters in the light than in the dark when mitochondrial dysfunction is induced by the mitochondria-specific inhibitor myxothiazol, overlap in the promoter sites bound by ANAC017 and a variety of clock regulators, and altered expression of MDR genes in clock mutants (Zhu et al., 2022).
In addition to the NAC transcription factors, several other transcription factors also link mitochondrial and chloroplast retrograde signaling. Sumoylation of MYB DOMAIN PROTEIN 30 (MYB30) positively upregulates the expression of AOX1a in Arabidopsis and leads to salt tolerance (Gong et al., 2020). MYB30 binds directly to the AOX1a promoter and rescues the salt-sensitive phenotype of a myb30 mutant. MYB30 also regulates systematic ROS signaling under high light but not other stress treatments, like wounding or heat stress (Fichman et al., 2020). Importantly, MYB30 is required to link the ROS wave to systematic acclimation. In addition to its role in the high-light systematic response, MYB30 also regulates photomorphogenesis by positively regulating expression of PHYTOCHROME INTERACTING FACTOR 4 and 5 under light conditions (Yan et al., 2020). The ANAC017 MRS pathway is also under diurnal control, with several diurnal and clock regulators having overlapping binding sites with those of ANAC017 (Zhu et al., 2022). In addition, multiple MRS marker genes and genes regulated by ANAC017 show altered expression in a variety of clock mutants. Thus, in addition to the well-characterized metabolite exchange between chloroplasts and mitochondria, it is now apparent that the alternative respiratory pathway that is part of the MDR and required for efficient photosynthesis is co-regulated with a variety of photosynthetic components by clock and diurnal regulators.
Cyclin-dependent kinase E1 (CDKE1) is associated with the Mediator complex, which is present in eukaryotes and links bound transcription factors to RNA polymerase II to regulate the rate of transcript initiation (Poss et al., 2013). The Mediator complex has a central role in many stress responses (He et al., 2021). CDKE1 was identified as a transcriptional and post-transcriptional regulator of AOX1a in Arabidopsis (Ng et al., 2013b), and subsequent studies have revealed that responses to chloroplast perturbation are also altered in a corresponding cdke1 mutant (Blanco et al., 2014). CDKE1 interacts with SNF1-RELATED KINASE 1 (SnRK1), a central mediator of energy signaling in plant cells (Crepin and Rolland, 2019). SnRK1 differs from the components outlined above in that it acts in anterograde regulation of organelle function (Wurzinger et al., 2018). It has been linked to a variety of stress responses and specifically to signaling of low energy status, i.e., extended darkness and hypoxia, in relation to chloroplasts and mitochondria. SnRK1 regulates organelle energy metabolism by phosphorylation of cytosolic enzymes such as nitrate reductase and sucrose–phosphate synthase. Also, several chloroplast and mitochondrial proteins show altered phosphorylation patterns in mutant backgrounds of SnRK1 (reviewed in Wurzinger et al., 2018). It also directly phosphorylates the bZIP63 transcription factor (Mair et al., 2015), which regulates electron transfer flavoprotein/ubiquinone oxidoreductase (Mair et al., 2015; Pedrotti et al., 2018). Dynamic subcellular localization/compartmentalization of SnRK1 is proposed to mediate the diverse processes in which it participates, with an interaction with TOR (target of rapamycin) at the ER to balance energy and stress signaling (Blanco et al., 2019; Gutierrez-Beltran and Crespo, 2022).
Dual targeting of proteins to coordinate chloroplast and mitochondrial function
In general, nuclear-encoded proteins have unique cellular localizations to fulfill specific functions within the cell. Yet in plants, over 250 proteins are targeted to two different cellular compartments (Carrie et al., 2009a; Krause et al., 2012; Baudisch et al., 2014). The frequency of dual targeting for mitochondria and chloroplasts is considerably higher than that observed between other organelles, as more than 100 proteins have been proposed to be dual targeted to mitochondria and chloroplasts (Carrie et al., 2009b).
However, many dual-targeted proteins appear to function primarily in only one organelle. This was demonstrated by hemi-complementation, i.e., the targeting of the corresponding protein to only one organelle, in mutant lines for dual targeted proteins, e.g., for Arabidopsis BT1 (mitochondrial carrier family protein) and RNA polymerase (Bahaji et al., 2011; Tarasenko et al., 2016). Thus, we will not discuss dual-targeted proteins unless they have demonstrated co-regulatory activities in both organelles, bearing in mind that roles in both organelles may be observed only under specific growth conditions and may go unnoticed under standard, non-limiting growth conditions.
The adenosine bisphosphate phosphatase enzyme SAL1 and Prolyl-tRNA synthetase 1 are both dual targeted to chloroplasts and mitochondria and play a role in regulation of mitochondrial and chloroplast function (Pesaresi et al., 2006; Estavillo et al., 2011). SAL1 produces monophosphate PAP from 3′-phosphoadenosine 5′-phosphosulfate and regulates the expression of a variety of nuclear genes in response to abiotic and biotic challenges (Estavillo et al., 2011; Chan et al., 2016). PAP is most extensively characterized as a mobile retrograde signal from chloroplasts (Chan et al., 2016). However, partial complementation of the mutant by targeting SAL1 to exclusively one location showed that although mitochondrial or nuclear targeting led to essentially full complementation, targeting to the chloroplast was less effective. Although protein stability may impact the interpretation of these results, they do suggest that SAL1 may have distinct functions in each organelle. Alternatively, as with ANAC017, SAL1 may be involved in supporting chloroplast function but have its primary role in mitochondria and the nucleus. Sulphotransferase 12, which transfers the sulfuryl group from 3′-phosphoadenosine 5′-phosphosulfate to a number of secondary metabolites, is directly regulated by ANAC017, further linking PAP chloroplast signaling to mitochondrial signaling (He et al., 2022). Dual-targeted Prolyl-tRNA synthetase 1 is involved in translation in both organelles, which has a synergetic effect on signaling compared with a defect in either organelle (Pesaresi et al., 2006). A variety of other dual-targeted components are involved with organelle gene expression, proteases, and ETC biogenesis (Figure 3). It is unclear whether they are regulatory.
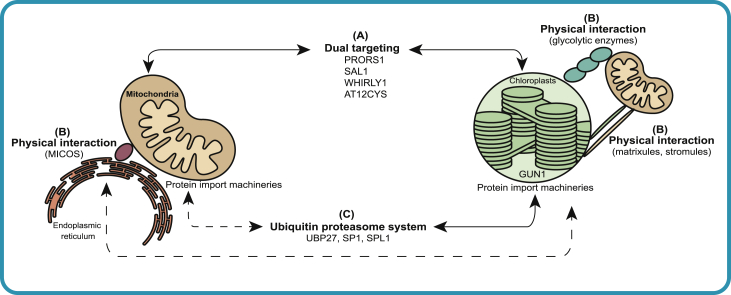
Figure 3.
Communication between mitochondria and chloroplasts at the post-translational level.
(A) The dual-targeted proteins SAL1, PRORS1, and WHIRLY1 have demonstrated roles in organelle retrograde signaling. The dual-targeted protein AT12CYS affects the function of both organelles.
(B) Mitochondria and chloroplasts physically interact through organellar extensions (matrixules and stromules) and glycolytic enzymes. Mitochondria also physically interact with the endoplasmic reticulum (ER) through the mitochondrial contact site and cristae organizing system (MICOS), but how the ER mediates interactions between the MICOS and chloroplasts is still unknown.
(C) Ubiquitin proteasome system regulates functions and biogenesis of mitochondria and chloroplasts by mediating the degradation of protein import machineries.
PRORS1, prolyl-tRNA synthetase 1; UBP27, ubiquitin-specific protease 27; SP1, suppressor of ppi1 locus 1; SPL1, SP1-like 1; GUN 1, genomes uncoupled 1.
A variation on dual-targeting of proteins is the retro-translocation of proteins from the chloroplast to the nucleus or between each organelle. Retro-translocation of WHIRLY1 from chloroplasts to the nucleus has been shown to influence several pathways, notably SA signaling (Krupinska et al., 2020), although it is difficult to demonstrate that such pathways are authentic and do not represent artifacts. In transplastomic Nicotiana tabacum (tobacco) transformed with WHIRLY1, the protein translocates from chloroplasts to the nucleus, providing strong evidence that this pathway is an authentic communication route (Isemer et al., 2012). The fact that WHIRLY proteins display dual targeting to mitochondria and chloroplasts suggests that retro-translocation from one organelle can directly impact the other by altered gene expression. Another notable example is the mitochondrial intermembrane space protein At12Cys. Its localization in cytosol, chloroplasts, and mitochondria was observed in a variety of genetic mutants with disrupted mitochondrial complex I biogenesis and activity (Wang et al., 2016). Depletion of At12Cys protein not only affects biogenesis/activity of the mitochondrial respiratory chain but also increases the protein abundance of Fe superoxide dismutase 1 (Wang et al., 2016), a chloroplastic, cytoplasmic, and nuclear enzyme that performs an antioxidant function in a copper-dependent manner (Dvořák et al., 2020; Melicher et al., 2022). These findings indicate that At12Cys protein not only affects respiratory chain biogenesis in mitochondria but also may impact ROS homeostasis in the chloroplasts, cytoplasm, and nucleus by affecting the abundance of Fe superoxide dismutase 1. The mechanism for alteration of At12Cys location could be inhibition of protein import into mitochondria or retro-translocation from mitochondria to the cytosol, with subsequent import into chloroplasts. The full extent of protein retro-translocation from organelles is probably underestimated, and it is a challenge to fully understand how organelles communicate.
Physical organelle interaction
Physical interactions between chloroplasts and mitochondria were observed over one hundred years ago and have more recently been analyzed for their function (Frederick and Newcomb, 1969; Sage and Sage, 2009; Hanson and Hines, 2018). The behavior of mitochondria in leaf palisade mesophyll cells was examined by confocal laser scanning microscopy. These observations revealed that mitochondria adopt different shapes during interactions with peroxisomes and chloroplasts, e.g., elongated morphology in the light and spherical in the dark (Oikawa et al., 2015). The proximity between mitochondria, chloroplasts, and peroxisomes increased after high-light exposure, a process proposed to be facilitated by reorganization of the ER (Jaipargas et al., 2016). Chloroplasts in nectary parenchyma cells from Citharexylum myrianthum form protrusions to connect with mitochondria (Machado et al., 2018), and elongated mitochondria dynamically interact with chloroplasts in Arabidopsis pavement cells (Barton et al., 2018). These interactions often occur through transient tubular extensions of mitochondria, chloroplasts, and peroxisomes, named matrixules, stromules, and peroxules, respectively (Köhler and Hanson, 2000; Scott et al., 2007) (Figure 3). While continuously changing shape and dynamically interacting with their surroundings, the extensions could enhance the exchange of metabolites and fluorescent proteins, and this process has been well reviewed (Hanson and Hines, 2018; Mathur, 2021). Exchange of other types of molecules such as RNA has also been postulated (Hanson and Hines, 2018).
Although microdomains on organellar outer membranes seem to be critical for organelle interaction (Schwarzländer and Fuchs, 2017; Sparkes, 2018), the specific components involved and how they regulate the interaction remain unknown. In plants, glycolytic enzymes are physically and dynamically associated with mitochondria, depending on the respiratory demand (Giegé et al., 2003; Graham et al., 2007). A specific glycolytic multi-enzyme complex (phosphoglycerate mutase/enolase/pyruvate kinase) plays a moonlighting role in mediating the interaction between chloroplasts and mitochondria (Zhang et al., 2020) (Figure 3). Genetic mutation of these enzymes resulted in a significant decrease in association between the two organelles (Zhang et al., 2020). Another putative pathway that regulates the organellar interaction is mediated by the MICOS. In plants, two components of the MICOS have been characterized, AtMic60 and AtDGS1 (Michaud et al., 2016; Li et al., 2019). Genetic mutation of the two components led to disordered lipid trafficking between mitochondria and chloroplasts, resulting in abnormal organelle morphology (Michaud et al., 2016; Li et al., 2019). However, there is no evidence that MICOS mediates a direct physical interaction between mitochondria and chloroplasts. Analysis of a quantitative, high-density genetic interaction map in yeast revealed that MICOS complex genes share a strong genetic interaction with ER–mitochondrial encounter structure (ERMES) components, indicating that there is a functional connection between the MICOS and ERMES in terms of lipid homeostasis, mtDNA transmission, and mitochondrial dynamics (Kornmann et al., 2009; Hoppins et al., 2011). Plus, the chloroplast–ER physical contact sites are directly involved in the lipid exchange (Tan et al., 2011; Block and Jouhet, 2015). We therefore postulate that perhaps the co-regulation of chloroplasts and mitochondria can be mediated by interactions between the MICOS and ERMES (Figure 3).
Regulation through the ubiquitin proteasome system
In yeast and mammals, the ubiquitin–proteasome system (UPS) directly regulates mitochondrial biogenesis and function (Mishra and Chan, 2014; Franz et al., 2015). A similar UPS seems to exist in plant systems, but its extent is unclear. Arabidopsis UBIQUITIN-SPECIFIC PROTEASE 27 (UBP27) is embedded in the mitochondrial outer membrane (Pan et al., 2014). Overexpression of UBP27 results in abnormal mitochondrial morphology and loss of association between mitochondria and DYNAMIN-RELATED PROTEIN 3, a component of the mitochondrial fission machinery. Thus, UBP27 has been proposed to affect mitochondrial morphology by modulating the function of mitochondrial division proteins. SUPPRESSOR OF PPI1 LOCUS 1 (SP1), a RING-type ubiquitin E3 ligase, is another UPS component that regulates organelle biogenesis in plants. Two independent studies have revealed that SP1 is a novel regulator of plastid and peroxisome biogenesis (Ling and Jarvis, 2015; Pan et al., 2016). The studies demonstrated that SP1 can attach to the organelle membrane, mediate ubiquitination of protein import components, and promote their degradation. The SP1 homolog SP1-like 1 (SPL1) plays a positive role in peroxisome biogenesis (Pan et al., 2018). Although still under some debate, both SP1 and SPL1 have also been observed to localize to mitochondria (Pan et al., 2018). Thus, the role of SP1 and/or SPL1 in the regulation of mitochondrial biogenesis in plants is of interest given that the UPS has been widely reported to have this role in yeast and mammals (Mishra and Chan, 2014; Franz et al., 2015). Based on their function in regulating multi-organelle biogenesis and localization, we postulate that SP1, SPL1, and/or other UPS components orchestrate the biogenesis and functional relationships of chloroplasts and mitochondria (and peroxisomes) (Figure 3). A recent study of GUN1 is consistent with this hypothesis. The study revealed that GUN1 regulates protein import from the cytosol to chloroplasts and that a gun1 clpc1 double mutant induces the cytosolic protein quality control system, including many components of the 26S proteasome (Wu et al., 2019). This indicates that GUN1 not only triggers retrograde signaling at the transcriptional level via ANAC017 targeting of unfolded protein response regulators (as discussed above) but also triggers retrograde signaling through the UPS that may also be linked with mitochondria.
Hormonal regulation of mitochondrial and chloroplast function
Given the central role of chloroplasts and mitochondria in primary and secondary metabolism, which provide the energy and building blocks for growth, it is not surprising that organelle signaling interacts with hormonal control of growth and development (Fàbregas and Fernie, 2021). Although this has long been recognized, the mechanisms of these interactions are only now being elucidated. It has been shown that both MRS via the ANAC pathway and chloroplast retrograde signaling (CRS) via the MEcPP pathway are antagonistic to auxin signaling. Two independent studies showed that perturbation of auxin signaling alone was sufficient to activate MRS and that mitochondrial and auxin signaling were antagonistic (Ivanova et al., 2014; Kerchev et al., 2014). Characterization of several genes involved in auxin homeostasis as direct targets of ANAC017, notably URIDINE DIPHOSPHATE GLYCOSYLTRANSFERASE 74E2, which glycosylates the auxin indole-3-butyric acid to reduce auxin levels, and the auxin biosynthetic enzyme YUCCA5, provides a mechanism for how this is achieved (Kerchev et al., 2014; He et al., 2022). It was also demonstrated that CRS via MEcPP specifically reduced auxin abundance and signaling (Jiang et al., 2018). Thus, both CRS and MRS converge to alter auxin signaling, coordinating their response to environmental conditions. Both these pathways also converge to interact with another phytohormone, ethylene, which is not surprising given the well-characterized interactions between auxin and ethylene (Muday et al., 2012). Biotic and abiotic (high-light) stress induces the synthesis of MEcPP leading to the phosphorylation of MPK3/6 and a systematic stress response (Zeng et al., 2022). ANAC017 activation by mitochondrial dysfunction leads to stimulation of the MITOGEN-ACTIVATED PROTEIN KINASE KINASE 9–MPK3/6 pathway involved in ethylene biosynthesis (He et al., 2022). Also, phosphorylation of the auxin signaling repressor IAA15 by MPK3/6 to repress root growth provides a direct link between organelle signaling and inhibition of auxin-mediated growth (Kim et al., 2022). ANAC017 also links to gibberellic acid signaling, as it is a gibberellin-suppressing factor (Chen et al., 2019), leading to the inactivation of gibberellic acid (GA) and suppression of genes for its biosynthesis. GA has been linked to repression of photomorphogenesis when levels are low in early plant development (Shanmugabalaji et al., 2018), and low levels of GA are also associated with dark-induced senescence (Zhang et al., 2018). A note of caution for an ANAC017-dependent role of GA is a lack of GA-related genes targeted by ANAC017 as identified by chromatin immunoprecipitation sequencing (He et al., 2022) and an age- and/or development-dependent epigenetic regulation of ANAC017 (Bui et al., 2020).
SA is another plant hormone that links chloroplast and mitochondrial function, although the molecular components involved have not been dissected as for other hormones as outlined above. It appears that plastid retrograde signals decrease SA synthesis in chloroplasts, and in some mutant backgrounds (i.e., plastid protein import 2, salicylic acid induction deficient 2, lesions stimulating disease resistance 1), addition of SA induces the expression of photosynthesis-associated proteins via transcriptional and post-transcriptional routes (Hirosawa et al., 2021). Silencing of thylakoid-located ascorbate oxidase also resulted in increased levels of SA (Maruta et al., 2012). More directly, in the var2 mutant lacking a functional FtsH protein, photosystem II damaged protein proteolysis and EXECUTER 1 signaling pathways are impaired. Accumulation of damaged proteins in the chloroplast appears to induce the synthesis of SA as a retrograde signal (Dogra and Kim, 2019). In mitochondria, it is well known that SA induces the expression of AOX1a, a marker of mitochondrial retrograde regulation (Rhoads and McIntosh, 1993), but this is likely because it is both an inhibitor and an uncoupler of the mitochondrial electron transport chain (Norman et al., 2004). The mitochondrial OUTER MEMBRANE PROTEIN OF 66 kDa (previously referred to as BIOSYNTHESIS OF CYTOCHROME C1) is dramatically induced upon SA treatment, and this induction differs in kinetics and sensitivity in different defense-signaling mutant backgrounds compared with AOX1a (Ho et al., 2008). Given that OUTER MEMBRANE PROTEIN OF 66 kDa is induced after treatments that alter mitochondrial function, it appears that SA does play a role in signaling. As outlined above, a disrupted in stress response mutant (dsr1) that maps to a gene encoding a subunit of mitochondrial succinate dehydrogenase shows altered SA gene-mediated expression and pathogen susceptibility (Gleason et al., 2011). Thus, it appears that SA is involved in both CRS and MRS, but the molecular pathways remain to be defined.
Abscisic acid (ABA) is clearly an important hormone in plant abiotic stress response, and given the role of organelle signaling in such responses, it is likely that organelle and ABA signaling converge. The role of ABA in organelle signaling has become complex, with reports refuting a role for the transcription factor ABA INSENSITIVE 4 (ABI4) in GUN1-mediated CRS (Kacprzak et al., 2019). On the other hand, ABI4 has been defined as a negative regulator of AOX1a in Arabidopsis based on a) analysis of two abi4 mutant backgrounds in which AOX1a was de-repressed; b) ABI4 binding to the promoter region of AOX1a in an electrophoretic mobility shift assay and a yeast two-hybrid system; and c) deletion of the ABI4 binding site(s) leading to a large increase in GUS reporter activity driven by the AOX1a promoter (Giraud et al., 2009). In addition, ABI4 is part of the hormonal cross talk between ABA and ethylene (Chandrasekaran et al., 2020). Finally, the CRS component PAP is a second messenger in the ABA-dependent regulation of stomatal closure and germination. Together, these results provide strong evidence for the interaction of organellar and ABA signaling pathways (Pornsiriwong et al., 2017).
Outstanding questions
-
•
The MRS and CRS pathways have largely been studied separately. The distinction between the two has become more blurred with the identification of molecular components that have a role in both pathways, e.g., SAL1-PAP and ANAC017. How signaling specificity is achieved (Møller and Sweetlove, 2010), if indeed a variety of components are shared, is an important question.
-
•
Dual targeting and retro-translation are a powerful means of coordinating organelle function and achieving specificity. Resolving how both are regulated is technically difficult, and this seems to be an underrepresented issue.
-
•
The cytosolic ubiquitin proteasomal system is emerging as a global system to coordinate organelle biogenesis and signaling. A link to the ER unfolded protein response also provides transcriptional and post-transcriptional routes for organelle communication.
-
•
The cell-specific nature of organelle signaling has yet to be explored. There are hints that such specificity exists (Beltrán et al., 2018), with possible different branches of pathways enriched in cell types, based on hormonal specificity (Berkowitz et al., 2021). The emergence of single-cell and spatial transcriptomes will reveal tissue and cell specificity of organelle signaling and interactions.
Funding
J.W. is supported by Australian Research Council Discovery grant DP21010325.
Acknowledgments
No conflict of interest is declared.
Published: November 26, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Bahaji A., Ovecka M., Bárány I., et al. Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol. 2011;52:597–609. doi: 10.1093/pcp/pcr019. [DOI] [PubMed] [Google Scholar]
- Balk J., Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011;16:218–226. doi: 10.1016/j.tplants.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Balparda M., Armas A.M., Estavillo G.M., Roschzttardtz H., Pagani M.A., Gomez-Casati D.F. The PAP/SAL1 retrograde signaling pathway is involved in iron homeostasis. Plant Mol. Biol. 2020;102:323–337. doi: 10.1007/s11103-019-00950-7. [DOI] [PubMed] [Google Scholar]
- Barreto P., Dambire C., Sharma G., Vicente J., Osborne R., Yassitepe J., Gibbs D.J., Maia I.G., Holdsworth M.J., Arruda P. Mitochondrial retrograde signaling through UCP1-mediated inhibition of the plant oxygen-sensing pathway. Curr. Biol. 2022;32:1403–1411.e4. doi: 10.1016/j.cub.2022.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K.A., Wozny M.R., Mathur N., Jaipargas E.A., Mathur J. Chloroplast behaviour and interactions with other organelles in Arabidopsis thaliana pavement cells. J. Cell Sci. 2018;131:jcs202275. doi: 10.1242/jcs.202275. [DOI] [PubMed] [Google Scholar]
- Baudisch B., Langner U., Garz I., Klösgen R.B. The exception proves the rule? Dual targeting of nuclear-encoded proteins into endosymbiotic organelles. New Phytol. 2014;201:80–90. doi: 10.1111/nph.12482. [DOI] [PubMed] [Google Scholar]
- Belt K., Huang S., Thatcher L.F., et al. Salicylic Acid-Dependent Plant Stress Signaling via Mitochondrial Succinate Dehydrogenase. Plant Physiol. 2017;173:2029–2040. doi: 10.1104/pp.16.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán J., Wamboldt Y., Sanchez R., LaBrant E.W., Kundariya H., Virdi K.S., Elowsky C., Mackenzie S.A. Specialized plastids trigger tissue-specific signaling for systemic stress response in plants. Plant Physiol. 2018;178:672–683. doi: 10.1104/pp.18.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O., Xu Y., Liew L.C., Wang Y., Zhu Y., Hurgobin B., Lewsey M.G., Whelan J. RNA-seq analysis of laser microdissected Arabidopsis thaliana leaf epidermis, mesophyll and vasculature defines tissue-specific transcriptional responses to multiple stress treatments. Plant J. 2021;107:938–955. doi: 10.1111/tpj.15314. [DOI] [PubMed] [Google Scholar]
- Bittner A., Cieśla A., Gruden K., et al. Organelles and phytohormones: a network of interactions in plant stress responses. J. Exp Bot. 2022;73:7165–7181. doi: 10.1093/jxb/erac384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco N.E., Guinea-Díaz M., Whelan J., Strand Å. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130231. doi: 10.1098/rstb.2013.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco N.E., Liebsch D., Guinea Díaz M., Strand Å., Whelan J. Dual and dynamic intracellular localization of Arabidopsis thaliana SnRK1.1. J. Exp. Bot. 2019;70:2325–2338. doi: 10.1093/jxb/erz023. [DOI] [PubMed] [Google Scholar]
- Block M.A., Jouhet J. Lipid trafficking at endoplasmic reticulum-chloroplast membrane contact sites. Curr. Opin. Cell Biol. 2015;35:21–29. doi: 10.1016/j.ceb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Bui L.T., Shukla V., Giorgi F.M., Trivellini A., Perata P., Licausi F., Giuntoli B. Differential submergence tolerance between juvenile and adult Arabidopsis plants involves the ANAC017 transcription factor. Plant J. 2020;104:979–994. doi: 10.1111/tpj.14975. [DOI] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Whelan J. Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- Carrie C., Kühn K., Murcha M.W., Duncan O., Small I.D., O'Toole N., Whelan J. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009;57:1128–1139. doi: 10.1111/j.1365-313X.2008.03745.x. [DOI] [PubMed] [Google Scholar]
- Castro B., Citterico M., Kimura S., Stevens D.M., Wrzaczek M., Coaker G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants. 2021;7:403–412. doi: 10.1038/s41477-021-00887-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee A., Alber N.A., Dahal K., Vanlerberghe G.C. The complementary roles of chloroplast cyclic electron transport and mitochondrial alternative oxidase to ensure photosynthetic performance. Front. Plant Sci. 2021;12:748204. doi: 10.3389/fpls.2021.748204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.X., Mabbitt P.D., Phua S.Y., Mueller J.W., Nisar N., Gigolashvili T., Stroeher E., Grassl J., Arlt W., Estavillo G.M., et al. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proc. Natl. Acad. Sci. USA. 2016;113:E4567–E4576. doi: 10.1073/pnas.1604936113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran U., Luo X., Zhou W., Shu K. Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Commun. 2020;1:100040. doi: 10.1016/j.xplc.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.I., Li P.F., Yang C.H. NAC-like gene GIBBERELLIN SUPPRESSING FACTOR regulates the gibberellin metabolic pathway in response to cold and drought stresses in Arabidopsis. Sci. Rep. 2019;9:19226. doi: 10.1038/s41598-019-55429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford T., Lehotai N., Strand Å. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018;69:2783–2795. doi: 10.1093/jxb/erx481. [DOI] [PubMed] [Google Scholar]
- Crepin N., Rolland F. SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 2019;51:29–36. doi: 10.1016/j.pbi.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Dahal K., Vanlerberghe G.C. Improved chloroplast energy balance during water deficit enhances plant growth: more crop per drop. J. Exp. Bot. 2018;69:1183–1197. doi: 10.1093/jxb/erx474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal K., Martyn G.D., Alber N.A., Vanlerberghe G.C. Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J. Exp. Bot. 2017;68:657–671. doi: 10.1093/jxb/erw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I., Vermeirssen V., Van Aken O., Vandepoele K., Murcha M.W., Law S.R., Inzé A., Ng S., Ivanova A., Rombaut D., et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell. 2013;25:3472–3490. doi: 10.1105/tpc.113.117168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra V., Kim C. Chloroplast protein homeostasis is coupled with retrograde signaling. Plant Signal. Behav. 2019;14:1656037. doi: 10.1080/15592324.2019.1656037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Ruiz-Sola M.Á., Couso A., Veciana N., Monte E. Red and blue light differentially impact retrograde signalling and photoprotection in rice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190402. doi: 10.1098/rstb.2019.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák P., Krasylenko Y., Ovečka M., Basheer J., Zapletalová V., Šamaj J., Takáč T. FSD1: developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant Cell Environ. 2020 doi: 10.1111/pce.13773. pce.13773. [DOI] [Google Scholar]
- Eisenhut M., Roell M.S., Weber A.P.M. Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol. 2019;223:1762–1769. doi: 10.1111/nph.15872. [DOI] [PubMed] [Google Scholar]
- Estavillo G.M., Crisp P.A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N., Fernie A.R. The interface of central metabolism with hormone signaling in plants. Curr. Biol. 2021;31:R1535–R1548. doi: 10.1016/j.cub.2021.09.070. [DOI] [PubMed] [Google Scholar]
- Fichman Y., Zandalinas S.I., Sengupta S., Burks D., Myers R.J., Jr., Azad R.K., Mittler R. MYB30 orchestrates systemic reactive oxygen signaling and plant acclimation. Plant Physiol. 2020;184:666–675. doi: 10.1104/pp.20.00859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Hanke G. ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J. 2022;111:642–661. doi: 10.1111/tpj.15856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A., Kevei É., Hoppe T. Double-edged alliance: mitochondrial surveillance by the UPS and autophagy. Curr. Opin. Cell Biol. 2015;37:18–27. doi: 10.1016/j.ceb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Frederick S.E., Newcomb E.H. Microbody-like organelles in leaf cells. Science. 1969;163:1353–1355. doi: 10.1126/science.163.3873.1353. [DOI] [PubMed] [Google Scholar]
- Fuchs P., Bohle F., Lichtenauer S., Ugalde J.M., Feitosa Araujo E., Mansuroglu B., Ruberti C., Wagner S., Müller-Schüssele S.J., Meyer A.J., Schwarzländer M. Reductive stress triggers ANAC017-mediated retrograde signaling to safeguard the endoplasmic reticulum by boosting mitochondrial respiratory capacity. Plant Cell. 2022;34:1375–1395. doi: 10.1093/plcell/koac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes D., Meneses M., Nunes-Nesi A., Araújo W.L., Tapia R., Gómez I., Holuigue L., Gutiérrez R.A., Fernie A.R., Jordana X. A deficiency in the flavoprotein of Arabidopsis mitochondrial complex II results in elevated photosynthesis and better growth in nitrogen-limiting conditions. Plant Physiol. 2011;157:1114–1127. doi: 10.1104/pp.111.183939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P., Heazlewood J.L., Roessner-Tunali U., Millar A.H., Fernie A.R., Leaver C.J., Sweetlove L.J. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15:2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 2009;150:1286–1296. doi: 10.1104/pp.109.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman N.P., Marshall R.S., Lee K.H., Vierstra R.D. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell. 2016;28:1279–1296. doi: 10.1105/tpc.15.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C., Huang S., Thatcher L.F., Foley R.C., Anderson C.R., Carroll A.J., Millar A.H., Singh K.B. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA. 2011;108:10768–10773. doi: 10.1073/pnas.1016060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers C.M.M., Ruiz-Sola M.Á., Ayats A., Pereira L., Pujol M., Monte E. GENOMES UNCOUPLED1-independent retrograde signaling targets the ethylene pathway to repress photomorphogenesis. Plant Physiol. 2021;185:67–76. doi: 10.1093/plphys/kiaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q., Li S., Zheng Y., Duan H., Xiao F., Zhuang Y., He J., Wu G., Zhao S., Zhou H., Lin H. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J. 2020;102:1157–1171. doi: 10.1111/tpj.14689. [DOI] [PubMed] [Google Scholar]
- Graham J.W.A., Williams T.C.R., Morgan M., Fernie A.R., Ratcliffe R.G., Sweetlove L.J. Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell. 2007;19:3723–3738. doi: 10.1105/tpc.107.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.W. Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol. Biol. Cell. 2017;28:1285–1287. doi: 10.1091/mbc.E16-07-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Okazaki Y., Zhang R., Saito K., Nikolau B.J. Dual-localized enzymatic components constitute the fatty acid synthase systems in mitochondria and plastids. Plant Physiol. 2020;183:517–529. doi: 10.1104/pp.19.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E., Crespo J.L. Compartmentalization, a key mechanism controlling the multi-tasking role of SnRK1. J. Exp. Bot. 2022;73:7055–7067. doi: 10.1093/jxb/erac315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.R., Hines K.M. Stromules: probing formation and function. Plant Physiol. 2018;176:128–137. doi: 10.1104/pp.17.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Liew L.C., Yin L., Lewsey M.G., Whelan J., Berkowitz O. The retrograde signalling regulator ANAC017 recruits the MKK9-MPK3/6, ethylene, and auxin signalling pathways to balance mitochondrial dysfunction with growth. Plant Cell. 2022;34:3460–3481. doi: 10.1093/plcell/koac177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Denecker J., Van Der Kelen K., Willems P., Pottie R., Phua S.Y., Hannah M.A., Vertommen D., Van Breusegem F., Mhamdi A. The Arabidopsis mediator complex subunit 8 regulates oxidative stress responses. Plant Cell. 2021;33:2032–2057. doi: 10.1093/plcell/koab079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosawa Y., Tada A., Matsuura T., Mori I.C., Ogura Y., Hayashi T., Uehara S., Ito-Inaba Y., Inaba T. Salicylic acid acts antagonistically to plastid retrograde signaling by promoting the accumulation of photosynthesis-associated proteins in Arabidopsis. Plant Cell Physiol. 2021;62:1728–1744. doi: 10.1093/pcp/pcab128. [DOI] [PubMed] [Google Scholar]
- Ho L.H.M., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 2008;147:1858–1873. doi: 10.1104/pp.108.121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A.U. Citrate valve integrates mitochondria into photosynthetic metabolism. Mitochondrion. 2020;52:218–230. doi: 10.1016/j.mito.2020.04.003. [DOI] [PubMed] [Google Scholar]
- Isemer R., Mulisch M., Schäfer A., Kirchner S., Koop H.U., Krupinska K. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 2012;586:85–88. doi: 10.1016/j.febslet.2011.11.029. [DOI] [PubMed] [Google Scholar]
- Ivanova A., Law S.R., Narsai R., Duncan O., Lee J.H., Zhang B., Van Aken O., Radomiljac J.D., van der Merwe M., Yi K., Whelan J. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates alternative Oxidase1a expression in Arabidopsis. Plant Physiol. 2014;165:1233–1254. doi: 10.1104/pp.114.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaipargas E.A., Mathur N., Bou Daher F., Wasteneys G.O., Mathur J. High light intensity leads to increased peroxule-mitochondria interactions in plants. Front. Cell Dev. Biol. 2016;4:6. doi: 10.3389/fcell.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Rodriguez-Furlan C., Wang J.Z., de Souza A., Ke H., Pasternak T., Lasok H., Ditengou F.A., Palme K., Dehesh K. Interplay of the two ancient metabolites auxin and MEcPP regulates adaptive growth. Nat. Commun. 2018;9:2262. doi: 10.1038/s41467-018-04708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak S.M., Mochizuki N., Naranjo B., Xu D., Leister D., Kleine T., Okamoto H., Terry M.J. Plastid-to-Nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 2019;179:18–23. doi: 10.1104/pp.18.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev P.I., De Clercq I., Denecker J., Mühlenbock P., Kumpf R., Nguyen L., Audenaert D., Dejonghe W., Van Breusegem F. Mitochondrial perturbation negatively affects auxin signaling. Mol. Plant. 2014;7:1138–1150. doi: 10.1093/mp/ssu071. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Bahk S., Nguyen N.T., Pham M.L.A., Kadam U.S., Hong J.C., Chung W.S. Phosphorylation of the auxin signaling transcriptional repressor IAA15 by MPKs is required for the suppression of root development under drought stress in Arabidopsis. Nucleic Acids Res. 2022;50:10544–10561. doi: 10.1093/nar/gkac798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B., Branca R.M.M., Murcha M.W., Lehtiö J., Glaser E., Teixeira P.F. A common peptidolytic mechanism for targeting peptide degradation in mitochondria and chloroplasts. Mol. Plant. 2018;11:342–345. doi: 10.1016/j.molp.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Kmiecik P., Leonardelli M., Teige M. Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 2016;67:3793–3807. doi: 10.1093/jxb/erw136. [DOI] [PubMed] [Google Scholar]
- Köhler R.H., Hanson M.R. Plastid tubules of higher plants are tissue-specific and developmentally regulated. J. Cell Sci. 2000;113:81–89. doi: 10.1242/jcs.113.1.81. [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K., Oetke S., Krupinska K. Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int. J. Mol. Sci. 2012;13:11085–11101. doi: 10.3390/ijms130911085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinska K., Blanco N.E., Oetke S., Zottini M. Genome communication in plants mediated by organelle-n-ucleus-located proteins. Philos Trans R Soc Lond B Biol Sci. 2020;375:20190397. doi: 10.1098/rstb.2019.0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.P., Elsässer M., Fuchs P., Fenske R., Schwarzländer M., Millar A.H. The versatility of plant organic acid metabolism in leaves is underpinned by mitochondrial malate-citrate exchange. Plant Cell. 2021;33:3700–3720. doi: 10.1093/plcell/koab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Piecing the puzzle together: the central role of reactive oxygen species and redox hubs in chloroplast retrograde signaling. Antioxid. Redox Signal. 2019;30:1206–1219. doi: 10.1089/ars.2017.7392. [DOI] [PubMed] [Google Scholar]
- Lewis W.H., Lind A.E., Sendra K.M., Onsbring H., Williams T.A., Esteban G.F., Hirt R.P., Ettema T.J.G., Embley T.M. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 2020;37:524–539. doi: 10.1093/molbev/msz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Lavell A., Meng X., Berkowitz O., Selinski J., van de Meene A., Carrie C., Benning C., Whelan J., De Clercq I., Wang Y. Arabidopsis DGD1 SUPPRESSOR1 is a subunit of the mitochondrial contact site and cristae organizing system and affects mitochondrial biogenesis. Plant Cell. 2019;31:1856–1878. doi: 10.1105/tpc.18.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Kim C. Chloroplast ROS and stress signaling. Plant Commun. 2022;3:100264. doi: 10.1016/j.xplc.2021.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.L., Voon C.P., Guan X., Yang Y., Gardeström P., Lim B.L. In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD(+) fluorescent protein sensors. Nat. Commun. 2020;11:3238. doi: 10.1038/s41467-020-17056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q., Jarvis P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 2015;25:2527–2534. doi: 10.1016/j.cub.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado S.R., Gregório E.A., Rodrigues T.M. Structural associations between organelle membranes in nectary parenchyma cells. Planta. 2018;247:1067–1076. doi: 10.1007/s00425-018-2844-7. [DOI] [PubMed] [Google Scholar]
- Mackenzie S.A., Mullineaux P.M. Plant environmental sensing relies on specialized plastids. J. Exp. Bot. 2022;73:7155–7164. doi: 10.1093/jxb/erac334. [DOI] [PubMed] [Google Scholar]
- Mair A., Pedrotti L., Wurzinger B., Anrather D., Simeunovic A., Weiste C., Valerio C., Dietrich K., Kirchler T., Nägele T., et al. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife. 2015;4:e05828. doi: 10.7554/eLife.05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiuchi T., Nozaki T. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 2014;100:3–17. doi: 10.1016/j.biochi.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Martin W., Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Maruta T., Noshi M., Tanouchi A., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 2012;287:11717–11729. doi: 10.1074/jbc.M111.292847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J. Organelle extensions in plant cells. Plant Physiol. 2021;185:593–607. doi: 10.1093/plphys/kiaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros D.B., Aarabi F., Martinez Rivas F.J., Fernie A.R. The knowns and unknowns of intracellular partitioning of carbon and nitrogen, with focus on the organic acid-mediated interplay between mitochondrion and chloroplast. J. Plant Physiol. 2021;266:153521. doi: 10.1016/j.jplph.2021.153521. [DOI] [PubMed] [Google Scholar]
- Melicher P., Dvořák P., Krasylenko Y., Shapiguzov A., Kangasjärvi J., Šamaj J., Takáč T. Arabidopsis iron superoxide dismutase FSD1 protects against methyl viologen-induced oxidative stress in a copper-dependent manner. Front. Plant Sci. 2022;13:823561. doi: 10.3389/fpls.2022.823561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Li L., De Clercq I., Narsai R., Xu Y., Hartmann A., Claros D.L., Custovic E., Lewsey M.G., Whelan J., Berkowitz O. ANAC017 coordinates organellar functions and stress responses by reprogramming retrograde signaling. Plant Physiol. 2019;180:634–653. doi: 10.1104/pp.18.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M., Gros V., Tardif M., Brugière S., Ferro M., Prinz W.A., Toulmay A., Mathur J., Wozny M., Falconet D., et al. AtMic60 is involved in plant mitochondria lipid trafficking and is part of a large complex. Curr. Biol. 2016;26:627–639. doi: 10.1016/j.cub.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielecki J., Gawroński P., Karpiński S. Retrograde signaling: understanding the communication between organelles. Int. J. Mol. Sci. 2020;21:6173. doi: 10.3390/ijms21176173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I.M., Sweetlove L.J. ROS signalling--specificity is required. Trends Plant Sci. 2010;15:370–374. doi: 10.1016/j.tplants.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Muday G.K., Rahman A., Binder B.M. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 2012;17:181–195. doi: 10.1016/j.tplants.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Nagashima Y., Mishiba K.I., Suzuki E., Shimada Y., Iwata Y., Koizumi N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 2011;1:29. doi: 10.1038/srep00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Giraud E., Duncan O., Law S.R., Wang Y., Xu L., Narsai R., Carrie C., Walker H., Day D.A., et al. Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 2013;288:3449–3459. doi: 10.1074/jbc.M112.416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Ivanova A., Duncan O., Law S.R., Van Aken O., De Clercq I., Wang Y., Carrie C., Xu L., Kmiec B., et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell. 2013;25:3450–3471. doi: 10.1105/tpc.113.113985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H. Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 2016;171:1581–1592. doi: 10.1104/pp.16.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C., Howell K.A., Millar A.H., Whelan J.M., Day D.A. Salicylic acid is an uncoupler and inhibitor of mitochondrial electron transport. Plant Physiol. 2004;134:492–501. doi: 10.1104/pp.103.031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Matsunaga S., Mano S., Kondo M., Yamada K., Hayashi M., Kagawa T., Kadota A., Sakamoto W., Higashi S., et al. Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat. Plants. 2015;1:15035. doi: 10.1038/nplants.2015.35. [DOI] [PubMed] [Google Scholar]
- Pan R., Kaur N., Hu J. The Arabidopsis mitochondrial membrane-bound ubiquitin protease UBP27 contributes to mitochondrial morphogenesis. Plant J. 2014;78:1047–1059. doi: 10.1111/tpj.12532. [DOI] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Hu J. E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2016;113:E7307–E7316. doi: 10.1073/pnas.1613530113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Chen C., Hu J. The E3 ubiquitin ligase SP1-like 1 plays a positive role in peroxisome biogenesis in Arabidopsis. Plant J. 2018;94:836–846. doi: 10.1111/tpj.13900. [DOI] [PubMed] [Google Scholar]
- Pedrotti L., Weiste C., Nägele T., Wolf E., Lorenzin F., Dietrich K., Mair A., Weckwerth W., Teige M., Baena-González E., Dröge-Laser W. Snf1-RELATED KINASE1-controlled C/S(1)-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell. 2018;30:495–509. doi: 10.1105/tpc.17.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Masiero S., Eubel H., Braun H.P., Bhushan S., Glaser E., Salamini F., Leister D. Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell. 2006;18:970–991. doi: 10.1105/tpc.105.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri S., Sukiran N.A., Jacobs B.C.I.C., Knight M.R. Chloroplast calcium signalling regulates thermomemory. J. Plant Physiol. 2021;264:153470. doi: 10.1016/j.jplph.2021.153470. [DOI] [PubMed] [Google Scholar]
- Pornsiriwong W., Estavillo G.M., Chan K.X., Tee E.E., Ganguly D., Crisp P.A., Phua S.Y., Zhao C., Qiu J., Park J., et al. A chloroplast retrograde signal, 3'-phosphoadenosine 5'-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. Elife. 2017;6:e23361. doi: 10.7554/eLife.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss Z.C., Ebmeier C.C., Taatjes D.J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.M., McIntosh L. The salicylic acid-inducible alternative oxidase gene aox1 and genes encoding pathogenesis-related proteins share regions of sequence similarity in their promoters. Plant Mol. Biol. 1993;21:615–624. doi: 10.1007/bf00014545. [DOI] [PubMed] [Google Scholar]
- Sage T.L., Sage R.F. The functional anatomy of rice leaves: implications for refixation of photorespiratory CO2 and efforts to engineer C4 photosynthesis into rice. Plant Cell Physiol. 2009;50:756–772. doi: 10.1093/pcp/pcp033. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Fuchs P. Plant mitochondrial membranes: adding structure and new functions to respiratory physiology. Curr. Opin. Plant Biol. 2017;40:147–157. doi: 10.1016/j.pbi.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Scott I., Sparkes I.A., Logan D.C. The missing link: inter-organellar connections in mitochondria and peroxisomes? Trends Plant Sci. 2007;12:380–381. doi: 10.1016/j.tplants.2007.08.010. author reply 381-383. [DOI] [PubMed] [Google Scholar]
- Shanmugabalaji V., Chahtane H., Accossato S., Rahire M., Gouzerh G., Lopez-Molina L., Kessler F. Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 2018;28:2616–2623.e5. doi: 10.1016/j.cub.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A., Vainonen J.P., Hunter K., Tossavainen H., Tiwari A., Järvi S., Hellman M., Aarabi F., Alseekh S., Wybouw B., et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife. 2019;8:7554. doi: 10.7554/eLife.43284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.R., Dupont C.L., McCarthy J.K., Broddrick J.T., Oborník M., Horák A., Füssy Z., Cihlář J., Kleessen S., Zheng H., et al. Evolution and regulation of nitrogen flux through compartmentalized metabolic networks in a marine diatom. Nat. Commun. 2019;10:4552. doi: 10.1038/s41467-019-12407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I. Lessons from optical tweezers: quantifying organelle interactions, dynamics and modelling subcellular events. Curr. Opin. Plant Biol. 2018;46:55–61. doi: 10.1016/j.pbi.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Tan X., Wang Q., Tian B., Zhang H., Lu D., Zhou J. A Brassica napus lipase locates at the membrane contact sites involved in chloroplast development. PLoS One. 2011;6:e26831. doi: 10.1371/journal.pone.0026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko V.I., Katyshev A.I., Yakovleva T.V., Garnik E.Y., Chernikova V.V., Konstantinov Y.M., Koulintchenko M.V. RPOTmp, an Arabidopsis RNA polymerase with dual targeting, plays an important role in mitochondria, but not in chloroplasts. J. Exp. Bot. 2016;67:5657–5669. doi: 10.1093/jxb/erw327. [DOI] [PubMed] [Google Scholar]
- Van Aken O., Zhang B., Carrie C., Uggalla V., Paynter E., Giraud E., Whelan J. Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant. 2009;2:1310–1324. doi: 10.1093/mp/ssp053. [DOI] [PubMed] [Google Scholar]
- Vogel M.O., Moore M., König K., Pecher P., Alsharafa K., Lee J., Dietz K.J. Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell. 2014;26:1151–1165. doi: 10.1105/tpc.113.121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J., Xiao Y., Wang J.Z., Baidoo E.E., Keasling J.D., Shen Z., Briggs S.P., Dehesh K. Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2015;112:6212–6217. doi: 10.1073/pnas.1504828112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lyu W., Berkowitz O., Radomiljac J.D., Law S.R., Murcha M.W., Carrie C., Teixeira P.F., Kmiec B., Duncan O., et al. Inactivation of mitochondrial complex I induces the expression of a twin cysteine protein that targets and affects cytosolic, chloroplastidic and mitochondrial function. Mol. Plant. 2016;9:696–710. doi: 10.1016/j.molp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Wu G.Z., Meyer E.H., Richter A.S., Schuster M., Ling Q., Schöttler M.A., Walther D., Zoschke R., Grimm B., Jarvis R.P., Bock R. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants. 2019;5:525–538. doi: 10.1038/s41477-019-0415-y. [DOI] [PubMed] [Google Scholar]
- Wu J., Sun Y., Zhao Y., Zhang J., Luo L., Li M., Wang J., Yu H., Liu G., Yang L., et al. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Res. 2015;25:621–633. doi: 10.1038/cr.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzinger B., Nukarinen E., Nägele T., Weckwerth W., Teige M. The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 2018;176:1085–1094. doi: 10.1104/pp.17.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Li C., Dong X., Li H., Zhang D., Zhou Y., Jiang B., Peng J., Qin X., Cheng J., et al. MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell. 2020;32:2196–2215. doi: 10.1105/tpc.19.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Wang J.Z., He X., Ke H., Lemos M., Gray W.M., Dehesh K. A plastidial retrograde signal potentiates biosynthesis of systemic stress response activators. New Phytol. 2022;233:1732–1749. doi: 10.1111/nph.17890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Z., Wang X., Wang J., Fan K., Li Z., Lin W. DELLA proteins negatively regulate dark-induced senescence and chlorophyll degradation in Arabidopsis through interaction with the transcription factor WRKY6. Plant Cell Rep. 2018;37:981–992. doi: 10.1007/s00299-018-2282-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sampathkumar A., Kerber S.M.L., Swart C., Hille C., Seerangan K., Graf A., Sweetlove L., Fernie A.R. A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat. Commun. 2020;11:4509. doi: 10.1038/s41467-020-18234-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Luo L., Xu J., Xin P., Guo H., Wu J., Bai L., Wang G., Chu J., Zuo J., et al. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 2018;28:448–461. doi: 10.1038/s41422-018-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yu H., Zhou J.M., Smith S.M., Li J. Malate circulation: linking chloroplast metabolism to mitochondrial ROS. Trends Plant Sci. 2020;25:446–454. doi: 10.1016/j.tplants.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Narsai R., He C., et al. Coordinated regulation of the mitochondrial retrograde response by circadian clock regulators and ANAC017. Plant Commun. 2022;3:100501. doi: 10.1016/j.xplc.2022.100501. [DOI] [PMC free article] [PubMed] [Google Scholar]